Abstract
Background
Joubert syndrome (JS) is a recessive neurodevelopmental disorder characterized by hypotonia, ataxia, cognitive impairment, abnormal eye movements, respiratory control disturbances, and a distinctive mid-hindbrain malformation. JS demonstrates substantial phenotypic variability and genetic heterogeneity. This study provides a comprehensive view of the current genetic basis, phenotypic range and gene-phenotype associations in JS.
Methods
We sequenced 27 JS-associated genes in 440 affected individuals (375 families) from a cohort of 532 individuals (440 families) with JS, using molecular inversion probe-based targeted capture and next generation sequencing. Variant pathogenicity was defined using the Combined Annotation Dependent Depletion (CADD) algorithm with an optimized score cut-off.
Results
We identified presumed causal variants in 62% of pedigrees, including the first B9D2 mutations associated with JS. 253 different mutations in 23 genes highlight the extreme genetic heterogeneity of JS. Phenotypic analysis revealed that only 34% of individuals have a “pure JS” phenotype. Retinal disease is present in 30% of individuals, renal disease in 25%, coloboma in 17%, polydactyly in 15%, liver fibrosis in 14% and encephalocele in 8%. Loss of CEP290 function is associated with retinal dystrophy, while loss of TMEM67 function is associated with liver fibrosis and coloboma, but we observe no clear-cut distinction between JS-subtypes.
Conclusion
This work illustrates how combining advanced sequencing techniques with phenotypic data addresses extreme genetic heterogeneity to provide diagnostic and carrier testing, guide medical monitoring for progressive complications, facilitate interpretation of genome-wide sequencing results in individuals with a variety of phenotypes, and enable gene-specific treatments in the future.
Keywords: Joubert syndrome, ciliopathy, genotype-phenotype, targeted sequencing, genetic heterogeneity, next generation sequencing
INTRODUCTION
Joubert syndrome (JS, OMIM 213300) is a recessive neurodevelopmental disorder characterized by abnormal eye movements, respiratory control disturbances, cognitive impairment, hypotonia and ataxia.[1–4] Diagnosis of JS relies on a pathognomonic combination of imaging findings on axial MRI: cerebellar vermis hypoplasia, thickened and horizontally oriented superior cerebellar peduncles and a deep interpeduncular fossa (the ”Molar Tooth Sign” - MTS).[5] In addition to these core central nervous system (CNS) features, subsets of individuals with JS have ocular (chorioretinal coloboma and progressive retinal dystrophy), kidney (nephronophthisis), liver (spectrum of ductal plate malformation and fibrosis), and/or skeletal (dystrophy and polydactyly) involvement. JS overlaps genetically and phenotypically with the more severe Meckel syndrome, often defined by co-occurrence of occipital encephalocele, cystic-dysplastic kidney disease, liver fibrosis, and perinatal lethality.[6] Care of individuals with JS is complex, requiring surveillance for progressive complications and input from multiple medical subspecialists.
JS can be caused by recessive mutations in more than 27 genes, all of which encode proteins localizing to the primary cilium or basal body.[3, 7] Primary cilia are microtubule-based organelles projecting from the surface of most differentiated cells where they serve as environmental sensors, transducing sensory, chemical or mechanical input, as well as signaling pathways (such as hedgehog) during development and homeostasis.[8] Given the key role of this organelle in such a wide variety of processes, it is not surprising that its dysfunction leads to a number of human diseases collectively named “ciliopathies”.[9] These disorders are unified not only by the underlying pathophysiology and shared genetic causes, but also by a wide array of overlapping phenotypes including cognitive dysfunction, central nervous system malformations, fibrocystic kidney disease, retinal degeneration, skeletal and craniofacial abnormalities, polydactyly and defects in left-right asymmetry.[10]
Ciliopathies in general, and JS in particular, display prominent genetic heterogeneity, i.e. biallelic mutations in many different genes cause the same disorder, albeit with variable severity. Clinically, identifying the genetic causes and understanding gene-phenotype correlations are essential for providing diagnostic testing, prognostic information and treatment recommendations; however, until recently, it has not been possible to identify the genetic cause in the majority of affected individuals. The advent of next-generation sequencing has revolutionized the study of Mendelian disorders by accelerating novel gene discovery.[11] Using JS as a paradigm, we highlight how next generation sequencing combined with extensive phenotypic data can inform prognosis leading to improved medical monitoring in rare disorders, generate insights into the differential tolerance of genes to mutation, and aid in interpreting genome-wide sequencing results in individuals with diverse phenotypes. Understanding the genetic architecture of Mendelian disorders is also leading to gene-specific treatments and improved patient care.
METHODS
Subject ascertainment and phenotypic data
Participants were referred to the University of Washington (UW) Joubert Syndrome Research Program by the Joubert Syndrome and Related Disorders Foundation and clinical collaborators internationally (see Acknowledgements). All participants have clinical findings of JS (intellectual impairment, hypotonia, ataxia and/or oculo-motor apraxia) and diagnostic or supportive brain imaging findings (MTS or cerebellar vermis hypoplasia), or they have a sibling with JS. Clinical data were obtained by direct examination of participants, review of medical records, and structured questionnaires. All participants or their legal representatives provided written informed consent for the study which was approved by the Institutional Review Boards at the UW and Seattle Children’s Hospital. Neurologically Normal Caucasian Control Panels (Coriell panels NDPT020 and NDPT090 - http://ccr.coriell.org) were sequenced as controls.
Mutation identification
Using Molecular Inversion Probes (MIPs),[12] all exons in genes associated with JS or the allelic disorder Meckel syndrome (AHI1, ARL13B, B9D1, B9D2, C2CD3, C5ORF42, CC2D2A, CEP290, CEP41, CSPP1, IFT172, INPP5E, KIF7, MKS1, NPHP1, OFD1, RPGRIP1L, TCTN1, TCTN2, TCTN3, TMEM138, TMEM216, TMEM231, TMEM237, TMEM67, TTC12B and ZNF423;[13–36] details in Supplementary Table S1) were captured using 100 ng of genomic DNA isolated from blood or saliva. Captured DNA was PCR amplified and sequenced on either the Illumina HiSeq or MiSeq platform. Sequence reads were mapped using the Burrows-Wheeler Aligner (BWA v.0.5.9). Variants were called using the Genome Analysis Tookit (GATK v2.5-2) and annotated with SeattleSeq (http://snp.gs.washington.edu/SeattleSeqAnnotation138/). We also included data previously generated by Sanger sequencing of individual genes in subsets of samples. We used the Combined Annotation Dependent Depletion (CADD) algorithm to estimate the deleteriousness of variants (version 1.1),[37] and considered all nonsense, frameshift and canonical splice-site mutations to be deleterious, regardless of CADD score. We defined a cause as the presence of ≥2 rare. deleterious variants (RDVs) or a homozygous RDV in one gene in an affected individual. RDVs that were of high quality (Depth ≥25, Quality by Depth >5, and Heterozygous Allele Balance <0.8) were not confirmed by Sanger sequencing based on the previously demonstrated high sensitivity and specificity of the MIPs method for well-covered variants [12]; however, in affected individuals with one high quality RDV, we did perform Sanger sequencing to confirm second RDVs that did not meet the above-mentioned quality criteria.
Statistical analysis
We tested the significance of associations between clinical features, as well as between features and genetic causes, using the Chi-square or Fisher’s exact tests (SAS, version 9.4; SAS Institute, Inc., Cary, NC). We present odds ratios and 95% confidence intervals as measures of these correlations. The Bonferroni method was used to correct for multiple hypothesis testing.
