ABSTRACT
Macroautophagy/autophagy plays an important role against pathogen infection in mammals and plants. However, little has been known about the role of autophagy in the interactions of insect vectors with the plant viruses, which they transmit. Begomoviruses are a group of single-stranded DNA viruses and are exclusively transmitted by the whitefly Bemisia tabaci in a circulative manner. In this study, we found that the infection of a begomovirus, tomato yellow leaf curl virus (TYLCV) could activate the autophagy pathway in the Middle East Asia Minor 1 (MEAM1) species of the B. tabaci complex as evidenced by the formation of autophagosomes and ATG8-II. Interestingly, the activation of autophagy led to the subsequent degradation of TYLCV coat protein (CP) and genomic DNA. While feeding the whitefly with 2 autophagy inhibitors (3-methyladenine and bafilomycin A1) and silencing the expression of Atg3 and Atg9 increased the viral load; autophagy activation via feeding of rapamycin notably decreased the amount of viral CP and DNA in the whitefly. Furthermore, we found that activation of whitefly autophagy could inhibit the efficiency of virus transmission; whereas inhibiting autophagy facilitated virus transmission. Taken together, these results indicate that TYLCV infection can activate the whitefly autophagy pathway, which leads to the subsequent degradation of virus. Furthermore, our report proves that an insect vector uses autophagy as an intrinsic antiviral program to repress the infection of a circulative-transmitted plant virus. Our data also demonstrate that TYLCV may replicate and trigger complex interactions with the insect vector.
KEYWORDS: autophagy, begomovirus, Bemisia tabaci, interaction, Tomato yellow leaf curl virus, virus transmission, whitefly
Introduction
The whitefly Bemisia tabaci is a species complex with more than 35 cryptic species.1,2 One of the cryptic species, Middle East Asia Minor 1 (MEAM1, previously referred to as the B “biotype”) has invaded many regions of the world and caused extensive crop damage through direct feeding and transmission of plant viruses.3,4 More than 200 species of plant viruses are exclusively transmitted by the whitefly B. tabaci, mostly begomoviruses.5,6 The epidemics of begomoviruses are usually associated with the outbreak of whiteflies7,8 and have caused a number of economically important diseases in the tropical and subtropical regions.9 During the past 30 years, the tremendous economic impact of begomovirus and the rapid spread of begomovirus disease worldwide has triggered a large body of research about plant-virus-insect relationship, epidemiology, disease management and breeding for resistance plants.6,10
The relationships between begomoviruses and whiteflies are complex.11,12 Viral particles are thought to be ingested along with phloem sap of infected plants through the stylet and enter the midgut. Virions are subsequently transported through the hemocoel into the salivary glands and from there they are finally excreted with the saliva during feeding.13,14 During the circulative transmission of begomoviruses, viral particles interact with various whitefly proteins.15,16 As demonstrated for tomato yellow leaf curl virus (TYLCV) which is a destructive pathogen of tomato crops, the whitefly GroEL protein seems to bind and protect TYLCV from degradation in the haemolymph. Disturbing the GroEL-TYLCV association leads to the degradation of the virus and to a marked decrease in virus transmission efficiency.17 TYLCV can be transmitted vertically and horizontally by whiteflies suggesting that the virus has invaded the reproductive system of whitefly.18-20 In addition, compared to healthy whiteflies, the longevity and fecundity of viruliferous whiteflies were reduced by 27% and 36% respectively, and the proportion of adults with detectable viral DNA maintained at 50% after feeding on cotton for 960 h.8,21 Luan et al.22 further demonstrate that TYLCV infection can activate the immune response of whiteflies, which might result in the degradation of viral particles within the body of viruliferous whiteflies. These results indicate that TYLCV infection has deleterious effects on whiteflies by inducing physiological changes and immune responses. However, the detailed mechanisms underlying the immune responses of the whitefly to begomoviruses are poorly understood.
Autophagy is a self-degradation process essential for cell survival, differentiation, development and homeostasis.23 There are at least 3 main forms of autophagy: chaperone-mediated autophagy, microautophagy and macroautophagy.24,25 The entire process of macroautophagy/autophagy encompasses the inclusion of cargos within the phagophore, the delivery of cargo to lysosomes and its subsequent breakdown and release of the resulting macromolecules back into the cytosol.23,26 Original studies in the yeast Saccharomyces cerevisiae have identified a group of autophagy-related (Atg) genes that can regulate autophagosome formation and other functions.27 Accumulating evidence indicates that many Atg genes are functionally conserved from yeast to mammals,25 and the ability of autophagy to protect against pathogen infection in mammals had been reported.28,29 For example, many viruses can be observed inside of autophagic compartments, including herpes simplex virus 1 and Sindbis virus.30 Moreover, autophagy plays a direct antiviral role against the mammalian viral pathogen vesicular stomatitis (VSV) in the model organism Drosophila.31 However, little is known about the role of autophagy in the interaction of plant viruses and their insect vectors.
Here we show that the infection of TYLCV is correlated with macroautophagy activation in the MEAM1 species of the B. tabaci complex. Within 48 h postinfection (hpi), the viral genomic DNA and coat protein (CP) were rapidly accumulated in whiteflies and reached their peak. Following the activation of autophagy at 48 hpi, the viral genomic DNA and CP were gradually degraded. Activation of autophagy significantly decreased the viral load and viral gene expression in whiteflies, while inhibition of autophagy resulted in higher viral concentration in whiteflies. These observations suggest that the autophagy pathway plays critical roles in the complex interactions between the MEAM1 whitefly and TYLCV.
Results
The autophagy pathway is activated in whiteflies, on TYLCV-infected tomato plants
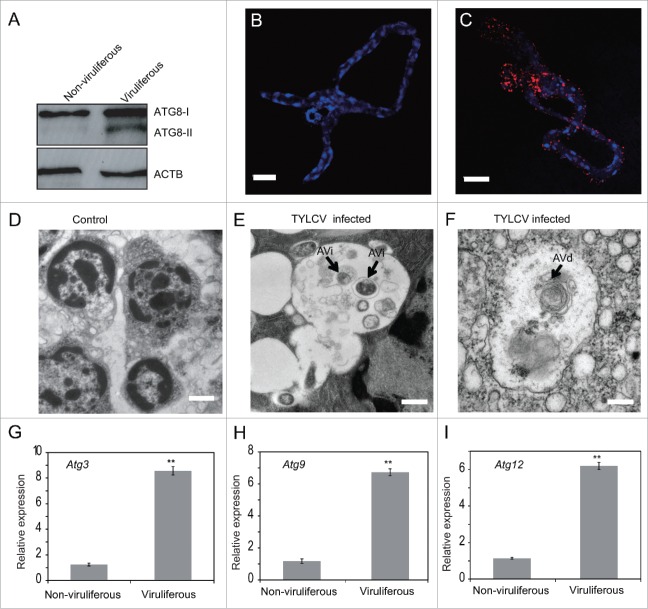
ATG8 is a ubiquitin-like protein involved in cargo recruitment into phagophores and the biogenesis of autophagosomes. Since ATG8 is selectively enclosed within autophagosomes, its breakdown allows measurement of the autophagic rate.31 In the transcriptome of whitefly MEAM1,32 only one Atg8 gene was found and it shows 79% identity with the Drosophila Atg8b gene (data not shown). To determine whether autophagy is triggered upon TYLCV infection, western blot with an antibody to ATG8 was used to analyze the level of ATG8-II in viruliferous and nonviruliferous MEAM1 whiteflies. Our results showed that the level of ATG8-II increased notably in whiteflies that fed on TYLCV infected tomato plants for 48 h (Fig. 1A). In addition, after 24 h and 72 h feeding on TYLCV infected tomato plants, the amount of ATG8-II was markedly upregulated as well (Fig. S1A). To further verify that the autophagy was activated by TYLCV infection, the formation of autophag-osome in the whitefly midgut was examined using immunofluorescence.33 While no autophagic signal was detected in uninfected whiteflies (Fig. 1B), specific ATG8-II puncta (red) was found in guts of TYLCV-infected whiteflies (Fig. 1C). Next, we performed electron microscopy analysis on viruliferous and nonviruliferous whiteflies; and found that the average number of autophagosomes in TYLCV-infected whiteflies is significantly higher than that in the healthy whiteflies (Fig. S1B). The autophagic vacuoles in infected midguts showed double-membrane with cytoplasmic contents or membranous contents indicative of autophagic compartment (Fig. 1E and F). The initial autophagic vacuole (AVi)/autophagosome with double membranes could be identified (Fig. 1E) and the degradative autophagic vacuoles (AVd)/autolysosomes with multimembrane structures were observed as well (Fig. 1F).34 In addition, a number of autophagosomes with different organelles, such as lysosome and mitochondrion were also found in the viruliferous whiteflies (Fig. S1C to H). Furthermore, the expression of 3 autophagy-related genes (Atg3, Atg9 and Atg12) was monitored. ATG3 conjugates ATG8 to phosphatidylethanolamine; ATG9 may act as a lipid carrier for expansion of the phagophore; and ATG12 can form a covalent bond with ATG3.34 Real-time PCR results showed that the expression of the 3 genes increased significantly in TYLCV-infected whiteflies as well (Fig. 1G to I). Taken together, these data indicate that the presence of TYLCV in whiteflies may activate autophagy.
