Abstract
A disease trait often can be characterized by multiple phenotypic measurements that can provide complementary information on disease etiology, physiology or clinical manifestations. Given that multiple phenotypes may be correlated and reflect common underlying genetic mechanisms, the use of multivariate analysis of multiple traits may improve statistical power to detect genes and variants underlying complex traits. The literature, however, has been unclear as to the optimal approach for analyzing multiple correlated traits. In this study, heritability and linkage analysis was performed for six Obstructive Sleep Apnea Hypopnea Syndrome (OSAHS) related phenotypes, as well as principal components of the phenotypes and principal components of the heritability (PCHs) using the data from Cleveland Family Study, which include both African and European American families. Our study demonstrates that principal components generally result in higher heritability and linkage evidence than individual traits. Furthermore, the PCHs can be transferred across populations, strongly suggesting that these PCHs reflect traits with common underlying genetic mechanisms for OSAHS across populations. Thus, PCHs can provide useful traits for using data on multiple phenotypes and for genetic studies of trans-ethnic populations.
Introduction
In genetic studies, multiple correlated phenotypes are often measured to better characterize a complex disease. Correlation among different measurements may be induced by common underlying genetic mechanisms or environmental factors. A number of analytical approaches have been suggested to improve statistical power in detecting genetic variants underlying complex traits[Stephens 2013] [Solovieff, et al. 2013] [Zhu, et al. 2015], although most current genome wide association studies perform analysis for each trait individually. The optimal method for analyzing multiple traits for genetic studies is still under debate [Aschard, et al. 2014]. Multivariate analysis offers one way to model genetic effects to multiple phenotypes simultaneously [Obrien 1984]. Multivariate regression analysis of modeling the correlation matrix among the traits has the flexibility of testing a variety of parameters without losing any information but can be computationally intensive. On the other hand, principal components (PC) analysis, based on a dimension reduction technique summarizing information across multiple measured traits has been suggested for use in linkage and association analysis [Comuzzie, et al. 1997] [Klei, et al. 2008] [Gu, et al. 2008] with the potential of losing important information when only the top PCs are analyzed [Aschard, et al. 2014]. The traditional PCs are the linear combination of traits calculated by maximizing the variance with the assumption of independent subjects, which we termed PCVs. When family data are available, the linear combinations of traits can be obtained by maximizing heritability, which we termed PCHs [Ott and Rabinowitz 1999]. The top PCHs have the potential to extract more information reflecting the genetic components of traits than may be derived from PCVs. Furthermore, when the correlation among phenotypes is driven by a common genetic mechanism, it is reasonable that the top PCVs and PCHs likely capture such effects. However, this hypothesis has to be examined using real data.
Obstructive sleep apnea-hypopnea syndrome (OSAHS) is a complex disorder characterized by the occurrence of repetitive episodes of complete or partial upper airway obstruction during sleep associated with snoring, intermittent hypoxemia, and daytime sleepiness [Mbata and Chukwuka 2012]. The public health importance of the disorder relates to its high prevalence and its impact on a wide range of health outcomes. Epidemiological studies indicate that OSAHS affects approximately 2 to 6% of children and more than 15% of adults [Badran, et al. 2015; Capdevila, et al. 2008; Rosen, et al. 2003]. The recurrent episodes of upper airway obstruction that occur with OSAHS lead to disruptive snoring, sleep fragmentation and daytime sleepiness, which negatively impact quality of life and daytime performance [Mbata and Chukwuka 2012]. OSAHS also results in overnight hypoxemia and heightened sympathetic activation, which adversely affect blood pressure, metabolism, and vascular health [Mbata and Chukwuka 2012]. Indeed, research over the last two decades has identified OSAHS as an independent risk factor for the development of hypertension, cardiovascular disease, stroke and premature mortality [Badran, et al. 2015].
There is large variation in the clinical expression of OSAHS. Although OSAHS is significantly more common in men and individuals who are obese [Badran, et al. 2015], male gender and obesity only explain a portion of the variation in the trait distribution. The mechanisms that account for individual susceptibility are not well understood but likely include anatomic and physiological factors that influence upper airway size and collapsibility and stability of ventilatory control during sleep [Redline and Tishler 2000; Young, et al. 2004]. Understanding the genetic etiology of OSAHS, including the genetic bases for intermediate mechanisms, may provide a means of better understanding its pathogenesis, with the goal of improving preventive strategies, diagnostic tools and therapies.
Although a significant genetic basis for the disorder is supported by family studies and emerging candidate gene association analyses, molecular studies of OSAHS genetics has lagged those of other chronic diseases [Redline, et al. 1995] [Palmer, et al. 2003] [Larkin, et al. 2008]. To date, the chief quantitative metric that has been used in genetic analysis is the apnea-hypopnea index (AHI), which averages the number of complete and partial airway occlusions that occur hourly during sleep. The advantages of using the AHI include its simplicity, high night-to-night reproducibility and widespread clinical use. However, the AHI does not provide information on attributes of OSAHS that may reflect intermediate pathways or reflect specific subtypes of OSAHS that my represent different underlying intermediate pathways.
