ABSTRACT
Cannabinoids and related drugs generate profound behavioral effects (such as analgesic effects) through activating CNR1 (cannabinoid receptor 1 [brain]). However, repeated cannabinoid administration triggers lysosomal degradation of the receptor and rapid development of drug tolerance, limiting the medical use of marijuana in chronic diseases. The pathogenic mechanisms of cannabinoid tolerance are not fully understood, and little is known about its prevention. Here we show that a protein involved in macroautophagy/autophagy (a conserved lysosomal degradation pathway), BECN2 (beclin 2), mediates cannabinoid tolerance by preventing CNR1 recycling and resensitization after prolonged agonist exposure, and deletion of Becn2 rescues CNR1 activity in mouse brain and conveys resistance to analgesic tolerance to chronic cannabinoids. To target BECN2 therapeutically, we established a competitive recruitment model of BECN2 and identified novel synthetic, natural or physiological stimuli of autophagy that sequester BECN2 from its binding with GPRASP1, a receptor protein for CNR1 degradation. Co-administration of these autophagy inducers effectively restores the level and signaling of brain CNR1 and protects mice from developing tolerance to repeated cannabinoid usage. Overall, our findings demonstrate the functional link among autophagy, receptor signaling and animal behavior regulated by psychoactive drugs, and develop a new strategy to prevent tolerance and improve medical efficacy of cannabinoids by modulating the BECN2 interactome and autophagy activity.
KEYWORDS: analgesia, autophagy inducer, BECN2, behavior, cannabinoid, cannabinoid receptor 1, drug tolerance, intracellular trafficking
Introduction
Cannabinoids and related drugs, such as the marijuana-derived ingredients, generate profound behavioral effects (such as analgesic effects) that are therapeutic in many pathological conditions, including neurodegeneration, digestive disorders, spasticity, and chronic and cancer-related pain.1-4 However, long-term administration of cannabinoids for either medical or recreational purposes induces rapid development of tolerance (a demonstration of physical dependence), which is a limitation and concern of its medical use and may lead to addiction and withdrawal symptoms.5,6 Clinical data showed that 9% of adult cannabis users, and 17% of adolescent users, develop dependence and addiction after repeated dosage,6 which is not trivial given the widespread usage of illicit cannabinoids in many countries. Yet the pathogenic mechanisms of cannabinoid tolerance are not fully understood, and little is known about its prevention methods. Consequently, only a very small number of cannabinoid therapeutics have been approved and used clinically in the market in limited regions; for example, the cannabis medication for spasticity, Sativex, is prescribed as an oromucosal spray to ensure slow blood delivery and is administered at relatively low doses.
The effects of cannabinoids are mediated by binding and activation of a G protein-coupled receptor (GPCR), CNR1/CB1 (cannabinoid receptor 1 [brain]), and chronic exposure to cannabinoids results in lysosomal trafficking and degradation of CNR1, which causes diminished cellular responses and requires higher doses to produce the same effects.7 Therefore, retaining the functionality of CNR1 may play an important role in the prevention of cannabinoid tolerance, especially in clinical applications. A recently identified autophagy gene, Becn2 (beclin 2), which belongs to the Beclin (coiled-coil, myosin-like BCL2-interacting protein) family, may link CNR1-regulated cell signaling and animal behavior to the autophagy pathway.8 Autophagy is an essential catabolic process that breaks down damaged or unnecessary structures in lysosomes, and the resulting metabolites are recycled and reused for new protein synthesis and energy production. Autophagy is intensely induced by physiological stimuli or stress, such as starvation9 and physical exercise,10 and malfunction of autophagy has been implicated in a variety of diseases, including neurodegeneration, cardiovascular diseases, cancer and metabolic disorders.11-13 In addition to a role in autophagy, BECN2 is also essential for agonist-induced lysosomal degradation of a group of specific GPCRs, including CNR1, DRD2 (dopamine receptor D2) and OPRD1 (opioid receptor, delta 1).8 In vitro biotin protection degradation data suggest that BECN2 mediates the degradation of these GPCRs by binding to GPRASP1/GASP1 (G protein-coupled receptor associated sorting protein 1),8 a receptor protein that degrades GPCRs independently of ubiquitination and certain components of the canonical endosomal sorting complex required for transport (ESCRT) machinery.14-18
It is unclear whether BECN2 regulates the downstream events of these GPCRs in response to chronic agonist exposure, including receptor resensitization, signaling cascades and drug-responsive behaviors in vivo, which are important questions especially when many of the GPCRs in this specific group are targets of psychoactive drugs, such as CNR1. In addition, the function, genetic basis and molecular mechanism of autophagy in the regulation of drug tolerance and dependence after repeated usage remain mysterious. It is also unknown how the autophagy pathway crosstalks with drug-responsive GPCR signaling and behavioral regulation, and whether BECN2 plays a role in the process. Here we found that BECN2 is essential for the development of analgesic tolerance after chronic cannabinoid exposure by preventing CNR1 recycling and resensitization in mice. We discovered that activation of autophagy restores CNR1 function and regulates cannabinoid analgesia through BECN2, and also identified novel synthetic and natural agents to prevent cannabinoid tolerance by modulating the BECN2 interactome and autophagy activity. Our findings demonstrated for the first time a new functional link among autophagy, drug responses and behaviors regulated by psychoactive substances, and opened up a new direction to potentially improve medical efficacy of cannabinoids in many circumstances such as pain management.
Results
Becn2+/− heterozygous knockout mice are resistant to analgesic tolerance induced by repeated cannabinoid dosage
We hypothesized that in response to prolonged cannabinoid exposure, BECN2 mediates the degradation and rapid deactivation of CNR1. To test this hypothesis, we sought to use the Becn2+/− heterozygous knockout (KO) mice, because becn2−/− homozygotes rarely survive the embryonic development period. We treated Becn2+/− mice and wild-type (WT) littermates daily for 14 d with WIN55,212–2 (WIN), a synthetic cannabinoid drug and CNR1 agonist, and analyzed the levels of CNR1 in the brain. Compared to WT mice, Becn2+/− mice maintained a significantly higher level of brain CNR1 after 14 d of chronic cannabinoid treatment (Fig. 1A), in addition to a trend of increased steady-state CNR1 levels under normal conditions (with DMSO treatment), which suggests that BECN2 loss-of-function increases the availability of CNR1 after repeated agonist dosage.
Figure 1.
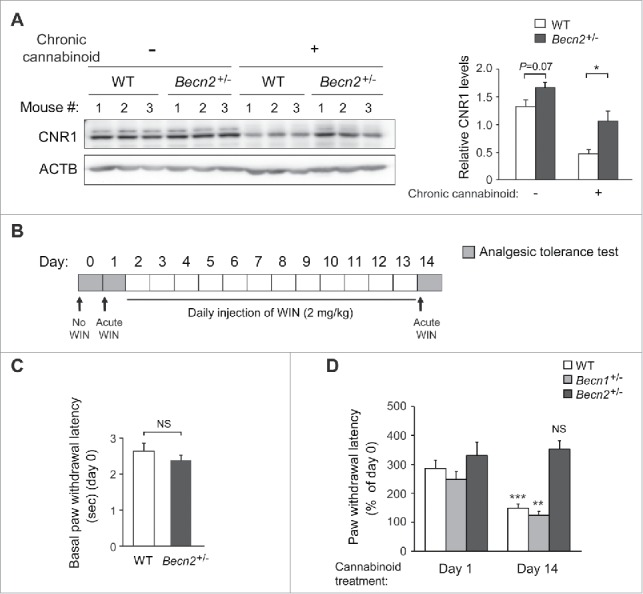
Loss of BECN2 confers resistance to analgesic tolerance induced by chronic usage of cannabinoid drugs. (A) Brain samples from Becn2+/− heterozygous knockout (KO) and WT mice were collected after 14 d of prolonged treatment of vehicle or the synthetic cannabinoid drug WIN55,212–2 (WIN). CNR1 levels were analyzed by Western blot of brain lysates (left) and quantified (right). Representative images of 3 mice in each group are shown. (B) Chronic cannabinoid treatment scheme, described in Materials and methods. Briefly, mice were injected with WIN for 14 d, and analgesic tolerance was measured without WIN treatment on day 0, and after 1-h WIN treatment on d 1 and d 14. (C) Baseline pain sensitivity of WT and Becn2+/− mice before chronic WIN treatment (day 0, agonist-free). N ≥ 16 mice/group. (D) Becn2+/− mice show resistance to analgesic tolerance to chronic WIN treatment. Becn2+/−, Becn1+/− and WT mice were treated with daily WIN for 14 d, and the analgesic effect of WIN was analyzed as in (B). Statistics are comparing the same genotype on day 1 and day 14. N = 9–13 mice/group. Results represent mean ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (t-test).
