Abstract
With the increasing anthropogenic CO2 concentration, ocean acidification (OA) can have dramatic effects on coral reefs. However, the effects of OA on coral physiology and the associated microbes remain largely unknown. In the present study, reef-building coral Acropora gemmifera collected from a reef flat with highly fluctuating environmental condition in the South China Sea were exposed to three levels of partial pressure of carbon dioxide (pCO2) (i.e., 421, 923, and 2070 μatm) for four weeks. The microbial community structures associated with A. gemmifera under these treatments were analyzed using 16S rRNA gene barcode sequencing. The results revealed that the microbial community associated with A. gemmifera was highly diverse at the genus level and dominated by Alphaproteobacteria. More importantly, the microbial community structure remained rather stable under different pCO2 treatments. Photosynthesis and calcification in A. gemmifera, as indicated by enrichment of δ18O and increased depletion of δ13C in the coral skeleton, were significantly impaired only at the high pCO2 (2070 μatm). These results suggest that A. gemmifera can maintain a high degree of stable microbial communities despite of significant physiological changes in response to extremely high pCO2.
Rising CO2 in the atmosphere elevates the partial pressure of carbon dioxide (pCO2) in seawater and reduces the global oceanic pH and carbonate ion concentrations, which is called ocean acidification (OA). It has been suggested that OA has profound effects on marine organisms and ecosystems, particularly calcifying organisms such as reef-building corals1,2,3. With the increasing OA associated with ocean warming due to the rising global CO2 emissions2, there is an urgent need to understand and predict the tolerance and response of corals to future climate change.
Reef-building corals are commonly referred to as holobionts, which comprise coral host and associated microorganisms including endosymbiotic photosynthetic algae, bacteria, and archaea, among others. These complex microbial partners play pivotal roles in coral health and holobiont function in carbon, nitrogen and sulfur cycles. The future fate of coral reefs largely depends on the capacity of corals and their symbionts to acclimatize or adapt to climate change2,4,5,6,7. There is emerging evidence that corals can adapt to climate change8, although coral photosynthesis and growth can be negatively impacted by OA9. Moreover, flexible coral-algal symbiosis may facilitate the acclimatization/adaptation of the holobiont through algal shuffling or switching10,11.
Shifting in the composition of coral-associated microbiota has been observed following environmental disturbances (e.g., elevated temperature) and has often been linked to impaired host health12,13,14, although it has also been hypothesized to mediate holobiont resistance to environmental perturbations6,15. Coral-associated microbial communities may be affected directly or indirectly by OA and would subsequently compromise holobiont fitness (i.e., changes in photosynthesis or calcification) and survival, possibly due to a shift in the functional roles of microbial associations12,16,17,18. However, to date it is not clear how microbial communities change in coral in response to OA, or whether theses changes also alter host physiology. Preliminary laboratory-based investigations have revealed a remarkable impact of increased pCO2 or reduced pH on coral microbial communities12,16,18. In contrast, no significant changes were observed in the microbial communities of transplanted corals in natural CO2 vents19 and associated with two Pacific corals after 8 weeks of exposure to increased pCO220. These contradictory findings underscore the need for further research. Most recently, microbial communities associated with coral and sponge from natural CO2 seeps have demonstrated species-specific acclimatization to their habitats21. Therefore, the potential of natural microbial communities in corals to acclimatize/adapt to OA cannot be overlooked.
Natural fluctuations in seawater pH/pCO2 are common, especially diel pH/pCO2 fluctuations in shallow water coral reefs22 and these fluctuations affect the abundance and distribution of marine organisms23. The fauna have been suggested to be locally acclimatized/adapted to the variable pH environment as an evolutionary mechanism to cope with future acidification24,25,26. However, the adaptation of the coral holobionts to OA remains largely unexplored and is worth careful investigation.
The Luhuitou fringing reef (18°12′N, 109°28′E) is located in the southern Hainan Island, South China Sea (see Supplementary Fig. S1) and used to have a high coverage of living coral, which has declined by 80% since the 1960s27. The diurnal and seasonal variation of the reef flat seawater pH/pCO2 is high28,29,30, and the recorded extreme level is even lower than the value currently predicted at the end of this century (see Supplementary Table S1). The rapidly-growing branching coral Acropora, which is distributed worldwide, is an ecologically important genus in this reef flat. In the present study, A. gemmifera colonies collected from this reef flat were exposed to three pCO2 levels to test our hypothesis that both the coral physiology and the microbial communities associated with this coral species are stable and resistant to OA exposure.
Results and Discussion
Overview of the microbial communities
After quality filtering, 308,591 reads were used for the downstream analyses. The number of operational taxonomic units (OTUs), Chao1 estimation of species richness, and Shannon index were obtained at a dissimilarity of 3% (Table 1). The rarefaction analyses revealed that the sequencing effort for each sample was sufficient to reflect the microbial diversity, and the rank-abundance curve showed that most OTUs had an abundance lower than 0.1%, which demonstrated that the microbial communities were occupied by rare species (see Supplementary Fig. S2). The prevalence of rare species has been widely demonstrated elsewhere, yet the ecological and functional roles of these rare species remain unknown31. There were no significant differences in beta diversity among the pCO2 treatments, which is in contrast to the findings of a previous report showing an increase in coral microbial diversity with decreasing pH, possibly caused by an intermediate disturbance16.
