Abstract
Ionotropic glutamate receptors (iGluRs) transduce signals derived from release of the excitatory neurotransmitter glutamate from pre-synaptic neurons into excitation of post-synaptic neurons on a millisecond time-scale. In recent years, the elucidation of full-length iGluR structures of NMDA, AMPA and kainate receptors by x-ray crystallography and single particle cryo-electron microscopy has greatly enhanced our understanding of the interrelationships between receptor architecture and gating mechanism. Here we briefly review full-length iGluR structures and discuss the similarities and differences between NMDA receptors and non-NMDA iGluRs. We focus on distinct conformations, including ligand-free, agonist-bound active, agonist-bound desensitized and antagonist-bound conformations as well as modulator and auxiliary protein-bound states. These findings provide insights into structure-based mechanisms of iGluR gating and modulation which together shape the amplitude and time course of the excitatory postsynaptic potential.
Introduction
Glutamate, the most abundant excitatory neurotransmitter in the vertebrate brain, targets two receptor families: ionotropic glutamate receptors (iGluRs) and metabotropic glutamate receptors (mGluRs). The iGluRs are responsible for both synaptic transmission and synaptic plasticity, are central to molecular mechanisms of learning and memory, and form tetrameric ligand-gated channel pores that allow the influx of Na+ and Ca2+ when glutamate binds to the extracellular ligand-binding domain (LBD). iGluRs can be further subdivided into N-methyl-D-aspartate (NMDA) receptors, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors and kainate receptors (Traynelis et al., 2010). Nearly three decades of functional characterization, including probes of biophysical and electrophysiological properties, pharmacology, and allosteric regulation, have increased our knowledge of iGluR function with respect to both physiological and pathological pathways (Paoletti et al., 2013; Traynelis et al., 2010). The absence of full-length iGluR structures in all physiologically-relevant conformations, however, has limited our understanding of the ion channel gating mechanism. Within the recent half decade, technical breakthroughs in structural biology comprising membrane protein x-ray crystallography and single-particle cryo-electron microscopy (cryo-EM), have enabled us to explore more deeply the structure-function relationship of intact iGluRs at near atomic resolution (Mayer, 2016). By elucidating the ‘molecular mechanics’ of iGluRs, we will be better able to understand the inner workings of the chemical synapse, a space in the synaptic cleft that is 20–40 nm wide and which is largely occupied by the extracellular domains of iGluRs accompanied by several families of pre synaptic targeting and synapse assembly proteins. At present, it is not clear why such large multi-domain, multi-layer molecular machines are needed to carry out excitatory synaptic transmission, compared to the much simpler architecture of receptors for acetylcholine, glycine and GABA.
Overall architecture and a gallery of iGluR structures
In 2009, the x-ray crystal structure of the intact rat GluA2 homomeric AMPA receptor provided the first atomic view of its global architecture and domain organization, together with the definition of an overall 2-fold symmetry and local, non aligned, axes of symmetry (Sobolevsky et al., 2009). The structure was captured in a closed channel, inhibited state bound with a competitive antagonist at 3.6 Å resolution. Five years later, two laboratories independently reported the x-ray crystal structures of the GluN1/GluN2B NMDA receptor in complex with agonists, an allosteric modulator and an ion channel blocker (Karakas and Furukawa, 2014; Lee et al., 2014). These milestone works revealed several unforeseen features. Globally, iGluRs form a dimer-of-dimers assembly, with individual subunits arranged in three main layers: the distal amino-terminal domain (ATD) on the ‘top’, the ligand-binding domain (LBD) sandwiched in the ‘middle’ and the transmembrane domain (TMD), harboring the ion channel, on the ‘bottom’. The large extracellular domains display an unanticipated feature: a non-equivalent subunit arrangement of the ATDs and LBDs, which together are composed of four clamshell-like modules (Sobolevsky et al., 2009) (Figure 1). On the ATD layer, the A/B and C/D subunits are coupled as local dimers, whereas on the LBD layer, the A/D and B/C subunits are coupled together as local dimers, respectively, an intertwining of subunits that is accomplished by the swapping of subunit-subunit interactions between the ATD and LBD layers (Sobolevsky et al., 2009). As a consequence of this subunit swapping, the A/C and B/D subunits, even in homomeric AMPA receptors, adopt two distinct conformations (Sobolevsky et al., 2009).
