ABSTRACT
In contrast to normal tissue, cancer cells display profound alterations in protein synthesis and degradation. Therefore, proteins that regulate endoplasmic reticulum (ER) homeostasis are being increasingly recognized as potential therapeutic targets. The ubiquitin-proteasome system and autophagy are crucially important for proteostasis in cells. However, interactions between autophagy, the proteasome, and ER stress pathways in cancer remain largely undefined. This study demonstrated that withaferin-A (WA), the biologically active withanolide extracted from Withania somnifera, significantly increased autophagosomes, but blocked the degradation of autophagic cargo by inhibiting SNARE-mediated fusion of autophagosomes and lysosomes in human pancreatic cancer (PC) cells. WA specifically induced proteasome inhibition and promoted the accumulation of ubiquitinated proteins, which resulted in ER stress-mediated apoptosis. Meanwhile, the impaired autophagy at early stage induced by WA was likely activated in response to ER stress. Importantly, combining WA with a series of ER stress aggravators enhanced apoptosis synergistically. WA was well tolerated in mice, and displayed synergism with ER stress aggravators to inhibit tumor growth in PC xenografts. Taken together, these findings indicate that simultaneous suppression of 2 key intracellular protein degradation systems rendered PC cells vulnerable to ER stress, which may represent an avenue for new therapeutic combinations for this disease.
KEYWORDS: autophagy, ER stress, proteasome, SNAREs, withaferin A
Introduction
Pancreatic cancer (PC) has one of the worst prognoses among malignant tumors. Despite advances in early diagnosis and treatment of PC, the 5-year survival rate is less than 5% and the median survival is only 6 mo.1 This neoplasm is often resistant to available therapeutic approaches such as radiotherapy and chemotherapy, in part because of frequent genomic alterations.2 Currently, gemcitabine is the mainstay of therapy; however, response rates are less than 20%.3 Thus, this lack of effective therapeutic strategies urgently raises the need for novel systemic treatments.
The endoplasmic reticulum (ER) is an important organelle that plays a critical role in protein metabolism. Accumulation of misfolded proteins in the ER initiates a specialized response known as the unfolded protein response (UPR), which is the major protective mechanism during ER stress.4,5 The UPR activates an array of ER-located sensors, including ERN1 (endoplasmic reticulum to nucleus signaling 1), EIF2AK3 (eukaryotic translation initiation factor 2 α kinase 3), and ATF6 (activating transcription factor 6), which are normally inactivated through interaction with HSPA5 (heat shock protein family A (Hsp70) member 5). The main functions of the UPR are to reduce the amount of protein that enters the ER and to increase the folding capacity of the ER.5 Moreover, if proteins cannot be folded correctly in the ER, they are exported to the cytoplasm and degraded by the ubiquitin-proteasome system (UPS), through a process called ER-associated degradation (ERAD).6 However, when these adaptation strategies fail, the same system will trigger cell death via induction of pro-apoptotic transcription factors such as DDIT3/CHOP (DNA damage inducible transcript 3).7,8 Thus, using a proteasome inhibitor may interfere with clearance of misfolded proteins through the ERAD system, which appears to induce ER stress-mediated apoptosis.9 In addition, unlike normal tissues, tumor cells are exposed to chronic metabolic stress conditions that favor the activation of ER stress.10 Therefore, ER homeostasis is increasingly recognized as a promising target for cancer therapy.11
Besides the UPS, autophagy is another evolutionarily conserved intracellular degradation system. Macroautophagy/autophagy is characterized by the formation of functionally double-membrane compartments, phagophores, that sequester long-lived, misfolded proteins as well as damaged organelles, which are subsequently degraded following fusion of the resulting autophagosomes with lysosomes.12,13 In contrast to the specific degradation of ubiquitinated short-lived proteins by the proteasome, autophagy is considered to be a nonselective degradation system for long-lived proteins. Nevertheless, evidence suggests that these 2 degradation pathways are complementary and interlinked. Ubiquitinated proteins can be degraded by selective autophagy mediated via receptor proteins (e.g., SQSTM1/p62),14 whereas impairment of the UPS stimulates autophagy, which acts as a compensatory mechanism to remove protein aggregates.15 Furthermore, recent findings indicate that ER stress is a potent inducer of autophagy, which acts to remove toxic and damaged proteins, and abate ER stress.16-18 However, the reciprocal interactions between these pathways and their influence on therapeutic outcomes in cancer are largely unclear.
Withaferin-A (WA), a purified steroidal lactone isolated from the medicinal plant Withania somnifera, exhibits pro-apoptotic and antiproliferative activities in several cancer models.19,20 WA targets the tumor proteasome, which leads to ubiquitinated protein accumulation and induction of ER stress in human cancer cells.21,22 WA also has effects on autophagy, although the role of autophagy in the anticancer effects of WA remains to be determined.23,24 In this study, the effect of WA on protein homeostasis in PC cells was examined. Our data indicate that WA induces incomplete autophagy by suppressing the fusion of autophagosomes and lysosomes. Furthermore, WA inhibits proteasome activity, which results in ER stress-mediated apoptosis. Importantly, WA-mediated obstruction of autophagy and the proteasome significantly increased the cytotoxic effect of ER stress aggravators in vitro and in vivo.
Results
WA induces incomplete autophagy in PC cells
As indicated in Fig. 1A, WA decreased PC cell (Panc-1, SW1990, MIAPaCa-2, AsPC-1 and BxPc-3) survival in a dose-dependent manner. In contrast, treatment of human pancreatic ductal epithelial cells (HPDE) resulted in minimal loss of viable cell when exposed to identical concentrations of WA for a similar period (Fig. 1A). To determine whether WA induced autophagy, we first monitored autophagic alterations by analyzing the abundance of MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3 β). Generally, the covalent conjugation of a soluble form of LC3B (LC3B-I) with phosphatidylethanolamine to form a nonsoluble form (LC3B-II) is a hallmark of autophagy.13 As shown in Fig. 1B and C, WA treatment of Panc-1 and MIAPaCa-2 cells increased the amount of LC3B-II protein in a dose- and time-dependent manner. Interestingly, all cell lines tested displayed similar trends in LC3B-II protein levels following WA treatment (Fig. S1). To monitor autophagosome formation, the lentivirus vector encoding the GFP-LC3B fusion protein was utilized. As expected, treatment with WA increased GFP-LC3B puncta in a dose- and time-dependent manner, indicating that there was a cumulative increase in autophagosomes (Fig. 1D, Fig. S2). Furthermore, transmission electron microscopy revealed an increase in autophagic vacuoles in the cytoplasm of Panc-1 and MIAPaCa-2 cells exposed to WA (Fig. 1E). Collectively, these data clearly demonstrate that WA causes the accumulation of autophagosomes in PC cells.
Figure 1.
WA specifically initiates autophagy but blocks the degradation of SQSTM1 in PC cells. (A) Panc-1, SW1990, MIAPaCa-2, AsPC-1, BxPc-3 and HPDE cells were treated with increasing concentrations of WA (0.5–5 μM) for 48 h; viable cells were quantified using the MTS assay. Data are presented as mean ± SD from 3 independent experiments. (B and C) Panc-1 and MIAPaCa-2 cells were treated for 24 h with the indicated concentrations of WA, or cells were treated with 2.5 μM WA for the indicated times. Levels of protein expression were analyzed by Western blot using antibodies against LC3B and GAPDH. (D) Panc-1 cells transfected with GFP-LC3B were treated with the indicated concentrations of WA for 24 h, or treated with 2.5 μM WA for the indicated period of time. The number of GFP-LC3B dots in each cell was quantified, and at least 50 cells were included for each group. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Scale bar: 20 μm. (E) Panc-1 and MIAPaCa-2 cells treated with DMSO (<0.1%) or WA (2.5 μM) for 24 h were imaged by transmission electron microscopy. Representative images of cells are shown. A magnified view of the electron photomicrograph shows a characteristic autophagosome. Arrowhead, autophagic vacuoles; N, nuclear. Quantification of the number of autophagic vacuoles from at least 20 randomly selected areas is shown (***, p < 0.001). Scale bar: 500 nm. (F) Panc-1 and MIAPaCa-2 cells were either untreated or treated with WA (1–2.5 μM) for 24 h in the absence or presence of Baf-A1 (100 nM), CQ (10 μM) or E64D together with pepstatin (P/E; 10 μg/ml). The indicated protein levels were analyzed by western blot. (G) Panc-1 cells were treated with DMSO (<0.1%), CQ (10 μM) or WA (2.5 μM) for 24 h followed by immunostaining with an anti-SQSTM1 antibody. Nuclei are counterstained with DAPI (blue). Scale bar: 20 μm. (H) Panc-1 cells were pretreated with Rap (100 nM) for 30 min, followed by treatment with 2.5 μM WA or 10 μM CQ for another 24 h, and then the indicated protein levels were analyzed by Western blot.
Autophagosome accumulation in cells is an intermediate process within the autophagic flux, which reflects a balance between their rate of formation and degradation. Thus, accumulation of autophagosomes in WA-treated cells could, in principle, be explained by 3 possibilities: (1) WA induces complete autophagy, (2) WA suppresses basic autophagic flux, or (3) WA induces incomplete autophagy. To examine this, a series of lysosomal inhibitors, bafilomycin A1 (Baf-A1), chloroquine (CQ), or E64D together with pepstatin, were used. As indicated in Figure 1F, the addition of these agents dramatically increased the accumulation of LC3B-II after WA (1–2.5 μM) treatment in both Panc-1 and MIAPaCa-2 cells, suggesting that WA induces autophagy (either complete autophagy or incomplete autophagy). Interestingly, the abundance of SQSTM1, an LC3B binding protein and receptor that is degraded via autophagy,25 was markedly increased in cells treated with WA (Fig. 1F and G). In addition, when CQ was combined with a high concentration of WA (5 μM), no difference in LC3B-II levels was observed (Fig. S3A), indicating that a high concentration of WA saturates the ability of CQ to block autophagic flux.