Table 1. Sample information and summary of microbial communities in corals and seawaters.
| Sample ID | Treatment | Number of qualified reads | Total OTUs | Chao1 Ave. | Observed species Ave. | Shannon Ave. |
|---|---|---|---|---|---|---|
| HA.1 | High pCO2 treatment | 37600 | 1301 | 1678.248 | 707.2 | 5.420 |
| HA.2 | 19098 | 1115 | 1826.916 | 874.8 | 5.494 | |
| HA.3 | 29468 | 1687 | 2102.160 | 1087 | 6.333 | |
| MA.1 | Medium pCO2 treatment | 27489 | 1108 | 1545.223 | 707.2 | 5.360 |
| MA.2 | 27946 | 1295 | 1780.889 | 822.9 | 5.316 | |
| MA.3 | 26650 | 1074 | 1528.017 | 709.3 | 5.431 | |
| CA.1 | Control pCO2 treatment | 24616 | 1065 | 1629.961 | 716.8 | 5.258 |
| CA.2 | 21224 | 989 | 1576.258 | 727.8 | 5.527 | |
| CA.3 | 28163 | 1244 | 1688.842 | 783.5 | 5.450 | |
| HW.1 | High pCO2 treatment | 14358 | 1237 | 1901.201 | 1152.6 | 5.252 |
| MW.1 | Medium pCO2 treatment | 15688 | 1203 | 1795.261 | 1075.2 | 4.860 |
| CW.1 | Control pCO2 treatment | 12463 | 1127 | 1886.261 | 1126.7 | 5.908 |
“H”, “M” and “C” refer to high, medium and control pCO2 treatment, respectively. “A” and “W” refer to coral and seawater samples, respectively. The number following the letter indicates a replicate. Chao1, Observed species and Shannon index were determined at 3% dissimilarity after normalizing the full 16S dataset (including bacterial and archaeal sequences) to 12,459 sequences per sample.
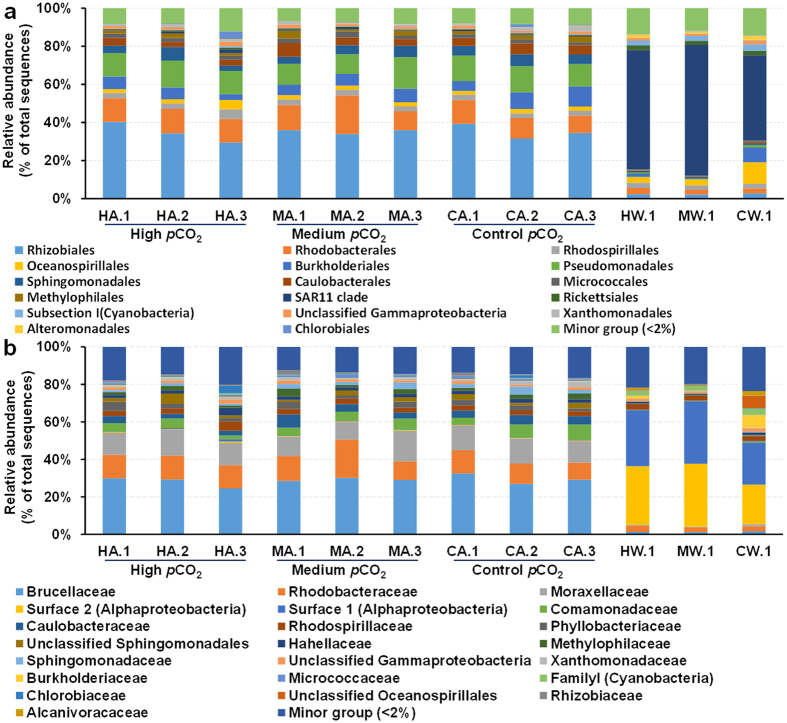
In total, 24 bacterial and 2 archaeal phyla were detected in the coral and seawater samples, including Proteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Thaumarchaeota and Euryarchaeota (see Supplementary Fig. S3). The relative abundance of archaea made up less than 0.1% of the seawater samples, with most OTUs belonging to autotrophic ammonia-oxidizing archaea (AOA) within the phylum Thaumarchaeota, which was even less abundant in the coral samples. It has been suggested that archaea may play prominent roles in corals and reefs5, although their abundance in both corals and reef water is much lower than that of bacteria32. Notably, the bacterial communities in both A. gemmifera and seawater were dominated by Proteobacteria and the most abundant class was Alphaproteobacteria (56–80%), among which the majority were assigned to the family Brucellaceae in the order Rhizobiales (Fig. 1) followed by Gammaproteobacteria (9–26%). Both Alphaproteobacteria and Gammaproteobacteria are commonly highly abundant in corals, but their relative abundance varies among species33.
Figure 1.
Coral and seawater microbial communities at the order (a) and family (b) level. The minor group represents the sum of all orders or families representing <2% in all samples. “H”, “M” and “C” refer to high, medium and control pCO2 treatment, respectively. “A” and “W” refer to coral and seawater samples, respectively. The number following the letter indicates a replicate.