RESULTS
UW Joubert syndrome cohort
The study cohort comprised 532 affected participants from 440 families, 79 families having >1 affected individual. Participants were recruited from 29 countries, the majority (59%) residing in North America. 19% of the families reported consanguinity. The mean age of the affected participants at the time of the analysis was 13.1 years (S.D. 9.1), with 34% of individuals <10 years of age and 30% 10-20 years of age. 56 % percent were male (Table 1). The large size of the cohort and world-wide ascertainment based on brain imaging and neurologic findings provide a relatively unbiased spectrum of the disorder.
Table 1.
Demographic characteristics of the UW JS cohort
| Characteristic | N | %° |
|---|---|---|
| Current Age (years) | ||
| 0-9 | 178 | 33.5 |
| 10-19 | 157 | 29.5 |
| 20-29 | 65 | 12.2 |
| 30-39 | 24 | 4.5 |
| ≥ 40 | 6 | 1.1 |
| Unknown age | 42 | 7.9 |
| Deceased | ||
| Terminations of pregnancy | 11 | 2.1 |
| Other deaths* | 49 | 9.2 |
| Total | 532 | 100 |
| Continent of residence | ||
| North America | 316 | 59.4 |
| Europe | 51 | 9.6 |
| Australia | 23 | 4.3 |
| South America | 17 | 3.2 |
| Asia (Middle East=88) | 125 | 23.5 |
| Families with known consanguinity° | 84 | 19.1 |
| Male | 295 | 55.5 |
| Families with ≥ 1 affected child° | 79 | 17.9 |
Includes one in utero demise.
Percentages are calculated by individual for all variables except consanguinity and > 1 affected child
Multi-organ involvement is common and the “pure JS” phenotype occurs in a minority of individuals
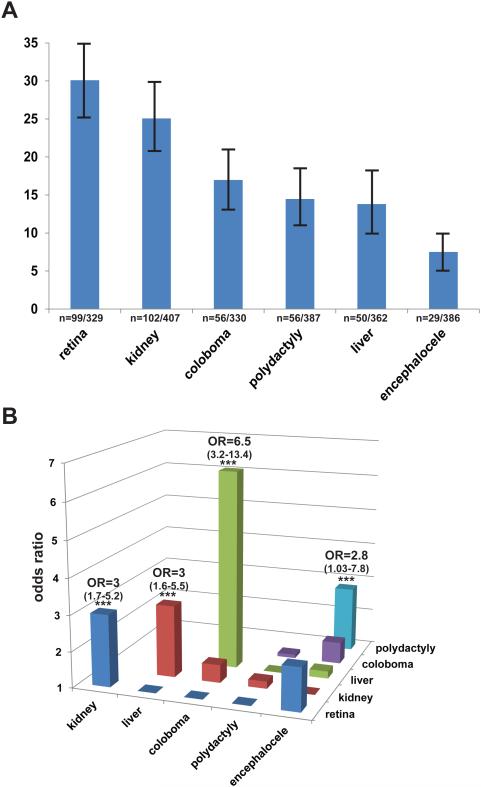
In addition to the core diagnostic features for JS (MTS, hypotonia, ataxia, cognitive dysfunction, abnormal breathing pattern, and oculo-motor apraxia) which were part of the inclusion criteria, several extra-CNS features are commonly described in JS. Based on the presence of these features, various subtypes of JS have been proposed: [2] “pure” JS (core diagnostic features only), JS plus retinal dystrophy, JS plus cystic kidney disease, JS plus retinal-renal involvement, JS plus liver fibrosis and JS plus oral-facial-digital features. Therefore, we systematically assessed the relevant features (Supplementary Table S2) in the cohort. As a consequence of the world-wide recruitment required to collect a large cohort for a rare disorder, the ascertainment of clinical features was variable. To be conservative in calculating the prevalence of each feature, we restricted our analysis to individuals for whom definite positive or negative information was available for a given feature; consequently, the denominator for calculating the frequency of individual features varies accordingly. Retinal dystrophy (n=99/329, 30%) and renal disease (n=102/407, 25%) were the most common associated features, followed by coloboma (n=56/330, 17%), polydactyly (n=56/387, 15%), liver fibrosis (n=50/362, 14%) and encephalocele (n=29/386, 8%) (Figure 1A). When considering only the individuals for whom definite information was available for all six associated features (n=201), only 68 (33.8%) had the “pure JS” phenotype (Supplementary Table S3).
Figure 1. Phenotypic analysis of a large JS cohort.
(A) Bar graph indicating the prevalence of major associated features. Absolute numbers are indicated below each bar and 95% confidence intervals are presented. Information about each feature was not available in every subject, so the denominators are different for each variable. (B) Odds ratios for the association between pairs of features. Hepatic disease and coloboma are highly associated with each other while encephalocele and polydactyly, retinal and renal disease, and hepatic and renal disease are less strongly associated with each other. Precise odds ratios with 95% confidence intervals are indicated for the four statistically significant (***) associations. Detailed odds ratios and confidence intervals for all pairwise possible associations are presented in Supplementary Table S4.
We next evaluated whether any of the major features were associated with each other. Liver fibrosis and coloboma were strongly associated (Odds Ratio (O.R.) 6.5; 95% Confidence Interval (C.I.) 3.2-13.4), i.e. the likelihood of having liver fibrosis in individuals with coloboma was 6.5 times the likelihood of having liver fibrosis in individuals without coloboma. Retinal dystrophy and kidney disease (O.R. 3.0; 95% C.I. 1.7-5.2), liver fibrosis and kidney disease (O.R. 3.0; 95% C.I. 1.6-5.5) and polydactyly and encephalocele (O.R. 2.8; 95% C.I. 1.03-7.8) were more weakly associated with each other (Figure 1B and Supplementary Table S4). In addition, we observed multiple combinations of features in subsets of individuals, often precluding categorization into one of the proposed sub-types (Supplementary Table S3). For example, individuals presenting with the combination of liver fibrosis and kidney disease could be categorized as either “JS plus kidney disease” or “JS plus liver disease”. While the most frequent associations of features are consistent with the proposed JS subtypes, the broad range of additional combinations observed indicates that no clear-cut distinction exists between subtypes.
Multiple additional clinical features
A variety of other clinically important features were documented in medical records and by families but were not systematically queried across the entire cohort (Table 2). Additional brain abnormalities were identified in 91 individuals, most commonly ventriculomegaly, and more rarely heterotopia, agenesis of the corpus callosum and polymicrogyria. This is likely an under-ascertainment compared to prior studies,[38] since detailed review of the brain imaging studies was not part of this study. Additional eye findings were also commonly reported in our cohort, including strabismus and ptosis in 167 and 104 individuals, respectively. Seizures were described in 55 individuals. Other, less common, features included scoliosis (n=28), cleft palate (n=20), hearing loss (n=16), tongue tumors (n=17), oral frenulae (n=9), heart defects (n=7), and a variety of mental health problems such as anxiety, aggression, depression and autism (total n=47). Since these features were not systematically assessed across the cohort, only minimum prevalence estimates can be calculated.
Table 2.
