Abstract
Aneuploidy, an imbalanced chromosome number, is associated with both cancer and developmental disorders such as Down syndrome (DS). To determine how aneuploidy affects cellular and organismal physiology, we have developed a system to evaluate aneuploid cell fitness in vivo. By transplanting hematopoietic stem cells (HSCs) into recipient mice after ablation of recipient hematopoiesis by lethal irradiation, we can directly compare the fitness of HSCs derived from a range of aneuploid mouse models with that of euploid HSCs. This experimental system can also be adapted to assess the interplay between aneuploidy and tumorigenesis. We hope that further characterization of aneuploid cells in vivo will provide insight both into the origins of hematopoietic phenotypes observed in DS individuals as well as the role of different types of aneuploid cells in the genesis of cancers of the blood.
Aneuploidy is a state in which cells harbor a chromosome number that is not a whole number multiple of the haploid chromosome complement. It is the cause of developmental disorders such as Down syndrome (DS; trisomy 21). Aneuploidy is also a feature of cancer cells and has been hypothesized to drive cancer cell proliferation (Boveri 1914). In recent years, research evaluating the effects of aneuploidy on cell physiology and investigating its role in cancer have greatly increased our understanding of this complicated cellular state. A number of model systems have been developed to study cells with increased rates of chromosome mis-segregation— chromosomal instability (CIN)—and the result of such chromosome mis-segregation events—aneuploidy—in tumorigenesis. However, the precise role of aneuploidy in cancer still remains unclear, as it has been shown to both promote and inhibit tumorigenesis in different model systems (Pfau and Amon 2012).
By studying aneuploidy per se, we have learned that whole chromosome gains and losses have a profound effect on cells and organisms (Santaguida and Amon 2015). Such large genomic alterations affect a multitude of cellular processes, from transcription and translation, to protein folding and degradation. Cells respond with a characteristic stress response, proliferating slowly and up-regulating protein quality control pathways. Because of its severe impact on cell physiology and development, aneuploidy is often lethal in multicellular organisms, and the few aneuploidies that do not cause lethality lead to reduced growth and developmental abnormalities.
To date, much of the work characterizing aneuploid cells has been performed in cell culture or in animal models of DS or CIN. While these systems have revealed some features of the aneuploid cell condition, they did not provide a complete picture of the effects of aneuploidy on cell physiology in vivo. Cell culture systems are limiting in that cells are studied outside the natural context of the organism. Mouse models of DS only permit study of the amplification of genes present on human chromosome 21. Mouse models of CIN do not solely examine the effects of aneuploidy but also evaluate events associated with chromosome mis-segregation such as DNA damage (Janssen et al. 2011; Crasta et al. 2012). Because of these limitations, it is important to develop additional model systems for the study of aneuploidy to better understand how aneuploidy affects cell physiology and to dissect the relationship between aneuploidy and cancer. Towards this goal, we have established an experimental system to study aneuploid cells in vivo. By utilizing transplantation, the fitness of a range of aneuploid hematopoietic stem cells (HSCs) can be evaluated in the context of an otherwise euploid organism. Further, this system has great potential to directly assess how aneuploidy affects the kinetics of tumorigenesis in leukemia and lymphoma.
HEMATOPOIETIC RECONSTITUTIONS TO STUDY ANEUPLOIDY IN VIVO
Study of the effects of aneuploidy on cellular fitness in vivo is difficult because most autosomal aneuploidies are embryonic lethal in mammals (Gropp 1982; Torres et al. 2008). We can circumvent this complication in mice by performing transplantation experiments with HSCs derived from aneuploid embryos, a procedure called an adoptive transfer. This model system is convenient for studying aneuploidy in vivo because HSCs can be isolated from embryos and transplanted into euploid adult recipient mice that have undergone lethal irradiation, which eliminates recipient hematopoiesis. The blood from recipients of aneuploid HSCs can then be evaluated over time to determine the potential of trisomic cells in this in vivo context.
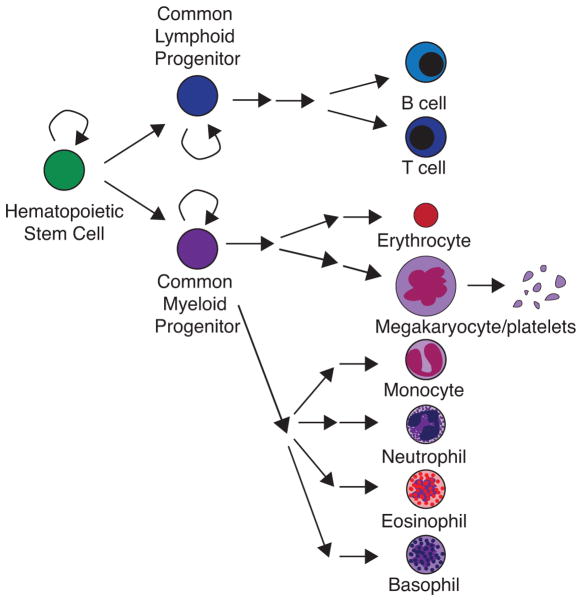
Blood contains many cell types, yet all blood cells are derived from a common precursor, the HSC. Hematopoiesis, the process by which blood is made, is a hierarchical differentiation pathway whereby HSCs divide asymmetrically to generate progenitor cells, which subsequently divide to amplify and also differentiate to give rise to all downstream cell types (Figure 1). HSCs are found in the bone marrow in adult mice and humans, where they are mostly quiescent (Sun et al. 2015; Busch et al. 2015); however, HSCs undergo several migrations during embryonic development to reach the bone marrow (Figure 2).