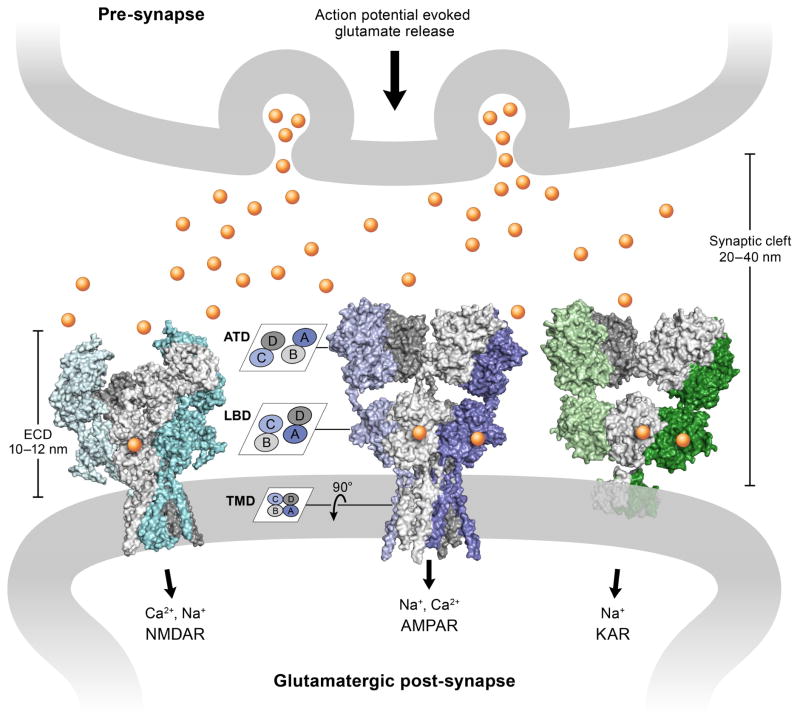
Figure 1. iGluRs at glutamatergic synapses.
The action potential evokes release of the excitatory neurotransmitter glutamate (shown as yellow cycles) from pre-synaptic vesicles. Structures of three main iGluRs (surface representation) are shown in the post-synaptic membrane. From left to right: NMDA receptor in the glycine/glutamate bound state (PDB: 5IOU), AMPA receptor in the competitive antagonist MPQX bound state (PDB: 5KK2) and kainate receptor in the presence of the agonist 2S,4R-4-methylglutamate (PDB: 4UQQ). iGluRs are heteromeric (NMDA receptors) and/or homomeric (AMPA/kainate receptors) dimer-of-dimers assemblies, with each subunit arranged in three layers defined by the amino-terminal domain (ATD), the ligand-binding domain (LBD) and the transmembrane domain (TMD). The subunit cross-talk on the ATD and LBD layers is illustrated in the cartoon on the left. The lengths of the synaptic cleft and the extracelluar domain (ECD) of the iGluRs are also marked.
Within the past six years, there has been a further exploration of iGluR structures using intact or nearly intact receptor constructs. Crystal structures of the homotetrameric GluA2 AMPA receptors have been determined for several states: the apo state (Dürr et al., 2014), the partial agonist-bound state (Yelshanskaya et al., 2014), the partial agonist/modulator bound pre-open state (Dürr et al., 2014), the agonist-bound desensitized state (Dürr et al., 2014) and the cone snail toxin bound state (Chen et al., 2014). Meanwhile, the cryo-EM technique has greatly advanced the structural biology of iGluRs. Moreover, cryo-EM structures of homomeric GluA2 AMPA receptors have been captured in perhaps more physiologically-relevant conformations by trapping the protein in vitreous ice (Dürr et al., 2014) (Meyerson et al., 2014). Interestingly, the cryo-EM structure of the heteromeric GluA2/GluA3 AMPA receptor with disulfide cross-linked cysteine mutations, in the nominally ligand-free state, exhibited a substantial compression between the ATD and LBD layers (Herguedas et al., 2016). Recently, two groups independently visualized the cryo-EM complex of the AMPA receptor and its auxiliary protein TARP (transmembrane AMPA-receptor regulatory protein) γ2 and elucidated their stoichiometry (Zhao et al., 2016) (Twomey et al., 2016), showing how the TARP is poised underneath the LBD ‘clamshells’ and in an appropriate position to modulate the ion channel gating.
Cryo-EM structures of NMDA receptors represent various conformations, including an agonist-bound inactive or perhaps desensitized-like state, an antagonist-bound inhibited state, an agonist/modulator bound state (Zhu et al., 2016) and an agonist-bound activated state (Tajima et al., 2016). Kainate GluK2 receptors were first captured in both the resting and the agonist-bound desensitized states by cryo-electron tomography at ~20 Å resolution (Schauder et al., 2013). Later, the resolution of the desensitized state was improved to 7.6 Å by single-particle cryo-EM (Meyerson et al., 2014) (Figure 1). Together, these structural studies deepen our understanding of receptor mechanism by showing how the isolated iGluR domains, some of which have been studied for nearly 20 years (Polhlsgaard et al., 2011), are arranged and undergo conformational changes in the context of intact receptors.
