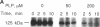Abstract
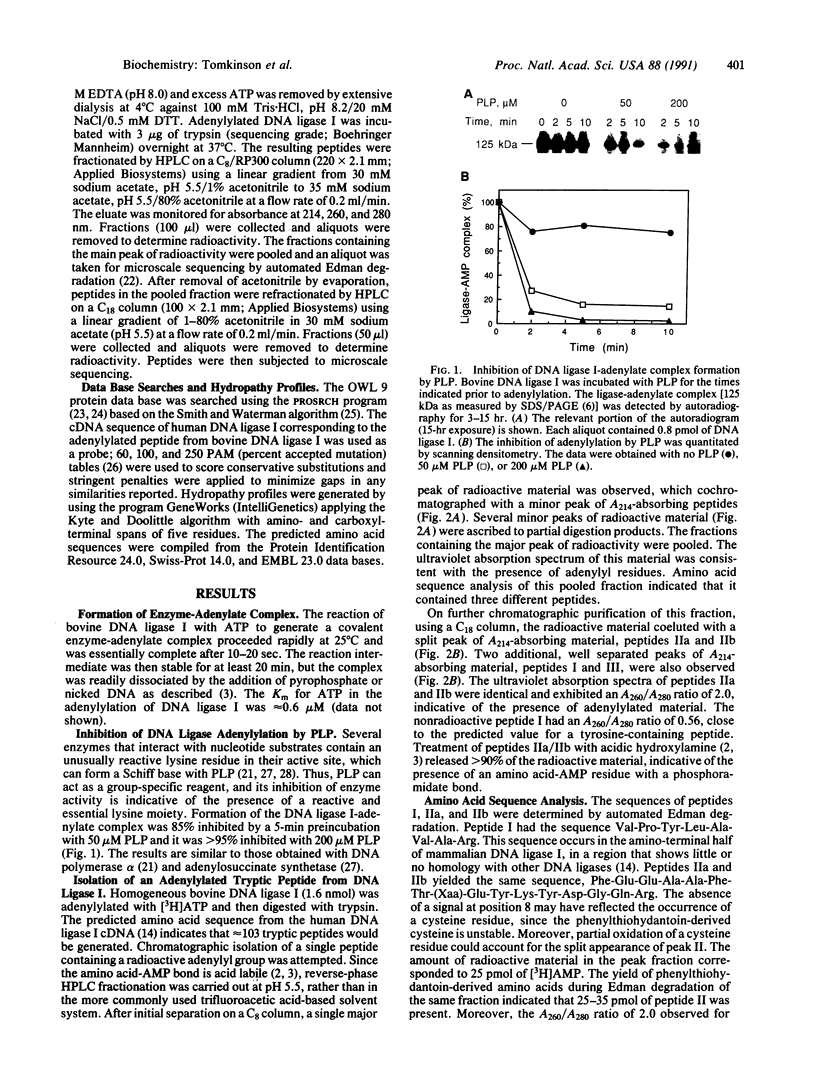
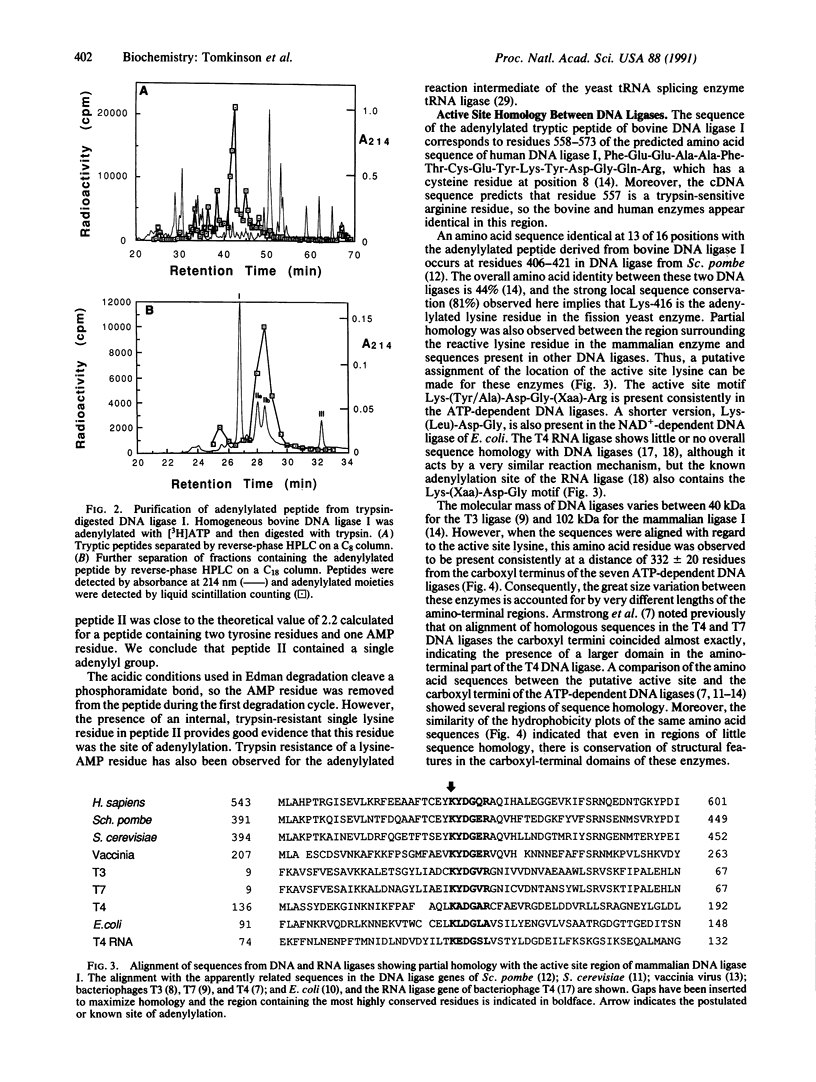
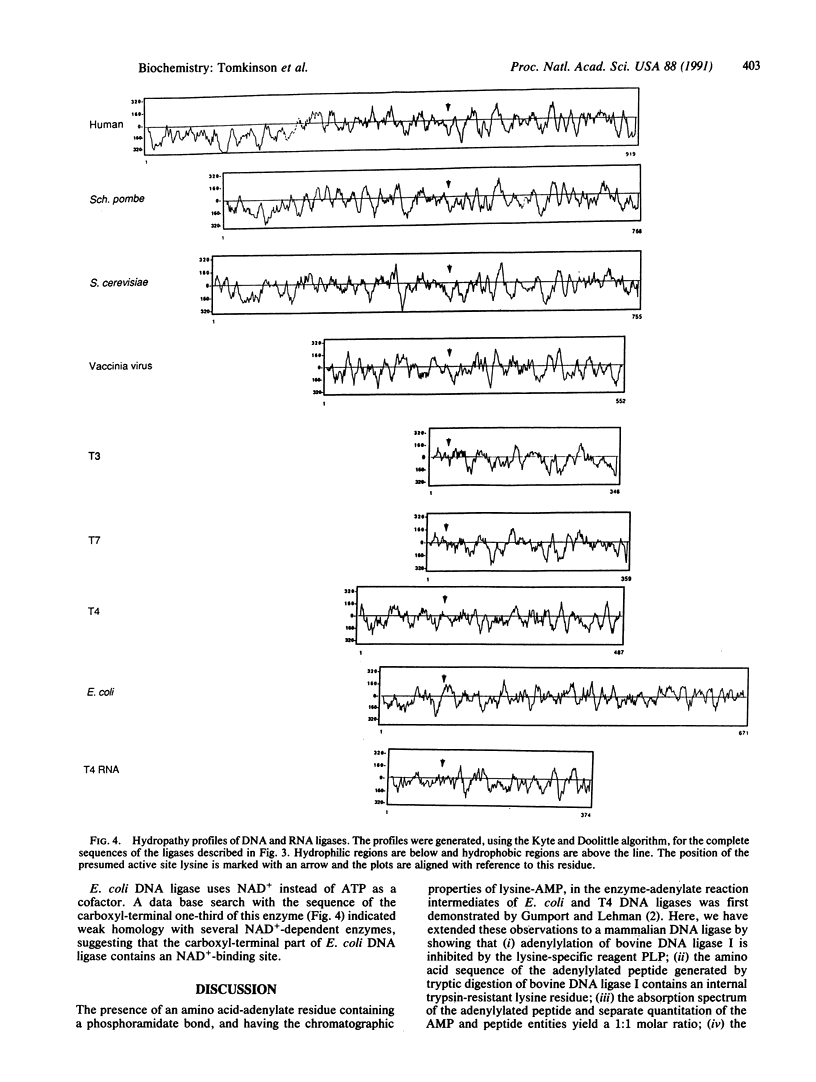
The enzyme-AMP reaction intermediate of the 102-kDa bovine DNA ligase I was digested with trypsin, and the adenylylated peptide was isolated by chromatography under conditions that maintain the acid-labile phosphoramidate bond. Microsequencing of the peptide showed that it contains an internal trypsin-resistant lysine residue, as expected for the site of adenylylation. Inhibition of DNA ligase I activity by pyridoxal 5'-phosphate also indicated the presence of a reactive lysine residue in the catalytic domain of the enzyme. Comparison of the known primary structures of several other DNA ligases with the adenylylated region of mammalian DNA ligase I allows their active sites to be tentatively assigned by sequence homology. The ATP-dependent DNA ligases of mammalian cells, fission yeast, budding yeast, vaccinia virus, and bacteriophages T3, T4, and T7 contain the active site motif Lys-Tyr/Ala-Asp-Gly-(Xaa)-Arg, with the reactive lysine residue flanked by hydrophobic amino acids. The distance between the postulated adenylylation site and the carboxyl terminus of the polypeptide is very similar in these ATP-dependent DNA ligases, whereas the size of the amino-terminal region is highly variable.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J., Brown R. S., Tsugita A. Primary structure and genetic organization of phage T4 DNA ligase. Nucleic Acids Res. 1983 Oct 25;11(20):7145–7156. doi: 10.1093/nar/11.20.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. Molecular characterisation of the DNA ligase gene, CDC17, from the fission yeast Schizosaccharomyces pombe. Eur J Biochem. 