Abstract
Chronic pain is a complex disease with limited treatment options. Several profiling efforts have been employed with the aim to dissect its molecular underpinnings. However, generated results are often inconsistent and nonoverlapping, which is largely because of inherent technical constraints. Emerging data-independent acquisition (DIA)-mass spectrometry (MS) has the potential to provide unbiased, reproducible and quantitative proteome maps - a prerequisite for standardization among experiments. Here, we designed a DIA-based proteomics workflow to profile changes in the abundance of dorsal root ganglia (DRG) proteins in two mouse models of chronic pain, inflammatory and neuropathic. We generated a DRG-specific spectral library containing 3067 DRG proteins, which enables their standardized quantification by means of DIA-MS in any laboratory. Using this resource, we profiled 2526 DRG proteins in each biological replicate of both chronic pain models and respective controls with unprecedented reproducibility. We detected numerous differentially regulated proteins, the majority of which exhibited pain model-specificity. Our approach recapitulates known biology and discovers dozens of proteins that have not been characterized in the somatosensory system before. Functional validation experiments and analysis of mouse pain behaviors demonstrate that indeed meaningful protein alterations were discovered. These results illustrate how the application of DIA-MS can open new avenues to achieve the long-awaited standardization in the molecular dissection of pathologies of the somatosensory system. Therefore, our findings provide a valuable framework to qualitatively extend our understanding of chronic pain and somatosensation.
Although acute pain is an evolutionary adaptive response, chronic pain is considered a pathology of the nervous system (1, 2). Treatment of chronic pain presents a big challenge as current therapies target molecules with key physiological functions throughout the body resulting in minimal safety profiles. Hence, the identification of proteins specifically involved in chronic pain states is of paramount importance for designing optimized interventions.
In vertebrates, noxious insults are detected by specialized peripheral sensory neurons (so-called nociceptors) such as those of dorsal root ganglia (DRG)1. These DRG neurons innervate the skin, muscles and all organs where they serve as primary cellular detectors of noxious stimuli of various qualities (chemical, thermal or mechanical) including tissue injury and inflammation (3, 4). Ultimately, their activity shapes the afferent message transmitted to the brain where the sensation of pain is generated. The function of nociceptors relies on dedicated protein networks (3, 4). During the last decade enormous progress has been made regarding the identification of ion channels, receptors and signaling proteins involved in nociception and pain (4–6). However, a systematic and quantitative comparison of protein dynamics across different pain states remains a major challenge.
Several -omics approaches have been developed to study pain-related changes in gene expression (7, 8) or of proteins (9–14). Deep-sequencing transcriptomics (RNA-Seq) is able to quantify the majority of the transcriptome (7, 8), but only half of mRNA changes account for protein variability (15) limiting the ability to relate results to protein function. On the other hand, proteome profiling schemes using data-dependent acquisition (DDA; commonly referred to as shotgun mass spectrometry) are semi-stochastic and biased toward detecting the most abundant proteins in complex samples, all of which leads to well-documented effects of undersampling (16–19). Consequently, an average shotgun mass spectrometry (MS) experiment fails to detect 84% of peptides present in a given sample (16). Furthermore, as many as 30–60% of sampled peptides may differ between technical replicates because of limited reproducibility (20) precluding reliable proteome profiling. Besides other factors related to experimental design, these technical limitations are largely responsible for the fact that the pain research community has accumulated inconsistent and nonoverlapping lists of regulated proteins during pain (9–14).
A potential solution might be offered by emerging alternative mass spectrometry-based methods such as data-independent acquisition mass spectrometry (DIA-MS) (21–23). In contrast to random selection and fragmentation of only the most abundant precursor ions in shotgun approaches, one major hallmark of DIA-MS lies in the concurrent selection and fragmentation of nearly all precursor ions irrespective of their abundance (as long as their abundance is above the detection limit of the mass spectrometer). Precursor ion selection is solely dependent on user-defined mass windows spanning several hundreds of mass-to-charge ratios (m/z). Consequently, one implementation of this technique is called Sequential Windowed Acquisition of all THeoretical mass spectra (SWATH) (21–23). However, recorded fragment ion spectra are of chimeric nature as they may derive from several coeluting precursor ions, which does not permit the application of classical database searches to decipher protein identities. Instead, a prevalent approach for DIA-MS data analysis is the use of reference spectral libraries containing information on retention time and fragmentation spectra for all included peptides (21–23). Thus, DIA-MS is capable to quantify thousands of proteins present in a reference spectral library across multiple samples in a highly reproducible and experimenter-independent fashion (21–23). Ergo, this technique is especially suited to globally interrogate pathology-related proteome alterations (23–25); however, DIA-MS has so far not been applied to decipher the protein signature of chronic pain.
Here, we present a multi-layer workflow consisting of mouse behavior, biochemistry and DIA-MS for standardized detection and quantification of the changes in the DRG proteome in two mouse models of chronic pain. We reproducibly quantified the same set of 2526 proteins in all biological and technical replicates, and revealed 141 differentially regulated candidates, of which dozens have not been characterized in the somatosensory system. The biological significance of our data is reflected by our functional validation experiments suggesting the involvement of selected candidates in nociceptive signaling and pain. In summary, our extensive data set establishes a valuable framework to qualitatively extend our understanding of chronic pain and somatosensation.
EXPERIMENTAL PROCEDURES
Animals
Adult C57Bl/6J mice (6–8 weeks old at the time of surgery or injection) were in-house bred and kept in a temperature- and humidity-controlled environment under a 12 h (h) light/dark cycle with food and water provided ad libitum. All animal experiments were approved and carried out in compliance with institutional guidelines and guidelines of the Landesamt für Verbraucherschutz und Lebensmittelsicherheit of Lower Saxony, Germany (33.9-42502-04-14/1638 and 3392-42502-04-13/1077).
Pain Paradigms and Measurement of Mechanical Hypersensitivity
Chronic Inflammatory Pain
Ten microliters of Complete Freund's Adjuvant (CFA, 1.0 mg/ml, Sigma-Aldrich, Carlsbad, CA) were injected into the plantar surface of one hind paw. Control mice were injected with 10 μl of phosphate-buffered saline (PBS; Veh; Life Technologies). Mechanical hypersensitivity was measured with a dynamic aesthesiometer (Ugo Basile) 24 h after injection according to standard procedures (26) and the manufacturer′s manual (Ugo Basile, Monvalle VA, Italy). In brief, mice were acclimatized for 2 h and paw withdrawal latencies were determined upon application of a graded force (0–10 g in 40 s) to the plantar surface of each hind paw. Paw withdrawal latencies were averaged from at least four readings per paw. In order to minimize stress (and associated potential alterations of the proteome) because of behavioral testing (27, 28), we intended to separate behavioral testing as much as possible from tissue collection in time. Therefore, we did not test mechanical hypersensitivity in CFA- and Veh-injected mice destined for tissue collection for subsequent experiments (calcium imaging, biochemistry and mass spectrometry). Instead, we used a separate cohort of mice to assess mechanical hypersensitivity. This separate mouse cohort was CFA-injected in parallel (control: Veh injection) and subsequently tested for mechanical hypersensitivity. Separate mouse cohorts consisted of at least 7 animals per DIA-MS replicate. All of them exhibited pronounced mechanical hypersensitivity as described previously (26, 29) and shown in Fig. 1B. Moreover, all CFA-injected animals in this study showed pronounced paw edema (26), which was absent in Veh-injected mice.
