Abstract
Primary cultures of rodent sensory neurons are widely used to investigate the cellular and molecular mechanisms involved in pain, itch, nerve injury, and regeneration. However, translation of these preclinical findings may be greatly improved by direct validation in human tissues. We have developed an approach to extract and culture human sensory neurons in collaboration with a local organ procurement organization. Here we describe the surgical procedure for extraction of human dorsal root ganglia (hDRG) and the necessary modifications to existing culture techniques to prepare viable adult human sensory neurons for functional studies. Dissociated sensory neurons can be maintained in culture for >10 days, and are amenable to electrophysiological recording, calcium imaging, and viral gene transfer. The entire process of extraction and culturing can be completed in less than 7 hours, and can be performed by trained graduate students. This approach can be applied at any institution with access to organ donors consenting to tissue donation for research and provides an invaluable resource for improving translational research.
Keywords: human sensory physiology, sensory physiology, chronic pain, itch, optogenetics, human neuron culture, viral gene therapy, neuron, calcium imaging, electrophysiological recording, sensory neuron, human dorsal root ganglia, hDRG
Introduction
Primary cultures of dissociated rodent sensory neurons are widely used in preclinical studies of pain, itch, nerve injury, regeneration, and axonal transport. Many candidate molecular targets and genes have been identified using this approach, yet few of these findings have directly translated into effective and safe clinical treatments1–5. Several notable failures in translation have raised questions about possible differences in fundamental biological mechanisms between humans and rodents6–12. For example, NK1 receptor antagonists failed to provide significant analgesia in humans, despite strong preclinical effects in rodents6. More recently, the Mas-related G protein-coupled receptor (Mrgpr) subtypes a3 and c11 were discovered to be key mediators of acute and chronic itch in mice, but are not conserved in humans, which instead have four MRGPRX receptors (MRGPRX1-4). Human MRGPRX1 can be activated by both Mrgpra3 and Mrgprc11 agonists, although key differences in the pharmacological profiles of these receptors demonstrate distinct molecular mechanisms in mice versus humans13–17. In support of these fundamental biological differences, GABAA receptor-mediated currents in cultured human sensory neurons were recently found to exhibit distinct biophysical features from those in rodents, demonstrating the importance of this type of preclinical approach18–21.
Using this protocol we have obtained DRG from multiple spinal levels and have used this opportunity to culture from half of the ganglia, while preserving the remaining tissue for mRNA and immunohistochemical analysis. This permits parallel functional and gene expression studies from the same donor with the ability to correlate observations with patient medical information19,21,22. We have also transduced cultured hDRG with viral vectors for expression of light-sensitive proteins for optogenetic applications, enabling the manipulation of sensory neuron activity with light23–25. We anticipate that by adapting these approaches, preclinical mechanisms of altered sensory processing can be directly verified in human neurons, facilitating the identification of novel therapeutic targets and clinically effective treatments.
Development of the protocol
Several recent studies have identified species differences and validated molecular mechanisms using human sensory neurons, but few have provided a detailed description of extraction or culturing methodologies18–21. Here, we provide a detailed surgical extraction protocol for post-mortem access to human dorsal root ganglia (hDRG) immediately after organ procurement from consented donors. We have used this protocol previously to extract and culture hDRG for studies investigating mGlu2 function in human sensory neurons.22. Our ventral approach through the abdominal cavity eliminates the need to move the donor during the procedure and preserves structural integrity of the body, thus significantly reducing post-mortem interval for DRG retrieval and resulting in minimal interference with other surgical procedures. Maintaining a sterile intraoperative field during the extraction allows subsequent tissue procurement of bone and skin, ensuring maximal utilization of donor tissues by organ procurement organizations (OPOs). Importantly, this approach maintains the overall integrity of the donor body for funeral purposes, allowing access to all donors who consent to tissue donation for research.
The culturing protocol we describe is adapted from our previously published rodent studies with a number of key modifications: a specifically designed preservation solution for transport, new instructions for tissue preparation, changes to enzyme digestion duration, and amount of time in culture prior to functional recordings26–32. Our studies using hDRG show that these adaptations maintain neuronal viability in transport and address several issues that arise from dissociation of substantially larger tissues with more abundant connective tissue than rodent DRG.22 Notable differences between this protocol and those previously described include the specific types of enzymes used (papain versus trypsin and DNase18, or proprietary enzyme mixture19,21) and the duration of enzymatic digestion (up to 3.5 hours in some studies). The type of culture medium and use of neurotrophic factors has also varied between studies, with several studies utilizing DMEM/F12-based media and all studies supplementing with varying concentrations of NGF and/or GDNF. The successful culturing of human cells in a variety of different culture media and following different digestion methodologies suggests that individual labs may be able to adapt this protocol. Producing viable cultures of adult human sensory neurons using conditions similar to those used in previous studies of rodent neurons should reduce sources of variation and allow for direct comparison between rodent and human sensory neurons.
Recent advances in induced pluripotent stem cell (iPSC) methods have produced nociceptor-like cells derived from human fibroblasts that express TRP channels and can be sensitized by inflammatory mediators such as PGE2.33,34 Unlike sensory neurons obtained from organ donors, such methods are not restricted by donor availability because they do not rely on post-mortem tissue collection and can be further strengthened by a living donor’s developing medical history. However, the iPSC approach is still significantly limited by low nociceptor conversion rates (<5%) and the use of varying transcription factor combinations to produce nociceptor-like populations. While both approaches can be informative, direct studies in human sensory neurons may more accurately recapitulate the complex overlapping sensory neuron subpopulations and downstream signaling cascades involved in sensory processing.
Applications of the method
Cultured human sensory neurons can serve as a valuable tool in functional and genetic studies to validate the numerous molecular targets identified in animal models. Using patch-clamp recordings, calcium imaging, immunohistochemistry, gene knock-down, and viral gene transfer, the fundamental physiological properties of human sensory neurons can be more accurately characterized. These approaches could lead to the discovery of novel target receptors or channels and allow for in-depth characterization of pharmacological agents in native human cells. Whole ganglia or cultured neurons can help identify human biomarkers for chronic pain and itch through advanced approaches in RNA and whole genome sequencing, which can be further strengthened by available donor medical history. Furthermore, the potential of novel therapeutic interventions involving gene therapies, and optogenetic and chemogenetic manipulations of cellular activity can be verified directly in human sensory neurons.
Limitations
A significant challenge to obtaining viable hDRG cultures is the identification of a local transplant service or OPO that is able to provide immediate access to their operating room, once vital organs are removed for transplantation. Minimizing the post-mortem interval for DRG procurement is crucial for optimal neuron health, but will be directly dependent on the duration of organ procurement. The frequency of suitable donor availability will vary depending on the institution and inclusion criteria. Factors related to donor medical history, age, sex, ethnic background, and body mass index may introduce variability and should be considered during analysis when averaging data from multiple donors. Primary cultures of human sensory neurons likely present the same caveats inherent to all dissociated primary cultures, the most notable of which is axotomy. During the dissociation process, the peripheral and central processes of the neurons are severed, resulting in the activation of injury- and regeneration-associated cascades. To address this potential confound, the expression pattern of proteins or genes of interest can be directly compared between intact ganglia and cultures of neurons from the same donor and spinal level. The specific culturing protocol we describe can produce live adult sensory neurons that are amenable to physiological and biochemical measures and are similar to those studied in our previous publications.21,22 However, an extensive characterization of specific neuronal subpopulations is not provided. Despite these limitations, dissociated cultures of human sensory neurons are a valuable resource that enables the precise manipulation of external factors to study neuronal physiology specific to humans.
