Abstract
Objective:
To characterize the clinical and molecular effect of mutations in the sortilin-related receptor (SORL1) gene.
Methods:
We performed whole-exome sequencing in early-onset Alzheimer disease (EOAD) and late-onset Alzheimer disease (LOAD) families followed by functional studies of select variants. The phenotypic consequences associated with SORL1 mutations were characterized based on clinical reviews of medical records. Functional studies were completed to evaluate β-amyloid (Aβ) production and amyloid precursor protein (APP) trafficking associated with SORL1 mutations.
Results:
SORL1 alterations were present in 2 EOAD families. In one, a SORL1 T588I change was identified in 4 individuals with AD, 2 of whom had parkinsonian features. In the second, an SORL1 T2134 alteration was found in 3 of 4 AD cases, one of whom had postmortem Lewy bodies. Among LOAD cases, 4 individuals with either SORL1 A528T or T947M alterations had parkinsonian features. Functionally, the variants weaken the interaction of the SORL1 protein with full-length APP, altering levels of Aβ and interfering with APP trafficking.
Conclusions:
The findings from this study support an important role for SORL1 mutations in AD pathogenesis by way of altering Aβ levels and interfering with APP trafficking. In addition, the presence of parkinsonian features among select individuals with AD and SORL1 mutations merits further investigation.
Alzheimer disease (AD) is the leading cause of dementia in the elderly.1 Multiple genes have been implicated in risk for both late-onset Alzheimer disease (LOAD; onset >65 years of age) and early-onset Alzheimer disease (EOAD; onset <65 years of age)2 including the sortilin-related receptor (SORL1) gene. Located on chromosome 11q23.2-q24.2, SORL1 influences the differential sorting of the amyloid precursor protein (APP) and regulation of β-amyloid (Aβ) production, making it biologically plausible for AD risk.3–9
Compelling evidence for the involvement of SORL1 in AD comes from a large meta-analysis of >30,000 individuals, which confirmed that variants in SORL1 are associated with AD risk.10 Furthermore, whole-exome sequencing (WES) has identified potentially damaging SORL1 mutations in patients with both EOAD and LOAD.11,12 Of note, a WES study of a large EOAD cohort found a greater frequency of predicted damaging missense SORL1 variants in cases vs controls, with this effect enriched among cases with a positive family history.13 Clearly, rare coding variants in SORL1 are tied to risk for EOAD and LOAD. Finally, while SORL1 mutations have been reported in multiple patients with AD, there has been little investigation of clinical phenotypes beyond dementia and age at onset (AAO) among these individuals.
For this study, we examined well-characterized EOAD families using WES to discover AD risk genes. Our efforts focused on clinical characterization of individuals with SORL1 alterations and investigation of the functional effect of the identified SORL1 alterations in a series of gene overexpression experiments.
METHODS
Standard protocol approvals, registrations, and patient consents.
All participants ascertained for this study gave written informed consent prior to their inclusion. If an individual was not competent to give consent, the immediate next of kin or a legal representative provided written consent on their behalf. All participants were ascertained using a protocol that was approved by the appropriate Institutional Review Board. Oversight of this study falls under the University of Miami Institutional Review Board #20070307.
Sources of participants.
EOAD families were ascertained as part of a larger study on AD genetics whose participants were enrolled under protocols previously described.14,15 Individuals were ascertained for this study after they provided informed consent at the John P. Hussman Institute for Human Genomics (HIHG) at the University of Miami Miller School of Medicine (Miami, FL). The majority of these families were self-reported non-Hispanic whites (N = 47); the remaining families were self-reported African Americans (N = 3). Clinical data from cognitively impaired individuals, including any that changed affection status, were evaluated by the HIHG AD clinical staff which includes a psychiatrist, neurologist, and neuropsychologist. Familial EOAD cases were defined as AAO <65 years of age. As reported in previous studies, AAO was defined as the age at which an individual or family historian reported onset of significant cognitive problems that interfered with normal activity, or the AAO of problems as documented in the medical record.15 All affected individuals met the internationally recognized standard NINCDS-ADRDA criteria.16,17 The cognitive status of participants was measured using either the Mini-Mental State Examination18 or the Modified Mini-Mental State.19
Patients with LOAD (N = 151) were part of a study investigating coding mutations in SORL1 in AD.11 These participants were drawn from a larger study of AD genetics restricted to Caribbean Hispanics. All affected individuals were of Caribbean Hispanic ancestry. All participants were assessed using standard clinical examinations and cognitive testing as described elsewhere.20 For this study, we reviewed the clinical records of participants who had SORL1 mutations to assess for possible features of Parkinson disease (PD) or more broadly, parkinsonism.
