Abstract
The capacity to integrate transgenes into the host cell genome makes retroviral vectors an interesting tool for gene therapy. Although stable insertion resulted in successful correction of several monogenic disorders, it also accounts for insertional mutagenesis, a major setback in otherwise successful clinical gene therapy trials due to leukemia development in a subset of treated patients. Despite improvements in vector design, their use is still not risk-free. Lentiviral vector (LV) integration is directed into active transcription units by LEDGF/p75, a host-cell protein co-opted by the viral integrase. We engineered LEDGF/p75-based hybrid tethers in an effort to elicit a more random integration pattern to increase biosafety, and potentially reduce proto-oncogene activation. We therefore truncated LEDGF/p75 by deleting the N-terminal chromatin-reading PWWP-domain, and replaced this domain with alternative pan-chromatin binding peptides. Expression of these LEDGF-hybrids in LEDGF-depleted cells efficiently rescued LV transduction and resulted in LV integrations that distributed more randomly throughout the host-cell genome. In addition, when considering safe harbor criteria, LV integration sites for these LEDGF-hybrids distributed more safely compared to LEDGF/p75-mediated integration in wild-type cells. This approach should be broadly applicable to introduce therapeutic or suicide genes for cell therapy, such as patient-specific iPS cells.
Introduction
The capacity to integrate transgenes into the host cell genome makes retroviral vectors (RV) an interesting tool for gene therapeutic applications as stable insertion of transgenes into the genome ensures long-term expression. Use of RV-mediated gene transfer resulted in successful cure of several monogenic, primary immunodeficiency disorders [1–3]. Yet, stable insertion occasionally altered endogenous gene regulation resulting in insertional mutagenesis. Due to this major setback 5 out of 19 treated patients developed leukemia in otherwise successful clinical gene therapy trials for X-SCID and 2 out of 2 patients treated for X-CGD acquired myelodysplastic syndrome [3–6]. Both trials employed murine leukemia virus (MLV)-based gammaretroviral vectors (γRV) that integrate in close proximity to gene regulatory regions [7–9] and resulted in transcriptional deregulation due to up-regulated LMO2 expression [10–13]. Similar reports on insertional mutagenesis were published after integration of γRV near CCDN2, BMI1 and EVI1 [14,15]. Despite improvements in vector design (e.g. self-inactivating (SIN) vectors) their use is still not risk-free [3,4,6,14–16], which shifted attention from yRV towards HIV-derived lentiviral vectors (LV). Even though LV display a more favorable integration pattern, induction of aberrant splicing [17,18] and insertional mutagenesis remain a major concern, as clonal expansion was observed in a gene therapy trial for β-thalassemia [19]. In addition, two recent independent studies revealed clonal expansion in HIV-1 infected patients on antiretroviral therapy due to HIV-1 virus triggered insertional mutagenesis [20,21]. Retroviral integration is a non-random process which is, depending on the viral genus, associated with specific chromatin marks and genomic features [22–24]. yRV predominantly integrate in the vicinity of gene regulatory regions, whereas LV preferably target the body of active transcription units [10,25]. Integration is catalyzed by the viral integrase (IN), whereas integration site choice bias is attributed to the cellular chromatin readers that are co-opted by the viral IN. Whereas the bromodomain and extra-terminal domain (BET) family of proteins (BRD2, 3 and 4) guide MLV integration [26–28], LV integration is directed by Lens epithelium-derived growth factor p75 (LEDGF/p75) [29,30]. Both function as molecular tethers in the cell, combining a chromatin-binding and a protein-interacting region (reviewed in [31]). For LEDGF/p75 (Fig 1A), the chromatin-binding part contains an N-terminal Pro-Trp-Trp-Pro (PWWP) epigenetic reader domain (aa 1–93), recognizing H3K36me3 chromatin marks [32–36], and a set of DNA-binding motifs (Fig 1A, [37,38]). Together, these elements allow LEDGF/p75 to explore the chromatin in a dynamic scan-and-lock fashion [39]. Even though its cellular role is not fully understood, it is clear that LEDGF/p75 acts as a molecular hub for a variety of endogenous proteins next to the lentiviral integrase (Fig 1A) [40,41],[42],[43],[44]. All these proteins, including the lentiviral integrase, bind the C-terminal Integrase-Binding Domain (IBD, aa 347–429; Fig 1A) of LEDGF/p75. We and others showed that replacement of the N-terminal LEDGF/p75 DNA-binding region (aa 1–325) with alternative DNA-binding domains retargets LV integration towards genomic loci bound by these domains [35,45–47]. Fusion of the heterochromatin binding Chromobox protein homolog 1 (CBX1) to the IN-binding C-terminal end of LEDGF/p75 shifted LV integration into the cognate H3K9mex-marked chromatin environment, pericentric heterochromatin and intergenic regions [46]. Despite integration in regions enriched in epigenetic marks associated with gene silencing, transgene expression remained efficient and resulted in successful phenotypic correction in a cell model for X-CGD [48].
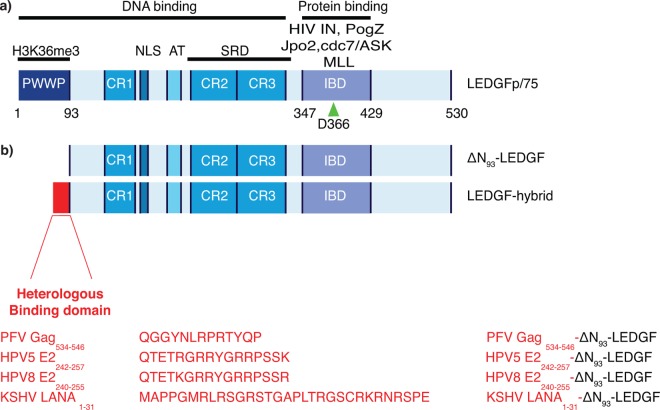
Fig 1. Schematic representation of the LEDGF/p75 domain structure and artificial LEDGF-hybrids.
(a) LEDGF/p75 contains a C-terminal protein-binding domain, coined Integrase Binding Domain (IBD) responsible for HIV-IN interaction. Several endogenous proteins like Jpo2, PogZ and MLL bind to the same interface. At its N-terminal end carries multiple chromatin interacting domains, the PWWP domain, the AT hook-like domain (AT) and three charged regions (CR1, 2, 3). D366 is a pivotal amino acid involved in HIV-IN interaction (arrowhead). Mutation to Asn (D366N) abolishes HIV-IN interaction. The lower panel (b) depicts the different LEDGF-hybrids, PFV Gag534-546-ΔN93-LEDGF, HPV5 E2242-257-ΔN93-LEDGF, HPV8 E2240-255-ΔN93-LEDGF and LANA1-31-ΔN93-LEDGF respectively. Numbers indicate the different amino acid residues. AT, AT-Hook; CR, Charged Region; SRD, Supercoiled Recognition Domein; IBD, Integrase Binding Domain; PWWP, Pro-Trp-Trp-Pro Domain; PFV, Prototype foamy virus; LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; LEDGF, Lens epithelium-derived growth factor; NLS, Nuclear localization signal.
Here we aimed at developing a LEDGF-based tether that results in a more random integration pattern to reduce the overall risk of insertional mutagenesis [11,49–51]. First, we truncated LEDGF/p75 by deleting the N-terminal chromatin-reading PWWP domain that binds H3K36me3 marks directing LEDGF/p75 into the body of active transcription units (Fig 1A and 1B). In addition, we replaced the PWWP-domain with three alternative viral protein domains and motifs, described in literature as pan-chromatin recognition peptides since they bind cellular chromatin without sequence specificity (Fig 1B; Fig 2). Several viruses reside as an episomal DNA genome in host cells, and evolved strategies to persist during mitosis through defined chromatin binding motifs. The spumavirus, Prototype Foamy Virus (PFV), contains a 13-amino acid motif in the group-specific antigen (Gag) binding the H2A/H2B core nucleosome [52–54]. Likewise, the Kaposi Sarcoma-associated Herpes Virus (KSHV) genome is tethered to the nucleosomal core via a chromatin binding sequence (CBS) at the N-terminal end of the latency-associated nuclear antigen protein (LANA) [55]. Finally, in the Beta-Papillomaviruses (PV) a conserved motif in the E2 hinge promotes binding to chromatin and mitotic chromosomes of the invaded cell [56–58]. Following the generation of stable cells lines, we monitored LV integration preferences and evaluated integration sites based on safe harbor region criteria [59] and determined a genotoxicity profile.
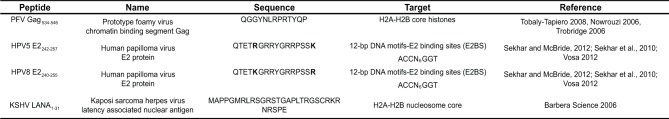
Fig 2. Peptide characteristics.
Fig showing the acronyms, aa-sequences and binding characteristics of the peptides used to generate the artificial LEDGF tethers. PFV, Prototype foamy virus; HPV, Human papilloma virus; KSHV, Kaposi’s sarcoma herpes virus; LANA, Latency associated nuclear antigen.
Materials and Methods
Generation of stable cell lines
SIV-based vector transfer plasmids (pGAE) were a kind gift of D. Nègre (Laboratoire de Vectorologie Rétrovirale et Thérapie Génique, INSERM U412, IFR 74, Ecole Normale Supérieure de Lyon, Lyon, France). A lentiviral vector carrying CMV promoter driving a Zeocin resistance gene and a LEDGF specific miRNA-based shRNA was described earlier [60] and used to generate stable LEDGFKD cells. All LEDGF/p75 hybrid expression constructs were cloned into the pGAE backbone and cloning steps sequence verified.