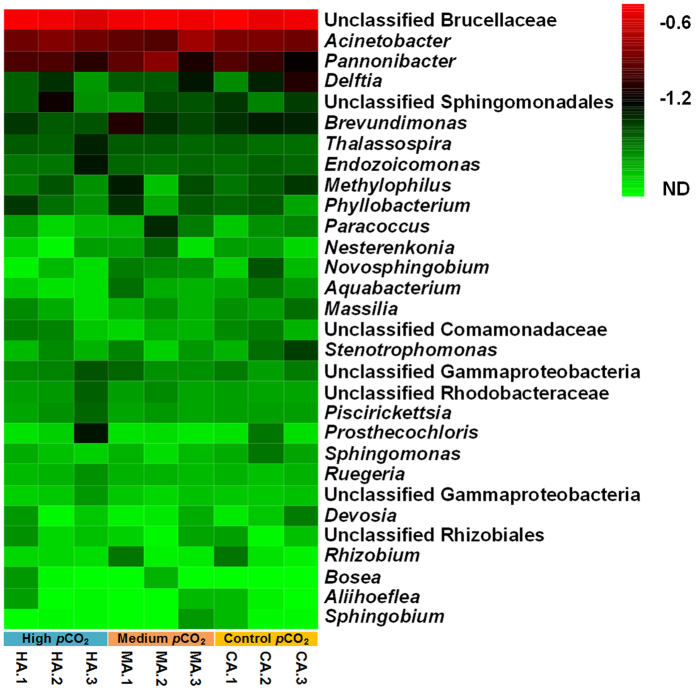
Taxonomic assignment at the genus level was summarized, and genera with an abundance of greater than 1% in at least one sample are shown in a heat map (Fig. 2). In the present study, the unclassified Brucellaceae (>24%), Acinetobacter (>9%) and Pannonibacter (>5%) were the most abundant genera in coral, regardless of the pCO2 treatment. Diazotrophs within the order Rhizobiales have been found in other coral species and were considered to be important for coral holobiont in nitrogen-limited waters5. It has been shown that copiotrophic taxa including Brucellaceae were enriched in algal-dominated environment34. Diverse algal communities on the Luhuitou fringing reef35 might contribute to the dominance of unclassified Brucellaceae in A. gemmifera. Acinetobacter spp. have also been commonly reported in bleached and healthy corals36. Therefore, it is reasonable to suggest that the dominant genera, including Acinetobacter and the unclassified Brucellaceae, play critical roles in A. gemmifera. Interestingly, the putatively endosymbiotic Endozoicomonas37 was detected at a very low level in all coral samples (<2%). The photosynthetic Cyanobacteria assigned to the genus Synechococcus have been reported in sponge and coral21 and were also detected at a very low abundance (0.2%) in A. gemmifera. However, the functions of bacteria and archaea and their interactions in the coral holobiont remain largely unclear.
Figure 2. Heat map showing the abundance of microbial assignments in coral at the genus level.
Genera abundance >1% at least in one sample are shown. The abundance values are log10(x + 0.01)-transformed for plotting. For the heat map scale, “ND, −1.2, and −0.6” indicate relative abundance “0, 5%, and 24%”, respectively. The heat map was generated with R (version 3.1.3, R Development Core Team, 2015). “H”, “M” and “C” refer to high, medium and control pCO2 treatment, respectively. “A” refer to coral samples. The number following the letter indicates a replicate.
Stability of microbial communities in A. gemmifera
As estimated by Adonis analysis at all taxonomic levels (Adonis test, p > 0.05) and nMDS ordination (see Supplementary Fig. S4), there were no significant differences in microbial community compositions in A. gemmifera among the different pCO2 treatments even after a 4-week exposure. Additionally, results from the SIMPER analysis showed that the dissimilarity of microbial communities among pCO2 treatments was very small (see Supplementary Fig. S5). Taken together, these findings suggest that the A. gemmifera microbiome was not significantly affected by elevated pCO2 and could remain relatively stable (Fig. 2). This result is inconsistent with the findings of some previous studies in which the coral microbiome shifted under higher pCO2 or lower pH treatments over treatment periods ranging from days to months16,38. However, our finding is consistent with some other studies. For example, there were no differences in the microbial community structure in coral between pH 7.7 (pCO2 = 1187 μatm) and 7.5 (pCO2 = 1638 μatm) whereas a significant difference was observed between pH 8.1 (pCO2 = 464 μatm) and 7.9 (pCO2 = 822 μatm) after 6 weeks of CO2 exposure18. Moreover, Meron et al.19 observed no significant changes in microbial communities associated with two Mediterranean coral species that were transplanted along natural pH gradients. A recent study reported that the microbial communities of two Pacific coral species were tolerant to reduced pH 7.9 (pCO2 738–835 μatm)20. These inconsistent results might reflect that some coral-microbial associations are more resistant to increases in pCO2/decreases in pH than others, but these findings could also be partially attributed to differences in the experimental conditions (e.g., field vs laboratory, pCO2 or pH level, among others), and the exposure duration.
In most cases, microbial communities are dynamic and can rapidly respond to OA in seawater39, biofilms40 and other associated systems41. The genus Acropora is among the most sensitive coral to environmental change42. The potential for coral acclimatization or adaptation to climate change has been studied43, and the physiological and molecular mechanisms responsible for OA resistance have recently been proposed8. Although there is limited evidence for biological adaptation to climate change in coral microbial symbionts, the adaptive power to climate change has been well documented in the photosymbiotic Symbiodinium10,11,44. The shallow habitat of the coral A. gemmifera sampled in the present study has been experiencing regular large diurnal and seasonal variations in pH/pCO2 (see Supplementary Table S1), which are mainly driven by biological activities of the reef28,29,30. Therefore, it is most likely that microbial communities harbored by the natural population of A. gemmifera are resistant to the increased pCO2, due to long-term acclimatization/adaptation to the highly dynamic pH conditions within the reef flat. Thus, there may be a resilient relationship between coral and microbial partners that can help corals overcome the fluctuations in seawater pH/pCO2. However, we note that the stability of the coral microbiome is based on only one species colleting from a fluctuating environment. The application of variable pCO2 conditions and controls from stable pH/pCO2 environments in highly replicated culture experiments with consideration of tank effects could further confirm this assumption in future studies.