In this paper, we postulate that additional measures of OSAHS- including features extracted from the overnight sleep study and associated symptoms – may be heritable, and when used alone or in combination with other metrics, may improve the ability to detect genetic signals. We evaluated PCVs and PCHs obtained using six OSAHS traits that captured a range of features of this disorder: the frequency of apneas/hypopneas (AHI), the duration of respiratory disturbance events during sleep (apnea or hypopnea duration), measures of nocturnal hypoxemia, and self-reported snoring and sleepiness. In a sample of European and African-American families from the Cleveland Family Study (CFS), we compared the heritability estimates of the PCVs, PCHs and individual traits as well as the transferability of PCs across ethnic populations. We studied whether a common genetic mechanism can be captured by the top PCs. We further investigate the impact of PCs on identifying linkage signals in linkage analysis of measures of OSAHS.
Methods
Cleveland Family Study (CFS)
Analyses were performed using data collected from the CFS, a family-based longitudinal study comprised of index cases with laboratory diagnosed sleep apnea, their family members and neighborhood control families. Individuals have been followed on as many as four occasions over a period of 16 years, and have completed sleep apnea, anthropometry, and other related measurements as well as standardized questionnaires, as detailed previously [Mehra, et al. 2010]. Table 1 presents the characteristics of 139 European and 147 African American families in CFS, including 645 Europeans (EAs) and 656 African Americans (AAs), respectively.
Table 1.
Sample characteristics of African Americans and European Americans in CFS
| African Americans | European Americans | |||
|---|---|---|---|---|
|
| ||||
| Total Subjects (Families) | 656 (147) | 645 (139) | ||
|
| ||||
| No. of Males (%) | 285 (43.4) | 302 (46.8) | ||
|
| ||||
| HABSNORE (%) | ||||
| 0 | 106 (16.2) | 137 (21.2) | ||
| 1 | 98 (14.9) | 132 (20.5) | ||
| 2 | 151 (23.0) | 139 (21.6) | ||
| 3 | 95 (14.5) | 88 (13.6) | ||
| 4 | 206 (31.4) | 149 (23.1) | ||
|
| ||||
| EXCSLPDY (%) | ||||
| 0 | 229 (34.9) | 171 (26.4) | ||
| 1 | 161 (24.5) | 181 (28.1) | ||
| 2 | 143 (21.8) | 165 (25.6) | ||
| 3 | 58 (8.8) | 81 (12.6) | ||
| 4 | 65 (9.9) | 47 (7.3) | ||
|
| ||||
| Mean ± Standard Deviation | Median with Interquartile ranges | Mean ± Standard Deviation | Median with Interquartile ranges | |
|
| ||||
| Age | 38.2 ± 19.2 | 39.0 (20.8–51.3) | 41.1 ± 19.5 | 42.2 (24.1–54.7) |
|
| ||||
| BMI | 31.8 ± 9.6 | 30.8 (24.8–37.2) | 30.0 ± 8.6 | 28.8 (24.3–34.4) |
|
| ||||
| AHI | 17.6 ± 26.8 | 5.6 (1.4–21.1) | 15.2 ± 23.1 | 4.6 (1.5–17.7) |
|
| ||||
| AVGO2 | 94.5 ± 3.7 | 95.3 (93.9–97.0) | 93.9 ± 3.4 | 94.8 (93.0–96.0) |
|
| ||||
| PER90 | 4.8 ± 13.5 | 0.23 (0.0–2.0) | 4.1 ± 12.2 | 0.2 (0.0–1.4) |
|
| ||||
| AVGDUR | 20.4 ± 5.6 | 20.0 (16.3–24.0) | 20.6 ± 6.2 | 20.0 (16.1–23.9) |
Abbreviations are as follows: AVGO2: Average nocturnal oxygen saturation; PER90: percentage of sleep time at oxygen saturation < 90%; EXCPLSPY: excessive daytime sleepiness; HABSNORE: habitual snoring; AHI: Apnea Hypopnea Index; AVGDUR: Average duration of apnea and hypopneas
Phenotypes
Analysis in this study was performed based on the measures from the last available examination for each participant. Body mass index (BMI) was defined as weight divided by the square of height derived from the same examination as the AHI. Sleep apnea was assessed using overnight sleep studies that consisted of either Type 3 polygraphy (measuring oxygen saturation, body position, airflow using thermistry, chest wall effort, and heart rate; Eden Trace; Eden Prairie, MN) which was performed in participants studied prior to 2000, or by 14-channel polysomnography (Compumedics E series, Abottsford, AU) performed after 2000. The AHI derived from each type of assessment were highly correlated, as described previously [Redline, et al. 2003].
The AHI was computed as the average number of apneas and hypopneas, each associated with a 3% desaturation, per hour of sleep. We also assessed the following sleep traits that each describes specific aspects of the disorder:
Average nocturnal oxygen saturation (AVGO2) and percentage of sleep time at oxygen saturation levels < 90% (PER90), two readily obtainable measures from the sleep study that quantify degree of overnight hypoxemia; extracted from the oximetry signal recorded during the overnight sleep study after manually excluding periods of artifact.
Average respiratory disturbance event duration (hypopnea and apnea; AVGDUR), a measure of the propensity to arouse and terminate a hypopnea or apnea [Berry and Gleeson 1997];
Habitual snoring (HABSNORE), based on self-report of average snoring frequency over the prior month, and reported on a 5-point Likert scale (never, rarely, sometimes, frequently, and almost always or always);
Excessive daytime sleepiness (EXCSLPDY), based on the report of whether the individual experienced “excessive (too much) sleepiness during the day” over the prior month, recorded on a 5-point likert scale (as above).
