Abstract
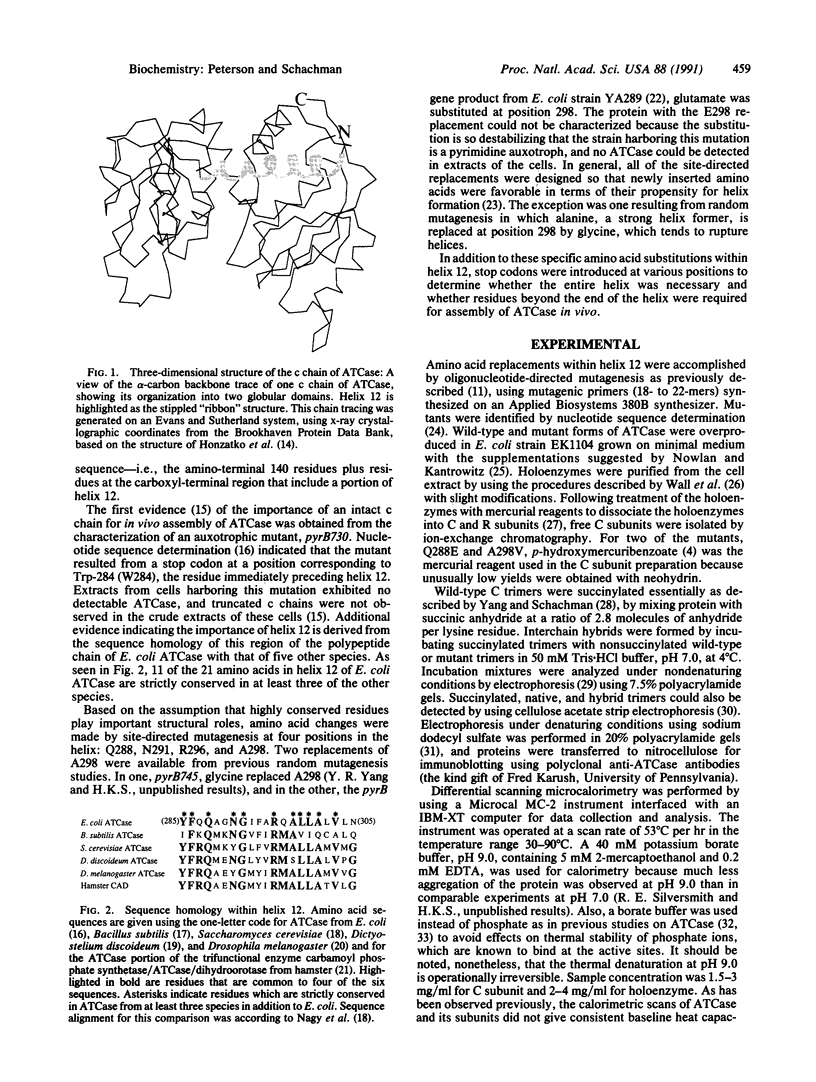
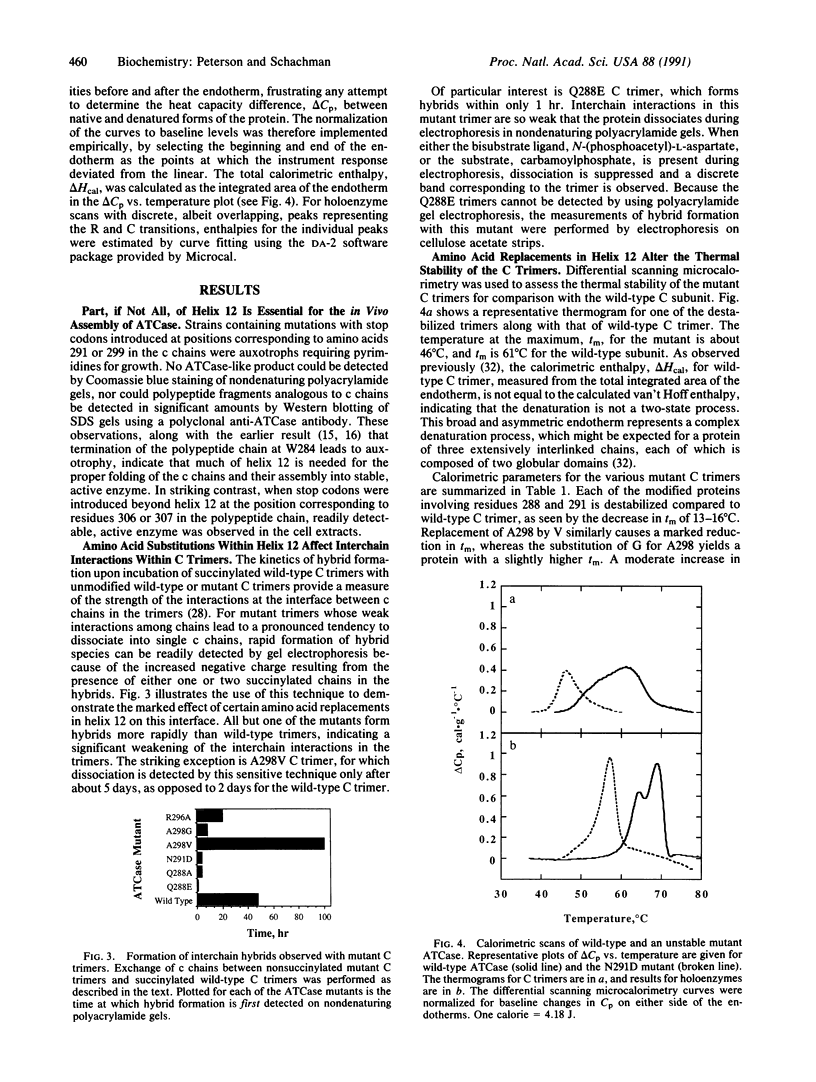
The six individual catalytic polypeptide chains within the two catalytic trimers of Escherichia coli aspartate transcarbamoylase (ATCase; EC 2.1.3.2) are folded into two discrete structural domains interconnected in part by helix 12, which comprises residues 285-305 and is located near the carboxyl terminus of the chain. The essential role of this helix in folding of the chains and their assembly into ATCase was demonstrated by introducing a stop codon at the position corresponding to amino acid 284, 291, or 299. Cells containing these mutations are pyrimidine auxotrophs lacking ATCase-like protein in cell extracts. In contrast, stable active enzyme is formed from chains truncated at position 306 or 307, showing that all 310 amino acids are not required for assembly. Replacements of Gln-288, Asn-291, Arg-296, and Ala-298 were introduced to assess the effect of alterations within helix 12 on protein stability. Stability of the trimers was measured both by differential scanning microcalorimetry and by the rate of exchange of chains at 4 degrees C when mutant trimers were incubated with succinylated wild-type trimers. Melting temperatures of the mutant trimers spanned a range of more than 20 degrees C, with a few higher and others lower than that of wild-type trimers. Large changes in interchain interaction energies were observed for the trimers, but there was no direct correlation between the ease of dissociation of the trimers and their thermal stability. Calorimetry on the mutant holoenzymes revealed alterations in the interactions between trimers and regulatory subunits within the intact enzymes. The striking changes in stability of both trimers and holoenzymes demonstrated that effects of relatively localized amino acid replacements in helix 12 are manifested by indirect global alterations propagated throughout the structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allewell N. M. Escherichia coli aspartate transcarbamoylase: structure, energetics, and catalytic and regulatory mechanisms. Annu Rev Biophys Biophys Chem. 1989;18:71–92. doi: 10.1146/annurev.bb.18.060189.000443. [DOI] [PubMed] [Google Scholar]
