ABSTRACT
Iron is an essential micronutrient for both pathogens and their hosts, which restrict iron availability during infections in an effort to prevent microbial growth. Successful human pathogens like the yeast Candida glabrata have thus developed effective iron acquisition strategies. Their regulation has been investigated well for some pathogenic fungi and in the model organism Saccharomyces cerevisiae, which employs an evolutionarily derived system. Here, we show that C. glabrata uses a regulation network largely consisting of components of the S. cerevisiae regulon but also of elements of other pathogenic fungi. Specifically, similarly to baker’s yeast, Aft1 is the main positive regulator under iron starvation conditions, while Cth2 degrades mRNAs encoding iron-requiring enzymes. However, unlike the case with S. cerevisiae, a Sef1 ortholog is required for full growth under iron limitation conditions, making C. glabrata an evolutionary intermediate to SEF1-dependent fungal pathogens. Therefore, C. glabrata has evolved an iron homeostasis system which seems to be unique within the pathogenic fungi.
IMPORTANCE
The fungus Candida glabrata represents an evolutionarily close relative of the well-studied and benign baker’s yeast and model organism Saccharomyces cerevisiae. On the other hand, C. glabrata is an important opportunistic human pathogen causing both superficial and systemic infections. The ability to acquire trace metals, in particular, iron, and to tightly regulate this process during infection is considered an important virulence attribute of a variety of pathogens. Importantly, S. cerevisiae uses a highly derivative regulatory system distinct from those of other fungi. Until now, the regulatory mechanism of iron homeostasis in C. glabrata has been mostly unknown. Our study revealed a hybrid iron regulation network that is unique to C. glabrata and is placed at an evolutionary midpoint between those of S. cerevisiae and related fungal pathogens. We thereby show that, in the host, even a successful human pathogen can rely largely on a strategy normally found in nonpathogenic fungi from a terrestrial environment.
INTRODUCTION
Iron is an essential micronutrient for all living organisms. Usually incorporated into heme or bound in iron sulfur clusters, it acts as an important cofactor for various cellular processes, including the tricarboxylic acid (TCA) cycle, DNA replication, chromatin remodeling, mitochondrial respiration, and detoxification of reactive oxygen species (ROS) (1). Its biological functions derive from its redox properties, based on the switch between the ferric state (Fe3+) and the ferrous state (Fe2+). This switch can, however, also become detrimental to the cell. While Fe3+ is poorly soluble under alkaline conditions, Fe2+ can become toxic by promoting the production of ROS via the Fenton reaction (2). Thus, uptake, distribution, utilization, and storage of iron have to be tightly regulated. In terrestrial environments, the poor solubility of Fe is the main reason for its low bioavailability (1). During infection, pathogenic microbes similarly face iron limitation mainly due to “nutritional immunity”—a process by which the host restricts iron to prevent proliferation of invading pathogens (3). In the human body, for example, iron is sequestered by various carrier and storage proteins such as hemoglobin, transferrin, and ferritin, and there is virtually no freely bioavailable iron (4). Therefore, successful pathogens had to develop sophisticated mechanisms to exploit host iron sources.
One of these is Candida glabrata, an opportunistic fungal pathogen that colonizes epithelial surfaces like the gastrointestinal tract as part of the normal human microbiota (5). However, under certain predisposing conditions, like immunosuppression or long-term hospitalization, the fungus can shift from commensalism to a pathogenic lifestyle. C. glabrata-induced diseases can range from superficial oral infections to systemic infections with high mortality rates (6). Among Candida species, C. glabrata ranks second to Candida albicans in isolation frequency in the United States and most of Europe, and together, those two species are responsible for 65% to 75% of all life-threatening systemic candidiasis (5, 7). Still, these two pathogens differ remarkably in their lifestyle, genetic makeup, and morphology (8). In fact, C. glabrata is much more closely related to the normally nonpathogenic baker’s yeast Saccharomyces cerevisiae than to C. albicans (9), and their respective pathogenicity strategies must have evolved independently.
A widespread strategy of pathogens, and an important virulence attribute for several fungal pathogens, is efficient and tightly controlled iron utilization. Fungi as diverse as C. albicans, Histoplasma capsulatum, Aspergillus fumigatus, and Cryptococcus neoformans harbor complex iron uptake and homeostasis mechanisms for their survival in the host (10–13). Frequently, high-affinity (HA-) uptake systems allow transport via the fungal membrane under iron starvation conditions (14), and siderophores are produced by many fungi, but not by Candida spp., which instead possess an uptake system for xenosiderophores (1, 15). Finally, host iron sources can be exploited: C. albicans, for example, uses many host iron sources like hemoglobin, ferritin, or transferrin (15–18), but no such uptake systems are known in C. glabrata (or S. cerevisiae) (15, 17, 18). In C. albicans, as in most other fungi, regulation of iron homeostasis is tightly controlled and mediated by two reciprocally acting transcriptional repressors, a GATA-type factor, Sfu1, and a CCAAT-binding factor, Hap43 (19). Under iron restriction conditions, the Cys6Zn2 transcriptional activator Sef1 induces iron uptake genes, while Hap43 represses iron-consuming processes as well as transcription of Sfu1. Under iron-replete conditions, lack of Sef1 activation leads to the absence of Hap43 and hence to an induction of Sfu1, which represses iron uptake (20). S. cerevisiae employs an iron regulatory system completely different from those employed by other fungi. Here, Aft1 is the major activator of iron uptake and recycling under iron restriction conditions (21, 22), together with its paralog Aft2, which contributes in a largely redundant manner (23). Additionally, posttranscriptional degradation of specific mRNAs ensures the downregulation of iron-consuming processes (24). Although an ortholog of Sef1 is present in S. cerevisiae, no iron-related function has been described for it. Both HAP43 and SFU1 are absent in the yeast genome.
In the present study, we analyzed C. glabrata mutants with deletions of potential orthologs of iron regulatory systems of both C. albicans and S. cerevisiae. Our experiments identified Aft1 as the main regulator for iron uptake and recycling under iron deprivation conditions and the mRNA degrading protein Cth2 as a posttranscriptional inhibitor of iron consumption processes. Surprisingly, the SEF1 transcript was found to be a novel target of Cth2 with importance for growth under iron limitation conditions in C. glabrata. We show that C. glabrata employs a unique iron regulatory system, containing elements of both S. cerevisiae and C. albicans and differing from other pathogens.
RESULTS
Screening of a mutant library under iron restriction conditions identified C. glabrata transcription factors Aft1 and Sef1.
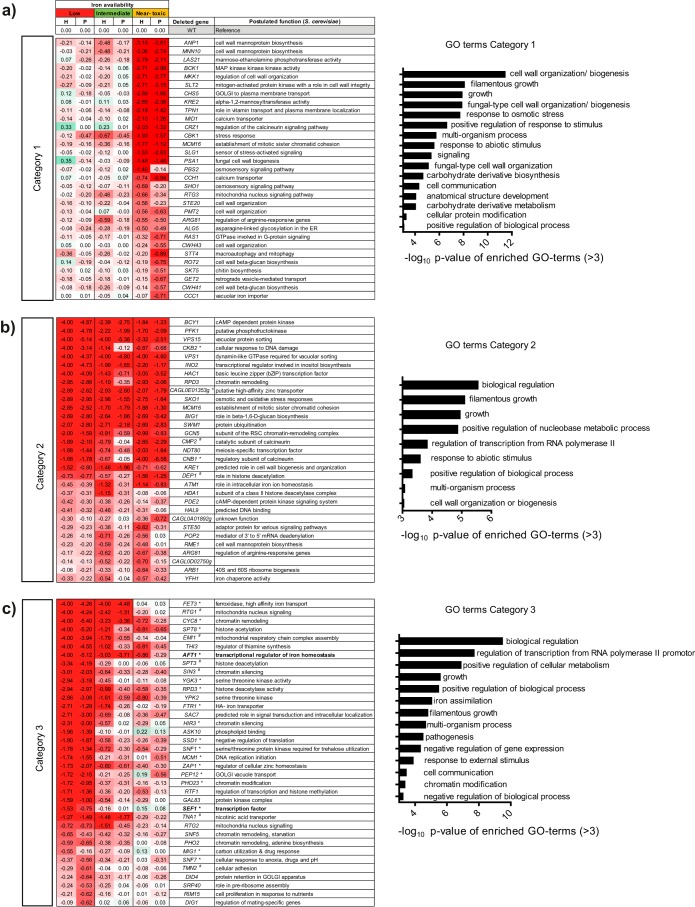
To identify factors important for iron homeostasis, an extensive deletion mutant library (Schwarzmüller et al. [25] and mutants generated in this work) was screened for iron-dependent growth defects. In total, 649 strains were analyzed for their growth behavior under conditions of different iron concentrations, and 100 of them displayed an iron-dependent phenotype under at least one set of conditions. These strains were divided into three different categories according to their growth behavior (Fig. 1). Category 1 contained 31 mutants with growth defects under specifically near-toxic iron conditions (Fig. 1a). Gene ontology (GO)-term analysis revealed an enrichment of processes associated with cell wall organization and integrity in this category. A mutant lacking Ccc1, a transporter with a possible role in vacuolar iron sequestration (26), also appeared in this category, while its supposed regulator Yap5 (27) seemed to play no role (see Dataset S1 in the supplemental material). Category 2 contained another 31 mutants with general growth defects that were independent of iron supplementation (Fig. 1b), including deletions of known and iron-associated genes (YFH1, ATM1) conserved in most fungi such as C. albicans (28, 29) and S. cerevisiae (30, 31). Additionally, category 1 and 2 contained strains lacking genes encoding components of calcium signaling or the stress-associated calcineurin pathway (CCH1, MID1, CMP2, CNB1, CRZ1). Most relevant to this work, category 3 contained 36 mutants with growth defects under low-iron conditions that were rescued by iron addition (Fig. 1c). GO-term analysis revealed an enrichment of genes involved in transcription regulation, chromatin remodeling (CYC8, PHO23), and—as expected—iron assimilation. These included the iron acquisition genes FTR1 and FET3 as well as AFT1 and SEF1, encoding orthologs of activators of iron acquisition in S. cerevisiae and C. albicans, respectively.
