Significance
Box C (RUGAUGA)/D (CUGA) and H (ANANNA)/ACA small nucleolar RNAs (snoRNAs) are important for the modification and processing of rRNA during ribosome biogenesis in eukaryotes. However, the molecular role of snoRNAs throughout the multiple steps of pre-rRNA processing remains poorly understood. This study shows that an uncharacterized C/D box snoRNA, HIDDEN TREASURE 2 (HID2), functions as a prominent player in the monitoring of efficient pre-rRNA processing, which, in turn, is essential for accurate ribosome assembly in Arabidopsis. Our data explore a link between a spatially regulated snoRNA and the complexity and the precise control of ribosome biogenesis. Further, the conservation of HID2’s signature motif and function highlights its importance in multicellular organisms. hid2 appears to be a representative snoRNA mutant exhibiting pleiotropic developmental defects in plants.
Keywords: C/D box small nucleolar RNA, pre-rRNA processing, ribosome biogenesis
Abstract
Ribosome production in eukaryotes requires the complex and precise coordination of several hundred assembly factors, including many small nucleolar RNAs (snoRNAs). However, at present, the distinct role of key snoRNAs in ribosome biogenesis remains poorly understood in higher plants. Here we report that a previously uncharacterized C (RUGAUGA)/D (CUGA) type snoRNA, HIDDEN TREASURE 2 (HID2), acts as an important regulator of ribosome biogenesis through a snoRNA–rRNA interaction. Nucleolus-localized HID2 is actively expressed in Arabidopsis proliferative tissues, whereas defects in HID2 cause a series of developmental defects reminiscent of ribosomal protein mutants. HID2 associates with the precursor 45S rRNA and promotes the efficiency and accuracy of pre-rRNA processing. Intriguingly, disrupting HID2 in Arabidopsis appears to impair the integrity of 27SB, a key pre-rRNA intermediate that generates 25S and 5.8S rRNA and is known to be vital for the synthesis of the 60S large ribosomal subunit and also produces an imbalanced ribosome profile. Finally, we demonstrate that the antisense-box of HID2 is both functionally essential and highly conserved in eukaryotes. Overall, our study reveals the vital and possibly conserved role of a snoRNA in monitoring the efficiency of pre-rRNA processing during ribosome biogenesis.
The eukaryotic ribosome responsible for protein synthesis is composed of a 60S large ribosomal subunit and a 40S small ribosomal subunit. To date, ribosome biogenesis has been studied most extensively in yeast. The generally conserved process, which encompasses a number of highly orchestrated steps, is pivotal to all organisms and requires the generation of mature rRNAs, ribosomal proteins with hundreds of processing and assembly factors, and many small nucleolar RNAs (snoRNAs) (1, 2). In plants, mutants in genes encoding essential ribosomal biogenesis factors display broad abnormal growth and developmental phenotypes and altered responses to stress stimuli (3). Likewise, a variety of human diseases have been shown to be caused by defects in ribosome assembly (4).
In plants, ribosome biogenesis begins in the nucleolus when 45S rRNA transcription is initiated by RNA polymerase I (5). The primary long transcript is comprised of three ribosomal RNAs, 18S, 5.8S, and 25S rRNA, which are separated by two internal transcribed spacers (ITS1 and ITS2) and flanked by external transcribed spacers (5′- and 3′-ETS) (6). After the initial splicing steps, the resulting 35S rRNA primarily follows the “ITS1-first” pathway, which is common among metazoans, to produce mature rRNAs. This pathway starts with an early cleavage of ITS1 at the A3 site, followed by complete elimination of the 5′-ETS (7, 8). Recently a minor pathway, known as “5′ ETS-first pathway,” also has been reported to coexist in plants and has been shown to resemble the strict processing pathway in yeast (9, 10). Each of the processes of rRNA transcription, processing, and modification has been shown to require snoRNAs (11).
Plants possess an abundance of snoRNAs, which are primarily transcribed in polycistronic gene clusters (12). snoRNAs ranging from 70 to 250 nt have been shown to work in concert with ribonucleoproteins (RNPs) during the RNA-modification process (13). C (RUGAUGA)/D (CUGA) box and H (ANANNA)/ACA box snoRNAs constitute the two main classes of snoRNAs, with a number of the former group directing the 2′-O-methylation of the ribose and a number of the latter group guiding pseudouridination (14, 15). Both types of small nucleolar RNP (snoRNP) recognize their target modification site via specific sequence base-paring.
At present, few snoRNAs involved in rRNA processing and/or rRNA folding have been explored. The U3 C/D box snoRNP shown to be universally conserved across eukaryotes is the core component of the small subunit processome essential for 18S rRNA processing (16, 17). In plants, it has been reported that the U3 snoRNP forms a stable complex with nucleolin protein 1 (NUC1), which binds nascent pre-RNA at the 5′ ETS and specifically cleaves pre-rRNA at the P site (18–20). Moreover, NUC1-U3 snoRNP binds rDNA before its interaction with pre-rRNA (20, 21). A few other snoRNAs also have been reported to be involved in pre-rRNA processing. For instance, in yeast U14, snR30, and snR10 are required for 18S rRNA processing, and in metazoans U8 and U24 have been demonstrated to be involved in large-subunit processing (22–25). Overall, however, with the exception of U3 snoRNA, little is known regarding the roles of plant snoRNAs in the regulation of rRNA processing.
Our previous work has annotated dozens of snoRNA gene clusters, as well as single snoRNAs, in both rice and Arabidopsis (26, 27). Here, taking advantage of our established collection of sequenced Arabidopsis snoRNAs, we identify and characterize HIDDEN TREATURE 2 (HID2), an Arabidopsis C/D box snoRNA that we show to be enriched in proliferating tissues known to be active in rRNA processing. Knocking down HID2 results in an increase of both 45S rRNA and pre-rRNA processing intermediates, overaccumulation of 27SB rRNA (5.8S-ITS2-25S) aberrant products, which most likely impede ribosomal assembly, and the occurrence of pleiotropic developmental defects. Thus, by elucidating the regulation of pre-rRNA processing pathways by snoRNAs, our study of HID2 provides insights into ribosome synthesis.
Results
Reduction in Arabidopsis HID2 Expression Impairs Normal Growth and Development.
To identify snoRNAs that serve as prominent players in Arabidopsis growth and development, we screened a collection of Agrobacterium transferred DNA (T-DNA) insertion mutants (27). The hid2 mutant identified in this screen defectively expressed five snoRNAs organized in a polycistronic gene cluster and exhibited retarded and pointed leaf-growth phenotypes (Fig. S1).
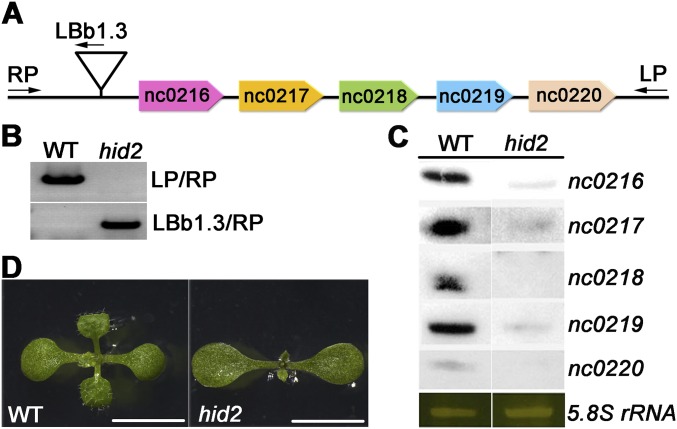
Fig. S1.
Molecular and phenotypic analysis of a T-DNA insertion mutant, hid2. (A) Schematic diagram showing the T-DNA insertion site in hid2. The primers used for genotyping are indicated by black arrows. (B) PCR-based genotyping of hid2 homozygous mutant and WT plants. (C) Northern blot analysis showing the expression of nc0216–nc0220 in WT and hid2 seedlings. 5.8S rRNA was used as the loading control. (D) Phenotypes of aerial parts of WT and hid2 seedlings. (Scale bars: 5 mm.)