A recent study supports this interpretation. Morrow et al.21 found that microbial communities associated with coral and sponge originally from natural volcanic CO2 seeps were distinct from the nearby control sites, reflecting the acclimatization of the host-symbiont to the high pCO2 environment. Local acclimatization/adaptation to environmental variations in pCO2, temperature and nutrients, among others, has revealed the capabilities of marine organisms including reef-building corals and symbiotic algae, to adapt to future climate change8,24,25,44,45. However, in general, the species-specific response of marine organisms to OA remains poorly understood1,23,46. Thus, it is rather premature to conclude whether we can extrapolate the adaptive power of coral and its associated microbes documented in the present study to other coral species living in highly fluctuating reef environments.
Skeletal isotopic response to ocean acidification
During our experiments, all blocks of A. gemmifera exposed to the different pCO2 treatments grew, survived and formed new skeleton (see Supplementary Fig. S6), even at the high pCO2 (pH reduced to 7.47). When the fast-growing coral A. gemmifera skeletal δ13C and δ18O were compared among the different pCO2 treatments, the skeletal δ13C values in A. gemmifera were significantly different between any two pCO2 treatments except between the control and the medium (Fig. 3). Skeletal δ13C values were depleted by 1.10‰ and 1.04‰ for the control vs. high pCO2 and for the medium vs. high pCO2, respectively (one-way ANOVA, Tukey test, p < 0.05). Skeletal δ18O values in A. gemmifera were enriched with increased pCO2; they were 0.55‰ and 0.38‰ heavier in response to high pCO2 than those in the control and medium, respectively (one-way ANOVA, Tukey test, p < 0.05). Compared with the previous data47, skeletal δ18O values revealed greater depletion in fast-growing coral A. gemmifera, while the skeletal δ13C values remained within range. In addition, the relationship between skeletal δ13C and δ18O in the non-photosynthetic coral Tubastrea sp. deviated the most from those both in the photosynthetic corals A. gemmifera and Pavona sp. (Fig. 3).
Figure 3. Skeletal isotopic response of Acropora gemmifera cultured under the control (421 μatm) and increased (923 μatm and 2070 μatm) pCO2 conditions.
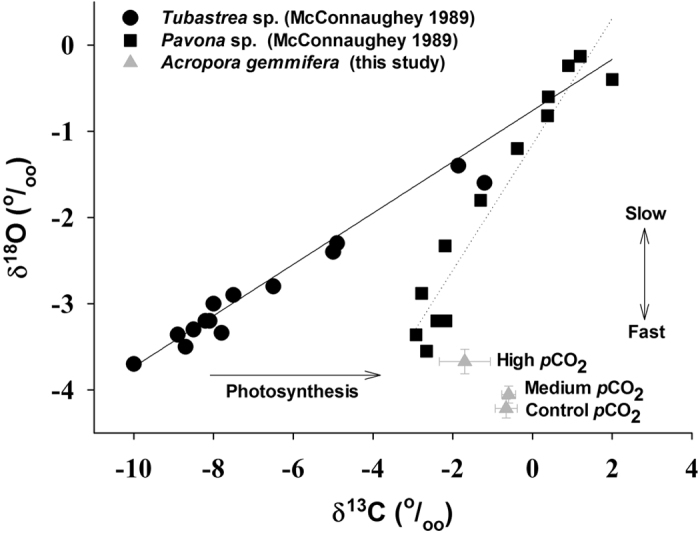
Skeletal isotopic composition of non-photosynthetic coral Tubastrea sp. and photosynthetic coral Pavona sp. reported in a previous study (McConnaughey 1989) are plotted for comparison. Photosynthesis indicated the carbon isotopic offset due to respiration and photosynthesis. “Slow” and “Fast” indicated slow and fast coral calcification rates. Fast growing Acropora corals are expected to have more enriched δ13C but depleted δ18O values.
The isotopic composition of the coral skeleton can be affected by metabolic isotope effects (e.g., photosynthesis and respiration) and kinetic isotope effects (e.g., the calcification process)48. The coral skeletal δ13C and δ18O were generally used as an effective proxy to study photosynthesis, respiration and calcification processes48,49. In general, photosynthesis and calcification can enrich skeletal δ13C but deplete skeletal δ18O due to isotope fractionation47,49. Compared non-photosynthetic (Tubastrea sp.) and photosynthetic (Pavona sp.) corals47, the relationship of δ13C vs. δ18O in Pavona sp. and A. gemmifera was different from that in Tubastrea sp. (non-photosynthetic coral) due to active photosynthesis. In addition, more δ18O deviation was observed in A. gemmifera than Tubastrea sp. and Pavona sp., mostly due to the highest growth rate in A. gemmifera (Fig. 3). Skeletal δ13C values in A. gemmifera were significantly depleted at the high pCO2, suggesting that the photosynthetic rates were much lower at the high pCO2 than at the control pCO2. The variation in skeletal δ18O values of A. gemmifera was consistent with the findings of a previous study demonstrating an enrichment of δ18O in the coral skeleton in response to elevated pCO249. The coral calcification rate decreases under reduced pH conditions9, which corresponds to heavier skeletal δ18O, whereas low pCO2 and higher pH lead to species with lighter δ18O because HCO3− is isotopically heavier than CO32,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49. Consequently, the significantly enriched δ18O and more depleted δ13C in A. gemmifera observed herein may reflect slight reductions in photosynthesis and calcification at the high pCO2. It should be noted that A. gemmifera skeletal δ13C and δ18O values did not vary significantly at the medium pCO2, potentially because this stress level did not exceed its acclimatization range. These findings indicate that the coral A. gemmifera is able to acclimate to an acidifying ocean, even in the presence of a dramatically increasing atmospheric CO2 concentration.