FIGURE 1. Hematopoiesis.
Hematopoiesis is initiated by a hematopoietic stem cell that can either divide to self-renew or to produce a cell that will become a common lymphoid progenitor or a common myeloid progenitor. These common progenitors further divide to self-renew or to differentiate into several types of committed progenitor cells that will ultimately give rise to all the differentiated cell types in the blood.
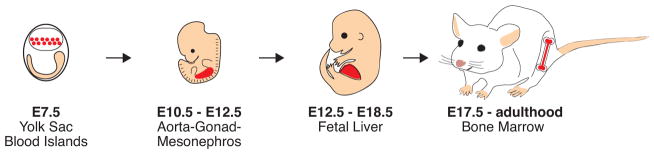
FIGURE 2. Ontogeny of the hematopoietic stem cell in mice.
Primitive HSCs arise around E7.5 in yolk sac blood islands (highlighted in red). These cells do not contribute to the adult HSC pool. Definitive HSCs are largely formed around E10.5 in the aorta-gonad-mesonephros (AGM) region (highlighted in red). These cells migrate to the fetal liver (highlighted in red), where they rapidly proliferate for several days before migrating to the bone marrow (highlighted in red) during late embryonic development. HSCs largely remain in the bone marrow throughout adulthood.
Primitive hematopoiesis is the process of forming blood cells that help sustain the embryo during development but do not ultimately contribute to the adult blood after birth. Primitive hematopoiesis begins around embryonic day 7.5 (E7.5) with the emergence of yolk sac blood islands, which primarily produce primitive erythroid cells (Baron et al. 2012; Mikkola and Orkin 2006). This production of red blood cells allows for proper oxygenization of embryonic tissues (Orkin and Zon 2008). Definitive hematopoiesis is the process of forming blood cells that both sustain the developing embryo and contribute to adult hematopoiesis after embryonic development. While there is evidence that HSCs in the yolk sac can also contribute to definitive hematopoiesis (Moore and Metcalf 1970; Palis et al. 1999; Samokhvalov et al. 2007), the majority of definitive HSCs are thought to arise from the aorta-gonad-mesonephros (AGM) region around E10.5 (Müller et al. 1994). These cells then migrate to the developing fetal liver around E11.5, where they rapidly expand for several days (Morrison et al. 1995) before gradually entering circulation around E15.5 (Christensen et al. 2004). HSCs then “home” to the bone marrow beginning around E17.5 (Christensen et al. 2004), where they are recruited to colonize the adult hematopoietic niche by interactions with niche cells (Lapidot 2005).
Much of what has been learned about HSCs has been gleaned from transplantation experiments. Following whole body irradiation, HSCs apoptose (Meng et al. 2003) or senesce (Wang et al. 2006), eliminating HSC function. Most mice die within 14 days after exposure to a lethal dose of irradiation due to bone marrow failure (Storer 1966). However, when non-irradiated donor HSCs are supplied by injection of fetal liver or bone marrow cell suspensions into the bloodstream, these HSCs home to the bone marrow and rapidly proliferate to reconstitute the blood and promote long-term survival of the irradiated recipient mouse (Figure 3). This “transplantability” of HSCs can be exploited experimentally, allowing the evaluation of HSCs of interest in promoting recipient survival and contributing to long-term recipient hematopoiesis after irradiation.
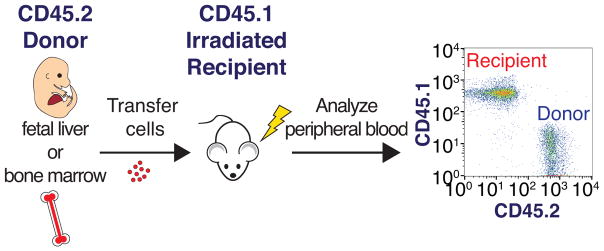
FIGURE 3. Adoptive transfer of fetal liver or bone marrow cells.
Donor cells containing HSCs are isolated from either the fetal liver or bone marrow of mice expressing the CD45.2 cell surface antigen. These cells are transferred by injection into irradiated recipient mice that express the CD45.1 cell surface antigen. Donor cells can later be detected in the peripheral blood of the recipient by flow cytometry using antibodies that distinguish between CD45.1 and CD45.2 expressing cells.
Donor- and recipient- derived blood cells can be distinguished by isoform-specific antibodies against the pan-leukocyte cell surface marker CD45 (Figure 3), which is present in two isoforms in laboratory mice (Morse et al. 1987). Further, using antibodies against cell surface markers on blood cells, an immunophenotypic identification scheme has been delineated for identifying HSCs and their differentiated progeny, enabling effective evaluation of donor HSC contribution to recipient peripheral blood. Thus, this system has not only been useful for identifying HSCs, but also for evaluating the fitness of HSCs derived from different genetic models, as in (Park et al. 2003).
Taken together, HSCs provide a tractable cellular system to study the effects of aneuploidy in vivo for several reasons. First, HSCs can be isolated from both embryos and adults from the fetal liver and the bone marrow, respectively, allowing for a range of aneuploid models to be tested. Second, the fitness of different aneuploid HSCs can be evaluated by comparing recipient survival and performing immunophenotyping analyses of recipient peripheral blood over time. Finally, aneuploid HSCs can be transplanted into euploid recipient mice, thus offering a unique model for studying aneuploid cells in the context of a euploid environment, a setting more typical of sporadic human cancers in vivo.