FIG 1 .
Screen-revealed C. glabrata deletion mutants with iron-dependent growth defects. Growth of iron-prestarved cells was assayed by measuring the OD600 under low-iron conditions (10 µM FeCl3), intermediate-iron conditions (500 µM FeCl3), and near-toxic iron conditions (40 mM FeCl3) in Chelex-treated SD (pH 5.8). Growth was evaluated compared to the WT by assessing the time of half-maximal growth (H) and the height of the plateau (P) after 50 h at 37°C. Zero corresponds to wild-type (WT) growth (shaded white), positive numbers indicate improved growth (shaded green), and negative numbers indicate growth defects (shaded red). Means of results of triplicate experiments are shown with growth equal to or less than a set cutoff of −0.5 for at least one evaluated parameter. Enriched GO-terms (–log10 P value of >3 displayed) are shown. Selected mutants were tested separately for growth on solid medium containing the iron chelator BPS. Of 27 tested mutants, 19 showed a growth defect (*), while 8 grew as well as the WT (#). (a) List of mutants with a growth defect under near-toxic iron conditions (category 1). (b) List of mutants with general growth defects under all tested iron conditions (category 2). (c) List of mutants with an iron-dependent growth defect that can be partially rescued by iron addition (category 3). Potential iron regulators are labeled in bold. cAMP, cyclic AMP; RSC, remodels the structure of chromatin; MAP, mitogen-activated protein kinase.
AFT1 and SEF1, but no other known genes encoding iron regulator orthologs, are required under iron limitation conditions.
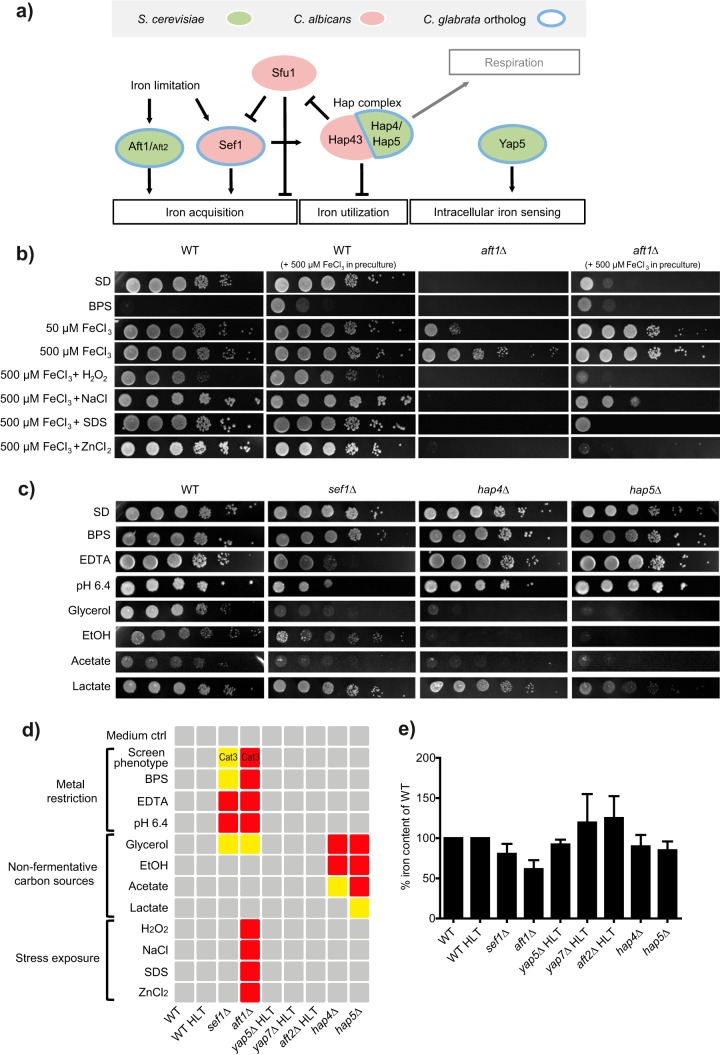
The identified regulators Aft1 and Sef1 are known key components in independent and distinct iron regulation networks of the baker’s yeast species S. cerevisiae and the pathogen C. albicans, respectively. Since these findings suggest overlapping contributions, we decided to analyze both networks in more detail. Therefore, we screened the C. glabrata genome in silico for potential orthologs of the known iron-associated regulators in C. albicans and S. cerevisiae and identified seven candidates. These comprised mostly orthologs from the S. cerevisiae iron network, including Aft1, its functionally redundant paralog Aft2 (32), and the fungus-specific bZIP transcription factor Yap5, an intracellular iron sensor (33). C. glabrata Yap5, along with Yap7, has recently been shown to regulate iron sulfur cluster biogenesis and heme biosynthesis in a global chromatin immunoprecipitation (ChIP) and transcriptome analysis approach (34). Additionally, members of the Hap complex were tested, e.g., Hap4 and Hap5, known to contribute to Hap43-regulated iron homeostasis in C. albicans (35) but to respiratory gene expression in S. cerevisiae (36) and with an unclear role in C. glabrata. Sef1 was chosen as the only direct ortholog from the C. albicans iron regulation network present in C. glabrata; a SFU1 ortholog was not found. Figure 2a summarizes these species-specific iron regulatory networks and the C. glabrata orthologs. These candidate regulator knockout strains were included in the growth screen along with aft1Δ and sef1Δ mutants but did not show any growth defects under low-iron conditions (see Dataset S1 in the supplemental material). In contrast, the aft1Δ mutant showed a severe iron-dependent growth defect and also showed increased sensitivity to oxidative, osmotic, high metal, and cell membrane stress (Fig. 2b). Growth under various metal restriction conditions, such as the presence of the general metal chelator ethylenediaminetetraacetic acid (EDTA) or of the iron chelator bathophenanthrolinedisulfonic acid (BPS), or under alkaline pH conditions, which reduce iron solubility, revealed a moderate defect for the sef1Δ mutant but not for the hap4Δ, hap5Δ, yap5Δ, yap7Δ, or aft2Δ mutant (Fig. 2c and d). Instead, growth of the hap4Δ and hap5Δ mutants was impaired on the nonfermentative carbon sources glycerol, lactate, ethanol, and acetate, which require respiration for energy generation, indicating a role for these genes in respiration similar to that in S. cerevisiae. Finally, we determined the intracellular iron content for all C. glabrata regulator knockout strains and found decreased iron content only in the aft1Δ and sef1Δ strains (Fig. 2e). Taking the data together, the C. glabrata phenotypes seen upon deletion of AFT1, HAP4, or HAP5 resemble the S. cerevisiae phenotypes, while the SEF1 phenotype is more reminiscent, at least in part, of those of C. albicans and other pathogenic fungi.
FIG 2 .
Iron regulators Aft1 and Sef1 are needed for C. glabrata growth under iron restriction conditions. (a) Scheme of the distinct iron regulation networks (black connections) in C. albicans (red) and S. cerevisiae (green) and the orthologs present in the C. glabrata genome (circled in blue). (b) Growth of the aft1Δ mutant on solid media (SD) under conditions of iron restriction (BPS) and a combination of iron supplementation in the preculture (500 µM FeCl3) and in the plate (50 µM FeCl3 or 500 µM FeCl3) and with oxidative stress (H2O2), osmotic stress (NaCl), cell membrane stress (SDS), and high metal stress (ZnCl2). (c) Growth of regulator deletion strains on solid media (SD) in the presence of iron-associated stressors (BPS, EDTA [pH 6.4]) and on nonfermentative carbon sources (glycerol, EtOH, acetate, lactate). (d) Growth levels of all regulator deletion strains are displayed schematically for all tested stressors as follows: no growth defect, gray box; moderate growth defect, yellow box; severe growth defect, red box. Cat3, category 3 iron screen phenotype; ctrl, control. (e) Intracellular iron content of C. glabrata regulator deletion strains of YPD-grown log-phase cells shown as a percentage of the content determined for the corresponding WT strain ± SEM. An unpaired two-tailed t test was performed.
The iron-dependent phenotype of Sef1 is specific to C. glabrata and not present in S. cerevisiae.
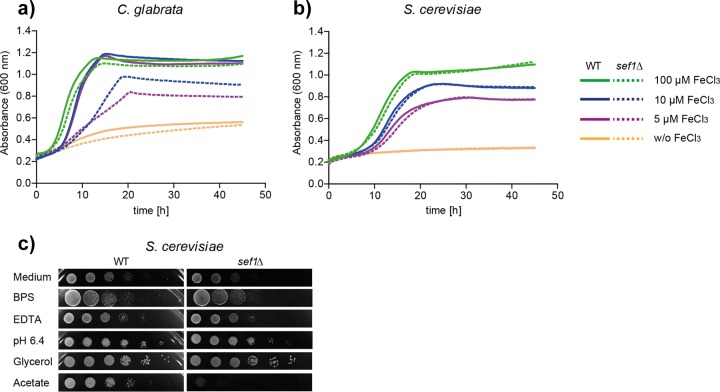
Sef1 is known to play a fundamental role in the C. albicans iron regulation network. Our findings likewise suggest an—albeit less pronounced—iron-related role in C. glabrata. The function of Sef1 in S. cerevisiae is thus far unknown, and so we investigated a possible connection to iron homeostasis similar to that seen with C. glabrata and C. albicans. Growth curve analysis showed growth representative of a defect under low-iron conditions (5 µM and 10 µM FeCl3) for a C. glabrata sef1Δ mutant which was restored to wild-type (WT) levels by addition of 100 µM FeCl3 (Fig. 3a). In contrast, the S. cerevisiae sef1Δ mutant results were indistinguishable from the WT results under all conditions (Fig. 3b). Similarly, exposure to growth conditions that included the presence of iron-limiting stressors such as BPS, EDTA, or alkaline, as well as of glycerol as a sole carbon source, caused no growth defects of S. cerevisiae sef1Δ (Fig. 3c), in clear contrast to C. glabrata sef1Δ (Fig. 2c). Instead, S. cerevisiae sef1Δ, but not C. glabrata sef1Δ, was strongly attenuated in growth on the nonfermentative carbon source acetate. Taken together, these data suggest a species-specific role for Sef1 which in C. glabrata resembles the function of Sef1 in C. albicans more than in S. cerevisiae.