WES and variant calling.
All samples were prepared using DNA extracted from the blood. Genomic DNA was then sheared and processed using the SureSelect Human All Exon 50 Mb v4 capture kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol at the HIHG Center for Genome Technology. After capture, the DNA was tested for uniform enrichment of targets via quantitative PCR. Sequencing was then performed on the Illumina HiSeq2000 at 2× 150 bp paired-end cycles at 40–50× on target depth. Exomes were sequenced to sufficient depth to achieve a minimum threshold of 80% of coding sequence covered with at least 15 reads, based on UCSC hg19 “known gene” transcripts. The mean depth of coverage across SORL1 was 68.
Sequencing data from the Illumina HiSeq2000 were processed using an established semiautomated pipeline. Initial image files were processed using the Firecrest module (Illumina, San Diego, CA) to determine cluster intensities and noise. After initial quality control, BWA-ELAND and CASAVA v1.9 were used for realignment to the human genome version hg19. Results from BWA and CASAVA are then fed into additional software packages (CLC Genomics Workbench and GenomeStudio) for secondary analysis, visualization of the called variants, and browsing of consensus reads.21,22 Genotype calling was performed with GATK Unified Genotyper. Variants were then normalized using BCFTools.23 Single nucleotide polymorphisms with read depth <6, variant quality score log odds ratio <0, and Phred-scaled likelihood score <100 were removed from further analysis. Variants were filtered to identify alterations that were likely to be damaging (missense, splicing, stop-gain, stop-loss, and insertion/deletions) in Gencode v19, NCBI RefSeq, or Ensembl gene annotations.24,25 Variants were screened to determine whether they occurred in a known or suspected EOAD gene (APP, GRN, MAPT, PSEN1, PSEN2, SORL1, and TREM2). Minor allele frequencies were obtained from the Exome Aggregation Consortium.26
Cloning of SORL1 variants.
Site-directed mutagenesis was used to generate the SORL1 T588I and SORL1 T2134IM mutation constructs using human SORL1-MYC pcDNA3.1 as a backbone according to the manufacturer's instructions as previously published.3,11,27–30 Sequencing was used to verify mutant constructs. Cell culture and transfection followed previously described standard protocols.3,11,27–30
Aβ, Western blot, and co-immunoprecipitation assays.
Aβ assays were measured by sandwich ELISA assay in culture medium from stably transfected HEK293 cells expressing the Swedish APP mutant (APPsw) and either wild-type SORL1 or mutant SORL1 as previously described.3,11,27–30 Cell surface biotinylation was performed using 1 mg/mL Sulfo-NHS-LC-Biotin (Sigma-Aldrich, St. Louis, MO) for 20 minutes at 4°C to prevent internalization. Cells were then washed and lysed, and biotinylated proteins were precipitated with NeutrAvidin beads (Thermo Fisher Scientific, Waltham, MA). Western blot band intensities were measured with ImageJ software and samples normalized to the wild-type control. Co-immunoprecipitation was performed after cell lysis in 1% CHAPSO buffer,3 using G Plus beads with 2 μg mouse monoclonal anti-c-MYC antibody for the immunoprecipitation of SORL1-myc, immunoblotted with anti-C-terminal APP antibody (Ab365), and anti-C-terminal SORL1 (S9200). Western blot band intensities were measured with ImageJ software. Full-length (FL) APP coprecipitated with c-MYC antibody was quantified and normalized to the amount of immunoprecipated SORL1 as previously described.3,11,27–30
Statistical analyses.
Statistical analyses were performed using Graphpad statistical software (graphpad.com/guides/prism/5/user-guide/prism5help.html?using_tour_overview.htm; GraphPad Prism 5). Analysis of variance and t tests were used to analyze statistical difference, followed by Bonferroni correction (*p < 0.05; **p < 0.01; and ***p < 0.001).
RESULTS
SORL1 variants in EOAD families.