Although the mechanisms by which extremely high pCO2 induces decreased photosynthesis and calcification efficiencies in A. gemmifera are unknown, several potential mechanisms have been proposed, such as photoinhibition and suppression of the carbon concentrating process3,9. Photosynthesis, calcification and other physiological processes in reef-building corals can be influenced by their microbial partners or vice versa under OA17. However, the microbial communities associated with A. gemmifera remained unchanged as a consequence of host physiological changes, further supporting our hypothesis that highly stable microbial associations are likely driven by local acclimatization/adaptation to the fluctuating environment. Alternatively, host physiological costs might result from a potentially increasing energy demand to maintain stable microbial assemblages at the extremely high pCO2 that exceeds its tolerance level.
It has also been proposed that physiological differences among symbiotic algal phylotypes may influence the stable isotopic composition of coral skeleton50. Furthermore, the distinct mechanisms used to concentrate carbon by different Symbiodinium phylotypes and their physiological responses to OA are phylotype- specific51. For example, Symbiodnium community shifts may occur in response to environmental stresses10,11. In the present study, we did not investigate Symbiodinium phylotypes associated with A. gemmifera. A recent study found no changes in Symbiodinium phylotypes associated with corals among different pH conditions19, suggesting the presence of stable Symbiodinium assemblages in corals in response to OA. In general, a stable microbial partnership to maintain key metabolic functions can improve coral holobiont acclimatization or adaptation to environmental stresses5. However, the interactions between microbial communities and coral physiology remain far from clarified.
Conclusions
In this study, the tropical fast-growing coral A. gemmifera from a shallow habitat with natural pH/pCO2 fluctuations was selected as a representative species and was exposed to a 4-week CO2 treatment. The microbial communities and skeletal isotopic compositions were examined simultaneously. We found that the microbial communities in A. gemmifera remained remarkably stable. In contrast, neither photosynthesis nor calcification in the coral were impacted under medium pCO2 but were both negatively affected under extremely high pCO2, as demonstrated by an enrichment of δ18O and increased depletion of δ13C in the skeleton under extremely high CO2 stress. The present findings indicate that some reef-building corals may be more tolerant to OA in pH/pCO2 fluctuating environments and have a high degree of host-symbiont fidelity, despite the observed impairment of host physiological processes in response to high CO2 stress. This study also contributes to our understanding of the variability of OA resistance among coral-microbial associations. Because coral reefs are facing other environmental stresses in addition to OA, the synergistic effects of multiple stressors on the coral microbiome must be carefully examined to understand the persistence of the coral holobiont and coral reefs in the future ocean.
Methods
Experimental design and sample collection
The experiment was conducted in an outdoor seawater flow-through system at the Tropical Marine Biological Research Station in Hainan (TMBRS) near the Luhuitou fringing reef. Seawater was pumped directly from a depth of 6 m in the front of the TMBRS into three 2000-L header tanks in which the designated pCO2 level was adjusted with CO2 gas. Three pCO2 treatments were projected current pCO2 levels, those at the end of the present century, and double the end of the present century: 421 μatm, 923 μatm, and 2070 μatm with pH values of 8.07, 7.76 and 7.47, respectively. The well-mixed seawater from the header tank continually flowed into three aquaria at a rate of 0.5 l min−1. Each aquaria was equipped with a submerged pump to drive the water flow, and all aquaria were maintained under a natural light–dark cycle to mimic the field condition.
Six healthy colonies of A. gemmifera were collected from the Luhuitou fringing reef flat at a depth of ~2 m in May 2014 and divided into small pieces. After acclimation for two weeks in large aquaria with running water, one coral nubbin was randomly selected and was suspended using fine nylon strings in three aquaria for exposure to each of the three pCO2 treatments. The coral nubbins from same colony were evenly distributed among the pCO2 treatments to avoid any possible sampling bias. A total of 9 coral nubbins were maintained for further analysis during the experiment.
At the end of the experiment (i.e., 4-week exposure), 1 L of seawater from each treatment was filtered through 0.2 μm polycarbonate (PC) membrane filters and stored at −20 °C for further analyses. One coral nubbin from each treatment aquarium was sampled and divided into two aliquots. One was rinsed three times and then preserved in 70% ethanol and stored at −20 °C for DNA extraction; the other was used for stable isotope analyses.
Determination of environmental parameters
Photosynthetically active radiation (PAR) in each aquarium was recorded every 30 min below the seawater surface using the Hobo® logger (Onset, USA). The average diurnal variations in PAR during the 4-week period are shown in Fig. S7. The seawater pH and temperature were measured daily in each aquarium using a pH meter (Orion Star™) and the total alkalinity was also determined weekly using an automated titration system (Metrohm 877 Titrino plus, Switzerland). Carbonate system parameters, including [HCO3−], [CO32−], pCO2 and aragonite saturation state (ΩA), were calculated from the measured pH and total alkalinity values using the CO2SYS program52 (Table 2).
