ABSTRACT
Spermiogenesis is a complex and highly ordered spermatid differentiation process that requires reorganization of cellular structures. We have previously found that Atg7 is required for acrosome biogenesis. Here, we show that autophagy regulates the round and elongating spermatids. Specifically, we found that Atg7 is required for spermatozoa flagella biogenesis and cytoplasm removal during spermiogenesis. Spermatozoa motility of atg7-null mice dropped significantly with some extra-cytoplasm retained on the mature sperm head. These defects are associated with an impairment of the cytoskeleton organization. Functional screening revealed that the negative cytoskeleton organization regulator, PDLIM1 (PDZ and LIM domain 1 [elfin]), needs to be degraded by the autophagy-lysosome-dependent pathway to facilitate the proper organization of the cytoskeleton. Our results thus provide a novel mechanism showing that autophagy regulates cytoskeleton organization mainly via degradation of PDLIM1 to facilitate the differentiation of spermatids.
Keywords: Atg7, autophagy, cytoskeleton organization, PDLIM1, spermatid differentiation
Introduction
Spermiogenesis is a complex and highly ordered spermatid differentiation process that lasts from the end of the meiosis to the release of the mature spermatozoa.1 During spermiogenesis, the spermatids undergo a structural reorganization including the generation of the acrosome, the condensation of the nuclear chromatin, the rearrangement of the mitochondria, the assembly of the sperm flagella, and the removal of unnecessary cytoplasm to facilitate spermatozoa motility.2 The above events are achieved by the cooperation of different cellular processes. Any failures of these processes in the spermatids differentiation could result in abnormalities in the sperm's morphology, motility, and function.3,4 However, due to the lack of in vitro study systems, the molecular mechanism underlying spermatids differentiation remains largely unknown.
Macroautophagy (hereafter referred to as autophagy) is the primary intracellular catabolic mechanism for degrading and recycling long-lived proteins and organelles. Once autophagy is induced, the cytoplasmic components and organelles are enclosed by a double-membrane vesicle called an autophagosome. The autophagosome moves along the cytoskeletal structures and fuses with the lysosome, forming the autolysosome, which is then degraded and recycled by the lysosome.5 In mammals, autophagy is initiated by the activation of LC3 by ATG7, which is homologous to the ubiquitin-activating enzyme E1 (UBA1). The activated LC3 is then transferred to an E2-like enzyme (ATG3) and is finally conjugated to the lipid-containing membrane. The LC3-lipid-containing membrane works as a scaffold to build the early autophagy element called the phagophore, which grows into an enclosed autophagosome. The last step is the fusion of the autophagosome with the lysosome, generating the autolysosome where the substrates are entirely degraded.6
The primary function of autophagy is to allow the lower eukaryotic organisms, such as yeast, to survive nutrient starvation conditions by recycling their cellular components in response to extracellular or intracellular stress conditions. In addition to its well-established primary function, autophagy is recently reported to have conserved roles in the differentiation and development processes because it can drive the rapid cellular changes necessary for proper differentiation and/or development.7 In mammals, autophagy is essential to many physiological and pathological processes, such as preimplantation development,8 exocytosis of the granules from the Paneth cells,9 polarized secretion of the lysosomal content in osteoclasts,10 biogenesis of melanosomes,11 cell differentiation during erythropoiesis,12 lymphopoeisis,13 and adipogenesis.14 Accordingly, it is believed that autophagy could “kill 2 birds with one stone“ during cellular differentiation because, in addition to eliminating the pre-existing materials, it provides support for the subsequent creation of new components.5
Our previous study reveals the requirement of Atg7 for the biogenesis of the acrosome during spermiogenesis.15 In addition to the acrosome biogenesis defect, here, the severe defects of motility and morphology in atg7-null spermatozoa were identified. These defects were found to be associated with the disorganization of the “9+2” structure of the flagella and other cytoskeletal components in spermatozoa. In a further effort to address the relationship between autophagy and cytoskeleton organization, we found that a negative regulator of cytoskeleton organization, PDLIM1, needs to be degraded by the autophagy-lysosome pathway to maintain the proper cytoskeleton networks during spermiogenesis, thus assuring the spermatids differentiation process. Our study uncovers a novel mechanism explaining the functional role of autophagy during some cellular differentiation processes, which involves the degradation of some negative regulators to maintain the proper cytoskeleton dynamics required to renovate the entire cellular system.
Results
LC3 and ATG7 are mainly localized in the elongating spermatids
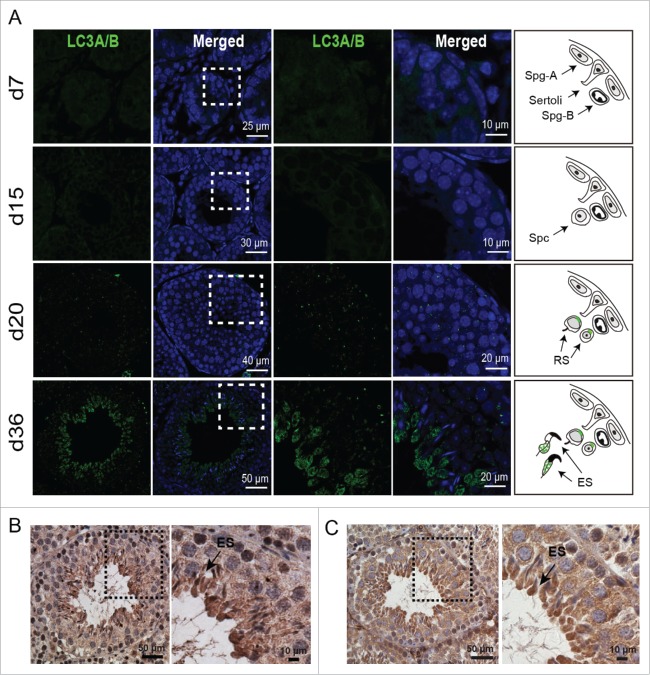
To further investigate the potential roles of autophagy in spermiogenesis, immunofluorescent staining was used to assess the expression and localization of MAP1LC3A/LC3A (microtubule-associated protein 1 light chain 3 α) and LC3B, an autophagy marker, in the seminiferous tubules at various developmental stages. The testes were collected at 4 typical developmental stages based on a previous report,16 in detail, the spermatogonium stage at postnatal d 7, the spermatocyte stage at d 15, the round spermatid stage at d 20, and the mature stage at d 36, when the elongating spermatids can be found in testes. Our results showed the absence of LC3A/B signals at the spermatogonium (d 7) and spermatocyte stages (d 15) (Fig. 1A, upper 2 panels). Consistent with our previous results, LC3A/B punctum signal was observed in the perinuclear region of the round spermatid in testes from d 20 mice (Fig. 1A, third panel). Surprisingly, robust LC3A/B staining was found in the central region of the seminiferous lumen in the d 36 testes (Fig. 1A, fourth panel), the enlarged images highlighting that LC3A/B was mainly expressed and distributed in the elongating spermatids. This result was further confirmed by immunohistochemical staining for LC3A/B in 8-wk-old mouse testes (Fig. 1B). The weak LC3A/B signal was observed in the round spermatids, while the robust staining of LC3A/B was found around the seminiferous lumen in the elongating spermatids (Fig. 1B).
Figure 1.
LC3A/B and ATG7 are mainly expressed and localized in the elongating spermatids. (A) Immunofluorescent staining (IF) of LC3A/B protein in testes of mice at postnatal d 7, d 15, d 20, and d 36. Blue, DAPI; green, LC3A/B. The enlarged images originated from the dotted squares highlighting green LC3A/B expression and localization in the elongating spermatids. The right panel model shows the schematic of LC3A/B expression in the testes of the indicated age, green markers LC3A/B localization. (B) Immunohistochemistry (IHC) of LC3A/B in 8-wk-old mouse testes. (C) The IHC of ATG7 in 8-wk-old mouse testes. The framed area was magnified. Sertoli, Sertoli cells; Spg-A, spermatogonium A; Spg-B, spermatogonium B; Spc, spermatocyte; RS, round spermatid; ES, elongating spermatid.
ATG7 is an E1-like activating enzyme playing an essential role in the 2 ubiquitin-like conjugation systems of autophagy. It is essential for ATG12 conjugation and for LC3 association with the membrane.5 To test whether ATG7 possesses a similar expression and distribution as LC3A/B, we detected its expression by immunohistochemistry (IHC) staining. Our results showed that ATG7 was ubiquitously expressed in the seminiferous tubules, and highly expressed in the elongated spermatids (Fig. 1C). This expression pattern further suggests that autophagy might also be involved in some processes in the elongating/elongated spermatids.
Alpl/Tnap-atg7 −/− mice produce spermatozoa with low motility
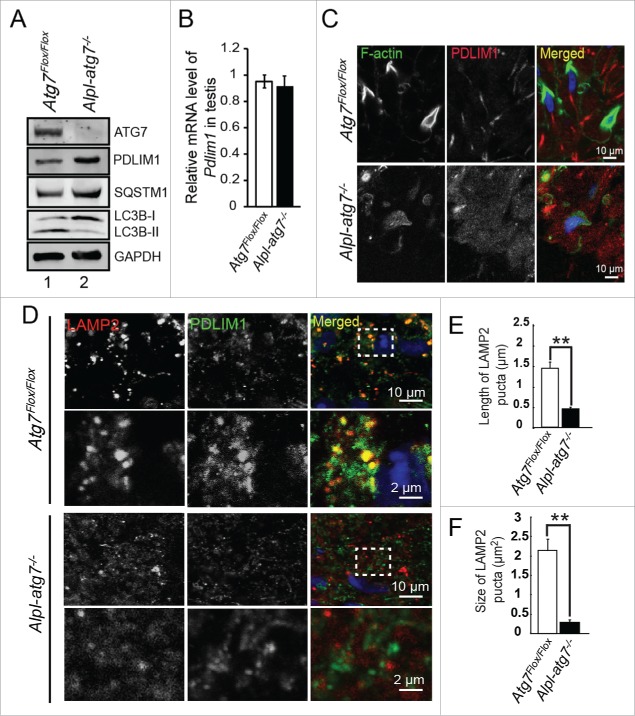
To address whether autophagy plays a distinct role in spermiogenesis in addition to the acrosome biogenesis, we constructed a germ cell-specific Atg7 knockout mouse model—the Atg7Flox/Flox; Alpl-Cre mice (hereafter described as Alpl-atg7−/− mice) (Fig. S1A),15 where Cre recombinase is under control of the Alpl/Tnap promoter, and the knockout efficiency was confirmed by ATG7 IHC staining (Fig. S1B). First, the sperm motility of Atg7Flox/Flox mice and Alpl-atg7−/− mice were measured by computer-assisted semen analysis. We found that the sperm motility of the Alpl-atg7−/− mice was severely damaged, with the significantly decreased motility rate compared to that of the Atg7Flox/Flox mice. Indeed, only 15.67 ± 2.19% of the atg7-null spermatozoa were motile, whereas the percentage of motile spermatozoa in the Atg7Flox/Flox mice was 83.5 ± 4.77% (P < 0.01, Fig. 2A).
Figure 2.
