Abstract
Familial hypertrophic cardiomyopathy (FHC) is associated with mild to severe cardiac problems and is the leading cause of sudden death in young people and athletes. Although the genetic basis for FHC is well-established, the molecular mechanisms that ultimately lead to cardiac dysfunction are not well understood. To obtain important insights into the molecular mechanism(s) involved in FHC, hearts from two FHC troponin T models (Ile79Asn [I79N] and Arg278Cys [R278C]) were investigated using label-free proteomics and metabolomics. Mutations in troponin T are the third most common cause of FHC, and the I79N mutation is associated with a high risk of sudden cardiac death. Most FHC-causing mutations, including I79N, increase the Ca2+ sensitivity of the myofilament; however, the R278C mutation does not alter Ca2+ sensitivity and is associated with a better prognosis than most FHC mutations. Out of more than 1200 identified proteins, 53 and 76 proteins were differentially expressed in I79N and R278C hearts, respectively, when compared with wild-type hearts. Interestingly, more than 400 proteins were differentially expressed when the I79N and R278C hearts were directly compared. The three major pathways affected in I79N hearts relative to R278C and wild-type hearts were the ubiquitin-proteasome system, antioxidant systems, and energy production pathways. Further investigation of the proteasome system using Western blotting and activity assays showed that proteasome dysfunction occurs in I79N hearts. Metabolomic results corroborate the proteomic data and suggest the glycolytic, citric acid, and electron transport chain pathways are important pathways that are altered in I79N hearts relative to R278C or wild-type hearts. Our findings suggest that impaired energy production and protein degradation dysfunction are important mechanisms in FHCs associated with poor prognosis and that cardiac hypertrophy is not likely needed for a switch from fatty acid to glucose metabolism.
Familial hypertrophic cardiomyopathy (FHC)1 is the most common monogenically inherited heart disease, estimated to affect one in 500 people (1, 2). It is characterized by thickening of the left ventricle, contractile dysfunction, heart failure, and a high incidence of potentially lethal arrhythmias (3). Over a thousand mutations in more than twenty genes have been identified that cause FHC, most of which encode proteins of the sarcomere; however, the pathological mechanisms of this disease are not well understood (1, 3, 4). Mutations in cardiac troponin T (TnT), encoded by TNNT2, carry a particularly poor prognosis for the patient and are associated with a high risk of sudden cardiac death (SCD) (5–8). Most FHC-causing mutations alter how the sarcomere responds to Ca2+ by increasing the myofilament Ca2+ sensitivity, which changes its contractile properties and affects Ca2+ handling in the cell (9, 10). Proposed pathological mechanisms include altered cardiac contractility, changes in Ca2+ handling, and altered energy homeostasis (1, 11). Current treatments include β-adrenergic receptor blockers, Ca2+ channel antagonists, myocardial reduction by surgical or septal ablation, and implanted defibrillators (2). Currently available pharmacological treatments can provide relief from symptoms and improve patient quality of life, but they do not target FHC-specific pathways and have not been shown to slow disease progression (1). An improved understanding of the molecular mechanisms of FHC is needed to develop specific therapies that address the underlying causes of the disease (2).
Alterations of proteins and pathways that are comorbid with heart diseases like dilated cardiomyopathy, congestive heart failure, and myocardial infarction have been identified by protein mass spectrometry (12). To gain a better understanding of the pathways affected in TnT-related FHC, we utilized a label-free proteomic approach to investigate how hearts from mice with two different TnT mutations (Ile79Asn [I79N] and Arg278Cys [R278C]) are affected at the protein level. These mice (wild-type (WT), R278C, and I79N) all express human TnT at ∼50% of the total TnT with the endogenous mouse TnT accounting for the other 50% (13, 14). Transgenic mice expressing human TnT are a useful tool for investigating the disease, as mice with the R278C or I79N mutation show similar features to humans; humans with the I79N mutation are at a high risk of sudden cardiac death, and those with the R278C mutation show a milder phenotype (7, 15–17). To determine changes at the metabolite level, a time-of-flight gas chromatography mass spectrometer system (GC-TOF) was utilized to determine metabolite levels in the different transgenic hearts. A major advantage of this investigation is the comparison of two different FHC models: hearts containing the myofilament Ca2+-sensitizing I79N mutation that is associated with poor prognosis and SCD, and the R278C mutation, which does not alter myofilament Ca2+ sensitivity and is associated with mild, late-onset cardiac effects and no SCD (7, 15–17).
No proteomic or metabolomic comparisons between mild and severe models of FHC related to sarcomeric mutations have been reported. Our data reveal that although metabolic pathways and stress responses were affected in both R278C and I79N hearts, a major protein complex affected differently by the two mutations was the proteasome, a large, ∼2 MDa complex that is conserved in eukaryotes and responsible for degrading the majority of cytosolic, nuclear, and nascent membrane proteins following their modification with the small protein ubiquitin (18). Through targeted degradation of proteins that are no longer required by the cell, the proteasome plays a role in virtually all cellular processes (19), and particularly in cardiomyocyte proteostasis, as these cells are prone to protein damage due to continual mechanical and metabolic stresses (20, 21). The heart has very limited capacity for self-renewal, meaning that cell death, which may result from impaired proteasome function, can be highly detrimental to the health of the organ (20, 22). A growing body of evidence indicates proteasome dysfunction is an important contributor to the pathogenesis of many cardiac diseases. In FHC caused by certain mutations in the thick filament-associated protein cardiac myosin binding protein-C (MyBP-C), the mutated protein is preferentially degraded by the proteasome, competitively inhibiting breakdown of other proteasome substrates (23–26). However, some single residue MyBP-C mutants do not seem to affect ubiquitin-dependent proteasomal proteolysis, and the proteasome has not been implicated in any other non-MyBP-C FHC model (23). To investigate the effect of TnT mutations on proteasomal activity, expression and activity of the proteasome was measured, revealing increased proteasome subunit expression in I79N hearts relative to R278C hearts but decreased constitutive proteasome and immunoproteasome activities in I79N mice relative to WT and R278C hearts. This is the first report of immunoproteasome activity being measured in heart tissue from any animal. Combined proteomic, metabolomic, immunological, and biological results all suggest that pathways associated with energy metabolism, protein degradation, and oxidant defense are major pathways that are altered. These studies show increased levels of enzymes associated with energy metabolism in I79N hearts and suggest I79N hearts favor glucose as a substrate for energy metabolism whereas R278C hearts favor fatty acids. Thus our findings suggest altered metabolism and proteasome function are likely important molecular mechanisms involved in TnT-related FHC.
EXPERIMENTAL PROCEDURES
Materials
Urea, DTT, triethylphosphine, iodoethanol, and ammonium bicarbonate (NH4HCO3) were purchased from Sigma-Aldrich (St. Louis, MO). LC-MS grade 0.1% formic acid in ACN and 0.1% formic acid in water (H2O) were purchased from Burdick & Jackson (Muskegon, MI). Modified sequencing grade porcine trypsin was obtained from Princeton Separations (Freehold, NJ).
Experimental Animals
All experiments involving animals were approved by the University of California, Davis Institutional Animal Care and Use Committee. Transgenic TnT mouse lines generated using the background mouse strain BL6SJF1/J were used (13, 14). Transgenic male mice were bred with B6SJLF1/J female mice purchased from the Jackson Laboratory (Sacramento, CA). Mice were fed standard chow and water ad libitum and maintained on a 12 h dark/light cycle. Male offspring were used in all studies. DNA was isolated from tail snips taken from mice at the time of weaning or sacrifice for genotyping. Transgenic mice were identified by performing a PCR reaction with primers to the 630-base pair 3′-untranslated region from the human growth hormone transcript that is included in the transgene insert (forward primer CTC CTG GCC CTG GAA GTT; reverse primer CTG GCC AAT ATG GTG AAA CC) (13, 14).
Tissue Collection
Three month old transgenic TnT mice were used in these studies, an age equivalent to young adulthood in humans. Mice were euthanized with isoflurane (3%), followed by cervical dislocation, and hearts were immediately exercised and briefly washed with ice-cold PBS, weighed, and flash frozen. Hearts prepared using this method has been previously shown to be excellent for subsequent proteomic investigation (27, 28).
Experimental Design and Statistical Rationale
Label-Free Proteomics
To 20 mg of pulverized aged-matched heart muscle each from 3 WT, 4 R278C, and 4 I79N mice 300 μl of 8 m urea was added. As described by Sengupta et al. (29), a minimum of triplicate biological samples with a significance level of 0.05, a 30% standard deviation, and large effect size, is needed to give a power of 0.69. Each tissue was treated by light sonication and mixing for 1 h followed by centrifugation at 13,000 rpm for 10 min. The protein concentration was measured by Bradford assay (30). 20 μl (100 μg) was removed and 20 μl 100 mm ammonium carbonate, pH 10.8 and 1 ng chicken lysozyme (to serve as an internal standard) were added to the samples. 40 μl of reduction/alkylation mixture (97.5% ACN, 2% iodoethanol and 0.5% triethylphosphine) was added to each sample, and the samples were incubated in a 37 °C incubator for 1.5 h. The samples were speed vacuumed to dryness overnight, and the dry pellets were resuspended in 50 μl ammonium bicarbonate. 2.5 μg trypsin in 100 μl ammonium bicarbonate was added to each sample, and they were incubated at 37 °C for 4 h. 2.5 μg trypsin in 50 μl ammonium bicarbonate was then added to each sample, and they were incubated at 37 °C overnight.
LC-MS/MS
The digested samples were analyzed using a Thermo Scientific Orbitrap Velos Pro mass spectrometer coupled with a Surveyor autosampler and MS HPLC system (Thermo Fisher Scientific, Waltham, MA). Each biological sample was run two independent times. Tryptic peptides (K/R cleavages) were injected onto a C18 reversed phase column (TSKgel ODS-100V, 3 μm, 1.0 mm × 150 mm) at a flow rate of 50 μl/min. The mobile phases A and B were LC-MS grade H2O with 0.1% formic acid and ACN with 0.1% formic acid, respectively. The gradient elution profile was as follows: 5% B for 5 min, 10–35% B for 150 min, 35–80% B for 10 min, 80% B for 10 min, and 5% B for 5 min. The data were collected in the “Data dependent MS/MS” mode of FT-IT (MS-MS/MS) with the ESI interface using normalized collision energy of 35% (CID). Dynamic exclusion settings were set to repeat count 1, repeat duration 30 s, exclusion duration 120 s, and exclusion mass width 10 ppm (low) and 10 ppm (high).
Peptide/Protein Identification and Quantification
The acquired data were searched against the UniProt protein sequence database of MOUSE (released on 03/19/2014) using X!Tandem algorithms in the Trans-Proteomic Pipeline (TPP, v. 4.6.3) (http://tools.proteomecenter.org/software.php). The Uniprot database used contained 102,778 entries including the decoy sequences (51,389 entries without decoys). General parameters were set to: parent monoisotopic mass error set as 10 ppm, cleavage semi set as yes, missed cleavage sites set at 2, static modification set as + 44.026215 Da on cysteine and no variable modifications. Although not including modifications reduced the number of peptides detected it also prevents ambiguous modification site assignments. A mass tolerance of 0.8 Da for fragment ion was used and only trypsin as a known contaminant was excluded in the analysis. The threshold score/expectation value for accepting individual spectra was the default value in the X!Tandem program. The searched peptides and proteins were validated by PeptideProphet (31) and ProteinProphet (32) in the TPP. Only proteins and peptides with protein probability ≥0.9000 and peptide probability ≥0.8000 were reported. Protein quantification was performed using a label-free quantification software package, IdentiQuantXLTM (33). Using this IdentiQuantXLTM method complex biological samples containing identical amounts of spiked protein standard showed a coefficient of variation (CV) of 8.6% across eight injections between two groups (33). This program individually aligns the retention time of each peptide and applies multiple filters for exclusion of unqualified peptides to enhance label-free protein quantification. Briefly, the protein ID, sequence, charge, m/z, scan time, injection, sample, and group information are extracted and collected for each peptide, and peptides with identical sequence and charge are considered as a single peptide entry for further analysis (33). After peptide frequency is calculated, the first filter removes peptides that were not identified in at least three different biological replicates. Alignment to determine peptide retention time was carried out using clustering. Peptide intensity extraction was carried out using “Acquisition time (minutes)” and “Limit the search to only custom m/z values (ignoring auto-fragmented m/z's)” under Custom SIC Options in the program MASIC (33). The CV for each peptide in each sample was calculated, and Pearson's correlation (for ≥3 variables) and uncentered Pearson's correlation (for 2 variables) methods were utilized to determine the correlation coefficient of peptides that correspond to a unique protein ID. The average CV of all the peptides in this investigation was 14.9%. The intensity of each peptide was compared with all other peptides, and the average was used as the final correlation coefficient value. The mass spectrometry data used in this manuscript have been deposited in the MassIVE (MSV000079538) and ProteomeXchange (PXD003703) databases.
Protein abundance was calculated using Ap = ∑i = 1n(IpFp)i.
Where Ap = protein abundance, Ip = peptide intensity, and Fp = frequency of peptide sharing. When a peptide was shared by different proteins, the intensity of this peptide (Ip) was divided by the sharing frequency (Fp), which decreases the impact of shared peptides (33). The quantification values were averaged over technical replicates, and the resulting values were then averaged over biological replicates. Comparisons between WT, R278C, and I79N samples were carried out using one-way ANOVA (supplemental Table S2). A value of p < 0.05 was considered statistically significant. False discovery rate (FDR) was estimated by a nonparametric concatenated randomized target-decoy database search (34). For this experiment and the TPP settings used, protein identification FDR was ≤ 2.35%.
