The global spread of ESBL-E. coli has been driven in large part by pandemic sequence type 131 (ST131). A recent study suggested that, within E. coli ST131, certain sublineages have disseminated worldwide with little association with their geographical origin, highlighting the complexity of the epidemiology of this pandemic clone. ST131 bacteria have also been classified into four virotypes based on the distribution of certain virulence genes. Information on virotype distribution in Asian ST131 strains is limited. We conducted whole-genome sequencing of ESBL-E. coli ST131 strains collected in Nepal and Japan, two Asian countries with a high and low prevalence of ESBL-E. coli, respectively. We systematically compared these ST131 genomes with those reported from other regions to gain insights into the molecular epidemiology of their spread and found the distinct phylogenetic characteristics of the spread of ESBL-E. coli ST131 in these two geographical areas of Asia.
KEYWORDS: CTX-M, Escherichia coli, ST131, antimicrobial resistance, extended-spectrum beta-lactamase, whole-genome sequence
ABSTRACT
The global spread of extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli (ESBL-E. coli) has largely been driven by the pandemic sequence type 131 (ST131). This study aimed to determine the molecular epidemiology of their spread in two Asian countries with contrasting prevalence. We conducted whole-genome sequencing (WGS) of ESBL-E. coli ST131 strains collected prospectively from Nepal and Japan, two countries in Asia with a high and low prevalence of ESBL-E. coli, respectively. We also systematically compared these genomes with those reported from other regions using publicly available WGS data for E. coli ST131 strains. Further, we conducted phylogenetic analysis of these isolates and all genome sequence data for ST131 strains to determine sequence diversity. One hundred five unique ESBL-E. coli isolates from Nepal (February 2013 to July 2013) and 76 isolates from Japan (October 2013 to September 2014) were included. Of these isolates, 54 (51%) isolates from Nepal and 11 (14%) isolates from Japan were identified as ST131 by WGS. Phylogenetic analysis based on WGS suggested that the majority of ESBL-E. coli ST131 isolates from Nepal clustered together, whereas those from Japan were more diverse. Half of the ESBL-E. coli ST131 isolates from Japan belonged to virotype C, whereas half of the isolates from Nepal belonged to a virotype other than virotype A, B, C, D, or E (A/B/C/D/E). The dominant sublineage of E. coli ST131 was H30Rx, which was most prominent in ESBL-E. coli ST131 isolates from Nepal. Our results revealed distinct phylogenetic characteristics of ESBL-E. coli ST131 spread in the two geographical areas of Asia, indicating the involvement of multiple factors in its local spread in each region.
IMPORTANCE The global spread of ESBL-E. coli has been driven in large part by pandemic sequence type 131 (ST131). A recent study suggested that, within E. coli ST131, certain sublineages have disseminated worldwide with little association with their geographical origin, highlighting the complexity of the epidemiology of this pandemic clone. ST131 bacteria have also been classified into four virotypes based on the distribution of certain virulence genes. Information on virotype distribution in Asian ST131 strains is limited. We conducted whole-genome sequencing of ESBL-E. coli ST131 strains collected in Nepal and Japan, two Asian countries with a high and low prevalence of ESBL-E. coli, respectively. We systematically compared these ST131 genomes with those reported from other regions to gain insights into the molecular epidemiology of their spread and found the distinct phylogenetic characteristics of the spread of ESBL-E. coli ST131 in these two geographical areas of Asia.
INTRODUCTION
The emergence of extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli (ESBL-E. coli) is a global problem. However, its prevalence and epidemiology differ significantly depending on its geographical location. According to a recent report from the World Health Organization (WHO) (1), the prevalences of E. coli resistance to third-generation cephalosporins were 14% in Europe, 27% in the Americas (15% in the United States), and 30% in Southeast Asia. The prevalence of E. coli resistance to these cephalosporins was the highest in developing countries in Asia (38% in Nepal, 68% in Myanmar, and 16% to 95% in India) while relatively low in more-developed Asian countries, including Japan (17%) (1).
The global spread of ESBL-E. coli has been driven in large part by pandemic sequence type 131 (ST131); E. coli ST131 bacteria are often resistant to multiple drugs (2). A recent study suggested that, within E. coli ST131, certain sublineages have disseminated worldwide with little association with their geographical origin, highlighting the complexity of the epidemiology of this pandemic clone (3). ST131 has also been classified into four virotypes based on the distribution of certain virulence genes, and virotype C has been reported to be globally disseminated (4). However, information pertaining to virotype distribution among Asian ST131 strains is limited.
In this study, we conducted whole-genome sequencing (WGS) of ESBL-E. coli ST131 strains collected prospectively in Nepal (5) and Japan. Although both countries are located in Asia, the socioeconomic statuses of Japan and Nepal are different and they also have distinct prevalences of ESBL-E. coli (Japan has a low prevalence, and Nepal has a high prevalence). We systematically compared these ST131 genomes with those reported from other regions to gain insights into the epidemiology of their spread.
RESULTS AND DISCUSSION
Characteristics of ESBL-E. coli ST131 isolates and E. coli ST131 genomes.
During the study period, 105 and 76 unique ESBL-E. coli isolates were identified from Nepal and Japan, respectively. Of these isolates, 54 (51%) isolates from Nepal and 11 (14%) isolates from Japan were identified as ST131 by WGS based on sequence typing and included in further analyses. Forty (38.1%) of 105 ESBL-E. coli isolates from Nepal and 19 (25%) of 76 ESBL-E. coli isolates from Japan were isolated from outpatients. Nineteen (35.2%) of 54 ESBL-E. coli ST131 isolates from Nepal and 2 (18.2%) of 11 of ESBL-E. coli ST131 isolates from Japan were from outpatients. This suggests that ESBL-E. coli ST131 bacteria were isolated from outpatients almost twice as often in Nepal as in Japan.
The median age of patients with ESBL-E. coli ST131 was significantly lower in Nepal (39 years [interquartile range {IQR}, 25 to 61 years]) than in Japan (83 years [IQR, 73 to 86 years]) (P < 0.001). The numbers of male patients with ESBL-E. coli ST131 were similar in the two countries (19 [35%] in Nepal and 4 in Japan [36%]). The majority of isolates were collected from urine samples (54 [100%] in Nepal and 8 [73%] in Japan). Pregnant female patients with ESBL-E. coli ST131 were more frequent in Nepal (n = 9 [26%]) than in Japan (n = 0) (P < 0.001). Comorbid conditions, such as malignancy (n = 2 [3.7%] in Nepal; n = 3 [27.3%] in Japan) and underlying urological conditions (e.g., benign prostatic hyperplasia, urolithiasis, obstructive urinary diseases) (n = 10 [18.5%] in Nepal; n = 6 [54.5%] in Japan) were more common in patients with ESBL-E. coli in Japan than in patients in Nepal (P = 0.031 and P = 0.02, respectively). ESBL-E. coli isolates from Nepal and Japan had distinct clinical characteristics.
