Summary
Bacteria that inhabit the rhizosphere of agricultural crops can have a beneficial effect on crop growth. One such mechanism is the microbial‐driven solubilization and remineralization of complex forms of phosphorus (P). It is known that bacteria secrete various phosphatases in response to low P conditions. However, our understanding of their global proteomic response to P stress is limited. Here, exoproteomic analysis of Pseudomonas putida BIRD‐1 (BIRD‐1), Pseudomonas fluorescens SBW25 and Pseudomonas stutzeri DSM4166 was performed in unison with whole‐cell proteomic analysis of BIRD‐1 grown under phosphate (Pi) replete and Pi deplete conditions. Comparative exoproteomics revealed marked heterogeneity in the exoproteomes of each Pseudomonas strain in response to Pi depletion. In addition to well‐characterized members of the PHO regulon such as alkaline phosphatases, several proteins, previously not associated with the response to Pi depletion, were also identified. These included putative nucleases, phosphotriesterases, putative phosphonate transporters and outer membrane proteins. Moreover, in BIRD‐1, mutagenesis of the master regulator, phoBR, led us to confirm the addition of several novel PHO‐dependent proteins. Our data expands knowledge of the Pseudomonas PHO regulon, including species that are frequently used as bioinoculants, opening up the potential for more efficient and complete use of soil complexed P.
Introduction
Phosphorus (P) is an essential macroelement for all living biota. In soil, microorganisms and plants compete for P. It is therefore essential that a sufficient amount of P is available for agricultural crops to sustain their yields. Plants acquire P as inorganic orthophosphate (Pi) from the soil solution (Vance et al., 2003; White and Hammond, 2008). The concentration of Pi in the soil solution is controlled by chemical and biological processes which fix and release Pi through complex interactions between the soil, soil microorganisms and plant roots (Richardson et al., 2009; Shen et al., 2011). To overcome these limitations in agricultural systems, inorganic fertilizers, derived from non‐renewable Pi rocks, are supplied to crops and pastures (Vance et al., 2003; López‐Arredondo et al., 2014). Over 85% of mined P is used in food production (Heffer et al., 2006) and consumption of this non‐renewable resource could lead to a peak P scenario (akin to peak oil; Raven, 2008; Cordell et al., 2009). It is, therefore, likely that there will be increasing pressures on Pi fertilizer availability and, consequently, cost in the future. These pressures will be exacerbated by increasing demand on food production systems as the human population increases and by fluctuation in oil prices (Cordell et al., 2009). Inappropriate use of inorganic Pi fertilizers can also perturb the nutrient balance of natural ecosystems and reduce biodiversity (White and Hammond, 2008, 2009). Hence, it is desirable to increase the efficiency by which plants can access the many forms of unavailable P that reside within soil, thus, reducing the requirements for Pi fertilizer application (Richardson et al., 2009; Stutter et al., 2012).
Plant‐growth promoting rhizobacteria (PGPR) are bacteria that can enhance crop yields through a variety of mechanisms, including P mobilization, and their identification has led to the notion that bacteria are an integral part of the plant‐root interface (rhizosphere) (Lugtenberg and Kamilova, 2009; Miller et al., 2010). One advantage of utilizing P‐liberating bacteria in agricultural systems is that they can have synergistic beneficial effects, for example, pathogen suppression (Vassilev et al., 2006). The majority of research into P mobilization by PGPR has focused on the solubilization of inorganic P through acidification of the surrounding soil via the release of organic acids, namely gluconic acid (Rodrı'guez and Fraga, 1999; Miller et al., 2010; Oteino et al., 2015). The genus Pseudomonas represents one soil bacterial group that is frequently associated with plant‐growth promotion, including P solubilization (Miller et al., 2010). Various Pseudomonas strains can also degrade organic P compounds, such as phytate, phosphonates and phosphites (Ternan and Quinn, 1998; White and Metcalf, 2004, 2007). Three Pseudomonas strains, Pseudomonas putida BIRD‐1, Pseudomonas fluorescens SBW25 and Pseudomonas stutzeri DSM4166 (hereafter, BIRD‐1, SBW25 and DSM4166 respectively) are three examples of PGPR (Naseby et al., 2001; Hass and Keel, 2003; Preston, 2004; Yu et al., 2011; Roca et al., 2013). SBW25 inhabits the rhizosphere of Pea plants and is antagonistic towards the pathogen Pythium ultimum (Naseby et al., 2001), whereas DSM4166, an ‘unusual’ nitrogen‐fixing bacterium, was isolated from a cultivar of Sorghum nutans (Yu et al., 2011). BIRD‐1, a P‐solubilising bacterium, has been previously utilized as a bioinoculant, since it can significantly improve the germination rates, growth and yields of various agricultural crops (Roca et al., 2013). BIRD‐1 can remineralize Pi from the plant P‐storage compound phytate, a major source of organic P in some soils (up to 50%) (Stutter et al., 2012). Furthermore, when BIRD‐1 was used as an inoculant, phosphatase activity was greater in the rhizosphere compared with bulk soil (Roca et al., 2013). Although Pseudomonas have been implicated in plant‐growth promotion, partially attributed to their effect on P mobilization, the precise mechanisms behind this process remain largely unknown.
The majority of Bacteria studied can undergo a physiological response to Pi depletion controlled by a two‐component regulatory system (PhoBR) encoded by phoBR. PhoBR regulates a large set of genes (the PHO regulon) in response to low P concentrations (Baek and Lee, 2006; Monds et al., 2006; Su et al., 2007). The majority of studies have focused on specific functions/mechanisms that are regulated by PhoBR, for example, the Pi specific transport (Pst) system, motility and swarming, and expression of alkaline phosphatases (APases), acid phosphatases or secondary metabolites (Rittmann et al., 2005; Monds et al., 2006; Sola‐Landa et al., 2008; Furtwängler et al., 2010; Zavaleta‐Pastor et al., 2010). Fewer studies have experimentally confirmed the global PHO regulon using recently developed ‘omics’ techniques. Microarrays were employed to identify genes regulated at the transcriptional level in Pseudomonas aeruginosa (Bains et al., 2012), E. coli (Baek and Lee, 2006) and Synechococcus sp. WH8102 (Tetu et al., 2009; Ostrowski et al., 2010) while traditional 2D‐gel electrophoresis was performed to make a qualitative assessment of the Bacillus subtilis proteome in response to low Pi (Antelmann et al., 2000). These studies identified a number of genes/proteins involved in Pi scavenging underlining the importance of performing ‘omics’ to study Pi acquisition. In Pseudomonas, it has been shown that phosphate binding proteins (PBPs), APases, and virulence factors are associated with phosphate‐stress and regulated by PhoBR (Monds et al., 2006; Bains et al., 2012; Putker et al., 2013: Santos‐Beneit, 2015).
Exoproteomics captures the extracellular protein fraction that results from active secretion, cell lysis or leakage during cell division (Armengaud et al., 2012; Ebner et al., 2016). Removing the cellular fraction prior to protein extraction can help to identify exoproteins that are involved in the interaction of microorganisms with their environment (Christie‐Oleza et al., 2012, 2015). The majority of exoproteins detected are associated with nutrient acquisition, motility, cell attachment, defence, communication as well as antagonism (Christie‐Oleza et al., 2012). Therefore, exoproteomics is the ideal method of choice to study the bacterial mechanisms for scavenging extracellular P. Due to the complexity and technical challenges associated with metaproteomics (Muth et al., 2015), this study aimed to identify Pi‐responsive proteins that can be used as markers for future studies investigating Pi mobilization within the rhizosphere. Therefore, exoproteomic analyses were performed on three plant‐associated Pseudomonas strains grown under Pi‐deplete conditions (50 μM). We hypothesized that Pseudomonas strains harbour a number of common Pi‐scavenging enzymes that would be expressed during Pi depletion. In reality, clear evidence for intra‐genus‐level heterogeneity in their exoproteomes was observed and a number of novel PHO‐regulon members specifically linked with the PHO regulon in Pseudomonas putida BIRD‐1 were also determined.
Results
Effects of P‐limitation on the growth of Pseudomonas strains
We investigated the effect of Pi stress on three strains, DSM4166, SBW25) and BIRD‐1 by comparing growth under Pi‐replete (1.4 mM) or Pi‐deplete (50 µM) conditions (n = 3). Both the growth rates and growth yields of all three Pseudomonas strains showed a significant decrease (t‐test score, P < 0.01) under Pi‐deplete growth conditions (Fig. 1A–C). As expected, Pi‐deplete cultures of all three strains demonstrated a significant increase in the level of APase activity (Fig. 1D) due to the induction of the PHO regulon (Monds et al., 2006; Putker et al., 2013).
Figure 1.
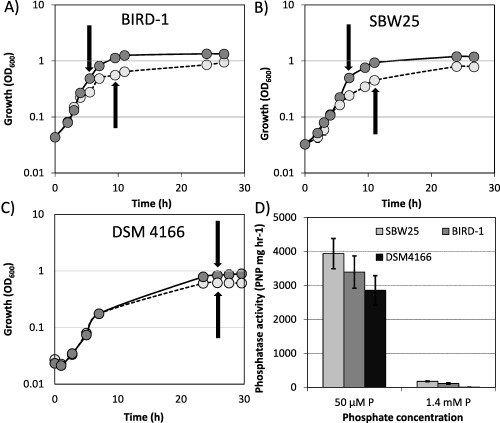
A–C. Growth of the three Pseudomonas strains under either Pi replete (1.4 mM) or Pi deplete (50 μM) growth conditions. Arrows denote sampling points for exoproteomics. D. During the growth experiments, alkaline phosphatase activity was quantified as a proxy for determining the activation of the PHO regulon. The values shown represent a given time point when the maximal alkaline phosphatase activity detected for each strain was obtained. Results presented are the mean of triplicate cultures. Error bars denote standard deviation.