FIG 3 .
Sef1 performs an iron-related role in C. glabrata but not in S. cerevisiae. (a) Growth of the C. glabrata WT and sef1Δ strains under conditions of iron restriction (SD+200 µM BPS) and iron supplementation (5 µM, 10 µM, or 100 µM FeCl3). The generation times (means ± standard deviations) of the WT and sef1Δ strains differed significantly in the presence of 5 µM FeCl3 (4.8 ± 1.14 h versus 13.2 ± 3.79 h; P < 0.05) and 10 µM FeCl3 (3.7 ± 0.35 h versus 9.6 ± 1.67 h; P < 0.01) but not in the presence of 100 µM FeCl3 (2.8 ± 0.09 h versus 3.1 ± 0.22 h). Data from a representative replicate are displayed (n = 3). (b) Growth levels of S. cerevisiae WT and sef1Δ as described for panel a showed no significant differences. A representative replicate is displayed. n = 3. (c) Growth of S. cerevisiae WT and sef1Δ in the presence of the stressors BPS, EDTA (pH 6.4), glycerol, and acetate.
Aft1 and Sef1 have different regulatory functions under iron starvation conditions.
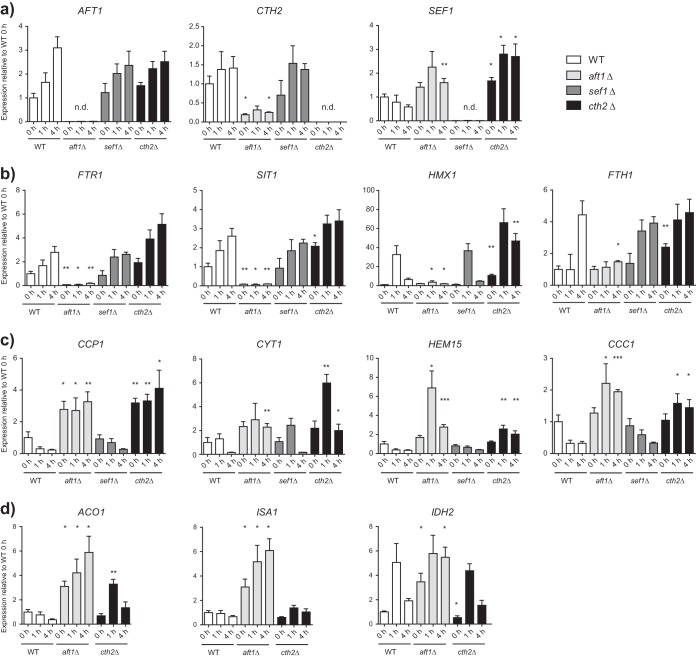
Our results indicate roles for both Aft1 and Sef1 in the regulation of iron homeostasis in C. glabrata. To gain insight into the target genes of these transcription factors, we compared the transcription profiles of the wild-type, aft1Δ, and sef1Δ strains under iron-restricted conditions. An ftr1Δ strain, deficient for the main high-affinity iron transporter, was included as a control. At 4 h after a shift from 5 µM FeCl3 to iron-free medium, genes involved in iron uptake processes (FTR1, FET3, SIT1) and iron recycling processes (FTH1, HMX1, SMF3) were strongly upregulated in the WT (Fig. 4a). However, these genes were significantly less transcribed in the aft1Δ mutant than in the WT after 4 h. A consensus-binding motif for Aft1 (PyPuCACCCPu) in S. cerevisiae is known (21). We found many genes associated with iron uptake or recycling containing one or more copies of this binding motif in their promoter regions (marked with an asterisk [*] in Fig. 4a). The data in Dataset S2 in the supplemental material indicate all C. glabrata ORFs (open reading frames) with at least one detectable upstream Aft1-binding motif as possible regulation targets. Remarkably, there was a large set of genes involved in iron-consuming processes (e.g., CYC1, COX6, CCP1, CCC1, HEM15), and those genes were strongly downregulated in the WT cells upon iron starvation but not in aft1Δ cells. Despite its phenotypic effects on growth and iron content, deletion of SEF1 had only subtle effects on the transcription profile under iron deprivation conditions. Genes involved in the TCA cycle and in glutamate biosynthesis in a broader sense (ACO1, IDH1, IDH2), as well as genes with iron sulfur cluster-dependent functions (ACO1, ISA1), were transcribed to a lesser degree under iron starvation conditions upon SEF1 deletion. Transcriptional downregulation was verified by quantitative reverse transcription-PCR (qRT-PCR) for the aconitase gene ACO1, the isocitrate dehydrogenase gene IDH2, and the gene encoding the iron sulfur cluster protein Isa1. These data indicate a potential role for Sef1 in TCA cycle regulation (Fig. 4b). Under replete-iron conditions, interestingly, some genes coding for iron-requiring enzymes (CCP1, CYT1, HEM15, CCC1, ACO1) showed a tendency of increased transcription in the sef1Δ mutant compared to the WT (see Dataset S3c in the supplemental material), pointing to a possible role of Sef1 in the “fine-tuning” of iron consumption processes. In summary, our data suggest a dual role of Aft1: as the major transcriptional activator of iron uptake and recycling processes and as a repressor of iron consumption processes. Additionally, Sef1 seems to regulate specific iron-associated processes such as the TCA cycle under iron restriction conditions.
FIG 4 .
Comparative transcriptional profiling shows major dysregulation of iron metabolism-associated genes in the aft1Δ mutant and reveals that Cth2 is a potential posttranscriptional regulator. (a) Relative mRNA levels of iron-associated genes in the WT strain compared to the aft1Δ, sef1Δ, and ftr1Δ mutants under iron starvation conditions. Microarray data are displayed in a normalized log2 scale for downregulation (negative numbers, shaded red), upregulation (positive numbers, shaded green), or no detected expression (n.d.) of target genes. Asterisks (*) indicate target genes that carry one or multiple copies of the Aft1 consensus-binding site PyPuCACCCPu in the 5′ UTR. (b) qRT-PCR expression of the following genes affected by SEF1 deletion relative to the levels seen with the WT at 0 h: ACO1 (encoding mitochondrial aconitase), ISA1 (encoding an iron sulfur cluster assembly), and IDH2 (encoding isocitrate dehydrogenase). Data represent means of results of biological triplicate experiments ± SEM (nd, no expression detected). Data are from an unpaired Student’s t test and represent results of comparisons to WT results from the same time point (*, P value < 0.05; **, P value < 0.01). (c) The postulated target consensus motif of the Cth2 mRNA-degrading protein (5′-UUAUUUAUU-3′) is found in the downstream sequences of 36 (of 106) genes normally downregulated in the WT strain upon iron starvation (0 h to 4 h) (volcano plot; 2-fold downregulation; P value = 0.05; false-discovery-rate [FDR] corrected) but is artificially upregulated in the aft1Δ strain (4 h against WT) (volcano plot; 2-fold upregulation; P value = 0.05; FDR corrected). Motifs (red box, forward strand; blue box, reverse strand) are shown for the previously displayed striking microarray genes in the annotated 3′ intergenic region (lower gray line) or the predicted 3′ UTR of the target gene (upper gray line) in proportion to the actual length (1 dot represents 4 bp; // indicates a discontinued display). (d) Growth of the cth2Δ mutant under low-iron conditions (10 µM FeCl3), intermediate-iron conditions (500 µM FeCl3), and near-toxic iron conditions (40 mM FeCl3) in Chelex-treated SD (pH 5.8) compared to the WT strain (zero, shaded white) determined by assessing the time of half-maximal growth (H) and the plateau height (P). Positive numbers indicate improved growth (shaded green), and negative numbers indicate growth defects (shaded red). Data represent means of results from triplicate experiments. (e) Growth of the cth2Δ mutant in the presence of the iron chelator BPS (50 µM) and under alkaline conditions (pH 6.4) on solid media.
CTH2 is induced under iron starvation conditions via Aft1 to downregulate iron consumption and SEF1.
According to our transcription data, iron-consuming processes were downregulated in the WT but not in the aft1Δ mutant during iron starvation (Fig. 4a). To investigate the underlying mechanism, we therefore selected genes downregulated in the WT at 4 h of starvation but with at least 2-fold higher expression in the aft1Δ mutant (aft1Δ mutant at 4 h versus the WT at 4 h). These 106 ORFs were analyzed for a common motif in their flanking regions using the bioinformatics tool FIMO (37). We found the motif 5′-TTATTTATT-3′ to be located in the 3′ untranscribed region (UTR) of 36 of these ORFs (E value 1.7 × 10−5) in single or multiple copies. A search for this motif in the intergenic regions of all known C. glabrata ORFs (see Dataset S4 in the supplemental material) detected additional iron metabolism-associated genes such as AFT1, SEF1, ACO1, and CCC1 (Fig. 4c). In fact, most of these genes were then found to be deregulated in the aft1Δ mutant and were originally excluded only because of our stringent threshold. Interestingly, this motif is known in S. cerevisiae as the nonamer consensus recognition motif of mRNA-degrading protein Cth2 (also known as Tis11). S. cerevisiae Cth2 (ScCth2), along with its paralog Cth1, was found specifically to target mRNA of iron-dependent genes for degradation via binding to this motif (22, 24), whereas the corresponding Cth2 protein in C. glabrata was still uncharacterized. Interestingly, C. glabrata has no ortholog of CTH1. To elaborate the role of Cth2 in C. glabrata, we constructed deletion mutants and performed growth assays. Growth of a mutant lacking CTH2 was strongly impaired under low-iron conditions in liquid culture (Fig. 4d), and the mutant showed susceptibility to BPS and alkaline growth conditions (Fig. 4e). In agreement with this, CTH2 transcription was highly upregulated upon iron starvation in the WT but not in the aft1Δ mutant (Fig. 4a). Since the CTH2 promoter also carries the Aft1 recognition motif, we postulated CTH2 to be a target gene of Aft1. In fact, qRT-PCR showed that lack of AFT1 abolished expression of CTH2 (Fig. 5a). Furthermore, expression of SEF1 was elevated in both aft1Δ and cth2Δ strains of C. glabrata. In silico analysis revealed that the S. cerevisiae ortholog of SEF1, in contrast, lacks the nonamer-binding site for Cth2 (2.5 kb up- and downstream of the ORF), reflecting its lack of iron-related function in this fungus. Taking the results together, we have identified C. glabrata Cth2 as a potential Aft1-induced mediator of mRNA degradation of iron-associated genes, a group which includes SEF1.
