Abstract
Kallistatin is an endogenous protein that regulates differential signaling pathways and biological functions. Our previous studies showed that kallistatin gene therapy inhibited angiogenesis, tumor growth and metastasis in mice, and kallistatin protein suppressed Wnt-mediated growth, migration and invasion by blocking Wnt/β-catenin signaling pathway in breast cancer cells. In this study, we show that kallistatin reduced cell viability, and increased apoptotic cell death and caspase-3 activity in MDA-MB-231 breast cancer cells. Kallistatin also induced cancer cell autophagy, as evidenced by increased LC3B levels and elevated Atg5 and Beclin-1 expression; however, co-administration of Wnt or PPARγ antagonist GW9662 abolished these effects. Moreover, kallistatin via its heparin-binding site antagonized Wnt3a-induced cancer cell proliferation and increased PPARγ expression. Kallistatin inhibited oncogenic miR-21 synthesis associated with reduced Akt phosphorylation and Bcl-2 synthesis, but increased BAX expression. Kallistatin via PKC-ERK activation reduced miR-203 levels, leading to increased expression of suppressor of cytokine signaling 3 (SOCS3), a tumor suppressor. Conversely, kallistatin stimulated expression of the tumorigenic suppressors miR-34a and p53. Kallistatin’s active site is essential for suppressing miR-21 and miR-203, and stimulating miR-34a and SOCS3 expression. This is the first study to demonstrate that kallistatin’s heparin-binding site is essential for inhibiting Wnt-mediated effects, and its active site plays a key role in regulating miR-21, miR-203, miR-34a and SOCS3 synthesis in breast cancer cells. These findings reveal novel mechanisms of kallistatin in inducing apoptosis and autophagy in breast cancer cells, thus inhibiting tumor progression by regulation of Wnt/PPARγ signaling, as well as miR-21, miR-203 and miR-34a synthesis.
Keywords: Kallistatin, MicroRNAs, Breast cancer, Wnt signaling, Apoptosis, Autophagy
1. Introduction
Breast cancer is a common malignancy and the second leading cause of cancer mortality in women [1]. The major treatment for breast cancer is surgery and postoperative adjuvant chemotherapy. However, the prognosis remains poor for a significant population of patients due to the adverse side effects of chemotherapy and drug resistance [2–4]. Recently, numerous evidences have indicated that microRNAs (miRNAs) are key players in cancer biology [5]. miRNAs are endogenous non-coding RNA molecules that regulate one-third of all human protein-coding genes and affect a wide variety of biological processes, including proliferation, differentiation, cell fate determination, apoptosis, organ injury and cancer [6,7]. miR-21 is one of the most common miRNAs associated with cancer patient outcome and has emerged as a novel molecular target for cancer therapy [8]. Indeed, the oncogenic miR-21 is overexpressed in many types of tumors [9]. In breast cancer, miR-21 is significantly increased and associated with patients’ poor survival [10,11]. Bcl-2, a key regulator of apoptosis in many types of human tumors, is positively regulated by miR-21 [12]. Moreover, miR-203 was found to be overexpressed in human breast cancer, while knockdown of miR-203 sensitized MCF-7 breast cancer cells to cisplatin-mediated apoptotic cell death by up-regulation of suppressor of cytokine signaling 3 (SOCS3) [13]. Conversely, miR-34a is a critical tumor suppressor in many types of cancers, including breast cancer [14–17]. miR-34a, which is regulated by the p53 network, is remarkably diminished in many types of human cancers [14,18]. Overexpression of miR-34a in breast cancer cells induced cell apoptosis and inhibited cell proliferation and migration by targeting Bcl-2 and various cyclins [17–19]. These findings indicate that miR-21, miR-203 and miR-34a play a vital role in cancer progression, either as an oncogene or a tumor suppressor.
Kallistatin, discovered in human plasma, is a tissue kallikrein-binding protein (KBP) and a unique serine proteinase inhibitor [20–22]. Plasma levels of kallistatin are reduced in patients with sepsis, liver disease and obesity [23,24]. Kallistatin gene or protein administration exerts multi-factorial properties by inhibiting inflammation, angiogenesis, oxidative stress, apoptosis, tumor growth and metastasis in animal models and cultured cells [25–32]. Local administration of the human kallistatin gene reduced breast tumor growth and angiogenesis in nude mice via antagonizing VEGF-mediated cell proliferation, migration and invasion of cultured endothelial cells [26,27]. Systemic injection of lenti-virus carrying the human kallistatin gene dramatically decreased cancer metastasis into lungs in association with reduced angiogenesis and inflammation, and enhanced survival of tumor-bearing mice [32]. SPTBN1 (β II-spectrin) has been shown to suppress the progression of hepatocellular carcinoma and Wnt signaling by regulation of kallistatin [33]. Moreover, kallistatin antagonizes the Wnt3a signaling pathway by forming a complex with Wnt co-receptor low-density lipoprotein receptor 6 (LRP6) in breast cancer cells [34]. Thus, kallistatin is capable of inhibiting Wnt-induced motility and invasion of breast cancer cells [34]. Furthermore, kallistatin (KBP) induced apoptotic cell death in human colorectal cancer cells [35]. As miR-21, miR-203 and miR-34a have been shown to play crucial roles in promoting or inhibiting tumor development, the aim of this study was to investigate potential mechanisms of kallistatin in programmed cell death by modulation of these miRNAs in breast cancer cells.
2. Materials and methods
2.1. Purification and characterization of recombinant human kallistatin
Recombinant human kallistatin was secreted into the serum-free medium of cultured human embryonic kidney cells (HEK293T). Cultured medium was then concentrated by ammonium sulfate precipitation followed by nickel-affinity chromatography [36]. Recombinant wild-type, heparin-binding site mutant and active site mutant kallistatin were expressed in Escherichia coli and purified as described [37]. The purity and identity of human kallistatin were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot using a specific monoclonal antibody [23,36].
2.2. Cell culture and treatment
Human MDA-MB-231 breast cancer cells were maintained at 37°C in a humidified atmosphere of 5% CO2 in 95% air, and grown in 1640 medium supplemented with 100 U/mL of penicillin, 100 g/mL of streptomycin and 10% fetal bovine serum. For protein expression studies, cells were starved overnight and treated with 1 µM kallistatin or media alone for 24 h. Phospho-Akt (Cell Signaling, Danvers, MA) was determined by western blot. For mRNA and miRNA expression, cells were starved overnight and treated with 1 µM kallistatin or media alone for 12 h. Total RNA was then extracted. The mRNA levels of Bcl-2, BAX, p53, Atg5 and Beclin-1, and miRNA levels of miR-21 and miR-34a were measured by real-time reverse transcription-polymerase chain reaction (RT-PCR). To determine the role of Wnt3a or PPARγ antagonist GW9662 (R&D systems, Minneapolis, MN) on kallistatin-mediated effect on PPARγ or autophagy-related gene expression, cells were pretreated with Wnt3a (200 ng/ml) or PPARγ antagonist GW9662 (10 µM) for 30 min and further incubated with 1 µM kallistatin or media alone for 24 h. Total RNA was then extracted. The mRNA levels of PPARγ, Atg5 and Beclin-1 were measured by real-time RT-PCR. To determine the effect of chelerythrine, a protein kinase C (PKC) inhibitor, on kallistatin-mediated phospho-ERK activation and levels of SOCS3 and miR-203, cells were starved overnight and pretreated with chelerythrine (1 µM) for 30 min followed by stimulation with 1 µM kallistatin for 12 or 24 h. Levels of SOCS3 and miR-203 were measured by real-time RT-PCR. Phospho-ERK was determined by western blot. The role of kallistatin’s active site in kallistatin-mediated apoptosis-related gene expression was also evaluated by starving cells overnight and incubating with 2 µM wild-type kallistatin, heparin-binding site mutant kallistatin or active site mutant kallistatin in the presence of polymyxin B (10 µg/ml) for 12 h. Total RNA was extracted and levels of PPARγ, miR-21, miR-203, miR-34a and SOCS3 were measured by real-time RT-PCR.