Genotyping
All the individuals were genotyped using the IBC array [Keating, et al. 2008]. The IBC array includes approximately 50,000 SNPs in cardiovascular, pulmonary, hematological and sleep-related disorders. In addition, 1900 Ancestry Informative Markers (AIMs) were also included in the IBC array for adjusting for the population structure between African and European, and within sub-European populations. Standard QC, including a Mendelian inconsistency check and Hardy-Weinberg equilibrium for each SNP, were performed.
Statistical Analysis
Principal Component Analysis maximizing variance and heritability
Since the traits were skewed, rank normal transformation was first performed for each of the six traits, including both continuous traits (AHI, AVGO2, PER90 and AVGDUR) and two ordinal traits (EXCSLPDY and HABSNORE) in the European and African-American cohorts separately. It has been known the population structure between the European-American and African American are different. One of our goals is to investigate whether the loadings of PCs can be transferred from one population to another. Thus, we did not perform rank normal transformation on European-American and African-American cohorts together. Two PC approaches were performed for the six rank normalized traits: PCV, which maximizes variance without consideration of family structure; and PCH, which maximizes heritability using family data, as introduced by Ott and Rabinowitz [Ott and Rabinowitz 1999]. The PCH was original developed for sib pair data [Ott and Rabinowitz 1999]. Here we applied the method by Wang et al. [Wang, et al. 2007], which is an extension of the method by Ott and Rabinowitz. Briefly, the PCH maximizes the ratio of the family-specific variance to the total variance: where β is the loading vector of PCs, Σg is calculated as the within-family variance-covariance matrix and Σg is calculated as the between-family variance-covariance matrix. It should be cautioned that this PCH will not differentiate variance components attributable to common environmental factors from polygenic variance, therefore only approximately maximizing the heritability.
Heritability Estimation
Heritability ( h2 ) estimates based on individual traits and PCVs and PCHs were determined using Statistical Analysis for Genetic Epidemiology (S.A.G.E.) ASSOC program (v6.1.0), which assumes a linear mixed model with polygenic and random error components. For each trait, PCV or PCH, two models for adjusting for covariates were investigated. Model 1 included the covariates age, age2, gender, age × gender, reflecting non-linear age associations and an age-gender interactions, and two principal components calculated from AIMs for correcting for population stratification [Zhu, et al. 2008]. Model 2 included all the covariates in Model 1 with the addition of BMI and BMI2. BMI and BMI2 were used in model 2 given the high, but non-linear correlation of BMI with indices of OSAHS severity. The corresponding residuals were used for estimating heritability of a trait, PCH or PCV.
Linkage Analysis
Variance components linkage analysis of the rank normalized traits, PCHs and PCVs was performed using SNPs on the IBC array using the software Merlin [Abecasis, et al. 2002]. In this analysis, the total covariance matrix of the residuals after adjusting for covariates was decomposed into three variance components: the variance due the major quantitative trait locus ( ), the variance due to the random polygenic effect ( ), and the variance due to the random environmental effect ( ),
The elements of Π matrix are the identical by descent probability between two related individuals i and j. Φ is the kinship matrix and I is the identity matrix. In particular, when i=j, the total variance of a trait is .
The null hypothesis of no linkage is and the alternative hypothesis is . Merlin uses the likelihood ratio test to test the null hypothesis. Again, we performed linkage analysis for each trait by adjusting for two sets of covariates as demonstrated in Models 1 and 2 above.
Results
PCHs and PCVs to combine six phenotypes
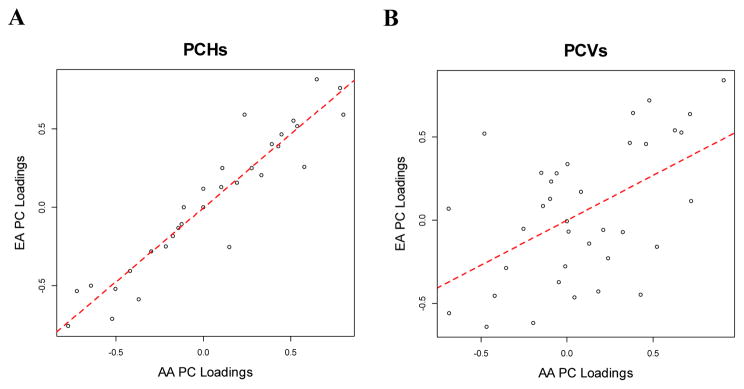
Descriptive statistics for the covariates and phenotypes in the AA and EA samples are shown in Table 1. The loadings of each of the PCVs and PCHs are presented in Table 2. The loadings of the corresponding PCH and PCV are substantially different, suggesting that a PC with the largest variances does not correspond to a PC with the largest heritability. In fact, PCH1, the first PCH, has the largest correlation with PCV2, the second PCV in AAs, while PCH1 has the largest correlation with PCV3 from EAs (Supplemental Table 1). The loadings of the PCHs are highly correlated between EAs and AAs with the correlation coefficient 0.94 (Figure 1A). In contrast, the correlation coefficient of the loadings from PCVs between EAs and AAs is 0.53 (Figure 1B).
Table 2.