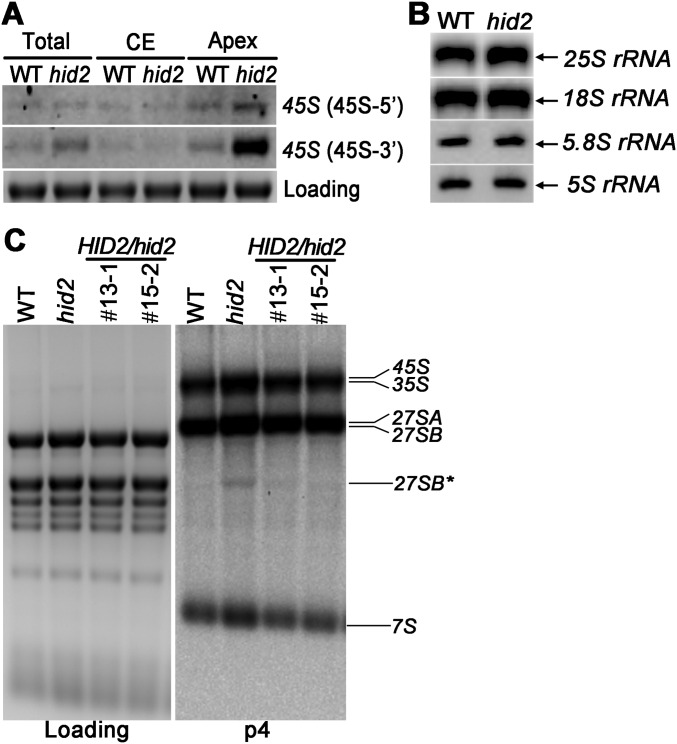
To determine whether the leaf phenotypes observed in the hid2 mutant were a direct result of the decrease in snoRNAs, we transformed a genomic DNA fragment encoding all five snoRNAs (nc0216–nc0220) under the control of their endogenous promoter into an hid2 mutant background (Fig. 1A). The resulting transgenic plants (pHID2: ALL/hid2) expressed all five snoRNAs at levels similar to or higher than their levels in WT plants and showed a complete rescue of the hid2 phenotype (Fig. 1B), indicating that the absence of these specific snoRNAs was responsible for the hid2 phenotypes observed at the seedling stage. Next, to determine which of these snoRNAs was the predominant player, we expressed the first snoRNA in the cluster, nc0216, under regulation of its endogenous promoter (pHID2:HID2) in hid2 mutants (Fig. 1A). Interestingly, the expression of nc0216 alone in hid2 mutants resulted in a dosage-dependent recovery of the mutant phenotype in our two independent transgenic lines (Fig. 1B). Furthermore, we found that the pleiotropic phenotypes observed in the hid2 mutant, including delayed seed germination, retarded root growth, and narrow, pointed leaves at the adult stage, were all complemented to WT in the pHID2:HID2/hid2 transgenes (Fig. S2). In contrast, transgenic lines expressing the other four snoRNAs (pHID2:ΔHID2/hid2) at WT or higher levels were observed to exhibit the hid2 mutant phenotype (Fig. 1B). These results suggest that nc0216 is physiologically important in hid2 mutants. We also analyzed the expression of genes located at either end of the nc0216-coding sequence in WT, hid2, and pHID2:HID2/hid2 plants using qRT-PCR (Fig. S3). Our results revealed that the expression levels of the neighboring genes were either comparable in all three genotypes or were not correlated with the levels of nc0216 expression. Together, our data indicate that nc0216 is a major driver of hid2, and we therefore refer to nc0216 as “HID2” hereafter. hid2 is a representative snoRNA mutant exhibiting developmental defects in plants. HID2 was previously annotated as a 90-nt C/D box snoRNA (27).
Fig. 1.
Down-regulation of HID2 results in developmental defects. (A) Schematic illustration of constructs containing all the members of the snoRNA gene cluster (pHID2:ALL), nc0216 only (pHID2:HID2), and nc0217–nc0220 (pHID2:ΔHID2) driven by the endogenous HID2 promoter. (B, Upper) Phenotypes of WT, hid2, and the indicated transgenic seedlings. (Scale bar: 5 mm.) (Lower) Northern blot analysis showing the expression levels of nc0216–nc0220, with 5.8S rRNA as the loading control.
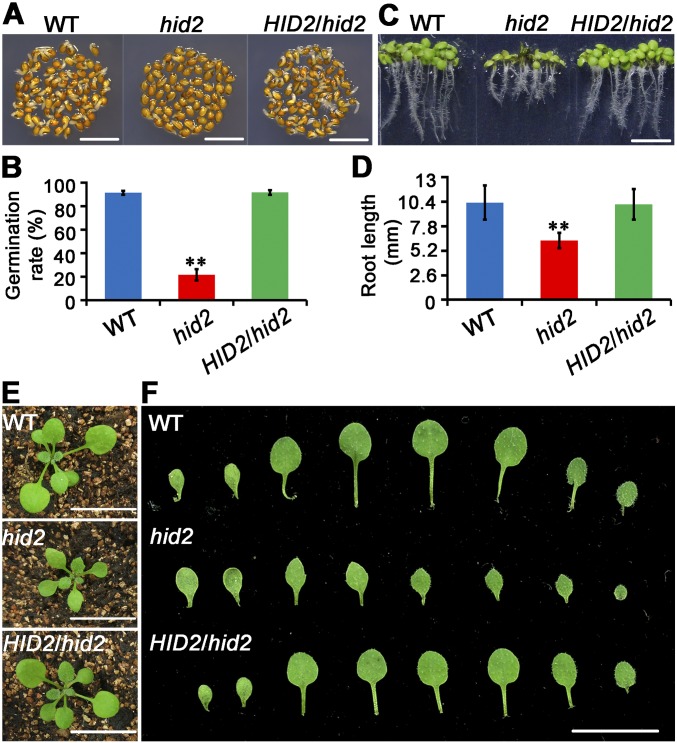
Fig. S2.
Pleiotropic developmental defects of hid2 mutants. (A and B) Delayed germination of hid2. The seed germination rates were analyzed in WT, hid2, and pHID2:HID2/hid2 seeds grown on Murashige and Skoog medium for 48 h after stratification. Data are shown as mean ± SD. Three independent replicates were performed; n = 105 for each replicate. P < 0.01 (t test). (Scale bars: 2 mm.) (C and D) Retarded root growth of hid2. Data are mean ± SD. Three biological replicates were performed; n = 25 for each replicate. P < 0.01 (t test). (Scale bar: 5 mm.) (E) Phenotypes of 15-d-old WT, hid2, and pHID2:HID2/hid2 plants grown on soil. (F) Narrow and pointed leaves of hid2 plants. (Scale bars: 10 mm.)
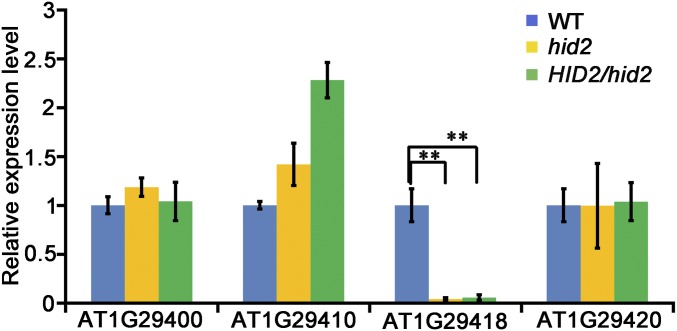
Fig. S3.
The expression levels of neighboring genes are not affected by HID2. qRT-PCR analysis showing the expression of HID2 neighboring genes in WT, hid2, and pHID2:HID2/hid2 seedlings. UBQ10 was used as the internal control. P < 0.01 (t test).
The Expression of HID2, Driven by a Site II Cis Element, Is Enriched in Actively Proliferating Tissues.
To gain initial insight into the developmental control of HID2, we first characterized the promoter activity of HID2 by introducing a pHID2:GUS reporter gene into WT plants. In shoots, β-glucuronidase (GUS) activity was significantly enriched in the shoot apex but not in the cotyledon. This result was also confirmed by Northern blot analysis using a HID2-specific antisense probe (Fig. 2 A and B). We further examined the expression pattern of HID2 at the shoot apical meristem using in situ hybridization. We detected high HID2 expression in the leaf primordia and growing young leaves and weak expression in the vascular tissues (Fig. 2 C and D). In addition, our FISH analysis validated the localization of HID2 in the nucleolus (Fig. 2E). Thus, our results indicate that nucleolus-localized HID2 is preferentially expressed in the rapidly proliferating tissues of Arabidopsis.
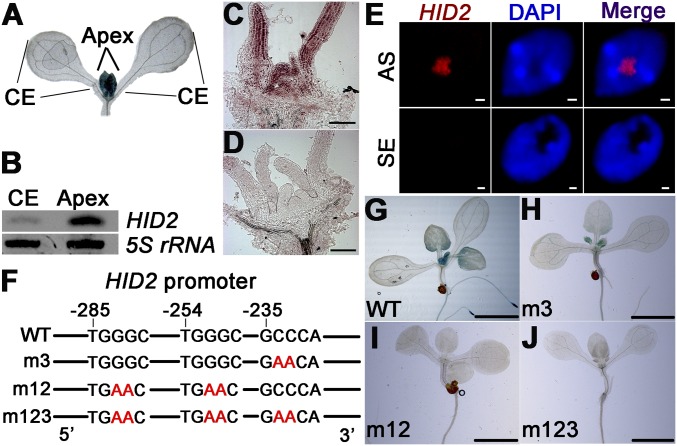
Fig. 2.
Nucleolus-localized HID2 is highly expressed in cell proliferating regions. (A) Histochemical staining showing the expression pattern of pHID2:GUS in 8-d-old WT seedlings. The apex and cotyledon (CE) are indicated. (B) Expression of HID2 in the cotyledon and apex, with 5S rRNA as the loading control. (C and D) In situ hybridizations of the apex in WT seedlings with the HID2 antisense (C) and sense (D) probes. (Scale bars: 100 μm.) (E) Nucleolar localization of HID2. FISH using the HID2 antisense (AS) and sense (SE) probes compared with nuclear staining with DAPI. (Scale bars: 1 μm.) (F) Schematic illustration showing the positions of site II and related motifs in the HID2 promoter. (G–J) GUS staining of pHID2WT:GUS (G) and pHID2mut:HID2 (H–J) transgenic seedlings. (Scale bars: 2 mm.)