2.3. Cell viability assay
Cell viability was determined using 3-[4,5-dimethylthiazol-2-yl]-2,5-dephenyl tetrazolium bromide (MTT; Sigma Chemical Co., St Louis, MO) assay. Cells were seeded in 96-well plates and treated with different concentrations of kallistatin (0.5, 1 and 2 µM) for 72 h. Each well was supplemented with 20 µl of 5 mg/mL MTT and incubated for 4 h. The medium was then removed and the MTT formazan was solubilized with 150 µl DMSO. The optical density was measured at 490 nm using a microplate ELISA reader. The experiment was repeated twice and each experiment had five replicate wells. For 5-bromo-20-deoxyuridine (BrdU) assay, MDA-MB-231 cells were serum-deprived, pretreated with 2 µM wild-type kallistatin, heparin-binding site mutant kallistatin or active site mutant kallistatin in the presence of polymyxin B (10 µg/ml) for 30 min, and then cultured with Wnt3a (200 ng/ml) for another 24 h. BrdU colorimetric assay was performed according to the manufacturer’s instructions (Roche Applied Science, Indianapolis, IN).
2.4. Apoptosis assay by Hoechst 33342 and TUNEL staining
MDA-MB-231 cells were treated with different concentrations of kallistatin (1 and 2 µM) for 48 h. Following treatment, the cells were incubated with 1 µg/mL Hoechst 33342 for 15 min at 37 °C and examined by fluorescence microscopy. TUNEL staining was performed using a TUNEL kit according to the manufacturer’s instructions (Roche, Indianapolis, IN).
2.5. Caspase-3 activity assay
Caspase-3 activity was determined with a caspase-3 colorimetric activity assay kit (EMD Millipore Corporation, Temecula, CA) according to the manufacturer’s instructions. Briefly, cells were treated with 1 µM kallistatin for 72 h and lysed in lysis buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 0.1% CHAPS, 1 mM dithiothreitol, 0.1 mM EDTA) on ice for 10 min, then centrifuged for collection of the supernatants. Supernatants were transferred to a fresh tube and the protein concentration for each sample was measured. Equal volumes of the lysates were incubated with assay mixture (including colorimetric substrate) at 37 °C for 1 h. The fluorescent intensity (405 nm) was recorded.
2.6. Immunofluorescence staining
Primary antibody for LC3B (Cell Signaling, Danvers, MA) was used to detect its protein levels with fluorescence microscopy. Briefly, cells were grown and treated with kallistatin (1 µM) for 48 h. At the end of treatment, cells were fixed with 4% formaldehyde diluted in PBS for 15 min at room temperature. Cells were then permeabilized with 0.2% Triton X-100 and blocked with 3% BSA in PBS for 1 h at room temperature, followed by incubation with primary antibody overnight at 4 °C. At the end of incubation, cells were rinsed three times with PBS and incubated with fluorochrome-conjugated secondary antibody (1:400) for 2 h at room temperature in the dark. Cells were then washed with PBS and observed under a fluorescence microscope.
2.7. Real-time RT-PCR
Total RNA was extracted from cells with TRIzol Reagent (Invitrogen, Calsbad, CA) following the manufacturer’s protocol. cDNA was synthesized with High Capacity cDNA Reverse Transcription Kit or MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Quantitative real-time PCR was performed by Prism 7300 Real Time PCR System (Applied Biosystems) using TaqMan Gene Expression Master Mix (Applied Biosystems) in a final reaction volume of 20 µl with each primer. The following human primers were purchased from Applied Biosystems: 18S (Hs 99999901_sl), tp53 (Hs01034249_m1), Bcl-2 (Hs00236808_s1), BAX (Hs00180269_m1), Atg5 (Hs00169468_m1), Beclin-1 (Hs00387943_m1), PPARγ (Hs00234592_m1), SOCS3 (Hs02330328_s1), U6 snRNA (001973), hsa-miR-203b-3p (464535-mat), hsa-miR-34a-3p (002316), and hsa-miR-21-3p (002438). A negative control without cDNA did not produce any amplicons. Data were analyzed with 2−ΔΔCt value calculation using 18S or U6 RNA for normalization.
2.8. Western blot analysis
Cells were lysed with ice-cold RIPA lysis buffer. All lysed samples were kept on ice for 30 min and centrifuged for 10 min at 4 °C at 12,000 g. The supernatant was collected and stored at −20 °C until further analysis. Cell lysates were subjected to 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The membranes were blocked with 7% milk in TBST (20 mM Tris, 500 mM NaCl, and 0.1% Tween 20) for 1 h. After washing with TBST twice, membranes were incubated with primary antibody overnight at 4 °C. The following primary antibodies were used: polyclonal anti-phospho-p44/42 MAPK (ERK1/2), polyclonal anti-p44/42 MAPK (ERK1/2), monoclonal anti-phospho-Akt, and monoclonal anti-Akt (Cell Signaling, Boston, MA). The membranes were washed twice with TBST and incubated with HRP conjugated secondary antibody in blocking buffer for 1 h. After washing three times with TBST, immunoreactive bands were visualized by incubation with ECL plus detection reagents (GE Healthcare, Waukesha, WI) for 5 min and exposed to BioMax light film (Thermo Scientific, Waltham, MA). The densitometry of bands was quantified with ImageJ2 software.
2.9. Statistical analysis
Data are expressed as mean ± standard error of the mean (SE). Statistical significance was determined by analysis of variance (ANOVA) with Fisher’s probability least-squares difference test or Student’s t-test using GraphPad Prism software. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. Kallistatin reduces viability and increases apoptotic cell death in breast cancer cells
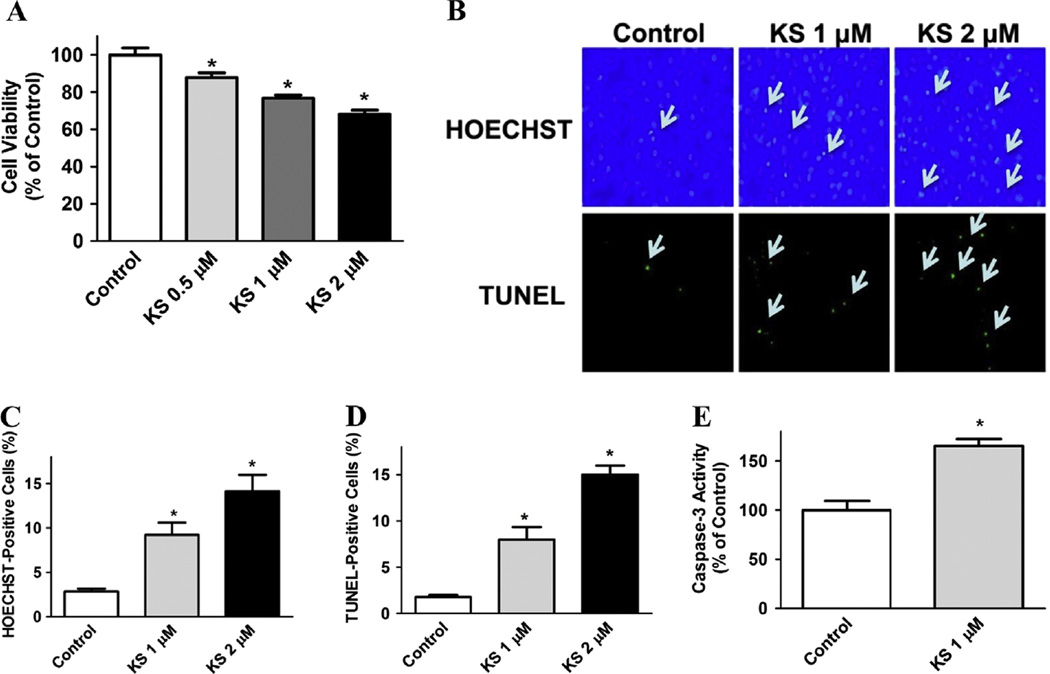
Human kallistatin protein inhibited the growth of MDA-MB-231 breast cancer cells, as determined by MTT assay. Kallistatin at concentrations at 0.5, 1, and 2 µM dose-dependently reduced cell viability to 87.8 ± 2.5%, 76.7 ± 1.7% and 68.1 ± 2.1% of the control group, respectively (P < 0.05; Fig. 1A). Representative images by Hoechst 33342 and TUNEL staining showed that kallistatin at 1 µM and 2 µM increased apoptotic cell death of breast cancer cells (Fig. 1B). Quantitative analysis confirmed these findings (Fig. 1C and D). Moreover, kallistatin treatment (1 µM) significantly augmented caspase-3 activity (1.7 ± 0.1 fold, n = 3, P < 0.05; Fig. 1E). These results indicate that kallistatin is capable of reducing the viability and inducing apoptosis of breast cancer cells.