1987 Feb 2;162(3):659–667. doi: 10.1111/j.1432-1033.1987.tb10688.x. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. E., Johnston L. H., Kodama K., Tomkinson A. E., Lasko D. D., Lindahl T. Human DNA ligase I cDNA: cloning and functional expression in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Collins J. F., Coulson A. F., Lyall A. The significance of protein sequence similarities. Comput Appl Biosci. 1988 Mar;4(1):67–71. doi: 10.1093/bioinformatics/4.1.67. [DOI] [PubMed] [Google Scholar]
- Diffley J. F. Affinity labeling the DNA polymerase alpha complex. I. Pyridoxal 5'-phosphate inhibition of DNA polymerase and DNA primase activities of the DNA polymerase alpha complex from Drosophila melanogaster embryos. J Biol Chem. 1988 Oct 15;263(29):14669–14677. [PubMed] [Google Scholar]
- Diffley J. F. Affinity labeling the DNA polymerase alpha complex. Identification of subunits containing the DNA polymerase active site and an important regulatory nucleotide-binding site. J Biol Chem. 1988 Dec 15;263(35):19126–19131. [PubMed] [Google Scholar]
- Dong Q., Fromm H. J. Chemical modification of adenylosuccinate synthetase from Escherichia coli by pyridoxal 5'-phosphate. Identification of an active site lysyl residue. J Biol Chem. 1990 Apr 15;265(11):6235–6240. [PubMed] [Google Scholar]
- Downward J., Yarden Y., Mayes E., Scrace G., Totty N., Stockwell P., Ullrich A., Schlessinger J., Waterfield M. D. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984 Feb 9;307(5951):521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol. 1981 Jun 5;148(4):303–330. doi: 10.1016/0022-2836(81)90178-9. [DOI] [PubMed] [Google Scholar]