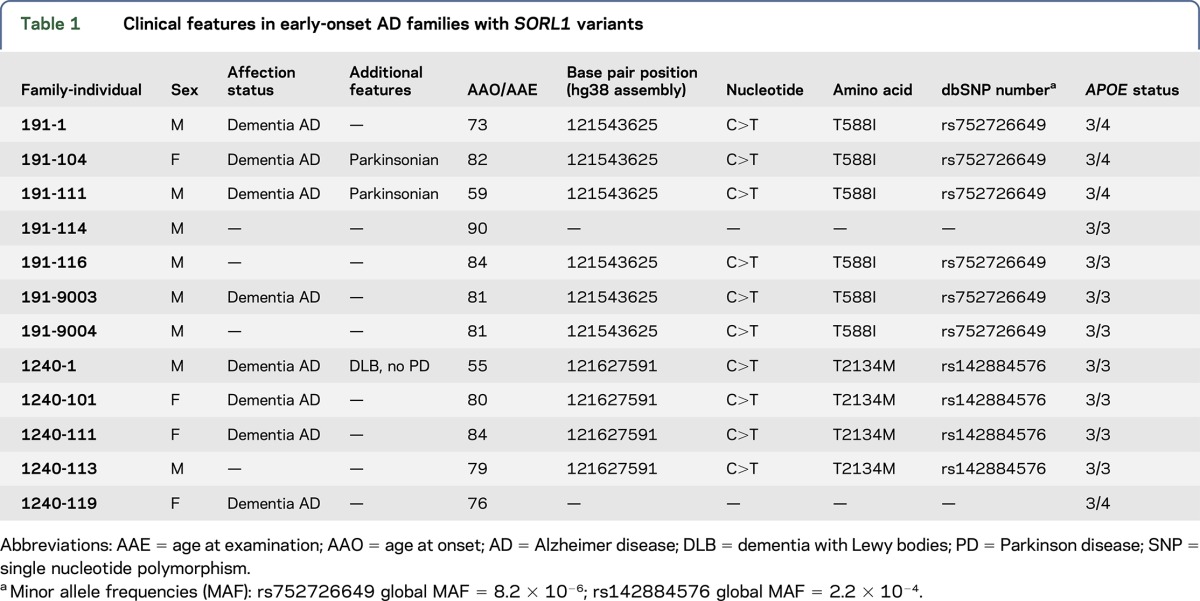
WES identified 10 individuals with SORL1 mutations in 2 unrelated EOAD families (table 1, figure 1). Neuropathology results were available for 1 affected individual. The first family, number 191, has 6 individuals with the predicted damaging SORL1 T588I mutation (rs752726649; C>T); all 4 affected individuals for whom DNA was available were found to carry this variant. These 4 affected individuals had AAOs that ranged from 59 to 82 years. While the progressive cognitive decline of each individual was consistent with dementia, individuals 104 and 111 had also parkinsonian features. Individual 104 began to show cognitive impairment at age 82. On examination, he demonstrated tremor at rest, hypophonia, micrographia, masked facial expression, smaller steps on gait, and overall bradykinesia. Chart review indicated that these symptoms were levodopa/carbidopa responsive. Imaging revealed white matter changes and moderate cerebral atrophy, and EEG was remarkable for a loss of alpha waves. Individual 111 had the earliest AAO in the family 191 at age 59, with diminished memory function in all domains, clinically judged to most likely represent EOAD. When seen by research staff at age 70, the individual was noted to exhibit parkinsonian features. This presentation was confounded by several years of treatment with haloperidol, a typical antipsychotic agent that can cause parkinsonian side effects. Two unaffected individuals in family 191 also carried the SORL1 T588I mutation. These individuals were last examined at ages 81 and 84 years, respectively. Individuals 116 and 9004 demonstrated a normal cognitive and physical examination.
Table 1.
Clinical features in early-onset AD families with SORL1 variants
Figure 1. Pedigrees of the early-onset Alzheimer disease families and SORL1 protein diagram.
(A and B) Pedigrees of the early-onset Alzheimer disease (EOAD) families and SORL1 protein diagram. Affected individuals are solid black while those presenting with parkinsonian features are patterned. Below each individual number is either the age at onset (AAO, for affected individuals) or the age at last examination (AAE, for unaffected individuals). For family 191, the SORL1 variant is present in all affected individuals examined. In family 1240, the variant occurs in 3 of 4 cases evaluated. (C) Diagram of SORL1 protein (2214 total amino acids) indicating the location of principal domains and the variants identified in the EOAD families (T588I and T2134M) and the late-onset Alzheimer disease individuals (A528T and T947M). AD = Alzheimer disease; VPS10 = vesicular protein sorting 10 domain; CR = complement type repeat domains; EGF = epidermal growth factor; FNIII = fibronectin type III repeats; TM = transmembrane region; DLB = dementia with Lewy bodies; SORL1 = sortilin-related receptor.