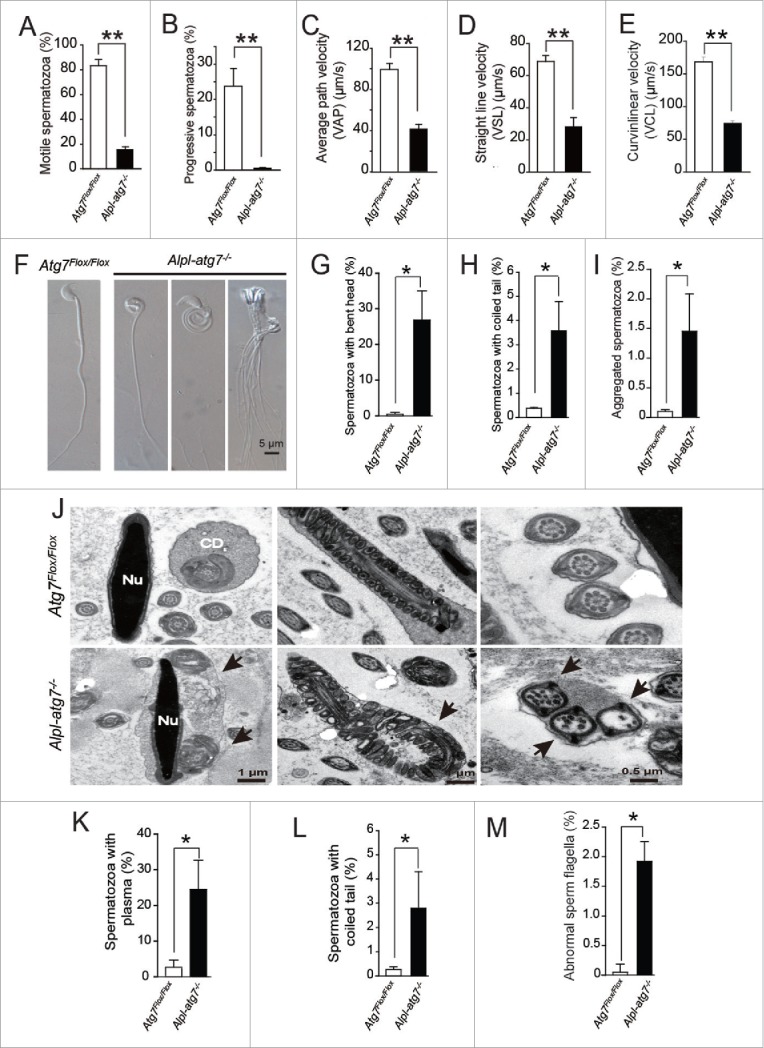
The defects of the Alpl-atg7−/− mouse spermatozoa. (A) The motile spermatozoa in the Atg7Flox/Flox mice (white column, 83.5 ± 4.77%) and in the Alpl-atg7−/− mice (black column, 15.67 ± 2.19%). (B) The progressive spermatozoa in the Atg7Flox/Flox mice (white column, 23.75 ± 4.99%) and in the Alpl-atg7−/− mice (black column, 0.66 ± 1.15%). (C) The average path velocity (VAP) of the spermatozoa from the Atg7Flox/Flox mice (white column, 99.42 ± 11.28 μm/s) and from the Alpl-atg7−/− mice (black column, 41.7 ± 7.64 μm/s). (D) The VSL of the spermatozoa from the Atg7Flox/Flox mice (white column, 68.9 ± 7.24 μm/s) and from the Alpl-atg7−/− mice (black column, 28.43 ± 9.47 μm/s). (E) The VCL of the spermatozoa from the Atg7Flox/Flox mice (white column, 168.35 ± 14.79 μm/s) and from the Alpl-atg7−/− mice (black column, 74.36 ± 5.73μm/s). (F) Morphology of the spermatozoa from Atg7Flox/Flox mice and from the Alpl-atg7−/− mice. (G) The percentage of spermatozoa with bent head the in Atg7Flox/Flox mice (white column) and in the Alpl-atg7−/− mice (black column). (H) The percentage of spermatozoa with coiled tail in the Atg7Flox/Flox mice (white column) and in the Alpl-atg7−/− mice (black column). (I) The percentage of aggregated spermatozoa in the Atg7Flox/Flox mice (white column) and in the Alpl-atg7−/− mice (black column). (J) The ultrastructural analysis of the spermatozoa from the Atg7Flox/Flox cauda epididymis (upper panel) and from the Alpl-atg7−/− cauda epididymis (lower panel). Lane 1, bent head spermatozoa, arrow indicates the undetached cytoplasm. Lane 2, spermatozoa severely coiled tail, arrow indicates coiled sperm tail. Lane 3, aggregated spermatozoa, arrow indicates the abnormal “9+2” structure of sperm flagella. (K) The percentage of spermatozoa with undetached cytoplasm in the Atg7Flox/Flox mouse (white column) and in the Alpl-atg7−/− mouse (black column) cauda epididymis by TEM analysis. (L) The percentage of spermatozoa with coiled tail in the Atg7Flox/Flox mouse (white column) and in the Alpl-atg7−/− mouse (black column) cauda epididymis by TEM analysis. (M) The percentage of abnormal sperm flagella in the Atg7Flox/Flox mouse (white column) and in the Alpl-atg7−/− mouse (black column) cauda epididymis by TEM analysis. Nu, nucleus; CD, cytoplasmic droplet.
In addition to the motile spermatozoa percentage, all other parameters of the atg7-null spermatozoa revealed a severe defect in motility. The progressive spermatozoa of the Alpl-atg7−/− mice were only 0.66 ± 1.15%, much lower than the 23.75 ± 4.99% found in the Atg7Flox/Flox mice (Fig. 2B). The average path velocity (VAP), straight-line velocity (VSL), and curvilinear velocity (VCL) are 3 important parameters to assess spermatozoa motility. The VAP of the atg7-null spermatozoa was 41.7 ± 7.64 μm/s, which was even less than half of the Atg7Flox/Flox spermatozoa's VAP of 99.42 ± 11.28 μm/s (Fig. 2C). The VSL of the Atg7Flox/Flox spermatozoa was 68.9 ± 7.24 μm/s, which is over 2-fold more than the atg7-null spermatozoa's VSL of 28.43 ± 9.47 μm/s (Fig. 2D). A sharp contrast was also found for the VCL, which was 168.35 ± 14.79 μm/s for the Atg7Flox/Flox spermatozoa vs. 74.36 ± 5.73 μm/s for the atg7-null spermatozoa (Fig. 2E). Collectively, our data revealed the severe damage of sperm motility in Alpl-atg7−/− mice, which suggest that Atg7 is required to maintain a high level of sperm motility and may be involved in spermatid differentiation.
The low motility of the atg7-null spermatozoa is associated with abnormal spermatozoa fine structures
The successful movement of spermatozoa is achieved by the frequent beating of a well-assembled tail using the energy provided by the mitochondria.17 The morphology of spermatozoa was further compared for Atg7Flox/Flox and Alpl-atg7−/− mice. Our data revealed a high percentage of abnormal spermatozoa in the mutant mouse epididymis. In addition to the acrosome abnormality that we have reported previously,15 we also identified striking morphological defects in spermatozoa with the abnormal sperm head and tail in Alpl-atg7−/− mice (Fig. 2F). These abnormal spermatozoa could be classified into 3 groups, the spermatozoa with bent head, the spermatozoa with coiled tail, and the aggregated spermatozoa (Fig. 2F). The statistical analysis revealed the significant higher percentage of these 3 kinds of defective spermatozoa in Alpl-atg7−/− mice, compared to those from Atg7Flox/Flox mice, respectively (Fig. 2G to I).
Transmission electron microscopy (TEM) analysis revealed the impaired fine structures of those abnormal spermatozoa. For the spermatozoa with bent head, a large portion of undetached cytoplasm remained and connected the bent head and the tail, thus inhibiting its beating (Fig. 2J, left panel). For the spermatozoa with coiled tail, the sperm tail was severely coiled with mislocalized and poorly condensed mitochondria (Fig. 2J, middle panel). The aggregated spermatozoa displayed the most severe defect, which was characterized by the presence of aggregation in which several sperm tails were clustered and wrapped by membrane and some cytoplasm (Fig. 2J, right panel). Notably, almost all of the clustered sperm flagella displayed a loss of their proper “9+2” microtubule structures in the axonemes, indicating microtubules misassembly during spermiogenesis. The percentages of the above abnormal ultrastructures were similar to those of the spermatozoa morphological abnormalities observed under light microscope (Fig. 2K to M). Collectively, these results reveal that the sharply decreased motility of the atg7-null spermatozoa is mainly caused by the mis-assembled “9+2” structure in the sperm flagellum and by the impaired cytoplasm removal.
The cytoskeleton of the atg7-null spermatids was disorganized during spermiogenesis
After meiosis, the nucleus of the haploid spermatid begins to condense and elongate. During this process, the most part of spermatid's cytoplasm is transported toward the sperm tail and is finally sloughed off from the spermatid. To investigate the precise mechanism by which Atg7 regulates cytoplasm removal, we dissected the spermatid maturation process step by step via TEM analysis.
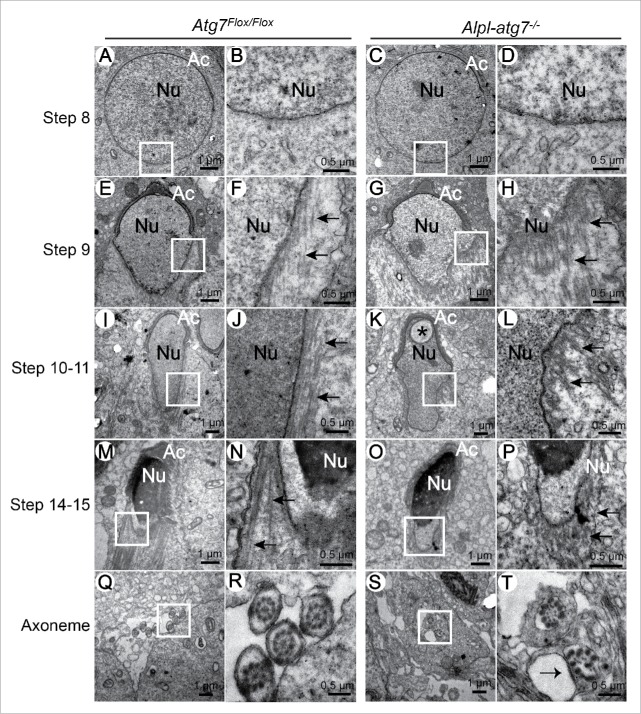
Since the spermatids begin to elongate from step 8, our investigation started from this stage. We did not found any clear differences between Atg7Flox/Flox (Fig. 3A and B) and atg7-null spermatids (Fig. 3C and D) at step 8. In the early elongating spermatids of the step 9, bundles of microtubules were assembled to form the manchette structure in the Atg7Flox/Flox testes, and the manchette was tightly arranged along the elongating nucleus (Fig. 3E and F). However, in the Alpl-atg7−/− testes, the enriched microtubules of the manchette were loosened and lost their normal orientation toward the centrosome (Fig. 3G and H). In the following elongating spermatids of steps 10 to 11, a well-assembled manchette consisting of long entire microtubule filaments could be observed in the Atg7Flox/Flox testes (Fig. 3I and J), whereas in the Alpl-atg7−/− testes, the manchette was disrupted, only some short microtubules could be observed, and the nucleus could not be condensed properly with a vesicle inside it (Fig. 3K and L). In the late elongating spermatids of steps 14 to 15, the manchette of elongated spermatids in Atg7Flox/Flox testes was well assembled, the microtubule bundles were finely arranged along the condensed nucleus (Fig. 3M and N), while in the same stage of Alpl-atg7−/− testes, only short and disordered microtubules were observed (Fig. 3O and P). In addition, abnormal flagella axonemes can also be found in the developing spermatids in Alpl-atg7−/− testes (Fig. 3S and T, arrow), but not in Atg7Flox/Flox testes (Fig. 3Q and R), meaning that the axoneme development was destroyed in Alpl-atg7−/− testes.
