Abstract
We report a high-throughput platform for delivering large cargo into 100,000 cells in 1 min. An array of micro-cavitation bubbles explode in response to laser pulsing, forming pores in adjacent cell membranes, and immediately thereafter, pressurized flows drive slow diffusing cargo through these pores into cells. The platform delivers large cargo including bacteria, enzymes, antibodies, and nanoparticles into diverse cell types with high efficiency and cell viability. We used this platform to explore the intracellular lifestyle of Francisella novicida and discovered that the iglC gene is unexpectedly required for intracellular replication even after phagosome escape into the cell cytosol.
INTRODUCTION
Heretofore, reliable methodologies for introducing large-sized cargo into mammalian cells at high throughput have not existed1. Biological approaches such as viruses can provide high transfer efficiency2, 3 but are restricted to kb-sized nucleic acids. Chemical methods that utilize lipids4, cationic polymers5, or insoluble precipitates vary in delivery efficiency, are highly cell type dependent, and frequently show undesirable endosome trapping of delivered cargo. Most physical approaches can bypass cell endocytosis processes, and the delivery typically involves two-steps: 1) disruption of the plasma membrane to create transient pores and 2) cargo delivery across transient pores before they reseal. To create transient pores, electroporation utilizes electrostatic forces to disrupt cell membranes6–8; sonoporation9, 10 generates acoustic pressure to trigger cavitation bubbles with strong fluid flows to induce membrane permeability; optoporation11–13 utilizes nonlinear optical absorption triggered by a short laser pulse to break down cell membranes; and microfluidic channels14 utilize narrow structures to squeeze cell membranes. These mechanisms are limited to small cargo delivery since they rely on thermal diffusion for cargo crossing at transient membrane pores. Slowly diffusing large cargo has little chance to transit pores before they reseal. Nanochannel electroporation8 can induce convective injection by electrophoresis, but it is only suitable for nanosized cargo. Molecules adhering onto nanomechanical pillars15 can be introduced into cells directly. However, sharp tips coated with large cargo may lose the ability to penetrate cell membranes. Micropipette-based approaches provide active pressure to drive large cargo into the cell cytosol16–19. These methods, however, are low throughput and may suffer from complications, such as clogging or traumatic cell lysis, as cargo size increases beyond ~500 nm.
To overcome the bottleneck for high throughput large cargo delivery, we developed a massively parallel photothermal platform, termed BLAST (Biophotonic Laser Assisted Surgery Tool), that can deliver up to micron-sized cargo into ~100,000 cells in 1 min, providing 5 orders of magnitude higher throughput than prior microcapillary based approaches19. A wide range of cargo including live bacteria, enzymes, antibodies, and functional nanoparticles have been successfully delivered into a variety of cell lines, including three types of primary cells [human peripheral blood monocyte-derived macrophages (PB-MDMs), primary normal human dermal fibroblasts (NHDFs), and human primary renal proximal tubule epithelial cells (RPTECs)] and one cancer cell line (HeLa), at high efficiency and high cell viability. We have also shown that BLAST delivers cargo directly into the cell cytosol, avoiding cargo entrapment in endosomes and maintaining their functionalities after delivery.
RESULTS
The Structure and Operating Principle of BLAST
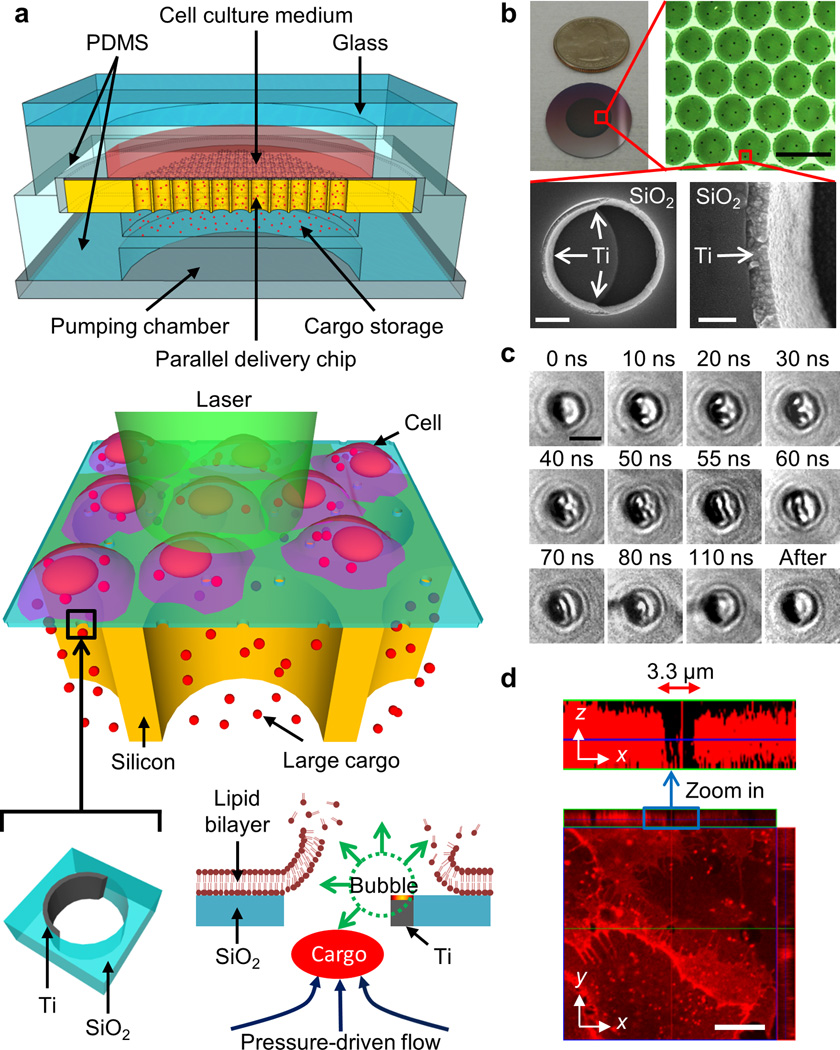
The BLAST platform consists of a silicon chip with a thin porous SiO2 membrane on top providing an array of micron-wide, trans-film holes, whose sidewalls are asymmetrically coated with crescent-shaped titanium thin films (Fig. 1a). Underneath the porous SiO2 membrane is an array of short, vertical silicon channels mechanically supporting the fragile porous membrane as well as providing fluid passage for cargo delivery (Supplementary Fig. 1 for detailed fabrication process). The procedure for cargo delivery on a BLAST platform is as follows: Step 1: Cells are cultured or made to adhere on a silicon chip. Step 2: The chip is assembled with a microliter chamber loaded with the cargo to be delivered. Step 3: A nanosecond pulse laser is triggered to scan rapidly across the entire chip to generate membrane pores in cells in contact with the titanium thin films, and immediately thereafter the elastic chamber is pressurized to deliver cargo into cells through these transient pores (Supplementary Note for detailed delivery mechanistic information). Laser pulsing and cargo pumping (Step 3) take 10 s. BLAST delivers cargo into cells in a batch mode. Each batch can deliver cargo into 100,000 cells, depending on chip area (currently 1 cm2). To deliver cargo to more cells, as many chips as desired can be prepared and Steps 2 and 3 repeated. Each batch delivery takes about 1 min.