Arrangement of LBD gating ring
All iGluRs play roles in evoking excitatory postsynaptic potentials, but each subfamily has diverse distributions due, at least in part, to their distinct gating kinetics (Attwell and Gibb, 2005). AMPA receptors largely participate in triggering the rapid excitatory synaptic current, leading to the depolarization of the postsynaptic membrane. By contrast, NMDA receptors mediate the slow component of synaptic transmission and contribute mainly to synaptic plasticity. Kainate receptors show similar fast activation as AMPA receptors, but modulate synaptic currents both pre-synaptically and post-synaptically. Whole-cell voltage-clamp recordings from native and recombinant receptors show that AMPA and kainate receptors exhibit pronounced and rapid desensitization occurring within a millisecond time window after activation by glutamate, while NMDA receptors desensitize much more slowly, with desensitization rates highly dependent upon the subunit composition. Thus, according to gating kinetics, iGluRs can be classified into a slow gating class represented by the NMDA receptors and a fast gating non-NMDA class by AMPA/kainate receptors (Figure 2) (Traynelis et al., 2010).
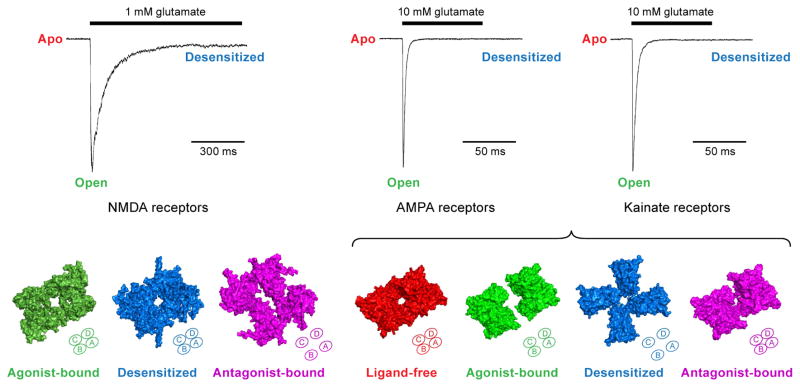
Figure 2. Conformational changes of the LBD gating ‘ring’ during gating cycle.
Top panel: Patch-clamp recordings of iGluR current, where NMDA receptors show slow desensitization kinetics and AMPA/kainate receptors display fast gating kinetics. Bottom panel: Top-down views of different ligand-binding domain (LBD) structures in physiologically-relevant conformations that correspond to the agonist-bound desensitized state (PDB: 5IOU) and antagonist-bound closed state (PDB: 5IPS) of the NMDA receptor; the ligand-free state (PDB: 4U2P), agonist-bound open state (PDB: 4UQK), antagonist-bound closed state (PDB: 3KG2) of the AMPA receptor; and the agonist-bound desensitized state (PDB: 4UQQ) of the kainate receptor. The tetrameric arrangement of the LBD gating ring in each state is also illustrated in diagrams at the bottom right corner of each structure.
The iGluR gating cycle during synaptic transmission can be sorted into three major states: the ligand-free (apo) state, the agonist-bound activated (open) state and the agonist-bound closed (desensitized) state. The structural elements of the gating machinery primarily involve the LBD and TMD regions (Mayer, 2016). Here we summarize a gallery of LBD conformations derived from full-length receptors trapped in distinct, gating-relevant states. The glycine/glutamate-bound active conformation of the GluN1/GluN2B NMDA receptor shows back-to-back interface within the LBD heterodimer, and also interactions between the two adjacent dimers. Unfortunately the TMD density has not yet been resolved (Tajima et al, 2016). The glycine/glutamate-bound inactive or desensitized LBDs in NMDA receptors resemble the LBD conformation in the ATD modulator phenylethanolamine-bound receptors, suggesting that the ATD negative allosteric modulator inhibits gating activity by stabilizing the LBD in a desensitized or inactive state (Zhu et al. 2016; Karakas et al, 2011; Tajima et al, 2016). Conversely, the desensitization process in AMPA/kainate receptors is accompanied by a remarkable disruption of the dimer interface, followed by the LBD conformational transition from two-fold to approximate four-fold symmetry (Meyerson et al. 2014; Durr et al, 2014). Interestingly, the four subunits do not rotate in a consistent manner or degree, with two clamshells ‘swinging’ ~120 degree clockwise and the other two turning ~10 degree clockwise (Figure 2). More strikingly, competitive antagonist-bound NMDA receptors somewhat resemble the desensitized AMPA/kainate receptors, with the LBD gating ring shifting into a pseudo four-fold arrangement, derived from the major motion of the GluN2-LBDs (Zhu et al, 2016). In contrast, antagonist-bound AMPA receptors maintain the same dimer-of-dimer arrangement as the apo state (Sobolevsky et al, 2009; Meyerson et al. 2014; Durr et al, 2014) (Figure 2). Taken together, these structures show how the LBD dimer is an essential element of the gating machinery and how clamshell closure, within the dimer, is coupled to ion channel gating. Disruption of the LBD dimer, either by rupture of the intradimer interface or by perturbation of dimer-dimer interactions, is associated with receptor inactivation. At present there is not a clear sense of the conformation of the LBD layer in a truly activated state due to the fact that a bona fide open channel structure has yet to be determined.
