Abstract
Background
Results from DNA microarray experiments have shown that the expression of miR-34s undergoes significant changes following spinal cord injury (SCI). The present study was designed to detect changes in the expression of miR-34s and its target genes during the acute and sub-acute stages of SCI.
Material/Methods
Luxol fast blue (LFB) staining for myelin was used to observe the differences in the general morphology of the spinal cord after SCI in a contusion model in rats. qPCR was carried out to determine the expression variation of miR-34s and its target genes during the acute and sub-acute stages of SCI. The mimic technique was used to further confirm the regulatory effect of miR-34a on the potential target genes.
Results
The expression level of miR-34a decreased immediately after SCI and persisted for 21 days after SCI. The expression level of miR-34c began decreasing at day 1 after SCI and persisted until day 14. The expression level of miR-34b did not undergo significant change after SCI. The results of double immunofluorescence and in-situ hybridization suggested that miR-34a was highly expressed in spinal cord neurons. Based on our bioinformatics analysis, we postulated that miR-34a might participate in post-SCI cell apoptosis by regulating the target gene Notch1, and likely participated in the inflammatory response and glial scar formation by regulating the candidate genes Csf1r and PDGFRα, respectively. The expression levels of the candidate genes Csf1r and PDGFRα were consistent with Notch1 after SCI. The mimic technique further confirmed the regulatory effect of miR-34a on the aforementioned target genes.
Conclusions
We postulate that miR-34a and miR-34c might participate in multiple aspects of cytobiological activities following SCI. MiR-34a in particular may participate in cell apoptosis, inflammatory response, and glial scar formation by regulating the target gene Notch1 and candidate target genes Csf1r and PDGFRα respectively.
MeSH Keywords: MicroRNAs; Receptor, Macrophage Colony-Stimulating Factor; Receptor, Notch1; Receptor, Platelet-Derived Growth Factor alpha; Spinal Cord Injuries
Background
Spinal cord injury (SCI) is one of the most common devastating central nervous system (CNS) injuries worldwide, often resulting in permanent disabilities such as paralysis, and loss of movement, sensation, or autonomic control below the affected level. The treatment of SCI remains one of the greatest challenges in basic science research and clinical practice despite the use of various nerve growth factors and cell implantation strategies used in an attempt to provide a permissive environment for neuron survival, improving the endogenous potentiality of regenerating axons, and hindering the progression of secondary damage [1]. However, identifying an effective therapeutic molecular target is a formidable task in clinical work. MicroRNAs (miRNAs) are a newly discovered class of gene expression regulators that may represent a novel class of therapeutic targets to promote neuron repair and regeneration [2].
MiRNAs are a class of ~22 nucleotides endogenous non-coding small RNAs. Mature miRNAs bind to the complementary sequences in the 3′-untranslated region (3′-UTR) of target mRNAs and negatively regulate these targets by destabilizing the mRNA or repressing its translation. A number of miRNAs have been identified in the mammalian central nervous system (CNS), including the brain and spinal cord, and are known to play key roles in neurodevelopment and regulation of neural plasticity [2]. Previous studies have uncovered alterations in the temporal expression of a large set of miRNAs following contusive spinal cord injuries in adult rats by performing microarray analysis [3,4]. Additionally, cluster analysis have shown that a large number of miRNAs are involved in pathophysiological events secondary to SCI, such as inflammation, oxidation, and apoptosis. Thus, miRNAs could become attractive novel therapeutic targets for the treatment of SCI. MiR-142-3p has been predicted as a potential therapeutic target for sensory function recovery after SCI via regulating cAMP and promoting neurite growth [5]. Previous studies found that the expression of miR-34a in the medulla oblongata, pons and spinal cord was significantly higher than that in other regions of the CNS [6]. Downregulation of miR-34a after SCI has been identified in several studies [3,4]. However, few studies have explored the precise expression variations and responses of miR-34a, miR-34b, and miR-34c following SCI.
In this study, we aimed to examine changes in miR-34s expression after SCI in a rat spinal cord contusion model in vivo, and to further explore the effects of expression changes of miR-34a and its potential targets on cell apoptosis, inflammatory response, and glia scar formation, in an attempt to gain deep insights into the function of miR-34a and its use as a potential therapeutic target for the treatment of SCI.
Material and Methods
Construction of a contusion model of spinal cord injury
A total of 48 adult female Sprague-Dawley (SD) rats weighing 200–220 g (Experimental Animal Company of Shanghai SlAC, China) were subjected to a contusive SCI. Each animal was anesthetized by an intraperitoneal injection of complex narcotics. Laminectomy of the ninth thoracic vertebra was performed to expose the dorsal surface of the T10 spinal cord and induce a T10 contusive SCI using an IH-0400 Spinal Cord Impactor (Precision Systems and Instrumentation, Fairfax Station, VA, USA) with a 2.5 mm tip with 145 Kdynes of force. Animals in the contusion groups were sacrificed at day 1, 3, 7, 14 and 21. Animals in sham control group only received laminectomy. All surgical interventions and postoperative animal care were performed in accordance with the Institutional Animal Care guidelines and approved ethically by the Administration Committee of Jiangsu Province of China.
Cell culture and transfection with miR-34a mimics
SH-SY5Y cells (Shanghai Institutes for Biological Sciences, China) were cultured in (DMEM/F12) (Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS). Cells were transfected with the miR-34a mimic and negative control oligomer (Biomics, Nantong, China) by using Lipofectamine™ 2000 in Opti-MEM® I Reduced Serum Medium (Invitrogen, USA) according to the manufacturer’s protocol.