Loadings of PCV and PCH in analyzing six OSAHS traits in CFS African American (AA) and European American (EA) cohorts
| PCH | PCV | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | |
| AA | AHI | 0.536 | 0 | 0.101 | 0 | 0.799 | 0.235 | −0.3540 | −0.1503 | 0.4261 | 0.2383 | 0.4779 | 0.6636 |
| AVGDUR | 0.191 | −0.64 | 0.648 | −0.333 | −0.143 | 0 | 0.9098 | −0.2549 | 0.1288 | 0.0803 | −0.1409 | 0.0086 | |
| EXCSLPDY | 0.277 | −0.37 | −0.722 | −0.513 | 0 | 0 | −0.0002 | 0.6256 | 0.3250 | 0.5229 | −0.0989 | −0.1975 | |
| PER90 | 0.445 | 0.427 | 0 | −0.108 | −0.52 | 0.576 | −0.0108 | −0.4203 | −0.4669 | 0.7160 | −0.6907 | 0.0413 | |
| HABSNORE | 0.389 | −0.417 | −0.175 | 0.773 | −0.215 | 0 | 0.2111 | 0.4594 | −0.6853 | −0.0600 | 0.1805 | 0.0006 | |
| AVGO2 | −0.501 | −0.299 | −0.112 | 0.123 | 0.149 | 0.781 | −0.0485 | 0.3652 | −0.0925 | 0.3836 | −0.4819 | 0.7203 | |
| EA | AHI | 0.517 | 0.118 | 0.129 | 0 | 0.591 | 0.59 | −0.287 | 0.283 | −0.447 | −0.227 | 0.719 | 0.5242 |
| AVGDUR | 0.157 | −0.5 | 0.814 | 0.204 | −0.131 | 0 | 0.837 | −0.053 | −0.142 | 0.169 | 0.085 | −0.068 | |
| EXCSLPDY | 0.251 | −0.584 | −0.535 | 0.553 | 0 | 0 | −0.007 | 0.537 | −0.071 | −0.161 | 0.127 | −0.617 | |
| PER90 | 0.464 | 0.389 | 0 | 0.249 | −0.71 | 0.258 | −0.277 | −0.455 | −0.641 | 0.636 | 0.068 | −0.463 | |
| HABSNORE | 0.401 | −0.405 | −0.182 | −0.757 | −0.25 | 0 | −0.058 | 0.455 | −0.558 | 0.281 | −0.429 | 0.336 | |
| AVGO2 | −0.518 | −0.281 | 0 | −0.107 | −0.254 | 0.759 | −0.371 | 0.463 | 0.23 | 0.641 | 0.52 | 0.113 | |
Abbreviations are as follows: AA: African American; EA: European American; AVGO2: Average nocturnal oxygen saturation; PER90: percentage of sleep time at oxygen saturation < 90%; EXCPLSPY: excessive daytime sleepiness; HABSNORE: habitual snoring; AHI: Apnea Hypopnea Index; AVGDUR: Average duration of apnea and hypopneas
Figure 1.
Plots between 36 PC loadings between EAs and AAs in PCH (A) and PCV (B).
Heritability of Individual Traits and the PCHs and PCVs
The estimated heritability for individual traits, PCHs and PCVs are presented in Figure 2 and Supplemental Table 2. In the individual trait analysis, heritability of the AHI, which is the traditional metric for sleep apnea, was 0.272 and 0.311 in AAs and EAs respectively for the age, gender and BMI-adjusted model. All individual traits were heritable other than HABSNORE in the EAs. Of the significantly heritable traits (P<0.05) in the EAs, heritability ranges from 0.261 (EXCSLPDY) to 0.401 (AVGDUR) and 0.409 (AVGO2) for BMI-adjusted models (Supplemental Table 2). In AAs, all traits are significantly heritable, ranging from 0.218 (HABSNORE) to 0.608 (AVGDUR) for BMI-adjusted models (Supplemental Table 2). Overall, heritability estimates are similar in BMI-adjusted and unadjusted models.
Figure 2. Heritability estimated for six individual traits (HABSNORE, EXCSLPDY, AHI, AVGO2, PER90 and AVGDUR) and six PCVs and six PCHs in models with and without BMI and BMI2 as covariates.
A. Heritability of each individual trait in EAs. B. Heritability of each individual trait in AAs. C. Heritability of PCVs in EAs. D. Heritability of PCVs in AAs. E. Heritability of PCHs in EAs. F. Heritability of PCHs in AAs.
For PCVs, the maximum heritability is observed for PCV2 in both EAs and AAs (0.513 and 0.538, respectively) (Supplemental Table 2). Heritability is not substantively changed with or without BMI adjustment. In contrast, for PCHs, the maximum heritability for EAs and AAs is observed for PCH1 with estimated heritability 0.512 and 0.657 respectively (Supplemental Table 2).