The second family, number 1240 (table 1 and figure 1), contains 3 affected individuals with the SORL1 T2134M mutation (rs142884576; C>T). These 3 affected individuals had AAOs that ranged from 55 to 84 years. While the clinical examinations revealed no motor abnormalities, there was autopsy evidence for Lewy bodies in individual 1, with the earliest AAO in the family at 55 years. Neuropathologic diagnosis of individual 1 was indicative of Braak & Braak stage IV tangles and limbic Lewy bodies. In addition, 1 individual (119) demonstrated progressive cognitive decline consistent with AD without the T2134M SORL1 mutation. This individual had an AAO of 76 years. Finally, there was 1 unaffected individual (113) with this T2134M SORL1 mutation who was last examined at 79 years of age.
Parkinsonian features in patients with LOAD with SORL1 variants.
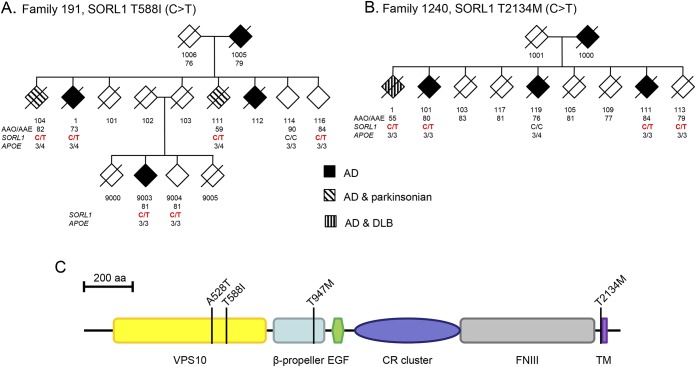
Given the clinical results from these 2 EOAD families, we examined in greater depth the clinical status of previously reported patients with SORL1 changes.11 Review of clinical history and physical examination data identified 4 additional AD individuals, all with LOAD (no neuropathology results were available), and who had evidence of parkinsonian features (table 2). The SORL1 mutations in these 4 individuals were distinct from those identified in the first 2 families. Specifically, 3 individuals which we previously reported carry a common variant at A528T (rs2298813A>G). Clinically, these individuals were diagnosed with both AD and PD and had ages of AD onset ranging from 78 to 84 years. The fourth individual had a different previously reported missense T947M variant (rs143571823, C>T). This individual had a clinical diagnosis of AD and parkinsonism with an age of AD onset at 90 years.
Table 2.
Clinical features in late-onset AD individuals with SORL1 variants
SORL1 variants alter Aβ levels and APP trafficking.
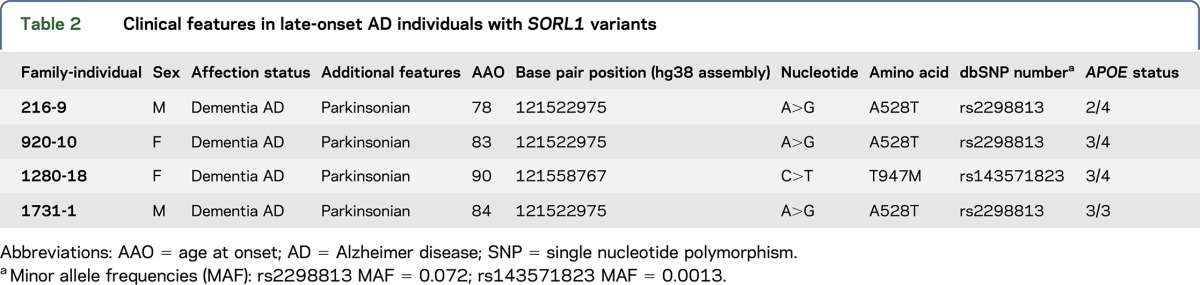
Next, we examined the functional consequences of the SORL1 T588I and T2134M alterations identified in the EOAD families; the variants identified in the LOAD individuals (A528T and T947M) were previously assessed and reported.11 To determine the effects on Aβ production by these SORL1 variants, Aβ42 and Aβ40 levels were measured in conditioned media collected from cultured HEK293 cells expressing equivalent levels of wild-type SORL1 protein, SORL1 T588I, or SORL1 T2134M. Both mutants increased Aβ42 secretion compared with the control (T588I: 113% ± 1.6% and T2134M: 117% ± 5.1%, p < 0.05, figure 2A). Overexpression of SORL1 T2134M also increased Aβ40 secretion (167% ± 9.9%, p < 0.001, figure 2B). While the SORL1 T588I alteration trended toward an increase of Aβ40 secretion in cells, it did not reach statistical significance (131% ± 17.6%, figure 2B).