Additional features observed in individuals with JS
| Characteristic | N | Minimum prevalence (%)* |
|---|---|---|
| Nervous system | ||
| Agenesis of the corpus callosum | 16a | 3.0 |
| Heterotopia | 15 | 2.8 |
| Polymicrogyria | 7 | 1.3 |
| Ventriculomegaly | 53 | 10.0 |
| Seizures | 55 | 10.3 |
| Mouth | ||
| Cleft Palate | ||
| Hard Palate | 13 | 2.4 |
| Soft Palate | 7 | 1.3 |
| Tongue Tumors | 17 | 3.2 |
| Oral Frenulae | 9 | 1.7 |
| Eye | ||
| Strabismus | 167 | 31.4 |
| Ptosis | 104 | 19.5 |
| Other | ||
| Hearing Loss | 16 | 3.0 |
| G-Tube | 43 | 8.1 |
| Scoliosis | 28 | 5.3 |
| Heart | 7b | 1.3 |
| Endocrine | ||
| Panhypopituitarism | 5 | 0.9 |
| Hypothyroidism | 4 | 0.8 |
| Micropenis | 10 | 1.9 |
| Other | 11c | 2.1 |
| Laterality defects | 3d | 0.8 |
| Mental Health Issues | 47e | 8.8 |
assumes that the feature is absent when the feature is not documented to be present. Denominator=532 individuals.
includes complete (13) and partial (3) agenesis of the corpus callosum
includes atrial septal defect (3), coarctation of aorta (2), bicuspid aortic valve and aortic stenosis (1), narrowing of aortic arch (1)
includes Hashimoto’s disease (1), type I diabetes mellitus (2), unknown type diabetes (1), ovarian failure (1), polycystic ovarian syndrome (1), growth hormone deficiency (3), elevated parathyroid hormone (1), and absence of pituitary bright spot, premature puberty, and borderline diabetes (1)
includes dextrocardia (1), situs inversus (2)
includes anxiety (6), ADHD/ADD (8), autism spectrum disorder (16), depression/bipolar disorder (5), aggression (2), obsessive compulsive disorder (2), borderline personality disorder (1), Anorexia Nervosa (1), non-specified behavioral problems (6)
Comprehensive sequencing identifies the presumed genetic cause in 62% of JS families
We sequenced 27 JS-associated genes in 428 affected individuals from 363 families for whom DNA was available using Molecular Inversion Probe (MIP) targeted capture followed by next generation sequencing. We previously demonstrated, using a subset of this cohort, that this method has 99.5% sensitivity and 98% positive predictive value for variant detection at covered basepairs compared to Sanger sequencing.[12] The MIP target included all coding positions and neighboring intronic basepairs (Supplementary Table 1), and >89% of basepairs were adequately covered (≥8X) for all genes except INPP5E (75% covered) (Supplementary Figure S1). We also included previous Sanger sequencing data, as well as sequencing data from clinical testing when available (n=12), bringing the total number of affected individuals with sequencing data to 440 from 375 families. Based on the estimated prevalence of JS (~1/80,000 Northern Europeans,[3]) and the genetic heterogeneity of the disease, we excluded variants with a MAF >0.2% in the Exome Variant Server (http://evs.gs.washington.edu/EVS/). We considered all nonsense, frameshift and canonical splice site mutations to be deleterious. We assessed the predicted deleteriousness of missense, synonymous and intronic variants using the CADD score algorithm,[37] which considers multiple available prediction techniques including conservation across species and protein function, and has the advantage of providing a score for all possible variants on a single scale. We selected the CADD score cutoff (11) for defining rare deleterious variants (RDVs) by maximizing the number of affected individuals with genes harboring two rare variants (or a homozygous rare variant), while minimizing the number of controls with genes harboring similar variants, an approach akin to generating a Receiver Operating Characteristic curve (Supplementary Figure S2). For missense variants, using the CADD score identified more presumed causes in the JS cohort compared to Polyphen2 without increasing the false-positive rate in controls (data not shown).
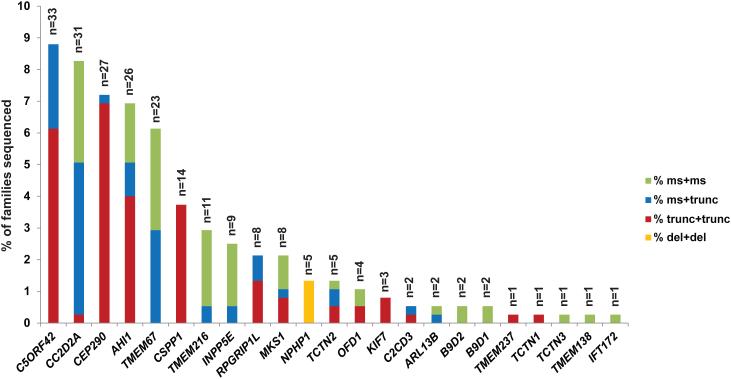
We defined a cause as the presence of ≥2 RDVs (or a homozygous RDV) in one gene in an affected individual. Using this definition and all available sequencing data, we identified the presumed genetic cause in 279 individuals from 232/375 families (62%) overall (Figure 2), 77% in consanguineous families, and 76% in families with >1 affected individual. The higher rate in the consanguineous families is likely due to the higher probability of calling a single homozygous variant, compared to the probability of calling two different heterozygous variants in the non-consanguineous families. Similarly, in 9% of families for whom we were able to sequence >1 affected individual, we identified two high quality RDVs in only one of the affected individuals. This likely accounts for the higher solve rate in multiplex families compared to families with only one affected child. In contrast to the results in affected individuals, 5/182 unrelated control individuals carried ≥2 RDVs in one of the known genes (Supplementary Table S5). In 68/70 (97%) families for which parental DNA was available, we confirmed that the identified compound heterozygous RDVs are in trans, excluding 2 samples from further analysis. We did not sequence parents of children with homozygous or hemizygous RDVs (90 families).. Parental samples were not available for controls.
Figure 2. Genetic causes in a large JS cohort.
Bar graph indicating the proportion of individuals with JS carrying two rare deleterious variants (RDVs) in each gene. Each bar is broken down to illustrate the relative frequency of the observed mutations in each gene: red indicates two truncating mutations (including nonsense, frameshift and canonical splice-site mutations), blue indicates one truncating and one missense mutation (including small in-frame indels), green indicates 2 missense mutations or small in-frame indels, and orange represents larger deletions.
Despite satisfying our criteria (MAF<0.2%, CADD>11), the variants in 12 families did not meet ACMG variant interpretation categories 1, 2, or 3.[39] In 8 of these12 families, one of the RDVs is a splice variant beyond +/-2 base pairs from the intron-exon junction, for which the functional effect on splicing has not been assessed. In 4/12 families, one RDV is a synonymous variant whose functional effect has not been evaluated. Therefore, we list these families separately in Supplemental Table S5 and excluded them from gene-phenotype analyses.
In addition, we identified 5 families with pairs of RDVs in each of two genes (Supplementary Table S6). In 3/5, the variants in one gene appeared much more likely to be causal than the variants in the second gene (e.g. a homozygous frameshift mutation in C5ORF42 versus two missense variants in CSPP1, which harbors exclusively truncating mutations in our cohort). In these three families, the more likely cause was retained for the subsequent analyses. In the other two families, we could not determine the cause and excluded them from the subsequent genetic analyses. Of note, based on the clinical information available, the phenotypic severity in these individuals was not substantially different from the rest of the cohort.
Including only the families with conservatively called genetic causes, five genes (C5ORF42, CC2D2A, AHI1, CEP290, TMEM67) each account for JS in ~6-9% of JS, three genes (CSPP1, TMEM216 and INPP5E) for ~3% each, and six genes for ~1-2%, while the remaining nine genes each account for JS in only 1-2 families. We also identified B9D2 mutations as the genetic cause in two families, further extending the known genetic overlap between JS and the more severe Meckel syndrome. The detailed phenotypic description of the two individuals with B9D2 mutations is presented in Supplementary Table S7. CEP41, TMEM138, TMEM231, and ZNF423 do not harbor ≥2 or homozygous RDVs in any affected individuals. A single affected individual carries one synonymous and one missense variant in TTC21B; however, this individual also carries a homozygous nonsense variant in C2CD3 that is predicted to truncate the protein near the N-terminus (Supplementary Table S6).