Fig. 1.
Kallistatin induces cytotoxicity and apoptosis in MDA-MB-231 cells. (A) Cell viability was determined by MTT assay. Kallistatin exhibited a dose-dependent growth inhibition in MDA-MB-231 cells. (B) Hoechst 33342 (apoptotic cells stain bright blue) and TUNEL (apoptotic cells stain green) staining were employed separately to investigate the effect of kallistatin on apoptosis. The representative images are shown at × 100 magnification. Arrows indicate apoptotic cells. (C and D) Quantitative analysis showed that kallistatin significantly increased apoptosis (n = 5–8). (E) Kallistatin treatment also significantly enhanced caspase-3 activity (n = 3). Data are expressed as means ± SE from at least two independent experiments. *P < 0.05 vs. Control group.
3.2. Kallistatin induces autophagy in breast cancer cells
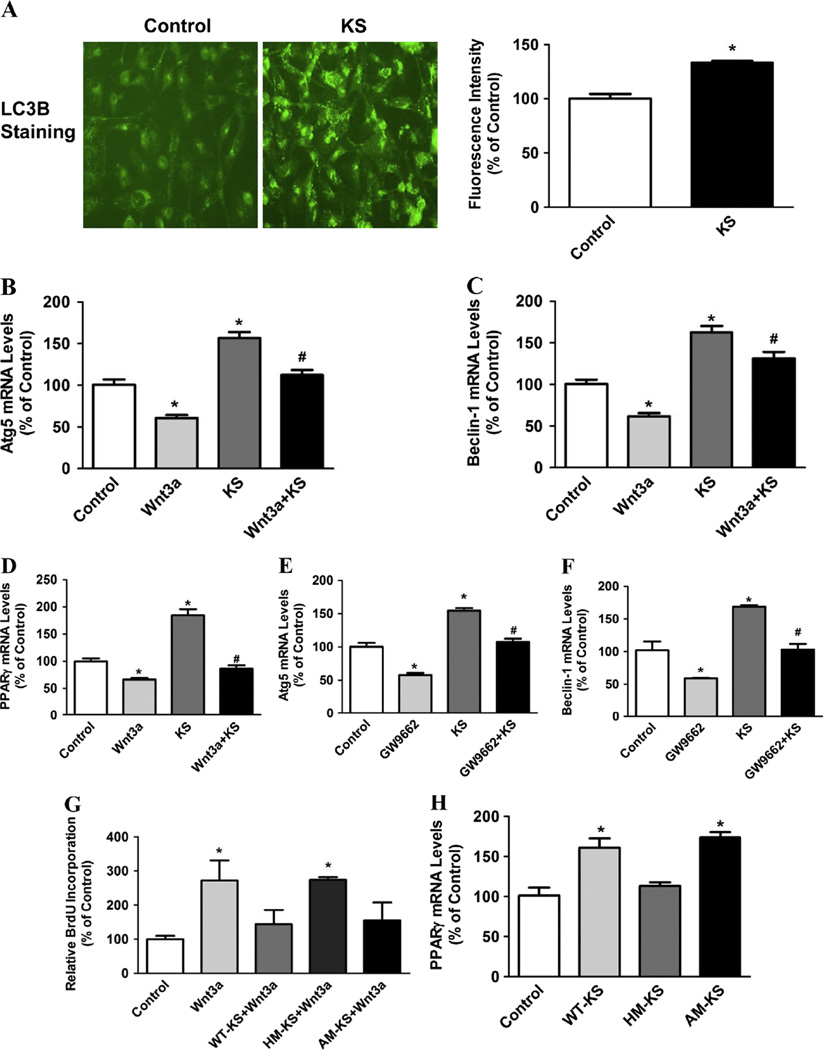
Since recent evidences have highlighted the important role of autophagy in breast cancer development [38], we next determined the effect of kallistatin on autophagy in MDA-MB-231 cells. Representative immunostaining showed that kallistatin (1 µM) significantly increased levels of the autophagy marker LC3B, which was confirmed by quantitative analysis (Fig. 2A). Moreover, kallistatin significantly increased expression of the autophagy gene markers Atg5 and Beclin-1 (1.6 ± 0.1 and 1.6 ± 0.1 fold, respectively; n = 3–6, P < 0.05), but the effect was blocked by pretreatment with Wnt3a (200 ng/ml; n = 3–6, P < 0.05; Fig. 2B and C). Wnt3a alone was found to inhibit Atg5 and Beclin-1 expression (39.2 ± 3.4% and 38.4 ± 4.0% reduction, respectively; n = 3–6, P < 0.05; Fig. 2B and C). PPARγ activation has been shown to induce autophagy in breast cancer cells [39]. Therefore, we investigated the role of PPARγ in kallistatin-mediated autophagy. Our results showed that kallistatin dramatically increased PPARγ expression (1.8 ± 0.1 fold, n = 3, P < 0.05; Fig. 2D) in MDA-MB-231 cells, while pretreatment with Wnt3a abolished this effect. Wnt3a alone was found to decrease PPARγ expression (34 ± 2.9% reduction, n = 3, P < 0.05; Fig. 2D). Furthermore, pretreatment with PPARγ antagonist GW9662 blocked kallistatin-induced Atg5 and Beclin-1 expression (n = 3, P < 0.05; Fig. 2E and F), whereas GW9662 alone markedly decreased Atg5 and Beclin-1 expression (n = 3, P < 0.05; Fig. 2E and F). Wnt3a-induced breast cancer cell proliferation was significantly blocked by wild-type kallistatin (n = 3, P < 0.05) and active-site mutant kallistatin (n = 3, P < 0.05), but not heparin-binding site mutant kallistatin (n = 3, P > 0.05; Fig. 2G). Moreover, wild-type kallistatin (n = 3, P < 0.05) and active-site mutant kallistatin (n = 3, P < 0.05), but not heparin-binding site mutant kallistatin (n = 3, P > 0.05; Fig. 2H), increased PPARγ expression. These composite findings demonstrate for the first time that kallistatin via its heparin-binding site induces autophagy by antagonizing Wnt signaling, thus increasing PPARγ expression in MDA-MB-231 cells.
Fig. 2.
Kallistatin induces autophagy in MDA-MB-231 cells. (A) LC3B immunostaining was performed, and quantitative analysis showed that kallistatin treatment significantly increased LC3B levels (n = 6). The representative images are shown at × 200 magnification. Kallistatin also significantly up-regulated (B) Atg5 and (C) Beclin-1 expression, which were blocked by Wnt3a pretreatment, as indicated by real-time PCR (n = 3–6). (D) Kallistatin significantly increased PPARγ expression, which was blocked by Wnt3a pretreatment, as indicated by real-time PCR (n = 3). PPARγ antagonist GW9662 abolished kallistatin-induced (E) Atg5 and (F) Beclin-1 expression (n = 3). (G) Wild-type kallistatin and active site mutant kallistatin, but not heparin-binding site mutant kallistatin, significantly inhibited Wnt3a-induced MDA-MB-231 cell proliferation (n = 3), as determined by BrdU assay. (H) Wild-type kallistatin and active site mutant kallistatin, but not heparin-binding site mutant kallistatin, significantly increased PPARγ expression (n = 3). Data are expressed as means ± SE from at least two independent experiments. *P < 0.05 vs. Control group. #P < 0.05 vs. KS alone group.