➢ Cloning of ΔN93-LEDGF and ΔN93-LEDGFD366N controls for the LEDGF ΔN93-LEDGF fusions
pGAE_SFFV_ZnF4_ ΔN93_BC_I_BsdR_WPRE cl.9 and pGAE_SFFV_ZnF4_ ΔN93 BC_D366N_I_BsdR_WPRE cl.3 were digested using BglII & XhoI. Ligation of the synthetic adaptor Ad_BglIIKO_AgeI_kozak generated pGAE_SFFV_ ΔN93_BC_I_BsdR_WPRE cl. E and pGAE_SFFV_ ΔN93_BC_D366N_I_BsdR_WPRE cl. 5. We further refer to the controls as ΔN93-LEDGF and ΔN93–LEDGFD366N respectively.
➢ Cloning of LEDGF ΔN93-LEDGF and ΔN93-LEDGFD366N hybrids
pGAE_SFFV_ZnF4_ ΔN93_BC_I_BsdR_WPRE cl.9 and pGAE_SFFV_ZnF4_ ΔN93 BC_D366N_I_BsdR_WPRE cl.3 were digested using BglII & XhoI. Ligation of the synthetic adaptors (for adaptor sequences see S1 Fig) LANA31, PFVCBS13, HPV5E2_16 and HPV8E_216 generated
pGAE_SFFV_LANA1-31_ ΔN93_BC_I_BsdR_WPRE cl.A9
pGAE_SFFV_LANA1-31_ ΔN93_BC_I_BsdR_WPRE cl.H
pGAE_SFFV_ PFVCBS13_ ΔN93_BC_I_BsdR_WPRE cl.13
pGAE_SFFV_ PFVCBS13_ ΔN93_BC_I_BsdR_WPRE cl.4
pGAE_SFFV_ HPV5E2_16 _ ΔN93_BC_I_BsdR_WPRE cl.19
pGAE_SFFV_ HPV5E2_16 _ ΔN93_BC_I_BsdR_WPRE cl.8
pGAE_SFFV_ HPV8E2_16 _ ΔN93_BC_I_BsdR_WPRE cl.23
pGAE_SFFV_ HPV8E2_16 _ ΔN93_BC_I_BsdR_WPRE cl.11
All cloning steps were confirmed by restriction digest and sequencing.
Cell culture
All cells were grown in a humidified atmosphere containing 5% CO2 at 37°C. HeLaP4 310 LEDGF/p75 depleted cells ([46], further referred to as LEDGFKD cells) were grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-BRL, Merelbeke, Belgium) supplemented with 5% v/v heat inactivated fetal calf serum (FCS; Sigma-Aldrich, Bornem, Belgium), 0.005% w/v gentamicin (GIBCO), 0.05% w/v geneticin (GIBCO) and 0.01% w/v zeocin (Life Technologies, Ghent, Belgium). These cells are monoclonal LEDGFKD cells, derived from HeLaP4 cells (gift from P. Charneau, Institut Pasteur, Paris, France). HelaP4 cells were grown on DMEM (GIBCO) supplemented with 5% v/v heat inactivated fetal calf serum (FCS; Sigma-Aldrich), 0.005% w/v gentamicin (GIBCO) & 0.05% w/v geneticin (GIBCO). HEK 293T cells (gift from O. Danos, Evry, France) were cultured in DMEM medium (GIBCO) with 8% v/v heat inactivated FCS (Sigma-Aldrich) and 0.005% w/v gentamicin (GIBCO). SupT1 cells were cultured in Roswell Park Memorial Institutes medium (RPMI, GIBCO-BRL, Merelbeke, Belgium) supplemented with 10% v/v heat inactivated fetal calf serum FCS (Sigma-Aldrich, Bornem,Belgium) and 0.005% w/v gentamicin (GIBCO). Nalm pre-B cells were cultured in RPMI (GIBCO) with 10% v/v heat inactivated FCS (Sigma-Aldrich) and 0.005% w/v gentamicin (GIBCO).
Retroviral vector production (SIV-based) and transduction
Lentiviral vector production was performed as described earlier [61]. Briefly, for the generation of vesicular stomatitis virus glycoprotein (VSV-G) pseudo-typed SIV-based lentiviral vectors, HEK 293T cells were transfected with the packaging plasmid specific for SIV (pAd_SIV3+; gift from D. Nègre, Lyon, France), the envelope plasmid encoding VSV-G (pLP-VSVG #646 B, from Invitrogen) and respective transfer plasmids, using polyethylenimine (PEI; Polysciences, Amsterdam, The Netherlands). After collecting the supernatant, the medium was filtered using a 0.45 μm filter (Corning Inc., Seneffe, Belgium) and concentrated using a Vivaspin 15 50,000 MW column (Vivascience, Bornem, Belgium). The vector containing concentrate was then aliquoted per 50 μl and stored at -80°C. Stable cell lines expressing a LEDGF hybrid were generated by transduction of polyclonal LEDGF/p75KD cells with SIV-based vectors and subsequent selection with 0,0003% w/v blasticidin (Invitrogen). For lentiviral transduction experiments (LV eGFP T2A fLuc) cells were transduced ON. 72 hours post-transduction cells were harvested when 90% confluent and used for eGFP FACS-analysis or luciferase activity. The remainder of the transduced cells was further cultivated for at least 20 days to eliminate non-integrated DNA and submitted for integration site sequencing.
Immunocytochemistry and Laser scanning microscopy
Cells were transfected using Lipofectamine 2000 (Life Technologies, Merelbeke, Belgium) as described earlier [62]. LEDGF-hybrids were detected with the primary polyclonal rabbit anti-LEDGF480-530 antibody (A300-848a; 1/500; Bethyl Laboratories-Imtec Diagnostics N.V., Antwerpen, Belgium) and secondary polyclonal goat anti-rabbit antibody (1/500 in PBS, goat-αRb488; Bethyl Laboratories-Imtec Diagnostics N.V., Antwerpen, Belgium). Confocal images were acquired using an LSM 510 META imaging unit (Carl Zeiss, Zaventem, Belgium). Alexa-488 was excited at 488 nm (AI laser), mRFP at 543 (HeNe laser) and DAPI at 790 nm (Spectra-physics Mai Tai laser; Spectra Physics, Mountain View CA). After the main beam splitter (HFT KP 700/543 for mRFP, HFT UV/488/543/633 for eGFP, and HFT KP650 for DAPI) a secondary dichroic beam splitter was used to divide the fluorescence signal (NFT 490 for eGFP, NFT 545 for mRFP). Distinct signals were directed to different detectors and data analysis was performed with the LSM image browser. Overlay images were obtained using ImageJ freeware.
Western Blot
Protein concentration of 1% SDS (AppliChem, Leuven, Belgium) protein extracts sheared with a 27 G needle (Terumo, Leuven, Belgium) was determined using a bicinchoninic acid (BCA) protein assay (Pierce, Aalst, Belgium). Proteins were separated on a 12.5% w/v SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (PVDF; BioRad) using an XCell SureLock electrophoresis system (Invitrogen). LEDGF-hybrids were detected using 1/2.000 polyclonal rabbit anti-LEDGF480–530 antibody (A300-848a; Bethyl Laboratories-Imtec Diagnostics N.V., Antwerpen, Belgium) and 1/5 000 secondary antibody (polyclonal goat anti-rabbit antibody coupled with horse radish peroxidase (HRP); Dako). Chemiluminescence was measured using a ECL plus western blotting detection kit (Amersham Biosciences, Roosendaal, The Netherlands). Equal loading was verified with a primary monoclonal antibody directed to α-tubulin (mouse, 1/10 000, 1 h at room temperature; T5168, Sigma-Aldrich) and secondary antibody in blocking buffer (1/10 000, polyclonal goat-anti mouse labelled with HRP; Dako). Visualization was done by chemiluminescence (Pierce ECL Western Blotting Substrate, Thermo scientific).
Luciferase assay
Cells were transduced with LV eGFP T2A fLuc and lysed with 70 μl of lysis buffer (50 mmol/l Tris pH 7.5, 200 mmol/l NaCl, 0.2% NP40, 10% glycerol). FLuc activity was determined using the ONE-glo luciferase assay system according to the manufacturers protocol (Promega, Leiden, The Netherlands) and normalized to the total protein concentration in order to correct for differences in metabolic state. The total protein concentration was measured in parallel using a bicinchoninic acid (BCA) protein assay (Pierce, Aalst, Belgium).
Flow cytometric analysis
Cells were transduced with LV eGFP T2A fLuc and harvested when 95% confluent. eGFP/YFP fluorescence was monitored by Flow cytometric analysis (FACS, Fluorescence activated cell sorting) using a FACSCalibur flow cytometer (BD Biosciences, Erembodegem, Belgium). Data analysis was performed with the CellQuest Pro software (BD Biosciences, Erembodegem-Aalst, Belgium). The percentage of eGFP positive cells (% of gated cells) multiplied by the mean fluorescence intensity (MFI) is further referred to as overall transduction efficiency.
