Abstract
Maintenance of the hematopoietic stem cell (HSC) compartment depends on the ability to metabolize exogenously and endogenously generated toxins, and to repair cellular damage caused by such toxins. Reactive aldehydes have been demonstrated to cause specific genotoxic injury, namely DNA interstrand cross-links. Aldehyde dehydrogenase 2 (ALDH2) is a member of a 19 isoenzyme ALDH family with different substrate specificities, subcellular localization, and patterns of expression. ALDH2 is localized in mitochondria and is essential for the metabolism of acetaldehyde, thereby placing it directly downstream of ethanol metabolism. Deficiency in ALDH2 expression and function are caused by a single nucleotide substitution and resulting amino acid change, called ALDH2*2. This genetic polymorphism affects 35–45% of East Asians (about ~560 million people), and causes the well-known Asian flushing syndrome, which results in disulfiram-like reactions after ethanol consumption. Recently, the ALDH2*2 genotype has been found to be associated with marrow failure, with both an increased risk of sporadic aplastic anemia and more rapid progression of Fanconi Anemia. This review discusses the unexpected interrelationship between aldehydes, ALDH2 and hematopoietic stem cell biology, and in particular its relationship to Fanconi anemia.
Keywords: Fanconi Anemia, aldehydes, hematopoiesis, hematopoietic stem cell, aplastic anemia, aldehyde dehydrogenase, ALDH2
Introduction
A central problem in hematology is the limited lifespan of mature non-lymphoid cells, which necessitates constant production of new blood cells. In humans, the estimated lifespan of circulating red blood cells (RBCs) is 120 days, polymorphonuclear neutrophils (PMNs) 2 days, and platelets 5–10 days. To maintain the blood system, approximately 2 × 1011 RBCs, 1.6 × 1011 PMNs, and 1011 platelets are produced daily [1–4]. The evolutionary resolution of this problem in mammals is based on a hierarchy of self-renewing multipotent hematopoietic stem cells (HSC), which give rise to non-self-renewing, lineage committed hematopoietic progenitor cells (HPC) capable of massive expansion and differentiation into mature blood cells.
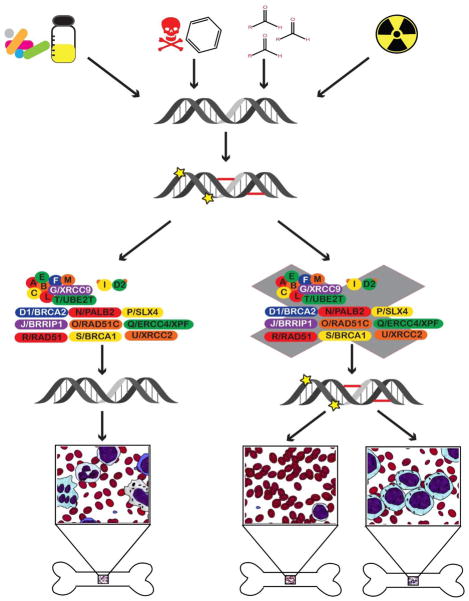
The dependence of blood cell production on a limited number of HSC and HPC (hereafter collectively referred to as HSPC) means that protection from potential toxins is essential for maintenance of the hematologic system (See Figure 1). For example, the generation of blood cells within the intramedullary marrow space of heavily calcified bones probably protects cells from typical doses of external ionizing radiation. Immature HSPC express a number of proteins which appear to protect them from toxicological injury. For example, the ABC transporter protein MDR1 expressed by HSC increases the export of various xenobiotics, including some chemotherapy drugs[5].
Figure 1. Genomic Instability and Bone Marrow Failure.
The genome is subject to a variety of different insults, including chemotherapy drugs, benzene and other toxic substances, radiation, and aldehydes (RCH=O). These can induce different types of DNA damage, including adduct formation, single and double strand breaks, and interstrand crosslinks. When the damage is appropriately repaired by DNA repair pathways, one of which is the FA pathway (depicted), there is no damage to HSPC in the bone marrow (depicted by a normal mixture of cells in the left inset). When the damage is not adequately repaired by DNA repair pathways, or by the FA pathway specifically, loss of HSPC can occur, leading to bone marrow failure (aplastic anemia, shown in the middle inset), or acute myelogenous leukemia (AML) shown on the right.
Because genotoxic injury is inevitable, an important protective mechanism is the expression of various proteins involved in DNA repair pathways [6]. These pathways include those for repair of single strand breaks, such as base excision repair (BER), nucleotide excision repair (NER), and mismatch repair (MMR). To repair double strand breaks, cells express pathways for non-homologous end joining (NHEJ), homologous recombination (HR), or microhomology-mediated end joining (MMEJ). Additionally, cells can use tolerance methods to continue to replicate DNA around lesions, called translesion synthesis (TLS). Interstrand crosslinks (ICL), another type of DNA lesion, are recognized and repaired by the Fanconi anemia pathway. Each of these means of DNA repair depend on both the type of DNA lesion, as well as the state of the cell cycle, as to whether a template will be available to make the repair, as in the case of a double strand break. Additionally, each repair pathway has its own unique set of proteins involved in recognizing, excising the damaged areas, and repairing the lesion, as well as proteins that are involved in multiple pathways, resulting in overlap and potentially crosstalk[7, 8].
Genomic instability and Fanconi anemia
Genetic diseases involving impaired repair of DNA damage, such as Fanconi Anemia (FA), have profound hematologic consequences. FA results from mutations of the FA pathway genes specifically required for repair of ICL[8]. Clinically, FA patients have congenital structural anomalies (birth defects); progressive exhaustion of their HSC pool leading to marrow failure (aplastic anemia); damage to HSPC, resulting in increased risk of leukemia, particularly acute myelogenous leukemia (AML) and increased susceptibility to other cancers, especially squamous cell carcinomas of the skin, upper airway and head and neck[9]. Hematopoietic stem cell transplant (HSCT) can cure aplastic anemia and AML in FA patients, but increased susceptibility to cancer in other somatic cells persists, resulting in high rates of post-transplant mortality.
FA is a syndrome caused by mutations in one of at least 20 genes, which comprise the Fanconi complementation (FANC) groups.[8, 10, 11] The proteins in the FA complex are essential for resolving DNA damage due to ICL. The FANC A, B, C, E, F, G, L, M, and T gene products form the Fanconi Anemia Core Complex, an ubiquitin E3 ligase which ubiquitinates the ID2 complex, which is comprised of the FANCI and FANCD2 subunits. [8]. Monoubiquinated ID2 recruits other proteins to excise ICLs, followed by recruitment of additional proteins to repair the lesions by homologous recombination (HR) [8, 10]. FA patients are homozygotes or compound heterozygotes for at least one of the genes encoding the FA complex, and are hypersensitive to DNA bifunctional alkylating agents (which cause crosslinks) and radiation. In the absence of functional FA pathway activation, the presence of ICL is deleterious to cells in several ways, notably interference with transcription and blocking of the replication fork in dividing cells. In addition, the inability to activate HR to repair the lesions results in activation of alternative DNA repair pathways, e.g., the error-prone non-homologous end-joining (NHEJ) repair. Activation of NHEJ likely causes mutations. The hypersensitivity to DNA damaging agents is demonstrated in vitro by induction of numerous cytogenetic abnormalities, such as radial chromosomes after exposure to alkylating agents, like diepoxybutane (DEB) or mitomycin C[12]. This hypersensitivity has been used to screen patients or family members for suspected FA. The hypersensitivity of FA cells to DNA damage is manifested clinically in several ways. Patients receiving chemotherapy for treatment of leukemia or other cancers, or as pre-transplant conditioning to ablate host immune cells before hematopoietic stem cell transplant or chemotherapy for leukemia, develop excessive toxicity and require reduced doses of chemotherapy or radiation to prevent fatal side effects. Secondly, even after successful cure of aplastic anemia, FA patients develop secondary malignancies at an alarming rate. Approximately one-half of transplanted patients develop cancer in the first 15 years after successful HSCT [13].