3.3. Kallistatin inhibits miR-21 and Bcl-2 synthesis and Akt phosphorylation, but increases BAX expression
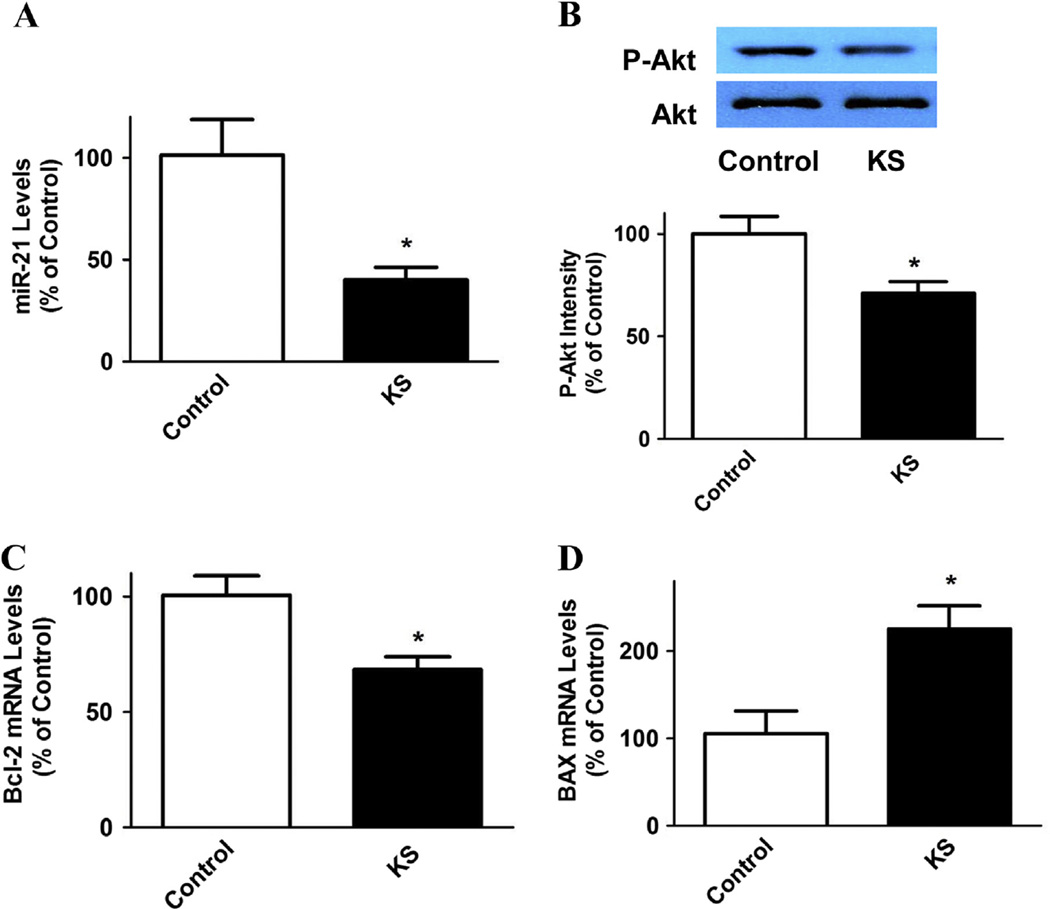
Oncogenic miR-21 and Bcl-2 levels were shown to be up-regulated in breast cancer patients [10,11,40]. Our results demonstrate that kallistatin (1 µM) significantly inhibited miR-21 expression (60 ± 6.1% reduction, n = 3, P < 0.05; Fig. 3A) in MDA-MB-231 cells. Kallistatin also reduced Akt phosphorylation (29 ± 5.6% reduction, n = 3, P < 0.05; Fig. 3B). Moreover, Bcl-2 expression was significantly decreased (32.1 ± 5.5% reduction, n = 3, P < 0.05; Fig. 3C), whereas BAX expression was increased after kallistatin treatment (2.1 ± 0.3 fold, n = 3, P < 0.05; Fig. 3D). These results indicate that kallistatin inhibits the miR-21-Akt pathway, leading to reduced Bcl-2 and increased BAX synthesis.
Fig. 3.
Kallistatin decreases miR-21 expression, Akt phosphorylation and Bcl-2 expression, but increases BAX synthesis in MDA-MB-231 cells. (A) miR-21 expression levels were significantly reduced by kallistatin, as indicated by real-time PCR (n = 3). (B) Western blot analysis showed that Akt phosphorylation was decreased after kallistatin treatment (n = 3). Kallistatin also significantly (C) down-regulated Bcl-2 expression, and thus (D) increased BAX synthesis, as evidenced by real-time PCR (n = 3). Data are expressed as means ± SE from at least two independent experiments. *P < 0.05 vs. Control group.
3.4. Kallistatin via PKC activation modulates miR-203 and SOCS3 synthesis, and stimulates ERK phosphorylation
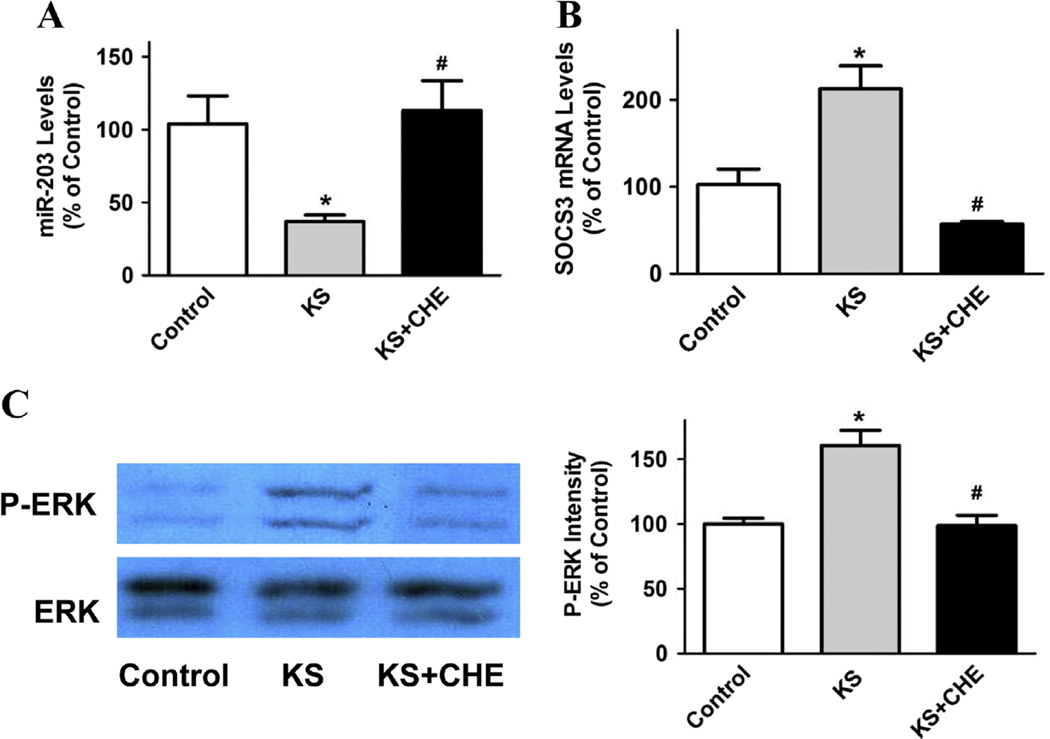
Kallistatin (1 µM) markedly reduced miR-203 expression (64 ± 4.2% reduction, n = 3, P < 0.05) in MDA-MB-231 cells, but the effect was blocked by chelerythrine (PKC inhibitor, 1 µM; n = 3, P < 0.05; Fig. 4A). Conversely, kallistatin induced SOCS3 expression (2.1 ± 0.3 fold, n = 3, P < 0.05), which was also reversed by chelerythrine (1 µM; n = 3, P < 0.05; Fig. 4B). In addition, kallistatin significantly increased ERK1/2 phosphorylation (1.6 ± 0.1 fold, n = 3, P < 0.05), and chelerythrine abolished the effect (1 µM; n = 3, P < 0.05; Fig. 4C). The present findings indicate that kallistatin activates a PKC-ERK signaling pathway, leading to down-regulation of miR-203 and up-regulation of SOCS3 expression.
Fig. 4.
Kallistatin enhances SOCS3 expression through activation of a PKC-ERK-miR-203 signaling pathway in MDA-MB-231 cells. (A) Kallistatin treatment significantly inhibited miR-203 expression, as shown by real-time PCR. The inhibitory effect of kallistatin on miR-203 expression was abolished by chelerythrine (CHE, PKC inhibitor). Kallistatin also induced (B) SOCS3 mRNA synthesis and (C) ERK phosphorylation, but the effects were reversed by chelerythrine. Data are expressed as means ± SE from at least two independent experiments (n = 3–4). *P < 0.05 vs. Control group; #P < 0.05 vs. KS alone group.