Fig. 1.
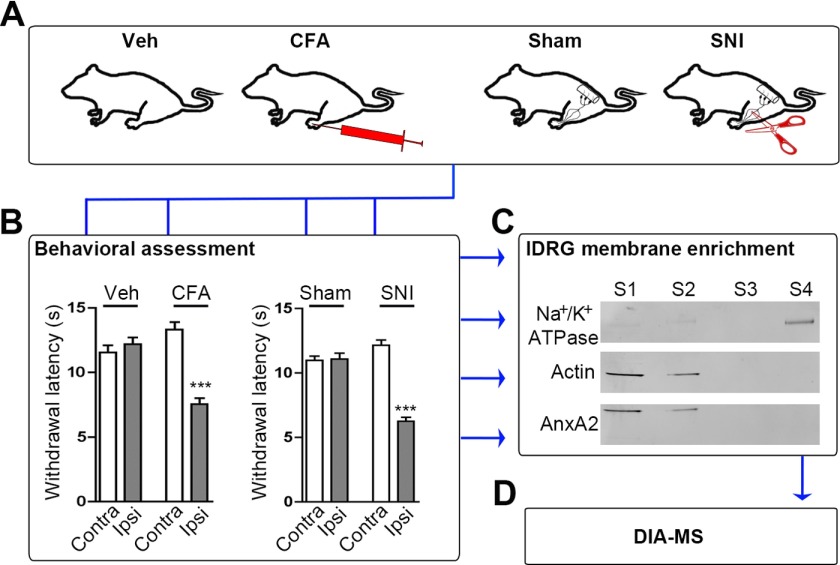
Experimental workflow. A, Representation of the four behavioral groups used in this study: (from left to right) Vehicle (Veh) and CFA injection (CFA) into one hind paw, exposed but not transected (Sham) and exposed as well as transected branches of the sciatic nerve (SNI). B, Representative assessment of hypersensitivity to mechanical stimulation reveals pronounced mechanical allodynia upon induction of inflammatory and neuropathic pain. Mechanical allodynia is manifested by decreased paw withdrawal latencies of the treated side (ipsilateral; Ipsi) in CFA and SNI animals (24 h after CFA-injection and 28 days after SNI). Paw withdrawal latencies were unaltered in Veh-injected (control for CFA-injection) and Sham animals (control for SNI) and contralateral paws (Contra) of all groups (***, p < 0.001 by one-way ANOVA followed by Bonferroni′s multiple comparison test; n = 21–24 animals/condition from three independent cohorts). C, Representative Western blot demonstrates enrichment for the membrane marker Na+/K+-ATPase and decrease of the cytosolic markers Actin and AnnexinA2 (AnxA2) in membrane-enriched fractions (S4). As expected, opposing results were obtained in cytosolic-enriched fractions (S1 and S2; n = 3 independent preparations). D, Membrane-enriched samples (S4) were analyzed by DIA-MS.
Neuropathic Pain
The spared nerve injury (SNI) paradigm was induced according to standard protocols (30). Mice were anesthetized with isoflurane (4% induction, 1.8% maintenance) and the sciatic nerve and its branches, the common peroneal, tibial and sural nerves, were exposed. Common peroneal and tibial nerves were ligated with a 6.0 silk suture (Braun) and transected distal to the ligation, removing 2–3 mm of each nerve. Care was taken to avoid damage to the sural nerve. Sham surgery was performed in a similar manner without ligating and transecting the nerves. Mechanical sensitivity was assessed 2 days before and 7, 14, 21, and 26 days after surgery, as described above, but applying the force to the lateral side of the plantar surface of both hind paws. Only mice that exhibited evident mechanical hypersensitivity (manifested as reduced withdrawal latencies) at all tested time points (90% of all mice receiving SNI) were included in the study and used for tissue collection 28 days after surgery. As explained above, tissue collection (at day 28 post surgery) was separated in time from behavioral testing (at day 26 post surgery) for the sake of stress-reduction.
Injection of Rotenone and PACMA 31
Ten microliters of CFA was unilaterally injected into the plantar surface of a hindpaw of mice. After 24 h, mice were acclimatized for 1–2 h, and injected into the plantar surface of the same hind paw with either 5 μl of Veh (0.63% DMSO/PBS) or a suspension of 5 μl of 2.5 mm Rotenone (Sigma-Aldrich) in 0.63% DMSO/PBS. Mice were acclimatized for another 15 min (min) and mechanical sensitivity determined by a plantar aesthesiometer (Ugo Basile) as described above up to 1 h after injection of Rotenone/Veh. Maximally 200 μl of PACMA 31 (Tocris, Bristol, UK; 20 mg/kg body weight) or Veh (6.9% DMSO in sterile PBS) were injected intraperitoneally. Mice were then acclimatized for 2 h and mechanical sensitivity determined by a plantar aesthesiometer (Ugo Basile) as described above. Withdrawal latencies were measured up to 6 h after injection of PACMA 31/Veh. At least two independent mouse cohorts were tested for each agent. In line with other studies (31–33), neither Rotenone nor PACMA 31 affected general health or motor coordination.
Membrane-enriched Fractions of DRG Lysates
Chemicals were obtained from Roth, unless stated otherwise. 24 h after induction of inflammatory pain and 28 days after induction of neuropathic pain mice were euthanized with CO2, decapitated, ipsilateral lumbar DRG were dissected, immediately snap-frozen in liquid nitrogen and stored at −80 °C until further use. To prepare membrane-enriched fractions we used a protocol modified from Lu and colleagues (34). Lumbar DRG of 10–13 mice per condition were homogenized in high salt buffer (2 m NaCl, 10 mm Tris/HCl, pH 7.5, complete protease inhibitor mixture, Roche), and centrifuged at 600 × g for 10 min, 4 °C. Supernatants were centrifuged at 16,000 × g for 15 min, 4 °C, and the pellets resuspended twice in 100 μl carbonate buffer (0.1 mm Na2CO3, pH 11.3), followed by centrifugation at 16,000 × g for 15 min, 4 °C. Pellets were then resuspended in 100 μl of 1% RapiGest (Waters, Eschborn, Germany) in ammonium bicarbonate buffer (50 mm (NH4)HCO3, pH 8.5), incubated overnight, 4 °C, and centrifuged at 16,000 × g for 15 min, 4 °C. For acetone-precipitation, 500 μl of ice-cold acetone were added to the supernatants and samples were incubated for 2 h, −20 °C, and centrifuged at 14,000 × g for 30 min. Pellets were washed with ice-cold 80% EtOH and centrifuged at 14,000 × g for 30 min. Supernatants were taken off and pellets air-dried. For the in-solution digest, 10 μl of 1% RapiGest in ammonium bicarbonate buffer were added followed by 10 μl of 50 mm dithiothreitol (Calbiochem) in 25 mm ammonium bicarbonate buffer, and samples were incubated at 850 rpm for 1 h, 56 °C on a thermoshaker. Next, 10 μl of 100 mm iodoacetamide (Sigma-Aldrich, Munich, Germany) in 25 mm ammonium bicarbonate buffer were added and samples were incubated at 850 rpm for 1 h, 37 °C. Seventy microliters of trypsin (Promega, Mannheim, Germany) solution in 25 mm ammonium bicarbonate buffer were added (1:20 enzyme to protein ratio) and samples were incubated overnight, 37 °C. Twenty microliters of 5% trifluoroacetic acid (Roth, Mannheim, Germany) were added, samples were incubated for 2 h, 37 °C, and centrifuged at 16000 × g for 30 min, room temperature. Supernatants were lyophilized and stored at −20 °C until further analysis.