Further examination of the sequence data revealed variation in the types of mutations across the different genes. Considering all nonsense, frameshift and canonical splice-site mutations as truncating, we observed that CEP290, CSPP1 and C5ORF42 mostly harbor a combination of two truncating mutations, CC2D2A and TMEM67 tend to have ≥1 missense mutation, and TMEM216 and INPP5E have mainly two missense mutations. All individuals with JS caused by mutations in NPHP1 (n=5) harbor the previously described deletion [24] in a homozygous state, and no causal point mutations were identified in this gene. The differences in mutation types across the genes were statistically significant (Supplementary Figure S3).
While the majority of RDVs were unique, we identified a subset of RDVs present in ≥3 families not known to be related (Supplementary Table S8). TMEM216 R73L is common in families of Ashkenazi Jewish descent,[34] and accounts for most of the families with TMEM216 mutations. Two C5ORF42 RDVs (p.Gly2663Alafs*40 and W2593*) were found homozygous in six families of Saudi Arabian descent. The p.Gly2663Alafs*40 variant has been previously associated with both JS and Meckel syndrome in Saudi Arabian families.[40, 41] One CC2D2A RDV (P1122S) was found homozygous in three families of Saudi Arabian descent. In three unrelated Brazilian families, the same combination of two CSPP1 RDVs was identified, suggesting that they might in fact be related.[20] None of the other recurring RDVs appeared to be associated with specific ethnic groups, so they may represent mutation hotspots (such as CEP290 G1890* identified in 10 unrelated families from 3 continents).
Gene-phenotype correlations
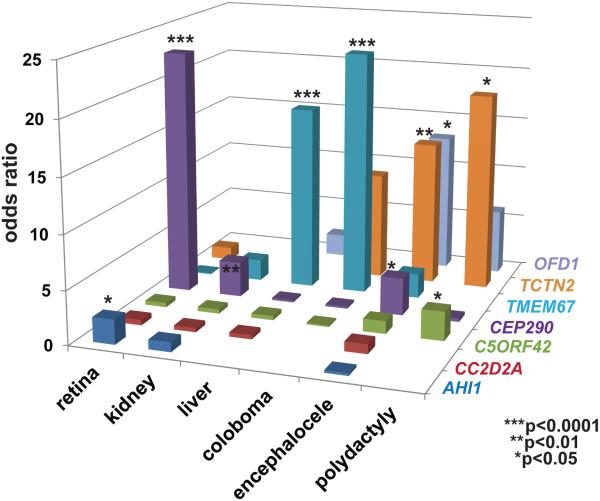
We next examined associations between the non-CNS features of JS and each genetic cause (Figure 3, Supplementary Table S9), and observed several significant gene-phenotype correlations: CEP290 mutations with retinal dystrophy (O.R. 22.9, C.I.6.7-78.4; p<0.0001) and cystic kidney disease (O.R. 3.3, C.I. 1.6-7.1; p=0.001); TMEM67 with liver fibrosis (O.R. 17.3, C.I. 7.2-42.0; p<0.0001) and coloboma (O.R. 22.9, C.I. 8.6-61.1; p<0.0001); C5ORF42 with polydactyly (O.R. 2.7, C.I. 1.2-5.9; p=0.01); OFD1 with encephalocele (O.R. 13.1, C.I: 1.8-97.0; p=0.03); TCTN2 with encephalocele (O.R. 13.6, C.I. 2.6-70.8; p=0.007) and polydactyly (O.R. 18.7, C.I. 1.9-182.9 p=0.01). Even after Bonferroni correction for multiple hypothesis testing, the associations between TMEM67 and liver disease and coloboma, and those between CEP290 and retinal dystrophy remained statistically significant (p<0.0001). In addition, a negative correlation was observed between TMEM67 mutations and retinal disease (O.R 0.1, C.I. 0.01-0.8; p=0.006), indicating that individuals with TMEM67 mutations are less likely to be diagnosed with retinal disease than those without mutations in this gene. When counseling families, the absolute prevalence of clinical features may be more useful than odds ratios, so this information is provided in Supplementary Figure S4.
Figure 3. Gene-Phenotype correlation in JS.
Bar graph indicating for each of the more frequently involved genes, and for two genes with significant phenotypic associations, the odds ratio for each of the six commonly associated features: retinal disease, renal disease, hepatic disease, coloboma, polydactyly and encephalocele. Statistically significant odds ratios (Fisher’s exact test or Chi-square test) are marked with an asterisk (***). Confidence intervals are omitted for clarity but are listed in Supplementary Table S9.
Although we cannot test the statistical significance of genetic associations with non-systematically assessed clinical features, several possible associations are notable. Both individuals with C2CD3 mutations had oral features including oral frenulae and/or cleft palate, suggesting C2CD3 mutations may lead to an OFD-like phenotype.[2] However, among the individuals with oral features (n=46), the majority did not have mutations in C2CD3 (or OFD1). Likewise, two of three individuals with KIF7 mutations had agenesis of the corpus callosum (while the status of the corpus callosum in the third individual was unknown), consistent with a KIF7-related “acro-callosal” subtype of JS. Again however, the majority of individuals with agenesis of the corpus callosum (n=14) had mutations in other genes without a clear predominance of one genetic cause. None of the 55 individuals with seizures had causal CEP290 mutations, despite CEP290 loss of function being the third most common cause of JS, suggesting a negative association.
DISCUSSION
Presumed genetic cause of JS identified in 62% of families
Just over ten years ago, the first genetic causes of JS were identified.[13, 24] Now, we can determine the presumed genetic cause in 62% of individuals with JS, using the highly efficient MIP capture technique, next generation sequencing, and an optimized CADD score cutoff to identify causal variants in 27 JS/Meckel genes. Five genes (C5ORF42, CC2D2A, CEP290, AHI1 and TMEM67) account for the majority of affected individuals, while nine genes are mutated in <15 families, and nine more genes are mutated in only 1-2 families. In two families with JS, we identified causal mutations in the Meckel-associated gene B9D2, further expanding the allelism between JS and Meckel syndrome. Not surprisingly, B9D2 is part of a transition zone sub-complex (with MKS1 and B9D1) that regulates protein trafficking in and out of the cilium.[42]
These findings illustrate the extreme genetic heterogeneity of JS. Therefore, given that no single gene predominates as a cause for JS, the most efficient method for clinical diagnostic testing is next-generation sequencing of all known JS genes through targeted gene panels or whole exome sequencing. The advantage of the MIP capture technique lies in its low cost and flexibility, allowing easy addition of newly identified JS genes to the target. For laboratories without a specific interest in JS, whole exome sequencing might be more practical, since it does not require any specialized set-up.
The genetic cause remains unidentified in 38% of families in our cohort. This may be due to mutations in genes not yet associated with JS, or variants in the known genes that were missed by our current techniques, either because they are inadequately covered in our data, located in non-coding regions, not called using our analysis pipeline, or not recognized as deleterious. Given the high coverage obtained for all but one gene (INPP5E) and the efficiency of MIP capture for identifying variants in the target regions,[12] it is likely that a sizeable fraction of the missed variants lie in non-coding regions that affect gene expression level, splicing or translation. Identifying these variants and understanding their significance will require integrating data from variant rating algorithms like CADD, global assessments of chromatin structure and regulatory elements from projects such as ENCODE,[43] and targeted functional assays in affected cell lines, animal models, or in vitro systems.