FIG 5 .
Expression analysis of iron-associated target genes by qRT-PCR. Data represent gene expression of target genes for the WT, aft1Δ, cth2Δ, and sef1Δ strains at 0 h, 1 h, and 4 h. After 4 h of preincubation under low-iron conditions (5 µM FeCl3), the 0-h sample was obtained. Residual cells were then incubated without iron for 1 h or 4 h. Target gene expression was normalized to that of housekeeping genes EFB1 and EFT2. Expression is shown relative to that of the WT at 0 h in biological triplicate experiments ± SEM (n.d., no expression detected). Data are from an unpaired Student’s t test and represent results of comparisons to WT results from the same time point (*, P < 0.05; **, P < 0.01; ***, P < 0.005). (a) Expression of the genes encoding regulators of iron metabolism: AFT1 and SEF1 (encoding transcription factors) and CTH2 (encoding an mRNA-degrading protein). (b) Expression of the iron acquisition genes FTR1 (encoding an HA-iron transporter), SIT1 (encoding a siderophore transporter), HMX1 (degrading heme oxidase), and FTH1 (encoding a vacuolar HA-iron exporter) under iron deprivation conditions. (c) Expression of the iron consumption genes CCP1 (encoding mitochondrial cytochrome c peroxidase), CYT1 (encoding cytochrome c), HEM15 (encoding heme synthesis), and CCC1 (encoding a vacuolar HA-iron importer) under iron deprivation conditions. (d) Expression of the following genes affected by SEF1 deletion: ACO1 (encoding mitochondrial aconitase), ISA1 (encoding an iron sulfur cluster assembly), and IDH2 (encoding isocitrate dehydrogenase).
Cth2 is required for limiting iron consumption and plays a minor role in iron uptake regulation.
Microarray transcriptional profiling analysis of the aft1Δ mutant had indicated a lack of induction of iron acquisition genes under iron deprivation conditions, which we confirmed by qRT-PCR for FTR1, SIT1, HMX1, and FTH1 (Fig. 5b). As the mRNA levels of all of these genes were also consistently elevated in the cth2Δ mutant, Cth2 seems to exert its posttranscriptional mRNA degradation on mRNA transcripts of highly expressed iron acquisition genes under iron starvation conditions. Consistently, lack of CTH2 did not influence their already low expression levels under iron-replete conditions (see Dataset S3b in the supplemental material). We similarly analyzed expression levels of genes associated with iron consumption (Fig. 5c), including respiration-associated genes such as CCP1 (cytochrome c peroxidase) and CYT1 (heme-dependent electron transport), HEM15 (heme synthesis), and CCC1 (vacuolar iron import). Consistent with our previous results, the mRNA levels of these genes were downregulated upon iron deprivation in the WT, but transcript levels were much higher in the aft1Δ mutant. Similarly, lack of CTH2 resulted in a significant and comparable increase in mRNA levels. Under iron-replete conditions, the iron consumption processes were mainly upregulated in the WT. Lack of CTH2, on the other hand, either slightly elevated the levels of respiration-associated genes or decreased them for other iron consumption processes (see Dataset S3c). Taking the results together, Cth2 is strictly needed for downregulating iron consumption processes under iron restriction conditions. Additionally, our data point to a continuous degradation of highly transcribed mRNAs involved in iron uptake processes.
C. glabrata mutants lacking AFT1 and SEF1 display diminished survival in human blood.
We have shown that Aft1, Cth2, and Sef1 are important for the regulation of iron metabolism in vitro. To elaborate whether these factors also play a role in the host, we analyzed the survival of C. glabrata aft1Δ, cth2Δ, and sef1Δ strains in an ex vivo human blood model mimicking, to some extent, the dissemination phase of systemic fungal infections (Fig. 6). There, the aft1Δ mutant showed a clear and significant reduction in early survival in blood. Survival of the sef1Δ mutant was likewise significantly attenuated early during exposure to blood (at 1 h and with the tendency already seen at 0.5 h), whereas, remarkably, deletion of CTH2 did not affect survival at any time point. A reduction of aft1Δ survival was observable also at later time points, albeit without reaching the statistical significance levels of the earlier samples. Hence, both Aft1 and Sef1 seem to be required for full survival in blood, especially during the early interaction. Assuming iron-scarce conditions during the initial immune defense, this would be in good agreement with the proposed roles of both transcription factors in iron homeostasis of C. glabrata.
FIG 6 .
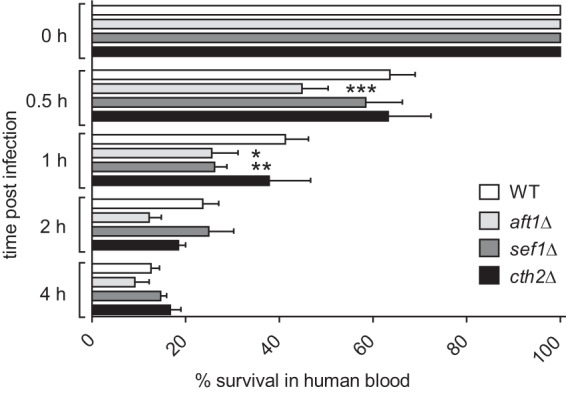
Aft1 and Sef1 are needed for survival in human blood. Human blood was infected with the C. glabrata WT, aft1Δ, sef1Δ, and cth2Δ strains, and survival at 37°C was determined by CFU plating after the indicated time points (0 h, 0.5 h, 1 h, 2 h, and 4 h). Data are shown as percent survival of the WT at 0 h (means ± standard deviation). Data represent results of a paired one-way analysis of variance (ANOVA) of comparisons to WT results from the same time point (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
Iron homeostasis networks have been studied in a variety of pathogenic fungi, and they are known to play important roles in virulence (38). Although transcriptomics data have been obtained during changes in environmental iron levels for C. glabrata (39), our knowledge of genes which are essential for iron homeostasis, of their integration into the complex regulatory networks, and of their role in virulence remained limited. We therefore employed a screen of deletion mutants where we mimicked both iron deprivation and iron overload to detect genes involved in iron homeostasis. Using this setup, we identified 100 deletion mutants with relevant growth defects. We were able to confirm the important roles of various iron-associated genes described previously in C. glabrata, such as FTR1, FET3, and YFH1 (40), or in C. albicans and S. cerevisiae, such as ATM1 (29, 31). However, most of our candidate genes have not been associated previously with iron homeostasis in C. glabrata. A shortage of iron represents a common challenge in most host niches of C. glabrata (41, 42); hence, we considered mutant growth defects under low-iron conditions especially informative for the identification of possible virulence factors. Strikingly, we identified Aft1 as an essential transcription factor of C. glabrata necessary for growth under low-iron conditions via activation of iron uptake and intracellular recycling. aft1, known as the major transcriptional activator of the iron regulon in S. cerevisiae (21, 22, 31), was presumed an essential gene in C. glabrata by Srivastava et al. (40), since a knockout strain could not be obtained. Here we show that AFT1 is not essential, or, more precisely, is conditionally essential, as it is required only under conditions of low to moderate iron concentrations and not under conditions of highly elevated concentrations. Hence, we were able to show that C. glabrata, like its closest relative—the baker’s yeast S. cerevisiae—uses an Aft1-dominated system for iron homeostasis. To our knowledge, C. glabrata is the first known pathogen engaging an Aft1-dependent system as the main iron regulation strategy. Considering that levels of iron availability and sources of iron, as well as the physical conditions (temperature, pH, oxygen levels), differ greatly between the terrestrial environment of S. cerevisiae and a pathogen’s animal host niches, it is astonishing that C. glabrata utilizes the Aft1-dependent system. Thus, our findings point to a rather unusual regulation strategy for C. glabrata compared to other fungal pathogens.
Interestingly, according to our transcription data, C. glabrata reacts to iron deprivation in a time-dependent manner and first activates the uptake mechanisms (FTR1, SIT1), followed by the mobilization of vacuolar storages (FTH1), and intermediately and transiently resorts to heme recycling (HMX1). These observations indicate a tightly controlled and timed response to iron deprivation, probably depending on the increasing magnitude of intracellular iron deficiency. In addition to the upregulation of iron acquisition genes, we also observed a marked downregulation of genes involved in iron consumption processes.
In S. cerevisiae, the Aft1 regulon is known to recruit the mRNA-degrading protein Cth2 (also known as Tis11), which posttranscriptionally reduces the abundance of iron consumption-related mRNAs under conditions of iron deprivation (22, 24). We were able to identify a functional homolog of CTH2 in C. glabrata which is also positively regulated by Aft1. As in S. cerevisiae, we found the highly conserved motif 5′-UUAUUUAUU-3′ in the 3′ UTR of target mRNAs of Cth2. In both species, these are transcripts coding especially for proteins with a role in iron consumption (24). Additionally, we have also observed a moderate effect of Cth2 on the mRNA levels of iron acquisition genes in C. glabrata. In contrast to S. cerevisiae (24), the Cth2-binding motif is present in FTR1, FET3, and SIT1 transcripts, pointing to a novel role in posttranscriptional fine-tuning of iron uptake likely via an increased turn-over of mRNA, which could be advantageous for survival in niches with often rapidly shifting iron levels.