In a study that identified ESBL-E. coli from travelers who returned from India, 47% of 17 ESBL-E. coli isolates collected from 2004 to 2006 were ST131 (6). The prevalence of ST131 among ESBL-E. coli bacteria isolated in Nepal in our study is in line with the previous data, considering the geographical proximity and traffic of visitors between Nepal and India. The ST131 prevalence of 14% found among ESBL-E. coli isolates from Japan is also similar to that reported in a previous study where 21% of 130 ESBL-E. coli isolates collected between 2002 and 2003 from Japan were ST131 (7). However, the prevalence was lower than that reported in a more recent study conducted in Japan, in which 37% of 581 ESBL-E. coli isolates collected from 2001 to 2010 were identified as ST131 (8). This discrepancy might be attributed to the differences in the year of collection, geographical locations in the country, and methods used to identify ST131 (PCR-based screening versus WGS).
fimH alleles and H30Rx sublineages in E. coli ST131.
fimH alleles and H30Rx sublineages in E. coli ST131 were determined and compared among various geographical regions (Table 1). fimH30, especially fimH30Rx comprised the majority of the fimH alleles in various geographical regions, followed by fimH41 and fimH22. fimH27 was observed only in E. coli ST131 from Africa. We found that all 54 ESBL-E. coli isolates from Nepal belonged to the H30Rx sublineage. There is little data on the prevalence of the H30Rx sublineage in South Asia. A previous study that included only a few ST131 strains from India revealed that all of them belonged to clade C (i.e., H30) and produced CTX-M-15 (3). Our result is in accordance with the high prevalence of CTX-M-15-producing ESBL-E. coli reported among travelers returning from the Indian subcontinent (6). However, the limited isolate collection period in one facility might have affected the clonal distribution of ESBL-E. coli ST131 isolates from Nepal in our study. H30R (non-Rx) were more prevalent among ESBL-E. coli ST131 isolates from Japan compared to isolates from Nepal, although more than 60% of ESBL-E. coli ST131 isolates from Japan still belonged to H30Rx. A previous report from Japan suggested a higher prevalence of H30R (non-Rx) and H22 among ESBL-E. coli ST131 isolates collected from 2001 to 2012 in 10 Japanese acute-care hospitals located in Kyoto, Shiga area, which is more than 460 km (286 miles) away from the hospital in this study (9). The relatively high prevalence of H30R (non-Rx) and H22 observed in this study in the United States and Europe, respectively, were consistent with previous reports (3, 10).
TABLE 1 .
Comparison of fimH alleles and H30Rx and H30R sublineages in Escherichia coli ST131
| fimH allele or sublineage | Total no. of isolates (n = 132) | No. of isolates (%) from: |
P valued | |||||
|---|---|---|---|---|---|---|---|---|
| Nepal (n = 54) | Japan (n = 12)a | Other Asian countries (n = 3)b | USA (n = 29)b | Europe (n = 29)b,c | Africa (n = 5)b | |||
| H30 | 110 | 54 (100) | 11 (91.7) | 3 (100) | 27 (93.1) | 12 (41.4) | 3 (60) | <0.001 |
| H30Rx | 88 | 54 (100) | 8 (66.7) | 2 (66.7) | 16 (55.2) | 6 (20.7) | 2 (40) | <0.001 |
| H30R (non-Rx) | 19 | 0 (0) | 3 (25) | 1 (33.3) | 11 (37.9) | 3 (10.3) | 1 (20) | 0.006 |
| H22 | 9 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 9 (31) | 0 (0) | <0.001 |
| H27 | 2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (40) | <0.001 |
| H41 | 11 | 0 (0) | 1 (8.3) | 0 (0) | 2 (6.9) | 8 (27.6) | 0 (0) | 0.001 |
Includes one publicly available sequence of E. coli ST131.
Publicly available sequence data for E. coli ST131.
One isolate from Austria belonged to H31.
The P values are comparing the prevalence of each antibiotic resistance gene among geographical regions. P values in boldface type represent statistically significant results.
Antimicrobial susceptibility and antibiotic resistance genes.
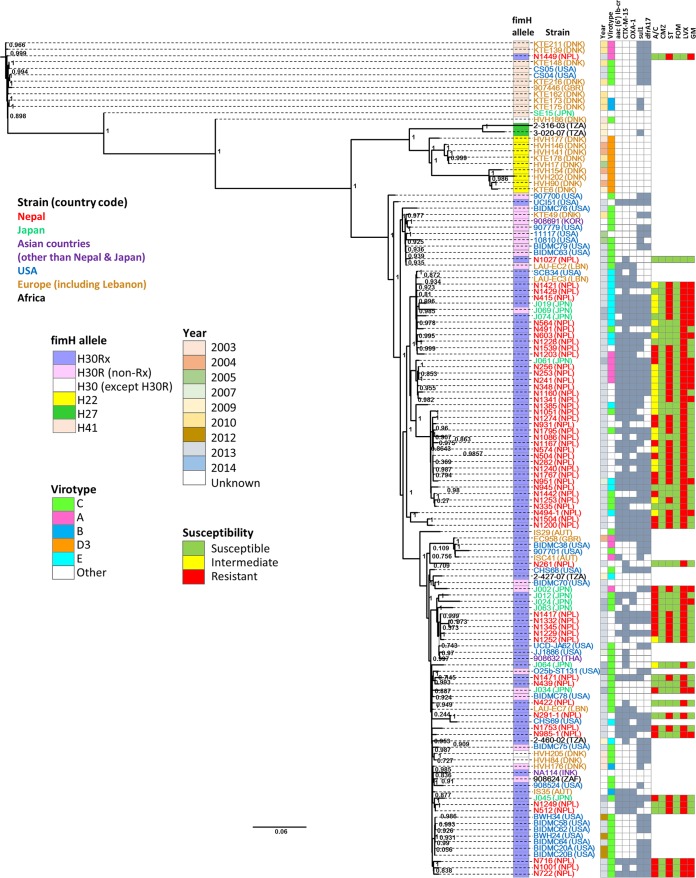
Antimicrobial susceptibility data were available for 54 and 11 ESBL-E. coli ST131 isolates from Nepal and Japan, respectively (Fig. 1). The susceptibilities of the 105 ESBL-E. coli isolates (including 54 ESBL-E. coli ST131 isolates described in this study) from Nepal were described elsewhere (5). The susceptibilities to trimethoprim-sulfamethoxazole, gentamicin, and levofloxacin were not significantly different in isolates from Nepal and Japan. All ESBL-E. coli ST131 isolates from Nepal and Japan were susceptible to cefmetazole and fosfomycin.
FIGURE 1 .