To study whether BECN2 functions in CNR1-regulated behavioral responsiveness following chronic cannabinoid treatment, we analyzed the anti-nociceptive (i.e., anti-pain) effect of WIN as a readout by analgesic tolerance tests (Fig. 1B), using WT mice and mice heterozygous for Becn2, or for Becn1, another Beclin family member that does not bind GPRASP1 or regulate GPCR degradation as a control. There were no genotype differences either in basal pain sensitivity to infrared heat-generated pain (agonist-free, day 0) (Fig. 1C, Video S1-S3), or in analgesic effects of acute WIN treatment prior to chronic WIN administration (day 1) (Fig. 1D, Video S4-S6). However, 14 d of repeated WIN dosage markedly decreased its analgesic effect in WT and Becn1+/− mice, but not in Becn2+/− mice (day 14) (Fig. 1D, Video S7-S9), suggesting that Becn2+/− mice are resistant to analgesic tolerance induced by prolonged cannabinoid exposure. Notably, body weight of mice of all genotypes remained constant during chronic WIN treatment (Fig. S1). Thus, these data demonstrate that BECN2 downregulates the level of brain CNR1 after prolonged WIN treatment, and BECN2 loss of function protects animals from developing analgesic tolerance to repeated exposure of cannabinoid drugs.
CNR1 is resensitized at the cell surface upon BECN2 loss
We next sought to investigate the cellular mechanism underlying the behavioral protection against tolerance upon loss of BECN2. Published data have shown that BECN2 is required for agonist-induced lysosomal transport of OPRD1, another GPRASP1-bound GPCR, which is relocalized to the plasma membrane upon loss of BECN2.8 However, it is unknown whether in the absence of BECN2 these recycled receptors are functional, or whether BECN2 also plays a role in the trafficking, recycling and signaling of CNR1 after prolonged agonist exposure. Here we proposed that BECN2 depletion stabilizes CNR1 levels by promoting its recycling and resensitization at the cell surface, and tested this hypothesis from 3 aspects. We first followed the intracellular trafficking of CNR1 in response to agonists, by examining its colocalization with endosome (EEA1) or lysosome (LAMP1) markers in the absence of BECN2. By pulse-labeling cell-surface CNR1 with antibody, we found that compared to basal conditions (0 min), in a significantly higher number of cells treated with control siRNA, CNR1 was transported to endosomes after 30 min of WIN treatment and to lysosomes after 60 min, whereas in cells transfected with Becn2 siRNA,8 instead of being transported to lysosomes for degradation, internalized CNR1 was trapped in endosomal structures even after 60 min of WIN treatment (Fig. 2A, Fig. S2A). In addition, in Becn2 siRNA-treated cells the endocytosed CNR1 relocalized to the cell surface after 30 min of agonist removal and antagonist treatment (Fig. 2B). Thus, these data demonstrate that loss of BECN2 leads to accumulation of CNR1 in the endosomes and its eventual recycling to the plasma membrane.
Figure 2.
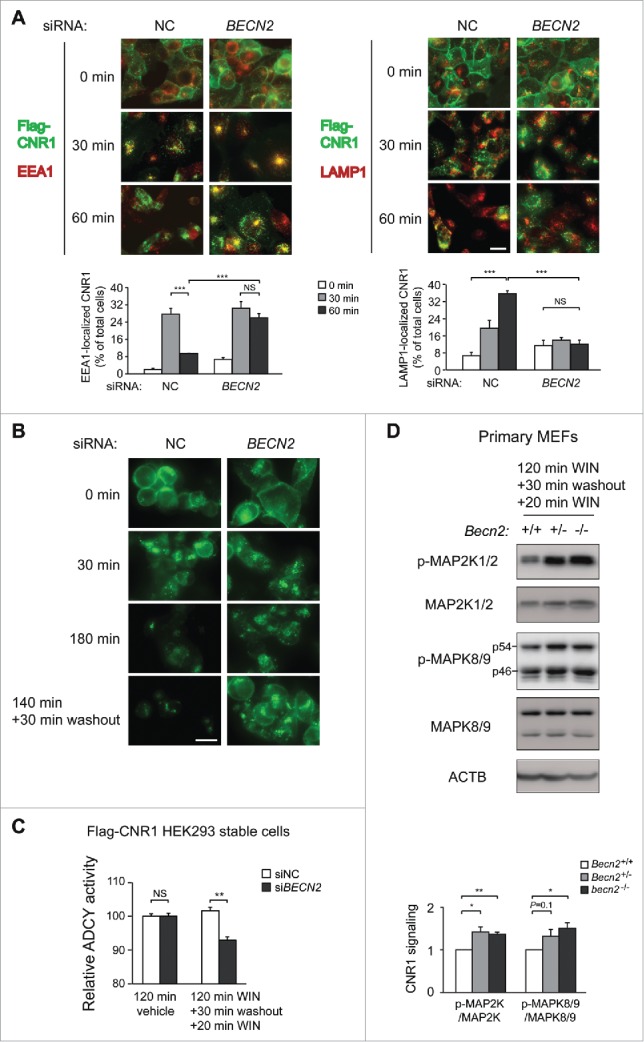
BECN2 depletion triggers resensitization of endocytosed CNR1 at the plasma membrane. (A) Immunofluorescence imaging (upper) and quantification (lower) of the effects of nontargeting control (NC) or Becn2 siRNA on the fate of endocytosed CNR1. HEK293 cells stably expressing FLAG-CNR1 were fed with anti-FLAG antibody, treated with the agonist WIN for 30 or 60 min and immunostained as in Materials and mMethods. Green, FLAG-CNR1; red, EEA1 or LAMP1. Percentages of cells with CNR1-EEA1 or CNR1-LAMP colocalization in >200 cells per experiment were quantified from 4 independent experiments. (B) Becn2 knockdown promotes CNR1 recycling to the cell surface after prolonged agonist exposure, analyzed by immunofluorescence imaging of FLAG-CNR1. HEK293 FLAG-CNR1 cells were fed with anti-FLAG antibody, and treated with WIN for 30 or 180 min or with WIN for 140 min followed by agonist washout and antagonist (rimonabant) treatment for 30 min. Scale bar: 20 μm. (C) Becn2 knockdown suppresses the ADCY/adenylyl cyclase activity downstream of CNR1 after prolonged agonist exposure. HEK293 FLAG-CNR1 cells transfected with the indicated siRNAs were treated either with vehicle for 120 min, or with WIN for 120 min to induce CNR1 internalization and degradation, followed by agonist washout and antagonist (rimonabant) treatment for 30 min to trigger CNR1 recycling and another 20 min treatment of WIN to activate any CNR1 at the cell surface. Results represent mean±s.e.m of 3 independent experiments. (D) BECN2 depletion increases cannabinoid-induced CNR1 signaling after prolonged exposure. Becn2+/+, Becn2+/− or becn2−/− primary MEFs were treated as in (C). Phosphorylation of MAP2K and MAPK8/9 downstream of CNR1 was analyzed by Western blot (upper) and quantified from 3 independent experiments (lower). Results represent mean±s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (t-test).