Integration site amplification and sequencing
Transduced HeLaP4 cells were further cultivated for at least 20 days to eliminate non-integrated DNA. Cells were harvested when ca. 90% confluent. Genomic DNA was extracted using the GenElute Mammalian Genomic DNA miniprep kit (Sigma-Aldrich). Integration sites were amplified by linker-mediated PCR as described previously [30]. Genomic DNA was digested using MseI and linkers were ligated (S1 Fig). Proviral-host junctions were amplified by nested PCR using Barcoded primers, generating 454 libraries. This enabled pooling of PCR products into one sequencing reaction. Products were gel-purified and sequenced using 454/Roche pyrosequencing (Titanium technology, Roche) on the 454 GS-FLX-instrument at the University of Pennsylvania. Reads were filtered based on perfect match to the LTR linker, Barcode and flanking LTR. All sites were mapped to the human genome requiring a perfect match within 3bp of the LTR end. Three random control sites were computationally generated and matched with respect to the distance to the nearest MseI Cleavage site for each experimental site (matched random control, MRC). A more detailed explanation can be found in the supplementary guidelines of [63]. Normalization of experimental HIV-derived lentiviral vector sites to those of the MRC sites functions as a control for recovery bias due to cleavage by restriction enzymes. Analysis was performed as described previously and genomic heat maps generated using the INSIPID software (Bushman Lab, University of Pennsylvania). [30]. A detailed guide to interpret the heat maps presented can be found in [63]. The computation of DNase I site density was based on a table of DNase I sites obtained from [64]. Datasets used in the safe harbor analysis were retrieved from ENSEMBLE and/or UCSC (TxDB knownGenes, miRNA biotype, UCR; hg19) using BioMART [65]. The Allonco-list was used for oncogenes as published in [66]
Results
Generation of LEDGF-hybrids and stable cell lines
In an effort to distribute lentiviral vector integration more randomly over the genome, we modified LEDGF/p75, the cellular tether of the HIV Pre-Integration complex (PIC), by deleting the chromatin-reading PWWP-domain (ΔN93-LEDGF, Fig 1A and 1B) [32–34] relying on the remaining non-specific DNA-interacting regions in LEDGF93-325, such as the AT-hook domains and the CRs (Fig 1A; [39]). In addition, we generated LEDGF-hybrids where the PWWP-domain was exchanged with a set of alternative pan-chromatin recognition peptides of viral origin (Fig 1B, and Fig 2). Prototype Foamy Virus chromatin binding segment of Gag534-546 (PFV Gag534-546) [52–54], Human Papilloma Virus serotype 5 E2242-257, Human Papilloma Virus serotype 8 E2240-255 (HPV5 E2242-257 and HPV8 E2240-255, respectively)[56–58] and Kaposi’s Sarcoma Herpes Virus Latency Associated Nuclear Antigen1-31 (KSHV LANA1-31) [55] were used to replace the PWWP domain, generating ΔN93-LEDGF, PFV Gag534-546-ΔN93-LEDGF, HPV5 E2242-257-ΔN93-LEDGF, HPV8 E2240-255-ΔN93-LEDGF and KSHV LANA1-31-ΔN93-LEDGF fusions, respectively. All above-mentioned LEDGF-hybrids were used to complement LEDGF/p75-depleted cells (HeLaP4 (LEDGFKD) [60] and Nalm (LEDGFKO) cells [67]) employing SIV-based lentiviral vectors. As a positive control, cells were complemented with WT LEDGF/p75 (referred to as LEDGF/p75 back complementation (LEDGFBC)). In order to control for non-specific effects resulting from the expression of the fusion proteins we also generated stable cell lines expressing the respective chimeras carrying a D366N mutation in the LEDGF/p75 part, which abrogates the interaction with lentiviral integrase (IN) [68]. Protein integrity was corroborated by western blot analysis, with all LEDGF-hybrids migrating at the predicted molecular weights (S2 Fig). Of note, protein levels of PFV Gag534-546-ΔN93-LEDGF were lower in all experiments. Viability and growth rates of all cell lines were comparable to the parental HeLaP4 cells (data not shown).
LEDGF-hybrids locate to the nucleus and display a distinct subnuclear distribution
In a first step, we evaluated the subcellular localization of the truncated ΔN93-LEDGF and the respective ΔN93-LEDGF-hybrids by immunocytochemistry (Fig 3). Complementation of LEDGF-depleted HeLaP4 cells (LEDGFKD) with LEDGFBC resulted in a typical pattern of dense, fine speckles in the nucleoplasm excluded from the nucleoli during interphase (Fig 3C), phenocopying the endogenous LEDGF/p75 pattern (Fig 3A), which is in line with earlier reports [46]. Contrary, LEDGF/p75 lacking the chromatin-reading PWWP-domain exhibited a more diffuse nuclear distribution and located to the nucleoli as well (ΔN93-LEDGF, Fig 3D). In addition, all ΔN93-LEDGF peptide-fusions located to the nucleus (Fig 3E–3H), displaying a unique sub-nuclear distribution: the PFV Gag534-546- and the KSHV LANA1-31-fusion to ΔN93-LEDGF showed a punctate appearance in the nucleus and were excluded from nucleoli (Fig 3E and 3H), contrary to both HPV5 E2242-257- and HPV8 E2240-255-ΔN93-LEDGF fusions that were enriched in the nucleoli (Fig 3F and 3G). Similar subcellular distributions were observed for the respective cognate LEDGFD366N-hybrids (data not shown).
Fig 3. Subcellular localization of LEDGF/p75-hybrids in interphase cells.
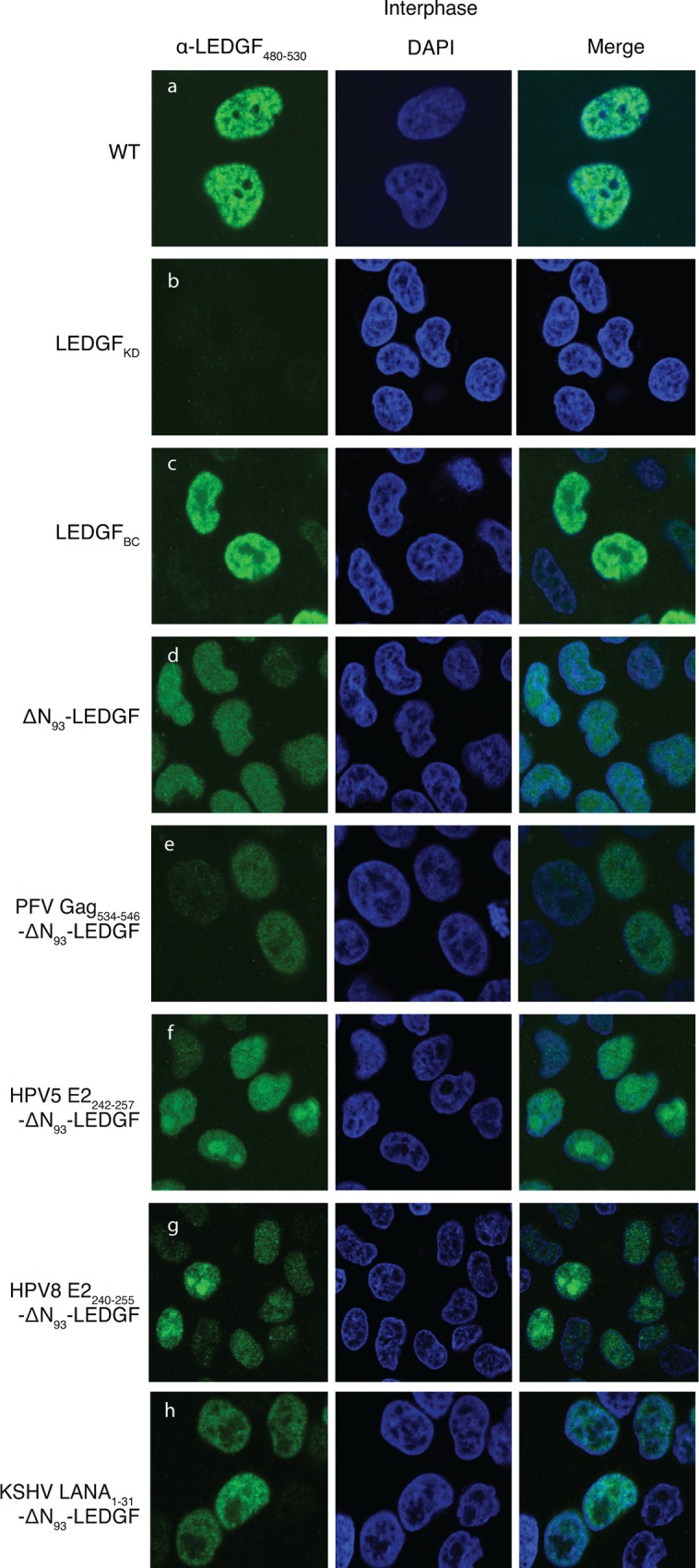
LEDGF/p75 depleted cell lines were complemented with the respective LEDGF-fusions. Laser scanning confocal images of HelaP4 cells, stained using an Ab recognizing LEDGF480-530, are shown in green. Nuclei were stained using Dapi (shown in blue). A merge of green and blue fluorescence is shown. Data depicted are representative for the respective cell lines. PFV, Prototype foamy virus; LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; LEDGF, Lens epithelium-derived growth factor; DAPI, 4',6-Diamidino-2-Phenylindole.