Figure 3.
Ultrastructural analysis of the spermiogenesis in Atg7Flox/Flox and Alpl-atg7−/− mice during different developmental stages. (A to D) The ultrastructures of step8 round spermatids from Atg7Flox/Flox mouse testis (A, B) and Alpl-atg7−/− mouse testis (C, D), showing that there were no differences between Atg7Flox/Flox and Alpl-atg7−/− mouse testis before manchette assembly. (E to H) The ultrastructures of step 9 spermatids from Atg7Flox/Flox mouse testis (E, F) and Alpl-atg7−/− mouse testis (G, H); arrows indicate the microtubule-based manchette. (I to L) The ultrastructures of step 10 to 11 spermatids from Atg7Flox/Flox mouse testis (I, J) and Alpl-atg7−/− mouse testis (K, L); arrows indicate the microtubule-based manchette. (M to P) The ultrastructures of step 14-15 spermatids from Atg7Flox/Flox mouse testis (M, N) and Alpl-atg7−/− mouse testis (O, P); arrows indicate the microtubule-based manchette. (Q to T) The ultrastructures of developing axonemes from Atg7Flox/Flox mouse testis (Q, R) and Alpl-atg7−/− mouse testis (S, T); the arrow indicates the destroyed axoneme in Alpl-atg7−/− mouse testis. The framed area was magnified, asterisk indicates the vesicle in the nucleus. Nu, nucleus; Ac, acrosome.
Because the F-actin bundles and the manchette structures are essential for nuclear remodeling, cargo transport, and spermatid elongation,18 the disrupted F-actin and microtubules in the Alpl-atg7−/− testes might be responsible for the observed spermatozoa structure abnormalities. Collectively, these results provide evidences indicating that ATG7 may affect spermatozoa maturation by regulating the cytoskeleton dynamics during spermatid differentiation.
ATG7 participates in cytoskeleton organization in mouse embryonic fibroblasts (MEFs)
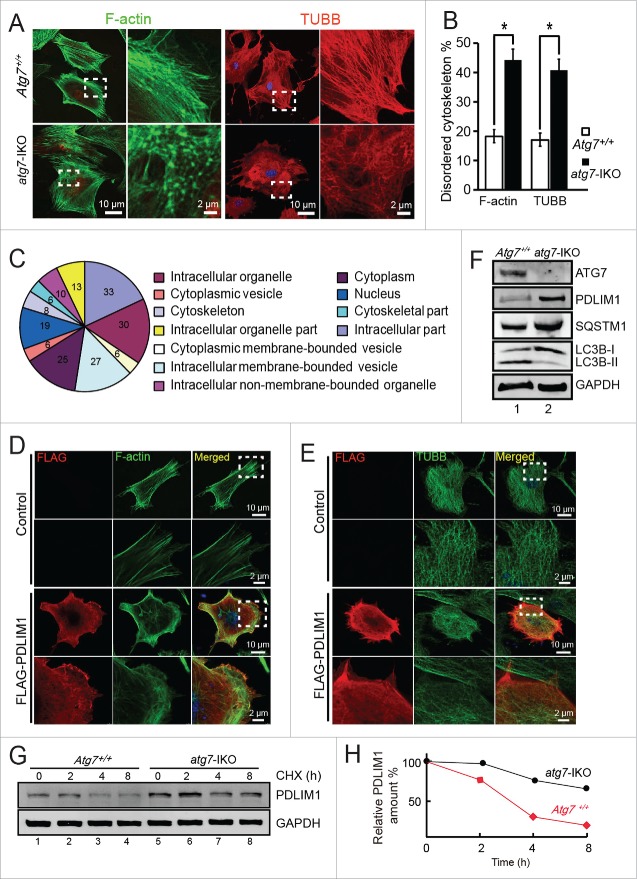
It is well known that the induction of autophagy depends on the proper organization of the cytoskeleton. For example, the autophagy marker LC3 is a microtubule-associated protein,19 and the assembly of F-actin within the phagophore may be required for shaping the autophagosome.20 However, how autophagy modulates cytoskeleton organization still remains unknown. Recently, Zhuo et al. have conducted a comparative mass spectrometry analysis between WT and atg7−/− MEFs and find that the F-actin networks are disrupted in the atg7−/− MEFs,21 suggesting that autophagy and cytoskeleton might mutually affect each other. To study the regulatory function of ATG7 on cytoskeleton organization, we prepared the Atg7Flox/Flox; Tamoxifen-cre MEF cells, in which the Cre recombinase activity could be induced by 4-OH-tamoxifen to delete the floxed Atg7 alleles, thus generating the Atg7 induced knockout MEF cells (hereafter described as atg7-IKO MEFs), and the untreated cells were described as Atg7+/+ MEFs. Then the microtubule and F-actin networks were examined in Atg7+/+ MEFs and atg7-IKO MEFs via immunofluorescent staining of TUBB/β-tubulin and F-actin. Consistent with the situations in the testis TEM analyses, the microtubule networks were disrupted in the atg7-IKO MEFs compared with that of the Atg7+/+ MEFs. The microtubules in the Atg7+/+ MEFs were well organized in parallel bundles, while the atg7-IKO MEFs displayed loosened and randomly distributed microtubules (Fig. 4A, right panel, Fig. 4B). The F-actin networks of the Atg7+/+ MEFs were made of clean and strong bundles composed of parallel F-actin filaments. However, the F-actin networks in the atg7-IKO MEFs were disordered, with F-actin signals accumulated into foci rather than the well-arranged filaments structures observed in the Atg7+/+ MEFs (Fig. 4A, left panel, Fig. 4B). To confirm that it is Atg7 depletion that results in the disorganization of the cytoskeleton, we reintroduced Atg7 and the control plasmids into Atg7+/+ and atg7-IKO MEFs respectively. The expression of exogenous ATG7 significantly reduced the disorganization frequencies of the cytoskeletons (Fig. S2A, lower panel green dotted area, S2C). Taken together, all these results suggest that ATG7 indeed is required for proper cytoskeleton assembly in MEFs.
Figure 4.
PDLIM1 is degraded via an ATG7-mediated autophagy-lysosome pathway to maintain the proper organization of the cytoskeleton. (A) IF of F-actin and TUBB in Atg7+/+ (upper panel) and atg7-IKO (lower panel) MEFs. The enlarged images originated from the dotted squares. (B) The percentage of disordered F-actin and TUBB cytoskeleton in (A). (C) Cluster analysis of upregulated proteins in atg7-IKO MEFs. (D, E) Co-IF of F-actin (D) and TUBB (E) with overexpressed FLAG-PDLIM1 in MEF cells, Framed area was magnified. Note that the overexpression of PDLIM1 impaired the disorganization of cytoskeleton. (F) Immunoblotting analysis of ATG7, PDLIM1, SQSTM1 and LC3B in both Atg7+/+ and atg7-IKO MEFs. (G) Cycloheximide (CHX) chase assay of PDLIM1 in Atg7+/+ and atg7-IKO MEFs. Samples were taken at 0, 2, 4, 8 h after the addition of CHX and PDLIM1 level was tested by immunoblotting. (H) Quantification of PDLIM1 level in the CHX assay using the ratio of PDLIM1:GAPDH.
ATG7 regulates cytoskeleton organization by degrading PDLIM1
The disorganization of the cytoskeleton in the Atg7 knockout cells (including atg7-null spermatids and atg7-IKO MEFs) could result from the accumulation of some negative regulators of cytoskeleton organization. In other words, some negative regulators of cytoskeleton organization may need to be eliminated by the autophagy-lysosome pathway to preserve the proper dynamics of the cytoskeleton. To test the aforementioned hypothesis, we analyzed the comparative mass spectrometry results of Zhuo et al. In that paper, they compared the protein levels between WT and atg7−/− MEFs by quantitative mass spectrometry, and among the 1,234 proteins they identified, 114 proteins were significantly altered, in which 66 proteins were downregulated and 48 proteins were upregulated.21 We found that the upregulated proteins could be grouped into approximately 10 clusters, according to their localizations and functions. The categories of “cytoskeleton-associated proteins” and “gene expression-related proteins” were 2 highly enriched clusters highlighted via gene ontology enrichment analyses. In addition, these genes were found to have multiple relationships with the downregulated proteins, according to the interaction network analysis calculated by STRING and visualized by Cytoscape 2.8.3. (Fig. 4C, Fig. S3A and S3B). To identify the potential negative regulators of cytoskeleton organization involved in our phenotype, 7 genes from the 2 aforementioned clusters were selected and their cDNAs were cloned. These FLAG-tagged genes were overexpressed in HeLa cells and the cells transfected with the empty vector were set as control. And the untransfected cells within the same dish could also serve as internal control for the transfected cells. This small-scale functional screening demonstrated that the overexpression of PDLIM1 (PDZ and LIM domain 1 [elfin]) and STMN1 (stathmin 1) lead to the disorganization of the cytoskeleton, including the microtubules and F-actin networks (Fig. S4).
Because we did not observe any expression of STMN1 in the testes (data not shown), we focused on PDLIM1 in our subsequent analyses. To further confirm the effect of PDLIM1 on MEF cytoskeleton organization, we overexpressed FLAG-PDLIM1 in WT MEFs, and similar disorganization of cytoskeleton was observed (Fig. 4D and 4E). PDLIM1 is a member of the PDZ and LIM protein family, which associates with F-actin via an interaction with ACTN1/α-actinin 1 and ACTN4.22 To clarify whether the disorganization of the cytoskeleton in the atg7-IKO MEFs was caused by PDLIM1 accumulation, the protein level of PDLIM1 was examined by immunoblotting and immunofluorescent staining in the Atg7+/+ and atg7-IKO MEFs. Our results showed that PDLIM1 was obviously accumulated in the atg7-IKO MEFs compared with the Atg7+/+ MEFs (Fig. 4F), which was consistent with the previously reported MS data.21 The accumulation of LC3B-I and autophagy substrate SQSTM1 suggested that the autophagic flux was blocked in these cells. Our IF staining of PDLIM1 with TUBB or F-actin showed that the accumulation of PLDIM1 was positively associated with disordered microtubule or F-actin networks in atg7-IKO MEFs, but these were not observed in Atg7+/+ MEFs (Fig. S5A and S5B). High level of PDLIM1 expression may be resulted from either upregulated expression or downregulated degradation, we first tested the relative mRNA level of Pdlim1 in both Atg7+/+ and atg7-IKO MEFs, but no difference in its mRNA level was found (Fig. S5C), suggesting that the elevated PDLIM1 might be a result of the failure of its degradation.
To prove that ATG7 was crucial for the degradation of PDLIM1, cycloheximide (CHX) chase assays were performed in Atg7+/+ MEFs and atg7-IKO MEFs. The results showed that the PDLIM1 degradation was strongly delayed in atg7-IKO MEFs compared with Atg7+/+ MEFs (Fig. 4G and H). Moreover, the reintroduction of ATG7 into atg7-IKO MEFs significantly restore PDLIM1 level and the autophagy flux (Fig. S2B). To determine whether ATG7 alone was involved in PDLIM1 degradation or if the whole autophagy-lysosome pathway was also involved, we used 2 lysosome acidic inhibitors, ammonium chloride (NH4Cl) and chloroquine (CQ), to treat WT MEFs for 24 h. A clear accumulation of PDLIM1 was observed in lysosome inhibitor-treated MEFs (Fig. S6). All these results suggest that PDLIM1 is mainly eliminated in an autophagy-dependent manner.