Phylogenetic trees of ST131 strains. ML phylogenetic trees were estimated using PHYML 3.0 and SH statistics for branch support. Antibiotics are abbreviated as follows: A/C, amoxicillin and clavulanate; CMZ, cefmetazole; ST, trimethoprim-sulfamethoxazole; FOM, fosfomycin; LVX, levofloxacin; GM, gentamicin. The countries are shown in parentheses after the strain name and abbreviated as follows: DNK, Denmark; NPL, Nepal; GBR, Great Britain; JPN, Japan; TZA, Tanzania; KOR, South Korea; LBN, Lebanon; AUT, Austria; THA, Thailand. Susceptibility data are available only for the 11 ESBL-E. coli ST131 isolates from the National Center for Global Health and Medicine, Japan, and 54 ESBL-E. coli ST131 isolates from Tribhuvan University, Nepal. For isolates from Japan, ampicillin-sulbactam was used instead of amoxicillin-clavulanate. For resistance genes, a filled square means positive for the resistance gene and an open square means negative for the resistance gene.
The frequency of resistance genes among the E. coli ST131 isolates is summarized in Table 2. More than 94% (n = 51) and 66% (n = 8) of ESBL-E. coli ST131 isolates were positive for blaCTX-M-15 in Nepal and Japan, respectively. Overall, 50 (70%) of the 71 blaCTX-M-15-positive isolates in the entire cohort were also positive for blaOXA-1 and aac(6′)-Ib-cr (encoding an aminoglycoside/fluoroquinolone acetyltransferase); of these 71 isolates, 38 isolates were obtained from Nepal, 5 isolates from Japan, and 6 isolates from Europe.
TABLE 2 .
Comparison of resistance genes in Escherichia coli ST131
| Antibiotic(s) and antibiotic resistance gene | fimH allele or sublineage | Total no. of isolates (n = 132) | No. of isolates (%) from: |
P valued | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nepal (n = 54) | Japan (n = 12)a | Other Asian countries (n = 3)b | USA (n = 29)b | Europe (n = 29)b,c | Africa (n = 5)b | ||||
| Aminoglycoside | |||||||||
| aac(3)-IIa | Total | 26 | 15 (27.8) | 5 (41.7) | 0 | 3 (10.3) | 3 (10.3) | 0 | 0.05 |
| H30Rx | 25 | 15 (27.8) | 4 (33.3) | 3 (10.3) | 3 (10.3) | ||||
| H30R | 1 | 1 (8.3) | |||||||
| aac(3)-IId | Total | 17 | 1 (1.9) | 2 (16.7) | 1 (33.3) | 5 (17.2) | 5 (17.2) | 3 (60) | 0.003 |
| H30Rx | 3 | 1 (1.9) | 1 (3.4) | 1 (20) | |||||
| H30R | 6 | 2 (16.7) | 1 (33.3) | 2 (6.9) | 1 (20) | ||||
| H27 | 1 | 1 (20) | |||||||
| H41 | 7 | 2 (6.9) | 5 (17.2) | ||||||
| aadA1 | Total | 5 | 0 | 0 | 0 | 2 (6.9) | 3 (10.3) | 0 | 0.213 |
| H30Rx | 1 | 1 (3.4) | |||||||
| H30R | 3 | 2 (6.9) | 1 (3.4) | ||||||
| H41 | 1 | 1 (3.4) | |||||||
| aadA2 | Total | 11 | 8 (14.8) | 0 | 0 | 2 (6.9) | 1 (3.4) | 0 | 0.329 |
| H30Rx | 8 | 8 (14.8) | |||||||
| H30R | 3 | 2 (6.9) | 1 (3.4) | ||||||
| aadA5 | Total | 68 | 31 (57.4) | 6 (50) | 0 | 21 (72.4) | 9 (31) | 1 (20) | 0.007 |
| H30Rx | 51 | 31 (57.4) | 4 (33.3) | 13 (44.8) | 2 (6.9) | 1 (20) | |||
| H30R | 9 | 2 (16.7) | 6 (20.7) | 1 (3.4) | |||||
| H41 | 8 | 2 (6.9) | 4 (13.8) | ||||||
| strA | Total | 45 | 20 (37) | 2 (16.7) | 1 (33.3) | 13 (44.8) | 6 (20.7) | 3 (60) | 0.211 |
| H30Rx | 28 | 20 (37) | 1 (8.3) | 6 (20.7) | 1 (20) | ||||
| H30R | 7 | 1 (8.3) | 1 (33.3) | 5 (17.2) | |||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 8 | 2 (6.9) | 6 (20.7) | ||||||
| strB | Total | 49 | 20 (37) | 2 (16.7) | 1 (33.3) | 15 (51.7) | 7 (24.1) | 4 (80) | 0.054 |
| H30Rx | 30 | 20 (37) | 1 (8.3) | 8 (27.6) | 1 (20) | ||||
| H30R | 3 | 1 (8.3) | 1 (33.3) | 1 (20) | |||||
| H22 | 5 | 5 (17.2) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 9 | 2 (6.9) | 7 (24.1) | ||||||
| Aminoglycoside and fluoroquinolone | |||||||||
| aac(6')-Ib-cr | Total | 55 | 39 (72.2) | 6 (50) | 0 | 3 (10.3) | 7 (24.1) | 0 | <0.001 |
| H30Rx | 53 | 39 (72.2) | 5 (41.7) | 3 (10.3) | 6 (20.7) | ||||
| H30R | 2 | 1 (8.3) | 1 (3.4) | ||||||
| Beta-lactams | |||||||||
| blaCTX-M-14 | Total | 2 | 0 | 0 | 0 | 0 | 2 (6.9) | 0 | 0.205 |
| H41 | 2 | 2 (6.9) | |||||||
| blaCTX-M-15 | Total | 71 | 51 (94.4) | 8 (66.7) | 1 (33.3) | 4 (13.8) | 6 (20.7) | 1 (20) | <0.001 |
| H30Rx | 69 | 51 (94.4) | 7 (58.3) | 1 (33.3) | 4 (13.8) | 5 (17.2) | 1 (20) | ||
| H30R | 2 | 1 (8.3) | 1 (3.4) | ||||||
| blaCTX-M-27 | Total | 3 | 2 (3.7) | 0 | 0 | 1 (3.4) | 0 | 0 | 0.874 |
| H30Rx | 2 | 2 (3.7) | |||||||
| H30R | 1 | 1 (3.4) | |||||||
| blaCTX-M-55 | Total | 4 | 0 | 3 (25) | 1 (33.3) | 0 | 0 | 0 | <0.001 |
| H30Rx | 1 | 1 (8.3) | |||||||
| H30R | 3 | 2 (16.7) | 1 (33.3) | ||||||
| blaOXA-1 | Total | 54 | 38 (70.4) | 7 (58.3) | 0 | 3 (10.3) | 6 (20.7) | 0 | <0.001 |
| H30Rx | 52 | 38 (70.4) | 6 (50) | 3 (10.3) | 5 (17.2) | ||||
| H30R | 2 | 1 (8.3) | 1 (3.4) | ||||||
| blaTEM-1B | Total | 61 | 20 (37) | 3 (25) | 1 (33.3) | 16 (55.2) | 16 (55.2) | 5 (100) | 0.036 |
| H30Rx | 33 | 20 (37) | 1 (8.3) | 6 (20.7) | 4 (13.8) | 2 (40) | |||
| H30R | 13 | 2 (16.7) | 1 (33.3) | 8 (27.6) | 1 (3.4) | 1 (20) | |||
| H22 | 1 | 1 (3.4) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 10 | 2 (6.9) | 8 (27.6) | ||||||