Notably, Cth2 has a partially redundant paralog, Cth1, in S. cerevisiae. Although there, both CTH1 and CTH2 are activated by Aft1 under iron deprivation conditions, CTH1 is expressed only transiently early during iron deficiency, while CTH2 expression persists during prolonged iron limitation conditions. Additionally, autoregulation and trans-regulation have been proposed for those two mRNA degradation mediators, since both carry a 5′-UUAUUUAUU-3′ recognition motif (24, 43, 44). C. glabrata is lacking a second copy of CTH2, indicating that while it seems to rely on the Cth system for downregulation, its specific implementation differs from that used by S. cerevisiae. Since Cth2 carries its own recognition motif, an autoregulatory loop seems likely. Remarkably, C. albicans also possesses a single Cth2 ortholog called Zfs1. However, Zfs1 target transcripts are not related to iron homeostasis but rather to biofilm formation (45). The only other fungus with a functionally described Cth2 ortholog, Schizosaccharomyces pombe, also displays no transcriptional alterations of its iron homeostasis genes when this gene is deleted (46, 47). These observations indicate a mode of recognition by Cth2 that is evolutionarily conserved between C. glabrata and S. cerevisiae, with a large set of overlapping target genes (24), while the sets of regulated target genes differ greatly between all other investigated species. To our knowledge, C. glabrata is thus the first example of a pathogenic fungus using its Cth2 ortholog to downregulate iron homeostasis-associated transcripts. Further factors may play a role in this iron-level-dependent transcript degradation process, as most but not all decreases in the mRNA level were dependent on the presence of Cth2.
In contrast, other pathogenic fungi such as Aspergillus fumigatus, Cryptococcus neoformans, and C. albicans generally rely on iron-dependent regulation via GATA factor orthologs and the CCAAT-binding complex (12, 48–51). Although this system is broadly conserved among ascomycetes and partly in basidiomycetes, there are a few species-specific modifications—for instance, the incorporation of Sef1 orthologs into this network (49). In C. albicans, Sef1 is a pivotal activator of iron uptake genes and thus plays an essential role during iron deficiency and for virulence in mice (19). In this study, we showed that, surprisingly, the C. glabrata Sef1 ortholog has an iron-related function although the fungus is not using the GATA factor and CCAAT-binding complex for iron-dependent regulation. A Sef1 ortholog can also be found in S. cerevisiae, but, in contrast to C. glabrata, we have found no connection of Sef1 to iron metabolism in baker’s yeast. These findings strongly suggest a novel hybrid iron regulation system for C. glabrata which combines features from the networks of S. cerevisiae (Aft1) and C. albicans (Sef1). The Candida and Saccharomycetaceae clade diverged evolutionarily from each other, with the latter partly undergoing a whole-genome duplication (WGD) event (52). During evolution, a complete rewiring of the iron regulation network from the likely more ancient GATA/CCAAT-binding complex system to Aft1/Cth2 must have evolved in the Saccharomycetaceae branch, accompanied by the loss of the GATA factor and the iron-associated CCAAT-binding factor function. Although the components of the CCAAT-binding complex (e.g., Hap4, Hap5) are still present in S. cerevisiae, they are now involved in the regulation of aerobic respiration (36). C. glabrata, as part of the post-WGD Saccharomycetaceae clade (53), underwent the same process but apparently has maintained a residual Sef1 function, resulting in a hybrid form of iron regulation with major features derived from the S. cerevisiae system and minor features retained from the system used by C. albicans and other pathogenic fungi.
The full role of Sef1 in the C. glabrata iron network still eludes us; however, we found important functional characteristics of Sef1 during iron starvation. Growth defects of the sef1Δ mutant under conditions of iron deprivation and of alkaline pH (with its concomitant low iron solubility), as well as the in silico analysis, place it firmly in the C. glabrata iron homeostasis regulatory network. Surprisingly, despite its phenotype, the transcriptome of the sef1Δ mutant did not differ strongly from that of the WT. This is especially striking in comparison to the large effect that SEF1 deletion has on C. albicans iron-dependent transcriptional responses, where Sef1 is a major activator of iron uptake-related genes (reference 49 and own unpublished observations). In the process of the change to an Aft-dependent system, most of these interactions seem to have been lost within the Saccharomycetaceae branch. However, at least in C. glabrata, very specific changes in gene expression were observed for genes associated with the (iron-requiring) TCA cycle (ACO1, IDH1, IDH2) as well as for genes encoding known iron sulfur cluster-containing proteins (ACO1, ISA1).
Interestingly, mammalian Aco1 has dual functions that depend on iron availability. Under conditions of low iron availability—when it is known as IRP1—it acts as an RNA-binding protein and inhibits translation initiation of mRNAs related to iron-consuming and -storing processes, such as ferritin mRNA (54, 55). On the other hand, IRP1 binding can also stabilize target RNAs under iron limitation conditions, e.g., the mRNA coding for the transferrin receptor, preventing degradation to enhance the transferrin-mediated iron uptake pathway (56). However, under high-iron conditions, IRP1 acquires an active iron sulfur cluster and is converted to a fully functional cytosolic aconitase (57–59). Although plants and fungi seem to lack this iron regulatory switch in function for Irp1/Aco1 (60, 61), the mitochondrial aconitase remains a crucial iron-dependent protein of the TCA cycle in fungi (29, 62).
Furthermore, Isa proteins have been proposed to function in the maturation of aconitase-like iron sulfur cluster proteins in S. cerevisiae (63, 64). Since we observed less expression of Isa1 and Aco1 in the sef1Δ mutant, the two proteins might be part of the same process, suggesting a need for Sef1 for the assembly of specific iron sulfur cluster proteins, such as Aco1 via Isa1. In S. cerevisiae, various iron sulfur proteins have been implicated in iron sensing, where the status of iron loading indicates the cellular lack or presence of iron (63, 65, 66). Since Sef1 is needed to sustain basal expression of ISA1 and ACO1 according to our data, it might be necessary to ensure the presence of specific iron sulfur cluster proteins under iron starvation conditions, which may then act as iron sensors, signaling iron deprivation to the cell by lack of iron binding. Overall, it seems that Sef1 has a very specialized function in the hybrid iron homeostasis regulation of C. glabrata, with the Aft1/Cth2 system bearing the brunt of transcriptional regulation under iron starvation conditions and Sef1 acting on specific pathways.
We were also interested in the role of Aft1 and Sef1 under conditions resembling a human infection. Establishment of systemic disease caused by Candida spp. requires (i) entry into the bloodstream by overcoming epithelial barriers or by colonizing of intravenous catheters (67) and (ii) initial dissemination via the bloodstream. These processes frequently lead to sepsis with significant associated morbidity and mortality and to high health care-associated costs (68, 69). To survive in the blood and successfully disseminate, Candida spp. evidently have to undergo drastic changes of metabolism. Metal availability is especially tightly restricted in blood to facilitate nutritional immunity. Free iron availability is very low (10−24 M Fe3+) (19) and below the minimal concentration required for microbial growth (10−6 to 10−7 M Fe3+) (70). Especially for C. glabrata, which (in contrast to C. albicans) is not known to use hemoglobin or transferrin for iron acquisition (8, 15), survival in this environment is likely dependent on tight and precise regulation of the balance between iron uptake and consumption. In C. albicans, Sef1 is crucial for iron uptake in bloodstream infections, as shown by severely decreased virulence and recovery from murine organs of a sef1Δ/Δ mutant (49). Here, we have shown that Sef1 is a transcriptional regulator required for early survival in human blood in C. glabrata as well, while an even more pronounced decrease in survival was observed for the aft1Δ (main regulator) mutant. We found that both genes are thus needed for early survival in blood, while at later time points, killing by immune cells probably reaches a level which starts to occlude the iron-dependent effects. It is noteworthy, however, that during dissemination, fungi are likely exposed only briefly to the bloodstream before they enter the target organs and hence that early survival may be crucial for successful systemic infections (71). These data hence underline the prominent role of Aft1 and Sef1 in C. glabrata iron homeostasis and suggest a role of these factors in human virulence.
To summarize, we propose a model for the regulation of iron metabolism in C. glabrata under conditions of iron deprivation (Fig. 7). Aft1 acts as the major transcriptional activator of iron acquisition processes and of CTH2 expression. Subsequently, Cth2 mediates posttranscriptional degradation of specific transcripts carrying a conserved recognition motif. Potential targets include predominantly iron consumption-related genes and SEF1, which itself regulates a subset of genes. Remarkably, this kind of iron regulation system has not been described in any other fungus and seems to be unique to the opportunistic pathogen C. glabrata. It mostly resembles the regulation system of the baker’s yeast S. cerevisiae, whereas some components from C. albicans are retained. These network adaptions might be of advantage for virulence of C. glabrata, contributing to its successful life as a pathogen despite its close evolutionary relations to S. cerevisiae.
FIG 7 .
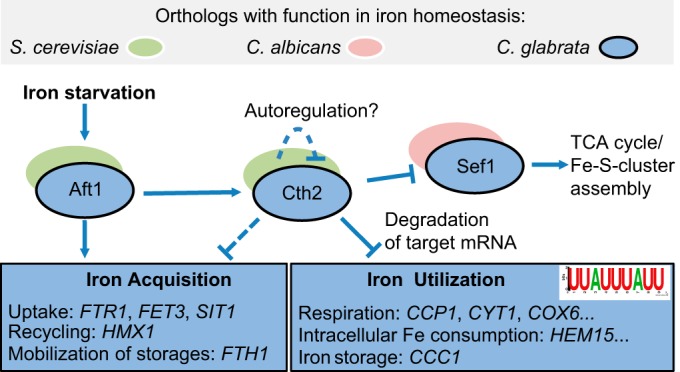
Model of iron metabolism regulation in C. glabrata. In C. glabrata (blue), similarly to S. cerevisiae (green), Aft1 acts as an activator of iron acquisition genes under iron starvation conditions (FTR1, FET3, SIT1), while it also induces expression of CTH2. Cth2, a mediator of posttranscriptional mRNA degradation, targets transcripts carrying the recognition motif 5′-UUAUUUAUU-3′, predominantly associated with iron consumption processes (HEM15, CCP1, ACO1). Additional postulated functions of Cth2 (dashed lines) also include the negative regulation of iron acquisition in a moderate manner and an autoregulatory role. The transcription factor SEF1, which is important in the C. albicans iron regulon (red), is also a Cth2 target gene in C. glabrata which has a specific impact on the TCA cycle and on iron sulfur cluster (Fe-S cluster) assembly.