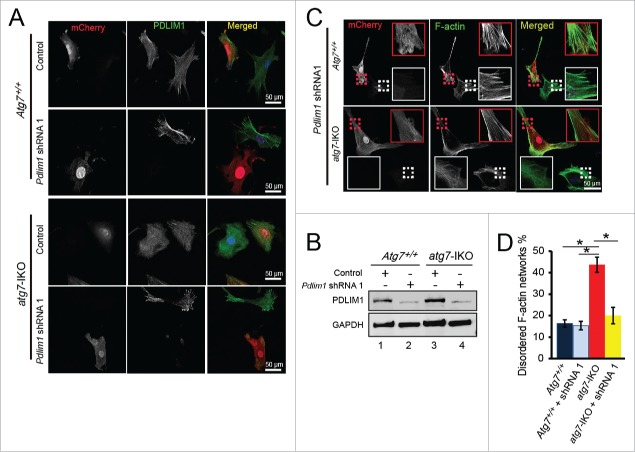
Next, we tested whether PDLIM1 is the major substrate for autophagy to facilitate cytoskeleton organization. If the answer is yes, then knockdown of Pdlim1 in atg7-IKO MEFs might at least partially rescue the cytoskeleton disorder phenotype of the cells. We then knocked down Pdlim1 by shRNA using pSicoR-mCherry lentiviral vector.23 We found that 2 shRNAs reduced PDLIM1 protein level effectively (Fig. S7), especially for shRNA No. 1 (shRNA1). We then chose shRNA1 for the following experiment. The IF staining results indicated that Pdlim1 shRNA1 inhibited PDLIM1 expression very efficiently in both Atg7+/+ MEFs and atg7-IKO MEFs (Fig. 5A, 5B), but this was not the case with the empty vector. We found that atg7-IKO MEF cells infected by Pdlim1 shRNA1 virus displayed relatively normal cytoskeleton networks (Fig. 5C lower panel, red dotted area), but the uninfected cells in the same dish still showed some disorganized F-actin networks (Fig. 5C lower panel, white dotted area). Statistical results showed that the percentage of cells with disordered cytoskeleton reduced significantly compare with that of the atg7-IKO cells (Fig. 5D). These results suggest that the accumulation of PDLIM1 in atg7-IKO MEFs might be causative to the cytoskeletal disorder in these cells. Collectively, these results demonstrate that PDLIM1 is degraded via an ATG7-mediated autophagy-lysosome pathway to maintain or facilitate proper cytoskeleton organization.
Figure 5.
Knockdown of Pdlim1 in atg7-IKO MEFs partially rescues the cytoskeleton organization defect. (A) Infection of Pdlim1 shRNA1 virus inhibited the expression of PDLIM1 protein in both Atg7+/+ and atg7-IKO MEFs. The uninfected cells in the same dish served as an internal control. (B) The knockdown efficiency of Pdlim1 by shRNA, detected by immunoblotting. (C) Knockdown of Pdlim1 by shRNA1 in atg7-IKO MEFs partially rescued the disordered F-actin networks (lower panel, red dotted area), but has no influence on Atg7+/+ MEFs (upper panel red dotted). The uninfected cells in the same dish served as an internal control (white dotted areas). (D) Quantification of the rescue efficiency of Pdlim1 knockdown.
The formation of autophagosomes and autolysosomes are impaired in atg7 knockout spermatids
To address whether the same mechanism applied to spermatozoa, we turned back to the germ cells. First, we tested whether the autophagy-lysosome system was impaired in the atg7-null spermatozoa. The substrates degraded by the autophagy-lysosome pathway are first engulfed by the autophagosome, which then fuses with the lysosome to generate a relatively large autolysosome for hydrolysis.6 To study the effect of Atg7 knockout on autophagosome formation, we examined the LC3A/B signals in seminiferous tubules at 4 typical developmental stages in both Atg7Flox/Flox and Alpl-atg7−/− testes, ie. stage I to III, stage IV to VI, stage VII to VIII and stage IX to XII. LC3A/B could form relatively large and bright puncta in the cytoplasm of elongating spermatids of the Atg7Flox/Flox testes, especially in stage IV to VI seminiferous tubule (Fig. S8A). Note that the large LC3A/B plaques next to the mature spermatozoa nucleus in stage VII to VIII seminiferous tubule was residual body of the elongated spermatids, which had been sloughed off. LC3A/B signals can also be detected in Alpl-atg7−/− seminiferous tubules of the corresponding 4 stages, but the LC3A/B distributed evenly in the cytoplasm of the elongating spermatids, few obvious puncta could be found (Fig. S8B). These results indicate that ATG7 is required for autophagosome formation in the elongating spermatids, and implying a potential role of autophagy in spermatid differentiation.
The autolysosomes can be detected using the same markers as for the lysosomes, namely LAMP1 (lysosomal-associated membrane protein 1) or LAMP2,24 but are distinguished from the lysosomes by their larger size, both in length and size.25 Therefore, we tested autolysosome formation in the Atg7Flox/Flox and Alpl-atg7−/− mouse testes using immunofluorescent staining for LAMP1. In the Atg7Flox/Flox mouse testes, LAMP1 signals were dominantly distributed around the seminiferous lumen (Fig. 6A, upper panel), with a similar distribution pattern as LC3A/B and ATG7. The LAMP1-positive puncta appeared large and bright in the Atg7Flox/Flox mouse testes, with an average length of 1.8 ± 0.14 μm (Fig. 6D, white column) and an average size of 2.36 ± 0.26 μm2 (Fig. 6E, white column). In contrast, in the Alpl-atg7−/− mouse testes, the LAMP1-positive puncta appeared much smaller and smeared (Fig. 6A, lower panel), with an average length of 0.67 ± 0.03 μm (Fig. 6D, black column) and an average size of 0.43 ± 0.03 μm2 (Fig. 6E, black column). These results indicate that the generation of relatively larger autolysosomes was impaired in the Alpl-atg7−/− mouse spermatids.
Figure 6.
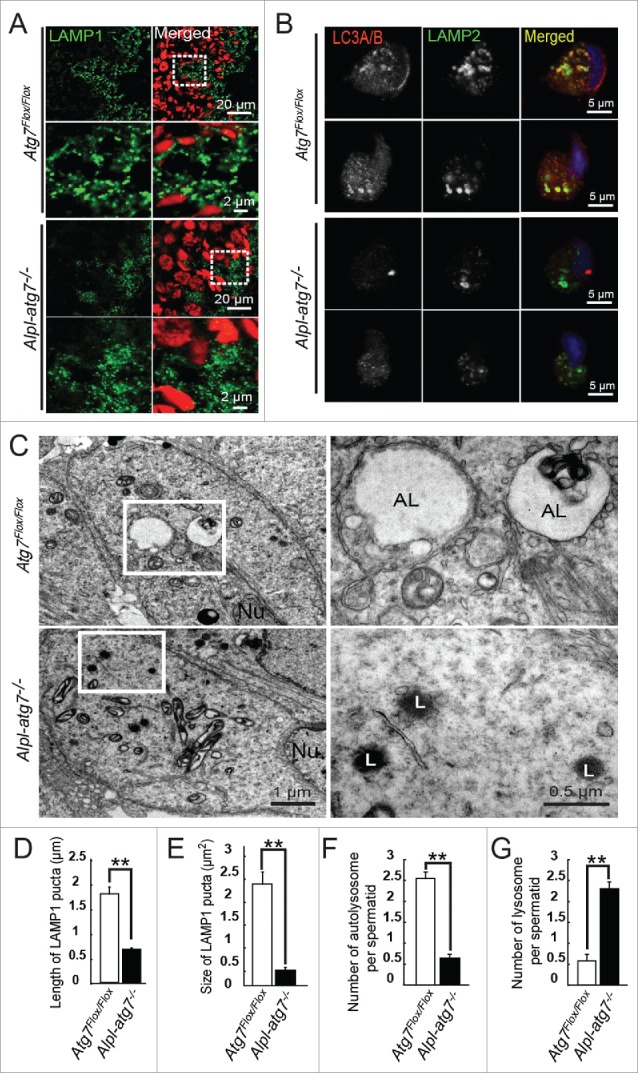
Impairment of autolysosome formation in atg7-null spermatids. (A) IF of LAMP1 in Atg7Flox/Flox mouse testis (upper 2 panels) and Alpl-atg7−/− mouse testis (lower 2 panels). Red, propidium iodide. (B) Co-IF of LC3A/B and LAMP2 in Atg7Flox/Flox spermatids (upper 2 panels) and atg7-null spermatids (lower 2 panels), 2 representative results are shown in each group. (C) Ultrastructural analysis of Atg7Flox/Flox spermatids (upper) and atg7-null spermatids (lower). (D) Quantification of the length of the LAMP1-positive puncta in Atg7Flox/Flox spermatids (white column, 1.8 ± 0.14 μm) and atg7-null spermatids (black column, 0.67 ± 0.03 μm). (E) Quantification of the size of the LAMP1-positive puncta in Atg7Flox/Flox spermatids (white column, 2.36 ± 0.26 μm2) and atg7-null spermatids (black column, 0.43 ± 0.03 μm2). (F) Quantification of the average number of autolysosomes in Atg7Flox/Flox (white column, 2.51 ± 0.33) and atg7-null elongating spermatids (black column, 0.69 ± 0.12) by TEM analysis. (G) Quantification of the average number of lysosomes in Atg7Flox/Flox (white column, 0.61 ± 0.19) and atg7-null elongating spermatids (black column, 2.23 ± 0.31) by TEM analysis. AL, autolysosome; L, lysosome; Nu, nucleus.
The colocalization of LC3A/B and LAMP1 refers to the fusion of autophagosome and lysosome, because the antibodies against LC3A/B and LAMP1 were both generated in rabbit, we chose another lysosome marker (LAMP2) for the costaining analyses with LC3A/B. LAMP1 and LAMP2 are both lysosome membrane-integrated proteins with a single transmembrane domain and they display a similar distribution pattern. First, we measured the LAMP2 immunofluorescence staining puncta in the Atg7Flox/Flox and Alpl-atg7−/− testes. Similar to the LAMP1 staining, the LAMP2 puncta in the Alpl-atg7−/− testes were smaller and smeared compared to those from Atg7Flox/Flox testes, indicating an impairment of the autolysosome generation (Fig. S9). To clearly identify autophagosome and lysosome, we used testis smear for staining of them. We found that, in Atg7Flox/Flox testes smear, LC3A/B and LAMP2 colocalized to each other in relatively large puncta in elongating spermatids of different stages (Fig. 6B, upper panel). Whereas in Alpl-atg7−/− testes, large LC3A/B puncta disappeared, and LAMP2 can only be found in small puncta which were not colocalized with LC3A/B (Fig. 6B, lower panel), confirming an autophagosome and lysosome fusion defect in the Alpl-atg7−/− spermatids.
To further confirm this defect, we analyzed the testes via TEM. Similar to the LAMP1 immunofluorescent staining results, large vacuoles filled with cargos were found in the Atg7Flox/Flox mouse testes (Fig. 6C, upper panel, AL), which was the typical appearance of autolysosomes.26,27 However, in the Alpl-atg7−/− mouse testes, only small electron dense lysosomes surrounded with some unfused small vesicles were observed (Fig. 6C, lower panel, L), few autophagosome was found in these spermatids.28 The average number of autolysosomes per spermatid on a single section dropped significantly and the lysosomes increased correspondingly in Alpl-atg7−/− mouse testes (Fig. 6F and 6G). Taken together, these results provide strong evidences showing that the depletion of ATG7 disrupts the generation of autolysosomes and blocks substrate degradation and cytoplasm component removal through the autophagy-lysosome pathway.