Next, dual treatment with CQ and WA plus rapamycin, which induces autophagy by inhibiting MTOR (mechanistic target of rapamycin [serine/threonine kinase]), resulted in a significant increase in LC3B-II levels (Fig. 1H, Fig. S3B) as well as the increased appearance of GFP-LC3B puncta (Fig. S3C). Conversely, SQSTM1 levels decreased under rapamycin treatment, but were markedly increased by exposure to the CQ and WA combination (Fig. 1H, Fig. S3B). Moreover, phosphorylated (p)-RPS6 levels (a surrogate measure of MTOR activity) were dramatically reduced by rapamycin treatment, but not obviously affected by WA or CQ (Fig. 1H). In addition, WA treatment prevented the starvation-induced degradation of LC3B-II and SQSTM1 protein levels in both Panc-1 and MIAPaCa-2 cells (Fig. S4). Together, these results indicate that WA is a potent autophagic flux inhibitor; WA-induced autophagosome accumulation is due to impaired autophagic degradation rather than promoting autophagic flux in PC cells.
WA inhibits the fusion of lysosomes and autophagosomes
Because autophagosome maturation depends on its fusion with a lysosome, inhibition of the fusion process or the activation of lysosomal proteases impairs autophagic degradation. To confirm whether WA suppressed the maturation of autophagy, a tandem labeled GFP-mRFP-LC3B construct was used. The GFP of this tandem autophagosome reporter is sensitive to pH and quenched in the acidic environment of the lysosome, whereas the mRFP is resistant. Therefore, the fusion of autophagosomes with lysosomes results in the loss of yellow puncta and the appearance of red-only puncta.26 As shown in Figs. 2A and S5, in rapamycin-treated cells, only parts of the LC3B-positive puncta were yellow. Conversely, CQ and WA inhibited the maturation of autophagy, resulting in predominantly autophagosomes (yellow) in cells.
Figure 2.
WA inhibits the fusion of lysosomes with autophagosomes, but has no effect on lysosomal function and degradation of endocytic cargo. (A) Panc-1 cells were transiently transfected with GFP-mRFP-LC3B for 48 h and subsequently treated with CQ (10 μM), WA (2.5 μM), or Rap (100 nM) for 12 h, and then observed for the change of both green and red fluorescence using a confocal microscope. Scale bar: 20 μm (white) and 2 μm (blue). Lower panel, the numbers of acidified autophagosomes (GFP− RFP+) versus neutral autophagosomes (GFP+RFP+) per cell in each condition were quantified. Data are presented as mean ± SD from 3 independent experiments (N.S, not significant; *, p < 0.05; ***, P < 0.001). (B) Western blot analysis of autophagy-related protein levels after Panc-1 and MIAPaCa-2 cells were treated with WA for 24 h at the indicated concentrations. (C) Panc-1 and MIAPaCa-2 cells were either untreated or treated with WA (2.5 μM) for 24 h in the absence or presence of Baf-A1 (100 nM), then stained with acridine orange (1 mg/ml for 15 min). Representative results of 2 independent experiments are shown. Scale bar: 100 μm. (D) FACS analysis of LysoTracker Red after Panc-1 and MIAPaCa-2 cells were treated without or with WA (2.5 μM) in the absence or presence of Baf-A1 (100 nM) for 24 h. (E) Enzymatic activity of CTSB and CTSD in WA-treated Panc-1 cells. Cells were treated with WA for 24 h at the indicated concentrations. Enzymatic activity was analyzed using fluorogenic kits. Data are presented as mean ± SD from 3 independent experiments. (F) Panc-1 cells were serum-starved overnight, and incubated without or with WA (2.5 μM) for 12 h before stimulating with 100 ng/ml EGF for 0, 0.5, 1 and 2 h. EGFR protein levels were analyzed by Western blot.
To address whether WA affects autophagosome-lysosome fusion, we examined the colocalization of GFP-LC3B and LysoTracker Red, a specific dye for live cell lysosome labeling. As a positive control, rapamycin treatment induced a remarkable increase of GFP-LC3B puncta, which were well colocalized with LysoTracker Red. In contrast, GFP-LC3B puncta were greatly increased in WA-treated cells and did not colocalize with LysoTracker Red (Fig. S6A). The intensity of LysoTracker Red dye can be changed by pH alteration; therefore we examined the colocalization of GFP-LC3B and LAMP2 (lysosomal-associated membrane protein 2), a marker for endosomal and lysosomal membranes. As shown in Figure S6B, WA-treated cells exhibited separation of GFP-LC3B puncta and LAMP2. In contrast, GFP-LC3B was well colocalized with LAMP2 during rapamycin treatment. These findings indicate that WA blocks the fusion of autophagosomes with lysosomes.
Subsequent experiments were performed to ascertain the mechanism(s) by which WA causes impaired autophagy. First, expression of several autophagy-related (ATG) proteins was investigated. Paradoxically, WA markedly decreased expression levels of BECN1, whereas it increased expression levels of ATG14 (Fig. 2B). Interestingly, downregulation of BECN1 did not attenuate the accumulation of LC3B-II or appearance of GFP-LC3B puncta induced by WA, whereas autophagosome accumulation elicited by WA was significantly abrogated after knockdown of ATG5 or ATG7 (Fig. S7), suggesting WA-induced autophagosome accumulation was autophagy-dependent. Because RAB5 (early endosome marker), RAB7 (late endosome marker), RAB11 (recycling endosome marker), and LAMP1 are critical for lysosome fusion with endosomes and autophagosomes,27 we examined the effects of WA on the expression of these proteins by western blot analysis. Interestingly, treating cells with WA increased the levels of RAB5 and RAB7 in a dose-dependent manner (Fig. S8A). However, no apparent change was detected in RAB11 or LAMP1 expression (Fig. S8A). These data suggested that the mechanism by which WA blocked autophagosome-lysosome fusion was not due to reduced expression of these proteins.
As autophagosome-lysosome fusion depends on the pH in acidic compartments,28 we first used acridine orange (AO; a dye that accumulates in intracellular acidic vesicles) staining to evaluate lysosomal pH. As shown in Figure 2C, treating cells with WA resulted in a significant increase in acidic vesicles compared to the control. In contrast, treating with Baf-A1, a selective inhibitor of the vacuolar-type H+-translocating ATPase that can raise the pH of acidic compartments, resulted in a considerable decrease in acidic vesicles. LysoTracker Red manifests red fluorescence in a pH-dependent manner in the lysosome. Our measurements also indicated that the LysoTracker Red fluorescent intensities were significantly enhanced in WA-treated cells, but were dramatically decreased in Baf-A1-treated cells (Fig. 2D). Thus, our data indicated that impaired autophagic flux induced by WA was not due to inhibition of lysosomal acidification.
Autophagy is a process involved in the proteolytic degradation of cellular macromolecules in lysosomes, which requires the activity of proteases. Cathepsins are the most studied lysosomal proteases that participate in autophagic degradation.29 We next investigated whether WA treatment affects the expression and maturation process of 2 main cathepsins, CTSB (cathepsin B) and CTSD (cathepsin D). As shown in Fig. S8B, WA had no affect on CTSB and CTSD protein levels in Panc-1 and MIAPaCa-2 cells, including both precursors and mature forms. Consistently, the enzymatic activity of CTSB and CTSD had not changed upon WA treatment (Fig. 2E). Finally, we used EGFR (epidermal growth factor receptor) as an endogenous substrate to monitor lysosomal activity. EGFR is a typical member of the receptor tyrosine kinase family, which after ligand binding-induced activation is endocytosed and delivered to lysosomes for degradation. In this assay, Panc-1 cells were treated with EGF for 0.5, 1 or 2 h in the presence or absence of WA, and the rate of EGFR degradation was assessed by western blot analysis. Consistent with our results from the cathepsin activity assay, no defect in ligand-induced EGFR degradation was observed after WA treatment (Fig. 2F).
Taken together, these data demonstrate that, although WA blocks the fusion of autophagosomes with lysosomes, this blockade has no effect on lysosomal functions or the degradation of endocytic cargo.
WA disrupted the function of the SNAREs
Recently, an elegant study demonstrated that fusion of autophagosomes with lysosomes is mediated by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), which are composed of STX17 (syntaxin 17), SNAP29 (synaptosome associated protein 29kDa), and VAMP8 (vesicle-associated membrane protein 8).30 Thus, we sought to determine whether WA blocks the fusion of autophagosomes with lysosomes by disrupting the function of SNAREs. As shown in Fig. 3A, levels of STX17 and SNAP29 decreased in a dose-dependent manner in WA-treated cells, whereas the abundance of SQSTM1 was markedly increased. Consistent with previous results,30 knockdown of STX17 or SNAP29 caused dramatic accumulation of LC3B-II and SQSTM1 in cells, even under normal conditions (Fig. 3B). In agreement with this finding, transient transfection of Panc-1 cells with GFP-mRFP-LC3B led to the formation of many yellow LC3B-positive puncta after knockdown of these SNARE proteins (Fig. 3C), suggesting that loss of function of these SNAREs inhibits autophagosome maturation. Furthermore, SNAP29 siRNA alone further increased LC3B-II levels under treatment with a low concentration of WA (1 μM) (Fig. 3D). Conversely, in cells exposed to high WA concentrations (5 μM), LC3B-II levels did not increase further, regardless of SNAP29 knockdown (Fig. 3D), which was supportive of high WA concentrations (5 μM) being sufficient to block the fusion of lysosomes and autophagosomes.
Figure 3.