| blaTEM-1C | Total | 3 | 0 | 0 | 0 | 0 | 3 (10.3) | 0 | 0.05 |
| H22 | 3 | 3 (10.3) | |||||||
| blaKPC-3 | Total | 6 | 0 | 0 | 0 | 6 (20.7) | 0 | 0 | <0.001 |
| H30Rx | 6 | 6 (20.7) | |||||||
| Macrolide | |||||||||
| mphA | Total | 69 | 46 (85.2) | 5 (41.7) | 0 | 10 (34.5) | 8 (27.6) | 0 | <0.001 |
| H30Rx | 56 | 46 (85.2) | 3 (25) | 5 (17.2) | 2 (6.9) | ||||
| H30R | 7 | 2 (16.7) | 4 (13.8) | 1 (3.4) | |||||
| H41 | 4 | 1 (3.4) | 3 (10.3) | ||||||
| Chloramphenicol | |||||||||
| catA1 | Total | 4 | 0 | 0 | 0 | 0 | 2 (6.9) | 2 (40) | <0.001 |
| H30Rx | 1 | 1 (20) | |||||||
| H27 | 1 | 1 (20) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| catB3 | Total | 51 | 37 (68.5) | 6 (50) | 0 | 2 (6.9) | 6 (20.7) | 0 | <0.001 |
| H30Rx | 48 | 37 (68.5) | 5 (41.7) | 1 (3.4) | 5 (17.2) | ||||
| H30R | 3 | 1 (8.3) | 1 (3.4) | 1 (3.4) | |||||
| Sulfonamide | |||||||||
| sul1 | Total | 82 | 39 (72.2) | 6 (50) | 0 | 22 (75.9) | 13 (44.8) | 2 (40) | 0.01 |
| H30Rx | 60 | 39 (72.2) | 4 (33.3) | 13 (44.8) | 3 (10.3) | 1 (20) | |||
| H30R | 11 | 2 (16.7) | 7 (24.1) | 2 (6.9) | |||||
| H27 | 1 | 1 (20) | |||||||
| H41 | 8 | 2 (6.9) | 6 (20.7) | ||||||
| sul2 | Total | 52 | 20 (37) | 2 (16.7) | 1 (33.3) | 18 (62.1) | 8 (27.6) | 3 (60) | 0.041 |
| H30Rx | 32 | 20 (37) | 1 (8.3) | 9 (31) | 1 (3.4) | 1 (20) | |||
| H30R | 9 | 1 (8.3) | 1 (33.3) | 7 (24.1) | |||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 9 | 2 (6.9) | 7 (24.1) | ||||||
| Trimethoprim | |||||||||
| dfrA12 | Total | 8 | 8 (14.8) | 0 | 0 | 0 | 0 | 0 | 0.031 |
| H30Rx | 8 | 8 (14.8) | |||||||
| dfrA17 | Total | 67 | 31 (57.4) | 6 (50) | 0 | 20 (69) | 9 (31) | 1 (20) | 0.014 |
| H30Rx | 50 | 31 (57.4) | 4 (33.3) | 12 (41.3) | 2 (6.9) | 1 (20) | |||
| H30R | 9 | 2 (16.7) | 6 (20.7) | 1 (3.4) | |||||
| H41 | 6 | 2 (6.9) | 4 (13.8) | ||||||
| dfrA1 | Total | 5 | 0 | 0 | 0 | 1 (3.4) | 4 (13.8) | 0 | 0.054 |
| H30Rx | 1 | 1 (3.4) | |||||||
| H30R | 1 | 1 (3.4) | |||||||
| H22 | 1 | 1 (3.4) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| Tetracycline | |||||||||
| tet(A) | Total | 86 | 40 (74.1) | 8 (66.7) | 1 (33.3) | 18 (62.1) | 15 (51.7) | 4 (80) | 0.292 |
| H30Rx | 61 | 40 (74.1) | 7 (58.3) | 11 (37.9) | 2 (6.9) | 1 (20) | |||
| H30R | 10 | 1 (8.3) | 1 (33.3) | 5 (17.2) | 2 (6.9) | 1 (20) | |||
| H22 | 4 | 4 (13.8) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 9 | 2 (6.9) | 7 (24.1) | ||||||
Includes one publicly available sequence of E. coli ST131.
Publicly available sequence data for E. coli ST131.
Three isolates from Europe were from Lebanon.
The P values are comparing the prevalence of each antibiotic resistance gene among geographical regions. P values in boldface type represent statistically significant results.
The aforementioned study from Japan (9) revealed higher prevalence of blaCTX-M-14 and blaCTX-M-27 in addition to blaCTX-M-15 among ESBL-E. coli isolates collected from another area in Japan. A study from Korea on E. coli isolates collected from 2012 to 2013 suggested that the majority of H30Rx isolates harbored blaCTX-M-15, whereas about half of the H30 non-Rx isolates harbored blaCTX-M-14 or blaCTX-M-27 (11). Geographical differences and different times of these studies may account for these discrepancies.
Virotypes and virulence-associated genes.
The distribution of virotypes and virulence-associated genes among the E. coli ST131 strains is summarized in Table 3. The predominant virotypes differed across the geographical regions. The majority of E. coli ST131 isolates from the United States and half of those from Japan belonged to virotype C, whereas almost half of the isolates from Nepal belonged to virotypes other than A, B, C, D, and E. Two-thirds of E. coli ST131 isolates from Europe belonged to either virotype C or D. The virulence-associated genes such as iha, sat, fyuA, traT, ompT, and malT were highly prevalent among isolates from most geographical regions. The papGII gene was frequently observed only among isolates from Nepal and Tanzania, and hlyA and cnf1 were common only in E. coli ST131 isolates obtained from Africa. Most of the virotypes obtained in our study belonged to virotype C (Table 1). Our finding that virotype C is the most prevalent virotype among E. coli ST131 isolates is consistent with previous reports on virotype distribution in ESBL-E. coli ST131 (4, 12). As previously suggested (4, 12), our results also suggested the association between virotype C and H30Rx sublineage. In our study, more isolates were identified as “other” virotype than previous studies (4, 12), which might be due to the different methodology used (e.g., WGS versus PCR). The majority of the ESBL-E. coli ST131 isolates (n = 28 [52%]) from Nepal were positive for papGII and sat but were negative for hlyA, and thus, were not categorized into any of the previously described virotypes (A to E) (13).