Figure 1.
TYLCV infection induces autophagy in whiteflies. (A) Immunoblot analysis of whiteflies infected with TYLCV for 24 h and transferred to cotton for 120 h. ATG8-I (16 kDa) is observed in both viruliferous and nonviruliferous whiteflies, and ATG8-II (14 kDa) is induced only in the viruliferous whiteflies. Midguts of nonviruliferous (B) and viruliferous (C) whiteflies were fixed and immunofluorescence labeled with anti-ATG8 antibody and secondary antibody conjugated to Dylight 549 (red). Blue indicates DAPI staining of the nuclei. Twenty midguts of viruliferous whiteflies were measured and 95% of them were positive. A representative image is shown, Bar: 50 μm. TYLCV infection induces autophagosome formation as measured by electron microscopy (D to F). Representative images are shown for nonviruliferous (D, Bar: 1 μm) and viruliferous whiteflies (E and F, Bar: 0.5 μm). The initial autophagic vacuole (AVi)/autophagosome can be identified by its rough endoplasmic reticulum, and a double membrane (E). The multimembrane structure in the degradative autophagic vacuole (AVd)/autolysosome can be observed as well (F). Relative expression level of Atg3 (G), Atg9 (H) and Atg12 (I) were tested by qRT-PCR and ACTB was used as the internal control (*, P < 0.05; **, P < 0.01).
Time-course of autophagy activation in response to TYLCV infection
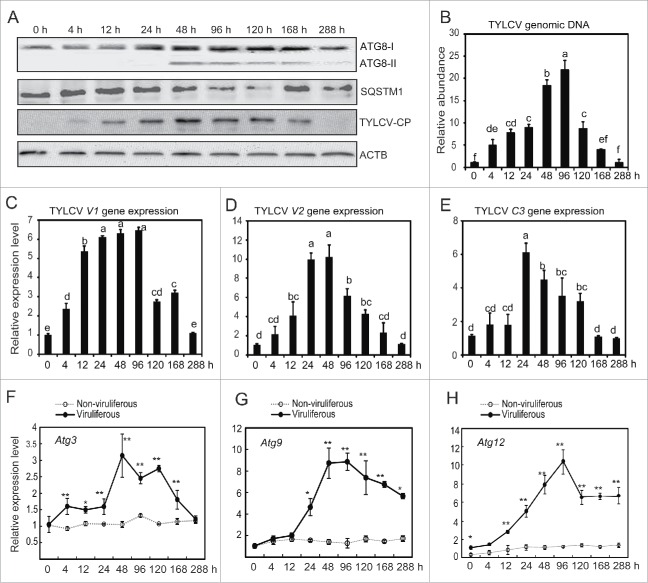
The virus-infected tomato may differ from the healthy plants in nutrition, plant volatiles and defense responses. Therefore, except viral infection, the physiological change of TYLCV-infected tomato plants may affect the whitefly autophagy pathway as well.35 To rule out the possible effect of infected tomato plants on insects, whiteflies were fed on virus-infected tomato plants for a 6-h acquisition access period (AAP) and then transferred onto cotton, a nonhost plant of TYLCV and very suitable for whiteflies.22,36,37 The activation of autophagy at different time points was monitored by western blot. The ATG8-II protein could hardly be observed for the first 24 h, but clearly increased at 48 h and then gradually decreased (Fig. 2A). While the viral CP was nearly undetectable for the first 4 h on cotton, a substantial amount of CP was accumulated after 12 h and peaked at 48 h (Fig. 2A). Interestingly, after 48 h, TYLCV CP began to decrease, corresponding to the time when autophagy was activated. Furthermore, except at 168 h, there was a clear correlation between the increase in ATG8-II and the decrease of SQSTM1/p62 from 24 h to 288 h after virus infection (Fig. 2A), suggesting the activation of autophagic flux.34 For whiteflies that were fed on healthy tomato plants for 6 h and then transferred onto cotton, no ATG8-II could be detected throughout all the time points (Fig. S2A). It suggests that virus infection but not host-plant transfer is critical for the activation of autophagy. To verify that changing the host from tomato to cotton plants is not a stress to whiteflies, the activation of a number of stress-related pathways in whiteflies was monitored by western blot. As shown in Figure S2B, transferring the whitefly from tomato to cotton plants had no effect on MAPK/ERK, MAPK/ p38, apoptosis and cell cycle pathways. Furthermore, we used another 2 tomato varieties Zheza 502 and Zhefen 701 which are resistant to TYLCV infection38 to test the activation of autophagy after transferring the viruliferous and healthy whiteflies from Hezuo903 to the resistant tomato cultivars. As shown in Figure S2C, the autophagy pathway was also activated after transferring the viruliferous whiteflies on these 2 tomato varieties in different times. In contrast, transferring healthy whiteflies from Hezuo903 to Zheza 502 and Zhefen 701 has no effect on autophagy (Fig. S2C). Collectively, these results confirm that the TYLCV infection but not the host plant shift activates the whitefly autophagy pathway.
Figure 2.
Time course of autophagy activation in response to TYLCV infection. (A) Accumulation of TYLCV CP and autophagy activation in whiteflies that were infected for 6 h and transferred to cotton for 0 to 288 h. The autophagic flux were also monitored by detecting the turnover of the autophagic receptor and substrate SQSTM1/p62. (B) TYLCV DNA was detected by qPCR (Significant differences are indicated with different letters, P < 0.05). (C to E) TYLCV genes (V1, V2 and C3) were detected by qRT-PCR (Significant differences are indicated with different letters, P < 0.05). (Fto H) Relative expression levels of 3 autophagy genes (Atg3, Atg9 and Atg12) were detected by qRT-PCR (*, P < 0.05; **, P < 0.01).
Then, virus genomic DNA abundance was determined using quantitative PCR (qPCR). Initially, the viral genome was undetectable in whiteflies. Similar to viral CP, the level of virus genomic DNA increased steadily and peaked at 48 to 96 h and then gradually decreased (Fig. 2B). The expression of TYLCV V1, V2 and C3 genes was monitored with quantitative reverse transcription PCR (qRT-PCR). The relative expression levels of TYLCV gene transcripts (V1, V2 and C3) also significantly increased during the initial phase of feeding on cotton and then gradually decreased (Fig. 2C to E). The expression levels of 3 autophagy-related genes (Atg3, Atg9 and Atg12) were also examined using both the ΑΤCB and ribosomal protein L29 (RPL29) as internal controls (Figs. 2F to H and S1I to K). The expression of these Atg genes followed a similar trend with that of TYLCV. When whiteflies were fed on virus-infected tomato plants for 6 h and then transferred onto cotton, the genomic DNA, CP and gene transcription of TYLCV all increased significantly within 48 h (Fig. 2). As cotton is a nonhost plant of TYLCV, these results suggest that TYLCV may accumulate (or replicate) within the whitefly.39,40 The accumulation of TYLCV at the early phase activates the whitefly autophagy, which may lead to the subsequent degradation of TYLCV (Fig. 2).
Dynamics of virus and activation of autophagy in the gut of whiteflies
Next, we measured the dynamics of TYLCV and autophagy at different time points in whitefly guts using confocal immunofluorescence. Both the viral CP (Fig. 3A) and the autophagosome (Fig. 3B) were undetectable for the first 4 h on cotton. With the accumulation of CP in whiteflies, the autophagosome was notably increased and peaked at 96 h. After the activation of autophagy, the viral load began to decrease. At 288 h, when CP became undetectable again, autophagosome signal disappeared as well (Fig. 3A and B). However, for nonviruliferous whiteflies, no autophagosome and TYLCV signals could be detected throughout all the time points (data not shown). We also performed colocalization experiments with CP and ATG8 antibodies on the same sample to show that autophagy is activated where virus is present (Fig. 3C). These observations confirm that the activation of autophagy is strongly correlated with the presence of virus.
Figure 3.

Dynamics of TYLCV and autophagy in the gut of the whitefly. Whiteflies were infected with TYLCV for 6 h on tomato and then transferred onto cotton for 0 to 288 h. Guts of viruliferous whiteflies were dissected, fixed and labeled with anti-CP (A) and anti-ATG8 (B) antibodies. Bar: 20 μm. Blue indicates DAPI staining of the nuclei. For each time point, 20 midguts were dissected and similar trend was observed. (C) Localization of CP and ATG8 on the same sample.