Figure 1. Schematic of a massively parallel photothermal platform for large cargo delivery.
(a) The BLAST platform consists of an array of trans-membrane holes patterned on a 1.5 µm thick SiO2 film. Crescent-shaped titanium films are asymmetrically coated on the sidewalls of these holes to harvest laser pulse energy. Rapid pulsed laser scanning of the entire active photothermal region on the chip triggers cavitation bubbles in all holes that disrupt contacting cell membranes. Following membrane opening, an external pressure source is applied to deform the bottom flexible polydimethylsiloxane (PDMS) storage chamber to push cargo into the cytosol of cells via these transient membrane pores. (b) Images of a BLAST chip for delivery. SEM images show a 100 nm thick titanium film coated on the inner sidewall of a 3 µm hole on the SiO2 membrane through a lift-off process using the e-beam evaporation method. Scale bars, 100 µm (upper right), 1 µm (lower left), 200 nm (lower right). (c) Time-resolved images capturing rapidly expanding cavitation bubbles triggered in a hole from 10 to 110 ns after laser pulsing. At 10–50 ns, two bubbles form and enlarge at the upper and lower poles of the hole; at 55 ns, the bubbles coalesce; and by 110 ns, the bubbles collapse. Scale bars, 3 µm. (d) Confocal z-axis scanning images show that the cell membrane of a fixed cell on the platform is disrupted and reveals a micron-sized pore after laser pulsing.
Mechanism of Opening Transient Cell Membrane Pores
BLAST utilizes laser energy harvested by the metallic titanium thin films in each trans-film hole (Fig. 1b) to induce rapid heating and vaporization of adjacent water layers to trigger cavitation bubbles that cause disruption of a contacting cell membrane20–24. Cavitation bubbles in each SiO2 hole initiate at the two tips of the crescent shaped titanium film, where hot spots are located. Local electric field enhancement occurs near the tips due to the lightning-rod effect. Scanning the chip with laser pulses triggers cavitation bubbles that grow, coalesce, and collapse within 110 ns (Fig. 1c). The size of a cavitation bubble is highly dependent upon laser fluence (Supplementary Fig. 2). This rapid bubble ‘explosion’ induces strong fluid flows that can disrupt an adjacent plasma membrane19. A confocal fluorescence image shows a pore formed in a plasma membrane of a chemically fixed cell on the platform (Fig. 1d). This reveals that cell membrane cutting by BLAST is highly localized.
Delivery Efficiency and Membrane Resealing Time
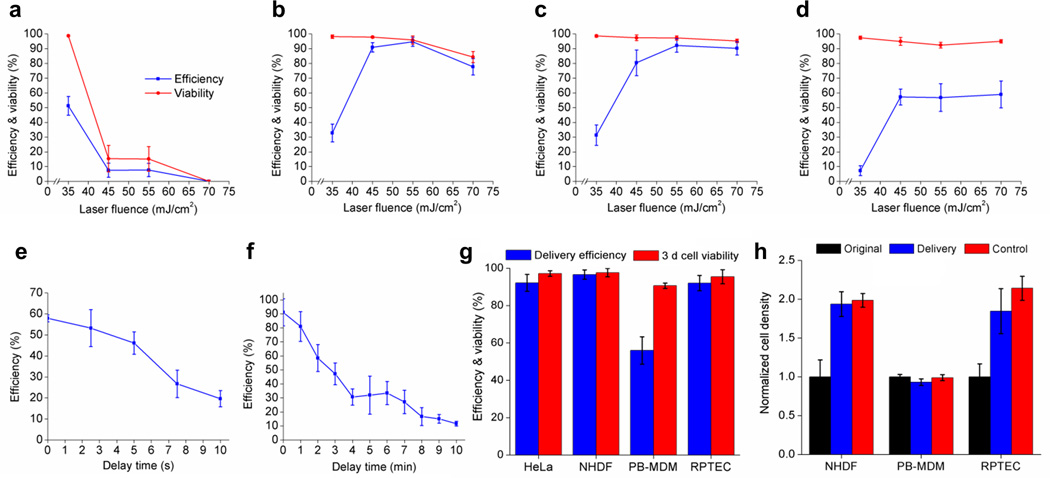
Cell viability and membrane opening efficiency by BLAST depends on the SiO2 hole density and laser pulse energy. Higher hole densities generate more transient pores per cell. Optimal conditions for high membrane opening efficiency with high cell viability (>90%) for HeLa cells has been experimentally determined as ~1-2 holes/cell with a laser fluence of 55 mJ/cm2 (Fig. 2a-d).
Figure 2. Cell membrane pore opening efficiency, cell viability, and pore resealing time.
(a-d) Plots show laser fluence and hole density are critical in determining optimal delivery conditions. The hole density on the chip varies as follows: (a) 1.15 (b) 0.28 (c) 0.18 or (d) 0.12 holes/10 µm2. Pore opening efficiency is determined by calcein (622 Da) dye uptake for HeLa cells. Error bars, s.d. (n = 2,031; 1,483; 1,601; 1,975 for a; b; c; d) (cell density: 1,138 cells/mm2). (e,f) Characterization of the transient window for large cargo and small molecule delivery. (e) Live bacteria, F. novicida, were delivered into HeLa cells at different delay time between laser pulsing and fluid pumping. Error bars, s.d. (n = 1,926; 2,062; 2,335; 2,406; 2,106 for 0; 2.5; 5; 7.5; 10 s). (f) Calcein was delivered at different time delays. The cell membrane pores remain permeable for longer than 10 min for small molecule delivery. Error bars, s.d. (n = 5,891 for all tests). (g) FITC-dextran (40 kDa) delivery efficiency and 3 d cell viability for different kinds of primary mammalian cells. Error bars, s.d. (n = 2,030; 1,512; 1,464; 532 for HeLa; NHDFs; PB-MDMs; RPTECs). (h) FITC-dextran (40 kDa) delivered primary cells were cultured on chips for 3 d and compared with their original cell densities. Control cells were seeded on chips at the same time without laser pulsing and fluid pumping. Error bars, s.d. (n = 3,062; 3,017; 1,149 for NHDFs; PB-MDMs; RPTECs) (initial cell densities: 265; 534; 98 cells/mm2 for NHDFs; PB-MDMs; RPTECs).