Figure 2. SORL1 mutants' overexpression increases β-amyloid secretion.
(A–C) Secreted β-amyloid 40 (Aβ40), Aβ42, and amyloid precursor protein soluble β-secretase (APPsβ) were measured from culture medium in stable HEK293 cells expressing the APP Swedish mutant (HEKsw) together with either wild-type SORL1 or mutant SORL1. Error bars represent standard error of the mean (SEM). ***p < 0.001, *p < 0.05, ns, not significant, n = 3 independent replications. (D) Western blot was performed to detect APP soluble α-secretase (APPsα) from cultured media. Bar graphs were normalized to control. **p < 0.01, n = 3 independent replications, and error bars represent the SEM. Aβ = β-amyloid; FL-APP = full-length APP; SORL1 = sortilin-related receptor.
SORL1 has been proposed to modulate the posttranslational biology of APP at several intracellular sites including during transport out of the Golgi and during re-entry and recycling from the cell surface. To examine further the effect of these SORL1 mutants on APP trafficking, we measured APP soluble β-secretase (APPsβ) secretion in a conditioned medium.31 Both mutations caused an increase in APPsβ secretion compared with the wild-type SORL1 (T588I: 132% ± 6.3%, p < 0.05; T2134M: 140% ± 9.4%, p < 0.05, figure 2C). Both mutations also increased production of the soluble α-secretase cleavage product compared with control cells (T588I: 207% ± 15.8%, p < 0.01; T2134M: 223% ± 29.6%, p < 0.05, figure 2D). These observations suggest that in the presence of these SORL1 mutants, APP is neither retained efficiently in the Golgi nor effectively retrieved from the cell surface into recycling pathways. This could result in additional APP lingering at the cell surface. This hypothesis was supported by surface biotinylation experiments which revealed that both SORL1 mutants increased the amount of surface APP compared with the control (T588I: 143% ± 13.1%, p < 0.05; T2134M: 138% ± 7.5%, p < 0.05, figure e-1 at Neurology.org/ng).
SORL1 variants decrease APP binding.
To understand the mechanism by which these SORL1 mutants might alter APP trafficking at the cell surface, we next measured levels of SORL1 protein at the cell surface. The T588I variant showed essentially normal levels of SORL1 both at the cell surface and in total cell lysates (∼87% ± 13.1% of control value, figure e-1). However, while the T2134M mutant showed normal levels of total cellular SORL, there were decreased amounts of surface SORL1 (∼25%, p < 0.05, figure e-1B).
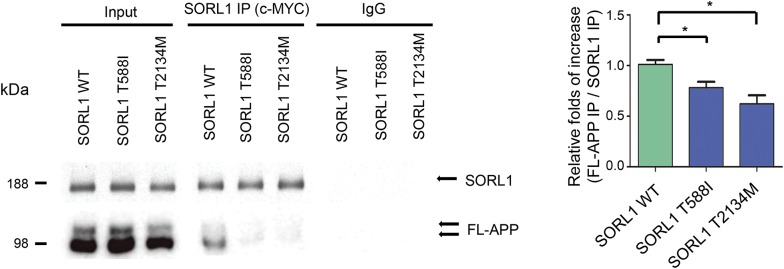
Previous work by us and others have demonstrated that SORL1 directly binds APP and regulates its sorting into secretory, endocytic, or recycling pathways.3,4,11,27,32–38 To assess whether the SORL1 T588I and T2134M mutations might alter the binding affinity of SORL1 to APP, we immunoprecipitated SORL1 from whole cell lysates using an anti-myc antibody directed to the myc epitope on the exogenous SORL1 protein. This strategy circumvents possible risk that the SORL1 mutants might alter binding affinity of anti-SORL1 antibodies, or that endogenous SORL1 might be pulled down in addition, to overexpressed SORL1 in the mutant APPsw cell lines. We then measured the amount of FL-APP that co-immunoprecipitated with the myc-tagged SORL1 proteins and expressed the binding as a normalized ratio of the abundance of coprecipitated FL-APP relative to the abundance of immunoprecipitated SORL1. Both mutations caused reductions in APP binding (T588I: ∼77.1% ± 5.8%, p < 0.05; T2134M: ∼61.5% ± 8.3%, p < 0.05, figure 3).