Sample Preparation, DIA-MS and Data Analysis
All solvents were HPLC-grade from Sigma-Aldrich and all chemicals were obtained from Sigma-Aldrich if not stated otherwise. All steps of DIA-MS and its analysis were performed by Biognosys AG (Zuerich, Switzerland) essentially as described (23); in short:
Sample Preparation
The 12 samples (3 biological replicates per experimental group) were prepared for mass spectrometric analysis. The peptides were re-suspended in 8 m urea/0.1 M ammonium bicarbonate at room temperature (RT). Peptides were cleaned up for mass spectrometric analysis using a C18 Ultra MicroSpin Column (The Nest Group, Southborough, MA) according to the manufacturer's instructions. The peptide concentration was determined using a SpectrostarNano spectrometer and was in the expected range of 1 μg/μl for all samples. Peptides were dried down to complete dryness and resuspended in liquid chromatography (LC) solvent A (40 μl of 1% acetonitrile in water with 0.1% formic acid).
Following, samples were analyzed for spectral library generation and Hyper Reaction Monitoring (HRM)-MS profiling. The LC gradient was 5–35% solvent B in 120 min followed by 35–100% B (3% water in acetonitrile with 0.1% formic acid) in 2 min and 100% B for 8 min (total gradient length was 240 min). For shotgun analyses on the Q Exactive a modified TOP12 method from Olsen et al. was used (35). Prior to mass spectrometric acquisition, the HRM Calibration Kit (Biognosys) was spiked into the samples at the manufacturer's instructions (36).
Spectral Library Generation
To generate the spectral library, we used one sample for each of the four experimental groups and pooled the results. Thus, 4 h gradient LC-MS/MS acquisitions were performed in shotgun proteomics mode using a Q Exactive mass spectrometer. The LC-MS/MS mass spectrometric data were analyzed using Maxquant 1.4 software (37) with the following settings: Trypsin/P as protease; maximal two missed cleavages; carbamidomethyl (C) as fixed modification; oxidation (M) and acetylation (at protein N-term) as variable modifications; false discovery rate on peptide and protein level set to 1%; mass tolerance for precursor ions 20 ppm and fragment ions (MS/MS) 20 ppm. A mouse Uniprot FASTA database (state 05.06.2014) was used for the search engine. The final spectral library contained 16,971 peptide assays including charge states and modifications (of which 15,850 were proteotypic) corresponding to 3067 proteins (of which 2530 were defined by at least one proteotypic peptide). On average, there were 5.5 peptides per protein.
DIA-MS and Data Analysis
The 12 samples of the four conditions and three replicates were acquired in HRM mode in a replicate blocking manner to avoid performance bias in the acquisition (supplemental Fig. S3). The HRM method consisted of loops of 1 survey scan and 19 SWATHs (i.e. isolation windows) spanning 400 to 1200 m/z. The LC gradient was 2 h. This resulted in on average 8 data points per peak, sufficient for quantitative analysis. The 12 HRM measurements were analyzed using the Spectronaut™ 5.0 software from Biognosys AG (Zuerich, Switzerland). The spectral library generated in this project was used for the targeted analysis approach. The false discovery rate (FDR) on peptide level was set to 1%. The resulting data were filtered using row based extraction, for a peptide identified in one measurement; all quantitative values were extracted in all measurements resulting in full profiles. The resulting data tables were normalized using local normalization controlling for loading differences and spray bias (supplemental Fig. S4).
Quality control was performed by a control sample (human blood plasma) measured before and after profiling MS measurements in shotgun proteomics. The analysis of the two control measurements revealed that the performance stayed constant over the time of acquisition (data not shown). To identify significantly differential expressed proteins, a state comparison analysis was performed comparing either CFA versus Veh or SNI versus Sham using a pairwise t test. The resulting p values were corrected for multiple testing using the Benjamini-Hochberg and q-value method to control the overall false discovery rates (38). State comparison analysis and the analysis of the detection of fold change were performed using MSstats 2.3.2 (39) essentially as described (23).
All data have been deposited to Peptide Atlas (http://www.peptideatlas.org, No. PASS00826, the username is PASS00826 and the password is ZE5945at).
The metadata analysis to compare all 141 regulated candidates with proteins and genes previously shown to be associated with pain states in humans and rodents was done manually by extensive literature search using Pubmed (NCBI) and the web-resource Pain Networks (painnetworks.org) (40). For the purpose of determining the amount of novel proteins identified in our approach (i.e. proteins that have not been investigated in the context of pain before), we disregarded all genes/proteins that (1) were shown to be involved in nociception and pain elsewhere, (2) are included in Pain Networks as pain-related genes or (3) are considered members of the mitochondrial electron transport chain (ETC) as inhibitors of mitochondrial ETC have been demonstrated to be involved in nociception and pain (32, 41). Gene ontology (GO) analysis was performed by uploading mouse gene IDs into the search GO data engine (42). To determine enriched GO terms we used the DAVID functional annotation tool to assign genes with their affiliate terms and to order them by enrichment (43, 44). Only significantly enriched GO terms (p < 0.05 after Benjamini-Hochberg correction) are reported. To visualize molecular interaction networks and predict direct and indirect associations among all significantly regulated proteins in each pain model we uploaded accession numbers (Uniprot; mus musculus) of differentially regulated proteins into STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) (45). STRING settings: Confidence view; confidence level 0.4; clustering algorithm MCL set to 2. Of note, we only display “connected proteins,” i.e. associations among significantly regulated proteins (Supplemental Table 3). As disconnected nodes are hidden for the purpose of visualizing molecular interaction networks, not all significantly regulated proteins are included in the presented protein networks (Fig. 4).
Fig. 4.
Association networks and functional clustering among significantly altered proteins (q-value < 0. 05; Fig. 2B, C) in each pain model. Links between (A) 40 out of 64 regulated proteins during inflammatory pain and (B) 56 out of 77 regulated proteins during neuropathic pain were discovered. Of note, only connected nodes, i.e. significantly regulated proteins (supplemental Table S3), which exhibit reported associations among each other are displayed in the depicted networks (number of observed interactions versus expected interactions in a random protein set: inflammatory pain: 111 versus 22; neuropathic pain: 162 versus 27; assessed by STRING). Association networks were generated by STRING and depict the “confidence view,” i.e. thicker lines represent stronger interactions (for details please see Experimental Procedures). Application of the MCL algorithm (STRING) highlights protein clusters by connecting protein nodes with continuous lines. Colors of protein nodes denote protein clusters involved in mitochondrial function, cytoskeleton and myelin maintenance, protein biosynthesis and maturation and others, respectively. Intercluster edges are displayed by dashed-lines.