Cell membrane resealing starts immediately after transient pores are generated25. Large cargo delivery is challenging since these transient pores shrink quickly, impeding the diffusion of large sized cargo through the pores, a relatively slow process. Therefore active, not diffusion based, cargo delivery is timed to occur before the cell membrane reseals. To measure the transient opening timeframe for large cargo delivery, we delivered a micron-sized bacterium, green fluorescence protein (GFP) - expressing Francisella novicida, into HeLa cells at various delay time intervals between laser pulsing and active fluid pumping. As expected, delivery efficiency decreased with increasing delay time. With no delay, the average delivery efficiency across the entire chip was ~58%, whereas with a 10 s delay, the efficiency dropped to ~20% (Fig. 2e). The delivery of small cargo can tolerate a longer delay time. For example, high efficiency delivery can be maintained for up to several minutes for membrane impermeable calcein dye (Fig. 2f), suggesting that a micron-sized pore opened by a cavitation bubble quickly shrinks to exclude the delivery of large cargo but remains permeable to small molecules. Complete membrane resealing takes longer than 10 min. Of note, the 40–50% average efficiency for delivering intracellular bacteria is partially due to the slow laser scanning. In our current setup, it takes about 10 s to scan the entire 1 cm2 chip. Many cells in the initially scanned regions have shrunk their pores by the time the pressured delivery is applied, reducing bacterial delivery efficiency.
BLAST delivery also works well for primary mammalian cells. FITC-dextran (40 kDa) was delivered into normal human dermal fibroblasts (NHDFs), peripheral blood monocyte-derived macrophages (PB-MDMs) and renal proximal tubule epithelial cells (RPTECs), and the cells incubated on the platform for 3 d to check viability and proliferation. All three primary cell lines retained high cell viability (>90%) 3 d after delivery (Fig. 2g). The delivery efficiencies for NHDFs and RPTECs were higher than 90%. PB-MDMs only approached 60% efficiency, likely because of their smaller size and reduced contact area with the platform compared to much larger NHDF and RPTEC cells (Supplementary Fig. 3). We quantified the proliferation of primary cells after cargo delivery by cell density determinations for cells with or without (control, no laser pulsing or fluid pumping) FITC-dextran delivery and found that BLAST delivery does not affect cell growth (Fig. 2h). PB-MDMs, which are terminally differentiated cells that do not divide, maintained their original cell density.
In addition to live intracellular bacteria, a variety of additional large cargo, including antibodies, micron-sized polystyrene beads, and magnetic beads have also been successfully delivered into cells using BLAST (Supplementary Fig. 4a-c). To study the effect of cargo size on delivery efficiency, we delivered an increasing series of five different sizes of polystyrene beads (20 nm, 200 nm, 500 nm, 1 µm, and 2 µm). The efficiency decreased with cargo size from 93% for 20 nm beads to 62% for 2 µm beads; cell viability in all cases was maintained above 90% (Supplementary Fig. 4d). Multiple payloads can be co-delivered at the same time. In fact, combinatorial delivery is one major advantage of BLAST since it offers less size-diffusion variance by providing active pressure driven flows during delivery (Supplementary Fig. 4e,f). Evaluation of stress levels of HeLa cells after BLAST delivery shows no major increase in heat shock HSPA6 gene expression, a general cell stress sensor (Supplementary Fig. 5). All cell and cargo types, delivery efficiencies, and cell viabilities are summarized (Supplementary Table 1).
Direct Delivery into the Cytosol Bypasses Endocytosis
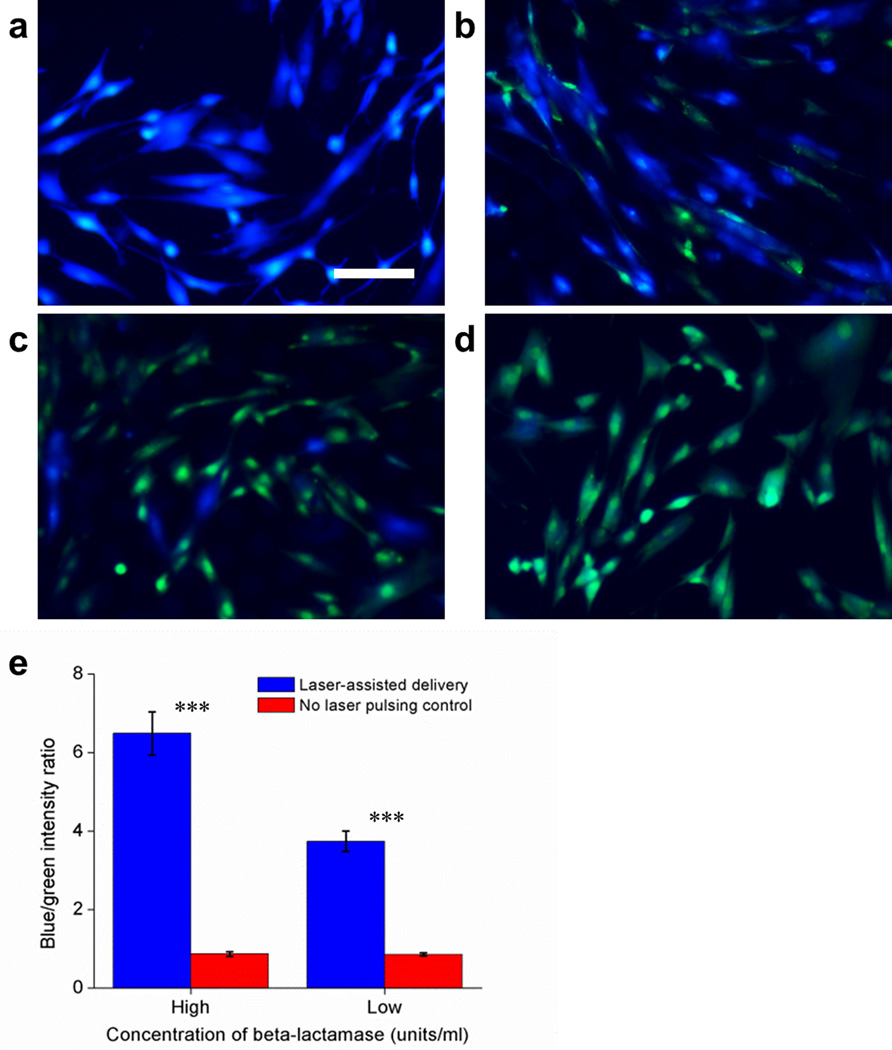