WA inhibits the fusion of lysosomes and autophagosomes by disrupting the function of SNAREs. (A) Western blot analysis of STX17, SNAP29, VAMP8 and SQSTM1 protein levels after Panc-1 and MIAPaCa-2 cells were treated with WA for 24 h at the indicated concentrations. (B) Panc-1 and MIAPaCa-2 cells were transfected with STX17 siRNA or SNAP29 siRNA for 48 h, and then the indicated protein levels were analyzed by Western blot. (C) Panc-1 cells were transfected with STX17 siRNA or SNAP29 siRNA for 48 h and then transiently transfected with a construct encoding GFP-mRFP-LC3B for an additional 48 h for colocalization assay. Representative fluorescence images are shown. Scale bar: 20 μm. (D) Panc-1 cells were transfected with SNAP29 siRNA for 48 h, and then cells were treated with 1 μM or 5 μM WA for an additional 24 h. The indicated protein levels were analyzed by Western blot. (E) Panc-1 cells stably expressing FLAG-STX17 or FLAG-SNAP29, or combinations thereof were either untreated or treated with WA (1–2.5 μM) for 24 h. The indicated protein levels were analyzed by Western blot. An asterisk indicates degradation products of transfected FLAG-STX17 and FLAG-SNAP29. (F) Panc-1 cells stably expressing FLAG-BECN1 were either untreated or treated with WA (1–2.5 μM) for 24 h. The indicated protein levels were analyzed by Western blot. (G) Panc-1 cells were either mock infected or infected with lentiviral vectors expressing STX17 plus SNAP29, and then untreated or treated with WA (2.5 μM) for 24 h followed by conventional electron microscopy analysis. Representative images of cells are shown. N, nuclear; arrows, autolysosomes; arrowhead, autophagosomes. Quantification of the number of autolysosomes from at least 20 randomly selected areas is shown (N.S, not significant; ***, p < 0.001). Scale bar: 500 nm.
To confirm that downregulation of STX17 and SNAP29 is the leading cause of WA-induced autophagy inhibition, Panc-1 and MIAPaCa-2 cells were either mock infected or infected with lentiviral vectors carrying the genes for STX17, SNAP29, or STX17 plus SNAP29, and then treated with WA (1–2.5 μM) or DMSO. As shown in Fig. 3E and S9A, compared with cells overexpressing STX17 or SNAP29 only, co-overexpression of STX17 and SNAP29, which had no significant effect on BECN1 expression, cooperatively reversed WA-induced LC3B-II and SQSTM1 accumulation. In contrast, BECN1 overexpression did not alter the expression of LC3B-II, SQSTM1, STX17 or SNAP29 affected by WA (Fig. 3F; Fig. S9B). Furthermore, transmission electron microscopy was used to observe the cellular ultrastructures. High magnification images clearly showed accumulation of autophagic vacuoles in the cytoplasm of mock-infected cells exposed to WA (Fig. 3G; Fig. S9C). Of note, most of these accumulated autophagic vacuoles contained intact cytoplasmic material without any features of degradation. More remarkably, WA-treated cells with co-overexpression of STX17 and SNAP29 exhibited numerous autolysosomes as well as hybrid autolysosomes fused with early endosomes or late endosomes, compared with the control. This observation indicates that co-overexpression of STX17 and SNAP29 accelerates autophagosome maturation under WA treatment. From these results, we conclude that WA inhibits the fusion of lysosomes and autophagosomes by disrupting the function of SNAREs.
WA inhibits proteasome activity and induces ER stress-related apoptosis in PC cells
Increasing evidence suggests that the UPS and autophagy are interdependent,14 whereas it has been reported that the tumor proteasome is a target of WA.21 Thus, we sought to determine whether the incomplete autophagy induced by WA was associated with proteasome inhibition. As shown in Fig. 4A, WA progressively inhibited the proteasomal chymotrypsin-like activity in a dose-dependent manner in Panc-1 and MIAPaCa-2 cells. Meanwhile, the level of ubiquitinated proteins dose-dependently increased (Fig. 4B), suggesting WA inhibited proteasome activity. It is generally thought that inhibition of autophagy can damage bulk protein degradation by lysosomes, leading to protein aggregation.14 Unexpectedly, the autophagy inhibitor CQ caused a slight elevation in the level of ubiquitinated proteins in Panc-1 cells, even at lethal concentrations (Fig. S10), suggesting WA-induced ubiquitinated protein accumulation primarily through proteasome inhibition.
Figure 4.
WA inhibits proteasome activity and induces ER stress-related apoptosis in PC cells. (A and B) WA affected ubiquitin-proteasomal activity in PC cells. Panc-1 and MIAPaCa-2 cells were treated with either DMSO (<0.1%) or the indicated concentrations of WA for 24 h, followed by measuring inhibition of the proteasomal chymotrypsin (CT)-like activity using a cell-based assay (A) and Western blot analysis using specific antibodies to ubiquitin (B). Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001). (C) Panc-1 and MIAPaCa-2 cells were treated with DMSO (<0.1%) or WA (2.5 μM) for 24 h followed by immunostaining with an anti-CANX antibody. Nuclei are counterstained with DAPI (blue). Arrows indicate dilated ER cavities. Scale bar: 10 μm. (D) Western blot analysis of ER stress-related protein levels after Panc-1 and MIAPaCa-2 cells were treated with the indicated concentrations of WA or tunicamycin (TM, 10 μg/ml) for 24 h. (E) Apoptotic cells were detected by flow cytometry using an ANXA5-FITC staining assay after cells were treated as described in (A). Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001). (F) Western blot analysis of PARP1, CASP9, CASP8 and cleaved (C)-CASP3 levels after cells were treated as described in (A). (G) Panc-1 cells were pretreated or not with CHX (1 mg/ml) for 1 h, followed by treatment with the indicated concentrations of WA for 24 h. The ubiquitinated protein levels (upper panel) and the cell viability (lower panel) were determined by Western blot and MTS assay, respectively. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05). (H) Panc-1 and MIAPaCa-2 cells were pretreated with TUDCA (1 mM) for 30 min, and then exposed to WA (2.5 μM) for 24 h. The indicated protein levels (upper panel) and the apoptotic cells (lower panel) were determined by Western blot and ANXA5-FITC staining assay, respectively. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05). (I) Panc-1 cells were transfected with DDIT3 siRNAs (#1 and #2) for 48 h and then cells were treated with WA (2.5 μM) for an additional 24 h. The indicated protein levels (upper panel) and the apoptotic cells (lower panel) were determined by Western blot and ANXA5-FITC staining assay, respectively. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05).
To verify whether ER stress was involved in WA-induced proteasome inhibition, cells were stained with the ER-specific marker CANX (calnexin; a calcium-binding protein embedded in the ER membrane). In untreated cells, the ER had a reticular pattern, whereas following WA treatment, numerous cytoplasmic vacuoles appeared (Fig. 4C). Furthermore, WA dose-dependently increased the levels of ERN1, p-EIF2A, HSPA5, and DDIT3, and decreased the levels of ATF6/p90 as distinctively as the typical ER stress inducer tunicamycin (TM) (Fig. 4D). These results indicate that WA induced ER stress.
We next determined whether WA-induced ER stress was related to apoptosis. First, ANXA5/annexin V-FITC staining assays revealed that WA increased cellular apoptosis in a dose-dependent manner (Fig. 4E). Moreover, WA effectively induced cleavage of PARP1 (poly[ADP-ribose] polymerase 1), CASP9 (caspase 9), CASP8 (caspase 8) and CASP3 (caspase 3) (Fig. 4F). Pretreatment of cells with zVAD-fmk (a pan-caspase inhibitor) almost completely blocked WA-induced cell death, whereas the RIPK1 (receptor interacting serine/threonine kinase 1) inhibitor necrostatin-1, which is an inhibitor of programmed necrotic cell death,31 was unable to block cell death in this system (Fig. S11). These data suggest that WA causes caspase-dependent apoptosis in PC cells. Subsequently, it was found that cycloheximide (CHX), an inhibitor of protein synthesis, significantly reduced WA-induced ubiquitinated protein accumulation and delayed the cell death response to WA, suggesting that proteotoxicity is important for the effect of WA (Fig. 4G). Moreover, the ER stress inhibitor tauroursodeoxycholic acid (TUDCA), which significantly suppressed WA-induced ER stress, strongly reduced PARP1 and CASP3 cleavage in WA-treated cultures concomitant with decreased WA-induced apoptosis (Fig. 4H). Interestingly, TUDCA attenuated WA-elicited formation of LC3B-II, although LC3B-II expression was not depressed to the basal level (Fig. 4H). However, TUDCA-treatment had no apparent effect on WA-induced downregulation of STX17 and SNAP29 levels. DDIT3 is generally considered to be a key pro-apoptotic factor during ER stress.7,8 Similarly, knockdown of DDIT3 markedly attenuated WA-induced apoptosis and LC3B-II formation (Fig. 4I). Together, these results indicate that WA inhibited proteasome activity and triggered ER stress, which subsequently induced apoptosis and autophagy in PC cells.
Simultaneous inhibition of autophagy and the proteasome triggers cell death through elevating ER stress
One purpose for upregulation of autophagy by ER stress is to remove misfolded protein aggregates, which in turn ameliorates ER stress.16-18 Indeed, confocal microscopy indicated that the GFP-LC3B puncta colocalized with ubiquitinated aggregates in WA-treated cells (Fig. 5A). However, WA-induced autophagy becomes impaired at the late stage of autophagy, which hinders autophagic function as a compensatory mechanism to reduce proteotoxicity. Moreover, coincubation with WA and CQ failed to augment the accumulation of ubiquitinated proteins and expression of HSPA5 and DDIT3 compared with WA alone (Fig. 5B and C). In contrast, compared with control cells, there was a significant reduction in the cleavage of PARP1 as well as expression of HSPA5 and DDIT3 in STX17- and SNAP29-co-overexpressing cells after treatment with WA (Fig. 5D). However, pretreatment of the cells with CQ almost completely blocked the effect of STX17 and SNAP29 co-overexpression (Fig. 5D). This result suggested that reversal of the impaired autophagy induced by WA was likely a cytoprotective effect. Interestingly, there was a pronounced increase in the cleavage of CASP3 and PARP1 in BECN1-overexpression cells after treatment with WA (Fig. S12), indicating that BECN1 exerts an autophagy-independent tumor suppressive effect in PC cells upon exposure to WA.
