Abstract
Reactivation of tumor suppressor genes (TSGs) involved in carcinogenesis by nontoxic bioactive food component represents a promising strategy for cancer chemoprevention. Recently, curcumin has been demonstrated to inhibit a bacterial DNA methyltransferase (M. Sss I) activity, induce global DNA hypomethylation in leukemia cells, and reactivate several hypermethylation silenced genes in lung and prostate cancer cells. Herein, we demonstrated that curcumin can enhance the mRNA and protein levels of ras-association domain family protein 1A (RASSF1A), 1 hypermethylation-silenced TSG, and decrease its promoter methylation in breast cancer cells. Mechanistic study demonstrated that curcumin can decrease DNA methylation activity of nuclear extract and downregulate the mRNA and protein levels of DNMT1 in MCF-7 cells, which may be associated with curcumin-induced disruption of NF-κB/Sp1 complex bound to the promoter region of DNMT1. Altogether, this study reveals a novel molecular mechanism of curcumin as a chemo-preventive agent for breast cancer through hypomethylation reactivation of RASSF1A.
INTRODUCTION
Breast cancer remains the second leading cause of mortality in American women. Recent cancer statistics from the American Cancer Society have estimated 32% incidence (269,000 new breast cancer cases) and 21% mortality (40,410 breast cancer related deaths) rate to breast cancer in the United States in 2011. Many factors contribute to the pathogenesis of breast cancer. These factors include family history, nutrition, age, and epigenetic changes including DNA methylation.
DNA methylation is a fundamental physiological process to convert cytosine to 5-methylcytosine in the context of the sequence 5’-cytosine-guanosine (CpG), which is mediated by DNA methyltransferases (DNMTs) (1). Cumulative evidence suggests that DNA methylation associated with nutrition, age, and environment exposure appears to be an early event in the etiology of breast carcinogenesis, resulting in the activation of many oncogenes and silencing of tumor suppressors, consequently promoting proliferation and metastasis of abnormal cells (2,3). In contrast to genetic mutation, DNA methylation is susceptible to perturbations by these diets and is reversible; therefore, it represents a promising target to identify dietary DNA methylation inhibitors for chemotherapy and chemoprevention (4,5).
Accumulative evidence suggested that regional hypermethylation represents one of the main mechanisms for early stages of breast cancer (6). Recently, genome-wide analysis has defined a list of hypermethylated genes, such as cadherin 1 (CDH1), death-associated protein kinase 1 (DAPK1), glutathione S-transferase pi hyper (GSTP1), high in normal-1 (HIN-1), ras-association domain family protein 1A (RASSF1A), tissue inhibitor of metalloproteinases-3 (TIMP-3), O6-methylguanine methyltransferase (MGMT), p16INK4a in various breast cancer cells and tumor tissues (7–9. Notably, the unique combination of methylated CpG islands correlating with breast cancer stage has been proposed to be a diagnostic marker of breast cancer (7). Among them, RASSF1A is shown to be the most frequently methylated tumor suppressor gene (TSG) in breast cancer (10,11). Methylated alleles are present to a similar extent in intraductal papillomas and epithelial hyperplasias, as well as in ductal carcinoma in situ, but never in normal breast tissue (11), which may indicate that RASSF1A methylation is an early event in breast tumorigenesis and may be used as a surrogate endpoint in chemoprevention trials. Notably, loss of RASSF1A expression is also one of the most common events in human cancers reported in various tumor types, including colon and lung cancers, which renders RASSF1A a prevalent target for chemoprevention of these cancers (10,11).
Curcumin, a yellow pigment extracted from turmeric, is widely consumed as a coloring and flavoring dietary additive in a variety of foods, including saffron and various beverages (12). Curcumin has also been historically used for the treatment of inflammation, skin wounds, as well as certain tumors (12–14). As such, there is a growing interest to study the cancer chemoprevention and chemotherapeutic properties of curcumin. The anticancer activities of curcumin have been the subject of hundreds of papers and thoroughly reviewed in several recent articles (15–18,); many favorable results have been documented in preclinical setting and to a lesser extent in clinical setting. Because of its extremely poor oral bioavailability, the in vivo molecular mechanism of curcumin remains largely unknown despite multiple targets were proposed for its anticancer activities. Recently, we have found that curcumin is a potent DNA methylation inhibitor via a potential mechanism that differs from that of nucleosides and other nonnucleoside DNA methylation inhibitors (19). Therefore, in this article, we evaluated whether curcumin can reactivate RASSF1A in an early stage breast cancer MCF-7 cell line and an advanced stage breast cancer MDA-MB-231 cell line. Also, the molecular mechanism of its hypomethylation activity was investigated in MCF-7 cells.
MATERIALS AND METHODS
Materials
Curcumin, methanol, acetonitrile (HPLC grade), ammonium formate, ammonium acetate, ammonium bicarbonate, decitabine, 5-methyl-2-deoxycytidine (5mdC), 2-deoxyguanosine, nucleophosphatase, snake venom phosphatase, and alkaline phosphatase, deoxynucleotide triphosphate (2.5 mM), Ampli-TaqGold polymerase and 10× PCR buffer were all purchased from Sigma (St. Louis, MO). The primers for amplification of RASSF1A, and its bisulfite-converted promoter region were purchased from Integrated DNA technology (IDT, Coralville, IA). M. SssI methylase, s-adenosyl-methionine (3.2 mM), 10× incubation buffer were purchased from New England Biolab Inc. (Beverly, MA). The HPLC-grade water (>18 mΩ) was obtained from an E-pure water purification system (Barnstead, Dubuque, IA).