Aldehyde-mediated DNA damage in Fanconi anemia
Although the components of the FA pathway have been identified and their functions have been elucidated, a number of questions about the pathophysiology of FA remain unanswered. First, efforts to link the FA genotype, i.e., which FANC group is mutated and the type of mutation (missense, nonsense, etc.), with time of onset of either marrow failure or cancer, or with the phenotype of congenital anomalies have been largely unsuccessful, suggesting the presence of modifying genes or unknown environmental factors that influence the course of FA. Second, the causes of widespread ICL in FA patients have not been defined. Although exposure to basal levels of radiation or chemotherapy like cytotoxic drugs is possible, a more attractive idea is that ICL form in response to more common toxicological events, e.g., ubiquitous substances in the environment such as airborne or ingested molecules.
The FA field has been considerably advanced by the identification of acetaldehyde and formaldehyde as important contributors to both ICL formation and protein modification, and both aldehyde dehydrogenase (ALDH2) and alcohol dehydrogenase 5 (ADH5) as the enzymes responsible for their respective detoxification in murine models. ALDH2 was first demonstrated to modify Fanconi Anemia in a study from the Patel laboratory that showed that the development of aplastic anemia is rapidly accelerated in aldehyde dehydrogenase 2 (ALDH2)−/− FANCD2−/− mice compared to ALDH2+/+ FANCD2−/− mice [14]. This group also demonstrated that ALDH2−/− or ALDH2+/− mothers could not give birth to ALDH2−/− FANC−/− progeny - the embryos were either resorbed or embryonic lethal with severe developmental defects. ALDH2−/− FANCA−/− pups could be rescued from embryonic lethality, if they were transferred at the 2-cell embryonic stage to ALDH2+/+ mothers, though with developmental defects and bone marrow failure later in life [15]. Thus, ALDH2 modulates the severity of both developmental and HSPC phenotypes in murine models of Fanconi anemia. Later work identified ADH5 as an additional modifier of the Fanconi anemia phenotype. Formaldehyde is present endogenously, as it is produced by oxidative demethylation reactions and other metabolic reactions, and is detectable at micromolar levels in the bloodstream [16, 17]. ADH5 is expressed in many tissues and is necessary for metabolism of formaldehyde, which is oxidized in a glutathione dependent manner [16, 18]. The number of formaldehyde-induced N2-hydroxymethyl-deoxyguanine DNA modifications is increased after mice were fed methanol, which is directly metabolized to formaldehyde [16]. This study found that ADH5−/− FANCD2−/− mice had decreased viability, numbers of nucleated cells in the bone marrow, and pancytopenia as well as liver and kidney defects. Competitive repopulation experiments, where equal amounts of bone marrow from ADH5−/− FANCD2−/− and wild type mice were transplanted into lethally irradiated recipients showed that ADH5−/−, FANCD2−/− marrow was selectively unable to repopulate the marrow, as these cells contributed to only 0.1% of the repopulated bone marrow.[16] Together, ALDH2 and ADH5 are probably two long sought modifying genes in FA. Of note for human health and disease, both ALDH2 and ADH5 polymorphisms associated with loss of function have been identified. While ALDH2 mutation is common and has been proven to be deleterious to health, ADH5 mutation is less prevalent and disease association has not been proven [19, 20]. Because ALDH2 is immediately downstream of alcohol dehydrogenases (including ADH5), it also is directly linked to potential common environmental impacts on the FA phenotype, e.g., impact of ethanol and its metabolite, acetaldehyde, as well as exposure to aldehydes from other sources, and is the focus of the rest of this review.
Speculation about the relationship of the FA developmental phenotype to aldehyde metabolism can be drawn from the similar patterns of malformation seen in FA and fetal alcohol syndrome (FAS). Children with FAS have congenital and neurodevelopmental abnormalities, and prenatal aldehyde exposure rather than ethanol itself has been hypothesized to be the relevant teratogen [21]. Both FA and FAS patients display microcephaly (small heads), developmental delays, microphthalmia (small eyes), ptosis (drooping of upper eyelid), strabismus (abnormal eye alignment), ventricular sepal defect (VSD) of the heart, horseshoe kidney, and finger and toe abnormalities [22–25] (see Figure 2). A unifying hypothesis to explain the similarities between FA and FAS is that aldehydes alter the development of various susceptible somatic stem and progenitor cell populations, resulting in a common pattern of malformation (reviewed in [26], along with alternative hypotheses for FAS, based on the protective role of Gli2 and Sonic Hedgehog (SHH) on fetal damage from ethanol [27].
Figure 2. Comparison of Birth Defects in Fanconi Anemia[28] and Fetal Alcohol Syndrome[29].
Images reproduced with permission, and are originally found in the works cited above.
Acetaldehyde
One of the most common environmental aldehydes is acetaldehyde (CH2CHO). Acetaldehyde, which is highly volatile, has been classified as a Group I human carcinogen by the International Agency for Research on Cancer[30]. Aldehydes can form DNA adducts, including ring-open forms of crotonaldehyde propanodeoxyguanosines, which can be generated from crotonaldehyde or two successive reactions with individual acetaldehyde molecules [31, 32]. These ring-open forms can dimerize with other guanosine residues, resulting in interstrand crosslink (ICL) formation between DNA strands [32–34]. There are other effects of aldehydes on cells, including adducts covalently added to proteins, especially by 4-hydroxy-nonenal (4-HNE)[34]. ICL crosslinks can lead to genotoxic effects due to DNA damage and protein adducts can disrupt cellular signaling and result in protein aggregation and protein unfolding responses. These effects have been implicated in pathological processes such as myocardial infarction and atherosclerosis [35–39].
Intracellular aldehydes are derived from both exogenous and endogenous sources (see Table 1). The major exogenous sources of aldehydes are likely to be direct exposure to environmental aldehydes including acrolein, formaldehyde and acetaldehyde. These aldehydes can be present in burned oil, industry glue and new buildings, as well as fossil fuel and fragrances [30, 31, 33, 40–42]. More sources of aldehydes in the environment as well as those generated endogenously are detailed in Table 1.
Table 1.