TABLE 3 .
Comparison of virotypes and virulence-associated traits and genes in Escherichia coli ST131
| Virotype or virulence-associated trait and gene | fimH allele or sublineage | Total no. of isolates (n = 132) | No. of isolates (%) from: |
P valued | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nepal (n = 54) | Japan (n = 12)a | Other Asian countries (n = 3)b | USA (n = 29)b | Europe (n = 29)b,c | Africa (n = 5)b | ||||
| Virotypes | |||||||||
| A | Total | 12 | 5 (9.3) | 2 (16.7) | 0 | 1 (3.4) | 4 (13.8) | 0 | 0.619 |
| H30Rx | 9 | 5 (9.3) | 1 (8.3) | 1 (3.4) | 2 (6.9) | ||||
| H30R | 1 | 1 (8.3) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| B | Total | 4 | 0 | 0 | 0 | 0 | 4 (13.8) | 0 | 0.012 |
| H30Rx | 1 | 0 | 1 (3.4) | ||||||
| H30R | 1 | 1 (3.4) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| C | Total | 54 | 13 (24.1) | 6 (50) | 2 (66.7) | 23 (79.3) | 9 (31) | 1 (20) | <0.001 |
| H30Rx | 33 | 13 (24.1) | 5 (41.7) | 1 (33.3) | 12 (41.4) | 2 (6.9) | |||
| H30R | 14 | 1 (8.3) | 1 (33.3) | 9 (31) | 2 (6.9) | 1 (20) | |||
| H41 | 4 | 2 (6.9) | 2 (6.9) | ||||||
| De | Total | 9 | 0 | 0 | 0 | 0 | 9 (31) | 0 | <0.001 |
| H22 | 9 | 9 (31) | |||||||
| E | Total | 18 | 10 (18.5) | 3 (25) | 0 | 2 (6.9) | 1 (3.4) | 2 (40) | 0.09 |
| H30Rx | 17 | 10 (18.5) | 2 (16.7) | 2 (6.9) | 1 (3.4) | 2 (40) | |||
| H30R | 1 | 1 (8.3) | |||||||
| Other | Total | 35 | 26 (48.1) | 1 (8.3) | 1 (33.3) | 3 (10.3) | 2 (6.9) | 2 (40) | <0.001 |
| H30Rx | 28 | 26 (48.1) | 1 (33.3) | 1 (3.4) | |||||
| H30R | 2 | 2 (6.9) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 3 | 1 (8.3) | 2 (6.9) | ||||||
| Virulence-associated traits and genes | |||||||||
| Adhesin | |||||||||
| papGII | Total | 46 | 35 (64.8) | 3 (25) | 1 (33.3) | 2 (6.9) | 1 (3.4) | 4 (80) | <0.001 |
| H30Rx | 43 | 35 (64.8) | 2 (16.7) | 1 (33.3) | 2 (6.9) | 1 (3.4) | 2 (40) | ||
| H30R | 1 | 1 (8.3) | |||||||
| H27 | 2 | 2 (40) | |||||||
| iha | Total | 119 | 53 (98.1) | 10 (83.3) | 3 (100) | 27 (93.1) | 24 (82.8) | 2 (40) | 0.001 |
| H30Rx | 86 | 53 (98.1) | 8 (66.7) | 2 (66.7) | 16 (55.2) | 6 (20.7) | 1 (20) | ||
| H30R | 16 | 2 (16.7) | 1 (33.3) | 9 (31) | 3 (10.3) | 1 (20) | |||
| H22 | 4 | 4 (13.8) | |||||||
| H41 | 10 | 2 (6.9) | 8 (27.6) | ||||||
| hra | Total | 33 | 11 (20.4) | 3 (25) | 0 | 8 (27.6) | 0 | 1 (20) | 0.085 |
| H30Rx | 11 | 3 (25) | 8 (27.6) | ||||||
| H27 | 1 | 1 (20) | |||||||
| Toxin | |||||||||
| hlyA | Total | 20 | 10 (18.5) | 3 (25) | 0 | 2 (6.9) | 1 (3.4) | 4 (80) | <0.001 |
| H30Rx | 7 | 2 (16.7) | 2 (6.9) | 1 (3.4) | 2 (40) | ||||
| H30R | 1 | 1 (8.3) | |||||||
| H27 | 2 | 2 (40) | |||||||
| cnf1 | Total | 19 | 9 (16.7) | 3 (25) | 0 | 2 (6.9) | 1 (3.4) | 4 (80) | <0.001 |
| H30Rx | 7 | 2 (16.7) | 2 (6.9) | 1 (3.4) | 2 (40) | ||||
| H30R | 1 | 1 (8.3) | |||||||
| H27 | 2 (40) | ||||||||
| sat | Total | 118 | 54 (100) | 10 (83.3) | 3 (100) | 26 (89.7) | 22 (75.9) | 3 (60) | 0.004 |
| H30Rx | 33 | 8 (66.7) | 2 (66.7) | 15 (51.7) | 6 (20.7) | 2 (40) | |||
| H30R | 16 | 2 (16.7) | 1 (33.3) | 9 (31) | 3 (10.3) | 1 (20) | |||
| H22 | 4 | 4 (13.8) | |||||||
| H41 | 8 | 2 (6.9) | 6 (20.7) | ||||||
| Siderophore | |||||||||
| iroN | Total | 11 | 0 | 0 | 0 | 0 | 11 (37.9) | 0 | <0.001 |
| H30Rx | 1 | 1 (3.4) | |||||||
| H30R | 1 | 1 (3.4) | |||||||
| H22 | 7 | 7 (24.1) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| fyuA | Total | 130 | 53 (98.1) | 12 (100) | 3 (100) | 29 (100) | 28 (96.6) | 5 (100) | 0.911 |
| H30Rx | 87 | 53 (98.1) | 8 (66.7) | 2 (66.7) | 16 (55.2) | 6 (20.7) | 2 (40) | ||
| H30R | 19 | 3 (25) | 1 (33.3) | 11 (37.9) | 3 (10.3) | 1 (20) | |||
| H22 | 9 | 9 (31) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 11 | 1 (8.3) | 2 (6.9) | 8 (27.6) | |||||
| ireA | Total | 3 | 2 (3.7) | 0 | 1 (33.3) | 0 | 0 | 0 | 0.009 |
| H30Rx | 3 | 2 (3.7) | 1 (33.3) | ||||||
| Protectins | |||||||||
| kfiA | Total | 43 | 13 (23.6) | 4 (33.3) | 0 | 13 (44.8) | 9 (31) | 4 (80) | 0.066 |
| H30Rx | 28 | 13 (23.6) | 2 (16.7) | 9 (31) | 2 (6.9) | 2 (40) | |||
| H30R | 8 | 2 (16.7) | 4 (13.8) | 1 (3.4) | 1 (20) | ||||
| H22 | 4 | 4 (13.8) | |||||||
| H27 | 1 | 1 (20) | |||||||
| iss | Total | 9 | 0 | 0 | 1 (33.3) | 0 | 8 (27.6) | 0 | <0.001 |
| H30Rx | 1 | 1 (33.3) | |||||||
| H30R | 1 | 1 (3.4) | |||||||
| H22 | 5 | 5 (17.2) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| Invasin | |||||||||
| ibeA | Total | 9 | 0 | 0 | 0 | 0 | 9 (31) | 0 | <0.001 |
| H22 | 9 | 9 (31) | |||||||
| Miscellaneous | |||||||||