General characteristics of the exoproteomes of the three Pseudomonas species
Based on qualitative 1D SDS‐PAGE analysis, there was visible evidence for heterogeneity in the profiles of Pseudomonas exoproteomes in response to Pi‐depletion (Fig. 2). Samples were subsequently processed for peptide identification using LC‐MS/MS. Proteins were considered present based on a minimum of at least two unique peptides. Exoproteins were identified by the presence of a signal peptide sequence (IMG/JGI). Proteins detected in the exoproteome that did not possess a signal peptide sequence were further analysed using SecretomeP and LipaseP to determine if they are secreted in a non‐classical manner (Christie‐Oleza and Armengaud, 2010; Christie‐Oleza et al., 2015). In Pi‐deplete cultures, ‘predicted’ exoproteins comprised 89.9%, 68.3% or 97.6% of the top‐60 most abundant proteins detected in the exoproteomes of BIRD‐1, SBW25 and DSM4166 respectively (Supporting Information Fig. S1), while the remaining fraction was comprised of cytoplasmic proteins. In BIRD‐1, SBW25 and DSM4166 29, 52 and 54 proteins were significantly enriched (t‐test, P value ≤ 0.05, fold‐change (log2) ≥ 1.5) in response to Pi‐depletion respectively (Supporting Information Tables S1–S3). In all three Pseudomonas exoproteomes a suite of previously characterized PHO‐dependent proteins (Santos‐Beneit, 2015) were enriched in their exoproteomes during growth under Pi depletion. These included the high affinity periplasmic substrate binding protein (SBP) subunits of the high affinity Pi transporter (PstS) and phosphonate transporter (PhnD), APases (PhoX, PhoD), 5'‐nucleotidase (UshA) and glycerolphosphodiesterase (GlpQ) (Figs. 3 and 4 and Supporting Information Fig. S2). However, the genomic content (Table 1) and thus exoproteomic response to Pi depletion varied between the three Pseudomonas strains (Fig. 3), revealing significant inter‐genus level heterogeneity. We should point out though that the exoproteome of DSM4166 was harvested after a longer period (DSM4166, 25 h; BIRD‐1 & SBW25, 7–11 h respectively) of Pi stress (Fig. 1) and as a result a higher percentage of its exoproteome was associated with Pi‐scavenging compared with either BIRD‐1 or SBW25 (Fig. 4).
Figure 2.
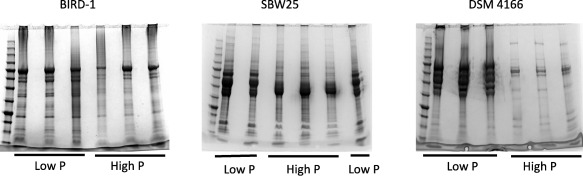
A qualitative assessment, using 1D‐SDS PAGE, of the exoproteomes of all three Pseudomonas strains examined prior to HPLC 2D‐MS/MS. Each gel lane represents 20 ml culture supernatant. For both Pi deplete and Pi replete growth conditions, three biological replicates were performed.
Figure 3.
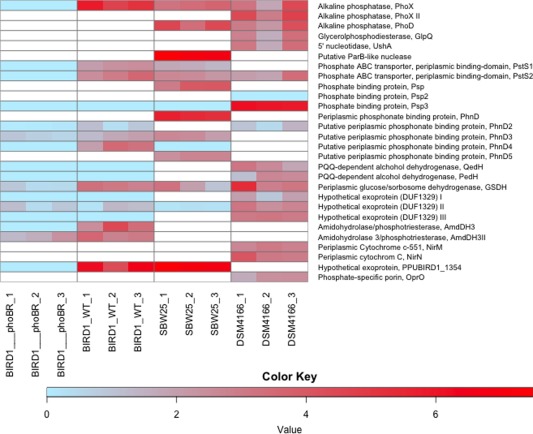
Protein expression analyses in response to Pi depletion of the three Pseudomonas exoproteomes in addition to the exoproteome of the phoBR mutant of P. putida BIRD‐1. White spaces represent the absence of genes encoding the corresponding proteins from their genomes. Each individual biological replicate is displayed. The colour key represents Log2 transformations of protein fold change.
Figure 4.
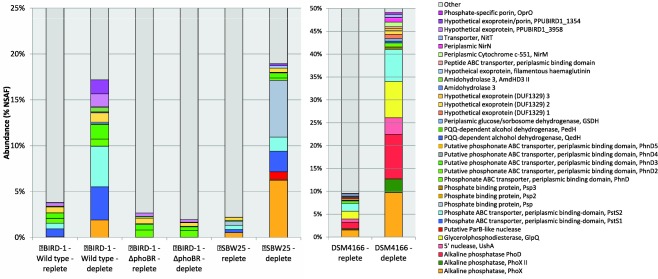
The relative abundance of Pi scavenging proteins detected in the exoproteomes of the three Pseudomonas strains and the phoBR mutant grown in both Pi replete and Pi deplete growth conditions. The normalized spectral abundance factor (NSAF) was calculated using Scaffold 4. Values displayed are the mean of triplicate cultures.
Table 1.
Comparative genomic analysis of selected proteins involved in the recycling of P among Pseudomonas isolates.
| P. aeruginosa PAO1 | P. fluorescens A506 | P. fluorescens F113 | P. fluorescens Pf0‐1 | P. fluorescens SBW25 | P. putida BIRD‐1 | P. putida GB‐1 | P. putida KT2440 | P. putida W619 | P. stutzeri A1501 | P. stutzeri DSM4166 | P. stutzeri DSM10701 | P. syringae CC1557 | P. syringae pv. actinidia ICMP 9617 | P. syringae B728a | P. syringae D C3000 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pi transport | ||||||||||||||||
| PstS1 | ♦ | ♦ | ♦ | ♦ | ♦ | |||||||||||
| PstS2 | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |
| LapA/LapB | ♦ | ♦ | ||||||||||||||
| Psp | ♦ | ♦ | ||||||||||||||
| Psp2 | ♦ | ♦ | ||||||||||||||
| Psp3 | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |
| Psp4 | ||||||||||||||||
| Po scavenging | ||||||||||||||||
| PhnF‐M | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||||||||||
| PhnXW | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||||||||
| PhnWAY | ||||||||||||||||
| PhnCDE | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||||||||||
| PhnC2D2E2 | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||
| PhnD3 | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||||
| PhnD4 | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | |||||||||
| PhnD5 | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||||||||||
| PalA | ||||||||||||||||
| PhoA | ♦ | |||||||||||||||
| PhoX | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |

|
♦ | ♦ | ♦ | ♦ | ♦ |
| PhoD | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |

|
|||||
| GlpQ | ♦ | ♦ | ♦ | ♦ | ||||||||||||
| Phytase | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | |||||||
| UshA |

|

|
♦ | |||||||||||||
| Lipid renovation | ||||||||||||||||
| PlcP | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | |||
| DagK | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | |||
| OlsA | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |
| OlsB | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |
| OlsF | ||||||||||||||||
| SqdBCD | ||||||||||||||||
| Cfa | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||
| BtaAB |
BLASTP was performed using the IMG/JGI database on selected strains whose genome was marked as ‘finished’. Multiple diamonds indicate two or more homologs.
Abbreviations are the same as in Figure 2 as well as: LapA/B, low molecular weight phosphatase; PhoT5, Flavobacterial putative Pi‐binding protein form V; PhnF‐M, C‐P lyase; PhnXW, phosphonatase; PhnWAY, alternative 2‐aminoethylphosphonatase; PhnCDE, phosphonate transporter I, PhnC2D2E2, putative phosphonate transporter II; PalA, phosphonopyruvate hydrolase; PhoA, alkaline phosphatase; PlcP, intracellular phospholipid phosphodiesterase; DagK, diacylglycerol kinase; OlsA, Lyso‐ornithine lipid:acyl‐ACP O‐acyltransferase; OlsB, Ornithine:acyl‐ACP N‐acyltransferase; OlsF, bifunctional ornithine acyltransferase; SqdBCD, sulfolipid biosynthesis; Cfa, cyclo‐propane fatty acid synthase; BtaAB, diacylglycerol trimethylhomoserine biosynthesis.
Pseudomonads show heterogeneity in their resource allocation towards organic P scavenging
Bacteria possess a number of different APases (PhoA, PhoD, PhoX) with different phosphomonoesterase and phosphodiesterase activities (Brickman and Beckwith, 1975; Scott and Wu, 2005). Again, there is genomic and thus exoproteomic variation between the three Pseudomonas strains with respect to the catabolism of organic P (Fig. 3 and Table 1). For example, BIRD‐1 possesses PhoX but lacks PhoD whereas DSM4166 and SBW25 possess both exoenzymes. Furthermore, PhoD was the second most abundant protein in DSM4166 while it was ranked 355th in the exoproteome of SBW25. In addition, DSM4166 also harbours a distinctive PhoX homolog that is duly expressed under Pi depletion (Fig. 4). DSM4166 also expressed homologs of GlpQ (Larson et al., 1983) and UshA (Zalkin and Nygaard, 1996; Rittmann et al., 2005; Pinchuk et al., 2008) while BIRD‐1 and SBW25 do not possess either of these exoenzymes. BIRD‐1 did possess a gene encoding a predicted exoprotein containing the same domains as UshA (Pfam00149 – metallophos; Pfam02872 – 5_nucleotid_C), but did not secrete this protein in response to low Pi.