Acetaldehyde and other aldehyde sources
| Type | Source | Aldehyde |
|---|---|---|
| Exogenous-Environmental[31, 34, 40–42] | Factory byproducts | Acetaldehyde |
| Linoleum, carpeting | Acetaldehyde, formaldehyde | |
| Plywood | Formaldehyde | |
| Pollution | Acetaldehyde, formaldehyde, acrolein, valeraldehyde, hexanal, heptanal, nonanal, dodecyl aldehyde, crotonaldehyde, methacrolein, benzaldehyde, m-tolualdehyde, 2,5-dimethylbenzaldehyde, 3-hydroxybenzaldehyde, gloxal, glycoaldehyde, gloxylic acid | |
| Fossil Fuel | ||
| Cigarette Smoke | Acrolein, crotonaldehyde, formaldehyde, diacetyl, methylglyoxal | |
| Fireplaces/Forest fires | Crotonaldehyde | |
| Crematoriums/Mortuaries | Formaldehyde | |
| Fragrances/Perfumes | Benzaldehyde, anisaldehyde, vanillin, cinnamaldehyde, citral, safranal | |
| Cosmetic/Hair Salons | Formaldehyde | |
| Therapeutic drugs | Acrolein, chloroacetaldehyde, glyoxal, atropalaldehyde, abacavir aldehyde, tribroacetaldyde, trichloroacetaldehyde | |
| Exogenous-Food[30, 31] | Coffee | Acetaldehyde |
| Ethanol (metabolite of) | Acetaldehyde | |
| Ripe Fruit | Acetaldehyde | |
| Vinegar | Acetaldehyde | |
| Yogurt | Acetaldehyde | |
| Pickled Foods | Acetaldehyde | |
| Fermented Foods | Acetaldehyde | |
| Aspartame (metabolite of) | Formaldehyde | |
| Cinnamon | Cinnamaldehyde | |
| Almonds | Benzaldehyde | |
| Cherries | Benzaldehyde | |
| Endogenous-Cell Metabolism[31] | β-oxidation of fatty acids | Acetaldehyde, crotonaldehyde, glyoxyal,4-hydroxynonenal (4-HNE) |
| Carbohydrate metabolism | Methylglyoxal | |
| Myeloperoxidase in neutrophils | Acetaldehyde | |
| Endogenous-microbiome[43] | Gut bacteria | Acetaldehyde |
Direct inhalation or contact with acetaldehyde can occur as an industrial hazard, since acetaldehyde is also important for production of a number of industrial chemicals, including acetic acid. Non-work related inhalation exposure to acetaldehyde occurs via breathing of indoor air, as acetaldehyde is released from building materials, laminates, cleaning products and varnished woods.
Besides exposure via industrial or environmental inhalation or contact, acetaldehyde exposure occurs through ingestion. This can occur by consumption of acetaldehyde directly (as in foods containing acetaldehyde), by drinking ethanol, or by ethanol produced by the gut microbiota. Acetaldehyde is directly ingested by consuming ripe fruits, coffee, and bread, which derive their fruity aroma from acetaldehyde. Vinegar and foods pickled with vinegar also contain acetaldehyde, as well as fermented foods such as yogurt[30]. An important source of acetaldehyde is from ethanol metabolism [44]. Ingested ethanol is oxidized mainly in the liver to acetaldehyde by alcohol dehydrogenase, which is coupled to reduction of NAD+ to NADH. Aldehyde dehydrogenase (ALDH) further oxidizes acetaldehyde to acetic acid, again in a reaction coupled to NAD+ reduction. The gut microbiome ferments sugars to form ethanol that is then converted to acetaldehyde[43].
Finally, endogenous production of acetaldehyde can occur as a result of several metabolic processes (reviewed in O’Brien PJ, 2005)[31], for example under inflammatory conditions[45]. Myeloperoxidase in neutrophils and myeloid progenitors generates acetaldehyde as a byproduct of hypochlorous acid and tyrosyl radical production. In times of oxidative stress, when the mitochondria are unable to generate enough ATP, deoxyribose phosphate aldolase can metabolize 2-deoxy-D-ribose-5 phosphate to glyceraldehyde-3-phosphate and acetaldehyde, which allows cells to use nucleotides to replenish ATP levels[46]. Other potential sources of endogenously generated acetaldehyde are products of lipid peroxidation, carbohydrate/ascorbate autoxidation, carbohydrate metabolism, amine oxidases and cytochrome P-450 catalyzed metabolism[31].
Acetaldehyde has been found to induce ICL in both mammalian and rodent cell lines, and to activate the FA pathway [47–49]. Acetaldehyde-induced DNA damage colocalizes with RAD51/FANCR [47], and increases FANCD2 monoubiquitination and BRCA1/FANCS activation via phosphorylation [48, 49]. These studies demonstrate that acetaldehyde may be a critical aldehyde in activation of the FA pathway.
The Aldehyde Dehydrogenase (ALDH) Enzyme Family
In humans, ALDH enzymes comprise a 19 isoenzyme family of proteins with different patterns of substrate specificity, tissue and cell distribution and subcellular localization [50]. Given the widespread sources of exogenous and endogenous aldehydes as well as the ability of these compounds to deleteriously modify both DNA and proteins, the expression of aldehyde dehydrogenase (ALDH) enzymes is necessary in all tissues. ALDH oxidize aldehydes to form respective acidic derivatives of their substrates. Because of the variety of reactive aldehydes, ALDH isoforms with different specificities are needed. The substrate specificity and redundancy of these isoenzymes has not been fully characterized (see review [51]). The focus of this review will be on ALDH2, the enzyme which preferentially detoxifies acetaldehyde, as denoted by its Km being 900-fold lower than that of ALDH1, another enzyme that can detoxify acetaldehyde[52]. ALDH2 is encoded by nuclear DNA, but is localized inside mitochondria, and is important in reducing oxidative stress[52]. ALDH2 uses NAD+ as co-enzyme and oxidizes acetaldehyde to acetic acid [53]. ALDH2 expression in mice is similar to that of humans, where it is present in liver, esophagus, lung, pancreas, brain and heart [54–56]. ALDH2 is a homotetrameric molecule localized in the mitochondrial matrix. ALDH2 synthesis is continuously required, because some reactive aldehyde substrates, like 4-HNE, bind to and inactivate the enzyme, thus rendering it unable to catalyze further reactions [57].
Asian Flushing Syndrome and ALDH2
A subset of the East Asian population exhibit facial flushing and experience heart palpitations and headaches upon ethanol consumption. This disorder, which was termed “Asian Flushing Syndrome”, was identified in 1979 as a deficiency in ALDH2 activity. Although it was first thought that this may be due to gene deletion [58, 59], later work demonstrated that in proteins extracted from livers of Asian patients with alcohol intolerance, there was a protein that cross reacted in an immunologically identical way as the mitochondrial ALDH2 protein from livers of those with normal alcohol tolerance. However, this cross-reactive immunological material (CRIM) isoform of ALDH2 had no measurable aldehyde dehydrogenase activity [60, 61]. ALDH2 is synthesized as an immature 517 amino acid protein, after which the 17 amino acid mitochondrial targeting sequence is cleaved from the newly translated protein as it enters the mitochondria[52, 62]. When the native ALDH2 protein was first isolated and sequenced, it was found to have a single amino acid substitution of lysine for glutamine at amino acid position 487 of the mature protein (E487K) in individuals who exhibit the flushing syndrome relative to those who do not[63–65].
The single nucleotide E504K polymorphism, which is denoted ALDH2*2, decreases both the enzyme’s stability and activity, thus resulting in a much slower rate of aldehyde metabolism. The ALDH2*2 mutant has ~ 200-fold higher Km for NAD and a 10-fold lower Kcat, as compared to wild-type (wt) ALDH2[66]. Because the KmNAD+ is 15-fold higher than the in vivo concentration of the NAD+ and the mutant enzyme is less stable, ALDH2*2 heterozygotes have 25–40% and homozygotes only 1–4% of the wt ALDH2 activity. The mutant effect is semidominant, because the E504K substitution destabilizes the subunit interface, thus decreasing the activity of adjacent wild type subunits [65, 67]. The ALDH2*2 protein has both lower activity, stability in tetrameric form, and half-life[67]. The crystal structure of the ALDH2*2 protein displays a disordered alpha-helix near the subunit interface, away from the catalytic site or the tetramer interface, which is well ordered in the region of the ALDH2 protein[52] (see Figure 3). The ALDH2*2 genotype is found in 35–45% of East Asians (about 560 million people)[52]. Because ALDH2*2 is so prevalent in the world (it is the most common known human mutation), it is possible that ALDH2 deficiency provides a yet uncharacterized evolutionary advantage.
