Significance
A high nuclear Notch homolog 1, translocation-associated (Notch1) intracellular domain level distinguishes high-risk prostate cancer and castration-resistant prostate cancer from benign and low/intermediate-risk prostate cancer. Chronic activation of Notch1 cooperates with multiple oncogenic pathways altered in early prostate cancer, including AKT, Myc, and Ras/Raf/MAPK, to promote progression to androgen ablation-resistant prostate adenocarcinoma.
Keywords: prostate, cancer, Notch1
Abstract
Metastatic castration-resistant prostate cancer (CRPC) is the primary cause of prostate cancer-specific mortality. Defining new mechanisms that can predict recurrence and drive lethal CRPC is critical. Here, we demonstrate that localized high-risk prostate cancer and metastatic CRPC, but not benign prostate tissues or low/intermediate-risk prostate cancer, express high levels of nuclear Notch homolog 1, translocation-associated (Notch1) receptor intracellular domain. Chronic activation of Notch1 synergizes with multiple oncogenic pathways altered in early disease to promote the development of prostate adenocarcinoma. These tumors display features of epithelial-to-mesenchymal transition, a cellular state associated with increased tumor aggressiveness. Consistent with its activation in clinical CRPC, tumors driven by Notch1 intracellular domain in combination with multiple pathways altered in prostate cancer are metastatic and resistant to androgen deprivation. Our study provides functional evidence that the Notch1 signaling axis synergizes with alternative pathways in promoting metastatic CRPC and may represent a new therapeutic target for advanced prostate cancer.
The first line of treatment for men with advanced prostate cancer is androgen deprivation therapy. However, the disease commonly relapses to a lethal metastatic form referred to as “castration-resistant prostate cancer” (CRPC) (1). Current therapies for CRPC include second-generation androgen inhibitors (enzalutamide and abiraterone acetate), chemotherapeutic agents (docetaxel), and immunotherapy (Sipuleucel-T). Unfortunately, these agents improve median overall survival by only 4 months (2, 3). There is an urgent need to define the pathways that drive metastatic CRPC, possibly gaining insights into new therapeutic strategies for targeting the advanced disease.
The Notch family of receptors regulates cell-fate determination throughout development in many organ systems, including the prostate (4–7). In neonatal and adult mouse prostate tissues, disruption of Notch signaling inhibits prostate epithelial cell differentiation (4). Notch homolog 1, translocation-associated (Notch1) was also shown to promote mouse luminal prostate cell survival and proliferation through the activation of the prosurvival NF-κB pathway (5). Elevated expression of the Notch ligand Jagged1 has been associated with metastatic prostate cancer (8, 9), and down-regulation of Notch1 and Jagged1 in human prostate cancer cell lines decreases in vitro invasion and cell growth (10). In addition, chemoresistance in human prostate cancer cells has been linked to the activation of Notch2 receptors (11, 12). Despite these studies, direct evaluation of Notch receptors as drivers of aggressive prostate cancer and the relationship of Notch receptors with other commonly altered pathways in prostate tumorigenesis remain unclear.
The canonical Notch pathway is activated through binding of Notch ligands (Jagged1/2 and Delta-like 1/3/4) to Notch receptors (1/2/3/4) (13–19). Ligand binding induces Notch-receptor cleavage through regulated intramembrane proteolysis (13–21), a multistep process carried out by members of the A Disintegrin and Metalloprotease (ADAMs) family within the extracellular region and by the γ-secretase complex within the transmembrane domain of Notch cell-surface receptors. Upon cleavage, the Notch intracellular domain (NICD) is released and translocates to the nucleus, where it is referred to as “activated Notch” (13–21). NICD serves as a transcriptional coactivator that in turn regulates a set of genes involved in proliferation, self-renewal, survival, and differentiation (20, 21).
Components of the Notch signaling pathway are altered in multiple cancers (21–32). Interestingly, Notch signaling previously has been reported to function as both a tumor suppressor and an oncogene (21–32). The dependency of Notch1 function in cancer may be tissue specific and context dependent (21–32). Loss-of-function mutations in Notch receptors support their tumor-suppressive role in multiple malignances, including bladder cancer and squamous cell carcinoma (22–24). Constitutive activation of the Notch receptors through gene rearrangements or mutations leads to Notch receptors' oncogenic function in 55–60% of patients with T-cell acute lymphoblastic leukemia (26, 27). An oncogenic role of Notch1 also has been demonstrated in chronic lymphocytic leukemia and in solid tumors such as lung adenocarcinoma and others (28–31). Mutations within Notch1 receptor are rare (3% frequency) in metastatic prostate cancer (32, 33).
Here we report significantly elevated levels of nuclear NICD1 in hormone-naive high-risk prostate cancer and nearly all metastatic CRPC specimens but not in benign tissues or low- and intermediate-risk localized prostate cancer. Although overexpression of NICD1 alone was not sufficient to drive prostate tumorigenesis, NICD1 in combination with components of pathways commonly altered in early prostate cancer, such as myristoylated AKT, Myc, and Ras/Raf/MAPK, promoted the development of aggressive prostate adenocarcinoma and progression to CRPC. Consistent with their aggressiveness, these tumors displayed an epithelial-to-mesenchymal (EMT) transition phenotype, high self-renewal, and the potential for metastatic colonization. Tumors driven by NICD1 in combination with myristoylated AKT, Myc, and the Ras/Raf/MAPK pathway are also resistant to androgen deprivation. Our results indicate that Notch1 receptor signaling plays a central role in the development and progression of prostate cancer and may serve as a rational therapeutic target in high-risk prostate cancer and metastatic CRPC.