Proteolytic Activity Assays
Powdered mouse cardiac tissue was homogenized in ice cold 1X homogenization buffer (50 mm Tris, 1 mm EDTA, 150 mm NaCl, 5 mm MgCl2, 0.5 mm DTT, pH 7.5) using 25 strokes with a glass dounce homogenizer. The homogenates were centrifuged for 30min at 12,000 × g at 4 °C, and the supernatants removed, quantified with a Nanodrop (Thermo Scientific), and normalized to equal protein concentrations. A portion of the samples normalized to the same concentration were used for SDS-PAGE to verify that the samples were equal in protein amount. The proteolytic activities of the different proteasome and calpain proteases were quantified by measuring the degradation of specific fluorogenic substrates using a kinetic protocol in the presence or absence of a specific inhibitor. Unless otherwise noted, all reactions were carried in 96-well flat black plates, run for 75 min at 37 °C, and measured using excitation and emission wavelengths of 390 nm and 460 nm respectively.
26S Proteasome Activity
Protein sample (25 μg) was combined with 1× homogenization buffer, bortezomib (bort) or an equal volume of DMSO, and 100 μm ATP, and incubated for 20 min at room temperature. The reaction was initiated by adding the specific substrate at a final concentration of 100 μm. Final concentrations of bort and specific fluorogenic substrates for each proteolytic proteasome subunit, respectively were: β1 - 100 μm bort, Z-Leu-Leu-Glu-MCA (Peptide Institute, 3179-v), β5 - 10 μm bort, Suc-Leu-Leu-Val-Tyr-AMC (Bachem, Bubendorf, Switzerland, I-1395). These assays have been previously described (35, 36).
20S Proteasome Activity
25 μg protein sample was combined with 1× Chemicon Buffer (25 mm HEPES, 0.5 mm EDTA, 0.05% Nonidet P-40, 0.001% SDS, pH 7.5), bort or an equal volume of DMSO, and water and incubated for 20 min at room temperature. The reaction was initiated by adding Suc-Leu-Leu-Val-Tyr-AMC at a final concentration of 100 μm to measure β5 activity. The final concentrations of bort and Suc-Leu-Leu-Val-Tyr-AMC were the same as described in the 26S proteasome activity section.
Immunoproteasome Activity
Protein sample (10 μg), Immunoproteasome Buffer (50 mm Tris, 5 mm MgCl2, 20 mm KCl, 1 mm DTT, pH 7.5), and inhibitor or an equal volume of DMSO were combined and incubated for 20 min at room temperature. The reactions were initiated by the addition of 25 μm fluorogenic substrate. The reaction was run at 37 °C for 60 min, and the fluorescence intensity was measured every 5 mins (498 excitation wavelength, 520 excitation wavelength). The β1i inhibitor and substrate, respectively, were: 50 μm bortezomib and (Ac-Pro-Ala-Leu)2-R110 (AAT Bioquest, Sunnyvale, CA, Cat. No. 13467). The β5i inhibitor and substrate, respectively, were: 20 μm ONX-0914 (Abmole Bioscience Inc., Houston, TX, Cat. No. M2071) and (Ac-Ala-Asn-Trp)2-R110 (AAT Bioquest, Cat. No. 13455). ONX-0914 is a β5i inhibitor.
Calpain Activity
Protein sample (50 μg) was combined with calpain activation buffer (25 mm Tris, 0.5 mm EDTA, 5 mm CaCl2, 75 mm NaCl, 0.25 mm DTT), 25 μm calpain inhibitor IV (Calbiochem San Diego, CA) or an equal volume of DMSO, and incubated for 20 min at room temperature. The reaction was initiated by adding the fluorogenic substrate Suc-Leu-Leu-Val-Tyr-MCA at a final concentration of 50 μm (36).
Western Blot Analysis
Preparation of Cytosolic Cardiac Homogenates
4× SDS Sample Buffer (8% SDS, 40% glycerol, 0.4% bromophenol blue, 5% β-mercaptoethanol, 240 mm Tris, pH 6.8) was added to cytosolic cardiac homogenates (prepared as described in the proteasome assays section), and the homogenates were boiled for 4 mins at 95 °C.
Preparation of Total Cardiac Homogenates
1 ml of 1× Laemmli sample buffer with freshly added β-mercaptoethanol (Bio-Rad, Hercules, CA, Cat. # 161–0737) was added to 20 μg powdered cardiac tissue. The tissue was mixed, incubated at room temperature for 5 mins, and boiled for 5 mins at 95 °C. The protein concentrations were determined using the RC DC Protein Assay (Bio-Rad, Cat. # 500–0119).
Electrophoresis and Western Blotting
Homogenates (30 μg) and 5 μl Spectra multicolor broad range protein ladder (product # 26634, Thermo Scientific) were separated on 4–20% 18-well TGX Precast Gels (Cat. # 567–8094 for Stain-Free gels, Cat. # 567–1094 for conventional gels, Bio-Rad) at 150V, 500mA maximum current. Stain-Free gels were activated for 1 min by UV transillumination. The proteins were transferred to a nitrocellulose or PVDF membrane (Trans-Blot Turbo Midi Nitrocellulose, #170–4159/PVDF Transfer Pack, #170–4157, Bio-Rad) using the Trans-Blot Turbo Transfer System (# 170–4155, Bio-Rad). The membranes were then probed with the respective antibodies (supplemental Table S1). All Western blotting procedures were carried out at room temperature with agitation except when stated otherwise. Unless otherwise noted, membranes were blocked for one hour in 3% blotting grade nonfat milk (Cat. # 170–6404, Bio-Rad) in Tris Buffered Saline (50 mm Tris, 150 mm NaCl, pH 7.5) containing 0.05% Tween-20 (TBST) at room temperature, then incubated in primary antibody diluted in 1% milk/TBST for two hours at room temperature and washed three times for 5 mins each in TBST. The membranes were incubated in peroxidase-conjugated rabbit anti-mouse or anti-rabbit IgG secondary antibody (Sigma-Aldrich, anti-mouse Cat. # A9044, anti-rabbit Cat. # A9169 or #A0545, Sigma-Aldrich) diluted 1:5000 in 1% milk/TBST. The membranes were then washed three times for 5 mins each in TBST. Blots were incubated with Clarity Western ECL Substrate (Cat. # 170–5061, Bio-Rad) and imaged using the ChemiDoc MP (Cat. # 170–8280, Bio-Rad) with ImageLab Version 5 (Bio-Rad). When conventional gels were used, the membrane was stained for total protein with Coomassie Blue to serve as a loading control following the Western blotting procedure.
OxyBlot
Homogenates were transferred from a Stain-Free gel to a PVDF membrane. The membrane was immersed in 100% methanol for 15 s, dried for 5 mins, and equilibrated in 20% methanol/TBS for 5 mins. The membrane was then washed in 2N HCl for 5 mins, and incubated in 10 mm 2,4-Dinitrophenylhydrazine (DNPH) (Sigma-Aldrich, Cat. # D199303) prepared in 2N HCl for 5 mins. The membrane was washed three times, 5 mins each in 2N HCl and five times, 5 mins each in 100% methanol. The membrane was then blocked in 5% milk/TBST for one hour and incubated in anti-DNP primary antibody (supplemental Table S1) for 2 h, followed by three 5 min washes in TBST. The membrane was then incubated in peroxidase-conjugated anti-rabbit secondary antibody diluted 1:5000 in 5% milk/TBST for 1 h, washed three times in TBST for 5 min per wash, incubated with Clarity Western ECL Substrate, and imaged as described in the previous section.
Ubiquitin Blot
Homogenates were transferred from a Stain-Free gel to a nitrocellulose membrane and washed three times for 2 min each with water, then incubated in 0.5% glutaraldehyde in PBS for 20 min. The membrane was washed three times for 5 min each with PBS and blocked in 5% milk/TTBS (0.1% Tween was used for all steps) for 30 min. The membrane was incubated overnight at 4 °C with mouse anti-ubiquitin primary antibody (supplemental Table S1) in 1% milk/TBST. The membrane was washed three times, 5 mins each with TBST, then incubated for one hour in peroxidase-conjugated anti-mouse secondary antibody diluted 1:5000 in 1% milk/TBST. The membrane was washed three times, 5 min each with TBST, incubated with Clarity Western ECL Substrate, and imaged.
Antioxidant Capacity Assay
The antioxidant capacity assay was used to measure the antioxidant activity of cardiac cytosolic homogenates based on the oxidation rate of ABTS (2,2′-azino-di-(3-ethyl-benzthiazoline-6-sulfonic acid) by the ferryl metmyoglobin radical. 10 mg of powdered heart tissue was homogenized using the Qiagen (Hilden, Germany) TissueLyser LT (30 oscillations/s for 3 mins at 4 °C) with a metal bead in 100 μl cold 1× Antioxidant Assay Buffer (5 mm potassium phosphate, 0.9% NaCl, 0.1% glucose, pH 7.4). The homogenates were centrifuged at 12,000 × g for 20 min at 4 °C. The supernatants were removed and diluted to equal protein concentrations. In a clear 96-well plate, 0.19 μm myoglobin and 130.18 mm ABTS were combined with 24 μg homogenate or increasing amounts of Trolox standard (0, 0.05, 0.1, 0.15 mm Trolox final concentration). 84 μm hydrogen peroxide was added to start the reaction. The reaction was incubated for 45 min, and the amount of ABTS radical was quantified by measuring absorbance at 415 nm. Trolox equivalent units were calculated based on the Trolox standard curve (37).
Metabolomics
Sample Preparation for GC-MS
Hearts (3 month old WT, R278C, and I79N) were pulverized in liquid nitrogen with a mortar and pestle and kept as 20 mg aliquots at −80 °C. Degassed extraction solvent (1 ml) was added to 20 mg of powdered heart tissue sample (three biological and two technical replicates for each transgenic line) and homogenized for 45 s using a Turex mini homogenizer. The extraction solvent was composed of ACN, isopropanol and water in proportions 3:3:2. The samples were then centrifuged at 2500 rpm for 5 min and the supernatant removed and aliquoted into two 500 μl aliquots. One aliquot was evaporated to complete dryness in a Labconco (Kansas City, MO) Centrivap cold trap concentrator. The dried aliquot was then resuspended with 500 μl 50% degassed ACN, centrifuged for 2 min at 14,000 × g, and the supernatant was transferred to a new tube. The supernatant was evaporated to dryness and the pellet used for derivatization.
GC-TOF
GC-TOF was carried out by the UC Davis Metabolomics core facility. Samples were derivatized with methoxyamine and MSTFA to trimethylsilation. Fatty acid methyl esters internal standards were added to the sample, and 500 μl of sample was injected into an Agilent 7890A GC- Pegasus HT TOF MS (Santa Clara, CA). The 7890A GC is equipped with a Gerstel automatic liner exchange system (Linthicum, MD). Injection volume was 0.5 μl with 10 μl/s injection speed on a splitless injector with a purge time of 25 s. After each injection the 10 μl injection syringe was washed three times with 10 μl ethyl acetate. Chromatography was carried out using a 30 m long, 0.25 mm i.d. Rtx-5Sil MS column (0.25 μm 95% dimethyl 5% diphenyl polysiloxane film) with an additional 10 m integrated guard column (Restek, Bellefonte, PA). Helium (99.9999% purity) with a built-in purifier (Airgas, Radnor, PA) was set at constant flow of 1 ml/min and the oven temperature held constant at 50 °C for 1 min and then increased at 20 °C/min to 330 °C and then held constant for 5 min. Mass spectrometry of the metabolites were carried out using a Leco Pegasus HT time of flight mass spectrometer controlled by Leco ChromaTOF software V4.50.8.0 (St. Joseph, MI). The transfer line temperature between the gas chromatograph and mass spectrometer was 280 °C. Electron impact ionization at 70V was employed with an ion source temperature of 250 °C. Acquisition rate was 17 spectra/second with a scan mass range of 85–500 Da.
The identities of all metabolites were validated through a multi-tiered matching algorithm with the UC Davis in-house BinBase database system (38, 39) using retention index information in addition to mass spectral matching, unique ion characteristics, peak purity, and signal/noise metadata (38). The quantification values were averaged over two technical replicates, and the resulting values were then averaged over biological replicates. Comparisons between WT, R278C and I79N samples were carried out using one-way ANOVA. Results are presented as mean ± standard deviation (S.D.), and a value of p < 0.05 was considered statistically significant.
RESULTS
A label-free proteomics approach was used to uncover changes in R278C and I79N mouse hearts. Proteomic analysis revealed that there were more changes in R278C than I79N mice relative to WT, indicating that the changes in I79N mice are likely to be more profound in nature, because the I79N hearts show a more severe phenotype. Of ∼1200 proteins identified, a total of 76 proteins were found to be statistically altered (p < 0.05 by ANOVA) when WT and R278C hearts were compared, whereas 53 proteins were altered in I79N hearts compared with WT hearts (Table I and supplemental Tables S2 and S3). Proteome analysis of complex biological samples remains challenging mainly due to the expansive range of protein concentrations. Because of the large protein concentration range all unfractionated as well as many fractionated samples suffer an intrinsic bias against low abundance proteins (40). When R278C hearts were compared with I79N hearts more than 400 proteins were found to be significantly altered. This is the first report of proteomic and metabolomic comparisons between mutations associated with mild (R278C) and severe (I79N) phenotypes in any sarcomeric gene. An advantage of this investigation is that both mutations are well characterized and occur in the same gene (TnT), which reduces the potential for artifacts when interpreting the results. Another advantage of these studies is that the I79N and R278C mutations are not associated with hypertrophy in patients or in the mouse models (7, 13–17). This allowed us to investigate the disease mechanisms independently of cardiac hypertrophy.