Effects of autophagy inhibitors on TYLCV infection
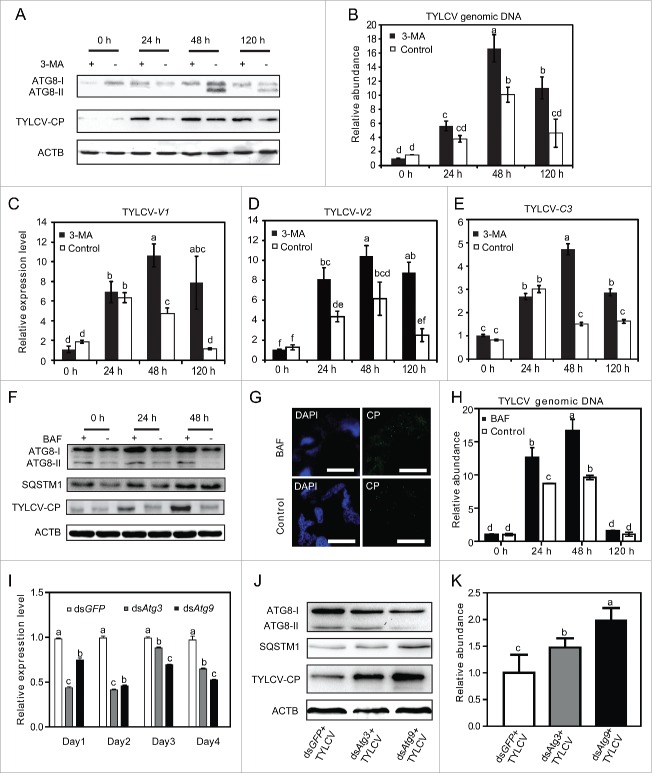
To observe the role of autophagy in TYLCV infection, whiteflies were treated with 3-methyladenine (3-MA) for 24 h, which inhibits autophagy due to suppression of the class III PtdIns 3-kinase.41 After 3-MA treatment, whiteflies were allowed to feed on virus-infected tomato for 6 h and then transferred onto cotton plant. The whitefly were collected after transferred onto cotton plant for 0, 24, 48 and 120 h; and the effects of 3-MA on autophagy activity were examined using confocal microscopy and western blot. Without 3-MA, TYLCV infection led to the conversion of ATG8-II at 48 h and 120 h (Fig. 4A, lanes with -). After treatment with 3-MA, the ATG8-II formation was nearly completely lost (Fig. 4A, lanes with +), suggesting that treatment with 3-MA resulted in obvious inhibition of TYLCV-induced autophagy. Confocal microscopy also showed that after 3-MA treatment the level of autophagic signal was obviously decreased in the TYLCV-infected whitefly (Fig. S2D). Interestingly, the accumulation of viral CP was substantially increased when insects were treated with 3-MA, as compared to that of the untreated insects, especially at 24 and 120 h (Fig. 4A). Next, TYLCV genomic DNA was detected by qPCR to determine whether viral genome accumulation in whiteflies was affected by autophagy inhibition. After 6 h AAP on infected tomato and transferred onto cotton (0 h), the TYLCV genomic DNA has no significant difference between the treatment and control groups, suggesting that 3-MA treatment did not affect the efficiency of virus acquisition (Fig. 4B). However, after 48 and 120 h on cotton, the accumulation of TYLCV genomic DNA in 3-MA treated whiteflies was substantially higher than that of the untreated ones (Fig. 4B). In addition, viral gene expression was measured by qRT-PCR. No significant difference of gene transcription was observed at 0 h. However, from 24 to 120 h, viral RNA in whiteflies treated with inhibitors was significantly higher than that in the control group (Fig. 4C to E). Next, bafilomycin A1 (BAF) which can block the fusion of autophagosomes with the vacuole was used to treat the whitefly. Our results showed that after being treated with BAF (lanes with +), the amount of ATG8-II slightly increased compared with the untreated group (lanes with -) (Fig. 4F). In addition, the level of SQSTM1/p62 was elevated after BAF treatment, suggesting successful inhibition of the late phase of autophagy. Accordingly, the accumulation of viral CP was substantially much higher when insects were treated with BAF, as compared with that of the untreated insects, especially at 48 h (Fig. 4F and G). Next, TYLCV genomic DNA was detected by qPCR. After 6 h AAP on infected tomato and transferred onto cotton (0 h), the TYLCV genomic DNA had no significant difference between the treatment and control groups, suggesting that BAF treatment did not affect the efficiency of virus acquisition (Fig. 4H). However, the accumulation of TYLCV genomic DNA in BAF treated whiteflies was substantially higher than that of the untreated ones at 24 and 48 h (Fig. 4H).
Figure 4.
Effects of inhibiting autophagy on TYLCV. (A) TYLCV CP and ATG8 of whiteflies treated with 3-MA (+) or without 3-MA (−) were detected by western blot. (B) TYLCV DNA was detected by qPCR (Significant differences are indicated with different letters, P < 0.05). (Cto E) TYLCV gene expression (V1, V2 and C3) were also detected by qRT-PCR (Significant differences are indicated with different letters, P < 0.05). (F) TYLCV CP, ATG8 and SQSTM1/p62 in whiteflies treated with (+) and without BAF (−) were detected by western blot. (G) The effect of BAF treatment on the accumulation of TYLCV CP at 48 h was detected by immunofluorescence. (H) The effect of BAF treatment on the accumulation of the TYLCV genome. TYLCV DNA was detected by qPCR at different time points (significant differences are indicated with different letters, P < 0.05). (I) Silencing of whitefly Atg3 and Atg9 by feeding double-strand RNA (dsRNA). The expression of Atg3 and Atg9 genes was monitored by qRT-PCR (significant differences are indicated with different letters, P < 0.05). (J) The level of ATG8, SQSTM1/p62 and TYLCV CP was then analyzed by immunoblot. (K) The effect of silencing Atg3 and Atg9 on the accumulation of the TYLCV genome. TYLCV DNA was detected by qPCR (Significant differences are indicated with different letters, P < 0.05).
Silencing Atg3 and Atg9 inhibits whitefly autophagy
To further examine the relationship between TYLCV and autophagy pathway, we silenced the expression of Atg3 and Atg9 genes by RNAi and then measured the formation of ATG8-II and the level of TYLCV CP in whiteflies. ATG3 is a ubiquitin-conjugating enzyme analog that conjugates ATG8 to phosphatidylethanolamine and ATG9 is the only integral membrane ATG protein essential for autophagosome formation. First, the whiteflies were fed with 200 ng/μl Atg3 and Atg9 dsRNA for different time intervals. We found that after feeding dsRNA for 48 h, the expression of Atg3 and Atg9 was significantly suppressed in whiteflies as revealed by qRT-PCR (Fig. 4I). Therefore, for subsequent RNAi experiments, whiteflies were always fed with dsRNA for 2 d. As shown in Figure 4J, the depletion of Atg3 and Atg9 reduced the formation of ATG8. In addition, the SQSTM1/p62 levels were noticeably increased, indicating successful inhibition of autophagic flux. Accordingly, silencing Atg3 and Atg9 strongly increased the amounts of virus coat protein (Fig. 4J) and genomic DNA (Fig. 4K) in whiteflies.
Effects of autophagy induction on TYLCV infection
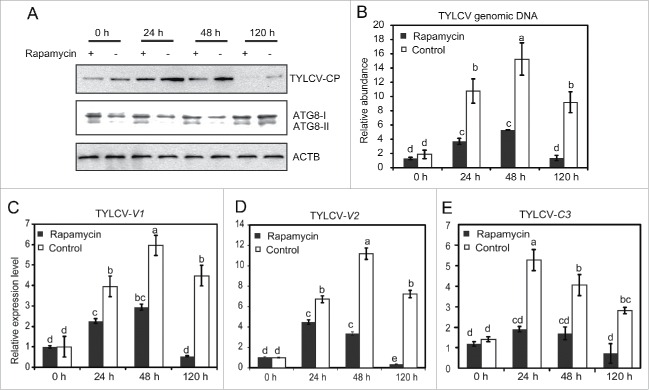
To further observe whether autophagy plays an antiviral role in whiteflies, we examined whether TYLCV DNA accumulation and protein expression could be modulated by inducing autophagy with rapamycin, which has been known to cause the activation of autophagy.42 After rapamycin treatment, whiteflies were allowed to feed on virus-infected tomato for 6 h and then transferred onto cotton plant. As presented in Figure S2D, feeding whiteflies with rapamycin activated the formation of autophagosome in both the viruliferous and nonviruliferous whiteflies. For whiteflies treated with rapamycin, ATG8-II was detected once they were transferred to cotton for 0 h, suggesting that autophagy was induced 24 h after rapamycin treatment. The amount of the viral CP in rapamycin-treated whiteflies was significantly lower than that of untreated whiteflies, indicating that viral CP synthesis was inhibited by autophagy (Fig. 5A). Likewise, TYLCV DNA in rapamycin-treated whiteflies was significantly lower than that in control group at 24 to 120 h, while no significant difference was observed at 0 h on cotton (after 6 h AAP on infected tomato). For the control group, the level of TYLCV DNA increased significantly from 24 to 48 h indicating the accumulation of TYLCV in whiteflies. But no significant difference was detected at 24 h and 48 h when whiteflies were treated with rapamycin (Fig. 5B). These results suggest that rapamycin treatment inhibits the viral accumulation and protein synthesis via activation of autophagy. Similar to TYLCV DNA, qRT-PCR analysis showed that, at all-time points, the expression of TYLCV-V1, V2 and C3 genes in whiteflies treated with rapamycin were lower than that in the untreated group (Fig. 5C to E). Taken together, our data suggest that activation of autophagy can lead to degradation of viral DNA, gene transcripts and CP.