LEDGF-peptide fusions rescue lentiviral vector transduction
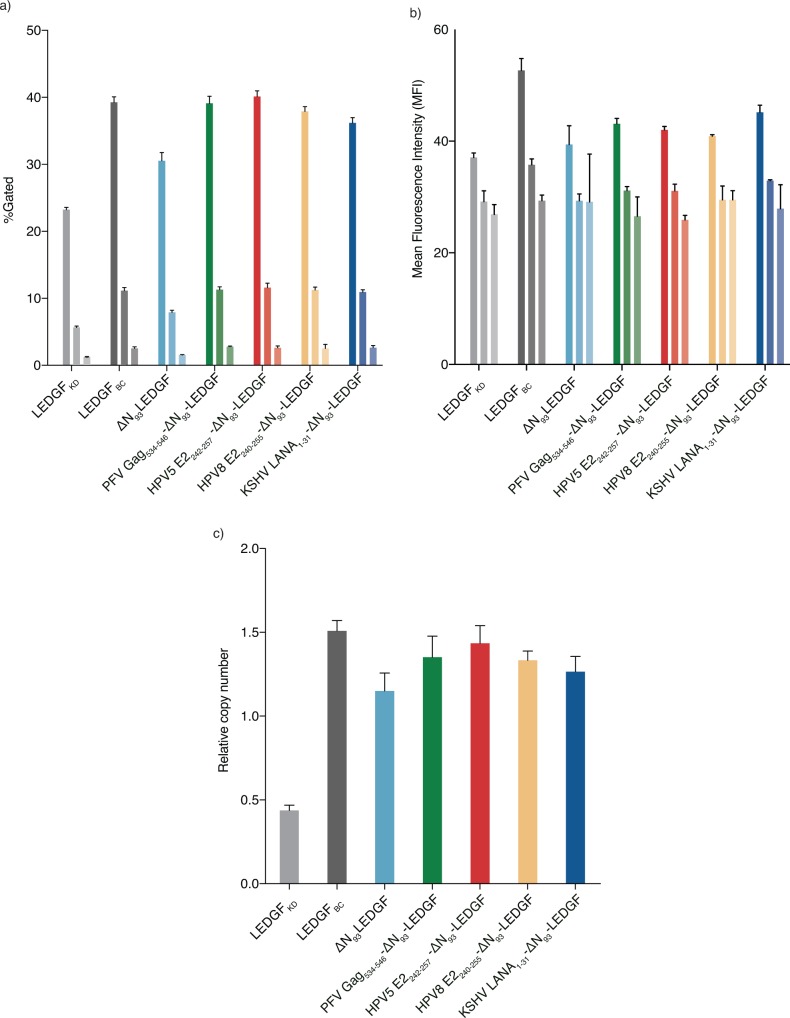
Next, we assessed whether the ΔN93-LEDGF-hybrids supported lentiviral vector transduction by complementing LEDGF-depleted cells (LEDGFKD) and employing wild-type LEDGF/p75 complemented cells (LEDGFBC) as control. The respective HeLaP4 cell lines were challenged with a dilution series of a lentiviral vector (multiplicity of infection (MOI) = 1, 0.2 or 0.04 (indicated in lighter colors)) encoding enhanced green fluorescent protein (eGFP) and firefly luciferase (fLuc) reporters [61]. Transduction efficiencies were determined by flow cytometry monitoring eGFP fluorescence (Fig 4A and 4B, showing transduction efficiency (eGFP positive cells; %Gated) and Mean Fluorescence Intensity (MFI), respectively). Complementation of LEDGF-depleted cells with LEDGFBC restored transduction efficiency (Fig 4A) (***, p<0.005; two-tailed t-test relative to LEDGFKD), in line with earlier reports [46,69]. Complementation of LEDGFKD cells with ΔN93-LEDGF, lacking the chromatin interacting PWWP-domain, partially rescued lentiviral transduction (78% compared to LEDGFBC) (Fig 4A, ***, p<0.005 compared to KD, two-tailed t-test). Addition of chromatin binding peptides to replace the PWWP domain displayed a significantly improved transduction relative toΔN93-LEDGF (***, p<0.005; two-tailed t-test), reaching efficiencies comparable to LEDGFBC (Fig 4A). Similar results were obtained for different vector dilutions (Fig 4A) or when assessing complemented LEDGF/p75 knock-out cells (Nalm-/-, data not shown) [67] or when evaluating fLuc as a reporter (data not shown). Looking at Mean Fluorescence intensities all LEDGF-peptide fusions were about 20% lower than LEDGFBC (Fig 4B). In addition to transduction efficiencies, we also determined the number of integrated copies (Fig 4C). Reintroduction of LEDGF/p75 (LEDGFBC) significantly improved vector integration (±-3.5-fold compared to LEDGFKD (***, p<0.005, two-tailed t-test).
Fig 4. Rescue of lentiviral vector transduction by artificial LEDGF-hybrids.
LEDGF-fusions were evaluated for their ability to support lentiviral vector transduction. LEDGF-depleted HelaP4-based cell lines stably complemented with LEDGF-hybrids were challenged with a VSV-G pseudo-typed lentiviral reporter vector encoding enhanced Green Fluorescent Protein (eGFP). Fluorescence was measured by fluorescence activated cell sorting and the different variables plotted:. (a) Percentage eGFP positive cells (transduction efficiency) and (b) Mean Fluorescence Intensity (MFI). Data are compiled for a representative experiment and depict averages of 3 replicates for 3 different vector dilutions (mean ± SD). (c) Lentiviral integrated proviral copies were determined by Q-PCR analysis on genomic DNA extracts of cells transduced with an MOI = 1. Data are represent the mean of 3 replicates ± SD. Statistical significance is calculated using a two-tailed t-test relative to LEDGFKD or ΔN93-LEDGF. PFV, Prototype foamy virus; LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; LEDGF, Lens epithelium-derived growth factor.
Likewise, ΔN93-LEDGF and all LEDGF-peptide fusions restored vector integration (2.5-fold more compared to LEDGFKD, ***, p<0.005, two-tailed t-test), albeit still to a lesser extent (reaching 68–74% of LEDGFBC, Fig 4B). The increased transduction efficiencies (%Gated) closely correlate with an increase in integrated viral vector copies (Fig 4C). Complementation with ΔN93-LEDGF alone, lacking any additional chromatin-tether, resulted in lower integrated copy numbers than ΔN93-LEDGF fused to chromatin engaging peptides (p-values<0.005, two-tailed t-test relative to ΔN93-LEDGF; 66.7% of LEDGFBC), supporting the notion that the chromatin-reading PWWP domain of LEDGF/p75 is not an absolute requirement for efficient integration.
LEDGF-peptide fusions efficiently redistribute lentiviral integration
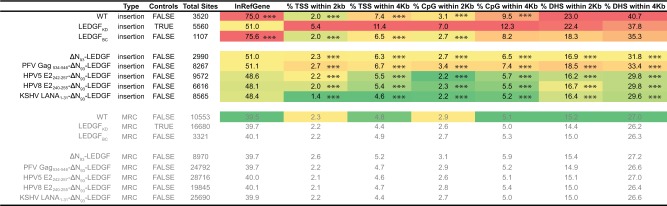
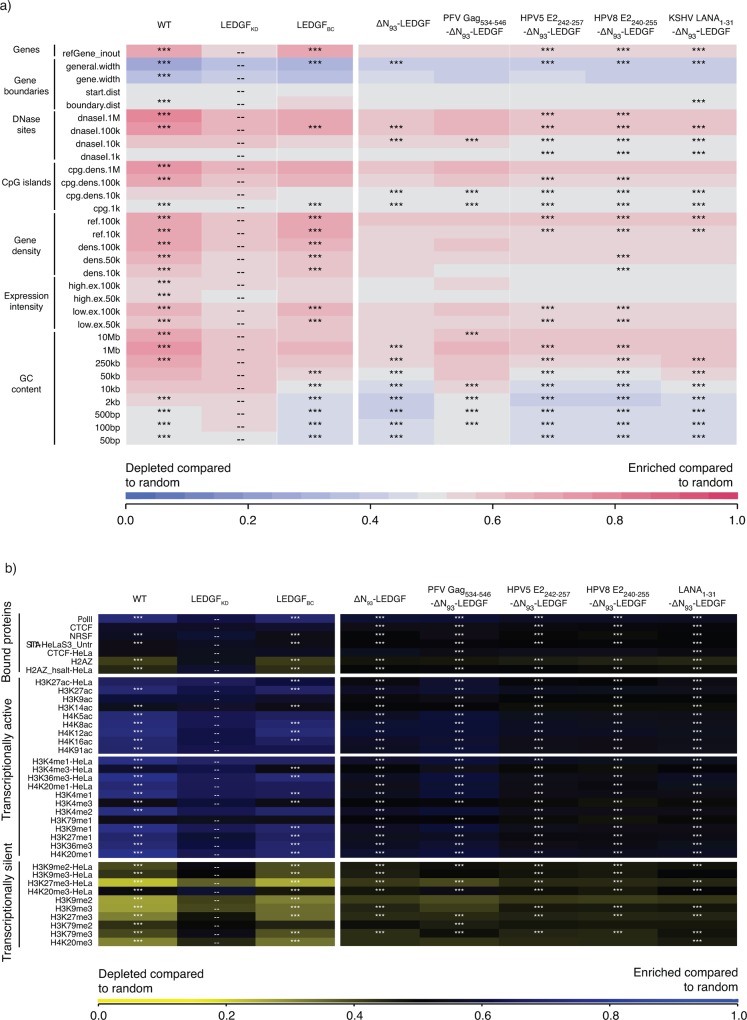
After showing that complementation of LEDGF/p75-depleted cells with ΔN93-LEDGF or any of the LEDGF-hybrids rescued vector integration, we determined the integration profiles in the respective cell lines. HIV-based viral vector integration sites were amplified and sequenced as described earlier [30,46], yielding a total of 62670 unique integration sites and their computationally generated matched random control (MRC) sites. Note that SIV-based viral vectors were used to complement LEDGF/p75-depleted cells, in order to avoid interference with the HIV-based viral vector integration site amplification and analysis. First, we analysed integration relative to a set of defined genomic features (Fig 5, Fig 6). Lentiviral vector integration in wild-type HeLaP4 cells (endogenous LEDGF/p75, Fig 6) is traditionally enriched in the body of transcription units (75.0% in RefSeq genes; Fig 5) but disfavoured transcription start sites (TSS) and promoter regions (2.0% within 2kb of the 5’ of a RefSeq gene and 3.1% within 2kb of a CpG island) [10,25]. LEDGF-depletion results in a more random integration site distribution, characterized by reduced integration into genes (51.0% in RefSeq genes) and increased integration close to TSS (5.4%) and CpG islands (7.0%), in line with previous work [30,46,70]. This phenotype was fully reverted upon LEDGF/p75 complementation (LEDGFBC; 75.6% in RefSeq genes). Comparable data were obtained for larger window sizes (only 2kb and 4kb are shown in Fig 5). Integration site distributions in cells expressing the respective LEDGFD366N mutants were not different from LEDGFKD cells (n = 16473; data not shown). Interestingly, the mere ablation of the PWWP domain (ΔN93-LEDGF) resulted in an overall more random distribution compared to LEDGFKD cells, with decreased integration near retrovirus-specific features like gene bodies, TSS and promoter regions (*** p<0.001; χ2 test compared to LEDGFKD; Fig 5). Complementation of LEDGF-depleted cells with LEDGF-peptide fusions resulted in a comparable more randomized distribution (*** p<0.001; χ2 test compared to LEDGFKD; Fig 5). In a more elaborate analysis, we analysed global integration preferences and included a wide selection of genomic features, depicted as a genomic heatmap (Fig 6), comparing integration site data sets obtained from HeLaP4 LEDGFKD cells to those of cells complemented with the respective LEDGF-hybrids. Tile color depicts the correlation for an integration dataset with the respective genomic feature (left) relative to matched random controls, as indicated by the colored receiver operating characteristic (ROC) curve area scale at the bottom of the panel. LEDGF/p75 depletion shifts integration out of transcriptionally active regions wich is reverted upon complementation with LEDGF/p75 (compare LEDGFKD and LEDGFBC; shown in Fig 6), in line with previous data [30,46,70]. Cells complemented with ΔN93-LEDGF displayed an more randomly distributed integration profile, with tiles overall coloring less red or blue compared to LEDGFKD, integrating less near DNase sensitive regions, CpG-islands and GC-rich regions compared to LEDGFKD (*** p<0.001, Wald statistics). Introduction of the heterologous HPV E2 and LANA1-31-peptide fragments to replace the PWWP-domain resulted in a ΔN93-LEDGF-like integration profile when compared to LEDGFKD (p<0.001), whereas integration for PFV Gag534-546-ΔN93-LEDGF was less random. When displaying statistics relative to ΔN93-LEDGF (S3A Fig) integration frequencies near these genomic features is not significantly different between ΔN93-LEDGF and the respective ΔN93-LEDGF peptide-fusions, except for PFV Gag534-546-ΔN93-LEDGF (Fig 6). The reproducibility of the data observed for HPV5 E2242-257-ΔN93-LEDGF and HPV8 E2240-255-ΔN93-LEDGF complemented