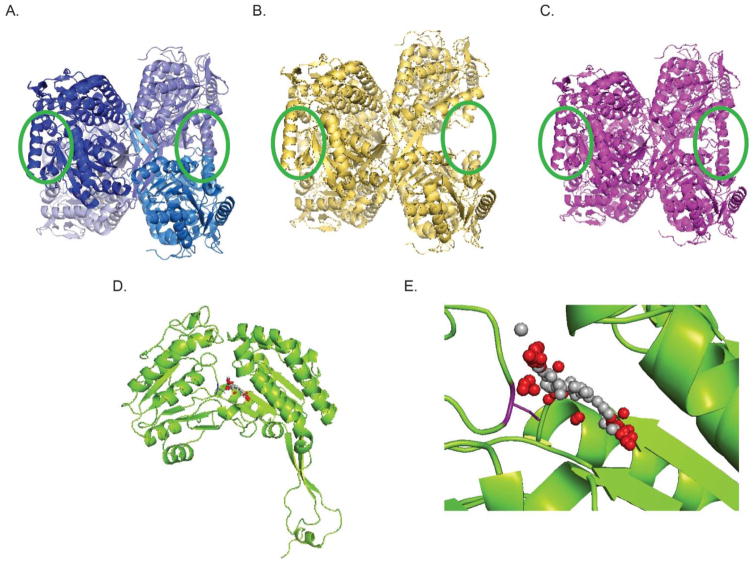
Figure 3. Examples of small molecule activators of ALDH2 and ALDH3A1.
Cartoon representations of homotetrameric structures of ALDH2, ALDH2*2, and ALDH3A1. A. Cartoon of ALDH2, each shade of blue represents a different monomer subunit within the tetramer. Note circled areas in green, which show two alpha helices pointing into the page on the left and 2 alpha helices aligned vertically with the page on the right. B. ALDH2*2, note absence of helices in circled areas. C. ALDH2*2 after binding of Alda-1, note structural helices are now present. D. Monomeric structure of ALDH3A1. E. Zoom in of the catalytic pocket of ALDH3A1. Binding area of Alda-89 is shown in red and grey spheres. Catalytic cysteine 243 is shown in purple.
Increased risk of other malignancies associated with ALDH2 deficiency
The ALDH2*1/ALDH2*2 (heterozygous) genotype is associated with an up to 70-fold increased risk of esophageal cancer depending on the amount of ethanol consumed [44], presumably due to exposure to high levels of acetaldehydes in these individuals. Although the high levels of acetaldehyde found in ALDH2*2 individuals after ethanol exposure deter many heterozygote individuals from imbibing alcoholic beverages, due to social pressure many East Asian individuals heterozygote for this mutation continue to consume alcohol[44]. Because the negative consequences of alcohol consumption in ALDH2*2/ALDH2*2 homozygote individuals, multiple studies have shown that this group does not drink alcohol, and is protected from esophageal cancers[68]. People with the ALDH2*2 genotype should be educated of the elevated cancer risk associated with alcohol ingestion [69].
ALDH isoforms and hematopoiesis
A critical role for ALDH in HSC has been known since the early 1980’s, when strategies to purge bone marrow of leukemia cells by in vitro treatment with the alkylating agent 4-hydroperoxy-cyclophosphamide (4-HC) were tested in clinical trials[70]. The metabolism of 4-HC by ALDH was found to be essential for the differential survival of HSC after 4-HC treatment, but at the time, the specific ALDH isoform needed for 4-HC metabolism had not been identified. A fluorescent ALDH substrate, Aldefluor (BODIPY aminoacetaldehyde), has been used as a viable stain for immature HSPC, and thus has been used to sort HSC for basic research[71] and for clinical trials of HSC transplantation[72]. Although Aldefluor activity was attributed to ALDH1A1, a cytosolic ALDH isoform, and was thought to be critical to hematopoiesis, an analysis of ALDH1A1 knockout mice has demonstrated that Aldefluor staining does not require ALDH1A1 and that ALDH1A1 is, in fact, dispensable for hematopoiesis[73]. The findings in this study show that Aldefluor is not specific to ALDH1A1, and that signal may instead be from activity of other ALDH isoforms including ALDH2, ALDH3A2, and ALDH9A1[73]. In another study, a general inhibitor of aldehyde dehydrogenases, diethylaminobenzaldehyde (DEAB) was found to promote the expansion of HSCs in mice. This group found that siRNA knockdown of ALDH1A1 could duplicate this effect, suggesting that ALDH1A1 is the key isoform required [74, 75]. These seemingly contradictory pieces of evidence for the importance of ALDH1A1 in hematopoiesis suggest the possibility that ALDH1A1 is involved in both metabolism of exogenous aldehydes and regulation of an endogenous signaling pathway. A likely candidate for the hematopoietic effects of ALDH1A1 is retinoic acid signaling. Retinoids are important in early development and embryogenesis, as well as hematopoiesis[76]. ALDH1A1 oxidizes retinaldehyde or retinal, to generate retinoic acid [73]. This molecule acts as a signaling molecule via the Wnt/β-catenin pathway to activate self-renewal, and when it is inhibited, leads to cellular differentiation [77, 78].
ALDH2*2 and Aplastic Anemia
Although aplastic anemia can be caused by environmental affects and drugs, such as benzene or pesticide exposure, chemotherapy, radiation, and other toxins[79, 80], sporadic aplastic anemia is thought to be frequently due to a dysregulated immune system, which is responsible for killing bone marrow stem cells[80]. Cytotoxic T-cells (CTL) secreting interferon-γ are thought to be the cause of this effect, and T regulatory cells, which normally act to suppress the activity of CTL, are reduced in most patients with aplastic anemia [81, 82]. Thus, the primary therapy for patients with newly diagnosed aplastic anemia is immunosuppressive therapy. Besides the immunologic mechanism for the development of aplastic anemia, direct damage to the HSC compartment is also a known cause, as mentioned above. For example, radiation exposure and certain chemical toxins like benzene can cause marrow failure. Likewise, aldehydes are a known source of genomic damage to the HSC compartment, suggesting that ALDH2 may play an important role in HSC maintenance and survival. Since 20–45% of the East Asian population are ALDH2 deficient, aplastic anemia in this population may be exacerbated by ALDH2 deficiency.
Indeed, the ALDH2*2 genotype is correlated with a worse prognosis in children with idiopathic aplastic anemia [83]. The distribution of aplastic anemia is biphasic, with two peaks of incidence, one occurring in children and young adults (10–25 years of age), and another occurring in older adults (above 60 years of age)[84]. In East Asia, the proportion of aplastic anemia that occurs in young adults is much higher than in Western countries [85–87].
Furthermore, the incidence of sporadic aplastic anemia is 2–3 fold higher in East Asia [80, 85, 88]; in Western countries the incidence is close to 2 cases per million people, while in East Asia, the prevalence ranges from 4 per million in Thailand to 7 per million in China[86]. It is therefore possible that the ALDH2*2 and reduced ALDH2 activity increases susceptibility to marrow failure.
ALDH2*2 and Fanconi Anemia
Fanconi Anemia (FA) is a disease characterized by birth defects and anemia leading to bone marrow failure in children. The ALDH2*2 genotype has been recently associated with a faster progression to marrow failure in children with FA and a younger age of onset of sporadic marrow failure in children who do not have FA. In Japanese children with FA, the median time to development of aplastic anemia was 72 months in homozygous wild-type ALDH2*1 allele, 28 months in heterozygous ALDH2*1/ALDH2*2, and 0 months (range 0–7) in homozygous for ALDH2*2[89]. In children with aplastic anemia not due to FA, both the age of onset and likelihood of failure-free survival are reduced in children homozygous for ALDH2*2. The median age of diagnosis for these patients was 9 years for homozygous ALDH2*1 and heterozygous ALDH2*1/ALDH2*2, and only 2 years if homozygous for ALDH2*2[83]. These studies highlight the possible role of ALDH2*2 in human FA and bone marrow failure, consistent with the previously discussed research in murine models [14–16, 21].