Western Blots
Membrane-enriched fractions of lumbar DRG of 12 mice per condition were obtained as described above. Instead of resuspending pellets in RapiGest (Waters), pellets were resuspended in 30 μl PBS/lithium dodecyl sulfate sample buffer (Life Technologies) and incubated for 1.5 h, 4 °C, while shaking. 1× NuPAGE sample reducing agent (Life Technologies) was added and equal amounts of protein separated by 1D NuPAGE. Protein gels of membrane-enriched fractions were blotted on PVDF membranes using the iBlot System (Life Technologies) and blocked for 30 min in 5% milk/PBS. Western blots were incubated for 1 h, RT, with primary antibodies to β-Actin (mouse, 1:500, Sigma-Aldrich #A1978), to Annexin A2 (rabbit; 1:100; Abcam #ab41803, Cambridge, UK), to sodium-potassium ATPase alpha2 (rabbit, 1:100, Millipore, Darmstadt, Germany #07–674) and Serca1 (rabbit, 1:100, Alomone, Jerusalem, Israel #ACP011) in 1% milk/PBS, and additionally overnight, 4 °C, at a three times lower concentration. Blots were incubated for 2 h at RT with secondary antibodies conjugated to Alexa Fluor 680 or 790 (1:8000, Life Technologies, #A10038/A10043) in 1% milk/PBS, washed extensively and imaged using infrared imaging (Odyssey, LI-COR, Bad Homburg, Germany). For our orthogonal validation experiments (Fig. 3) Western blots were cut to be able to separately probe for high (Serca1) and low molecular weight proteins (β-Actin) (please see supplemental Fig. S7 for full blots). Western blots were repeated two times. Only for presentation purposes size, brightness, and contrast levels of paired blots (i.e. experiments done in parallel and probed with the same antibody) were adjusted using Photoshop CS6 (Adobe).
Fig. 3.
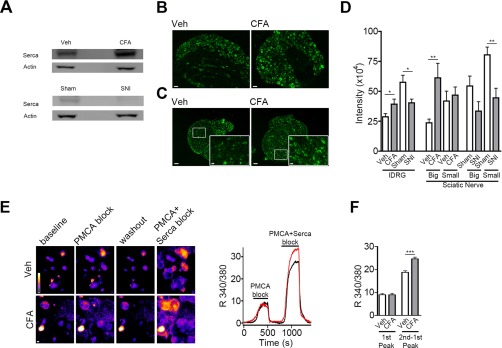
Correlation of proteomics results with altered Serca expression and function. A, Representative Western blot of tissue pooled from 12 animals per condition illustrates the regulation of Serca during inflammatory (Veh versus CFA: 70% increase) and neuropathic pain (Sham versus SNI: 65% decrease). The cytosolic marker β-Actin was unchanged and served as loading control (n = 2 independent preparations). B–D, Representative immunohistochemistry for Serca in somata of lDRG neurons (B) and sciatic nerves (C) of Veh- and CFA-injected animals. Insets in (C) show a magnified image of indicated regions. Scale bars: 50 μm and 10 μm (for insets). D, Quantification of Serca abundance (represented by staining intensity in arbitrary units) in somata of lDRG neurons as well as in big and small fascicles of sciatic nerves of all four experimental groups (*, p < 0.05 and **, p < 0.01 by two-tailed unpaired t test for pairwise comparison of CFA with Veh and SNI with Sham; n = 20–40 images analyzed from n = 3–4 independent experiments each). Serca was increased in big fascicles (corresponding to the tibial and common peroneal nerve) of the sciatic nerve upon CFA-injection whereas its down-regulation was more pronounced in small, noninjured (i.e. spared) fascicles in the SNI model (please refer to Experimental Procedures for a description of this model). The percentage of Serca-positive neurons was not changed in any condition (data not shown). Of note, in experiments (A–D) Veh-injected animals served as controls for CFA-injection and Sham animals served as controls for SNI. A comparison of Veh and Sham animals was not performed because of different treatment (injection versus surgery) and age (Sham animals are 4 weeks older). E, Representative ratiometric calcium-imaging in cultured lDRG neurons from Veh- and CFA-injected animals. Left, pseudocolored calcium oscillations upon functional blocking of PMCA followed by simultaneous block of PMCA and Serca (see Experimental Procedures for assay description). Scale bar: 10 μm. Right, representative traces showing changes in intracellular calcium (expressed by the 340/380 fluorescence ratio in arbitrary intensity units) in cultured lDRG neurons of a Veh- (black line) and CFA- (red line) injected animal. F, Quantification of the relative calcium increase (expressed by the 340/380 fluorescence ratio in arbitrary intensity units) upon PMCA block (measured at the maximum of the first peak) or Serca inhibition (measured after subtracting the first from the second peak); ***, p < 0.001 by two-tailed unpaired t test; n > 450 neurons analyzed from n = 4 independent experiments.
Immunohistochemistry
Mice were euthanized with CO2, decapitated and lumbar DRG and sciatic nerves carefully dissected, collected in 4% PFA/PBS, and fixed overnight, 4 °C. After overnight cryoprotection in 30% sucrose/PBS, tissue was frozen in optimal cutting temperature medium (Sakura, Staufen, Germany), 10 μm-thick step serial sections were cut with a cryostat (Leica, Wetzlar, Germany), mounted on SuperFrost Plus slides (Thermo Scientific, Waltham, MA), and stored at −80 °C. We prepared cross sections of DRG and longitudinal as well as cross sections of sciatic nerves. Frozen slides were thawed at RT for 60 min, blocked for 30 min in 0.4% Triton X-100/PBS containing 5% donkey serum (Dianova, Hamburg, Germany), and incubated overnight, 4 °C, with primary antibodies to Serca1 (rabbit, 1:100, Alomone #ACP011) or Peripherin (chicken, 1:100, Abcam #39374) in 0.1% Triton X-100/PBS containing 1% donkey serum. After extensive washing in PBS, sections were incubated for 2 h, RT, with secondary antibodies conjugated to Alexa Fluor 488, 546 and 647 (1:250, Thermo Fisher) or Alexa Fluor 488-conjugated-Tubulin III antibody (Tuj1, directed against neuron-specific class III β-Tubulin, 1:500, HISS Diagnostics #A488–435L) in 0.1% Triton X-100/PBS containing 1% donkey serum. Some sections were then incubated with Fluoromyelin™ Red Fluorescent Myelin Stain for 20 min at RT (1:300, Thermo Fisher #F34652) and all were mounted in SlowFade Gold reagent (Thermo Fisher). At least three independent stainings (from at least three independent animal cohorts) per experimental paradigm were performed.
Image Acquisition and Analysis of Immunohistochemistry
Stained cryosections were imaged at a Zeiss Axio Observer Z1 inverted epifluorescence microscope. Images for all experimental groups were taken using identical acquisition parameters. For each experimental group, at least six pictures were randomly analyzed. Raw images were analyzed using NIH ImageJ (46). Signal was considered positive when higher than mean background fluorescence plus three times the standard deviation, measured from at least 10 random unstained tissue spots. Mean background fluorescence intensity was subtracted from the mean surface fluorescence intensity. Signal particle size threshold was set at 15 square pixels to account for nonspecific staining. Values were averaged among experiments. As the SNI model involves transection/injury of two major branches of the sciatic nerve (30) we separately analyzed big fascicles (corresponding to branches of the tibial and common peroneal nerves) and small fascicles (corresponding to the sural nerve and smaller branches of the tibial and common peroneal nerves, e.g. articular branch) in cross sections of sciatic nerves (47). To be consistent, we applied the same procedure to cross sections obtained from Sham animals as well as CFA- and Veh-injected animals. The analysis of fluorescence intensity was only performed on cross sections. Only for presentation purposes brightness and contrast of paired images were adjusted using Photoshop CS6 (Adobe).