ROBERTSONIAN CHROMOSOMES AS A TOOL TO GENERATE ANEUPLOID HSCs
Because we are particularly interested in evaluating the effects of aneuploidy on cellular fitness in vivo, we sought to utilize this transplantation system in particular to study trisomic HSCs. Although trisomy is usually embryonic lethal in mice—precluding the analysis of animals with constitutional aneuploidies—some trisomic mice survive to E14.5, enabling isolation of HSCs from the fetal liver and thus permitting analysis of trisomic HSCs in adoptive transfers.
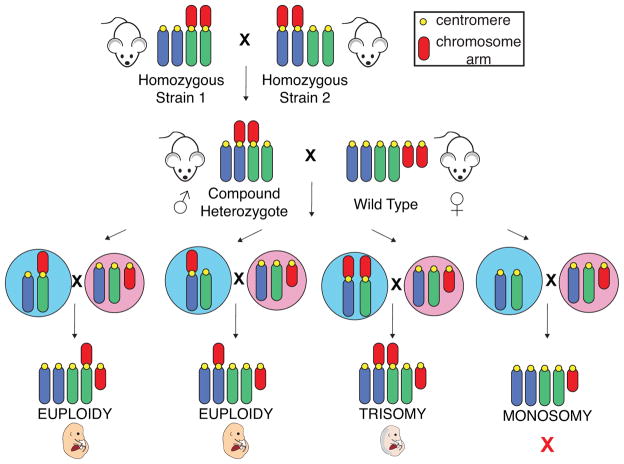
We employ a genetic system that relies on Robertsonian translocations to generate trisomic embryos (Figure 4; Williams et al. 2008). The breeding scheme to generate these trisomic animals was originally developed by Alfred Gropp and colleagues (Gropp et al. 1972). Gropp and colleagues discovered that feral house mice caught in Switzerland have 26 chromosomes instead of the 40 chromosomes previously found in mice (Gropp et al. 1969; Hsu and Benirschke 1970). This mouse, called the “tobacco mouse” because of its dark brown coat color, was initially thought to be a different species because it had fewer chromosomes and showed reduced fertility when mated with laboratory strains. However, further analysis revealed that the genome of these mice had the same complement of chromosomes as laboratory mice but that it was comprised in part by 7 Robertsonian translocations (Figure 5; Gropp et al. 1972; Hsu and Benirschke 1970). A number of other Robertsonian translocations have since been isolated from feral mouse populations in Europe, Africa and Asia (Capanna and Castiglia 2004), enabling the isolation of most combinations of Robertsonian translocations for laboratory use.
FIGURE 4. Breeding schematic for generating trisomic mice.
Mice homozygous for a Robertsonian translocation of two chromosomes are crossed to mice homozygous for a different Robertsonian translocation. Importantly, one chromosome is shared in each of the two Robertsonian translocations. The resulting mice, which are heterozygous for both of these Robertsonian translocations, are called “compound heterozygotes.” When meiotic nondisjunction occurs in the germline of compound heterozygotes and both Robertsonian chromosomes are segregated into the same gamete, a trisomic embryo is produced upon fertilization. Male compound heterozygotes were utilized in our studies to permit repeated matings. Potential male gametes are shown in blue, and potential female gametes are shown in pink. Monosomic embryos are not observed, as they die in a very early stage of embryonic development.
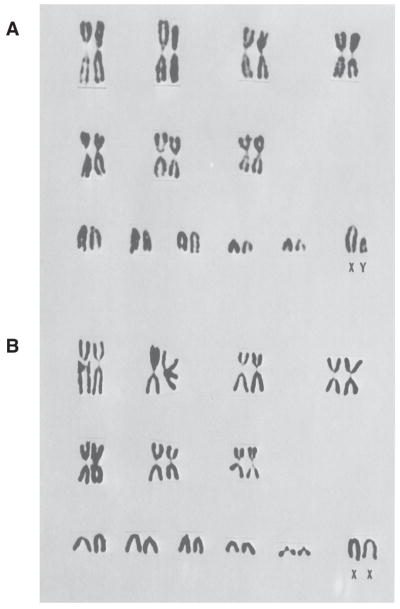
FIGURE 5. Tobacco mouse karyogram.
Chromosome spreads of (A) male and (B) female mice reveals the karyotype of the tobacco mouse. This feral mouse contains 7 Robertsonian fusion chromosomes and 19 acrocentric chromosomes, for a total of 26 chromosomes. Reprinted with permission from (Hsu and Benirschke 1970).
To generate trisomic mice (Figure 4), mice homozygous for a single Robertsonian translocation of two chromosomes (e.g. Rb 13.16) are crossed to mice homozygous for another Robertsonian translocation that includes one chromosome found in the first Robertsonian translocation fused with another chromosome (e.g. Rb 16.17). Male mice that are heterozygous for these Robertsonian translocations—“compound heterozygotes”—though genetically euploid themselves, give rise to trisomic embryos upon mating with wild type females when meiotic nondisjunction occurs in the male germline. The frequency of non-disjunction is increased in this system because more than two chromosomes will recombine during meiosis I, creating multivalents and hence causing mis-segregation to occur more often. The corresponding monosomic embryos are not observed, as they die in a very early stage of embryonic development (Gropp 1982).