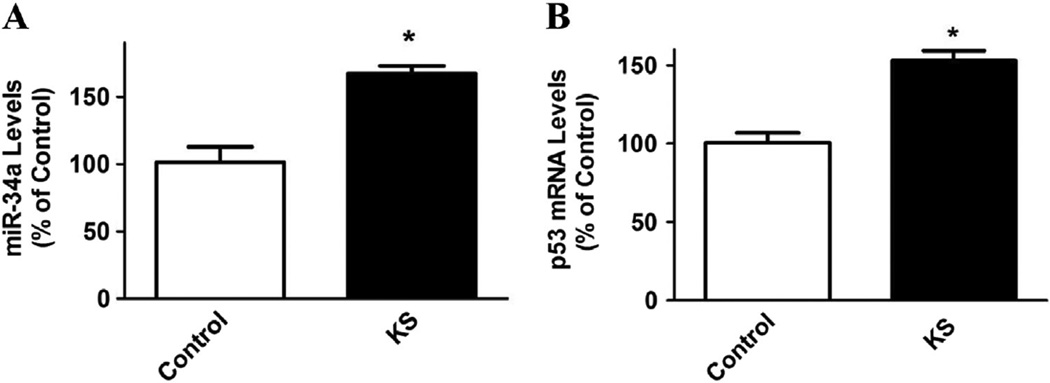
3.5. Kallistatin increases miR-34a and p53 expression
miR-34a has been widely reported as a critical tumor suppressor, which can be regulated by the p53 network in human cancer [14–17]. In contrast to suppression of miR-21 and miR-203 synthesis, kallistatin (1 µM) up-regulated miR-34a and p53 expression in MDA-MB-231 cells (1.6 ± 0.1 and 1.5 ± 0.1 fold, respectively; n = 3, P < 0.05; Fig. 5A and B). These findings reveal that kallistatin’s anti-tumor effect is partly mediated by stimulating miR-34a and p53 synthesis.
Fig. 5.
Kallistatin increases miR-34a and p53 expression in MDA-MB-231 cells. Kallistatin significantly up-regulated (A) miR-34a and (B) p53 expression, as determined by real-time PCR. Data are expressed as means ± SE from at least two independent experiments (n = 3). *P < 0.05 vs. Control group.
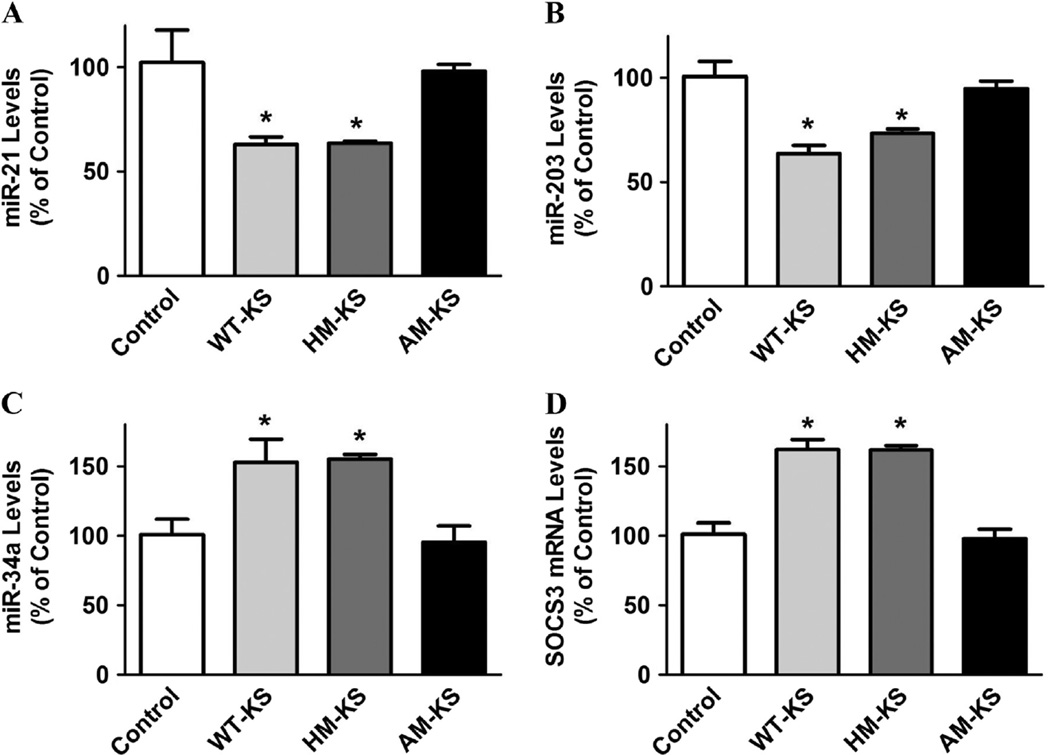
3.6. Kallistatin via its active site suppresses miR-21 and miR-203, and stimulates miR-34a and SOCS3 expression
Kallistatin contains two important structural elements: an active site and a heparin-binding domain [37,41]. We therefore examined the role of kallistatin’s structural elements in regulating miRNA synthesis. miR-21 and miR-203 expression was significantly decreased by wild-type kallistatin (38.5 ± 3.6% and 36.7 ± 3.9% reduction, respectively; n = 3, P < 0.05) and heparin-binding site mutant kallistatin (37.8 ± 1.1% and 27.1 ± 2.1% reduction, respectively; n = 3, P < 0.05), but not by active site mutant kallistatin (n = 3, P > 0.05; Fig. 6A and B). Moreover, miR-34a synthesis was significantly increased by wild-type kallistatin (1.5 ± 0.2 fold, n = 3, P < 0.05) and heparin-binding site mutant kallistatin (1.5 ± 0.1 fold n = 3, P < 0.05), but not active site mutant kallistatin (n = 3, P > 0.05; Fig. 6C). SOCS3 expression was also significantly increased by wild-type kallistatin (1.6 ± 0.1 fold, n = 3, P < 0.05) and heparin-binding site mutant kallistatin (1.6 ± 0.1 fold, n = 3, P < 0.05), but not active site mutant kallistatin (n = 3, P > 0.05; Fig. 6D). These results indicate that kallistatin's active site plays a crucial role in suppressing miR-21 and miR-203, and stimulating miR-34a and SOCS3 expression in breast cancer cells.
Fig. 6.
Kallistatin’s active site is essential for down-regulation of miR-21 and miR-203 expression, and up-regulation of miR-34a and SOCS3 expression in MDA-MB-231 cells. Wild-type kallistatin and heparin-binding site mutant kallistatin, but not active site mutant kallistatin, significantly decreased (A) miR-21 and (B) miR-203 expression, as determined by real-time PCR. (C) miR-34a and (D) SOCS3 synthesis were significantly increased by wild-type kallistatin and heparin-binding site mutant kallistatin, but not active site mutant kallistatin. Data are expressed as means ± SE from at least two independent experiments (n = 3). *P < 0.05 vs. Control group.