Next, to study whether the recycled CNR1 caused by BECN2 depletion is resensitized and functional, we examined the signaling pathways downstream of CNR1 at both biochemical and cellular levels. CNR1 is coupled to the GNAI/Gi/o protein, and CNR1 activation inhibits the ADCY/adenylyl cyclase activity and cyclic adenosine monophosphate (cAMP) production, while activating the mitogen-activated protein kinase (MAPK) pathways, including MAP2K1 (mitogen-activated protein kinase 1)/MEK1-MAP2K2/MEK2 and MAPK8/JNK1-MAPK9/JNK2 cascades.7,19 Thus, we measured cannabinoid-induced cAMP suppression (Fig. 2C) and MAPK cascade phosphorylation (Fig. 2D) in cell lines and primary MEFs (mouse embryonic fibroblasts) following prolonged agonist treatment. We found that the intracellular cAMP level was reduced in Becn2 siRNA-treated cells compared to control cells (Fig. 2C), which suggests an increased level of functional CNR1 at the cell surface and is an indication of CNR1 resensitization upon loss of BECN2. Furthermore, we detected a higher level of cannabinoid-induced phosphorylation of MAP2K1/2 and MAPK8/9 in Becn2 KO primary MEFs (Fig. 2D), as well as a trend of higher MAP2K1/2 and MAPK8/9 phosphorylation in the brain of Becn2+/− mice (Fig. S2B), than in WT MEFs or brain, suggesting higher CNR1 functionality after chronic agonist exposure in the absence of BECN2. Overall, these data suggest that loss of BECN2 leads to CNR1 resensitization.
Competitive recruitment model of BECN2 and identification of novel brain-penetrable autophagy inducers
Accordingly, an important translational question is how to reduce BECN2 activity to maintain CNR1 sensitivity and prevent drug tolerance induced by repeated cannabinoid exposure. To avoid the technical difficulty and risk of directly deleting Becn2 in vivo (such as by injecting shRNAs or CRISPR constructs), which may disrupt BECN2-regulated autophagy, we developed and tested a novel and more convenient strategy to achieve the same goal via modulating the BECN2 interactome, which will not affect the essential autophagy function of BECN2 (Fig. 3A). As previously reported,8 BECN2 is present in 2 distinct protein complex pools, the BECN2-class III phosphatidylinositol 3-kinase complex for autophagy, and the BECN2-GPRASP1-GPCR complex in which BECN2-GPRASP1 binding is required for GPCR degradation. Thus, we hypothesized that BECN2 is a limiting factor that regulates the balance between the 2 lysosomal degradation pathways, and that competitive recruitment of BECN2 to the autophagy machinery by activating autophagy prevents it from functioning in CNR1 downregulation and thus restores CNR1 functionality and reverses drug tolerance. To test this hypothesis, we first examined whether autophagy activation by brain-penetrable inducers inhibits the BECN2-GPRASP1 interaction. From a small-molecule library screen20 and a phytochemical screen (Y. Fan, A. Rocchi, N. Wang, W. Zhang, R. Vassar, Y. Zhou and C. He, manuscript in preparation), we identified 2 novel brain-penetrable autophagy-inducing compounds: ML246 (also named metarrestin based on its anti-metastasis effects in cancer,20 Fig. 3B), a synthetic small molecule derived from a high-throughput screen against a pan-cancer cellular marker; and Rg2,21 a 785 Da natural steroid glycoside compound isolated from Panax ginseng by analytical chemistry approaches.
Figure 3.
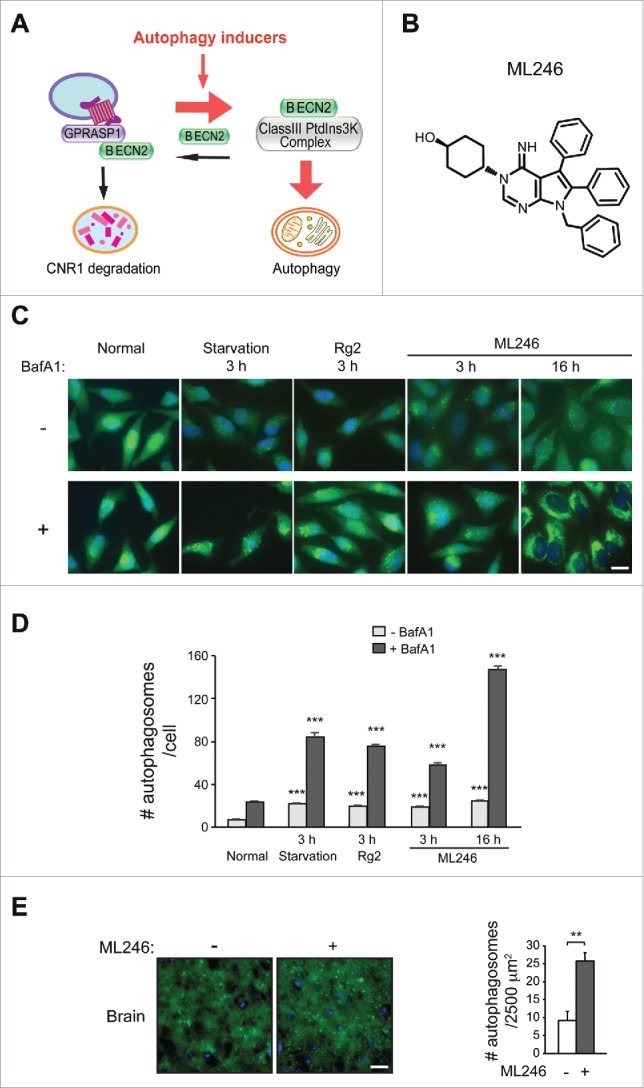
Identification of novel brain-penetrable autophagy-inducing compounds. (A) Competitive recruiting model of BECN2 by the 2 lysosomal degradation pathways: inducing autophagy shunts BECN2 away from the BECN2-GPRASP1 complex, and thus attenuates lysosomal degradation of CNR1 and maintains its responsiveness. Red arrows, pro-autophagy; black arrows, pro-CNR1 downregulation. PtdIns3K, phosphatidylinositol 3-kinase. (B) Chemical structure of ML246. (C and D) Representative images (C) and quantification (D) of GFP-LC3 puncta in HeLa cells stably expressing GFP-LC3 cultured for 3 h in normal or starvation medium, or treated with Rg2 or ML246 in normal medium for 3 h or 16 h, in the presence or absence of bafilomycin A1 (BafA1). Results represent mean ± s.e.m. Statistics are comparing the indicated value with or without BafA1 to the one under the normal condition. Blue, DAPI; scale bar: 20 μm. (E) Representative images (left) and quantification (right) of GFP-LC3 puncta in the brain of GFP-LC3 transgenic mice injected with vehicle or ML246. Results represent mean ± s.d. Scale bar: 25 μm. N>100 cells or N ≥ 4 mice. **, P < 0.01; ***, P < 0.001 (t-test).