Results
Nuclear NICD1 Is Highly Elevated in High-Risk Prostate Canter and CRPC.
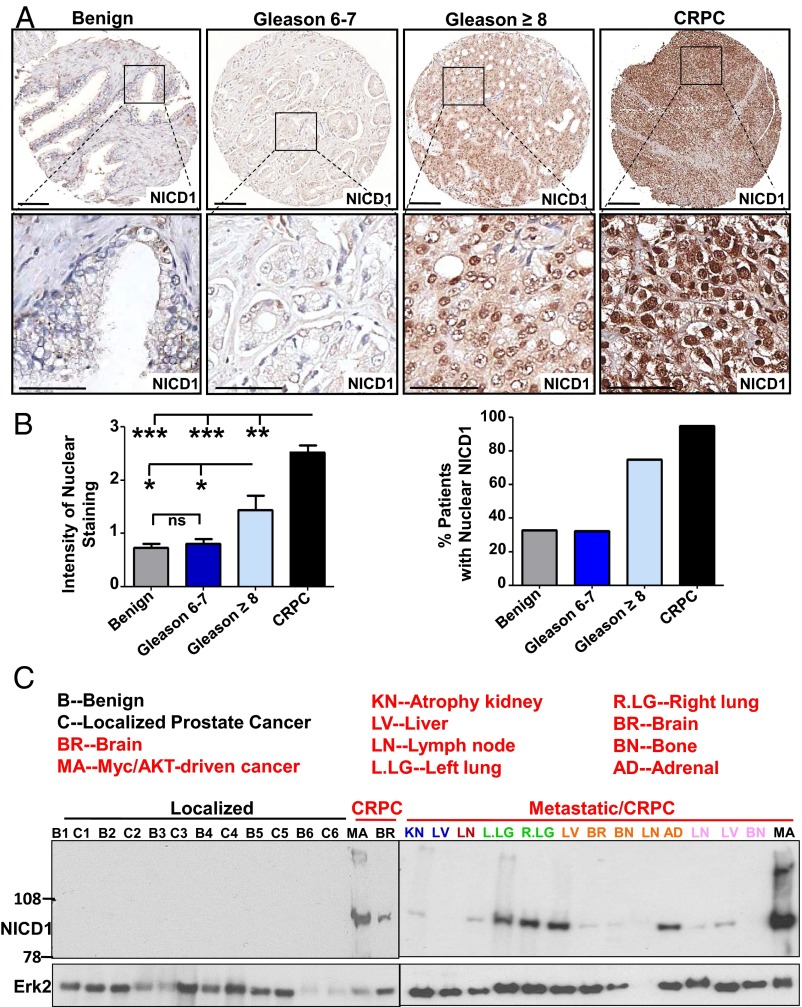
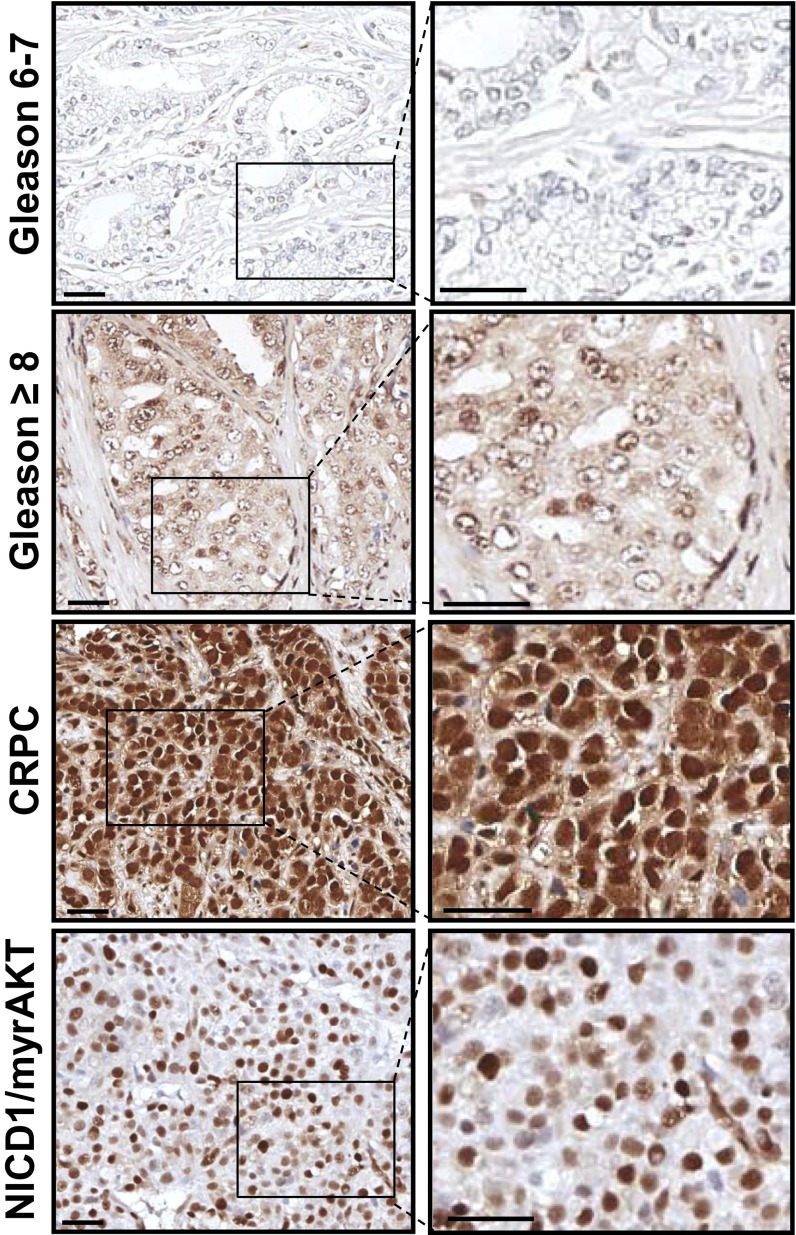
To determine if alterations in the Notch signaling axis are associated with prostate tumorigenesis, we first assessed the levels of Notch1 in benign prostate tissue and different stages of human prostate cancer including low- to intermediate-risk (Gleason score 6 and 7) and high-risk (Gleason score 8–10) prostate cancer and CRPC. Immunohistochemical analysis of human prostate tissue microarrays (TMAs) showed a significant (more than two-fold) increase in the average intensity of nuclear NICD1 staining in high-risk prostate cancer in comparison with benign prostate tissue or localized low- to intermediate-risk prostate cancers (Fig. 1 A and B and Fig. S1). The average intensity of the staining was even higher in CRPC samples and showed a dramatic increase, by more than threefold, compared with benign prostate tissue or localized low- to intermediate-risk prostate cancers (Fig. 1 A and B and Fig. S1). Analysis of NICD1 subcellular localization indicated the presence of nuclear NICD1 in 75% of samples from high-risk prostate cancers (n = 23) and in 95% of CRPC samples (n = 19) (Fig. 1 A and B). In contrast, nuclear NICD1 was found in 32% of benign prostate tissue samples (n = 221) and in 33% of localized low- to intermediate-risk prostate cancers (n = 207) but at low intensity (Fig. 1 A and B). Similar results were observed through Western blot analysis of benign tissue, localized low- to intermediate-risk prostate cancers. and CRPC from distinct metastatic sites. Prostate tissues designated as low-risk localized prostate tumor, benign tissue adjacent to the tumor, or CRPC from distinct metastatic sites obtained from rapid autopsies performed in eight different patients (34, 35) were subjected to Western blot analysis (Fig. 1C and Fig. S2). As a positive control, we used a human CRPC model initiated in primary human cells and driven by the combination of Myc and myristoylated/activated AKT (myrAKT) oncogenes that express high levels of NICD1 (36). NICD1 (100 kDa) was observed only in the human metastatic CRPC samples and the Myc/myrAKT human CRPC model (36) but not in the benign tissue or in localized low- to intermediate-risk prostate cancers (Fig. 1C and Fig. S2). Cleavage of Notch1 at valine1744 in human metastatic CRPC was confirmed by Western blot with antibody against cleaved Notch1 (Fig. S3). These results demonstrate that high levels of nuclear NICD1 distinguish low- to intermediate-risk prostate cancer from high-risk prostate cancer and metastatic CRPC and prompted us to investigate the functional role of NICD1 in prostate tumorigenesis.
Fig. 1.
The Notch1 receptor intracellular domain is highly elevated in advanced human prostate cancer. (A) Human prostate TMAs were stained with an antibody against NICD1 (Novus Biologicals). Representative images are shown. (Scale bars: 100 microns in Upper Row and 50 microns in Lower Row.) (B) NICD1 expression was scored from 0–3 in benign tissue (n = 221), localized low- to intermediate-risk prostate cancer (Gleason score 6 or 7) (n = 207), localized high-risk prostate cancer (Gleason score 8–10) (n = 23), and CPRC (n = 19) specimens. (Left) The intensity of nuclear staining was plotted. ***P < 0.0001. **P < 0.005; *P < 0.05; ns, nonsignificant; one-way ANOVA. (Right) Percentage of patients exhibiting nuclear NICD1. (C) Western blot analyses were performed with anti-NICD1 antibody (Epitomics/Abcam) and anti-Erk2 used as loading control using the following specimens. (i) Human specimens were separated into regions of benign tissue and low- to intermediate-risk prostate cancer by a urologic pathologist. (ii) Metastatic CRPC tissues were obtained from the rapid autopsy program at the University of Michigan. (iii) The Myc/myrAKT CRPC model initiated in primary human cells. Metastatic CRPC tissues were obtained from total of eight different patients (including the samples presented in Fig. S1). Twenty-three distinct metastatic sites are shown. Each distinct patient is indicated by a different color.
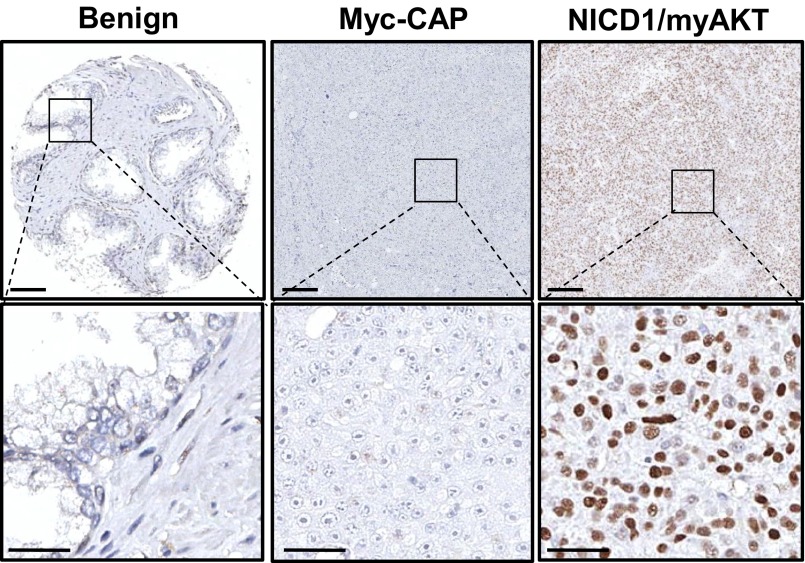
Fig. S1.
Nuclear NICD1 is elevated in advanced human prostate cancer. The figure shows negative and positive controls for the human prostate TMA stained with an antibody against the intracellular domain of Notch1 presented in Fig. 1A. (Left) Benign human tissue; epithelial cells are negative for NICD1. (Center) Previously described mouse Myc-CAP xenografts (41). Myc-CAP xenografts are negative for nuclear NICD1. (Right) As a positive control we used tumors driven by the combination of NICD1 overexpression and active AKT overexpression (myrAKT). (Scale bars: 100 microns in the upper row; 50 microns in the lower row.)
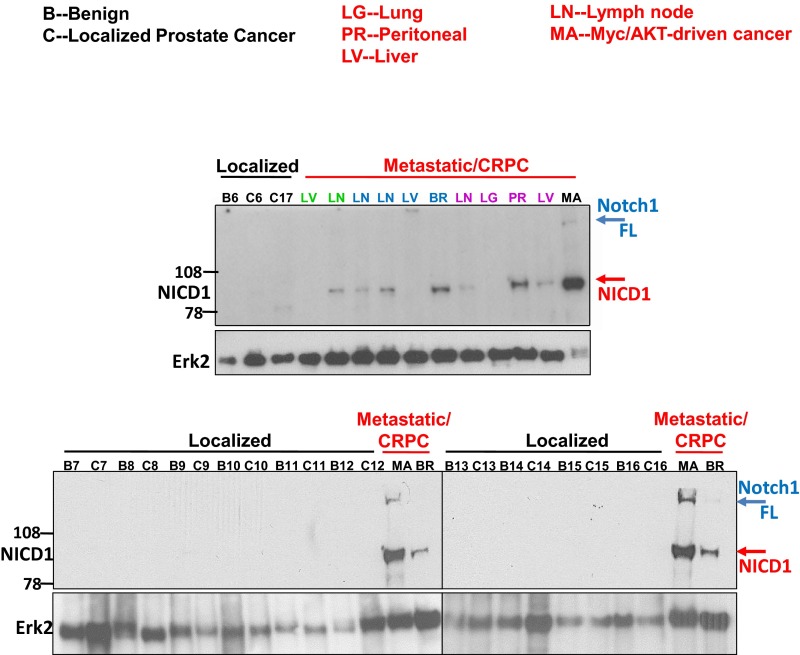
Fig. S2.
NICD1 is highly expressed in metastatic prostate cancer but not in benign prostate tissues and low- or intermediate-risk prostate cancer. Western blot analyses were performed using anti-NICD1 antibody (Epitomics; currently Abcam), and anti-Erk2 was used as a loading control. For the Western blot analyses we used (i) human prostate tissues separated into benign (B) and low- to intermediate-risk prostate cancer (T) regions; (ii) metastatic CRPC tissues obtained from the rapid autopsy program at the University of Michigan; and (iii) the Myc/myrAKT CRPC model initiated in primary human cells that express high levels of full-length Notch1 (Notch1 FL, ∼300 kDa) and NICD1 (∼100 kDa). Distinct patients with metastatic disease are shown in different colors.
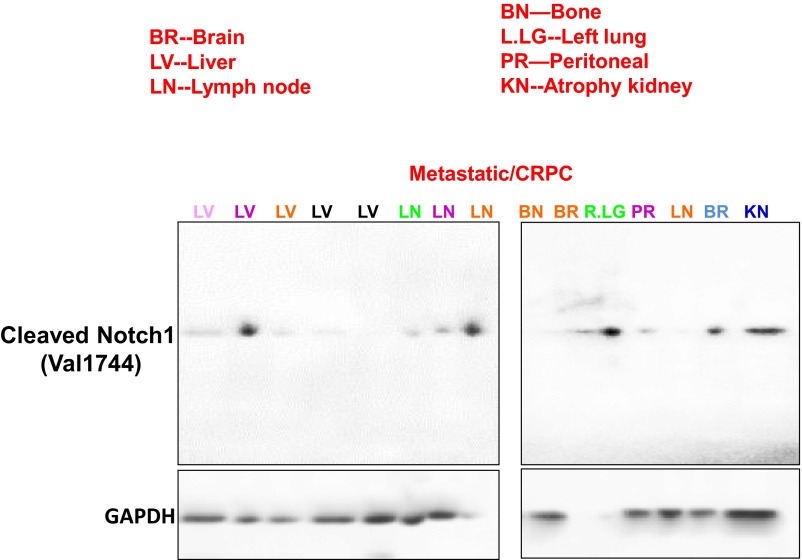
Fig. S3.