Figure 3. Both SORL1 mutants have a reduced binding affinity to APP.
SORL1 was immunoprecipitated from cell lysates with a c-MYC antibody, and the amount of coprecipitated full-length amyloid precursor protein (FL-APP) was measured by densitometry of the anti-APP immunoreactive bands on the Western blot of the SORL1 immunoprecipitation products. *p < 0.05, **p < 0.01, n = 3 replications, and error bars represent the SEM. IgG = immunoglobulin G; IP = immunoprecipitated; SORL1 = sortilin-related receptor.
DISCUSSION
In this study, we identified SORL1 alterations in EOAD families thus confirming previously reported studies showing a role for SORL1 in risk for EOAD. Furthermore, we presented functional evidence that these SORL1 alterations are pathogenic.
Evidence for functional consequences of SORL1 mutations is scant. However, the evidence shown here suggests that the variants identified in the EOAD families, SORL1 T588I and T2134, weaken the interaction of SORL1 with FL-APP. This can culminate in excessive APP accumulating at the cell surface either due to failure of the mutant SORL1 to slow trafficking of APP to the cell surface39 or failure of mutant SORL1 to retrieve FL-APP into the retromer-recycling endosome pathway.3,4,11,27,32–38 Our result agrees with prior work which suggests that some SORL1 mutants cause reduced trafficking of the mutant SORL1 protein from the endoplasmic reticulum (ER)/Golgi network to the cell surface.11 The resulting misdirection of more APP into the late endosome pathway exposes the APP to β-secretase and γ-secretase cleavage, with the consequent increase in Aβ production, especially Aβ42. Intriguingly, but consistent with prior work, our data suggest that the molecular mechanisms underlying this common overall effect differ between the 2 variants. Thus, the T2134M mutant, which is located close to the transmembrane domain (figure 1), appears to disrupt trafficking of SORL1 to the cell surface, presumably due to its removal from the ER-Golgi secretory pathway by the ER quality control systems which remove misfolded proteins. In contrast, the T588I mutant survives the ER quality control mechanisms, but appears to be less efficient than wild-type SORL1 in binding to APP. The molecular mechanism for the reduced binding of T588I is unclear, but may relate to subtle changes in the fold of the extracellular domain of SORL1 such that putative APP-binding sites in VPS10 and/or in complement type repeat domains.39,40 Crucially, while they may have different underlying molecular mechanisms, the net effect of both mutations is the same.
A secondary finding in our study was the observation of additional clinical features beyond AD among select individuals with SORL1 alterations. These clinical findings, based on extensive clinical reviews, included clinical Parkinson-related features and neuropathology-proven Lewy bodies without clinical parkinsonism. While these findings point to a potential association between SORL1 alterations and a broader spectrum of neurodegenerative disorders, it is important to note that these clinical features were not present in all individuals with SORL1 alterations and may simply represent features of coincidental sporadic PD.
The results from this study demonstrate that select SORL1 variants present in EOAD and LOAD alter Aβ levels and interfere with APP trafficking. In addition, we observed parkinsonian features among some EOAD/LOAD individuals with SORL1 alterations. These clinical findings should be viewed cautiously but suggest the need for exploration of the additional phenotype consequences of SORL1 alterations beyond dementia.
Supplementary Material
ACKNOWLEDGMENT
The authors are grateful to the families and staff who participated in this study. In addition, they acknowledge Larry Deon Adams for his assistance in data management and Anthony Griswold for his help in bioinformatics.
