Abstract
The fidelity of RNA splicing is maintained by a network of factors, but the molecular mechanisms that govern this process have yet to be fully elucidated. We previously found that TDP-43, an RNA-binding protein implicated in neurodegenerative disease, utilizes UG microsatellites to repress nonconserved cryptic exons and prevent their incorporation into mRNA. Here, we report that two well characterized splicing factors, polypyrimidine tract-binding protein 1 (PTBP1) and polypyrimidine tract-binding protein 2 (PTBP2), are also nonconserved cryptic exon repressors. In contrast to TDP-43, PTBP1 and PTBP2 utilize CU microsatellites to repress both conserved tissue-specific exons as well as nonconserved cryptic exons. Analysis of these conserved splicing events suggests that PTBP1 and PTBP2 repression is titrated to generate the transcriptome diversity required for neuronal differentiation. Together, we establish that PTBP1 and PTBP2 are members of a family of cryptic exon repressors.
Graphical abstract
Introduction
RNA splicing in higher eukaryotes is a complex but efficient process that uses only ~170 spliceosomal factors (Wahl et al., 2009) to reliably identify hundreds of thousands of exon and intron boundaries with nucleotide precision (Chen and Manley, 2009; Fu and Ares, 2014; Huelga et al., 2012). This delicate system is often disrupted in human disease (Brinegar and Cooper, 2016; Scotti and Swanson, 2015) and has been a major focal point for interpreting mutations that are identified through whole-genome medicine (Xiong et al., 2015). Currently, our ability to predict exon-intron junctions from the primary genomic sequence is limited, in part due to the loose consensus sequence of most splicing proteins. Coupling next-generation sequencing technology with a complete characterization of individual splicing factors will help reveal the mechanisms that underlie splice site selection. Here, we report a model for the function of two well-studied splicing factors, polypyrimidine tract-binding protein 1 (PTBP1) and polypyrimidine tract-binding protein 2 (PTBP2) (Gil et al., 1991; Markovtsov et al., 2000; Patton et al., 1991; Polydorides et al., 2000).
PTBP1 (also known as PTB or hnRNP I) and its paralog PTBP2 (also known as nPTB or brPTB) are RNA-binding proteins that, in addition to roles involving mRNA regulation and gene expression, have been extensively characterized as trans-acting splicing repressors (Kafasla et al., 2012; Keppetipola et al., 2012; Licatalosi et al., 2012; Xue et al., 2009). PTBP1 and PTBP2 both utilize four highly conserved RNA-recognition motifs (RRMs) to bind to their consensus sequences, the CU-rich pyrimidine tract (Oberstrass et al., 2005). Interestingly, the expression patterns of PTBP1 and PTBP2 are mutually exclusive, since PTBP1 downregulates PTBP2 through an alternative splicing event that leads to nonsense-mediated decay (NMD) (Boutz et al., 2007). PTBP1 is highly expressed and PTBP2 is kept low in most tissues except for certain organs such as brain, where the opposite is true (Lilleväli et al., 2001). During neuronal differentiation, PTBP1 is downregulated and PTBP2 is upregulated to compensate (Li et al., 2014; Makeyev et al., 2007). Thus, it is thought that PTBP2 controls an alternative splicing program that is critical for neuronal maturation (Li et al., 2007; Raj and Blencowe, 2015).
We recently found that TDP-43—a splicing factor implicated in the pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia—binds to UG microsatellites to repress nonconserved cryptic exons (Ling et al., 2015). TDP-43 loss of function results in the incorporation of nonconserved cryptic exons that often induce NMD of the associated mRNA. Since PTBP1 and PTBP2 are splicing repressors that bind to CU repeats, we reasoned that these two splicing factors might perform a similar function.
We now report that PTBP1 and PTBP2 repress nonconserved cryptic exons. Interestingly, while short pentamer pyrimidine stretches have long been postulated to be a consensus binding motif for PTBP1 (Ashiya and Grabowski, 1997; Pérez et al., 1997; Xue et al., 2009), we find that highly repressed PTBP1/2 exons are flanked by long CU microsatellites (often >20bp in length). Furthermore, our analysis of unannotated splicing events reveals a subset of conserved exons that, while repressed in undifferentiated cells, become actively spliced in differentiated neurons. Previous studies have characterized a PTBP1-specific alternative splicing program for early neuronal differentiation (Linares et al., 2015) and a late program that is PTBP2-specific (Li et al., 2014). The conserved exons identified in our work, however, belong to the set of targets that are regulated by both PTBP1 and PTBP2. Our findings suggest a potential bridge between the early and late programs via the titration of CU-repeat associated splicing repression. Thus, PTBP1 and PTBP2 are precisely coordinated to both repress deleterious nonconserved cryptic exons and to generate the alternative splicing complexity required for differentiated tissue.