Figure 5.
Reversal of the impaired autophagy induced by WA attenuates ER stress and cell death. (A) Colocalization of the ubiquinated protein aggregates with GFP-LC3B puncta was examined by confocal microscopy. Scale bar: 10 μm. (B and C) Panc-1 and MIAPaCa-2 cells were treated with WA (2.5 μM) for 24 h in the absence or presence of CQ (10 μM). The levels of ubiquitinated proteins (B) and ER stress-related proteins (C) were assessed by Western blot. (D) Panc-1 and MIAPaCa-2 cells were either mock infected or infected with lentiviral vectors expressing STX17 plus SNAP29, and then untreated or treated with the indicated concentration of WA in the absence or presence of CQ (10 μM) for 24 h. The indicated protein levels were analyzed by Western blot.
To further verify the above hypothesis, the canonical proteasome inhibitor bortezomib was used. It has been reported that bortezomib induces autophagy in PC cells.32 Consistent with a previous report, bortezomib treatment of Panc-1 and MIAPaCa-2 cells increased LC3B-II protein levels, whereas the level of SNARE proteins was unaltered (Fig. S13). Moreover, CQ dramatically increased the accumulation of LC3B-II after bortezomib exposure, indicating that autophagic flux was enhanced (Fig. 6A). Furthermore, knockdown of ATG5, ATG7 or both, simultaneously augmented bortezomib-induced expression of HSPA5 and DDIT3, as well as PARP1 cleavage and ubiquitinated protein accumulation in Panc-1 cells (Fig. 6B and C), indicating a protective role for autophagy in bortezomib-induced proteotoxicity. Similar results were generated in MIAPaCa-2 cells (data not shown). Additionally, CQ and WA potentiated bortezomib-induced ER stress and cell death (Fig. 6D-F). Interestingly, while both CQ and WA were able to sensitize cells to bortezomib when applied alone, the addition of CQ together with WA had no additional sensitizing effect on bortezomib-induced toxicity compared to WA alone (Fig. 6F; Fig. S14). This indicates that CQ and WA sensitize cells to bortezomib largely though the same mechanism. Collectively, these results indicate that autophagy has a protective role against bortezomib-induced proteotoxicity, and simultaneous inhibition of autophagy and the proteasome triggers cell death by elevating ER stress.
Figure 6.
Inhibition of autophagy augments proteasome inhibitor-induced cell death through elevating ER stress in PC cells. (A) Panc-1 and MIAPaCa-2 cells were either untreated or treated with Bor (100 nM) for 24 h in the absence or presence of CQ (10 μM). The indicated protein levels were analyzed by Western blot. (B and C) Panc-1 cells were transfected with ATG5 or ATG7 siRNA, or combinations thereof for 48 h, and then cells were treated with Bor (100 nM) for an additional 24 h. The indicated protein levels were analyzed by Western blot. (D and E) Panc-1 cells were treated with Bor (100 nM) for 24 h in the absence or presence of CQ (10 μM) or WA (2.5 μM). The indicated protein levels were analyzed by Western blot. (F) Cell viability (MTS) of Panc-1 cells treated with Bor (100 nM) for 24 h in the absence or presence of CQ (10 μM) or/and WA (2.5 μM). Data are presented as mean ± SD from 3 independent experiments (N.S, not significant; *, p < 0.05; #, p < 0.05).
The combination of WA with ER stress aggravators causes synergistic enhancement of apoptosis in PC cells
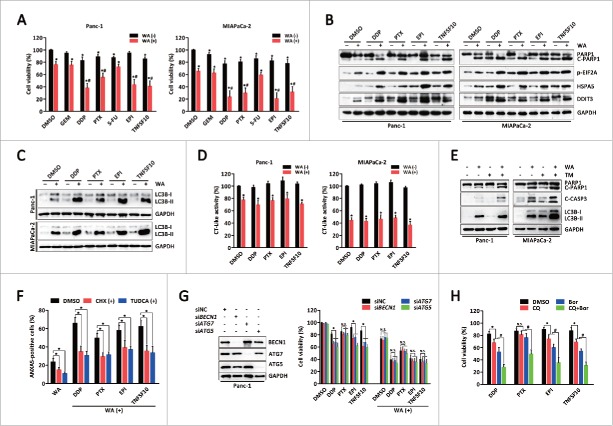
Because WA and bortezomib are both proteasome inhibitors, the ability of WA to sensitize PC cells to other cytotoxic agents, including DNA-damaging agents (cisplatin and epirubicin), anti-metabolite agents (gemcitabine and 5-fluorouracil), an anti-mitotic agent (paclitaxel), and a death receptor agonist (TNFSF10/TRAIL), was examined. Combined treatment with WA and cisplatin, paclitaxel, epirubicin or TNFSF10 synergistically reduced viability and induced PARP1 cleavage in Panc-1 and MIAPaCa-2 cells (Fig. 7A and B). Morphological and isobologram analyses demonstrated that these combination treatments acted synergistically (Fig. S15). Conversely, the combination of either gemcitabine or 5-fluorouracil with WA did not exhibit synergistic effects (Fig. 7A).
Figure 7.
WA sensitizes PC cells to multiple cytotoxic agents by augmenting ER stress. (A) Panc-1 and MIAPaCa-2 cells were pretreated with WA (2.5 μM), and then exposed to gemcitabine (GEM, 50 μM), cisplatin (DDP, 20 μM), paclitaxel (PTX, 200 nM), 5-fluorouracil (5-FU, 50 μM), epirubicin (EPI, 200 nM) or TNFSF10 (50 ng/ml) for 24 h. Cell viability was measured by MTS assay. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05; and synergistic [#, CI < 1] effect are indicated). (Band C) Panc-1 and MIAPaCa-2 cells were pretreated with WA (2.5 μM), and then exposed to cisplatin (DDP, 20 μM), paclitaxel (PTX, 200 nM), epirubicin (EPI, 200 nM) or TNFSF10 (50 ng/ml) for 24 h. After treatment, the indicated protein levels were analyzed by western blot. (D) Panc-1 and MIAPaCa-2 cells were treated as described in (B). The proteasomal chymotrypsin (CT)-like activity was measured by a cell-based assay. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05). (E) Panc-1 and MIAPaCa-2 cells were treated with WA (2.5 μM) and/or tunicamycin (TM, 10 μg/ml) for 24 h. The indicated protein levels were analyzed by western blot. (F) Panc-1 cells were pretreated with CHX (1 mg/ml) or TUDCA (1 mM) for 30 min, and then cells were exposed to the combination treatment as described in (B) for 24 h. The apoptotic cells were determined by ANXA5-FITC staining assay. Data are presented as mean ± SD from 3 independent experiments (*, p < 0.05). (G) Panc-1 cells were transfected with ATG5, ATG7, or BECN1 siRNA for 48 h, and then exposed to cisplatin (DDP, 20 μM), paclitaxel (PTX, 200 nM), epirubicin (EPI, 200 nM) or TNFSF10 (50 ng/ml) in the absence or presence of WA (2.5 μM) for an additional 24 h. Cell viability was measured by MTS assay. Data are presented as mean ± SD from 3 independent experiments (N.S, not significant; *, p < 0.05). (H) Panc-1 cells were treated with cisplatin (DDP, 20 μM), paclitaxel (PTX, 200 nM), epirubicin (EPI, 200 nM) or TNFSF10 (50 ng/ml) for 24 h in the absence or presence of CQ (10 μM) or Bor (100 nM) or both. Cell viability was measured by MTS assay. Data are presented as mean ± SD from 3 independent experiments (N.S, not significant; *, p < 0.05; #, p < 0.05).
Interestingly, combined treatment of WA with cisplatin, paclitaxel, epirubicin or TNFSF10 resulted in a significant increase in ER stress-associated proteins compared with either agent alone (Fig. 7B). Of note, treatment of cells with cisplatin, paclitaxel, epirubicin or TNFSF10 alone (for 24 h) also induced accumulation of ER stress-associated proteins (Fig. 7B). However, gemcitabine or 5-fluorouracil alone did not induce ER stress nor enhance WA-induced ER stress (Fig. S16). In addition, combination of WA with these ER stress aggravators caused further elevations in LC3B-II levels (Fig. 7C), whereas they had no synergistic effects on proteasomal chymotrypsin-like activity (Fig. 7D). These data suggest that WA may synergize with cytotoxic agents by augmenting ER stress. To enhance ER stress, TM was applied with WA, leading to, as expected, a significant increase in PARP1 and CASP3 cleavage as well as LC3B-II levels compared with either agent alone (Fig. 7E). Conversely, pretreatment with CHX or TUDCA attenuated the cytotoxicity induced by combination treatment (Fig. 7F).
Because these data suggested that WA inhibits autophagic flux, it was investigated whether WA sensitizes these ER stress aggravators though the same mechanism. As shown in Fig. 7G, suppression of autophagy by BECN1, ATG7 or ATG5 knockdown augmented cell death induced by the majority of ER stress aggravators, except for paclitaxel; however, none of the single drugs achieved the effect of combining with WA. Moreover, knockdown of BECN1, ATG7 or ATG5 neither enhanced WA-induced cell death nor augmented the effects of the combination of WA and ER stress aggravators. Furthermore, while both CQ and bortezomib alone sensitized cells to the ER stress aggravators, the addition of CQ to bortezomib had an additional sensitizing effect on inducing toxicity compared with either agent alone (Fig. 7H). Taken together, these data demonstrate that simultaneous inhibition of the proteasome and autophagy renders PC cells vulnerable to ER stress.