GLOSSARY
- AAO
age at onset
- Aβ
β-amyloid
- AD
Alzheimer disease
- APP
amyloid precursor protein
- APPsβ
APP soluble β-secretase
- APPsw
Swedish APP mutant
- EOAD
early-onset Alzheimer disease
- ER
endoplasmic reticulum
- FL-APP
full-length APP
- HIHG
John P. Hussman Institute for Human Genomics
- LOAD
late-onset Alzheimer disease
- PD
Parkinson disease
- SORL1
sortilin-related receptor
- WES
whole-exome sequencing
Footnotes
Supplemental data at Neurology.org/ng
AUTHOR CONTRIBUTIONS
M.L.C., R.M.C., B.W.K., and M.A.P.-V. conceived and designed the experiments. M.L.C., R.M.C., R.M., and M.A.P.-V. acquired and assessed participants. P.L.W. and H.N.C. performed custom capture sequencing and exome sequencing. B.W.K., H.N.C., and M.A.P.-V. analyzed the sequencing data. B.W.K. and M.A.P.-V. performed the statistical analysis. Y.Z., C.B., and P.S.G.-H. cloned the SORL1 variants, performed all assays, and analyzed all resulting data. M.L.C. and R.M.C. drafted the manuscript. M.L.C., R.M.C., Y.Z., C.B., B.W.K., H.N.C., P.S.G.-H., and M.A.P.-V. edited the manuscript. The authors jointly discussed the experimental results over the course of the study. All authors read and approved the final manuscript.
STUDY FUNDING
This research was supported by grants from the National Institutes of Health (R01 AG027944 and R01 AG028786 to M.A.P.-V.; R01 AG019085 to J.L.H.; P20 MD000546 and R01 AG28786-01A1 to G.S.B.; U01-AG032984 and RC2-AG036528 to G.D.S., U24 AG026395, U24 AG026390, R01AG037212, R37 AG015473, and R01AG015473 to R.M.; R01 AG009029 to L.A.F.; U01-AG016976 to W.A.K.; U24-AG021886 to T.M.F.; R01AG009956 and RC2 AG036650 to K.H.; UO1 AG06781 and UO1 HG004610 to E.L.; 5R01AG20688 to M.D.F.; P50 AG005133 and AG030653 to I.K.; R01 AG1101, R01 AG030146, and RC2 AG036650 to D.A.E.; P30AG10161, R01AG15819, R01AG30146, R01AG17917, and R01AG15819 to D.A.B.; R01AG028786 to J.J.M.; R01AG22018 and P30AG10161 to L.L.B.; P50AG16574, R01 032990, and KL2 RR024151 to N.E.T. and N.R.G.R.; AG005138 to J.D.B.; P50 AG05681, P01 AG03991, and P01 AG026276 to A.M.G.; U24-AG041689 to L.S.W.; and U19 AG047133 and UF1AG047133); the Department of Defense (W81XWH-12-1-0013 to M.A.P.-V.); a joint grant from the Alzheimer's Association (SG-14-312644) and the Fidelity Biosciences Research Initiative to M.A.P.-V.; and the BrightFocus Foundation (A2011048 to M.A.P.-V.).
DISCLOSURE
Dr. Cuccaro has served on the editorial board of Child Psychiatry & Human Development. Dr. Carney, Dr. Zhang, Dr. Bohm, and Dr. Kunkle report no disclosures. Dr. Vardarajan has served on the scientific advisory board of, been a consultant of, and received research support from the Immuneering Corporation. Ms. Whitehead reports no disclosures. Dr. Cukier has served on the scientific advisory board of the General Direction for Scientific Research and Health Innovation (Italian Ministry of Health); and has received research support from NARSAD, the Hussman Foundation, and the Alzheimer's Association. Dr. Mayeux has received research support from the following grants: RO1 AG037212 Epidemiology of Biomarkers of Risk and Progression in Late onset Alzheimer's Disease; and RF1 AG015473 Genetic Studies of Alzheimer's Disease in Caribbean Hispanics. Dr. St. George-Hyslop has served on the scientific advisory board of the University of Lille (basic AD program) and has received research support from the Canadian Institutes of Health, the Medical Research Council, and Wellcome Trust. Dr. Pericak-Vance reports no disclosures. Go to Neurology.org/ng for full disclosures.