Overview of the Procedure
The key stages of this protocol are extraction of hDRG from an organ donor, cleaning of ganglia, enzymatic and mechanical dissociation, and isolation and plating of dissociated neurons. Dorsal root ganglion extraction takes place in the operating room of the OPO or transplant service, where access to tissue can be obtained once vital organs have been removed from consented organ donors. After procurement, hDRG are stored in N-methyl-D-glucamine (NMDG) artificial cerebrospinal fluid (aCSF) solution for transport, cleaning and enzymatic digestion. NMDG-based aCSF solutions reduce neuronal permeability to sodium and calcium, reduce oxidative stress, and are very beneficial for producing adult rodent brain and spinal cord slices for electrophysiological recordings35–38. Upon return to the laboratory, DRG are cleaned of connective tissue and fat, dura mater is removed, and nerve roots and rami are trimmed. The remaining ganglion is finely cut and enzymatically digested prior to mechanical dissociation. Sensory neurons are isolated from large debris by filtration and centrifugation, and then plated on poly-D-lysine and collagen-coated glass coverslips.
Experimental design
Access to human donor tissue
This protocol provides investigators seeking to establish partnerships with transplant services or OPOs a clear roadmap and the specific details for extraction of human sensory neurons. After establishing contact with a local OPO or transplant service, investigators will need to obtain the appropriate regulatory documentation and agreements, including institutional review board waivers and documentation of donor or family consent to tissue donation for research. The logistics in establishing a network for donor notification, the timing for the research team preparation and access, and the surgical tools to be used must be coordinated with the OPO/transplant service. Since establishing this approach at our institution, we have had access to donor tissue approximately once per month. When a donor becomes available and consent has been obtained for tissue donation to research, we are contacted by our collaborator Mid-America Transplant, generally 3–8 hours prior to organ procurement.
Cell yield
The total number of coverslips obtained for functional studies will depend on the number of ganglia cultured and cell density needed for the particular study. We have found that one DRG can produce approximately 10–12 coverslips plated at a density of 500 cells/coverslip. However, this can vary based on several factors, including DRG size (L1 is smaller than L2–4), degree of dissociation, and general health of the tissue. The density of 500 cells/coverslip provides an optimal number of cells for calcium imaging studies (~20–30 cells per viewing field at a 10× magnification) without excessive debris. Alternatively, plating density can be adjusted to fit investigator needs. For example, plating at a lower density of 200 cells/coverslip was suitable for electrophysiological recordings and immunocytochemical techniques, but did not provide robust numbers for calcium imaging studies.
Growth factors (optional)
Human nerve growth factor (hNGF, 25 ng/mL), and human glial cell line-derived neurotrophic factor (hGDNF, 25 ng/mL), may be included in the medium based on the experimental design of the investigator21. We have found that cells cultured without growth factors survive for more than 10 days and can be used for electrophysiological recordings and calcium imaging, without measurable effects on membrane properties (unpublished observation). The ability to maintain these neurons in culture in the absence of growth factors allows for future studies to determine the acute effects of growth factors on receptor expression or sensitization.
Culturing methods
This culturing protocol follows our prior published studies very closely, utilizing the same coverslip coating solution, digestive enzymes, and culture medium26–32. Following these procedures will produce viable dissociated cultures of hDRG neurons, but recent publications indicate that it may also be possible to culture human sensory neurons in the presence of other types of culture media18,19,21. Investigators may decide to incorporate the same enzymatic digestion, plating protocol, or culture media they have previously used in animal model studies if the goal is to make direct comparisons to those prior studies. In such cases, our protocol can provide a positive control to confirm the viability of neurons as investigators adopt the extraction and dissection procedures in their own laboratories.
Materials
REAGENTS
Donor tissue from human organ donors Tissue should be extracted as soon as possible to minimize the post-mortem interval. We are generally able to extract DRG 1.5–3 hours post-mortem, immediately after the surgical resection of donor organs for transplantation. However, we have obtained viable neuron cultures from tissues extracted up to 4 hours post-mortem. Caution: Any experiments involving human tissue must conform to relevant institutional and national regulations. Extraction procedures were approved by Mid-America Transplant and IRB waiver was obtained from the Human Research Protection Office at Washington University in St. Louis.
70% (vol/vol) ethanol
Collagen from rat tail (10 mg, Sigma, cat. no. C7661-10MG)
Collagenase from Clostridium histolyticum (100 mg, Sigma, cat. no. C6885-100MG)
Poly-D-lysine (Sigma, P7405-5MG)
B27 Supplement (10 ml, Gibco, 17504-044)
Papain (Suspension, 25 mg, Worthington, cat. no. LS003124)
L-cysteine (Sigma, cat. no. C7352-25G)
Hank’s balanced salt solution (HBSS) without calcium and magnesium (Corning, cat. no. 21-021-CM)
Neurobasal A-medium 1× (500 ml, Invitrogen (Gibco), cat. no. 10888-022)
GlutaMAX (2 mM, Life Technologies, cat. no. 35050-61)
Heat-inactivated fetal bovine serum (FBS) (Gibco/Life Technologies, cat. no. 26140-079)
Penicillin/streptomycin (10,000 I.U. Penicillin, 10,000 μg/ml Streptomycin, Corning/CellGro, cat. no. 30-002-CI)
NaOH (0.5 M, Fisher, cat. no. BP359-500)
Sterile water
N-Methyl-D-glucamine (NMDG) (Sigma, cat. no. M2004-500G)
HCl (12 N, Fisher, cat. no. A144-212) Caution Corrosive and can be fatal if swallowed. Proper ventilation, a fume hood and personal protection (gloves, lab coat and safety goggles) must be used when handling concentrated HCl solutions.
KCl (Sigma, cat. no. P5405-250G)
NaH2PO4 (Sigma, cat. no. S5011-100G)
NaHCO3 (Sigma, cat. no. S5761-1KG)
Hepes (Sigma, cat. no. H4034-500G)
D-(+)-Glucose (Sigma, cat. no. G8270-1KG)
L-Ascorbic acid (Sigma, cat. no. A5960-25G)
Thiourea (Sigma, cat. no. T8656-50G) Caution Potential carcinogen and may cause irritation to the eyes, skin and respiratory tract. Wear gloves, lab coat and safety goggles when handling thiourea solution.
Na+ pyruvate (Sigma, cat. no. P2256-25G)
MgSO4 (2 M, Fisher, cat. no. BP213-1)
CaCl2 dihydrate (Sigma, cat. no. C7902-500G)
N-acetylcysteine (Sigma, cat. no. A7250-50G)
Carbogen gas (95% O2, 5% CO2, Airgas)
(optional) Human β-nerve growth factor (hNGF) (Cell Signaling Technology, cat. no. 5221)
(optional) Recombinant human glial cell line-derived neurotrophic factor (hGDNF) (Peprotech, cat. no. 450-10)
EQUIPMENT
Straight osteotome (50 mm tip, Aesculap, cat. no. FL662R)
Curved osteotome (38 mm tip, Aesculap, cat. no. FL790R)
Orthopedic mallet (Aesculap, cat. no. MB630R or similar)
(optional) Stryker Autopsy Saw Model 811 (not autoclavable, cat. no. 0811-000-000) with spinal column arbor blade (cat. no. 1105)
(optional) Stryker Micro Oscillating Saw (autoclavable, cat. no. 5100-31 or similar, requires CORE console) and blade (cat. no. 2296-003-108 or similar)
Scalpel handle with #20 or #10 blades (Aesculap, cat. no. BB073R or similar)
DeBakey pick-ups (8 in, Aesculap, cat. no. FB402R or similar)
Curved dissecting scissors (Aesculap, cat. no. BC575R or similar)
12 mm glass coverslips (Fisher Scientific, cat. no. 12-545-80)
24-well tissue culture plate (BD Falcon, cat. no. 353226)
50 ml conical tubes (Corning, cat. no. 430290)
2 glass Pasteur pipettes (fire polished to a tip diameter of 2 mm and 3 mm, VWR, cat. no. 53283-911)
100 μm Falcon cell strainer (VWR, cat. no. 21008-950)
150 ml sterile filter (0.22 μm, Corning, cat. no. 431153)
60 mm tissue culture dish (BD Falcon, cat. no. 353002)
14 ml snap-cap tube (VWR, cat. no. 60819-499)
Dissection microscope (Olympus, SZ51)
2 pairs of Dumont #5 forceps (WPI, cat. no. 500342)
1 pair of large spring scissors (WPI, cat. no. 15905)
Scalpel handle and #10 blades (WPI, cat. no. 500236, or similar)
Parafilm
BSL II-certified tissue culture hood/biosafety cabinet with UV light sterilization (any standard model)
REAGENT SETUP
Poly-D-lysine (PDK) solution
Dissolve 5 mg PDK in 50 ml sterile water and aliquot 1 ml (0.1 mg/ml) per tube, store at −20°C. Can be used for up to 1 year.