MATERIALS AND METHODS
Strains.
The mutants screened in this study comprised the strains described in Schwarzmüller et al. (25) as well as C. glabrata gene knockout strains generated in this study, constructed in the ATCC 2001 wild-type strain using the same gene deletion strategy. The latter included orthologs of S. cerevisiae or C. albicans high- and low-affinity iron uptake systems (fet3Δ, ftr1Δ, and fet4Δ mutants) and transcriptional regulators (aft1Δ, sef1Δ, hap4Δ, and hap5Δ mutants) (Table 1). Briefly, the gene of interest (GOI) was replaced via homologous recombination by a cassette containing constant flanking regions (U1 and D1), specific up- and downstream bar codes, and the nourseothricin resistance marker NAT1. Cassettes were fused with 500-bp to 1,000-bp target gene-specific 5′ and 3′ flanks and integrated into XbaI-linearized pUC19 vector using an Infusion HD cloning kit (Clontech). The insertion was PCR amplified (with primers GOI fwd and GOI rev) and used for C. glabrata transformation by a modified heat shock method (72) (with 45°C heat shock for 15 min). The transformants were plated onto yeast extract-peptone-dextrose (YPD; 1% yeast extract, 2% Bacto peptone, 2% glucose) agar containing 250 µg/ml nourseothricin, supplemented where necessary with 500 µM FeCl3 (strains ftr1Δ and aft1Δ). Knockout strains were verified by sequencing and Southern blotting (NAT1-specific probe; primers NAT fwd South and Nat rev South). All primers are listed in Dataset S5 in the supplemental material.
TABLE 1 .
Strains used in this studya
| Strain name | Description | Source or reference |
|---|---|---|
| C. glabrata | ||
| WT | C. glabrata WT strain ATCC 2001 | American Type Culture Collection |
| WT HLT | C. glabrata WT strain for collection ATCC 2001; his3Δ::FRT leu2Δ::FRT trp1Δ::FRT | 25 |
| Deletion mutant collection (GOIΔ HLT) | ATCC 2001; goiδ::NAT1 his3Δ::FRT leu2Δ::FRT trp1Δ::FRT | 25 |
| aft1Δ | ATCC 2001; CAGL0H03487gΔ::NAT1 | This study |
| ftr1Δ | ATCC 2001; CAGL0I06743gΔ::NAT1 | This study |
| sef1Δ | ATCC 2001; CAGL0A04455gΔ::NAT1 | This study |
| fet3Δ | ATCC 2001; CAGL0F06413gΔ::NAT1 | This study |
| fet4Δ | ATCC 2001; CAGL0F00187gΔ::NAT1 | This study |
| cth2Δ | ATCC 2001; CAGL0E01243gΔ::NAT1 | This study |
| hap4Δ | ATCC 2001; CAGL0K08624gΔ::NAT1 | This study |
| hap5Δ | ATCC 2001; CAGL0K09900gΔ::NAT1 | This study |
| S. cerevisiae | ||
| BY4741 | S. cerevisiae WT strain; MAT a; his3Δ 1; leu2Δ 0; met15Δ 0; ura3Δ 0 | EUROSCARFb |
| YBL066cΔ/ sef1Δ | BY4741; Mat a; his3Δ 1; leu2Δ 0; met15Δ 0; ura3Δ 0; YBL066c::kanMX4 | EUROSCARFb |
GOI (gene of interest); HLT (auxotrophy for histidine, leucine, and tryptophan); WT (wild type).
Screen for iron-dependent growth defects.
C. glabrata cultures were grown in 96-well plates (TPP) at 37°C and 180 rpm in three consecutive overnight cultures by 1:50 transfers: first, in rich medium (YPD); second, in synthetic defined medium (SD; 0.67% yeast nitrogen base [YNB], 2% glucose, 0.079% complete supplement mix [CSM; Formedium]); and third, in citrate-buffered SD (pH 5.8) supplemented with 200 µM bathophenanthrolinedisulfonic acid (BPS). The optical density at 600 nm (OD600) of the last culture was adjusted to 0.1 in iron-free water for growth in iron-free citrate-buffered SD (pH 5.8). This medium was pretreated overnight with Chelex 100 (Sigma) to remove metals, the Chelex 100 beads were removed by filtration, and noniron metals were restored (100 mg/ml CaCl2, 500 mg/ml MgSO4 ⋅ 7H2O, 400 µg/ml MnSO4, 40 µg/ml CuSO4, 400 µg/ml ZnSO4 ⋅ 7H2O). Defined amounts of FeCl3 were added to obtain low-iron conditions (10 µM FeCl3), intermediate iron conditions (500 µM FeCl3), or near-toxic iron conditions (40 mM FeCl3). Then, 20 µl of the C. glabrata inoculum was added to 180 µl of each test medium. Growth was followed by determination of the OD600 at 37°C in a Tecan Infinite 200 enzyme-linked immunosorbent assay (ELISA) reader (with intermittent shaking) for 2 days. The relative growth levels of the mutants were determined by the time of half-maximal OD600 (H) and the final plateau height of the stationary phase (P) relative to those determined for the wild type. The values were normalized to the wild-type control values, and the log2 of this ratio was calculated; hence, negative numbers indicate growth defects. The screen was performed once for all 648 mutants and repeated twice to obtain triplicate measures whenever a possible phenotype was seen.
Serial dilution growth tests.
Precultures were prepared in SD supplemented with CSM and optionally with 500 µM FeCl3. Sensitivity of C. glabrata strains to stressors was tested by spotting 1:10 serial dilutions on agar containing H2O2 (SD, 12 mM), NaCl (SD, 1.1 M), sodium dodecyl sulfate (SDS) (SD, 60 µg/ml), ZnCl2 (SD, 12 mM), BPS (YPD, 50 µM), or EDTA (SD, 8 µM). The alternative carbon sources used instead of glucose were glycerol (3%), sodium lactate (2%), ethanol (2%), and sodium acetate (4%). Growth at a more alkaline pH was tested by buffering SD agar with phosphate buffer to pH 6.4.
Cultivation of yeast for RNA isolation.
For RNA isolation, the strains were cultured overnight in SD, harvested by centrifugation, and washed four times in iron-free water. All steps were carried out with acid-washed iron-free equipment. Cells were adjusted to 1 × 107 cells/ml in iron-free citrate-buffered SD (pH 5.8) supplemented with 5 µM FeCl3 and grown for 4 h at 37°C and 180 rpm. The 0-h sample was immediately frozen in liquid nitrogen and stored at −80°C. The remaining cultures were harvested by centrifugation and washed four times with iron-free SD. The start of the time series was defined by the first contact with iron-free SD. Further samples were taken at 0.5 h, 1 h, 2 h, and 4 h. To obtain iron-replete samples, 100 µM FeCl3 was added following the 4-h time point and a sample was taken after an additional 0.5 h. RNA was isolated with an RNeasy minikit (Qiagen), and quality was verified using an Agilent 2100 Bioanalyzer Nanochip system according to the manufacturer’s protocol. The concentration was determined using a Nanodrop 2000 instrument.
Microarray.
For microarray analysis, 1 µg high-quality RNA was used to generate cRNA fluorescently labeled with Cy5 CTP (GE Healthcare, United Kingdom) using a one-color Quick Amp labeling kit (Agilent). For a common reference experimental design, cRNA from mid-log-phase YPD-grown C. glabrata was labeled with Cy3 CTP (GE Healthcare). After cleanup with an RNeasy minikit (Qiagen), sufficiency of dye incorporation (specific activity, >7 pmol/µg) was checked by NanoDrop. C. glabrata arrays were purchased from Agilent Technologies (8-by-15K format) (GEO accession number for the C. glabrata Agilent Array, GPL10713 [73]), hybridized using a Gene Expression Hybridization kit (Agilent) as described previously (73), and scanned using a GenePix 4200AL scanner (software, GenePix Pro 6.1; automatically calibrated photomultiplier tube [PMT] wavelengths, 635 and 594 nm; pixel size, 5 µm). Data were extracted with AgilentFE and imported into GeneSpring 12 for data analysis. The experiment was conducted in biological triplicate to cover biological variation. The microarray data are available at the NCBI GEO microarray repository (see below).
qRT-PCR.
For qRT-PCR, 600 ng high-quality RNA was treated with DNase (Epicentre) and reverse transcribed into cDNA using oligo(dT) primers and Superscript III (Invitrogen). One microliter of cDNA (1:20 dilution) was used for gene expression analysis in a C1000 thermocycler (Bio-Rad; CFX96 Realtime system) using an EvaGreen system (Bio & Sell). The expression rates were determined in biological triplicate and normalized to expression of the housekeeping genes EFB1 and EFT2 using Bio-Rad CFX Manager 3.1. All primers are listed in Dataset S5 in the supplemental material.
Determination of cellular iron content.