PDLIM1 is accumulated in the atg7-null spermatids
Next, we assessed the PDLIM1 protein levels in the testes by immunoblotting. Similar to the results obtained in MEFs, upon Atg7 depletion, the autophagic flux was blocked and the PDLIM1 level in the Alpl-atg7−/− testes was strongly increased compared with the Atg7Flox/Flox testes (Fig. 7A), but the mRNA level of Pdlim1 remains unchanged (Fig. 7B). To show the correlation between PDLIM1 and cytoskeleton organization in the testes directly, PDLIM1 and F-actin immunofluorescent staining was performed in the Atg7Flox/Flox and Alpl-atg7−/− testes. In the Atg7Flox/Flox testes, a clear F-actin signal indicating the presence of F-actin bundles was detected in the elongating spermatids, but the PDLIM1 signal was relatively weak (Fig. 7C, upper panel). In contrast, in the Alpl-atg7−/− testes, the F-actin signal was disordered suggesting a disruption in the organization of the cytoskeleton, and the PDLIM1 signal was clearly increased (Fig. 7C lower panel). These results suggest that Atg7 depletion in the spermatid results in the accumulation of PDLIM1, which finally causes the disorganization of the cytoskeleton.
Figure 7.
PDLIM1 is degraded within the autolysosome in an ATG7-dependent manner. (A) Immunoblotting analysis of ATG7, PDLIM1, SQSTM1, and LC3B in Atg7Flox/Flox and Alpl-atg7−/− testes. (B) The mRNA expression level of Pdlim1 in Atg7Flox/Flox and Alpl-atg7−/− testes. (C) IF of F-actin and PDLIM1 in Atg7Flox/Flox (upper panel) and Alpl-atg7−/− testes (lower panel). Framed area was magnified. (D) IF of LAMP2 and PDLIM1 in Atg7Flox/Flox spermatids (upper 2 panels) and atg7-null spermatids (lower 2 panels). Framed area was magnified. (E) Quantification of the length of LAMP2-positive puncta in Atg7Flox/Flox spermatids (white column, 1.42 ± 0.13 μm) and atg7-null spermatids (black column, 0.46 ± 0.08 μm). (F) Quantification of the area of the LAMP2-positive puncta in Atg7Flox/Flox spermatids (white column, 2.16 ± 0.14 μm2) and atg7-null spermatids (black column, 0.31 ± 0.04 μm2).
PDLIM1 is degraded within the autolysosome in spermatozoa
If PDLIM1 was degraded via an ATG7-mediated autophagy-lysosome pathway, it might be targeted to the autolysosome in spermatids. PDLIM1 could be detected mainly in the haploid spermatids at different stages in testis, and formed puncta-like structures at stages VII to VIII (Fig. S10). We then examined the relationship between PDLIM1 and LAMP2 at VII to VIII stage by coimmunofluorescent staining in the Atg7Flox/Flox and Alpl-atg7−/− testes. As shown in the Figure 7D, PDLIM1 was colocalized with large LAMP2 puncta in the Atg7Flox/Flox testes (Fig. 7D, upper 2 panels), which means that, in the presence of functional ATG7, PDLIM1 could be targeted by the autophagy pathway and delivered to the autolysosomes for degradation. In contrast, the LAMP2 puncta were relatively smaller in the Alpl-atg7−/− testes (Fig. 7E and F), and PDLIM1 did not colocalize with the LAMP2 puncta (Fig. 7D, lower 2 panels), demonstrating that, in the absence of Atg7 in spermatozoa, PDLIM1 cannot be transported into the autolysosomes. Collectively, these results suggest that PDLIM1 was degraded by an ATG7-mediated autophagy-lysosome pathway in spermatids to maintain a proper level to facilitate cytoskeleton organization.
Discussion
In the current work, we found that autophagy mainly works in 2 types of germ cells, the round spermatids and the elongating spermatids. Consistent with the LC3A/B and ATG7 expression patterns, the germ cell-specific atg7 knockout in mice results in multiple defects in their spermatozoa. In addition to the acrosome biogenesis defect, we have found an abnormal “9+2” structure of the sperm flagellum and an inefficient cytoplasm removal in atg7-null spermatozoa (Fig. 2). The efficient cytoplasm removal is thought to be critical for the generation of functional and motile spermatozoa, but the mechanisms underlying this process remain poorly understood.29 For a long time, the elimination of residual bodies was thought to be a passive process, and they were thought to be engulfed by the Sertoli cells.30 However, the role of the spermatids themselves is largely ignored. Recently, the notion of a proper cytoplasmic removal under a genetic control was well established by the functional study of Spem1.31 SPEM1 is directly involved in the regulation of the cytoplasm removal from the late elongated spermatids in mice, and the lack of SPEM1 causes spermatozoa deformation and male infertility. Actually, cytoplasmic removal defects have been reported in mice deficient in the function of 3 types of genes. The first group includes mice deficient in nuclear packaging and condensation during the late stage of spermiogenesis, harboring defects in either Tnp1/Tnp2,32 Prm1/Prm2,33 H1fnt/H1t2),34 or Camk4.35 The second group includes mice deficient in acrosome biogenesis,15 with defects in Csnk2a2, Gopc, Gba2, Zpbp1, Pick1, or Spaca1. Spem1 itself stands alone as the third group because its knockout does not affect either of the 2 processes. These results suggest that the regulation of DNA packaging, acrosome biogenesis, and cytoplasmic removal during spermiogenesis might be partially interconnected. For the DNA packaging deficient mice, the cytoplasmic remnants left on spermatozoa seem less than those on either the acrosome biogenesis defect or Spem1 knockout mice, suggesting that the cytoplasmic remnants may be left as a side effect of improperly packed nuclear. In contrast to the DNA packaging deficient mice, usually the cytoplasmic remnants on spermatozoa with acrosome defect or Spem1 deficiency are relatively larger, suggesting that acrosome biogenesis and cytoplasmic removal maybe 2 tightly coupled processes.
During normal spermiogenesis, the cytoplasm of the apical head is compressed by the close contact between the cytoplasm membrane and the outer acrosomal membrane,36 while the bulk of the cytoplasm moves distally along the manchette.37 In the elongating spermatids, the F-actin based acroplaxome provides a docking site for the acrosome development, and anchors it to the spermatid nucleus.38 Thus, due to lack of proper anchor, the nucleus could not be efficiently pushed out from the cytoplasm when there were no acrosomes or with abnormal acrosomes, finally results in the presence of excessive perinuclear cytoplasm.
Actually, it is cytoskeleton that couples the acrosome biogenesis, cytoplasm removal and sperm flagella development during the spermiogenesis. It has been well established that the transport of the proacrosomal vesicles requires microtubule-based and F-actin-based motor proteins (Kinesin/dynein and MYO5A/myosin Va, respectively).38 Meanwhile, with the initiation of the spermatid nucleus elongation, a microtubule- and actin-containing manchette begins to assemble. Most cargos and macromolecules are transported to the centrosome along the manchette, and at the same time, the nucleus is pushed out of the cytoplasm.39 In the atg7-null spermatids, the assembly of a functional manchette was affected, thus inhibiting the transportation of the cargos toward the direction of sperm tail and the proper development of the sperm tail along with the microtubules of the manchette. In addition, we found that the axonemes of the atg7-null aggregated sperm flagella had lost their regular “9+2” structure. Instead, several microtubule filaments were distributed randomly. Actually, plenty of genes have been reported to be essential for normal sperm flagella development and for the head-tail connection, such as Spag6,40 Pacrg,41 Odf2,42 Hook1,43 Trip11/Gmap210 and Ift88.44 Most importantly, a recent work brought autophagy and primary ciliogenesis together.45 It has been reported that a ciliopathy protein, OFD1 (oral-facial-digital syndrome 1 gene homolog [human]), is degraded by autophagy to promote primary ciliogenesis.45 Although we currently cannot test this hypothesis directly due to the absence of a good antibody to detect OFD1 during mice spermiogenesis, as a specialized cilium, autophagy may also participate in sperm flagella biogenesis by degrading OFD1. Therefore, all of these defects in atg7-null spermatozoa were found to be associated with cytoskeletal disorganizations.
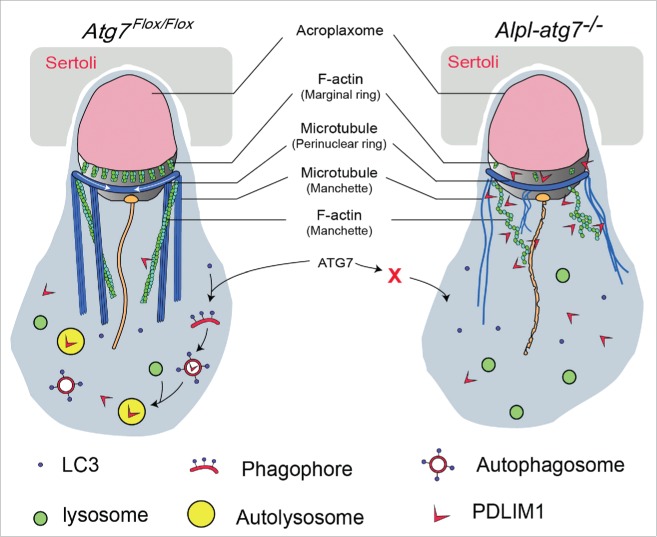
Based on our data, we propose a model in which actin-associated protein PDLIM1 was a mediator between autophagy and cytoskeleton organization during spermiogenesis. Under normal conditions, PDLIM1 is targeted to the autolysosome for degradation in an ATG7-dependent manner to maintain a proper dynamics of the cytoskeleton network. However, after Atg7 knockout, LC3 could not be recruited to the membrane of the phagophore and the autophagic flux is blocked. Consequently, the autophagosome and autolysosome do not engulf their substrates, such as PDLIM1 and OFD1, causing their accumulation in the cytoplasm. This leads to the formation of stress fibers-like structures and disrupts the assembly of the spermatozoa flagella. The ablation of Atg7 also disrupts the parallel orientation of the TUBB- and F-actin-containing cytoplasmic manchettes and some other cytoskeleton-based structures, impairs acrosome formation, decreases sperm motility and prevents the complete cytoplasm removal, and finally results in sterility (Fig. 8).
Figure 8.
Model for the functional role of Atg7 in spermatid differentiation. In the presence of ATG7 (left), excessive PDLIM1 could be engulfed by the LC3-labeled phagophore, and then be delivered to the autolysosome for degradation. F-actin-based structures and microtubule-based structures are assembled properly. In the absence of ATG7 (right), autophagy cannot be initiated, and PDLIM1 accumulates inside the cytosol. The accumulated PDLIM1 results in the disruption of the cytoskeletons in spermatids.