WA enhances the therapeutic efficacy of ER stress aggravators in PC xenografts
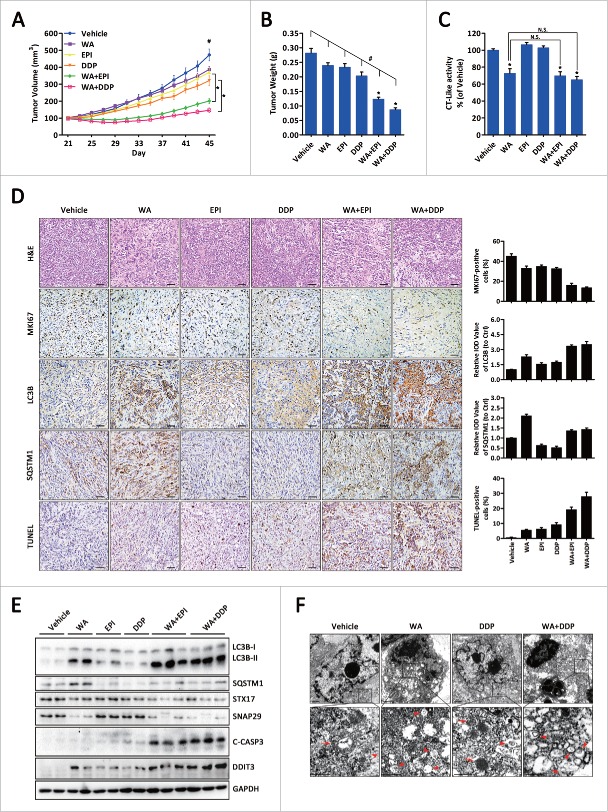
To translate the above results to an in vivo setting, Panc-1 cell pancreatic tumor xenograft models were established. At 21 d post-cell injection, mice with tumors of 100 mm3 were randomly assigned to vehicle, WA alone, epirubicin alone, cisplatin alone, WA + epirubicin, or WA + cisplatin. All treatments were administered for 24 d. As depicted in Fig. 8A, there were minimal effect on tumor volume after WA or epirubicin administration compared with vehicle group at d 45 (p = 0.052; p = 0.047). As expected, the WA and epirubicin or WA and cisplatin combinations significantly reduced tumor volume (p < 0.001). Consistent with the tumor volumes, mean tumor weights were substantially reduced in the combination groups compared with the single-drug groups (Fig. 8B). Notably, mice receiving epirubicin and cisplatin appeared to be sick, with loss of appetite and weight loss; however, no other significant toxicity in terms of progressive weight loss was observed in the combination groups (Fig. S17). In addition, although there was no difference, WA alone or the combination treatment caused inhibition of the proteasomal chymotrypsin-like activity in vivo (Fig. 8C).
Figure 8.
Combination of WA plus ER stress aggravators showed significant tumor growth inhibition in a xenograft model. (A) Panc-1 cells were inoculated subcutaneously and when tumors reached 100 mm3, mice were treated with vehicle, WA, epirubicin, cisplatin, WA + epirubicin, or WA + cisplatin as described in Materials and Methods. Data are presented as mean ±S.E.M. from each group (*, p < 0.01; #, p < 0.001, treatment vs. control by one-way ANOVA). (B) Tumor weight of each group at d 45 collected immediately after sacrificing the animals. Data are presented as mean ± SEM from each group (*, p < 0.01, monotherapy versus combination; #, p < 0.001, treatment vs. control by one-way ANOVA). (C) Tissue proteins exacted from tumor xenografts were subjected to the proteasomal chymotrypsin (CT)-like activity assay. Data are presented as mean ±S.E.M. from each group (N.S, not significant; *, p < 0.01, treatment versus control by t-test). (D) H&E and immunohistochemical analysis for MKI67, LC3B, SQSTM1 and TUNEL in tumor specimens harvested from mice (original magnification, ×200). Scale bar: 100 μm. Specimens were scored and estimated in relative integrated optical density (IOD) value or in percentage of positive cells. (E) Total proteins were extracted from the xenograft tumors and the indicated protein levels were analyzed by western blot. (F) Ultrastructure of tissue samples exacted from tumor xenografts shown by electron microscopy. Arrows, autolysosomes; arrowhead, autophagosomes. Scale bar: 500 nm.
As shown in Fig. 8D, immunohistochemical hematoxylin and eosin (H&E) staining of samples from mice treated in the combination group demonstrated that cell density was lower than in the single-drug groups. MKI67 staining confirmed a pronounced reduction in cell proliferation, whereas TUNEL staining revealed a significant increase in the number of apoptotic cells after combination treatment compared with either drug alone. Expression levels of LC3B and SQSTM1 were assessed as a measure of autophagy, with the finding that vehicle-treated control tumors had low expression levels of LC3B and SQSTM1, whereas epirubicin or cisplatin slightly increased LC3B expression and decreased SQSTM1 expression, implying that autophagy was activated. Conversely, WA administration significantly increased the expression of LC3B and SQSTM1, which was further enhanced by combination therapy, indicating that WA inhibits epirubicin- or cisplatin-induced autophagy in vivo. To confirm these results, Western blot and electron microscopy analyses were carried out. As shown in Fig. 8E, tissue lysates from harvested tumors revealed that WA treatment reduced the protein levels of STX17 and SNAP29 and induced LC3B-II and SQSTM1 accumulation even in combined treatments. Electron microscopy showed accumulation of autophagosomes containing cytoplasmic material without degradation after WA treatment alone or in combination with chemotherapeutic drugs (Fig. 8F), indicating that WA also induced incomplete autophagy in vivo.
Thus, these findings corroborate the in vitro data, verifying the synergistic antitumor activity of the combination of WA with ER stress aggravators in human PC.
Discussion
Here, we report that WA inhibited proteasome activity and triggered ER stress, resulting in apoptosis induction and autophagy in PC cells. However, WA-induced autophagy impairment at a late stage was likely due to blocking SNARE-mediated fusion of autophagosomes with lysosomes, whereas overexpression of SNAREs corrected the autophagosome fusion blockage in WA-treated cells. Furthermore, WA significantly increased the cytotoxic effect of ER stress aggravators in vitro and in vivo, suggesting that simultaneous inhibition of the ubiquitin-proteasome system and autophagy rendered PC cells vulnerable to ER stress.
UPS-mediated proteolysis consists of 2 steps: ubiquitination and proteasomal degradation, which is involved in the regulation of cell proliferation, differentiation, survival, and apoptosis.33 Thus, targeting of its pathway has emerged as effective antitumor approach.34 Moreover, proteasome inhibition appears to prevent the clearance of misfolded proteins through the ERAD system, possibly triggering ER stress-mediated apoptosis. Indeed, it has been widely reported that ER stress is implicated in antitumoral effects of proteasome inhibitors.34-36 Consistent with previous research,21,22 the present study demonstrated that WA inhibited proteasome activity and accumulation of ubiquitinated proteins in PC cells. Subsequent investigations revealed that CHX attenuated WA-induced cell death, indicating that proteotoxicity is important for the effect of WA. Furthermore, pretreatment with the ER stress inhibitor TUDCA or DDIT3 siRNA dramatically attenuated WA-induced apoptosis, leading to the suggestion that ER stress is directly involved in WA-induced apoptosis. Similarly, WA promotes ER stress-induced apoptosis in human renal carcinoma cells.37
Accumulating data indicate that ER stress is a potent trigger of autophagy.16-18 In these cases, ER stress-induced autophagy counterbalances ER expansion, removes aggregated proteins, and has a cytoprotective function. In this study, we also found that WA treatment of PC cells stimulated the formation of autophagic vacuoles, and promoted GFP-LC3B redistribution and the accumulation of LC3B-II, all of which confirmed that autophagy was activated. Similar to our findings, WA has also been demonstrated to have effects on autophagy, although the role of autophagy in the anticancer effects of WA remains to be determined.23,24 Moreover, pretreatment with TUDCA partially reduced WA-induced LC3B-II accumulation, suggesting that ER stress precedes WA-induced autophagy. Previous studies reported that major transducers of the UPR, EIF2AK3, EIF2A, ATF4 and DDIT3, are able to induce autophagy through ER stress.38,39 In accordance with this, accumulation of LC3B-II was partially ameliorated in DDIT3-depleted cells, which further supports the idea that ER stress occurs upstream of WA-induced autophagy.
Inhibition of autophagy leading to the accumulation of autophagy substrates and receptors may lie upstream of proteasomal dysfunction in certain cases.40 Under these conditions, detection of SQSTM1 levels concomitant with LC3B conversion has been proposed to be useful in monitoring autophagic flux.25 The present findings clearly indicate that WA treatment blocks autophagic flux in PC cells, while SQSTM1 failed to be degraded. In addition, results obtained using either LysoTracker Red, LAMP2 (a marker of endosomal and lysosomal membranes), or a tandem-labeled GFP-mRFP-LC3B construct demonstrated that autophagosomes remained separate from lysosomes for the duration of WA treatment. Thus, autophagy did not occur normally and was impaired at the latter stages. Interestingly, although WA enlarged the lysosomal compartment, which retained the ability to be stained by LysoTracker Red, no changes in lysosomal pH or a reduction in proteolytic activity were observed after WA treatment. These results demonstrate that WA does not impair endolysosomal or lysosomal activities. These data are in conflict with another report indicating that an azido-derivative of withaferin A (3-azido withaferin A) induces autophagy coupled with gradual degradation of SQSTM1.24 Unexpectedly, consistent with our results, this report showed that treating cells with 3-azido withaferin A induces ER stress to trigger apoptosis and ER stress-associated autophagy. It is possible that withanolide compounds function differently in distinct cellular processes including autophagy. Further investigations are needed to resolve this controversy.