| traT | Total | 107 | 47 (87) | 10 (83.3) | 1 (33.3) | 17 (58.6) | 29 (100) | 3 (60) | <0.001 |
| H30Rx | 68 | 47 (87) | 7 (58.3) | 6 (20.7) | 6 (20.7) | 2 (40) | |||
| H30R | 16 | 3 (25) | 1 (33.3) | 9 (31) | 3 (10.3) | ||||
| H22 | 9 | 9 (31) | |||||||
| H27 | 1 | 1 (20) | |||||||
| H41 | 10 | 2 (6.9) | 8 (27.6) | ||||||
| ompT | Total | 129 | 54 (100) | 12 (100) | 2 (66.7) | 28 (96.6) | 28 (96.6) | 5 (100) | 0.01 |
| H30Rx | 87 | 54 (100) | 8 (66.7) | 1 (33.3) | 16 (55.2) | 6 (20.7) | 2 (40) | ||
| H30R | 18 | 3 (25) | 1 (33.3) | 10 (34.5) | 3 (10.3) | 1 (20) | |||
| H22 | 9 | 9 (31) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 11 | 1 (8.3) | 2 (6.9) | 8 (27.6) | |||||
| malX | Total | 131 | 54 (100) | 12 (100) | 3 (100) | 29 (100) | 28 (96.6) | 5 (100) | 0.911 |
| H30Rx | 88 | 54 (100) | 8 (66.7) | 2 (66.7) | 16 (55.2) | 6 (20.7) | 2 (40) | ||
| H30R | 19 | 3 (25) | 1 (33.3) | 11 (37.9) | 3 (10.3) | 1 (20) | |||
| H22 | 9 | 9 (31) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 11 | 1 (8.3) | 2 | 8 (27.6) | |||||
Includes one publicly available sequence of E. coli ST131.
Publicly available sequence data for E. coli ST131.
Three isolates from Europe were from Lebanon.
The P values are comparing the prevalence of each antibiotic resistance gene among geographical regions. P values in boldface type represent statistically significant results.
All isolates of virotype D were identified as D3.
Plasmid replicon types.
The distribution of plasmid replicon types was similar across geographical regions except that FIA was prevalent in Nepal and the United States and considerably less prevalent in Europe; other geographical regions showed intermediate prevalence (Table 4). FII was commonly found among ESBL-E. coli ST131 isolates from Nepal, whereas it was considerably less prevalent in Japan. IncP was detected in 40% of E. coli ST131 isolates from Africa, but it was very rarely found or not found in isolates from other areas.
TABLE 4 .
Comparison of plasmid replicon types of Escherichia coli ST131
| Replicon type | fimH allele or sublineage | Total no. of isolates | No. of isolates (%) from: |
P valued | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nepal (n = 54) | Japan (n = 12)a | Other Asian countries (n = 3)b | USA (n = 29)b | Europe (n = 29)b,c | Africa (n = 5)b | ||||
| IncFIA | Total | 84 | 38 (70.4) | 6 (50) | 2 (66.7) | 25 (86.2) | 10 (34.5) | 3 (60) | 0.002 |
| H30Rx | 62 | 38 (70.4) | 4 (33.3) | 1 (33.3) | 14 (48.3) | 3 (10.3) | 2 (40) | ||
| H30R | 17 | 2 (16.7) | 1 (33.3) | 11 (37.9) | 2 (6.9) | 1 (20) | |||
| H41 | 2 | 2 (6.9) | |||||||
| IncFIB | Total | 89 | 40 (74.1) | 8 (66.7) | 1 (33.3) | 14 (48.3) | 23 (79.3) | 3 (60) | 0.09 |
| H30Rx | 48 | 40 (74.1) | 5 (41.7) | 2 (6.9) | 1 (3.4) | ||||
| H30R | 18 | 3 (25) | 1 (33.3) | 10 (34.5) | 3 (10.3) | 1 (20) | |||
| H22 | 9 | 9 (31) | |||||||
| H27 | 2 | 2 (40) | |||||||
| H41 | 10 | 2 (6.9) | 8 (27.6) | ||||||
| IncFIC | Total | 3 | 0 | 0 | 0 | 0 | 3 (10.3) | 0 | 0.053 |
| H22 | 1 | 1 (3.4) | |||||||
| H41 | 2 | 2 (6.9) | |||||||
| IncFII | Total | 95 | 43 (79.6) | 4 (33.3) | 1 (33.3) | 23 (79.3) | 21 (72.4) | 3 (60) | 0.017 |
| H30Rx | 60 | 43 (79.6) | 2 (16.7) | 11 (37.9) | 2 (6.9) | 2 (40) | |||
| H30R | 17 | 2 (16.7) | 1 (33.3) | 10 (34.5) | 3 (10.3) | 1 (20) | |||
| H22 | 8 | 8 (27.6) | |||||||
| H41 | 8 | 2 (6.9) | 6 (20.7) | ||||||
| IncI1 | Total | 9 | 3 (5.6) | 2 (16.7) | 1 (33.3) | 1 (3.4) | 2 (6.9) | 0 | 0.29 |
| H30Rx | 5 | 3 (5.6) | 1 (8.3) | 1 (3.4) | |||||
| H30R | 2 | 1 (8.3) | 1 (33.3) | ||||||
| H41 | 2 | 2 (6.9) | |||||||
| IncFrepB | Total | 40 | 13 (24.1) | 3 (25) | 0 | 15 (51.7) | 7 (24.1) | 2 (40) | 0.091 |
| H30Rx | 32 | 13 (24.1) | 2 (16.7) | 12 (41.4) | 3 (10.3) | 2 (40) | |||
| H30R | 4 | 1 (8.3) | 3 (10.3) | ||||||
| H22 | 4 | 4 (13.8) | |||||||
| IncB/O | Total | 1 | 0 | 0 | 0 | 1 (3.4) | 0 | 0 | 0.611 |
| H30R | 1 | 1 (3.4) | |||||||
| IncY | Total | 2 | 0 | 0 | 0 | 1 (3.4) | 1 (3.4) | 0 | 0.763 |
| H30Rx | 1 | 1 (3.4) | |||||||
| H30R | 1 | 1 (3.4) | |||||||
| IncN | Total | 8 | 0 | 2 (16.7) | 0 | 2 (6.9) | 4 (13.8) | 0 | 0.092 |
| H30Rx | 6 | 1 (8.3) | 2 (6.9) | 3 (10.3) | |||||
| H30R | 1 | 1 (8.3) | |||||||
| H22 | 1 | 1 (3.4) | |||||||
| IncP | Total | 3 | 0 | 0 | 0 | 0 | 1 (3.4) | 2 (40) | <0.001 |
| H30R | 1 | 1 (3.4) | |||||||
| H27 | 2 | 2 (40) | |||||||
| IncA/C | Total | 2 | 0 | 0 | 0 | 2 (6.9) | 0 | 0 | 0.205 |
| H30R | 2 | 2 (6.9) | |||||||
Includes one publicly available sequence of E. coli ST131.