Pseudomonads harbour a number of phosphate binding proteins (PBPs) that are enriched in their exoproteome in response to Pi‐depletion
In total, the three Pseudomonas strains increased the secretion of four different Pi binding proteins (PBP) containing the Pfam01249 domain, in response to Pi‐depletion (Figs. 3 and 4). All PBPs found in the genomes of the three Pseudomonas strains contain the key residues associated with Pi binding (Supporting Information Fig. S3) (Berna et al., 2008; Liebschner et al., 2009). DSM4166 has a single operon encoding Pst (pstSCAB), whereas BIRD‐1 and SBW25 have two operons encoding two separate Pst systems (Fig. 5A). All PstS homologs were secreted in large quantities during Pi depletion (Fig. 4). The two operons were named pst1 and pst2. PstS1 is closely related to PstS found in E. coli (Wanner, 1990) and Synechocystis sp. PCC6803 (herein, Synechocystis) (Pitt et al., 2010), whereas PstS2 is phylogenetically distinct and closely related to Vibrio PstS (Pratt et al., 2010) (Fig. 5B). Interestingly, pstSCAB1 is not present in all Pseudomonads whereas pstSCAB2 is (Table 1).
Figure 5.
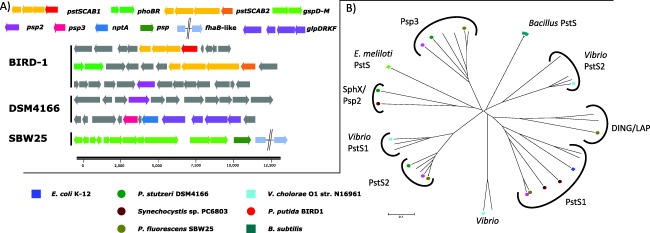
Genomic analyses of the Pi binding proteins found in the three Pseudomonas strains (A) The genetic neighbourhood profiles of the different Pi binding proteins located in the three Pseudomonas strains (B) The diversity of proteins that contain the Pfam12849 domain using a number of genome‐sequenced soil bacteria with the inclusion of characterized Pi binding proteins. Abbreviations; pstSCAB1/2, Pi‐specific ABC transporter; psp, DING‐family Pi binding protein; psp2/3, uncharacterized Pi binding protein; nptA, NA+/Pi co‐transporter; glpD, glycerol 3‐phosphate dehydrogenase; glpR, transcriptional regulator; glycerol kinase; glpK, glycerol uptake facilitator; fhaB‐like, putative filamentous haemagglutinin; phoBR, two component regulator; gsp, type II secretion system.
In SBW25, another PBP (encoded by PFLU2427), hereafter referred to as Psp, was heavily secreted in Pi‐deplete cultures (Fig. 4) alongside a large hypothetical exoprotein (encoded by PFLU2428) that contains a domain related to an adhesion virulence factor (Inatsuka et al., 2005). All Pseudomonas screened encode Psp3. However, only DSM4166 appeared to secrete this exoprotein (PSTAA_2217) in response to Pi stress. Psp3 contains an outer membrane protein A (OmpA) domain implying that this protein may be located in the outer membrane.
Acquisition of phosphonates in phosphate‐depleted Pseudomonas cells
To date, PhnD, which is located in an operon with genes encoding the promiscuous C‐P lyase (PhnF‐M), is the only characterized phosphonate transporter (Baker et al., 1998). A number of putative SBPs responsible for phosphonate transport were identified in the Pseudomonas strains genomes (Fig. 6A) and detected in their exoproteomes, with almost all showing a positive response to Pi‐depletion (Supporting Information Fig. S4). Phylogenetic analysis categorized these homologs into five groups, hereafter referred to as PhnD, PhnD2, PhnD3, PhnD4 and PhnD5. However, none of the three Pseudomonas strains possesses all five homologs in their genomes (Fig. 6B and Table 1). PhnD and PhnD2 both contain the Pfam012974 domain (phosphonate‐binding domain) and these two homologs are mutually exclusive with one another among the genomes of Pseudomonas strains. Only SBW25 possesses and secreted PhnD, a known PHO‐regulon member (Baker et al., 1998), during Pi‐depletion (Fig. 6). BIRD‐1 and DSM4166 both possess PhnD2, which showed a modest increase in abundance in both strains under Pi depletion (Supporting Information Fig. S4). The abundance of PhnD3 also increased in Pi‐deplete SBW25 and BIRD‐1 exoproteomes. PhnD4 is closely related to a SBP (SM_b21540) located upstream of genes (phnWAY) encoding an alternative pathway for the degradation of 2‐AEP in Sinorhizobium meliloti (Fig. 6A) (Borisova et al., 2011) and was only detected in Pi‐deplete BIRD‐1 cultures, albeit at a low abundance.
Figure 6.
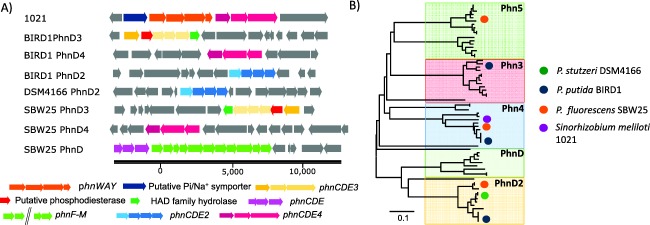
Genomic and proteomic analyses of the phosphonate binding proteins found in the three Pseudomonas strains (A) The genetic neighbourhood profiles of the different phosphonate binding proteins located in the three Pseudomonas strains (BIRD‐1, SBW25, DSM4166) as well as Sinorhizobium meliloti 1021 (1021). (B) Neighbour‐joining phylogenetic analysis of the different phosphonate binding proteins detected in the Pseudomonas strains outlined in Table 1 with the addition of various Burkholderia and Flavobacteria strains. Bootstrap values (500 runs) have been omitted for clarity. IMG accession numbers have been included as a reference. Abbreviations: phnWAY, alternative 2‐aminoethylphosphonate degradation pathway; phnDCE1/2/3/4, phosphonate ABC transporter; phnF‐M, C‐P lyase.
Organic acid production
The Pi solubilization potential of the three Pseudomonads showed marked differences at both the genomic and exoproteomic level. Only DSM4166 induced secretion of two extracellular proteins annotated as pyroloquinoline quinone (PQQ)‐dependent alcohol dehydrogenases (QedH, PSTAA_2299; PedH, PSTAA_2293) in response to Pi stress (Fig. 3). Although BIRD‐1 does possess homologs of both of these proteins neither were detected in its exoproteome. All three strains expressed and secreted an exoprotein under Pi‐depletion that contained the Pfam07995 domain (glucose/sorbosone dehydrogenase) and this enzyme may represent a novel mechanism for extracellular organic acid production.
Identification of novel Pi‐responsive exoproteins in Pseudomonas strains
In response to Pi‐depletion BIRD‐1 and SBW25 expressed a hypothetical outer membrane protein (PPUBIRD1_1354, PFLU4536) containing the Pfam16930 domain which is linked to porin structure (Figs. 3 and 4). In BIRD‐1 this was one of the most abundant proteins in the exoproteome of P‐stressed cells (Fig. 3). P‐stressed SBW25 cells secreted an uncharacterized lipoprotein containing a ParB‐like nuclease domain (Pfam08857), which may have a similar function to UshA (Rittmann et al., 2005) (Fig. 3). BIRD‐1 also secreted a putative extracellular phosphotriesterase/amidohydrolase (AmdDH3 II, PPUBIRD1_5046) that has the potential to cleave the C‐O‐P bond of certain pesticides (Sun et al., 2004). In DSM4166, three hypothetical exoproteins containing the domain of unknown function (DUF) 1329 increased in abundance in response to low Pi. However, these proteins did not increase in abundance in the exoproteomes of either of the other two Pseudomonas isolates. Finally, a number of putative extracellular proteases were enriched in the exoproteome of SBW25 in response to Pi depletion (Supporting Information Table S2).
Analysis of the Pi‐responsive whole‐cell proteome in BIRD‐1
To gain a deeper understanding of the Pi‐responsive proteome, the whole‐cell proteome of BIRD‐1, which included both cytosolic and membrane protein fractions, was analysed. The majority of abundant proteins detected in the proteome of BIRD‐1 were housekeeping and central metabolism proteins (e.g. GyrB, DnaK, GroEL, RpoD, RpoB, FusA, IleS, GuaA, Tuf, SdhA, SdhB, AtpA, AtpB, RpsA, RpsC, SucC, SucA) whose abundance was not significantly affected by differing Pi regime (Supporting Information Table S4). A total of 267 proteins were significantly enriched [t‐test, P value ≤ 0.05, fold‐change (log2) ≥ 1.5] during Pi depletion and there was concordance between the two datasets. These enriched proteins included the transmembrane and ATP‐binding domains of the Pst system (PstC, PstA, PstB), a 2‐aminoethylphosphonate (2‐AEP)–specific phosphonatase (PhnX, PhnW) (Jiang et al., 1995; Baker et al., 1998; White and Metcalf, 2007), proteins involved in lipid remodelling (PlcP, DagK, OlsA, OlsB, Cfa, TauD) (Liu and Hulett, 1998; Antelmann et al., 2000; Zavaleta‐Pastor et al., 2010; Carini et al., 2015; Sebastian et al., 2016), a putative intracellular phosphatase (UxpA) and the twin‐arginine translocation (TAT) pathway (Putker et al., 2013) (Table 2). Proteins for both starch (MalQ, GlgE, GlgX, GlpA and GlpB) and polyhydroxyalkanoic acid (PhaA, PhaG, PhaC) biosynthesis (carbon storage) were also enriched during Pi stress (Supporting Information Table S4). The abundance of a cytoplasmic glucose‐6‐phosphate dehydrogenase (Zwf), as well as two distinct membrane‐bound (PPUBIRD1_4115, PPYBiRD1_2225) glucose dehydrogenases (Gcd, GcdII respectively), all of which are known to play a role in Pi solubilization through gluconic acid production (Miller et al., 2010; Roca et al., 2013), was also greater in Pi‐deplete cells. Finally, another HAD‐family phosphatase (encoded by PPUBIRD1_3492), similar to PhnX was only detected in the proteome of Pi‐deplete cells (Table 2).