RNA extraction and qPCR
Spinal cord tissue (10 mm) centered on the injury site was obtained at day 1, 3, 7, 14 and 21 after injury and used for RNA extraction with miRNeasy Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. For detection of miR-34s or U6 snRNA as endogenous miRNA control, qPCR was performed with a TaqMan Universal Master Mix II using a specific FAM-dye labeled TaqMan MicroRNA Assay in Applied Biosystem sequence detection system AB7500 (Applied Biosystems, USA). Total RNA from cultured SH-SY5Y cells was extracted with miRNeasy Mini kit (Qiagen, Germany). A total of 500 ng RNA was used for generating cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo, Lithuania). The qPCR was performed with the Fast Start Universal SYBR Green Master/Probe Master (Rox) Mix, (Roche Diagnostics GmbH, Mannheim, Germany) in Bio-Rad CFX-96 (Bio-Rad, USA). The following primers were used: GAPDH forward GAGGTAGTTATGGCGTAGTGC, reverse CTGGTTTCT GGAGGATGG; Notch1 forward CACCCATGACCACTACCCAGTT, reverse CCTCGGACCAATCAGAGATGTT; Csf1r forward ACTGGTGA AGGATGGATACCAAA, reverse GGACTGCATGATGCTGTATATGTTC; pdgfra forward ACCTTGCACAATAACGGGAG, reverse CAGTTTGA TGGACGGGAGTT. Analyses of gene expression were carried out by the 2−ΔΔCT method. All data were expressed as means ±SD.
MiRNA potential target prediction and bioinformatics analysis
Target genes of miR-34s were predicted in according to miRDB online database (http://mirdb.org/miRDB/). GO-Analysis was performed with Gene Ontology (GO, http://geneontology.org) for indicating the biological functional coherence of target genes. Fisher’s exact test and multiple comparison test were used to calculate p-values and the false discovery rate (FDR) of differential genes after acquiring annotation functions of miR targets. Pathway-Analysis was performed with Kyoto Encyclopedia of Genes and Genomes (KEGG), and screened significance function of differential genes using Fisher’s exact test and the chi-squared according to p value <0.05. Based on prior literature reports, potential target genes involved in SCI associated biological processes, such as inflammation, oxidation and apoptosis, were selected preferentially for further study. In addition, the portion of the potential target genes that have been validated as being regulated by miR-34a in other tissues, were also preferentially selected for further study.
Luxol Fast Blue (LFB) staining
Briefly, sections were stained overnight at 56°C in 0.1% Luxol Fast Blue (LFB) (Sigma, USA) in acidified 95% ethanol, and then rinsed in 95% ethanol and differentiated in 0.05% Li2CO3 solution followed by 70% ethanol. Differentiation was terminated by washing in distilled water until the unmyelinated tissue looked white.
In-situ hybridization with subsequent immunofluorescence
Locked nucleic acid (LNA) hybridization probes complementary to miR-34a (5′-3′/5DigN/ACAACCAGCTAAGACACTGCCA) were provided from Exiqon (Vedbaek, Denmark). A negative non-hybridizing control named scramble miRNA (5′-3′/5DigN/GTGTAACACGTCTATACGCCCA) and a LNA U6 small nuclear RNA positive control probe (5′-3′/5DigN/CACGAATTTGCGTGTCATCCTT) were also purchased from Exiqon. Frozen tissue sections were prepared following the description of MicroRNA Protocol for In-situ Hybridization on Frozen Sections (Exiqon, Denmark). Briefly, tissue sections were fixed in 4% PFA and subsequently incubated with 15 μg/mL proteinase K for 10 minutes at 37°C. Hybridization with 20 nM miR-34a, 20 nM scramble-miR, or 4 nM U6 was respectively performed at 55°C, 57°C, or 54°C for 2 hours in hybridization buffer. After stringent washes with saline-sodium-citrate buffer, blocking was performed for 15 minutes with 1× blocking buffer (DIG Wash and Block Buffer Set, Roche Diagnostics GmbH, Germany). After blocking, sections were incubated for two hours at room temperature in a humidified chamber with Anti-Digoxigenin-AP, Fab fragments (1: 300, Roche Diagnostics GmbH). Sections were visualized with NBT/BCIP, and incubated for two hours at 37°C in the dark. Finally, sections were incubated with Nuclear Fast Red™ (Sigma, USA) for one minute for nuclear counter staining, then rinsed and mounted. After in-situ hybridization, subsequent immunofluorescence was carried out. After incubation with Anti-Digoxigenin-AP, sections were blocked in 10% NGS/3% BSA in PBS and incubated with the following primary antibodies overnight at 4°C: mouse anti-neurofilament 200 (NF200, 1: 100, Boster, Wuhan, China). Subsequently, sections were rinsed in PBS and conjugated with goat anti-mouse IgG (H+L)-FITC (1: 100, Biowold, China) dissolved in PBS-Tween 0.1% (PBST) for two hours at room temperature. Immunological detection of Anti-Digoxigenin-AP antibody was performed with the HNPP Fluorescent Detection Set (Roche Diagnostics GmbH).
Statistical analysis
Statistical analysis was performed with GraphPad Prism 6.0 software. All data are presented as mean ±SD. For all comparisons between untreated and treated cells, student’s unpaired t-test was used. For comparison of transcripts expressed after SCI, one-way analysis of variance (ANOVA) was used to determine whether or not a significant interaction was present. Positive results were followed up with Tukey’s post hoc t test for comparison within groups. A p value <0.05 was deemed significant.