Results
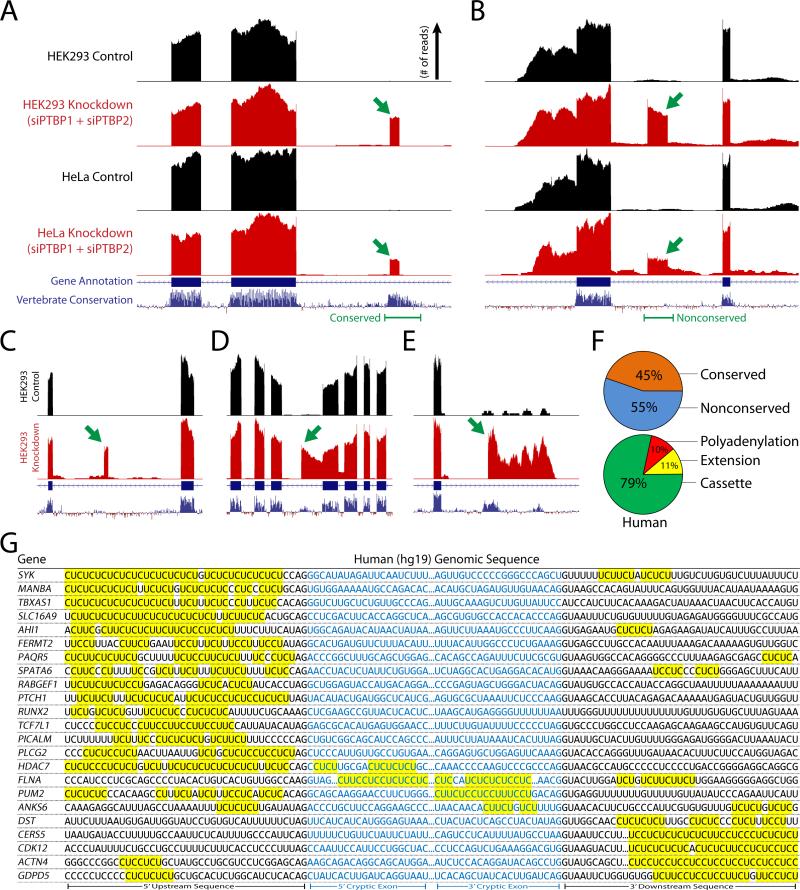
Recent studies have used high-throughput RNA-sequencing technology (RNA-seq) to study the alternative splicing events that are regulated by PTBP1 and PTBP2 (Gueroussov et al., 2015; Li et al., 2014). To identify cryptic exons associated with PTBP1 and PTBP2, we reanalyzed these published datasets in addition to RNA-seq datasets generated from concurrent knockdown of PTBP1 and PTBP2 in HeLa cells. As previously described (Ling et al., 2015), unannotated splicing events were identified in HEK293 (Gueroussov et al., 2015) and HeLa cells, revealing numerous repressed exons within these PTBP1 and PTBP2 knockdown datasets (Fig. 1A and B; 183 total, Supplemental Excel File).
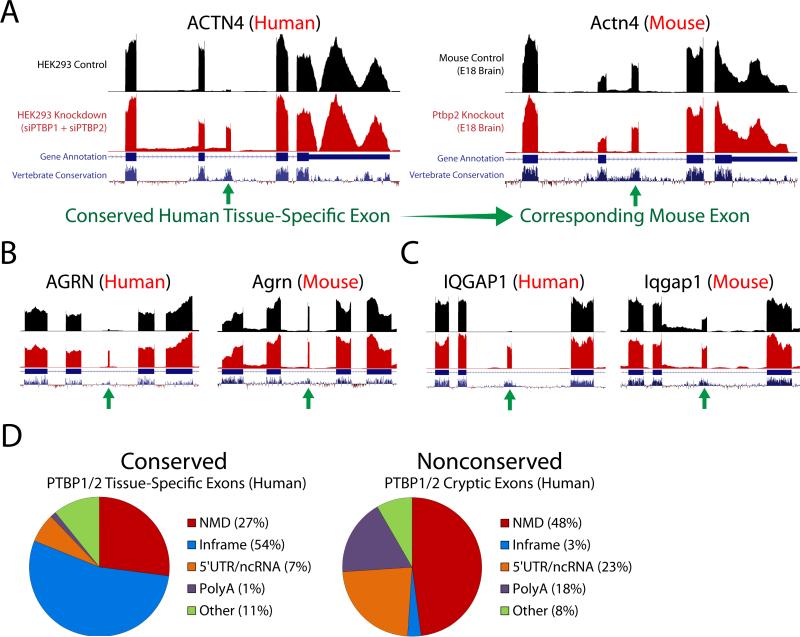
Figure 1. Identification of human nonconserved cryptic exons and conserved tissue-specific exons repressed by PTBP1 and PTBP2.
(A and B) Analysis of RNA-seq data from concurrent knockdown of PTBP1 and PTBP2 in HEK293 (Gueroussov et al., 2015) and HeLa cells reveals unannotated conserved [FERMT2, (A)] and nonconserved [ANKS6, (B)] repressed exons (green arrows). (C–E) PTBP1/2 repressed exons can be classified as standard cassette exons [SPATA6, (C)], exon extensions [FLNA, (D)] and premature polyadenylation sites [RUNX2, (E)]. (F) Approximately 45% of PTBP1/2 repressed exons are conserved, while 55% are nonconserved. The majority of exons are cassette exons (79%) while exon extensions (11%) and polyadenylation sites (10%) constitute a smaller fraction. (G) Repressed exons are flanked by CU microsatellites in the upstream, downstream, or internal sequence. Sequences that are missing the 3’ splice site ‘AG’ or 5’ splice site ‘GU’ dinucleotide reflect repressed exons that are premature polyA sites or exon extensions where a 5’ or 3’ splice junction is absent.
PTBP1/2 repressed exons could be classified as standard cassette exons (Fig. 1C), alternative splice site selections leading to extensions of conserved exons (Fig. 1D), or premature polyadenylation sites due to alternative 3’ exon splicing (Fig. 1E). The majority of repressed exons were cassette exons (79%), while extensions (11%) and polyadenylation sites (10%) contributed a smaller fraction (Fig. 1F). Furthermore, ~45% of these human PTBP1/2 repressed exons resided in conserved domains of the genome, while ~55% were found in nonconserved regions (Fig. 1F). Indeed, many of the nonconserved cryptic exons were predicted to introduce frameshifts or stop codons (~48%), which lead to NMD and the downregulation of associated transcripts (Supplemental Fig. 1). Sequence analysis of PTBP1/2 repressed exons further confirmed the presence of adjacent CU microsatellites (Fig. 1G). These microsatellites reside in the canonical polypyrimidine tract of the 3’ splice site, although certain CU microsatellites are also found within the repressed exon itself or downstream of the 5’ splice site.