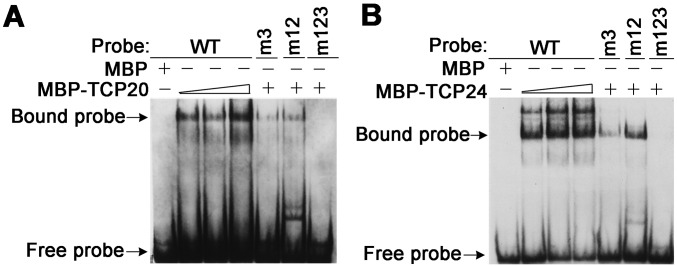
To understand better the spatial control of HID2’s expression pattern, we analyzed the HID2 promoter sequence using MEME software that enabled us to identify the specific motifs driving HID2 expression in vivo. We found two site II (TGGGC) motifs and one site II-related (GCCCA) motif in the HID2 promoters (Fig. 2F). Site II motifs have been shown to be present in promoters of some, but not all, snoRNA genes. They also have been shown to be present in ribosomal genes (28–30). To examine the relation between this cis element and the expression of HID2 in planta, we constructed pHID2WT:GUS and pHID2mut:GUS and compared their activity. We observed that mutation of either the site II (m12) or site II-related (m3) motifs reduced the GUS reporter activity (Fig. 2 H and I), and mutation of all three motifs (m123) abolished GUS activity (Fig. 2J). Taken together, these results indicate that site II cis elements are essential drivers of HID2 expression in vivo. Given that the site II element can be recognized by TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (TCP), a family of key transcriptional factors known to regulate cell proliferation in planta (31, 32), we next tested the binding of selected TCPs to the WT and mutated HID2 promoter by gel-shift analysis. Both TCP20 and TCP24 were found to bind the HID2 WT promoter, but not the mutant HID2 promoter m123, directly in vitro (Fig. S4). This finding suggests that TCP proteins may be important determinants of HID2 transcription.
Fig. S4.
Site II and related motifs are required for TCP’s binding on HID2 promoter in vitro. Shown are EMSAs of recombinant TCP proteins and HID2 WT and mutated promoter regions. (A) MCP-TCP20. (B) MCP-TCP24.
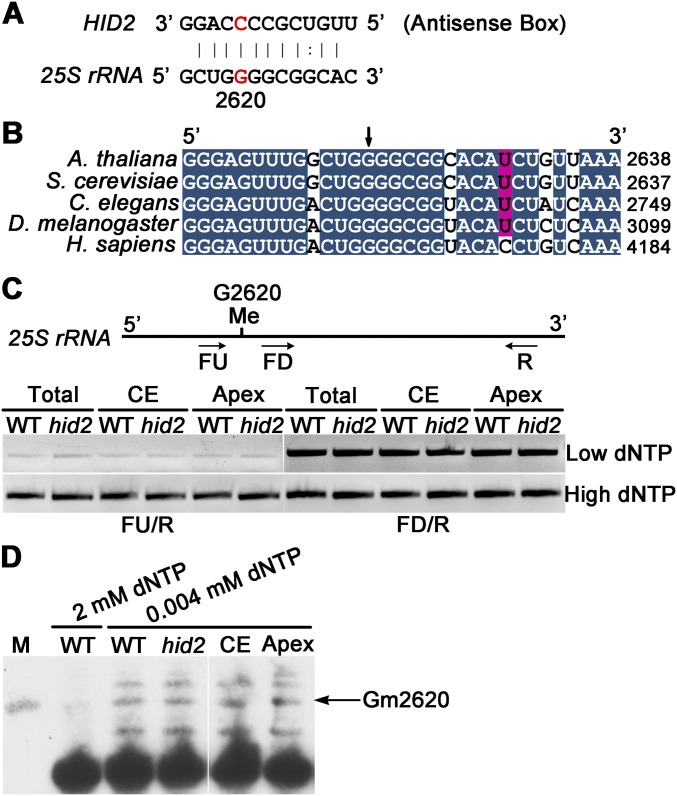
2′-O-Methylation on the 25S rRNA G2620 Apparently Is Not Affected in hid2.
To gain insight into HID2 function, we first searched for canonical targets of HID2 in Arabidopsis. We found that the HID2 antisense-box element uniquely matched a conserved fragment in 25S rRNA. Within this fragment, the G2620 site is predicted to undergo 2′-O-methylation (Fig. S5 A and B). To test the connection between the 2′-O-methylation of 25S rRNA G2620 and HID2, we performed reverse transcription at low deoxy-ribonucleotide triphosphate concentrations (RTL-P) followed by PCR in WT and hid2 mutants (33). Our data support the presence of a 2′-O-methylated site within the tested region. However, no obvious difference in band intensity was observed in either the cotyledon or apex of the WT and hid2 samples (Fig. S5C), and this result was further supported using primer extension analysis (Fig. S5D).
Fig. S5.
The levels of 2′-O-ribose methylation are not affected in hid2. (A) Base-pairing between the HID2 antisense box and a region in 25S rRNA. (B) Sequence alignment of a 25S/26S/28S rRNA region that is complementary to the antisense box of HID2 in five model organisms. The positions of each base at the 3′ end relative to the 5′ end of mature 25S/26S/28S rRNAs are indicated. (C, Upper) Schematic illustration showing the putative 2′-O-methylation site on 25S rRNA G2620. (Lower) RTL-P using total seedlings (Total), cotyledons (CE), and apices (Apex) of WT and hid2 plants. Primers are indicated by black arrows. (D) Primer extension analysis of 25S rRNA G2620 methylation in WT and hid2 mutant seedlings. M, molecular marker with 31 nt.
A Decrease in HID2 Expression Causes an Increase in the Transcription Rate of rDNA in Arabidopsis.
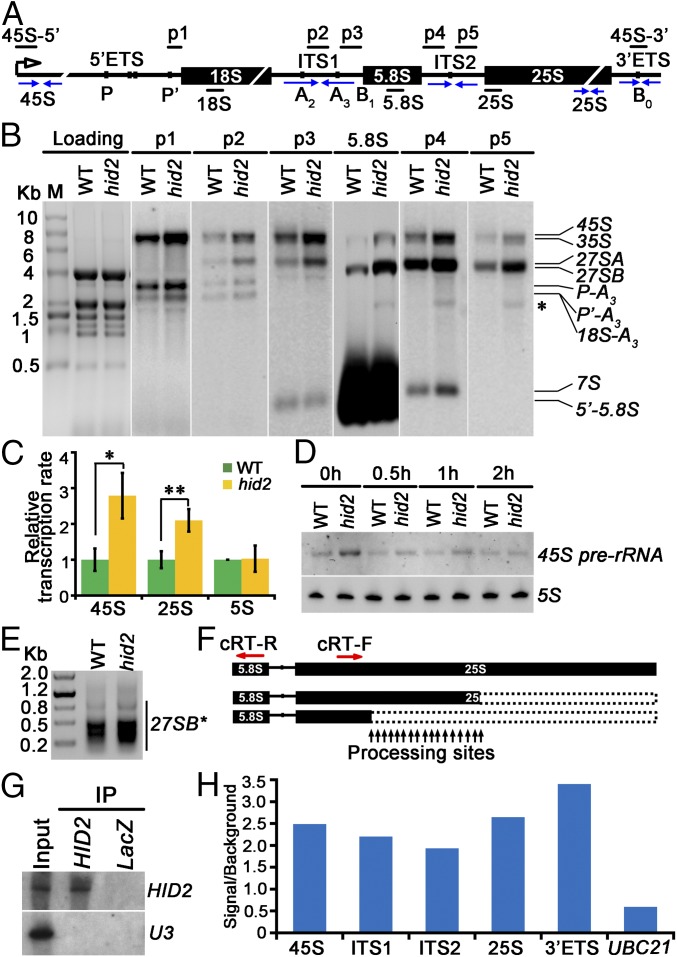
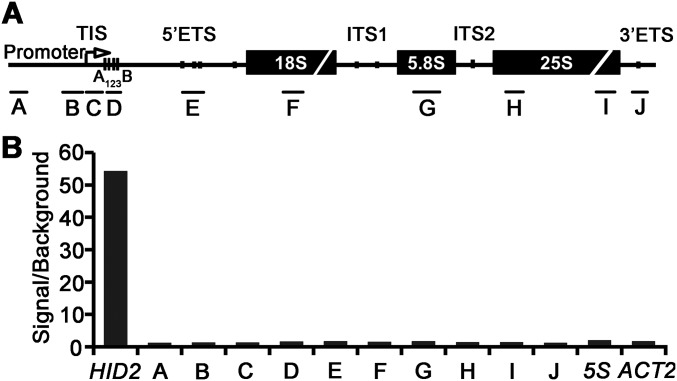
To detect the steady-state levels of pre-rRNA precursors and intermediates in each processing step, Northern blot analyses were performed, showing an accumulation of 45S and 35S pre-rRNA precursors in hid2 mutants (Fig. 3 A and B and Fig. S6A). To examine whether rDNA transcription was increased in hid2 mutants, we conducted a nuclear run-on (NRO) transcription analysis using WT and hid2 seedling samples. Our run-on results revealed an increased 45S rDNA transcription rate in hid2 (Fig. 3C). Selective inhibition of DNA-dependent RNA polymerase I activity by actinomycin D treatment suggested that the degradation rate of 45S rRNA in hid2 is not reduced compared with WT (Fig. 3D). Next, to determine whether HID2 directly affects the transcription of rDNA, we used chromatin isolation by RNA purification (ChIRP) to purify HID2 RNA and subsequently investigated whether it interacts with 45S rDNA. Following retrieval of HID2-interacting chromatin using anti-sense DNA tilling probes, we could not detect the 45S rDNA locus but clearly detected the HID2 locus, indicating that HID2 may not interact with 45S rDNA (Fig. S7). Thus, HID2 may modulate rDNA transcription indirectly.