NICD1 is highly expressed in metastatic prostate cancer. Western blot analyses were performed on metastatic CRPC tissues obtained from the rapid autopsy program at the University of Michigan using anti-cleaved Notch1 V1744 antibody (Cell Signaling Technology). GAPDH was used as a loading control. Distinct patients with metastatic disease are shown in different colors. Different colors and metastatic sites match the patients’ samples analyzed in Fig. 1C and Fig. S2.
NICD1 Synergizes with the myrAKT, Myc, and Ras/Raf/MAPK Pathways to Promote the Development of Aggressive Prostate Adenocarcinoma.
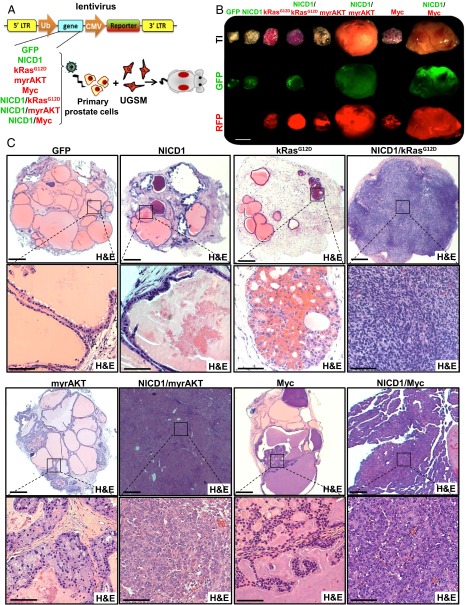
High levels of nuclear NICD1 are observed predominantly in high-risk prostate cancer and CRPC, suggesting a role for the pathway in prostate cancer progression. We sought to determine whether NICD1 promotes prostate tumorigenesis through collaboration with early genetic alterations. Deletion of the PTEN tumor suppressor is observed in up to 70% of prostate cancers and results in the activation of AKT (37, 38). Other common alterations in advanced human prostate cancer include elevated expression of the Myc oncogene and activation of the Ras/Raf/MAPK signaling pathway (38, 39). To mimic the loss of PTEN, we used myrAKT. Activation of Ras/Raf/MAPK was achieved through overexpression of mutant kRasG12D. Dissociated mouse prostate cells can regrow prostate-like structures in vivo when combined with urogenital sinus mesenchyme (UGSM) followed by implantation under the kidney capsule of SCID mice (Fig. 2A) (40). This prostate regeneration model allows testing the functional role of single genes or combinations of genes in vivo (Fig. 2A) (40). Overexpression of a single oncogene such as myrAKT, kRasG12D, or Myc initiates prostate intraepithelial neoplasia (PIN) in our model but is not sufficient to drive prostate adenocarcinoma. Therefore these genes are suitable for addressing the role of NICD1 in promoting tumor progression (Fig. 2).
Fig. 2.
NICD1 synergizes with multiple oncogenic pathways to drive prostate cancer. (A, Left) Schematic representation of the in vivo mouse tissue regeneration assay. Lentiviral transduction was used to overexpress GFP alone (GFP), human NICD1 and GFP (NICD1), mutant kRasG12D and RFP (kRasG12D), myristoylated/activated AKT and RFP (myrAKT), or Myc and RFP (Myc), alone or in combination with NICD1 (NICD1/kRasG12D, NICD1/myrAKT, NICD1/Myc). (Right) Transduced epithelial cells were combined with UGSM and implanted under the kidney capsule of SCID mice. Twelve weeks later the grafts were evaluated histologically. (B) Representatives of the grafts recovered 12 wk after implantation are shown for each condition. (Scale bar: 5 mm.) One of the five experiments performed is shown. (C) The recovered grafts from each condition were stained with H&E and were evaluated histologically. Representatives of the recovered grafts are shown. (Scale bars: 500 microns in upper panels; 100 microns in lower panels.)
Overexpression of NICD1 alone or in combination with kRasG12D (NICD1/kRasG12D), myrAKT (NICD1/myrAKT), or Myc (NICD1/Myc) was investigated for its ability to drive prostate tumorigenesis using the in vivo mouse prostate regeneration model (Fig. 2A). NICD1 alone was not sufficient to drive prostate cancer initiation, because we observed normal prostate tubular structures similar to the GFP-expressing control cells (Fig. 2). In contrast, NICD1 strongly synergized with kRasG12D, myrAKT, and Myc, giving rise to prostate adenocarcinoma (Fig. 2 B and C). RFP and GFP signals in the recovered tissues were used as a control for infection efficiency (Fig. 2B). Importantly, the level of NICD1 overexpression in our in vivo regeneration assay was comparable to the levels observed in human CRPC specimens (Fig. S4). Oncogene expression or pathway activity was confirmed through immunohistochemical analysis (Fig. 3 and Fig. S5).
Fig. S4.
Expression levels of NICD1 in tumors driven by NICD1 in combination with kRasG12D, myrAKT, or Myc resemble levels of nuclear NICD1 in human CRPC specimens. Levels of nuclear NICD1 overexpression mimic the levels of nuclear NICD1 in human CRPC. Immunohistochemical analyses of 4- μm sections of paraffin-embedded human low- to intermediate-risk prostate cancer (Gleason score 6 or 7), localized high-risk prostate cancer (Gleason score 8–10), CRPC specimens, and NICD1/myrAKT tumors with antibodies against NICD1 (Novus Biologicals) are shown. (Scale bars: 100 microns in left panels; 50 microns in right panels.)
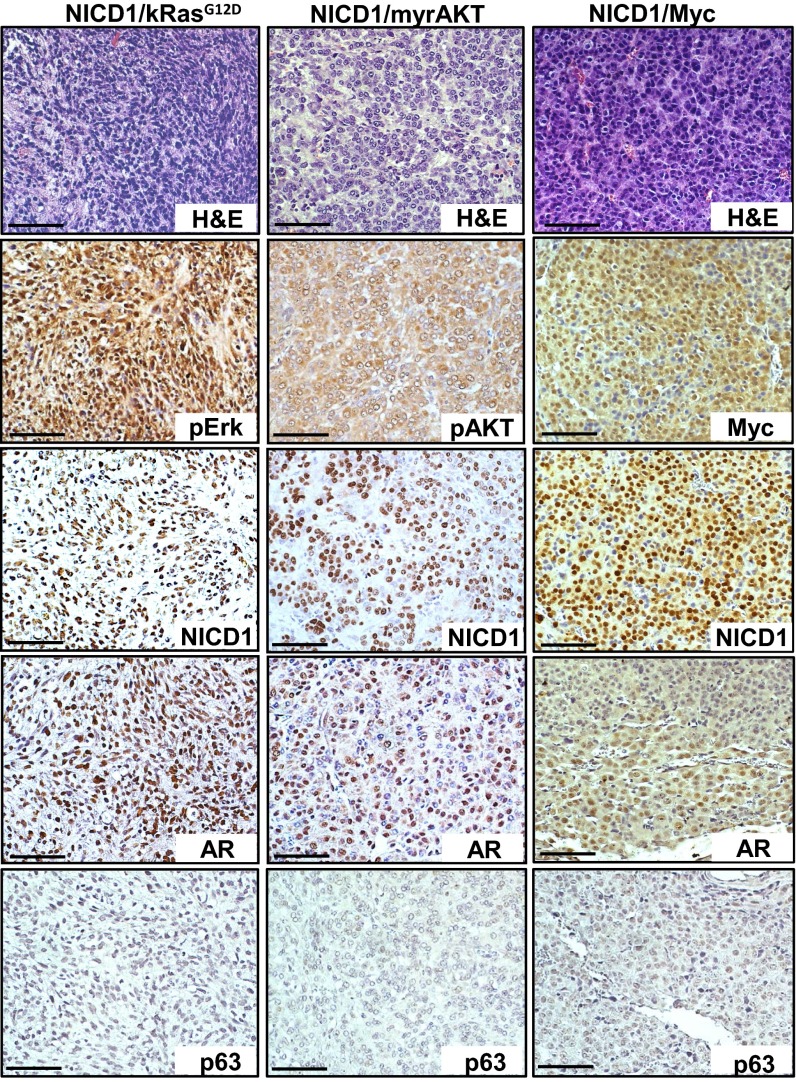
Fig. 3.
Characterization of NICD1/myrAKT, NICD1/Myc, and NICD1/kRasG12D tumors. Immunohistochemical analysis was performed on 4-μm sections of paraffin-embedded NICD1/myrAKT, NICD1/Myc, and NICD1/kRasG12D tumors stained with H&E or with antibodies against NICD1, pErk, pAKT, Myc, p63, or AR. (Scale bars: 100 microns.)
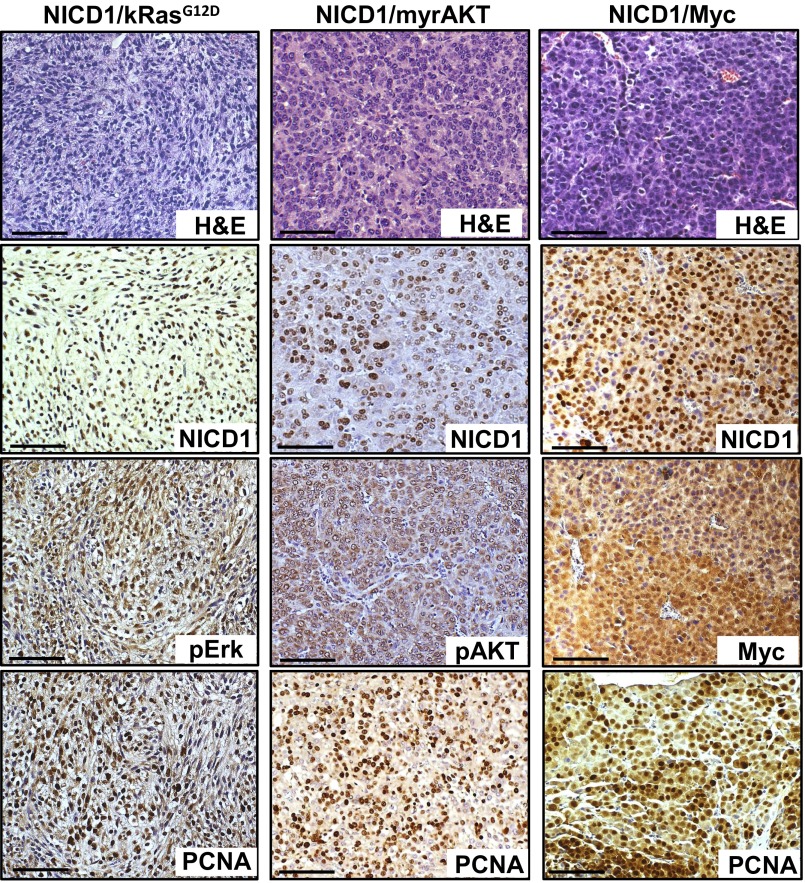
Fig. S5.