Initial characterization of all possible trisomic mice that could be generated using the available Robertsonian translocations revealed that trisomy is generally embryonic lethal in mice, with different trisomies causing lethality at different developmental stages (Gropp 1982). However, Gropp and coworkers did observe live birth of some trisomic animals, including those trisomic for chromosomes 13, 16 and 19. This phenotypic heterogeneity is likely due to the fact that feral mice harboring Robertsonian chromosomes were outcrossed to laboratory strains to isolate specific fusion chromosomes. Such mating procedures typically result in “hybrid vigor,” or heterosis, a condition of increased fitness due to heterozygosity of alleles at many loci (Birchler et al. 2006). While this system makes studying trisomic embryos easier because of increased fitness, it can make further genetic studies of the progeny of these strains difficult to interpret because of a wide range of genetic variability (Simpson et al. 1997). Indeed, matings of these feral strains with different laboratory strains led to strain-specific differences in trisomic embryo survival (Gropp et al. 1983).
To reduce this genetic variability for research purposes, inbred syngeneic strains generated by breeding siblings for hundreds of generations are used (Silver 1995). Inbreeding results in homozygosity of all alleles in the genome but also causes decreased fitness, known as an inbreeding depression (Charlesworth and Charlesworth 1987). This decreased fitness is thought to be due to factors such as fixation of deleterious alleles in the strain background and a reduced ability to respond to environmental challenges. Thus, crossing of mice with an established inbred strain for many filial generations reduces genetic variability of strains – making phenotypes easier to interpret – but also results in decreased fitness.
BACKCROSSING REDUCES TRISOMIC EMBRYO FITNESS
Our lab undertook the effort of backcrossing a number of mouse strains harboring Robertsonian translocations into the inbred mouse strain C57BL/6J. The generation of backcrossed strains was especially important for adoptive transfer experiments to prevent graft rejection. Conditions such as graft-versus-host disease occur during HSC transplantation due to histoincompatibility (Allen et al. 1999). Graft-versus-host disease occurs when donor-derived T cells recognize the recipient as “non-self” and attack the recipient cells (Shlomchik 2007), often resulting in death or a chronic autoimmune-like condition following transplantation. The use of inbred mouse strains eliminates such complications.
Histoincompatibility likely affected previous adoptive transfers with trisomic HSCs. HSCs derived from mouse embryos trisomic for six different chromosomes were shown to be capable of repopulating the bone marrow of lethally irradiated mice in adoptive transfers (Herbst and Winking 1991a). Several hematopoietic phenotypes were observed in the recipients in this study. Mice reconstituted with trisomy 16 fetal liver cells showed decreased survival, anemia—decreased red blood cell counts—and leukopenia—decreased white blood cell counts. Mice reconstituted with trisomy 19 HSCs also showed leukopenia, and mice reconstituted with trisomy 13 HSCs showed decreased survival. However, because this study utilized non-backcrossed animals, the results were likely affected by graft rejection factors such as delayed graft-versus-host disease (Lowenberg 1977; Gropp et al. 1983), as only 70% of control animals survived at 9 weeks after adoptive transfers (Herbst and Winking 1991b). This high incidence of lethality in even control recipient mice makes the observed differences in survival and peripheral blood counts between trisomic and euploid reconstitutions difficult to interpret. Backcrossing the Robertsonian strains into an inbred line both reduces lethality due to histoincompatibility and ensures the phenotypes observed are due to differences in the donor HSCs and not a consequence of delayed graft rejection in the recipients.
We initially selected three trisomic strains, trisomy 13, 16 and 19, to perform adoptive transfer experiments because they span a distribution of mouse chromosome sizes: chromosomes 13 and 16 are intermediate sized chromosomes and chromosome 19 is the smallest mouse autosome. Additionally, these trisomic embryos were shown to survive to late stages of embryogenesis in initial characterizations in mixed backgrounds (Gropp 1982). We backcrossed mice harboring the Robertsonian chromosomes required to generate these trisomies for 10 generations into the C57BL/6J inbred background, leading to an estimated homozygosity of 99.9%. To evaluate the impact of backcrossing on trisomic embryo fitness, we documented the results of efforts to isolate these trisomic embryos. Our breeding data indicate that backcrossing these Robertsonian chromosomes has likely led to reduced embryonic survival for all three trisomies (Table 1).
TABLE 1.
Backcrossing reduces trisomic embryo fitness
| Trisomy | Percentage pregnant after evidence of mating plug (number of plugs observed) | Percentage of pregnancies yielding trisomic embryo | Overall percentage of trisomic embryos | Average litter size | Average number of resorptions per litter (range) |
|---|---|---|---|---|---|
| WT | 95.6 (266) | -- | -- | ~7.5 | ~1 |
| 13 | 28.5 (56) | 5.3 | 0.8 | 7.3 | 1.83 (1–5) |
| 16 | 65.4 (188) | 40.5 | 11.9 | 6.6 | 2.1 (1–6) |
| 19 | 58.5 (53) | 55 | 15.7 | 7.3 | 2.25 (1–5) |
Data obtained from embryos between ages E12.5 and E15.5 for trisomy 13 and E13.5 and E15.5 for WT (Holinka et al. 1979), trisomies 16 and 19.