Clinical utility of gene-phenotype correlations and phenotypic associations
Gene-phenotype correlations in well-characterized, comprehensively sequenced cohorts translate directly into improved prognostic information and medical management for individuals with JS. For instance, results from this study indicate that individuals with JS harboring causal mutations in TMEM67 have a higher risk of developing liver fibrosis, necessitating closer monitoring to allow early diagnosis and treatment of portal hypertension. Likewise, individuals with causal mutations in CEP290 require closer surveillance for retinal dystrophy. Our findings validate prior results from smaller cohorts focused on single genes,[44,45,46] and also identify additional positive and negative correlations. For example, individuals with causal mutations in TMEM67 appear less likely to develop retinal disease and may require less frequent monitoring for this complication. Even when the genetic cause is unknown, phenotypic associations can also guide management and surveillance; for example, individuals with JS and retinal dystrophy should be monitored more closely for renal dysfunction, and those with coloboma should be monitored more closely for liver fibrosis.
While the strongest phenotypic associations observed in this cohort are consistent with previously described JS-subtypes such as COACH syndrome,[45,46] and the retinal-renal form of JS,[44] we did not observe clear-cut distinctions between phenotypic subgroups corresponding to specific genetic causes. The molar tooth sign provides a unifying feature for all affected individuals in our cohort, but the distribution of associated phenotypes highlights the phenotypic variability and overlap with other ciliopathies. This is particularly well illustrated by the individuals with mutations in the OFD-associated genes C2CD3 or OFD1 who have oral features, consistent with an OFD-like JS subtype; however, most individuals with oral features in our cohort harbor mutations in other genes. Therefore, phenotypic sub-typing is of limited clinical value for guiding molecular genetic testing. Fortuitously, next-generation sequencing panels now preclude the need for prioritizing single gene tests. Nonetheless, grouping individuals by genetic cause or clinical phenotype retains value for determining their risk of developing progressive features and guiding clinical management as described above.
Gene-specific mutation patterns provide insights into gene function
The observed gene-phenotype correlations, along with the gene-specific mutation distributions provide information about the function of the different genes. Genes associated preferentially with particular phenotypes suggest a specific or more important role for these genes in the affected organ systems. For instance, the association of CEP290 mutations with retinal dystrophy in JS and Leber congenital amaurosis,[47] confirms the importance of CEP290 function in the human retina, as seen in animal models.
The distribution of mutation types harbored by each gene also reveals information about gene function. For instance, the near-absence of biallelic truncating mutations in some genes suggests that full loss of function for these genes is poorly tolerated in humans, leading to more severe phenotypes, such as Meckel syndrome or early fetal lethality. In support of this hypothesis, fetuses with Meckel syndrome tend to carry two truncating mutations in CC2D2A and TMEM67 compared to individuals with JS who usually carry at least one missense mutation as previously described.[48-50] Likewise, biallelic truncating mutations in TMEM216 and INPP5E have not been previously identified in individuals with JS and are not found in our cohort.[22, 33, 34, 51] In contrast, virtually all individuals with JS due to mutations in CSPP1 or CEP290 harbor two truncating variants in these genes, indicating that severe loss-of-function is required to cause JS. This type of gene-specific information should be considered when interpreting the significance of newly identified sequence variants, in combination with allele frequency in controls, deleteriousness prediction algorithms, and the phenotype of the affected individual. For example, missense mutations in CEP290 or CSPP1 detected by targeted or genome-wide clinical sequencing are less likely to be clinically significant than missense mutations in TMEM216 or INPP5E. A further consequence of the gene-specific distribution of mutation types lies in the development of potential specific therapies: genes harboring a majority of nonsense mutations such as CEP290 may be amenable to read-through therapies,[52] while this therapeutic direction would be less valuable for genes harboring mainly missense mutations.
Limitations
While larger than previously published studies, our analysis is still limited by the small number of individuals with two RDVs in several genes associated with JS, precluding statistically significant gene-phenotype correlations for these genetic causes. This is an inherent limitation to the study of rare disorders with prominent genetic heterogeneity. Similarly, the relative rarity of JS necessitates the world-wide enrollment of study participants; consequently, phenotypic assessment is inhomogeneous and some features, especially neurodevelopmental outcome, are difficult to assess at a distance. This is currently a universal problem in the field of rare disorder genetics, where, for the first time, genetic data is more easily available than phenotypic data. In this study, we made every effort to use conservative assumptions for tests of statistical significance; however, until validated by other studies, these results should be translated into clinical practice with caution.
Impact of next-generation sequencing on diagnosis and treatment of Mendelian disorders
In summary, this work illustrates how applying advanced DNA sequencing technologies and improved functional prediction algorithms to large, well-characterized cohorts, is enhancing our understanding of the genetic architecture and gene-phenotype correlations in rare Mendelian disorders. Identifying the genetic cause empowers individuals with JS and their families to make family planning decisions, and gene-phenotype correlations provide more reliable prognostic information leading to individually-tailored, organ-specific surveillance, thereby improving the health and longevity of affected individuals while conserving healthcare costs. In parallel, identifying the genetic causes of Mendelian disorders is required for developing and applying gene-specific treatments. Similar to recent breakthroughs in cancer treatment based on genomic information (reviewed in Sameek et al,2014),[53] understanding the genetic causes of Mendelian disorders will inform future gene-specific treatments and is a major step toward personalized medicine for affected individuals.
Supplementary Material
Acknowledgements:
We greatly appreciate the participation of all of the individuals with JS and their families. We also thank the Joubert Syndrome and Related Disorders Foundation and the innumerable clinicians who have referred participants to us over many years, including Jumana Al-Aama, Beth Allen, Ala’a Arafat, Mutluay Arslan, Louis Bartoshesky, Erawati Bawle, Anastashia Boli, Carsten Bönnemann, Sarah Bowdin, Steven Braddock, John Carey, Umran Cetincelik, Alicia Chan, David Chitayat, Juan Chemke, Krystyna Chrzanowska, Brian Chung, Robin Clark, Vida Culic, Duygu Cura, Cynthia Curry, Sumita Danda, Stephen Deputy, Charu Deshpande, William Dobyns, Emily Doherty, Lisa Ewans, Michael Gabbett, Özlem Giray, Himanshu Goel, Elaine Goh, Crystal Grimsley, Sachin Gupta, Jonathan Holt, Niels Illum, Olafur Indridason, A. Micheil Innes, Louise Izatt, Sujatha Jagadeesh, Usha Kini, Jarkko Kirjavainen, Edwin Kirk, Antonie Kline, Kristleifur Kristjannson, Mark Labinksy, Yves Lacassie, Amparo Lopez Lafuente, James Lemons, Richard Leventer, Jan Liebeit, Shawn Lipinski, Sally Lynch, Alan Ma, Richard Macias, Sahar Mansour, Bernard Maria, Isabelle Maystadt, Carole McKeown, Scott McLean, Nancy Mendelsohn, Markella Mikkelsen, Vinod Misra, Sheela Nampoothiri, Vinodh Narayanan, David Neubauer, Ann Olney, Wendy Osterling, Alex Paciorkowski, Shashidhar Pai, Ivan Pavkovic, Joan Pellegrino, Ruthann Pfau, Joseph Pinter, Kate Pope, Gerald Raymond, Miriam Regev, James Reggin, Janet Rennie, Anne Ronan, Alan Shanske, Lisa Sharf, Lori Skallerud, Diana Smith, Rhonda Spiro, Zornitza Stark, Lois Starr, Helen Stewart, Anne Summers, Krzysztof Szczaluba, David Tilstra, John Tolmie, Priya Verghese, Alain Verloes, Julie Vogt, Richard Webster, Kara Weisiger, Mark Wells, Kevin White, Sue White, Dorota Wicher, Robert Wildin, Denise Williams, Meredith Wilson, Barry Wolf, Lisa Worgan, Grace Yoon, Takehito Yokoi, Marc Yudkoff, Elaine Zackai. We thank Karen Barnett for help establishing the cohort, Elizabeth Blue for comments on the manuscript, and Stephan Neuhauss and Anita Rauch for their support for R.B.-G.