Linkage analysis of Individual Traits and PCs from PCV and PCH
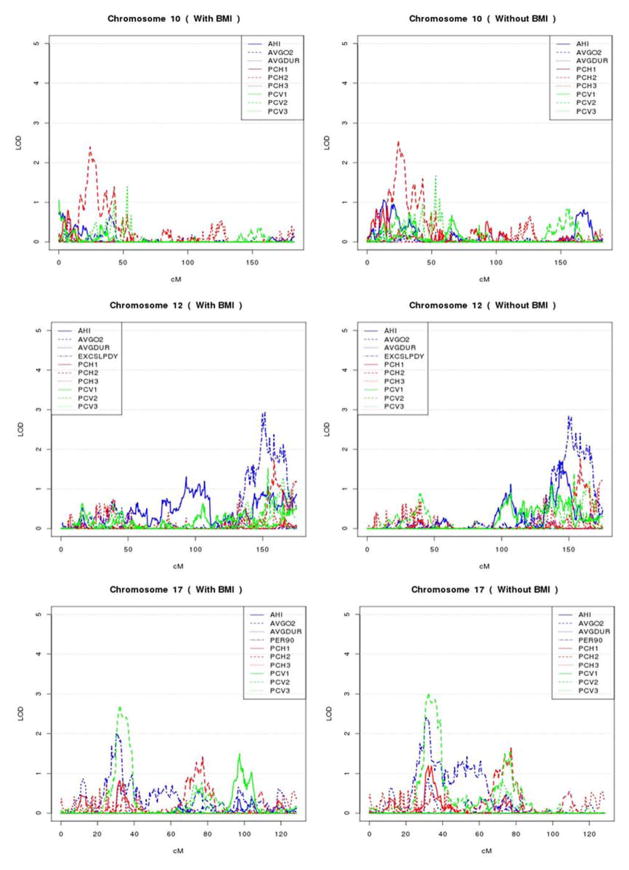
We performed linkage analysis using individual traits, PCHs and PCVs. There are 6 and 8 regions with LOD score > 2.0 for at least one trait in each of the AA and EA populations, respectively, which are summarized in Table 3 and Figures 3 and 4. Among these 14 linkage regions, both PCH and PCV analyses have the largest LOD scores six times and individual traits analyses have the largest LOD scores two times. Among the 14 linkage regions, 10, 9 and 7 regions have LOD scores larger than 2.0 for PCH, PCV and individual trait analysis, respectively. In general, the linkage evidence identified by individual trait analysis could also be identified by PCH or PCV analysis, except for the two regions on chromosome 12 at 150 cM and chromosome 8 at 48 cM that were identified for EXCSLPDY and PER90, respectively. In contrast, 7 linkage regions identified by PCH or PCV analysis would have been missed by individual trait analyses. Overall, PCH and PCV analysis resulted in a greater number of linkage peaks with LOD scores > 2.0 as well as larger LOD scores than those from an individual trait analysis (Table 3). The phenomena are also true when examining EAs or AAs separately. The two most improved linkage peaks by PCH and PCV analysis are at 16.2 cM and 21.2 cM on chromosome 8, with maximum LOD scores 4.6 and 4.83 identified by PCH3 and PCV1 respectively in EAs, in contrast to the maximum LOD score of 3.23 and 4.71 for AVGO2 and PER90. These two peaks may reflect the same peak since they are only 5 cM away. Comparing PCH and PCV analysis, 7 linkage regions could be identified by both PCH and PCV analysis, although they may correspond to different PCs. Interestingly, all the regions identified by PCHs and PCVs are restricted to the first three PCs.
Table 3.
Summaries of linkage regions with linkage peak LOD > 2 in both EAs and AAs.
| Chr | Region in cM | Region in BP | Maximum LOD(Trait) | Traits with LOD>2 | |
|---|---|---|---|---|---|
| AAs | 2 | 143.502~145.725 | 121881498~123834583 | 2.221 (PCH1) | PCH1 |
| 4 | 195.304~214.765 | 181408817~188517935 | 2.849 (PCV2) | PCV2 | |
| 5 | 180.937~182.707 | 168147329~168571866 | 2.487 (PCV2) | PCV2; PCH1 | |
| 10 | 22.934~28.636 | 10414194~12797758 | 2.561 (PCH2) | PCH2 | |
| 12 | 148.622~165.362 | 124851420~129867979 | 2.914 (EXCSLPDY) | EXCSLPDY | |
| 17 | 30.235~38.844 | 11393896~14290156 | 3.002 (PCV2) | PCV2; PER90 | |
| EAs | 6 | 61.753~67.369 | 41552005~44211867 | 2.606 (PCH1) | PCH1; PCV2 |
| 8 | 16.142~16.591 | 6263083~6424907 | 4.603 (PCH3) | PCV1; PCH3 AVGO2 | |
| 8 | 18.165~22.662 | 6828813~10888096 | 4.83 (PCV1) | PCV1; PCH3; AVGO2; PER90; | |
| 8 | 36.014~54.054 | 17157677~27399518 | 3.52 (PER90) | PER90 | |
| 8 | 94.429~102.106 | 77437702~90802045 | 2.894 (PCH2) | PCH2 | |
| 14 | 44.458~60.275 | 51251157~65573162 | 2.417 (PCH3) | PCH3; PCV1 | |
| 14 | 96.296~101.303 | 95439228~ 97601602 | 3.429 (PCV1) | PCH3; PCV1; AHI; | |
| 14 | 105.457~112.999 | 99588940~103588515 | 3.289 (PCV1) | PCH3; PCV1; AHI; |
Figure 3. Variance component linkage analysis of chromosome 2q, 4q, 5q, 10q, 12q, and 17q in African Americans.
Genetic distance in cM is plotted against the multipoint variance component LOD score for different individual traits, PCVs and PCHs with BMI adjusted or unadjusted.