Another intriguing observation from the present study was that WA disrupts the function of SNAREs. Recent studies demonstrated that STX17 is inserted into completed autophagosomes via its unusual C-terminal hairpin-like structure, and mediates autophagosome-lysosome fusion by binding to its partner SNAP29 and VAMP8 in mammalian cells, all of which is indispensable for the fusion of autophagosomes with autolysosomes.30 Surprisingly, WA substantially decreased levels of STX17 and SNAP29 in PC cells, whereas it had no effect on VAMP8. Furthermore, knockdown of SNAP29 caused a dramatic accumulation of LC3B-II and SQSTM1 in PC cells under normal conditions, and did not further increase LC3B-II levels above those induced by a high concentration of WA, suggesting that WA is sufficient to block autophagosome-lysosome fusion. In contrast, co-overexpression of STX17 and SNAP29 reversed the autophagosome fusion blockade in WA-treated cells in which BECN1 overexpression had no effect. The mechanism underlying the destabilization of BECN1, STX17 and SNAP29 in WA-treated cells remains unclear, since 2 major protein degradation pathways, the ubiquitin-proteasome pathway and the autophagy-lysosome pathway, are inhibited. It is likely that nonlysosomal cysteine proteases (CAPN/calpain, etc.,) mediate the degradation of these proteins. It has been reported that CAPN is involved in proteasome inhibitor-induced androgen receptor breakdown,41,42 and the autophagic proteins BECN1 is cleaved by CAPN.43 The involvement of CAPN and other proteases in autophagic SNARE protein breakdown will be further explored in future studies. Indeed, although the underlying molecular mechanisms for WA's effects on the SNAREs require further investigation, this is the first report describing inhibition of the SNAREs by a natural compound.
The development of resistance to antineoplastic agents is one of the primary obstacles for PC treatments. An increasing number of studies have reported the use of WA as an adjunct agent for enhancing the antitumor activity of chemotherapy drugs.44,45 However, the detailed molecular mechanisms are not yet fully understood. Interestingly, dramatic synergistic effects were noted when WA was combined with cisplatin, epirubicin, paclitaxel or TNFSF10, all of which induce ER stress, whereas no synergism was found with gemcitabine or 5-fluorouracil. Importantly, combined treatment with WA yielded a synergistic increase in ER stress, whereas gemcitabine or 5-fluorouracil alone did not induce ER stress nor enhance WA-induced ER stress. Furthermore, suppression of autophagy augmented ER stress aggravator-induced cell death; however, the effect of all single agents was inferior to that achieved by combined treatment with WA. Of note, although inhibition of autophagy has little effect on the antitumor activity of paclitaxel, WA is able to sensitize paclitaxel and cause a synergistic increase in ER stress. Therefore, it was speculated that WA sensitizes ER stress aggravators through simultaneous suppression of autophagy and the UPS, which rendered PC cells vulnerable to ER stress. Indeed, WA also sensitized PC cells to TM (an ER stress inducer). Conversely, pretreatment with CHX or TUDCA attenuated the cytotoxicity induced by all the combination treatments. Similar to these findings, it was previously reported that combining inhibitors of proteasome and autophagy elevates ER stress and enhances anticancer activity in myeloma cells.46 Thus, in the context of ER stress loading, targeting intracellular protein degradation pathways appears to be an important factor for determining sensitivity to chemotherapy.
In summary, the findings suggest that disrupting ER homeostasis by simultaneously blocking 2 major protein degradation systems is a promising therapeutic strategy to enhance the cytotoxicity of ER stress aggravators. These data provide a basis for future clinical trials to explore proteasome inhibitors, autophagy inhibitors, and ER stress aggravators as combinatory therapeutic approaches for the treatment of PC.
Materials and methods
Antibodies and reagents
All commercial antibodies and chemicals were purchased from the following resources: anti-LC3B (3868), anti-SQSTM1 (8025; for western blot), anti-SQSTM1 (7695; for immunofluorescence/immunohistochemistry), anti-ATG7 (2631), anti-ATG5 (8540), anti-BECN1 (3495), anti-DDIT3 (2895), anti-HSPA5 (3177), anti-ERN1 (3294), anti-EIF2A (5324), anti-CANX (2679), anti-ubiquitin (3936), anti-PARP1 (9542), anti-CASP9 (9502), anti-cleaved (C)-CASP3 (9664), anti-FLAG (14793) antibodies and paclitaxel (PTX; 9807) were from Cell Signaling Technology; anti-GAPDH (60004-1-Ig), anti-STX17 (17815-1-AP), anti-ATG14 (19491-1-AP) and anti-CASP8 (13423-1-AP) antibodies were from Proteintech Group; anti-p-RPS6 (2268-1), anti-SNAP29 (6700-1), anti-VAMP8 (2379-1), anti-ATF6 (T3355), anti-p-EIF2A (1090-1) and anti-EGFR (2116-1) antibodies were from Epitomics; anti-MKI67/Ki67 (sc-15402) antibody, bortezomib (Bor; sc-217785), epirubicin (EPI; sc-203041) and withaferin A (WA; sc-200381) were from Santa Cruz Biotechnology; horseradish peroxidase-conjugated secondary antibodies (anti-rabbit or anti-mouse) were purchased from Beyotime (A0208 and A0216); Cy3-conjugated secondary antibodies (anti-rabbit or anti-mouse) were purchased from Jackson ImmunoResearch (111-165-003 and 115-165-003); bafilomycin A1 (Baf-A1; B1793), chloroquine (CQ; C6628), rapamycin (Rap; R0395), pepstatin A (77170), E-64d (E8640), acridine orange (AO; 01662), human EGF (E9644), cycloheximide (CHX; C7698), cisplatin (DDP; C479306), gemcitabine (GEM; G6423), recombinant human TNFSF10/TRAIL (T9701) and 5-fluorouracil (5-FU; F6627) were from Sigma-Aldrich; LysoTracker Red DND-99 (L-7528) was purchased from Molecular Probes; tauroursodeoxycholic acid (TUDCA; 580549), zVAD-FMK (627610) and tunicamycin (TM; 654380) were obtained from Calbiochem. The chemicals were dissolved in either appropriate media solution or dimethyl sulfoxide (DMSO) and then treated at the required working dilution. All chemicals were handled in accordance with the supplier's recommendations.
Cell cultures
Human pancreatic cancer cell lines Panc-1, SW1990, MIAPaCa-2, AsPC-1 and BxPc-3 were purchased from ATCC (CRL-1469, CRL-2172, CRL-1420, CRL-1682 and CRL-1687). The immortalized human pancreatic ductal epithelial cell line HPDE was obtained from North Carolina Chuanglian Biotechnology research institute (BNCC338284). All cells were maintained in DMEM or RPMI-1640 medium (Gibco, 12100-046 and 31800-089) supplemented with 10% fetal bovine serum (Gibco, 10438-026), 2 mM glutamine, 100 units/ml of penicillin and 100 mg/ml of streptomycin in a 5% CO2 atmosphere at 37°C. All cell lines used in this study were regularly authenticated by morphological observation and routinely tested for mycoplasma contamination.
Cell viability assay
The cell viability was detected by using the CellTiter96® Aqueous Non-Radioactive Cell Proliferation Assay kit as described previously.47 Briefly, cells (3,000 per well) were plated in 96-well plates. After 24 h, cells were treated with the chemical agents as indicated in the figure legends. DMSO was used as vehicle. 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) solution (Promega, G5430) was added to each well and the cells were incubated at 37°C with 5% CO2 for 1 h. Absorbance at 490 nm was then measured with a microplate reader (Bio-tek Instruments, VT, USA). To investigate the synergistic effect between WA and the chemotherapy agents, cells were exposed to drugs at a fixed concentration ratio and a combination index (CI) was calculated using CalcuSyn software (Biosoft). CI < 1, CI = 1 and CI > 1 indicate synergism, additive effect and antagonism, respectively.
Acridine orange staining
Cell staining with acridine orange (AO) was performed according to the protocol from the manufacturer. Following the different treatments, the cells were stained with acridine orange (1 mg/ml) in phosphate-buffered saline (PBS; Beyotime, C0221A) at 37°C for 15 min. After washing with PBS, the cells were immediately analyzed under an inverted fluorescence microscope (OLYMPUS).
LysoTracker red staining
Cells from different treatments were collected and then incubated in the dark for 30 min at 37°C with LysoTracker Red DND-99. After washing with PBS, stained cells were assessed by FACS-Calibur flow cytometry (Becton Dickinson). Data analysis was performed with FlowJo software.
Cathepsin activity assay
The catalytic activities of cathespins were determined by CTSB and CTSD activity fluorometric assay kits (BioVision, K140-100 and K143-100,) according to the manufacturer's protocol. Briefly, cells were harvested after treatment and lysed in 200 μl of cell lysis buffer. Then 50 μl of cell lysate was transferred into 96-well plates, mixed with reaction buffer and substrate, and incubated at 37°C for 2 h. The samples were read in a fluorometer and the activity was normalized with the samples' protein concentration.
Detection of cell death
In this study, cell death was quantitatively and qualitatively analyzed by the following approaches: (i) morphological analysis under phase-contrast microscopy; (ii) ANXA5-FITC staining assay using the ANXA5/Annexin V-FITC/PI apoptosis detection kit (Invitrogen, V13242) according to the manufacturer's protocol; (iii) levels of cleaved CASP3 and PARP1 were determined by immunoblotting.