cell lines and the pronounced redistribution towards more random relative to LEDGFBC (S3B Fig) underscores the effectiveness of LEDGF-based artificial tethers for retargeting of LV integration. Next to integration relative to genomic features, we also analyzed integration site densities near epigenetic features (Fig 6B). The epigenetic heat map displays yellow and blue tiles, with blue tiles indicating that integration frequency is enriched near these marks relative to MRC, whereas yellow tiles indicate that integration is disfavored compared to MRC. A near random distribution would result in a black tile. As reported previously, lentiviral integration correlates with histone marks associated with open and transcriptionally active chromatin (H3K4 mono-, di- and tri methylation, H3K14 and H4 acetylation, as well as acetylation and monomethylation of H3K9/K27/K79, H4K20 and H2BK5, …): [8] while disfavoring integration in transcriptionally silent regions or heterochromatin (H3K27me3, H3K9me3 or H4K20me3 and H3K79, respectively): [8] (WT; Fig 6). Depletion of LEDGF/p75 (LEDGFKD) resulted in a more random distribution (with tiles displaying a less pronounced blue or yellow color, and shifting towards black). This tendency was more outspoken for ΔN93-LEDGF and the HPV E2 and LANA1-31-peptide fusions compared to LEDGFKD (Fig 6B), ΔN93-LEDGF (S4A Fig) or LEDGFBC (S4B Fig), potentially because integration in LEDGF-depleted cells, at least in part, is tethered by HRP-2 [67].
Fig 5. Integration frequency near genomic features.
Fig showing the percentage HIV-derived lentiviral vector integration sites relative to features specific for integration viral vectors such as integration into the body of genes (Refseq genes, InRefGene), integration within 2kb-4kb windows near Transcription Start Sites (X5-end of genes, TSS), midpoint of CpG islands or DNase I-hypersensitive sites (DHS), counted in both the 5’ and 3’ direction. Dataset details are described in the MM section. Asterisks depict a significant deviation from LEDGF KD (two-tailed Chi-square test; ***, p-values <0.001). TSS, Transcription start sites; DHS, DNase I-hypersensitive sites; PFV, Prototype foamy virus; HPV, Human papilloma virus; KSHV, Kaposi’s sarcoma herpes virus; LANA, Latency associated nuclear antigen; MRC, matched random control.
Fig 6. LEDGF-hybrids retarget lentiviral integration towards a more randomized pattern.
(a) Genomic heat map comparing integration site data sets obtained from HeLaP4 LEDGF/p75 KD cells overexpressing different artificial LEDGF-hybrids to genomic features. Tile color depicts the correlation for an integration dataset with the respective genomic feature (left) relative to matched random controls, as indicated by the colored receiver operating characteristic (ROC) curve area scale at the bottom of the panel. Statistical significance (asterisks, ***p<0.001 ranked Wald tests) is shown relative to LEDGF KD population (double dash). Columns indicate different data sets, while rows indicate different genomic features analyzed (described in [63]). LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; PFV, Prototype foamy virus; a; LEDGF, Lens epithelium-derived growth factor. (b) Epigenetic heat map comparing integration site data sets obtained from HeLaP4 LEDGF/p75 KD cells overexpressing different artificial LEDGF-hybrids to epigenetic features. Tile color depicting a positive or negative correlation to the respective epigenetic feature (10kb windows), relative to MRC, as indicated by the receiver operating characteristic (ROC) curve area scale at the bottom of the panel. Statistical significance (asterisks, ***p<0.001, ranked Wald tests) is shown relative to LEDGF KD population (dashed). Columns indicate different data sets while rows indicate different epigenetic features analyzed. Included features were limited to those identified in high-throughput studies HeLaP4 and primary CD4+ T-cells. Detailed information on epigenetic marks and their roles can be found in [87,88]. LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; PFV, Prototype foamy virus; a; LEDGF, Lens epithelium-derived growth factor.
Artificial peptide-LEDGF/p75 hybrids result in a safer integration profile
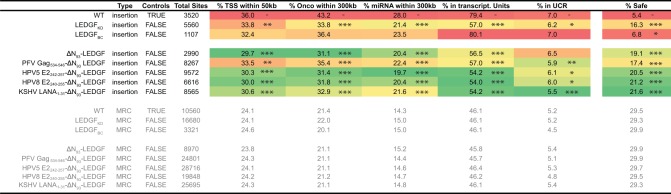
Together, the presented above data indicate that lentiviral vector integration preferences are defined by LEDGF/p75 as a cellular tether, and are mostly dictated by the N-terminal PWWP-domain. The mere deletion of this domain, or replacement with alternative chromatin-interacting modules redistributes vector integration sites in a more random fashion. The question remains whether redistribution of proviral integration sites obtained for our LEDGF-hybrids also translated in a safer therapy, with a lower chance on insertional mutagenesis. In an effort to get a better view on the safety profile, we calculated integration frequencies near a specific set of previously defined criteria [59,66], such as transcription start sites (<50kb), oncogenes (<300kb) or miRNA coding regions (<300kb), transcription units and ultraconserved elements to define potentially unsafe integration events. The large window sizes impose a very stringent selection for lentiviral integration events away from these features, which in turn can thus be considered as more safe [59]. For each data set we evaluated the percentage of unsafe integrations (Fig 7 and S5 Fig) and in addition determined the percentage of safe sites (events not captured in any of the other criteria; Fig 7, % safe). When calculating the percentage in the parental cell line only 5.4% of all LV integration sites may be considered safe. LEDGF/p75-depletion results in a shift to 16.3% safe sites (p-value <0.005, Pearsons Chi-square compared to the LEDGFWT control condition), a phenotype that was fully reverted upon LEDGF/p75BC complementation (5.4%, no significant difference compare to LEDGFWT). Ablation of the N-terminal PWWP-domain again boosted the percentage safe integration events to 19.7% (p-value <0.005, Pearsons Chi-square compared to the LEDGFWT control condition). Addition of heterologous peptide fragments KSHV LANA1-31 and HPV8 E2240-255 to the N-terminal end of LEDGF93-530 slightly increased the % safe integrations relative to ΔN93-LEDGF (S5A Fig) with complementation up to 20.2% for HPV5 E2242-257-ΔN93-LEDGF, 21.2% for HPV8 E2240-255-ΔN93-LEDGF and 21.6% for KSHV LANA1-31-ΔN93-LEDGF when considering these criteria (p-value <0.05, Pearsons Chi-square compared to the ΔN93-LEDGF control condition). Relative to the LEDGFBC condition our LEDGF-chimera increased the percentage of safe sites more than 3 fold (p-value <0.005, Pearsons Chi-square, S5B Fig). Of note, for the MRC conditions, we obtained a maximum of 30% integrations in safe harbors.
Fig 7. Integration frequency near safe harbor criteria.
Fig showing the percentage HIV-derived lentiviral vector integration frequencies near features (TSS, Oncogenes [66], miRNA encoding regions, Transcription units and ultra conserved regions) that, when hit, are considered to be UNsafe as defined in [59] (Dataset details are described in the MM section). As such these features are used to define safe harbors as regions that fall outside these criteria. Percentages depict the fraction of integrations falling within the corresponding range relative to the criteria. The % of integrations negatively associated with these 5 features is used to calculate a safety profile. (*, p-value <0.5; **, p-value <0.05;***, p-value <0.005, Pearsons Chi-square compared to LEDGFWT control). TSS, Transcription start sites; UCR, Ultra conserved regions; PFV, Prototype foamy virus; HPV, Human papilloma virus; KSHV, Kaposi’s sarcoma herpes virus; LANA, Latency associated nuclear antigen; MRC, matched random control.