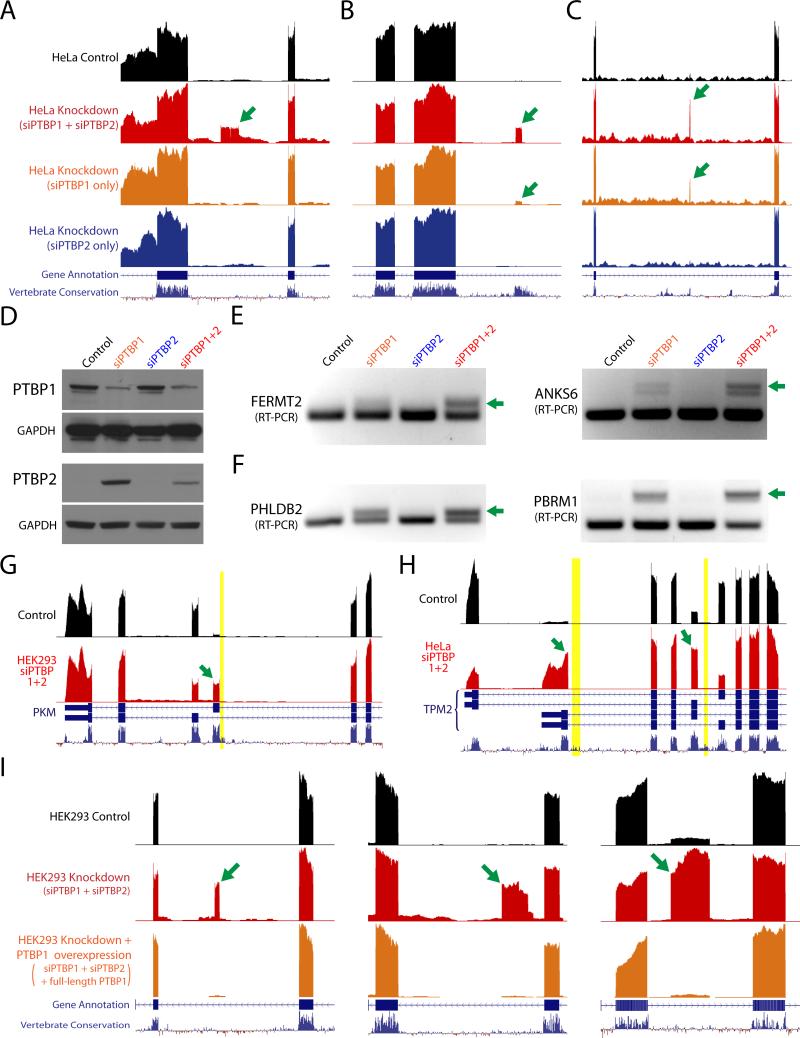
We then analyzed RNA-seq data from knockdown of either PTBP1 or PTBP2 alone to verify that loss of both proteins is required for conserved and nonconserved exon incorporation. Due to compensation by PTBP2, many exons were still repressed when only PTBP1 was reduced (Fig. 2A). For some exons, however, PTBP2 was not sufficient for complete repression (Fig. 2B and C). We also confirmed with immunoblot analysis that knockdown of PTBP1 alone led to increased levels of PTBP2 (Fig. 2D) and validated several repressed exons by reverse transcription-polymerase chain reaction (RT-PCR) analysis (Fig. 2E and F).
Figure 2. Concurrent knockdown of both PTBP1 and PTBP2 is required for cryptic exon activation.
(A–C) RNA-seq data from control HeLa cells, double knockdown of PTBP1 and PTBP2 (red), single knockdown of PTBP1 (orange), and single knockdown of PTBP2 (blue). While some repressed exons (green arrows) do not appear unless both PTBP1 and PTBP2 are knocked down [ANKS6, (A)], other exons are revealed in single PTBP1 knockdown [FERMT2, PHLDB2, (B and C)], supporting previous observations that PTBP2 may be a weaker repressor than PTBP1 in certain contexts (Keppetipola et al., 2016; Markovtsov et al., 2000). (D) Immunoblot analysis of PTBP1 and PTBP2 knockdown; PTBP2 levels increase after knockdown of PTBP1. (E and F) Certain repressed exons are strongly spliced only after double knockdown of PTBP1 and PTBP2 while other exons (F) appear after single knockdown of PTBP1, albeit at lower levels. (G and H) PTBP1 and PTBP2 have been previously identified to regulate alternatively spliced exons in genes such as PKM (G) and TPM2 (H) (David et al., 2010; Xue et al., 2009). Numerous CU microsatellites (yellow bars) can be seen directly upstream of upregulated alternatively spliced exons (green arrows). (I) Overexpression of full-length PTBP1 after double knockdown of PTBP1 and PTBP2 results in the restoration of exon repression (Gueroussov et al., 2015).
To establish that repressed exons were direct targets of PTBP1, we used the Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP) method to map transcriptome-wide binding sites (Hafner et al., 2010). As expected, intronic peaks corresponding to direct binding by PTBP1 could be found adjacent to repressed exons (Supplemental Fig. 2). Many reads also aligned to the more abundant exonic regions, supporting the notion that PTBP1 has additional roles in mRNA processing and translation (Kafasla et al., 2012; Keppetipola et al., 2012). PAR-CLIP reads also mapped PTBP1 to the alternatively spliced exons of PKM and TPM2, sites which have been previously identified as under the control of PTBP1 (David et al., 2010; Xue et al., 2009). Numerous CU microsatellites can be found directly upstream of these alternative exons, strengthening the model of CU-repeat associated splicing repression (Fig. 2G and H). Indeed, PTBP1 and PTBP2 may utilize the CU microsatellite upstream of PKM exon 9 to regulate an alternative splicing event that plays an important role in the metabolism of many cancers (David et al., 2010; Israelsen et al., 2013). Finally, to determine whether expression of PTBP1 could restore exon repression, we also analyzed RNA-seq data obtained from PTBP1/2 knockdown cells expressing full-length PTBP1 (Gueroussov et al., 2015) and observed that exons were again repressed (Fig. 2I, Supplemental Fig. 3).