Table 2.
P‐responsive proteins in BIRD‐1 that are under the control of the PHO regulator, PhoBR.
| Identified proteins | Accession number | Locus tag | Fold change – WT* | Fold change – mutant* | |
|---|---|---|---|---|---|
| Proteins silenced in the phoBR mutant | |||||
| Hypothetical protein, conserved | ADR57845 | PPUBIRD1_0136 | 4.12 | ND | |
| Taurine dioxygenase | tauD | ADR57960 | PPUBIRD1_0256 | 2.31 | ND |
| Phosphate‐specific methyl‐accepting chemotaxis protein | ctpL | ADR58302 | PPUBIRD1_0612 | 4.95 | ND |
| Dehydratase | ADR58320 | PPUBIRD1_0630 | 2.32 | ND | |
| HlyD family type I secretion membrane fusion protein | ADR58534 | PPUBIRD1_0849 | 3.42 | ND | |
| ABC transporter related protein | ADR58535 | PPUBIRD1_0850 | 4.34 | ND | |
| lyso‐ornithine lipid acyltransferase | olsA | ADR58658 | PPUBIRD1_0974 | 2.18 | ND |
| ornithine‐acyl[acyl carrier protein] N‐acyltransferase | olsB | ADR58659 | PPUBIRD1_0975 | 4.14 | ND |
| L‐serine dehydratase | ADR58719 | PPUBIRD1_1037 | 2.05 | ND | |
| Arginine/ornithine antiporter | arcD | ADR58734 | PPUBIRD1_1052 | 3.04 | ND |
| General secretion pathway protein K | tatA | ADR58773 | PPUBIRD1_1091 | 3.34 | ND |
| Alkaline phosphatase | phoX | ADR58775 | PPUBIRD1_1093 | 5.14 | ND |
| 2',3'‐cyclic‐nucleotide 2'‐phosphodiesterase | uxpA | ADR58776 | PPUBIRD1_1094 | 3.24 | ND |
| General secretion pathway protein G | tatG | ADR58781 | PPUBIRD1_1099 | 4.60 | ND |
| Hypothetical protein, conserved | ADR59032 | PPUBIRD1_1354 | 5.10 | ND | |
| Metallophosphoesterase | plcP | ADR59065 | PPUBIRD1_1390 | 4.59 | ND |
| Gluconate 2‐dehydrogenase acceptor subunit | ADR59804 | PPUBIRD1_2165 | 3.15 | ND | |
| 2Fe‐2S iron‐sulfur cluster binding domain | ADR59806 | PPUBIRD1_2167 | 4.47 | ND | |
| Probable quinate dehydrogenase | gcdII | ADR59861 | PPUBIRD1_2225 | 3.26 | ND |
| UDP‐glucose 4‐epimerase | ADR60229 | PPUBIRD1_2604 | 2.14 | ND | |
| Response regulator | ADR60354 | PPUBIRD1_2733 | 3.33 | ND | |
| UDP‐glucose 6‐dehydrogenase | tuaD | ADR60422 | PPUBIRD1_2809 | 5.63 | ND |
| Putative cyclopropane fatty acid synthase A | cfa2 | ADR60552 | PPUBIRD1_2940 | 2.93 | ND |
| Hypothetical protein, conserved | ADR60627 | PPUBIRD1_3016 | 4.96 | ND | |
| Phosphate ABC transporter, ATP‐binding domain | pstB1 | ADR60628 | PPUBIRD1_3017 | 6.70 | ND |
| Phosphate ABC transporter, transmembrane domain | pstA1 | ADR60629 | PPUBIRD1_3018 | 4.52 | ND |
| Phosphate ABC transporter transmembrane domain | pstC1 | ADR60630 | PPUBIRD1_3019 | 3.91 | ND |
| Phosphate ABC transporter, periplasmic binding domain | pstS1 | ADR60631 | PPUBIRD1_3020 | 5.08 | ND |
| 2‐aminoethylphosphonate–pyruvate transaminase | phnW | ADR61037 | PPUBIRD1_3442 | 3.87 | ND |
| Phosphonoacetaldehyde hydrolase | phnX | ADR61038 | PPUBIRD1_3443 | 4.76 | ND |
| Hypothetical protein, conserved (HAD‐like domain) | ADR61085 | PPUBIRD1_3492 | 3.081 | ND | |
| Cation/acetate symporter actP | actP | ADR61460 | PPUBIRD1_3874 | 2.74 | ND |
| GntR family transcriptional regulator | ADR61476 | PPUBIRD1_3890 | 3.57 | ND | |
| ABC transporter ATP‐binding protein | ADR61481 | PPUBIRD1_3895 | 4.72 | ND | |
| Metallopeptidase, zinc binding protein | ADR61927 | PPUBIRD1_4353 | 2.26 | ND | |
| Fe3+ ABC transporter, periplasmic binding domain | ADR62476 | PPUBIRD1_4925 | 2.04 | ND | |
| Fe3+ ABC transporter, ATP‐binding domain | ADR62478 | PPUBIRD1_4927 | 2.85 | ND | |
| Arylesterase, putative | ADR62594 | PPUBIRD1_5047 | 3.47 | ND | |
| Winged helix family regulator | phoB | ADR62659 | PPUBIRD1_5112 | 4.38 | ND |
| Phosphate regulon sensor protein | phoR | ADR62660 | PPUBIRD1_5113 | 4.64 | ND |
| Phosphate ABC transporter, ATP‐binding domain | pstB2 | ADR62665 | PPUBIRD1_5118 | 5.16 | ND |
| Phosphate ABC transporter, transmembrane domain | pstA2 | ADR62666 | PPUBIRD1_5119 | 4.93 | ND |
| Phosphate ABC transporter, transmembrane domain | pstC2 | ADR62667 | PPUBIRD1_5120 | 6.46 | ND |
| Phosphate ABC transporter, periplasmic binding domain | pstS2 | ADR62668 | PPUBIRD1_5121 | 3.55 | ND |
| Proteins that were down‐regulated compared with the WT | |||||
| Taurine ABC transport, periplasmic binding domain | tauA | ADR57963 | PPUBIRD1_0259 | 3.10 | −1.14 |
| Quinoprotein glucose dehydrogenase A | gcd | ADR61697 | PPUBIRD1_4115 | 2.03 | 0.86 |
| Methyl‐accepting chemotaxis sensory transducer | ADR61928 | PPUBIRD1_4354 | 1.99 | −0.38 | |
| Phosphate transport regulator | phoU | ADR62664 | PPUBIRD1_5117 | 2.07 | −0.32 |
| Proteins whose expression was not affected by PhoBR | |||||
| Cyclopropane‐fatty‐acyl‐phospholipid synthase | cfa1 | ADR57745 | PPUBIRD1_0035 | 2.71 | 4.55 |
| Phosphonate ABC transporter, periplasmic binding domain | phnD2 | ADR58557 | PPUBIRD1_0873 | 0.62 | 1.16 |
| ABC‐type Fe3+ transport system periplasmic binding domain | phnD3 | ADR61477 | PPUBIRD1_3891 | 2.69 | 2.31 |
| Exopolyphosphatase | ppX | ADR62560 | PPUBIRD1_5012 | 0.44 | 0.66 |
| Polyphosphate kinase | ppK | ADR62561 | PPUBIRD1_5013 | 1.34 | 0.89 |
| Amidohydrolase 3 | amdhd3II | ADR62593 | PPUBIRD1_5046 | 4.45 | 3.97 |
| PhoH family protein | phoH | ADR61846 | PPUBIRD1_4270 | 1.38 | 2.78 |
ND, not detected; WT, wild type.
A number of P‐responsive proteins of interest that are not regulated by PhoBR are also listed. The abundance of these proteins within the Proteome of BIRD‐1 is shown in Table S4. The accession number shown refers to the Uniprot database. * represents Log2 transformation of fold change values that are the mean of triplicate cultures. All proteins displayed in the Table with a Log2 value ≥ 1.5, were also statistically significantly enriched under Pi depletion (t‐test, P value ≤ 0.05).
Identification of PHO‐regulated Pi‐responsive proteins in BIRD‐1
To assess how the Pi‐responsive proteome and exoproteome in BIRD‐1 is regulated we disrupted the genes encoding the master regulator of the PHO regulon, phoBR. Compared with the wild type, the phoBR mutant showed a substantial reduction in final growth yield when grown under Pi deplete, but not Pi replete conditions (Supporting Information Fig. S5). As with the wild type strain, we performed exoproteome and whole‐cell proteome analysis for the phoBR strain. In the phoBR mutant, three categories of Pi responsive proteins were determined: (1) proteins that were absent (below detection level) in the mutant and observed in the wild type (2) proteins that no longer increased in abundance under Pi stress and (3) proteins whose abundance in both Pi‐replete and Pi‐deplete growth conditions was similar to the wild type. In the whole‐cell proteome, PstSCAB1&2, PhoX, UxpA, PhoBR, PhoU, TatADG, PhnXW, PlcP, OlsAB, TuaD were all absent during Pi‐deplete growth of the mutant (Table 2). With respect to Pi solubilization, GcdII was also absent, whereas Gcd was not enriched in Pi‐deplete cells, unlike the wild type. However, the abundance of Zwf was unaffected by mutation of phoBR. The putative HAD‐like phosphatase (PPUBIRD‐1_3492), as well as the hypothetical outer membrane protein PPBUBIRD1_1354, were also absent in the mutant, suggesting that these two proteins are novel members of the pho regulon (Table 2). Interestingly, AmdHd3II was still enriched during Pi‐stress suggesting that it is not regulated by phoBR. The abundance of several other proteins was also not affected by mutation of phoBR (Table 2 and Supporting Information Table S5) indicating that a PHO‐independent response to Pi stress occurs in BIRD‐1. For example, all the carbon storage proteins and proteins linked with biofilm formation (encoded by PPUBIRD1_2591‐2608) still showed a PHO‐independent response to Pi stress.