Figure 5.
Effect of rapamycin on TYLCV DNA, CP and gene expression. (A) TYLCV CP and ATG8 of whiteflies induced with rapamycin (+) or without rapamycin (−) were detected by western blot. (B) TYLCV DNA was detected by qPCR (Significant differences are indicated with different letters, P<0.05). (C to E) TYLCV gene expression (V1, V2 and C3) were also detected by qRT-PCR (significant differences are indicated with different letters, P < 0.05).
Acquisition, retention and transmission of TYLV by whiteflies treated with 3-MA and rapamycin
To investigate the effect of autophagy on the acquisition of TYLCV by whiteflies, we treated the insects with 3-MA and rapamycin respectively before transferring them onto virus-infected tomato plants for viral acquisition. PCR was then deployed to detect TYLCV DNA in each insect at different time points. We could amplify TYLCV DNA in 60% to 70% of the individuals tested as early as 3 h after access to the infected plant (Table 1). For whiteflies treated with 3-MA, the frequency of detection increased as the length of the AAP increased and reached 100% after 12 h AAP to the infected plants and thereafter consistently remained at 100% (Table 1). Next, the efficiency of TYLCV retention in whiteflies treated with 3-MA or rapamycin were examined (Fig. 6A). After feeding on cotton plants for 15 d, viral DNA can only be detected in 30% of whiteflies treated with rapamycin; while in the untreated and 3-MA treated ones, 90% of the whiteflies remained with detectable TYLCV DNA even after 30 d. These results suggested that 3-MA had no effect on the efficiency of TYLCV acquisition and retention by the whitefly, while rapamycin substantially reduced the retention of virus by the insect. To demonstrate whether inducing or inhibiting autophagy might affect the efficiency of TYLCV transmission, whiteflies with and without rapamycin and 3-MA treatment were used to infect susceptible tomato plants and the virus transmission frequency were determined by PCR (Table 2). Our results showed that rapamycin-treatment markedly reduced the rate of virus transmission; and 3-MA treatment considerably increased the number of plants with virus infection. To further confirm our observation, the amount of virus in each tomato plant was also measured by qPCR. As shown in Figure 6B, the average level of TYLCV also significantly higher than the control group and rapamycin-treated group. It demonstrates that regulating autophagy pathway in the whitefly can affect the efficiency of virus transmission.
Table 1.
Efficiency of TYLCV acquisition in rapamycin or 3-MA treated whiteflies.
| Insects with TYLCV DNA (%) |
|||
|---|---|---|---|
| Acquisition access period (h)a | Control | Rapamycin | 3-MA |
| 0 | 0 | 0 | 0 |
| 3 | 60 | 60 | 70 |
| 6 | 70 | 60 | 70 |
| 12 | 100 | 80 | 100 |
| 24 | 100 | 80 | 100 |
| 48 | 100 | 80 | 100 |
Note.
Ten whiteflies were collected at each of the time points as shown in the table.
Figure 6.
TYLCV retention in whiteflies treated with 3-MA or rapamycin. (A) The whiteflies treated with 3-MA or rapamycin feeding on TYLCV-infected tomato plants for 48 h and then transferred to cotton plant. Ten whiteflies were collected at every time point as shown in the figure after transferring to cotton plants. PCR analyses showed that percentage of samples with viral DNA (significant differences are indicated with different letters, P < 0.05). (B) The amount of virus in each tomato plant. The level of TYLCV is each tomato plant was measured by qPCR and normalized with tomato actin. Statistical analysis was done with the Mann-Whitney test. The horizontal line depicts the medians. Significant differences are indicated with different letters, P < 0.05.
Table 2.
Rate of TYLCV transmission after rapamycin or 3-MA treatment.
| Treatment | No. of infected plants /Total No. of plantsa |
|---|---|
| Control | 10/20 |
| 3-MA | 16/20 |
| Rapamycin | 5/20 |
Note.
For each group, 20 tomato plants were used for virus transmission and the numerator is the number of plants detected with virus, by PCR.
Tomato yellow leaf curl China virus (TYLCCNV) infection activates autophagy as well
Tomato yellow leaf curl China virus (TYLCCNV) is another begomovirus closely related to TYLCV; however, unlike TYLCV which is a monopartite virus, TYLCCNV has bipartite genomes, referred to as DNA-A and DNA-B.43 To investigate whether TYLCCNV can activate the whitefly autophagy pathway as well, the level of ATG8-II in TYLCCNV-infected whiteflies was monitored using western blot. Similar to previous descriptions, whiteflies were allowed to feed on TYLCCNV-infected plants for 6 h and then transferred onto cotton, a nonhost plant of TYLCCNV. While the TYLCCNV CP was undetectable for the first 4 h on cotton, substantial amount of CP was accumulated after 12 h and peaked at 48 h (Fig. 7A), indicating that the TYLCCNV CP may accumulate in the whitefly as well. Similar to TYLCV infection, the whitefly autophagy pathway was clearly activated at 24 and 48 h. After the activation of autophagy, the amount of TYLCCNV CP suddenly decreased (Fig. 7A). Similar to the change of CP, the level of TYLCCNV genomic DNA also increased gradually, peaked at 48 h and then decreased (Fig. 7B). Immunofluorescence showed that autophagosomes were also induced at 12 h, peaked at 48 h and then decreased at 72 h (Fig. 7C) in the gut. This profile is highly consistent with the dynamics of TYLCCNV genome DNA and CP as well, indicating that the presence of TYLCCNV is responsible for the activation of autophagy.
Figure 7.

Autophagy activation in response to TYLCCNV infection. (A) Accumulation of TYLCCNV CP and autophagy activation in whiteflies which were infected for 6 h and transferred to cotton for 0 to 120 h. (B) TYLCCNV DNA was detected by qPCR (significant differences are indicated with different letters, P < 0.05). (C) Midguts of viruliferous whiteflies were dissected, fixed and labeled with anti-ATG8 antibodies. The formation of autophagosomes (red) was monitored in TYLCCNV-infected guts from 0 to 120 h.
Activation of the autophagy pathway is triggered by viral replication
To further prove that the activation of autophagy is a common phenomenon for viral infection, we infected whiteflies with another monopartite begomovirus, Papaya leaf curl China virus (PalCuCNV),44 and monitored the activation of autophagy pathway. Similar to the previous experiments, whiteflies were first fed on PalCuCNV-infected tomato plants for 6 h to acquire the virus and then transferred onto cotton. To our surprise, no ATG8-II was found by western blot throughout all the time points (Fig. 8A) and neither the autophagic signal could be detected by immunofluorescence (Fig. 8B). It indicates that, unlike TYLCV and TYLCCNV, PalCuCNV infection cannot activate the whitefly autophagy pathway. Interestingly, qPCR results showed that the genomic DNA of PalCuCNV rapidly decreased and remained at a very low level thereafter, suggesting that this virus cannot replicate in the whitefly (Fig. 8C). These observations indicate that virus replication may be required to activate the autophagy pathway.
Figure 8.
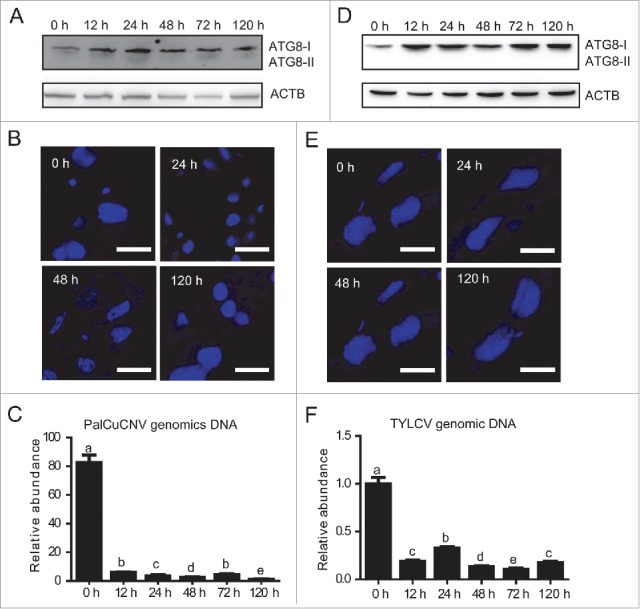
Viral replication and activation of the autophagy pathway. (A) Autophagy activation in whiteflies which were infected for 6 h with PalCuCNV and transferred onto cotton for 0 to 120 h. (B) Formation of autophagosomes in guts of PalCuCNV-infected whiteflies was monitored from 0 to 120 h. (C) PalCuCNV DNA was detected by qPCR (significant differences are indicated with different letters, P < 0.05). (D) Autophagy activation in greenhouse whiteflies which were infected with TYLCV for 6 h and transferred onto cotton for 0 to 120 h. (E) The formation of autophagosomes in guts of TYLCV-infected greenhouse whiteflies from 0 to 120 h. (F) The amount of TYLCV DNA in greenhouse whiteflies was detected by qPCR (significant differences are indicated with different letters, P < 0.05).