Fig. 3.
HID2 interacts with 45S rRNA and promotes efficient pre-rRNA processing. (A) Schematic illustration of the rDNA locus encoding 45S pre-rRNA. (B) Northern blots showing the accumulated pre-rRNAs and aberrant processing products (asterisk) in WT and hid2 seedlings. The SYBR Green II-stained gel image is shown as a loading control. Probes (black bars) are indicated in A. (C) NRO assay showing increased rDNA transcription in hid2 seedlings compared with WT. *P < 0.05; **P < 0.01 (t test). Primers are indicated by blue arrows in A. (D) Northern blots showing the transcript levels of 45S pre-rRNA in WT and hid2 seedlings treated with Actinomycin D (50 ng/mL) for the indicated time. 5S rRNA was used as a control. (E) Identification of 27SB* by circular RT-PCR. (F) Schematic illustration of 27SB*. The primers for RT-PCR are indicated by red arrows. (G) Northern blots showing that ChIRP enriches HID2. (H) qRT-PCR analysis of HID2-ChIRP RNA shows the retrieval of 45S rRNA. Amplicons are indicated by facing arrows in A. Data represent one of three biological replicates.
Fig. S6.
HID2 is required for proper pre-rRNA processing. (A) Northern blots showing the levels of 45S pre-rRNA in WT and hid2 total seedlings (Total), cotyledons (CE), and apices (Apex) detected by the probes 45S-5′ and 45S-3′. (B) Northern blots showing the mature rRNA levels in WT and hid2 seedlings. Probes are indicated in Fig. 3A by black bars. (C) Northern blots showing the levels of pre-rRNAs in WT, hid2, and pHID2:HID2/hid2. Pre-rRNAs were detected by the probe p4 shown in Fig. 3A, with SYBR Green II-stained gel as a loading control.
Fig. S7.
HID2 is not likely to associate with rDNA. (A) Schematic diagram of the rDNA locus showing primer pairs (black bars) used for HID2-ChIRP qRT-PCR. (B) qRT-PCR analysis of HID2-ChIRP. ACT2 was used as a negative control.
HID2 Is Required for Efficient Processing of Pre-rRNA.
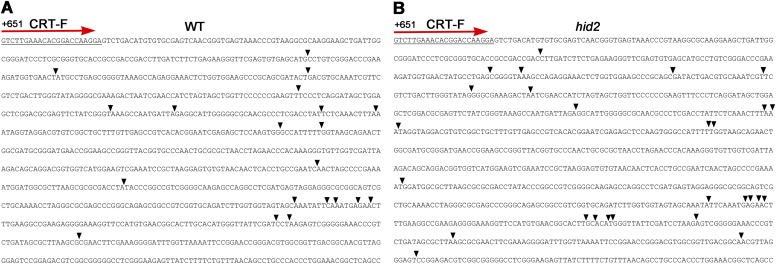
Using Northern blot analysis, we determined that P-A3 and 27S intermediate pre-rRNAs were accumulated in hid2 mutants, whereas mature rRNAs were expressed at their normal levels (Fig. 3B and Fig. S6B). This finding suggests that pre-rRNA processing is delayed in hid2 mutants. Notably, the appearance of a significantly sized band around 1.8 Kb when both 5.8S and ITS2 probes were used (Fig. 3B, asterisk) drove us to determine the precise 5′ and 3′ ends of the undetermined transcript via circular RT-PCR (Fig. 3B). Most intriguingly, our sequencing results demonstrated that the overproduced band in hid2 mutants was a series of 3′- truncated 27SB intermediates (27SB* in short) that had been cleaved within the 25S region (Fig. 3 E and F and Fig. S8). By analyzing 50 clones each for the WT and hid2 mutant circular RT-PCR products, we found the frequency of cleavage inside the 25S region of 27SB to be increased in the hid2 mutant (Fig. S8). It is worth noting that the observed increase in 45S and 35S rRNA and 27SB* accumulation is recovered in the pHID2:HID2/hid2 transgenes (Fig. S6C), suggesting that HID2 plays an important role in both the efficiency and accuracy of rRNA processing.
Fig. S8.
Sequencing results of 27SB aberrant products by circular RT-PCR. The sequence of 25S rRNA is shown from position +651 to +1729 with respect to the 5′ end of mature 25S rRNA. The forward primer for PCR amplification is indicated. Arrowheads indicate the positions of cutting sites. (A) WT. (B) hid2.
To understand the molecular basis of HID2 function further, we isolated HID2-interacting RNA by ChIRP. Our results demonstrated that HID2 was efficiently purified (Fig. 3G) and that amplicons on 45S pre-rRNA were equally enriched (Fig. 3H). This finding, in turn, suggests that HID2 is able to bind 45S pre-rRNA.
hid2 Mutants Confer an Imbalanced Ribosome Profile and Altered Responses to Specific Antibiotics.
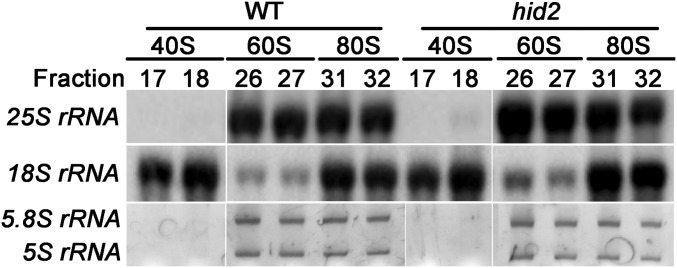
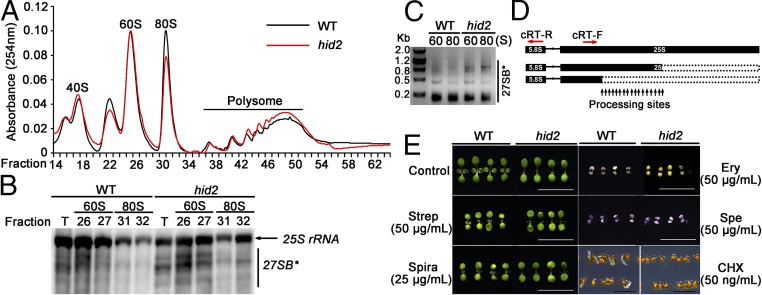
To evaluate further the potentially impaired ribosome assembly state caused by defects of pre-rRNA processing in the hid2 mutants, we performed a polysome profiling assay. Lysates from WT and hid2 seedlings were fractionated through sucrose gradients to separate the ribosomal subunits and the 80S monosome (Fig. S9). It appeared that the hid2 mutation caused a reduction in the 80S/40S ratio and an increased ratio of polysomes (Fig. 4A). Further insight regarding the association between each ribosomal particle and rRNAs was gained by using Northern blot analyses using a 25S rRNA-specific probe. The results showed an accumulation of prominent bands smaller than the mature 25S rRNA that cofractionated with 60S and 80S of hid2 mutants (Fig. 4B). Through circular RT-PCR and sequencing, we found these bands to be 27SB* (Fig. 4 C and D), raising the possibility that the overaccumulation of 27SB* in hid2 mutants may have an impact on the lower 80S/40S ratio.
Fig. S9.
rRNA detection in the ribosome fractions. RNA was extracted from equal aliquots of each indicated fraction. 25S rRNA and 18S rRNA were detected by Northern blots; 5.8S rRNA and 5S rRNA were detected by SYBR Gold staining.
Fig. 4.
hid2 mutants confer an imbalanced ribosome profile and altered responses to specific antibiotics. (A) Quantitative sucrose density gradient analysis showing the imbalanced ribosome profile in hid2 mutants. (B) Northern blots showing prominent aberrant products detected by 25S antisense probe that were cosedimented with 60S and 80S ribosomal fractions in hid2. T, total ribosomal extracts. Fraction numbers are indicated above each lane. (C) Identification of 27SB* by circular RT-PCR. The primers are indicated by red arrows in D. (D) Schematic illustration of 27SB*. (E) Phenotypes of WT and hid2 seedlings treated with antibiotics and cycloheximide. CHX, cycloheximide; Ery, erythromycin; Spe, spectinomycin; Spira, spiramycin; Strep, streptomycin. (Scale bars: 10 mm.)