PDLIM1 is a member of the PDZ and LIM protein family, which contains an N-terminal PDZ domain and a C-terminal LIM domain.46 Most interactions between the PDZ-LIM proteins and the cytoskeleton have been identified in striated muscle. In contrast, PDLIM1 interacts with the F-actin-based stress fibers in nonmuscle tissues via an association with nonmuscle ACTN1 and ACTN4.22 It has been reported that PDLIM1 is involved in cytoskeletal reorganization and signal transduction in a variety of cells.47 The reduction of PDLIM1 expression leads to the loss of stress fibers in BeWo48 and F2408 cells,49 but not in SAOS-2 cells.50 This difference in the function of PDLIM1 might originate from various partners in the cells because PDLIM1 works as a scaffold of cytoskeleton organization and an adaptor for the recruitment of several factors. It has been reported that a spermatid-enriched kinase, Clik1, is recruited to the actin stress fibers by PDLIM1.51 In addition, an actin regulatory protein named PALLD/palladin is concentrated along the tubulin- and F-actin-containing cytoplasmic manchette.52 Because PDLIM1 might form a triple complex with ACTN1 and PALLD on stress fibers,53 the localization of PALLD in the elongating spermatids might also be mediated by PDLIM1, hinting that PDLIM1 and ACTN1 may also localize on the manchette. In striated muscle, ACTN1 cross-links in an antiparallel manner to the F-actin at the Z disk, whereas in nonmuscle cells, ACTN1 is a highly flexible cross-linker,54 it can cross-link parallel,55 antiparallel bundles,56 and other kinds of actin filaments.57 Therefore, the accumulation of PDLIM1 might lead to the formation of a stress fibers-like structure, and finally result in the random orientation of the filamentous bundles and manchette in either MEFs or spermatids.
There remains one question to be answered, which is why the knockout of Atg7 in germ cells results in the disorder of microtubules. Actually, as the 2 major systems of cytoskeleton, F-actin and microtubules interact with and influence each other mutually,58 and adaptors such as DBNL/drebin and MAPRE3/EB3 have been identified to crosslink the F-actin and microtubules directly.59 What is more, besides the acroplaxome, F-actin also participates in manchette, suggesting that the microtubule based and F-actin based structure and transport systems do interact with each other during spermiogenesis.60 So, as an F-actin associated factor, the accumulation of PDLIM1 may further influence microtubules organization. An alternative possibility is that another negative regulator of microtubules, which is still totally unknown, needs to be degraded by the autophagy-lysosome pathway. All these possibilities still need further experimental data to be distinguished in the future.
The actin stress fibers form a dynamic organelle that is important in some processes such as cell migration, cell polarity, and cytokinesis.61 However, some of these structures are less suited for cell motility and more suited to static contraction. It has been reported that stress fibers are more prominent in stationary cells,62 F-actin-based stress fibers are poorly organized in migrating cells,63 suggesting that they might inhibit cell migration.64 The reduction of ACTN expression by the transfection of an antisense construct leads to increased motility in 3T3 cells.65 When PDLIM1 and its paralog PDLIM4/RIL are suppressed, the F2408 cells not only display a reduction of stress fiber formation but also the generation of lamellipodia with extensive ruffling activity and a dramatic increase of both directional and random migration.49 Along with the migration toward the lumen of the elongated spermatids, the cytoplasm is removed to generate mature spermatozoa.
The current study found that once Atg7 was knocked out, PDLIM1 accumulated in the cytoplasm of the spermatids, the accumulation of PDLIM1 disrupted the proper dynamics of the cytoskeleton and finally resulted in the inefficient cytoplasm removal during spermiogenesis. We have to point out that PDLIM1 might be just one case of these negative regulators, and multiple kinds of negative regulators, such as RIL and OFD1, may also need to be eliminated by autophagy-lysosome pathway to ensure efficient cytoplasm removal during spermiogenesis. In fact, the autophagy-dependent regulation of cytoskeleton such as stress fibers has been hinted at more than 2 decades ago by Ridley and Hall.66 They show that once Swiss 3T3 fibroblasts are starved by serum deprivation for a few hours, the pre-existing stress fibers and focal adhesions disappear, suggesting that they might be degraded by the autophagy-lysosome pathway. Therefore, our work, along with other previous studies, uncovered a novel crosstalk between autophagy and the cytoskeleton, suggesting that autophagy and the cytoskeleton mutually modulate each other during the differentiation of some cell types.
Materials and methods
Animals
The Atg7Flox/Flox mouse strain (RBRC02759)67 were purchased from the RIKEN Bio Resource Center with permission from Dr. Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science). The Atg7Flox/Flox; Alpl/Tnap-cre mice were bred by crossing the Atg7Flox/Flox mice and Alpl/Tnap-Cre mice (Jackson Laboratory, 008569). The Cre-recombinase is driven by the promoter of Tnap (tissue nonspecific alkaline phosphatase) or Alpl (alkaline phosphatase, liver/bone/kidney ) which allows it to delete the floxed Atg7 alleles in primordial germ cells between E9.5 to E10.5.68 All of the animal experiments were approved by the Animal Research Panel of the Committee on Research Practice of the University of the Chinese Academy of Science.
Antibodies
The rabbit anti-LC3A/B polyclonal antibody (ab58610) was purchased from Abcam. The rabbit anti-SQSTM1 polyclonal antibody (5114) was purchased from Cell Signaling Technology. The mouse anti-ATG7 monoclonal antibody (SAB4200304) for immunoblotting, and the rabbit anti-LC3B polyclonal antibody (L7543) for immunoblotting were purchased from Sigma Aldrich. The mouse anti-TUBB/β-tubulin monoclonal antibody (M20005) was purchased from Abmart, FITC-conjugated phalloidin (40735ES75) and TRITC-conjugated phalloidin (40734ES75) were purchased from Yeason. The goat anti-rabbit FITC (ZF-0311), donkey anti-rat TRITC, and goat anti-mouse TRITC (ZF-0313) -conjugated secondary antibodies, as well as the goat anti-mouse (ZB-2305), goat anti rabbit (ZB-2301)-horseradish peroxidase-conjugated secondary antibodies were purchased from Zhong Shan Jin Qiao. The Alexa Fluor 680-conjugated goat anti-mouse (A21057) and Alexa Fluor 800-conjugated goat anti-rabbit (A21109) secondary antibodies for immunoblotting were purchased from Invitrogen. The rabbit anti-PDLIM1 antibody (11674-1-AP) was purchased from Proteintech Group. The rabbit anti-LAMP1 antibody (SAB3000285) was purchased from Sigma-Aldrich. The rat anti-LAMP2 antibody (ab13524, Abcam) was kindly provided by Dr. Li Yu (Tsinghua University).
Plasmids, cell culture and transfection
Genes encoding FLAG-tagged PDLIM1, STMN1 (stathmin 1), PPP4R2 (protein phosphatase 4, regulatory subunit 2), NUDC (nudC nuclear distribution protein), MAP6 (microtubule-associated protein 6), YBX1 (Y box protein 1), and CD2AP (CD2-associated protein) were cloned into the SalI and NotI site of the modified pRK5 vector (AddGene, 71868; deposited by David Sabatini). The plasmids were transfected using Lipofectamine 2000 (Invitrogen, 11668019) according to the manufacturer's instructions. FLAG-tagged ATG7 was cloned into the XbaI and XmaI site of pHIV-EGFP lentiviral vector (AddGene, 21373; deposited by Bryan Welm) and a stop codon was inserted before the open reading frame of the EGFP. The Pdlim1 shRNA1 (5′-CGAAGTGCCATGCCATTTA-3′), or shRNA2 (5′-GTTAGTCATGCGCAATAAA-3′) were cloned into the HpaI and XhoI site of pSicoR-mCherry lentiviral vector (AddGene, 21907; deposited by Miguel Ramalho-Santos). The lentiviral plasmid was cotransfected into 293T cells with the packaging plasmids pMD2.G (AddGene, 12259; deposited by Didier Trono) and psPAX2 (AddGene, 12260; deposited by Didier Trono),69 then the virus was collected 48 h and 60 h after transfection, and then used to infect the cells along with 4 mg/ml polybrene (Sigma, H9268).
Sperm motility assays
The sperm motility assays were performed as described previously.70 Briefly, the cauda epididymis was dissected from adult mice. Spermatozoa were squeezed out from the cauda epididymis and incubated for 30 min at 37°C under a 5% CO2-containing atmosphere. 10-μl aliquots from the sample were taken and placed into 80-μm deep glass cell chambers (Hamilton Thorne, 80 micron 2X-CEl). The chambers were imaged using an Olympus BX51 microscope (Olympus, Tokyo, Japan) through a 20X phase objective, while being maintained at 37°C on a heated platform. The viewing areas on each chamber were captured using a CCD camera. The samples were analyzed via computer-assisted semen analysis using the Animal Motility system (Hamilton Thorne, Beverly, MA, USA). Different sperm motility parameters were analyzed, including the total motility, average path velocity (VAP), VSL, and the VCL. Unfixed sperm were spread onto precoated slides for morphological observation.
Testis smear
Atg7Flox/Flox and Alpl-atg7−/− mice (8-wk old) were killed by cervical dislocation. The testes were surgically removed and the tunica albunigea was removed from the testes. Then, the testes were digested by 1mg/ml collagenase and 1mg/ml hyaluronidase. Cells were dissociated by gently pipetting, filtered through a 70-μm filter and then pelleted by centrifugation at 500 g for 10 min. Cells were suspended in 1 ml phosphate-buffered saline (PBS; Gibco, C14190500BT) and fixed with 4% paraformaldehyde (PFA) solution, washed with PBS and finally spread onto polylysine-coated slides for staining.
Real-time quantitative PCR (qPCR) analyses
Total RNAs were isolated from MEF cells or testis as previously described.71 cDNA was synthesized by the PrimeScriptTM RT Reagent Kit (TaKaRa, RR037A), and primer sets for Pdlim1 (forward:5′- CCACATCCTTCCTGGTTCTG3′ and reverse: 5′- TGGTGATCCC TCAGCTTCAC-3′) and Gapdh (forward:5′-GGTGGTGCTAAGCGTGTTAT-3′ and reverse: 5′-ACCTCTGTCATCTCTCCACA-3′) were used. Real-time PCR was carried out with the Roche Light Cycler® 480II System (WA, USA), and the results were analyzed using the LightCycle480 SW 1.5.1.
Immunoblotting
Tissue or cell extracts were prepared using a Dounce Tissue Grinders (Corning, 7722 15) in cold RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40 [Sigma, 74385], 1% sodium deoxycholate [Amresco, D0613], 0.1% sodium dodecyl sulfate [Amresco, 0227]) supplemented with 1 mM phenylmethylsulfonyl fluoride (Amresco, M145) and a protein inhibitor cocktail (Roche Diagnostics, 04693132001). The homogenates were centrifuged at 13,200 g for 15 min, and the protein concentrations were determined by the Bio-Rad protein assay (500-0006). The protein lysates (25 μg) were separated by SDS-PAGE and electrotransferred onto a nitrocellulose membrane. The membrane was incubated with corresponding primary antibodies and detected by Alexa Fluor 680-conjugated goat anti-mouse or Alexa Fluor 800-conjugated goat anti-rabbit secondary antibody, and finally scanned using the ODYSSEY Sa Infrared Imaging System (LI-COR biosciences, USA).
Transmission electron microscopy
The adult testes were fixed with 2.5% glutaraldehyde in 0.2 M cacodylate buffer overnight. After washing them in 0.2 M PBS, the tissues were cut into small pieces of approximately 1 mm3, and immersed in 1% OsO4 in 0.2 M cacodylate buffer for 2 h at 4°C. Then, the samples were dehydrated through a graded ethanol series and embedded in resin. Ultrathin sections were cut on an ultramicrotome, stained with uranyl acetate and lead citrate, and observed using a JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan).