Western blot analysis
Upon treatment, cell pellets were lysed on ice in RIPA buffer (150 mM NaCl, 1% Triton X-100 [Sigma, X100], 0.5% deoxycholate [Sigma, D6750], 0.1% SDS [Sigma, 74255], 50 mM Tris, pH 7.5, protease inhibitor cocktail [Roche, 04693116001]). Protein concentration was determined using the bicinchoninic acid protein assay kit (Beyotime, P0009). Equivalent amounts of protein (20 µg) from each sample were subjected to electrophoresis on a SDS-polyacrylamide gel, and then transferred onto polyvinylidene difluoride membranes. Target antigens were detected with primary antibodies and subsequently secondary antibodies. Immunoreactive bands were then developed using an enhanced chemiluminescence reagent (Pierce, 32132), according to the manufacturer's instructions.
Transmission electron microscopy
After designated treatments, cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer and stored at 4°C until embedding. Then, they were postfixed with 1% OsO4 in 0.1 M cacodylate buffer (pH 7.2) containing 0.1% CaCl2 for 1 h at 4°C. After rinsing with cold distilled water, cells were dehydrated through a graded series of ethanol (30%–100%). The samples were embedded in Embed-812 (EMS, 14120). After polymerization of the resin at 60°C for 36 h, serial sections were cut using an ultramicrotome (Leica) and mounted on formvar-coated slot grids (EMS, GA300-Cu). Sections were stained with 4% uranyl acetate and lead citrate, and examined under a Tecnai G2 F20 S-TWIN transmission electron microscope (FEI).
Proteasome activity assay
Proteasome activity was determined by Proteasome-Glo™ Chymotrypsin-Like Cell-Based Assay kit (Promega, G8661). Briefly, cells (6,000 per well) were plated in 96-well plates. After 24 h, cells were treated with the chemical agents as indicated in the figure legends. Equal volumes of Proteasome-Glo reagent were then added and the luminescence signal was measured using a microplate reader (F200/M200, TECAN).
Immunofluorescence and confocal laser-scanning microscopy
After transfection with GFP-LC3B or GFP-mRFP-LC3B, cells were grown on glass coverslips. Following designated treatments, the fluorescent autophagy marker GFP-LC3B or GFP-mRFP-LC3B were observed using a confocal microscope LSM710 (Carl Zeiss, Germany). The average number of GFP-LC3B dots per cell was determined from 3 independent experiments. Ten random fields representing 200 cells were counted on each coverslide.
To detect fusion of lysosomes and autophagosomes, GFP-LC3B-expressing cells were grown on glass coverslips. Following designated treatments, the cells were then stained with 50 nM LysoTracker Red DND-99 in DMEM medium at 37°C for 30 min. After washing with PBS, the cells were immediately fixed with 4% paraformaldehyde for 10 min and observed under a confocal LSM710 microscope (Carl Zeiss, Germany).
For immunofluorescence, cells were seeded on sterile cover glasses placed in the 6-well plates. After designated treatments, cells were fixed with 4% formaldehyde for 20 min followed by permeabilization with 0.5% Triton X-100 (Sigma-Aldrich, X100) for 20 min. Fixed cells were washed and blocked with 2% BSA (Roche, 10735086001) for 30 min, then incubated with primary antibodies against SQSTM1, LAMP2, CANX or ubiquitin at 4°C overnight. After washing twice with PBS, antibodies were visualized with Cy3-conjugated secondary antibodies. Subsequently, cells were counterstained with DAPI (Sigma-Aldrich, D8417) and observed under a laser scanning confocal microscope LSM710 (Carl Zeiss, Germany).
Transfection
A lentiviral vector containing GFP-LC3B reporter, 3 × FLAG-STX17, 3 × FLAG-SNAP29, or 3 × FLAG-BECN1 were constructed by GenePharma (Shanghai, China) and transfection was carried out according to the manufacturer's instructions. The tandem labeled GFP-mRFP-LC3B plasmid were purchased from Addgene (#21074, depositing Dr. Tamotsu Yoshimori's lab). For transfection experiments, cells were seeded into 6-well plates overnight and transiently transfected using Lipofectamine 2000 (Invitrogen, 11668019) according to the manufacturer's instructions. After 48 h of transfection, the cells were incubated with the indicated reagents for further experiments.
RNA interference
The specific nucleotide RNAs and scrambled siRNA were synthesized chemically from Ribobio (Guangzhou, China) and resuspended in RNase-free water to a concentration of 20 μM. For siRNA transfection, cells were seeded into 6-well or 12-well plates in antibiotic-free media and transfected at 30–50% confluency with 50 nM siRNA in Opti-MEM (Invitrogen, 22600050) using Lipofectamine 2000 (Invitrogen, 11668019) according to the manufacturer's instructions. Cells were used 48 h after transfection and siRNA effects were monitored by Western blot analysis with suitable antibodies. The effective sequences of siRNAs used in experiments were as follows: ATG5 (5′-GUGAGAUAUGGUUUGAAUA-3′), ATG7 (5′-CAGCUA UUGGAACACUGUA-3′), BECN1 (5′-GGUCUAAGACGUCCAACAA-3′), STX17 (5′-CCGAAAGGAUGACCUAGUA-3′), SNAP29 (5′-AGACAGAAAUUGAGGAGCA-3′), DDIT3-1 (5′-GCCUGGUAUGAGGACCUGC-3′) and DDIT3-2 (5′-GAACCAGCAGAGGUCACAA-3′).
Xenograft experiments
The Institutional Animal Care and Treatment Committee of Huazhong University of Science and Technology approved all studies herein. Female athymic BALB/c nude mice (HFK Bioscience, China), 6- to 8-wk old, were injected subcutaneously with Panc-1 cells (5 × 106 cells/mouse). When the tumors became palpable (∼100 mm3) at d 21, the mice were randomized into the following treatment groups (n = 6): i) intraperitoneal (i.p.) injection of vehicle every other day; ii) i.p. injection of WA 4 mg/kg every other day; iii) i.p. injection of epirubicin 2 mg/kg twice a wk; iv) i.p. injection of cisplatin 4 mg/kg twice a wk; v) a combination of treatments ii + iii; or vi) a combination of treatments ii + iv. WA was dissolved in vehicle (10% DMSO, 40% cremophor:ethanol (3:1), and 50% PBS).48 Tumor size and body weight were measured every 2 d from day 21 after implantation. The tumor volume was calculated by the formula: volume (mm3) = (length × width2)/2. Animals were sacrificed and tumors were segregated and weighed by d 45 after treatment. Samples were prepared for proteasome activity assay, immunohistochemistry, immunoblotting and transmission electron microscopy.
Immunohistochemistry and TUNEL analysis
The tumor samples were embedded in paraffin, cut into 4-μm sections, and stained with H&E or incubated with primary antibodies for MKI67, LC3B and SQSTM1 using the ElivisionTM plus Polyer HRP IHC Kit (Maxim, KIT-9901). TUNEL assays for tissue sections were performed using an ApopTag® Plus Peroxidase Apoptosis Detection Kit (Roche, S7101) according to the manufacturer's instructions.
Statistical analysis
Data are presented as mean ± SD or mean ± SEM. Levels of significance were evaluated by 2-tailed Student ttest between 2 groups or by one-way ANOVA for multiple groups using GraphPad Prism 5 (GraphPad Software). P < 0.05 was considered statistically significant.
Supplementary Material
Abbreviations
- AO
acridine orange
- ATF6
activating transcription factor 6
- ATG
autophagy related
- Baf-A1
bafilomycin A1
- CHX
cycloheximide
- CI
combination index
- CQ
chloroquine
- CTSB
cathepsin B
- CTSD
cathepsin D
- DDIT3
DNA damage inducible transcript 3
- EIF2A
eukaryotic translation initiation factor 2A
- ERN1
endoplasmic reticulum to nucleus signaling 1
- ER
endoplasmic reticulum
- H&E
hematoxylin and eosin
- HPDE
human pancreatic ductal epithelial
- GFP
green fluorescent protein
- LAMP
lysosomal-associated membrane protein
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 β
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- PC
pancreatic cancer
- RAB
member RAS oncogene GTPases
- RFP
red fluorescent protein
- SNAREs
soluble N-ethylmaleimide-sensitive factor attachment protein receptors
- SNAP29
synaptosome associated protein 29kDa
- SQSTM1
sequestosome 1
- STX17
syntaxin 17
- TM
tunicamycin
- TUDCA
tauroursodeoxycholic acid
- TUNEL
terminal deoxynucleotidyl transferase-mediated nick end labeling
- UPR
unfolded protein response
- UPS
ubiquitin-proteasome system
- VAMP8
vesicle associated membrane protein 8
- WA
withaferin A
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Pei Zhang from the core facility and technical support, Wuhan Institute of Virology, for her help with producing EM micrographs.
Funding
This study was supported by The National Natural Science Foundation of China (No. 81272659 to R.Y.Q., No. 81502633 to X.L., No. 81101621 to M.W., No. 81160311 to J.X.J, No. 81372353 to X. W., No. 81172064 to M.S., No. 81301860 to C.J.S., No. 81402443 to P.F. and No. 81101870 to J.H.); Research Fund of Young Scholars for the Doctoral Program of Higher Education of China (No. 20110142120014 to R.Y.Q., No 20120142110055 to M.W., No 20130142120102 to D.W.Y.); and International Science & Technology Cooperation Program of China (No. 2014DFA31420 to C.Y.S.).