Collagen solution
Dissolve collagen in sterile water for a final concentration of 1 mg/ml, store at 4°C and use within 2–3 months.
PDK/Collagen solution
Combine 1 ml of 0.1 mg/ml PDK, 2 ml of 1 mg/ml collagen, and 7 ml sterile water. Final concentration: 0.01 mg/ml PDK, 0.2 mg/ml collagen. Store at 4°C and use within 2–3 months.
L-cysteine
Dissolve 20 mg L-cysteine into 1 ml sterile HBSS to make 1 mg/50μl aliquots. Store at 4°C. Use within 6 months.
Papain
Calculate the number of active units (U) per ml of enzyme solution and aliquot 45U per tube. Store at 4°C for up to 6 months.
Collagenase
Dissolve 100 mg collagenase in 4.5 mL HBSS. Aliquot 200 μl of enzyme solution per tube (4.5 mg) and store at −20°C. Can be used up to 6 months.
DRG media (100 ml)
Combine 1 ml of penicillin/streptomycin, 2 ml of B27 supplement, 5 ml FBS, 1 ml GlutaMAX, and 91 ml Neurobasal A. Sterile filter the media using a vacuum pump and aliquot into 50ml conical tubes. Store at 4°C protected from light for up to 2 weeks.
NMDG-artificial cerebrospinal fluid (aCSF) solution
Combine the reagents listed in table below in 450 ml Milli-Q H2O in the order listed. Adjust pH to 7.4 with NMDG/HCl and osmolarity to 300–310 mOsm, if necessary, with H2O or sucrose35,36.
| Reagent | Final Concentration (mM) | MW or Concentration of stock | 500 ml |
|
| |||
| NMDG | 93 | 195.2 | 9.08 g |
| HCl | 12.1 N | 3.5 ml | |
| KCl | 2.5 | 2.5 M | 500 μl |
| NaH2PO4 | 1.25 | 1.25 M | 500 μl |
| NaHCO3 | 30 | 84.0 | 1.26 g |
| HEPES | 20 | 2 M | 5 ml |
| Glucose | 25 | 2.5 M | 5 ml |
|
| |||
| Ascorbic acid | 5 | 176.1 | 0.44 g |
| Thiourea | 2 | 76.1 | 0.08 g |
| Na+ pyruvate | 3 | 110.0 | 0.17 g |
|
| |||
| MgSO4 | 10 | 2 M | 2.5 ml |
| CaCl2 | 0.5 | 1 M | 250 μl |
|
| |||
| N-acetylcysteine | 12 | 163.2 | 0.98 g |
|
| |||
CRITICAL NMDG-aCSF solution should always be made fresh and bubbled with carbogen gas (95% O2, 5% CO2) for at least 15 min prior to use to achieve stable pH and saturate with oxygen.
Papain solution
CRITICAL Add 0.5 μl of 0.5 M NaOH to 1mg L-cysteine aliquot. Combine 45U papain with L-cysteine solution in 3 ml of NMDG-aCSF in a 14 ml snap cap tube. Make fresh in sterile culture hood and keep on ice. 15 min prior to use, warm the solution to 37°C in incubator.
CRITICAL 3 ml papain solution is suitable for 1–2 ganglia. Scale up or down as needed.
Coating of coverslips
Place glass coverslips in individual wells of 24-well plate. Wash with 500 μl 70% ethanol, aspirate, and allow to completely dry while uncovered (approx. 15–30 minutes). UV-sterilize in tissue culture hood for 10 min. Tap 24-well plate to ensure all coverslips lay flat at the bottom of each well. Pipet 80μl PDK/collagen solution onto the center of each coverslip, carefully allowing the surface tension to hold the solution on the coverslip without spilling off onto the bottom of the well. Incubate for 1–4 hours at 37°C. Aspirate the PDK/collagen solution off each coverslip and allow them to completely air dry uncovered in the laminar flow tissue culture hood for >30 min. PDK/collagen-coated coverslips can be stored wrapped in aluminum foil at 4°C for up to 4 weeks. Ensure that the 24-well plate remains closed and the aluminum foil is thoroughly cleaned with 70% ethanol to maintain sterility of the coverslips. Ensure that the plates remain upright to prevent flipping of coverslips (PDK-coated surface must remain face up).
CRITICAL This setup must be performed in a sterile tissue culture hood. Do not allow the PDK/collagen solution to flow off the glass coverslip into the rest of the well. Coverslips should be completely dry to prevent this. If at the end of incubation, some coverslips are found with the PDK/collagen solution leaking off of the coverslip, do not use them for cell plating as they will not provide good cell adherence.
See TROUBLESHOOTING Table 1 for further guidance.
TABLE 1.
TROUBLESHOOTING
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| Equipment setup | Few neurons found on coverslip | Coverslips are not properly coated; bad or expired PDK/collagen | Verify that PDK/collagen was made properly and that it did not spill off coverslip for entire duration of coating procedure |
| 15–23 | Over-trituration; too much enzymatic digestion | Triturate with larger tip diameter pipette; triturate for less time; Decrease enzyme incubation times. | |
| 15–23 | Under-trituration; not enough enzymatic digestion | Triturate with a smaller tip diameter pipette and/or for longer; cut tissue into smaller pieces; increase enzyme incubation times. | |
| 15–33 | Unhealthy cells due to prolonged dissociation | Expedite culturing procedure and reduce time outside incubator | |
| 4–6 | Anterior spinal column cannot be removed | Vertebral bodies or pedicles are not fully transected, there are remaining bone or connective tissue attachments | Apply greater force when using osteotomes and mallet to ensure bone connecting to the anterior spinal column is fully detached |
| 6–7 | Cannot localize DRG after spinal column is removed and dura is torn | Tearing due to the pulling of dura and DRG by the posterior longitudinal ligament | Take more time to ensure dura is completely detached from the posterior longitudinal ligament during column removal; check posterior portion of spinal column for DRG that may be attached |
| 24 | Cell strainer clogs, few cells in suspension after filtration | Too much debris has blocked dissociated cells from passing through cell strainer | Cut tissue into smaller pieces, use more than one cell strainer to filter less volume at one time |
| 31–32 | Cell suspension spills off of coverslips into well bottom during plating | Coverslips are not dry prior to plating | Dry coverslips in uncovered dish in tissue culture hood for longer period of time prior to cell plating; vacuum several times to remove any remnants of PDK/collagen solution (see Equipment setup). |
| 34 | Bacterial/fungal contamination | Contamination of cells during dissection, trituration, or incubation | Perform dissection under sterile conditions (blow out hood, BSL2 tissue culture hood) using sterilized tools; ensure penicillin/streptomycin is fresh; clean and sterilize incubator |
PROCEDURE
Removal of hDRG from spinal column TIMING 20–30 min
CRITICAL The following steps can be performed with the internal organs retracted, but the procedure is considerably easier if all viscera are removed. The dissection tools (with the exception of the Stryker autopsy bone saw) used below were made available for this extraction protocol in the operating room. Investigators should discuss with their OPO or transplant service whether they need to provide their own instruments.
! CAUTION Sterile operating room procedures need to be followed for tissue extraction. Exercise proper safety precautions (e.g. gloves, eye protection, scrubs, lab coat) when working with human tissues.
-
1)
Prepare several 40 ml aliquots (6–10) of NMDG-aCSF solution in air-tight 50 ml conical tubes wrapped in parafilm and maintain on ice for hDRG transport after extraction. Continue to bubble the remaining NMDG-aCSF solution on ice for hDRG dissection.