To confirm that BLAST delivers biologically active and undamaged cargo directly into the cell cytosol, we delivered the bacterial enzyme β-lactamase (29 kDa) by BLAST into NHDFs, and evaluated its delivery and functional activity by incubating the cells with the esterified β-lactamase substrate, CCF4-AM. When CCF4-AM enters the cytosol, endogenous esterases convert it to CCF4, which can be detected by fluorescence resonance energy transfer (FRET). Excitation of CCF4 at 408 nm wavelength leads to efficient FRET and emission of green fluorescence at 530 nm wavelength. Bacterial β-lactamase that has been delivered by BLAST into the cell cytosol cleaves CCF4 into two separate fluorophores and the FRET effect is lost, with the emission fluorescence changing from green to blue at 460 nm wavelength. CCF4-loaded cells at 5 h after β-lactamase delivery are shown (Fig. 3). NHDFs into which more enzyme was delivered show a higher blue-to-green fluorescence intensity ratio (Supplementary Fig. 6 for lower magnification images covering a larger area). This result shows that the enzyme remains functional in the cytosol after BLAST delivery.
Figure 3. BLAST delivered β-lactamase enzyme is functional inside NHDFs.
(a-d) β-lactamase at 50 units/ml (a, c) or 1 unit/ml (b, d) in PBS were delivered into NHDFs using BLAST under the condition of fluid pumping but with (a, b) or without (c, d) laser pulsing. Scale bar, 100 µm. (e) Blue and green fluorescent intensities for all 4 groups were measured 5 h after β-lactamase delivery. A higher blue-to-green fluorescence ratio means more β-lactamase was delivered into cells and remained functional, and thus able to cleave CCF4 molecules. Error bars, s.d. (n = 976; 577; 1,005; 353 for 50 units/ml delivery; 50 units/ml control; 1 unit/ml delivery; 1 unit/ml control, *** p < 0.0001, t-test). The experiment was conducted 3 times with the same results.
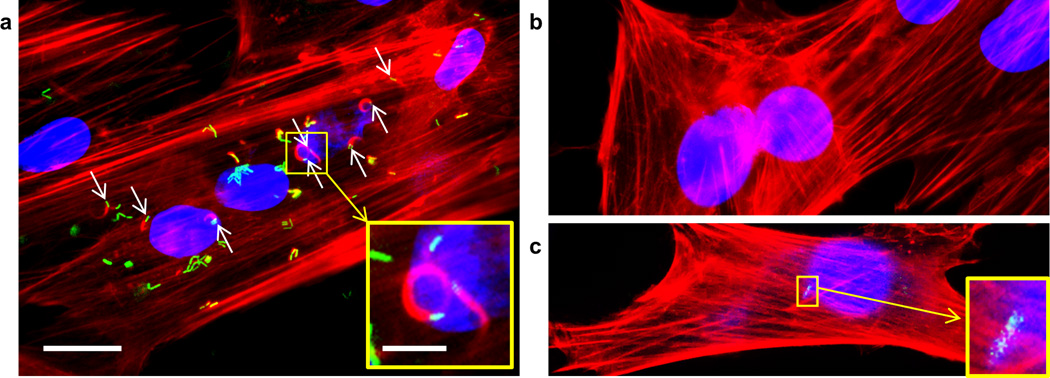
We also delivered an escape-incompetent strain of Listeria monocytogenes deficient in listeriolysin O and phospholipase C26 into NHDFs (strains used are described in Supplementary Table 2). This L. monocytogenes mutant is well established as being unable to escape its vacuole to recruit actin and thereby form actin comet tails in the cytosol26. Formation of actin tails (visualized by red fluorescent rhodamine-phalloidin staining, Fig. 4a) by these bacteria after BLAST delivery is strong direct evidence that they have been delivered directly into the cell cytosol and remain alive and metabolically active (Fig. 4a). In the absence of laser pulsing, these escape incompetent bacteria are taken up poorly and never form actin tails (Fig. 4b, c). Because NHDFs are not phagocytic, uptake of the bacteria by NHDFs in the absence of laser pulsing is extremely inefficient, and as a result, considerably more bacteria are observed in the laser pulsed cells (Fig. 4a) than in non-laser pulsed control cells (Fig. 4b, c) (p < 0.0001 by t-test for difference in the number of bacteria delivered with versus without laser pulsing). We have obtained the same results after BLAST delivery of these bacteria into HeLa cells (data not shown) and have confirmed that they are unable to form actin tails after phagocytic uptake and endolysosomal entrapment by THP-1 macrophages (Supplementary Fig. 7).
Figure 4. Escape-incompetent Listeria monocytogenes, deficient in listeriolysin O and broad range phospholipase C, nucleate actin after BLAST delivery directly into the cytosol of NHDFs.
L. monocytogenes-GFP Δhly ΔplcB (green), strain 10403S, was delivered by BLAST with laser pulsing into the cytosol of NHDFs (a) or, as a sham control, in the absence of laser pulsing (b, c). Nuclei, stained with DAPI, fluoresce blue and F-Actin, stained by phalloidin-rhodamine, fluoresces red. A higher magnification of a green fluorescent bacterium and its actin comet tail after BLAST delivery with laser pulsing is shown (a, inset). In the absence of laser pulsing, intracellular bacteria are rare and comet tails are not seen (b, c, inset). Scale bars, 20 µm and 5 µm for zoom-in images. The experiment was conducted twice with the same results each time.