Current therapies for metabolic diseases
Inborn errors of metabolism caused by deficiency of particular enzymes due to loss of function mutations or deletions have been treated by decreasing the intake of the enzymatic substrate, supplementation with an essential cofactor or vitamin, increasing metabolism by enzyme replacement therapy (ERT), or more recently, by gene therapeutic approaches[90]. For some diseases, substrate intake can be decreased by dietary modification. The most advanced example of this approach has been phenylketonuria, which is managed by newborn diagnosis and institution of a restricted diet that avoids food proteins and dietary supplements, e.g., the artificial sweetener aspartame, which are rich in toxic phenylalanine. In some metabolic diseases of defective carboxylation, e.g., biotinidase deficiency, or holocarboxylase synthetase deficiency, defects in the biotinylation of protein substrates can be treated by lifelong supplementation with biotin. Enzyme replacement therapy in which purified or recombinant enzyme has been used to replace adenosine deaminase (ADA) in some types of severe combined immunodeficiency syndrome (SCIDS), glucocerebrosidase in Gaucher disease, acid alpha-glucosidase in Pompe disease, and alpha-galactosidase A in Fabry disease have all been used[91]. Efforts to increase enzymatic activity by gene therapy are still experimental, but offer the promise of permanent correction of metabolic defects. Early strategies have focused on the use of viral or non-viral vectors to introduce wild-type copies of a defective enzyme into affected cells, either by transplantation of a patient’s own cells after in vitro gene transduction or by in vivo injection. The recent discovery and rapid development of the CRISPR/Cas9 system as a method to modify the mutant gene sequence, thereby normalizing it, make gene editing a real possibility.
ALDH activators—a new strategy for treatment of metabolic disease
One avenue that is still in the exploratory phase is the activation of defective enzymes to improve their stability and catalytic function. The use of small molecules to do this offers the promise of a low cost therapy that could be applied to a wide variety of different enzyme disorders. The ALDH2*2 mutation lends itself to this approach because the defect causes the expression of a full-length ALDH2 protein with altered conformation, subunit interactions, and enzymatic activity. Therefore, small molecule activators could function as molecular chaperones to induce normalized conformation of the ALDH2*2 protein, with increased subunit interactivity and higher enzymatic activity.
The Mochly-Rosen laboratory has developed a series of small molecule activators for several members of the ALDH family of enzymes. These small molecules, called Aldas, were designed to increase enzymatic activity of specific ALDH isoforms. One such activator of ALDH2 enzymatic activity, is Alda-1 [N-(1,3-benzodioxol-5-ylmethyl)-2,6-dichlorobenzamide], a chemical chaperone which corrects the disturbed helix in the mutant enzyme (Figure 3). Alda-1 increases the stability of both the wild-type and mutant ALDH2 tetramer. In addition, Alda-1 prevents ALDH inactivation by reactive aldehydes and acts as an allosteric agonist, increasing the catalytic activity of the enzyme [66]. Alda-1 appears to be safe, as it has been used in many animal studies and shows no toxicity [36, 39, 92] and one Alda was found to be safe in a small phase 1 clinical study (unpublished data).
A strategy that can be used as an alternative to or complementary to Alda-1 treatment is to recruit other members of the ALDH family of enzymes to take over the role of ineffective or mutant ALDH2 to increase the detoxification of genotoxic aldehydes, like acetaldehyde. Alda-89 was developed by the Mochly-Rosen lab to direct ALDH3A1 (another member of the ALDH family of enzymes) to metabolize acetaldehyde, which is not normally a substrate for ALDH3A1 [93]. Using computer simulated docking, Alda-89 was found to bind in the catalytic pocket of ALDH3A1, near catalytic cysteine 243, where it is available to direct acetaldehyde to the active site[93]. Using enzyme activation and recruitment strategies like these may increase the number of targets available to treat genetic metabolic diseases. Potential clinical application of ALDH2 activators, in FA include both delaying or preventing marrow failure or AML as well as preventing post-BMT incidence of secondary malignancies. Because ALDH3A1 levels are high in the airways, mouth and upper digestive system, which is where FA patients tend to have high incidence of solid tumors as secondary malignancies, Alda-89 or other ALDH3A1 activating compounds may be a cancer chemopreventive in these patients.
Aldehyde sensors to monitor substrate levels
Previous efforts to ameliorate metabolic diseases frequently relied on biochemical methods which could measure the levels of toxic substrates in cells or bodily fluids (blood and serum or plasma, urine, cerebrospinal fluid). Therapeutic efforts to decrease exposure to toxic reactive aldehydes are likely to be advanced by developing methods for measurement of these substrates Recently, a general sensor of reactive aldehydes and specific sensors of formaldehyde have been developed [94–96]. The challenges for use of these sensors include the volatility of the aldehydic substrates, and the need for reactions to occur at physiological pH and temperature if they are to be applied to viable cells. We are presently validating the dark hydrazone sensor [96] for measurement of overall reactive aldehydes in HSC, with the goal of using this technology to detect differences in intracellular aldehyde levels in HSC or other cells from ALDH2*1 and ALDH2*2 individuals and to monitor the effects of ALDH2 activators on these levels, in anticipation of clinical trials.
Conclusions and Future Directions
ALDH2 may be an important regulator of HSC function and maintenance as evidenced by its connection to both aplastic anemia and Fanconi Anemia. In the future, it may prove important to consider ALDH2 genotype in donors for bone marrow transplant, as ALDH2 deficiency may negatively impact engraftment and survival of donor-derived HSC. Alda-1 and other ALDH family activators may be useful in treating diseases of the bone marrow. Additionally, because FA patients are subject to secondary malignancies including leukemia, and head and neck cancers, and are sensitive to the traditional chemotherapy and radiation used to treat them, non-genotoxic Aldas may be useful in prevention of these cancers.
The work described above suggests a scenario in which the activation of the ALDH2 enzyme would provide a general health benefit in terms of cancer prevention to the estimated 560 million people carrying the ALDH2*2 genotype. However, cancer prevention research is technically very difficult and expensive due to having to follow cohorts for many years and to tease out an association with treatment from all other confounding factors, as well as chronic delivery of a drug. An alternative way to address the ability of ALDH2 activators to prevent cancer would be in a more specific scenario studying children with Fanconi anemia, who are a combination of ALDH2*1/*1, ALDH2*1/*2 and ALDH2*2/*2 genotype. Infants identified to have Fanconi Anemia could be treated to see if aplastic anemia and AML could be delayed, since these patients are known to develop either of these conditions within the first few years of life [89]. Additionally, the rate of progression to secondary malignancies could be studied, as the likelihood of developing secondary malignancy after marrow transplant to cure aplastic anemia or acute myelogenous leukemia (AML) is as much as 50-fold greater than that of the general population, and for head and neck cancers, 100–1000-fold greater[97]. The concept of pharmacologic ALDH2 activation also raises the possibility that other important enzymes in alcohol and aldehyde metabolism,. e.g., ADH5, could be similarly activated.
Additionally, the use of surrogate measures or biomarkers to quantify aldehydic load in peripheral blood or HSPC would be a way to measure the effectiveness of ALDH2 activators in reducing DNA-damaging aldehydic load. Such surrogate measures could be in incorporated into larger cancer prevention studies and could shorten the time scale.
Finally, the applicability of an activator of ALDH2 to such a large population suggests a problem in which people with the ALDH2*2 genotype may consume more alcohol, as ALDH2 activators may be able to reduce the effects of Asian Flushing syndrome. Additionally, if people who had traditionally avoided alcohol because of the disulfiram-like effects or concern about cancer susceptibility, now ingest ethanol with the assumption that the ALDH2*2 activators protect from cancer, this change in behavior may reduce the benefit of the drug. A social discussion of the impact of such a drug on human behavior must be addressed before beginning the large scale use of ALDH2*2 activators.