To determine autophagy activation by the 2 compounds, we analyzed several markers of autophagy induction, including formation of autophagosomes in cells and transgenic mice expressing GFP-tagged MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3; an autophagosome marker),9 degradation of SQSTM1/p62 (sequestosome 1; an autophagy receptor and substrate protein), and conversion of LC3 from the non-lipidated form (LC3-I) to the phagophore- and autophagosome-associated lipidated form (LC3-II). Both compounds markedly increased numbers of GFP-LC3 puncta (representing autophagosomes) and decreased levels of SQSTM1 in HeLa cells and in mouse brain, as potently as starvation (Fig. 3C-E, Fig. S3A-C), although as previously reported22 we did not detect significant changes in LC3-II conversion in brain. In addition, when cotreated with the lysosomal inhibitor bafilomycin A1, ML246 or Rg2 led to more accumulation of GFP-LC3 puncta, LC3-II and SQSTM1 compared to normal conditions (Fig. 3C-D, Fig. S3D), suggesting an increased level of autophagic flux induced by either compound. Therefore, both agents are effective autophagy inducers in vitro and in vivo.
Activation of autophagy pharmacologically or physiologically attenuates BECN2-GPRASP1 binding, dependent on the early initiation step of autophagy
Based on our model, we subsequently examined the effect of the ML246 and Rg2 inducers on the BECN2-GPRASP1 interaction in vitro and in mouse brain. We found that autophagy induction either pharmacologically with ML246 or physiologically with starvation medium for 3 h potently blocked co-immunoprecipitation of endogenous GPRASP1 by BECN2 in HEK293 cells (Fig. 4A). Importantly, early initiation of autophagy is required for the sequestration of BECN2 from GPRASP1 binding by autophagy inducers, as the BECN2-GPRASP1 complex remained intact during starvation or ML246 treatment after knockdown of any member in the upstream ATG13-RB1CC1/FIP200-ULK1 kinase complex essential for autophagy induction (Fig. 4A).23-26 Furthermore, injection of ML246 or Rg2 in vivo to WT mice, as well as 48-h starvation, dramatically decreased the binding between endogenous BECN2 and endogenous GPRASP1 in mouse brain (Fig. 4B). Overall, these data suggest that both physiological and novel pharmacological autophagy inducers attenuate BECN2-GPRASP1 binding, which is dependent on the early initiating step of autophagy and supports our competitive recruitment model of BECN2. Thus, autophagy inducers serve as new candidates in the prevention of receptor downregulation and cannabinoid tolerance to prolonged exposure.
Figure 4.
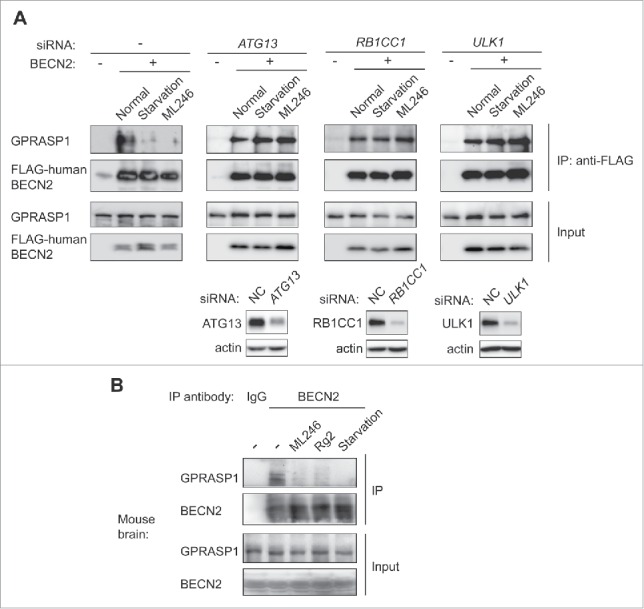
Autophagy induction by synthetic, natural or physiological inducers disrupts the BECN2-GPRASP1 interaction in vitro and in vivo, dependent on the upstream ULK1 kinase complex. (A) Co-immunoprecipitation (co-IP) of endogenous human GPRASP1 with FLAG-human BECN2 in HEK293 cells treated with normal or starvation medium, or normal medium with ML246 for 3 h. The same co-IPs were also performed in HEK293 cells transfected with siRNAs against Atg13, Rb1cc1 or Ulk1. The lower panel shows the siRNA knockdown efficiency of each gene. (B) Co-IP of endogenous GPRASP1 with endogenous BECN2 using brain lysates from WT mice treated with vehicle, ML246 or Rg2, or with 48-h starvation. Lysates in each group were pooled from 3 mice.
Synthetic, natural or physiological autophagy inducers prevent WT mice from analgesic tolerance after chronic cannabinoid usage
We then tested the efficacy of ML246 and Rg2 in the maintenance of cannabinoid analgesia in WT mice. We found that compared with vehicle injection, cotreatment of either ML246 or Rg2 with WIN potently rescued the pain-relieving effect of WIN in WT mice on day 14 to a day 1-like level (i.e., before repeated dosage) (Fig. 5A, Video S10-S12), suggesting that the autophagy-inducing compounds prevent analgesic tolerance induced by chronic WIN administration. The cotreatment regimen did not alter body weight of the mice (Fig. S4A). To confirm the findings with pharmacological inducers, we further asked whether autophagy induction by physiological methods also rescues cannabinoid tolerance. We adopted 2 methods that activate autophagy in mouse brain:22,27 the first is daily voluntary exercise by use of running wheels, which allows mice to run for approximately 1 km/night; and the second is a “2-day on/1-day off” periodic starvation protocol, in which mice were subjected to cycles of 48-h fasting followed by 24-h feeding that allowed them to return to normal weight in each cycle (Fig. 5B, Fig. S4B).
Figure 5.
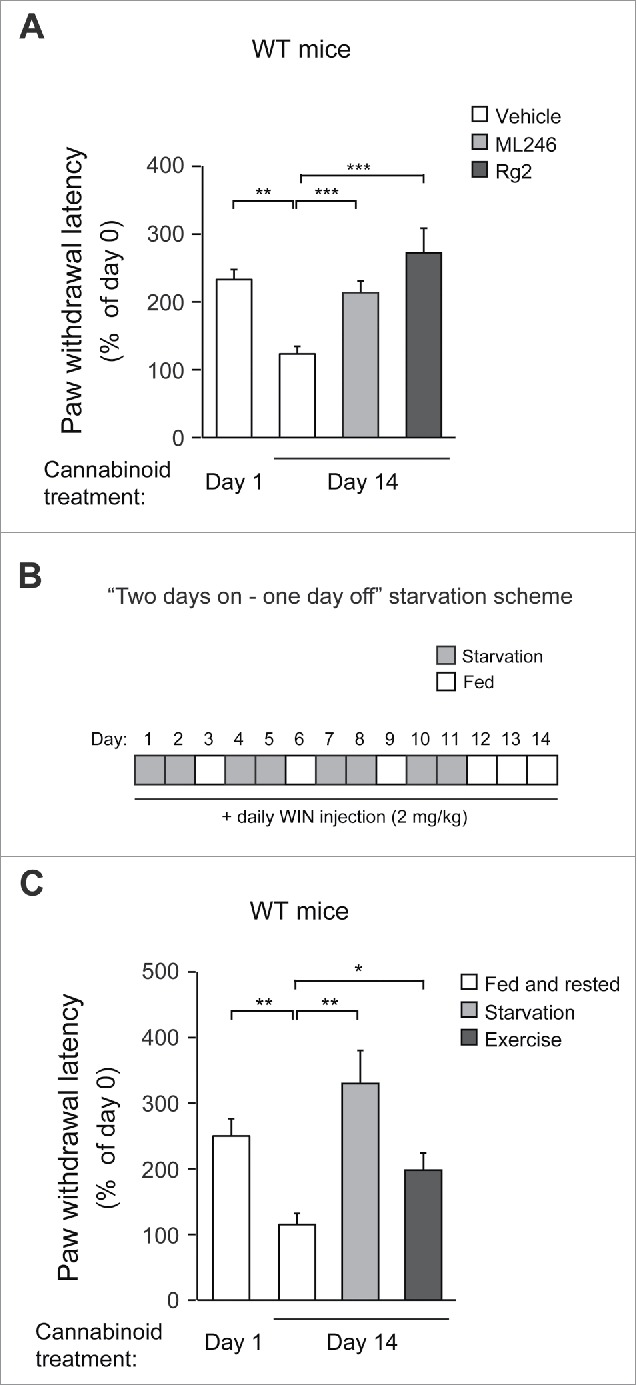
Autophagy activation either pharmacologically or physiologically prevents WT mice from developing behavioral tolerance to repeated cannabinoid dosage. (A) Analgesic tolerance of WT mice simultaneously treated with daily WIN and vehicle, ML246, or Rg2 for 14 d. (B) Periodic fasting scheme. WT mice underwent 48-h fasting followed by 24-h feeding cycles during chronic WIN treatment. (C) Analgesic tolerance of WT mice housed under normal conditions or subjected to either “2 days on–1 day off” periodic starvation or running-wheel exercise, with daily WIN treatment for 14 d. N = 8–14 mice/group. Results represent mean ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t-test).