Study of Intracellular Lifestyle of a Pathogen using BLAST
BLAST enables massively parallel, high efficiency, and nearly simultaneous delivery of large cargo into mammalian cells under uniform physiological conditions. This feature allows reliable statistical analysis of delivered cargo and their interactions with cells over time. Here, as an example, we utilize BLAST to examine the role of iglC, a gene within the Francisella Pathogenicity Island (FPI), in intracellular replication of Francisella.
Francisella is a highly virulent Gram-negative facultative intracellular bacterium that causes a fatal zoonotic disease, tularemia. Francisella subverts host cell trafficking by preventing phagolysosomal fusion and escapes into the host cell cytosol, where it replicates27. Essential to its virulence and its capacity to escape into the cytosol and replicate intracellularly is a cluster of 17 genes, the FPI, thought to encode a Type 6 Secretion System (T6SS)28–30. Disruption of almost any of the FPI genes prevents phagosome escape and intracellular replication31. Because vacuolar escape precedes and appears to be required for intracellular bacterial replication of Francisella, it has been impossible to determine whether the FPI is required only for vacuolar permeabilization and bacterial escape into the cytosol or is additionally required for intracytosolic activities such as replication following escape.
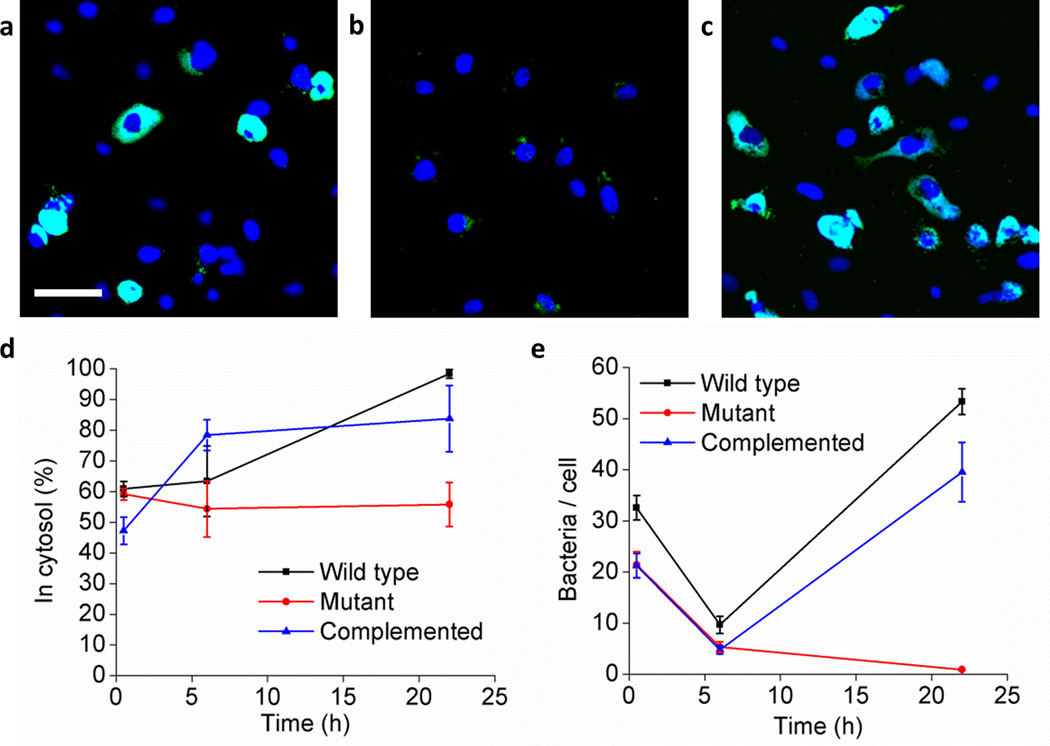
Three strains of F. novicida – the wild-type, a ΔiglC mutant in which the iglC gene has been deleted, and a ΔiglC mutant complemented with the iglC gene - were delivered into HeLa cells (Supplementary Fig. 8). HeLa cells are non-phagocytic and inefficiently internalize F. novicida, but using BLAST, the F. novicida strains were delivered directly into the cell cytosol. After the HeLa cells recovered, they were harvested from the BLAST platform for study of bacterial intracellular trafficking and multiplication over time. Bacterial replication at 22 h after these three strains were delivered into HeLa cells is shown in fluorescence images (Fig. 5a-c). We used a modification of the differential digitonin permeabilization assay32 to identify cytosolic versus vacuolar bacteria, confirming extensive cytosolic localization of the bacteria (Fig. 5d and Supplementary Fig. 9). Approximately 50–60% of these three types of F. novicida delivered on the platform were found in the cytosol 1 h after delivery, suggesting that 40–50% of bacteria were re-packaged into vacuoles. The wild-type and iglC complemented F. novicida show a higher ratio in the cytosol with increasing incubation time (p < 0.001 and p < 0.01, respectively, 2-way ANOVA with Bonferroni post-test correction). We assayed bacterial intracellular growth by counting the number of bacteria in HeLa cells over time (Fig. 5e). The Francisella wild-type strain replicated extensively within the cytosol as evidenced by large areas of green fluorescence within the HeLa cells at 22 h (Fig. 5a, e), whereas, unexpectedly, the Francisella with a deleted iglC gene33 was unable to replicate intracellularly even after direct delivery into the cytosol (Fig. 5b, e). Complementation of the iglC gene from a plasmid restored the capacity of the ΔiglC mutant to multiply within HeLa cells after cytosolic delivery (Fig. 5c, e), indicating that iglC is required for bacterial cytosolic replication even after vacuolar escape. Indeed, growth curves revealed that numbers of ΔiglC mutant bacteria per HeLa cell decrease over time, suggesting that host defense mechanisms of HeLa cells were able to kill and destroy the mutant bacteria (Fig. 5e). In contrast, wild-type bacteria, which replicate intracellularly, appear immune to these host defenses. At 22 h, wild-type and complemented bacteria/cell were both significantly greater than mutant bacteria/cell (p < 0.001, 2-way ANOVA with Bonferroni post-test correction). Previously, the iglC gene has been known to be critically important in phagosome permeabilization, but our data indicate that it is also required for Francisella cytosolic replication even after phagosome escape.
Figure 5. The iglC gene of F. novicida is required for intracellular multiplication after cytosolic delivery.