Table I. Proteins which are differentially expressed in hearts from 3 month old wild-type, R278C and I79N mice, grouped according to gene ontology (GO) biological process. Details of protein expression are shown in supplemental Table S2. Abbreviations: mito, mitochondrial.
| I79N versus WT |
R278C versus WT |
||||
|---|---|---|---|---|---|
| Protein ID | Gene Name | Protein Name | Protein ID | Gene Name | Protein Name |
| Metabolic Process | Metabolic Process | ||||
| H7BX88 | Crat | Carnitine O-acetyltransferase ↑ | Q8R0F8 | Fahd1 | Acylpyruvase FAHD1, mito. ↑ |
| Q04447 | Ckb | Creatine kinase B-type ↑ | O35143 | Atpif1 | ATPase inhibitor, mito. ↓ |
| Q9DCW5 | Cox6a1 | Cytochrome c oxidase subunit 6A, mito. ↑ | Q8VCT4 | Ces1d | Carboxylesterase 1D ↑ |
| Q9DCW5 | Cox6a2 | Cytochrome c oxidase subunit 6A2, mito. ↑ | Q99LC5 | Etfa | Electron transfer flavoprotein subunit alpha, mito. ↑ |
| P10518 | Alad | Delta-aminolevulinic acid dehydratase ↑ | Q9DCW4 | Etfb | Electron transfer flavoprotein subunit beta ↑ |
| Q9DCW4 | Etfb | Electron transfer flavoprotein subunit beta ↑ | P42125 | Eci1 | Enoyl-CoA delta isomerase 1, mito. ↑ |
| P42125 | Eci1 | Enoyl-CoA delta isomerase 1, mito. ↑ | P16858 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase ↓ |
| Q05816 | Fabp5 | Fatty acid-binding protein, epidermal ↑ | Q9ET01 | Pygl | Glycogen phosphorylase, liver form ↑ |
| B0R1E3 | Hint1 | Histidine triad nucleotide-binding protein 1 ↑ | B8JK32 | Hnrnpm | Heterogeneous nuclear ribonucleoprotein M ↓ |
| D3Z636 | Ppa2 | Inorganic pyrophosphatase 2, mito. ↑ | Q9D0S9 | Hint2 | Histidine triad nucleotide-binding protein 2, mito. ↓ |
| Q9D6R2 | Idh3a | Isocitrate dehydrogenase [NAD] subunit alpha, mito. ↑ | Q9D819 | Ppa1 | Inorganic pyrophosphatase ↓ |
| P16125 | Ldhb | l-lactate dehydrogenase B chain ↑ | Q8BWT1 | Acaa2 | 3-ketoacyl-CoA thiolase, mito. ↑ |
| Q9CXT8 | Pmpcb | Mitochondrial-processing peptidase subunit beta ↑ | P53395 | Dbt | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mito. ↑ |
| O88492 | Plin4 | Perilipin-4 ↑ | P16125 | Ldhb | l-lactate dehydrogenase B chain ↑ |
| P17751 | Tpi1 | Triosephosphate isomerase ↑ | Q9EQ20 | Aldh6a1 | Methylmalonate-semialdehyde dehydrogenase [acylating], mito. ↑ |
| Biological Adhesion | Q9JLZ3 | Auh | Methylglutaconyl-CoA hydratase, mito. ↑ | ||
| Q04857 | Col6a1 | Collagen alpha-1(VI) chain ↑ | Q9CR61 | Ndufb7 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 ↓ |
| P08122 | Col4a2 | Collagen alpha-2(IV) chain ↑ | Q8BMF3 | Me3 | NADP-dependent malic enzyme, mito. ↑ |
| Q99K47 | Fga | Fibrinogen, alpha polypeptide ↑ | Q8R5J9 | Arl6ip5 | PRA1 family protein 3 ↓ |
| E9PWQ3 | Col6a3 | Protein Col6a3 ↑ | P62192 | Psmc1 | 26S protease regulatory subunit 4 (Rpt2) ↓ |
| Multicellular Organismal Process | Q9R1P4 | Psma1 | Proteasome subunit alpha type-1 (α6) ↓ | ||
| Q6ZWQ9 | Myl12a | Myosin, light chain 12A, regulatory, non-sarcomeric ↑ | Q9DB15 | Mrpl12 | 39S ribosomal protein L12, mito. ↑ |
| Developmental Process | P60867 | Rps20 | 40S ribosomal protein S20 ↓ | ||
| Q8VDQ1–2 | Ptgr2 | Prostaglandin reductase 2, Isoform 2 | Q62417–4 | Sorbs1 | Sorbin and SH3 domain-containing protein 1, Isoform 4 ↓ |
| Biological Regulation | P35235–2 | Ptpn11 | Tyrosine-protein phosphatase non-receptor type 11, | ||
| Isoform 2 ↓ | |||||
| Q99PT1 | Arhgdia | Rho GDP-dissociation inhibitor 1 ↑ | Biological Adhesion | ||
| Q60854 | Serpinb6 | Serpin B6 ↑ | P11087 | Col1a1 | Collagen alpha-1(I) chain ↓ |
| Apoptotic Process | Q01149 | Col1a2 | Collagen alpha-2(I) chain ↓ | ||
| D3YX27 | Htra2 | Serine protease HTRA2, mito. ↑ | Multicellular Organismal Process | ||
| Cellular Process | Q3UH68–2 | Limch1 | LIM and calponin homology domains-containing protein 1, Isoform 2 ↓ | ||
| Bsg | Basigin ↑ | Q62000 | Ogn | Mimecan ↓ | |
| P05132–2 | Prkaca | cAMP-dependent protein kinase catalytic subunit α, Isoform 2 ↑ | Developmental Process | ||
| D3Z630 | Mylk3 | Myosin light chain kinase 3 ↑ | Q80X90 | Flnb | Filamin-B ↓ |
| Q91YZ8 | Pabpc4 | Poly(A) binding protein, cytoplasmic 4 ↑ | A2AEX7 A2AEX8 | Fhl1 | Four and a half LIM domains protein 1 ↓ |
| P67778 | Phb | Prohibitin ↑ | O70433 | Fhl2 | Four and a half LIM domains protein 2 ↓ |
| P26039 | Tln1 | Talin-1 ↑ | Q9JK92 | Hspb8 | Heat shock protein beta-8 ↓ |
| Cellular Component Organization or Biogenesis | Biological Regulation | ||||
| P26040 | Ezr | Ezrin ↑ | Q06890 | Clu | Clusterin ↓ |
| P68433 | Hist1h3a | Histone H3.1 ↑ | Q9JKS4–2 | Ldb3 | LIM domain-binding protein 3, Isoform 2 ↓ |
| P14733 | Lmnb1 | Lamin-B1 ↑ | Localization | ||
| Q62261–2 | Sptbn1 | Spectrin beta chain, non-erythrocytic 1, Isoform 2 ↑ | Q9DCC8 | Tomm20 | Mitochondrial import receptor subunit TOM20 homolog ↓ |
| Immune System Process | Cellular Process | ||||
| P01029 | C4b | Complement C4-B ↑ | P84078 | Arf1 | ADP-ribosylation factor 1 ↓ |
| P63028 | Tpt1 | Translationally-controlled tumor protein ↑ | Q9CXJ4 | Abcb8 | ATP-binding cassette sub-family B member 8, mito. ↓ |
| No GO Biological Process | Q9D7X Q3V2Y9 | Dusp3 | Dual specificity protein phosphatase 3 ↑ | ||
| P08226 | Apoe | Apolipoprotein E ↑ | P16045 | Lgals1 | Galectin-1 ↓ |
| Q91WS0 | Cisd1 | CDGSH iron-sulfur domain-containing protein 1 ↑ | Q61011 | Gnb3 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-3 ↓ |
| E9Q4B5 | Ccdc58 | Coiled-coil domain-containing protein 58 ↑ | Q6URW6–2 | Myh14 | Myosin-14, Isoform 2 ↓ |
| Q9CPQ1 | Cox6c | Cytochrome c oxidase subunit 6C ↑ | Q3UIZ8–2 | Mylk3 | Myosin light chain kinase 3, Isoform 2 ↓ |
| P05063 | Aldoc | Fructose-bisphosphate aldolase C ↑ | P26645 | Marcks | Myristoylated alanine-rich C-kinase substrate ↓ |
| Q9QUH0 | Glrx | Glutaredoxin-1 ↑ | P68254 | Ywhaq | 14–3-3 protein theta ↓ |
| P10649 | Gstm1 | Glutathione S-transferase Mu 1 ↑ | P61982 | Ywhag | 14–3-3 protein gamma ↓ |
| Q91V64 | Isoc1 | Isochorismatase domain-containing protein 1 ↑ | F6UIT9 | Ppp1r12b | Protein phosphatase 1 regulatory subunit 12B ↓ |
| P99029–2 | Prdx5 | Isoform cytoplasmic+peroxisomal of Peroxiredoxin-5, mito. ↑ | P62071 | Rras2 | Ras-related protein R-Ras2 ↓ |
| P20029 | Hspa5 | 78 kDa glucose-regulated protein ↑ | Q8CC35–2 | Synpo | Synaptopodin, Isoform 2 ↓ |
| P34884 | Mif | Macrophage migration inhibitory factor ↑ | P21107–2 | Tpm3 | Tropomyosin alpha-3 chain, Isoform 2 ↓ |
| E9Q7L0 | Ogdhl | Oxoglutarate dehydrogenase-like ↑ | O70373 | Xirp1 | Xin actin-binding repeat-containing protein 1 |
| Q9D1G1 | Rab1b | Ras-related protein Rab-1B ↑ | Cellular Component Organization or Biogenesis | ||
| E9Q3Y1 E9Q0P9 | Serpinb6a | Serpin B6 ↑ | P61161 | Actr2 | Actin-related protein 2 ↓ |
| O54988–2 | Slk | STE20-like serine/threonine-protein kinase, Isoform 2 ↑ | E9Q557 | Dsp | Desmoplakin ↓ |
| P80314 | Cct2 | T-complex protein 1 subunit beta ↑ | P47754 | Capza2 | F-actin-capping protein subunit alpha-2 ↓ |
| Q9CQN6 | Tmem14c | Transmembrane protein 14C ↑ | F8WIX8 | Hist1h2al | Histone H2A ↓ |
| E0CZ27 | H3f3a | Histone H3 ↓ | |||
| P68433 | Hist1h3a | Histone H3.1 ↑ | |||
| P43274 | Hist1h1e | Histone H1.4 ↓ | |||
| E9PUM4 Q71LX4 | Tln2 | Talin-2 ↓ | |||
| No GO Biological Process | |||||
| Q9CQR4 | Acot13 | Acyl-coenzyme A thioesterase 13 ↑ | |||
| Q9DCX2 | Atp5h | ATP synthase subunit d, mito. | |||
| Q8VBT1 | Txlnb | Beta-taxilin ↓ | |||
| Q5SXR6 | Cltc | Clathrin heavy chain ↓ | |||
| G3UX44 | H2-Ke6 | Estradiol 17-beta-dehydrogenase 8 ↑ | |||
| Q9DCM2 | Gstk1 | Glutathione S-transferase kappa 1 ↑ | |||
| K3W4S6 | Gyg | Glycogenin-1 ↓ | |||
| P17879 | Hspa1b | Heat shock 70 kDa protein 1B ↑ | |||
| Q9ESB3 | Hrg | Histidine-rich glycoprotein ↓ | |||
| Q8K310 | Matr3 | Matrin-3 ↓ | |||
| Q9QYA2 | Tomm40 | Mitochondrial import receptor subunit TOM40 homolog ↓ | |||
| P63030 | Mpc1 | Mitochondrial pyruvate carrier 1 | |||
| Q99JI4 | Psmd6 | 26S proteasome non-ATPase regulatory subunit 6 (Rpn7) ↓ | |||
| Q9D1G1 | Rab1b | Ras-related protein Rab-1B ↓ | |||
| P53994 | Rab2a | Ras-related protein Rab-2A ↓ | |||
| P61027 | Rab10 | Ras-related protein Rab-10 ↓ | |||
| Q4VAE3 | Tmem65 | Transmembrane protein 65 ↑ | |||
Metabolic Processes
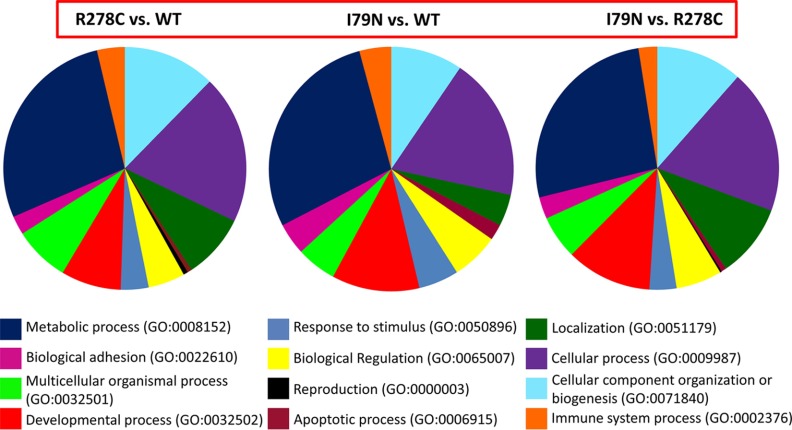
Mass spectrometry results analyzed using the Panther program (http://pantherdb.org/) showed that when comparing WT and R278C or I79N, proteins associated with metabolic processes (GO:0008152) were the most common process altered when differentially expressed proteins were grouped by biological process (Fig. 1). However, the relative proportion of proteins altered that were part of metabolic processes were larger for I79N (35.3%) than for R278C (29.3%). This is consistent with the largest proportion of proteins altered when comparing R278C with I79N (26.4%) also being involved in metabolic processes. The next two biological processes with the most proteins altered when comparing WT and R278C or I79N were cellular processes (GO:0009987) and cellular component organization or biogenesis (GO:0071840) (Fig. 1). One biological process that appears to be affected to a lesser degree in R278C versus WT hearts than I79N versus WT hearts is the apoptosis process (GO:0006915), which accounted for 2.1% of the altered proteins in I79N versus WT, and only 0.6% of altered proteins in R278C versus WT. When comparing protein changes based on molecular function, the greatest proportion of altered proteins were associated with catalytic activity (GO:0003824) for all comparisons (WT versus R278C or I79N and R278C versus I79N) (supplemental Fig. S1).
Fig. 1.
Summary of results for the proteomic analysis of wild-type, R278C, and I79N mouse hearts. Biological processes associated with the differentially expressed proteins are shown as identified using the gene ontology (GO) biological processes classification. Most proteins are associated with multiple biological functions; they are grouped according to the biological function they are most often associated with.