Viability Study
MCF-10A, MCF-7, or MDA-MB-231 breast carcinoma cell line was purchased from ATCC and passages 9–25 of these breast cancer cells maintained in Dulbecco’s Modifed Eagle’s Medium (DMEM; Mediatech, Herndon, VA) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco, Grand Island, NY) and 1% (v/v) penicillin/streptomycin (Invitrogen Life Technologies, Carlsbad, CA) antibiotic solution at 37°C supplemented with 5% CO2. The effects of curcumin on cell viability were determined by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT; Sigma, St. Louis, MO) colorimetric assay. Briefly, 3 × 103 cells in 100 µL the aforementioned media were plated in 96-well plates in triplicate and allowed to attach overnight. The media was replaced with DMEM supplemented with 5% FBS and 1% P/S antibiotics and the indicated concentrations of curcumin and allowed to incubate for 72 h.
Immunoblotting and the Methylation Activity of Nuclear Extracts
Fresh cells (106–108 cells) in culture medium or frozen cell pellets, thawed on ice and resuspended in 1 mL ice-cold PBS, were centrifuged (<1,000 g) at 4°C for 5 min and the supernatant was removed and discarded. The pellet was resuspended in 100 to 200 µL ice-cold lysis buffer (20 mM pH 7.0 HEPES, 150 mM NaCl, 0.1% NP40 supplemented with 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM NaF, 1 mM benzimedin, and 1 mM phenylmethylsulfonyl fluoride) with protease inhibitors (Protease Inhibitor cocktail Set III; Calbiochem-Novabiochem Corporation, La Jolla, CA) and incubated on ice for 40 min. The lysate was centrifuged at 1000 g for 15 min at 4°C. The supernatants were frozen in liquid nitrogen and stored at −80°C. Equal amounts of protein for each sample were then separated on 5%–15% SDS-polyacrylamide gels as confirmed by probing with β-actin. The DNMT1, RASSF1A, and β-actin levels were determined using appropriate antibodies. The blots were blocked in TBST (10 mM Tris-HCl, pH8.0, 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat milk and subsequently incubated with anti-DNMT1, and RASSF1A, and β-actin (New England Biolabs; 1:2000). Protein recognized by the antibody was detected using the Chemiluminescent Detection Kit (Pierce, Rockford, IL). To test the methylation activity of nuclear extract from MCF-7 cells, NE was extracted from MCF-7 cells using a NE extraction kit (Active Motif, Carlsbad, CA). The methylation activity was measured using a EpiQuik™DNMT assay kit (Epigentek Group Inc., Brooklyn, NY), according to the manufacturer’s instruction.
DNA Extraction and Hydrolysis
For global DNA methylation study, genomic DNA was isolated from 1 million MCF-7 breast carcinoma cells treated with indicated concentration of curcumin at specific time points using DNeasy Tissue kit (Qiagen, Minneapolis, MN), according to the manufacturer’s instruction. DNA hydrolysis was performed as previously described (20). Briefly, 200 ng of genomic DNA was first denatured by heating at 100°C for 3 min and then chilled on ice. After adding a 1/10 volume of 0.1 M ammonium acetate (pH 5.3) and 2 units of nuclease P1 (US Biological, Swampscott, MA), the mixture was incubated at 45°C for 2 h. A 1/10 volume of 1 M ammonium bicarbonate and 0.002 unit of venom phosphodiesterase I (Sigma Chemical Co., Louis, MO) was then added, and the mixture was incubated at 37°C for 1 h. Next, 0.5 unit of alkaline phosphatase (Sigma Chemical Co., St. Louis, MO) was added, and the mixture was incubated at 37°C for 1 h.
Instrumentation
For quantification, the LC-MS system used consisted of a Perkin-Elmer Sciex API 3000 triple-quadrupole mass spectrometer (Thornhill, Ontario, Canada) coupled to a Shimadzu HPLC system (Shimadzu, Columbia, MD). The HPLC system was equipped with an SCL-10A system controller, a LC-10AD pump, and a SIL-10A auto-sampler (Shimadzu, Columbia, MD).
HPLC Chromatographic and Mass Spectrometric Conditions
2dC, 5mdC, and the internal standard 5,6-dihydro-5-azacytidine were separated on a 250 × 2.1 mm Hypersil Aquasil C18 5 µm stainless steel column (Thermo Hypersil-Keystone, Bellefonte, PA), which was coupled to a 2-µm Aquasil pre-column (Thermo Hypersil-Keystone, Bellefonte, PA), using the mobile phase consisting of 30% methanol in 10 mM ammonium formate at a flow rate of 0.2 mL/min. The mass spectrometer was operated under electrospray ionization with an ion-spray voltage of +4700 V. The positive ion multiple reaction-monitoring mode analysis was performed using nitrogen as the collision gas. A dwell time of 200 ms and a pause time of 5 ms between scans were used to monitor the following precursor/product ion pair at m/z 247.1/115.2 for internal standard and m/z 242.1/126.1 for 5 mdC. The mass spectrometer was tuned to its optimum sensitivity and mass accuracy by infusion of a fresh standard solution of 5 mdC at 500 ng/mL. Data acquisition was performed using the Analyst (version 1.4).