In the exoproteome of the BIRD‐1 phoBR mutant, PstS1, PhoX, the hypothetical exoprotein/porin (PPUBIRD1_1354), PhnD4 were all absent from either growth condition. Meanwhile, the abundance of PstS2 PhnD2, PhnD3, GSDH, AmdHd3, AmdHdII and the hypothetical exoprotein (PPUBIRD1_3958) were all reduced in Pi deplete cultures compared with the wild type, but were still detected (Figs. 3 and 4).
Discussion
Characterizing the exoproteomes, and thus the functional entities associated with environmental interactions (Armengaud et al., 2012), of various Pseudomonas strains allowed us to deepen our understanding of Pi‐regulated protein expression in this genus (Figs. 3 and 4). While, genomic comparisons, based on known proteins in the literature, allowed us to access the heterogeneity in their P‐mobilizing and P‐scavenging ‘potential’, proteomic analyses revealed both differences in their global regulatory networks and also helped to identify novel Pi‐responsive proteins, which may be of further biotechnological interest. For example, a novel Pi‐responsive extracellular nuclease in SBW25 was discovered that was not identified through our comparative genomic analysis. Importantly, the genes (PPUBIRD1_5077, PPUBIRD1_0727, PPUBIRD1_2395, PPUBIRD1_0951, PPUBIRD1_0932) identified in BIRD‐1, based solely on in silico annotation (Roca et al., 2013), were not members of the PHO regulon, highlighting the need for auxiliary studies to confirm genomic annotation. Furthermore, the strong secretion of exoproteins, such as PstS and PhoX, may serve as markers for characterizing complex communities in soil/rhizosphere to enable identification of the key microbial taxa involved in P recycling.
Soil organic P exists in many forms and frequently accounts for 30%–65% of total P in soils (Harrison, 1987) and its mineralization to Pi can have a great impact on total P bioavailability (Turner et al., 2002; Shen et al., 2011). From the genomic comparison of each strain, it appears that DSM4166 has the greatest ability to degrade organic P compounds as it contains two distinct PhoX homologs as well as PhoD, UshA and GlpQ (Larson et al., 1983; Antelmann et al., 2000; Rittmann et al., 2005; Monds et al., 2006; Pinchuk et al., 2008; Putker et al., 2013). We also identified an UshA‐like homolog and a GlpQ‐like homolog (PFLU4789) in the genomes of BIRD‐1 and SBW25, respectively, but in contrast to DSM4166, neither or these homologs were secreted in response to Pi depletion. SBW25 also heavily secreted Psp, a phosphate‐binding protein (Scott and Wu, 2005), which is closely related to the low molecular weight phosphatases, LapA and LapB, in P. aeruginosa (Tan and Worobec, 1993; Ball et al., 2002). Psp may, therefore, be the exoenzyme responsible for the unnaccounted APase activity detected in P. fluorescens Pf0‐1 (Monds et al., 2006). Interestingly, it was SBW25 that elicited the strongest APase activity towards pNPP (phosphomonoesterase activity). To date, little is known about the natural substrate range of these promiscuous enzymes in soil and it is likely that differences occur between the different PhoX homologs. In support of this hypothesis, SBW25 PhoX is phylogenetically distinct (Supporting Information Fig. S6) from either BIRD‐1 or DSM4166 homologs.
Although P solubilization through organic acid production has been well studied in Pseudomonas (Rodrı'guez and Fraga, 1999; Miller et al., 2010) several putatively new P solubilizing proteins likely involved in this process were still identified at the genomic level, e.g. QedH, PedH, and an alternative PHO‐regulated Gcd, GcdII (Fig. 2; Table 2). However, a similar discordance between genomic prediction and proteomic abundance was observed for Pi‐solubilizing enzymes. For example, QedH and PedH were detected in Pi‐depleted DSM4166 cultures, but not in BIRD‐1. GSDH, another previously uncharacterized protein with respect to the PHO regulon, and not identified in our genomic assessment, was secreted in all three strains in response to Pi‐depletion and likely has a role in Pi‐solubilization though organic acid release. The lower abundance of Pi‐solubilizing proteins may have resulted from an absence of glucose in the growth medium the precursor substrate for these Pi‐solubilizing enzymes.
Although the three Pseudomonas strains can grow on phytate as a source of P (Lim et al., 2007; Roca et al., 2013), there was no evidence that either the known phytase of DSM4166 and SBW25, or the predicted phytase, responsible for the growth of BIRD‐1 on this substrate (Roca et al., 2013) are regulated by PhoBR. Considering that phytate is usually ubiquitous in soils (Stutter et al., 2012), Pseudomonads have likely adapted to express their respective phytases solely in response to the presence of this compound and may explain why these Pi‐mobilizing enzymes are unexpectedly not part of the Pseudomonas PHO regulon.
The existence of two distinct Pst systems in both BIRD‐1 and SBW25 is similar to that of Synechocystis, Vibrio cholerae and the Archaeon, Halobacterium salinarium R1 (Furtwängler et al., 2010; Pitt et al., 2010; Mudrak and Tamayo, 2012). As Pst2 is present in all Pseudomonas strains, while Pst1 appears in only a few, we hypotheize that Pst2 is essential for efficient uptake of Pi in Pseudomonas. However, Pst1 must clearly have a role in Pi uptake as we detected PstS1 as well as PstS2 in both BIRD‐1 and SBW25 exoproteomes. Furthermore, although disruption of PstS1 in SBW25 did not affect growth on low Pi in isolation, it did confer a fitness reduction in the presence of the wild type (Zhang et al., 2007). In Synechocystis, Vibrio and H. salinarium, Pst1 and Pst2 either have different kinetic parameters for the uptake of Pi (Furtwängler et al., 2010; Pitt et al., 2010) or are expressed during different growth phases (planktonic v biofilm) (Pratt et al., 2010; Mudrak and Tamayo, 2012). The data presented in this study favours the hypothesis that they have different kinetic parameters as both were expressed in BIRD‐1 and SBW25 during planktonic growth.
Only BIRD‐1 and SBW25 have the genetic potential to catabolize phosphonates (Table 1), and in BIRD‐1, phosphonatase (PhnWX) was PHO‐regulated. Based on our data, we cannot rule out the possibility that DSM4166 can also grow on phosphonates as a source of P for two reasons: (i) Although phosphonate degradation has been well documented in recent years (Jiang et al., 1995; White and Metcalf, 2007; Villarreal‐Chiu et al., 2012; McGrath et al., 2013), bacteria capable of growing on phosphonates that do not possess any of the characterized genes/proteins have been isolated, demonstrating alternative pathways for phosphonate degradation must exist in nature (Fox and Mendz, 2006); (ii) DSM4166 possesses and expressed a number of putative phosphonate transporters (Table 1 and Fig. 6). Characterizing these putative transporters will surely enhance our knowledge regarding phosphonate degradation in Pseudomonas strains and may provide new molecular markers for investigating the in situ cycling of these compounds (Mauchline et al., 2006; Christie‐Oleza and Armengaud, 2010; Lidbury et al., 2014).
Bacteria that can remodel their lipid membranes in order to reduce their ratio of P‐containing:non P‐containing lipids (Zavaleta‐Pastor et al., 2010; Carini et al., 2015; Sebastian et al., 2016) are desirable to use as PGPR as their requirement for P is reduced. We found genes encoding for the key proteins required for lipid remodelling (PlcP, DagK, OlsA and OlsB) in the genomes of all Pseudomonas strains scrutinised (Gao et al., 2004; Zavaleta‐Pastor et al., 2010). Furthermore, in BIRD‐1 these proteins were expressed in a PHO‐dependent manner. Interestingly, in BIRD‐1 UDP‐glucose 6‐phosphate dehydrogenase (TuaD) was also PHO‐regulated. In B. subtilis, TuaD is encoded by the tua operon that is involved in the production of teichuronic acid lipids during Pi‐depletion (Liu and Hulett, 1998; Antelmann et al., 2000). The rest of the genes required for teichuronic acid were absent, therefore, making the role of TuaD somewhat unclear in BIRD‐1. As all of these proteins were silenced in the phoBR mutant, it is likely that remodelling the lipid membrane accounts for the gross difference observed in growth between this strain and the wild type when grown under Pi‐deplete conditions (Fig. 7).
The observed heterogeneity in the Pi responsive portion of the proteome of the three Pseudomonas strains in this study highlights how the utilization of different PGPR can have potentially different effects in the rhizosphere. For example, based on our genomic and proteomics data, differences in the ability of the strains to degrade phospholipids, nucleic acids and organopesticides (Singh and Walker, 2006; Bigley and Raushel, 2013), as well as a likely difference in their broad organic P substrate range may have marked effects on their ability to mobilize P for a plant host under certain environmental conditions. For example, the addition of a strain comparable to DSM4166, expressing UshA and GlpQ, may increase the acquisition of P in plants when using manure as the nutrient source, which is rich in nucleic acids and phospholipids (Turner and Leytem, 2004; Shen et al., 2011). Furthermore, DSM4166 is a known nitrogen fixer and in certain soils (nitrogen‐limited) employing this strain over that of either BIRD‐1 or SBW25 may provide more efficient plant‐growth promotion. Likewise, in soils contaminated with phosphorus‐containing organopesticides, it may be more suitable to deploy a strain similar to SBW25, which contains the promiscuous C‐P lyase, capable of degrading these compounds, or BIRD‐1 which possesses a Pi‐responsive putative phosophotriesterase.