Figure 4. Variance component linkage analysis of chromosome 6q, 8q, and 14q in European Americans.
Genetic distance in centimorgans is plotted against the multipoint variance component LOD score for different individual traits, PCHs and PCVs with BMI adjusted or unadjusted.
Discussion
The comparison of heritability estimation and linkage analysis for multiple sleep apnea traits in this study has important inferences. These analyses supported the utility of PCHs in genetic analyses of family data and the PCHs can be transferred across populations. Principal components generally result in higher heritability and linkage evidence with an increased number of linkage peaks identified in comparison to individual trait analysis.
In calculating PCHs and PCVs between AAs and EAs, we observed a high correlation among the loadings of the PCHs between the two populations of AAs and EAs (Figure 1). In contrast, the loadings of PCVs were less strongly correlated. This result suggests that common genetic mechanisms can contribute the top PCHs and these genetic mechanisms are transferable between African Americans and European Americans. In comparison, PCVs are less likely to capture the underlying common genetic mechanisms, possibly due to different environmental factors across populations. However, whether this conclusion is generalizable for other complex traits in other populations needs further analysis.
In general, the PCHs have higher heritability estimates than PCVs. Individual traits analysis resulted in the lowest heritability estimates. It is not surprising that a PCV may not correspond to a PCH, as suggested by our results. For example, the first PCH is most strongly correlated to the second PCV in AAs with R2 = 0.61 and the third PCV in EAs with R2 =0.59. For PCHs, heritability always decreases across PCH1 to PCH6, while PCV2 has the maximum heritability in both EAs and AA. This result suggests that PCV1 may not have an advantage over the other PCVs in detecting genetic factors, which is consistent with the conclusion by Aschard et al. [Aschard, et al. 2014]. In both PCH and PCV analysis, the first three PCs capture most of trait heritability, and all three PCs result in higher heritability estimates than individual trait analysis, suggesting an advantage of using PCs in genetic analyses.
In the linkage analysis using individual traits as well as PCHs and PCVs, we also observed that PCs usually resulted in higher LOD scores and more linkage peaks with LOD scores > 2 than individual traits (Table 3), suggesting that PC analysis has advantages for searching for genetic variants for complex traits. In particular, most of the linkage peaks from individual trait analysis could also be identified by PC analysis. Specifically, only two linkage peaks (chromosome 8 for PER90 in EAs and 12 for EXCSLPDY in AAs) from the individual trait analysis could not be detected by either PCHs or PCVs. In contrast, the linkage peaks identified by PCHs or PCVs usually were not identified by individual trait analysis. For example, 7 linkage regions identified by PCHs or PCVs could not been detected by individual trait analysis. In particular, we observed a substantial improvement of linkage evidence on chromosome 8 at 16.2 cM using PCH or PCV analysis over the individual trait analysis, with the LOD score for the highest individual trait (AVGO2) of 3.23 compared to a LOD score of greater than or equal to 4.6 for the PC traits. In a combined linkage and association analysis of AVGO2, we identified that the ANGPT2 gene in this linkage region potentially contributes to the observed linkage evidence of AVGO2 (data not shown). Furthermore, all the linkage peaks identified in both PCHs and PCVs are from the first three PCs, suggesting that an approach that focuses on the top PCs may be advantageous; however, the number of PCs which should be analyzed remains a question for future studies. In practice, we suggest to examine the eigenvalues of the variance-covariance matrix. For example, we can only study the top PCs which account for over 90% total trait variability. Limiting to the top PCs could substantially reduce the penalty accrued due to multiple comparisons. In addition, analysis of linkage peaks can enhance the detection of rare variants [Zhu, et al. 2010].
Multivariate linkage analysis employs a multivariate variance components approach which can test multiple parameters, such as pleiotropic effect, and has been suggested to have greater power to identify genetic loci with small effects than a single trait analysis [Iturria et al. 2000, Turner et al. 2004]. However, for many complex diseases, genetically relevant disease definition is not very clear and people tend to collect large numbers of phenotypes related directly or indirectly to the disease. If combining all the traits, multivariate linkage analysis usually have difficult computational task burden [Oualkacha et al. 2012]. Alternatively, principal components of heritability (PCH), as a dimension reduction technique, capture the familiar information across traits by calculating linear combinations of trait that maximize heritability. Thus, top PCHs capturing most of trait heritability can be used to do linkage analysis, which is advantageous.
Finally, it should also be noted that an analysis of multiple phenotypes provides opportunities to discover genetic signals that may not emerge with single trait analysis. For example, recent studies in the psychiatric literature indicate that analysis of multiple phenotypes that likely reflect pleiotropy have allowed discovery of genetic variants that are common to several related disorders [Cross-Disorder Group of the Psychiatric Genomics 2013]. For hypertension, an analysis combining systolic blood pressure, diastolic blood pressure and hypertensive status identified four genes which were missed by individual trait analysis [Zhu, et al. 2015]. Combining data across traits may also enhance the reliability of measurement, as has been proposed for studies of inflammatory pathways [Fibrinogen Studies 2009]. Use of data from several related traits may provide an enriched description of the phenotype, which may improve its association with genetic variants that influence such features. For example, for OSAHS, the AHI is usually used clinically and in genetic studies. However, as a simple count of the number of respiratory disturbances per hour of sleep, it does not provide information on key physiological processes that likely are heritable-such as ventilatory arousability and propensity for oxygen desaturation and sleepiness. Notably, we found that respiratory event duration (AVGDUR) was among the most heritable individual traits. Although this trait has never been analyzed in genetic epidemiological studies of OSAHS, recent data suggest that this trait better predicts mortality than does the AHI [Butler, et al. 2015]. Furthermore, ventilatory arousability is considered to be a fundamental risk factor for OSAHS. AVGDUR loads on PCV1 and PCH2 and PCH3, which may improve the heritability estimate of these PCs. Other factors loading on the top 3 PCs include AHI, EXCSLPDY, AVGO2 for PCH and AHI, EXCSLPDY, HABSNORE for PCV, and suggest that OSAHS is better described when considering multiple aspects of physiological sleep disturbance.