-
2)
Expose the lumbar spinal column using a scalpel to remove abdominal vasculature and connective tissue located anterior to the spine in the retroperitoneal space (This includes the abdominal aorta, inferior vena cava, lymphatics, etc.).
-
3)
Using a scalpel, cut and retract or remove psoas major and psoas minor muscles from their origin and expose the lateral aspects of the spinal column and the roots of the lumbar plexus.
-
4)
Using a mallet and straight osteotome, transect L1 and L5 vertebral bodies, stopping at the spinal canal (Fig. 1a).
-
5)
Transect the pedicles bilaterally between L1 and L5, staying close to the posterior edge of the vertebral bodies, but anterior to the spinal canal (Fig. 1b). If using a sterile mallet and curved osteotome, use a scalpel or dissecting scissors to detach the dura mater from the posterior longitudinal ligament to minimize tearing and pulling of the DRG when the vertebral bodies are removed. If using an autopsy bone saw the blade is wide enough to cut the posterior longitudinal ligament, preventing tearing of the dura mater in step 12 without the need to use a scalpel.
CRITICAL STEP If using a sterile mallet and curved osteotome to perform this step, the sterility of the surgical field is maintained, but they do not offer the more precise cutting of a bone saw and require significantly more manual power. The autopsy saw allows for more rapid and precise identification of spinal levels however it cannot be sterilized, therefore it will compromise the sterile surgical field. Therefore its use is limited by OPO/transplant service procedures for tissue retrieval following hDRG extraction (Alternatively, surgical grade micro-saws which can be sterilized may be used, but are considerably more expensive).
-
6)
Manually remove the entire anterior portion of the vertebral column between L1 and L5, lift carefully to minimize tearing of structures in the intervertebral foramina and simultaneously cut any dural attachments to the posterior longitudinal ligament. Ensure that the transverse processes, laminae, and spinous processes remain in place (Fig. 1c).
CRITICAL STEP It is essential to leave the dorsal portion of the vertebral column intact to maintain the integrity of the body for further tissue procurement and funeral service purposes. This is a major concern for potential partnering transplant services or organ procurement organizations.
? TROUBLESHOOTING
-
7)
Gently tug on the dura mater to visualize DRG and dissect each ganglion away from surrounding connective tissue and bone using DeBakey pick-ups and dissecting scissors. Cut the spinal roots and rami of peripheral nerves to completely free the DRG (Fig. 1d).
? TROUBLESHOOTING
-
8)
Rinse DRG in 40 ml of ice-cold NMDG-aCSF, and place in 40 ml of fresh ice-cold NMDG-aCSF in air-tight 50ml conical tubes. Maintain on ice for transport.
Figure 1. Surgical extraction of hDRG using ventral approach.
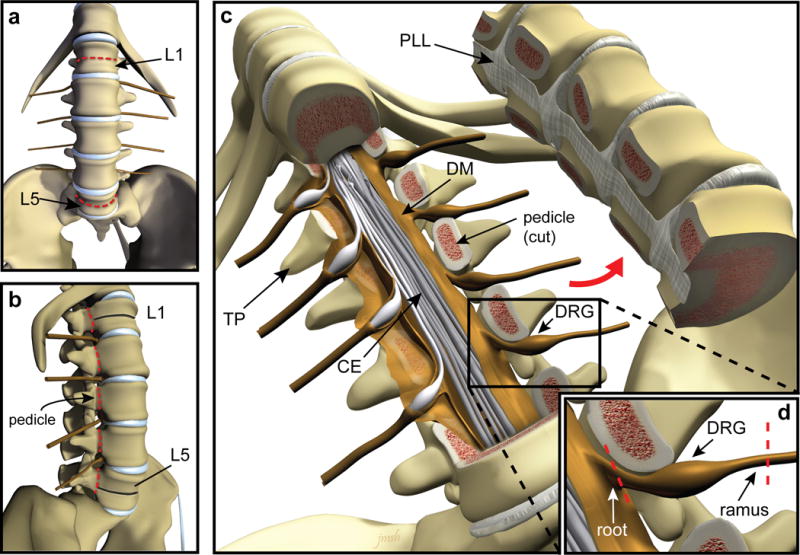
(a–b) Illustration depicting the ventral (a) and lateral (b) views of the spinal column with lumbar vertebral bodies L1 and L5 indicated by arrows. Red dashed lines indicate the location of bone transection. After the spinal column is visualized, lumbar vertebrae can be correctly identified by counting up from L5, which is located directly above the sacrum (a,b). Using a straight osteotome and surgical mallet, the L1 and L5 vertebral bodies are transected, stopping at the spinal canal (red dashed lines in panel a, black lines indicate transected bone in panel b). Using a curved osteotome and mallet or autopsy saw, the pedicles of each vertebrae are transected bilaterally between L1 and L5 (red dashed lines in panel b). (c) Illustration showing the anterior portion of the vertebral column removed to expose the spinal canal where the cauda equina and DRG are located (TP: transverse process, CE: cauda equina, PLL: posterior longitudinal ligament, DM: dura mater, DRG: dorsal root ganglion). (d) Each DRG is dissected away from surrounding bone and connective tissue and the nerve roots and rami are cut to completely free each ganglion (red dashed lines). CAUTION: Donor consent for tissue donation for research purposes must be obtained by OPO or transplant service prior to tissue extraction.
Cleaning of DRG TIMING 30–90 min
! CAUTION Exercise proper safety precautions when working with and disposing of materials containing human tissues. All procedures need to be performed using sterile technique in biosafety cabinets and tools must be properly cleaned and sterilized after every use.
-
9)
Place individual DRG in a 60 mm petri dish filled with 15–20 ml of ice cold NMDG-aCSF to fully submerge the ganglion, keeping the petri dish on ice for the duration of cleaning and dissection.
-
10)
Using large spring scissors and forceps, remove remaining fat and connective tissue surrounding DRG (Fig. 2a).
-
11)
Under a dissection microscope, use Dumont #5 forceps and large spring scissors to identify the edge of the dura along the severed ends of the central and peripheral nerve processes. Cut the dura longitudinally from the peripheral to the central endings.
-
12)
Remove dura by continually pulling away from the ganglion and cutting along the edge where the dura meets the DRG and nerves (Fig. 2b).
-
13)
Trim the remaining central and peripheral nerve processes with a scalpel, so that only the ganglion body remains (Fig. 2c).
-
14)
Optional: If desired, the remaining DRG can be saved for immunohistochemistry or RNA sequencing studies. Once the ganglion is cleaned, cut the DRG with a scalpel into small pieces and freeze tissue in RNAlater for RNA sequencing/RT-PCR or post-fix in 4% paraformaldehyde solution for intact ganglion histology. For direct comparisons from the same spinal level, culture one DRG and use the contralateral DRG for histology/RNA purposes.
Figure 2. Cleaning of hDRG prior to dissociation.
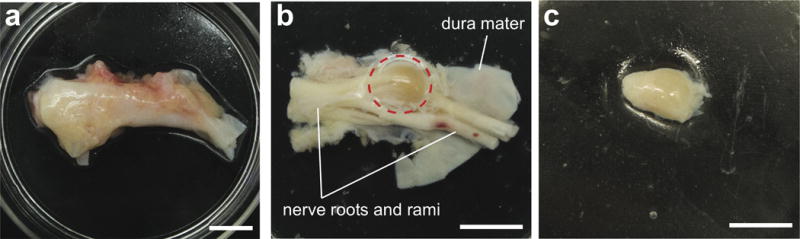
(a) Extracted ganglia are first cleaned of all visible fat and connective tissue. (b) Dura mater is then removed to expose underlying ganglion and nerve roots and rami. (c) Nerve roots are trimmed so that only the ganglion remains for dissociation. Scale bars represent 1 cm. The human tissue extraction procedures were approved by Mid-America Transplant and IRB waiver was obtained from the Human Research Protection Office at Washington University in St. Louis.