Table 2. Carbonate chemistry parameters of seawater for each treatment.
| Treatment | pHNBS | Alkalinity (μmol kg−1) | HCO3− (μmol kg−1) | CO32− (μmol kg−1) | pCO2 (μatm) | Ωara |
|---|---|---|---|---|---|---|
| Control pCO2 | 8.07 ± 0.02 | 2233 ± 22 | 1701 ± 35 | 214 ± 15 | 421 ± 49 | 3.6 ± 0.25 |
| Medium pCO2 | 7.76 ± 0.02 | 2223 ± 11 | 1912 ± 36 | 125 ± 11 | 923 ± 113 | 2.1 ± 0.19 |
| High pCO2 | 7.47 ± 0.02 | 2230 ± 13 | 2089 ± 25 | 67 ± 6 | 2070 ± 259 | 1.1 ± 0.01 |
Values are means ± SE. Seawater pH (NBS scale), and salinity (31.5) were measured daily during the experiment (n = 28). Total alkalinity (TA) was measured at a specific time point every week (n = 4). The remaining parameters for carbonate seawater chemistry were calculated using the CO2SYS program.
DNA extraction and 16S rDNA amplicon sequencing
A preserved piece of coral was homogenized in liquid nitrogen and then the total DNA from the resulting coral powder and filtered seawater samples was subsequently extracted using the Fast DNA® SPIN Kit for Soil (MP Biomedicals, Irvine, CA) according to the company’s protocol. The DNA samples were amplified by PCR using barcoded primers targeting the hypervariable region V3-V4 of the 16S rRNA gene of Bacteria and Archaea: 341F (5′-CCTAYGGGRBGCASCAG-3′) and 802R (5′-TACNVGGGTATCTAATCC-3′)53. The PCR amplification was performed using a thermocycle controller (MJ Research Inc., Bio-Rad) with the following program: an initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 5 min. All PCR products were purified using the Qiagen Agarose Gel DNA Purification Kit (Qiagen, Germany) and quantified using a NanoDrop device (Thermo Scientific, USA). All amplicon products were mixed at equal concentrations and sequenced on an Illumina Miseq platform using 2 × 300 bp mode at Novogene (Beijing, China). The raw reads were submitted to the NCBI Sequence Read Archive under accession number SRP066229 (SRR2917919).
Sequence data processing
Overlapping paired-end reads were merged to obtain full-length 16S V3-V4 fragments using PEAR54. After de-multiplexing and quality control, the downstream bioinformatics analysis was performed with QIIME1.5.0 pipelines55. Briefly, OTUs with 97% similarity were defined after the qualified reads were clustered using Uclust56. Representative sequences for each OTU were assigned to different taxa using the Ribosomal Database Project (RDP) classifier version 2.257 against the SILVA108 database58 with a 50% cut-off threshold. Representatives assigned to eukaryotes, chloroplasts and mitochondria were filtered out. The taxon and abundance were summarized at the phylum, class, order, family, and genus levels. The species diversity, Shannon index, rarefaction curves and rank-abundance curves were determined using the QIIME pipeline.
Stable isotope analyses
Skeleton fragments of A. gemmifera were soaked in 30% hydrogen peroxide to remove coral tissue and then sonicated for 4 min at 20 °C. Skeletons were subsequently washed several times with double-distilled water and dried overnight at 50 °C. The newly grown part was scalped and ground into powder. Coral skeletal δ13C and δ18O data were obtained using a Finnigan MAT 253 Isotope Ratio Mass Spectrometer coupled to a Kiel Carbonate Device IV at the South China Sea Institute of Oceanology, Chinese Academy of Sciences, China. δ13C and δ18O were determined by repeated measurements of the international reference standard NBS-18. δ13C and δ18O values were presented as the per mil (‰) deviation of 13C/12C and 18O/16O, respectively.
Statistical analyses
To test the effect of pCO2 treatments on microbial community compositions of coral samples, after normalizing all sequence reads for taxonomic analysis to the lowest sequencing depth (19,098 reads), pairwise dissimilarities among coral samples were calculated based on the Bray–Curtis index for ‘Adonis’, which is a non-parametric multivariate analysis of variance. In the Adonis analysis, the distance matrix was the response variable with pCO2 treatment as independent variable. Non-metric multidimensional scaling (nMDS) was also performed to visualize the dissimilarities. Genera making the greatest contribution to dissimilarity among the pCO2 treatments were further investigated through similarity percentage (SIMPER) analysis. To compare coral skeletal δ13C and δ18O among pCO2 treatments, one-way ANOVA and Tukey’s test were employed. All analyses were conducted using the package vegan within the R statistical environment (version 3.1.3, R Development Core Team, 2015).
Additional Information
How to cite this article: Zhou, G. et al. Changes in microbial communities, photosynthesis and calcification of the coral Acropora gemmifera holobiont in response to ocean acidification. Sci. Rep. 6, 35971; doi: 10.1038/srep35971 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. Yuyang Zhang and Mr. Shize Zhang for laboratory assistance, and the Administration of Sanya Coral Reef National Nature Reserve for providing sampling permits. This work was supported by grants from the National Natural Science Foundation of China (U1301232, 41206140) and the National Key Technology R&D Program (2012BAC19B08).
Footnotes
Author Contributions G.W.Z. wrote the manuscript with all authors commenting. G.W.Z., T.Y., L.C. and L.J. performed the experiment and undertook carbonate analyses. W.P.Z., R.M.T., H.Y.T. and X.C.Y. participated in data analyses. G.W.Z., S.L., H.H. and P.Y.Q. designed the research.