Pharmacology and auxiliary proteins
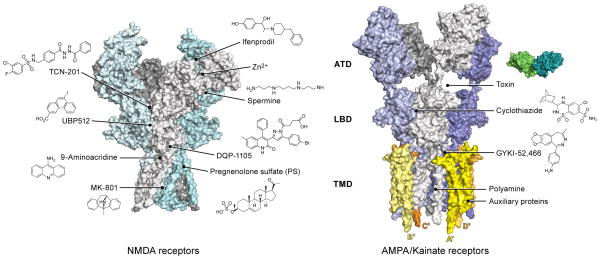
Due to the important role iGluRs play in neurophysiology and their deep involvement in learning and memory, tremendous efforts have been made in the past two decades to develop therapeutic small molecules that can precisely modulate iGluR channel activities (Paoletti et al, 2007; Traynelis et al, 2010; Zhu et al, 2015; Karakas et al, 2015). In addition to the orthosteric sites for binding of full agonists, partial agonists and competitive antagonists, iGluRs harbor multiple binding sites for ions and molecules acting as channel blockers and subunit-selective negative or positive allosteric modulators (Figure 3). In NMDA receptors, binding pockets for negative allosteric modulators such as the phenylethanolamines, are present in the upper lobe of the ATD heterodimeric interface (Perin-Dureau et al., 2002) (Karakas et al., 2011), for Zn2+ in the GluN2A-ATD (Paoletti et al., 1997) and for polyamines within the bottom lobe of the ATD heterodimeric interface (Zhu et al., 2013). Within the LBD heterodimeric interface there are binding sites for a broad range of allosteric modulators that include the TCNs, UBPs and GNEs (Hackos and Hanson, 2016) (Hackos and Hanson, 2016), as well as for DQP-1105 (Acker et al., 2011). The TMD is the binding site for well known small molecules that include the ion channel blockers MK-801 and ketamine, as well as the open channel blocker, 9-amino acridine (Traynelis et al., 2010). By contrast with NMDA receptors, AMPA/kainate receptors have fewer binding sites for small compounds. These binding sites are mainly located at the LBD dimeric interface and modulate desensitization (cyclothiazide), are near the linker region between the S1 and M1 segment (GYKI-52,466, perampanel), or are within the ion channel (polyamines, philanthotoxin and related spider toxins) (Traynelis et al., 2010).
Figure 3. Pharmacological properties and auxiliary proteins.
iGluRs harbor multiple binding sites for small molecules acting as allosteric modulators and for auxiliary proteins that modulate receptor trafficking and activity. The left panel shows how NMDA receptors have binding pockets on ATD, LBD and TMD layers. The right panel shows that AMPA/kainate receptors have binding cavities between the ATD-LBD layer, within the LBD dimer interface, and within the TMDs for both small molecules and proteins.
In addition to small molecules, receptor-protein complexes have also been reported with iGluRs. The crystal structure of the GluA2 AMPA receptor in complex with a cone snail toxin (Chen et al., 2014) was recently reported, showing how the toxin can bind to the large groove between the ATD and LBD layers of the AMPA receptor and allosterically potentiate channel activity by stabilizing the LBD gating ring in an active-like conformation. The Gouaux lab also reported the first cryo-EM structure of the AMPA receptor fully occupied with TARPs, illustrating how four TARP γ2 subunits surround the ion channel pore. They observed that the A′/C′ subunits and B′/D′ subunits of the γ2 TARP associate with the tetramer in an asymmetric manner (Zhao et al., 2016) (Figure 3). At the same time, the Sobolevsky lab reported the cryo-EM structures of the antagonist-bound AMPA receptor in combination with one or two γ2 auxiliary protein subunits (Twomey et al., 2016). These complex structures illustrate that the auxiliary proteins form extensive interactions with the TMDs and also interact with a conserved sequence in the D2 lobe of the LBD to modulate the gating behavior of AMPA receptors.