For the determination of cellular iron content, a modified ferrozine-based colorimetric method (74) was used. C. glabrata cells grown overnight in YPD were inoculated into fresh YPD medium and grown to logarithmic phase (OD600, ~1). From that, 100 ml was harvested, washed with iron-free water, resuspended in 600 µl 0.5 M NaOH, and disrupted after addition of 300 mg iron-free acid-washed glass beads using a Precellys cell homogenizer (6,500 × g; 3 homogenizations at 45 s per homogenization with a 15-s pause between homogenizations). Beads and cell debris were removed by centrifugation, and 200 µl of the cell supernatant or 0.5 M NaOH (negative control) was used for detection. A 1:2 serial dilution (300 µM to 4.68 µM) of FeCl3–0.5 M NaOH was used as a standard. To free heme-bound iron, 100 µl freshly prepared iron-releasing agent (equal volumes of 1.4 M HCl and 4.5% [wt/vol] KMnO4) was added to the samples, followed by incubation for 2 h at 60°C and 300 rpm and cooling for 5 min with open lids. Finally, 30 µl of detection mix (6.5 mM ferrozine, 6.5 mM neocuproine to suppress copper signals, 2.5 M ammonium acetate, 1 M ascorbic acid; prepared fresh in iron-free water) was added and samples were incubated for 30 min at room temperature. The iron content was determined by measurement of the OD568 normalized against the cell number (initial OD600). The experiment was performed in biological triplicate.
Whole-blood killing assay.
Venous blood of healthy volunteers was collected in hirudin Monovettes (Sarstedt). Overnight C. glabrata YPD cultures were harvested and washed twice with sterile water. Yeasts (1 × 106 in 10 µl) were added to 990 µl freshly drawn human blood and incubated using gentle rolling at 37°C. The initial inoculum (0 h) was determined, and after 30, 60, 120, and 240 min, 10 µl of infected blood sample was diluted and plated onto YPD agar (or SD supplemented with 500 µM in FeCl3 for the aft1Δ mutant) for CFU determination in technical triplicate. This assay was performed independently with three healthy blood donors.
In silico analysis and statistics.
Information about gene orthologs and intergenic regions was obtained from the Candida Genome Database (CGD—http://www.candidagenome.org), the Candida Gene Order Browser (CGOB—http://cgob.ucd.ie/), and the Saccharomyces Genome Database (SGD—http://www.yeastgenome.org/). GO terms were obtained from the CGD and processed using the online tool Revigo (http://revigo.irb.hr/). GO terms with a negative log10 P value higher than 3 (P < 0.001) are shown. The 3′ UTR of ORFs was predicted as described by Linde et al. (75) and downloaded from CGD. MEME (37) (http://meme-suite.org) was used to identify enriched motifs in a list of genes or to find genes carrying a target motif via MEME/FIMO (http://meme-suite.org/tools/fimo). For analysis of statistics, GraphPad Prism 5 (GraphPad Inc.) was used. All data are reported as means ± standard errors of the means (SEM) or standard deviation where appropriate, and the two-tailed, unpaired Student’s t test was performed, if not stated otherwise. Similarity (nonsignificant differences) of variance data was tested in Prism 5 for all tests. Statistically significant results were marked as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Ethics statement.
Human peripheral blood was collected from healthy volunteers after written informed consent was obtained. The study was conducted in accordance with the Declaration of Helsinki, and all protocols were approved by the Ethics Committee of the University Hospital Jena (permission number 2207-01/08).
Accession number(s).
The microarray data determined in this work are available at the NCBI GEO microarray repository (accession number GSE84816).
SUPPLEMENTAL MATERIAL
Iron screen results for all tested C. glabrata mutants. Download
C. glabrata genes with ScAft1-binding motif PyPuCACCCPu in 5′ intergenic region (IR) identified by FIMO. Download
Expression analysis of iron-associated target genes by qRT-PCR under iron-replete conditions. Data represent expression of target genes for the WT, cth2Δ, and sef1Δ strains at 0 h and 4 h (under iron deprivation conditions) and at +0.5 h (under iron supplementation conditions). After 4 h preincubation under low-iron conditions (50 µM FeCl3), the 0-h sample was obtained. Residual cells were then incubated without iron for 4 h before 100 µM FeCl3 was supplemented (+0.5 h). Target gene expression was normalized to that of housekeeping genes EFB1 and EFT2. Expression is shown relative to that of the WT at 0 h in biological triplicate experiments (standard deviation ± SEM; n.d., no expression detected). Statistical significance data were determined by an unpaired Student’s t test (*, P value < 0.05; **, P value < 0.01; ***, P value < 0.001). (a) Expression of the regulators of iron metabolism AFT1 and SEF1 (encoding transcription factors) and CTH2 (encoding mRNA-degrading protein). (b) Expression of iron acquisition genes FTR1 (encoding an HA-iron transporter), SIT1 (encoding a siderophore transporter), HMX1 (degrading heme oxidase), and FTH1 (encoding a vacuolar HA-iron exporter). (c) Expression of iron consumption genes CCP1 (encoding mitochondrial cytochrome c peroxidase), CYT1 (encoding cytochrome c), HEM15 (encoding heme synthesis), and CCC1 (encoding a vacuolar HA-iron importer). (d) Expression of the following genes affected by SEF1 deletion: ACO1 (encoding mitochondrial aconitase), ISA1 (encoding an iron sulfur cluster assembly), and IDH2 (encoding isocitrate dehydrogenase). Download
C. glabrata ORFs with Cth2 nonamer recognition motif and degenerated variants in 3′ intergenic region (IR) with FIMO. Download
Primers used in this study. Download
ACKNOWLEDGMENTS
F.G., P.K., and B.H. are members of the excellence graduate school Jena School for Microbial Communication (JSMC) and the International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS). We thank Oliver Kurzai and Kerstin Hünniger (Fungal Septomics, Jena, Germany) for their help with the blood experiments, Ilse D. Jacobsen (MI, HKI, Jena, Germany), Katja Graf, and Daniel Fischer (MPM, HKI, Jena, Germany) for their valuable input, and our reviewers for their time and helpful suggestions. We also thank all voluntary blood donors.
This work was partially supported via the Centre for Sepsis Control and Care (CSCC; http://www.cscc.uniklinikum-jena.de/cscc/en/CSCC.html, grant 01EO1002; B.H.) by the German Federal Ministry of Education and Health (BMBF), and by the German Research Foundation (DFG) within the priority program SPP1580 (http://www.spp1580.uni-bonn.de, grant HU 528/16-2; L.K. and B.H.) within the Collaborative Research, Centre/Transregio 124 FungiNet (http://www.funginet.de/, subproject INF; J.L.), and by the DACH program (DFG HU 528/17-1; F.G. and B.H.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Gerwien F, Safyan A, Wisgott S, Hille F, Kaemmer P, Linde J, Brunke S, Kasper L, Hube B. 2016. A novel hybrid iron regulation network combines features from pathogenic and nonpathogenic yeasts. mBio 7(5):e01782-16. doi:10.1128/mBio.01782-16.
REFERENCES
- 1.Haas H, Eisendle M, Turgeon BG. 2008. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol 46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- 2.Ehrensberger KM, Bird AJ. 2011. Hammering out details: regulating metal levels in eukaryotes. Trends Biochem Sci 36:524–531. doi: 10.1016/j.tibs.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins HL. 2008. Withholding iron as a cellular defence mechanism—friend or foe? Eur J Immunol 38:1803–1806. doi: 10.1002/eji.200838505. [DOI] [PubMed] [Google Scholar]
- 5.Perlroth J, Choi B, Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol 45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- 6.Kaur R, Domergue R, Zupancic ML, Cormack BP. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol 8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20(Suppl 6):5–10. doi: 10.1111/1469-0691.12539. [DOI] [PubMed] [Google Scholar]
- 8.Brunke S, Hube B. 2013. Two unlike cousins: Candida albicans and C. Glabrata infection strategies. Cell Microbiol 15:701–708. doi: 10.1111/cmi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML. 2004. Genome evolution in yeasts. Nature 430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 10.Noble SM. 2013. Candida albicans specializations for iron homeostasis: from commensalism to virulence. Curr Opin Microbiol 16:708–715. doi: 10.1016/j.mib.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman SL, Smulian AG. 2013. Iron uptake and virulence in Histoplasma capsulatum. Curr Opin Microbiol 16:700–707. doi: 10.1016/j.mib.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jöchl C, Moussa TA, Wang S, Gsaller F, Blatzer M, Werner ER, Niermann WC, Brakhage AA, Haas H. 2010. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog 6:e01782-16. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kronstad JW, Hu G, Jung WH. 2013. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol 21:457–465. doi: 10.1016/j.tim.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philpott CC. 2006. Iron uptake in fungi: a system for every source. Biochim Biophys Acta 1763:636–645. doi: 10.1016/j.bbamcr.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Nevitt T, Thiele DJ. 2011. Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog 7:e01782-16. doi: 10.1371/journal.ppat.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, Turano P, Becker J, Lewinson O, Kornitzer D. 2014. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog 10:e01782-16. doi: 10.1371/journal.ppat.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4:e01782-16. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight SA, Vilaire G, Lesuisse E, Dancis A. 2005. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect Immun 73:5482–5492. doi: 10.1128/IAI.73.9.5482-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Pande K, French SD, Tuch BB, Noble SM. 2011. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu PC, Yang CY, Lan CY. 2011. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell 10:207–225. doi: 10.1128/EC.00158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi-Iwai Y, Dancis A, Klausner RD. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J 14:1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell 15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR. 2001. A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci U S A 98:14322–14327. doi: 10.1073/pnas.261381198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 25.Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, Ames L, Firon A, Green B, Cabral V, Marcet-Houben M, Jacobsen ID, Quintin J, Seider K, Frohner I, Glaser W, Jungwirth H, Bachellier-Bassi S, Chauvel M, Zeidler U, Ferrandon D, Gabaldon T, Hube B, d’Enfert C, Rupp S, Cormack B, Haynes K, Kuchler K. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10:e01782-16. doi: 10.1371/journal.ppat.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Chen OS, McVey Ward D, Kaplan J. 2001. Ccc1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Bagley D, Ward DM, Kaplan J. 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos R, Buisson N, Knight SA, Dancis A, Camadro JM, Lesuisse E. 2004. Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Mol Microbiol 54:507–519. doi: 10.1111/j.1365-2958.2004.04281.x. [DOI] [PubMed] [Google Scholar]