Proteasome Dysfunction
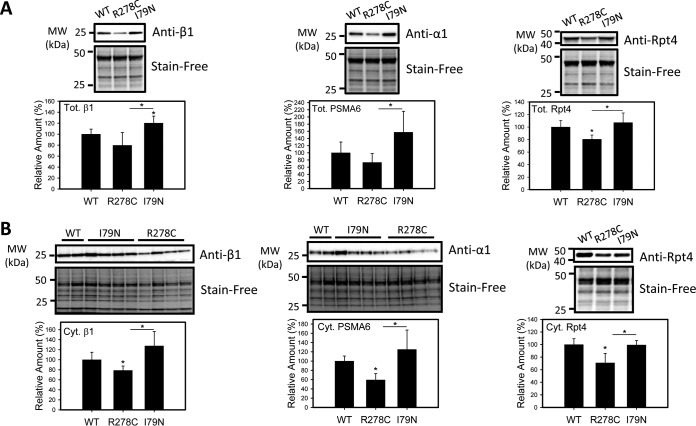
One large protein complex associated with metabolic function is the proteasome. The proteasome metabolizes many cellular proteins, and has been shown to play a role in lipid metabolism (41, 42). The Ubiquitin Proteasome System (UPS) has also been shown to regulate 5′-AMP-activated protein kinase (AMPK), a key enzyme in regulating cellular energy homeostasis (43). A total of 23 proteasome subunits were identified in this investigation. Four proteasome subunits showed a significant decrease in R278C mice relative to WT hearts (supplemental Table S3). When these 23 proteasome subunits were compared between R278C and I79N hearts, 9 subunits were found to be significantly higher in I79N hearts than R278C hearts. Expression of 3 proteasome subunits was also lower in R278C hearts than WT hearts. Hence the results from the label-free proteomic approach suggest that the proteasome is differentially regulated in R278C and I79N mouse hearts. To confirm the differences in proteasome levels detected by mass spectrometry, we examined the expression of a subset of proteasome subunits in R278C and I79N hearts (Fig. 2). Expression of two subunits of the 20S proteasome, α1 (PSMA6) and β1, was measured in total and cytosolic cardiac homogenates (Fig. 2). In total homogenates, β1 expression was 20% higher in I79N hearts than WT hearts, and expression of both subunits was higher in I79N hearts than R278C hearts (Fig. 2). Hearts from R278C mice showed a trend toward lower total α1 and β1 levels relative to WT; however, these differences were not statistically significant (p = 0.217 and 0.156 respectively). Expression of these subunits in cytosolic cardiac homogenates was also investigated, yielding similar results. Expression of α1 and β1 was higher in I79N cytosolic heart fractions than R278C cytosolic heart fractions, and decreased in R278C hearts relative to WT hearts by 41 and 21%, respectively (Fig. 2). A trend toward increased expression of α1 and β1 in I79N hearts relative to WT hearts was observed; however these differences were not statistically significant (p = 0.289 and 0.139 respectively). Expression of the 20S subunit β2 was also examined in cytosolic heart fractions, showing lower expression in R278C hearts and higher expression in I79N than R278C hearts (Fig. 3).
Fig. 2.
Validation of increased proteasome subunit expression in hearts from I79N mice relative to R278C hearts by Western blotting. Western blots of proteasome levels (α1/PSMA6, β1, Rpt4) and total protein (Stain-Free, used as loading controls) and corresponding semiquantitative bar graphs are shown. A, Determination of proteasome amount in total cardiac homogenates (n = 4). B, Determination of proteasome amount in cytosolic cardiac homogenates (n = 4). *, p < 0.05.
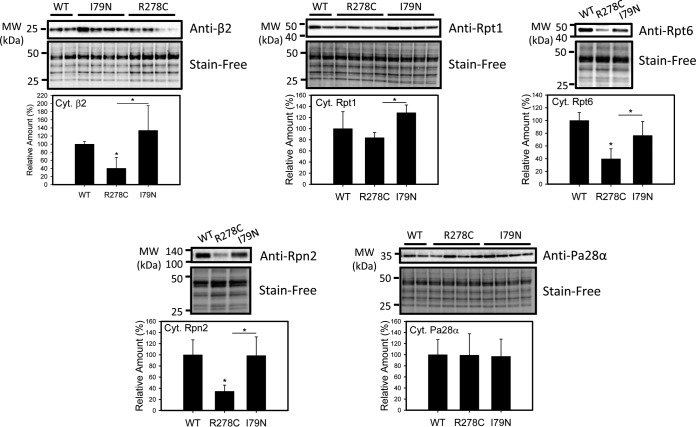
Fig. 3.
Determination of proteasome amount in cytosolic fractions from wild-type, R278C, and I79N mouse hearts. Western blots of proteasome levels (β2, Rpt1, Rpt6, Rpn2, PA28α) and total protein (Stain-Free, used as loading controls) and corresponding semi-quantitative bar graphs are shown (n = 3–4). *, p < 0.05.
The expression of four subunits of the 19S regulatory particle was also examined: Rpt1, Rpt4, Rpt6, and Rpn2, as well as PA28α, a subunit of the 11S regulatory particle (Figs. 2 and 3). A similar trend to 20S expression was observed in 19S subunit expression; expression of three out of the four subunits (Rpt4, Rpt6, and Rpn2) was decreased in R278C hearts compared with WT hearts, and expression of all four subunits examined was higher in I79N hearts than R278C hearts (Figs. 2 and 3). PA28α levels were not affected in R278C hearts or I79N hearts, in agreement with MS data (Fig. 3). Overall, expression of proteasome subunits was higher in I79N hearts than R278C hearts as judged by Western blotting, which is consistent with the proteomic data (supplemental Table S4).
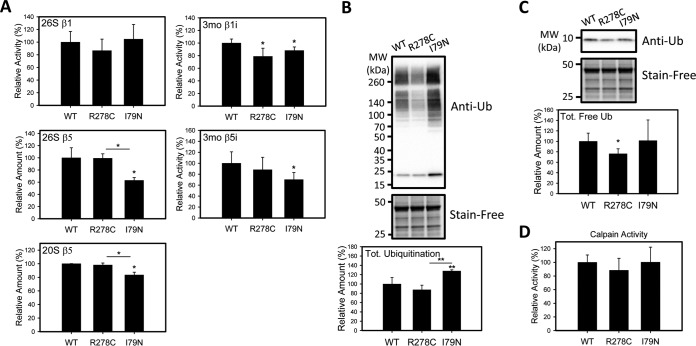
To further explore the effect of the altered proteasome levels in the transgenic hearts, the proteolytic activities of the 26S proteasomes were measured (Fig. 4). 26S proteasome activities are ATP dependent, in contrast to 20S activities, and the assays are conducted in the presence of ATP to optimize measurement of the 26S activity only. The 26S and 20S β5 (chymotrypsin-like) activities of the proteasome in the I79N hearts were significantly lower than in WT and R278C hearts, suggesting proteasome dysfunction (Fig. 4). The 26S and 20S β5 activities of the R278C hearts were similar to WT hearts. The β5 activity is the main proteolytic activity of the proteasome, so it is likely that other components of the proteasome pathway would be affected. The 26S β1 (caspase-like) activity of the proteasome was also measured and found to be similar between the three groups of transgenic hearts. Because we have previously shown that the immunoproteasome is important for the removal of oxidized proteins, the immunoproteasome β1i and β5i activities were determined (35, 36) (Fig. 4). Although the immunoproteasome β1i activity was decreased in both I79N and R278C hearts relative to control hearts, the β5i activity was only decreased in the I79N hearts. The β1i activity was the only type of proteasome activity that we determined to be decreased in R278C hearts. To determine if other cytosolic proteolytic enzymes such as calpain were also affected in I79N hearts, calpain activity was determined and found to be similar between all hearts investigated (Fig. 4).
Fig. 4.
Characterization of proteolytic activity and ubiquitination in wild-type, R278C, and I79N mouse hearts. A, 26S proteasome activity of lysates of I79N, R278C, and wild-type mouse hearts (n = 3). B, 20S proteasome activity of lysates of wild-type, R278C, and I79N mouse hearts (n = 3). C, Western blot of free ubiquitin levels and ubiquinated proteins in transgenic mouse hearts (n = 3). D, Calpain activity (n = 3). *, p < 0.05. **, p < 0.001.
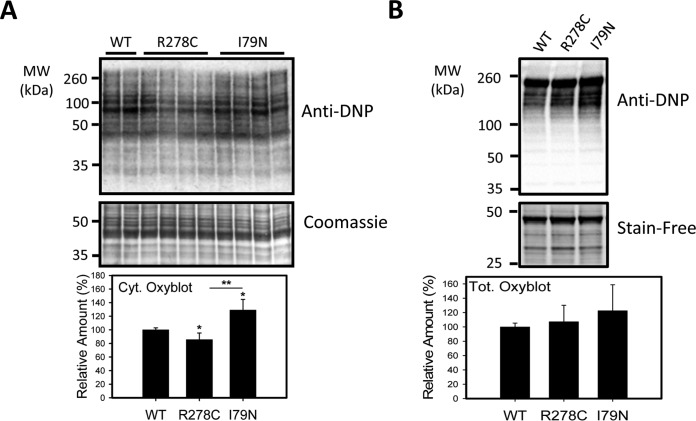
In addition to its roles in signaling and intracellular trafficking, ubiquitination is important as a signal for protein degradation, and decreases in proteasome function and/or dysregulation of the ubiquitination pathway have been linked to up-regulation of stress related proteins, increases in polyubiquinated proteins, and increased oxidative stress (44, 45). To determine if impaired proteasome function is associated with increased ubiquitination, heart lysates were electrophoresed and probed with an anti-ubiquitin antibody. Levels of ubiquitinated proteins in total homogenates and oxidized proteins in cytosolic homogenates (as determined using the OxyBlot procedure, which detects carbonylated proteins) were increased in I79N mice relative to WT and R278C mice, consistent with impaired UPS function (Figs. 4 and 5). Interestingly, the levels of oxidized proteins in cytosolic homogenates were lower in R278C hearts than in WT and I79N hearts (Fig. 5).
Fig. 5.
Characterization of levels of oxidized proteins in wild-type, R278C, and I79N mouse hearts. A, Oxidized protein levels in cytosolic heart lysates from 3 month old hearts were determined by the Oxyblot method. Carbonyl residues were derivatized with DNPH and subsequently detected by Western blotting using anti-DNP antibody (n = 4). B) Oxidized proteins in total heart lysates from 3 month old hearts (n = 4). *, p < 0.05. **, p < 0.001.
Antioxidant Defense System
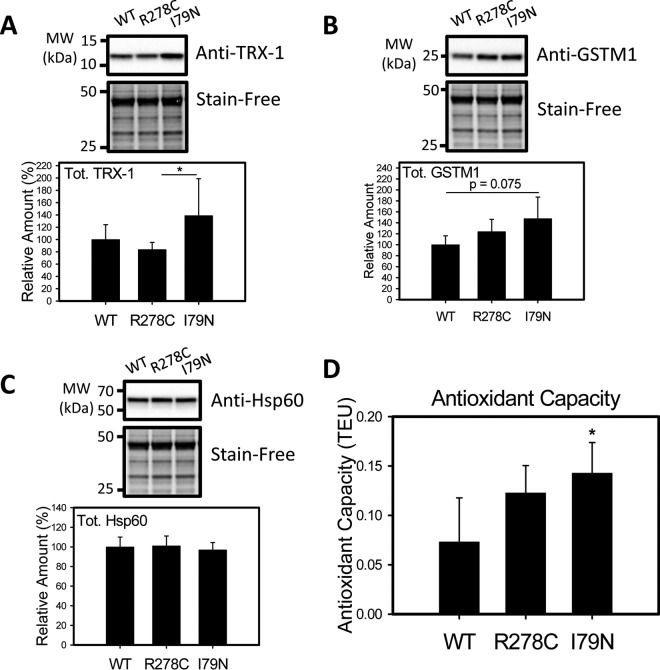
The proteomic data also suggests that several antioxidant enzymes including thioredoxin-1 (TRX-1) and glutathione S-transferase mu 1 (GSTM1) were up-regulated in I79N hearts compared with R278C hearts, and GSTM1 and Isoform cytoplasmic+peroxisomal of Peroxiredoxin-5, mitochondrial were up-regulated in I79N compared with WT hearts (supplemental Table S5). To further investigate the expression of stress-related proteins that may be affected in R278C or I79N hearts, TRX-1, GSTM1 and one chaperone [heat shock protein 60 (Hsp60)] were selected for Western blotting. TRX-1 is a mainly cytosolic antioxidant enzyme that reduces oxidized proteins, and thus plays an important role in protecting against oxidative stress (46). Oxidative stress and other types of cellular stresses have been shown to upregulate thioredoxin expression (47–49). Western blotting showed that TRX-1 was significantly up-regulated in I79N hearts relative to R278C hearts, consistent with the proteomic results (Fig. 6). GSTM1 is an antioxidant enzyme that also plays a role in coping with oxidative stress by conjugating the antioxidant glutathione to target proteins (50). GSTM1 expression was found to be increased in I79N hearts when compared with WT and R278C hearts by proteomic analysis. Western blotting showed a trend toward increased GSTM1 expression in I79N hearts relative to WT hearts, but this increase was not statistically significant (p = 0.075). Hsp60 is a mitochondrial chaperone that is implicated in mitochondrial import, proper folding of imported proteins, and promoting refolding of improperly folded proteins under stress conditions (51, 52). Protein expression of Hsp60 was not altered in R278C or I79N hearts, in agreement with the proteomic data (Fig. 6).
Fig. 6.
Characterization of levels of oxidative stress-related proteins and antioxidant capacity in wild-type, R287C, and I79N mouse hearts. Western blot of enzymes associated with oxidative stress, A, Thioredoxin-1 (TRX-1) and B, Glutathione S-transferase mu 1 (GSTMI). C, Western blot of heat shock protein 60 (HSP60), which was not found to be differentially expressed by proteomic methods. D, Antioxidant capacity of cardiac homogenates were carried out using 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS)/metmyoglobin. Antioxidant activity is measured relative to the antioxidant Trolox and reported in Trolox Equivalent Units (n = 4). *, p < 0.05.