Immunofluorescence
The indicated male C57BL/6 mice were euthanized according to the guidelines of the Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences. The tissues were immediately embedded in an optimum cutting temperature compound (OCT; Tissue-Tek, 4583) and cut in 8-µm sections using a microtome-cryostat (Leica CM1950, Wetzlar, Germany). Each section was fixed with 4% PFA and rinsed 3 times in PBS (pH 7.4). Then, the primary antibody was added to the sections and incubated at 4°C overnight, followed by incubation with the secondary antibody. The F-actin networks were stained with FITC or TRITC-conjugated phalloidin. The nuclei were stained with propidium iodide or DAPI. The images were taken immediately using a LSM 780/710 microscope (Zeiss, Oberkochen, Germany) or SP8 microscope (Leica, Wetzlar, Germany). For cellular immunofluorescence, 1×105 cells were plated on cover glass slips in 35-mm dish and culture in DMEM with 10% fetal bovine serum for 24 h, the cells were rinsed 3 times with PBS, fixed, and stained as described above. For protein overexpression and PDLIM1 knockdown, cells were stained 48 h after plasmid transfection or virus infection.
Immunohistochemistry
The paraffin sections were fixed with 4% PFA and rinsed 3 times in PBS (pH 7.4). Then, the sections were boiled for 15 min in sodium citrate buffer for antigen retrieval. Next, 3% H2O2 was added to the sections to eliminate the internal peroxidase activity. After blocking with 5% bovine serum albumin (Sigma, V900933), each section was incubated with the primary antibody at 4°C overnight, followed by staining with the horseradish peroxidase-conjugated secondary antibody. The negative controls were prepared without the primary antibody. Finally, the sections were stained with 3, 3′-diaminobenzidine, and the nuclei were stained with hematoxylin. The images were acquired using a Nikon 80i invert microscope equipped with a CCD camera (Nikon, Tokyo, Japan).
Generation of atg7-IKO MEFs
The atg7-IKO MEFs were constructed using a 4-OH-tamoxifen inducible Cre recombinase, briefly, the Cre recombinase is fused to the mutated human ER LBD (estrogen receptor ligand-binding domain) containing a glycine at amino-acid 400 and an arginine at amino-acid 521, so it is also called Cre-ER(GR). This fused recombinase is strongly activated in the presence of the ER antagonists 4-hydroxytamoxifen (OHT) and ICI 182,780 (ICI), but not ER.72
Atg7Flox/Flox; Tamoxifen-cre mice were generated from the breeding of Atg7Flox/Flox mice with Tamoxifen-cre mice. When the female mice showed plugs and were pregnant on d 12, they were sacrificed and the embryos were isolated as previously described.73 For the knockout of Atg7 in the MEFs, 4-OH-tamoxifen (Sigma-Aldrich, H7904) was added to the medium at a final concentration of 0.1 mM to induce the gene deletion for at least 3 d and the 4-OH-tamoxifen induced Atg7 knockout MEFs are called atg7-IKO MEFs. The cells were incubated at 37°C in a 5% CO2-containing atmosphere. The knockout efficiency was confirmed by immunoblotting.
Bioinformatics analyses
The information of up- and downregulated proteins in atg7-IKO MEFs was obtained from a published paper,21 and the DAVID platform was used to annotate their biological themes. DAVID version 6.7 (http://david.abcc.ncifcrf.gov/) enables the generation of specific functional annotations of biological processes affected by a treatment from the target gene lists produced from the high-throughput experiments.74 We selected the cellular components and biological processes showing a P value < 0.1 and a Count > 5 as the significance cutoff for the terms. Moreover, the gene symbols corresponding to these regulated proteins were also sent to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING version 9. One;75 http://string.embl.de/), to build a network using edge information from 3 separate form of evidence: databases, experiments, and text mining. We used 0.4 (medium confidence) as the value for the edge confidence provided by STRING.
Cycloheximide (CHX) chase assay
Atg7+/+ and atg7-IKO MEFs were plated one d before the experiment and cycloheximide (R750107, Sigma-Aldrich) was added to the culture at 100 μg/ml to block new protein synthesis. Samples were taken at 0, 2, 4, 8 h after the addition of CHX and protein levels were determined by immunoblotting. The quantification of PDLIM1 was obtained as the ratio of PDLIM1 intensity to GAPDH intensity in each sample.
Quantification of IF and TEM
The IF images were obtained using a LSM 780/710 microscope (Zeiss, Oberkochen, Germany) or SP8 microscope (Leica, Wetzlar, Germany). The quantification of IF was counted up to 100 cells in randomly selected microscopy fields from the slides, and each experiment was repeated for 3 times. Data were analyzed and expressed as the percentage of cells with disordered cytoskeleton in each group. The TEM images were observed using a JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan). The quantification of TEM was counted up to 50 cells in randomly selected microscopy fields, and repeated for 3 times.
Statistical analysis
All data were presented as the means ± SEM. The statistical significance of the differences between the mean values for the different genotypes was measured by the Student t test with a paired, 2-tailed distribution. The data were considered significant when the P value was less than 0.05 (*) or 0.01 (**).
Supplementary Material
Abbreviations
- Ac
acrosome
- AL
autolysosome
- Atg7
autophagy-related 7
- CD
cytoplasmic droplet
- CHX
cycloheximide
- CQ
chloroquine
- IF
immunofluorescent staining
- IHC
immunohistochemistry
- L
lysosome
- LAMP1
lysosomal-associated membrane protein 1
- LAMP2
lysosomal-associated membrane protein 2
- MAP1LC3A/LC3A
microtubule-associated protein 1 light chain 3 α
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 β
- MEF
mouse embryonic fibroblast
- MS
mass spectrometry
- OFD1
oral-facial-digital syndrome 1 gene homolog (human)
- PDLIM1
PDZ and LIM domain 1 (elfin)
- PFA
paraformaldehyde
- SQSTM1
sequestosome 1
- STMN1
stathmin 1
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TEM
transmission electron microscopy
- VAP
average path velocity
- VCL
curvilinear velocity
- VSL
straight-line velocity
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Quan Chen, Li Yu and Qingyuan Sun for their critical reading of the manuscript. We are grateful to Masaaki Komatsu for providing the Atg7Flox/Flox mice, Li Yu for providing anti-LAMP2 antibody.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 91519317, 31471277) and the Major Basic Research Program (Grant No. 2012CB944404).
References
- [1].Parvinen M. Regulation of the seminiferous epithelium. Endocr Rev 1982; 3:404-17; PMID:6295753; http://dx.doi.org/ 10.1210/edrv-3-4-404 [DOI] [PubMed] [Google Scholar]
- [2].Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc Res Tech 2010; 73:241-78; PMID:19941293; http://dx.doi.org/ 10.1002/jemt.20783 [DOI] [PubMed] [Google Scholar]
- [3].Calvel P, Rolland AD, Jegou B, Pineau C. Testicular postgenomics: targeting the regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci 2010; 365:1481-500; PMID:20403865; http://dx.doi.org/ 10.1098/rstb.2009.0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med 2008; 14:1197-213; PMID:18989307; http://dx.doi.org/ 10.1038/nm.f.1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; http://dx.doi.org/ 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- [6].Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 2009; 335:1-32; PMID:19802558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823-30; PMID:20811354; http://dx.doi.org/ 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008; 321:117-20; PMID:18599786; http://dx.doi.org/ 10.1126/science.1154822 [DOI] [PubMed] [Google Scholar]
- [9].Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al.. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 2009; 136:308-21; PMID:19167332; http://dx.doi.org/ 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
- [10].Nollet M, Santucci-Darmanin S, Breuil V, Al-Sahlanee R, Cros C, Topi M, Momier D, Samson M, Pagnotta S, Cailleteau L, et al.. Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy 2014; 10:1965-77; PMID:25484092; http://dx.doi.org/ 10.4161/auto.36182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim ES, Chang H, Choi H, Shin JH, Park SJ, Jo YK, Choi ES, Baek SY, Kim BG, Chang JW, et al.. Autophagy induced by resveratrol suppresses α-MSH-induced melanogenesis. Exp Dermatol 2014; 23:204-6; PMID:24499351; http://dx.doi.org/ 10.1111/exd.12337 [DOI] [PubMed] [Google Scholar]
- [12].Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A 2010; 107:832-7; PMID:20080761; http://dx.doi.org/ 10.1073/pnas.0913170107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al.. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 2008; 4:309-14; PMID:18188005; http://dx.doi.org/ 10.4161/auto.5474 [DOI] [PubMed] [Google Scholar]
- [14].Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 2009; 119:3329-39; PMID:19855132; http://dx.doi.org/ 10.1172/JCI35541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, Yang L, Tang H, Zhang X, Duan E, et al.. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res 2014; 24:852-69; PMID:24853953; http://dx.doi.org/ 10.1038/cr.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 1993; 225:84-113; PMID:8231890; http://dx.doi.org/ 10.1016/0076-6879(93)25009-Q [DOI] [PubMed] [Google Scholar]
- [17].MacLean JA, Wilkinson MF. Gene Regulation in Spermatogenesis. 2005; 71:131-97; PMID:16344105 [DOI] [PubMed] [Google Scholar]
- [18].AL K. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch Histol Cytol 2004; 67:271-84; PMID:15700535; http://dx.doi.org/ 10.1679/aohc.67.271 [DOI] [PubMed] [Google Scholar]
- [19].Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem 1994; 269:11492-7; PMID:7908909 [PubMed] [Google Scholar]
- [20].Mi N, Chen Y, Wang S, Chen M, Zhao M, Yang G, Ma M, Su Q, Luo S, Shi J, et al.. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol 2015; 17:1112-23; PMID:26237647; http://dx.doi.org/ 10.1038/ncb3215 [DOI] [PubMed] [Google Scholar]
- [21].Zhuo C, Ji Y, Chen Z, Kitazato K, Xiang Y, Zhong M, Wang Q, Pei Y, Ju H, Wang Y. Proteomics analysis of autophagy-deficient Atg7−/− MEFs reveals a close relationship between F-actin and autophagy. Biochem Biophys Res Commun 2013; 437:482-8; PMID:23850690; http://dx.doi.org/ 10.1016/j.bbrc.2013.06.111 [DOI] [PubMed] [Google Scholar]
- [22].Vallenius T, Luukko K, Makela TP. CLP-36 PDZ-LIM protein associates with nonmuscle α-actinin-1 and α-actinin-4. J Biol Chem 2000; 275:11100-5; PMID:10753915; http://dx.doi.org/ 10.1074/jbc.275.15.11100 [DOI] [PubMed] [Google Scholar]
- [23].Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al.. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 2009; 460:863-8; PMID:19587682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 2009; 10:623-35; PMID:19672277; http://dx.doi.org/ 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- [25].Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gerland LM, Genestier L, Peyrol S, Michallet MC, Hayette S, Urbanowicz I, Ffrench P, Magaud JP, Ffrench M. Autolysosomes accumulate during in vitro CD8+ T-lymphocyte aging and may participate in induced death sensitization of senescent cells. Exp Gerontol 2004; 39:789-800; PMID:15130673; http://dx.doi.org/ 10.1016/j.exger.2004.01.013 [DOI] [PubMed] [Google Scholar]