Funding: R.B.-G. was supported by a Swiss NSF grant Ambizione-SCORE PZ00P3_142404/1. The following authors received support from the National Institutes of Health: M.A.P. K23NS45832, I.A.G. K24HD046712, D.D R01NS064077. D.D. also received funding from a March of Dimes Basil O’Connor Starter Scholar Research Award, The Arc of Washington Trust Fund, and private donations from families of children with Joubert syndrome. The work was also supported by the University of Washington Intellectual and Developmental Disabilities Research Center Genetics Core (National Institutes of Health U54HD083091). Sequencing was provided by the University of Washington Center for Mendelian Genomics (UW CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant 1U54HG006493 to Drs. Debbie Nickerson, Jay Shendure and Michael Bamshad.
Footnotes
Supplemental Data: This section contains 9 tables and 4 figures.
Contributors RBG, JCD, IGP, PC, MAP, IG, JS, and DD participated in the design of the study. JCD, IGP, BJO, DMK, GEI, CRI, NG, JA, EAB, DO, AA, ARD, LL, CL, LM, AGC, HO, GH, BT, MT, MAP, UWCMG and DD collected and/or generated the data. RBG, JCD, IGP, BJO, TCR, EAB, NdL, UWCMG, and DD analyzed and interpreted the data. RBG, JCD, IGP, and DD drafted the manuscript. All co-authors read and approved the final manuscript.
REFERENCES
- 1.Joubert M. Familial agenesis of the cerebellar vermis. A syndrome of episodic hyperpnea, abnormal eye movements, ataxia, and retardation. Neurology. 1969;19(9):813–825. doi: 10.1212/wnl.19.9.813. [DOI] [PubMed] [Google Scholar]
- 2.Brancati F, Dallapiccola B, Valente E. Joubert Syndrome and related disorders. Orphanet J Rare Dis. 2010;5(1):20. doi: 10.1186/1750-1172-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parisi MA, Glass I. Joubert syndrome and related disorders 2003. doi: 10.1038/sj.ejhg.5201648. Available at: http://www.ncbi.nlm.nih.gov/books/NBK1325/ [DOI] [PubMed]
- 4.Boltshauser E. Joubert syndrome: episodic hyperpnea, abnormal eye movements, retardation and ataxia, associated with dysplasia of the cerebellar vermis. Neuropadiatrie. 1977;8(1):57–66. doi: 10.1055/s-0028-1091505. IW. [DOI] [PubMed] [Google Scholar]
- 5.Poretti A, Boltshauser E, Valente EM. The Molar Tooth Sign is Pathognomonic for Joubert Syndrome! Pediatr Neurol. 2014;50(6):e15. doi: 10.1016/j.pediatrneurol.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Barker AR, Thomas R, Dawe HR. Meckel-Gruber syndrome and the role of primary cilia in kidney, skeleton, and central nervous system development. Organogenesis. 2014;10(1):96–107. doi: 10.4161/org.27375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romani M, Micalizzi A, Valente EM. Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol. 2013;12(9):894–905. doi: 10.1016/S1474-4422(13)70136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364(16):1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badano JL, Mitsuma N, Beales PL, Katsanis N. The Ciliopathies: An Emerging Class of Human Genetic Disorders. Annu Rev Genomics Hum Genet. 2006;7(1):125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 11.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 12.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O’Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J. Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders. Science. 2012;338(6114):1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-Nouri D, Al-Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart Y. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36(9):1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 14.Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attié-Bitach T, Holden KR, Dobyns WB, Traver D, Al-Gazali L, Ali BR, Lindner TH, Caspary T, Otto EA, Hildebrandt F, Glass IA, Logan CV, Johnson CA, Bennett C, Brancati F, Valente EM, Woods CG, Gleeson JG. Mutations in the Cilia Gene ARL13B Lead to the Classical Form of Joubert Syndrome. Am J Hum Genet. 2008;83(2):170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romani M, Micalizzi A, Kraoua I, Dotti M, Cavallin M, Sztriha L, Ruta R, Mancini F, Mazza T, Castellana S, Hanene B, Carluccio M, Darra F, Mate A, Zimmermann A, Gouider-Khouja N, Valente EM. Mutations in B9D1 and MKS1 cause mild Joubert syndrome: expanding the genetic overlap with the lethal ciliopathy Meckel syndrome. Orphanet J Rare Dis. 2014;9(1):72. doi: 10.1186/1750-1172-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srour M, Schwartzentruber J, Hamdan FF, Ospina LH, Patry L, Labuda D, Massicotte C, Dobrzeniecka S, Capo-Chichi J-M, Papillon-Cavanagh S, Samuels ME, Boycott KM, Shevell MI, Laframboise R, Désilets V, Maranda B, Rouleau GA, Majewski J, Michaud JL. Mutations in C5ORF42 Cause Joubert Syndrome in the French Canadian Population. Am J Hum Genet. 2012;90(4):693–700. doi: 10.1016/j.ajhg.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorden NT, Arts HH, Parisi MA, Coene KL, Letteboer SJF, van Beersum SEC, Mans DA, Hikida A, Eckert M, Knutzen D, Alswaid AF, Özyurek H, Dibooglu S, Otto EA, Liu Y, Davis EE, Hutter CM, Bammler TK, Farin FM, Dorschner M, Topçu M, Zackai EH, Rosenthal P, Owens KN, Katsanis N, Vincent JB, Hildebrandt F, Rubel EW, Raible DW, Knoers NVAM, Chance PF, Roepman R, Moens CB, Glass IA, Doherty D. CC2D2A Is Mutated in Joubert Syndrome and Interacts with the Ciliopathy-Associated Basal Body Protein CEP290. Am J Hum Genet. 2008;83(5):559–571. doi: 10.1016/j.ajhg.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38(6):674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 19.Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, Ikegami K, Schlossman AM, Merriman B, Attie-Bitach T, Logan CV, Glass IA, Cluckey A, Louie CM, Lee JH, Raynes HR, Rapin I, Castroviejo IP, Setou M, Barbot C, Boltshauser E, Nelson SF, Hildebrandt F, Johnson CA, Doherty D, Valente EM, Gleeson JG. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet. 2012;44(2):193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuz K, Bachmann-Gagescu R, O’Day DR, Hua K, Isabella CR, Phelps IG, Stolarski AE, O’Roak BJ, Dempsey JC, Lourenco C, Alswaid A, Bönnemann CG, Medne L, Nampoothiri S, Stark Z, Leventer RJ, Topçu M, Cansu A, Jagadeesh S, Done S, Ishak GE, Glass IA, Shendure J, Neuhauss SCF, Haldeman-Englert CR, Doherty D, Ferland RJ. Mutations in CSPP1 Cause Primary Cilia Abnormalities and Joubert Syndrome with or without Jeune Asphyxiating Thoracic Dystrophy. Am J Hum Genet. 