REFERENCES
- 1.Karch CM, Goate AM. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 2015;77:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer's Association. 2015 Alzheimer's disease facts and figures. Alzheimers Dement 2015;11:332–384. [DOI] [PubMed] [Google Scholar]
- 3.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 2007;39:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Offe K, Dodson SE, Shoemaker JT, et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci 2006;26:1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Barral S, Reitz C. The neuronal sortilin-related receptor gene SORL1 and late-onset Alzheimer's disease. Curr Neurol Neurosci Rep 2008;8:384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng Y, Lee JH, Cheng R, St George-Hyslop P, Mayeux R, Farrer LA. Association between SORL1 and Alzheimer's disease in a genome-wide study. Neuroreport 2007;18:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolsch H, Jessen F, Wiltfang J, et al. Association of SORL1 gene variants with Alzheimer's disease. Brain Res 2009;1264:1–6. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology 2008;70:887–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin RH, Yu JT, Tan L. The role of SORL1 in Alzheimer's disease. Mol Neurobiol 2015;51:909–918. [DOI] [PubMed] [Google Scholar]
- 10.Reitz C, Cheng R, Rogaeva E, et al. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch Neurol 2011;68:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vardarajan BN, Zhang Y, Lee JH, et al. Coding mutations in SORL1 and Alzheimer disease. Ann Neurol 2015;77:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pottier C, Hannequin D, Coutant S, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry 2012;17:875–879. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas G, Charbonnier C, Wallon D, et al. SORL1 rare variants: a major risk factor for familial early-onset Alzheimer's disease. Mol Psychiatry 2016;21:831–836. [DOI] [PubMed] [Google Scholar]
- 14.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carney RM, Slifer MA, Lin PI, et al. Longitudinal follow-up of late-onset Alzheimer disease families. Am J Med Genet B Neuropsychiatr Genet 2008;147B:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology 1984;34:939–944. [DOI] [PubMed] [Google Scholar]
- 17.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association Workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011;7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 19.Teng EL, Chui HC. The modified mini-mental state (3MS) examination. J Clin Psychiatry 1987;48:314–318. [PubMed] [Google Scholar]
- 20.Vardarajan BN, Faber KM, Bird TD, et al. Age-specific incidence rates for dementia and Alzheimer disease in NIA-LOAD/NCRAD and EFIGA families: National Institute on Aging Genetics Initiative for Late-Onset Alzheimer Disease/National Cell Repository for Alzheimer Disease (NIA-LOAD/NCRAD) and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA). JAMA Neurol 2014;71:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raney BJ, Dreszer TR, Barber GP, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC genome browser. Bioinformatics 2014;30:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reitz C, Tokuhiro S, Clark LN, et al. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer's disease risk. Ann Neurol 2011;69:47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohm C, Seibel NM, Henkel B, Steiner H, Haass C, Hampe W. SorLA signaling by regulated intramembrane proteolysis. J Biol Chem 2006;281:14547–14553. [DOI] [PubMed] [Google Scholar]
- 29.Hasegawa H, Sanjo N, Chen F, et al. Both the sequence and length of the C terminus of PEN-2 are critical for intermolecular interactions and function of presenilin complexes. J Biol Chem 2004;279:46455–46463. [DOI] [PubMed] [Google Scholar]
- 30.Yu G, Nishimura M, Arawaka S, et al. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature 2000;407:48–54. [DOI] [PubMed] [Google Scholar]
- 31.Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999;286:735–741. [DOI] [PubMed] [Google Scholar]
- 32.Jacobsen L, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem 2001;276:22788–22796. [DOI] [PubMed] [Google Scholar]
- 33.Andersen OM, Reiche J, Schmidt V, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci USA 2005;102:13461–13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodson SE, Andersen OM, Karmali V, et al. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer's disease. J Neurosci 2008;28:12877–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermey G. Targeting amyloid precursor protein. Ann Neurol 2011;69:8–10. [DOI] [PubMed] [Google Scholar]
- 36.Fjorback AW, Seaman M, Gustafsen C, et al. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci 2012;32:1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane RF, Steele JW, Cai D, Ehrlich ME, Attie AD, Gandy S. Protein sorting motifs in the cytoplasmic tail of SorCS1 control generation of Alzheimer's amyloid-beta peptide. J Neurosci 2013;33:7099–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt V, Sporbert A, Rohe M, et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem 2007;282:32956–32964. [DOI] [PubMed] [Google Scholar]
- 39.Mehmedbasic A, Christensen SK, Nilsson J, et al. SorLA complement-type repeat domains protect the amyloid precursor protein against processing. J Biol Chem 2015;290:3359–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gustafsen C, Glerup S, Pallesen LT, et al. Sortilin and SorLA display distinct roles in processing and trafficking of amyloid precursor protein. J Neurosci 2013;33:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.