Results
General morphology of the spinal cord following SCI
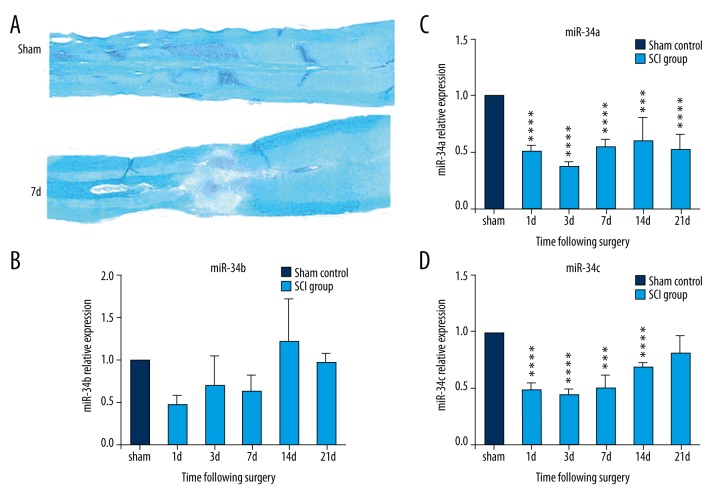
LFB staining of the normal spinal cord showed the entire structure with a distinct dividing line between grey matter and white matter with tight myelin. As shown by LFB staining, myelin in the spinal cord was mostly restricted to the white matter, the integrity of the spinal cord was destroyed, a greater proportion of the tissue was loosened, and the neural conduct tract was blocked following SCI as compared with the sham control group (Figure 1A). Large amounts of cell debris, degenerated axons, and cavities were observed at the injury center on day 7 following SCI due to abundant cell death and demyelination.
Figure 1.
Luxol fast blue (LFB) staining of the spinal tissue and miR-34s expression change after spinal cord injury (SCI). (A) LFB staining of the spinal cord. (B–D) miR-34s expression change after SCI by qPCR. (B) The trend of miR-34a expression change. (C) The trend of miR-34b expression change. (D) The trend of miR-34c expression change, where the horizontal ordinate represents different time points after SCI, and the vertical ordinate represents the relative expression level of miRNA in the spinal tissue (compared with sham control group, *** p<0.001; **** p<0.0001).
Aberrant expression of miR-34s in the injured spinal cord
The result of qPCR demonstrated that the expression levels of miR-34a and miR-34c were significantly declined after SCI. MiR-34a expression was declined in both acute and sub-acute phases. Compared with the sham control, miR-34a expression decreased to 50.30%, 37.12%, 54.54%, 59.98% and 52.20% on day 1, day 3, day 7, day 14 and day 21 after injury, respectively (p<0.001) (Figure 1C). The miR-34c expression level decreased to 49.15% on day 1, 44.93% on day 3, 50.99% on day 7 and 69.50% on day 14 after SCI (p<0.001), and almost restored to the baseline level on day 21 after injury (Figure 1D). Although the expression level of miR-34b decreased after SCI, the expression difference did not reach the significant threshold compared with the sham control group (Figure 1B) (p>0.05).
KEGG pathway and GO enrichment analysis of predicted target genes
Given the significant expression changes after SCI, miR-34a and miR-34c were selected for further study. To explore the roles of miR-34a and miR-34c in SCI, potential downstream targets of miR-34a and miR-34c were predicted by the miRDB databases. A total of 163 and 157 potential downstream targets were identified for miR-34a and miR-34c, respectively. The KEGG pathway analysis was performed to identify the functional categories and biological processes that were most prominent within the potential downstream targets of miR-34a and miR-34c. Several significantly enriched pathways of potential targets for miR-34a were identified as potential therapeutic targets after SCI, including MAPK signaling pathway (p=0.01), PPAR signaling pathway (p=0.02), and Wnt signaling pathway (p=0.03). However, the potential downstream targets of miR-34c showed moderate enrichment: MAPK signaling pathway (p=0.07), and fail to enrich in the Wnt signaling pathway (Table 1). GO-Analysis revealed that the potential targets of miR-34a were significantly enriched in several GO functions, such as cell division, mitosis, negative regulation of cell growth, metanephric mesenchyme development, apoptotic process, cell adhesion, and positive regulation of cell migration. It was noted that Csf1r and PDGFRα were enriched in several common GO functions, such as protein autophosphorylation, positive regulation of cell migration, and phosphatidylinositol-mediated signaling (Table 2). Consequently, we further confirmed the regulatory effect of miR-34a on Csf1r and PDGFRα using the mimics technique. In addition, Notch1 was also selected for further study, as it has been demonstrated to be the direct target of miR-34a in several studies [7–10] and involved in proliferation, growth, differentiation, and apoptosis of neural precursors [11].
Table 1.
KEGG pathway enrichment analysis for potential downstream targets of miR-34a and miR-34c.