Although the increase in autophagosome formation was not significant in the brain (frontal cortex) of GFP-LC3 mice after a single bout of 48-h fasting as previously reported using fluorescence microscopy (Fig. S4C),9 we observed a cumulative effect on autophagy induction in the same brain region after multiple rounds of alternating fasting and feeding, demonstrated by a significant induction of GFP-LC3 puncta in the frontal cortex after 4 cycles of “2-day on/1-day off” starvation (Fig. S4C). Although the exact mechanism of this additive effect is currently unclear, we propose that it may be due to a relatively stable glucose supply and low metabolism in the brain (compared to muscle and liver), leading to slow formation/turnover of autophagosomes that can be detected after repeated induction. This hypothesis is supported by our observation that skeletal muscle, which has high metabolic activity, does not show much cumulative effect with regard to autophagy induction by periodic starvation cycles (Fig. S4C). At the end of chronic WIN treatment, similar to mice treated with autophagy-inducing compounds, mice undergoing daily running or intermittent fasting showed significantly higher sensitivity to the analgesic effects of WIN (Fig. 5C, Video S13-S15), suggesting that physiological autophagy inducers, such as exercise and starvation, prevent analgesic tolerance as well. Altogether, these data suggest that both pharmacological and physiological autophagy inducers prevent cannabinoid tolerance to repeated dosage.
Autophagy induction preserves brain CNR1 level and activity in response to chronic cannabinoids
To investigate whether restoration of CNR1 signaling underlies the behavioral regulation by autophagy activation, we analyzed the level and functionality of CNR1 in mouse brain after co-administration of chronic cannabinoids and autophagy inducers. Consistent with behavioral sensitization to WIN, after chronic WIN treatment we detected higher levels of CNR1 in the brain of mice concurrently treated with ML246 or Rg2 (Fig. 6A), or fasted or exercised (Fig. 6B), which were comparable to the brain CNR1 level prior to chronic cannabinoid exposure (Fig. 1A). To determine whether the increased level of CNR1 represents CNR1 resensitization in the brain, we analyzed the agonist-induced phosphorylation of signaling kinases downstream of CNR1 after chronic WIN exposure. We found that repeated WIN treatment decreased activation of CNR1 signaling induced by agonists (Fig. 6C), whereas autophagy induction, either pharmacologically by ML246 or Rg2, or physiologically by starvation or exercise, rescued the CNR1 signaling in spite of repeated WIN dosage, demonstrated by increased phosphorylation of MAP2K and MAPK8/9 (Fig. 6D-E). Thus, these findings suggest that activating autophagy pharmacologically or physiologically effectively improves CNR1 signaling after chronic cannabinoid exposure.
Figure 6.
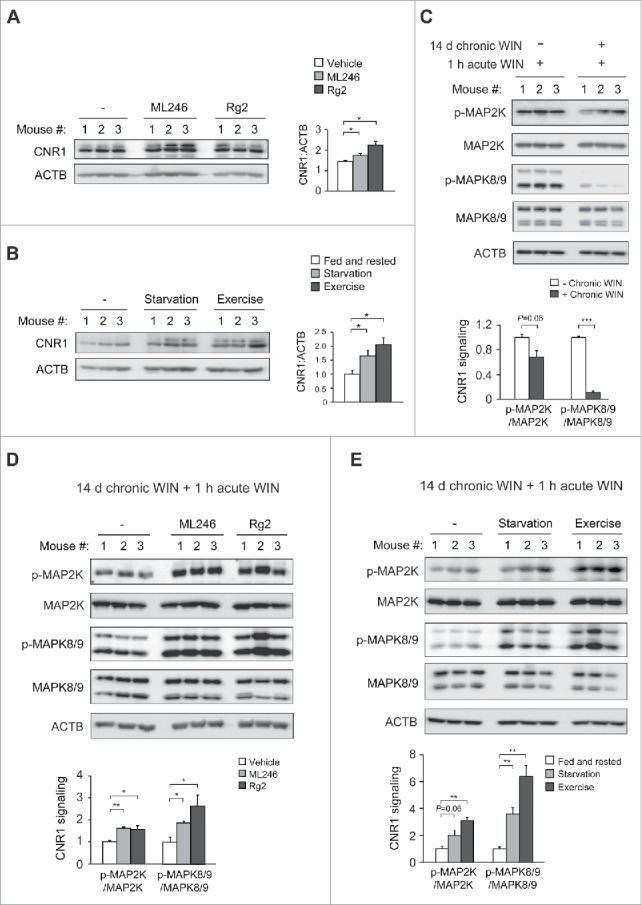
Autophagy activation prevents chronic cannabinoid-induced CNR1 reduction and dysfunction in the brain. (A) Brain CNR1 levels of WT mice simultaneously treated with daily WIN and vehicle, ML246 or Rg2 for 14 d. (B) Brain CNR1 levels of WT mice housed under normal conditions or subjected to either “2 days on–1 day off” periodic starvation or running-wheel exercise, with daily WIN treatment for 14 d. Western blot images (left) and quantification (right) of 3 mice in each group are shown. (C) Decreased CNR1 signaling after chronic WIN treatment. WT mice were treated with either daily WIN or vehicle for 14 d, and then subjected to an acute dosage of WIN 1 h prior to collection of brain samples. Western blot images (upper) and quantification (lower) of 3 mice in each group are shown. (D and E) Pharmacological (ML246 or Rg2) and physiological (starvation or exercise) inducers of autophagy maintain CNR1 signal transduction in mouse brain after chronic agonist exposure. (D) WT mice were treated as in (A), and then received an acute dosage of WIN 1 h prior to western blot analyses on brain lysates. (E) WT mice were treated as in (B), and injected with acute WIN 1 h prior to Western blot analyses on brain lysates. Western blot images (upper) and quantification (lower) of 3 mice in each group are shown. Results represent mean ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t-test).