GFP-expressing F. novicida wild-type (a), ΔiglC mutant (b), and ΔiglC mutant complemented with iglC (c) in the cytosol of HeLa cells at 22 h after delivery. The bacteria fluoresce green and cell nuclei are stained blue with DAPI. Scale bar, 50 µm. (d) Percentage of bacteria in the cytosol was assessed by the differential digitonin permeabilization assay. Error bars, s.d. (n = 712 for all tests). (e) Growth of F. novicida wild-type, ΔiglC mutant and complemented ΔiglC mutant in HeLa cells after cytosolic delivery. Error bars, s.e.m. (n = at least 7 randomly selected microscopic fields of view per data point with at least 74 cells evaluated per data point). The experiment was conducted twice with similar results.
DISCUSSION
Potential applications of BLAST are broad. In addition to the delivery of small and medium sized cargo, such as quantum dots, gold nanospheres, functional enzymes, and DNA and RNA delivered in combination, BLAST may enable new fields by providing a high-throughput tool to deliver ultra large cargo into cells, which was not possible prior to this invention. Examples include delivery of whole mitochondria for study of diseases of mutant mitochondrial DNA, whole chromosomes for cell engineering, and bacteria for dissecting the pathogenesis of infectious organisms, etc.
BLAST provides a physical approach for reliably puncturing mammalian cell membranes in contact with metallic nanostructured thin films using short laser pulse illumination. Our data show that a similar pulse energy efficiently opens pores in the plasma membranes of a variety of mammalian cells. Cell-type independent membrane opening capability, cargo-type independent pressured flow driven delivery, and direct cytosol delivery that avoids cargo entrapment in endosomes are unique features that render BLAST a powerful and versatile platform for delivering a variety of cargo up to a few micrometers in size into a wide spectrum of mammalian cell types.
Massively parallel and nearly simultaneous delivery of cargo into cells under the same physiological conditions is another unique advantage of BLAST. The transient membrane pore resealing time studies presented in this work take advantage of this unique capability (Fig. 2). A laser beam programmed to scan across the entire chip to open cell membranes sequentially with known delay time is followed by uniform pressured delivery across the entire chip. The delivery efficiencies in different zones on the chip show the effect of delay time between laser pulsing and fluid pumping under the same delivery conditions. A single experiment using a single chip can produce sufficient data for statistical analysis.
The study of the intracellular lifestyle of bacteria in live cells shown in this manuscript was made possible with BLAST. That bacteria can be delivered into 100,000 host cells at one time allows separation of the infected cells into multiple subgroups for study of various subsequent phenomena over time after delivery, such as bacterial localization and intracellular multiplication performed in this study. Although the bacterial delivery experiment was intended only for demonstrating the utility of BLAST, this study unexpectedly discovered that the iglC gene is not only important for phagosome permeabilization, but also required for cytosolic replication after phagosome escape. Such a study is nearly impossible using conventional pipette-based delivery approaches since they do not provide the throughput required for reliable statistical analysis and the cells are not infected simultaneously, making it difficult to perform studies over time for fast occurring events, such as the re-packaging of bacteria into vacuoles after delivery.
Online Methods
Experimental setup
The silicon-based delivery chip was loaded onto a custom-built membrane pump for laser pulsing. The laser scanning system includes a Q-switched Nd:YAG laser (Minilite I, Continuum) with a wavelength at 532 nm, a pulse duration of 6 ns, and a beam diameter of 1 mm and is coupled with a 2D scanning mirror (Thorlabs) for rapid scanning across the entire active 1 cm2 photothermal delivery area. After laser scanning, an external synchronized pressure source (FemtoJet, Eppendorf) is applied to deform the PDMS membrane to transfer cargo in the storage chamber into all cells on the platform. The setup for time-resolved imaging characterization is the same as Wu’s research19.
Cell culture on platform chips prior to BLAST
HeLa (ATCC), NHDF (Lonza) and RPTEC (Lonza) cells were routinely tested for mycoplasma using the Universal Mycoplasma Detection Kit (ATCC, 30-1012K) according to the manufacturer's protocol. Cells were seeded onto a clean chip 24 h before the BLAST procedure. Chips with HeLa and NHDF cells were placed in Dulbecco’s modified essential medium (DMEM, Corning) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Thermo Scientific) and 1% penicillin/streptomycin (Mediatech) and kept in an incubator at 37°C and 5% CO2. In the case of PB-MDMs, 0.01% poly-L-lysine (Sigma) was coated on chips to improve cell adhesion. Peripheral blood mononuclear cells were isolated on Ficoll-Hypaque gradients, adjusted to 6 × 106 cells/ml in RPMI-1640 (Corning) supplemented with 200 mM L-glutamine, penicillin/streptomycin, and 20% autologous serum, and incubated for 3 d in sterile screw-cap Teflon wells (Savillex Corp. Minnetonka, MN) at 37°C, 5% CO2. The PB-MDMs were washed, resuspended in RPMI 1640 with glutamine, penicillin/streptomycin and 10% (vol/vol) autologous serum and seeded onto the chip. After 90 min incubation at 37°C and 5% CO2, the chip was washed to remove the non-adherent cells and fresh culture medium added for overnight incubation prior to use in experiments. These cells were obtained from a healthy donor and showed no evidence of mycoplasma infection by DAPI or Hoechst staining. Chips with RPTEC cells were placed in renal epithelial cell basal medium (REBM, Lonza) supplemented with 0.5% FBS and incubated at 37°C and 5% CO2.
Delivery materials
The total volume of the cargo storage chamber is 10 µL. F. novicida were grown in trypticase soy broth supplemented with 0.2% cysteine (TSBC), washed once with Hank’s Balanced Salt Solution (HBSS, Mediatech) and resuspended in HBSS to a final OD of 10.0 at 600 nm. L. monocytogenes were grown in brain heart infusion medium (BHI) at 30°C to O.D. 1.0, washed once with HBSS and resuspended to O.D. 1.0. Green fluorescent polystyrene beads (20 nm, 200 nm, 500 nm, 1 µm and 2 µm) modified by carboxylate (FluoSpheres, Invitrogen) were suspended in phosphate-buffered saline (PBS) (Corning) at 1014, 1011, 1011, 1010 and 1010 beads/ml, respectively. 200 nm red fluorescent magnetic beads (Chemicell) modified by streptavidin were diluted to 2×1010 beads/ml in PBS. Mouse anti-α tubulin with Alexa Fluor 488 (abcam) was prepared to 100 µg/ml in PBS. β-lactamase enzyme from Enterobacter cloacae (Sigma) was dissolved in PBS at 50 units/ml.