Real-Time RT-PCR Assays
Quantitative RT-PCR was used to quantify the expression of RASSF1A and DNMT1. Untreated cells were used as a negative control. RT-PCR was performed using 2 µg of total RNA extracted with Trizol reagent (Invitrogen, CA) according to the manufacturer instruction and reverse transcribed by Moloney murine leukemia virus reverse transcriptase (Invitrogen, CA). Conditions for Sybergreen real-time RT-PCR were stage 1, 50°C 2 min; stage 2, 95°C 3 min; stage 3, 95°C 15 s, 60°C 30 s, 72°C 30 s with 40 cycles. Real-time RT-PCR reactions were performed with an ABI prism 7700 sequence detector (Taqman; PE Applied Biosystems), and data were analyzed with the Sequence Detector version 1.6 software to establish the PCR cycle at which the fluorescence exceeded a cycle threshold (CT) for each sample. Data were analyzed according to the comparative CT method using the internal control (18S) transcript levels to normalize differences in sample loading and preparation. Results represent the n-fold difference of transcript levels between different samples and are expressed as the mean ± SD of triplicate reaction wells.
Electrophoretic Mobility-Shift Assays (and Antibody-Supershift Assays
Nuclear extracts were prepared using Nuclear and Cytoplasmic Extraction Reagent (Pierce, Rockford, IL). Electrophoretic mobility-shift assays (EMSA) with nuclear extracts and 32p-labeled oligonucleotides containing the Sp1 motif and DNMT1 promoters’ oligos were performed as previously described (21). Briefly, for antibody competition assay, nuclear extracts were preincubated with designated antibodies at 4°C for 16 h before adding the probes. Three pairs of oligonucleotides derived from human DNMT1 promoter regions containing putative Sp1 binding sites were chemically synthesized, complementary oligos annealed, and labeled with 32P-dCTP by Klenow fragment.
Xenograft Animal Model Study
Female athymic nu/nu mice (4–6 wk old, 18–22 g) were obtained from Charles River Laboratory (Wilmington, MA) and acclimated for 1 wk in a pathogen-free enclosure before start of study. Animals were given sterile rodent chow and water ad libitum and were housed in sterile filter-top cages with 12-h light/dark cycles. All experiments were conducted in accordance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International. Prior to the inoculation of MCF-7 cells, an estradiol pellet was inset under the skin of each nude mouse. MCF-7 Cells (5 × 106 cells per mouse) were suspended with cell culture medium (Becton Dickinson) and subcutaneously implanted into the right flank of the athymic nu/nu mice. The tumor size was then measured by external caliper and the greatest longitudinal diameter (length, L) and the greatest transverse diameter (width, W) were determined. Tumor volume based on caliper measurements were calculated as V = 0.52 * L * W * W. When tumors were grown between 100 to 200 mm3, treatments were initiated. Mice were randomly assigned into 2 cohorts for the antitumor growth activity studies, with 5 mice in the control group and 8 mice in the treatment group. Curcumin was given as a solution interperitoneal at the dose of 100 mg/kg 5 days/wk and the placebo formulation was used as a control for 4 wk.
RESULTS
Curcumin Increased the Protein Level of RASSF1A in MCF-7 and MDA-MB-231 Cells In Vitro and Enhanced its mRNA Levels in MCF-7 Cells In Vitro and In Vivo
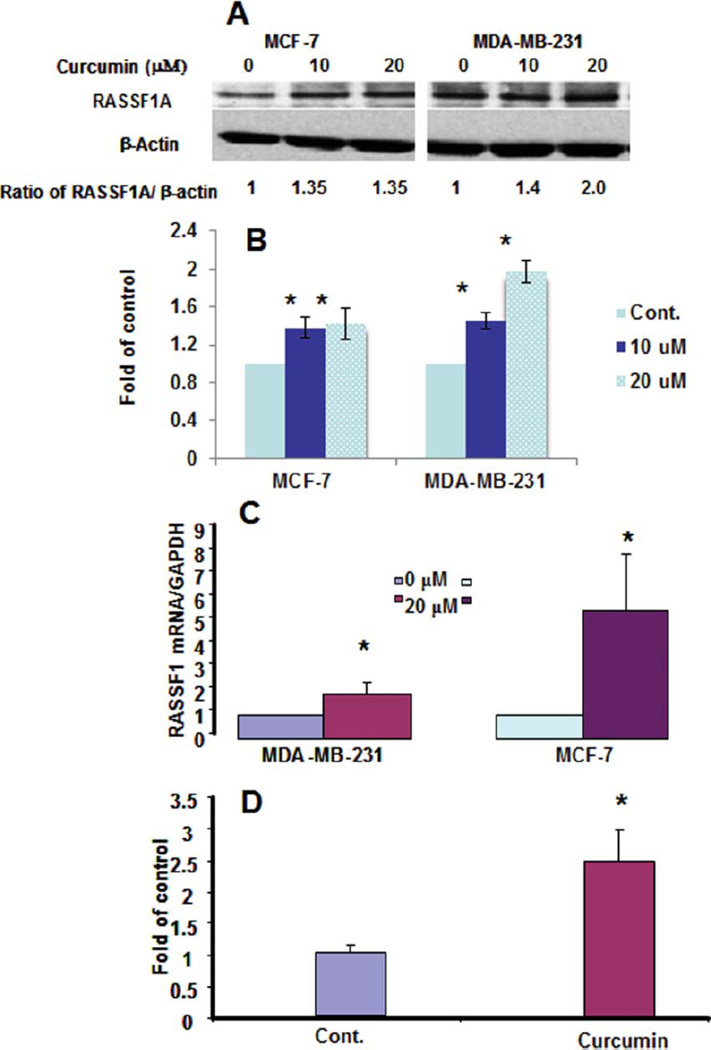