Ratiometric Calcium Imaging and Serca Inhibition
CFA- and Veh-injected littermate mice were sacrificed 24 h post injection by CO2 inhalation followed by decapitation. Ipsilateral lumbar DRG were dissected and plated as previously described (48) with the following modifications: growth medium (Hams F12/DMEM 1:1 ratio with l-glutamine; Life Technologies) was supplemented with 10% horse serum (Life Technologies) and 100 ng/ml NGF, 50 ng/ml GDNF, 50 ng/ml BDNF, 50 ng/ml NT-3, and 50 ng/ml NT-4 (all R&D Systems, Minneapolis, MN). Neurons were plated on coverslips coated with Poly-d-lysine (1.0 mg/ml, Millipore), followed by Laminin (20 μg/ml, Life Technologies).
Experiments were performed 24 h after plating. Assay buffer consisted of regular Tyrodes's (in mm): NaCl 140, KCl 4, CaCl2 2, Glucose 10, MgCl2 2, HEPES 10, with an osmolality of 295–300 mOsm and pH 7.4. Neuronal cultures were incubated with 2.5 μm Fura2-AM (Life Technologies) and 0.04% Pluronic F-127 (Life Technologies) in Tyrode's, for 30 min, 37 °C, washed and mounted on a recording chamber constantly superfused by a gravity-driven system. To block PMCA activity, HEPES was replaced by 10 mm Trizma and pH adjusted to 8.8 (49). Block of Serca activity was obtained by further addition of 150 nm Thapsigargin (TG; Sigma-Aldrich). At this concentration TG has been shown to be specific for Serca while not affecting voltage-gated calcium currents in mouse DRG neurons (50). The fluorophore was excited alternately at 340 nm and 380 nm, and emission acquired at 510 nm, using the Zeiss Axio Observer Z1. Intracellular calcium concentration is expressed as the 340/380 ratio. All experimental groups to be compared were processed in parallel using the same culture preparation. At least two coverslips from three independent culture preparations were analyzed as described (48). The mean intensity value was assessed with ImageJ following background subtraction and particle analysis as described above.
Experimental Design and Statistical Rationale
All DIA-MS experiments were performed in triplicates, i.e. 3 biological replicates were analyzed. Experimental conditions were CFA-treated animals and animals with spared nerve injury (SNI). Control conditions involved Veh-treated animals (control for CFA-treated animals) and animals receiving Sham surgery (control for SNI). DIA-MS experiments involved pooled tissue from 10–13 animals/replicate/condition. To identify significantly differential expressed proteins, a state comparison analysis was performed comparing either CFA versus Veh or SNI versus Sham using a pairwise t test. The resulting p values were corrected for multiple testing using the Benjamini-Hochberg and q-value method to control the overall false discovery rates. State comparison analysis and the analysis of the detection of fold change were performed using MSstats 2.3.2 (39) essentially as described (23).
For statistical analysis of all other experiments, two-tailed unpaired t test was used for single comparisons and one-way ANOVA was used for multiple comparisons. Statistical analysis was performed with GraphPad Prism if not stated otherwise. No statistical methods were used to predetermine sample sizes for any experiment, but our sample sizes are similar to those generally employed in the field. Compiled data are expressed as mean ± sem; in Fig. Legends n indicates the sample number, N indicates the number of independent experiments, P denotes the significance (*, p < 0.05, **, p < 0.01, ***, p < 0.001) and refers to the respective control in each experimental group if not noted otherwise, and NS indicates “not significant.”
RESULTS
Overview of Our Integrated Workflow: From Murine Chronic Pain Models to DIA-MS Proteome Profiling
In order to study proteins relevant for somatosensory signaling by DIA-MS, we generated a customized spectral library (22, 51, 52) from DRG isolated under pain and control conditions, respectively. To this end, we employed two well-established mouse models of chronic pain with different etiology (26, 53) (Fig. 1A): inflammatory pain induced by injection of Complete Freund's Adjuvant (CFA; Vehicle (Veh) injections served as controls) and neuropathic pain using the spared nerve injury model, in which the tibial and common peroneal branch of the sciatic nerve are transected (SNI; control groups received Sham surgery; please refer to Experimental Procedures for further details). Both models mimic clinically relevant features of chronic pain patients including mechanical hypersensitivity (54, 55). Therefore, we assessed hypersensitivity to innocuous punctuate mechanical stimulation (allodynia) by the plantar aesthesiometer test (26) in treated (ipsilateral) hindpaws compared with the contralateral side as well as to Veh and Sham surgery controls, respectively (Fig. 1B and supplemental Fig. S1). Only mouse cohorts showing prominent mechanical allodynia were included in our study (Fig. 1B; please refer to Experimental Procedures for further details). Animals were sacrificed for dissection of ipsilateral lumbar DRG (lDRG) 24 h after induction of inflammatory pain and 28 days after induction of neuropathic pain (and respective controls). The time points were chosen based on previous publications reporting robust mechanical allodynia (26, 29, 30) and our assessment of mechanical hypersensitivity (Fig. 1B and supplemental Fig. S1).
The function and interplay of proteins expressed in sensory neurons and other cell types of DRG (e.g. peripheral glial cells and various immune cells) shape nociceptive signaling (56, 57). Especially transmembrane and membrane-associated proteins (hereafter summarized as “membrane proteins”) are critically involved in stimulus detection, neuronal activity and consequently, the sensation of pain (4–6, 58–61). Hence, they represent attractive targets for pharmacotherapy (62, 63). Yet, the experimental analysis of membrane proteins is hampered by their low abundance and pronounced hydrophobicity (64). As a corollary to these difficulties, the membrane proteome is traditionally understudied despite its paramount importance for diverse (patho-) physiological processes including pain (4–6, 58–61, 63). For these reasons we aimed at assessing the potential of DIA-MS to profile a membrane-enriched proteome of DRG. To this end we performed biochemical sub-fractionation and obtained membrane-enriched lDRG lysates of each experimental and control condition (Fig. 1C). These were subsequently used for DIA-MS-based profiling (Fig. 1D).
A DRG-specific Spectral Library
We prepared three independent biological replicates per pain paradigm (CFA versus Veh and SNI versus Sham) each consisting of membrane-enriched lDRG lysates from 10–13 mice. One replicate per condition was used to generate the spectral library on a Q Exactive mass spectrometer. Data were analyzed using Maxquant 1.4 software (37), setting the false discovery rate on peptide and protein level to 1%. A mouse Uniprot FASTA database (state 05.06.2014) was used for the search engine. The final spectral library contained 16,971 peptide assays (of which 15,850 were proteotypic) corresponding to 3067 proteins (of which 2530 were defined by at least one proteotypic peptide) (supplemental Table S1). To examine the representation of cellular compartments and functions in our spectral library, we systematically analyzed gene ontology (GO) terms with the Database for Annotation, Visualization and Integrated Discovery (DAVID; david.ncifcrf.gov). Spectral library components broadly covered diverse cellular compartments and molecular functions (supplemental Fig. S2). As expected, this analysis confirmed the enrichment of membrane-associated proteins (including membrane-passing, i.e. transmembrane proteins) in our spectral library (supplemental Fig. S2). Among the latter were several ion channels with known roles in somatosensation and pain, most of which have not been identified in a previous proteomic study of DRG membranes (65), e.g. Piezo2 and TRPA1 channels (supplemental Table S2). Further comparison with published proteomes of neuromas, sciatic nerve, DRG, and spinal cord (harboring presynaptic terminals of DRG neurons) demonstrates, that our spectral library constitutes the largest compendium of DRG proteins reported to date (9, 10, 12–14, 65). The generated resource is publically available in PeptideAtlas (66) and constitutes an optimized resort for unbiased, consistent and highly accurate protein quantification in future DIA-MS experiments of any mouse sample of interest.