Timed matings were employed to harvest trisomic embryos at E13.5 or E14.5, when HSCs could be readily isolated from fetal livers. Compound heterozygous males were mated with wild type C57BL/6J females, and embryos were harvested from pregnant females 13 or 14 days after successful mating. Trisomic embryos were initially distinguished from euploid embryos by gross morphological characteristics: trisomic embryos are smaller and generally paler than their wild type littermates and exhibit nuchal edema, a build-up of fluid on the spine (Gropp et al. 1983; Williams et al. 2008). However, the karyotype of embryos deemed trisomic was later confirmed by chromosome spreads and qPCR.
Embryos that die mid-gestation are resorbed into the maternal circulation over the course of a few days and can be detected in the uterine horn during this time (Flores et al. 2014). A previous study detected low numbers of resorbed embryos between E13.5 and E15.5 in matings of 3–7 month old C57BL/6J females with C57BL/6J male mice (Table 1; Holinka et al. 1979).
Previous characterizations of trisomy 16 in outcrosses yielded about 20% trisomic progeny at E14 (Miyabara et al. 1982). Trisomy 19 embryos were obtained at an even higher frequency (White et al. 1974a; 1974b). These studies also showed that these trisomic strains frequently survived until at least E16, and sometimes even survived to birth depending on the strain background used. As shown in Table 1, backcrossing into the C57BL/6J background greatly reduced the frequency at which trisomic embryos were obtained with our Robertsonian lines. For trisomy 13, the number of successful matings was low and yielded only one trisomic embryo (0.8% of all embryos isolated). Crosses to obtain trisomy 16 and 19 embryos led to pregnancies in approximately half of attempted matings, and trisomic embryos represented only about 12 and 16%, respectively, of all embryos isolated (Table 1). This decreased yield in trisomic embryos makes the isolation of trisomic fetal liver cells challenging, especially given that trisomy 16 and 19 fetal livers on average yield about 50% and 70% fewer cells than the livers isolated from their wild type littermates, respectively (data not shown). These data suggest that trisomic embryos of the C57BL/6J background will not survive to late embryonic development as frequently as previously characterized trisomic embryos.
While backcrossing likely led to decreased recovery of trisomic embryos at this embryonic stage, there are a number of additional factors that could have also influenced this decrease in later term embryo survival. Variable rates of chromosome mis-segregation were observed in the original hybrid strains both for different Robertsonian translocations and in male and female mice ranging from 4 to 26% when mating two compound heterozgyotes (Gropp et al. 1974). We have not evaluated the rate of chromosome mis-segregation in strains carrying Robertsonian fusion chromosomes after backcrossing; however, the presence of multiple resorbed embryos in most litters (Table 1) suggests that trisomic embryos are being produced. Additionally, we chose to use male compound heterozygotes to generate trisomic embryos because they can be mated multiple times. However, chromosome mis-segregation was reported to be lower in male than in female compound heterozygotes (Gropp et al. 1974). Indeed, recent breedings with female compound heterozygotes harboring the 11.13 and 13.16 Robertsonian chromosomes have yielded several trisomic embryos at E13.5 (P. Hsu, personal communication), suggesting that low mis-segregation rate – or likely male sterility (Gropp et al. 1983) – in these compound heterozygotes may be an additional factor limiting embryo production.
It is important to note that we did not attempt to isolate embryos before or after E13.5 – E15.5. It is thus possible that more trisomic embryos could have been recovered earlier in development. Consistent with this idea is the observation that while only half of the pregnancies aimed at obtaining trisomy 16 and trisomy 19 yielded trisomic embryos when harvested between E13.5 and E15.5, nearly all contain resorbed embryos. Further, because we did not isolate embryos after E15.5, it is possible that embryos surviving to E15.5 may have survived longer. Thus, our data provide us with only an estimate for a range of embryonic survival after backcrossing. Regardless, these data demonstrate that backcrossing of the Robertsonian chromosomes has led to decreased embryonic fitness.
TOWARDS AN IN VIVO SYSTEM TO STUDY ANEUPLOID CELLS
Although backcrossing into an inbred line led to reduced organismal fitness of the trisomic embryos, we have focused on characterizing the in vivo fitness of HSCs from two of the trisomic mouse strains that could be successfully isolated, trisomy 19, the smallest mouse autosome, and trisomy 16, the closest whole-chromosomal mouse model for human DS. Using our backcrossed animals, we have begun flow cytometric analyses of trisomic blood lineages in vivo to quantitatively assess differences in reconstitution in recipient mice over an extended period of time. We are also utilizing improved HSC characterization methods (Kiel et al. 2005; Kim et al. 2006; Yilmaz et al. 2006) to better understand the source of potential hematopoietic defects.
We complement our studies of constitutional trisomic HSCs with the analysis of HSCs harboring random aneuploidies that are generated by mouse models with increased rates of chromosome mis-segregation. The mouse model we first utilized to characterize HSCs with CIN harbors a hypomorphic allele of the spindle assembly checkpoint protein BubR1 (BUB1BH/H; Baker et al. 2004). The spindle assembly checkpoint ensures proper chromosome segregation. Thus, when BubR1 function is compromised by this loss-of-function allele, chromosomes mis-segregate more frequently, resulting in a randomly aneuploid cell population containing cells with one or more whole chromosomal aneuploidies mixed with euploid cells. The BubR1 hypomorphic allele is better tolerated than constitutional trisomy, as these mice survive to adulthood with a mean life expectancy of 6 months; however these mice have a progeroid phenotype and become progressively more aneuploid with age as evaluated by metaphase spreads of stimulated splenocytes (Baker et al. 2004). The use of BubR1H/H cells enables us to differentiate between chromosome-specific effects and general aneuploidy effects. Furthermore, because this mouse model survives to adulthood, both fetal liver and bone marrow HSCs can be evaluated from BubR1H/H mice. Finally, having a mouse model of CIN enables us to determine whether observed phenotypes are graded depending on the severity of the aneuploidy present, as has been seen previously in yeast and murine tissue culture (Torres et al. 2007; Williams et al. 2008).