Publicly available sequence data for E. coli ST131.
Three isolates from Europe were from Lebanon.
The P values are comparing the prevalence of each antibiotic resistance gene among geographical regions. P values in boldface type represent statistically significant results.
The association of blaCTX-M-15 and IncF replicon has been reported previously (14), and the H30Rx sublineage was created by introduction of IncF (15). We previously found that a plasmid resembling pEC958 and harboring FIA and FII (16) was present in approximately 80% of the ESBL-E. coli ST131 isolates from Nepal (5). The high prevalence of IncF (IncFIA, FIB, and FII) observed among ESBL-E. coli ST131 isolates from Nepal is consistent with these previous reports and is reasonable considering the high prevalence of H30Rx among ESBL-E. coli ST131 isolates.
Phylogenetic analysis of E. coli ST131 in different geographical regions.
In the phylogenetic analysis based on WGS (Fig. 1), the majority of ESBL-E. coli ST131 isolates from Nepal clustered together, whereas those from Japan were more diverse. Thus, ESBL-E. coli ST131 may have been introduced into Japan more sporadically over time. Strict requirements for antibiotic prescription in Japan might have resulted in relatively low selective pressure, along with more advanced medical and social infrastructure. Carriers of ESBL-E. coli are usually placed under contact precaution in health care facilities in Japan, including the hospital in this study, to minimize transmission. These factors might explain at least in part the differences in the prevalence and clonality of ST131 between these two Asian countries. In contrast, prevalent clonal spread of ESBL-E. coli appears to have occurred in Nepal, where poor infection control practices and sanitation might facilitate dissemination of antimicrobial-resistant strains. In Nepal, antibiotics can be purchased in the community at general retail stores and pharmacies. According to recent reports, 80% of the drugs are purchased outside the government-supplied health system (17, 18). In addition, inappropriate prescription occurs in up to 40% of patients (17, 18).
E. coli ST131 isolates from the United States are distributed across the phylogeny, even those isolates that were collected in the same year. The wide diversity of population in the United States might explain in part this phylogenetic characteristic. E. coli ST131 isolates from Europe (including three E. coli ST131 isolates from Lebanon) could be roughly divided into four clusters, with one major cluster consisting mainly of E. coli ST131 isolates from Denmark and one cluster consisting of two E. coli ST131 isolates from Lebanon. E. coli ST131 isolates from Denmark predominantly consisted of E. coli ST131 from Europe (72%). Four E. coli ST131 isolates from Tanzania and one isolate from South Africa were included in E. coli ST131 from Africa. E. coli ST131 isolates from Africa were phylogenetically diverse. E. coli ST131 isolates from Asian countries other than Nepal and Japan included one isolate from South Korea, one isolate from India, and one isolate from Thailand. They did not belong to the same cluster.
There are several limitations to this study. Since ESBL-E. coli ST131 isolates from Nepal and Japan were collected during a relatively short period of time, epidemiologically related isolates may have been included in the analysis, and the trends across years could not be elucidated. However, as shown in Fig. 1, the distribution of ESBL-E. coli ST131 isolates in each region suggests that the contribution of such clonal isolates was unlikely to have been remarkable. Due to the small number of isolates from Japan, the power of the statistical comparisons between isolates from Nepal and Japan is limited. Also, only small numbers of E. coli ST131 isolates from some geographical regions such as Africa could be included due to limited publicly available data. Finally, the difference in the molecular analyses, i.e., WGS and PCR, used might have caused the difference in the identification of the target genes.
In conclusion, comparative analysis of ESBL-E. coli ST131 genomes from Japan and Nepal and those from other geographical regions revealed distinct phylogenetic characteristics of the spread of ESBL-E. coli ST131 in these two geographical areas of Asia. Multiple yet distinct factors might contribute to the local spread of ESBL-E. coli ST131 in each region.
MATERIALS AND METHODS
Isolates and susceptibility testing.
All ESBL-E. coli isolates were serially collected from the Tribhuvan University Teaching Hospital (444 beds), Kathmandu, Nepal, between 1 February 2013 and 31 July 2013 and from the National Center for Global Health and Medicine (NCGM) (781 beds), Tokyo, Japan, between 1 October 2013 and 30 September 2014; both centers are tertiary teaching hospitals. Some of the isolates from Nepal were included in our previous study (5).
The E. coli strains from both Nepal and Japan were identified, and their susceptibility was tested in accordance with the Clinical and Laboratory Standards Institute (CLSI) criteria (19) by using an automated broth microdilution system (MicroScan; Siemens AG, Germany) unless otherwise stated. The MIC of fosfomycin was determined using an NC6.11J panel (Siemens AG, Germany) which contains glucose-6-phosphate, and its susceptibility status was determined as previously reported (20). ESBL production was confirmed by disc diffusion tests in accordance with the 2009 CLSI criteria (19). If multiple ESBL-E. coli isolates were identified from a patient during the study period, only the first isolate was included.
Whole-genome sequencing and phylogenetic analysis.
Molecular analysis was conducted at the Department of Infectious Diseases, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan. The strains were cultured overnight in lysogeny broth (LB) (Nakarai Tesque, Kyoto, Japan), and genomic DNA was purified using a DNeasy blood and tissue kit (Qiagen, Venlo, Netherlands). The genomes of the isolates were then subjected to MiSeq sequencing by using Nextera XT library kits (Illumina, Inc., San Diego, CA), according to the manufacturer’s instructions. Approximately 1 million paired-end reads (301 bp × 2) were obtained from each genome and analyzed using the CLC Genomics Workbench software (CLC Bio, Aarhus, Denmark). The reads from each isolate were trimmed by screening for base quality (quality score limit of 0.05; reads that contained greater than two ambiguous nucleotides or that were less than 15 bp in length were removed) (21). The resulting sequencing data were registered with the DNA Data Bank of Japan (DDBJ) (DDBJ accession no. DRA003515). Velvet and Mummer (open source MUMmer 3.0) described previously were used for de novo assembly to prepare contigs and single nucleotide polymorphism (SNP) calling, respectively (21, 22).