NICD1/myrAKT, NICD1/Myc, and NICD1/kRasG12D tumors are highly proliferative. Immunohistochemical analysis of 4-μm sections of paraffin-embedded NICD1/myrAKT, NICD1/Myc, and NICD1/kRasG12D tumors stained with H&E or with antibodies against NICD1, pErk, pAKT, Myc, or PCNA. (Scale bars: 100 microns.)
NICD1/kRasG12D, NICD1/myrAKT, and NICD1/Myc tumors were highly proliferative as measured by proliferating cell nuclear antigen (PCNA), exhibit loss of basal cell marker p63, and express androgen receptors (ARs) (Fig. 3 and Fig. S5). These results demonstrate the functional role of NICD1 in synergizing with alternative pathways to promote the development of advanced prostate adenocarcinoma.
Gene-Expression Profiling of Tumors Driven by NICD1 in Combination with Alternative Pathways Reveals the EMT Signature.
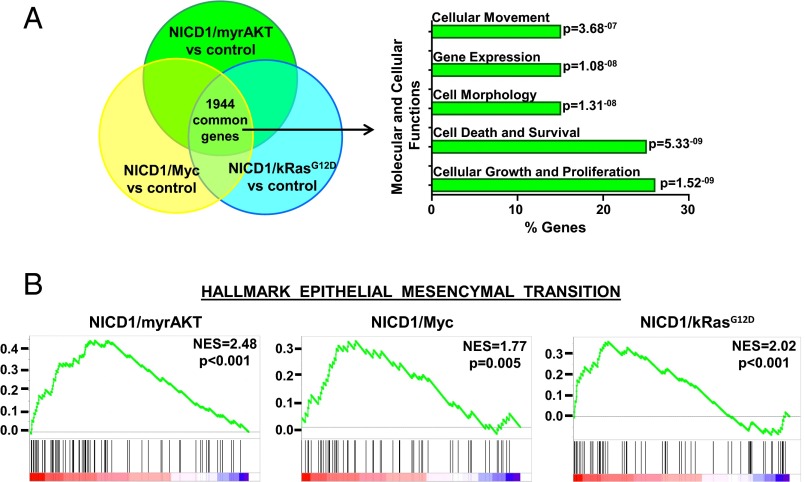
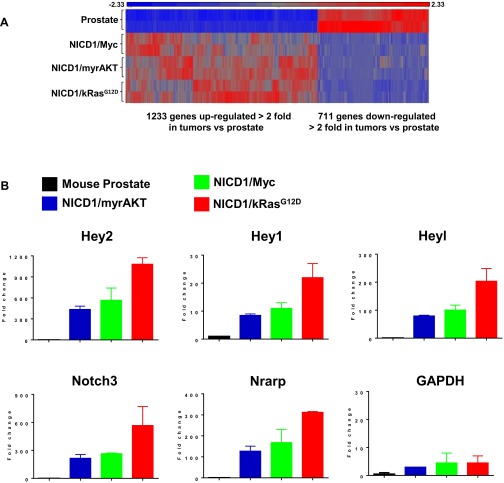
To gain insight into how NICD1 promotes advanced prostate cancer, we performed genome-wide transcriptome profiling of NICD1-driven tumors. Tumors initiated by the combination of NICD1/kRasG12D, NICD1/myrAKT, or NICD/Myc and normal mouse prostate were subjected to high-throughput RNA-sequencing and differential gene-expression analysis (Fig. 4A and Fig. S6A). We identified 4,238 genes significantly up-regulated or down-regulated in NICD1/kRasG12D tumors, 4,433 genes in NICD1/myrAKT tumors, and 3,502 genes in NICD1/Myc tumors compared with normal mouse prostate. Of the differentially expressed genes, 1,944 were common for all three tumors (Fig. 4A and Fig. S6A). Ingenuity pathway analysis identified “cell movement” and “migration” as one of the top regulatory networks across all NICD1-driven tumors (Fig. 4A). As expected, we observed a significant increase in direct transcriptional targets of Notch1 such as Hey2, Hey1, Heyl, Notch3, and Nrarp in NICD1/kRasG12D, NICD1/myrAKT, and NICD/Myc tumors compared with normal mouse prostate (Fig. S6B).
Fig. 4.
Expression profiling of NICD1-driven tumors reveals the EMT signature. (A) Venn diagram of genes differentially expressed in NICD1/kRasG12D, NICD1/myrAKT, and NICD1/Myc versus normal mouse prostate. A total of 1,944 common differentially expressed genes were analyzed for molecular and cellular functions using Ingenuity engine software. The top statistically significant molecular and cellular functions are presented with the corresponding P values. (B) The plots show the GSEA of genes differentially expressed in NICD1/myrAKT, NICD1/Myc, or NICD1/kRasG12D versus normal mouse prostate. NES, normalized enrichment score.
Fig. S6.
Tumors driven by NICD1 in combination with pathways altered in prostate cancer express high levels of Notch1 target genes. (A) Heat map of common genes differentially expressed in NICD1/Myc, NICD1/myrAKT, and NICD1/kRasG12D tissue versus normal mouse prostate. Red represents genes overexpressed, and blue represents genes down-regulated across all NICD1/Myc, NICD1/myrAKT, and NICD1/kRasG12D tissues versus normal mouse prostate. (B) The fold change in reads per kilobase per million mapped reads (RPKM) of transcript of Notch1 target genes (Hey2, Hey1, Heyl, Notch3, and Nrarp) and GAPDH from our RNA-sequencing analysis is shown. The average of RPKM of each gene in the mouse prostate is considered as 1.
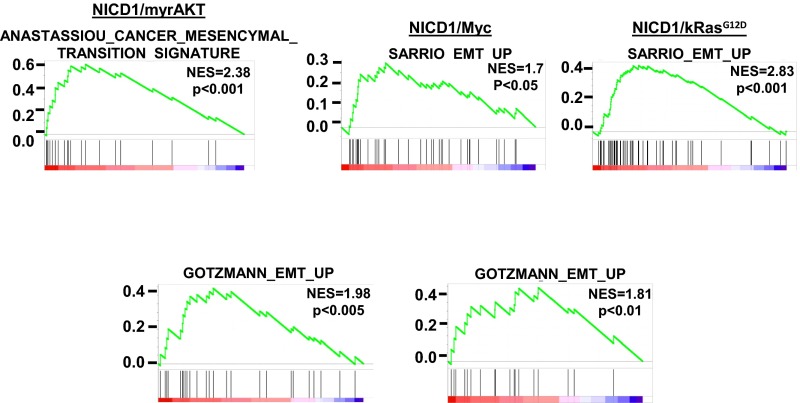
Gene set enrichment analysis (GSEA) (41) of differentially expressed genes across all NICD1-driven tumors revealed enrichment of previously published EMT signatures (Fig. 4B and Fig. S7). Our results demonstrate that tumors driven by NICD1 in combination with kRasG12D, myrAKT, and Myc exhibit EMT features, a phenotype that may characterize invasive, poorly differentiated carcinoma (42–45).
Fig. S7.
Tumors driven by NICD1 in combination with pathways altered in prostate cancer exhibit EMT characteristics. Plots representing GSEA of genes differentially expressed in NICD1/myrAKT, NICD1/Myc, and NICD1/kRasG12D versus normal mouse prostate are shown. NES, normalized enrichment score.
Tumors Driven by NICD1 in Combination with Pathways Altered in Prostate Cancer Exhibit High Self-Renewal Activity.
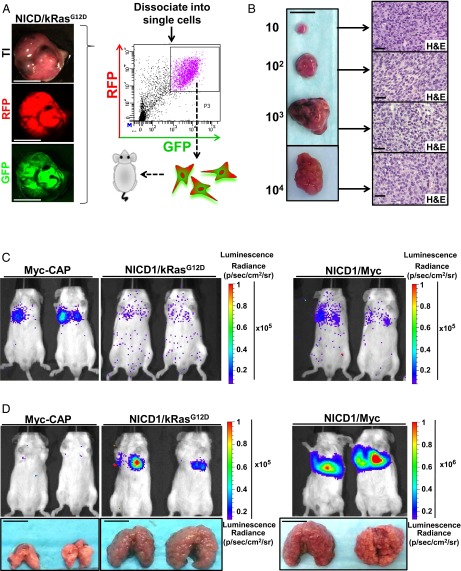
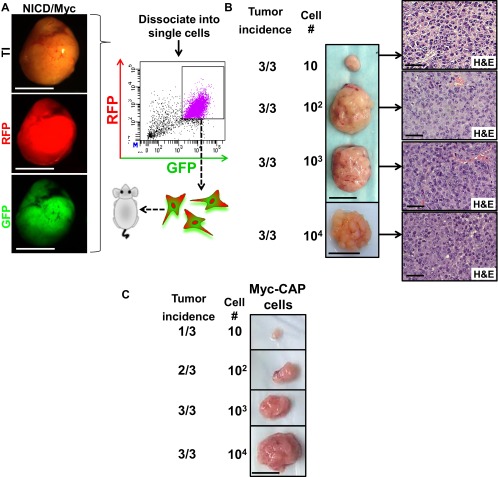
EMT is a morphological change in which epithelial cells acquire mesenchymal features and is commonly associated with self-renewal activity, an invasive tumor phenotype, and metastasis (46). EMT has been previously demonstrated to stimulate cancer stem cell self-renewal (47). We performed limiting dilution experiments to assess functionally the acquisition of cancer self-renewal activity and stem cell properties and to evaluate the minimum number of cells required to regenerate new tumors upon transplantation. Regenerated NICD1/kRasG12D and NICD1/Myc tumors were dissociated into single cells and subjected to FACS based on expression of RFP and GFP (Fig. 5A and Fig. S8A). The RFP color marker identified cells expressing kRasG12D or Myc, and GFP allowed the detection of NICD1-expressing cells. GFP/RFP double-positive cells were isolated, and 10, 100, 1,000, or 10,000 RFP/GFP double-positive cells were combined with Matrigel and transplanted s.c. into SCID mouse recipients (Fig. 5A and Fig. S8A). By 4 wk as few as 10 cancer cells were sufficient to regenerate the original tumor, demonstrating the high frequency of cancer cells with self-renewing activity within the tumors (Fig. 5B and Fig. S8 B and C). Histological analysis identified a phenotype consistent with the original NICD1/kRasG12D- and NICD1/Myc-driven tumors (Fig. 5B and Fig. S8B).