| Term | Count | % | P Value | Genes | Benjamini | FDR | |
|---|---|---|---|---|---|---|---|
| Targets of miR-34a* | rno05214: Glioma | 5 | 3.33 | 0.00 | MAP2K1, PDGFRA, TP53, PDGFRB, PRKCB | 0.13 | 1.71 |
| rno04010: MAPK signaling pathway | 8 | 5.33 | 0.01 | MAP2K1, ELK4, MRAS, PDGFRA, TP53, PDGFRB, MAP3K14, PRKCB | 0.23 | 6.50 | |
| rno05215: Prostate cancer | 5 | 3.33 | 0.01 | CCNE2, MAP2K1, PDGFRA, TP53, PDGFRB | 0.17 | 6.86 | |
| rno05200: Pathways in cancer | 8 | 5.33 | 0.02 | CCNE2, MAP2K1, PDGFRA, TP53, KITLG, PDGFRB, CSF1R, PRKCB | 0.28 | 15.55 | |
| rno05218: Melanoma | 4 | 2.67 | 0.02 | MAP2K1, PDGFRA, TP53, PDGFRB | 0.28 | 19.29 | |
| rno03320: PPAR signaling pathway | 4 | 2.67 | 0.02 | SCD1, ACSL1, ACSL4, PCK1 | 0.26 | 20.65 | |
| rno04810: Regulation of actin cytoskeleton | 6 | 4.00 | 0.03 | MAP2K1, MRAS, WASF1, PDGFRA, PDGFRB, RDX | 0.30 | 27.73 | |
| rno04540: Gap junction | 4 | 2.67 | 0.03 | MAP2K1, PDGFRA, PDGFRB, PRKCB | 0.27 | 27.91 | |
| rno05210: Colorectal cancer | 4 | 2.67 | 0.03 | MAP2K1, PDGFRA, TP53, PDGFRB | 0.27 | 27.91 | |
| rno04310: Wnt signaling pathway | 5 | 3.33 | 0.03 | TBL1XR1, PPP2R5A, TP53, DAAM1, PRKCB | 0.26 | 29.97 | |
| rno04666: Fc gamma R-mediated phagocytosis | 4 | 2.67 | 0.04 | MAP2K1, WASF1, PPAP2A, PRKCB | 0.27 | 33.35 | |
| rno04910: Insulin signaling pathway | 4 | 2.67 | 0.10 | MAP2K1, FLOT2, PRKCI, PCK1 | 0.48 | 67.10 | |
| Targets of miR-34c** | rno05214: Glioma | 4 | 2.76 | 0.01 | MAP2K1, PDGFRA, TP53, PRKCB | 0.72 | 14.41 |
| rno05200: Pathways in cancer | 8 | 5.52 | 0.02 | CCNE2, MAP2K1, PIAS3, PDGFRA, TP53, KITLG, BIRC3, PRKCB | 0.50 | 15.71 | |
| rno03320: PPAR signaling pathway | 4 | 2.76 | 0.02 | SCD1, ACSL1, ACSL4, PCK1 | 0.47 | 20.86 | |
| rno05222: Small cell lung cancer | 4 | 2.76 | 0.03 | CCNE2, PIAS3, TP53, BIRC3 | 0.51 | 29.72 | |
| rno05020: Prion diseases | 3 | 2.07 | 0.03 | NOTCH1, C9, MAP2K1 | 0.46 | 31.65 | |
| rno04666: Fc gamma R-mediated phagocytosis | 4 | 2.76 | 0.04 | MAP2K1, WASF1, PPAP2A, PRKCB | 0.43 | 33.66 | |
| rno05215: Prostate cancer | 4 | 2.76 | 0.04 | CCNE2, MAP2K1, PDGFRA, TP53 | 0.40 | 35.26 | |
Potential downstream targets of * miR-34a and ** miR-34c were analyzed to identify the significantly enriched pathways.
Table 2.
Top 20 GO function for potential targets of miR-34a.
| GO Name | Total count | Diff count | P Value | FDR | Gene |
|---|---|---|---|---|---|
| Regulation of transcription, DNA-dependent | 284 | 24 | 0.00000 | 0.00003 | Ahrr, Arid4b, Ccnc, Ctnnd2, Dmtf1, Dnmt1, E2f1, Gtf2b, Hipk1, Morf4l1, Ncoa3, Nlk, Nono, Osr1, Pias3, Pkn2, Ptov1, Rnf2, Spz1, Tceal8, Tle4, Txnip, Wt1, Zbtb10 |
| Cell cycle | 207 | 19 | 0.00000 | 0.00008 | Ccnf, Cdc26, Cep55, Clasp2, Dmtf1, Gnai3, Haus1, Msh2, Pafah1b1, Pkn2, Ppp6c, Rab11a, Stag3, Stk11, Tp53, Tsg101, Txnip, Wee1, Zfyve26 |
| Protein autophosphorylation | 39 | 8 | 0.00000 | 0.00036 | Aak1, Csf1r, Epha4, Map3k12, Mink1, Pdgfra, Pdgfrb, Stk11 |
| Cell division | 136 | 14 | 0.00000 | 0.00037 | Aurka, Ccne2, Ccnf, Cdc26, Cdk2, Cep55, Clasp2, Gnai3, Haus1, Mapre3, Pafah1b1, Pkn2, Tsg101, Wee1 |
| Mitosis | 80 | 10 | 0.00002 | 0.00133 | Aurka, Ccnf, Cdc26, Cdk2, Cep55, Clasp2, Haus1, Mapre3, Pafah1b1, Wee1 |
| Negative regulation of cell growth | 17 | 5 | 0.00003 | 0.00228 | Dcun1d3, Rbbp7, Stk11, Tp53, Wt1 |