- Gumport R. I., Lehman I. R. Structure of the DNA ligase-adenylate intermediate: lysine (epsilon-amino)-linked adenosine monophosphoramidate. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2559–2563. doi: 10.1073/pnas.68.10.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy S., Singh M., Gait M. J. Effect of single amino acid changes in the region of the adenylylation site of T4 RNA ligase. Biochemistry. 1987 Mar 24;26(6):1688–1696. doi: 10.1021/bi00380a030. [DOI] [PubMed] [Google Scholar]
- Hinz H. R., Kirley T. L. Lysine 480 is an essential residue in the putative ATP site of lamb kidney (Na,K)-ATPase. Identification of the pyridoxal 5'-diphospho-5'-adenosine and pyridoxal phosphate reactive residue. J Biol Chem. 1990 Jun 25;265(18):10260–10265. [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Tsunasawa S., Sakiyama F., Nakata A. Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol Gen Genet. 1986 Jul;204(1):1–7. doi: 10.1007/BF00330179. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Panasenko S. M., Alazard R. J., Lehman I. R. A simple, three-step procedure for the large scale purification of DNA ligase from a hybrid lambda lysogen constructed in vitro. J Biol Chem. 1978 Jul 10;253(13):4590–4592. [PubMed] [Google Scholar]
- Rand K. N., Gait M. J. Sequence and cloning of bacteriophage T4 gene 63 encoding RNA ligase and tail fibre attachment activities. EMBO J. 1984 Feb;3(2):397–402. doi: 10.1002/j.1460-2075.1984.tb01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. P., Beck P. J., Kearney C. A., Spence J. L., DiGiovanni D., Condreay J. P., Molineux I. J. Sequence of a conditionally essential region of bacteriophage T3, including the primary origin of DNA replication. J Mol Biol. 1987 Feb 5;193(3):479–495. doi: 10.1016/0022-2836(87)90261-0. [DOI] [PubMed] [Google Scholar]