The strengths of this study include its consideration of the influence of alternative approaches for analyzing multiple traits through heritability and linkage analysis and the availability of phenotype and genotype data for both European-American and African-American families. Each of the six sleep apnea traits underwent rank normalization before analysis ensuring that the trait distributions were normal, but reducing statistical power in linkage analysis. Since the goal of this study is to compare two PC approaches and individual traits in genetic analysis, including heritability estimating and linkage analysis, ensuring normality for each trait enhanced their comparability. Using both EA and AA families enabled us to make a comparison across populations. Finally we were able to compare the linkage results among different phenotype approaches as well as across populations. However, our study does have limitations. First, the heritability estimated by PC approaches may be over-estimated because within-family shared environmental effects that are confounded with genetic effects. In fact, this is a general problem for heritability estimation using family data. Second, we only analyzed sleep apnea traits from CFS in which a similar design was applied to European and African American families. It is still not clear whether our general conclusion of PCHs being better than individual traits in trans-ethnic analysis can be generalized to different complex diseases. However, it has already been suggested that the effect sizes of genetic variants detected in many GWAS are transferable, including hypertension related traits [Franceschini, et al. 2013]. Finally, our study sample sizes are relatively small and have limited power in linkage analysis. For the linkage peaks detected in PCHs in either European or African American families, we did not observe any peaks with a LOD score larger than 2 in the other population, which could be attributed to low statistical power. We did not obtain PCs when maximizing either linkage evidence or association evidence, as proposed in [Klei et al. 2008]. Therefore, the approaches used in the current study will not lead to biased inference in the subsequent association or linkage analysis. However, it should be pointed out that the method by Klei et al. 2008 should be more powerful than that first calculating PCVs or PCHs then performing association analysis. In this study, we have not studied how PCVs and PCHs affect family-based association analysis. Similar to linkage analysis, we expect PCVs and PCHs will improve statistical power but this is not always true, as suggested by Aschard et al. [2014].
In summary, we compared two PC approaches with individual trait analysis in genetic analysis of six sleep apnea traits collected from CFS data. Our study strongly suggests that PC based approaches, especially PCs calculated by maximizing trait heritability have multiple advantages in genetic studies. A common genetic mechanism of PCHs across ethnic populations may provide useful traits for trans-ethnic analysis and association analysis aimed to detect both rare and common genetic variants underlying complex traits.
Supplementary Material
Acknowledgments
The work was supported by the National Institutes of Health grants HG003054 from the National Human Genome Research Institute, HL113338 and HL046380 from the National Heart, Lung, Blood Institute, and RR024990. In addition, some of the results of this paper were obtained by using the software package S.A.G.E., which is supported by a U.S. Public Health Service Resource Grant (RR03655) from the National Center for Research Resources.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Aschard H, Vilhjalmsson BJ, Greliche N, Morange PE, Tregouet DA, Kraft P. Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies. Am J Hum Genet. 2014;94(5):662–76. doi: 10.1016/j.ajhg.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran M, Yassin BA, Fox N, Laher I, Ayas N. Epidemiology of Sleep Disturbances and Cardiovascular Consequences. Can J Cardiol. 2015 doi: 10.1016/j.cjca.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Berry RB, Gleeson K. Respiratory arousal from sleep: mechanisms and significance. Sleep. 1997;20(8):654–75. doi: 10.1093/sleep/20.8.654. [DOI] [PubMed] [Google Scholar]
- Butler MP, Emch JT, Rueschman M, Lasarev M, Wellman A, Shea SA, Redline S. Apnea Duration and Inter-apnea Interval as Predictors of Mortality in a Prospective Study. Sleep. 2015;38:A160. Abstract Supplement, 2015. [Google Scholar]
- Capdevila OS, Kheirandish-Gozal L, Dayyat E, Gozal D. Pediatric obstructive sleep apnea: complications, management, and long-term outcomes. Proc Am Thorac Soc. 2008;5(2):274–82. doi: 10.1513/pats.200708-138MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie AG, Mahaney MC, Almasy L, Dyer TD, Blangero J. Exploiting pleiotropy to map genes for oligogenic phenotypes using extended pedigree data. Genet Epidemiol. 1997;14(6):975–80. doi: 10.1002/(SICI)1098-2272(1997)14:6<975::AID-GEPI69>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibrinogen Studies C. Correcting for multivariate measurement error by regression calibration in meta-analyses of epidemiological studies. Stat Med. 2009;28(7):1067–92. doi: 10.1002/sim.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, Tayo BO, Sun YV, Gottesman O, Adeyemo A, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet. 2013;93(3):545–54. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu CC, Flores HR, de las Fuentes L, Davila-Roman VG. Enhanced detection of genetic association of hypertensive heart disease by analysis of latent phenotypes. Genet Epidemiol. 2008;32(6):528–38. doi: 10.1002/gepi.20326. [DOI] [PubMed] [Google Scholar]