| Metanephric mesenchyme development | 4 | 3 | 0.00005 | 0.00298 | Osr1, Pdgfrb, Wt1 |
| Apoptotic process | 144 | 12 | 0.00013 | 0.00722 | Bcl10, E2f1, Eaf2, Egln3, Ei24, Gzmb, Pak7, Pkn2, Prkcb, Sqstm1, Tox3, Tp53 |
| Cell adhesion | 237 | 16 | 0.00014 | 0.00661 | Cdh11, Cdh13, Cercam, Cntn2, Col12a1, Ctnnd2, Fat2, Flot2, Kitlg, Olr1, Pcdha4, Pcdha6, Pcdhac1, Pcdhac2, Pkn2, Ssx2ip |
| Positive regulation of cell migration | 15 | 4 | 0.00031 | 0.01354 | Csf1r, F2rl1, Pdgfra, Pdgfrb |
| Protein transport | 208 | 14 | 0.00036 | 0.01428 | Chmp4c, Nup35, Nxt2, Rab10, Rab35, Rab3b, Scamp1, Sec61a1, Serp1, Snx15, Snx27, Stxbp1, Trim3, Tsg101 |
| Phosphatidylinositol-mediated signaling | 7 | 3 | 0.00040 | 0.01442 | Csf1r, Pdgfra, Pdgfrb |
| Nervous system development | 100 | 9 | 0.00051 | 0.01713 | Arhgef7, Cln8, Cyb5d2, Dclk1, Epha5, Ina, Ndel1, Nptn, Pafah1b1 |
| Positive regulation of NF-kappaB transcription factor activity | 17 | 4 | 0.00052 | 0.01626 | Bcl10, Prkcb, Prkch, Prkci |
| Cardiac myofibril assembly | 2 | 2 | 0.00053 | 0.01319 | Pdgfra, Pdgfrb |
| Posterior mesonephric tubule development | 2 | 2 | 0.00053 | 0.01319 | Osr1, Wt1 |
| Regulation of Rap GTPase activity | 2 | 2 | 0.00053 | 0.01319 | Epha4, Sipa1l1 |
| Positive regulation of glial cell proliferation | 2 | 2 | 0.00053 | 0.01319 | Prkch, Prkci |
| Positive regulation of B cell receptor signaling pathway | 2 | 2 | 0.00053 | 0.01319 | Prkcb, Prkch |
| Retina vasculature development in camera-type eye | 2 | 2 | 0.00053 | 0.01319 | Pdgfra, Pdgfrb |
MiR-34a expression was reduced following SCI and relatively high expression in spinal cord neurons
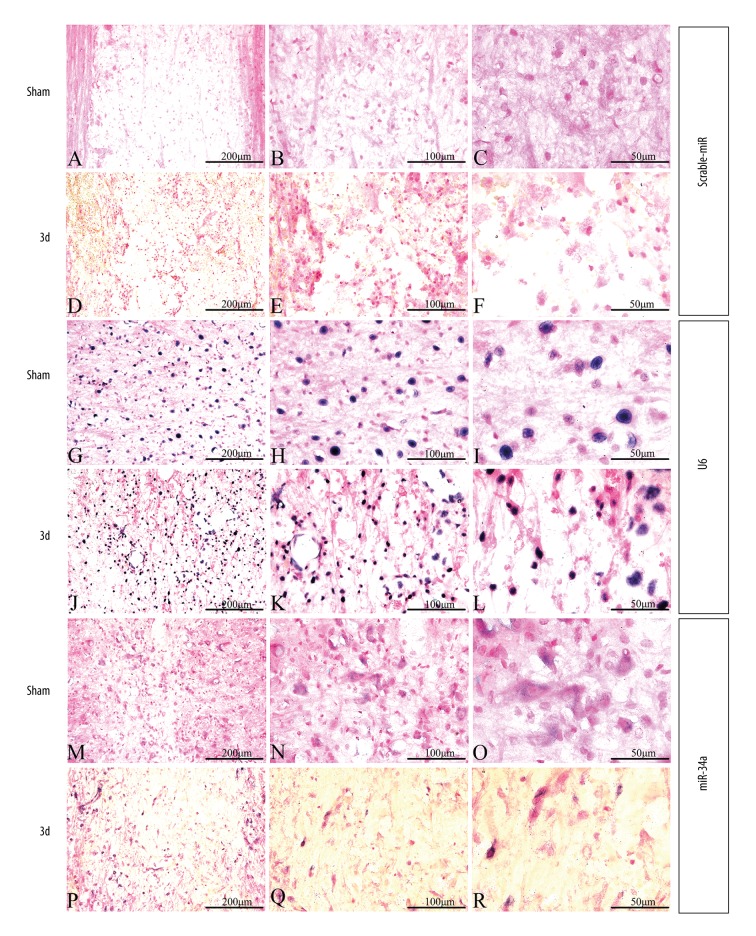
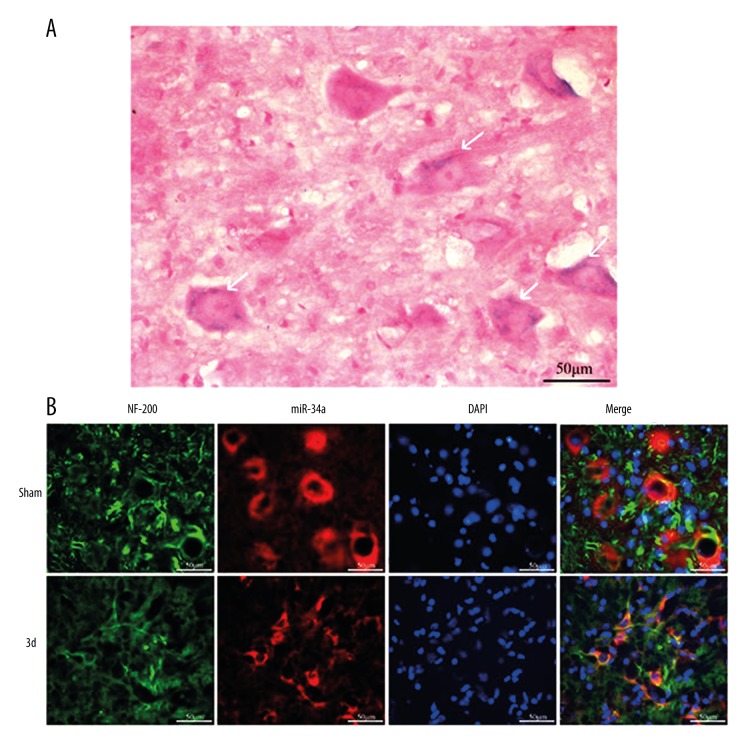
The result of qPCR showed that the expression of miR-34a declined to the lowest level on day 3 following SCI. In addition, changes in miR-34a expression were detected on day 3 after SCI by in-situ hybridization (Figure 2). Scramble-miR and U6 were used as a negative control and a positive control, respectively. The scramble control probe showed no significant staining in the sham control group and the SCI groups (Figure 2A–2F). U6 was evenly distributed in both injured and uninjured spinal cords (Figure 2G–2L), while miR-34a-expressing cells were mostly found in the gray matter (Figure 2M–2R). For miR-34a, relatively strong staining was observed in the uninjured spinal cord (Figure 2M–2O) and weakly expressed in cells at the lesion epicenter (Figure 2P–2R), as well as in spinal segments 400 μm rostral (Supplementary Figure 1) and caudal region (Supplementary Figure 2) to the injury lesion. The morphologic characteristics of miR-34a positive cells were similar to neurons with large cell bodies and round big nuclei (Figure 3A). To validate whether the miR-34a positive cells were neurons, co-expression and distribution of the neurofilament marker NF200 and miR-34a mRNA expression were investigated by combined immunohistochemistry and in-situ hybridization, respectively. Co-localization and distribution of NF200 were observed in miR-34a positive cells (Figure 3B). In addition, the expression pattern of NF200 was similar to that of miR-34a, which decreased on day 3 post-injury.