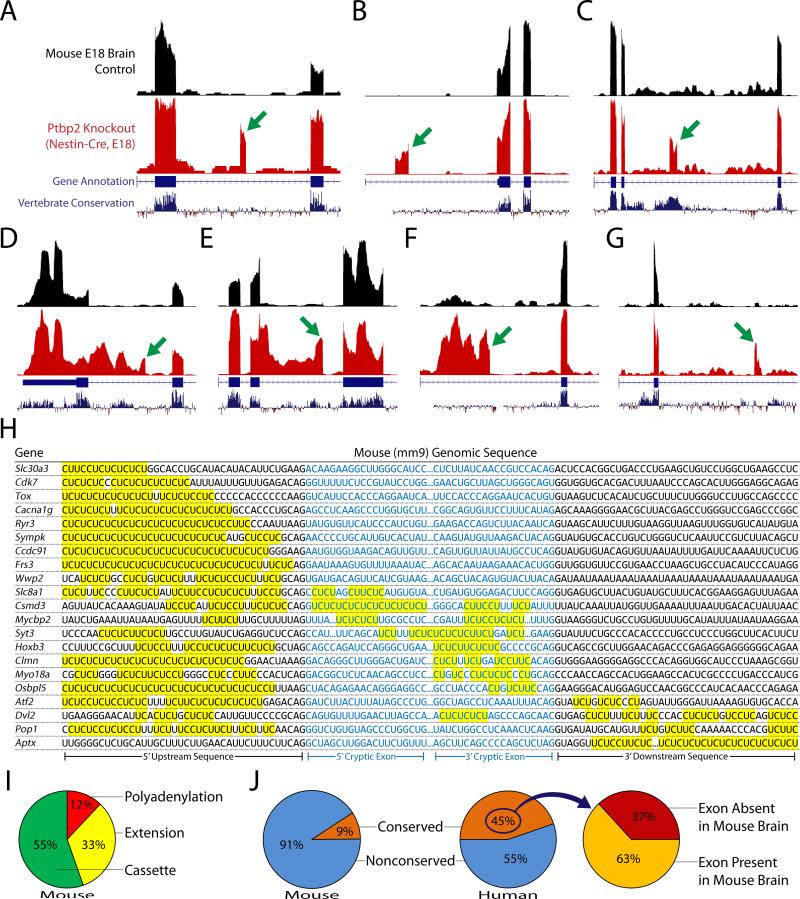
To validate that the nonconserved PTBP1/2 cryptic exons identified in human cells were distinct from other species, we examined RNA-seq data from mouse brain lacking Ptbp2 in neurons (Li et al., 2014). Analysis of these datatsets revealed cryptic exons that could be categorized as cassette exons (Fig. 3A to C), exon extensions (Fig. 3D and E), and polyadenylation sites (Fig. 3F and G) (166 total, Supplemental Excel File). Furthermore, robust CU microsatellites could be identified upstream, downstream, or internal to each cryptic exon (Fig. 3H). As expected, no overlap was found between human and mouse cryptic exons.
Figure 3. Identification of mouse nonconserved cryptic exons and conserved tissue-specific exons repressed by Ptbp1 and Ptbp2.
(A–G) Genetic deletion of PTBP2 in mouse neurons (Li et al., 2014)—where PTBP1 is already downregulated—reveals repressed exons that are analogous to those found in humans. The identified exons can also be classified as cassette exons [Ryr3, Sympk, Mycbp2, (A–C)], exon extensions [Slc30a3, Cacna1g, (D and E)] and polyadenylations sites [Cdk7, Wwp2, (F and G)]. H, Robust CU microsatellites can be found in the upstream, downstream, or internal sequences. Sequences that are missing the 3’ splice site ‘AG’ or 5’ splice site ‘GU’ dinucleotide reflect repressed exons that are premature polyA sites or exon extensions where a 5’ or 3’ splice junction is absent. (I) The majority of exons are cassette exons (55%), while polyadenylation sites (12%) comprise a smaller fraction. In contrast with humans, however, exon extensions (33%) are found nearly three times higher in frequency. (J) Furthermore, 91% of mouse repressed exons are nonconserved while only 9% are conserved—a striking difference when compared to humans where 45% of repressed exons are conserved. Cross referencing the conserved human exons (which are normally repressed in undifferentiated cells) to the mouse genome led to the discovery that many (~63%) of these exons are in fact actively spliced in mouse brain (Fig. 4).
Intriguingly, the vast majority of unannotated exons identified in the mouse neuron Ptbp2 knockout datasets were nonconserved: 91% in mouse compared with 55% in human (Fig. 3J). Upon further analysis, we observed that the majority (63%) of human conserved alternative exons—found to be repressed in non-neuronal cells—are normally spliced in at high levels in mouse neurons (Fig. 4A to C). Surprisingly, deletion of Ptbp2 has either no impact on many of these neuron-included exons (Fig. 4B, Supplemental Fig. 4) or only moderately increases exon inclusion (Fig. 4C, Supplemental Fig. 5). Alternative splicing analysis of the mouse Ptbp2 knockout datasets fails to identify the importance of these exons without the context of the human PTBP1 and PTBP2 double knockdown.
Figure 4. Conserved PTBP1/2 cryptic exons are activated in brain tissue.