- [27].Eskelinen EL, Reggiori F, Baba M, Kovacs AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 2011; 7:935-56; PMID:21566462; http://dx.doi.org/ 10.4161/auto.7.9.15760 [DOI] [PubMed] [Google Scholar]
- [28].Klionsky DJ, Eskelinen EL. The vacuole versus the lysosome: when size matters. Autophagy 2014; 10:185-7; PMID:24343261; http://dx.doi.org/ 10.4161/auto.27367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cheng CY, Mruk DD. The biology of spermatogenesis: the past, present and future. Philos Trans R Soc Lond B Biol Sci 2010; 365:1459-63; PMID:20403863; http://dx.doi.org/ 10.1098/rstb.2010.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carr I, Clegg EJ, Meek GA. Sertoli cells as phagocytes: an electron microscopic study. J Anat 1968; 102:501-9; PMID:5656139 [PMC free article] [PubMed] [Google Scholar]
- [31].Zheng H, Stratton CJ, Morozumi K, Jin J, Yanagimachi R, Yan W. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc Natl Acad Sci U S A 2007; 104:6852-7; PMID:17426145; http://dx.doi.org/ 10.1073/pnas.0701669104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod 2004; 71:1220-9; PMID:15189834; http://dx.doi.org/ 10.1095/biolreprod.104.029363 [DOI] [PubMed] [Google Scholar]
- [33].Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet 2001; 28:82-6; PMID:11326282 [DOI] [PubMed] [Google Scholar]
- [34].Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, et al.. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol 2005; 25:7107-19; PMID:16055721; http://dx.doi.org/ 10.1128/MCB.25.16.7107-7119.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet 2000; 25:448-52; PMID:10932193; http://dx.doi.org/ 10.1038/78153 [DOI] [PubMed] [Google Scholar]
- [36].Burgos MH, Fawcett DW. Studies on the fine structure of the mammalian testis. I. Differentiation of the spermatids in the cat (Felis domestica). J Biophys Biochem Cytol 1955; 1:287-300; PMID:13242594; http://dx.doi.org/ 10.1083/jcb.1.4.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fawcett DW, Anderson WA, Phillips DM. Morphogenetic factors influencing the shape of the sperm head. Dev Biol 1971; 26:220-51; PMID:5168310; http://dx.doi.org/ 10.1016/0012-1606(71)90124-2 [DOI] [PubMed] [Google Scholar]
- [38].Kierszenbaum AL, Tres LL, Rivkin E, Kang-Decker N, van Deursen JM. The acroplaxome is the docking site of Golgi-derived myosin Va/Rab27a/b- containing proacrosomal vesicles in wild-type and Hrb mutant mouse spermatids. Biol Reprod 2004; 70:1400-10; PMID:14724135; http://dx.doi.org/ 10.1095/biolreprod.103.025346 [DOI] [PubMed] [Google Scholar]
- [39].Kierszenbaum AL. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol Reprod Dev 2002; 63:1-4; PMID:12211054; http://dx.doi.org/ 10.1002/mrd.10179 [DOI] [PubMed] [Google Scholar]
- [40].Sapiro R, Kostetskii I, Olds-Clarke P, Gerton GL, Radice GL, Strauss IJ. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol Cell Biol 2002; 22:6298-305; PMID:12167721; http://dx.doi.org/ 10.1128/MCB.22.17.6298-6305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lorenzetti D, Bishop CE, Justice MJ. Deletion of the Parkin coregulated gene causes male sterility in the quaking(viable) mouse mutant. Proc Natl Acad Sci U S A 2004; 101:8402-7; PMID:15148410; http://dx.doi.org/ 10.1073/pnas.0401832101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rivkin E, Tres LL, Kierszenbaum AL. Genomic origin, processing and developmental expression of testicular outer dense fiber 2 (ODF2) transcripts and a novel nucleolar localization of ODF2 protein. Mol Reprod Dev 2008; 75:1591-606; PMID:18398819; http://dx.doi.org/ 10.1002/mrd.20911 [DOI] [PubMed] [Google Scholar]
- [43].Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, Neesen J. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet 2002; 11:1647-58; PMID:12075009; http://dx.doi.org/ 10.1093/hmg/11.14.1647 [DOI] [PubMed] [Google Scholar]
- [44].Kierszenbaum AL, Rivkin E, Tres LL, Yoder BK, Haycraft CJ, Bornens M, Rios RM. GMAP210 and IFT88 are present in the spermatid golgi apparatus and participate in the development of the acrosome-acroplaxome complex, head-tail coupling apparatus and tail. Dev Dyn 2011; 240:723-36; PMID:21337470; http://dx.doi.org/ 10.1002/dvdy.22563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 2013; 502:254-7; PMID:24089205; http://dx.doi.org/ 10.1038/nature12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bauer K, Kratzer M, Otte M, de Quintana KL, Hagmann J, Arnold GJ, Eckerskorn C, Lottspeich F, Siess W. Human CLP36, a PDZ-domain and LIM-domain protein, binds to α-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood 2000; 96:4236-45; PMID:11110697 [PubMed] [Google Scholar]
- [47].Miehe U, Kadyrov M, Neumaier-Wagner P, Bartz C, Rath W, Huppertz B. Expression of the actin stress fiber-associated protein CLP36 in the human placenta. Histochem Cell Biol 2006; 126:465-71; PMID:16609848; http://dx.doi.org/ 10.1007/s00418-006-0182-5 [DOI] [PubMed] [Google Scholar]
- [48].Tamura N, Ohno K, Katayama T, Kanayama N, Sato K. The PDZ-LIM protein CLP36 is required for actin stress fiber formation and focal adhesion assembly in BeWo cells. Biochem Biophys Res Commun 2007; 364:589-94; PMID:17964547; http://dx.doi.org/ 10.1016/j.bbrc.2007.10.064 [DOI] [PubMed] [Google Scholar]
- [49].Miyazaki K, Ohno K, Tamura N, Sasaki T, Sato K. CLP36 and RIL recruit α-actinin-1 to stress fibers and differentially regulate stress fiber dynamics in F2408 fibroblasts. Exp Cell Res 2012; 318:1716-25; PMID:22659164; http://dx.doi.org/ 10.1016/j.yexcr.2012.05.006 [DOI] [PubMed] [Google Scholar]
- [50].Maeda M, Asano E, Ito D, Ito S, Hasegawa Y, Hamaguchi M, Senga T. Characterization of interaction between CLP36 and palladin. FEBS J 2009; 276:2775-85; PMID:19366376; http://dx.doi.org/ 10.1111/j.1742-4658.2009.07001.x [DOI] [PubMed] [Google Scholar]
- [51].Vallenius T, Makela TP. Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J Cell Sci 2002; 115:2067-73; PMID:11973348 [DOI] [PubMed] [Google Scholar]
- [52].Niedenberger BA, Chappell VK, Kaye EP, Renegar RH, Geyer CB. Nuclear localization of the actin regulatory protein Palladin in sertoli cells. Mol Reprod Dev 2013; 80:403-13; PMID:23559268; http://dx.doi.org/ 10.1002/mrd.22174 [DOI] [PubMed] [Google Scholar]
- [53].Hasegawa T, Ohno K, Funahashi S, Miyazaki K, Nagano A, Sato K. CLP36 interacts with palladin in dorsal root ganglion neurons. Neurosci Lett 2010; 476:53-7; PMID:20381583; http://dx.doi.org/ 10.1016/j.neulet.2010.03.081 [DOI] [PubMed] [Google Scholar]
- [54].Courson DS, Rock RS. Actin cross-link assembly and disassembly mechanics for α-Actinin and fascin. J Biol Chem 2010; 285:26350-7; PMID:20551315; http://dx.doi.org/ 10.1074/jbc.M110.123117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Taylor KA, Taylor DW, Schachat F. Isoforms of α-actinin from cardiac, smooth, and skeletal muscle form polar arrays of actin filaments. J Cell Biol 2000; 149:635-46; PMID:10791977; http://dx.doi.org/ 10.1083/jcb.149.3.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Liu J, Taylor DW, Taylor KA. A 3-D reconstruction of smooth muscle α-actinin by CryoEm reveals two different conformations at the actin-binding region. J Mol Biol 2004; 338:115-25; PMID:15050827; http://dx.doi.org/ 10.1016/j.jmb.2004.02.034 [DOI] [PubMed] [Google Scholar]
- [57].Hampton CM, Taylor DW, Taylor KA. Novel structures for α-actinin:F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J Mol Biol 2007; 368:92-104; PMID:17331538; http://dx.doi.org/ 10.1016/j.jmb.2007.01.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou FQ, Waterman-Storer CM, Cohan CS. Focal loss of actin bundles causes microtubule redistribution and growth cone turning. J Cell Biol 2002; 157:839-49; PMID:12034775; http://dx.doi.org/ 10.1083/jcb.200112014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol 2008; 10:1181-9; PMID:18806788; http://dx.doi.org/ 10.1038/ncb1778 [DOI] [PubMed] [Google Scholar]
- [60].Kierszenbaum AL, Rivkin E, Tres LL. Acroplaxome, an F-actin-keratin-containing plate, anchors the acrosome to the nucleus during shaping of the spermatid head. Mol Biol Cell 2003; 14:4628-40; PMID:14551252; http://dx.doi.org/ 10.1091/mbc.E03-04-0226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci 2007; 120:3491-9; PMID:17928305; http://dx.doi.org/ 10.1242/jcs.018473 [DOI] [PubMed] [Google Scholar]
- [62].Couchman JR, Rees DA. The behaviour of fibroblasts migrating from chick heart explants: changes in adhesion, locomotion and growth, and in the distribution of actomyosin and fibronectin. J Cell Sci 1979; 39:149-65; PMID:575139 [DOI] [PubMed] [Google Scholar]
- [63].Badley RA, Couchman JR, Rees DA. Comparison of the cell cytoskeleton in migratory and stationary chick fibroblasts. J Muscle Res Cell Motil 1980; 1:5-14; PMID:7014630; http://dx.doi.org/ 10.1007/BF00711922 [DOI] [PubMed] [Google Scholar]
- [64].Burridge K. Are stress fibres contractile? Nature 1981; 294:691-2; PMID:7198718; http://dx.doi.org/ 10.1038/294691a0 [DOI] [PubMed] [Google Scholar]
- [65].Gluck U, Ben-Ze'ev A. Modulation of α-actinin levels affects cell motility and confers tumorigenicity on 3T3 cells. J Cell Sci 1994; 107(Pt 7):1773-82; PMID:7983147 [DOI] [PubMed] [Google Scholar]
- [66].Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992; 70:389-99; PMID:1643657; http://dx.doi.org/ 10.1016/0092-8674(92)90163-7 [DOI] [PubMed] [Google Scholar]
- [67].Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al.. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169:425-34; PMID:15866887; http://dx.doi.org/ 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hammond SS, Matin A. Tools for the genetic analysis of germ cells. Genesis 2009; 47:617-27; PMID:19548313; http://dx.doi.org/ 10.1002/dvg.20539 [DOI] [PubMed] [Google Scholar]
- [69].Zhu S, Wang H, Ding S. Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nat Protoc 2015; 10:959-73; PMID:26042385; http://dx.doi.org/ 10.1038/nprot.2015.059 [DOI] [PubMed] [Google Scholar]
- [70].Jimenez T, Sanchez G, Wertheimer E, Blanco G. Activity of the Na,K-ATPase alpha4 isoform is important for membrane potential, intracellular Ca2+, and pH to maintain motility in rat spermatozoa. Reproduction 2010; 139:835-45; PMID:20179187; http://dx.doi.org/ 10.1530/REP-09-0495 [DOI] [PubMed] [Google Scholar]
- [71].Yuan S, Stratton CJ, Bao J, Zheng H, Bhetwal BP, Yanagimachi R, Yan W. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc Natl Acad Sci U S A 2015; 112:E430-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 1997; 237:752-7; PMID:9299439; http://dx.doi.org/ 10.1006/bbrc.1997.7124 [DOI] [PubMed] [Google Scholar]
- [73].Conner DA. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr Protoc Mol Biol 2001; Chapter 23:Unit 23 2. [DOI] [PubMed] [Google Scholar]
- [74].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/ 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- [75].Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 2000; 28:3442-4; PMID:10982861; http://dx.doi.org/ 10.1093/nar/28.18.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.