Figure 2.
The result of in-situ hybridization in different groups. (A–C, G–I, M–O) In-situ hybridization in the normal control group. (D–F, J–L, P–R) In-situ hybridization in experimental groups 3 days after SCI. (A–F) In-situ hybridization of scramble-miR. (G–L) In-situ hybridization of U6; (M–P) In-situ hybridization of miR-34a. (A, D, G, J, M, P) Bar=200 μm. (B, E, H, K, N, Q) Bar=100 μm. (C, F, I, L, O, R) Bar=50 μm.
Figure 3.
The result of double immunofluorescence and in-situ hybridization suggest that miR-34a was highly expressed in spinal cord neurons. (A) Detection of miR-34a in spinal gray matter by in-situ hybridization (indicated with white arrows). (B) Fluorescence and in-situ hybridization double labeling (NF-200 labeled neurons, green; miR-34a, red; DAPI labeled nucleus, blue), Bar=50 μm.
The expression trends of Csf1r and PDGFRα were consistent with Notch1 after SCI
A single miRNA has the potential to target hundreds of distinct mRNAs, and one mRNA can be regulated by multiple miRNAs. Abnormal expression of miR-34a suggested that miR-34a participated in pathophysiological processes through targeting candidate genes after SCI. To determine the specific effect of miR-34a, three significant candidate genes (Notch1, Csf1r, and PDGFRα) were selected from predicted 163 target genes of miR-34a for the subsequent study. Several studies have demonstrated that Notch1 was directly regulated by miR-34a [7–10]. In our study, the expression levels of Notch1, Csf1r, and PDGFRα were conducted by qPCR following SCI. Notch1, PDGFRα, and Csf1r expression levels were downregulated on day 1 after SCI, reached the highest level on day 7 after SCI, and were upregulated until day 21 post-injury compared with the sham control group. The expression pattern of PDGFRα and Csf1ra was consistent with Notch1 after SCI (Figure 4A).
Figure 4.
Verification of Notch1, Csf1r, and PDGFRa as target genes of miR-34a and their expression changes after SCI. (A) qPCR detection of the trend of Notch1, Csf1r, and PDGFRa expression change in different stages after SCI. The horizontal coordinate represents the time points following SCI; the longitudinal coordinate represents the relative expression level of the target genes in the spinal tissue. (B) qPCR verification of changes in Notch1, Csf1r, and PDGFRa after miR-34a overexpression, compared with the control group, **** p<0.0001.
Validation of the downstream targets of miR-34a in vitro
MiR-34a was seen to be a regulator of the Notch pathway through its targeting of Notch1. To further investigate whether Csf1r and PDGFRα were targets of miR-34a, SH-SY5Y cells were transfected with miR-34a mimics to investigate the effect of overexpression of miR-34a. As expected, Notch1, Csf1r, and PDGFRα declined in the mimic-transfected cells compared with cells that were transfected with control mimics. The similar expression patterns of Notch1, Csf1r, and PDGFRα after the overexpression of miR-34a indicated that other than Notch 1, Csf1r and PDGFRα were also targets of miR-34a (Figure 4B).
Discussion
An increased numbers of studies have demonstrated that miRNA dysregulation in the CNS is associated with traumatic injury and significantly affects nerve regeneration and various neurodegenerative disorders, including Alzheimer, Parkinson, and Huntington diseases [3,12]. Recently, miRNAs have drawn high attention due to their potential as therapeutic targets through regulating related gene expression. Therefore, it is necessary to know the underlying mechanism of miRNA effects on SCI. Previous studies have identified a large number of dysregulated miRNAs after SCI. A marked increase in the number of downregulated microRNAs following moderate contusive SCI have been found when screened by microarray analysis [3,4]. The result of microarray analysis by Liu et al. [4] showed that more than 35% of miRNAs underwent significant changes on day 7 after SCI, and further bioinformatics analysis revealed that these miRNAs could be classified into several categories of target genes that respectively participated in regulating inflammatory response, apoptotic response, and oxidative stress. However, changes in miRNA-34s expression at different stages of SCI and the roles that they play in these stages remain unclear.
It was found in the present study that miR-34a and miR-34c underwent significant changes after SCI, which is consistent with several other study reports. For example, microarray analyses in BALB/c mouse traumatic brain injury (TBI) demonstrated that miR-34a and miR34b were downregulated at four hours post-TBI [13]. Microarray analysis by Yunta et al. [3] showed that miR-34a level was decreased on both day 3 and day 7 in a spinal cord contusion injury model. Similarly, Liu et al. [4] reported that miR-34a was downregulated at four hours and on day 1 and day 7 in a rat spinal cord traumatic injury model. These studies demonstrated that miR-34a was deregulated following spinal cord contusion injury, which is similar to our finding that miR-34a was aberrantly expressed after SCI. However, the result of our qPCR showed that the expression of miR-34a reached the lowest level on day 3 after SCI, and declined persisted until day 21 after SCI, while the expression of miR-34c began declining on day 1, continued to decline until day 14, and restored to the normal level on day 21. Although Liu et al. reported the abnormal expression of miR-34a and miR-34c after SCI, they did not mention whether the expression level of miR-34b was affected by SCI or not. It was found in our study that the expression of miR-34b did not undergo significant change after SCI as compared with the sham control group.