(A–C) Conserved repressed exons identified in the human, non-neuronal RNA-seq datasets that are actively spliced in mouse neurons. Many of these activated tissue-specific exons are surprisingly unaffected by PTBP2 deletion [(A and B), Supplemental Fig. 4] and would thus be undetected by alternative splicing software algorithms when analyzing the mouse datasets without the context of the human double knockdown. Some exons, however, do show a moderate increase in splicing frequency [(C), Supplemental Fig. 5]. (D) When conserved and nonconserved exons are functionally categorized, a clear difference emerges. Conserved tissue-specific exons are more likely to produce in-frame insertions into the protein sequence than nonconserved cryptic exons (54% vs. 3%). In contrast, nonconserved cryptic exons are more frequently not frame-preserving (i.e. NMD or premature polyadenylation).
What are the functional consequences of activating these repressed exons during neuronal differentiation? Many of these conserved sequences (54%) produce in-frame insertions that may alter the protein structure to perform roles necessary for neural development (Fig. 4D, Supplemental Table 1). In contrast, nonconserved cryptic exons are generally not frame-preserving; only 3% of nonconserved cryptic exons are in-frame (Fig. 4D). Some conserved PTBP1/2 exons, however, appear to downregulate non-neuronal transcripts through NMD (Supplemental Table 2). For example, SNAP23—a ubiquitous component of membrane fusion machinery—appears to be downregulated by NMD as PTBP1/2 repression decreases, allowing for the neuronal homolog SNAP25 to dominate.
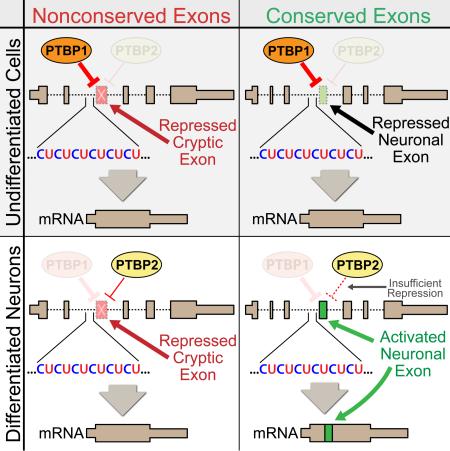
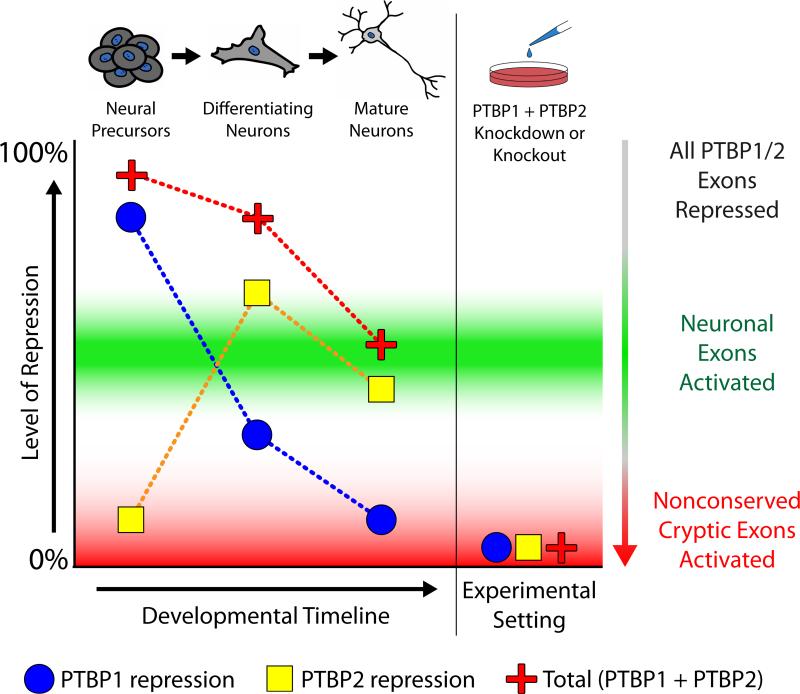
To explain these observations, we propose a splicing model that builds upon the current mechanistic understanding of PTBP1 and PTBP2 (Fig. 5). In undifferentiated neural precursor cells, PTBP1 expression is high and both neuronal exons and nonconserved cryptic exons are repressed. Through the process of neuronal differentiation, PTBP1 expression is reduced and PTBP2 expression is increased but because PTBP2 is a weaker splicing repressor (Keppetipola et al., 2016), neuronal exons become activated. Despite weaker repression, however, PTBP2 is still able to repress deleterious nonconserved cryptic exons. It is only under experimental conditions that we are able to reduce PTBP1 and PTBP2 to levels low enough for nonconserved cryptic exon activation. Thus, neurons may titrate down the levels of PTBP1/2 repression to activate conserved tissue-specific exons, thereby generating the transcriptome diversity required for neuronal differentiation.
Figure 5. PTBP1 and PTBP2 regulate conserved neuronal exons and nonconserved cryptic exons by titrating the level of CU-repeat associated repression.
A proposed model to merge the dual roles of PTBP1 and PTBP2 in neuronal differentiation and cryptic exon repression. Initially, levels of PTBP1 are high in undifferentiated cells. During neuronal differentiation, PTBP1 expression decreases and PTBP2 is increased. However, while the increase in PTBP2 expression is sufficient to repress nonconserved cryptic exons, it is insufficient to repress conserved neuronal exons. Thus, in mature neurons, total repression levels are reduced to activate exons that are important for neuronal differentiation, but not reduced enough to allow the incorporation of deleterious nonconserved cryptic exons.