HEMATOPOIESIS IN DS AND MOUSE MODELS OF DS
In addition to providing insights into the effects of aneuploidy on cellular fitness and tumorigenesis, the trisomy 16 mouse model is also relevant to DS, as DS individuals exhibit a number of hematopoietic phenotypes, including an increased risk of leukemia in the first few years of life (Satge et al. 1998). Most DS individuals who develop cancer show perturbed hematopoiesis before the onset of the disease. Neutrophilia, increased neutrophil counts, thrombocytopenia, decreased platelet counts, and polycythemia or increased hemoglobin concentration in the peripheral blood – manifest in 80, 66 and 34% of DS newborns, respectively (Choi 2008; Henry et al. 2007). These aberrations can also be detected in human trisomy 21 fetal livers. Trisomy 21 fetal livers harbor higher numbers of HSCs and megakaryocyte-erythroid progenitors and decreased numbers of B cell progenitors (Roy et al. 2012; Chou et al. 2008). Similar perturbations in the myeloid lineage were observed in HSCs derived from trisomy 21 induced pluripotent stem cells (Chou et al. 2012; Maclean et al. 2012). Furthermore, about 10% of DS infants develop transient myeloproliferative disorder (TMD), a condition caused by increased proliferation in the myeloid lineage that is usually apparent within the first few weeks of life and generally spontaneously resolves within 3 months (Choi 2008). This alteration in the myeloid lineage can be attributed to both trisomy 21 and mutations in the transcription factor GATA1 (Mundschau et al. 2003). In some cases, TMD will progress to acute myeloid leukemia (Alford et al. 2011). Additionally, while not present at birth (Henry et al. 2007), macrocytosis – increased cell volume – of red blood cells has been observed in children with DS analyzed between the ages of 2 and 15 (David et al. 1996).
Perturbations in blood lineages are also observed in mouse models of DS. The Ts65Dn mouse model – which harbors an extra copy of about two-thirds of the mouse genes orthologous to human chromosome 21 – shows sustained macrocytic anemia—increased red blood cell volume accompanied by decreased red blood cell counts and blood hemoglobin concentration—and increased numbers of megakaryocytes and common myeloid progenitors in the bone marrow (Kirsammer et al. 2008). The Ts1Cje mouse model – which contains about two-thirds of the genes amplified in the Ts65Dn mouse model – also exhibits macrocytic anemia throughout life; however, no differences in the myeloid lineage were observed, even after a loss-of-function allele of GATA1 was introduced into the line (Carmichael et al. 2009). The Tc1 mouse model – which harbors an exogenous copy of 80% of human chromosome 21– also showed macrocytic anemia throughout life (Alford et al. 2010). Introduction of a mutant, truncated form of GATA1 induced increased megakaryopoiesis in this model, but was not sufficient to induce leukemia (Alford et al. 2010).
In summary, there is evidence for perturbations of the hematopoietic lineages in both humans with DS and mouse models of DS. While the phenotypes observed are variable, there are some features frequently shared by humans and mice. For example, macrocytosis is observed both in some individuals with DS and in all DS mouse models studied. Notably, increased cell volume is also a feature of aneuploid cells in general (Torres et al. 2007; Williams et al. 2008). Further study of the effect of aneuploidy on HSCs using mouse models of trisomy and CIN will permit evaluation of these phenotypes in the context of different chromosomal amplifications. This evaluation will determine whether these phenotypes are specific to the particular amplification of the set of genes present on human chromosome 21 or if some of these phenotypes can be attributed to a general cellular response to aneuploidy in the blood.
ADOPTIVE TRANSFER AS A TOOL TO STUDY ANEUPLOID HEMATOPOIETIC MALIGNANCIES
It is still not well understood why cancer cells are aneuploid. However, adaptation of the system described here could provide a unique way to assess the effects of a range of aneuploid cells on tumorigenesis in vivo. HSCs can be transduced in vitro and then transplanted into irradiated recipients. If transduced with oncogenes, this system can be used to induce leukemias and lymphomas (Schmitt et al. 2000; 2002; Hemann et al. 2005). Additionally, leukemias and lymphomas are often transplantable and amenable to manipulation in culture, making this a potentially powerful system to assess the role of cellular aneuploidy in tumorigenesis in the future without requiring repeated isolations of trisomic fetal liver cells. This system will greatly expand the range of our in vivo analysis model, allowing us to assess the combinations of many aneuploidies and oncogenes on the kinetics of transformation in leukemia and lymphoma in mice and enabling us to test the effect of aneuploidy-targeting drugs (Tang et al. 2011) in aneuploid blood cancers.
CONCLUSIONS
Aneuploidy is a generally detrimental cellular state that exhibits a complicated relationship with cancer. We are working towards establishing a cellular system to better understand this relationship in vivo in mice by characterizing the fitness and differentiation potential of HSCs with trisomy or harboring mutations that cause CIN. Establishment and characterization of this system also permits future evaluation of the role of aneuploidy in tumorigenesis through the activation of oncogenes or inactivation of tumor suppressor genes before transplantation. We hope that our findings and future work using this system will help clarify the effects of aneuploidy in DS and provide insight into the precise role of aneuploidy in cancer.