Fig. 5.
Tumors driven by NICD1 in combination with pathways altered in prostate cancer exhibit the acquisition of self-renewal activity and metastatic potential. (A) Primary NICD/kRasG12D tumor was dissociated into single cells and subjected to FACS. (Scale bars: 1 cm.) GFP/RFP double-positive cells were sorted, and 10, 100, 1,000, or 10,000 cells were implanted into SCID recipient mice. (B, Left) Representative recovered tumors initiated from 10, 100, 1,000, or 10,000 cells. (Scale bar: 1 cm.) (Right) Histology of each tumor is shown. (Scale bar: 50 microns.) One of the three independent experiments is presented. (C) Tumors driven by NICD1 in combination with pathways altered in prostate cancer exhibit metastatic potential. Primary tumors driven by NICD1/kRasG12D and NICD1/Myc were dissociated into single cells, infected with lentivirus expressing luciferase and YFP, and cultured. One week after infection cells were sorted by FACS for YFP. As a negative control, we used Myc-CAP cells expressing luciferase with YFP. Cells (8 × 105) were introduced to immunocompromised mice via tail veil injection. Bioluminescent images taken on the day of injection are shown. One of the three independent experiments is shown. (D, Upper) Bioluminescent images taken day 28 after injection are shown. (Lower) Recovered lungs are presented. (Scale bars: 1 cm.) The size of the recovered lungs, histology of the lungs, and immunohistochemistry for NICD1, pErk, myc, and AR are presented in Fig. S9.
Fig. S8.
NICD1 in combination with alternative pathways exhibits high self-renewal activity and metastatic potential in vivo. (A) Dissociated single cells from a primary NICD1/Myc tumor were subjected to FACS. GFP/RFP double-positive cells were sorted. GFP/RFP cells were implanted into SCID recipient mice at limiting dilutions of 10, 100, 1,000, or 10,000. (Scale bars: 0.5 cm.) (B, Left) Representative tumors initiated from 10, 100, or 1,000 cells recovered 4 wk after implantation or from 10,000 cells recovered 3 wk after implantation. (Scale bars: 1 cm.) (Right) H&E staining of each tumor is shown. (Scale bars: 50 microns.) One of the two independent experiments is shown. (C) The Myc-CAP cell line (44) has lower self-renewal activity and was used as a control. Representative tumors initiated from 10, 100, or 1,000 Myc-CAP cells recovered 8 wk after implantation. (Scale bar: 1 cm.)
Tumors Driven by NICD1 in Combination with Pathways Altered in Prostate Cancer Exhibit High Metastatic Colonization Potential.
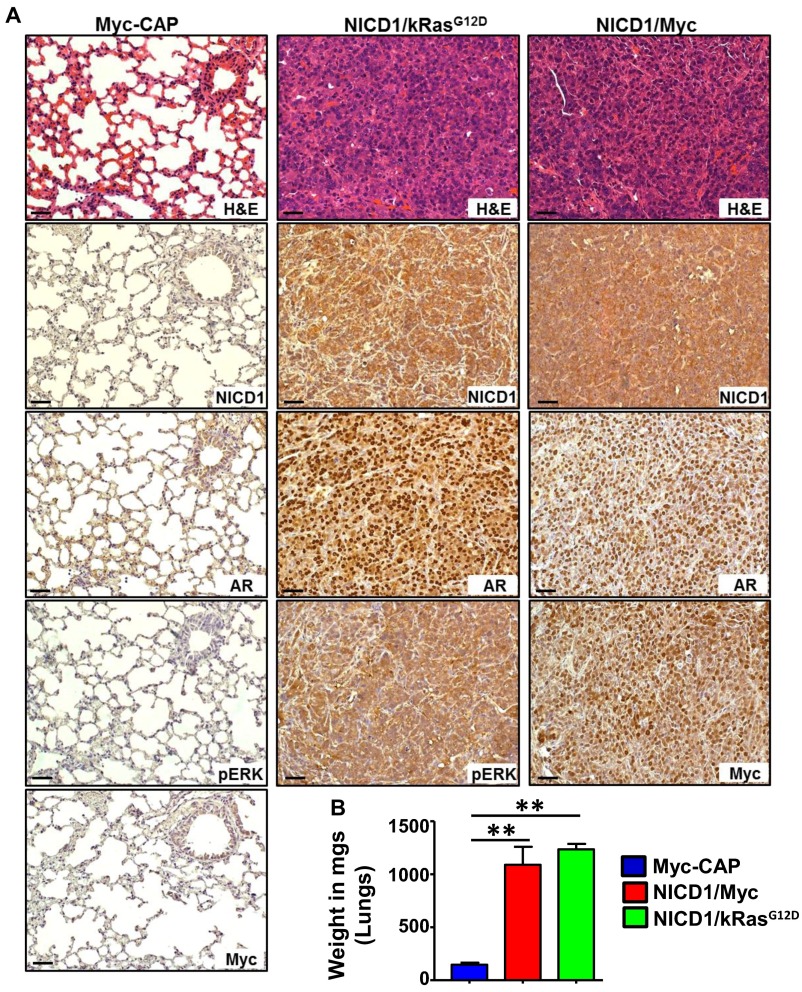
Self-renewal activity of tumor cells is a necessary property during metastasis that allows cancer cells to colonize tumors at new sites. The metastatic potential of tumors driven by NICD1 in combination with alternative pathways was assessed using an in vivo lung-colonization assay. Regenerated NICD1/kRasG12D and NICD1/Myc tumors were dissociated to single cells and infected with a luciferase-expressing lentivirus. As negative control, we used a previously characterized Myc-CAP cancer cell model derived from Myc transgenic mouse prostates (48) expressing luciferase. Immunocompromised mice were subjected to tail-vein injection with 8 × 105 cells. Twenty-eight days after injection we observed large lung tumors in animals injected with NICD1/kRasG12D and NICD1/Myc tumor cells but not in the control animals injected with Myc-CAP cells (Fig. 5 C and D and Fig. S9). Histological and immunohistochemical analyses showed that these tumors closely resembled the primary tumors and exhibited high levels of nuclear AR (Fig. S9A). These results demonstrate that NICD1/kRasG12D and NICD1/Myc tumor cells can survive and regrow tumors at a distant site, revealing the metastatic potential of NICD1 combination tumors.
Fig. S9.
Tumors driven by NICD1 in combination with other pathways altered in prostate cancer exhibit metastatic potential. (A) Immunohistochemistry analysis of paraffin-embedded lungs from the mice injected with NICD1/kRasG12D and NICD1/Myc cells shown in Fig. 5A stained with H&E or antibodies against NICD1, pErk, Myc, and AR. (Scale bars: 100 microns in left panels; 50 microns in center and right panels.) (B) Weight of the lungs recovered at day 28 after injection from the animals shown in Fig. 5C injected with Myc-CAP-Luciferase, NICD1/kRasG12D-Luciferase, or NICD1/Myc-Luciferase was measured in milligrams and plotted. **P < 0.005, one-way ANOVA.
Tumors Driven by NICD1 in Combination with Pathways Altered in Prostate Cancer Exhibit a Castration-Resistant Phenotype.
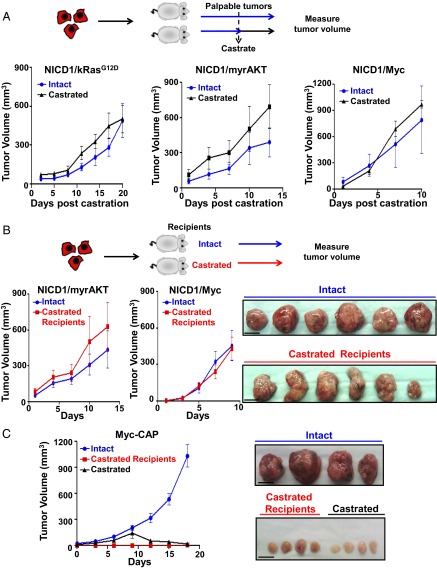
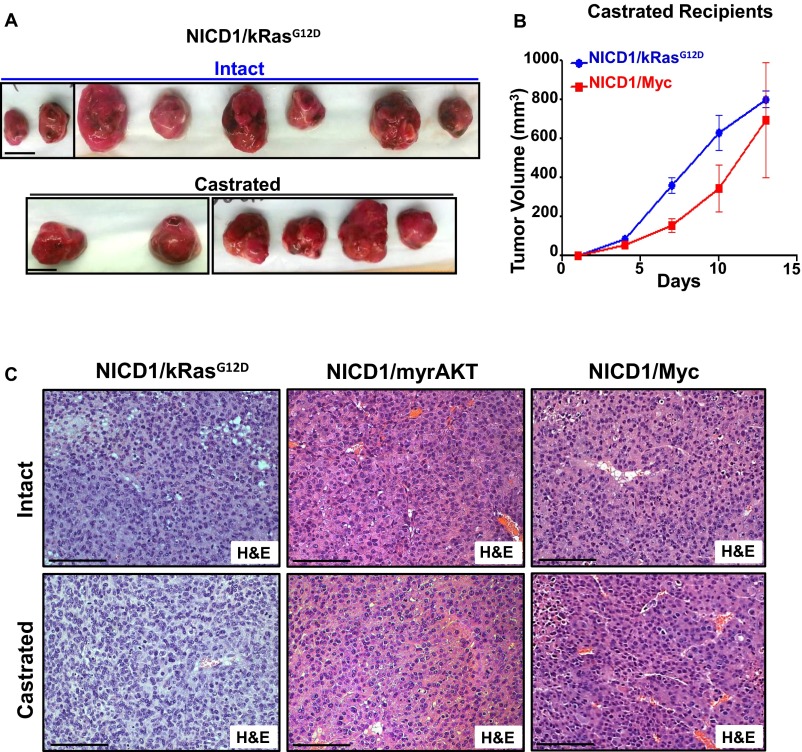
The frequent activation of NICD1 in clinical CRPC samples coupled with the high frequency of cancer stem cells and metastatic phenotype of NICD1-driven tumors led us to investigate the role of NICD1 in the development of castration resistance. To test if castration can affect tumor growth, we used cells derived from hormone-naive NICD1/kRasG12D, NICD1/myrAKT, and NICD1/Myc tumors (Fig. 6A and Fig. S10A). Myc-CAP mouse prostate cancer cells, previously demonstrated to be castration sensitive, were used as a control (Fig. 6C) (48). Tumor cells (6 × 105) were implanted s.c. into immunocompromised recipients. Upon detection of palpable tumors (average tumor volume, 50 mm3), recipients were subjected to surgical castration. Myc-CAP cells grew only in intact mice and failed to grow in castrated mice, whereas NICD1/kRasG12D, NICD1/myrAKT, and NICD1/Myc tumors continued to grow rapidly after androgen deprivation (Fig. 6 A and C and Fig. S10A).