Increases in antioxidant enzymes are typically associated with increased total antioxidant capacity of the tissue. Measuring the antioxidant capacity of the cardiac homogenates showed that the antioxidant capacity was nearly doubled in I79N hearts relative to WT hearts (increased by 95%), suggesting that antioxidant levels are up-regulated to compensate for increased oxidative stress (Fig. 6D). These findings all suggest that oxidative stress is higher in I79N hearts.
Energy Production Pathways
Proteomic results also suggest that several glycolytic, tricarboxylic acid cycle (TCA) and electron transport chain (ETC) enzymes are up-regulated in I79N hearts compared with WT and R278C hearts (supplemental Tables S2, S3, and S6). Some of the enzymes up-regulated in I79N hearts relative to WT and R278C hearts in glycolytic, TCA and ETC pathways include triosephosphate isomerase, fructose-bisphosphate aldolase C, isocitrate dehydrogenase [NAD] subunit alpha (mitochondrial), oxoglutarate dehydrogenase-like, and cytochrome c oxidase subunits 6A and 6C. To further confirm the importance and involvement of these pathways, the levels of metabolites (intermediates and products of metabolism) present in these hearts were determined by GC-TOF (Figs. 7 and 8, Table II, supplemental Table S8). Mass spectra of representative single peptides for which significant changes were observed are shown in supplemental Fig. S4.
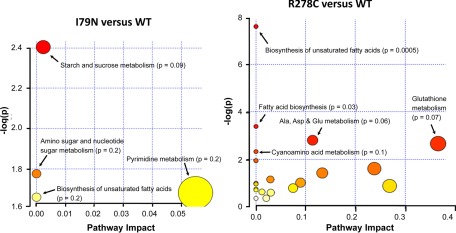
Fig. 7.
Pathway analysis of the metabolomic data comparing wild-type, R278C, and I79N hearts. Pathway analysis was carried out using MetaboAnalyst 2.0 (92).
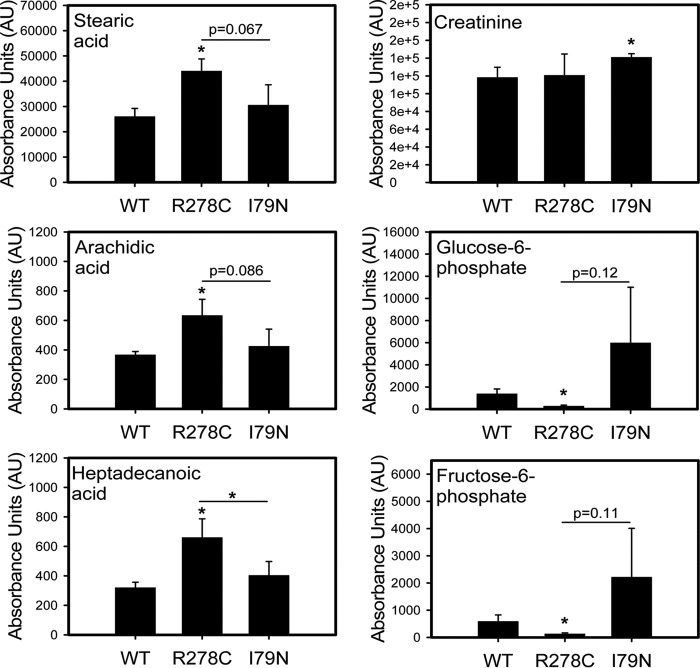
Fig. 8.
Effect of R278C and I79N mutations on metabolite levels in transgenic hearts. Changes in metabolite levels in WT, R278C, and I79N hearts. Bar charts for stearic acid, arachidic acid, heptadecanoic acid, creatinine, glucose-6-phosphate, and fructose-6-phosphate are shown. *, p < 0.05. **, p < 0.001.
Table II. Metabolites which are differentially expressed in hearts from 3 month old wild-type, R278C and I79N mice grouped according to type of metabolite. Details of metabolite expression are shown in supplemental Table S7.
| R278C vs. WT | I79N vs. WT | I79N vs. R278C |
|---|---|---|
| Metabolite | Metabolite | Metabolite |
| Fatty Acids | Fatty Acids | Fatty Acids |
| Arachidic acid ↑ | cis-gondoic acid ↑ | Heptadecanoic acid ↓ |
| Capric acid ↑ | Sugars | Sugars |
| cis-gondoic acid ↑ | 1,5-anhydroglucitol ↑ | 1,5-anhydroglucitol ↑ |
| Heptadecanoic acid ↑ | Fructose ↑ | Sugar Acids |
| Oleic acid ↑ | Sugar Alcohols | Pentonic acid ↑ |
| Palmitic acid ↑ | Ribitol ↑ | Ribonic acid ↑ |
| Pelargonic acid ↑ | Methylphenols | Vitamins/Derivatives |
| Stearic acid ↑ | p-cresol ↑ | Dehydroascorbic acid ↑ |
| Sugars | Guanidino Compounds | Pantothenic acid ↓ |
| Fructose-6-phosphate ↓ | Creatinine ↑ | Amino Acids |
| Glucose-6-phosphate ↓ | Nucleosides | Asparagine ↑ |
| Hexose-6-phosphate ↓ | Thymidine ↓ | Glycine ↑ |
| Maltotriose ↓ | Other Organic Acids | Other Organic Acids |
| Sugar Alcohols | Dehydroabietic acid ↑ | Uric acid ↑ |
| Beta-glycerolphosphate ↑ | Unknown Metabolites (13) | Hydrocarbons |
| Bisphosphoglycerol ↑ | 110 ↑ | Squalene ↓ |
| Xylitol ↑ | 257 ↑ | Unknown Metabolites (7) |
| Vitamins/Derivatives | 2061 ↑ | 1760 ↓ |
| Dehydroascorbic acid ↓ | 3083 ↑ | 3200 ↓ |
| Nicotinamide ↑ | 6104 ↑ | 31962 ↑ |
| Peptides | 16561 ↑ | 84544 ↑ |
| Glutathione ↓ | 32153 ↑ | 97584 ↓ |
| Amino Acids | 42424 ↑ | 100321 ↑ |
| Aspartic acid ↓ | 43021↑ | 102601 ↑ |
| Glycine ↓ | 84223↑ | |
| Other Organic Acids | 97326 ↑ | |
| Benzoic acid ↑ | 100584 ↑ | |
| Gamma-aminobutyric acid ↓ | 100730 ↑ | |
| Malic acid ↑ | ||
| Uric acid ↓ | ||
| Other Organic Alcohols | ||
| Methanolphosphate ↑ | ||
| Hydrocarbons | ||
| Squalene ↑ | ||
| Methylphenols | ||
| p-cresol ↑ | ||
| Unknown Metabolites (19) | ||
| 54 ↑ | ||
| 1064 ↑ | ||
| 1704 ↑ | ||
| 9320 ↑ | ||
| 17651 ↑ | ||
| 17954 ↑ | ||
| 18485 ↓ | ||
| 21665 ↑ | ||
| 21682 ↑ | ||
| 21683 ↑ | ||
| 26746 ↑ | ||
| 33989 ↑ | ||
| 42424 ↑ | ||
| 84223 ↑ | ||
| 84382 ↓ | ||
| 84682 ↑ | ||
| 97584 ↑ | ||
| 100321 ↓ | ||
| 102601 ↓ |
Metabolomic Results
The metabolome consists of all of the metabolites, including substrates, intermediates and products of metabolic reactions within the cell. As the end-products of metabolic reactions, the metabolome can be indicative of the overall health of the cell, and offer useful information on the dysregulation of certain pathways. Similar to the results of the proteomic study, more metabolites were differentially expressed in R278C hearts when compared with WT hearts (46 changed of 304 identified metabolites) than when I79N hearts were compared with WT hearts (21 changed of 304 identified), strongly suggesting that the relative number of proteins or metabolites differentially expressed were not major factors with respect to the resulting phenotype. When R278C and I79N hearts were directly compared, 17 metabolites were changed.
Pathway analysis of the metabolomics data using MetaboAnalyst 2.0, a comprehensive suite of web tools for metabolomic data processing, visualization and analysis (53), showed that two pathways were significantly affected in R278C hearts compared with WT hearts: biosynthesis of unsaturated fatty acids and fatty acid synthesis, and two other pathways bordered on significance (Fig. 7). Although several metabolites were altered in I79N hearts relative to WT hearts no pathway was statistically significantly affected. One pathway that bordered on significance in I79N versus hearts was starch and sucrose metabolism (Fig. 7). Comparison of I79N and R278C hearts revealed the greatest number of significant differences, with four pathways affected (supplemental Fig. S2). These were aminoacyl-tRNA biosynthesis, cyanoamino acid metabolism, nitrogen metabolism, and methane metabolism, which were up-regulated in R278C hearts relative to I79N hearts (supplemental Fig. S2). Many unknown metabolites were also detected and quantified and shown in the supplemental Table S8.
Metabolomic analysis showed that the levels of eight fatty acids were increased in R278C hearts relative to WT hearts, compared with one fatty acid that was decreased in I79N hearts compared with WT hearts. Increased levels of creatinine in I79N hearts relative to WT hearts (Fig. 8) as well as other changes in metabolites associated with energy metabolism (Table II) suggest impaired energy production in I79N hearts. R278C hearts showed decreases in glucose-6-phosphate, fructose-6-phosphate, and hexose-6-phosphate when compared with WT hearts, whereas the I79N hearts showed a trend toward increased levels of these metabolites as well as increased fructose levels in comparison to WT hearts (Fig. 8 and supplemental Fig. S3). This trend of increased levels of glycolytic metabolites in I79N hearts relative to both WT and R278C hearts is consistent with the proteomics findings (increased levels of glycolytic enzymes) which suggest that glycolysis may be accelerated in I79N hearts. A summary of the proteins and metabolites found to be differentially expressed in the glycolytic pathway are shown in Fig. 9.
Fig. 9.
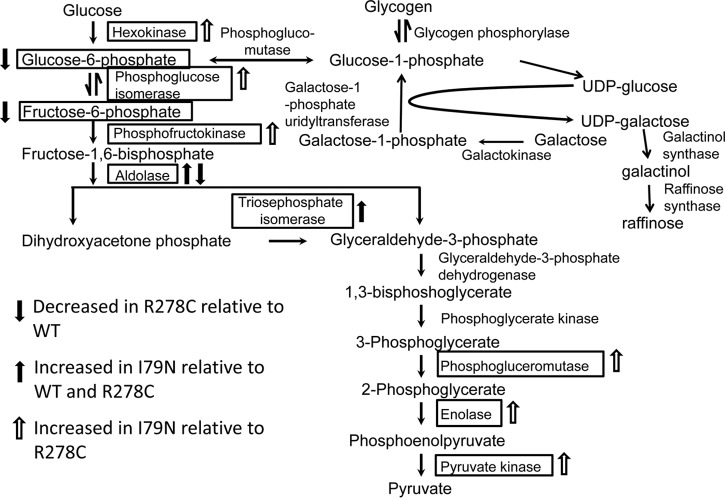
Changes in levels of metabolites and enzymes in the glycolytic pathway in R278C and I79N hearts. A solid black arrow indicates metabolite or enzyme is increased in I79N hearts relative to WT and R278C. An empty arrow indicates that the metabolite or enzyme is increased in I79N relative to R278C only. An arrow with gray fill indicates a decrease in R278C relative to WT. The galactose pathway is also shown (see Discussion and supplemental Fig. S3).
DISCUSSION
FHC-causing TnT mutations are associated with a high risk of sudden cardiac death, and the mechanisms underlying the disease are not well understood. Transgenic mice expressing the I79N mutation have low exercise tolerance and die upon isoproterenol treatment, whereas R278C mice are tolerant of exercise and isoproterenol treatment (54, 55). We used proteomic and metabolomic approaches to uncover pathways affected in TnT-related FHC. The only previous proteomic approach to investigate FHC models was using two-dimensional polyacrylamide gel electrophoresis (2DE) coupled with a Q-TOF Ultima MS (56). Mice predisposed to developing hypertrophic cardiomyopathy (HCM) due to overexpression of mutated (Gly203Ser [G203S]) troponin I (TnI) were studied by 2DE coupled with LC-MS/MS to identify protein spots that showed altered expression. This study identified 34 protein spots (corresponding to 22 different proteins) that showed at least a 2-fold change in G203S hearts relative to WT hearts (56). Altered proteins in the G203S hearts were involved in energy production and utilization (e.g. NADH dehydrogenase), calcium handling (e.g. S100 calcium binding protein A10), and cell structure and muscle contraction (e.g. myosin) (56). 2DE and mass spectrometry have also been used in studies on human patients and mouse models of dilated cardiomyopathy (DCM). A study using shotgun LC-MS/MS to compare protein expression in DCM mouse hearts to WT mouse hearts showed altered levels of proteins involved in the endoplasmic reticulum (ER) stress response, cytoskeletal remodeling, and apoptosis (57). Proteomic investigation of DCM by other groups found changes in expression of metabolic enzymes (58–61), stress response proteins (58, 60, 61), and proteins involved in cell structure and contraction (58, 59). Although 2DE is a powerful technique, it has several disadvantages including the low dynamic protein range and limitations on the isoelectric point and molecular weights of proteins that can be resolved and analyzed. The investigation of the I79N and R278C hearts using MS suggests that TnT-related cardiomyopathies are more complex than previous proteomic and metabolomic studies on cardiomyopathy hearts showed. More pathways were found to be affected, including pathways that have not previously been shown to be affected in any FHC model.