To test further whether the ribosome is structurally impaired in hid2 mutants, we analyzed the mutants’ response to an array of antibiotics known to target ribosomal locations. Although the hid2 mutants showed a response to spectinomycin similar to that observed in the WT, they showed mild resistance to erythromycin. The mutants also were found to be resistant to streptomycin and spiramycin, exhibiting green cotyledons (Fig. 4E). Streptomycin belongs to aminoglycoside class of antibiotics, which inhibit aminoacyl tRNA binding to position A during elongation. In contrast, spiramycin and erythromycin are macrolide antibiotics and inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes (34). The specific resistance of hid2 mutants to these antibiotics suggests that an altered ribosomal structure or an aberrant population of ribosomes in the mutants prohibits the proper binding of these antibiotics by the ribosomes. To assess further whether HID2 could affect de novo protein synthesis, we tested the effect of the protein synthesis inhibitor cycloheximide on the hid2 mutants (35). As expected, the hid2 mutant was more sensitive to cycloheximide than the WT, suggesting that the mutant has reduced translational activity (Fig. 4E). Taken together, our data reveal that HID2-regulated rRNA biogenesis is essential for the proper assembly and activity of the ribosome.
The Contribution of the HID2 Signature Motif to Its Function in Vivo.
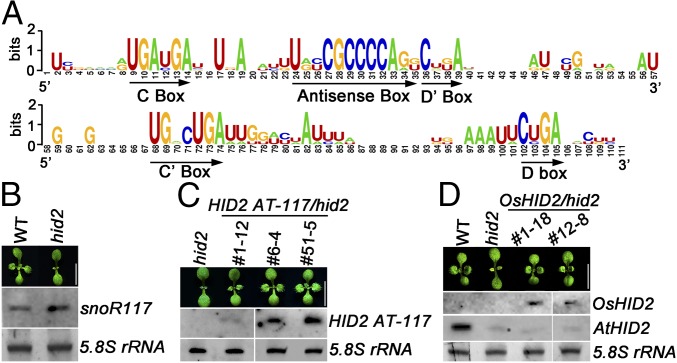
HID2 has three clearly identifiable signature motifs, the C/D box, the C′/D′ box, and an antisense box that is complementary to 25S rRNA. Our sequence comparison analysis revealed the conservation of those signature motifs in yeast, in model metazoans ranging from worm to human, and in monocot rice (Fig. 5A). To test the functional importance of the HID2 antisense box, we designed a chimeric construct that included a HID2 antisense-box sequence and a backbone sequence of snoR117, which is annotated to another classic C/D box snoRNA with an antisense-box sequence upstream of its D′ box (36). The expression of snoR117 was found to be comparable in the WT and hid2 mutant (Fig. 5B). We then transformed the chimeric construct into the hid2 mutant background under the regulation of the HID2 endogenous promoter (HID2 AT-117). Intriguingly, the expressed chimeric snoR117 successfully rescued the hid2 phenotype to WT (Fig. 5C). Moreover, we found the extent of phenotypic recovery to be correlated with the expression level of HID2 AT-117, confirming that the antisense box of HID2 is critical for its function in vivo.
Fig. 5.
The conserved antisense box of HID2 is essential for its function in plants. (A) The consensus signature motifs of Arabidopsis HID2 and its orthologs in Oryza sativa, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens are presented using the WebLogo software. (B–D, Upper) Phenotypes of WT, hid2, and the indicated transgenic seedlings (Scale bars: 5 mm). (Lower) Northern blots showing the expression levels of snoR117 (B), HID2 AT-117 (C), and AtHID2 and OsHID2 (D) in the indicated seedlings, with 5.8S rRNA as a loading control.
To evaluate the functional conservation of HID2 further, we transformed hid2 mutants with OsHID2:OsHID2, an HID2 ortholog in rice. Interestingly, the expression of OsHID2 completely rescued the phenotype of hid2 mutants without disturbing the expression of AtHID2 (Fig. 5D), suggesting that the function of HID2 is conserved in monocots and dicots. Moreover, our findings may imply a general role of HID2 throughout the eukaryotic kingdom.
Discussion
In this study, we show that HID2 encodes a C/D box snoRNA that is highly expressed in actively proliferative tissues and is essential for the normal growth and development of Arabidopsis. HID2 associates with 45S rRNA and acts as an important regulator of ribosome biogenesis, predominantly by promoting efficient pre-rRNA processing.
In plants, a large number of snoRNAs have been identified and annotated (26, 27, 36). However, to the best of our knowledge, no characteristic developmental defects have been previously connected to the function of a single snoRNA in plants, at least in part because of the higher diversity of the snoRNA genes in plants (12). The sequence redundancy of snoRNA genes also may make it difficult to obtain a single snoRNA mutant displaying apparent physiological defects (12, 26). Thus, the identification of our hid2 mutant represents a valuable opportunity to dissect and further our understanding of the role of snoRNA in regulating ribosome biogenesis.
A few snoRNAs have been found to regulate pre-rRNA processing in yeast and animals, but little is known about the function of snoRNAs in planta, with the exception of recent studies of the U3 snoRNP (18–21). As one of the core components in the small subunit processome, U3 snoRNP has been shown to have a conserved function in 18S rRNA biogenesis (8, 17, 23). Depletion of U3 snoRNA in yeast has been shown to lead to under-accumulation of mature 18S rRNA (37). Because the expression levels of mature 18S rRNA were unchanged in hid2 mutants (Fig. 3 and Fig. S6), we assume that HID2 has a function different from that of U3 snoRNA in pre-rRNA processing. Given that the antisense-box sequence of HID2 is functionally essential (Fig. S5 and Fig. 5) and that HID2 is associated with 45S rRNA in vivo (Fig. 3), we propose that HID2–rRNA base-pairing is required for HID2 to regulate pre-rRNA processing. In addition, given the disruptions in pre-rRNA processing documented in ribosome biogenesis factor mutants with distinct aberrant pre-rRNAs (3, 7, 9), it is conceivable that HID2 may serve as a safeguard responsible for promoting the processing and minimizing the production of aberrant rRNA species in the error-prone processes that require a large set of transacting factors. A complete model of HID2’s function has yet to be established in vivo, and, consequently, a number of important questions remain unanswered. For instance, it is still unclear whether the base-pairing between HID2 and 25S rRNA affects rRNA folding, facilitates the accessibility of HID2–RNP to the target site, or has a functional relevance to the binding and release of HID2–RNP.
Although the general scheme of pre-rRNA processing is conserved in eukaryotes, differences do occur in different species in the factors required at specific developmental stages to ensure the successful processing of pre-rRNA (1, 3). We suggest that HID2 may represent one of these factors that have plant-specific features. Our study showed that the site II motif in the promoter of HID2 is essential for its expression in actively proliferative tissues (Fig. 2), which rely on rapid ribosome biogenesis. However, we did not find similar cis elements in metazoans and yeast. Thus, regulation of the expression of HID2 orthologs in animals and yeast requires further investigation. In addition, to our knowledge, there has been no report on the functional analysis of HID2 orthologs in model animals, and whether these orthologs have a common or a divergent role in controlling ribosome biogenesis remains of great interest for future explorations. Furthermore, changes in snoRNA expression previously have been shown to affect the physiological conditions of cells and to correlate with increases in dysfunctional ribosome activity that, in turn, lead to various human diseases, including various forms of cancer (38–40). This insight into the control of snoRNA expression no doubt will contribute both to our understanding of how growth and development in eukaryotes is controlled and to the development of innovative diagnostic approaches and therapeutic agents for human disease.
Materials and Methods
The plant materials, growth conditions, plasmid construction, and transgenic plants generation procedures are described in SI Materials and Methods. The detailed procedures of Northern blot analysis, GUS staining, circular RT-PCR, in situ hybridization, FISH, EMSA, primer extension analysis, ChIRP, and the NRO assay are provided in SI Materials and Methods. The primers and probes used in this study are listed in Table S1. The genes mentioned in this article have been given the following accession numbers by the Arabidopsis Information Resource (TAIR) database: TCP20, AT3G27010; TCP24, T1G30210; UBQ10, AT4G05320; UBC21, AT5G25760; ACT2, AT3G18780.
Table S1.