- Blackburn M. N., Schachman H. K. Allosteric regulation of aspartate transcarbamoylase. Effect of active site ligands on the reactivity of sulfhydryl groups of the regulatory subunits. Biochemistry. 1977 Nov 15;16(23):5084–5091. doi: 10.1021/bi00642a022. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Hu C. Q., Lin L. N., Mos M. T. A simple model for proteins with interacting domains. Applications to scanning calorimetry data. Biochemistry. 1989 Oct 17;28(21):8588–8596. doi: 10.1021/bi00447a048. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Edge V., Allewell N. M., Sturtevant J. M. Differential scanning calorimetric study of the thermal denaturation of aspartate transcarbamoylase of Escherichia coli. Biochemistry. 1988 Oct 18;27(21):8081–8087. doi: 10.1021/bi00421a017. [DOI] [PubMed] [Google Scholar]
- Edge V., Allewell N. M., Sturtevant J. M. High-resolution differential scanning calorimetric analysis of the subunits of Escherichia coli aspartate transcarbamoylase. Biochemistry. 1985 Oct 8;24(21):5899–5906. doi: 10.1021/bi00342a032. [DOI] [PubMed] [Google Scholar]
- Eisenstein E., Markby D. W., Schachman H. K. Changes in stability and allosteric properties of aspartate transcarbamoylase resulting from amino acid substitutions in the zinc-binding domain of the regulatory chains. Proc Natl Acad Sci U S A. 1989 May;86(9):3094–3098. doi: 10.1073/pnas.86.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure M., Camonis J. H., Jacquet M. Molecular characterization of a Dictyostelium discoideum gene encoding a multifunctional enzyme of the pyrimidine pathway. Eur J Biochem. 1989 Feb 1;179(2):345–358. doi: 10.1111/j.1432-1033.1989.tb14560.x. [DOI] [PubMed] [Google Scholar]
- Freund J. N., Jarry B. P. The rudimentary gene of Drosophila melanogaster encodes four enzymic functions. J Mol Biol. 1987 Jan 5;193(1):1–13. doi: 10.1016/0022-2836(87)90621-8. [DOI] [PubMed] [Google Scholar]
- Gerhart J. C., Schachman H. K. Allosteric interactions in aspartate transcarbamylase. II. Evidence for different conformational states of the protein in the presence and absence of specific ligands. Biochemistry. 1968 Feb;7(2):538–552. doi: 10.1021/bi00842a600. [DOI] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Howlett G. J., Schachman H. K. Allosteric regulation of aspartate transcarbamoylase. Changes in the sedimentation coefficient promoted by the bisubstrate analogue N-(phosphonacetyl)-L-aspartate. Biochemistry. 1977 Nov 15;16(23):5077–5083. doi: 10.1021/bi00642a021. [DOI] [PubMed] [Google Scholar]