4. Discussion
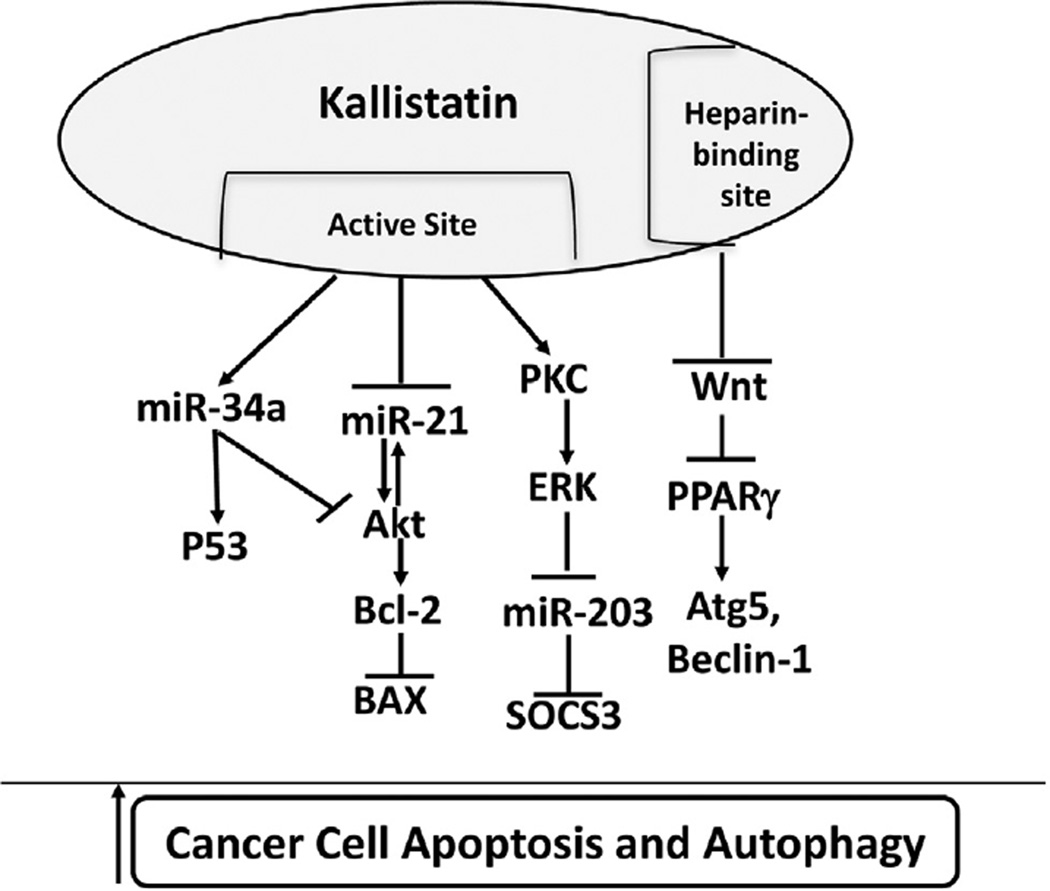
This is the first study to demonstrate that kallistatin inhibits tumor progression by promoting cancer cell apoptosis through suppression of oncogenic miR-21 and miR-203, and stimulation of tumor suppressor miR-34a in MDA-MB-231 breast cancer cells. Recent reports indicate that miR-21, miR-203 and miR-34a play important roles in tumor development, either as oncogenes or tumor suppressors [10,13,19]. Our previous studies have shown that kallistatin inhibits tumor growth and invasion in animal models and cancer cells [26,27,34,35]. These present findings further demonstrate that kallistatin can reduce cancer cell viability and induce programmed cell death by down-regulation of oncogenes (miR-21 and miR-203) and by up-regulation of a tumor suppressor gene (miR-34a). Kallistatin inhibited miR-21-Akt signaling, thereby suppressing Bcl-2 and stimulating BAX expression. Moreover, kallistatin via PKC-ERK activation inhibited miR-203 and increased SOCS3 expression. Conversely, kallistatin up-regulated the miR-34a/p53 axis, and may also modulate miR-34a-Akt-Bcl2 signaling. In particular, kallistatin’s active site was demonstrated to play a key role in suppressing miR-21 and miR-203, and stimulating miR-34a and SOCS3 expression. These new findings reveal new mechanisms of kallistatin in inducing breast cancer cell death (Fig. 7).
Fig. 7.
Proposed mechanism by which kallistatin induces breast cancer cell death. Kallistatin via its active site down-regulates miR-21 and miR-203 expression, and up-regulates miR-34a and SOCS3 expression. The heparin-binding domain is essential for kallistatin's ability to increase PPARγ expression via inhibition of Wnt signaling, and further induce autophagy. SOCS3, suppressor of cytokine signaling 3.
Oncogenic miR-21 levels are increased in many types of cancer, including gastric, colorectal, lung, pancreatic, ovarian and breast [9]. Elevation of miR-21 in breast cancer patients is associated with larger tumor size, higher stage, higher grade and lower overall survival [10,42]. Anti-miR-21 inhibitor was shown to down-regulate levels of the survival protein Bcl-2 in breast cancer [43]. Similarly, matrine, one of the major alkaloids extracted from Sophora flavescens, inhibited breast cancer cell growth by suppressing miR-21, which in turn dephosphorylated Akt and increased accumulation of Bad [44]. Akt activation also has an impact on miR-21 expression in cancer cells, as resveratrol reduced prostate cancer growth and metastasis by inhibiting the Akt-miR-21 pathway [45]. Herein, we showed that kallistatin treatment increased apoptotic cell death in conjunction with reduced miR-21 synthesis and phospho-Akt expression, which in turn inhibited Bcl-2 expression and elevated BAX synthesis in breast cancer cells. Kallistatin may also regulate miR-21 by suppressing the Akt-miR-21 signaling pathway. These new findings indicate that kallistatin-mediated breast cancer cell apoptosis is partly mediated by miR-21 through an Akt-Bcl-2 signaling pathway.
In addition to miR-21, miR-203 has also found to be increased in human breast cancer tissues [13]. Depletion of miR-203 enhances cisplatin chemosensitivity by increasing SOCS3, p53 and BAX expression in breast cancer cells [13]. SOCS3, a tumor suppressor, was found to inhibit breast cancer cell proliferation [46], and a MEK inhibitor was shown to increase miR-203 synthesis [47]. Our present study showed that kallistatin treatment significantly increased ERK phosphorylation and decreased miR-203, but increased SOCS3 expression; however, these effects were abolished by a PKC inhibitor. These results indicate a novel mechanism of kallistatin in inducing cancer cell death by up-regulation of SOCS3 expression through a PKC-ERK-miR-203 signaling pathway.
The tumor suppressor miR-34a is markedly reduced in breast cancer cell lines and clinical specimens [19]. miR-34a was reported to inhibit breast cancer cell proliferation by inhibiting the Akt signaling pathway [48]. Restoration of miR-34a reduces breast cancer proliferation and migration through regulation of Bcl-2 [19]. miR-34a, regulated by p53, was shown to suppress breast cancer invasion and metastasis [17]. Moreover, diallyl disulfide was reported to inhibit human breast cancer proliferation and metastasis through up-regulation of miR-34a [49]. In the present study, we showed that kallistatin significantly increased the expression of the tumor suppressors miR-34a and p53 in breast cancer cells (Fig. 5). As kallistatin is capable of inhibiting Akt activation (Fig. 3), kallistatin may also induce cancer cell death through miR-34a-mediated inhibition of Akt signaling, in addition to stimulation of the miR-34a/p53 axis (Fig. 7).
Kallistatin appears to have double-edged actions in apoptosis. Our previous studies showed that kallistatin gene delivery attenuated cardiovascular and renal injury in association with reduced cell death in rats with myocardial ischemia/reperfusion or salt-induced hypertension [28,50]. In addition, kallistatin treatment prevented apoptosis and inflammation in septic mice [36]. Moreover, kallistatin blocked TNF-α-induced apoptosis in cultured endothelial cells and endothelial progenitor cells [28,51]. However, SERPINA3K (KBP) was shown to promote apoptotic cell death in colorectal cancer cells [35]. Our present study also indicated that kallistatin is capable of inducing breast cancer cell death. Taken together, these combined findings indicate that kallistatin exerts both anti- and pro-apoptotic effects depending on pathological conditions and cell types.
Recent studies have highlighted the association between autophagy and breast cancer [38,52]. Autophagy is a morphologically distinctive mode of programmed cell death, and controlled by autophagy-related genes (ATGs) [52]. Beclin-1 and LC3B are two major markers of autophagy. The role of autophagy in cancer cell death is still controversial. However, increasing evidence has shown that autophagy can suppress tumor cell growth and is a promising strategy for the treatment of breast cancer [38,53]. Overexpression of Beclin-1 in MCF-7 breast cancer cells is associated with reduced cellular proliferation, in vitro clonigenicity, and tumorigenesis in nude mice [54]. Moreover, Beclin-1 expression was found to inhibit breast cancer development, and its expression was negatively correlated with Bcl-2 expression in human breast cancer tissue [55]. Our present study showed that kallistatin induces autophagy, as kallistatin significantly induced LC3B levels, Atg5 and Beclin-1 expression in MDA-MB-231 cells. Our previous report indicated that kallistatin antagonizes Wnt-β-catenin signaling via binding to LRP6 in MDA-MB-231 cells [34]. Resveratrol has been demonstrated to induce autophagy via suppressing Wnt/β-catenin signaling pathway in breast cancer stem-like cells [56]. In addition, β-catenin levels are negatively correlated with PPARγ expression in breast cancer patients [57]. Inhibition of LRP6 by RNAi leads to increased PPARγ expression in human mesenchymal stem cells [58], and PPARγ activation induces autophagy in human breast cancer cells [39,59]. Furthermore, high expression levels of PPARγ significantly correlate with long-term survival of breast cancer patients [57]. Consistent with these findings, our data showed that Wnt3a markedly decreased PPARγ expression, while kallistatin increased PPARγ synthesis, and kallistatin’s effect was blocked by Wnt3a pretreatment. Kallistatin alone induced Atg5 and Beclin-1 synthesis; however, co-administration of Wnt3a or PPARγ antagonist GW9662 abolished kallistatin's effects. Using purified human kallistatin mutants, we further showed that kallistatin’s heparin-binding site is essential for blocking Wnt-mediated proliferation and PPARγ expression in breast cancer cells. Our data demonstrated for the first time that kallistatin via its heparin-binding site induces autophagy by antagonizing Wnt signaling pathway, thus increasing PPARγ expression in breast cancer cells. These findings indicate that kallistatin increases programmed cancer cell death, in part, via induction of autophagy.