A wide array of hypermethylation-silenced TSGs have been documented in various human breast cancer cell lines and primary breast tumor tissues from patients with breast cancer. Among them, RASSF1A is a putative silenced TSG predominantly associated with its promoter hypermethylation; conversely, reactivation has been found to be associated with promoter demethylation by decitabine (22), a clinically used DNA hypomethylating agent, in breast cancer cells. Curcumin has been shown as a novel DNA methylation inhibitor in leukemia cells (19). To demonstrate whether curcumin can be used as a hypomethylating agent in breast cancer, first, we evaluated whether curcumin can reactivate RASSF1A in 2 breast cancer MCF-7 and MDA-MB-231 cells. As shown in Fig. 1A, at the previously demonstrated pharmacological effective (antiproliferation and apoptosis) concentrations, curcumin can significantly enhance the protein levels of RASSF1A in MCF-7 cells (about 1.3–1.5-fold, P < 0.05, 2 sample t-test, n = 3) and MDA-MB-231 cells (1.4–2.0-fold, P < 0.05, 2 sample t-test, n = 3). Notably, curcumin also enhanced RASSF1A mRNA level (5–7-folds) as shown in Fig. 1B. This result suggested that the curcumin-induced increased protein levels of RASSF1A may be associated with the enhanced mRNA level of RASSF1A in MCF-7 cells. Notably, the mRNA level of RASSF1A was also found to be significantly higher (Fig. 1C) in MCF-7 cell engrafted tumor tissue collected from tumor bearing nude mice treated with an i.p. administration of 100 mg/kg curcumin in reference to those treated with the vehicle. This result demonstrated that curcumin can increase the expression of RASSF1A in MCF-7 cell is in vitro and in vivo.
FIG. 1.
Reactivation of RASSF1A in Breast Cancer Cell Lines by Curcumin MCF-7 cells and MDA-MB-231 cells treated with 10 and 20 uM curcumin, the protein level of RASSF1A was measured by immunological anti-body of RASSF1A (western blot, WB) and showed significant increase in a typical WB (A) and densitometry analysis (B); the mRNA level of RASSF1A was quantified using q-RT-PCR and showed pronounced increase (C) in the curcumin-treated MCF-7 and MDA-MB-231 cells; and the in vivo enhancement of RASSF1A mRNA level in MCF-7 engrafted tumor in the nude mice treated with a daily i.p. administration of 100 mg/kg curcumin for 5 day/week for four weeks (D). (*, 0.01 < p < 0.05) (Color figure available online).
Curcumin Can Induce RASSF1A Promoter and Global DNA Hypomethylation and Decrease the DNA Methylation Activity of NE of MCF-7 Cells
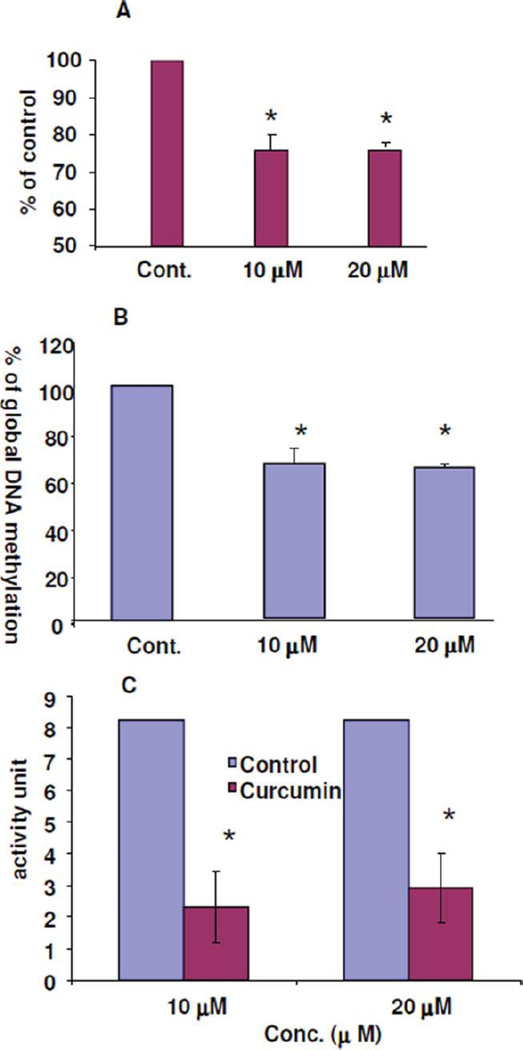
To evaluate whether the above-described RASSF1A reactivation is associated with its promoter hypomethylation, the promoter methylation level of RASSF1A was measured using our established LC-MS/MS method (22). Consistent with the enhanced expression level of RASSF1A, the promoter methylation level was decreased ~25%of the basal level ofMCF-7 cells (Fig. 2A). The global DNA methylation (GDM) level was also measured. It was found that GDM level in curcumin-treated MCF-7 cells decreased about 30%–35% when compared to untreated MCF-7 cells (Fig. 2B). To understand the mechanism of the hypomethylating activity of curcumin, the methylation activity of the nuclear extract from MCF-7 cells treated with curcumin or decitabine was determined. Consistent with its hypomethylation activity, the methylation activity of NE from MCF-7 cells treated with curcumin decreased about 70% relative to that of the control (Fig. 2C), whereas decitabine can completely inhibit the methylation activity of cell extracts. As a positive control, the methylation activity of NE from MCF-7 cells treated with decitabine is the lowest. These results implicate that the reactivation of RASSF1A is at least partially associated with its promoter demethylation through the inhibition of DNMTs’ methylation activity.