Highlights.
Aldehydes cause DNA damage
ALDH2*2 genotype exacerbates Fanconi Anemia (FA)
Small molecule activators of ALDH2 activity may prevent marrow failure in FA
Small molecule activators of ALDH2 activity could prevent cancer in FA
Acknowledgments
Supported in part by R37 NIAAA11147 to DM-R, Supported in part by the Stanford Child Health Research Institute (CHRI) Transdisciplinary Initiatives Program (TIP) The role of ALDH2 genetic variation and aldehyde metabolism in hematopoietic stem cell biology and the pathogenesis of bone marrow failure’ award to KIW, and by NIH grants T32 GM089626, T32 DK098132 to LDVW
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon MY, Lewis JL, Marley SB. Of mice and men...and elephants. Blood. 2002;100:4679–4680. doi: 10.1182/blood-2002-08-2517. [DOI] [PubMed] [Google Scholar]
- 2.Lieber JG, Webb S, Suratt BT, Young SK, Johnson GL, Keller GM, Worthen GS. The in vitro production and characterization of neutrophils from embryonic stem cells. Blood. 2004;103:852–859. doi: 10.1182/blood-2003-04-1030. [DOI] [PubMed] [Google Scholar]
- 3.Harker LA. The kinetics of platelet production and destruction in man. Clin Haematol. 1977;6:671–693. [PubMed] [Google Scholar]
- 4.Machlus KR, Italiano JE., Jr The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785–796. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Licht T, Pastan I, Gottesman M, Herrmann F. P-glycoprotein-mediated multidrug resistance in normal and neoplastic hematopoietic cells. Ann Hematol. 1994;69:159–171. doi: 10.1007/BF02215949. [DOI] [PubMed] [Google Scholar]
- 6.Sokolov M, Neumann R. Lessons learned about human stem cell responses to ionizing radiation exposures: a long road still ahead of us. Int J Mol Sci. 2013;14:15695–15723. doi: 10.3390/ijms140815695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes B, Saadat N, Myung B, Shekhar MP. Crosstalk between translesion synthesis, Fanconi anemia network, and homologous recombination repair pathways in interstrand DNA crosslink repair and development of chemoresistance. Mutat Res Rev Mutat Res. 2015;763:258–266. doi: 10.1016/j.mrrev.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mouw KW, D’Andrea AD. Crosstalk between the nucleotide excision repair and Fanconi anemia/BRCA pathways. DNA Repair (Amst) 2014;19:130–134. doi: 10.1016/j.dnarep.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Kutler DI, Patel KR, Auerbach AD, Kennedy J, Lach FP, Sanborn E, Cohen MA, Kuhel WI, Smogorzewska A. Natural history and management of Fanconi anemia patients with head and neck cancer: A 10 year follow-up. Laryngoscope. 2015 doi: 10.1002/lary.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 11.Park JY, Virts EL, Jankowska A, Wiek C, Othman M, Chakraborty SC, Vance GH, Alkuraya FS, Hanenberg H, Andreassen PR. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J Med Genet. 2016 doi: 10.1136/jmedgenet-2016-103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yabe M, Yabe H, Hamanoue S, Inoue H, Matsumoto M, Koike T, Ishiguro H, Morimoto T, Arakawa S, Ohshima T, Masukawa A, Miyachi H, Yamashita T, Katob S. In vitro effect of fludarabine, cyclophosphamide, and cytosine arabinoside on chromosome breakage in Fanconi anemia patients: relevance to stem cell transplantation. Int J Hematol. 2007;85:354–361. doi: 10.1532/IJH97.06191. [DOI] [PubMed] [Google Scholar]
- 13.Yabe M, Yabe H, Matsuda M, Hinohara T, Oh Y, Hattori K, Ishikawa K, Ohshima T, Yamamoto H, Kato S. Bone marrow transplantation for Fanconi anemia. Adjustment of the dose of cyclophosphamide for preconditioning. Am J Pediatr Hematol Oncol. 1993;15:377–382. [PubMed] [Google Scholar]
- 14.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 15.Oberbeck N, Langevin F, King G, de Wind N, Crossan GP, Patel KJ. Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol Cell. 2014;55:807–817. doi: 10.1016/j.molcel.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, Swenberg JA, Crossan GP, Patel KJ. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo W, Li H, Zhang Y, Ang CY. Determination of formaldehyde in blood plasma by high-performance liquid chromatography with fluorescence detection. J Chromatogr B Biomed Sci Appl. 2001;753:253–257. doi: 10.1016/s0378-4347(00)00552-1. [DOI] [PubMed] [Google Scholar]
- 18.Sanghani PC, Stone CL, Ray BD, Pindel EV, Hurley TD, Bosron WF. Kinetic mechanism of human glutathione-dependent formaldehyde dehydrogenase. Biochemistry. 2000;39:10720–10729. doi: 10.1021/bi9929711. [DOI] [PubMed] [Google Scholar]
- 19.Wang LL, Yang AK, Li Y, Liu JP, Zhou SF. Phenotype prediction of deleterious nonsynonymous single nucleotide polymorphisms in human alcohol metabolism-related genes: a bioinformatics study. Alcohol. 2010;44:425–438. doi: 10.1016/j.alcohol.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Ladeira C, Viegas S, Carolino E, Gomes MC, Brito M. The influence of genetic polymorphisms in XRCC3 and ADH5 genes on the frequency of genotoxicity biomarkers in workers exposed to formaldehyde. Environ Mol Mutagen. 2013;54:213–221. doi: 10.1002/em.21755. [DOI] [PubMed] [Google Scholar]
- 21.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 22.Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302:999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- 23.Jones KL, Smith DW, Hanson JW. The fetal alcohol syndrome: clinical delineation. Ann N Y Acad Sci. 1976;273:130–139. doi: 10.1111/j.1749-6632.1976.tb52873.x. [DOI] [PubMed] [Google Scholar]
- 24.Williams JF, Smith VC. A Committee On Substance, Fetal Alcohol Spectrum Disorders. Pediatrics. 2015;136:e1395–1406. doi: 10.1542/peds.2015-3113. [DOI] [PubMed] [Google Scholar]
- 25.Levy W. Aplastic anemia in siblings with multiple congenital anomalies (the Fanconi type) J Pediatr. 1952;40:24–41. doi: 10.1016/s0022-3476(52)80231-8. [DOI] [PubMed] [Google Scholar]
- 26.Eberhart JK, Parnell SE. The Genetics of Fetal Alcohol Spectrum Disorders. Alcohol Clin Exp Res. 2016;40:1154–1165. doi: 10.1111/acer.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kietzman HW, Everson JL, Sulik KK, Lipinski RJ. The teratogenic effects of prenatal ethanol exposure are exacerbated by Sonic Hedgehog or GLI2 haploinsufficiency in the mouse. PLoS One. 2014;9:e89448. doi: 10.1371/journal.pone.0089448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DW. Compendium on shortness of stature. J Pediatr. 1967;70:463–519. doi: 10.1016/s0022-3476(67)80355-x. [DOI] [PubMed] [Google Scholar]
- 29.Jones KL, Smith DW, Ulleland CN, Streissguth P. Pattern of malformation in offspring of chronic alcoholic mothers. Lancet. 1973;1:1267–1271. doi: 10.1016/s0140-6736(73)91291-9. [DOI] [PubMed] [Google Scholar]