To further support this splicing model, we analyzed RNA-seq data generated from the brain tissue of various vertebrate species (Barbosa-Morais et al., 2012; Merkin et al., 2012). As expected, conserved exons that become activated during neuronal differentiation are also present in brain tissues of other vertebrate species (Supplemental Fig. 6). In contrast, nonconserved cryptic exons are species-specific and not found in any other vertebrate brains (Supplemental Fig. 7). Finally, we also analyzed mouse RNA-seq data from various tissue types to determine whether the splicing of conserved exons was correlated with the expression of PTBP1/2. Indeed, conserved PTBP1/2 exons were found in brain and muscle—tissues that express low levels of PTBP1 (Lilleväli et al., 2001; Llorian et al., 2016)—but not found in colon, kidney, liver, lung, spleen, or testes (Supplemental Fig. 8).
Discussion
Together, our data establish that PTBP1 and PTBP2 are splicing factors that utilize CU repeats to repress conserved tissue-specific exons and nonconserved cryptic exons. These results expand upon our previous finding that TDP-43 targets UG repeats to repress nonconserved cryptic exons (Ling et al., 2015). In contrast to TDP-43, however, PTBP1 and PTBP2 regulate both conserved and nonconserved splicing events. This dichotomy can be clearly demonstrated when repressed exons are functionally categorized. For example, 54% of conserved exons are in-frame, while only 3% of nonconserved cryptic exons are in-frame, suggesting that conserved tissue-specific exons are far more likely to alter protein sequences in a productive manner. We also find that nonconserved cryptic exons—which only arise after PTBP1/2 depletion—are completely repressed under normal conditions across different tissue types and brains from various vertebrate species. Indeed, the nonconserved cryptic exons identified in brains of Ptbp2 knockout mice are not present in any RNA-seq datasets from normal mouse brain or rat brain, despite high concordance between mouse and rat genomes. These findings lead to the conclusion that conserved and nonconserved exons are differentially regulated by PTBP1/2 and functionally distinct.
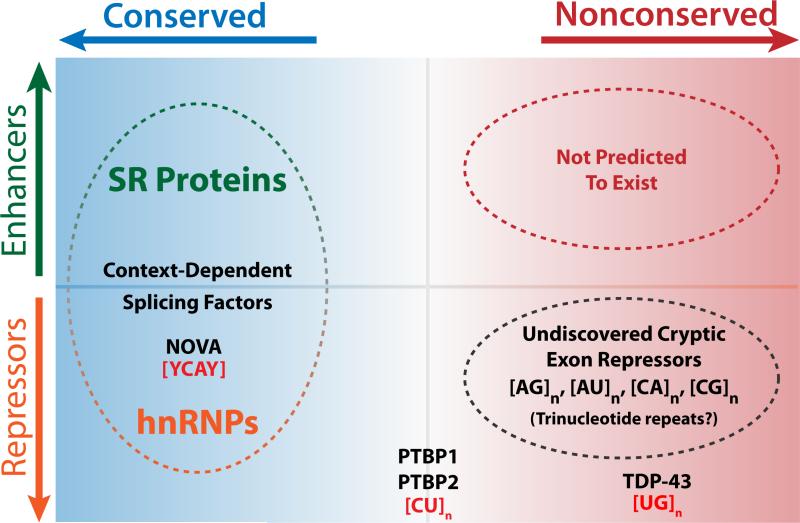
We therefore propose the existence of a conserved vs. nonconserved axis of RNA splicing (Fig. 6). Cells must actively repress nonconserved cryptic exons that would otherwise disrupt the fidelity of canonical mRNA splicing. In certain contexts, however, activating conserved exons by reducing repeat-associated repression may play an important role in generating transcriptome and proteome complexity (Eom et al., 2013). The conserved exons associated with PTBP1 and PTBP2 are expected to be important for neuronal differentiation and may even play a role in muscle differentiation as well (Supplemental Fig. 8). The mechanistic parallels between PTBP1, PTBP2, and TDP-43 suggest that these splicing factors may belong to a larger family of cryptic exon repressors. Given the diversity of microsatellites sequences within the genome, we predict that other cryptic exon repressors will be uncovered in the near future.
Figure 6. Conserved vs. nonconserved axis of the splicing landscape.
In the exon definition model of RNA splicing, conserved exons are governed by splicing factors that can act as enhancers or repressors, depending on context. Many splicing factors bind to loosely defined consensus sequences, allowing for the wide diversity in exonic sequences. However, this flexibility comes at a cost since cryptic “exon-like” sequences may stochastically appear within introns—a consequence of mutations that accumulate over evolutionary timescales. Certain microsatellite-binding splicing factors appear to serve as cryptic exon repressors, although their original functions prior to the evolution of higher order RNA-splicing remain unclear. Interestingly, while TDP-43 appears to primarily regulate nonconserved cryptic exons, PTBP1 and PTBP2 have gained additional roles in the regulation of conserved tissue-specific exons involved in neuronal maturation and cellular differentiation. Other splicing factors such as NOVA have been previously documented to regulate cryptic exons (Eom et al., 2013), although it remains to be seen whether NOVA regulates nonconserved splicing events. It is predicted that additional cryptic exon repressors exist to bind repetitive sequences beyond [CU]n and [UG]n.
Advances in whole-genome sequencing have greatly accelerated in recent years, but our ability to interpret and understand these vast amounts of data has lagged behind. This disparity is particularly apparent when comparing exonic and intronic regions of the genome. Mutations discovered in the exome reflect direct changes in the encoded protein but mutations that occur in the intron have a far more mysterious effect. Intronic sequences that are regulated by cryptic exon repressors may represent regions of susceptibility for neurodegeneration, cancer, and other conditions. Thus, characterizing the nonconserved cryptic exons within the human genome may couple with the advent of personalized genetic medicine and allow us to more fully interpret SNPs and somatic mutations that arise in human disease.