FIG. 2.
The hypomethylation activity of curcumin in MCF-7 cells The alteration of the promoter methylation level of RASSF1A (A), the global DNA methylation level (B), and the methylation activity of nuclear extract (C) from untreated and 10 or 20 M curcumin-treated MCF-7 cells for 72 hour. The promoter DNA methylation level and global DNA methylation level were measured by two tandem LC-MS/MS methods for global DNA methylation and regional DNA methylation as the molar ratio of 5mdC to 2dC and the methylation activity was measured using an EpiQuik TM DNMT1 assay Kit from Epigentek Group Inc. (*: 0.01 < p < 0.05) (Color figure available online).
Curcumin Can Downregulate DNMT1 Expression Levels in MCF-7 Cells
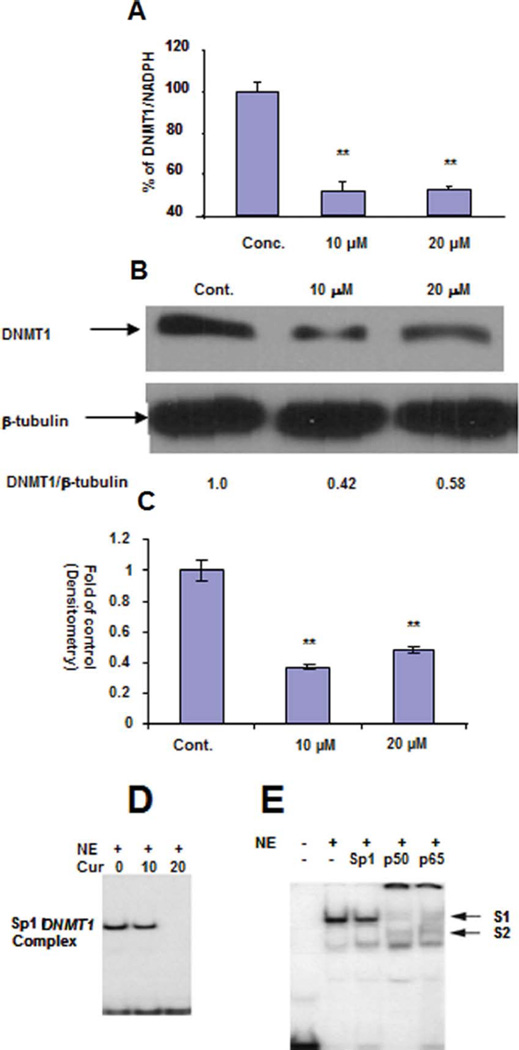
There are 2 putative mechanisms for the reduced enzymatic activity of NE of curcumin-treated MCF-7 cells: 1) chemical inhibition of DNMTs and/or 2) biological inhibition of DNMTs synthesis. We have shown that curcumin is a potent M. SssI inhibitor with an IC 50 of 30 nM (19); therefore, this above-documented inhibitory activity of curcumin on DNMTs’ methylation activity could be due to its chemical inhibition. Recently, we have defined Sp1/NF-κB as a complex binding to the human DNMT1 promoter region to transactivate its expression, which can be inhibited by a proteasomal inhibitor bortezomib (21) and NF-κB inhibitor parthenolide (23). Curcumin has been shown to be a potent NF-κB inhibitor; therefore we first check the protein levels of DNMT1 in curcumin-treated MCF-7 cells. As shown in Fig. 3A, DNMT1 protein levels were significantly decreased in 10 or 20 µM curcumin-treated cells. Consistent with the decreased protein level of DNMT1, the mRNA level of DNMT1 was also significantly downregulated in curcumin-treated MCF-7 cells as shown in Fig. 3B. Hence, inhibition of DNMT1 synthesis may be an alternative or complementary mechanism for curcumin’s overall inhibitory effect on DNA methylation of NE. The EMSA study has shown that curcumin can disrupt the complex of Sp1/NF-κB binding to the promoter of DNMT1 as shown in Fig. 3C. Antibody gel shift study has shown that the complex of Sp1 and the promoter of DNMT1 contained all 3 transcriptional factors—Sp1, p65, and p50—because the intensity of the complex was significantly decreased when they were incubated with these 3 respective antibodies as shown in Fig. 3D. The presence of p65 and p50 in this complex was further supported by showing the formation of novel slower-moving complex labeled as S1 and S2 in Fig. 3D.
FIG. 3.