- 29.Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol 53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 30.Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G. 2000. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am J Hum Genet 67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y. 2012. Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol Cell Biol 32:4998–5008. doi: 10.1128/MCB.00726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courel M, Lallet S, Camadro JM, Blaiseau PL. 2005. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol Cell Biol 25:6760–6771. doi: 10.1128/MCB.25.15.6760-6771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes L, Rodrigues-Pousada C, Struhl K. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17:6982–6993. doi: 10.1128/MCB.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merhej J, Thiebaut A, Blugeon C, Pouch J, Ali Chaouche Mel A, Camadro JM, Le Crom S, Lelandais G, Devaux F. 2016. A network of paralogous stress response transcription factors in the human pathogen Candida glabrata. Front Microbiol 7:645. doi: 10.3389/fmicb.2016.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh RP, Prasad HK, Sinha I, Agarwal N, Natarajan K. 2011. Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem 286:25154–25170. doi: 10.1074/jbc.M111.233569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schüller HJ. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 37.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding C, Festa RA, Sun TS, Wang ZY. 2014. Iron and copper as virulence modulators in human fungal pathogens. Mol Microbiol 93:10–23. doi: 10.1111/mmi.12653. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava VK, Suneetha KJ, Kaur R. 2015. The mitogen-activated protein kinase CgHog1 is required for iron homeostasis, adherence and virulence in Candida glabrata. FEBS J 282:2142–2166. doi: 10.1111/febs.13264. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava VK, Suneetha KJ, Kaur R. 2014. A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem J 463:103–114. doi: 10.1042/BJ20140598. [DOI] [PubMed] [Google Scholar]
- 41.Nevitt T. 2011. War-Fe-re: iron at the core of fungal virulence and host immunity. Biometals 24:547–558. doi: 10.1007/s10534-011-9431-8. [DOI] [PubMed] [Google Scholar]
- 42.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. 2012. Mechanisms of mammalian iron homeostasis. Biochemistry 51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puig S, Vergara SV, Thiele DJ. 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab 7:555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martínez-Pastor M, Vergara SV, Puig S, Thiele DJ. 2013. Negative feedback regulation of the yeast CTH1 and CTH2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation. Mol Cell Biol 33:2178–2187. doi: 10.1128/MCB.01458-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells ML, Washington OL, Hicks SN, Nobile CJ, Hartooni N, Wilson GM, Zucconi BE, Huang W, Li L, Fargo DC, Blackshear PJ. 2015. Post-transcriptional regulation of transcript abundance by a conserved member of the tristetraprolin family in Candida albicans. Mol Microbiol 95:1036–1053. doi: 10.1111/mmi.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuthbertson BJ, Liao Y, Birnbaumer L, Blackshear PJ. 2008. Characterization of zfs1 as an mRNA-binding and -destabilizing protein in Schizosaccharomyces pombe. J Biol Chem 283:2586–2594. doi: 10.1074/jbc.M707154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells ML, Huang W, Li L, Gerrish KE, Fargo DC, Ozsolak F, Blackshear PJ. 2012. Posttranscriptional regulation of cell-cell interaction protein-encoding transcripts by Zfs1p in Schizosaccharomyces pombe. Mol Cell Biol 32:4206–4214. doi: 10.1128/MCB.00325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baek YU, Li M, Davis DA. 2008. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell 7:1168–1179. doi: 10.1128/EC.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C, Noble SM. 2012. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog 8:e01782-16. doi: 10.1371/journal.ppat.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW. 2010. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog 6:e01782-16. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thön M, Kniemeyer O, Abt B, Seeber B, Werner ER, Kato M, Brakhage AA, Haas H. 2007. Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J 26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roetzer A, Gabaldón T, Schüller C. 2011. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol Lett 314:1–9. doi: 10.1111/j.1574-6968.2010.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabaldón T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, Arnaise S, Boisnard S, Aguileta G, Atanasova R, Bouchier C, Couloux A, Creno S, Almeida Cruz J, Devillers H, Enache-Angoulvant A, Guitard J, Jaouen L, Ma L, Marck C, Neuveglise C, Pelletier E, Pinard A, Poulain J, Recoquillay J, Westhof E, Wincker P, Dujon B, Hennequin C, Fairhead C. 2013. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics 14:623. doi: 10.1186/1471-2164-14-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eisenstein RS. 2000. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr 20:627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 55.Hentze MW, Muckenthaler MU, Andrews NC. 2004. Balancing acts: molecular control of mammalian iron metabolism. Cell 117:285–297. doi: 10.1016/S0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 56.Kühn LC. 2015. Iron regulatory proteins and their role in controlling iron metabolism. Metallomics 7:232–243. doi: 10.1039/c4mt00164h. [DOI] [PubMed] [Google Scholar]
- 57.Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K. 2006. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 58.Kühn LC. 2009. How iron controls iron. Cell Metab 10:439–441. doi: 10.1016/j.cmet.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Benjamin JA, Massé E. 2014. The iron-sensing aconitase B binds its own mRNA to prevent sRNA-induced mRNA cleavage. Nucleic Acids Res 42:10023–10036. doi: 10.1093/nar/gku649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hentze MW, Kühn LC. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci U S A 93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnaud N, Ravet K, Borlotti A, Touraine B, Boucherez J, Fizames C, Briat JF, Cellier F, Gaymard F. 2007. The iron-responsive element (IRE)/iron-regulatory protein 1 (IRP1)-cytosolic aconitase iron-regulatory switch does not operate in plants. Biochem J 405:523–531. doi: 10.1042/BJ20061874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farooq MA, Pracheil TM, Dong Z, Xiao F, Liu Z. 2013. Mitochondrial DNA instability in cells lacking aconitase correlates with iron citrate toxicity. Oxid Med Cell Longev 2013:493536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lill R, Dutkiewicz R, Elsässer HP, Hausmann A, Netz DJ, Pierik AJ, Stehling O, Urzica E, Mühlenhoff U. 2006. Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes. Biochim Biophys Acta 1763:652–667. doi: 10.1016/j.bbamcr.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 64.Mühlenhoff U, Richter N, Pines O, Pierik AJ, Lill R. 2011. Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J Biol Chem 286:41205–41216. doi: 10.1074/jbc.M111.296152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma AK, Pallesen LJ, Spang RJ, Walden WE. 2010. Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J Biol Chem 285:26745–26751. doi: 10.1074/jbc.R110.122218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rietzschel N, Pierik AJ, Bill E, Lill R, Mühlenhoff U. 2015. The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol Cell Biol 35:370–378. doi: 10.1128/MCB.01033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akbari F, Kjellerup BV. 2015. Elimination of bloodstream infections associated with Candida albicans biofilm in intravascular catheters. Pathogens 4:457–469. doi: 10.3390/pathogens4030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delaloye J, Calandra T. 2014. Invasive candidiasis as a cause of sepsis in the critically ill patient. Virulence 5:161–169. doi: 10.4161/viru.26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duggan S, Leonhardt I, Hünniger K, Kurzai O. 2015. Host response to Candida albicans bloodstream infection and sepsis. Virulence 6:316–326. doi: 10.4161/21505594.2014.988096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 71.Mavor AL, Thewes S, Hube B. 2005. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr Drug Targets 6:863–874. doi: 10.2174/138945005774912735. [DOI] [PubMed] [Google Scholar]
- 72.Sanglard D, Ischer F, Monod M, Bille J. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother 40:2300–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferrari S, Sanguinetti M, De Bernardis F, Torelli R, Posteraro B, Vandeputte P, Sanglard D. 2011. Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob Agents Chemother 55:1852–1860. doi: 10.1128/AAC.01271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. 2004. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem 331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 75.Linde J, Duggan S, Weber M, Horn F, Sieber P, Hellwig D, Riege K, Marz M, Martin R, Guthke R, Kurzai O. 2015. Defining the transcriptomic landscape of Candida glabrata by RNA-Seq. Nucleic Acids Res 43:1392–1406. doi: 10.1093/nar/gku1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Iron screen results for all tested C. glabrata mutants. Download
C. glabrata genes with ScAft1-binding motif PyPuCACCCPu in 5′ intergenic region (IR) identified by FIMO. Download
Expression analysis of iron-associated target genes by qRT-PCR under iron-replete conditions. Data represent expression of target genes for the WT, cth2Δ, and sef1Δ strains at 0 h and 4 h (under iron deprivation conditions) and at +0.5 h (under iron supplementation conditions). After 4 h preincubation under low-iron conditions (50 µM FeCl3), the 0-h sample was obtained. Residual cells were then incubated without iron for 4 h before 100 µM FeCl3 was supplemented (+0.5 h). Target gene expression was normalized to that of housekeeping genes EFB1 and EFT2. Expression is shown relative to that of the WT at 0 h in biological triplicate experiments (standard deviation ± SEM; n.d., no expression detected). Statistical significance data were determined by an unpaired Student’s t test (*, P value < 0.05; **, P value < 0.01; ***, P value < 0.001). (a) Expression of the regulators of iron metabolism AFT1 and SEF1 (encoding transcription factors) and CTH2 (encoding mRNA-degrading protein). (b) Expression of iron acquisition genes FTR1 (encoding an HA-iron transporter), SIT1 (encoding a siderophore transporter), HMX1 (degrading heme oxidase), and FTH1 (encoding a vacuolar HA-iron exporter). (c) Expression of iron consumption genes CCP1 (encoding mitochondrial cytochrome c peroxidase), CYT1 (encoding cytochrome c), HEM15 (encoding heme synthesis), and CCC1 (encoding a vacuolar HA-iron importer). (d) Expression of the following genes affected by SEF1 deletion: ACO1 (encoding mitochondrial aconitase), ISA1 (encoding an iron sulfur cluster assembly), and IDH2 (encoding isocitrate dehydrogenase). Download
C. glabrata ORFs with Cth2 nonamer recognition motif and degenerated variants in 3′ intergenic region (IR) with FIMO. Download
Primers used in this study. Download