References
- [1].Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin 2013; 63:318-48; PMID:23856911; http://dx.doi.org/ 10.3322/caac.21190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Paulson AS, Tran Cao HS, Tempero MA, Lowy AM. Therapeutic advances in pancreatic cancer. Gastroenterology 2013; 144:1316-26; PMID:23622141; http://dx.doi.org/ 10.1053/j.gastro.2013.01.078 [DOI] [PubMed] [Google Scholar]
- [3].Kindler HL, Niedzwiecki D, Hollis D, Sutherland S, Schrag D, Hurwitz H, Innocenti F, Mulcahy MF, O'Reilly E, Wozniak TF, et al.. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303). J Clin Oncol 2010; 28:3617-22; PMID:20606091; http://dx.doi.org/ 10.1200/JCO.2010.28.1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8:519-29; PMID:17565364 [DOI] [PubMed] [Google Scholar]
- [5].Boyce M, Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ 2006; 13:363-73; PMID:16397583 [DOI] [PubMed] [Google Scholar]
- [6].Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol 2005; 7:766-72; PMID:16056268; [DOI] [PubMed] [Google Scholar]
- [7].Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function and promotes cell survival in multiple mouse models of diabetes. J Clin Invest 2008; 118:3378-89; PMID:18776938; http://dx.doi.org/ 10.1172/JCI34587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011; 13:184-90; PMID:21364565; http://dx.doi.org/ 10.1038/ncb0311-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov 2013; 12:703-19; PMID:23989796; http://dx.doi.org/ 10.1038/nrd3976 [DOI] [PubMed] [Google Scholar]
- [10].Martinon F. Targeting endoplasmic reticulum signaling pathways in cancer. Acta Oncol 2012; 51:822-30; PMID:22686473; http://dx.doi.org/ 10.3109/0284186X.2012.689113 [DOI] [PubMed] [Google Scholar]
- [11].Schönthal AH. Pharmacological targeting of endoplasmic reticulum stress signaling in cancer. Biochem Pharmacol 2013; 85:653-66; PMID:23000916; http://dx.doi.org/ 10.1016/j.bcp.2012.09.012 [DOI] [PubMed] [Google Scholar]
- [12].Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al.. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http://dx.doi.org/ 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- [14].Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 2010; 584:1393-8; PMID:20040365; http://dx.doi.org/ 10.1016/j.febslet.2009.12.047 [DOI] [PubMed] [Google Scholar]
- [15].Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171:513-24; PMID:17620365; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem 2007; 282:4702-10; PMID:17135238; [DOI] [PubMed] [Google Scholar]
- [17].Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 2010; 6:239-47; PMID:20104019; [DOI] [PubMed] [Google Scholar]
- [18].Verfaillie T, Salazar M, Velasco G, Agostinis P. Linking ER Stress to Autophagy: Potential Implications for Cancer Therapy. Int J Cell Biol 2010; 2010:930509; PMID:20145727; http://dx.doi.org/ 10.1155/2010/930509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vyas AR, Singh SV. Molecular targets and mechanisms of cancer prevention and treatment by withaferin a, a naturally occurring steroidal lactone. AAPS J 2014; 16:1-10; PMID:24046237; http://dx.doi.org/ 10.1208/s12248-013-9531-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nishi M, Akutsu H, Kudoh A, Kimura H, Yamamoto N, Umezawa A, Lee SW, Ryo A. Induced cancer stem-like cells as a model for biological screening and discovery of agents targeting phenotypic traits of cancer stem cell. Oncotarget 2014; 5:8665-80; PMID:25228591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang H, Shi G, Dou QP. The tumor proteasome is a primary target for the natural anticancer compound Withaferin A isolated from “Indian winter cherry”. Mol Pharmacol 2007; 71:426-37; PMID:17093135 [DOI] [PubMed] [Google Scholar]
- [22].Khan S, Rammeloo AW, Heikkila JJ. Withaferin A induces proteasome inhibition, endoplasmic reticulum stress, the heat shock response and acquisition of thermotolerance. PLoS One 2012; 7:e50547; PMID:23226310; http://dx.doi.org/ 10.1371/journal.pone.0050547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hahm ER, Singh SV. Autophagy fails to alter withaferin A-mediated lethality in human breast cancer cells. Curr Cancer Drug Targets 2013; 13:640-50; PMID:23607597; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rah B, ur Rasool R, Nayak D, Yousuf SK, Mukherjee D, Kumar LD, Goswami A. PAWR-mediated suppression of BCL2 promotes switching of 3-azido withaferin A (3-AWA)-induced autophagy to apoptosis in prostate cancer cells. Autophagy 2015; 11:314-31; PMID:25803782; http://dx.doi.org/ 10.1080/15548627.2015.1017182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304 [DOI] [PubMed] [Google Scholar]
- [26].Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ 2014; 21:348-58; PMID:24440914; http://dx.doi.org/ 10.1038/cdd.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy 2007; 3:154-7; PMID:17204842; [DOI] [PubMed] [Google Scholar]
- [29].Appelqvist H, Wäster P, Kågedal K, Öllinger K. The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol 2013; 5:214-26; PMID:23918283; http://dx.doi.org/ 10.1093/jmcb/mjt022 [DOI] [PubMed] [Google Scholar]
- [30].Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256-69; PMID:23217709; http://dx.doi.org/ 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- [31].Park EJ, Min KJ, Lee TJ, Yoo YH, Kim YS, Kwon TK. β-Lapachone induces programmed necrosis through the RIP1-PARP-AIF-dependent pathway in human hepatocellular carcinoma SK-Hep1 cells. Cell Death Dis 2014; 5:e1230; PMID:24832602; http://dx.doi.org/ 10.1038/cddis.2014.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Min H, Xu M, Chen ZR, Zhou JD, Huang M, Zheng K, Zou XP. Bortezomib induces protective autophagy through AMP-activated protein kinase activation in cultured pancreatic and colorectal cancer cells. Cancer Chemother Pharmacol 2014; 74:167-76; PMID:24842158; http://dx.doi.org/ 10.1007/s00280-014-2451-7 [DOI] [PubMed] [Google Scholar]
- [33].Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochemistry (Mosc) 2009; 74:1411-42; PMID:20210701; [DOI] [PubMed] [Google Scholar]
- [34].Shen M, Schmitt S, Buac D, Dou QP. Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin Ther Targets 2013; 17:1091-108; PMID:23822887; http://dx.doi.org/ 10.1517/14728222.2013.815728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Obeng EA, Carlson LM, Gutman DM, Harrington WJ Jr, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 2006; 107:4907-16; PMID:16507771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fels DR, Ye J, Segan AT, Kridel SJ, Spiotto M, Olson M, Koong AC, Koumenis C. Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res 2008; 68:9323-30; PMID:19010906; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Choi MJ, Park EJ, Min KJ, Park JW, Kwon TK. Endoplasmic reticulum stress mediates withaferin A-induced apoptosis in human renal carcinoma cells. Toxicol In Vitro 2011; 25:692-8; PMID:21266191; http://dx.doi.org/ 10.1016/j.tiv.2011.01.010 [DOI] [PubMed] [Google Scholar]
- [38].Li T, Su L, Zhong N, Hao X, Zhong D, Singhal S, Liu X. Salinomycin induces cell death with autophagy through activation of endoplasmic reticulum stress in human cancer cells. Autophagy 2013; 9:1057-68; PMID:23670030; http://dx.doi.org/ 10.4161/auto.24632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shimodaira Y, Takahashi S, Kinouchi Y, Endo K, Shiga H, Kakuta Y, Kuroha M, Shimosegawa T. Modulation of endoplasmic reticulum (ER) stress-induced autophagy by C/EBP homologous protein (CHOP) and inositol-requiring enzyme 1α (IRE1α) in human colon cancer cells. Biochem Biophys Res Commun 2014; 445:524-33; PMID:24565834; http://dx.doi.org/ 10.1016/j.bbrc.2014.02.054 [DOI] [PubMed] [Google Scholar]
- [40].Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 2009; 33:517-27; PMID:19250912; http://dx.doi.org/ 10.1016/j.molcel.2009.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pelley RP, Chinnakannu K, Murthy S, Strickland FM, Menon M, Dou QP, Barrack ER, Reddy GP. Calmodulin-androgen receptor (AR) interaction: calcium-dependent, calpain-mediated breakdown of AR in LNCaP prostate cancer cells. Cancer Res 2006; 66:11754-62; PMID:17178871 [DOI] [PubMed] [Google Scholar]
- [42].Yang H, Murthy S, Sarkar FH, Sheng S, Reddy GP, Dou QP. Calpain-mediated androgen receptor breakdown in apoptotic prostate cancer cells. J Cell Physiol 2008; 217:569-76; PMID:18726991; http://dx.doi.org/ 10.1002/jcp.21565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, Rotiroti D, Morrone LA, Bagetta G, Corasaniti MT. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis 2011; 2:e144; PMID:21490676; http://dx.doi.org/ 10.1038/cddis.2011.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fong MY, Jin S, Rane M, Singh RK, Gupta R, Kakar SS. Withaferin A synergizes the therapeutic effect of doxorubicin through ROS-mediated autophagy in ovarian cancer. PLoS One 2012; 7:e42265; PMID:22860102; http://dx.doi.org/ 10.1371/journal.pone.0042265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cohen SM, Mukerji R, Timmermann BN, Samadi AK, Cohen MS. A novel combination of withaferin A and sorafenib shows synergistic efficacy against both papillary and anaplastic thyroid cancers. Am J Surg 2012; 204:895-900; PMID:23231932; http://dx.doi.org/ 10.1016/j.amjsurg.2012.07.027 [DOI] [PubMed] [Google Scholar]
- [46].Kawaguchi T, Miyazawa K, Moriya S, Ohtomo T, Che XF, Naito M, Itoh M, Tomoda A. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol 2011; 38:643-54; PMID:21174067; http://dx.doi.org/ 10.3892/ijo.2010.882 [DOI] [PubMed] [Google Scholar]
- [47].Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res 2009; 69:5820-8; PMID:19584296; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stan SD, Hahm ER, Warin R, Singh SV. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res 2008; 68:7661-9; PMID:18794155; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.