Dissociation of DRG for cell culture TIMING: 2.5–3 hours
CRITICAL Prepare Papain Solution as described in the Reagent setup and warm the solution to 37°C in incubator
-
15)
Use a scalpel to mince each DRG as finely as possible (pieces should be no larger than 3 mm in diameter).
-
16)
Place all tissue fragments into a 14 ml snap-cap tube containing 3 ml of pre-warmed (37°C) papain solution and incubate for 1 hour at 37°C and 5% CO2. Gently agitate every 20 min to mix tissue with solution. Warm remaining NMDG-aCSF to 37°C. [AU: Volume of papain solution correct?]
-
17)
15 min prior to the end of incubation, prepare collagenase solution by diluting 4.5 mg aliquot of collagenase in 3 ml warmed (37°C) NMDG-aCSF.
-
18)
Carefully wash the tissue 3 times with 3 ml of fresh NMDG-aCSF, warmed to 37°C.
CRITICAL STEP Avoid aspirating or pipetting tissue during the washing step. Allow the tissue to settle if it has been disturbed and is interfering with washes.
-
19)
Replace entire solution volume with 3 ml of NMDG-aCSF-collagenase solution and incubate tissue for 1 hour at 37°C and 5% CO2. Gently agitate every 20 min to mix the tissue with the solution.
-
20)
Prepare DRG medium as described in the Reagent set up and pre-warm ~30 ml per set of 2 ganglia in tissue culture incubator set at 37°C.
-
21)
Wash tissue 3 times with 3 ml of fresh NMDG-aCSF warmed to 37°C. Take care to avoid disturbing tissue pieces while pipetting.
-
22)
Remove most of the NMDG-aCSF solution, leaving ~100 μl, and add 2 ml of pre-warmed DRG medium.
-
23)
Gently triturate tissue through the fire-polished tip of a sterile glass Pasteur pipette until the solution becomes cloudy and little resistance remains (approximately 10–20 times). Tissue from younger donors will require less trituration than older donors.
CRITICAL STEP Do not over-triturate and take care not to generate bubbles during trituration.
? TROUBLESHOOTING.
Carefully pass the solution of dissociated neurons through a 100 μm sterile filter into a 50 ml conical tube. CRITICAL STEP hDRG neurons are approximately twice the diameter of dissociated sensory neurons from rodents, and filters traditionally used for rodent cultures will not be suitable.
? TROUBLESHOOTING
-
24)
Centrifuge the filtrate at room temperature (20–25°C) for 4 min at 180g.
-
25)
Remove supernatant containing debris and resuspend pellet (in same tube) in 1ml warmed DRG medium, mixing the solution by pipetting up and down several times.
-
26)
Centrifuge solution for 3 min at 180g.
-
27)
Remove supernatant and resuspend pellet in 1ml warmed DRG medium, mixing gently.
Plating on glass coverslips
TIMING: 1.5–2 h
-
28)
Calculate the density of the cell suspension using a standard hemocytometer.
-
29)
Dilute the cell suspension to 5,000 cells/ml with DRG medium (or adjust to other desired density).
-
30)
Carefully plate 100 μl of cell suspension onto the center of each PDK/collagen-coated glass coverslip for a final plating density of approximately 500 cells/coverslip.
CRITICAL STEP Take care to allow the surface tension to hold the solution onto the coverslip without spilling into the bottom of the well.
-
31)
Incubate at 37°C, 5% CO2 for 1–1.5 hours to allow neurons to adhere to the glass coverslips.
-
32)
Carefully fill each well with 900 μl warm DRG medium and maintain neurons in culture at 37°C, 5% CO2 in a humidified chamber until use. ? TROUBLESHOOTING
-
33)
Replace half of the culture medium with fresh warmed medium every 3 days.
-
34)
Culture neurons for 3–4 days to ensure that satellite glial cells peel off the neurons allowing membrane access to characterize the cells by further downstream analyses, as described in the anticipated results. CRITICAL STEP Cells can be cultured for over 10 days.
TIMING
REAGENT SETUP, preparation of NMDG-aCSF solution: 30 min
REAGENT SETUP, preparation of papain solution: 5 min
REAGENT SETUP, coating of coverslips: 1–4 h
Steps 1–8, hDRG surgical extraction: 20–30 min
Steps 9–14, cleaning of DRG: 30–90 min
Steps 15–28, dissociation of DRG for cell culture: 2.5–3 h
Steps 29–35, plating on glass coverslips: 1.5–2 h
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
ANTICIPATED RESULTS
Appearance of neurons in culture
While most rodent studies can be completed within 24 hours of cell plating, human sensory neurons are often tightly encased by satellite glia until the third or fourth day in vitro (DIV). During these first days in culture, glial cells peel off allowing membrane access for patch pipettes or direct fluorescence visualization (Fig. 3). More vigorous dissociation protocols can yield neurons free of supporting glia, but the health and survival of these neurons may be reduced21. The neurons produced with this extraction and dissociation protocol can be maintained in culture for more than 10 days22.
Figure 3. Dissociated hDRG neurons over time in vitro.
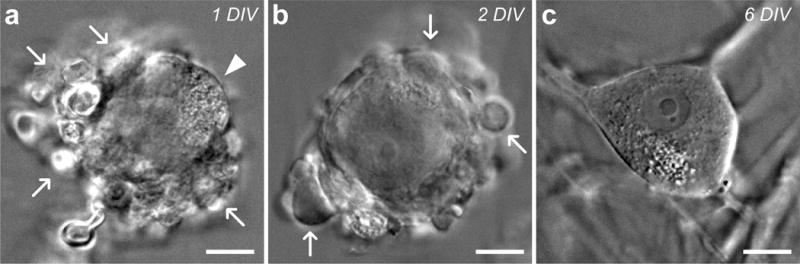
Infrared differential interference contrast microscopy (IR-DIC) images of cultured human sensory neurons. (a) Initially, most dissociated neurons are encased in glial cells (white arrows) after 1 day in vitro (DIV), but can be identified by partially visible plasma membrane (white arrowhead). (b) As time in culture progresses, glia (white arrows) continue to peel off and adhere to the coverslip, exposing more of the plasma membrane. (c) After 6 DIV, the plasma membrane of most neurons is completely exposed, leaving them amenable to patch-clamp recordings and calcium imaging studies. Scale bars represent 20 μm.
Immunocytochemical studies of human sensory neurons
Whole ganglia and dissociated neurons can be studied using validated antibodies and standard immunocytochemical techniques to characterize cell morphology and the localization and expression of ion channels, receptors, and intracellular proteins (Fig. 4)30,38. Importantly, the coverslips used for functional studies by patch-clamp21,24,31,32 and calcium imaging26,31 can be fixed immediately after the experiment to allow for immunocytochemical studies in the same functionally-characterized neurons.
Figure 4. Immunocytochemical analysis of cultured hDRG neurons.