The current study demonstrates that kallistatin promotes programmed cell death by differential regulation of miR-21, miR-203 vs. miR-34a in breast cancer cells. Kallistatin, an endogenous protein, exerts cancer suppression through multi-faceted pathways, including inhibition of angiogenesis, inflammation, tumor growth and metastasis, and induction of cancer cell death [25–33,35]. Kallistatin inhibits tumor growth by blocking VEGF-mediated angiogenesis and NF-κB signaling [26,27,60]. Moreover, kallistatin attenuates breast cancer cell migration and invasion via antagonizing a Wnt signaling pathway [34]. The present results provide new insights into the mechanisms by which kallistatin promotes breast cancer cell death: 1) inhibiting miR-21-Akt-Bcl-2 signaling; 2) suppressing miR-203 and increasing SOCS3 expression through PKC-ERK activation; 3) stimulating the miR-34a/p53 axis and/or miR-34a-mediated inhibition of Akt-Bcl-2 signaling; and 4) blocking Wnt signaling, inducing PPARγ expression and autophagy (Fig. 7). Taken together, these findings provide novel insights into the beneficial effects of kallistatin in tumor development by down-regulating miR-21 and miR-203 and up-regulating miR-34a synthesis.
Acknowledgments
This work was supported by National Institutes of Health grants HL-118516.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author contributions
Conceived and designed the experiments: PL, LC and JC. Performed the experiments: PL, YG, and ZY; Analyzed the data: PL, YG, ZY, GB and JC; Contributed reagents/materials/analysis tools: PL, YG, ZY and LC; Wrote the paper: PL, GB, JC, YG and LC. All authors have read and approved the manuscript.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 3.Crowder RJ, Phommaly C, Tao Y, Hoog J, Luo J, Perou CM, Parker JS, Miller MA, Huntsman DG, Lin L, Snider J, Davies SR, Olson JA, Jr, Watson MA, Saporita A, Weber JD, Ellis MJ. PIK3CA and PIK3CB inhibition produce synthetic lethality when combined with estrogen deprivation in estrogen receptor-positive breast cancer. Cancer Res. 2009;69:3955–3962. doi: 10.1158/0008-5472.CAN-08-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love RR, Young GS, Laudico AV, Van Dinh N, Uy GB, Quang le H, De La Pena AS, Dofitas RB, Bisquera OC, Jr, Siguan SS, Salvador JD, Mirasol-Lumague MR, Navarro NS, Jr, Linh ND, Jarjoura D. Bone mineral density following surgical oophorectomy and tamoxifen adjuvant therapy for breast cancer. Cancer. 2013;119:3746–3752. doi: 10.1002/cncr.28302. [DOI] [PubMed] [Google Scholar]
- 5.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 2013;5:111. doi: 10.1186/gm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng EK, Wong CL, Ma ES, Kwong A. MicroRNAs as new players for diagnosis, prognosis, and therapeutic targets in breast cancer. J. Oncol. 2009;2009:305420. doi: 10.1155/2009/305420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarver JE, Sperling EA, Nailor A, Heimberg AM, Robinson JM, King BL, Pisani D, Donoghue PC, Peterson KJ. miRNAs: small genes with big potential in metazoan phylogenetics. Mol. Biol. Evol. 2013;30:2369–2382. doi: 10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- 8.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Wang X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clin. Transl. Oncol. 2014;16:225–233. doi: 10.1007/s12094-013-1132-z. [DOI] [PubMed] [Google Scholar]
- 10.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 12.Dong J, Zhao YP, Zhou L, Zhang TP, Chen G. Bcl-2 upregulation induced by miR-21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa-2 pancreatic cancer cells. Arch. Med. Res. 2011;42:8–14. doi: 10.1016/j.arcmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Ru P, Steele R, Hsueh EC, Ray RB. Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer. 2011;2:720–727. doi: 10.1177/1947601911425832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, Chen H, Ding F, Wang X, Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294–4303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 18.Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, Xiao X, Yang Y, Sheng W, Wu Y, Zeng Y. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 20.Chao J, Tillman DM, Wang MY, Margolius HS, Chao L. Identification of a new tissue-kallikrein-binding protein. Biochem. J. 1986;239:325–331. doi: 10.1042/bj2390325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VC, Chao L, Chao J. Roles of the P1, P2, and P3 residues in determining inhibitory specificity of kallistatin toward human tissue kallikrein. J. Biol. Chem. 2000;275:38457–38466. doi: 10.1074/jbc.M005605200. [DOI] [PubMed] [Google Scholar]
- 22.Zhou GX, Chao L, Chao J. Kallistatin: a novel human tissue kallikrein inhibitor. Purification, characterization, and reactive center sequence. J. Biol. Chem. 1992;267:25873–25880. [PubMed] [Google Scholar]
- 23.Chao J, Schmaier A, Chen LM, Yang Z, Chao L. Kallistatin, a novel human tissue kallikrein inhibitor: levels in body fluids, blood cells, and tissues in health and disease. J. Lab. Clin. Med. 1996;127:612–620. doi: 10.1016/s0022-2143(96)90152-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Chao J, Kotak I, Guo D, Parikh SJ, Bhagatwala J, Dong Y, Patel SY, Houk C, Chao L. Plasma kallistatin is associated with adiposity and cardio-metabolic risk in apparently healthy African American adolescents. Metabolism. 2013;62:642–646. doi: 10.1016/j.metabol.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao L, Yin H, Smith R JS, Chao L, Chao J. Role of kallistatin in prevention of cardiac remodeling after chronic myocardial infarction. Lab. Investig. 2008;88:1157–1166. doi: 10.1038/labinvest.2008.85. [DOI] [PubMed] [Google Scholar]
- 26.Miao RQ, Agata J, Chao L, Chao J. Kallistatin is a new inhibitor of angiogenesis and tumor growth. Blood. 2002;100:3245–3252. doi: 10.1182/blood-2002-01-0185. [DOI] [PubMed] [Google Scholar]
- 27.Miao RQ, Chen V, Chao L, Chao J. Structural elements of kallistatin required for inhibition of angiogenesis. Am. J. Physiol. Cell Physiol. 2003;284:C1604–C1613. doi: 10.1152/ajpcell.00524.2002. [DOI] [PubMed] [Google Scholar]
- 28.Shen B, Gao L, Hsu YT, Bledsoe G, Hagiwara M, Chao L, Chao J. Kallistatin attenuates endothelial apoptosis through inhibition of oxidative stress and activation of Akt-eNOS signaling. Am. J. Physiol. Heart Circ. Physiol. 2010;299:H1419–H1427. doi: 10.1152/ajpheart.00591.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen B, Hagiwara M, Yao YY, Chao L, Chao J. Salutary effect of kallistatin in salt-induced renal injury, inflammation, and fibrosis via antioxidative stress. Hypertension. 2008;51:1358–1365. doi: 10.1161/HYPERTENSIONAHA.107.108514. [DOI] [PubMed] [Google Scholar]