The main reason for the irreversibility of neurological deficits from SCI lies in the difficulty in regenerating neurons. Significant efforts have been directed toward regenerative therapies that may facilitate neuronal repair. Recent work by Agostini et al. [14] found that miR-34a regulated neurite outgrowth, spinal morphology, and function. Combination of in-situ hybridization and immunofluorescence staining in our study demonstrated that miR-34a was predominant in neurons, indicating that miR-34a may also play a key role in neuronal degeneration and regeneration after SCI, although its function remains largely unknown. In the present study, a total of 163 potential downstream targets for miR-34a were predicted by miRDB databases. We further explore the effects of expression change of miR-34a and its potential targets, such as Notch1, Csf1r, and PDGFRα.
Several studies have validated that Notch1 was regulated by miR-34a in colorectal cancer [8], glioblastoma [9], medulloblastoma cells [15], non-small cell lung cancer cell lines [7], and pancreatic cancer [10]. During the development of the nervous system, Notch1 regulates the proliferation, growth, differentiation, and apoptosis of neural precursors, determines the cell fate, and affects the morphology of dendrites and axons in embryonic neurons [11]. Previous studies have demonstrated that hybridization signals for Notch-1 were not detectable in the intact spinal cord, and significantly increased after injury [16]. In our study, it was found that expression change of miR-34a after SCI exhibited a negative correlation with Notch1. Location of miR-34a by in-situ-hybridization and immunohistochemistry showed high expression of miR-34a in normal spinal neurons, and this high expression decreased significantly after SCI. Based on this finding, we postulated that the phenomenon of neuronal apoptosis after SCI was also associated with the downregulation of miR-34a, which weakens the downregulatory effect on Notch1 gene expression and induces cell apoptosis via the Notch signaling pathway, thus decreasing the number of residual cells, especially neurons, and making spinal cord neuron regeneration difficult. In addition, in vitro studies [11] showed that activation of the Notch1 signaling pathway inhibited the differentiation of neural precursors, whereas attenuating Notch1 signaling and enhancing Ngn2 could increase nerve generation. The in vitro study by Chen et al. [16] showed that neurons that survived SCI may most likely participate in spontaneous neural circuit reconstruction produced by the newly generated synapses through Notch1 expression change. Whether Notch1 expression change is associated with cell apoptosis or nerve reconstruction, or both needs further investigation.
The results of the present study showed that the trend of Csf1r and PDGFRα change after SCI was consistent with Notch1. To verify whether Csf1r and PDGFRα were truly regulated by miR-34a, we transfected them with miR-34a mimics to overexpress miR-34a, and found that the expression level of Notch1, the target gene of miR-34a, was decreased significantly, and at the same time the expression level of candidate gene Csf1r and PDGFRα was also decreased significantly, which further confirmed that the expression of Csf1r and PDGFRα was regulated by miR-34a.
CSF-1R is a receptor tyrosine kinase with two cognate ligands, CSF-1 and IL-34, that regulate microglial proliferation and neuronal apoptosis [17]. Previous studies demonstrated, in the rat SCI model, granulocyte-macrophage colony-stimulating factor (GM-CSF) reduced apoptosis in the injured spinal cord and improved neurological function [18]. In our study, we found significant expression change of Csf1r after SCI, which may be associated with the migration and infiltration of related inflammatory cells such as microglia and macrophages and their participation in inflammatory response; on the other hand, it may be also be related to cell self-regulation to protect neurons against SCI-induced apoptosis.
NG2 cells, as a fourth glial cell type, which is widely distributed in the adult brain and spinal cord, simultaneously expressed PDGFRα and NG2. In the healthy nervous tissue, NG2 cells produce oligodendrocytes, and display wide differentiation potential under pathological conditions, where they could give rise to reactive astrocytes, such as involvement in the formation of glial scar astrocytes following a surgical lesion [19]. Recently, sonic hedgehog (Shh) has been identified as an important regulator in the differentiation of NG2 cells into astrocytes [20]. In addition, previous studies have demonstrated that PDGFRα was the target gene of miR-34a during progression of glioblastoma [21] and lung tumor [22]. Although the aforementioned studies did not make direct analysis of PDGFRα after SCI, their observation and our analysis on PDGFRα expression changes allows us to postulate that PDGFRα may simultaneously participate in neuronal pathological changes and astrocyte activation. Its linear correlation with miR-34a expression changes further encouraged us to postulate that miR-34a may participate in glial scar formation by regulating PDGFRα after SCI.
Conclusions
The present study was intended to answer the question whether miR-34s participated in responses during acute and sub-acute stages of SCI. The result showed that miR-34a and miR-34c expression underwent changes during the acute and sub-acute stages of SCI, although miR-34b expression change was not statistically significant. In addition, high expression of miR-34a was located in spinal cord neurons. Based on bioinformatics analysis, we collected candidate target genes of miR-34a and found that Csf1r and PDGFRα (two newly screened candidate genes by using the in vivo SCI model and mimics technique in vitro) were regulated by miR-34a. We postulated that miR-34a may participate in cell apoptosis, inflammatory response, and glial scar formation by regulating the target gene Notch1, Csf1r, and PDGFRα respectively following SCI.
Supplementary materials
The result of spinal in-situ hybridization in different groups (SCI group: 250–550 μm from the head end of the contusion center). (A–C, G–I, M–O) In-situ hybridization in the normal control group. (D–F, J–L, P–R) In-situ hybridization in experimental groups 3 days after SCI. (A–F) In-situ hybridization of scramble-miR. (G–L) In-situ hybridization of U6; (M–P) In-situ hybridization of miR-34a. (A, D, G, J, M, P) Bar=200 μm. (B, E, H, K, N, Q) Bar=100 μm. (C, F, I, L, O, R) Bar=50 μm.