- Smith G. L., Chan Y. S., Kerr S. M. Transcriptional mapping and nucleotide sequence of a vaccinia virus gene encoding a polypeptide with extensive homology to DNA ligases. Nucleic Acids Res. 1989 Nov 25;17(22):9051–9062. doi: 10.1093/nar/17.22.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. Mammalian DNA ligases. Serological evidence for two separate enzymes. J Biol Chem. 1975 Nov 10;250(21):8438–8444. [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. Mammalian deoxyribonucleic acid ligase. Isolation of an active enzyme-adenylate complex. J Biol Chem. 1973 Jan 25;248(2):672–675. [PubMed] [Google Scholar]
- Teraoka H., Tsukada K. Eukaryotic DNA ligase. Purification and properties of the enzyme from bovine thymus, and immunochemical studies of the enzyme from animal tissues. J Biol Chem. 1982 May 10;257(9):4758–4763. [PubMed] [Google Scholar]
- Thøgersen H. C., Morris H. R., Rand K. N., Gait M. J. Location of the adenylylation site in T4 RNA ligase. Eur J Biochem. 1985 Mar 1;147(2):325–329. doi: 10.1111/j.1432-1033.1985.tb08753.x. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Lasko D. D., Daly G., Lindahl T. Mammalian DNA ligases. Catalytic domain and size of DNA ligase I. J Biol Chem. 1990 Jul 25;265(21):12611–12617. [PubMed] [Google Scholar]
- Xu Q., Teplow D., Lee T. D., Abelson J. Domain structure in yeast tRNA ligase. Biochemistry. 1990 Jul 3;29(26):6132–6138. doi: 10.1021/bi00478a004. [DOI] [PubMed] [Google Scholar]
- Yang S. L., Frey P. A. Nucleophile in the active site of Escherichia coli galactose-1-phosphate uridylyltransferase: degradation of the uridylyl-enzyme intermediate to N3-phosphohistidine. Biochemistry. 1979 Jul 10;18(14):2980–2984. doi: 10.1021/bi00581a011. [DOI] [PubMed] [Google Scholar]