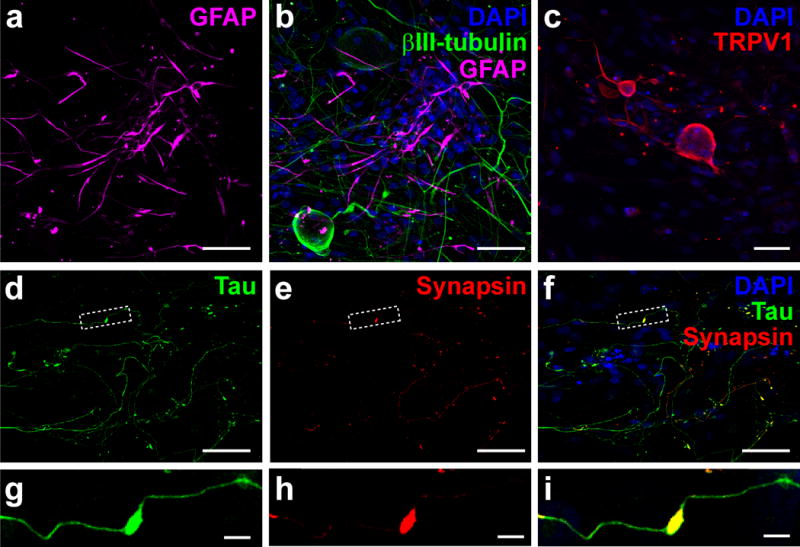
Collapsed confocal micrographs of immunolabeled neurons using standard techniques38. (a) GFAP+ cells (purple) – presumably satellite glia that initially encase dissociated hDRG neurons – that have migrated onto the coverslip after 8 DIV. (b) Merged image with neurons immunolabeled for the cytoskeletal protein βIII-tubulin (green) and nuclei labeled with DAPI (blue). (c) TRPV1 immunofluorescence (red), a common nociceptive marker in rodent DRG, was observed in subsets of cultured human sensory neurons. Nuclei are labeled with DAPI (blue). (d–f) Fluorescence images of cultured hDRG demonstrating extensive axonal process growth and branching marked by tau immunoreactivity (green) and a component of the synaptic vesicle release machinery, synapsin (red). Merged images are shown in (f) with DAPI-labeled nuclei (blue). (g–i) Cropped sections from the boxed areas in (d–f), showing an en passant-type presynaptic enlargement (synapsin, red) that formed along a sensory axon (tau, green). Scale bars represent 50 μm for panels (a–f), and 5 μm for the panels shown in (g–i). Antibodies used in immunocytochemistry experiments include: Rabbit anti-TRPV1 (1:800), custom-made serum directed against the TRPV1 C-terminus peptide CLKPEDAEVFKDSMVPGEK, specificity confirmed in TRPV1−/− mice30,43; Mouse anti-βIII-tubulin (1:2000) – EMD Millipore (cat. no. 05-559), species reactivity (hm/bv/rat/ms), Antibody Registry ID: AB_309804; Guinea pig anti-GFAP (1:1000) – Synaptic Systems (cat. no. 173 004), species reactivity (hm/rat/ms/ck), Antibody Registry ID: AB_1064116;. Mouse anti-synapsin 1 (1:2000) – Synaptic Systems (cat. no. 106 001), species reactivity (hm/ms/rat/avian/zebrafish), Antibody Registry ID: AB_887805; Guinea pig anti-tau (1:2000) – Synaptic Systems (cat. no. 314 004), species reactivity (hm/ms/rat), Antibody Registry ID: AB_1547385.
Calcium imaging to study functional subpopulations of hDRG neurons
Population-wide responses to chemical or thermal stimuli can be characterized using calcium imaging procedures we have previously published26,30,31. Primary cultures of dissociated human sensory neurons were loaded with the calcium indicator Fura2-AM (Life Technologies), and individual neuronal responses to several bath-applied stimuli were measured (Fig. 5). Waiting until the satellite glia have moved off the neurons to expose the soma (Fig. 3c) is important for obtaining neuron-specific calcium signals, as glia are also labeled by Fura2 dyes. Calcium imaging recordings on DIV4-7 demonstrate labeled neurons that are much larger than surrounding glial cells (Fig. 5a). An intracellular increase in calcium was visualized in subsets of neurons during application of the pruritogens histamine and chloroquine (Fig. 5a), and the algogens mustard oil, menthol, and capsaicin (Fig. 5b).
Figure 5. Calcium imaging of human sensory neurons for characterizing population responses to algogens and pruritogens.
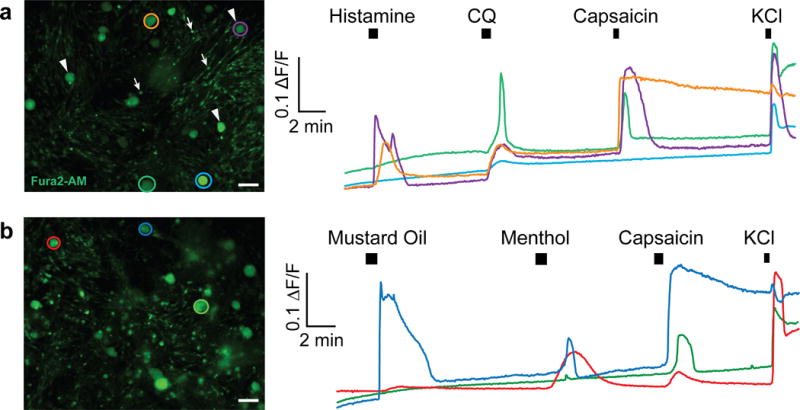
Cultured sensory neurons (DIV 4–7) were labeled with the fluorescent calcium indicator Fura2-AM (3μg/ml, Life Technologies) at 37°C for 45 min and then allowed to de-esterify for 15 min at 37°C in warmed external solution. (a) Left panel shows fluorescent Fura2-AM-labeled dissociated neurons (arrowheads), surrounded by non-neuronal cells (arrows) in culture. (a, right) Response traces corresponding to colored circles in (a, left panel) demonstrate different populations of neurons responding to the pruritogens histamine (100 μM) and chloroquine (1 mM) and the algogen capsaicin (200 nM). KCl (50 mM) was added at the end to confirm viability. (b) Example traces from a separate experiment demonstrates that dissociated neurons (b, left) also responded to the algogens mustard oil (100 μM), menthol (100 μM), and capsaicin (200 nM). Scale bars represent 100 μm.
Electrophysiological studies of hDRG neurons
Human sensory neurons obtained using this protocol can also be studied using patch-clamp electrophysiology techniques. Gigaohm seals were obtained from >95% of selected neurons. We were careful to only patch neurons with a clearly visible soma devoid of satellite glial cells and exhibited a “smooth” membrane (Fig. 3c). Whole-cell recordings were successful from >95% of these neurons, which included stable access resistance and a resting membrane potential hyperpolarized over −40 mV. We found that neurons could be cultured in the absence or presence of the growth factors NGF and GDNF (25ng/ml each), and that chronic treatment did not affect cellular excitability, resting membrane potential, or soma size (Fig. 6).
Figure 6. Long-term culturing with neurotrophic factors does not alter hDRG excitability.
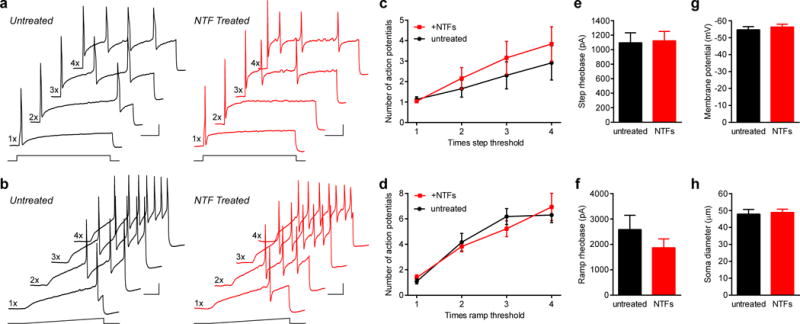
Human sensory neurons were grown in the absence (black traces) or presence (red traces) of the neurotrophic factors NGF and GDNF (NTFs, 25ng/ml each, added to wells chosen randomly from all spinal levels). Recordings were performed between 4–9 DIV and cells were excluded from further analysis if the resting membrane potential was more depolarized than −40 mV. (a,b) Voltage traces illustrating action potential firing to 1 s depolarizing step (a) or ramp (b) current injections of 1–4 times current threshold (rheobase) of neurons grown in the absence (black traces) or presence of NTFs (red traces). Scale bars are 20 mV and 200 ms. (c,d) Summary plots of the number of action potentials elicited in neurons in response to both step (c) and ramp (d) stimuli, which were not significantly different between the two culturing conditions (two-tailed Mann-Whitney test, P values range from 0.143 to 0.964). (e,f) Quantification of step (e) or ramp (f) current threshold to elicit an action potential. No differences were found between the two groups (two-tailed Mann-Whitney test, P=0.738 for panel e, and P=0.361 for panel f). (g) Summary graph of the resting membrane potential for cells cultured in the absence or presence of NTFs, which were not significantly different (two-tailed unpaired t-test; P=0.657). (h) Average soma diameters of neurons in both culturing conditions, which were not significantly different (two-tailed unpaired t-test; P=0.775). This suggests that the preferential survival of sub-populations of sensory neurons with different cell diameters was not influenced by either culturing condition. 19–30 cells were used for quantification per condition from four donors (4–30 cells for each measurement per donor), and all data are represented by the mean ± s.e.m of cells pooled across all donors.