Primers and probes used in this study
| Genotyping primers | |
| hid2-LP | AAAACCCAAACCAAACCAAAG |
| hid2-RP | GATAGCAGCGGAATTCACAAC |
| LBb1.3 | ATTTTGCCGATTTCGGAAC |
| Northern blot probes | |
| nc0216 (HID2) | ACAACCATCGTAATGGATGCAGATTAAGCCTGGGGCGACAATTTCATATCA |
| nc0217 | AGGATCAGAAAATATGGCAAATCAATCACGAAGATCAGACTGGGTGTCACAAAACATTA |
| nc0218 | TCAGATGGTAGTGCATGTCATTGTATCAACACTGTATTTCAGATTTGCAGACGTATGGA |
| nc0219 | AGCAATCTCAGCCGAAAGATGGTGATCTAGTGATTATAATAATCATTGTATAATCATTG |
| nc0220 | GTGTCGCAAAGCCTGAAATCACTTTCTGCCTAAGAAAAACGAAATGTCCAAACACAATT |
| 5S rRNA | TCGCCCAAGAACGCTTAACTGCGGAGTTCTGATGGGATCCGGTGCATTAGTGCTG |
| 5.8S rRNA | TCGATGGTTCACGGGATTCTGCAATTCACACCAAGTATCGCATTTCGCTACG |
| 18S rRNA | GACCAGGAGCGTATCGCCGACCGAAGGGACAAGCCGACCAATGCACACCA |
| 25S rRNA | CCTCCGCTTATTGATATGCTTAAACTCAGCGGGTAATCCCGCCTGACCTGGG |
| p1 | TCGATCACGGCAATTCCCCGCCACATCCTCTCAAACGCAATGGAAAGAGA |
| p2 | GATCCGGCGGGCAAGGAATCGGCTAAGAAACCGGCCCACCGAGAGTGGTG |
| p3 | AGACTTCAGTTCGCAGCACAGCATCCGCCCACACTCCGTCTCCGGGGAGG |
| p4 | TCCAGGCGTCCTTGGCTCGGATTTAGGCCAACCGCGTGCGGTAACACACG |
| p5 | GGACTTTGGGTCATCTACTGCTTCCGGACAAGAGCGACCGATAAAATGTAATGGATC |
| 45S-5′ | CGGTCGGTCATTCCTCGTGTCGATATCCGATACCATCCCTCGATCGCTACCCAAGTC |
| 45S-3′ | GTGTAGAAACACTTGTGTAGAATTGGGGATTGTTTTTTTTGGAGTGATTTAGGGGAGG |
| U3 | AGAAATCAAGGAAACAGAGGTACGAGCCTATAGAACAGATCCTGTTCAAGTAAGGTCGT |
| snoR117 | CAGAAAGTGGAGGTAATCGTCAAGGAGAAACTCAGATGGTACGGACAATCTCAAACA |
| HID2-AT-117 | CAGAAAGTGGAGGTAATCGTCAAGGAGAAACTCAGCCTGGGGCGACATCTCAAACA |
| OsHID2 | GAGTCAGATTTACATGTCAATCACCACACATCCGCGATTACAGCGGATCATAAGACT |
| Cloning primers | |
| pHID2:ALL-F | ACTGGATCCATCAGCTGCTCTGAGTATCGTGT |
| pHID2:ALL-R | ACTGTCGACCTACACCAAGAGAACAAGCCAAC |
| pHID2:HID2-F | ACTGGATCCATCAGCTGCTCTGAGTATCGTGT |
| pHID2:HID2-R | ACTGTCGACATATGGCAAATCAATCACGAAG |
| pHID2:ΔHID2-up-F | AGGGGTACCGATTGATAGCAGCGGAATTCAC |
| pHID2:ΔHID2-up-R | CGATTTGAGACTCAAAAACATTAAACGAATTACGAA |
| pHID2:ΔHID2-down-F | TTGAGTCTCAAATCGTTTATTATGATTCTG |
| pHID2:ΔHID2-down-R | CAACTGCAGCAAGCCAACAAATCCTGCACAAG |
| HID2-promoter-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTAAATCAGCTGCTCTGAGTATCGTG |
| HID2-promoter-R | GGGGACCACTTTGTACAAGAAAGCTGGGTACTCAAAAACATTAAACGAATTACG |
| HID2-promoter-m1-F | TCGAATTATGAACCTTTTGGATTTTA |
| HID2-promoter-m1-R | CCAAAAGGTTCATAATTCGATCGT |
| HID2-promoter-m2-F | GGATATAAATTGAACCTATAATAAACTAGG |
| HID2-promoter-m2-R | TAGTTTATTATAGGTTCAATTTATATCCAA |
| HID2-promoter-m3-F | TAAACTAGGAACATATATAAAGCGGTGG |
| HID2-promoter-m3-R | GCTTTATATATGTTCCTAGTTTATTATAGGT |
| TCP20-F | AATTCTAGAATGGATCCCAAGAACCTAAATCG |
| TCP20-R | AATAAGCTTTTAACGACCTGAGCCTTGAGAATC |
| TCP24-F | AATGGATCCATGGAGGTTGACGAAGACATTGA |
| TCP24-R | AATCTGCAGCTATCTCCTTTCCTTTGCCTTGTC |
| snoR117-F | AGGGGTACCAAGCAGGATGTCTCGATTTCTC |
| snoR117-R | CAACTGCAGGAAGTTTACATCAACGAGCGAA |
| HID2-AT-117-backbone-F | GTTTTTGAGAGCCTGTGATGTTTGAGATG |
| HID2-AT-117-backbone-R | GATTTGAGAAAGCCTCAGAAAGTGGAGGTA |
| HID2-AT-117-tail-F | CTGAGGCTTTCTCAAATCGTTTATTATGA |
| HID2-AT-117-tail-R | TCACAGGCTCTCAAAAACATTAAACGAAT |
| OsHID2-F | AGGGGTACCAACTCCGTACTAGCTAAAAGGCT |
| OsHID2-R | CAACTGCAGGAGTACAGTCGATGGAATCATGG |
| Primers for T7 in vitro transcription | |
| HID2-Sense-T7-F | TAATACGACTCACTATAGGGAGAGAAAGTGATGATATGAAATTGTCGC |
| HID2-Sense-T7-R | GAATCAGACAAAAATAGTCAATCACCA |
| HID2-Antisense-T7-F | GAAAGTGATGATATGAAATTGTCGC |
| HID2-Antisense-T7-R | TAATACGACTCACTATAGGGAGAGAATCAGACAAAAATAGTCAATCACCA |
| Primers for EMSA probes | |
| HID2 promoter-EMSA-F | GAGACGATCGAATTATG |
| HID2 promoter-EMSA-R | CCACCGCTTTATATATG |
| RTL-P primers | |
| 25S rRNA-G2620-FU | ACATTGTCAGGTGGGGAGTTTG |
| 25S rRNA-G2620-FD | AAGATAACGCAGGTGTCCTAAGATG |
| 25S rRNA-G2620-R | TCTGACACCTCTAGCTTCAAATTCC |
| Primers for primer extension assay | |
| Gm2620-R | 5′-DIG-CCTGCGTTATCTTTTAACAGATG |
| 31 nt-marker | GGGCGGCACATCTGTTAAAAGATAACGCAGG |
| qRT-PCR primers | |
| AT1G29400-RT-F | TGCTCATGGCGAGATCAAAGA |
| AT1G29400-RT-R | TGCAATCTCACACCTGTTTAATGC |
| AT1G29410-RT-F | GAATTTCTCCAAAACCTCCAAATC |
| AT1G29410-RT-R | TCCCAATGAAATCAGCACCAG |
| AT1G29418-RT-F | CTTAGGCAGAAAGTGATTTCAGGC |
| AT1G29418-RT-R | CAACAAATCCTGCACAAGACCAC |
| AT1G29420-RT-F | ATCCGATAGAATCCGCTTTGC |
| AT1G29420-RT-R | GGATGCCAAACTCTTCTTCGG |
| UBQ10-RT-F | CTATGTTTCCGTTCCTGTTATCTC |
| UBQ10-RT-R | GCTCAACACTTTCGCTACATCTATT |
| Biotinylated probes for ChIRP | |
| CHIRP-HID2 antisense-1 | CTGGGGCGACAATTTCATAT |
| CHIRP-HID2 antisense-2 | CAGATTAAGCCTGGGGCGAC |
| CHIRP-HID2 antisense-3 | TACAACCATCGTAATGGATG |
| CHIRP-HID2 antisense-4 | GAATCAGACAAAAATAGTCA |
| CHIRP-Lac Z antisense-1 | ATTAAGTTGGGTAACGCCAG |
| CHIRP-Lac Z antisense-2 | AATTCAGACGGCAAACGACT |
| CHIRP-Lac Z antisense-3 | TTTCGACGTTCAGACGTAGT |
| CHIRP-Lac Z antisense-4 | AGACGATTCATTGGCACCAT |
| CHIRP-qPCR primers | |
| 45S pre-rRNA-45S-F | TATAGGGGGGTGGGTGTTGAGGGA |
| 45S pre-rRNA-45S-R | CGGTCGGTCATTCCTCGTGTCGATATC |
| 45S pre-rRNA-25S-F | TTACTTACTCCGTGAATCGGAGG |
| 45S pre-rRNA-25S-R | GCTTTTACCCTTTTGTTCCACAC |
| 45S pre-rRNA-ITS1-F | CCAAAGATCACCACTCTCGGTG |
| 45S pre-rRNA-ITS1-R | GTTCGTTTGCATGTTCCTTGACA |
| 45S pre-rRNA-ITS2-F | TCCCTCACCATCCTTTGCTG |
| 45S pre-rRNA-ITS2-R | CGGACAAGAGCGACCGATAA |
| 45S pre-rRNA-3′ETS-F | ACAGCTCCTTAACGAATTCCCAA |
| 45S pre-rRNA-3′ETS-R | CTCACTATGCACCTTTGCGAGC |
| UBC21-mRNA-F | CTGCGACTCAGGGAATCTTCTAA |
| UBC21-mRNA-R | TTGTGCCATTGAATTGAACCC |
| HID2-DNA-F | AGGTGTTGTTGGTGTCTATGCTTCT |
| HID2-DNA-R | GTCAATCACCATGTTACAACCATCG |
| rDNA-CHIRP-A-F | GAATGCTCGACCAGGACGATG |
| rDNA-CHIRP-A-R | CGACGATTCCAACTTCTGGTCGA |
| rDNA-CHIRP-B-F | CAAGTTTGCATCAAATATGCCCA |
| rDNA-CHIRP-B-R | GCTTAATAGCCCTTTTCCCTCTTG |
| rDNA-CHIRP-C-F | TATAGGGGGGTGGGTGTTGAGGGA |
| rDNA-CHIRP-C-R | CGGTCGGTCATTCCTCGTGTCGATATC |
| rDNA-CHIRP-D-F | TCGGATATCGACACGAGGAATGAC |
| rDNA-CHIRP-D-R | ACGCCTCGAAGAACTAATGGCA |
| rDNA-CHIRP-E-F | CTAGGCTGTCCCGAAGGTATCTC |
| rDNA-CHIRP-E-R | TAGGTGGTAGGTAGTTCGATGCG |
| rDNA-CHIRP-F-F | GAACAACTGCGAAAGCATTTGC |
| rDNA-CHIRP-F-R | AAGTTTCAGCCTTGCGACCATAC |
| rDNA-CHIRP-G-F | CTCTCGGCAACGGATATCTCG |
| rDNA-CHIRP-G-R | TTGTGACACCCAGGCAGACG |
| rDNA-CHIRP-H-F | CCGACCGACCTTGATCTTCTG |
| rDNA-CHIRP-H-R | GATGGTTCGATTAGTCTTTCGCC |
| rDNA-CHIRP-I-F | TTACTTACTCCGTGAATCGGAGG |
| rDNA-CHIRP-I-R | GCTTTTACCCTTTTGTTCCACAC |
| rDNA-CHIRP-J-F | ACAGCTCCTTAACGAATTCCCAA |
| rDNA-CHIRP-J-R | CTCACTATGCACCTTTGCGAGC |
| rDNA-CHIRP-5S-F | GGATGCGATCATACCAGCACTAA |
| rDNA-CHIRP-5S-R | AGGGATGCAACACGAGGACTT |
| ACT2-RT-F | GAGAGATTCAGATGCCCAGAAGTC |
| ACT2-RT-R | TGGATTCCAGCAGCTTCCA |
| NRO qPCR primers | |
| 45S pre-rRNA-45S-F | TATAGGGGGGTGGGTGTTGAGGGA |
| 45S pre-rRNA-45S-R | CGGTCGGTCATTCCTCGTGTCGATATC |
| 45S pre-rRNA-25S-F | TTACTTACTCCGTGAATCGGAGG |
| 45S pre-rRNA-25S-R | GCTTTTACCCTTTTGTTCCACAC |
| 5S-NRO-F | GGATGCGATCATACCAGCACTAA |
| 5S-NRO-R | AGGGATGCAACACGAGGACTT |
| Circular RT-PCR primers | |
| cRT-F | GTCTTGAAACACGGACCAAGGA |
| cRT-R | CGAGATATCCGTTGCCGAGAGT |
SI Materials and Methods
Plant Materials and Growth Conditions.