Finally, we examined the effect of TYLCV infection on the autophagy of the greenhouse whitefly Trialeurodes vaporariorum. Previous studies have shown that TYLCV cannot penetrate the gut barrier of the greenhouse whitefly and only exists in the gut lumen.45 When adults of the greenhouse whitefly were fed on TYLCV-infected tomato plants for 6 h and transferred onto cotton, we could not detect the activation of autophagy pathway until 120 h either by western blot (Fig. 8D) or immunofluorescence (Fig. 8E). Accordingly, the genomic DNA of TYLCV never increased in the greenhouse whitefly (Fig. 8F). Taken together, these results confirmed that the presence of virus inside the whitefly tissue and viral replication is a prerequisite for the activation of the autophagy pathway.
Discussion
Autophagy is an evolutionarily conserved intracellular process by which bulk cytoplasm is enveloped inside a double-membrane vesicle and shuttled to lysosomes for degradation.31,46 Here we found that autophagy plays an important role against TYLCV and TYLCCNV infection in whiteflies. First, we investigated whether the TYLCV infection could induce autophagy. By immunoblotting and qRT- PCR, we found that the level of autophagosome-specific protein marker ATG8-II and the expression of autophagy-related genes (Atg3, Atg9 and Atg12) were significantly higher in viruliferous whiteflies than those of nonviruliferous whiteflies (Fig. 1). The western blotting results were further confirmed by the immunofluorescence staining of autophagic signals and electron microscopy of autophagosome formation in the midgut of viruliferous whiteflies (Fig. 1B to F). A previous study demonstrates that TYLCV localizes mostly to the filter chamber and the descending midgut of the whitefly digestive tract.47 Therefore, the activation of ATG8-II in midgut (Fig. 3) suggests that autophagy may play critical roles in the whitefly midgut, an organ shown to be a barrier to the transmission of some viruses.11,13,48 By modulating the autophagy pathway in vivo, either by inhibiting it with 3-MA and BAF or activating it with rapamycin, we demonstrated a reciprocal effect on viral accumulation: virus activated the autophagy, which was utilized by whiteflies to inhibit the viral infection (Figs. 4 and 5). Furthermore, we found that activation of whitefly autophagy could inhibit the efficiency of virus transmission; whereas inhibiting autophagy facilitated virus transmission (Table 2 and Fig. 6). Taken together, these results indicate that TYLCV infection can activate the whitefly autophagy pathway, which leads to the subsequent degradation of virus.
Interestingly, the anti-ATG8 antibody (Abgent, AP1802a) can detect both ATG8-I and ATG8-II in western blot (Fig. 2A). However, when we did immunofluorescence staining, no signal could be detected in the nonviruliferous whiteflies (Fig. 2B). Therefore, we think that this antibody can only recognize the cleaved ATG8-II in immunofluorescence analysis. To prove that this antibody can detect ATG8-II specifically in immunofluorescence, whiteflies were also subjected to chloroquine or starvation treatment. Our results showed that after chloroquine (30 μM) or starvation (24 h) treatment, activation of autophagy was clearly detected (Fig. S3A). In addition, we tried another 2 antibodies from Sigma (L8918) and Cell Signaling Technology (3868S). Interestingly, the ATG8 antibody from Sigma could detect the ATG8-I in the nonviruliferous whiteflies and the signal increased markedly in the viruliferous whiteflies. Conversely, the antibody from Cell Signaling Technology cannot recognize ATG8-I or ATG8-II from whiteflies (Fig. S3B). These results suggest that these antibodies perform differently in whiteflies. It is possible that the antibody from Abgent (AP1802a) can detect both ATG8-I and ATG8-II in the western blot, but it can only recognize ATG8-II during immunofluorescence staining. Another possible explanation for the lack of autophagic signal in the nonviruliferous whiteflies in immunofluorescence is that under normal conditions the ATG8-I protein is predominantly diffusely distributed throughout the cytoplasm. Under our experimental condition, these signals are rather weak and cannot be detected. Upon virus infection, there is a marked increase in the formation of ATG8-II puncta, which are detectable under confocal microscopy.
It has been postulated that TYLCV may accumulate and transcript in whiteflies.39,40 Accumulation of viral DNA in B. tabaci reared on a nonhost plant of TYLCV after first feeding on plants infected with TYLCV has been interpreted as multiplication of TYLCV in its vector.11 This phenomenon has been clarified by feeding whiteflies with TYLCV infected tomato plants for 1 h, and measuring the viral DNA by Southern blot hybridization after the insects were transferred to feed on artificial diet. The amount of TYLCV DNA steadily increased after a lag period of 8 h, reached the maximum level approximately after 16 h and decreased thereafter.39 These reports seem to be confirmed by our studies: well after whiteflies had a short access (6 h) to infected tomato plants and moved onto cotton, both TYLCV and TYLCCNV CP and DNA increased significantly and then decreased (Figs. 2 and 7). TYLCV gene transcription in its vector whiteflies was also assessed after feeding whiteflies on virus-infected tomato plants and subsequent transfer to cotton.11,39,40 The increase of virion (+) strand (V1 and V2) in mRNA level (Fig. 2) indicates active TYLCV transcription, which appears similar to the results of Sinisterra et al.40 In plants, the geminivirus genome (+) strand replicates in the nucleus of infected cells, via the rolling circle model.49 The genome complementary (−) strand is synthesized, forming a dsDNA replicative form and the (−) strand serves as template for the synthesis of genome (+) strands. So, we also assessed the presence of the (−) strand in the whiteflies (Fig. 2) and detected the expression of C3 gene from the viral complementary (−) strand. A similar finding has just been reported by Pakkianathan et al.50 Interestingly, these phenomena are not universal for all begomoviruses, because no increase of vial genome could be detected for PalCuCNV (Fig. 8A) and gene transcripts of Tomato mottle virus (ToMoV, a New World bipartite begomovirus) rapidly became undetectable in whiteflies following transfer from tomato to cotton.40 These observation further suggesting that the replication of TYLCV and TYLCCNV in insect vectors is not random. Taken together, the possibility that TYLCV and TYLCCNV replicate in whiteflies is appealing.39,40,50 Nevertheless, to further confirm viral replication, the expression of Rep protein and other begomoviral gene products involved in the putative replication of TYLCV in whiteflies should be investigated.
Following the end of the 6 h AAP, TYLCV DNA was detected throughout whitefly's lifetime, while the amount of TYLCV CP steadily decreased and became undetectable after 288 h. These results suggest a difference in the retention of viral DNA and CP in B. tabaci, an observation that seems to be supported by previous studies.51 It has been shown that following a 4 d AAP on infected abutilon plants, the Abutilon mosaic virus (AbMV) DNA remained associated with B. tabaci throughout the 15 d sampling period, while the CP was detectable only for up to 7 d.17 Similarly, TYLCV CP began to decrease after 48 h when autophagy was detected, while TYLCV DNA still increased from 48 h to 96 h (Fig. 2). The activation of autophagy was associated with a rapid decrease of the viral CP, which may be one of the reasons for shorter retention period of CP than DNA.17,51 Furthermore, we successfully induced the formation of autophagosomes by rapamycin and reduced the viral accumulation in whiteflies (Fig. 5). These results were also confirmed by inhibiting autophagy by RNAi and 2 chemical inhibitors (3-MA and BAF) (Fig. 4). These observations support our hypothesis that autophagy is a key process during viral infection. Moreover, our data suggest that 3-MA and rapamycin treatment do not affect TYLCV acquisition, because the amount of CP and viral genome was comparable after 6 h AAP on infected plants in both treated and untreated whiteflies (Figs. 4 and 5). Thereby, autophagy is a direct defense mechanism against the retention of TYLCV in whiteflies. During the submission of our manuscript, another study also showed that TYLCV proteins may be degraded by plant and whitefly protease, ubiquitin 26S proteasome and autophagy.52 Increased understanding of this process will greatly improve our knowledge of the pathogenesis of plant virus.53
Materials and methods
Source and maintenance of insects, plants and viruses
The whitefly B. tabaci is a species complex with more than 35 morphologically indistinguishable cryptic species and mitochondrial cytochrome oxidase I (mtCOI) has been widely used to distinguish these cryptic species. The MEAM1 species of the B. tabaci complex (mtCOI GenBank accession no. GQ332577) and the greenhouse whitefly T. vaporariorum (mtCOI GenBank accession number HM153751) were reared on cotton (Gossypium hirsutum cv. Zhe-Mian 1793). Cotton plants were sown into pots and were cultivated to 7 to 8 true leaf stage for experiments. Tomato plants (Lycopersicon esculentum cv Hezuo903) at 3 to 4 true leaf stage were inoculated with TYLCV by agroinoculation as previously described.43 All cultures were grown in cages held in a greenhouse at temperatures of 25 to 27°C, 60% relative humidity and 14 h light/10 h darkness. Clones of TYLCV isolate SH2 (GenBank accession number AM282874.1), TYLCCNV isolate Y10 (GenBank accession number AJ319675.1) with a DNA-B (GenBank accession number AJ421621.1) and PalCuCNV isolate HeNZM1 (GenBank accession number FN256260.1) were obtained from the Institute of Biotechnology, Zhejiang University and agroinoculated into tomato.