Discussion
Integration of retroviral vectors into the host cell genome makes them invaluable tools for gene therapeutic applications where life-long correction is key. Previous reports showed effective gene transfer enabling long-term gene correction (For a review see [71]). However, severe adverse events in these clinical studies (using full-LTR driven gamma-retrovirus vectors) raised serious concerns regarding the safety of gene therapy when using integrating vectors (derived from the family of retroviruses) [14,15]. The yRV preference for integration into enhancer regions and concomitant activation of proto-oncogenes led to malignant transformation of cells and clonal expansion [10–12]. Therefore, multiple studies have been triggered to increase the safety of the used retroviral vectors, which include the use of other subtypes (lenti or alpha instead of gammaretroviral), SIN-LTR design [72–75], tissue specific promoters [76], changing integration properties [45–47] and insulator sequences as enhancer and silencer blockers [77]. Meanwhile, lentivirus vectors became the mean of choice when using retroviruses for gene transfer and clinical gene therapy due to their safer integration profile and lower genotoxicity in preclinical models. As such, any successful modification avoiding an increased integration of these vectors into gene coding regions may be relevant for translation into the clinics. Stable integration however will always imply the intrinsic risk of vector-induced genomic perturbation, open reading frame-disruption, leading to loss of function or transcriptional deregulation of neighbouring genes as indicated by the report on SIN-LV affected splicing [78]. In addition, also LV integration may lead to clonal dominance as reported in the beta-thalassemia trial, which could be an indicator of upcoming malignant transformation [19]. Therefore it is important to gain additional mechanistic insights into the molecular mechanism of integration and integration site selection for LVs to be accepted for general therapeutic use. We and others substantially contributed to the elucidation of the role of LEDGF/p75 as a molecular tether of lentiviral vector integration. As a cellular cofactor of lentiviral integration, LEDGF/p75 orchestrates lentiviral integration preference by binding H3K36me3 in the body of active transcription units via its N-terminal PWWP domain, but it is the vector-encoded integrase that catalyzes the integration reaction. Depletion of LEDGF/p75 by knockdown or knockout strategies shifts lentiviral vector integration out of active genes, yet integration is not completely random [67,79], which at least in part can be explained by residual targeting via HRP-2 [67]. Here we set out to study whether different LEDGF-hybrids could be generated to distribute lentiviral integration sites more randomly. This line of vector development is based on the further increasing interest in new vector platforms displaying a close-to-random insertional profile potentially reducing the probability of proto-oncogene activation lowering the genotoxic potential [51,80,81]. In an effort to achieve a more random integration site distribution, we deleted the specific chromatin-binding PWWP module of LEDGF/p75 (aa 1–93), or we replaced it with alternative pan-chromatin binding modules. In case of LEDGF/p75, it is demonstrated that the PWWP domain recognizes H3K36me3, a chromatin mark that is particularly enriched in the body of active transcription units [32–36]. Complementation of LEDGF-depleted cells with a LEDGF/p75-protein that had its PWWP domain deleted (ΔN93-LEDGF) or replaced with alternative chromatin binding modules showed unique subnuclear distributions for each of the constructs, indicating that these deletion of the PWWP domain, or the replacements with any of the other peptides, resulted in a specific redistribution within the nuclear compartment of the artificial LEDGF chimera (Fig 3). The latter phenotype can be attributed to the AT-hook motifs and charged regions present in the N-terminal end of ΔN93-LEDGF, together with the specific peptides that replaced the PWWP domain. After working up integration sites, analysis showed that lentiviral integration preferences for most of the constructs resulted in a more random distribution than under LEDGF depleted conditions (genomic and the epigenetic heat map representations; Fig 6A and 6B), except for PFV Gag534-546-ΔN93-LEDGF. For example, in the latter cells LV integration was still enriched near epigenetic markers for transcriptionally active chromatin, albeit less outspoken than observed with LEDGFWT and LEDGFBC cells (S4B Fig). Interestingly, peptide addition was not required to obtain a more random distribution. Lentiviral integrations in ΔN93-LEDGF expressing cells were redistributed in a fairly random manner, with tile colors shifting to grey and black (for the genomic and the epigenetic heat map representations, respectively) indicating that integration frequencies for these features are not enriched nor depleted compared to the matched random integration site distribution. Comparison with LEDGF KD shows that integration is more randomly distributed than under LEDGF depletion (*** p<0.001, Wald statistics; Fig 6A and 6B). Fusion of short pan-chromatin binding peptides to the truncated ΔN93-LEDGF resulted in similar shifts towards a more randomized integration profile. The fact that all peptide fusions display a unique subnuclear location, suggest that their interaction with chromatin is different. Even though the overall integration frequencies are highly similar (considering the genomic and the epigenetic features analyzed), larger integration site datasets (>10e5 sites) would be required to allow more detailed analysis on the specific subsets. In an effort to estimate the effect of the more randomized distribution on safety, we calculated the frequency of integration relative to a set of safe harbor criteria for the individual integration site datasets [59]. This analysis showed that the more random distributions resulted in a lower genotoxic profile with 18–22% of integrations meeting safe harbor criteria for our LEDGF-chimera compared to only 5.4% for cells carrying wild-type LEDGF/p75, all LEDGF-chimera resulted in a safer distributions over the genome. Fully targeted integration towards safe harbor regions like the AAVS1 or CCR5 locus would be the ultimate solution to circumvent insertional mutagenesis [59,66,82]. Several methods for site-directed gene correction have been developed using genetic scissors based on Zinc-finger nucleases, transcription activator like effector nucleases or more recently RNA-guided nucleases (CRISPR/Cas9) (for a review [83]). However, site directed integration would no doubt impair transduction efficiencies. Our approach improves the therapeutic potential of lentiviral vectors by decreasing the risk/benefit ratio, still supporting high transduction efficiencies. The fact that integration can be directed to genomic regions that are not targeted under wild-type conditions nor LEDGF-depleted conditions, indicates that integration in these areas is disfavored due to the absence of a tether, rather than the presence of specific obstacles such as steric hindrance resulting from the condensed chromatin structure. As an alternative to the generation of stable cell lines as employed here, we demonstrated earlier that mRNA-electroporation ensures timely, high-level recombinant protein expression that is sufficient to retarget lentiviral vector integration [48]. When combined with IN mutant lentiviral vectors that selectively bind complementary LEDGF/p75 variants [84], this approach should be broadly applicable to introduce therapeutic or suicide genes for cell therapy, such as genetic modification of patient-specific iPS cells and improve safety of lentiviral vectors. With the occurrence of potential adverse effects being of multi-factorial nature [85] novel therapeutic approaches should be evaluated in relevant functional assays able to predictively assess the cytotoxicity observed in vivo [86], a continuous effort aiming at abolishing the risk of insertional mutagenesis will be required for gene therapy to become a broadly accepted treatment alternative.
Supporting Information
Fig depicting the different oligos used in this study together with their nucleotide sequence.
(EPS)
LEDGF depleted cell lines were complemented with the respective LEDGF-hybrids. Total cell lysates were prepared and separated on a 12,5% SDS gel. An antibody recognizing LEDGF325-530 was used for detection. β-tubuline detection was used as an equal loading control. WT, Wild type; KD, Knockdown; PFV, Prototype foamy virus; LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; LEDGF, Lens epithelium-derived growth factor.
(EPS)
Genomic heat maps comparing integration site data sets obtained from HeLaP4 LEDGF/p75 KD cells overexpressing different artificial LEDGF-hybrids to genomic features. Tile color depicting the nature of the correlation for an integration dataset with the respective genomic feature (left) relative to matched random controls, as indicated by the colored receiver operating characteristic (ROC) curve area scale at the bottom of the panel. Statistical significance (asterisks, ***p<0.001, ranked Wald tests) is shown relative to (a) ΔN93-LEDGF or (b) LEDGFBC, respectively (double dash). Columns show different data sets while rows indicate different genomic features analyzed (described in [63]). LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; PFV, Prototype foamy virus; LEDGF, Lens epithelium-derived growth factor.
(EPS)
Epigenetic heat map comparing integration site data sets obtained from HeLaP4 LEDGF/p75 depleted cells overexpressing different artificial LEDGF-hybrids to epigenetic features, generated using the INSIPID software (Bushman Lab, University of Pennsylvania). Tile color depicting a positive or negative correlation to the respective epigenetic feature (10 kb windows), relative to matched random controls, as indicated by the receiver operating characteristic (ROC) curve area scale at the bottom of the panel. Statistical significance (asterisks, ***p<0.001; ranked Wald tests) is shown relative to (a) ΔN93-LEDGF or (b) LEDGFBC, respectively (double dash). Significance is reached when p<0.001, compared to MRC. Columns indicate different data sets while rows indicate different epigenetic features analyzed. Included features were limited to those identified in high-throughput studies performed in HeLa and primary CD4+ T-cells. Detailed information on epigenetic marks and their roles can be found in [87,88]. LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; PFV, Prototype foamy virus; a; LEDGF, Lens epithelium-derived growth factor; MRC, matched random control.
(EPS)
Fig showing the percentage HIV-derived lentiviral vector integration frequencies near features (TSS, Oncogenes [66], miRNA encoding regions, Transcription units and ultra conserved regions) that, when hit, are considered to be unsafe as defined in [59] (Dataset details are described in the MM section). As such these features are used to define safe harbors as regions that fall outside these criteria. Percentages depict the fraction of integrations falling within the corresponding range relative to the criteria. The % integrations negatively associated with these 5 features is used to calculate a safety profile. (*, p-value <0.5; **, p-value <0.05;***, p-value <0.005, Pearsons Chi-square compared to (a) ΔN93-LEDGF or (b) LEDGFBC control condition). TSS, Transcription start sites; UCR, Ultra conserved regions; PFV, Prototype foamy virus; HPV, Human papilloma virus; KSHV, Kaposi’s sarcoma herpes virus; LANA, Latency associated nuclear antigen; MRC, matched random control.
(EPS)
Acknowledgments
We are grateful to Jan De Rijck and Dominique Van Looveren for critical reading and to Paulien Van de Velde for her technical assistance. Viral vector production was performed at the Leuven Viral Vector Core. L.V. is a doctoral fellow supported by the Flemish Fund for Scientific Research (FWO; Fonds voor Wetenschappelijk Onderzoek). Research at KU Leuven received financial support from the FWO, the KU Leuven Research Council (OT), the KU Leuven IDO program and the Belgian IAP Belvir (IDO/12/008, ZKB9996 SB/0881057 and FWO ZKC0523, ZKC3378).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
L.V. is a doctoral fellow supported by the Flemish Fund for Scientific Research (FWO; Fonds voor Wetenschappelijk Onderzoek). Research at KU Leuven received financial support from the FWO, the KU Leuven Research Council (OT), the KU Leuven IDO program and the Belgian IAP Belvir (IDO/12/008, ZKB9996 SB/0881057 and FWO ZKC0523, ZKC3378).