Viral gene transfer into hDRG neurons for functional analysis
We also determined that viral vectors carrying exogenous genes can be used to transduce cultured human sensory neurons. Herpes simplex viral (HSV) vectors have natural tropism for sensory neurons and have been used in rodent models of chronic pain and in human gene therapy trials39,40. We used these vectors to deliver optogenetic tools to bi-directionally manipulate human sensory neuron excitability using LED illumination24,25. (Figure 7, unpublished data). These approaches should also be amenable to gene knockdown strategies using RNAi or more advanced genome editing techniques including the CRISPR-Cas system41,42. In pilot studies, we have also had success using adeno-associated viral (AAV) vectors with attenuated lytic activity, which may significantly expand approaches to understand human sensory neuron physiology using viral transgene delivery38.
Figure 7. Viral transduction of human sensory neurons for expression of optogenetic tools to manipulate neuronal firing.
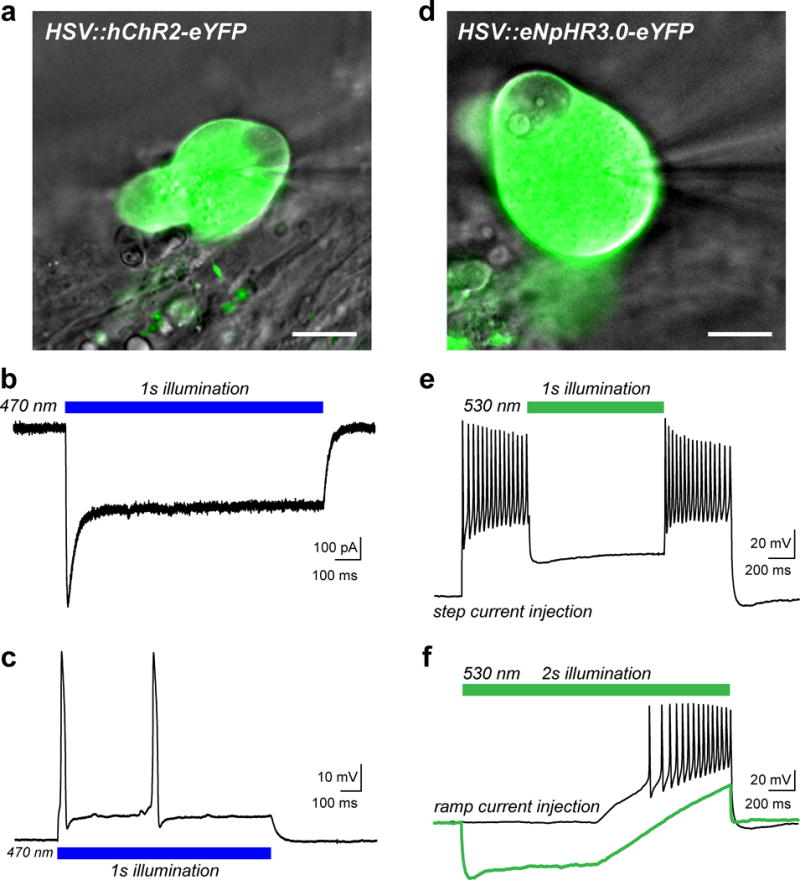
(a) Overlay of a DIC image of a human sensory neuron (gray) transduced with herpes simplex virus (HSV) carrying a transgene for the humanized excitatory opsin, channelrhodopsin-2 (hChR2-eYFP, green). Viruses were added at a concentration of 3.2×108 vector genomes/ml after 8 DIV and cells were examined 1–2 days later. The patch-pipette is visible on the right side of the image. (b) Representative inward photocurrent induced by illumination with blue LED light (blue bar, 10 mW/mm2) as previously described24. (c) Current-clamp recording showing action potential (AP) firing in response to blue light illumination as in (b). (d) DIC image (gray) of a sensory neuron that was transduced with HSV carrying the inhibitory Cl− pump halorhodopsin (eNpHR3.0-eYFP, green). (e) Voltage trace from a current-clamp recording in response to a 2s suprathreshold step current injection to elicit AP firing. Neuronal activity was acutely silenced by green LED illumination (green bar, 30 mW/mm2). (f) Current-clamp trace showing AP firing elicited by a ramp protocol of suprathreshold depolarizing current injection (black trace). Illumination with green LED light (30 mW/mm2) hyperpolarized the neuron and blocked AP discharge to the stimulus (green trace). Scale bars represent 20 μm. HSV vectors used include: CMV:ChR2(H134R)-eYFP and CMV:eNpHR3.0-eYFP, (both at 3.2×108 vector genomes/ml) from Rachael Neve at the MIT McGovern Viral Core Facility.
Acknowledgments
We sincerely thank the donors and their families for their generous donations to science. We thank Mid-America Transplant for providing access to donor tissue and their facilities, and especially Patrick Silva and John Lemen for their tremendous help coordinating and performing hDRG extractions. We thank Dr. Andrea Vannucci and Dr. Yiing Lin for helpful discussions and guidance in establishing the research collaboration with Mid-America Transplant. We thank Orlando Crisp from the Washington University Pathology Department for demonstrating DRG removal using the autopsy saw. We thank Janet Sinn-Hanlon for generating the illustrations in Figure 1. We thank Daniel Brenner for helpful discussions on the development of extraction procedures. This work was supported by National Institute of Neurological Disorders and Stroke grants R01NS042595 (R.W.G.), F31NS089130 (M.V.V.), F32NS076324 (S.D.), and T32GM108539 (B.A.C.) and T32GM007067-41 (M.Y.P.) from the National Institute of General Medical Sciences.
Footnotes
AUTHOR CONTRIBUTIONS
M.V.V., S.D., B.A.C., and K.D. developed the surgical approach. M.V.V., S.D., B.A.C., and T.D.S. optimized existing rodent culturing protocol to establish viable hDRG cultures. M.V.V., B.A.C., T.D.S., M.Y.P., and J.G.M. contributed to applications including immunocytochemistry, calcium imaging, electrophysiology, and optogenetic experiments. All authors contributed to writing and editing the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Contributor Information
Manouela V. Valtcheva, Email: mvaltcheva@wustl.edu.
Bryan A. Copits, Email: copitsb@anest.wustl.edu.
Steve Davidson, Email: davidsst@ucmail.uc.edu.
Tayler D. Sheahan, Email: tayler.sheahan@wustl.edu.
Melanie Y. Pullen, Email: melanie.pullen@wustl.edu.
Jordan G. McCall, Email: jordangmccall@wustl.edu.
Krikor Dikranian, Email: dikranik@wustl.edu.