Differential Regulation of Proteins During Inflammatory and Neuropathic Pain
We next used the generated spectral library for the targeted search of DIA-MS runs conducted with three biological replicates of each of the four conditions: induction of inflammatory pain by CFA versus Veh-injection and induction of neuropathic pain by SNI versus Sham (Fig. 1). The 12 samples were acquired in Hyper Reaction Monitoring (HRM) mode in a replicate blocking manner to avoid performance bias in the acquisition (23), which resulted in high quality raw data acquisition (supplemental Fig. S3). The spectral library was applied setting the false discovery rate (FDR) of detection at 1% on a peptide level. This analysis enabled the profiling of more than 14,500 peptides representing the same set of 2526 proteins across all replicates and samples (2581 proteins in all CFA and Veh replicates; 2600 proteins in all SNI and Sham replicates; supplemental Table S3), which reflects the high reproducibility of the presented workflow (Fig. 2A). The resulting data tables were normalized using local normalization in order to account for loading differences and spray bias (67) (supplemental Fig. S4). The analysis of the coefficient of variation of the quantified signals (which determines the variance from the biological pool, sample preparation and instrument performance) was in the expected range below 20% (68) indicating conserved and reliable quantification performance (supplemental Fig. S5).
Fig. 2.
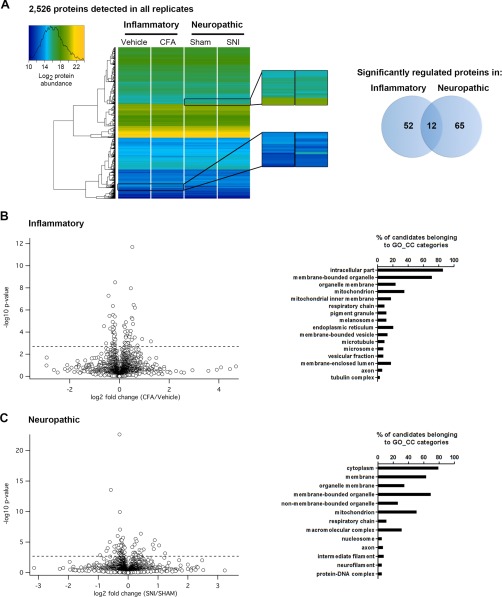
Results of DIA-MS proteomics of inflammatory and neuropathic pain in mice. A, Left, Heatmap upon unsupervised hierarchical clustering analysis of abundance levels of the same set of 2526 proteins quantified in all four experimental groups. Magnified regions show examples of changes in abundance (represented as changes in the color code) of specific proteins. Right, Venn diagram shows the number of specifically and commonly regulated proteins (intersection) detected in inflammatory and neuropathic pain models, respectively. B, Left, Volcano plot for all proteins profiled in inflammatory (CFA/Veh) pain. Dotted horizontal line represents the significance level, i.e. q-value = 0.05. Hence, all proteins above this dotted line exhibit a q-value < 0.05 and are considered to be significantly regulated. Right, Assigned Gene Ontology (GO) Cellular Component (CC) categories and the relative abundance of significantly regulated proteins during inflammatory pain. C, Left, Volcano plot for all proteins profiled in neuropathic (SNI/Sham) pain. Significance level q-value < 0.05 as in B. Right, Assigned Gene Ontology (GO) Cellular Component (CC) categories and the relative abundance of significantly regulated proteins during neuropathic pain.
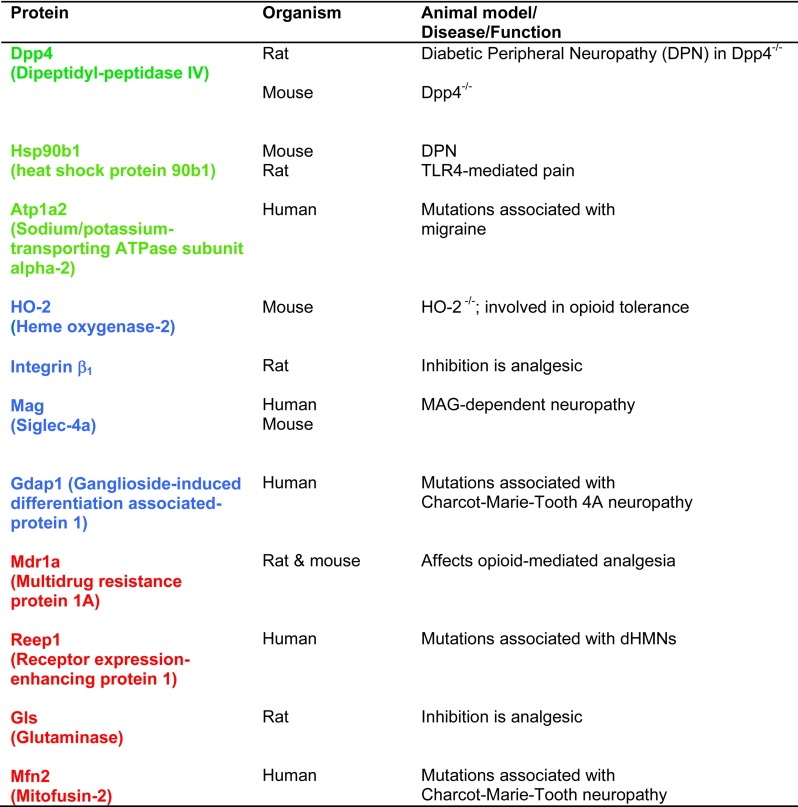
Upon statistical comparison of these data (corrected for multiple testing using the q-value (38); for complete data set please see supplemental Table S3), we observed little overall proteome changes as visualized in the heatmap and volcano plots (Fig. 2). Only around 3% of all quantified proteins showed significant up- and downregulations in each pain paradigm (64 and 77 proteins in inflammatory and neuropathic pain, respectively; Fig. 2A) and 12 proteins were commonly altered in both pain models, suggesting a high degree of pain model-specificity (Fig. 2A; supplemental Table S4). Gene Ontology (GO) analysis revealed that regulated proteins were expressed in different cellular compartments (Fig. 2B, 2C) and covered distinct as well as overlapping molecular functions (supplemental Fig. S6; supplemental Table S4). Intriguingly, several significantly regulated candidates of our data set have previously been associated with diverse pathologies involving the somatosensory system (including painful ones) in humans and rodents (examples are given in Table I), demonstrating the ability of our workflow to uncover meaningful candidates. For instance, Dipeptidylpeptidase 4 (Dpp4) was significantly up-regulated in both pain models (Table I and supplemental Tables S3 and S4). Dpp4 is a multifunctional cell surface glycoprotein with peptidase activity controlling the activity of numerous peptides and proteins (69). Interestingly, Dpp4−/− mice show enhanced nociceptive pain (70), whereas in vivo inhibition of Dpp4 produces analgesia in chronic diabetic neuropathy in rats (71). Similarly, Heat shock protein 90b1 (HSP90b1), commonly regulated in both pain models of our study (Table I), is implicated in diabetic neuropathy in rodents (72, 73). Other candidates in our study are involved in painful human pathologies such as familial hemiplegic migraine type II caused by mutations of the ATPase ATP1A2 (ATP1A2 is commonly regulated in our study, Table I) (74) or Charcot-Marie-Tooth 4A neuropathy linked to mutations of Ganglioside-induced differentiation associated-protein 1 (Gdap1) (Gdap1 is regulated during inflammatory pain in our study, Table I) (75).