Fig. 6.
NICD1 in combination with the Ras/Raf/MAPK, myrAKT, or Myc pathways drives CRPC. (A, Upper) Schematic representation of the experimental design. Primary tumors driven by NICD1/kRasG12D, NICD1/myrAKT, or NICD1/Myc were dissociated into single cells. NICD1/kRasG12D, NICD1/myrAKT, or NICD1/Myc tumor cells (6 × 105) were mixed with Matrigel and transplanted s.c. into six to eight mice per condition. Upon detection of palpable tumors (50 mm3), six to eight mice carrying a total of six to eight tumors were castrated (marked as day 1). Tumor length, width, and height were measured every 3 d for a total of 10–20 d. (Lower) Tumor volumes were calculated by multiplying length × width × height/2 and were plotted. One of the two independent experiments for each tumor type (NICD1/kRasG12D, NICD1/myrAKT, or NICD1/Myc) is shown. (B, Upper) The experimental design. NICD1/myrAKT or NICD1/Myc tumor cells (6 × 105) were mixed with Matrigel and transplanted s.c. into six castrated or six intact recipient mice. (Lower Left) Upon detection of palpable tumors (50 mm3), marked as day 1, tumor volumes were measured every 3 d. (Lower Right) Recovered NICD1/Myc secondary tumors from intact or castrated recipients are shown. (Scale bars: 1 cm.) One of the two independent experiments for each tumor type (NICD1/myrAKT or NICD1/Myc) is presented. (C, Left) Myc-CAP cells (6 × 105) were mixed with Matrigel and implanted s.c. into eight intact or four castrated immunodeficient recipient mice. Upon detection of palpable tumors, one group of intact mice (n = 4) was subjected to surgical castration. (Right) Images of the recovered tumors. (Scale bars: 1 cm.)
Fig. S10.
Tumors driven by NICD1 in combination with other pathways altered in prostate cancer are castration resistant. (A) Images of the transplanted tumors driven by NICD1/kRasG12D plotted in Fig. 6A. (Scale bars: 1 cm.) (B) NICD1/kRasG12D or NICD1/Myc tumor cells (6 × 105) were mixed with Matrigel and were transplanted s.c. into six castrated or six intact recipient mice. Upon detection of palpable tumors (50 mm3), tumors volumes were measured and plotted. (C) Histology of NICD1/kRasG12D, NICD/myrAKT, and NICD1/Myc tumors recovered from intact or castrated recipients. (Scale bars: 100 microns.)
To establish the capacity of NICD1/kRasG12D, NICD1/myrAKT, and NICD1/Myc tumors to resist androgen deprivation, cancer cells were implanted into castrated SCID mouse recipients. No significant difference in size and histology was seen in the NICD1/kRasG12D, NICD1/myrAKT, and NICD1/Myc secondary tumors recovered from intact or castrated recipients (Fig. 6B and Fig. S10 B and C). These results establish that NICD1 in combination with the activation of other pathways promotes the development of prostate cancer and the progression to CRPC.
Loss of Notch1 and Pharmacological Inhibition of γ-Secretase Delay Prostate Cancer Cell and Tumor Growth.
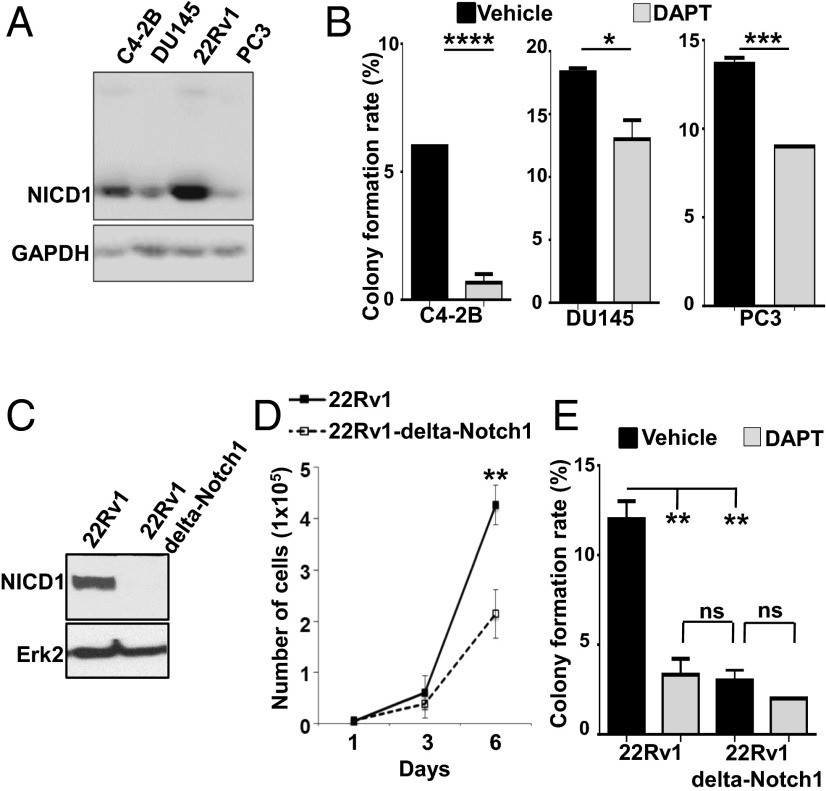
The γ-secretase complex cleaves Notch cell-surface receptors within the transmembrane domains, leading to the release of the NICD from the membrane to the nucleus, where it is referred to as “activated Notch” (13–21). To evaluate the therapeutic potential of Notch1 inhibition, we first used the γ-secretase inhibitor (GSI) (S)-tert-butyl 2-((S)-2-(2-(3,5-difluorophenyl)acetamido)propanamido)-2-phenylacetate (DAPT) (GSI-IX). We tested multiple prostate cancer cell lines for expression of NICD1 (Fig. 7A). The androgen-independent prostate cancer cell lines C4-2B, DU145, 22Rv1, and PC3 express different levels of NICD1 (Fig. 7A). All four prostate cancer cell lines were subjected to a colony-formation assay in the presence of DAPT or vehicle (Fig. 7B). Treatment of C4-2B, DU145, 22Rv1, and PC3 cells with DAPT significantly inhibited cell growth as measured by colony formation in vitro (Fig. 7 B and E). Additionally, we generated a 22Rv1 Notch1-knockout cell line via CRISPR/Cas9 to demonstrate the effect of Notch1 loss on cell growth and colony formation (Fig. 7C). Deletion of Notch1 in 22Rv1 cells (22Rv1 ΔNotch1) significantly delayed cell growth (Fig. 7D). Comparable to the effect of DAPT on colony formation, the deletion of Notch1 in 22Rv1 cells led to a significant decrease in colony-formation potential. DAPT had no significant effect on cell growth in ΔNotch1 22Rv1 cells, demonstrating the specificity of the Notch1 effect on prostate cancer cell growth (Fig. 7E).
Fig. 7.
Notch1 down-regulation and γ-secretase inhibition delay prostate cancer cell growth. (A) C4-2B, DU145, 22Rv1, and PC3 cells were lysed and subjected to Western blot analysis with antibodies against NICD1 and GAPDH. (B) C4-2B, DU145, and PC3 cells were subjected to a colony-formation assay. Plated cells were treated with DMSO (vehicle) or 25 μM GSI-IX (DAPT) every 48 h for 9 d. The mean colony-formation rate is plotted. Statistical analysis was performed by Student t test. ****P < 0.0001; *** P < 0.005; *P < 0.05; ns, nonsignificant. One of the two independent experiments is shown. (C) 22Rv1 and 22Rv1 ΔNotch1 cells were lysed and subjected to Western blot analysis with antibodies against NICD1 and Erk-2. (D) 22Rv1 or 22Rv1Δ Notch1 cells (1 × 104) were plated. Cell number was counted 1, 3, and 6 d after plating. Student t test was used for statistical analysis. **P < 0.01. (E) 22Rv1 and 22Rv1 ΔNotch1 cells were subjected a colony-formation assay. Cells were treated with DMSO (vehicle) or 25 μM GSI-IX (DAPT) every 48 h for 9 d. The mean colony-formation rate is shown. Student t test was used for statistical analysis. **P < 0.01; ns, nonsignificant.
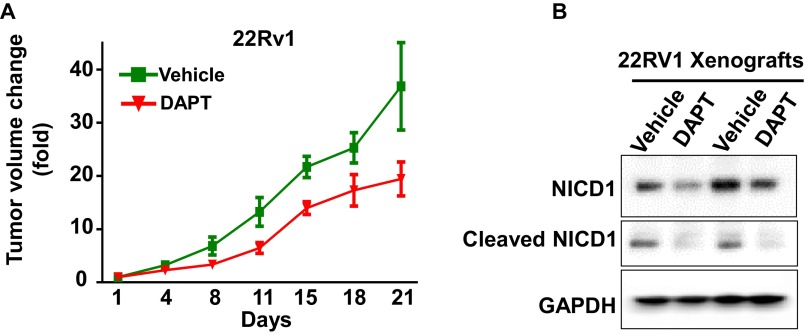
To evaluate the therapeutic potential of Notch1 inhibition in vivo, mice harboring androgen ablation-resistant 22Rv1 xenograft tumors were treated with either vehicle (n = 4) or DAPT (GSI-IX) (n = 4) at 50 mg/1 kg (Fig. S11A). Treatment was initiated upon the detection of palpable tumors, and tumor volume was assessed every 4 d after treatment induction. Treatment with DAPT delayed tumor growth of 22Rv1 xenografts. To confirm the on-target effect of DAPT in vivo, the 22Rv1 tumors were harvested and analyzed for NICD1 and cleaved Notch1 levels (Fig. S11B). These results demonstrate that loss or inhibition of Notch1 may represent a promising therapeutic strategy for CRPC.