Delivery efficiency and cell viability
Cells with cargo inside were counted as delivered cells. Delivery efficiency, expressed as a percentage, was calculated as the number of delivered cells divided by the total number of cells × 100. Cell viability, expressed as a percentage, was determined by propidium iodide (5 µg/ml) staining 90 min after delivery and calculated as the number of the cells lacking red fluorescence divided by the total number of cells × 100.
Reproducibility information
Data on optimizing the performance of BLAST chips are means of at least 5 randomly selected fields of view with at least 438 cells counted per data point; error bars represent s.d. (Fig. 2a-d). Data on testing the transient window for large cargo delivery are means of at least 4 randomly selected fields of view with at least 1,825 cells counted per data point; error bars represent s.d. (Fig. 2e). Data on testing the transient window for small molecule delivery are means of at least 3 randomly selected fields of view with at least 351 cells counted per data point; error bars represent s.d. (Fig. 2f). Data regarding the 3-day cell viability test are means of at least 5 randomly selected fields of view with at least 1,057 (HeLa), 737 (NHDFs), 1,464 (PB-MDMs) or 387 (RPTECs) cells counted per data point; error bars represent s.d. (Fig. 2g). Data on cell proliferation after cargo delivery are means of at least 5 randomly selected fields of view with at least 780 (NHDFs), 1,569 (PB-MDMs) or 576 (RPTECs) cells counted per data point; error bars represent s.d. (Fig. 2h). Data on enzyme delivery are means of at least 3 randomly selected fields of view with at least 353 cells counted per data point; error bars represent s.d. (Fig. 3). Data on the intracellular localization and growth of F. novicida after BLAST are means of at least 7 randomly selected fields of view with at least 74 cells counted per data point; error bars represent s.e.m. (Fig. 5). Data on delivery efficiency and cell viability after delivery of different polystyrene beads are means of at least 4 randomly selected fields of view with at least 562 cells counted per data point; error bars represent s.d. (Supplementary Fig. 4d).
Bacterial and cell culture
Francisella novicida Utah 112 was grown in TSBC or on chocolate agar. When needed, kanamycin and hygromycin were included in the F. novicida cultures at 20 µg/ml or 200 µg/ml, respectively. L. monocytogenes was grown in BHI medium. When needed, streptomycin and erythromycin were included in the L. monocytogenes cultures at 200 µg/ml or 5 µg/ml, respectively. Human monocytic cell line THP-1 (ATCC TIB202) was maintained in RPMI-1640 supplemented with 10% FBS and penicillin (100 IU)/streptomycin (100 µg/ml) and confirmed as being free of mycoplasma by using the Universal Mycoplasma Detection Kit (ATCC). Prior to infection, THP-1 cells were differentiated with 100 nM phorbol 12-myristate 13-acetate for 3 d. Bacteria were pre-opsonized by incubation in 10% human serum type AB in HBSS at 37°C for 10 min, spun onto THP-1 macrophages at 800 g for 30 min at 4°C, and incubated at 37°C for 30 min to allow uptake. The infected monolayers were treated with 10 µg/ml gentamicin in DMEM containing 10% FBS for 30 min, washed and incubated in DMEM containing 10% FBS and 0.1 µg/ml of gentamicin. At specified time points, the infected monolayers were lysed with 1% saponin in PBS for 5 min, and the serial diluted lysates were plated on chocolate agar for bacterial CFU enumeration.
Actin phalloidin staining
NHDFs were seeded onto BLAST chips as described above. L. monocytogenes-GFP Δhly ΔplcB (green), strain 10403S, was delivered by BLAST with (or as a sham control) without laser pulsing. The NHDF cells were washed, incubated at 37°C for 6 h in DMEM with 10% FBS and 5 µg/ml gentamicin (which kills any residual extracellular bacteria), and fixed in 4% paraformaldehyde in PBS. F-Actin was stained with phalloidin-rhodamine (Molecular Probes) according to the manufacturer’s protocol.
Generation of ΔiglC and green fluorescent F. novicida strains
F. novicida was chemically transformed with the suicide plasmid pMP590-ΔiglC-ExC to generate an unmarked in-frame deletion mutant of iglC by allelic exchange. The resulting kanamycin resistant clones with plasmid integration were counter-selected on chocolate agar containing 5% sucrose for loss of the integrated plasmid. The clones that were resistant to sucrose and sensitive to kanamycin were screened for deletion of the iglC gene by PCR and Western immunoblotting analyses. GFP expressing F. novicida strains were generated by introducing pMP633 carrying a codon optimized superfolder gfp (sfGFP; Dr. Francis Nano, University of Victoria) driven by the bacterioferritin (FTL_0617) promoter. For complementation of iglC, pMP633BC-iglC-sfGFP was introduced into F. novicida ΔiglC for bi-cistronic expression of iglC and sfGFP. Bacterial strains, plasmid constructs, and primers used in this study are listed (Supplementary Table 2).
Digitonin permeabilization assay
Vacuolar versus cytosolic localization of bacteria was assessed by a modification of the differential digitonin permeabilization method of Chercoun et. al.32, which permeabilizes plasma membranes but not bacterial vacuoles. After BLAST delivery of bacteria into HeLa cells, the monolayers were allowed to recover for 30 min; stained for 10 min with 20 µg/ml Alexa-Fluor 647 WGA (Molecular Probes); washed with HBSS; fixed for exactly 1 min with 4% paraformaldehyde in HBSS; permeabilized for exactly 1 min with 0.05 mg/ml digitonin in 110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2, and 15% sucrose (KHM with 15% sucrose); washed twice with KHM containing 15% sucrose; incubated with chicken anti-F. novicida antibody (1:1000 dilution, a gift from Dr. Denise Monack, Stanford University) in KHM buffer containing 15% sucrose and 0.1% BSA; washed three times with KHM containing 15% sucrose; fixed for 30 min in 4% paraformaldehyde in 75 mM sodium phosphate, pH 7.4; permeabilized thoroughly with 0.1% saponin in PBS; washed with PBS; and incubated with Texas Red-X conjugated goat anti-chicken antibody (1:50 dilution, Invitrogen) for 90 min at room temperature. Monolayers were incubated with DAPI (1 µg/ml), washed with PBS, and viewed by fluorescence microscopy. Green fluorescent bacteria within the WGA-stained host cell borders that stained by the red antibody probe were scored as being cytosolic and those that were not stained by the red fluorescent antibody probe were scored as being membrane bound (vacuolar) (Supplementary Fig. 9).