Table I. Examples of significantly regulated proteins in our study, which have been reported to be implicated in rodent pain models or vertebrate pathologies involving the somatosensory system. Of note, this table contains only examples of such proteins for which functional significance was demonstrated in vivo. In our study, blue-labeled proteins were significantly regulated during inflammatory pain, red-labeled proteins during neuropathic pain and green-labeled proteins in both pain models. dHMNs: distal hereditary motor neuropathies.
Conversely, a detailed literature search revealed that roughly 50% of significantly altered proteins identified by our approach have not been investigated in the context of nociception or pain before (please see Experimental Procedures for details on literature search). Hence, they represent potentially promising candidates for future studies on their role in painful conditions.
Correlation of proteomic changes with altered protein function and expression
Next, we intended to validate our results by orthogonal methods. To this end, we focused on sarco/endoplasmic reticulum calcium-ATPase (Serca) as it was differentially regulated in the two pain models we used for this study, i.e. Serca was up-regulated in CFA and downregulated in SNI animals (supplemental Table S4). Using independent mouse cohorts, we induced inflammatory and neuropathic pain and compared the abundance of Serca in DRG by Western blotting (Fig. 3A) as well as by immunohistochemistry (Fig. 3B–3D). Of note, animals were sacrificed for tissue dissection at the same time points as for the DIA-MS screen, i.e. 24 h after CFA-injection (controls: Veh-injection) and 28 days after SNI (controls: Sham surgery). In total accordance with our MS data, we recapitulated the differential and pain model-specific regulation of Serca not only in DRG (Fig. 3A, 3B, 3D; please see supplemental Fig. S7 for full blots), but also in distinct fascicles of the sciatic nerve (Fig. 3C, 3D; supplemental Fig. S8).
We next sought to investigate whether these changes in Serca abundance are indicative of altered Serca function. To this end, we cultured DRG neurons from CFA- and Veh-injected animals and performed ratiometric calcium imaging (Fig. 3E, 3F). Serca is a calcium-ATPase in the endoplasmic reticulum (ER) membrane and as such controls cytoplasmic calcium levels by active calcium transport from the cytosol to the ER. Hence, monitoring cytoplasmic calcium levels upon application of the Serca inhibitor thapsigargin can indirectly assess Serca activity. In order to do so without interference by the plasma membrane calcium ATPase (PMCA), we used an established protocol consisting of initial block of PMCA followed by coapplication of thapsigargin at a concentration shown to be specific for Serca (please see Experimental Procedures for details) (49, 50). Interestingly, application of both blockers revealed increased cytosolic calcium levels in DRG neurons from CFA-injected animals suggesting higher Serca activity compared with Veh-injected controls (Fig. 3E, 3F). In contrast, intracellular calcium levels were comparable when only PMCA was inhibited indicating no difference of PMCA activity among conditions (Fig. 3E, 3F). Importantly, and in line with our functional data, a retrospective in silico interrogation of our proteomics results found comparable PMCA levels in CFA or SNI animals and their respective controls (q-values for PMCA: CFA: 0.357; SNI: 0.645; supplemental Table S3). These data highlight the capability of DIA-MS profiling to predict functional alterations of distinct proteins within the same cellular pathway.
Major Cellular Signaling Networks are Altered in Both Chronic Pain Models
The high statistical power and reproducibility of our DIA-MS screen provided us with the opportunity to meaningfully interrogate the data set in respect to changes in protein networks. To this end, we tested for evidence of associations among all significantly regulated proteins in both pain models (q-value < 0.05, supplemental Table S3) using the web-based application STRING (45). Indeed, we found associations among 40 out of 64 proteins regulated during inflammatory pain and 56 out of 77 regulated proteins during neuropathic pain, respectively (Fig. 4A, 4B). Despite the certain degree of ambiguity intrinsic to functional categorization based on previously annotated information, this analysis underlined mutual interconnections among regulated proteins and associated functional processes (Fig. 4). For example, in the neuropathic pain model we observed the down-regulation of structural proteins involved in cytoskeleton organization and myelin maintenance (supplemental Table S3 and Fig. 4B; pink-colored protein nodes). Alterations in these proteins have been reported in another neuropathic pain model and are consistent with nerve injury and repair processes (11).
Moreover, our network analysis indicated differential alterations of multiple proteins implicated in mitochondrial function (Fig. 4A, 4B: represented by orange-brown-colored protein nodes) and protein biosynthesis/maturation (Fig. 4A, 4B: represented by green-colored protein nodes). Both cellular pathways are crucially interconnected determinants of apoptosis, energy metabolism as well as oxidative and ER stress (76–79). Furthermore, the notion is evolving that mitochondrial abnormality (33, 80–82) and ER stress (77, 79, 83) contribute to several forms of neuropathic pain. Mitochondrial dysfunction in painful conditions appears to be multifactorial as it may arise from changes in the activity of the electron transport chain (ETC) (33, 80–82), a concomitant increase of reactive oxygen species leading to oxidative stress (84) or mitochondrial calcium handling (85). Similarly, mitochondrial fission represents a crucial step for cellular homeostasis and has recently been implicated in painful neuropathies brought on by cancer chemotherapy and anti-HIV medication (41). ER stress represents another significant driver of pain in diverse neuropathic models such as diabetic neuropathy (77, 79, 83). ER stress may be caused by a disequilibrium in ER homeostasis regarding protein demand, synthesis and folding. Its pronounced impact on pain phenotypes has recently been demonstrated in healthy animals where initiation of ER stress signaling cascade generated lasting pain, which could be reversed by ER stress blockers (77).