References
- Kroeker K. J., Kordas R. L., Crim R. N. & Singh G. G. Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecol Lett 13, 1419–1434, 10.1111/j.1461-0248.2010.01518.x (2010). [DOI] [PubMed] [Google Scholar]
- Pandolfi J. M., Connolly S. R., Marshall D. J. & Cohen A. L. Projecting coral reef futures under global warming and ocean acidification. Science 333, 418–422, 10.1126/science.1204794 (2011). [DOI] [PubMed] [Google Scholar]
- Hofmann G. E. et al. The effect of ocean acidification on calcifying organisms in marine ecosystems: an organism-to-ecosystem perspective. Annu Rev Ecol Evol S, 41, 127–147, 10.1146/annurev.ecolsys.110308.120227 (2010). [DOI] [Google Scholar]
- Ainsworth T. D., Thurber R. V. & Gates R. D. The future of coral reefs: a microbial perspective. Trends Ecol Evol 25, 233–240, 10.1016/j.tree.2009.11.001 (2010). [DOI] [PubMed] [Google Scholar]
- Krediet C. J., Ritchie K. B., Paul V. J. & Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. P Roy Soc B-Biol Sci 280, 20122328, 10.1098/rspb.2012.2328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R. & Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5, 355–362, 10.1038/nrmicro1635 (2007). [DOI] [PubMed] [Google Scholar]
- Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J. & Wild C. Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol 23, 490–497, 10.1016/j.tim.2015.03.008 (2015). [DOI] [PubMed] [Google Scholar]
- Palumbi S. R., Barshis D. J., Traylor-Knowles N. & Bay R. A. Mechanisms of reef coral resistance to future climate change. Science 344, 895–898, 10.1126/science.1251336 (2014). [DOI] [PubMed] [Google Scholar]
- Anthony K. R., Kline D. I., Diaz-Pulido G., Dove S. & Hoegh-Guldberg O. Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc Natl Acad Sci USA 105, 17442–17446, 10.1073/pnas.0804478105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans R. & van Oppen M. J. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. P Roy Soc B-Biol Sci 273, 2305–2312, 10.1098/rspb.2006.3567 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse T. C. et al. Host-symbiont recombination versus natural selection in the response of coral-dinoflagellate symbioses to environmental disturbance. P Roy Soc B-Biol Sci 277, 2925–2934, 10.1098/rspb.2010.0385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R. V. et al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11, 2148–2163, 10.1111/j.1462-2920.2009.01935.x (2009). [DOI] [PubMed] [Google Scholar]
- Bourne D., Iida Y., Uthicke S. & Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J 2, 350–363, 10.1038/ismej.2007.112 (2008). [DOI] [PubMed] [Google Scholar]
- Roder C. et al. Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J 8, 31–39, 10.1038/ismej.2013.127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef L., Koren O., Loya Y., Zilber-Rosenberg I. & Rosenberg E. The coral probiotic hypothesis. Environ Microbiol 8, 2068–2073, 10.1111/j.1462-2920.2006.01148.x (2006). [DOI] [PubMed] [Google Scholar]
- Meron D. et al. The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J 5, 51–60, 10.1038/ismej.2010.102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien P. A., Morrow K. M., Willis B. & Bourne D. Implications of ocean acidification for marine microorganisms from the free-living to the host-associated. Front Mar Sci 3, 47, 10.3389/fmars.2016.00047(2016). [DOI] [Google Scholar]
- Webster N. S. et al. Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environ Microbiol Rep 5, 243–251, 10.1111/1758-2229.12006 (2013). [DOI] [PubMed] [Google Scholar]
- Meron D. et al. Changes in coral microbial communities in response to a natural pH gradient. ISME J 6, 1775–1785, 10.1038/ismej.2012.19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. et al. Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci Rep 6, 19324, 10.1038/srep19324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K. M. et al. Natural volcanic CO2 seeps reveal future trajectories for host–microbial associations in corals and sponges. The ISME journal 9, 894–908, 10.1038/ismej.2014.188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann G. E. et al. High-frequency dynamics of ocean pH: a multi-ecosystem comparison. PLoS One 6, e28983, 10.1371/journal.pone.0028983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius K. E. et al. Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat Clim Change 1, 165–169, 10.1038/nclimate1122 (2011). [DOI] [Google Scholar]
- Sunday J. M. et al. Evolution in an acidifying ocean. Trends Ecol Evol 29, 117–125, 10.1016/j.tree.2013.11.001 (2014). [DOI] [PubMed] [Google Scholar]
- Sanford E. & Kelly M. W. Local adaptation in marine invertebrates. Ann Rev Mar Sci 3, 509–535, 10.1146/annurev-marine-120709-142756 (2011). [DOI] [PubMed] [Google Scholar]
- Kelly M. W., Padilla-Gamino J. L. & Hofmann G. E. Natural variation and the capacity to adapt to ocean acidification in the keystone sea urchin Strongylocentrotus purpuratus. Glob Chang Biol 19, 2536–2546, 10.1111/gcb.12251 (2013). [DOI] [PubMed] [Google Scholar]
- Hughes T. P., Huang H. & Young M. A. The wicked problem of China’s disappearing coral reefs. Conserv Biol 27, 261–269, 10.1111/j.1523-1739.2012.01957.x (2013). [DOI] [PubMed] [Google Scholar]
- Zhang C. L. et al. Diurnal and seasonal variations of carbonate system parameters on Luhuitou fringing reef, Sanya Bay, Hainan Island, South China Sea. Deep-Sea Res Pt II 96, 65–74, 10.1016/j.dsr2.2013.02.013 (2013). [DOI] [Google Scholar]