References
- 1.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000. April 28;288(5466):669–72. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004. December;364(9452):2181–7. 10.1016/S0140-6736(04)17590-9 [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Kalle Von C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. American Association for the Advancement of Science; 2003. October 17;302(5644):415–9. 10.1126/science.1088547 [DOI] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. American Society for Clinical Investigation; 2008. September;118(9):3132–42. 10.1172/JCI35700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. American Society for Clinical Investigation; 2008. September;118(9):3143–50. 10.1172/JCI35798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nature Medicine. Nature Publishing Group; 2010. February;16(2):198–204. 10.1038/nm.2088 [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. American Association for the Advancement of Science; 2003. June 13;300(5626):1749–51. 10.1126/science.1083413 [DOI] [PubMed] [Google Scholar]
- 8.De Ravin SS, Su L, Theobald N, Choi U, Macpherson JL, Poidinger M, et al. Enhancers are major targets for murine leukemia virus vector integration. Journal of Virology. American Society for Microbiology; 2014. April;88(8):4504–13. 10.1128/JVI.00011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaFave MC, Varshney GK, Gildea DE, Wolfsberg TG, Baxevanis AD, Burgess SM. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Research. Oxford University Press; 2014. April;42(7):4257–69. 10.1093/nar/gkt1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell RS, Beitzel BF, Schroder ARW, Shinn P, Chen H, Berry CC, et al. Retroviral DNA Integration: ASLV, HIV, and MLV Show Distinct Target Site Preferences. Plos Biol. Public Library of Science; 2004;2(8):e234 10.1371/journal.pbio.0020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derse D, Crise B, Li Y, Princler G, Lum N, Stewart C, et al. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. Journal of Virology. American Society for Microbiology; 2007. June;81(12):6731–41. 10.1128/JVI.02752-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. American Society for Clinical Investigation; 2007. August;117(8):2225–32. 10.1172/JCI31659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007. September 15;110(6):1770–8. 10.1182/blood-2007-01-068759 [DOI] [PubMed] [Google Scholar]
- 14.Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Díez IA, Dewey RA, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010. November 11;363(20):1918–27. 10.1056/NEJMoa1003548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, et al. Gene therapy for Wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med. 2014. March 12;6(227):227ra33–3. 10.1126/scitranslmed.3007280 [DOI] [PubMed] [Google Scholar]
- 16.Maetzig T, Galla M, Baum C, Schambach A. Gammaretroviral vectors: biology, technology and application. Molecular Diversity Preservation International; 2011. June;3(6):677–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesana D, Sgualdino J, Rudilosso L, Merella S, Naldini L, Montini E. Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J Clin Invest. American Society for Clinical Investigation; 2012. May;122(5):1667–76. 10.1172/JCI62189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cesana D, Ranzani M, Volpin M, Bartholomae C, Duros C, Artus A, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther. Nature Publishing Group; 2014. April;22(4):774–85. 10.1038/mt.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. Nature Publishing Group; 2010. September 16;467(7313):318–22. 10.1038/nature09328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. American Association for the Advancement of Science; 2014. July 11;345(6193):179–83. 10.1126/science.1254194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. American Association for the Advancement of Science; 2014. August 1;345(6196):570–3. 10.1126/science.1256304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens SW, Griffith JD. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. Journal of Virology. 1996. September;70(9):6459–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA. National Acad Sciences; 2005. April 26;102(17):6103–7. 10.1073/pnas.0501646102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. Journal of Virology. American Society for Microbiology; 2005. April;79(8):5211–4. 10.1128/JVI.79.8.5211-5214.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002. August 23;110(4):521–9. [DOI] [PubMed] [Google Scholar]
- 26.Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci USA. National Acad Sciences; 2013. July 16;110(29):12036–41. 10.1073/pnas.1307157110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Rijck J, de Kogel C, Demeulemeester J, Vets S, Ashkar El S, Malani N, et al. The BET Family of Proteins Targets Moloney Murine Leukemia Virus Integration near Transcription Start Sites. CellReports. The Authors; 2013. November 27;5(4):886–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, et al. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. Journal of Virology. American Society for Microbiology; 2013. December;87(23):12721–36. 10.1128/JVI.01942-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. Journal of Biological Chemistry. American Society for Biochemistry and Molecular Biology; 2003. January 3;278(1):372–81. 10.1074/jbc.M209278200 [DOI] [PubMed] [Google Scholar]
- 30.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, et al. Role of PSIP1/LEDGF/p75 in Lentiviral Infectivity and Integration Targeting. Fox D, editor. PLoS ONE. Public Library of Science; 2007. December 19;2(12):e1340 10.1371/journal.pone.0001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debyser Z, Christ F, De Rijck J, Gijsbers R. Host factors for retroviral integration site selection. Trends Biochem Sci. 2015. February;40(2):108–16. 10.1016/j.tibs.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 32.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Research. Oxford University Press; 2010. October;38(18):6135–47. 10.1093/nar/gkq410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. Reik W, editor. PLoS Genet. Public Library of Science; 2012;8(5):e1002717 10.1371/journal.pgen.1002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Research. Oxford University Press; 2013. April 1;41(6):3924–36. 10.1093/nar/gkt074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gijsbers R, Vets S, De Rijck J, Ocwieja KE, Ronen K, Malani N, et al. Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2011. December 2;286(48):41812–25. 10.1074/jbc.M111.255711 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, et al. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin. BioMed Central Ltd; 2013;6(1):12 10.1186/1756-8935-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Research. Oxford University Press; 2006;34(5):1653–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsutsui KM, Sano K, Hosoya O, Miyamoto T, Tsutsui K. Nuclear protein LEDGF/p75 recognizes supercoiled DNA by a novel DNA-binding domain. Nucleic Acids Research. Oxford University Press; 2011. July;39(12):5067–81. 10.1093/nar/gkr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrix J, Gijsbers R, De Rijck J, Voet A, Hotta J- I, McNeely M, et al. The transcriptional co-activator LEDGF/p75 displays a dynamic scan-and-lock mechanism for chromatin tethering. Nucleic Acids Research. Oxford University Press; 2011. March;39(4):1310–25. 10.1093/nar/gkq933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. Journal of Cell Science. 2006. June 15;119(Pt 12):2563–71. 10.1242/jcs.02995 [DOI] [PubMed] [Google Scholar]
- 41.Bartholomeeusen K, De Rijck J, Busschots K, Desender L, Gijsbers R, Emiliani S, et al. Differential Interaction of HIV-1 Integrase and JPO2 with the C Terminus of LEDGF/p75. Journal of Molecular Biology. 2007. September;372(2):407–21. 10.1016/j.jmb.2007.06.090 [DOI] [PubMed] [Google Scholar]
- 42.Bartholomeeusen K, Christ F, Hendrix J, Rain J-C, Emiliani S, Benarous R, et al. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. Journal of Biological Chemistry. American Society for Biochemistry and Molecular Biology; 2009. April 24;284(17):11467–77. 10.1074/jbc.M807781200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes S, Jenkins V, Dar MJ, Engelman A, Cherepanov P. Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2010. January 1;285(1):541–54. 10.1074/jbc.M109.036491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008. July 8;14(1):36–46. 10.1016/j.ccr.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci USA. National Acad Sciences; 2010. February 16;107(7):3135–40. 10.1073/pnas.0914142107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, et al. LEDGF Hybrids Efficiently Retarget Lentiviral Integration Into Heterochromatin. Molecular Therapy. Nature Publishing Group; 2009. November 13;18(3):552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, et al. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Human Gene Therapy. Mary Ann Liebert, Inc. 140 Huguenot Street, 3rd Floor New Rochelle, NY 10801 USA; 2010. March;21(3):337–49. 10.1089/hum.2009.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vets S, De Rijck J, Brendel C, Grez M, Bushman F, Debyser Z, et al. Transient Expression of an LEDGF/p75 Chimera Retargets Lentivector Integration and Functionally Rescues in a Model for X-CGD. Mol Ther Nucleic Acids. Nature Publishing Group; 2013;2(3):e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatziandreou I, Siapati EK, Vassilopoulos G. Genetic correction of X-linked chronic granulomatous disease with novel foamy virus vectors. Exp Hematol. 2011. June;39(6):643–52. 10.1016/j.exphem.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 50.Kaufmann KB, Brendel C, Suerth JD, Mueller-Kuller U, Chen-Wichmann L, Schwäble J, et al. Alpharetroviral vector-mediated gene therapy for X-CGD: functional correction and lack of aberrant splicing. Mol Ther. Nature Publishing Group; 2013. March;21(3):648–61. 10.1038/mt.2012.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suerth JD, Maetzig T, Brugman MH, Heinz N, Appelt J- U, Kaufmann KB, et al. Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol Ther. Nature Publishing Group; 2012. May;20(5):1022–32. 10.1038/mt.2011.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobaly-Tapiero J, Bittoun P, Lehmann-Che J, Delelis O, Giron M-L, de Thé H, et al. Chromatin Tethering of Incoming Foamy Virus by the Structural Gag Protein. Traffic. Blackwell Publishing Ltd; 2008. October;9(10):1717–27. 10.1111/j.1600-0854.2008.00792.x [DOI] [PubMed] [Google Scholar]
- 53.Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, et al. Genome-wide mapping of foamy virus vector integrations into a human cell line. Journal of General Virology. Society for General Microbiology; 2006. May;87(Pt 5):1339–47. 10.1099/vir.0.81554-0 [DOI] [PubMed] [Google Scholar]