The molecular mechanism of hypomethylation activity of curcumin in MCF-7 cells down-regulation of the mRNA (A) and protein (B) level of DNMT1 and disruption of Sp1/DNMT1 complex in curcumin-treated MCF-7 cells (C) and the components of this complex (D). MCF-7 cells were incubated with 10 or 20 µM curucmin for 72 hour, DNMT1 mRNA level was detected by q-RT-PCR and protein level was detected by western blot. EMSA was performed by the incubation of Sp1-DNMT1-1 oligonucleotide probe with MCF-7 nuclear extract (NE) to form protein DNA complex and antibody gel supershift assays was performed by the pre-incubation of MCF-7 NE with Sp1 and NF-B (p65 and p50) antibodies before formation of protein-DNA complex. The novel slower moving complexes of S1 and S2 are of p65/p50-DNMT1-Sp1 concensus binding oligonucleotides and Sp1/p65/p50-DNMT1-Sp1 concensus binding oligonucleotides complex (*: 0.01 < p < 0.05, **: 0.001 < p < 0.01, ***: 0.0001 < p) (Color figure available online).
The Anticancer Activities of Curcumin on Breast Cancer In Vitro and in Vivo
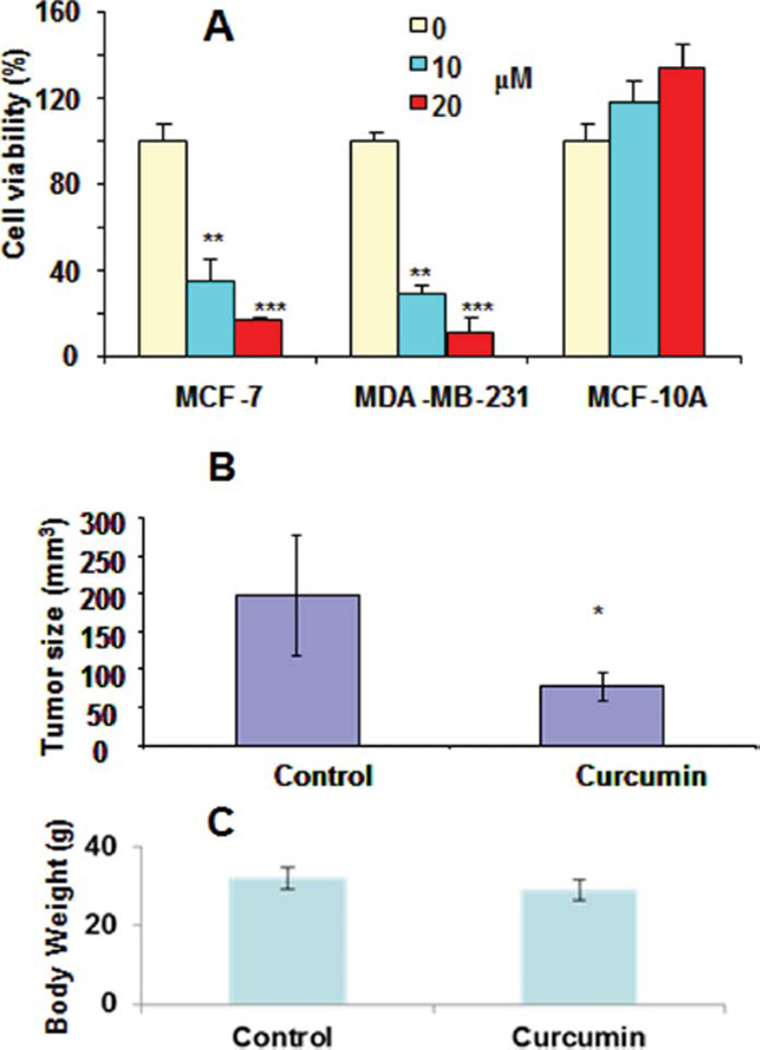
Functional analyses of silenced and overexpressed RASSF1A in several cancer cell lines have shown that RASSF1A is associated with the deregulated proliferation activity of some cancer cells (11,24). As shown above, both mRNA and protein levels of RASSF1A are upregulated in MCF-7 and MDA-MB-231 cells. To understand whether this reactivation is associated with the antiproliferation activity of curcumin, the cytotoxicity of curcumin has been evaluated in these 2 cell lines and a nontumoric immortalized breast epithelium cells MCF-10A. Curcumin has shown significant antiproliferation activity in MCF-7 and MDA-MB-231 cells with an IC50 <10 µM. The cell viability of MCF-7 cells is about 38%, 19% at 10 and 20 µM, and the cell viability ofMDA-MB-231 cells about 25%, and 13% at 10 and 20 µM, respectively; however, the cell viability of MCF-10A remains about 100% (Fig. 4A). These results implicate that curcumin is a selective cytotoxic agent for breast cancer cells while sparing the breast epithelium cells. Notably, this antibreast cancer cell growth and proliferation activity of curcumin was also documented in a MCF-7 cell engrafted tumor growth in nude mice as a 65% decrease of tumor size (Fig. 4B) without any observed cytotoxicity, because no apparent body-weight loss was observed in these mice treated with daily interperitoneal dose of 100 mg/kg curcumin 5 days/wk for 4 wk. Therefore, these observed RASSF1A reactivation may be associated with curcumin’s antiproliferation activity in vitro and antitumor growth activity in vivo.
FIG. 4.