Fig. S11.
γ-Secretase inhibition delays prostate cancer tumor growth. (A) 22Rv1 cells (1 × 106) were injected s.c. in NSG mice. Upon detection of palpable tumors, animals were treated with either vehicle (n = 4) or DAPT (GSI-IX) (n = 4) administered for 20 d according to the following schedule: 50 mg/kg DAPT for 3 d followed by 1 d off DAPT, then 50 mg/kg DAPT for 6 d and 1 d off DAPT. Day 1 is the day of treatment initiation. (B) Inhibition of Notch1 by DAPT in vivo was confirmed by Western blot for NICD1 and the cleaved Notch1 expression level.
Discussion
Defining the mechanisms that drive CRPC is critical for the development of new therapies for the lethal form of the disease. Here we demonstrate that NICD1 is localized to the nucleus in 70% of the high-risk prostate cancer and in 95% of CRPC samples analyzed. These findings suggest that nuclear NICD1 may distinguish between low- to intermediate-risk and high-risk prostate cancer and may predict prostate cancer that will recur as CRPC. Although activating mutations of Notch receptors are found in several cancers, there is no evidence of Notch1-activating mutations in advanced prostate cancer. Nuclear NICD1 in prostate cancer may arise as a consequence of Notch1 cleavage caused by elevated levels of Notch ligands, such as Jagged1, which has been reported to be overexpressed in metastasis (7). Mechanisms underlying the activation of Notch1 in advanced prostate cancer are yet to be elucidated.
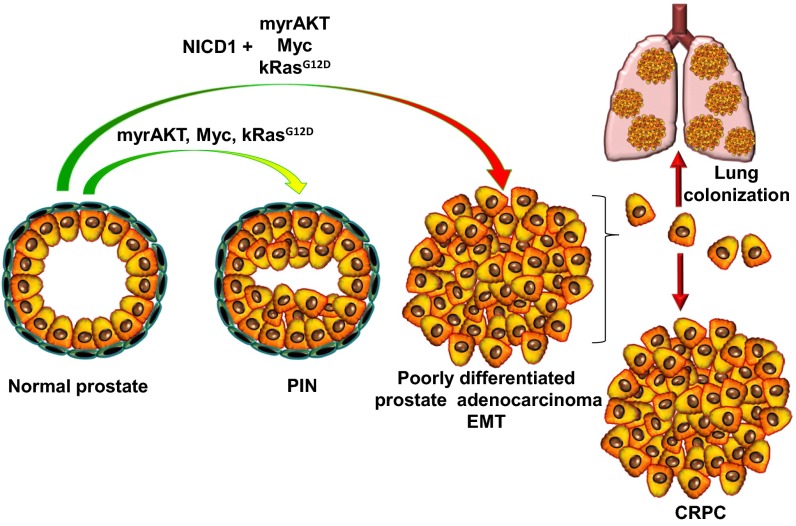
Because the nuclear localization of Notch receptors reflects their activation state, we devised a strategy to mimic the high expression of Notch1 observed in clinical metastatic CRPC by engineering the overexpression of the NICD in primary mouse prostate epithelium. NICD1 strongly synergized with a broad range of pathways commonly altered in prostate cancer, such as myrAKT, Myc, and Ras/Raf/MAPK. These combinations resulted in advanced prostate adenocarcinoma (Fig. 8). Our observation that NICD1 alone is not sufficient to drive prostate cancer initiation but instead synergizes with other pathways that can initiate only low-grade PIN lesions to drive prostate adenocarcinoma suggests that the Notch 1 receptor has a critical role in the progression of prostate cancer. These results are consistent with previous findings in osteogenic sarcoma and lung adenocarcinoma in which NICD1 strongly synergizes with p53 loss or Myc overexpression to accelerate tumor progression (49, 50).
Fig. 8.
NICD1 in combination with the myrAKT, Myc, or Ras/Raf/MAPK pathways promotes metastatic CRPC. The schematic representation summarizes our results.
Transcriptome profiling of tumors driven by NICD1 in combination with myrAKT, Myc, and Ras/Raf/MAPK revealed features of EMT. During development, Notch1 is known to drive EMT (51), a process by which epithelial cells lose their polarity and acquire mesenchymal features. These changes are accompanied by down-regulation of epithelial and adhesion genes such as E-cadherin and cytokeratins and elevated expression of mesenchymal markers such as vimentin and N-cadherin. Acquisition of an EMT phenotype is characteristic of invasive carcinoma, metastasis, and increased tumor cell self-renewal (47). In prostate cancer, down-regulation of E-cadherin is associated with poor prognosis and survival (40), whereas increased expression of the mesenchymal marker N-cadherin correlates with a high Gleason score and drives CRPC (42, 44). Consistent with the acquisition of EMT features, we found NICD1/myrAKT, NICD1/Myc, and NICD1/RasG12D tumors to be highly metastatic and capable of self-renewal activity. Notch signaling may serve a more general role in cancer stem cell maintenance in multiple tumor types (47).
Promising strategies targeting individual Notch receptors, ligand-receptor interactions, and Notch transcriptional activity have been developed and have demonstrated anticancer activity in animal models (52–54). Humanized monoclonal antibodies that block the ligand–receptor interaction between Notch and its ligand DLL4 demonstrate potent anticancer activity in patient-derived xenograft models (52). Another class of antibodies that targets individual Notch receptors, locking them in an inactive conformation, has also demonstrated promising therapeutic potential (53). Another strategy to inhibit Notch receptors is the use of GSIs that block regulated intramembrane proteolysis and subsequent Notch receptor activation (55). Our finding that Notch1 cooperates with a range of common genetic alterations in prostate cancer suggests that Notch1 inhibition may represent an effective therapy for advanced prostate cancer. Indeed, here we demonstrate that GSI and loss of Notch1 decrease prostate cancer cell growth. Additionally, GSI treatment delays the tumor growth of prostate cancer xenografts. Consistent with our findings, previous study has shown that the GSI PF-03084014 results in a significant decrease in tumor growth in two xenograft models of prostate cancer (PC3 and DU145) (12). PF-03084014 demonstrated an even greater antitumor effect in prostate cancer growth when combined with the chemotherapeutic agent docetaxel, which is currently in clinical use (12).
We provide functional evidence that NICD1 synergizes with multiple pathways in driving poorly differentiated prostate adenocarcinoma and metastatic CRPC in vivo (Fig. 8). Our results suggest that activation of Notch1 may serve as a biomarker to predict the potential benefit of Notch1 inhibition and as a therapeutic target for patients suffering from metastatic CRPC.
Materials and Methods
TMAs.
Seventy-five prostatectomy specimens from patients never treated with hormonal therapy were reviewed, and areas of normal prostate (indicated as benign tissue) and low- to intermediate-risk prostate cancer (Gleason patterns 6–7) were marked for sampling. Two cores per sample, each measuring 0.6 mm in diameter, were taken from selected regions in each paraffin block and transferred to a recipient paraffin block containing a total of 150 benign or cancer cores. Unstained sections of 4-μm thickness were used for immunohistochemical staining. Two additional TMAs were constructed containing (i) benign tissues and adjacent low- to intermediate-risk prostate cancer from 40 patients (three cores per sample) and (ii) benign tissues and adjacent cancer samples (low- to intermediate-risk and high-risk) from 115 patients. Additional CRPC TMAs were constructed from blocks containing transurethral resection tissues from 20 patients who failed androgen-ablation therapy (i.e., patients with CRPC) and who developed urinary obstruction. Two cores from each patient block were transferred to a new TMA block (CRPC TMA). A section from the TMA blocks was obtained and used for immunohistochemical staining.
Mouse Strains.
C57BL/6, CB17Scid/Scid, and NSG (NOD-SCID-IL2Rγ–null) mice were purchased from the Jackson Laboratory. Animals were housed at the University of California, Los Angeles animal facilities according to the regulations of the Division of Laboratory Animal Medicine.
Prostate Tissue Regeneration Assays.
Housing, breeding, and all surgical procedures were performed in agreement with the guidelines of the Division of Laboratory Animal Medicine of the University of California, Los Angeles. All experimental procedures were approved by the Division of Laboratory Animal Medicine of the University of California, Los Angeles. The details of the regeneration process have been explained previously (56).
Lung Colonization Assay.
NICD1/Myc and NICD1/RasG12D secondary tumors were excised, minced, and incubated in DMEM containing 10% FBS, 1 mg/mL collagenase, and 1 mg/mL Dispase for 1 h at 37 °C. To achieve single-cell dissociation, tumor chucks were spun down, further treated with 0.05% Trypsin-EDTA for 5 min at 37 °C, passed through a 20-gauge syringe, and filtered through a 70-μm filter mesh. Cells were cultured for 2 wk followed by infection with lentivirus-expressing luciferase and YFP. Mice were injected with 8 × 105 NICD1/Myc, NICD1/RasG12D, and Myc-CAP cells expressing luciferase and YFP via tail vein injection. Thirty minutes after injection, animals were injected i.p. with 150 μL of 15 mg/mL luciferin followed by bioluminescence imaging with the IVIS Lumina II imaging system.
RNA Sequencing.
RNA was extracted from mouse tumor tissue using the RNeasy Mini Kit from Qiagen. Libraries for RNA-sequencing were prepared with NuGen Ovation RNA-Seq System V2. The workflow consists of double-stranded cDNA generation using a mixture of random and poly (T) priming, fragmentation of double-stranded cDNA, end repair to generate blunt ends, A-tailing, adaptor ligation, and PCR amplification. Different adaptors were used for multiplexing samples in one lane. Sequencing was performed on an Illumina HiSeq 2500 for a pair-read 100-nt run. Data quality control was done on an Illumina Sequencing Analysis Viewer. Demultiplexing was performed with Illumina CASAVA 1.8.2. The reads were first mapped to the latest University of California, Santa Cruz transcript set using Bowtie2 version 2.1.0, and the gene-expression level was estimated using RSEM v1.2.15. TPM (transcript per million) was used as the normalized gene expression. Differentially expressed genes were identified using the DESeq package. Genes showing altered expression with a false-discovery rate P < 0.05 and more than twofold changes were considered differentially expressed.