- 30.Wang CR, Chen SY, Wu CL, Liu MF, Jin YT, Chao L, Chao J. Prophylactic adenovirus-mediated human kallistatin gene therapy suppresses rat arthritis by inhibiting angiogenesis and inflammation. Arthritis Rheum. 2005;52:1319–1324. doi: 10.1002/art.20991. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Gao L, Shen B, Chao L, Chao J. Kallistatin inhibits vascular inflammation by antagonizing tumor necrosis factor-alpha-induced nuclear factor kappaB activation. Hypertension. 2010;56:260–267. doi: 10.1161/HYPERTENSIONAHA.110.152330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiau AL, Teo ML, Chen SY, Wang CR, Hsieh JL, Chang MY, Chang CJ, Chao J, Chao L, Wu CL, Lee CH. Inhibition of experimental lung metastasis by systemic lentiviral delivery of kallistatin. BMC Cancer. 2010;10:245. doi: 10.1186/1471-2407-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhi X, Lin L, Yang S, Bhuvaneshwar K, Wang H, Gusev Y, Lee MH, Kallakury B, Shivapurkar N, Cahn K, Tian X, Marshall JL, Byers SW, He AR. betaII-Spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology. 2015;61:598–612. doi: 10.1002/hep.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Yang Z, Li P, Bledsoe G, Chao L, Chao J. Kallistatin antagonizes Wnt/beta-catenin signaling and cancer cell motility via binding to low-density lipoprotein receptor-related protein 6. Mol. Cell Biochem. 2013;379:295–301. doi: 10.1007/s11010-013-1654-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Y, Li L, Huang X, Gu X, Xu Z, Zhang Y, Huang L, Li S, Dai Z, Li C, Zhou T, Cai W, Yang Z, Gao G, Yang X. SERPINA3K induces apoptosis in human colorectal cancer cells via activating the Fas/FasL/caspase-8 signaling pathway. FEBS J. 2013;280:3244–3255. doi: 10.1111/febs.12303. [DOI] [PubMed] [Google Scholar]
- 36.Li P, Bledsoe G, Yang ZR, Fan H, Chao L, Chao J. Human kallistatin administration reduces organ injury and improves survival in a mouse model of polymicrobial sepsis. Immunology. 2014;142:216–226. doi: 10.1111/imm.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VC, Chao L, Chao J. Reactive-site specificity of human kallistatin toward tissue kallikrein probed by site-directed mutagenesis. Biochim. Biophys. Acta. 2000;1479:237–246. doi: 10.1016/s0167-4838(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 38.Zarzynska JM. The importance of autophagy regulation in breast cancer development and treatment. BioMed Res. Int. 2014;2014:710345. doi: 10.1155/2014/710345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J, Zhang W, Liang B, Casimiro MC, Whitaker-Menezes D, Wang M, Lisanti MP, Lanza-Jacoby S, Pestell RG, Wang C. PPARgamma activation induces autophagy in breast cancer cells. Int. J. Biochem. Cell Biol. 2009;41:2334–2342. doi: 10.1016/j.biocel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellemans P, van Dam PA, Weyler J, van Oosterom AT, Buytaert P, Van Marck E. Prognostic value of bcl-2 expression in invasive breast cancer. Br. J. Cancer. 1995;72:354–360. doi: 10.1038/bjc.1995.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen VC, Chao L, Pimenta DC, Bledsoe G, Juliano L, Chao J. Identification of a major heparin-binding site in kallistatin. J. Biol. Chem. 2001;276:1276–1284. doi: 10.1074/jbc.M005791200. [DOI] [PubMed] [Google Scholar]
- 42.Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic implications of micro-RNA-21 overexpression in invasive ductal carcinomas of the breast. J. Breast Cancer. 2011;14:269–275. doi: 10.4048/jbc.2011.14.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Bourguignon LY. Hyaluronan-CD44 interaction promotes c-Jun signaling and miRNA21 expression leading to Bcl-2 expression and chemoresistance in breast cancer cells. Mol. Cancer. 2014;13:52. doi: 10.1186/1476-4598-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li LQ, Li XL, Wang L, Du WJ, Guo R, Liang HH, Liu X, Liang DS, Lu YJ, Shan HL, Jiang HC. Matrine inhibits breast cancer growth via miR-21/PTEN/Akt pathway in MCF-7 cells. Cell Physiol. Biochem. 2012;30:631–641. doi: 10.1159/000341444. [DOI] [PubMed] [Google Scholar]
- 45.Sheth S, Jajoo S, Kaur T, Mukherjea D, Sheehan K, Rybak LP, Ramkumar V. Resveratrol reduces prostate cancer growth and metastasis by inhibiting the Akt/MicroRNA-21 pathway. PloS One. 2012;7:e51655. doi: 10.1371/journal.pone.0051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barclay JL, Anderson ST, Waters MJ, Curlewis JD. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int. J. Cancer. 2009;124:1756–1766. doi: 10.1002/ijc.24172. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y, Hu HY, Meng W, Jiang L, Zhang X, Sha JJ, Lu Z, Yao Y. MEK inhibitor effective against proliferation in breast cancer cell. Tumour Biol. 2014;35:9269–9279. doi: 10.1007/s13277-014-1901-5. [DOI] [PubMed] [Google Scholar]
- 48.Zhao G, Guo J, Li D, Jia C, Yin W, Sun R, Lv Z, Cong X. MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA Cell Biol. 2013;32:699–707. doi: 10.1089/dna.2013.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao X, Chen B, Liu X, Liu P, Zheng G, Ye F, Tang H, Xie X. Diallyl disulfide suppresses SRC/Ras/ERK signaling-mediated proliferation and metastasis in human breast cancer by up-regulating miR-34a. PloS One. 2014;9:e112720. doi: 10.1371/journal.pone.0112720. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Chao J, Yin H, Yao YY, Shen B, Smith RS, Jr, Chao L. Novel role of kallistatin in protection against myocardial ischemia-reperfusion injury by preventing apoptosis and inflammation. Hum. Gene Ther. 2006;17:1201–1213. doi: 10.1089/hum.2006.17.1201. [DOI] [PubMed] [Google Scholar]
- 51.Gao L, Li P, Zhang J, Hagiwara M, Shen B, Bledsoe G, Chang E, Chao L, Chao J. Novel role of kallistatin in vascular repair by promoting mobility, viability, and function of endothelial progenitor cells. J. Am. Heart Assoc. 2014;3:e001194. doi: 10.1161/JAHA.114.001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li JP, Yang YX, Liu QL, Zhou ZW, Pan ST, He ZX, Zhang X, Yang T, Pan SY, Duan W, He SM, Chen XW, Qiu JX, Zhou SF. The pan-inhibitor of Aurora kinases danusertib induces apoptosis and autophagy and suppresses epithelial-to-mesenchymal transition in human breast cancer cells. Drug Des. Dev. Ther. 2015;9:1027–1062. doi: 10.2147/DDDT.S74412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 55.Won KY, Kim GY, Kim YW, Song JY, Lim SJ. Clinicopathologic correlation of beclin-1 and bcl-2 expression in human breast cancer. Hum. Pathol. 2010;41:107–112. doi: 10.1016/j.humpath.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Fu Y, Chang H, Peng X, Bai Q, Yi L, Zhou Y, Zhu J, Mi M. Resveratrol inhibits breast cancer stem-like cells and induces autophagy via suppressing Wnt/beta-catenin signaling pathway. PloS One. 2014;9:e102535. doi: 10.1371/journal.pone.0102535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang Y, Zou L, Zhang C, He S, Cheng C, Xu J, Lu W, Zhang Y, Zhang H, Wang D, Shen A. PPARgamma and Wnt/beta-Catenin pathway in human breast cancer: expression pattern, molecular interaction and clinical/prognostic correlations. J. Cancer Res. Clin. Oncol. 2009;135:1551–1559. doi: 10.1007/s00432-009-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perobner I, Karow M, Jochum M, Neth P. LRP6 mediates Wnt/beta-catenin signaling and regulates adipogenic differentiation in human mesenchymal stem cells. Int. J. Biochem. Cell Biol. 2012;44:1970–1982. doi: 10.1016/j.biocel.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Rovito D, Giordano C, Plastina P, Barone I, De Amicis F, Mauro L, Rizza P, Lanzino M, Catalano S, Bonofiglio D, Ando S. Omega-3 DHA- and EPA-dopamine conjugates induce PPARgamma-dependent breast cancer cell death through autophagy and apoptosis. Biochim. Biophys. Acta. 1850;2015:2185–2195. doi: 10.1016/j.bbagen.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 60.Huang KF, Huang XP, Xiao GQ, Yang HY, Lin JS, Diao Y. Kallistatin, a novel anti-angiogenesis agent, inhibits angiogenesis via inhibition of the NF-kappaB signaling pathway. Biomed. Pharmacother. 2014;68:455–461. doi: 10.1016/j.biopha.2014.03.005. [DOI] [PubMed] [Google Scholar]