Discussion
Our findings characterized an autophagy gene as a novel regulator of drug-responsive behaviors, and linked autophagy for the first time to CNR1 sensitization and drug tolerance to cannabinoids, a substance that has emerged as a major medical and social challenge in recent decades. We demonstrated that BECN2 is a new target in the prevention of tolerance to repeated cannabinoid dosage, and developed the concept of activating autophagy as an anti-tolerance therapeutic method. Furthermore, we identified novel autophagy-inducing compounds that achieve this goal by biochemically manipulating the BECN2-GPRASP1 protein complex, which may serve as candidate drug compounds to strengthen the pain-relieving effects of cannabinoids for chronic usage. Notably, we also found that periodic starvation and daily exercise are effective to disrupt the BECN2-GPRASP1 interaction and to prevent mice from cannabinoid tolerance after repeated usage, although we cannot rule out that in the cases of daily exercise and alternating fasting/feeding, BECN2- or autophagy-independent mechanisms may also contribute to the anti-toleranceanti-tolerance effects or changes on CNR1 levels and signaling. To test this hypothesis, further studies using mutant mice deficient in early initiation of autophagy by starvation or exercise are required. Furthermore, to conclusively establish a causal relationship between BECN2-GPRASP1 binding and cannabinoid tolerance, it is necessary to directly disrupt the BECN2-GPRASP1 interaction or specifically perturb the BECN2 function in autophagy in vivo, by generation of knock-in mice containing loss-of-interaction mutations in BECN2 with either GPRASP1 or the autophagy machinery.
Cannabinoids can serve as promising therapeutics in many clinical occasions, such as alternative pain-relieving drugs for patients who develop tolerance to opioid medication. Yet cannabinoids' own potential of drug tolerance and dependence limits the medical use, which is sometimes a neglected issue due to the notion that cannabis may not cause as strong tolerance and dependence as some other drugs of abuse, such as opioids. Here our findings opened up many unanswered questions on analgesic tolerance to cannabinoids for further investigation. For example, drug tolerance is likely to develop at different kinetics to various CNR1 agonists of different efficacy;28 thus, in addition to WIN55,212–2, which is a highly potent CNR1 agonist, it will be essential to study whether the pharmacological or physiological autophagy inducers mediate the same effects on high doses of the natural cannabinoid THC (tetrahydrocannabinol) and other synthetic CNR1 agonists developed for therapeutic use. It is also important to analyze the role of BECN2 and autophagy in the regulation of analgesic tolerance to cannabinoids in disease models of inflammatory or neuropathic pain,29-31 as thermal pain generated in the analgesic tolerance tests may not exactly mimic these types of clinical pain. Thus, future studies will involve surgical- or chemical-stimulation of inflammatory or neuropathic pain using the Becn2 heterozygous KO mouse model. Finally, given that BECN2 regulates the degradation of several additional drug-responsive GPCRs besides CNR1,8 such as DRD2 (dopamine receptor D2) and OPRD1 (opioid receptor, delta 1), the current knowledge on cannabinoid tolerance may open up a new direction for studying autophagy in tolerance or dependence to drugs of abuse targeting many other BECN2-regulated receptors.
Materials and methods
Mice
All mice were in the C57BL/6J background and housed on a 12-h light/dark cycle. The global Becn2+/− and Becn1+/− heterozygous knockout mice have been described previously.8,32 All animal protocols were approved by the Northwestern University Institutional Animal Care and Use Committee (IACUC).
Cell lines
HEK293 and HeLa cell lines were obtained from ATCC, and stable FLAG-CNR1 HEK293 cells were from J. Whistler (University of California, San Francisco). Cells were cultured in Dulbecco's modified Eagle's medium (ThermoFisher Scientific, 11965) supplemented with 10% fetal bovine serum (ThermoFisher Scientific, 16140071).
Drug treatment
For in vivo use, WIN55,212–2 (Sigma-Aldrich, W102), ML246 (synthesized in-house) and Rg2 (purified in-house) were dissolved in a solvent containing 5% NMP (Sigma-Aldrich, 494496), 20% PEG400 (EMD Millipore, PX1286B) and 75% of a 10% solution of HP-β-CD (Tokyo Chemical Industry, H0979) in water, and injected intraperitoneally at the dosage of 2 mg/kg, 10 mg/kg, and 20 mg/kg body weight, respectively. For cell culture use, WIN55,212–2 and ML246 were dissolved in 100% DMSO (Fisher Scientific, BP231) and Rg2 was dissolved in 10% DMSO and used at the concentration of 1 µM, 0.5 μM and 100 μM, respectively. Bafilomycin A1 (Sigma-Aldrich, B1793) was dissolved in 100% DMSO and used at 100 nM in cells.
Chronic WIN55,212–2 treatment and analgesic tolerance test
The analgesic effect of the synthetic cannabinoid drug WIN55,212–2 was measured by the latency time taken by mice to feel the pain generated by an infrared beam and withdraw their paw using the Hargreaves apparatus. Paw withdrawal was detected and timed automatically by a fiber optic sensor. According to the paradigm shown in Figure 1B, 8-wk-old male mice were treated with WIN55,212–2 once per day at the dosage of 2 mg/kg body weight by intraperitoneal injection for 14 d continuously. On day 0, baseline pain sensitivity of the mice was measured without injection of WIN55,212–2. At the beginning (d 1) and the end (d 14) of chronic WIN55,212–2 treatment, an acute bout of 2 mg/kg WIN55,212–2 was injected into mice intraperitoneally 1 h prior to the analgesic tolerance test. Data are shown as the percentage of time taken by each mouse on day 0.
Cyclic AMP measurement
Intracellular cAMP levels were analyzed in HEK293 cells stably expressing FLAG-CNR1 by the cAMP-Glo assay kit (Promega, V1681). Briefly, 40,000 cells/well were cultured in a 96-well plate in triplicate, and treated with either cAMP stabilizers 0.5 mM IBMX (Sigma-Aldrich, I5879) and 1 µM forskolin (Sigma-Aldrich, F6886) alone for 120 min, or with 1 µM WIN55,212–2 plus IBMX and forskolin for 120 min, followed by a phosphate-buffered saline (PBS; Sigma-Aldrich, D8537) wash and 200 nM rimonabant (Cayman Chemical, 9000484) treatment for 30 min, and then another 20 min incubation with 1 µM WIN55,212–2 in the presence of IBMX and forskolin. The cells were subsequently lysed and cAMP production was measured following the manufacturer's instruction.
CNR1 trafficking and recycling by antibody pulse-chase assay
The antibody-feeding immunofluorescence internalization and recycling assays were performed essentially as previously described.33,34 Briefly, HEK293 cells stably expressing FLAG-CNR1 were grown on chamber slides, and incubated with 3.5 µg/ml M1 anti-FLAG antibody (Sigma-Aldrich, F3040) for 30 min. The cells were either directly fixed and immunostained as below as “0 min” control, or treated with the CNR1 agonist WIN55,212–2 (1 µM) for 30 min or 60 min and then washed with calcium-free PBS (Sigma-Aldrich, D8537) to strip off the residual noninternalized M1 antibody (M1-FLAG binding is calcium-dependent). The cells were subsequently fixed in 2% paraformaldehyde (PFA; Sigma-Aldrich, P6148), permeabilized with 0.5% Triton X-100 (Bio-Rad, 161–0407), and then immunostained with anti-EEA1 (ThermoFisher Scientific, MA5–14794) or anti-LAMP1 (Cell Signaling Technology, 9091) antibody for imaging. An Alexa Fluor 488-conjugated secondary antibody (ThermoFisher Scientific, A-21202) was used to detect FLAG and an Alexa Fluor 594-conjugated secondary antibody (ThermoFisher Scientific, A-11037) was used to detect EEA1 and LAMP1. For recycling experiments, after incubation with M1 anti-FLAG antibody, cells were either fixed directly as “0 min” control, or treated with WIN55,212–2 (1 µM) for 30 min, 140 min or 180 min, and then washed with calcium-free PBS to strip off the non-internalized M1 antibody. To detect recycled CNR1 (“140 min+30 min washout”), cells were treated with the antagonist rimonabant (200 nM) for 30 min following the 140 min-agonist treatment and M1 antibody strip-off. The cells were fixed and permeablized as described above.