Acquisition of viruliferous whiteflies from TYLCV-infected tomato plants
First, 300 mixed-sex adult whiteflies on cotton were separately transferred to virus-infected and uninfected tomato plants for 48 h. PCR analyses showed that nearly 100% of the whiteflies on virus-infected tomato carried viruses. The 2 groups of whitefly samples were referred to as the viruliferous and nonviruliferous whiteflies. Then, the activation of autophagy pathway in viruliferous and nonviruliferous whiteflies was analyzed by western blot, qRT-PCR, transmission electron microscopy and immunofluorescence.
Time-course of autophagy activation in response to viral infection
About 5,000 of mixed-sex whiteflies were separately collected and moved onto virus-infected and uninfected tomato plants in different cages. After 6-h feeding, they were transferred to cotton plants. Then, 300 nonviruliferous and viruliferous whiteflies were collected respectively after various time-points to determine if the viral infection can induce autophagy. The accumulation of viral genome and CP were measured by qPCR and western blot. The activation of autophagy at various time points after viral infection was analyzed by western blot, qRT-PCR and immunofluorescence.
Western blot
For western blot analyses, total protein was isolated using the loading buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 0.1% bromophenol blue, 1% β-mercaptoethanol and 2% SDS). Protein samples were separated by 12% or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidine difluoride membranes (MultiSciences Biotech, 4279NK). The membranes were blocked with 5% nonfat milk in phosphate-buffered saline (PBS; Sangon Biotech, SB0627) with 0.1% Tween 20 (BBI Life Sciences, 9005-64-5) and then incubated with the first antibody. After incubation with secondary antibody (MultiSciences Biotech, GAM007), signals were visualized with the ECL Plus Detection System (Bio-Rad, 170-5060). The antibodies for TYLCV CP and TYLCCNV CP were provided by the Institute of Biotechnology, Zhejiang University. The commercial anti-ATG8 (AP1802a) and anti-ACTB antibodies (sc-130656) were purchased from Abgent and Santa Cruz Biotechnology, respectively. The commercial anti-SQSTM1/p62 (8025s), anti-cyclin D (3741s), anti-MAPK/ERK (4370s), anti-MAPK/p38 (9215s) and anti-CASP3/caspase 3 (9662s) were purchased from Cell Signal Technology.
Virus DNA extraction and quantitative PCR (qPCR)
Total DNA was extracted using the AquaPure Genomic DNA Isolation kit (Bio-Rad, 732-6340). qPCR was performed on a CFX96™ Real-Time system (Bio-Rad, USA) with SYBR green detection (Takara, RR071A). The TYLCV genomic DNA (GenBank accession no. AM282874.1) abundance was determined using qPCR with the following primers, F (5′-GAAGCGACCAGGCGATATAA-3′) and R (5′-GGAACATCAGGGCTTCGATA-3′). The TYLCCNV genomic DNA (GenBank accession number AJ319675.1) was measured with the following primers: F (5′-CCTTCGAATTGGATGAGGAT-3′) and R (5′-GGAGGCACTTTCCCAAATTA-3′). The PalCuCNV genomic DNA (GenBank accession no. FN256260.1) was measured with the following primers: F (5′-ATGTTCCTCGTGGTTGTGAA-3′) and R (5′-TCCGACACACAAATGACCTT -3′). ACTB gene of B. tabaci (GenBank accession number AF071908) was measured in parallel as an internal control with the forward primer, 5′-TCTTCCAGCCATCCTTCTTG-3′ and reverse primer, 5′-CGGTGATTTCCTTCTGCATT-3′. RPL29 (GenBank accession no. EE596314) was measured in parallel as an internal control with the forward primer, 5′-TCGGAAAATTACCGTGAG-3′ and reverse primer, 5′-GAACTTGTGATCTACTCCTCTCGTG-3′. ACTB gene of greenhouse whitefly (GenBank accession number EU934242.1) was measured in parallel as an internal control with the forward primer, 5′-TCTTCCAGCCATCCTTCTTG-3′ and reverse primer, 5′-CGGTGATTTCCTTCTGCATT-3′.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR) analysis
Total RNA was isolated using the SV Total RNA isolation system (Promega, Z3100) according to the manufacturer's protocol. cDNA was synthesized using the SYBR PrimeScript reverse transcription-PCR (RT-PCR) kit II (Takara, RR037A). qRT-PCRs were carried out on the CFX96™ Real-Time system with SYBR green detection. The gene expression of Atg3 (GenBank accession no. HP660691) was determined using qRT-PCR with the following primers, F (5′-CCAGATTGTCTCCAGCAGCA-3′) and R (5′-CGTTTAAGGGAACAGCACTTG-3′). The gene expression of Atg9 (GenBank accession number HP662980) was determined with the following primers, F (5′-AGGGTTCCTGGTTCACGC-3′) and R (5′-TTGCCATCATTAACTTTCTGCT-3′); and Atg12 (GenBank accession no. HP659053) was measured by F (5′-TCAAAGCCACTGGAAACGC-3′) and R (5′-TCTGGTCTGGAGCAGGAGC-3′). The gene expression of TYLCV V1, V2 and C3 (GenBank accession number AM282874.1) were determined using the following pairs of primers: V1 (F: 5′-GAAGCGACCAGGCGATATAA-3′ and R: 5′-GGAACATCAGGGCTTCGATA-3′); V2 (F: 5′-TCTGTTCACGGATTTCGTTG-3′ and R: 5′- GCTGTCGAAGTTCAGCCTTC-3′); C3 (F: 5′-TGAGGCTGTAATGTCGTCCA-3′ and R: 5′-GCTCCTCAAGCAGAGAATGG-3′). As endogenous controls, the expression of ACTB and RPL29 was measured in parallel.22,54
Immunofluorescence
For immunofluorescence analyses, whitefly guts were dissected in TBS (10 mM Tris-HCl, 150 mM sodium chloride, pH 7.5) and fixed in 4% paraformaldehyde (MultiSciences Biotech, LK-F0001) for 2 h at room temperature. The guts were washed in TBS:0.1% Triton X-100 (Sangon Biotech, 9002-93-1) twice, and blocked in 1% BSA:TBST (TBS buffer with 0.05 % Tween 20) for 2 h. Primary antibody was diluted in TBST and incubated overnight at 4°C. The guts were washed twice in TBST and incubated with secondary antibody diluted in TBST for 2 h at room temperature. After washing twice in TBST, the dissected guts were mounted in fluoroshield mounting medium with DAPI (Abcam, ab104139) and imaged on Zeiss LSM710 confocal microscope (Zeiss, Germany).55 Experiments were performed at least 3 times. Whitefly guts were incubated with anti-CP MAb (3E10) at a 1:500 dilution and ATG8 antibody (Abgent, AP1802a) at a 1:50 dilution. Anti-CP binding was detected with the secondary antibody labeled with Dylight 488 (MultiSciences Biotech, LK-GAM4882) and ATG8 antibody binding was detected with the secondary antibody labeled with Dylight 549 (MultiSciences Biotech, LK-GAR5492).
Transmission electron microscopy
For transmission electron microscopy analyses, whitefly guts were first dissected and fixed with 2.5% glutaraldehyde (Sangon Biotech, A500484) in phosphate buffer (0.1 M, pH 7.0) for 4 h, and then postfixed with 1% OsO4 (TED PELLA INC, 4008-132207) in phosphate buffer for 2 h. After a series of ethanol dehydration, gut samples were transferred to acetone for 20 min and then embedded in Spurr resin (SPI-CHEM, 2386-87-0). After staining with uranyl acetate (SCRC, 6159-44-0) and alkaline lead citrate (Sigma, Sodium citrate tribasic hydrate, 25114; Sigma, Lead (II) nitrate, 203580), samples were sectioned and observed under a Hitachi H-7650 transmission electron microscope (Japan).
Effects of autophagy inhibitors on TYLCV infection
Autophagy inhibitors 3-methyladenine (3-MA; Sigma, M9281) and BAF (Enzo, BML-CM110-0100) were used to investigate the effects of autophagy on TYLCV infection. Whiteflies were first fed with 1 μM 3-MA or 10 nM BAF (dissolved in PBS), and mixed with 15% sucrose (SCR, 10021418) in 50-mm diameter cylindrical containers for 24 h. Whiteflies fed with 15% sucrose and PBS were referred to as control groups. Next, the whiteflies were treated as described above (Time-course of autophagy activation in response to TYLCV infection in whiteflies).