The ecotype of all WT Arabidopsis thaliana used in this study was Columbia-0 (Col-0). hid2 (SALK_138192) was ordered from the Arabidopsis Biological Resource Center. Surface sterilization and stratification of plant seeds and standard seedling growth experiments were performed as previously outlined (41). Ten-day-old light-grown seedlings were used for the experiments unless otherwise stated. The primers used for genotyping are listed in Table S1.
Plasmid Construction and Generation of Transgenic Plants.
To generate the pHID2: ALL and pHID2:HID2 constructs, a fragment containing 1,241 bp upstream of nc0216 followed by full-length nc0216–nc0220 coding sequences or full-length nc0216-only coding sequences were amplified from Col-0 genomic DNA and cloned into BamHI/SalI sites of pCAMBIA1300 (Cambia). For the pHID2:ΔHID2 construct, a fragment containing 541 bp upstream of nc0216 and full-length nc0217–nc0220 coding sequences was amplified from Col-0 genomic DNA. The chimeric fragment then was generated by overlapping PCR and cloned into KpnI/PstI sites of pCAMBIA1300.
To generate the pHID2WT:GUS and pHID2mut:GUS (m3, m12 and m123) reporter constructs, a fragment containing 1,241 bp upstream of the HID2 coding sequence or its mutated forms generated by PCR-based mutagenesis with attB sites was cloned into the Gateway pDONR221 vector (Invitrogen), as previously described (42). The inserts then were transferred into the Gateway binary vector pBGWFS7 by a recombination reaction between the attL and attR sites (LR reaction) (Invitrogen).
For the pHID2:HID2 AT-117 construct, the full-length snoR117 coding sequence was amplified from Col-0 genomic DNA. The antisense-box along with 12 nt upstream of the D′ box were replaced by the HID2 antisense-box sequence (TGTCGCCCCAGG) through overlapping PCR. The final chimeric fragment of pHID2:HID2 AT-117 driven by the HID2 endogenous promoter was cloned into the BamHI/SalI sites of pCAMBIA1300.
For the pOsHID2:OsHID2 construct, a fragment containing the full-length OsHID2 coding sequence and its 500-bp upstream and 246-bp downstream sequences was amplified from O. sativa spp. Japonica cv. Nipponbare genomic DNA and cloned into the KpnI/PstI sites of pCAMBIA1300.
The transgenic plants were generated as previously described (42). All primers are listed in Table S1.
Northern Blot Analysis.
For the detection of noncoding RNAs (ncRNAs) smaller than 300 nt, Northern blot analysis was performed as previously described (26). For the detection of pre-rRNA precursors, intermediates, and mature rRNAs, 1.8% (wt/vol) agarose/formaldehyde gels were used to separate total RNA. The subsequent procedure was performed essentially as previously reported (26, 42). The sequences used for probing ncRNAs in this study are listed in Table S1.
Histochemical GUS staining.
GUS staining was performed as previously described (43).
In Situ Hybridization.
We followed basic procedures for in situ hybridization as previously described (44). Digoxigenin (DIG)-labeled HID2 antisense and sense probes were transcribed using the AmpliScribe T7-Flash Transcription Kit (Illumina). Hybridization was performed at 50 °C overnight. Immunodetection was performed using an anti-DIG antibody coupled to alkaline phosphatase, according to the manufacturer’s instructions (Roche).
For FISH, 6-d-old seedlings were fixed in 4% paraformaldehyde, and the nuclei were prepared as previously described (45). Slides then were postfixed in 4% paraformaldehyde for 20 min, and hybridization was performed as previously described, with modifications (36). Immunofluorescence was performed using Alexa Fluor 594-conjugated streptavidin (1:200 dilution; Life Technologies), and samples were mounted in Vectashield mounting medium with DAPI (Vector H-1200). Nuclei were analyzed using a Zeiss LSM710 confocal laser-scanning microscope at 100× magnification.
EMSA.
To generate the MBP-TCP20 and MBP-TCP24 constructs, fragments containing the coding sequence of TCP20 or TCP24 were amplified from Col-0 cDNA and cloned into the XbaI/HindIII sites and BamHI/PstI sites of the pMal-C2X vector (New England Biolabs), respectively. MBP-TCP20 and MBP-TCP24 constructs were transformed into Escherichia coli BL21 (DE3)-CodonPlus cells. Expression of MBP fusion proteins was induced with isopropyl-b-d-thiogalactoside, and the proteins were purified using Amylose resin (New England Biolabs). EMSA was performed using the gel shift kit, according to the manufacturer’s instructions (Roche). The DIG-labeled probes were detected as previously described (26).
RTL-P Analysis.
RTL-P analysis was performed as previously reported with minor modifications (33). Briefly, the mixture containing 100 ng total RNA, 1 pmol 25S rRNA G2620-R primer (Table S1), and a low (1 μM) or high (1 mM) concentration of dNTPs was denatured at 70 °C and then chilled on ice. After annealing at 42 °C for 10 min, reverse transcription was performed at 42 °C for 30 min using M-MLV reverse transcriptase (Promega).
Primer Extension Analysis.
Total RNA was subjected to detection of 2′-O-methylation as previously reported, with some modifications (46). Briefly, 5 μg DNase I-digested total RNA was mixed with 5′ DIG-labeled Gm2620-R primer in AMV RT buffer (Promega) containing 2 mM or 4 μM dNTP at 58 °C for 20 min. Primer extension using 1 U AMV reverse transcriptase (Promega) was performed at 42 °C for 30 min.
NRO Assay.
The NRO assay was conducted as previously described with some modifications (47, 48). Briefly, nuclei were purified from WT and hid2 seedlings by Honda buffer and were washed with transcription buffer [125 mM Tris·HCl (pH 7.5), 500 mM KCl, 25 mM MgCl2, 10 mM DTT, 500 mM sucrose, 25% (vol/vol) glycerol]. To check the transcription rate of 45S pre-rRNA, 0.8 mM biotin-UTP (Roche) was used for NRO in 1× transcription buffer with 4 mM A/G/CTP and 1 U/μL RNase inhibitor at 30 °C for 10 min. The biotinylated RNA was purified using Streptavidin Sepharose beads (GE Healthcare), reverse transcribed by SuperScript III reverse transcriptase using a random hexamer primer (Life Technologies), and quantified by qPCR. Five independent replicates were performed and analyzed.
ChIRP.