Experimental Procedures
Cell Culture and Manipulation
HeLa cells were cultured in Dulbecco's Modified Eagle's Medium (Corning, 10-017-CV) supplemented with 1% GlutaMAX (Thermo Fisher Scientific, 35050-061), 10% FBS (Corning, 35-010-CV) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific, 15070-063). Knockdown of PTB1 and PTB2 was performed by transfecting using siRNA as previously described (Ling et al., 2015) (Sigma) while control was transfected with siRNA against GFP (Thermo Fisher Scientific). Transfection of siRNA was achieved using Lipofectamine 3000 (Thermo Fisher Scientific, L3000-008) following the manufacturer's protocol.
RT-PCR and qRT-PCR of Human Targets
cDNA was derived from transfected HeLa cell total RNA (1ug total RNA / 20ul first strand cDNA reaction) using ProtoScript II (NEB, E6560S). Primers were designed against cryptic exon targets and PCR reactions were performed using Phusion High-Fidelity DNA Polymerase (NEB, M0530S). qRT-PCR primers were designed against ANKS6, IQGAP1, LEPRE1 and LEPREL2. Primers were then used to amplify cDNA products from controls and PTBP1/2 knockdown, in triplicate, using the ABI Prism 7900 Sequence Detector System.
Primer sequences used for RT-PCR and qRT-PCR are listed in the Supplemental Experimental Procedures.
Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP) of PTBP1
HeLa cells were transfected with a plasmid expressing PTBP1 Tagged at the C-terminus with a dual 6His-biotin tag (Tagwerker, 2005). Cells were grown for 14 hrs in the presence of 4-thiouridine, media was then removed, and cells were irradiated for one minute with a 365nm UV-LED light source (Hamamatsu LC-L5). Cross-linked cells were removed by scraping in PBS, centrifuged; pellets were frozen at −80°C. Pellets from ten 15cm plates of cells were resuspended in 7.5 ml of 300 mM NaCl, 0.5% NP-40, 50 mM NaPO4 pH 7.2, 10 mM immidazole, 6 M Guanidine HCl, Protease inhibitor Cocktail VII (RPI) and the fusion protein was purified by tandem affinity chromatography and cross-linked RNA used for library construction and sequencing by Illumina HiSeq as previously described (Schaughency et al., 2014).
For hexamer motif analysis, reads were deduplicated using the FASTX-toolkit (Hannon Lab, Cold Spring Harbor Laboratory). Both deduplicated and original reads were, then, aligned to the human genome (hg19) using bowtie (v1.1.2) (Langmead et al., 2009) with the following options: -y --best --strata -n 3 -M 1. The output was then filtered for only reads that contained a T>C transversion using custom scripts (available upon request). Bedtools (Quinlan and Hall, 2010) was used to divide the reads into those that were exonic or intronic.
RNA Preparation and RNA-seq Analysis
RNA was extracted from cell culture samples using TRIzol (Thermo Fisher Scientific, 15596-026) and RNeasy Mini Kits (Qiagen, 74104). Total RNA for RNA-seq was then processed using the TruSeq Stranded Total RNA Library Prep Kit (Illumina) to construct 100-bp paired end stranded RNA-seq libraries. Sample libraries were then sequenced on a HiSeq 2500. Samples were then de-multiplexed and converted into fastq files.
Fastq files were aligned to mouse and human genomes using HISAT2 (Kim et al., 2015) and gene abundances were calculated using Cufflinks (Trapnell et al., 2012). To identify cryptic exons, HISAT2 generated BAM files were processed using StringTie (Pertea et al., 2015), a transcript assembly software. StringTie GTF outputs were then processed for unannotated exons identified in the PTBP1/2 knockdown datasets. Relative read coverage and splice junction coverage between control and knockdown conditions were then calculated for each unannotated exon using Cufflinks and SpliceMap (Au et al., 2010) and ranked by fold change accordingly. Data was then displayed on the UCSC Genome Browser to visualize RNA-seq coverage and each unannotated exon was manually curated for the presence of CU repeats, vertebrate conservation, and cryptic exon classification. Vertebrate conservation was determined using the “100 vertebrates Basewise Conservation by PhyloP” conservation track; positive likelihood ratio values were considered as conserved and negative values as nonconserved. Low quality alignments or misalignments, highly repetitive intergenic regions, intra-exonic splice junctions, and other false positives were manually discarded. Data visualization on the IGV genome browser was also used to ensure that cryptic exon alignments were strand-matched to the associated transcripts.
Antibodies
The following antibodies were used in this study: rabbit polyclonal against PTBP1 (1:1000; Cell Signaling, 8776); rabbit polyclonal against PTBP2 (1:500; Proteintech, 55186-1-AP); mouse monoclonal against GAPDH (1:10,000; Sigma-Aldrich, G8795) and rabbit monoclonal against GAPDH (1:10,000; Cell Signalling, D16H11).
Protein biochemistry
To obtain homogenate from treated cell cultures, cells were lysed and homogenized in RIPA buffer (Sigma-Aldrich) containing protease inhibitor (Roche). Cell debris and nuclei were removed by centrifugation at 3200 rpm for 10 min resulting in supernatant S1 (soluble fraction) and pellet P1 (membrane associated fraction). Protein concentration of S1 was determined by BCA. Twenty microgram protein was loaded onto 4-12% (w/v) SDS–polyacrylamide gels. Separated proteins were blotted onto PVDF membranes (Whatman, Maidstone, UK) and blocked with 5% (w/v) BSA/0.01% (v/v) Tween 20 in TBS for 1 h. After blocking, membranes were incubated overnight at 4 °C in blocking solution containing primary antibody. Blots were washed and incubated with the appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. Proteins were visualized and quantified using AFP imaging mini medical system (AFP imaging corporation).