Imaging
Fluorescent images were obtained using an inverted fluorescence microscope (Axio Observer.D1m, Carl Zeiss) with 10× and 40× objective lens. Scanning electron micrographs were taken by a field emission scanning electron microscope (Hitachi S4700). Confocal images were captured using a laser scanning confocal microscope (SP2 AOBS, Leica).
Supplementary Material
Acknowledgments
This work is supported by a University of California Discovery Biotechnology Award (178517), National Institute of Health grants AI065359, GM114188, and EB014456, and by NanoCav, LLC. This work is also funded by NSF CBET-1404080. The authors thank K. Niazi and S. Rabizadeh for helpful discussions and support, D. A. Portnoy for providing the L. monocytogenes strain DP-L2318, D. M. Monack for the F. novicida ΔFPI strain, K. E. Klose for the F. novicida U112 strain, F. E. Nano for the codon optimized superfolder gfp and J. S. Hong for the stress gene assay.
Footnotes
Author Contributions
Y.-C.W., T.-H.W. and P.-Y.C. had the idea for the platform. Y.-C.W. designed and fabricated the BLAST device. T.-H.W. built the experimental setup. Y.-C.W., T.-H.W., D.L.C. and B.-Y.L. performed the experiments and analyzed the data. B.-Y.L. prepared mutant and complemented Francisella strains. X.W. ran the numerical simulations. P.-Y.C., M.A.T. and M.A.H. advised on experiments, data analysis and paper writing. All authors discussed the experimental results and wrote the paper.
The authors do not have competing financial interests.
References
- 1.Rusk N. Seamless delivery. Nat. Methods. 2011;8:44–44. [Google Scholar]
- 2.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 3.Akin D, et al. Bacteria-mediated delivery of nanoparticles and cargo into cells. Nat. Nanotechnol. 2007;2:441–449. doi: 10.1038/nnano.2007.149. [DOI] [PubMed] [Google Scholar]
- 4.Felgner PL, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Smedt S, Demeester J, Hennink W. Cationic polymer based gene delivery systems. Pharm. Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 6.Somiari S, et al. Theory and in vivo application of electroporative gene delivery. Mol. Ther. 2000;2:178–187. doi: 10.1006/mthe.2000.0124. [DOI] [PubMed] [Google Scholar]
- 7.Guignet EG, Meyer T. Suspended-drop electroporation for high-throughput delivery of biomolecules into cells. Nat. Methods. 2008;5:393–395. doi: 10.1038/nmeth.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boukany PE, et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat. Nanotechnol. 2011;6:747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 9.Kim HJ, Greenleaf JF, Kinnick RR, Bronk JT, Bolander ME. Ultrasound-mediated transfection of mammalian cells. Hum. Gene Ther. 1996;7:1339–1346. doi: 10.1089/hum.1996.7.11-1339. [DOI] [PubMed] [Google Scholar]
- 10.Mitragotri S. Healing sound: the use of ultrasound in drug delivery and other therapeutic applications. Nat. Rev. Drug Discov. 2005;4:255–260. doi: 10.1038/nrd1662. [DOI] [PubMed] [Google Scholar]
- 11.Tirlapur UK, Konig K. Cell biology: Targeted transfection by femtos laser. Nature. 2002;418:290–291. doi: 10.1038/418290a. [DOI] [PubMed] [Google Scholar]
- 12.Tao W, Wilkinson J, Stanbridge EJ, Berns MW. Direct gene transfer into human cultured cells facilitated by laser micropuncture of the cell membrane. Proc. Natl. Acad. Sci. U. S.A. 1987;84:4180–4184. doi: 10.1073/pnas.84.12.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarty P, Qian W, El-Sayed MA, Prausnitz MR. Delivery of molecules into cells using carbon nanoparticles activated by femtos laser pulses. Nat. Nanotechnol. 2010;5:607–611. doi: 10.1038/nnano.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharei A, et al. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shalek AK, et al. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Yu L-C. Microinjection as a tool of mechanical delivery. Curr. Opin. Biotechnol. 2008;19:506–510. doi: 10.1016/j.copbio.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Hurtig J, Orwar O. Injection and transport of bacteria in nanotube-vesicle networks. Soft Matter. 2008;4:1515–1520. doi: 10.1039/b800333e. [DOI] [PubMed] [Google Scholar]
- 19.Wu T-H, et al. Photothermal nanoblade for large cargo delivery into mammalian cells. Anal. Chem. 2011;83:1321–1327. doi: 10.1021/ac102532w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartland GV. Optical studies of dynamics in noble metal nanostructures. Chem. Rev. 2011;111:3858–3887. doi: 10.1021/cr1002547. [DOI] [PubMed] [Google Scholar]
- 21.Link S, El-Sayed MA. Spectral properties and relaxation dynamics of surface plasmon electronic oscillations in gold and silver nanodots and nanorods. J. Phys. Chem. B. 1999;103:8410–8426. [Google Scholar]
- 22.Kotaidis V, Dahmen C, von Plessen G, Springer F, Plech A. Excitation of nanoscale vapor bubbles at the surface of gold nanoparticles in water. J. Chem. Phys. 2006;124:184702. doi: 10.1063/1.2187476. [DOI] [PubMed] [Google Scholar]
- 23.Lukianova-Hleb E, et al. Plasmonic nanobubbles as transient vapor nanobubbles generated around plasmonic nanoparticles. ACS Nano. 2010;4:2109–2123. doi: 10.1021/nn1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furlani EP, Karampelas IH, Xie Q. Analysis of pulsed laser plasmon-assisted photothermal heating and bubble generation at the nanoscale. Lab Chip. 2012;12:3707–3719. doi: 10.1039/c2lc40495h. [DOI] [PubMed] [Google Scholar]
- 25.Yamane D, et al. Electrical impedance monitoring of photothermal porated mammalian cells. J. Lab. Autom. 2014;19:50–59. doi: 10.1177/2211068213494390. [DOI] [PubMed] [Google Scholar]
- 26.Marquis H, Doshi V, Portnoy DA. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemens DL, Lee B-Y, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nano FE, Schmerk C. The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 29.Barker JR, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nano FE, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bruin OM, et al. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology. 2011;157:3483–3491. doi: 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golovliov I, Sjöstedt A, Mokrievich A, Pavlov V. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 2003;222:273–280. doi: 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.