- 30.Uebelacker M, Lachenmeier DW. Quantitative determination of acetaldehyde in foods using automated digestion with simulated gastric fluid followed by headspace gas chromatography. J Autom Methods Manag Chem. 2011;2011:907317. doi: 10.1155/2011/907317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien PJ, Siraki AG, Shangari N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit Rev Toxicol. 2005;35:609–662. doi: 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- 32.Brooks PJ, Zakhari S. Acetaldehyde and the genome: beyond nuclear DNA adducts and carcinogenesis. Environ Mol Mutagen. 2014;55:77–91. doi: 10.1002/em.21824. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1,n(2)-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem Res Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hecht SS, McIntee EJ, Wang M. New DNA adducts of crotonaldehyde and acetaldehyde. Toxicology. 2001;166:31–36. doi: 10.1016/s0300-483x(01)00436-x. [DOI] [PubMed] [Google Scholar]
- 35.Antoniak DT, Duryee MJ, Mikuls TR, Thiele GM, Anderson DR. Aldehyde-modified proteins as mediators of early inflammation in atherosclerotic disease. Free Radic Biol Med. 2015;89:409–418. doi: 10.1016/j.freeradbiomed.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Budas GR, Disatnik MH, Mochly-Rosen D. Aldehyde dehydrogenase 2 in cardiac protection: a new therapeutic target? Trends Cardiovasc Med. 2009;19:158–164. doi: 10.1016/j.tcm.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes KM, Bechara LR, Lima VM, Ribeiro MA, Campos JC, Dourado PM, Kowaltowski AJ, Mochly-Rosen D, Ferreira JC. Aldehydic load and aldehyde dehydrogenase 2 profile during the progression of post-myocardial infarction cardiomyopathy: benefits of Alda-1. Int J Cardiol. 2015;179:129–138. doi: 10.1016/j.ijcard.2014.10.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes KM, Campos JC, Bechara LR, Queliconi B, Lima VM, Disatnik MH, Magno P, Chen CH, Brum PC, Kowaltowski AJ, Mochly-Rosen D, Ferreira JC. Aldehyde dehydrogenase 2 activation in heart failure restores mitochondrial function and improves ventricular function and remodelling. Cardiovasc Res. 2014;103:498–508. doi: 10.1093/cvr/cvu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amiri A, Pryor E, Rice M, Downs CA, Turner-Henson A, Fanucchi MV. Formaldehyde exposure during pregnancy. MCN Am J Matern Child Nurs. 2015;40:180–185. doi: 10.1097/NMC.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 41.Kunjapur AM, Prather KL. Microbial engineering for aldehyde synthesis. Appl Environ Microbiol. 2015;81:1892–1901. doi: 10.1128/AEM.03319-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavoue J, Vincent R, Gerin M. Formaldehyde exposure in U.S. industries from OSHA air sampling data. J Occup Environ Hyg. 2008;5:575–587. doi: 10.1080/15459620802275023. [DOI] [PubMed] [Google Scholar]
- 43.de Medeiros IC, de Lima JG. Is nonalcoholic fatty liver disease an endogenous alcoholic fatty liver disease? - A mechanistic hypothesis. Med Hypotheses. 2015;85:148–152. doi: 10.1016/j.mehy.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 44.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6:e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zambelli VO, Gross ER, Chen CH, Gutierrez VP, Cury Y, Mochly-Rosen D. Aldehyde dehydrogenase-2 regulates nociception in rodent models of acute inflammatory pain. Sci Transl Med. 2014;6:251ra118. doi: 10.1126/scitranslmed.3009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salleron L, Magistrelli G, Mary C, Fischer N, Bairoch A, Lane L. DERA is the human deoxyribose phosphate aldolase and is involved in stress response. Biochim Biophys Acta. 2014;1843:2913–2925. doi: 10.1016/j.bbamcr.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Kotova N, Vare D, Schultz N, Gradecka Meesters D, Stepnik M, Grawe J, Helleday T, Jenssen D. Genotoxicity of alcohol is linked to DNA replication-associated damage and homologous recombination repair. Carcinogenesis. 2013;34:325–330. doi: 10.1093/carcin/bgs340. [DOI] [PubMed] [Google Scholar]
- 48.Marietta C, Thompson LH, Lamerdin JE, Brooks PJ. Acetaldehyde stimulates FANCD2 monoubiquitination, H2AX phosphorylation, and BRCA1 phosphorylation in human cells in vitro: implications for alcohol-related carcinogenesis. Mutat Res. 2009;664:77–83. doi: 10.1016/j.mrfmmm.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abraham J, Balbo S, Crabb D, Brooks PJ. Alcohol metabolism in human cells causes DNA damage and activates the Fanconi anemia-breast cancer susceptibility (FA-BRCA) DNA damage response network. Alcohol Clin Exp Res. 2011;35:2113–2120. doi: 10.1111/j.1530-0277.2011.01563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64:520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94:1–34. doi: 10.1152/physrev.00017.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Segura L, Ho KK, Perez-Miller S, Weiner H, Hurley TD. Catalytic contribution of threonine 244 in human ALDH2. Chem Biol Interact. 2013;202:32–40. doi: 10.1016/j.cbi.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oyama T, Isse T, Kagawa N, Kinaga T, Kim YD, Morita M, Sugio K, Weiner H, Yasumoto K, Kawamoto T. Tissue-distribution of aldehyde dehydrogenase 2 and effects of the ALDH2 gene-disruption on the expression of enzymes involved in alcohol metabolism. Front Biosci. 2005;10:951–960. doi: 10.2741/1589. [DOI] [PubMed] [Google Scholar]
- 55.Stewart MJ, Malek K, Crabb DW. Distribution of messenger RNAs for aldehyde dehydrogenase 1, aldehyde dehydrogenase 2, and aldehyde dehydrogenase 5 in human tissues. J Investig Med. 1996;44:42–46. [PubMed] [Google Scholar]
- 56.Harada S, Agarwal DP, Goedde HW. Isozyme variations in acetaldehyde dehydrogenase (e.c.1.2.1.3) in human tissues. Hum Genet. 1978;44:181–185. doi: 10.1007/BF00295411. [DOI] [PubMed] [Google Scholar]
- 57.Budas GR, Disatnik MH, Chen CH, Mochly-Rosen D. Activation of aldehyde dehydrogenase 2 (ALDH2) confers cardioprotection in protein kinase C epsilon (PKCvarepsilon) knockout mice. J Mol Cell Cardiol. 2010;48:757–764. doi: 10.1016/j.yjmcc.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goedde HW, Harada S, Agarwal DP. Racial differences in alcohol sensitivity: a new hypothesis. Hum Genet. 1979;51:331–334. doi: 10.1007/BF00283404. [DOI] [PubMed] [Google Scholar]
- 59.Teng YS. Human liver aldehyde dehydrogenase in Chinese and Asiatic Indians: gene deletion and its possible implications in alcohol metabolism. Biochem Genet. 1981;19:107–114. doi: 10.1007/BF00486141. [DOI] [PubMed] [Google Scholar]
- 60.Impraim C, Wang G, Yoshida A. Structural mutation in a major human aldehyde dehydrogenase gene results in loss of enzyme activity. Am J Hum Genet. 1982;34:837–841. [PMC free article] [PubMed] [Google Scholar]
- 61.Ikawa M, Impraim CC, Wang G, Yoshida A. Isolation and characterization of aldehyde dehydrogenase isozymes from usual and atypical human livers. J Biol Chem. 1983;258:6282–6287. [PubMed] [Google Scholar]