- Chen X. F. et al. Biological controls on diurnal variations in seawater trace element concentrations and carbonate chemistry on a coral reef. Mar Chem 176, 1–8, 10.1016/j.marchem.2015.06.030 (2015). [DOI] [Google Scholar]
- Yan H. et al. Seasonal variations of seawater pCO2 and sea‐air CO2 fluxes in a fringing coral reef, northern South China Sea. J Geophys Res: Oceans 121, 998–1008, 10.1002/2015JC011484 (2016). [DOI] [Google Scholar]
- Lynch M. D. & Neufeld J. D. Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13, 217–229, 10.1038/nrmicro3400 (2015). [DOI] [PubMed] [Google Scholar]
- Tout J. et al. Variability in microbial community composition and function between different niches within a coral reef. Microb Ecol 67, 540–552, 10.1007/s00248-013-0362-5 (2014). [DOI] [PubMed] [Google Scholar]
- Blackall L. L., Wilson B. & van Oppen M. J. Coral-the world’s most diverse symbiotic ecosystem. Mol Ecol 24, 5330–5347, 10.1111/mec.13400 (2015). [DOI] [PubMed] [Google Scholar]
- Haas A. F. et al. Global microbialization of coral reefs. Nat Microbiol 1, 16042, 10.1038/nmicrobiol.2016.42 (2016). [DOI] [PubMed] [Google Scholar]
- Tytlyanov E. A., Titlyanova T. V., Huang H. & Li X. Seasonal changes in benthic algal communities of the upper subtidal zone in Sanya Bay (Hainan Island, China). J Mar Biol Assoc UK 94, 51–64, 10.1017/S0025315413001112 (2014). [DOI] [Google Scholar]
- Ainsworth T. D. et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9, 2261–2274, 10.1038/ismej.2015.39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T. et al. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol 79, 4759–4762, 10.1128/AEM.00695-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L., Edwards R., Rodriguez-Brito B., Liu H. & Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9, 2707–2719, 10.1111/j.1462-2920.2007.01383.x (2007). [DOI] [PubMed] [Google Scholar]
- Krause E. et al. Small changes in pH have direct effects on marine bacterial community composition: a microcosm approach. PLoS One 7, e47035, 10.1371/journal.pone.0047035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt V., Wild C., Anthony K. R., Diaz-Pulido G. & Uthicke S. Effects of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ Microbiol 13, 2976–2989, 10.1111/j.1462-2920.2011.02571.x (2011). [DOI] [PubMed] [Google Scholar]
- Webster N. S., Uthicke S., Botte E. S., Flores F. & Negri A. P. Ocean acidification reduces induction of coral settlement by crustose coralline algae. Glob Chang Biol 19, 303–315, 10.1111/gcb.12008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya Y. et al. Coral bleaching: the winners and the losers. Ecol Lett 4, 122–131, 10.1046/j.1461-0248.2001.00203.x (2001). [DOI] [Google Scholar]
- Hughes T. P. et al. Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933, 10.1126/science.1085046 (2003). [DOI] [PubMed] [Google Scholar]
- Howells E. J. et al. Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat Clim Change 2, 116–120, 10.1038/nclimate1330 (2012). [DOI] [Google Scholar]
- Kelly L. W. et al. Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc Natl Acad Sci USA 111, 10227–10232, 10.1073/pnas.1403319111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker K. J. et al. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob Chang Biol 19, 1884–1896, 10.1111/gcb.12179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnaughey T. 13C and 18O isotopic disequilibrium in biological carbonates: I. Patterns. Geochim Cosmochim Ac 53, 151–162, 10.1016/0016-7037(89)90282-2 (1989). [DOI] [Google Scholar]
- Schoepf V. et al. Kinetic and metabolic isotope effects in coral skeletal carbon isotopes: A re-evaluation using experimental coral bleaching as a case study. Geochim Cosmochim Ac 146, 164–178, 10.1016/j.gca.2014.09.033 (2014). [DOI] [Google Scholar]
- Krief S. et al. Physiological and isotopic responses of scleractinian corals to ocean acidification. Geochim Cosmochim Ac 74, 4988–5001, 10.1016/j.gca.2010.05.023 (2010). [DOI] [Google Scholar]
- Carilli J. E., Charles C. D., Garren M., McField M. & Norris R. D. Baseline shifts in coral skeletal oxygen isotopic composition: a signature of symbiont shuffling? Coral Reefs 32, 559–571, 10.1007/s00338-012-1004-y (2013). [DOI] [Google Scholar]
- Brading P. et al. Differential effects of ocean acidification on growth and photosynthesis among phylotypes of Symbiodinium (Dinophyceae). Limnol Oceanogr 56, 927–938, 10.4319/lo.2011.56.3.0927 (2011). [DOI] [Google Scholar]
- Pierrot D., Lewis E. & Wallace D. MS excel program developed for CO2 system calculations. Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, U.S. Department of Energy, Oak Ridge, Tennessee, (2006).
- Cai L., Ye L., Tong A. H. Y., Lok S. & Zhang T. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PLoS ONE 8, 10.1371/journal.pone.0053649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. J., Kobert K., Flouri T. & Stamatakis A. PEAR: a fast and accurate Illumina Paired-End read mergeR. Bioinformatics 30, 614–620, 10.1093/bioinformatics/btt593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, 10.1038/nmeth.f.303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461, 10.1093/bioinformatics/btq461 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267, 10.1128/AEM.00062-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35, 7188–7196, 10.1093/nar/gkm864 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.