RNA interference
For RNA interference, about 100 adult whiteflies were put in 50-mm diameter cylindrical containers, allowing them to feed on a 15% sucrose diet with 200 ng/μl Atg3 or Atg9 dsRNA.56 The sucrose diet containing 200 ng/μl dsGFP was used as a negative control. The dsRNA was synthesized using AmpliScribeTM T7-Flash Transcription kit (Epicentre, ASF3507). For RNAi experiments, whiteflies were first fed on a 15% sucrose diet with dsRNA for 2 d and then transferred onto the virus-infected tomato for 6 h to acquire the virus. Subsequently, the whiteflies were transferred onto cotton for 120 h and at different time points the whiteflies were collected for western blot, confocal and qPCR analysis. All the treatments were replicated 3 times.
Effects of autophagy induction on TYLCV infection
The autophagy inducer rapamycin (LC Laboratories, R-5000) was used to investigate the effects of autophagy on TYLCV infection. Whiteflies were first fed with 10 μM rapamycin (dissolved in PBS) and mixed with 15% sucrose in 50-mm diameter cylindrical containers for 24 h. Whiteflies fed with 15% sucrose and PBS were referred to as control groups. Next, the whiteflies were treated as described above. Viral accumulation and autophagy activation were tested by western blot and qPCR.
Acquisition, retention and transmission of TYLCV by whiteflies treated with 3-MA and rapamycin
A total of 600 newly emerged (0 to 24 h) adult whiteflies were fed with 3-MA or rapamycin mixed with 15% sucrose in 50-mm diameter cylindrical containers. Then they were inoculated onto each of the 2 TYLCV-infected tomato plants, which were enclosed singly in insect-proof cages. Thereafter, 10 adults were randomly collected from the top second and third leaves of each of the 3 plants at different AAP on the plants (Table 1). The adults collected were stored at −80°C and assayed individually by PCR for TYLCV DNA. Two hundred adult whiteflies that had been fed on TYLCV-infected tomato plant for 48 h were placed on a healthy cotton plant. Following the initial placement, a group of 10 live adults were collected on the d 0, 15 and 30 and assayed individually by PCR for TYLCV DNA. For virus transmission experiment, whiteflies were first fed on diet with and without 10 μM rapamycin, 1 μM 3-MA for 24 h. Then approximately 200 whitefly adults were separately collected from the rapamycin-treated, 3-MA treated and control groups and allowed to feed on TYLCV-infected tomato plants. After 6 h, the viruliferous whitefies were transferred onto cotton for 120 h. Then one viruliferous whitefly was collected and transferred into a clip-cage on the third leaf from the bottom of an uninfected tomato plant at the 3-true leaf stage. After 96 h, the whiteflies were removed and the plants were sprayed with imidacloprid at a concentration of 50 mg/L to kill the eggs. For each group, 20 plants (replicates) were used for virus transmission. After 30 d, the leaves of each tomato plant were collected and subjected to PCR examination of virus infection rate. In addition, the amount of virus in each plant was also quntified by qPCR.
Statistical methods
The data of qPCR was calculated using the comparative CT method (2−ΔΔCt), and normalized against whitefly RPL29 or ACTB. A 2 × 3 test of independence was applied to compare frequencies of virus-infected versus uninfected plants among the 3 treatments. For the amount of virus in each tomato plant, statistical analysis was done with the Mann-Whitney test.
Supplementary Material
Abbreviations
- 3-MA
3-methyladenine
- AAP
acquisition access period
- ATG
autophagy related
- DAPI
4′,6-diamidino-2-phenylindole
- hpi
hours postinfection
- ATG8
microtubule-associated protein 1 light chain 3
- MEAM1
Middle East Asia Minor 1
- PBS
phosphate-buffered saline
- PalCuCNV
Papaya leaf curl China virus
- qPCR
quantitative PCR
- qRT-PCR
quantitative reverse transcription PCR
- ssDNA
single-stranded DNA
- TYLCV
tomato yellow leaf curl virus
- TYLCCNV
tomato yellow leaf curl China virus
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Yang Yue-Jian from Zhejiang Academy of Agricultural Science for kindly providing the tomato cultivars of Zheza 502 and Zhefen 701.
Funding
Financial support for this study was provided by the National Natural Science Foundation of China (31390421), National Basic Research Program of China (2014CB138404) and the National Natural Science Foundation of China (31321063).
References
- [1].De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: A statement of species status. Annu Rev Entomol 2011; 56:1-19; PMID:20690829; http://dx.doi.org/ 10.1146/annurev-ento-112408-085504 [DOI] [PubMed] [Google Scholar]
- [2].Boykin LM, P DB. A practical guide to identifying members of the Bemisia tabaci species complex: and other morphologically identical species. Front Ecol Evol 2014; 2:45; http://dx.doi.org/ 10.3389/fevo.2014.00045 [DOI] [Google Scholar]
- [3].Liu SS, De Barro PJ, Xu J, Luan JB, Zang LS, Ruan YM, Wan FH. Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 2007; 318:1769-72; PMID:17991828; http://dx.doi.org/ 10.1126/science.1149887 [DOI] [PubMed] [Google Scholar]
- [4].Oliveira MRV, Henneberry TJ, Anderson P. History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot 2001; 20:709-23; http://dx.doi.org/ 10.1016/S0261-2194(01)00108-9 [DOI] [Google Scholar]
- [5].Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 2008; 46:327-59; PMID:18680428; http://dx.doi.org/ 10.1146/annurev.phyto.022508.092135 [DOI] [PubMed] [Google Scholar]
- [6].Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S. Emergingvirus diseases transmitted by whiteflies. Annu Rev Phytopathol 2011; 49:219-48; PMID:21568700; http://dx.doi.org/ 10.1146/annurev-phyto-072910-095235 [DOI] [PubMed] [Google Scholar]
- [7].Colvin J, Omongo CA, Govindappa MR, Stevenson PC, Maruthi MN, Gibson G, Seal SE, Muniyappa V. Host-plant viral infection effects on arthropod-vector population growth, development and behaviour: management and epidemiological implications. Adv Virus Res 2006; 67:419-52; PMID:17027686; http://dx.doi.org/ 10.1016/S0065-3527(06)67011-5 [DOI] [PubMed] [Google Scholar]
- [8].Jiu M, Zhou XP, Tong L, Xu J, Yang X, Wan FH, Liu SS. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS One 2007; 2:e182; PMID:17264884; http://dx.doi.org/ 10.1371/journal.pone.0000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Czosnek H, Laterrot H. A worldwide survey of Tomato yellow leaf curl viruses. Arch Virol 1997; 142:1391-406; PMID:9267451; http://dx.doi.org/ 10.1007/s007050050168 [DOI] [PubMed] [Google Scholar]
- [10].Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 2011; 12:938-54; PMID:22017770; http://dx.doi.org/ 10.1111/j.1364-3703.2011.00752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Czosnek H, Ghanim M. Back to basics: Are begomoviruses whitefly pathogens? J Integrative Agriculture 2012; 11:225-34; http://dx.doi.org/ 10.1016/S2095-3119(12)60007-0 [DOI] [Google Scholar]
- [12].Gray S, Cilia M, Ghanim M. Circulative, “nonpropagative” virus transmission: an orchestra of virus-, insect-, and plant-derived instruments. Adv Virus Res 2014; 89:141-99; PMID:24751196; http://dx.doi.org/ 10.1016/B978-0-12-800172-1.00004-5 [DOI] [PubMed] [Google Scholar]
- [13].Czonek H, Ghanim M. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci-insights from studies with Tomato yellow leaf curl virus. Ann Appl Biol 2002; 140:215-31; http://dx.doi.org/ 10.1111/j.1744-7348.2002.tb00175.x [DOI] [Google Scholar]
- [14].Wei J, Zhao JJ, Zhang T, Li FF, Ghanim M, Zhou XP, Ye GY, Liu SS, Wang XW. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. J Virol 2014; 88:13460-8; PMID:25210181; http://dx.doi.org/ 10.1128/JVI.02179-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Kontsedalov S, Skaljac M, Brumin M, Sobol I, Czosnek H, Vavre F, Fleury F. The transmission efficiency of Tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J Virol 2010; 84:9310-7; PMID:20631135; http://dx.doi.org/ 10.1128/JVI.00423-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li JM, Ruan YM, Li FF, Liu SS, Wang XW. Gene expression profiling of the whitefly (Bemisia tabaci) Middle East-Asia Minor 1 feeding on healthy and Tomato yellow leaf curl China virus-infected tobacco. Insect Sci 2011; 18:11-22; http://dx.doi.org/ 10.1111/j.1744-7917.2010.01386.x [DOI] [Google Scholar]
- [17].Morin S, Ghanim M, Sobol I, Czosnek H. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology 2000; 276:404-16; PMID:11040131; http://dx.doi.org/ 10.1006/viro.2000.0549 [DOI] [PubMed] [Google Scholar]