Proteins Involved in Protein Degradation
Proteomic analysis revealed changes in expression of several proteasome subunits. Several of the proteasome subunits identified by mass spectrometry showed significantly higher levels in I79N mice than R278C mice (9 of 23) and some showed decreases in R278C relative to WT (3 of 23). Validation of these findings by Western blotting revealed a similar trend; expression of proteasome subunits was decreased in R278C hearts and increased in I79N hearts in one case, and was consistently higher in I79N than R278C hearts (Figs. 3 and 4). Further investigation of the proteasome revealed that β5 (chymotrypsin-like) proteasome activity was decreased in I79N mice relative to WT and R278C mice (Fig. 4). Thus, despite higher proteasome subunit expression in I79N than R278C hearts, both 20S and 26S chymotrypsin-like activities are decreased, indicating proteasome impairment. 20S proteasome activity was measured because the 20S proteasome has been shown to degrade oxidized proteins independently of ATP. Lower 20S activities would be expected to result in higher levels of oxidized proteins. The immunoproteasome activity was also affected; the β1i activity was decreased in R278C and I79N hearts, and β5i activity was decreased in I79N hearts. Immunoproteasome activity has not been previously measured in hearts, and these results suggest that the immunoproteasome function is impaired in I79N hearts relative to WT hearts.
Accumulation of ubiquitinated and oxidized proteins, typical indicators of proteasome impairment, was investigated to see if they were affected in I79N or R278C mice. Western blots showed a greater accumulation of ubiquitinated proteins in total homogenates and oxidized proteins in cytosolic fractions from I79N hearts relative to WT and R278C, consistent with impaired proteasome function (Figs. 4 and 5). The increased level of oxidized proteins is consistent with impaired immunoproteasome activity in I79N hearts. Increased levels of polyubiqutinated proteins are usually associated with increased levels of oxidized proteins, an independent indicator of increased oxidative stress. With up-regulated antioxidant enzymes and antioxidant capacity, it is likely that the intracellular homeostasis in I79N heart cells is significantly compromised. Increased levels of oxidized proteins suggest that oxidative stress in cytosolic cardiac homogenates is increased in I79N mice relative to WT and R278C hearts. Levels of oxidized proteins in total homogenates were similar in I79N, R278C and WT hearts, indicating that oxidative stress may be predominantly in the cytosol (Fig. 5). Overall, I79N hearts, which serve as a model of TnT-related cardiomyopathy with a poor prognosis, displayed higher levels of oxidative stress, increased ubiquitination, and an increase in proteasome subunit and TRX-1 expression relative to R278C hearts, the mild, late-onset disease model. Accumulation of ubiquitinated proteins, increases and decreases in proteasome activity, and changes in expression of the proteasome and associated proteins have been observed in several experimental and human heart diseases (62–65). These results suggest that proteasome dysfunction is an important difference between the I79N and WT or R278C hearts. Impaired proteasome function by itself could account for the poor prognosis in I79N FHC patients, because recent work with pigs showed that chronic proteasome inhibition led to functional and structural cardiomyopathic sequelae (66). Proteasome dysfunction has been previously shown to occur in some but not all hearts containing MyBPC mutants associated with FHC. This was because of some MyBPC mutants not being properly degraded by the proteasome, resulting in MyBPC aggregates and insufficient proteasome activity to degrade other substrates (23–26). This possibility needs to be tested with other thin and thick filament mutations.
Proteins Involved in Metabolism
Glucose Metabolism
Proteins involved in metabolism account for the largest group of biological processes affected when comparing R278C or I79N hearts to WT hearts (Fig. 1). Some proteins involved in glycolysis were up-regulated in I79N hearts, whereas expression of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase was decreased in R278C hearts (Fig. 9 and supplemental Table S6). Metabolomic analysis revealed several changes in key metabolites including altered levels of metabolites in the glycolytic pathway in R278C hearts (Table II and Fig. 9). Another affected pathway that seems to be important in cardiac physiology is the galactose pathway. Important components of the galactose pathway, glucose-1-phosphate and raffinose (a trisaccharide containing galactose), although not statistically significant, showed higher levels for all averaged biological replicates than WT or R278C averaged biological replicates (supplemental Table S2). Galactose is an epimer of glucose which can be converted to glucose-6-phosphate, at which point it may enter the glycolytic pathway. Impaired galactose metabolism is associated with hypertrophic cardiomyopathy and heart failure (67, 68). Additionally, mutations in α-galactosidase A (a glycoside hydrolase enzyme that catalyzes the hydrolysis of terminal alpha-galactosyl moieties from glycolipids and glycoproteins) have been shown to phenocopy hypertrophic cardiomyopathy caused by mutations in sarcomeric proteins (69–71). Further studies into the galactose pathway should be carried out in hearts with cardiomyopathy as this pathway may be important in FHC.
The increased protein expression of hexokinase, aldolase, and triosphosphate isomerase in I79N hearts relative to R278C (as determined by mass spectrometry) suggest accelerated glycolysis. Enhanced dependence on glucose and accelerated glycolysis have previously been reported in hypertrophied hearts (72). Several other previous studies have demonstrated altered energy metabolism in hypertrophic cardiomyopathy, and energy depletion due to inefficient sarcomeric ATP usage has been suggested as an important unifying factor in FHC (73). Previous studies also suggest that a switch in energy substrate utilization occurs during disease progression in cardiomyopathies (74).
Impaired energy metabolism has previously been suggested by several groups to be a major contributing factor to HCM. The majority of the 22 proteins that were altered in TnI FHC G203S hearts were involved in energy production (56). The G203S hearts showed up-regulation of several proteins involved in the tricarboxylic acid cycle (TCA) and the electron transport chain (ETC), which is consistent with the elevated TCA and ETC proteins in the I79N hearts. However, the I79N hearts also showed an up-regulated glycolytic pathway which was not detected in the TnI G203S hearts. The increased levels of enzymes in the glycolytic pathway may indicate that the cell is attempting to produce more ATP. Indeed, it has been demonstrated that energy metabolism is impaired in I79N hearts, and inefficient sarcomeric ATP usage is one factor thought to play a role in the pathogenesis of FHC (73, 75, 76). Interestingly, the ubiquitin-dependent proteolysis performed by the proteasome utilizes ATP, and a decrease in ATP levels may also decrease protein degradation (77, 78); therefore it is conceivable that one mechanism by which TnT mutations affect the proteasome is through ATP depletion.
Altered energetics has been demonstrated in various models of TnT-related FHC. After six months of daily exercise, most transgenic rats containing truncated human cardiac TnT, resulting from an intron 15 splice donor site mutation observed in FHC patients, showed ventricular tachycardia/fibrillation and myofibrillar disarray, whereas no transgenic rats expressing WT TnT showed ventricular tachycardia/fibrillation and myofibrillar disarray (79). These truncated TnT transgenic rats showed impaired cardiac metabolism, decreased phosphocreatine levels and decreased phosphocreatine to ATP ratios (-31%, p < 0.05) (80), although the ATP levels were not significantly affected. These changes in metabolites were observed even though the basal cardiac parameters (ejection fraction, end-diastolic volume etc.) of the transgenic truncated TnT rats were not significantly different from the control transgenic rats. Another study on energy utilization in TnT-FHC showed that transgenic hearts with R92W and R92L TnT mutations both showed greater ATP utilization during muscle contraction, with the R92W hearts showing more severe energetic abnormalities and greater contractile dysfunction than the R92L hearts (81). These hearts had higher intracellular ADP and Pi levels, as well as lower ATP and phosphocreatine levels (81). It was shown in our study that the levels of creatinine were higher in I79N hearts than WT hearts.
Fatty Acid Metabolism
In the heart, phosphocreatine is the main source of reserve energy; when ATP demand exceeds supply, as in acute pump failure or high wall stress, utilization of phosphocreatine by the creatine kinase system maintains constant ATP levels to meet the heart's energy needs (82). As pathogenesis progresses, the demand for ATP increases and the level of phosphocreatine is depleted, which necessitates the use of alternative energy substrates (76, 83). In the early compensatory stages of cardiomyopathy, the heart utilizes free fatty acids as an energy substrate, which yields more ATP than glucose but also requires more oxygen to metabolize (84). At more advanced stages of disease, as in heart failure and end-stage HCM, the heart has been shown to favor glucose over fatty acids as substrate (85). This may be due to cardiac growth without sufficient angiogenesis, resulting in a hypoxic state; data suggests that a mildly hypoxic state results in increased glucose utilization (86), and exposure of the developing heart to the oxygen-rich postnatal environment at birth correlates with a shift from glycolysis as the major source of ATP production to fatty acid and lactate oxidation (87). All of these findings support the notion that disease progression in the heart is correlated with a shift in energy substrate utilization, with an initially increased dependence on fatty acids and later an increased dependence on glucose. The R278C hearts show increased levels of mitochondrial 3-ketoacyl-CoA thiolase (involved in fatty acid synthesis) and acyl-coenzyme A thioesterase 13 and enoyl-CoA delta isomerase 1 (involved in fatty acid metabolism) relative to WT hearts (supplemental Table S7). Metabolomic analysis showed increased levels of eight fatty acids in R278C hearts relative to WT hearts, compared with only one increased in I79N hearts compared with WT hearts (Table II). Although it is possible that elevated levels of fatty acids may be due to decreased fatty acid usage, the increase in protein levels of enzymes involved in fatty acid metabolism suggest that the R278C hearts utilize increased levels of fatty acids. Hence, the 3 month old R278C hearts are likely in a compensatory mode while the I79N hearts show a shift to glucose utilization, suggesting that these hearts have already started to decompensate and that cardiac growth is not needed for a switch in metabolism, because these mice do not have cardiac growth. I79N hearts also show higher levels of creatinine, a breakdown product of phosphocreatine, relative to WT hearts, supporting the notion that phosphocreatine is used at an accelerated rate to meet the energy demands of I79N hearts.
Proteins Involved in Cellular Defense
Mass spectrometry also showed that levels of several antioxidants were increased in I79N hearts relative to R278C hearts, including glutathione, glutathione peroxidase, TRX-1, superoxide dismutase, and the glutathione S-transferases GSTM1, GSTM2, and GSTM7 (supplemental Table S5). Some of these findings were verified by Western blot; TRX-1 expression levels were higher in I79N hearts compared with R278C hearts. GSTM1 showed a trend toward increased levels relative to WT (p = 0.075), whereas glutathione amount was shown to be higher in metabolomic investigations (Table II). An assay to measure the antioxidant capacity revealed that antioxidant capacity is increased in I79N hearts relative to WT hearts, consistent with increased expression of antioxidants. Thus, it appears that I79N hearts are subject to more stress, as shown by increased ubiquitinated and oxidized protein levels. Expression of proteasome subunits and antioxidants may be up-regulated to cope with this stress.
Proteins Involved with Ca2+ Handling
Because previous reports have shown that increased Ca2+ sensitivity in I79N hearts results in increased intracellular Ca2+ levels, it is possible that these changes in Ca2+ levels would directly affect Ca2+-binding proteins. Although several proteins involved in calcium handling were up-regulated in I79N hearts compared with R278C, no Ca2+ binding or Ca2+ handling protein was altered in either I79N or R278C hearts when compared with WT hearts suggesting that altered protein expression levels of Ca2+ binding and handling proteins were not affected by the increased intracellular Ca2+ levels. This is different from the TnI G203S hearts, which show elevated levels of Ca2+ handling proteins S100A10, S110A11 and annexin A2 (56).
Link Between Different Cellular Pathways
No proteomic or metabolomic analyses of any TnT-FHC animal model have been previously performed. The proteomic results suggest that the two main pathways that differentiate the I79N hearts from the R278C hearts are metabolic processes (metabolism and protein degradation by the proteasome), and cellular processes (such as cellular defense). The increased levels of oxidized proteins suggest that many enzymes are likely to be oxidized and have lower enzymatic activity. Increased levels of oxidized proteins in I79N hearts could result from the decreased proteasome activity because the proteasome is involved in degrading oxidized proteins (36). It is also possible that the increased levels of oxidized proteins in these stressed hearts are due to increased energy production. Increased ATP production, as may be required in the I79N transgenic heart, would likely increase mitochondrial ROS production. A previous report has shown that increased ROS production from complex III in the mitochondria is proportional to respiration rate in adult guinea pig ventricular myocytes (88). Increased ROS levels could cause an increase in proteasome and antioxidant defense proteins to remove the increased levels of oxidized proteins and reduce the levels of ROS respectively. If the proteasome itself or activators of the proteasome are oxidized, then even with higher levels of proteasome, the activity of most proteasomes would be compromised resulting in lower proteasome activity. Independently the need for more ATP for contraction in I79N hearts would likely result in increased levels of enzymes involved in ATP production, as observed in the proteomic data. However, a trend toward higher levels of AMP in I79N hearts suggest that ATP levels may be lower, indicating impairment of the enzymes involved in ATP production occurs, possibly by oxidation of these enzymes. The R278C hearts also showed a trend toward increased levels of AMP suggesting that energy production may be compromised, though to a lesser extent in these hearts. R278C hearts also showed some signs of altering ATP production, as l-lactate dehydrogenase B chain was up-regulated in both I79N and R278C hearts relative to WT hearts (Table I). Although the R278C mutation is associated with a better prognosis, some patients with this mutation do present with dyspnea at ages above 65 years old (17). Hence R278C hearts may show more significant changes in energy metabolism in older hearts. Several other reports suggest that inefficient energy utilization may be a common and important molecular pathway associated with HCM caused by sarcomeric mutations (73, 75, 76, 89). Our results suggest that inefficient cellular ATP utilization (energetic alterations) and protein degradation in TnT-related FHC contributes to the pathogenesis of the disease.
In the previous report of a TnI G203S FHC model, 17 proteins were found to be up-regulated in the G203S hearts (56). Two of these proteins, cytochrome c oxidase subunit 6A and 39S ribosomal protein L12 (both mitochondrial), were found to be increased in I79N hearts relative to WT hearts. The ribosomal protein L12 was also found to be up-regulated in R278C hearts. Important structural proteins α cardiac actin, desmin and dynein light chain roadblock-type I which were found to be up-regulated in the TnI G203S hearts were not up-regulated in I79N or R278C hearts, suggesting that these proteins may be more important for structural changes associated with hypertrophy in the G203S hearts (56). Unlike transgenic hearts containing TnT I79N or R278C mutations, hearts containing TnI G203S showed left ventricular hypertrophy by age 21 weeks, and cardiomyocytes isolated from these hearts show cellular hypertrophy (56).