Conclusion
Recent structural studies of full-length iGluRs determined by cryo-EM and X-ray crystallography, together with expansive functional data, have greatly increased our understanding of the interrelationships between iGluR architecture, gating mechanism, biophysics and pharmacology. Moreover, these studies have begun to shed light on interactions with scaffold/auxiliary proteins, as well as on the differences between the NMDA receptors and non-NMDA iGluRs. With these efforts, more holistic views of the structure-function relationship of iGluRs are beginning to emerge.
Although remarkable progress has been achieved, future work remains. First, a structure capturing the open conformation of the ion channel pore will provide an important missing piece in the gating cycle and will also elucidate how the conformational changes of the LBD are coupled with movements of the transmembrane helix segments. Second, the manner in which the domains communicate and interact with each other within the large multi-domain multi-layer iGluR complex is still unclear. Third, structures of heteromeric iGluRs (e.g. triheteromeric NMDA, diheteromeric AMPA or kainate receptors) would offer more information about the physiological assemblies in native tissue, which would allow us to elucidate specific functions of iGluR subtypes in sub-brain regions. Finally, the precise way in which auxiliary proteins modulate the AMPA/kainate receptor gating on the synaptic membrane remains a puzzle. Capturing the distinct conformations of the TARP/iGluR complex would be helpful to visualize how protein interactions are involved in the modulation of synaptic transmission. Structural insights combined with functional validation will ultimately lead to the development of future iGluR-based therapeutics aimed at alleviating the deleterious effects of glutamatergic dysfunction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acker TM, Yuan H, Hansen KB, Vance KM, Ogden KK, Jensen HS, Burger PB, Mullasseri P, Synder JP, Liotta DC, Traynelis SF. Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-asparate receptor subunit-selective modulators. Mol Pharmacol. 2011;80:782–795. doi: 10.1124/mol.111.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D, Gibb A. Neuroenergetics and the kinetic design of excitatory synapses. Nature Rev Neurosci. 2005;6:841–849. doi: 10.1038/nrn1784. [DOI] [PubMed] [Google Scholar]
- Chen L, Dürr K, Gouaux E. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanisms. Science. 2014;345:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr K, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, Mchaourab HS, Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackos DH, Hanson JE. Diverse modes of NMDA receptor positive allosteric modulatioin: Mechanisms and consequences. Neuropharmacology. 2016 doi: 10.1016/j.neuropharm.2016.07.037. doi:10.1016j. [DOI] [PubMed] [Google Scholar]
- Herguedas B, Garcia-Nafria J, Cais O, Fernandez-Leiro R, Krieger J, Ho H, Greger IH. Structure and organization of heteromeric AMPA-type glutamate receptors. Science. 2016;352:352. doi: 10.1126/science.aad3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Furukawa H. Crystal structure of a heteromeric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2 NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Lu W, Carlisle Michel J, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML. Structural biology of glutamate receptor ion channel complexes. Curr Opin Struct Biol. 2016;41:119–127. doi: 10.1016/j.sbi.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Perin-Dureau F, Rachline J, Neyton J, Paoletti P. Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors. J Neurosci. 2002;22:5955–5965. doi: 10.1523/JNEUROSCI.22-14-05955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polhlsgaard J, Frydenvang K, Madsen U, Kastrup JS. Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology. 2011;60:135–150. doi: 10.1016/j.neuropharm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Schauder DM, Kuybeda O, Zhang J, Klymko K, Bartesaghi A, Borgnia MJ, Mayer ML, Subramaniam S. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proc Natl Acad Sci USA. 2013;110:5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–68. doi: 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science. 2016;353:83–86. doi: 10.1126/science.aaf8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Li M, Sobolevsky AI. Structure of an agonist-bound ionotropic glutamate receptor. Science. 2014;345:1070–1074. doi: 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E. Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature. 2016;536:108–111. doi: 10.1038/nature18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stein RA, Yoshioka C, Lee CH, Goehring A, Mchaourab HS, Gouaux E. Mechanism of NMDA receptor inhibition and activation. Cell. 2016;165:704–714. doi: 10.1016/j.cell.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol. 2013;20:477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]