- JOVIN T., CHRAMBACH A., NAUGHTON M. A. AN APPARATUS FOR PREPARATIVE TEMPERATURE-REGULATED POLYACRYLAMIDE GEL ELECTROPHORESIS. Anal Biochem. 1964 Nov;9:351–369. doi: 10.1016/0003-2697(64)90192-7. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Schachman H. K. Genetic characterization of the folding domains of the catalytic chains in aspartate transcarbamoylase. J Biol Chem. 1983 Mar 10;258(5):3266–3279. [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamoylase: the molecular basis for a concerted allosteric transition. Trends Biochem Sci. 1990 Feb;15(2):53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988 Aug 5;241(4866):669–674. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Lipscomb W. N., Cho Y. J., Honzatko R. B. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988 Dec 5;204(3):725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Pan Z. X., Honzatko R. B., Ke H. M., Lipscomb W. N. Structural asymmetry in the CTP-liganded form of aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1987 Aug 20;196(4):853–875. doi: 10.1016/0022-2836(87)90410-4. [DOI] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. 2.5 A structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J Mol Biol. 1987 Feb 5;193(3):527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerner C. G., Switzer R. L. Cloning and structure of the Bacillus subtilis aspartate transcarbamylase gene (pyrB). J Biol Chem. 1986 Aug 25;261(24):11156–11165. [PubMed] [Google Scholar]
- Meighen E. A., Pigiet V., Schachman H. K. Hybridization of native and chemically modified enzymes. 3. The catalytic subunits of aspartate transcarbamylase. Proc Natl Acad Sci U S A. 1970 Jan;65(1):234–241. doi: 10.1073/pnas.65.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M., Le Gouar M., Potier S., Souciet J. L., Hervé G. The primary structure of the aspartate transcarbamylase region of the URA2 gene product in Saccharomyces cerevisiae. Features involved in activity and nuclear localization. J Biol Chem. 1989 May 15;264(14):8366–8374. [PubMed] [Google Scholar]
- Nowlan S. F., Kantrowitz E. R. Superproduction and rapid purification of Escherichia coli aspartate transcarbamylase and its catalytic subunit under extreme derepression of the pyrimidine pathway. J Biol Chem. 1985 Nov 25;260(27):14712–14716. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachman H. K. Can a simple model account for the allosteric transition of aspartate transcarbamoylase? J Biol Chem. 1988 Dec 15;263(35):18583–18586. [PubMed] [Google Scholar]