In summary, this is the first study to use proteomic and metabolomic analysis to investigate mice with TnT-related cardiomyopathies, as well as the first study to compare mouse models of mild and severe FHC due to a sarcomeric gene mutation using proteomics or metabolomics. It was found that FHC was associated with altered expression of proteasome subunits and antioxidant enzymes, specifically higher levels in I79N versus R278C hearts. Proteasome activity was also decreased, and oxidative stress and ubiquitination were higher in I79N mice. The data suggest that I79N hearts are subject to more stress, and likely upregulate expression of antioxidants and proteasome subunits to cope with increased stress, but these changes in expression are insufficient for regaining intracellular homeostasis. It is likely that impaired metabolism in the cytoplasm and mitochondria leads to increased mitochondrial ROS production resulting in increased levels of oxidized proteins. In support of this possibility, an increased level of oxidized proteins was detected in I79N hearts using the OxyBlot procedure. Increased levels of ROS could also directly affect the proteasome, resulting in lower activity. As such, even with similar or higher levels of proteasome in I79N hearts compared with WT and R278C hearts, the overall proteasome activity would be decreased in I79N hearts. Further experiments will determine if the proteasome from I79N hearts is oxidatively modified.
In I79N hearts there is also metabolic dysregulation of the glycolytic pathway which is likely to contribute to the pathogenesis of the disease. It is interesting to note that these studies revealed more changes in R278C than I79N mice relative to WT, suggesting that the changes in I79N mice are likely more profound in nature, because the I79N mutation is associated with a more severe phenotype than the R278C mutation. A possible mechanism for the I79N mutation causing the observed effects may be due to the mutation triggering conformational changes in TnT which increase the binding of Ca2+ to TnC. The resulting increase in Ca2+ sensitivity of myofilament contraction affects intracellular Ca2+ homeostasis resulting in inefficient sarcomeric ATP utilization (55, 73, 90). The higher demand for ATP may be inducing stress on the cellular ATP producing pathways: glycolysis, critic acid cycle, and oxidative phosphorylation. A byproduct of oxidative phosphorylation is ROS production (91), and increased ATP production is likely to result in increased ROS production. Increased ROS production would result in increased levels of enzymes that reduce ROS levels as well as increased levels of oxidized proteins (which are both observed in the I79N hearts) which would strain the UPS. Because the levels of proteasome subunits in the I79N mice are not decreased it is possible that increased ROS levels may be directly oxidizing proteasome subunits and reducing proteasome activity. It is also possible that due to the increased levels of oxidized proteins in I79N hearts some of these oxidized proteins which interact with the proteasome (for degradation) are non-degradable or only slowly degradable, reducing the availability of the proteasome to act on other substrates, resulting in reduced overall proteasome activity. The impaired proteasome activity may result in altered expression of key signaling pathways enzymes that contribute to cardiac dysfunction. Proteasome function is vital for protein homeostasis in the heart because chronic decreased proteasome activity have been shown to directly cause cardiac dysfunction (66).
Overall, our findings reveal a new pathway (the ubiquitin-proteasome system) that was not previously known to be important in TnT-related cardiomyopathies. Our results also suggest that increases in structural proteins such as α-cardiac actin and desmin are likely to be associated predominantly with cell enlargement and that cardiac growth is not needed for switch from fatty acid to glucose metabolism. Further investigation of these pathways to determine their involvement in different FHC models as well as the effects of rectifying these pathways on diseased heart functions is needed. It is possible that modulating the proteasome activity in the heart will reduce the cardiac dysfunction observed during exercise.
Footnotes
Author contributions: A.V.G. designed research; J.E.G., X.L., F.A.W., and A.V.G. performed research; X.L., F.A.W., and A.V.G. contributed new reagents or analytic tools; J.E.G., X.L., F.A.W., and A.V.G. analyzed data; J.E.G., F.A.W., and A.V.G. wrote the paper.
* This research was supported by a National Institutes of Health Grant HL096819. JG is part of the BMCDB graduate group. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare no competing financial interests.
1 The abbreviations used are:
- FHC
- familial hypertrophic cardiomyopathy
- AMP
- adenosine monophosphate
- GC-TOF
- gas chromatography - time of flight mass spectrometry
- HCM
- hypertrophic cardiomyopathy
- LC-MS/MS
- tandem mass spectrometry
- MyBPC
- Myosin binding protein C
- SCD
- sudden cardiac death
- TnI
- troponin I
- TnT
- troponin T
- Ile79Asn
- I79N
- Arg278Cys
- R278C.
REFERENCES
- 1. Frey N., Luedde M., and Katus H. A. (2012) Mechanisms of disease: hypertrophic cardiomyopathy. Nat. Rev. Cardiology 9, 91–100 [DOI] [PubMed] [Google Scholar]
- 2. Maron B. J. (2002) Hypertrophic cardiomyopathy: a systematic review. JAMA 287, 1308–1320 [DOI] [PubMed] [Google Scholar]
- 3. Ashrafian H., McKenna W. J., and Watkins H. (2011) Disease pathways and novel therapeutic targets in hypertrophic cardiomyopathy. Circulation Res. 109, 86–96 [DOI] [PubMed] [Google Scholar]
- 4. Xu Q., Dewey S., Nguyen S., and Gomes A. V. (2010) Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. J. Mol. Cell. Cardiol. 48, 899–909 [DOI] [PubMed] [Google Scholar]
- 5. Gomes A. V., Barnes J. A., Harada K., and Potter J. D. (2004) Role of troponin T in disease. Mol. Cell. Biochem. 263, 115–129 [DOI] [PubMed] [Google Scholar]
- 6. Moolman J. C., Corfield V. A., Posen B., Ngumbela K., Seidman C., Brink P. A., and Watkins H. (1997) Sudden death due to troponin T mutations. J. Am. College Cardiol. 29, 549–555 [DOI] [PubMed] [Google Scholar]
- 7. Watkins H., McKenna W. J., Thierfelder L., Suk H. J., Anan R., O'Donoghue A., Spirito P., Matsumori A., Moravec C. S., Seidman J. G., and et al. (1995) Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. New Engl. J. Med. 332, 1058–1064 [DOI] [PubMed] [Google Scholar]
- 8. Pasquale F., Syrris P., Kaski J. P., Mogensen J., McKenna W. J., and Elliott P. (2012) Long-term outcomes in hypertrophic cardiomyopathy caused by mutations in the cardiac troponin T gene. Circulation Cardiovascular Genetics 5, 10–17 [DOI] [PubMed] [Google Scholar]
- 9. Wen Y., Pinto J. R., Gomes A. V., Xu Y., Wang Y., Potter J. D., and Kerrick W. G. (2008) Functional consequences of the human cardiac troponin I hypertrophic cardiomyopathy mutation R145G in transgenic mice. J. Biol. Chem. 283, 20484–20494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomes A. V., Venkatraman G., Davis J. P., Tikunova S. B., Engel P., Solaro R. J., and Potter J. D. (2004) Cardiac troponin T isoforms affect the Ca(2+) sensitivity of force development in the presence of slow skeletal troponin I: insights into the role of troponin T isoforms in the fetal heart. J. Biol. Chem. 279, 49579–49587 [DOI] [PubMed] [Google Scholar]
- 11. Gomes A. V., and Potter J. D. (2004) Molecular and cellular aspects of troponin cardiomyopathies. Ann. N. Y. Acad. Sci. 1015, 214–224 [DOI] [PubMed] [Google Scholar]
- 12. Cui Z., Dewey S., and Gomes A. V. (2011) Cardioproteomics: advancing the discovery of signaling mechanisms involved in cardiovascular diseases. Am. J. Cardiovascular Dis. 1, 274–292 [PMC free article] [PubMed] [Google Scholar]
- 13. Hernandez O. M., Szczesna-Cordary D., Knollmann B. C., Miller T., Bell M., Zhao J., Sirenko S. G., Diaz Z., Guzman G., Xu Y., Wang Y., Kerrick W. G., and Potter J. D. (2005) F110I and R278C troponin T mutations that cause familial hypertrophic cardiomyopathy affect muscle contraction in transgenic mice and reconstituted human cardiac fibers. J.Biol. Chem. 280, 37183–37194 [DOI] [PubMed] [Google Scholar]
- 14. Miller T., Szczesna D., Housmans P. R., Zhao J., de Freitas F., Gomes A. V., Culbreath L., McCue J., Wang Y., Xu Y., Kerrick W. G., and Potter J. D. (2001) Abnormal contractile function in transgenic mice expressing a familial hypertrophic cardiomyopathy-linked troponin T (I79N) mutation. J. Biol. Chem. 276, 3743–3755 [DOI] [PubMed] [Google Scholar]
- 15. Varnava A. M., Elliott P. M., Baboonian C., Davison F., Davies M. J., and McKenna W. J. (2001) Hypertrophic cardiomyopathy: histopathological features of sudden death in cardiac troponin T disease. Circulation 104, 1380–1384 [DOI] [PubMed] [Google Scholar]
- 16. Elliott P. M., D'Cruz L., and McKenna W. J. (1999) Late-onset hypertrophic cardiomyopathy caused by a mutation in the cardiac troponin T gene. N Engl. J. Med. 341, 1855–1856 [DOI] [PubMed] [Google Scholar]
- 17. Van Driest S. L., Ellsworth E. G., Ommen S. R., Tajik A. J., Gersh B. J., and Ackerman M. J. (2003) Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation 108, 445–451 [DOI] [PubMed] [Google Scholar]
- 18. Li J., Horak K. M., Su H., Sanbe A., Robbins J., and Wang X. (2011) Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J. Clin. Invest. 121, 3689–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Li J., Zheng H., Su H., and Powell S. R. (2011) Proteasome functional insufficiency in cardiac pathogenesis. American journal of physiology. Heart Circulatory Physiol. 301, H2207–H2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patterson C., Ike C., Willis P. W. t., Stouffer G. A., and Willis M. S. (2007) The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation 115, 1456–1463 [DOI] [PubMed] [Google Scholar]
- 21. Wang X., and Robbins J. (2006) Heart failure and protein quality control. Circulation Res. 99, 1315–1328 [DOI] [PubMed] [Google Scholar]
- 22. Wang X. (2013) Repeated intermittent administration of a ubiquitous proteasome inhibitor leads to restrictive cardiomyopathy. Eur. J. Heart Failure 15, 597–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bahrudin U., Morisaki H., Morisaki T., Ninomiya H., Higaki K., Nanba E., Igawa O., Takashima S., Mizuta E., Miake J., Yamamoto Y., Shirayoshi Y., Kitakaze M., Carrier L., and Hisatome I. (2008) Ubiquitin-proteasome system impairment caused by a missense cardiac myosin-binding protein C mutation and associated with cardiac dysfunction in hypertrophic cardiomyopathy. J. Mol. Biol. 384, 896–907 [DOI] [PubMed] [Google Scholar]
- 24. Sarikas A., Carrier L., Schenke C., Doll D., Flavigny J., Lindenberg K. S., Eschenhagen T., and Zolk O. (2005) Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovascular Res. 66, 33–44 [DOI] [PubMed] [Google Scholar]
- 25. Schlossarek S., Englmann D. R., Sultan K. R., Sauer M., Eschenhagen T., and Carrier L. (2012) Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res. Cardiol. 107, 235. [DOI] [PubMed] [Google Scholar]
- 26. Vignier N., Schlossarek S., Fraysse B., Mearini G., Kramer E., Pointu H., Mougenot N., Guiard J., Reimer R., Hohenberg H., Schwartz K., Vernet M., Eschenhagen T., and Carrier L. (2009) Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circulation Res. 105, 239–248 [DOI] [PubMed] [Google Scholar]
- 27. Gomes A. V., Young G. W., Wang Y., Zong C., Eghbali M., Drews O., Lu H., Stefani E., and Ping P. (2009) Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Mol. Cell. Proteomics 8, 302–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gomes A. V., Zong C., Edmondson R. D., Li X., Stefani E., Zhang J., Jones R. C., Thyparambil S., Wang G. W., Qiao X., Bardag-Gorce F., and Ping P. (2006) Mapping the murine cardiac 26S proteasome complexes. Circulation Res. 99, 362–371 [DOI] [PubMed] [Google Scholar]
- 29. Sengupta D., Byrum S. D., Avaritt N. L., Davis L., Shields B., Mahmoud F., Reynolds M., Orr L. M., Mackintosh S. G., Shalin S. C., and Tackett A. J. (2015) Quantitative Histone Mass Spectrometry identifies elevated Histone H3 Lysine 27 Trimethylation in Melanoma. Mol. Cell. Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Ma K., Vitek O., and Nesvizhskii A. I. (2012) A statistical model-building perspective to identification of MS/MS spectra with PeptideProphet. BMC Bioinformatics 13, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 33. Lai X., Wang L., Tang H., and Witzmann F. A. (2011) A novel alignment method and multiple filters for exclusion of unqualified peptides to enhance label-free quantification using peptide intensity in LC-MS/MS. J. Proteome Res. 10, 4799–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 35. Cui Z., Gilda J. E., and Gomes A. V. (2014) Crude and purified proteasome activity assays are affected by type of microplate. Anal. Biochem. 446, 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cui Z., Hwang S. M., and Gomes A. V. (2014) Identification of the immunoproteasome as a novel regulator of skeletal muscle differentiation. Mol. Cell. Biol. 34, 96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dewey S., Lai X., Witzmann F. A., Sohal M., and Gomes A. V. (2013) Proteomic analysis of hearts from akita mice suggests that increases in soluble epoxide hydrolase and antioxidative programming are key changes in early stages of diabetic cardiomyopathy. J. Proteome Res. 12, 3920–3933 [DOI] [PubMed] [Google Scholar]