Supplementary Material
Acknowledgments
We thank J. Nathans [Johns Hopkins Medical Institution (JHMI)] for valuable comments and suggestions. We also thank D. Ramos (JHMI) for assisting with qRTPCR analysis. Last, we thank the staff of the Next Generation Sequencing Center (JHMI) for their RNA sequencing service. This work was supported in part by the JHU Neuropathology Pelda fund and NIH grants P30-CA006973, P50-GM107632 (NIGMS subaward to S.J.W.), R01-GM066108 (J.L.C.), and R01-NS095969 (P.C.W.). R.C. is supported by the Excellence Initiative of the German Federal and State Governments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
All authors designed experiments and interpreted results. J.P.L. and R.C. performed in vitro cell culture, transfection and molecular biology experiments. J.D.M. and J.L.C. performed PAR-CLIP experiments and analysis. P.M.S. and S.J.W. assisted with PAR-CLIP mapping and motif analysis. J.P.L. analyzed RNA-seq data and characterized the cryptic exons. J.P.L. and P.C.W. wrote the paper and all authors approved the manuscript.
Accession Codes: raw sequencing files have been deposited at the NCBI Sequence Read Archive under SRP068862.
Conflicts of Interest: None declared.
References
- Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–8. doi: 10.1093/nar/gkq211. doi:10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–93. doi: 10.1126/science.1230612. doi:10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin C, Chawla G, Ostrow K, Shiue L, Ares M, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–52. doi: 10.1101/gad.1558107. doi:10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinegar AE, Cooper TA. Roles for RNA-binding proteins in development and disease. Brain Res. 2016:1–8. doi: 10.1016/j.brainres.2016.02.050. doi:10.1016/j.brainres.2016.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. doi:10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. doi:10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, Zhang C, Wang H, Lay K, Fak J, Noebels JL, Darnell RB. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife. 2013;2:e00178. doi: 10.7554/eLife.00178. doi:10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. doi:10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Sharp PA, Jamison SF, Garcia-Blanco MA. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–36. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- Gueroussov S, Gonatopoulos-Pournatzis T, Irimia M, Raj B, Lin Z-Y, Gingras A-C, Blencowe BJ. An alternative splicing event amplifies evolutionary differences between vertebrates. Science. 2015;349:868–73. doi: 10.1126/science.aaa8381. doi:10.1126/science.aaa8381. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. doi:10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TD, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, Ares M, Yeo GW. Integrative Genome-wide Analysis Reveals Cooperative Regulation of Alternative Splicing by hnRNP Proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. doi:10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, DePinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. doi:10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafasla P, Mickleburgh I, Llorian M, Coelho M, Gooding C, Cherny D, Joshi A, Kotik-Kogan O, Curry S, Eperon IC, Jackson RJ, Smith CWJ. Defining the roles and interactions of PTB. Biochem. Soc. Trans. 2012;40:815–20. doi: 10.1042/BST20120044. doi:10.1042/BST20120044. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. doi:10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola NM, Yeom K-H, Hernandez AL, Bui T, Sharma S, Black DL. Multiple determinants of splicing repression activity in the polypyrimidine tract binding proteins, PTBP1 and PTBP2. RNA. 2016 doi: 10.1261/rna.057505.116. doi:10.1261/rna.057505.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. doi:10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee J-A, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. doi:10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Li Q, Zheng S, Han A, Lin C-H, Stoilov P, Fu X-D, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. Elife. 2014;3:e01201. doi: 10.7554/eLife.01201. doi:10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. doi:10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilleväli K, Kulla A, Ord T. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech. Dev. 2001;101:217–20. doi: 10.1016/s0925-4773(00)00566-9. doi:S0925477300005669 [pii] [DOI] [PubMed] [Google Scholar]
- Linares AJ, Lin CH, Damianov A, Adams KL, Novitch BG, Black DL. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. Elife. 2015;4:1–25. doi: 10.7554/eLife.09268. doi:10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–5. doi: 10.1126/science.aab0983. doi:10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Gooding C, Bellora N, Hallegger M, Buckroyd A, Wang X, Rajgor D, Kayikci M, Feltham J, Ule J, Eyras E, Smith CWJ. The alternative splicing program of differentiated smooth muscle cells involves concerted non-productive splicing of post-transcriptional regulators. Nucleic Acids Res. 2016:gkw560. doi: 10.1093/nar/gkw560. doi:10.1093/nar/gkw560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-mRNA Splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. doi:10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman J. a, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. doi:10.1128/MCB.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–9. doi: 10.1126/science.1228186. doi:10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH-T. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. doi:10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Patton JG, Mayer SA, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–51. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Pérez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3’ splice site selection in vivo. RNA. 1997;3:764–78. [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–5. doi: 10.1038/nbt.3122. doi:10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6350–5. doi: 10.1073/pnas.110128397. doi:10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. doi:10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. doi:10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Schaughency P, Merran J, Corden JL. Genome-wide mapping of yeast RNA polymerase II termination. PLoS Genet. 2014;10:e1004632. doi: 10.1371/journal.pgen.1004632. doi:10.1371/journal.pgen.1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat. Rev. Genet. 2015;17:19–32. doi: 10.1038/nrg.2015.3. doi:10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C. A Tandem Affinity Tag for Two-step Purification under Fully Denaturing Conditions: Application in Ubiquitin Profiling and Protein Complex Identification Combined with in vivo Cross-Linking. Mol. Cell. Proteomics. 2005;5:737–748. doi: 10.1074/mcp.M500368-MCP200. doi:10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. doi:10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. doi:10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, Morris Q, Barash Y, Krainer AR, Jojic N, Scherer SW, Blencowe BJ, Frey BJ. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. doi:10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, Fu XD, Zhang Y. Genome-wide Analysis of PTB-RNA Interactions Reveals a Strategy Used by the General Splicing Repressor to Modulate Exon Inclusion or Skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. doi:10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.