- 62.Li H, Borinskaya S, Yoshimura K, Kal’ina N, Marusin A, Stepanov VA, Qin Z, Khaliq S, Lee MY, Yang Y, Mohyuddin A, Gurwitz D, Mehdi SQ, Rogaev E, Jin L, Yankovsky NK, Kidd JR, Kidd KK. Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann Hum Genet. 2009;73:335–345. doi: 10.1111/j.1469-1809.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A. 1984;81:258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu LC, Tani K, Fujiyoshi T, Kurachi K, Yoshida A. Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2. Proc Natl Acad Sci U S A. 1985;82:3771–3775. doi: 10.1073/pnas.82.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83:314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Miller S, Younus H, Vanam R, Chen CH, Mochly-Rosen D, Hurley TD. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17:159–164. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao Q, Weiner H, Crabb DW. The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J Clin Invest. 1996;98:2027–2032. doi: 10.1172/JCI119007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis SJ, Smith GD. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005;14:1967–1971. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 69.Gross ER, Zambelli VO, Small BA, Ferreira JC, Chen CH, Mochly-Rosen D. A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu Rev Pharmacol Toxicol. 2015;55:107–127. doi: 10.1146/annurev-pharmtox-010814-124915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith BD, Jones RJ, Lee SM, Piantadosi S, Vala MS, Fuller D, Gore SD, Noga SJ, O’Donnell PV, Braine H, Vogelsang GB, Fuchs EJ, Flinn IW, Brodsky RA, Ambinder RF, Miller CB. Autologous bone marrow transplantation with 4-hydroperoxycyclophosphamide purging for acute myeloid leukaemia beyond first remission: a 10-year experience. Br J Haematol. 2002;117:907–913. doi: 10.1046/j.1365-2141.2002.03530.x. [DOI] [PubMed] [Google Scholar]
- 71.Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, Lako M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 72.Chen Y, Yu D, Zhang C, Shang B, He H, Chen J, Zhang H, Zhao W, Wang Z, Xu X, Zhen Y, Shao RG. Lidamycin inhibits tumor initiating cells of hepatocellular carcinoma Huh7 through GSK3beta/beta-catenin pathway. Mol Carcinog. 2015;54:1–8. doi: 10.1002/mc.22069. [DOI] [PubMed] [Google Scholar]
- 73.Levi BP, Yilmaz OH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood. 2009;113:1670–1680. doi: 10.1182/blood-2008-05-156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muramoto GG, Russell JL, Safi R, Salter AB, Himburg HA, Daher P, Meadows SK, Doan P, Storms RW, Chao NJ, McDonnell DP, Chute JP. Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010;28:523–534. doi: 10.1002/stem.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Napoli JL. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim Biophys Acta. 1999;1440:139–162. doi: 10.1016/s1388-1981(99)00117-1. [DOI] [PubMed] [Google Scholar]
- 77.Ghiaur G, Yegnasubramanian S, Perkins B, Gucwa JL, Gerber JM, Jones RJ. Regulation of human hematopoietic stem cell self-renewal by the microenvironment’s control of retinoic acid signaling. Proc Natl Acad Sci U S A. 2013;110:16121–16126. doi: 10.1073/pnas.1305937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155:215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 79.Ishii K, Young NS. Anemia of Central Origin. Semin Hematol. 2015;52:321–338. doi: 10.1053/j.seminhematol.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young NS, Scheinberg P, Calado RT. Aplastic anemia. Curr Opin Hematol. 2008;15:162–168. doi: 10.1097/MOH.0b013e3282fa7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sloand E, Kim S, Maciejewski JP, Tisdale J, Follmann D, Young NS. Intracellular interferon-gamma in circulating and marrow T cells detected by flow cytometry and the response to immunosuppressive therapy in patients with aplastic anemia. Blood. 2002;100:1185–1191. doi: 10.1182/blood-2002-01-0035. [DOI] [PubMed] [Google Scholar]
- 82.Solomou EE, Rezvani K, Mielke S, Malide D, Keyvanfar K, Visconte V, Kajigaya S, Barrett AJ, Young NS. Deficient CD4+ CD25+ FOXP3+ T regulatory cells in acquired aplastic anemia. Blood. 2007;110:1603–1606. doi: 10.1182/blood-2007-01-066258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawashima N, Narita A, Wang X, Xu Y, Sakaguchi H, Doisaki S, Muramatsu H, Hama A, Nakanishi K, Takahashi Y, Kojima S. Aldehyde dehydrogenase-2 polymorphism contributes to the progression of bone marrow failure in children with idiopathic aplastic anaemia. Br J Haematol. 2015;168:460–463. doi: 10.1111/bjh.13122. [DOI] [PubMed] [Google Scholar]
- 84.Heimpel H. Epidemiology and etiology of aplastic anemia, Aplastic Anaemia; Pathophysiology and Treatment. 2000:97–116. [Google Scholar]
- 85.Kojima S. Aplastic anemia in the Orient. Int J Hematol. 2002;76(Suppl 2):173–174. doi: 10.1007/BF03165112. [DOI] [PubMed] [Google Scholar]
- 86.Young NS, Kaufman DW. The epidemiology of acquired aplastic anemia. Haematologica. 2008;93:489–492. doi: 10.3324/haematol.12855. [DOI] [PubMed] [Google Scholar]
- 87.Montane E, Ibanez L, Vidal X, Ballarin E, Puig R, Garcia N, Laporte JR A Catalan Group for Study of, A. Aplastic. Epidemiology of aplastic anemia: a prospective multicenter study. Haematologica. 2008;93:518–523. doi: 10.3324/haematol.12020. [DOI] [PubMed] [Google Scholar]
- 88.IARC Monogr Eval Carcinog Risks Hum; Dry cleaning, some chlorinated solvents and other industrial chemicals; Lyon, France. 7–14 February 1995; 1995. pp. 33–477. [PMC free article] [PubMed] [Google Scholar]
- 89.Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, Ogawa S, Matsuo K, Takata M, Yabe M. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122:3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ginocchio VM, Brunetti-Pierri N. Progress toward improved therapies for inborn errors of metabolism. Hum Mol Genet. 2016;25:R27–35. doi: 10.1093/hmg/ddv418. [DOI] [PubMed] [Google Scholar]
- 91.Todd AG, McElroy JA, Grange RW, Fuller DD, Walter GA, Byrne BJ, Falk DJ. Correcting Neuromuscular Deficits With Gene Therapy in Pompe Disease. Ann Neurol. 2015;78:222–234. doi: 10.1002/ana.24433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koda K, Salazar-Rodriguez M, Corti F, Chan NY, Estephan R, Silver RB, Mochly-Rosen D, Levi R. Aldehyde dehydrogenase activation prevents reperfusion arrhythmias by inhibiting local renin release from cardiac mast cells. Circulation. 2010;122:771–781. doi: 10.1161/CIRCULATIONAHA.110.952481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen CH, Cruz LA, Mochly-Rosen D. Pharmacological recruitment of aldehyde dehydrogenase 3A1 (ALDH3A1) to assist ALDH2 in acetaldehyde and ethanol metabolism in vivo. Proc Natl Acad Sci U S A. 2015;112:3074–3079. doi: 10.1073/pnas.1414657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roth A, Li H, Anorma C, Chan J. A Reaction-Based Fluorescent Probe for Imaging of Formaldehyde in Living Cells. J Am Chem Soc. 2015;137:10890–10893. doi: 10.1021/jacs.5b05339. [DOI] [PubMed] [Google Scholar]
- 95.Brewer TF, Chang CJ. An Aza-Cope Reactivity-Based Fluorescent Probe for Imaging Formaldehyde in Living Cells. J Am Chem Soc. 2015;137:10886–10889. doi: 10.1021/jacs.5b05340. [DOI] [PubMed] [Google Scholar]
- 96.Yuen LH, Saxena NS, Park HS, Weinberg K, Kool ET. Dark Hydrazone Fluorescence Labeling Agents Enable Imaging of Cellular Aldehydic Load. ACS Chem Biol. 2016 doi: 10.1021/acschembio.6b00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]