- 54.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem H- P, Kaul R, et al. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. National Acad Sciences; 2006. January 31;103(5):1498–503. 10.1073/pnas.0510046103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, et al. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. American Association for the Advancement of Science; 2006. February 10;311(5762):856–61. 10.1126/science.1120541 [DOI] [PubMed] [Google Scholar]
- 56.Sekhar V, Reed SC, McBride AA. Interaction of the betapapillomavirus E2 tethering protein with mitotic chromosomes. Journal of Virology. American Society for Microbiology; 2010. January;84(1):543–57. 10.1128/JVI.01908-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekhar V, McBride AA. Phosphorylation regulates binding of the human papillomavirus type 8 E2 protein to host chromosomes. Journal of Virology. American Society for Microbiology; 2012. September;86(18):10047–58. 10.1128/JVI.01140-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Võsa L, Sudakov A, Remm M, Ustav M, Kurg R. Identification and analysis of papillomavirus E2 protein binding sites in the human genome. Journal of Virology. American Society for Microbiology; 2012. January;86(1):348–57. 10.1128/JVI.05606-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LMS, et al. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. Nature Publishing Group; 2011. January;29(1):73–8. 10.1038/nbt.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osório L, Gijsbers R, Oliveras-Salvá M, Michiels A, Debyser Z, Van den Haute C, et al. Viral vectors expressing a single microRNA-based short-hairpin RNA result in potent gene silencing in vitro and in vivo. J Biotechnol. 2014. January;169:71–81. 10.1016/j.jbiotec.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 61.Ibrahimi A, Vande Velde G, Reumers V, Toelen J, Thiry I, Vandeputte C, et al. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Human Gene Therapy. Mary Ann Liebert, Inc. 140 Huguenot Street, 3rd Floor New Rochelle, NY 10801 USA; 2009. August;20(8):845–60. 10.1089/hum.2008.188 [DOI] [PubMed] [Google Scholar]
- 62.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, et al. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. Journal of Virology. American Society for Microbiology; 2006. December;80(23):11498–509. 10.1128/JVI.00801-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ocwieja KE, Brady TL, Ronen K, Huegel A, Roth SL, Schaller T, et al. HIV Integration Targeting: A Pathway Involving Transportin-3 and the Nuclear Pore Protein RanBP2. Cullen BR, editor. PLoS Pathog. 2011. March 10;7(3):e1001313–4. 10.1371/journal.ppat.1001313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. Public Library of Science; 2006. June;2(6):e60 10.1371/journal.ppat.0020060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford). Oxford University Press; 2011;2011(0):bar030–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nature Reviews Cancer. Nature Publishing Group; 2011. December 1;12(1):51–8. 10.1038/nrc3179 [DOI] [PubMed] [Google Scholar]
- 67.Schrijvers R, De Rijck J, Demeulemeester J, Adachi N, Vets S, Ronen K, et al. LEDGF/p75-Independent HIV-1 Replication Demonstrates a Role for HRP-2 and Remains Sensitive to Inhibition by LEDGINs. Krausslich H-G, editor. PLoS Pathog. Public Library of Science; 2012. March 1;8(3):e1002558 10.1371/journal.ppat.1002558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cherepanov P, Sun Z-YJ, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005. May 15;12(6):526–32. 10.1038/nsmb937 [DOI] [PubMed] [Google Scholar]
- 69.Vandekerckhove L, Christ F, Van Maele B, De Rijck J, Gijsbers R, Van den Haute C, et al. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. Journal of Virology. American Society for Microbiology; 2006. February;80(4):1886–96. 10.1128/JVI.80.4.1886-1896.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shun M-C, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. Cold Spring Harbor Lab; 2007. July 15;21(14):1767–78. 10.1101/gad.1565107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naldini L. Gene therapy returns to centre stage. Nature. 2015. October 15;526(7573):351–60. 10.1038/nature15818 [DOI] [PubMed] [Google Scholar]
- 72.Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. Nature Publishing Group; 2008. April;16(4):718–25. 10.1038/mt.2008.5 [DOI] [PubMed] [Google Scholar]
- 73.Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. American Society for Clinical Investigation; 2009. April;119(4):964–75. 10.1172/JCI37630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. Nature Publishing Group; 2009. November;17(11):1919–28. 10.1038/mt.2009.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newrzela S, Cornils K, Li Z, Baum C, Brugman MH, Hartmann M, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008. September 15;112(6):2278–86. 10.1182/blood-2007-12-128751 [DOI] [PubMed] [Google Scholar]
- 76.Antoniou MN, Skipper KA, Anakok O. Optimizing retroviral gene expression for effective therapies. Human Gene Therapy. Mary Ann Liebert, Inc. 140 Huguenot Street, 3rd Floor New Rochelle, NY 10801 USA; 2013. April;24(4):363–74. 10.1089/hum.2013.062 [DOI] [PubMed] [Google Scholar]
- 77.Emery DW. The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Human Gene Therapy. Mary Ann Liebert, Inc. 140 Huguenot Street, 3rd Floor New Rochelle, NY 10801 USA; 2011. June;22(6):761–74. 10.1089/hum.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trono D. Gene therapy: too much splice can spoil the dish. J Clin Invest. American Society for Clinical Investigation; 2012. May;122(5):1600–2. 10.1172/JCI63066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nature Medicine. 2005. November 27;11(12):1287–9. 10.1038/nm1329 [DOI] [PubMed] [Google Scholar]
- 80.Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z, et al. Hybrid Lentivirus-transposon Vectors With a Random Integration Profile in Human Cells. Nature. Nature Publishing Group; 2009. November 13;17(7):1205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moldt B, Miskey C, Staunstrup NH, Gogol-Döring A, Bak RO, Sharma N, et al. Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Mol Ther. Nature Publishing Group; 2011. August;19(8):1499–510. 10.1038/mt.2011.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Meth. 2011. August 21;8(10):861–9. [DOI] [PubMed] [Google Scholar]
- 83.Mussolino C, Cathomen T. RNA guides genome engineering. Nature Biotechnology. Nature Publishing Group; 2013. March;:208–9. 10.1038/nbt.2527 [DOI] [PubMed] [Google Scholar]
- 84.Wang H, Shun M-C, Li X, Di Nunzio F, Hare S, Cherepanov P, et al. Efficient Transduction of LEDGF/p75 Mutant Cells by Gain-of-Function HIV-1 Integrase Mutant Viruses. Mol Ther Methods Clin Dev. Nature Publishing Group; 2014. January 8;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. Nature Publishing Group; 2009. September;17(9):1537–47. 10.1038/mt.2009.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rothe M, Schambach A, Biasco L. Safety of gene therapy: new insights to a puzzling case. Curr Gene Ther. 2014;14(6):429–36. [DOI] [PubMed] [Google Scholar]
- 87.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007. May 18;129(4):823–37. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 88.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007. November 5;14(11):1025–40. 10.1038/nsmb1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig depicting the different oligos used in this study together with their nucleotide sequence.
(EPS)
LEDGF depleted cell lines were complemented with the respective LEDGF-hybrids. Total cell lysates were prepared and separated on a 12,5% SDS gel. An antibody recognizing LEDGF325-530 was used for detection. β-tubuline detection was used as an equal loading control. WT, Wild type; KD, Knockdown; PFV, Prototype foamy virus; LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; LEDGF, Lens epithelium-derived growth factor.
(EPS)
Genomic heat maps comparing integration site data sets obtained from HeLaP4 LEDGF/p75 KD cells overexpressing different artificial LEDGF-hybrids to genomic features. Tile color depicting the nature of the correlation for an integration dataset with the respective genomic feature (left) relative to matched random controls, as indicated by the colored receiver operating characteristic (ROC) curve area scale at the bottom of the panel. Statistical significance (asterisks, ***p<0.001, ranked Wald tests) is shown relative to (a) ΔN93-LEDGF or (b) LEDGFBC, respectively (double dash). Columns show different data sets while rows indicate different genomic features analyzed (described in [63]). LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; PFV, Prototype foamy virus; LEDGF, Lens epithelium-derived growth factor.
(EPS)
Epigenetic heat map comparing integration site data sets obtained from HeLaP4 LEDGF/p75 depleted cells overexpressing different artificial LEDGF-hybrids to epigenetic features, generated using the INSIPID software (Bushman Lab, University of Pennsylvania). Tile color depicting a positive or negative correlation to the respective epigenetic feature (10 kb windows), relative to matched random controls, as indicated by the receiver operating characteristic (ROC) curve area scale at the bottom of the panel. Statistical significance (asterisks, ***p<0.001; ranked Wald tests) is shown relative to (a) ΔN93-LEDGF or (b) LEDGFBC, respectively (double dash). Significance is reached when p<0.001, compared to MRC. Columns indicate different data sets while rows indicate different epigenetic features analyzed. Included features were limited to those identified in high-throughput studies performed in HeLa and primary CD4+ T-cells. Detailed information on epigenetic marks and their roles can be found in [87,88]. LANA, Latency associated nuclear antigen; HPV, Human papilloma virus; PFV, Prototype foamy virus; a; LEDGF, Lens epithelium-derived growth factor; MRC, matched random control.
(EPS)
Fig showing the percentage HIV-derived lentiviral vector integration frequencies near features (TSS, Oncogenes [66], miRNA encoding regions, Transcription units and ultra conserved regions) that, when hit, are considered to be unsafe as defined in [59] (Dataset details are described in the MM section). As such these features are used to define safe harbors as regions that fall outside these criteria. Percentages depict the fraction of integrations falling within the corresponding range relative to the criteria. The % integrations negatively associated with these 5 features is used to calculate a safety profile. (*, p-value <0.5; **, p-value <0.05;***, p-value <0.005, Pearsons Chi-square compared to (a) ΔN93-LEDGF or (b) LEDGFBC control condition). TSS, Transcription start sites; UCR, Ultra conserved regions; PFV, Prototype foamy virus; HPV, Human papilloma virus; KSHV, Kaposi’s sarcoma herpes virus; LANA, Latency associated nuclear antigen; MRC, matched random control.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.