- 38. Kind T., Wohlgemuth G., Lee do Y., Lu Y., Palazoglu M., Shahbaz S., and Fiehn O. (2009) FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 81, 10038–10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fiehn O., Wohlgemuth G., and Scholz M. (2005) Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. Proc. Lect. Notes Bioinformatics 3615, 224–239 [Google Scholar]
- 40. Ferguson P. L., and Smith R. D. (2003) Proteome analysis by mass spectrometry. Ann. Rev. Biophys. Biomol. Structure 32, 399–424 [DOI] [PubMed] [Google Scholar]
- 41. Dong S., Jia C., Zhang S., Fan G., Li Y., Shan P., Sun L., Xiao W., Li L., Zheng Y., Liu J., Wei H., Hu C., Zhang W., Chin Y. E., Zhai Q., Li Q., Jia F., Mo Q., Edwards D. P., Huang S., Chan L., O'Malley B. W., Li X., and Wang C. (2013) The REGgamma proteasome regulates hepatic lipid metabolism through inhibition of autophagy. Cell Metabolism 18, 380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yoshizawa T., Karim M. F., Sato Y., Senokuchi T., Miyata K., Fukuda T., Go C., Tasaki M., Uchimura K., Kadomatsu T., Tian Z., Smolka C., Sawa T., Takeya M., Tomizawa K., Ando Y., Araki E., Akaike T., Braun T., Oike Y., Bober E., and Yamagata K. (2014) SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metabolism 19, 712–721 [DOI] [PubMed] [Google Scholar]
- 43. Ronnebaum S. M., Patterson C., and Schisler J. C. (2014) Minireview: Hey U(PS): Metabolic and Proteolytic Homeostasis Linked via AMPK and the Ubiquitin Proteasome System. Mol. Endocrinol. 28, 1602–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Popovic D., Vucic D., and Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 [DOI] [PubMed] [Google Scholar]
- 45. Shang F., and Taylor A. (2011) Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radical Biol. Med. 51, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu J., and Holmgren A. (2014) The thioredoxin antioxidant system. Free Radical Biol. Med. 66, 75–87 [DOI] [PubMed] [Google Scholar]
- 47. Ohira A., Honda O., Gauntt C. D., Yamamoto M., Hori K., Masutani H., Yodoi J., and Honda Y. (1994) Oxidative stress induces adult T cell leukemia derived factor/thioredoxin in the rat retina. Lab. Invest. 70, 279–285 [PubMed] [Google Scholar]
- 48. Li K., Pasternak C., and Klug G. (2003) Expression of the trxA gene for thioredoxin 1 in Rhodobacter sphaeroides during oxidative stress. Arch. Microbiol. 180, 484–489 [DOI] [PubMed] [Google Scholar]
- 49. Turoczi T., Chang V. W., Engelman R. M., Maulik N., Ho Y. S., and Das D. K. (2003) Thioredoxin redox signaling in the ischemic heart: an insight with transgenic mice overexpressing Trx1. J. Mol. Cell. Cardiol. 35, 695–704 [DOI] [PubMed] [Google Scholar]
- 50. Eaton D. L., and Bammler T. K. (1999) Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol. Sci. 49, 156–164 [DOI] [PubMed] [Google Scholar]
- 51. Ostermann J., Horwich A. L., Neupert W., and Hartl F. U. (1989) Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 341, 125–130 [DOI] [PubMed] [Google Scholar]
- 52. Lee H. S., Bhagat L., Frossard J. L., Hietaranta A., Singh V. P., Steer M. L., and Saluja A. K. (2000) Water immersion stress induces heat shock protein 60 expression and protects against pancreatitis in rats. Gastroenterology 119, 220–229 [DOI] [PubMed] [Google Scholar]
- 53. Xia J., Psychogios N., Young N., and Wishart D. S. (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Knollmann B. C., Blatt S. A., Horton K., de Freitas F., Miller T., Bell M., Housmans P. R., Weissman N. J., Morad M., and Potter J. D. (2001) Inotropic stimulation induces cardiac dysfunction in transgenic mice expressing a troponin T (I79N) mutation linked to familial hypertrophic cardiomyopathy. J. Biol. Chem. 276, 10039–10048 [DOI] [PubMed] [Google Scholar]
- 55. Baudenbacher F., Schober T., Pinto J. R., Sidorov V. Y., Hilliard F., Solaro R. J., Potter J. D., and Knollmann B. C. (2008) Myofilament Ca2+ sensitization causes susceptibility to cardiac arrhythmia in mice. J. Clin. Investigation 118, 3893–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lam L., Tsoutsman T., Arthur J., and Semsarian C. (2010) Differential protein expression profiling of myocardial tissue in a mouse model of hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 48, 1014–1022 [DOI] [PubMed] [Google Scholar]
- 57. Gramolini A. O., Kislinger T., Alikhani-Koopaei R., Fong V., Thompson N. J., Isserlin R., Sharma P., Oudit G. Y., Trivieri M. G., Fagan A., Kannan A., Higgins D. G., Huedig H., Hess G., Arab S., Seidman J. G., Seidman C. E., Frey B., Perry M., Backx P. H., Liu P. P., MacLennan D. H., and Emili A. (2008) Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Mol. Cell. Proteomics 7, 519–533 [DOI] [PubMed] [Google Scholar]
- 58. Corbett J. M., Why H. J., Wheeler C. H., Richardson P. J., Archard L. C., Yacoub M. H., and Dunn M. J. (1998) Cardiac protein abnormalities in dilated cardiomyopathy detected by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis 19, 2031–2042 [DOI] [PubMed] [Google Scholar]
- 59. Buscemi N., Murray C., Doherty-Kirby A., Lajoie G., Sussman M. A., and Van Eyk J. E. (2005) Myocardial subproteomic analysis of a constitutively active Rac1-expressing transgenic mouse with lethal myocardial hypertrophy. Am. J. Physiol. 289, H2325–H2333 [DOI] [PubMed] [Google Scholar]
- 60. Knecht M., Regitz-Zagrosek V., Pleissner K. P., Jungblut P., Steffen C., Hildebrandt A., and Fleck E. (1994) Characterization of myocardial protein composition in dilated cardiomyopathy by two-dimensional gel electrophoresis. Eur. Heart J. 15 Suppl D, 37–44 [DOI] [PubMed] [Google Scholar]
- 61. Rosello-Lleti E., Alonso J., Cortes R., Almenar L., Martinez-Dolz L., Sanchez-Lazaro I., Lago F., Azorin I., Juanatey J. R., Portoles M., and Rivera M. (2012) Cardiac protein changes in ischaemic and dilated cardiomyopathy: a proteomic study of human left ventricular tissue. J. Cell. Mol. Med. 16, 2471–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mearini G., Schlossarek S., Willis M. S., and Carrier L. (2008) The ubiquitin-proteasome system in cardiac dysfunction. Biochim. Biophys. Acta 1782, 749–763 [DOI] [PubMed] [Google Scholar]
- 63. Day S. M. (2013) The ubiquitin proteasome system in human cardiomyopathies and heart failure. American journal of physiology. Heart Circulatory Physiol. 304, H1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schlossarek S., and Carrier L. (2011) The ubiquitin-proteasome system in cardiomyopathies. Current Opinion Cardiol. 26, 190–195 [DOI] [PubMed] [Google Scholar]
- 65. Weekes J., Morrison K., Mullen A., Wait R., Barton P., and Dunn M. J. (2003) Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics 3, 208–216 [DOI] [PubMed] [Google Scholar]
- 66. Herrmann J., Wohlert C., Saguner A. M., Flores A., Nesbitt L. L., Chade A., Lerman L. O., and Lerman A. (2013) Primary proteasome inhibition results in cardiac dysfunction. Eur. J. Heart Failure 15, 614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Salganik R. I., Solov'eva N. A., Nepomniashchikh L. M., and Semenov D. E. (1994) [Morphologic phenomena of hereditary hypertrophic cardiomyopathy in W/SSM line rats]. Biulleten' eksperimental'noi biologii i meditsiny 118, 203–207 [PubMed] [Google Scholar]
- 68. Solov'eva N. A., Salganik R. I., Grishaeva O. N., Dikalov S. I., and Semenov D. E. (1995) [Biochemical mechanisms of hereditary cardiomyopathy development in W/SSM rats]. Biulleten' eksperimental'noi biologii i meditsiny 120, 151–154 [PubMed] [Google Scholar]
- 69. O'Mahony C., and Elliott P. (2010) Anderson-Fabry disease and the heart. Progress Cardiovasc. Dis. 52, 326–335 [DOI] [PubMed] [Google Scholar]
- 70. von Scheidt W., Eng C. M., Fitzmaurice T. F., Erdmann E., Hubner G., Olsen E. G., Christomanou H., Kandolf R., Bishop D. F., and Desnick R. J. (1991) An atypical variant of Fabry's disease with manifestations confined to the myocardium. N Engl. J. Med. 324, 395–399 [DOI] [PubMed] [Google Scholar]
- 71. Perrot A., Osterziel K. J., Beck M., Dietz R., and Kampmann C. (2002) Fabry disease: focus on cardiac manifestations and molecular mechanisms. Herz 27, 699–702 [DOI] [PubMed] [Google Scholar]
- 72. Kolwicz S. C. Jr., and Tian R. (2011) Glucose metabolism and cardiac hypertrophy. Cardiovascular Res. 90, 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ashrafian H., Redwood C., Blair E., and Watkins H. (2003) Hypertrophic cardiomyopathy:a paradigm for myocardial energy depletion. Trends Genetics 19, 263–268 [DOI] [PubMed] [Google Scholar]
- 74. Harvey P. A., and Leinwand L. A. (2011) The cell biology of disease: cellular mechanisms of cardiomyopathy. J. Cell Biol. 194, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huke S., Venkataraman R., Faggioni M., Bennuri S., Hwang H. S., Baudenbacher F., and Knollmann B. C. (2013) Focal energy deprivation underlies arrhythmia susceptibility in mice with calcium-sensitized myofilaments. Circulation Res. 112, 1334–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Crilley J. G., Boehm E. A., Blair E., Rajagopalan B., Blamire A. M., Styles P., McKenna W. J., Ostman-Smith I., Clarke K., and Watkins H. (2003) Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J. Am. College Cardiol. 41, 1776–1782 [DOI] [PubMed] [Google Scholar]
- 77. Solomon V., and Goldberg A. L. (1996) Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem. 271, 26690–26697 [DOI] [PubMed] [Google Scholar]
- 78. Gronostajski R. M., Pardee A. B., and Goldberg A. L. (1985) The ATP dependence of the degradation of short- and long-lived proteins in growing fibroblasts. J. Biol. Chem. 260, 3344–3349 [PubMed] [Google Scholar]
- 79. Frey N., Franz W. M., Gloeckner K., Degenhardt M., Muller M., Muller O., Merz H., and Katus H. A. (2000) Transgenic rat hearts expressing a human cardiac troponin T deletion reveal diastolic dysfunction and ventricular arrhythmias. Cardiovascular Res. 47, 254–264 [DOI] [PubMed] [Google Scholar]
- 80. Luedde M., Flogel U., Knorr M., Grundt C., Hippe H. J., Brors B., Frank D., Haselmann U., Antony C., Voelkers M., Schrader J., Most P., Lemmer B., Katus H. A., and Frey N. (2009) Decreased contractility due to energy deprivation in a transgenic rat model of hypertrophic cardiomyopathy. J. Mol. Med. 87, 411–422 [DOI] [PubMed] [Google Scholar]
- 81. He H., Javadpour M. M., Latif F., Tardiff J. C., and Ingwall J. S. (2007) R-92L and R-92W mutations in cardiac troponin T lead to distinct energetic phenotypes in intact mouse hearts. Biophys. J. 93, 1834–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ingwall J. S., and Weiss R. G. (2004) Is the failing heart energy starved? On using chemical energy to support cardiac function. Circulation Res. 95, 135–145 [DOI] [PubMed] [Google Scholar]
- 83. Smith C. S., Bottomley P. A., Schulman S. P., Gerstenblith G., and Weiss R. G. (2006) Altered creatine kinase adenosine triphosphate kinetics in failing hypertrophied human myocardium. Circulation 114, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Neubauer S. (2007) The failing heart–an engine out of fuel. N Engl. J. Med. 356, 1140–1151 [DOI] [PubMed] [Google Scholar]
- 85. Davila-Roman V. G., Vedala G., Herrero P., de las Fuentes L., Rogers J. G., Kelly D. P., and Gropler R. J. (2002) Altered myocardial fatty acid and glucose metabolism in idiopathic dilated cardiomyopathy. J.e Am. College Cardiol. 40, 271–277 [DOI] [PubMed] [Google Scholar]
- 86. Huang Y., Hickey R. P., Yeh J. L., Liu D., Dadak A., Young L. H., Johnson R. S., and Giordano F. J. (2004) Cardiac myocyte-specific HIF-1alpha deletion alters vascularization, energy availability, calcium flux, and contractility in the normoxic heart. FASEB J. 18, 1138–1140 [DOI] [PubMed] [Google Scholar]
- 87. Lopaschuk G. D., Spafford M. A., and Marsh D. R. (1991) Glycolysis is predominant source of myocardial ATP production immediately after birth. Am. J. Physiol. 261, H1698–H1705 [DOI] [PubMed] [Google Scholar]
- 88. Cortassa S., Aon M. A., Winslow R. L., and O'Rourke B. (2004) A mitochondrial oscillator dependent on reactive oxygen species. Biophys. J. 87, 2060–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ferrantini C., Belus A., Piroddi N., Scellini B., Tesi C., and Poggesi C. (2009) Mechanical and energetic consequences of HCM-causing mutations. J. Cardiovasc. Translational Res. 2, 441–451 [DOI] [PubMed] [Google Scholar]
- 90. Guinto P. J., Haim T. E., Dowell-Martino C. C., Sibinga N., and Tardiff J. C. (2009) Temporal and mutation-specific alterations in Ca2+ homeostasis differentially determine the progression of cTnT-related cardiomyopathies in murine models. Am. J. Physiol. 297, H614–H626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen Q., Vazquez E. J., Moghaddas S., Hoppel C. L., and Lesnefsky E. J. (2003) Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem. 278, 36027–36031 [DOI] [PubMed] [Google Scholar]
- 92. Xia J., Mandal R., Sinelnikov I. V., Broadhurst D., and Wishart D. S. (2012) MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 40, W127–W133 [DOI] [PMC free article] [PubMed] [Google Scholar]