Co-immunoprecipitation (co-IP)
For co-IP in cell culture, cells were lysed in lysis buffer containing 50 mM Tris-HCl, pH 7.4 (Sigma-Aldrich, 252859), 150 mM NaCl (Fisher Scientific, S271), 1 mM EDTA (Fisher Scientific, BP2482), 0.9% Triton X-100, Halt protease inhibitor cocktail (ThermoFisher Scientific, 1861279) and Halt phosphatase inhibitor cocktail (ThermoFisher Scientific, 78427), and subjected to immunoprecipitation with anti-FLAG antibody (Sigma-Aldrich, F3165). Eluates were subjected to Western blot analyses with anti-FLAG-HRP (Sigma-Aldrich, A8592) and anti-GPRASP1 (Prestige Antibodies, HPA000161) antibodies. The Atg13, Rb1cc1 and Ulk1 siRNAs used for co-IP experiments were purchased from Cell Signaling Technology (M-020765-01-0005, M-021117-01-0005, and M-005049-00-0005, respectively). For co-IP in mouse brain, whole brain was dissected and homogenized in the above lysis buffer and subjected to immunoprecipitation with a polyclonal anti-mouse BECN2 antibody (custom made by GenScript Corporation). Eluates were separated and detected by anti-mouse BECN2 antibody and anti-GPRASP1 antibody using the ONE-HOUR Western Detection Kit (GenScript Corporation, L00204C) following the manufacturer's instruction.
Autophagy analyses
HeLa cells stably expressing GFP-LC3 were cultured in normal medium, starvation (Earle's balanced salt solution; Sigma-Aldrich, E7510) medium, or normal medium with the indicated drugs for 3 h or 16 h. The cells were fixed with 2% PFA for 20 min, and GFP-LC3 puncta were quantified by fluorescence microscopy.35 For assessment of autophagy in vivo, 8-wk-old male GFP-LC3 mice were subjected to 48-h fasting or 4 cycles of “2-day on/1-day off” fasting, or treated with the indicated drugs (5 mg/kg ML246 and 20 mg/kg Rg2) once daily for 3 d by intraperitoneal injection, and then anaesthetized and perfused with 4% PFA. Brain samples were fixed in 4% PFA overnight, 15% sucrose (Sigma-Aldrich, S0389) for 4 h and 30% sucrose overnight before frozen sections were prepared. The number of GFP-LC3 puncta per unit area of tissue was quantified by fluorescence microscopy.36 Autophagy in vivo was also analyzed by Western blot analysis of brain tissue extracts with antibodies against LC3 and SQSTM1 (see below for details).
Western blot analyses
Cell or mouse brain extracts were prepared in RIPA buffer (Sigma-Aldrich, R0278) containing Halt protease inhibitor cocktail and Halt phosphatase inhibitor cocktail, and subjected to western blot analysis with anti-human BECN2 (ProSci Inc., 7989), anti-LC3 (Novus Biologicals, NB100-2220), anti-SQSTM1 (Abnova, H00008878-M01), anti-CNR1 (Cayman Chemical, 101500), anti-MAP2K1/2 (Cell Signaling Technology, 8727), anti-phosphorylated-MAP2K1/2 (Cell Signaling Technology, 9154), anti-MAPK8/9 (Cell Signaling Technology, 9258), anti-phosphorylated-MAPK8/9 (Cell Signaling Technology, 4668), anti-ATG13 (Cell Signaling Technology, 13273), anti-RB1CC1 (Cell Signaling Technology, 12436), anti-ULK1 (Cell Signaling Technology, 8054) and anti-ACTB/β-actin-HRP (Santa Cruz Biotechnology, sc-47778 HRP) antibodies. Band intensity of Western blot gel lanes was measured by ImageJ.
Statistical analyses
P ≤ 0.05 was considered statistically significant in two-tailed, unpaired Student's t -tests. The investigator was blinded to the group allocation during experiments, except for the Becn1+/− heterozygous knockout mice, as they are distinguishable due to a brown coat color rather than black. No animals were excluded from the analyses.
Supplementary Material
Abbreviations
- ATG13
autophagy- related 13
- BECN1
beclin 1, autophagy related
- cAMP
cyclic adenosine monophosphate
- CNR1
cannabinoid receptor 1 (brain)
- CRISPR
clustered regularly-interspaced short palindromic repeats
- GPCR
G protein-coupled receptor
- GPRASP1
G protein-coupled receptor associated sorting protein 1
- KO
knockout
- MAP1LC3/LC3
microtubule-associated protein light chain 3
- MAPK
mitogen-activated protein kinase
- MAP2K1/2
mitogen-activated protein kinase kinase 1/2
- MEFs
mouse embryonic fibroblasts
- RB1CC1
RB1-inducible coiled-coil 1
- SQSTM1
sequestosome 1
- ULK1
unc-51 like kinase 1
- WIN
WIN55,212–2
- WT
wild-type
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Behavioral Phenotyping Core and the Mouse Histology and Phenotyping Laboratory at Northwestern University Feinberg School of Medicine for technical support and assistance, B. Levine (University of Texas Southwestern Medical Center) for providing Becn1+/− and Becn2+/− mice, N. Mizushima (University of Tokyo) for providing GFP-LC3 transgenic mice, and J. Whistler (University of California San Francisco) for providing HEK293/FLAG-CNR1 cells.
Funding
Synthesis of ML246 was supported by the University of Kansas Specialized Chemistry Center (Molecular Libraries Initiative grant U54HG005031 from National Institutes of Health, Jeffrey Aubé PI). Y. Z. was supported by the grant from National Natural Science Foundation of China (No. 31470798) and the Doctoral Fund of Ministry of Education of China (No. 20120043130001). S. H. was supported by the grant from National Institutes of Health (GM078555). K. K., N. W., Y. F., W. Z. and C. H. were supported by the startup funds from Northwestern University and the grant from National Institutes of Health (DK094980). Y. F. was also supported by the National Natural Science Foundation of China (Grant No. 31300287). N. W. was also supported by the National Natural Science Foundation of China (Grant No. 31171303).