Acknowledgments
We would like to thank Stacie Dodgson and Kristin Knouse for critical reading of this manuscript. This work is supported by the National Institute of Health grant GM056800 to A.A, the Kathy and the Curt Marble Cancer Research Fund and by an NSF Graduate Research Fellowship to S.J.P. A.A is also an investigator of the Howard Hughes Medical Institute and the Glenn Foundation for Biomedical Research.
References
- Alford KA, Reinhardt K, Garnett C, Norton A, Böhmer K, Neuhoff von C, Kolenova A, Marchi E, Klusmann J-H, Roberts I, et al. Analysis of GATA1 mutations in Down syndrome transient myeloproliferative disorder and myeloid leukemia. Blood. 2011;118:2222–2238. doi: 10.1182/blood-2011-03-342774. [DOI] [PubMed] [Google Scholar]
- Alford KA, Slender A, Vanes L, Li Z, Fisher EMC, Nizetic D, Orkin SH, Roberts I, Tybulewicz VLJ. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood. 2010;115:2928–2937. doi: 10.1182/blood-2009-06-227629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD, Dobkins JA, Harper JM, Slayback DL. Genetics of graft-versus-host disease, I. A locus on chromosome 1 influences development of acute graft-versus-host disease in a major histocompatibility complex mismatched murine model. Immunology. 1999;96:254–261. doi: 10.1046/j.1365-2567.1999.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nature Genetics. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baron MH, Isern J, Fraser ST. The embryonic origins of erythropoiesis in mammals. Blood. 2012;119:4828–4837. doi: 10.1182/blood-2012-01-153486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Unraveling the genetic basis of hybrid vigor. Proc Natl Acad Sci USA. 2006;103:12957–12958. doi: 10.1073/pnas.0605627103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Zur Frage der Entstehung Maligner Tumoren. Gustav Fischer; Jena: 1914. [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald H-R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Capanna E, Castiglia R. Chromosomes and speciation in Mus musculus domesticus. Cytogenetic and Genome Research. 2004;105:375–384. doi: 10.1159/000078210. [DOI] [PubMed] [Google Scholar]
- Carmichael CL, Majewski IJ, Alexander WS, Metcalf D, Hilton DJ, Hewitt CA, Scott HS. Hematopoietic defects in the Ts1Cje mouse model of Down syndrome. Blood. 2009;113:1929–1937. doi: 10.1182/blood-2008-06-161422. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annual review of ecology and systematics. 1987;18:237–268. [Google Scholar]
- Choi JK. Hematopoietic disorders in down syndrome. International journal of clinical and experimental pathology. 2008;1:387–395. [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Byrska-Bishop M, Tober JM, Yao Y, Vandorn D, Opalinska JB, Mills JA, Choi JK, Speck NA, Gadue P, et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proceedings of the National Academy of Sciences. 2012;109:17573–17578. doi: 10.1073/pnas.1211175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Opalinska JB, Yao Y, Fernandes MA, Kalota A, Brooks JSJ, Choi JK, Gewirtz AM, Danet-Desnoyers G-A, Nemiroff RL, et al. Trisomy 21 enhances human fetal erythro-megakaryocytic development. Blood. 2008;112:4503–4506. doi: 10.1182/blood-2008-05-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2:E75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O, Fiorucci GC, Tosi MT, Altare F, Valori A, Saracco P, Asinardi P, Ramenghi U, Gabutti V. Hematological studies in children with Down syndrome. Pediatr Hematol Oncol. 1996;13:271–275. doi: 10.3109/08880019609030827. [DOI] [PubMed] [Google Scholar]
- Flores LE, Hildebrandt TB, AAK, Drews B. Early detection and staging of spontaneous embryo resorption by ultrasound biomicroscopy in murine pregnancy. Reproductive Biology and Endocrinology. 2014;12:1–12. doi: 10.1186/1477-7827-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp A. Value of an animal model for trisomy. Virchows Arch A Pathol Anat Histol. 1982;395:117–131. doi: 10.1007/BF00429606. [DOI] [PubMed] [Google Scholar]
- Gropp A, Giers D, Kolbus U. Trisomy in the fetal backcross progeny of male and female metacentric heterozygotes of the mouse. i. Cytogenet Cell Genet. 1974;13:511–535. doi: 10.1159/000130304. [DOI] [PubMed] [Google Scholar]
- Gropp A, Tettenborn U, Lehmann von E. Chromosomenuntersuchungen bei der Tabakmaus (M. poschiavinus) und bei Tabakmaus-Hybriden. Cell Mol Life Sci. 1969;25:875–876. doi: 10.1007/BF01897931. [DOI] [PubMed] [Google Scholar]
- Gropp A, Winking H, Herbst EW, Claussen CP. Murine trisomy: developmental profiles of the embryo, and isolation of trisomic cellular systems. Journal of Experimental Zoology. 1983;228:253–269. doi: 10.1002/jez.1402280210. [DOI] [PubMed] [Google Scholar]
- Gropp A, Winking H, Zech L, Müller H. Robertsonian chromosomal variation and identification of metacentric chromosomes in feral mice. Chromosoma. 1972;39:265–288. doi: 10.1007/BF00290787. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature Cell Biology. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E, Walker D, Wiedmeier SE, Christensen RD. Hematological abnormalities during the first week of life among neonates with Down syndrome: Data from a multihospital healthcare system. Am J Med Genet. 2007;143A:42–50. doi: 10.1002/ajmg.a.31442. [DOI] [PubMed] [Google Scholar]