- Schachman H. K., Pauza C. D., Navre M., Karels M. J., Wu L., Yang Y. R. Location of amino acid alterations in mutants of aspartate transcarbamoylase: Structural aspects of interallelic complementation. Proc Natl Acad Sci U S A. 1984 Jan;81(1):115–119. doi: 10.1073/pnas.81.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigesada K., Stark G. R., Maley J. A., Niswander L. A., Davidson J. N. Construction of a cDNA to the hamster CAD gene and its application toward defining the domain for aspartate transcarbamylase. Mol Cell Biol. 1985 Jul;5(7):1735–1742. doi: 10.1128/mcb.5.7.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., Bothwell M. A., Gibbons I., Yang Y. R., Schachman H. K. Ligand-promoted weakening of intersubunit bonding domains in aspartate transcarbamolylase. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3777–3781. doi: 10.1073/pnas.74.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., Schachman H. K. Mechanism of disproportionation of asparate transcarbamoylase molecules lacking one regulatory subunit. J Biol Chem. 1980 Sep 10;255(17):8136–8143. [PubMed] [Google Scholar]
- Thornton J. M., Sibanda B. L. Amino and carboxy-terminal regions in globular proteins. J Mol Biol. 1983 Jun 25;167(2):443–460. doi: 10.1016/s0022-2836(83)80344-1. [DOI] [PubMed] [Google Scholar]
- Vickers L. P., Compton J. G., Wall K. A., Flatgaard J. E., Schachman H. K. Comparison of active mutants and wild-type aspartate transcarbamoylase of Escherichia coli. J Biol Chem. 1984 Sep 10;259(17):11027–11035. [PubMed] [Google Scholar]
- Vickers L. P., Donovan J. W., Schachman H. K. Differential scanning calorimetry of asparate transcarbamoylase and its isolate subunits. J Biol Chem. 1978 Dec 10;253(23):8493–8498. [PubMed] [Google Scholar]
- Wall K. A., Flatgaard J. E., Schachman H. K., Gibbons I. Purification and characterization of a mutant aspartate transcarbamoylase lacking enzyme activity. J Biol Chem. 1979 Dec 10;254(23):11910–11916. [PubMed] [Google Scholar]
- Yang Y. R., Kirschner M. W., Schachman H. K. Aspartate transcarbamoylase (Escherichia coli): preparation of subunits. Methods Enzymol. 1978;51:35–41. doi: 10.1016/s0076-6879(78)51007-0. [DOI] [PubMed] [Google Scholar]
- Yang Y. R., Schachman H. K. Hybridization as a technique for studying interchain interactions in the catalytic trimers of aspartate transcarbamoylase. Anal Biochem. 1987 May 15;163(1):188–195. doi: 10.1016/0003-2697(87)90111-4. [DOI] [PubMed] [Google Scholar]