Vector Production and Viral Packaging.
The third-generation lentiviral vectors FUCGW and FUCRW, derived from FUGW, were used for the construction of the human NICD1, kRasG12D, myrAKT, and Myc constructs.
Antibodies.
The following antibodies were used: anti-Notch1 [Novus Biologicals 3E12; Abcam ab52627; Cell Signaling Technology C37C7; Cell Signaling Technology (Val1744) (D3B8)]; anti-PCNA (Cell Signaling), anti–AR-N-20 (Santa Cruz), anti-p63 (Santa Cruz), and anti-AKT (Cell Signaling).
Immunohistochemistry.
Indicated tissues were fixed in 10% buffer and were embedded in formalin and paraffin. Four-micron sections were deparaffinized in xylene and rehydrated in 100, 95, and 70% ethanol. Antigen retrieval was performed with citrate buffer (pH 6.0) at 95 °C for 20 min. Sections were blocked using mouse-on-mouse blocking reagents (BMK-2202; Vector Labs). The sections were incubated with the indicated antibodies overnight. Slides were washed with 1× PBS and incubated with anti-mouse HRP or anti-rabbit HRP antibodies (DAKO) for 1 h, developed with HRP substrate (DAKO), and counterstained with hematoxylin.
Colony-Formation Assay.
Five hundred C4-2B, DU145, 22Rv1, and PC3 cells per well were plated in triplicate in a six-well plate. Cells were treated with DMSO (vehicle) or 25 μM DAPT (GSI-IX; Selleck Chemicals LLC) every 48 h. Nine days after plating colonies were fixed with ice-cold methanol for 30 min at −20 °C and were stained with 0.1% crystal violet in 1× PBS for 30 min at room temperature. Plates were washed for 1 h with water. Colonies were counted, and the colony formation rate (%) was calculated as the number of colonies per 500 cells × 100.
Generation of ΔNotch1 22Rv1 Cell Lines.
The 5′ UTR and first exons of Notch1 were used to design guide RNAs. Sequences were loaded on crispr.mit.edu. A sequence spanning the ATG was identified (guide #1 Notch1 CRISPR forward CACCGGGGAGGCATGCCGCCGCTCC; guide #1 Notch CRISPR reverse AAACGGAGCGGCGGCATGCCTCCCC). Overhangs appropriate for cloning onto BbsI sites were added. Oligos were annealed and cloned into the PX458 vector. pSpCas9(BB)-2A-GFP (PX458) (Addgene plasmid no. 48138) was a gift from Feng Zhang, Broad Institute of MIT and Harvard, Cambridge, MA; McGovern Institute for Brain Research, Cambridge, MA; and Departments of Brain and Cognitive Sciences and Biological Engineering, MIT, Cambridge, MA (57). 22RV1 cells (5 × 105 cells per well in 2 mL 10% DMEM in a six-well plate) were transfected with 5 μg DNA using Lipofectamine 2000. GFP+ cells were sorted by FACS 24 h after transfection and were plated in a 96-well plate at four cells per well. Cells were grown until wells with single clones were 50% confluent (4 wk). Loss of Notch1 was confirmed by Western blot.
In Vivo Studies with GSI.
NSG mice (8–10 wk old) were injected s.c. in the flank with 1 × 106 22Rv1 cells in Matrigel. When tumors reached palpable size (50 mm3), animals were randomly assigned to treatment with vehicle (control) or with DAPT (50 mg/kg GSI-IX). Vehicle and DAPT were dissolved in 10% ethanol and 90% corn oil and were administered by oral gavage on the following schedule: 3 d on, 1 d off, 6 d on, and 1 d off. The schedule was repeated two times. The length (L), width (W), and height (H) of tumors were measured every 4 d. Tumor volume was calculated by (L × W × H)/2.
Acknowledgments
We thank the University of California, Los Angeles (UCLA) Tissue Procurement Core Laboratories for tissue preparation. T.S. is supported by a Prostate Cancer Foundation Young Investigator Award and NIH/National Cancer Institute (NCI) K99/R00 Pathway to Independence Award 4R00CA184397. C.M.F. is supported by a UCLA Tumor Cell Biology Training Grant. S.L. is supported in part by the American Urological Association/Urology Care Foundation Research Scholar Award Program. B.A.S. is supported by UCLA Tumor Immunology Training Grant T32-CA009120. J.K.L. is supported by the Specialty Training and Advanced Research (STAR) Program at UCLA, a Prostate Cancer Foundation Young Investigator Award, and a UCLA Clinical and Translational Science Institute-KL2 Award KL2TR001882 (principal investigator: Mitchell Wong) from the NIH/National Center for Advancing Translational Sciences. J.M.D. is supported by Department of Defense Prostate Cancer Research Program Grant W81XWH-15-1-0236 and a Prostate Cancer Foundation Young Investigator Award. J.H. is supported by NIH/NCI Grant P50CA092131 [to the UCLA Specialized Program of Research Excellence (SPORE) in prostate cancer; principal investigator: R.E.R.], a Creativity Award from the Prostate Cancer Foundation (PCF) (principal investigator: M. Rettig), and NIH Grant 1R01CA158627 (principal investigator: L. Marks). A.S.G. is supported by the Department of Defense and a Prostate Cancer Foundation Young Investigator Award. O.N.W. is an Investigator of the Howard Hughes Medical Institute and is partly supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research. K.J.P. is supported by a PCF Challenge Award and by the NIH. R.E.R. is supported by NIH/NCI Grant P50CA092131 (to the UCLA SPORE in prostate cancer). A.S.G and O.N.W. are supported by a PCF Creativity Award (principal investigator: O.N.W.). A.S.G, J.H., and O.N.W. are supported by a PCF Challenge Award (principal investigator: O.N.W.) and the UCLA SPORE in prostate cancer (principal investigator: R.E.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE87448).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614529113/-/DCSupplemental.
References
- 1.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 2.Kantoff PW, et al. IMPACT Study Investigators Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363(5):411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 3.Tannock IF, et al. TAX 327 Investigators Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351(15):1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 4.Wang XD, et al. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol. 2006;290(1):66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Kwon OJ, et al. Increased Notch signalling inhibits anoikis and stimulates proliferation of prostate luminal epithelial cells. Nat Commun. 2014;5:4416. doi: 10.1038/ncomms5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valdez JM, et al. Notch and TGFβ form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell Stem Cell. 2012;11(5):676–688. doi: 10.1016/j.stem.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerhardt DM, et al. The Notch1 transcriptional activation domain is required for development and reveals a novel role for Notch1 signaling in fetal hematopoietic stem cells. Genes Dev. 2014;28(6):576–593. doi: 10.1101/gad.227496.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santagata S, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64(19):6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H, Zhou X, Redfield S, Lewin J, Miele L. Elevated Jagged-1 and Notch-1 expression in high grade and metastatic prostate cancers. Am J Transl Res. 2013;5(3):368–378. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109(4):726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 11.Domingo-Domenech J, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22(3):373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui D, et al. Notch pathway inhibition using PF-03084014, a γ-Secretase inhibitor (GSI), enhances the antitumor effect of docetaxel in prostate cancer. Clin Cancer Res. 2015;21(20):4619–4629. doi: 10.1158/1078-0432.CCR-15-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393(6683):382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 15.Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12(23):3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93(4):649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 17.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 18.Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19(6):781–791. doi: 10.1016/s1074-7613(03)00325-x. [DOI] [PubMed] [Google Scholar]
- 19.Allman D, Punt JA, Izon DJ, Aster JC, Pear WS. An invitation to T and more: Notch signaling in lymphopoiesis. Cell. 2002;109(Suppl):S1–S11. doi: 10.1016/s0092-8674(02)00689-x. [DOI] [PubMed] [Google Scholar]
- 20.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 21.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: A centennial for notch signaling. Cancer Cell. 2014;25(3):318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rampias T, et al. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20(10):1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 2011;108(43):17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 27.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 28.Girard L, et al. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10(15):1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 29.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westhoff B, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci USA. 2009;106(52):22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: It’s NOTCH what you think. J Exp Med. 2011;208(10):1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubin MA, et al. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6(3):1038–1045. [PubMed] [Google Scholar]
- 35.Mehra R, et al. Characterization of bone metastases from rapid autopsies of prostate cancer patients. Clin Cancer Res. 2011;17(12):3924–3932. doi: 10.1158/1078-0432.CCR-10-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoyanova T, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci USA. 2013;110(50):20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray IC, et al. Mutation and expression analysis of the putative prostate tumour-suppressor gene PTEN. Br J Cancer. 1998;78(10):1296–1300. doi: 10.1038/bjc.1998.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci USA. 2003;100(1) Suppl 1:11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaggi M, et al. N-cadherin switching occurs in high Gleason grade prostate cancer. Prostate. 2006;66(2):193–199. doi: 10.1002/pros.20334. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka H, et al. Monoclonal antibody targeting of N-cadherin inhibits prostate cancer growth, metastasis and castration resistance. Nat Med. 2010;16(12):1414–1420. doi: 10.1038/nm.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gravdal K, Halvorsen OJ, Haukaas SA, Akslen LA. A switch from E-cadherin to N-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin Cancer Res. 2007;13(23):7003–7011. doi: 10.1158/1078-0432.CCR-07-1263. [DOI] [PubMed] [Google Scholar]
- 45.Lang SH, et al. Enhanced expression of vimentin in motile prostate cell lines and in poorly differentiated and metastatic prostate carcinoma. Prostate. 2002;52(4):253–263. doi: 10.1002/pros.10088. [DOI] [PubMed] [Google Scholar]
- 46.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson PA, et al. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer Res. 2005;65(24):11565–11571. doi: 10.1158/0008-5472.CAN-05-3441. [DOI] [PubMed] [Google Scholar]
- 49.Tao J, et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell. 2014;26(3):390–401. doi: 10.1016/j.ccr.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res. 2011;71(18):6010–6018. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takebe N, Nguyen D, Yang SX. Targeting notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol Ther. 2014;141(2):140–149. doi: 10.1016/j.pharmthera.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444(7122):1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 56.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5(4):702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]