References
- 1.Lacroix-Fralish ML, Ledoux JB, Mogil JS. The Pain Genes Database: An interactive web browser of pain-related transgenic knockout studies. Pain. 2007;131:3 e1–4. doi: 10.1016/j.pain.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 2.Chizh BA, et al. The effects of the TRPV1 antagonist SB-705498 on TRPV1 receptor-mediated activity and inflammatory hyperalgesia in humans. Pain. 2007;132:132–141. doi: 10.1016/j.pain.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Kivitz AJ, et al. Efficacy and safety of tanezumab versus naproxen in the treatment of chronic low back pain. Pain. 2013;154:1009–1021. doi: 10.1016/j.pain.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Skljarevski V, et al. Efficacy of Duloxetine in Patients with Chronic Pain Conditions. Current drug therapy. 2011;6:296–303. doi: 10.2174/157488511798109592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wernicke JF, et al. A randomized controlled trial of duloxetine in diabetic peripheral neuropathic pain. Neurology. 2006;67:1411–1420. doi: 10.1212/01.wnl.0000240225.04000.1a. [DOI] [PubMed] [Google Scholar]
- 6.Hill R. NK1 (substance P) receptor antagonists–why are they not analgesic in humans? Trends in pharmacological sciences. 2000;21:244–246. doi: 10.1016/s0165-6147(00)01502-9. [DOI] [PubMed] [Google Scholar]
- 7.Taneja A, Di Iorio VL, Danhof M, Della Pasqua O. Translation of drug effects from experimental models of neuropathic pain and analgesia to humans. Drug discovery today. 2012;17:837–849. doi: 10.1016/j.drudis.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Mogil JS. Animal models of pain: progress and challenges. Nature reviews Neuroscience. 2009;10:283–294. doi: 10.1038/nrn2606. [DOI] [PubMed] [Google Scholar]
- 9.Contopoulos-Ioannidis DG, Ntzani E, Ioannidis JP. Translation of highly promising basic science research into clinical applications. The American journal of medicine. 2003;114:477–484. doi: 10.1016/s0002-9343(03)00013-5. [DOI] [PubMed] [Google Scholar]
- 10.Ergorul C, Levin LA. Solving the lost in translation problem: improving the effectiveness of translational research. Current opinion in pharmacology. 2013;13:108–114. doi: 10.1016/j.coph.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hug A, Weidner N. From bench to beside to cure spinal cord injury: lost in translation? Int Rev Neurobiol. 2012;106:173–196. doi: 10.1016/B978-0-12-407178-0.00008-9. [DOI] [PubMed] [Google Scholar]
- 12.Gereau RW, et al. A pain research agenda for the 21st century. The journal of pain: official journal of the American Pain Society. 2014;15:1203–1214. doi: 10.1016/j.jpain.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han L, et al. A subpopulation of nociceptors specifically linked to itch. Nature neuroscience. 2012 doi: 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, et al. The distinct roles of two GPCRs, MrgprC11 and PAR2, in itch and hyperalgesia. Science signaling. 2011;4:ra45. doi: 10.1126/scisignal.2001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–1365. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solinski HJ, Gudermann T, Breit A. Pharmacology and signaling of MAS-related G protein-coupled receptors. Pharmacological reviews. 2014;66:570–597. doi: 10.1124/pr.113.008425. [DOI] [PubMed] [Google Scholar]
- 17.Solinski HJ, Zierler S, Gudermann T, Breit A. Human sensory neuron-specific Mas-related G protein-coupled receptors-X1 sensitize and directly activate transient receptor potential cation channel V1 via distinct signaling pathways. The Journal of biological chemistry. 2012;287:40956–40971. doi: 10.1074/jbc.M112.408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XL, Lee KY, Priest BT, Belfer I, Gold MS. Inflammatory mediator-induced modulation of GABA currents in human sensory neurons. Neuroscience. 2015;310:401–409. doi: 10.1016/j.neuroscience.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. The Cancer Chemotherapeutic Paclitaxel Increases Human and Rodent Sensory Neuron Responses to TRPV1 by Activation of TLR4. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35:13487–13500. doi: 10.1523/JNEUROSCI.1956-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand U, et al. Angiotensin II type 2 receptor (AT2 R) localization and antagonist-mediated inhibition of capsaicin responses and neurite outgrowth in human and rat sensory neurons. European journal of pain. 2013;17:1012–1026. doi: 10.1002/j.1532-2149.2012.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson S, et al. Human sensory neurons: Membrane properties and sensitization by inflammatory mediators. Pain. 2014;155:1861–1870. doi: 10.1016/j.pain.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson S, et al. Group II mGluRs suppress hyperexcitability in mouse and human nociceptors. Pain. 2016 doi: 10.1097/j.pain.0000000000000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer SM, et al. Virally mediated optogenetic excitation and inhibition of pain in freely moving nontransgenic mice. Nature biotechnology. 2014;32:274–278. doi: 10.1038/nbt.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SI, et al. Soft, stretchable, fully implantable miniaturized optoelectronic systems for wireless optogenetics. Nature biotechnology. 2015;33:1280–1286. doi: 10.1038/nbt.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copits BA, Pullen MY, Gereau RW. Spotlight on pain: optogenetic approaches for interrogating somatosensory circuits. Pain. 2016 doi: 10.1097/j.pain.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang D, Gereau RW. Peripheral group II metabotropic glutamate receptors (mGluR2/3) regulate prostaglandin E2-mediated sensitization of capsaicin responses and thermal nociception. The Journal of neuroscience: the official journal of the Society for Neuroscience. 6660;22:6388–6393. doi: 10.1523/JNEUROSCI.22-15-06388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu HJ, Bhave G, Gereau RW. Prostaglandin and protein kinase A-dependent modulation of vanilloid receptor function by metabotropic glutamate receptor 5: potential mechanism for thermal hyperalgesia. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2002;22:7444–7452. doi: 10.1523/JNEUROSCI.22-17-07444.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X, Gereau RW. Acute p38-mediated modulation of tetrodotoxin-resistant sodium channels in mouse sensory neurons by tumor necrosis factor-alpha. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:246–255. doi: 10.1523/JNEUROSCI.3858-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D, Gereau RW. Group II metabotropic glutamate receptors inhibit cAMP-dependent protein kinase-mediated enhancemednt of tetrodotoxin-resistant sodium currents in mouse dorsal root ganglion neurons. Neuroscience letters. 2004;357:159–162. doi: 10.1016/j.neulet.2003.11.074. [DOI] [PubMed] [Google Scholar]
- 30.Valtcheva MV, Davidson S, Zhao C, Leitges M, Gereau RW. Protein kinase Cdelta mediates histamine-evoked itch and responses in pruriceptors. Molecular pain. 2015;11:1. doi: 10.1186/1744-8069-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valtcheva MV, Samineni VK, Golden JP, Gereau RW, Davidson S. Enhanced nonpeptidergic intraepidermal fiber density and an expanded subset of chloroquine-responsive trigeminal neurons in a mouse model of dry skin itch. The journal of pain: official journal of the American Pain Society. 2015;16:346–356. doi: 10.1016/j.jpain.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheahan TD, Copits BA, Golden JP, Gereau RW. Voluntary Exercise Training: Analysis of Mice in Uninjured, Inflammatory, and Nerve-Injured Pain States. PloS one. 2015;10:e0133191. doi: 10.1371/journal.pone.0133191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchard JW, et al. Selective conversion of fibroblasts into peripheral sensory neurons. Nature neuroscience. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wainger BJ, et al. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nature neuroscience. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S, et al. Cell type-specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nature methods. 2011;8:745–752. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ting JT, Daigle TL, Chen Q, Feng G. Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods in molecular biology. 2014;1183:221–242. doi: 10.1007/978-1-4939-1096-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hille B. The permeability of the sodium channel to organic cations in myelinated nerve. The Journal of general physiology. 1971;58:599–619. doi: 10.1085/jgp.58.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siuda ER, et al. Spatiotemporal control of opioid signaling and behavior. Neuron. 2015;86:923–935. doi: 10.1016/j.neuron.2015.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goverdhana S, et al. Regulatable gene expression systems for gene therapy applications: progress and future challenges. Molecular therapy: the journal of the American Society of Gene Therapy. 2005;12:189–211. doi: 10.1016/j.ymthe.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glorioso JC, Fink DJ. Herpes vector-mediated gene transfer in the treatment of chronic pain. Molecular therapy: the journal of the American Society of Gene Therapy. 2009;17:13–18. doi: 10.1038/mt.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhave G, et al. Protein kinase C phosphorylation sensitizes but does not activate the capsaicin receptor transient receptor potential vanilloid 1 (TRPV1) Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12480–12485. doi: 10.1073/pnas.2032100100. [DOI] [PMC free article] [PubMed] [Google Scholar]