- [18].Ghanim M, Czosnek H. Tomato yellow leaf curl geminivirus (TYLCV-Is) is transmitted among whiteflies (Bemisia tabaci) in a sex-related manner. J Virol 2000; 74:4738-45; PMID:10775612; http://dx.doi.org/ 10.1128/JVI.74.10.4738-4745.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ghanim M, Morin S, Zeidan M, Czosnek H. Evidence for transovarial transmission of Tomato yellow leaf curl virus by its vector, the whitefly Bemisia tabaci. Virology 1998; 240:295-303; PMID:9454703; http://dx.doi.org/ 10.1006/viro.1997.8937 [DOI] [PubMed] [Google Scholar]
- [20].Wang J, Zhao H, Liu J, Jiu M, Qian YJ, Liu SS. Low frequency of horizontal and vertical transmission of two begomoviruses through whiteflies exhibits little relevance to the vector infectivity. Ann Appl Biol 2010; 157:125-33; http://dx.doi.org/ 10.1111/j.1744-7348.2010.00403.x [DOI] [Google Scholar]
- [21].Jiu M, Zhou XP, Liu SS. Acquisition and transmission of two Begomoviruses by the B and a non-B Biotype of Bemisia tabaci from Zhejiang, China. J Phytopathol 2006; 154:587-91; http://dx.doi.org/ 10.1111/j.1439-0434.2006.01151.x [DOI] [Google Scholar]
- [22].Luan JB, Li JM, Varela N, Wang YL, Li FF, Bao YY, Zhang CX, Liu SS, Wang XW. Global analysis of the transcriptional response of whitefly to Tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J Virol 2011; 85:3330-40; PMID:21270146; http://dx.doi.org/ 10.1128/JVI.02507-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; http://dx.doi.org/ 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005; 118:7-18; PMID:15615779; http://dx.doi.org/ 10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jiang X. Animal genes identification and mTOR signaling reactivation in autophagy. Protein Cell 2010; 1:699-701; PMID:21203908; http://dx.doi.org/ 10.1007/s13238-010-0091-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang WP, Klionsky DJ. Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct 2002; 27:409-20; PMID:12576634; http://dx.doi.org/ 10.1247/csf.27.409 [DOI] [PubMed] [Google Scholar]
- [27].Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5:539-45; PMID:14536056; http://dx.doi.org/ 10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- [28].Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 2007; 7:767-77; PMID:17767194; http://dx.doi.org/ 10.1038/nri2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753-66; PMID:15607973; http://dx.doi.org/ 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- [30].Tallóczy Z, Virgin IV H, Levine B. PKR-dependent xenophagic degradation of herpes simplex virus type 1. Autophagy 2006; 2:24-9; PMID:Can't; http://dx.doi.org/ 10.4161/auto.2176 [DOI] [PubMed] [Google Scholar]
- [31].Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against Vesicular stomatitis virus. Immunity 2009; 30:588-98; PMID:19362021; http://dx.doi.org/ 10.1016/j.immuni.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang XW, Luan JB, Li JM, Su YL, Xia J, Liu SS. Transcriptome analysis and comparison reveal divergence between two invasive whitefly cryptic species. BMC Genomics 2011; 12:458; PMID:21939539; http://dx.doi.org/ 10.1186/1471-2164-12-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7:167-78; PMID:15296714; http://dx.doi.org/ 10.1016/j.devcel.2004.07.009 [DOI] [PubMed] [Google Scholar]
- [34].Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fang Y, Jiao X, Xie W, Wang S, Wu Q, Shi X, Chen G, Su Q, Yang X, Pan H, et al.. Tomato yellow leaf curl virus alters the host preferences of its vector Bemisia tabaci. Sci Rep 2013; 3:2876; PMID:24096821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xu J, Lin KK, Liu SS. Performance on different host plants of an alien and an indigenous Bemisia tabaci from China. J Appl Entomol 2011; 135:771-9; http://dx.doi.org/ 10.1111/j.1439-0418.2010.01581.x [DOI] [Google Scholar]
- [37].Xu HX, Hong Y, Zhang MZ, Wang YL, Liu SS, Wang XW. Transcriptional responses of invasive and indigenous whiteflies to different host plants reveal their disparate capacity of adaptation. Sci Rep 2015; 5:10774; PMID:26041313; http://dx.doi.org/ 10.1038/srep10774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jiao XY, Zhou XP, Yang YJ, Xie Y. Identification for resistance of tomato varieties against geminiviruses. ACTA Phytopathologica Sinica 2013; 43:655-8. [Google Scholar]
- [39].Czosnek H, Ghanim M, Morin S, Rubinstein G, Fridman V, Zeidan M. Whiteflies: vectors, and victims (?), of geminiviruses. Adv Virus Res 2001; 57:291-322; PMID:11680387; http://dx.doi.org/ 10.1016/S0065-3527(01)57006-2 [DOI] [PubMed] [Google Scholar]
- [40].Sinisterra XH, McKenzie CL, Hunter WB, Powell CA, Shatters RG. Differential transcriptional activity of plant-pathogenic begomoviruses in their whitefly vector (Bemisia tabaci, Gennadius: Hemiptera Aleyrodidae). J Gen Virol 2005; 86:1525-32; PMID:15831966; http://dx.doi.org/ 10.1099/vir.0.80665-0 [DOI] [PubMed] [Google Scholar]
- [41].Takatsuka C, Inoue Y, Matsuoka K, Moriyasu Y. Three-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol 2004; 45:265-74; PMID:15047874; http://dx.doi.org/ 10.1093/pcp/pch031 [DOI] [PubMed] [Google Scholar]
- [42].Tanemura M, Nagano H, Taniyama K, Kamiike W, Mori M, Doki Y. Role of rapamycin-induced autophagy in pancreatic islets. Am J Transplant 2012; 12:1067; PMID:22299600; http://dx.doi.org/ 10.1111/j.1600-6143.2011.03933.x [DOI] [PubMed] [Google Scholar]
- [43].Cui X, Tao X, Xie Y, Fauquet CM, Zhou X. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J Virol 2004; 78:13966-74; PMID:15564504; http://dx.doi.org/ 10.1128/JVI.78.24.13966-13974.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang H, Ma XY, Qian YJ, Zhou XP. Molecular characterization and infectivity of Papaya leaf curl China virus infecting tomato in China. J Zhejiang Univ Sci B 2010; 11:109-14; PMID:20104645; http://dx.doi.org/ 10.1631/jzus.B0900176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ohnishi J, Kitamura T, Terami F, Honda K. A selective barrier in the midgut epithelial cell membrane of the nonvector whitefly Trialeurodes vaporariorum to Tomato yellow leaf curl virus uptake. J Gen Plant Pathol 2009; 75:131-9; http://dx.doi.org/ 10.1007/s10327-009-0147-3 [DOI] [Google Scholar]
- [46].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ghanim M, Brumin M, Popovski S. A simple, rapid and inexpensive method for localization of Tomato yellow leaf curl virus and Potato leafroll virus in plant and insect vectors. J Virol Methods 2009; 159:311-4; PMID:19406154; http://dx.doi.org/ 10.1016/j.jviromet.2009.04.017 [DOI] [PubMed] [Google Scholar]
- [48].Czosnek H. Tomato yellow leaf curl virus disease: management, molecular biology, breeding for resistance. Springer; Dordrecht, The Netherlands, 2007. [Google Scholar]
- [49].Saunders K, Lucy A, Stanley J. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res 1991; 19:2325-30; PMID:2041773; http://dx.doi.org/ 10.1093/nar/19.9.2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pakkianathan BC, Kontsedalov S, Lebedev G, Mahadav A, Zeidan M, Czosnek H, Ghanim M. Replication of Tomato yellow leaf curl virus in its whitefly vector Bemisia tabaci. J Virol 2015; 89(19):9791-803; PMID:26178995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rubinstein G, Czosnek H. Long-term association of Tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J Gen Virol 1997; 78:2683-9; PMID:9349491; http://dx.doi.org/ 10.1099/0022-1317-78-10-2683 [DOI] [PubMed] [Google Scholar]
- [52].Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463; PMID:15068787; http://dx.doi.org/ 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- [53].Gorovits R, Fridman L, Kolot M, Rotem O, Ghanim M, Shriki O, Czosnek H. Tomato yellow leaf curl virus confronts host degradation by sheltering in small/midsized protein aggregates. Virus Res 2016; 213:304-13; PMID:26654789; http://dx.doi.org/ 10.1016/j.virusres.2015.11.020 [DOI] [PubMed] [Google Scholar]
- [54].Li R, Xie W, Wang S, Wu Q, Yang N, Yang X, Pan H, Zhou X, Bai L, Xu B, et al.. Reference gene selection for qRT-PCR analysis in the sweet potato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE 2013; 8:e53006; PMID:23308130; http://dx.doi.org/ 10.1371/journal.pone.0053006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang LL, Wei XM, Ye XD, Xu HX, Zhou XP, Liu SS, Wang XW. Expression and functional characterisation of a soluble form of Tomato yellow leaf curl virus coat protein. Pest Manag Sci 2014; 70:1624-31; PMID:24488592; http://dx.doi.org/ 10.1002/ps.3750 [DOI] [PubMed] [Google Scholar]
- [56].Asokan R, Rebijith KB, Roopa HK, Kumar NK. Non-invasive delivery of dsGST is lethal to the sweet potato whitefly, Bemisia tabaci (G.) (Hemiptera: Aleyrodidae). Appl Biochem Biotechnol 2015; 175:2288-99; PMID:25480347; http://dx.doi.org/ 10.1007/s12010-014-1437-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.