- Iturria SJ, Blangero J. An EM algorithm for obtaining maximum likelihood estimats in the multi-phenotype variance componnets linkage model. Ann Hum Genet. 2000;64(Pt 4):349–62. doi: 10.1017/S0003480000008228. [DOI] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3(10):e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klei L, Luca D, Devlin B, Roeder K. Pleiotropy and principal components of heritability combine to increase power for association analysis. Genet Epidemiol. 2008;32(1):9–19. doi: 10.1002/gepi.20257. [DOI] [PubMed] [Google Scholar]
- Larkin EK, Patel SR, Elston RC, Gray-McGuire C, Zhu X, Redline S. Using linkage analysis to identify quantitative trait loci for sleep apnea in relationship to body mass index. Ann Hum Genet. 2008;72(Pt 6):762–73. doi: 10.1111/j.1469-1809.2008.00472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbata G, Chukwuka J. Obstructive sleep apnea hypopnea syndrome. Ann Med Health Sci Res. 2012;2(1):74–7. doi: 10.4103/2141-9248.96943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Xu F, Babineau DC, Tracy RP, Jenny NS, Patel SR, Redline S. Sleep-disordered breathing and prothrombotic biomarkers: cross-sectional results of the Cleveland Family Study. Am J Respir Crit Care Med. 2010;182(6):826–33. doi: 10.1164/rccm.201001-0020OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrien PC. Applied Multivariate Statistical-Analysis - Johnson, Ra, Wichern, Dw. Journal of the American Statistical Association. 1984;79(385):231–231. [Google Scholar]
- Ott J, Rabinowitz D. A principal-components approach based on heritability for combining phenotype information. Hum Hered. 1999;49(2):106–11. doi: 10.1159/000022854. [DOI] [PubMed] [Google Scholar]
- Oualkacha K, Labbe A, Ciampi A, Roy MA, Maziade M. Principal components of heritablity for high dimension quantitative traits and general pedigrees. Stat Appl Genet Mol Biol. 2012;11(2) doi: 10.2202/1544-6115.1711. [DOI] [PubMed] [Google Scholar]
- Palmer LJ, Buxbaum SG, Larkin E, Patel SR, Elston RC, Tishler PV, Redline S. A whole-genome scan for obstructive sleep apnea and obesity. Am J Hum Genet. 2003;72(2):340–50. doi: 10.1086/346064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redline S, Schluchter MD, Larkin EK, Tishler PV. Predictors of longitudinal change in sleep-disordered breathing in a nonclinic population. Sleep. 2003;26(6):703–9. doi: 10.1093/sleep/26.6.703. [DOI] [PubMed] [Google Scholar]
- Redline S, Tishler PV. The genetics of sleep apnea. Sleep Med Rev. 2000;4(6):583–602. doi: 10.1053/smrv.2000.0120. [DOI] [PubMed] [Google Scholar]
- Redline S, Tishler PV, Tosteson TD, Williamson J, Kump K, Browner I, Ferrette V, Krejci P. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med. 1995;151(3 Pt 1):682–7. doi: 10.1164/ajrccm/151.3_Pt_1.682. [DOI] [PubMed] [Google Scholar]
- Rosen CL, Larkin EK, Kirchner HL, Emancipator JL, Bivins SF, Surovec SA, Martin RJ, Redline S. Prevalence and risk factors for sleep-disordered breathing in 8- to 11-year-old children: association with race and prematurity. J Pediatr. 2003;142(4):383–9. doi: 10.1067/mpd.2003.28. [DOI] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14(7):483–95. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. A unified framework for association analysis with multiple related phenotypes. PLoS One. 2013;8(7):e65245. doi: 10.1371/journal.pone.0065245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner ST, Kardia SL, Boerwinkle E, de Andrade M. Multivariate linkage analysis of blood pressure and body mass index. Genet Epidemiol. 2004;27(1):64–73. doi: 10.1002/gepi.20002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fang Y, Wang S. Clustering and principal-components approach based on heritability for mapping multiple gene expressions. BMC Proc. 2007;1(Suppl 1):S121. doi: 10.1186/1753-6561-1-s1-s121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T, Skatrud J, Peppard PE. Risk factors for obstructive sleep apnea in adults. JAMA. 2004;291(16):2013–6. doi: 10.1001/jama.291.16.2013. [DOI] [PubMed] [Google Scholar]
- Zhu X, Feng T, Li Y, Lu Q, Elston RC. Detecting rare variants for complex traits using family and unrelated data. Genet Epidemiol. 2010;34(2):171–87. doi: 10.1002/gepi.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, Smith JA, Yanek LR, Sun YV, Edwards TL, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am J Hum Genet. 2015;96(1):21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Li S, Cooper RS, Elston RC. A unified association analysis approach for family and unrelated samples correcting for stratification. Am J Hum Genet. 2008;82(2):352–65. doi: 10.1016/j.ajhg.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.