All genome sequence data for ST131 strains available up to January 2015 were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/genome/genomes/167?genome_assembly_id=group161531; accessed 26 September 2016) and included for phylogenetic analysis to determine the sequence diversity. SNP concatemers were prepared using a custom script and aligned using MAFFT (23). E. coli SE15 (accession no. NC_013654.1) was used as the reference to call SNPs of the isolates after mobile elements were removed from the chromosome. The appropriate evolutionary model (transversion model plus gamma distribution) was determined using jModelTest2 (24). Maximum likelihood (ML) phylogenetic trees were estimated using PHYML 3.0 (25). Branch support for nodes was assessed using the Shimodaira and Hasegawa (SH) test implemented in PHYML.
The contigs were subjected to further analyses by using the BLAST algorithm (26) and ResFinder (27) to identify whether virulence marker genes and drug resistance genes were present in the genomes (28, 29). An identification rate of more than 97% was considered positive for each target gene. Phylotypes (30), virulence genotypes (31), fimH alleles and H30R and H30Rx sublineages of ST131 (10), and distribution of acquired drug resistance genes were determined. The nucleotide sequences corresponding to the ST131 multilocus sequence typing (MLST) allelic profile (adk53, fumC40, gyrB47, icd13, mdh36, purA28, and recA29) were downloaded from the University of Warwick (http://mlst.warwick.ac.uk/mlst/) and compared by BLAST algorithm to the WGS data (3, 26). Virotypes were determined based on the virulence gene scheme (13). Sequence typing of plasmid replicons was performed as described elsewhere (32, 33).
We added one E. coli ST131 isolate from Japan for which sequence data were available publicly (http://www.ncbi.nlm.nih.gov/genome/genomes/167?genome_assembly_id=group161531; accessed 26 September 2016). SE15 is a completely sequenced ST131 representative strain, which was used as a reference in the current study and a previous study as well (3).
Statistical analysis.
All statistical analyses were performed using IBM-SPSS statistics 20 (2012). Bivariate analyses were performed using Fisher’s exact test or chi-square test for categorical variables and the Mann-Whitney U test for continuous variables. All P values were two sided. The percentage values included in this article are the “valid percentages,” which exclude the missing data.
Accession number(s).
Sequence data were deposited in the DNA Data Bank of Japan (DDBJ) under accession no. DRA003515.
ACKNOWLEDGMENTS
We thank Y. Sakurai for help in the preparation of the figure.
This work was supported in part by a grant in Clinical Epidemiology Research, St. Luke's International University, Tokyo, Japan (2016).
Funding Statement
This work was funded in part by St. Luke's International University, Tokyo, Japan.
REFERENCES
- 1.World Health Organization 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland: http://www.who.int/drugresistance/documents/surveillancereport/en/. [Google Scholar]
- 2.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 3.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernández V, de la Cruz F, Martínez-Martínez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, López-Cerero L, Pascual A, Rodríguez-Baño J, Spanish Group for Nosocomial Infections (GEIH). . 2013. Four main virotypes among extended-spectrum-beta-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51:3358–3367. doi: 10.1128/JCM.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherchan JB, Hayakawa K, Miyoshi-Akiyama T, Ohmagari N, Kirikae T, Nagamatsu M, Tojo M, Ohara H, Sherchand JB, Tandukar S. 2015. Clinical epidemiology and molecular analysis of extended-spectrum-beta-lactamase-producing Escherichia coli in Nepal: characteristics of sequence types 131 and 648. Antimicrob Agents Chemother 59:3424–3432. doi: 10.1128/AAC.00270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout JD, Campbell L, Church DL, Gregson DB, Laupland KB. 2009. Molecular characteristics of travel-related extended-spectrum-beta-lactamase-producing Escherichia coli isolates from the Calgary Health Region. Antimicrob Agents Chemother 53:2539–2543. doi: 10.1128/AAC.00061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 63:72–79. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 8.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group . 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-beta-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 67:2612–2620. doi: 10.1093/jac/dks278. [DOI] [PubMed] [Google Scholar]
- 9.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group . 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 10.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SY, Park YJ, Johnson JR, Yu JK, Kim YK, Kim YS. 2016. Prevalence and characteristics of Escherichia coli sequence type 131 and its H30 and H30Rx subclones: a multicenter study from Korea. Diagn Microbiol Infect Dis 84:97–101. doi: 10.1016/j.diagmicrobio.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Peirano G, van der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, Tchesnokova VL, Pitout JD. 2014. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother 58:3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mora A, Dahbi G, López C, Mamani R, Marzoa J, Dion S, Picard B, Blanco M, Alonso MP, Denamur E, Blanco J. 2014. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One 9:e87025. doi: 10.1371/journal.pone.0087025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcadé G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J Antimicrob Chemother 63:67–71. doi: 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- 15.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan MD, Forde BM, Peters KM, Sarkar S, Hancock S, Stanton-Cook M, Ben Zakour NL, Upton M, Beatson SA, Schembri MA. 2015. Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS One 10:e0122369. doi: 10.1371/journal.pone.0122369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alliance for the Prudent Use of Antibiotics (APUA) Nepal 2015. APUA Nepal. Alliance for the Prudent Use of Antibiotics, Boston, MA: http://www.tufts.edu/med/apua/intl_chapters/nepal.shtml. Accessed 20 September 2016. [Google Scholar]
- 18.GARP-Nepal National Working Group 2015. Situation analysis and recommendations. Antibiotic use and resistance in Nepal. Global Antibiotic Resistance Partnership (GARP), Center for Disease Dynamics, Economics & Policy, Washington, DC: http://cddep.org/publications/garp_nepal_situation_analysis#sthash.DKyQe07e.dpbs. Accessed 20 September 2016. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement. Approved standard M100-S19 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Kresken M, Pfeifer Y, Hafner D, Wresch R, Körber-Irrgang B, Working Party ‘Antimicrobial Resistance’ of the Paul-Ehrlich-Society for Chemotherapy . 2014. Occurrence of multidrug resistance to oral antibiotics among Escherichia coli urine isolates from outpatient departments in Germany: extended-spectrum beta-lactamases and the role of fosfomycin. Int J Antimicrob Agents 44:295–300. doi: 10.1016/j.ijantimicag.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 26.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JR, Gajewski A, Lesse AJ, Russo TA. 2003. Extraintestinal pathogenic Escherichia coli as a cause of invasive nonurinary infections. J Clin Microbiol 41:5798–5802. doi: 10.1128/JCM.41.12.5798-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JR, Johnston B, Clabots CR, Kuskowski MA, Roberts E, DebRoy C. 2008. Virulence genotypes and phylogenetic background of Escherichia coli serogroup O6 isolates from humans, dogs, and cats. J Clin Microbiol 46:417–422. doi: 10.1128/JCM.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]