2014;94(1):62–72. doi: 10.1016/j.ajhg.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halbritter J, Bizet AA, Schmidts M, Porath JD, Braun DA, Gee HY, McInerney-Leo AM, Krug P, Filhol E, Davis EE, Airik R, Czarnecki PG, Lehman AM, Trnka P, Nitschké P, Bole-Feysot C, Schueler M, Knebelmann B, Burtey S, Szabó AJ, Tory K, Leo PJ, Gardiner B, McKenzie FA, Zankl A, Brown MA, Hartley JL, Maher ER, Li C, Leroux MR, Scambler PJ, Zhan SH, Jones SJ, Kayserili H, Tuysuz B, Moorani KN, Constantinescu A, Krantz ID, Kaplan BS, Shah JV, UK10K Consortium. Hurd TW, Doherty D, Katsanis N, Duncan EL, Otto EA, Beales PL, Mitchison HM, Saunier S, Hildebrandt F. Defects in the IFT-B Component IFT172 Cause Jeune and Mainzer-Saldino Syndromes in Humans. Am J Hum Genet. 2013;93(5):915–925. doi: 10.1016/j.ajhg.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, Kayserili H, Swistun D, Scott LC, Bertini E, Boltshauser E, Fazzi E, Travaglini L, Field SJ, Gayral S, Jacoby M, Schurmans S, Dallapiccola B, Majerus PW, Valente EM, Gleeson JG. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41(9):1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dafinger C, Liebau MC, Elsayed SM, Hellenbroich Y, Boltshauser E, Korenke GC, Fabretti F, Janecke AR, Ebermann I, Nürnberg G, Nürnberg P, Zentgraf H, Koerber F, Addicks K, Elsobky E, Benzing T, Schermer B, Bolz HJ. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121(7):2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DWW, McDonald R, Eddy A, Chance PF, Glass IA. The NPHP1 Gene Deletion Associated with Juvenile Nephronophthisis Is Present in a Subset of Individuals with Joubert Syndrome. Am J Hum Genet. 2004;75(1):82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coene KLM, Roepman R, Doherty D, Afroze B, Kroes HY, Letteboer SJF, Ngu LH, Budny B, van Wijk E, Gorden NT, Azhimi M, Thauvin-Robinet C, Veltman JA, Boink M, Kleefstra T, Cremers FPM, van Bokhoven H, de Brouwer APM. OFD1 Is Mutated in X-Linked Joubert Syndrome and Interacts with LCA5-Encoded Lebercilin. Am J Hum Genet. 2009;85(4):465–481. doi: 10.1016/j.ajhg.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arts HH, Doherty D, van Beersum SEC, Parisi MA, Letteboer SJF, Gorden NT, Peters TA, Marker T, Voesenek K, Kartono A, Ozyurek H, Farin FM, Kroes HY, Wolfrum U, Brunner HG, Cremers FPM, Glass IA, Knoers NVAM, Roepman R. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39(7):882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, Garcia-Verdugo JM, Katsanis N, Hildebrandt F, Reiter JF. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43(8):776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas S, Legendre M, Saunier S, Bessières B, Alby C, Bonnière M, Toutain A, Loeuillet L, Szymanska K, Jossic F, Gaillard D, Yacoubi MT, Mougou-Zerelli S, David A, Barthez M, Ville Y, Bole-Feysot C, Nitschke P, Lyonnet S, Munnich A, Johnson CA, Encha-Razavi F, Cormier-Daire V, Thauvin-Robinet C, Vekemans M, Attié-Bitach T. TCTN3 Mutations Cause Mohr-Majewski Syndrome. Am J Hum Genet. 2012;91(2):372–378. doi: 10.1016/j.ajhg.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, Huntzicker EG, Sfakianos MK, Sandoval W, Bazan JF, Kulkarni P, Garcia-Gonzalo FR, Seol AD, O'Toole JF, Held S, Reutter HM, Lane WS, Rafiq MA, Noor A, Ansar M, Devi ARR, Sheffield VC, Slusarski DC, Vincent JB, Doherty DA, Hildebrandt F, Reiter JF, Jackson PK. Mapping the NPHP-JBTS-MKS Protein Network Reveals Ciliopathy Disease Genes and Pathways. Cell. 2011;145(4):513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srour M, Hamdan FF, Schwartzentruber JA, Patry L, Ospina LH, Shevell MI, Désilets V, Dobrzeniecka S, Mathonnet G, Lemyre E, Massicotte C, Labuda D, Amrom D, Andermann E, Sébire G, Maranda B, Consortium FC, Rouleau GA, Majewski J, Michaud JL. Mutations in TMEM231 cause Joubert syndrome in French Canadians. J Med Genet. 2012;49(10):636–641. doi: 10.1136/jmedgenet-2012-101132. [DOI] [PubMed] [Google Scholar]
- 31.Huang L, Szymanska K, Jensen VL, Janecke AR, Innes AM, Davis EE, Frosk P, Li C, Willer JR, Chodirker BN, Greenberg CR, McLeod DR, Bernier FP, Chudley AE, Müller T, Shboul M, Logan CV, Loucks CM, Beaulieu CL, Bowie RV, Bell SM, Adkins J, Zuniga FI, Ross KD, Wang J, Ban MR, Becker C, Nürnberg P, Douglas S, Craft CM, Akimenko M, Hegele RA, Ober C, Utermann G, Bolz HJ, Bulman DE, Katsanis N, Blacque OE, Doherty D, Parboosingh JS, Leroux MR, Johnson CA, Boycott KM. TMEM237 Is Mutated in Individuals with a Joubert Syndrome Related Disorder and Expands the Role of the TMEM Family at the Ciliary Transition Zone. Am J Hum Genet. 2011;89(6):713–730. doi: 10.1016/j.ajhg.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, Silhavy JL, Lee JE, Al-Gazali L, Thomas S, Davis EE, Bielas SL, Hill KJ, Iannicelli M, Brancati F, Gabriel SB, Russ C, Logan CV, Sharif SM, Bennett CP, Abe M, Hildebrandt F, Diplas BH, Attié-Bitach T, Katsanis N, Rajab A, Koul R, Sztriha L, Waters ER, Ferro-Novick S, Woods CG, Johnson CA, Valente EM, Zaki MS, Gleeson JG. Evolutionarily Assembled cis-Regulatory Module at a Human Ciliopathy Locus. Science. 2012;335(6071):966–969. doi: 10.1126/science.1213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valente EM, Logan CV, Mougou-Zerelli S, Lee JH, Silhavy JL, Brancati F, Iannicelli M, Travaglini L, Romani S, Illi B, Adams M, Szymanska K, Mazzotta A, Lee JE, Tolentino JC, Swistun D, Salpietro CD, Fede C, Gabriel S, Russ C, Cibulskis K, Sougnez C, Hildebrandt F, Otto EA, Held S, Diplas BH, Davis EE, Mikula M, Strom CM, Ben-Zeev B, Lev D, Sagie TL, Michelson M, Yaron Y, Krause A, Boltshauser E, Elkhartoufi N, Roume J, Shalev S, Munnich A, Saunier S, Inglehearn C, Saad A, Alkindy A, Thomas S, Vekemans M, Dallapiccola B, Katsanis N, Johnson CA, Attie-Bitach T, Gleeson JG. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat Genet. 2010;42(7):619–625. doi: 10.1038/ng.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edvardson S, Shaag A, Zenvirt S, Erlich Y, Hannon GJ, Shanske AL, Gomori JM, Ekstein J, Elpeleg O. Joubert Syndrome 2 (JBTS2) in Ashkenazi Jews Is Associated with a TMEM216 Mutation. Am J Hum Genet. 2009;86(1):93–97. doi: 10.1016/j.ajhg.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baala L, Romano S, Khaddour R, Saunier S, Smith UM, Audollent S, Ozilou C, Faivre L, Laurent N, Foliguet B, Munnich A, Lyonnet S, Salomon R, Encha-Razavi F, Gubler M, Boddaert N, de Lonlay P, Johnson CA, Vekemans M, Antignac C, Attié-Bitach T. The Meckel-Gruber Syndrome Gene, MKS3, Is Mutated in Joubert Syndrome. Am J Hum Genet. 