Conclusions
Observing the global exoproteomic response of just three Pseudomonas species revealed new insights into the P scavenging capabilities of this genus and has provided a number of markers (Muth et al., 2015) that can be utilized to investigate P‐mobilization directly in the rhizosphere. Given the enormous task of identifying proteins in situ from complex communities, the data presented in this paper will serve as a platform to investigate the key enzymes and microbial taxa involved in P‐mobilization at the level of functional entities (proteins) in situ. Meta‐exoproteomics has already identified differences between genomic and proteomic assessments of soil chintase‐degrading communites (Johnson‐Rollings et al., 2014) and should also shed light on the ‘black box’ concerning P‐mobilization in the rhizosphere.
Experimental procedures
Growth and maintenance of bacterial strains
All three Pseudomonas strains were maintained on Luria Bertani (LB) agar (1.5% w/v) medium at 30°C. To investigate the effect of Pi‐depletion on the three strains, each was grown (n = 3) in an adapted Minimal A medium comprising: Na‐Succinate 5.4 g l−1, NaCl 200 mg l−1, NH4Cl 450 mg l−1, CaCl2 200 mg l−1, KCL mg l−1 MgCl2 450 mg l−1, FeCl2 10 mg l−1, MnCl2 10 mg l−1, 10 mM 4‐(2‐hydroxyethyl)−1‐piperazineethanesulfonic acid (HEPES) pH 7.2, with KH2PO4 added to a final concentration of either 50 μM or 1.4 mM. Each strain was pre‐cultured in minimal A medium containing 400 μM Pi to ensure cells had adequate Pi while minimizing the potential for carry over of residual Pi into triplicate experimental cultures.
Quantification of alkaline phosphatase activity
A 0.5 ml culture (n = 3) was incubated with 20 μl para‐nitrophenyl phosphate (pNPP) (final conc. 4mM) and incubated at room temperature for 1 h or when colour development started to occur. The reaction was stopped using 25 μl NaOH (2 mM) and incubated for 10 min. Cell debris and precipitants were removed via centrifugation (2 min, 8,000 × g) prior to spectrophotometry (optical density 405 nm). A standard curve for para‐nitrophenol was generated using a range of known concentrations (0, 4, 8, 25, 50, 75, 100 mg ml−1).
Preparation of exoproteomes, trypsin in‐gel proteolysis, nano‐LC‐MS/MS analysis and peptide identification through MS/MS database searching
Exoproteomes were analysed using modified methods of Christie‐Oleza and Armengaud (2010). The recorded MS/MS spectra were searched against the protein sequence database (P. putida BIRD‐1, NC_017530.1; P. fluorescens SBW25, NC_012660.1; P. stutzeri DSM4166, NC_017532.1). Full details of the protocol and parameters used for peptide identification can be found in the supplementary materials and methods.
Quantification of detected proteins
The Normalized Spectral Abundance Factor (NSAF) values were calculated using SCAFFOLD v4.0 according to software defaults. For the exoproteomes, no further normalization was performed. For whole‐cell proteomics, 25 μg of protein was loaded onto SDS‐PAGE gels prior to identification. No further normalization was performed. However, we examined the abundance of several housekeeping proteins and central metabolic enzymes and did not observe substantial change in their abundance. For determining the proportion of proteins within the exoproteome, replicate cultures (n = 3) were averaged. The proteomics data has been deposited in the Proteomics Identification (PRIDE) database (Martens et al., 2005) with the following accession numbers: PXD004065, PXD004064, PXD003830, PXD003829, PXD003828, PXD003827, PXD003826.
Bioinformatics analysis of detected proteins and comparative genomics
The majority of analyses were performed using the Integrated Microbial Genomes Database at the Joint Genome Institute (IMG/JGI) server (http://img.jgi.doe.gov/). Please refer to supplementary information for a detailed summary of the bioinformatics approaches used. IMG/JGI was also used for comparative genomic analyses. BLASTP (expected value, e‐30, minimum identity = 20%) searches were performed using the proteins detected in the exoproteomes/proteome. In some cases other proteins identified from the literature known to be involved in Pi scavenging/recycling were used as queries for BLASTP analysis. For the Pi‐binding proteins (PBP), a function search using the IMG/JGI database was performed using the Pfam domain, 12849 as the query.
Genetic manipulation of P. putida BIRD‐1
To construct a phoBR mutant of P. putida BIRD‐1, the method outlined by Lidbury et al. (2014) was adapted. Please refer to the supplementary information for a detailed procedure.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Proportion of the extracellular and intracellular proteins detected in the four protein fractions extracted during this study. Only the top‐60 most abundant proteins were included in the analyses. The values displayed are taken from Pi‐deplete and Pi‐replete cultures. Results presented are the mean of triplicate cultures.
Fig. S2. The abundance of proteins in both the high Pi and low Pi exoproteomes of the three Pseudomonas strains. (A) Pseudomonas fluorescens SBW25, (B) Pseudomonas putida BIRD‐1, (C) Pseudomonas stutzeri DSM4166. Results are the mean of triplicate cultures and error bars denote standard deviation.
Fig. S3. Conservation of the key residues (highlighted in red) involved in phosphate binding among the periplasmic binding proteins containing the domain, Pfam12849‐ PBP. Locus tags are used as the identifier. Abbreviations: VP, Vibrio parahaemolyticus; VC/VCA, V. cholerae; VAA, V. anguillarum; V. harveyi MYO, Synechocystis sp. PCC6803; PFLU, P. fluorescens; PA, P. aeruginosa; PPUBIRD1, P. putida; PSTAA, P. stutzeri; Psyr, P. syringae; EcDH1, E. coli; P. Antarctica; Smc; Ensifer meliloti.
Fig. S4. Semi‐quantitative abundance analysis of the putative phosphonate substrate binding proteins detected in the exoproteomes of the three Pseudomonas strains. and the phoBR mutant. Results presented are the mean of triplicate cultures. Error bars denote standard deviation.
Fig. S5 Growth of the phoBR mutant strain of P. putida BIRD‐1. A. A comparison of the phoBR mutant grown under Pi‐replete (Black circles) and Pi‐deplete (Grey circles) conditions. Concentrations of Pi were the same as those used for the wild type. Black arrows indicated the times of sampling for proteomics and exoproteomics. The striped arrow indicates the addition of Pi (50 μM) to help generate enough biomass for sampling. B. Growth yields of either the wild type or phoBR mutant sampled after 48 hours grown on Pi‐replete or Pi‐deplete growth media. Results presented are the mean of triplicate cultures. Error bars denote standard deviation.
Fig. S6. Evolutionary relationships of PhoX‐like homologs. The evolutionary history was inferred using the Neighbor‐Joining method [1]. The optimal tree with the sum of branch length = 3.66263884 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p‐distance method [2] and are in the units of the number of amino acid differences per site. The analysis involved 27 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 842 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [3].
Table S1. A rank‐abundance profile of the identified proteins in the exoproteome of Pseudomonas putida BIRD‐1.
Table S2. A rank‐abundance profile of the identified proteins in the exoproteome of Pseudomonas fluorescens SBW25.
Table S3. A rank‐abundance profile of the identified proteins in the exoproteome of Pseudomonas stutzeri DSM4166.
Table S4. A rank‐abundance profile of the identified proteins in the proteome of P. putida BIRD‐1.
Table S5. A rank‐abundance profile of the identified proteins in the cellular proteome of the P. putida BIRD‐1 phoBR mutant.
Table S6. A rank‐abundance profile of the identified proteins in the exoproteome of the P. putida BIRD‐1 phoBR mutant.
Acknowledgements
A special thank you goes to Prof. Juan Luis Ramos and Dr. Andrew Spiers for their kind donations of P. putida BIRD‐1 and P. fluorescens SBW25, respectively. This study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) under the project code BB/L026074/1. We also acknowledge the WPH Proteomics RTP, specifically Cleidiane Zampronio, for mass spectrometry and proteomic analysis.