The result of spinal in-situ hybridization in different groups (SCI group: 250–550 μm from the tail end of the contusion center). (A–C, G–I, M–O) in-situ hybridization in the normal control group. (D–F, J–L, P–R) In-situ hybridization in experimental groups 3 days after SCI. (A–F) In-situ hybridization of scramble-miR. (G–L) In-situ hybridization of U6; (M–P) In-situ hybridization of miR-34a. (A, D, G, J, M, P) Bar=200 μm. (B, E, H, K, N, Q) Bar=100 μm. (C, F, I, L, O, R) Bar=50 μm.
Footnotes
Conflict of interest
The authors have no conflicts of interest to disclose.
Source of support: This work was supported by the grants from the Natural Science Foundation of China (numbers 81271721, 81200828 and 81501610), the Priority Academic Program Development of Jiangsu Higher Education Institutes (PAPD) and National Undergraduate Training Program for Innovation and Entrepreneurship (201510304035Z)
Statement
None of sources of support were directly involved in the design of the study, or in the collection and analysis of the data in this report.
References
- 1.Kabu S, Gao Y, Kwon BK, Labhasetwar V. Drug delivery, cell-based therapies, and tissue engineering approaches for spinal cord injury. J Control Release. 2015;219:141–54. doi: 10.1016/j.jconrel.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7(12):911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 3.Yunta M, Nieto-Diaz M, Esteban FJ, et al. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS One. 2012;7(4):e34534. doi: 10.1371/journal.pone.0034534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219(2):424–29. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Yuan W, Liu Y, et al. miR-142-3p is a potential therapeutic target for sensory function recovery of spinal cord injury. Med Sci Monit. 2015;21:2553–56. doi: 10.12659/MSM.894098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bak M, Silahtaroglu A, Moller M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–44. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang J, Kim E, Kim W, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem. 2013;288(38):27343–57. doi: 10.1074/jbc.M113.490482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–76. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia J, Duan Q, Ahmad A, et al. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets. 2012;13(14):1750–56. doi: 10.2174/138945012804545597. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto S, Nagao M, Sugimori M, et al. Transcription factor expression and Notch-dependent regulation of neural progenitors in the adult rat spinal cord. J Neurosci. 2001;21(24):9814–23. doi: 10.1523/JNEUROSCI.21-24-09814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu B, Zhou S, Yi S, Gu X. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Prog Neurobiol. 2015;134:122–39. doi: 10.1016/j.pneurobio.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Guo F, Pan C, et al. Effects of low temperatures on proliferation-related signaling pathways in the hippocampus after traumatic brain injury. Exp Biol Med (Maywood) 2012;237(12):1424–32. doi: 10.1258/ebm.2012.012123. [DOI] [PubMed] [Google Scholar]
- 14.Agostini M, Tucci P, Steinert JR, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA. 2011;108(52):21099–104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Antonellis P, Medaglia C, Cusanelli E, et al. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One. 2011;6(9):e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Leong SY, Schachner M. Differential expression of cell fate determinants in neurons and glial cells of adult mouse spinal cord after compression injury. Eur J Neurosci. 2005;22(8):1895–906. doi: 10.1111/j.1460-9568.2005.04348.x. [DOI] [PubMed] [Google Scholar]
- 17.Chitu V, Gokhan S, Nandi S, et al. Emerging roles for CSF-1 receptor and its ligands in the nervous system. Trends Neurosci. 2016;39(6):378–93. doi: 10.1016/j.tins.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha Y, Kim YS, Cho JM, et al. Role of granulocyte-macrophage colony-stimulating factor in preventing apoptosis and improving functional outcome in experimental spinal cord contusion injury. J Neurosurg Spine. 2005;2(1):55–61. doi: 10.3171/spi.2005.2.1.0055. [DOI] [PubMed] [Google Scholar]
- 19.Alonso G. NG2 proteoglycan-expressing cells of the adult rat brain: possible involvement in the formation of glial scar astrocytes following stab wound. Glia. 2005;49(3):318–38. doi: 10.1002/glia.20121. [DOI] [PubMed] [Google Scholar]
- 20.Honsa P, Valny M, Kriska J, et al. Generation of reactive astrocytes from NG2 cells is regulated by sonic hedgehog. Glia. 2016;64(9):1518–31. doi: 10.1002/glia.23019. [DOI] [PubMed] [Google Scholar]
- 21.Silber J, Jacobsen A, Ozawa T, et al. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One. 2012;7(3):e33844. doi: 10.1371/journal.pone.0033844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garofalo M, Jeon YJ, Nuovo GJ, et al. MiR-34a/c-dependent PDGFR-alpha/beta downregulation inhibits tumorigenesis and enhances TRAIL-induced apoptosis in lung cancer. PLoS One. 2013;8(6):e67581. doi: 10.1371/journal.pone.0067581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The result of spinal in-situ hybridization in different groups (SCI group: 250–550 μm from the head end of the contusion center). (A–C, G–I, M–O) In-situ hybridization in the normal control group. (D–F, J–L, P–R) In-situ hybridization in experimental groups 3 days after SCI. (A–F) In-situ hybridization of scramble-miR. (G–L) In-situ hybridization of U6; (M–P) In-situ hybridization of miR-34a. (A, D, G, J, M, P) Bar=200 μm. (B, E, H, K, N, Q) Bar=100 μm. (C, F, I, L, O, R) Bar=50 μm.
The result of spinal in-situ hybridization in different groups (SCI group: 250–550 μm from the tail end of the contusion center). (A–C, G–I, M–O) in-situ hybridization in the normal control group. (D–F, J–L, P–R) In-situ hybridization in experimental groups 3 days after SCI. (A–F) In-situ hybridization of scramble-miR. (G–L) In-situ hybridization of U6; (M–P) In-situ hybridization of miR-34a. (A, D, G, J, M, P) Bar=200 μm. (B, E, H, K, N, Q) Bar=100 μm. (C, F, I, L, O, R) Bar=50 μm.