ChIRP was performed as previously outlined with some modifications (49). Antisense DNA probes were designed against the full-length HID2 sequence and biotinylated at the 3′ end (Invitrogen). A set of probes against lacZ RNA was used as mock. WT seedlings (0.5 g) were cross-linked in 1% (vol/vol) formaldehyde (Sigma Aldrich) at room temperature for 20 min in a vacuum. Crosslinking then was quenched with 0.125 M glycine for 5 min. Nuclei were isolated as described in the NRO assay and were sonicated using a Bioruptor ultrasonicator (Diagenode). Chromatin was diluted in two volumes of hybridization buffer [750 mM NaCl, 1% SDS, 50 mM Tris·HCl (pH 7.0), 1 mM EDTA, 15% formamide, 0.1 mM PMSF, protease inhibitor, and RNase inhibitor] and was mixed gently. After preclearance with Streptavidin Sepharose beads (GE Healthcare), 100 pmol of probes were added and mixed by end-to-end rotation at 37 °C for 4 h. Washed/blocked Streptavidin Sepharose beads (20 μL) were added, and the reaction was rotated at 37 °C for 30 min. The beads were washed three times with high-salt wash buffer (2× SSC, 0.5% SDS, 1 mM DTT, and 1 mM PMSF) and three times with low-salt wash buffer (0.1× SSC, 0.5% SDS, 1 mM DTT, and 1 mM PMSF) for 5 min each at 37 °C. DNA was purified using the ChIP DNA Clean & Concentrator Kit (Zymo Research) and analyzed by qPCR. Four biological replicates were performed and produced similar results.
Circular RT-PCR.
Five micrograms of purified RNA were circulated by T4 RNA ligase I (New England Biolabs) at 25 °C for 2 h. The first-strand cDNA was generated by SuperScript III reverse transcriptase (Invitrogen) from ligated RNAs with the 5.8S-cRT-R primer. After amplification for 30 cycles by PCR, bands of the predicted size were cloned into the pMD20-T vector (Takara), and at least 50 clones were sequenced (9).
Polysome profiling.
Polysomes were fractionated over sucrose gradients as previously described with minor modifications (9, 50). A total of 6,000 A260 units of the supernatant were layered onto a linear 10–60% sucrose gradient poured with the Gradient Master 108 (BioComp Instruments). After ultracentrifugation in a Hitachi P40ST rotor at 217,000 × g for 4 h at 4 °C, the gradients were analyzed using the Piston Gradient Fractionator (BioComp Instruments) attached to a Model EM-1 Econo UV Monitor (Bio-Rad) for continuous measurement of the absorbance at 254 nm. All 64 fractions were collected using a Gilson 203B Fraction Collector, and several selected fractions were subjected to Northern blot analysis.
Acknowledgments
We thank Dr. Jian Lu, Dr. Weiqiang Qian, Dr. Shunong Bai, Dr. Ligeng Ma, and Dr. Qing Li for their helpful discussions and comments; Dr. Yu Zhang, Yue Huang, Yan Hu, Jiawei Pan, and Shengqian Dou for technical assistance; and Dr. Daniel A. Chamovitz, Dr. Sigal Rencus-Lazar, and Abigail Coplin for critical reading of the manuscript. This work was supported by National Key Basic Research Program Grant 2016YFA0500800, Major Program of National Natural Science Foundation of China Grant 91540105, General Program of National Natural Science Foundation of China Grant 31471155, and National Basic Research Program (973 Program) Grant 2012CB910900, and in part by the State Key Laboratory of Protein and Plant Gene Research at Peking University and the Peking-Tsinghua Center for Life Sciences. Y.W is supported in part by a Postdoctoral Fellowship from the Peking-Tsinghua Center for Life Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614852113/-/DCSupplemental.
References
- 1.Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6(2):225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol. 2015;22(1):11–19. doi: 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- 3.Weis BL, Kovacevic J, Missbach S, Schleiff E. Plant-specific features of ribosome biogenesis. Trends Plant Sci. 2015;20(11):729–740. doi: 10.1016/j.tplants.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23(1):63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Pontvianne F, et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013;27(14):1545–1550. doi: 10.1101/gad.221648.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Layat E, Sáez-Vásquez J, Tourmente S. Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 2012;53(2):267–276. doi: 10.1093/pcp/pcr177. [DOI] [PubMed] [Google Scholar]
- 7.Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010;38(13):4487–4502. doi: 10.1093/nar/gkq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004;10(6):942–953. doi: 10.1261/rna.5256704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hang R, et al. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc Natl Acad Sci USA. 2014;111(45):16190–16195. doi: 10.1073/pnas.1412697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández-Pevida A, Kressler D, de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev RNA. 2015;6(2):191–209. doi: 10.1002/wrna.1267. [DOI] [PubMed] [Google Scholar]
- 11.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3(3):397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 12.Rodor J, Letelier I, Holuigue L, Echeverria M. Nucleolar RNPs: From genes to functional snoRNAs in plants. Biochem Soc Trans. 2010;38(2):672–676. doi: 10.1042/BST0380672. [DOI] [PubMed] [Google Scholar]
- 13.Brown JW, Echeverria M, Qu LH. Plant snoRNAs: Functional evolution and new modes of gene expression. Trends Plant Sci. 2003;8(1):42–49. doi: 10.1016/s1360-1385(02)00007-9. [DOI] [PubMed] [Google Scholar]
- 14.Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85(7):1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 15.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89(5):799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 16.Dutca LM, Gallagher JE, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011;39(12):5164–5180. doi: 10.1093/nar/gkr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marmier-Gourrier N, Cléry A, Schlotter F, Senty-Ségault V, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 2011;39(22):9731–9745. doi: 10.1093/nar/gkr675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sáez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverría M. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol Cell Biol. 2004;24(16):7284–7297. doi: 10.1128/MCB.24.16.7284-7297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samaha H, et al. Identification of protein factors and U3 snoRNAs from a Brassica oleracea RNP complex involved in the processing of pre-rRNA. Plant J. 2010;61(3):383–398. doi: 10.1111/j.1365-313X.2009.04061.x. [DOI] [PubMed] [Google Scholar]
- 20.Pontvianne F, et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010;6(11):e1001225. doi: 10.1371/journal.pgen.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sáez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverría M. Characterization of a crucifer plant pre-rRNA processing complex. Biochem Soc Trans. 2004;32(Pt 4):578–580. doi: 10.1042/BST0320578. [DOI] [PubMed] [Google Scholar]
- 22.Dunbar DA, Baserga SJ. The U14 snoRNA is required for 2′-O-methylation of the pre-18S rRNA in Xenopus oocytes. RNA. 1998;4(2):195–204. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Wu C, Cai G, Chen S, Ye K. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016;30(6):718–732. doi: 10.1101/gad.274688.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peculis BA. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol. 1997;17(7):3702–3713. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu LH, et al. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23(14):2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu TT, et al. A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol Plant. 2013;6(3):830–846. doi: 10.1093/mp/sss087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Genomic features and regulatory roles of intermediate-sized non-coding RNAs in Arabidopsis. Mol Plant. 2014;7(3):514–527. doi: 10.1093/mp/sst177. [DOI] [PubMed] [Google Scholar]
- 28.Trémousaygue D, et al. Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 2003;33(6):957–966. doi: 10.1046/j.1365-313x.2003.01682.x. [DOI] [PubMed] [Google Scholar]
- 29.Qu G, et al. Promoter-based identification of novel non-coding RNAs reveals the presence of dicistronic snoRNA-miRNA genes in Arabidopsis thaliana. BMC Genomics. 2015;16:1009. doi: 10.1186/s12864-015-2221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102(36):12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hervé C, et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009;149(3):1462–1477. doi: 10.1104/pp.108.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Mao Y, Yang J, He Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Front Plant Sci. 2015;6:436. doi: 10.3389/fpls.2015.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong ZW, et al. RTL-P: A sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 2012;40(20):e157. doi: 10.1093/nar/gks698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 35.Abbasi N, et al. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010;64(6):960–976. doi: 10.1111/j.1365-313X.2010.04393.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim SH, et al. Plant U13 orthologues and orphan snoRNAs identified by RNomics of RNA from Arabidopsis nucleoli. Nucleic Acids Res. 2010;38(9):3054–3067. doi: 10.1093/nar/gkp1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valleron W, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26(9):2052–2060. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- 39.Su H, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014;33(11):1348–1358. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 40.Thorenoor N, Slaby O. Small nucleolar RNAs functioning and potential roles in cancer. Tumour Biol. 2015;36(1):41–53. doi: 10.1007/s13277-014-2818-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20(9):2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci USA. 2014;111(28):10359–10364. doi: 10.1073/pnas.1409457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell. 2008;20(6):1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu MF, Wagner D. RNA in situ hybridization in Arabidopsis. Methods Mol Biol. 2012;883:75–86. doi: 10.1007/978-1-61779-839-9_5. [DOI] [PubMed] [Google Scholar]
- 45.Jing Y, et al. SUVH2 and SUVH9 couple two essential steps for transcriptional gene silencing in Arabidopsis. Mol Plant. 2016;9(8):1156–1167. doi: 10.1016/j.molp.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Qu LH, Meng Q, Zhou H, Chen YQ. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001;29(7):1623–1630. doi: 10.1093/nar/29.7.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu F, Bakht S, Dean C. Cotranscriptional role for Arabidopsis DICER-LIKE 4 in transcription termination. Science. 2012;335(6076):1621–1623. doi: 10.1126/science.1214402. [DOI] [PubMed] [Google Scholar]
- 48.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340(6132):619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012;(61):3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]