References
- Antelmann, H. , Scharf, C. , and Hecker, M. (2000) Phosphate starvation‐inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J Bacteriol 182: 4478–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud, J. , Christie‐Oleza, J.A. , Clair, G. , Malard, V. , and Duport, C. (2012) Exoproteomics: exploring the world around biological systems. Expert Rev Proteomics 9: 561–575. [DOI] [PubMed] [Google Scholar]
- Bains, M. , Fernández, L. , and Hancock, R.E.W. (2012) Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa . Appl Environ Microbiol 78: 6762–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, A.S. , Ciocci, M.J. , Metcalf, W.W. , Kim, J. , Babbitt, P.C. , Wanner, B.L. , et al (1998) Insights into the mechanism of catalysis by the P−C bond‐cleaving enzyme phosphonoacetaldehyde hydrolase derived from gene sequence analysis and mutagenesis. Biochemistry 37: 9305–9315. [DOI] [PubMed] [Google Scholar]
- Ball, G. , Durand, É. , Lazdunski, A. , and Filloux, A. (2002) A novel type II secretion system in Pseudomonas aeruginosa . Mol Microbiol 43: 475–485. [DOI] [PubMed] [Google Scholar]
- Berna, A. , Bernier, F. , Chabrière, E. , Perera, T. , and Scott, K. (2008) DING proteins; novel members of a prokaryotic phosphate‐binding protein superfamily which extends into the eukaryotic kingdom. Int J Biochem Cell B 40: 170–175. [DOI] [PubMed] [Google Scholar]
- Bigley, A.N. , and Raushel, F.M. (2013) Catalytic mechanisms for phosphotriesterases. Biochim Biophys Acta 1834: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisova, S.A. , Christman, H.D. , Metcalf, M.E.M. , Zulkepli, N.A. , Zhang, J.K. , van der Donk, W.A. , and Metcalf, W.W. (2011) Genetic and biochemical characterization of a pathway for the degradation of 2‐aminoethylphosphonate in Sinorhizobium meliloti 1021. J Biol Chem 286: 22283–22290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman, E. , and Beckwith, J. (1975) Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol 96: 307–316. [DOI] [PubMed] [Google Scholar]
- Carini, P. , Van Mooy, B.A.S. , Thrash, J.C. , White, A. , Zhao, Y. , Campbell, E.O. , et al (2015) SAR11 lipid renovation in response to phosphate starvation. Proc Natl Acad Sci USA 112: 7767–7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , and Armengaud, J. (2010) In‐depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography‐tandem mass spectrometry: the Ruegeria pomeroyi DSS‐3 case‐study. Mar Drugs 8: 2223–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Piña‐Villalonga, J.M. , Bosch, R. , Nogales, B. , and Armengaud, J. (2012) Comparative proteogenomics of twelve Roseobacter exoproteomes reveals different adaptive strategies among these marine bacteria. Mol Cell Proteomics 11: M111.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Armengaud, J. , Guerin, P. , and Scanlan, D.J. (2015) Functional distinctness in the exoproteomes of marine Synechococcus . Environ Microbiol 17: 3781–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell, D. , Drangert, J.‐O. , and White, S. (2009) The story of phosphorus: global food security and food for thought. Global Environ Change 19: 292–305. [Google Scholar]
- Ebner, P. , Rinker, J. , and Götz, F. (2016) Excretion of cytoplasmic proteins in Staphylococcus is most likely not due to cell lysis. Curr Genet 62: 19–23 [DOI] [PubMed] [Google Scholar]
- Fox, E.M. , and Mendz, G.L. (2006) Phosphonate degradation in microorganisms. Enzyme Microb Technol 40: 145–150. [Google Scholar]
- Furtwängler, K. , Tarasov, V. , Wende, A. , Schwarz, C. , and Oesterhelt, D. (2010) Regulation of phosphate uptake via Pst transporters in Halobacterium salinarum R1. Mol Microbiol 76: 378–392. [DOI] [PubMed] [Google Scholar]
- Gao, J.‐L. , Weissenmayer, B. , Taylor, A.M. , Thomas‐Oates, J. , López‐Lara, I.M. , and Geiger, O. (2004) Identification of a gene required for the formation of lyso‐ornithine lipid, an intermediate in the biosynthesis of ornithine‐containing lipids. Mol Microbiol 53: 1757–1770. [DOI] [PubMed] [Google Scholar]
- Harrison, A.F. (1987) Soil Organic Phosphorus – A Review of the Literature. Wallingford, Oxon, UK: CAB International, p. 257 [Google Scholar]
- Hass, D. , and Keel, C. (2003) Regulation of antibiotic production in root‐colonising Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41: 117–153. [DOI] [PubMed] [Google Scholar]
- Heffer, P. (2006) Phosphorus fertilisation: issues and outlook : proceedings [of a] paper presented to the International Fertiliser Society at a conference in Cambridge, on 14th December 2006. York: International Fertiliser Society.
- Hwan Baek, J. , and Yup Lee, S. (2006) Novel gene members in the Pho regulon of Escherichia coli. FEMS Microbiology Letters 264: 104–109. [DOI] [PubMed]
- Inatsuka, C.S. , Julio, S.M. , and Cotter, P.A. (2005) Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci USA 102: 18578–18583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Metcalf, W.W. , Lee, K.S. , and Wanner, B.L. (1995) Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J Bacteriol 177: 6411–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson‐Rollings, A.S. , Wright, H. , Masciandaro, G. , Macci, C. , Doni, S. , Calvo‐Bado, L.A. , et al (2014) Exploring the functional soil‐microbe interface and exoenzymes through soil metaexoproteomics. ISME J 8: 2148–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, T.J. , Ehrmann, M. , and Boos, W. (1983) Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem 258: 5428–5432. [PubMed] [Google Scholar]
- Lidbury, I. , Murrell, J.C. , and Chen, Y. (2014) Trimethylamine N‐oxide metabolism by abundant marine heterotrophic bacteria. Proc Natl Acad Sci USA 111: 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner, D. , Elias, M. , Moniot, S. , Fournier, B. , Scott, K. , Jelsch, C. , et al (2009) Elucidation of the phosphate binding mode of DING proteins revealed by subangstrom X‐ray crystallography. J Am Chem Soc 131: 7879–7886. [DOI] [PubMed] [Google Scholar]
- Lim, B.L. , Yeung, P. , Cheng, C. , and Hill, J.E. (2007) Distribution and diversity of phytate‐mineralizing bacteria. ISME J 1: 321–330. [DOI] [PubMed] [Google Scholar]
- Liu, W. , and Hulett, F.M. (1998) Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho‐regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144: 1443–1450. [DOI] [PubMed] [Google Scholar]
- Lugtenberg, B. , and Kamilova, F. (2009) Plant‐growth‐promoting rhizobacteria. Annu Rev Microbiol 63: 541–556. [DOI] [PubMed] [Google Scholar]
- López‐Arredondo, D.L. , Leyva‐González, M.A. , González‐Morales, S.I. , López‐Bucio, J. , and Herrera‐Estrella, L. (2014) Phosphate nutrition: improving low‐phosphate tolerance in crops. Annu Rev Plant Biol 65: 95–123. [DOI] [PubMed] [Google Scholar]
- Martens, L. , Hermjakob, H. , Jones, P. , Adamski, M. , Taylor, C. , States, D. , et al (2005) PRIDE: the proteomics identifications database. Proteomics 5: 3537–3545. [DOI] [PubMed] [Google Scholar]
- Mauchline, T.H. , Fowler, J.E. , East, A.K. , Sartor, A.L. , Zaheer, R. , Hosie, A.H.F. , et al (2006) Mapping the Sinorhizobium meliloti 1021 solute‐binding protein‐dependent transportome. Proc Natl Acad Sci USA 103: 17933–17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, J.W. , Chin, J.P. , and Quinn, J.P. (2013) Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat Rev Microbiol 11: 412–419. [DOI] [PubMed] [Google Scholar]
- Miller, S.H. , Browne, P. , Prigent‐Combaret, C. , Combes‐Meynet, E. , Morrissey, J.P. , and O'Gara, F. (2010) Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ Microbiol Rep 2: 403–411. [DOI] [PubMed] [Google Scholar]
- Monds, R.D. , Newell, P.D. , Schwartzman, J.A. , and O'Toole, G.A. (2006) Conservation of the Pho regulon in Pseudomonas fluorescens Pf0‐1. Appl Environ Microbiol 72: 1910–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudrak, B. , and Tamayo, R. (2012) The Vibrio cholerae Pst2 phosphate transport system is upregulated in biofilms and contributes to biofilm‐induced hyperinfectivity. Infect Immun 80: 1794–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth, T. , Kolmeder, C.A. , Salojärvi, J. , Keskitalo, M. , Varjosalo, M. , Froukje J., et al (2015) Navigating through metaproteomics data – a logbook of database searching. Proteomics 15: 3439–3453. [DOI] [PubMed] [Google Scholar]
- Naseby, D.C. , Way, J.A. , Bainton, N.J. , and Lynch, J.M. (2001) Biocontrol of Phythium in the pea rhizosphere by antifungal metabolite producing and non‐producing Pseudomonas strains. J Appl Microbiol 90: 421–429. [DOI] [PubMed] [Google Scholar]
- Ostrowski, M. , Mazard, S. , Tetu, S.G. , Phillippy, K. , Johnson, A. , Palenik, B. , et al (2010) PtrA is required for coordinate regulation of gene expression during phosphate stress in a marine Synechococcus . ISME J 4: 908–921. [DOI] [PubMed] [Google Scholar]
- Oteino, N. , Lally, R.D. , Kiwanuka, S. , Lloyd, A. , Ryan, D. , Germaine, K.J. , and Dowling, D.N. (2015) Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol 6: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchuk, G.E. , Ammons, C. , Culley, D.E. , Li, S.‐M.W. , McLean, J.S. , Romine, M.F. , et al (2008) Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp.: ecological and physiological implications for dissimilatory metal reduction. Appl Environ Microbiol 74: 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt, F.D. , Mazard, S. , Humphreys, L. , and Scanlan, D.J. (2010) Functional characterization of Synechocystis sp. strain PCC 6803 pst1 and pst2 gene clusters reveals a novel strategy for phosphate uptake in a freshwater cyanobacterium. J Bacteriol 192: 3512–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, J.T. , Ismail, A.M. , and Camilli, A. (2010) PhoB regulates both environmental and virulence gene expression in Vibrio cholerae . Mol Microbiol 77: 1595–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, G.M. (2004) Plant perceptions of plant growth‐promoting Pseudomonas . Philos Trans R Soc Lond B Biol Sci 359: 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putker, F. , Tommassen‐van Boxtel, R. , Stork, M. , Rodríguez‐Herva, J.J. , Koster, M. , and Tommassen, J. (2013) The type II secretion system (Xcp) of Pseudomonas putida is active and involved in the secretion of phosphatases. Environ Microbiol 15: 2658–2671. [DOI] [PubMed] [Google Scholar]
- Raven, J.A. (2008) Phosphorus and the future. In: White PJ, Hammond JP, editors, The Ecophysiology of Plant‐Phosphorus Interactions. Dordrecht: Springer. pp. 271–283.