2007;80(1):186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, Muzny DM, Young AC, Wheeler DA, Cruz P, Morgan M, Lewis LR, Cherukuri P, Maskeri B, Hansen NF, Mullikin JC, Blakesley RW, Bouffard GG, NISC Comparative Sequencing Program. Gyapay G, Reiger S, Tönshoff B, Kern I, Soliman NA, Neuhaus TJ, Swoboda KJ, Kayserili H, Gallagher TE, Lewis RA, Bergmann C, Otto EA, Saunier S, Scambler PJ, Beales PL, Gleeson JG, Maher ER, Attié-Bitach T, Dollfus H, Johnson CA, Green ED, Gibbs RA, Hildebrandt F, Pierce EA, Katsanis N. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43(3):189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poretti A, Huisman TAGM, Scheer I, Boltshauser E. Joubert Syndrome and Related Disorders: Spectrum of Neuroimaging Findings in 75 Patients. Am J Neuroradiol. 2011;32(8):1459–1463. doi: 10.3174/ajnr.A2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 40.Shaheen R, Faqeih E, Alshammari MJ, Swaid A, Al-Gazali L, Mardawi E, Ansari S, Sogaty S, Seidahmed MZ, AlMotairi MI, Farra C, Kurdi W, Al-Rasheed S, Alkuraya FS. Genomic analysis of Meckel-Gruber syndrome in Arabs reveals marked genetic heterogeneity and novel candidate genes. Eur J Hum Genet. 2013;21(7):762–768. doi: 10.1038/ejhg.2012.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alazami AM, Alshammari MJ, Salih MA, Alzahrani F, Hijazi H, Seidahmed MZ, Abu Safieh L, Aldosary M, Khan AO, Alkuraya FS. Molecular characterization of Joubert syndrome in Saudi Arabia. Hum Mutat. 2012;33(10):1423–1428. doi: 10.1002/humu.22134. [DOI] [PubMed] [Google Scholar]
- 42.Dowdle WE, Robinson JF, Kneist A, Sirerol-Piquer MS, Frints SGM, Corbit KC, Zaghloul NA, Zaghloul NA, van Lijnschoten G, Mulders L, Verver DE, Zerres K, Reed RR, Attié-Bitach T, Johnson CA, García-Verdugo JM, Katsanis N, Bergmann C, Reiter JF. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89(1):94–110. doi: 10.1016/j.ajhg.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 44.Brancati F, Barrano G, Silhavy JL, Marsh SE, Travaglini L, Bielas SL, Amorini M, Zablocka D, Kayserili H, Al-Gazali L, Bertini E, Boltshauser E, D’Hooghe M, Fazzi E, Fenerci EY, Hennekam RCM, Kiss A, Lees MM, Marco E, Phadke SR, Rigoli L, Romano S, Salpietro CD, Sherr EH, Signorini S, Stromme P, Stuart B, Sztriha L, Viskochil DH, Yuksel A, Dallapiccola B, The International JSRD Study Group. Valente EM, Gleeson JG. CEP290 Mutations Are Frequently Identified in the Oculo-Renal Form of Joubert Syndrome–Related Disorders. Am J Hum Genet. 2007;81(1):104–113. doi: 10.1086/519026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doherty D, Parisi MA, Finn LS, Gunay-Aygun M, Al-Mateen M, Bates D, Clericuzio C, Demir H, Dorschner M, van Essen AJ, Gahl WA, Gentile M, Gorden NT, Hikida A, Knutzen D, Özyurek H, Phelps I, Rosenthal P, Verloes A, Weigand H, Chance PF, Dobyns WB, Glass IA. Mutations in 3 genes (MKS3, CC2D2A and RPGRIP1L) cause COACH syndrome (Joubert syndrome with congenital hepatic fibrosis) J Med Genet. 2010;47(1):8–21. doi: 10.1136/jmg.2009.067249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brancati F, Iannicelli M, Travaglini L, Mazzotta A, Bertini E, Boltshauser E, D'Arrigo S, Emma F, Fazzi E, Gallizzi R, Gentile M, Loncarevic D, Mejaski-Bosnjak V, Pantaleoni C, Rigoli L, Salpietro CD, Signorini S, Stringini GR, Verloes A, Zabloka D, Dallapiccola B, Gleeson JG, Valente EM. MKS3/TMEM67 mutations are a major cause of COACH Syndrome, a Joubert Syndrome related disorder with liver involvement. Hum Mutat. 2009;30(2):E432. doi: 10.1002/humu.20924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KEJ, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, Hoyng CB, van den Born LI, Rohrschneider K, Cremers FPM. Mutations in the CEP290 (NPHP6) Gene Are a Frequent Cause of Leber Congenital Amaurosis. Am J Hum Genet. 2006;79(3):556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachmann-Gagescu R, Ishak GE, Dempsey JC, Adkins J, O'Day D, Phelps IG, Gunay-Aygun M, Kline AD, Szczaluba K, Martorell L, Alswaid A, Alrasheed S, Pai S, Izatt L, Ronan A, Parisi MA, Mefford H, Glass I, Doherty D. Genotype–phenotype correlation in CC2D2A-related Joubert syndrome reveals an association with ventriculomegaly and seizures. J Med Genet. 2012;49(2):126–137. doi: 10.1136/jmedgenet-2011-100552. [DOI] [PubMed] [Google Scholar]
- 49.Mougou-Zerelli S, Thomas S, Szenker E, Audollent S, Elkhartoufi N, Babarit C, Romano S, Salomon R, Amiel J, Esculpavit C, Gonzales M, Escudier E, Leheup B, Loget P, Odent S, Roume J, Gérard M, Delezoide A, Khung S, Patrier S, Cordier M, Bouvier R, Martinovic J, Gubler M, Boddaert N, Munnich A, Encha-Razavi F, Valente EM, Saad A, Saunier S, Vekemans M, Attié-Bitach T. CC2D2A mutations in Meckel and Joubert syndromes indicate a genotype–phenotype correlation. Hum Mutat. 2009;30(11):1574–1582. doi: 10.1002/humu.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iannicelli M, Brancati F, Mougou-Zerelli S, Mazzotta A, Thomas S, Elkhartoufi N, Travaglini L, Gomes C, Luigi Ardissino G, Bertini E, Boltshauser E, Castorina P, D'Arrigo S, Fischetto R, Leroy B, Loget P, Bonnière M, Starck L, Tantau J, Gentilin B, Majore S, Swistun D, Flori E, Lalatta F, Pantaleoni C, Penzien J, Grammatico P, The International JSRD Study Group. Dallapiccola B, Gleeson JG, Attie-Bitach T, Valente EM. Novel TMEM67 mutations and genotype-phenotype correlates in meckelin-related ciliopathies. Hum Mutat. 2010;31(5):E1319. doi: 10.1002/humu.21239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacoby M, Cox JJ, Gayral S, Hampshire DJ, Ayub M, Blockmans M, Pernot E, Kisseleva MV, Compere P, Schiffmann SN, Gergely F, Riley JH, Perez-Morga D, Woods CG, Schurmans S. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat Genet. 2009;41(9):1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- 52.Schwarz N, Carr A, Lane A, Moeller F, Chen LL, Aguilà M, Nommiste B, Muthiah MN, Kanuga N, Wolfrum U, Nagel-Wolfrum K, da Cruz L, Coffey PJ, Cheetham ME, Hardcastle AJ. Translational read-through of the RP2 Arg120stop mutation in patient iPSC-derived retinal pigment epithelium cells. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sameek R, Chinnaiyan AM. Translating Genomics for Precision Cancer Medicine. Annu Rev Genomics Hum Genet. 2014;15(1):395–415. doi: 10.1146/annurev-genom-090413-025552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.