The anti-proliferative activity (A) and the anti-tumor growth activity (B) of curcumin on breast cancer cells MCF-7, MDA-MB-231, MCF-10 A cells were treated with 10 and 20 µM curcumin for 72 hour and the cytotoxicity of curcumin was evaluated using MTT assay (Triplicates); The MCF-7 cell engrafted tumor-bearing nude mice were treated with curcumin (n = 8) and placebo formulation (n = 5) daily 5 days/week for four weeks. The tumor size was measured (B) and the body weight was weighed (C) and recorded on day 29. (*: 0.01 < p < 0.05, **: 0.001 < p < 0.01, ***: 0.0001 < p) (Color figure available online).
DISCUSSION
Several lines of evidence suggest that curcumin may be an effective preventive and therapeutic agent for breast cancer. However, the molecular mechanism of action remains largely unknown. In this study, we first demonstrated that the most-tested curcumin concentrations (10 and 20 µM) can enhance the protein and mRNA levels of one omnipresent hypermethylation silenced TSG RASSF1A (Fig. 1) in both MCF-7 and MDA-MB-231 cells. In MCF-7 cells, RASSF1A reactivation is at least partially associated with its promoter hypomethylation (25% decreases). The hypomethylation activity of curcumin is possibly from dual functions of curcumin on DNMT1: the chemical inhibition of DNMT1 and the biological downregulation of DNMT1. Mechanistic study suggested that downregulation of DNMT1 could be associated with its disruption of a recently defined transactivating complex of Sp1 and NF-κB binding to its promoter region. Notably, we have demonstrated the antiproliferation activity of curcumin in 2 breast cancer cell lines:MCF-7 and MDA-MB-231 cells with an IC50 < 10 µM (Fig. 4A). A significant antitumor growth activity was also documented in MCF-7 engrafted tumor bearing nude mice with about 65% tumor size decrease (Fig. 4B). Taken together, curcumin is an effective DNA methylation inhibitor and has significant antitumor growth activity on breast cancer cell lines in vitro and in vivo.
This finding that curcumin inhibited DNMT1, thereby reactivating RASSF1A through its promoter hypomethylation, represents a novel molecular mechanism of its anticancer chemopreventive activity for breast cancer. Recently, Manson et al. reported that extended treatment with curcumin (≤3 µM) results in altered gene expression; for example, E-cadherin-11 and reduced growth and apoptosis of cancer cells in MDA-MB-231 cells, similar to that induced by nucleoside DNA methylation inhibitors (25). In addition, several hypermethylation silenced TSGs, for example, GSTP1 (26) and MGMT (27), have been reported to be reactivated in breast cancer or other cancer cells lines. Therefore, collective data suggest that curcumin can induce DNA hypomethylation and reactivate hypermethylation silenced genes.
Epigenetic chemotherapy has been demonstrated to be a novel therapeutic strategy for leukemia management (4); currently, 2 DNA hypomethylation agents, decitabine and 5-azacytidine collectively termed as azanucleosides have been approved for treatment of myelodysplatic syndrome (28). However, extrapolating this strategy to solid tumor including breast cancer meets an unprecedented challenge. Therefore, this discovery of curcumin as a hypomethylation agent may provide a novel epigenetic preventive treatment for breast cancer because curcumin is a nontoxic, long-term consumable, and widely used dietary supplement. Cell cycle study demonstrated that curcumin at concentrations showing significant hypomethylation activity can induce cell cycles at G2/M phase and significantly decrease S-phase cell population in these 2 breast cancer cells lines (29). Therefore, curcumin may have the advantage over S-phase dependent azanucleoside hypomethylation agents as exerting its hypomethylation activity without entailing the majority of breast cancer cells passing through S-phase, thereby inducing hypomethylation activities on nonreplicating subpopulations of breast cancer cells including stem cell, which are generally nonreplicating and may be particularly difficult populations of cells to be treated with azanucleoside hypomethylation agents.
CONCLUSION
Curcumin reactivates a silenced TSG RASSF1A at least partially due to its promoter hypomethylation in breast cancer MCF-7 and MDA-MB-231 cell lines. Consistent with its hypomethylation activity, curcumin downregulates mRNA and protein levels of DNMT1, thereby decreasing the methylating activity of nuclear extract, and global DNA methylation in MCF-7 cells. A pronounced in vitro antiproliferative and in vivo antitumor growth activity was documented in MCF-7 cells. Correlation of RASSF1A reactivation and plasma and tissue levels of curcumin and its metabolites is ongoing.
Acknowledgments
This work was supported by National Institute of Health (NIH) grants [R21 CA135478] (Zhongfa Liu), and Biomedical Mass Spectrometric Laboratory (Kenneth K. Chan and Zhongfa Liu) at the Ohio State University.
Footnotes
Full terms and conditions of use: http://www.tandfonline.com/page/terms-and-conditions
This article may be used for research, teaching, and private study purposes. Any substantial or systematic reproduction, redistribution, reselling, loan, sub-licensing, systematic supply, or distribution in any form to anyone is expressly forbidden.
Publisher's Disclaimer: The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae, and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand, or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
Contributor Information
Liping Du, College of Pharmacy, Ohio State University, Columbus, Ohio, USA.
Zhiliang Xie, College of Pharmacy and Comprehensive Cancer Center, Ohio State University, Columbus, Ohio, USA.
Lai-chu Wu, Department of Molecular & Cellular Biochemistry, the Ohio State University, Columbus, Ohio, USA.
Ming Chiu, College of Pharmacy, Ohio State University, Columbus, Ohio, USA.
Jiayuh Lin, Center for Childhood Cancer, The Research Institute at Nationwide Children’s Hospital, Columbus, Ohio.