References
- [1].Hill KP. Medical Marijuana for Treatment of Chronic Pain and Other Medical and Psychiatric Problems: A Clinical Review. Jama 2015; 313:2474-83; PMID:26103031; http://dx.doi.org/ 10.1001/jama.2015.6199 [DOI] [PubMed] [Google Scholar]
- [2].Gerich ME, Isfort RW, Brimhall B, Siegel CA. Medical marijuana for digestive disorders: high time to prescribe? Am J Gastroenterol 2015; 110:208-14; PMID:25199471; http://dx.doi.org/ 10.1038/ajg.2014.245 [DOI] [PubMed] [Google Scholar]
- [3].England TJ, Hind WH, Rasid NA, O'Sullivan SE. Cannabinoids in experimental stroke: a systematic review and meta-analysis. J Cerebral Blood Flow Metab 2015; 35:348-58; PMID:25492113; http://dx.doi.org/ 10.1038/jcbfm.2014.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Whiting PF, Wolff RF, Deshpande S, Di Nisio M, Duffy S, Hernandez AV, Keurentjes JC, Lang S, Misso K, Ryder S, et al.. Cannabinoids for Medical Use: A Systematic Review and Meta-analysis. Jama 2015; 313:2456-73; PMID:26103030; http://dx.doi.org/ 10.1001/jama.2015.6358 [DOI] [PubMed] [Google Scholar]
- [5].Panlilio LV, Goldberg SR, Justinova Z. Cannabinoid abuse and addiction: Clinical and preclinical findings. Clin Pharmacol Therapeutics 2015; 97:616-27; PMID:25788435; http://dx.doi.org/ 10.1002/cpt.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hall W, Degenhardt L. Adverse health effects of non-medical cannabis use. Lancet 2009; 374:1383-91; PMID:19837255; http://dx.doi.org/ 10.1016/S0140-6736(09)61037-0 [DOI] [PubMed] [Google Scholar]
- [7].Fratta W, Fattore L. Molecular mechanisms of cannabinoid addiction. Curr Opin Neurobiol 2013; 23:487-92; PMID:23490548; http://dx.doi.org/ 10.1016/j.conb.2013.02.002 [DOI] [PubMed] [Google Scholar]
- [8].He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, et al.. Beclin 2 functions in autophagy, degradation of g protein-coupled receptors, and metabolism. Cell 2013; 154:1085-99; PMID:23954414; http://dx.doi.org/ 10.1016/j.cell.2013.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15:1101-11; PMID:14699058; http://dx.doi.org/ 10.1091/mbc.E03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, et al.. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012; 481:511-5; PMID:22258505; http://dx.doi.org/ 10.1038/nature10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67-93; PMID:19653858; http://dx.doi.org/ 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; http://dx.doi.org/ 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- [13].Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823-30; PMID:20811354; http://dx.doi.org/ 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Whistler JL, Enquist J, Marley A, Fong J, Gladher F, Tsuruda P, Murray SR, Von Zastrow M. Modulation of postendocytic sorting of G protein-coupled receptors. Science 2002; 297:615-20; PMID:12142540; http://dx.doi.org/ 10.1126/science.1073308 [DOI] [PubMed] [Google Scholar]
- [15].Martini L, Waldhoer M, Pusch M, Kharazia V, Fong J, Lee JH, Freissmuth C, Whistler JL. Ligand-induced down-regulation of the cannabinoid 1 receptor is mediated by the G-protein-coupled receptor-associated sorting protein GASP1. FASEB J 2007; 21:802-11; PMID:17197383; http://dx.doi.org/ 10.1096/fj.06-7132com [DOI] [PubMed] [Google Scholar]
- [16].Tappe-Theodor A, Agarwal N, Katona I, Rubino T, Martini L, Swiercz J, Mackie K, Monyer H, Parolaro D, Whistler J, et al.. A molecular basis of analgesic tolerance to cannabinoids. J Neurosci 2007; 27:4165-77; PMID:17428994; http://dx.doi.org/ 10.1523/JNEUROSCI.5648-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol 2008; 48:537-68; PMID:18184106; http://dx.doi.org/ 10.1146/annurev.pharmtox.48.113006.094830 [DOI] [PubMed] [Google Scholar]
- [18].Marchese A, Paing MM, Temple BR, Trejo J. G protein-coupled receptor sorting to endosomes and lysosomes. Annu Rev Pharmacol Toxicol 2008; 48:601-29; PMID:17995450; http://dx.doi.org/ 10.1146/annurev.pharmtox.48.113006.094646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Howlett AC, Blume LC, Dalton GD. CB(1) cannabinoid receptors and their associated proteins. Curr Medicinal Chem 2010; 17:1382-93; PMID:20166926; http://dx.doi.org/ 10.2174/092986710790980023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Frankowski K, Patnaik S, Schoenen F, Huang S, Norton J, Wang C, Titus S, Ferrer M, Zheng W, Southall N, et al.. Discovery and Development of Small Molecules That Reduce PNC Prevalence. National Center for Biotechnology Information (US), 2010. http://www.ncbi.nlm.nih.gov/books/NBK143533/ [PubMed] [Google Scholar]
- [21].Gui FJ, Yang XW, Li LY, Tian JM. Simultaneous enantiomer determination of 20 (R)- and 20 (S)-ginsenoside-Rg2 in rat plasma after intravenous administration using HPLC method. J Chromatography B, Analytical Technologies Biomedical Life Sci 2007; 850:1-6; PMID:17113840; http://dx.doi.org/ 10.1016/j.jchromb.2006.11.008 [DOI] [PubMed] [Google Scholar]
- [22].He C, Sumpter R Jr, Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 2012; 8:1548-51; PMID:22892563; http://dx.doi.org/ 10.4161/auto.21327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181:497-510; PMID:18443221; http://dx.doi.org/ 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura SI, Natsume T, Takehana K, Yamada N, et al.. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20(7):1981-91; PMID:19211835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20(7):1992-2003; PMID:19225151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and essential autophagy. J Biol Chem 2009; 284:12297-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Alirezaei M, Kemball CC, Flynn CT, Wood MR, Whitton JL, Kiosses WB. Short-term fasting induces profound neuronal autophagy. Autophagy 2010; 6:702-10; PMID:20534972; http://dx.doi.org/ 10.4161/auto.6.6.12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Flores-Otero J, Ahn KH, Delgado-Peraza F, Mackie K, Kendall DA, Yudowski GA. Ligand-specific endocytic dwell times control functional selectivity of the cannabinoid receptor 1. Nat Commun 2014; 5:4589; PMID:25081814; http://dx.doi.org/ 10.1038/ncomms5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang LX, Wang ZJ. Animal and cellular models of chronic pain. Adv Drug Deliv Rev 2003; 55:949-65; PMID:12935939; http://dx.doi.org/ 10.1016/S0169-409X(03)00098-X [DOI] [PubMed] [Google Scholar]
- [30].Boyce-Rustay JM, Honore P, Jarvis MF. Animal models of acute and chronic inflammatory and nociceptive pain. Methods Mol Biol 2010; 617:41-55; PMID:20336412; http://dx.doi.org/ 10.1007/978-1-60327-323-7_4 [DOI] [PubMed] [Google Scholar]
- [31].Le Bars D, Gozariu M, Cadden SW. Animal models of nociception. Pharmacol Rev 2001; 53:597-652; PMID:11734620 [PubMed] [Google Scholar]
- [32].Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al.. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809-20; PMID:14638851; http://dx.doi.org/ 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bartlett SE, Enquist J, Hopf FW, Lee JH, Gladher F, Kharazia V, Waldhoer M, Mailliard WS, Armstrong R, Bonci A, et al.. Dopamine responsiveness is regulated by targeted sorting of D2 receptors. Proc Natl Acad Sci U S A 2005; 102:11521-6; PMID:16049099; http://dx.doi.org/ 10.1073/pnas.0502418102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Finn AK, Whistler JL. Endocytosis of the mu opioid receptor reduces tolerance and a cellular hallmark of opiate withdrawal. Neuron 2001; 32:829-39; PMID:11738029; http://dx.doi.org/ 10.1016/S0896-6273(01)00517-7 [DOI] [PubMed] [Google Scholar]
- [35].Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005; 1:46-52; PMID:16874027; http://dx.doi.org/ 10.4161/auto.1.1.1542 [DOI] [PubMed] [Google Scholar]
- [36].Qu X, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, et al.. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809-20; PMID:14638851; http://dx.doi.org/ 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