- Herbst EW, Winking H. Adoptive transfer of the hematopoietic system of trisomic mice with limited life span: stem cells from six different trisomies are capable of survival. Dev Genet. 1991a;12:415–422. doi: 10.1002/dvg.1020120606. [DOI] [PubMed] [Google Scholar]
- Herbst EW, Winking H. Adoptive transfer of the hematopoietic system of trisomic mice with limited life span: stem cells from six different trisomies are capable of survival. Dev Genet. 1991b;12:415–422. doi: 10.1002/dvg.1020120606. [DOI] [PubMed] [Google Scholar]
- Holinka CF, Tseng YC, Finch CE. Reproductive aging in C57BL/6J mice: plasma progesterone, viable embryos and resorption frequency throughout pregnancy. Biol Reprod. 1979;20:1201–1211. doi: 10.1095/biolreprod20.5.1201. [DOI] [PubMed] [Google Scholar]
- Hsu TC, Benirschke K. An Atlas of Mammalian Chromosomes. Springer; New York: 1970. Mus poschiavinus (Tobacco mouse) [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsammer G, Jilani S, Liu H, Davis E, Gurbuxani S, Le Beau MM, Crispino JD. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenberg B, de Zeeuw HM, Dicke KA, van Bekkum DW. Nature of the delayed graft-versus-host reactivity of fetal liver cell transplants in mice. J Natl Cancer Inst. 1977;58:959–966. doi: 10.1093/jnci/58.4.959. [DOI] [PubMed] [Google Scholar]
- Lapidot T. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- Maclean GA, Menne TF, Guo G, Sanchez DJ, Park I-H, Daley GQ, Orkin SH. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proceedings of the National Academy of Sciences. 2012;109:17567–17572. doi: 10.1073/pnas.1215468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng A, Wang Y, Brown SA, Van Zant G, Zhou D. Ionizing radiation and busulfan inhibit murine bone marrow cell hematopoietic function via apoptosis-dependent and -independent mechanisms. Exp Hematol. 2003;31:1348–1356. doi: 10.1016/j.exphem.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Mikkola HKA, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- Miyabara S, Gropp A, Winking H. Trisomy 16 in the mouse fetus associated with generalized edema and cardiovascular and urinary tract anomalies. Teratology. 1982;25:369–380. doi: 10.1002/tera.1420250314. [DOI] [PubMed] [Google Scholar]
- Moore MA, Metcalf D. Ontogeny of the haemopoietic system: yolk sac origin of in vivo and in vitro colony forming cells in the developing mouse embryo. Br J Haematol. 1970;18:279–296. doi: 10.1111/j.1365-2141.1970.tb01443.x. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci USA. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse HC, Shen FW, Hämmerling U. Genetic nomenclature for loci controlling mouse lymphocyte antigens. Immunogenetics. 1987;25:71–78. doi: 10.1007/BF00364270. [DOI] [PubMed] [Google Scholar]
- Mundschau G, Gurbuxani S, Gamis AS, Greene ME, Arceci RJ, Crispino JD. Mutagenesis of GATA1 is an initiating event in Down syndrome leukemogenesis. Blood. 2003;101:4298–4300. doi: 10.1182/blood-2002-12-3904. [DOI] [PubMed] [Google Scholar]
- Müller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Park I-KI, Qian DD, Kiel MM, Becker MWM, Pihalja MM, Weissman ILI, Morrison SJS, Clarke MFM. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pfau SJ, Amon AA. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Cowan G, Mead AJ, Filippi S, Bohn G, Chaidos A, Tunstall O, Chan JKY, Choolani M, Bennett P, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proceedings of the National Academy of Sciences. 2012;109:17579–17584. doi: 10.1073/pnas.1211405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov IM, Samokhvalova NI, Nishikawa S-I. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Satge D, Sommelet D, Geneix A, Nishi M, Malet P, Vekemans M. A tumor profile in Down syndrome. Am J Med Genet. 1998;78:207–216. [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Rosenthal CT, Lowe SW. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- Silver LM. Mouse genetics: concepts and applications. Oxford University Press; New York: 1995. [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nature Genetics. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Storer JB. Acute responses to ionizing radiation. In: Green EL, editor. The biology of the laboratory mouse. McGraw-Hill; New York: 1966. pp. 427–446. [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho Y-J, Klein A, Hofmann O, Camargo FD. Clonal dynamics of native haematopoiesis. Nature. 2015;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Schulte BA, LaRue AC, Ogawa M, Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Tjio JH, Van de Water LC, Crandall C. Trisomy 19 in the laboratory mouse. I. Frequency in different crosses at specific developmental stages and relationship of trisomy to cleft palate. Cytogenet Cell Genet. 1974a;13:217–231. doi: 10.1159/000130274. [DOI] [PubMed] [Google Scholar]
- White BJ, Tjio JH, Van de Water LC, Crandall C. Trisomy 19 in the laboratory mouse. II. Intra-uterine growth and histological studies of trisomics and their normal littermates. Cytogenet Cell Genet. 1974b;13:232–245. doi: 10.1159/000130275. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy Affects Proliferation and Spontaneous Immortalization in Mammalian Cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Kiel MJ, Morrison SJ. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood. 2006;107:924–930. doi: 10.1182/blood-2005-05-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]