- Richardson, A.E. , Barea, J.‐M. , McNeill, A.M. , and Prigent‐Combaret, C. (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321: 305–339. [Google Scholar]
- Rittmann, D. , Sorger‐Herrmann, U. , and Wendisch, V.F. (2005) Phosphate starvation‐inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl Environ Microbiol 71: 4339–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca, A. , Pizarro‐Tobías, P. , Udaondo, Z. , Fernández, M. , Matilla, M.A. , Molina‐Henares, M.A. , et al (2013) Analysis of the plant growth‐promoting properties encoded by the genome of the rhizobacterium Pseudomonas putida BIRD‐1. Environ Microbiol 15: 780–794. [DOI] [PubMed] [Google Scholar]
- Rodrı'guez, H. , and Fraga, R. (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17: 319–339. [DOI] [PubMed] [Google Scholar]
- Santos‐Beneit, F. (2015) The Pho regulon: a huge regulatory network in bacteria. Front Microbiol 6: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K. , and Wu, L. (2005) Functional properties of a recombinant bacterial DING protein: comparison with a homologous human protein. Biochim Biophys Acta Mol Cell Res 1744: 234–244. [DOI] [PubMed] [Google Scholar]
- Sebastian, M. , Smith, A.F. , Gonzalez, J.M. , Fredricks, H.F. , Van Mooy, B. , Koblizek, M. , et al (2016) Lipid remodelling is a widespread strategy in marine heterotrophic bacteria upon phosphorus deficiency. ISME J. 10: 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J. , Yuan, L. , Zhang, J. , Li, H. , Bai, Z. , Chen, X. , et al (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, B.K. , and Walker, A. (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30: 428–471. [DOI] [PubMed] [Google Scholar]
- Sola‐Landa, A. , Rodríguez‐García, A. , Apel, A.K. , and Martín, J.F. (2008) Target genes and structure of the direct repeats in the DNA‐binding sequences of the response regulator PhoP in Streptomyces coelicolor . Nucleic Acids Res 36: 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutter, M.I. , Shand, C.A. , George, T.S. , Blackwell, M.S.A. , Bol, R. , MacKay, R.L. , et al (2012) Recovering phosphorus from soil: a root solution? Environ Sci Technol 46:1977–1978. [DOI] [PubMed] [Google Scholar]
- Su, Z. , Olman, V. , and Xu, Y. (2007) Computational prediction of Pho regulons in cyanobacteria. BMC Genomics 8: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Dong, Y. , Zhou, Y. , Yang, M. , Zhang, C. , Rao, Z. , and Zhang, X. (2004) Crystallization and preliminary X‐ray studies of methyl parathion hydrolase from Pseudomonas sp. WBC‐3. Acta Crystallogr C Struct Chem D 60: 954–956. [DOI] [PubMed] [Google Scholar]
- Tan, A.S.P. , and Worobec, E.A. (1993) Isolation and characterization of two immunochemically distinct alkaline phosphatases from Pseudomonas aeruginosa . FEMS Microbiol Lett 106: 281–286. [DOI] [PubMed] [Google Scholar]
- Ternan, N.G. , and Quinn, J.P. (1998) Phosphate starvation‐independent 2‐aminoethylphosphonic acid biodegradation in a newly isolated strain of Pseudomonas putida, NG2. Syst Appl Microbiol 21: 346–352. [DOI] [PubMed] [Google Scholar]
- Tetu, S.G. , Brahamsha, B. , Johnson, D.A. , Tai, V. , Phillippy, K. , Palenik, B. , and Paulsen, I.T. (2009) Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J 3: 835–849. [DOI] [PubMed] [Google Scholar]
- Turner, B.L. , and Leytem, A.B. (2004) Phosphorus compounds in sequential extracts of animal manures: chemical speciation and a novel factionation procedure. Environ Sci Technol 38: 6101–6108. [DOI] [PubMed] [Google Scholar]
- Turner, B.L. , Richardson, A.E. , and Mullaney, E.J. (2002) Inositol phosphates in the environment. Philos Trans R Soc Lond B Biol Sci 357: 449–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance, C.P. , Uhde‐Stone, C. , and Allan, D.L. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447. [DOI] [PubMed] [Google Scholar]
- Vassilev, N. , Vassileva, M. , and Nikolaeva, I. (2006) Simultaneous P‐solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl Microbiol Biotechnol 71: 137–144. [DOI] [PubMed] [Google Scholar]
- Villarreal‐Chiu, J.F. , Quinn, J.P. , and McGrath, J.W. (2012) The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front Microbiol 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, B.L. (1990) Phosphorus assimilation and its control of gene expression in Escherichia coli In The Molecular Basis of Bacterial Metabolism. Hauska G., and Thauer R. (eds). Berlin, Heidelberg: Springer, pp. 152–163. [Google Scholar]
- White, A.K. , and Metcalf, W.W. (2004) The htx and ptx operons of Pseudomonas stutzeri WM88 are new members of the Pho regulon. J Bacteriol 186: 5876–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, A.K. , and Metcalf, W.W. (2007) Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol 61: 379–400. [DOI] [PubMed] [Google Scholar]
- White, P.J. , and Hammond, J.P. (2008) Phosphorus nutrition of terrestrial plants In The Ecophysiology of Plant‐Phosphorus Interactions. White P.J., and Hammond J.P. (eds). Dordrecht, Netherlands: Springer, pp. 51–81. [Google Scholar]
- White, P.J. , and Hammond, J.P. (2009) The sources of phosphorus in the waters of Great Britain. J Environ Qual 38: 13–26. [DOI] [PubMed] [Google Scholar]
- Yu, H. , Yuan, M. , Lu, W. , Yang, J. , Dai, S. , Li, Q. , et al (2011) Complete genome sequence of the nitrogen‐fixing and rhizosphere‐associated bacterium Pseudomonas stutzeri strain DSM4166. J Bacteriol 193: 3422–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin, H. , and Nygaard, P. (1996) Biosynthesis of purine nucleotides In Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F.C., Curtiss R, III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., et al (eds). Washington, DC: ASM Press, pp. 561–579. [Google Scholar]
- Zavaleta‐Pastor, M. , Sohlenkamp, C. , Gao, J.‐L. , Guan, Z. , Zaheer, R. , Finan, T.M. , et al (2010) Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc Natl Acad Sci USA 107: 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.‐X. , Scott, K. , Meffin, R. , and Rainey, P.B. (2007) Genetic characterization of psp encoding the DING protein in Pseudomonas fluorescens SBW25. BMC Microbiol 7: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Proportion of the extracellular and intracellular proteins detected in the four protein fractions extracted during this study. Only the top‐60 most abundant proteins were included in the analyses. The values displayed are taken from Pi‐deplete and Pi‐replete cultures. Results presented are the mean of triplicate cultures.
Fig. S2. The abundance of proteins in both the high Pi and low Pi exoproteomes of the three Pseudomonas strains. (A) Pseudomonas fluorescens SBW25, (B) Pseudomonas putida BIRD‐1, (C) Pseudomonas stutzeri DSM4166. Results are the mean of triplicate cultures and error bars denote standard deviation.
Fig. S3. Conservation of the key residues (highlighted in red) involved in phosphate binding among the periplasmic binding proteins containing the domain, Pfam12849‐ PBP. Locus tags are used as the identifier. Abbreviations: VP, Vibrio parahaemolyticus; VC/VCA, V. cholerae; VAA, V. anguillarum; V. harveyi MYO, Synechocystis sp. PCC6803; PFLU, P. fluorescens; PA, P. aeruginosa; PPUBIRD1, P. putida; PSTAA, P. stutzeri; Psyr, P. syringae; EcDH1, E. coli; P. Antarctica; Smc; Ensifer meliloti.
Fig. S4. Semi‐quantitative abundance analysis of the putative phosphonate substrate binding proteins detected in the exoproteomes of the three Pseudomonas strains. and the phoBR mutant. Results presented are the mean of triplicate cultures. Error bars denote standard deviation.
Fig. S5 Growth of the phoBR mutant strain of P. putida BIRD‐1. A. A comparison of the phoBR mutant grown under Pi‐replete (Black circles) and Pi‐deplete (Grey circles) conditions. Concentrations of Pi were the same as those used for the wild type. Black arrows indicated the times of sampling for proteomics and exoproteomics. The striped arrow indicates the addition of Pi (50 μM) to help generate enough biomass for sampling. B. Growth yields of either the wild type or phoBR mutant sampled after 48 hours grown on Pi‐replete or Pi‐deplete growth media. Results presented are the mean of triplicate cultures. Error bars denote standard deviation.
Fig. S6. Evolutionary relationships of PhoX‐like homologs. The evolutionary history was inferred using the Neighbor‐Joining method [1]. The optimal tree with the sum of branch length = 3.66263884 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p‐distance method [2] and are in the units of the number of amino acid differences per site. The analysis involved 27 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 842 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [3].
Table S1. A rank‐abundance profile of the identified proteins in the exoproteome of Pseudomonas putida BIRD‐1.
Table S2. A rank‐abundance profile of the identified proteins in the exoproteome of Pseudomonas fluorescens SBW25.
Table S3. A rank‐abundance profile of the identified proteins in the exoproteome of Pseudomonas stutzeri DSM4166.
Table S4. A rank‐abundance profile of the identified proteins in the proteome of P. putida BIRD‐1.
Table S5. A rank‐abundance profile of the identified proteins in the cellular proteome of the P. putida BIRD‐1 phoBR mutant.
Table S6. A rank‐abundance profile of the identified proteins in the exoproteome of the P. putida BIRD‐1 phoBR mutant.