Kenneth K. Chan, College of Pharmacy and Comprehensive Cancer Center, Ohio State University, Columbus, Ohio, USA
Shujun Liu, The Hormel Institute, University of Minnesota, Austin, Minnesota, USA.
Zhongfa Liu, College of Pharmacy and Comprehensive Cancer Center, Ohio State University, Columbus, Ohio, USA.
REFERENCES
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- 3.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 4.Kopelovich L, Crowell JA, Fay JR. The epigenome as a target for cancer chemoprevention. J Natl Cancer Inst. 2003;95:1747–1757. doi: 10.1093/jnci/dig109. [DOI] [PubMed] [Google Scholar]
- 5.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 6.Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004;68:1187–1197. doi: 10.1016/j.bcp.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal A, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol. 2007;20:711–721. doi: 10.1038/modpathol.3800822. [DOI] [PubMed] [Google Scholar]
- 8.Giacinti L, Claudio PP, Lopez M, Giordano A. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist. 2006;11:1–8. doi: 10.1634/theoncologist.11-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann U, Langer F, Feist H, Glockner S, Hasemeier B, et al. Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol. 2002;160:605–612. doi: 10.1016/S0002-9440(10)64880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krop IE, Sgroi D, Porter DA, Lunetta KL, LeVangie R, et al. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:9796–9801. doi: 10.1073/pnas.171138398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shishodia S, Chaturvedi MM, Aggarwal BB. Role of curcumin in cancer therapy. Curr Probl Cancer. 2007;31:243–305. doi: 10.1016/j.currproblcancer.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78:2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Singh S, Khar A. Biological effects of curcumin and its role in cancer chemoprevention and therapy. Anticancer Agents Med Chem. 2006;6:259–270. doi: 10.2174/187152006776930918. [DOI] [PubMed] [Google Scholar]
- 15.Hsu CH, Cheng AL. Clinical studies with curcumin. Adv Exp Med Biol. 2007;595:471–480. doi: 10.1007/978-0-387-46401-5_21. [DOI] [PubMed] [Google Scholar]
- 16.Lin JK. Molecular targets of curcumin. Adv Exp Med Biol. 2007;595:227–243. doi: 10.1007/978-0-387-46401-5_10. [DOI] [PubMed] [Google Scholar]
- 17.Anand P, Sundaram C, Jhurani S, Kunnumakkara AB, Aggarwal BB. Curcumin and cancer: an “old-age” disease with an “age-old” solution. Cancer Lett. 2008;267:133–164. doi: 10.1016/j.canlet.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Lazaro M. Anticancer and carcinogenic properties of curcumin, considerations for its clinical development as a cancer chemopreventive and chemotherapeutic agent. Mol Nutr Food Res. 2008;52(Suppl 1):S103–S127. doi: 10.1002/mnfr.200700238. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009;19:706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Liu S, Xie Z, Blum W, Perrotti D, et al. Characterization of in vitro and in vivo hypomethylating effects of decitabine in acute myeloid leukemia by a rapid, specific and sensitive LC-MS/MS method. Nucleic Acids Res. 2007;35:e31. doi: 10.1093/nar/gkl1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Liu Z, Xie Z, Pang J, Yu J, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008;111:2364–2373. doi: 10.1182/blood-2007-08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Wu J, Xie Z, Liu S, Fan-Havard P, et al. Quantification of regional DNA methylation by liquid chromatography/tandem mass spectrometry. Anal Biochem. 2009;391(2):106–113. doi: 10.1016/j.ab.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Liu S, Xie Z, Pavlovicz RE, Wu J, et al. Modulation of DNA Methylation by a Sesquiterpene Lactone Parthenolide. J Pharmacol Exp Ther. 2009;329(2):505–514. doi: 10.1124/jpet.108.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 25.Moiseeva EP, Almeida GM, Jones GD, Manson MM. Extended treatment with physiologic concentrations of dietary phytochemicals results in altered gene expression, reduced growth, and apoptosis of cancer cells. Mol Cancer Ther. 2007;6:3071–3079. doi: 10.1158/1535-7163.MCT-07-0117. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran C, Rodriguez S, Ramachandran R, Raveendran Nair PK, Fonseca H, et al. Expression profiles of apoptotic genes induced by curcumin in human breast cancer andmammary epithelial cell lines. Anticancer Res. 2005;25:3293–3302. [PubMed] [Google Scholar]
- 27.Niture SK, Velu CS, Smith QR, Bhat GJ, Srivenugopal KS. Increased expression of the MGMT repair protein mediated by cysteine prodrugs and chemopreventative natural products in human lymphocytes and tumor cell lines. Carcinogenesis. 2007;28:378–389. doi: 10.1093/carcin/bgl155. [DOI] [PubMed] [Google Scholar]
- 28.Blum W, Marcucci G. Targeting epigenetic changes in acute myeloid leukemia. Clin Adv Hematol Oncol. 2005;3:855–865. 882. [PubMed] [Google Scholar]
- 29.Prasad CP, Rath G, Mathur S, Bhatnagar D, Ralhan R. Potent growth suppressive activity of curcumin in human breast cancer cells: Modulation of Wnt/beta-catenin signaling. Chem Biol Interact. 2009;181:263–271. doi: 10.1016/j.cbi.2009.06.012. [DOI] [PubMed] [Google Scholar]