In addition to their impact on neuropathic pain, we observed a previously unreported regulation of proteins involved in mitochondrial function (orange-brown-colored nodes) and protein biosynthesis/maturation (green-colored nodes) upon CFA-induced inflammatory pain (Fig. 4A). Prompted by this finding, we set out to test whether manipulation of selected candidates in vivo may affect mouse pain behaviors upon CFA-injection. For this purpose, we looked for agents neither affecting general health nor motor function in naïve animals and at the same time being suitable to generally manipulate mitochondria and protein maturation in the ER in vivo (31, 32). For the first, we used the mitochondrial complex I inhibitor Rotenone (32, 33) as many of the regulated proteins are members of the respiratory electron chain complex I (e.g. Ndufs4, Ndufv2; Fig 4A). For the latter, we used PACMA 31, a nonselective inhibitor of protein disulfide isomerases (PDI) (31), of which three isoforms (PDIA1, PDIA3, and PDIA6) were strongly up-regulated during inflammatory pain (Fig. 4A and supplemental Table S3). Indeed unilateral injection of Rotenone into the ipsilateral paw of CFA-treated mice resulted in diminished hypersensitivity to innocuous mechanical stimulation delivered with a plantar aesthesiometer compared with Veh-injected controls (Fig. 5A). This finding considerably extends previous studies that reported the preventive action of modulating mitochondrial function by application of inhibitors before induction and chronification of inflammatory pain (32, 41). Likewise, we observed significantly less CFA-mediated mechanical allodynia upon intraperitoneal injection of PACMA 31 compared with Veh-injected mice (Fig. 5B).
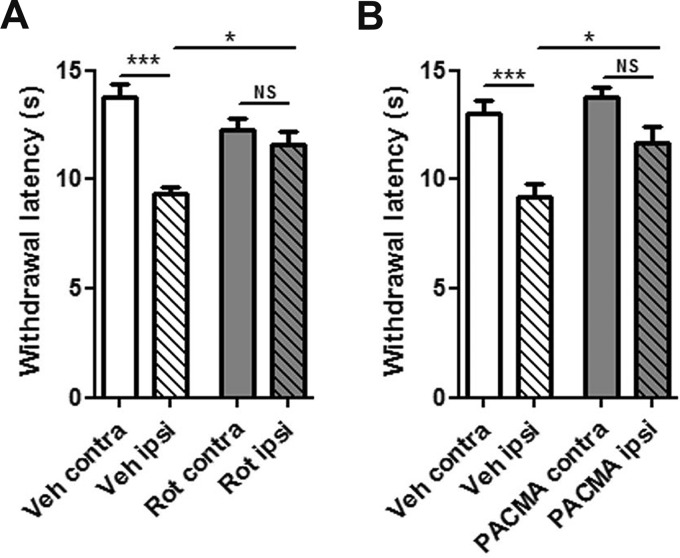
Fig. 5.
Mitochondrial and PDI activity play a role for chronic inflammatory pain in mice. A, Rotenone-mediated analgesia of established chronic inflammatory pain. Illustration of paw withdrawal latencies of CFA-injected animals, which received ipsilaterally (ipsi) either Veh or Rotenone (Rot) 24 h after CFA-application (***, p < 0.001 and *, p < 0.05 by one-way ANOVA followed by Bonferroni′s multiple comparison test; 17 animals per condition from 3 independent mouse cohorts). B, Inhibition of PDI by PACMA 31 attenuated established chronic inflammatory pain. Illustration of paw withdrawal latencies of CFA-injected animals, which received either Veh or PACMA 31 (PACMA) via intraperitoneal injection 24 h after CFA-application (***, p < 0.001 and *, p < 0.05 by one-way ANOVA followed by Bonferroni′s multiple comparison test; 10 animals per condition from 2 independent mouse cohorts). Importantly, withdrawal responses of contralateral (i.e. untreated) paws were unaffected arguing against unspecific motor effects upon intraperitoneal injection of PACMA 31.
Even though we used similar concentrations of Rotenone and PACMA 31 as described previously (31–33), we cannot unambiguously determine whether the observed analgesic action of Rotenone and PACMA 31 is solely mediated by inhibition of complex I and PDI in somatosensory neurons, respectively. Sensory neuron-specific conditional knockout mice would be required to critically interrogate potential off-target effects. These may derive from nonspecific action either on other proteins in sensory neurons and/or effects on nonneuronal cells in vivo (e.g. different cell types in the skin or DRG). The latter hypothesis is especially relevant to systemic injection of PACMA 31, yet neither general motor coordination nor withdrawal latencies of contralateral (noninjected) paws were affected (Fig. 5) arguing against major side-effects elicited by compound injection. These observations are in line with other studies using these compounds (31–33).
In summary, our results suggest an acute analgesic potential of mitochondrial complex I and PDI inhibitors in fully established inflammatory pain and provide a proof-of-principle that pathologically relevant proteins and protein networks can be uncovered with our profiling approach.
DISCUSSION
The unbiased characterization of protein signaling networks is key for understanding the molecular signature of health and disease. Here we present an integrated workflow combining mouse behavior and DIA-MS-based protein profiling to standardize the reproducible detection and quantification of several thousand proteins in two mouse models of chronic pain. We found substantial and pain model-specific alterations of several dozens of proteins, among which 50% have not been linked to nociception and pain before. Strikingly, many of those represent novel and functionally uncharacterized candidates. Comparison with known biology paired with our validation experiments strongly suggests that indeed proteins relevant for pain and somatosensory signaling were identified. Taken together, we present (1) the so far largest compendium of the DRG proteome and (2) a list of proteins specifically regulated during chronic pain states. These constitute valuable resources enabling further investigations of pathologies of the somatosensory system.
We focused our analysis on DRG isolated from male mice subjected to two different chronic pain states. As DRG harbor primary sensory neurons, which are the interface of the nervous system with external and internal environments, they are crucially implicated in the beginning of somatosensory pathways including nociception (3, 4, 86). This, together with their accessibility, renders DRG key cellular targets for analgesic therapies (2, 4, 87).
Because of the cellular complexity of DRG, we cannot assign the detected changes in protein abundance to specific cell types, at least at the level of mass spectrometry. However, we partially solved this limitation employing MS-independent and cell type-specific techniques for validation of our results. For example, we show that the regulation of Serca occurs in sensory neurons as revealed by calcium-imaging in primary DRG cultures as well as by immunohistochemistry on DRG and sciatic nerves.
The current profiling study recapitulates known biology regarding the identification of proteins previously linked to pain. In addition, we provide a proof-of-principle with orthogonal validation experiments that our DIA-MS workflow is capable to uncover functionally meaningful changes. Using various methods, we correlated our DIA-MS data with altered Serca function and expression in DRG neurons. Serca is crucially involved in neuronal calcium and ER homeostasis by governing ER calcium uptake and sequestration. Consequently, the activity of Serca requires precise control to enable adequate neuronal responses to different conditions. In fact, Serca-activity in DRG neurons is modulated by noxious substances (88), protein kinase C (89) and upon painful neuropathy (90). In turn, injection of Serca-inhibitors has recently been shown to elicit ER stress-mediated nociceptive responses in vivo (77). Our profiling data suggest diminished abundance of Serca in neuropathic mice that is in complete agreement with previous reports (77, 90). Strikingly, we observed the opposite regulation, i.e. augmented Serca levels and function specifically during inflammatory pain. Although the underlying mechanisms require further investigations, which are beyond the scope of this study, these results propose the existence of signaling mechanisms converging on Serca in a pain model-specific manner.
Integration of DIA-MS profiling together with protein network analysis offers another level of understanding the molecular signature of diseases. Using the web-based application STRING, we visualized associations among significantly regulated proteins (Fig. 4), which revealed that components of major signaling pathways are altered during chronic pain, e.g. mitochondrial function as well as protein synthesis and maturation. Although this notion is not new and many transcriptional profiling techniques provide a systems-wide view on cellular networks (7, 8, 29, 91), proteome profiling has so far made only minor contributions (92–94). Undersampling and limited reproducibility inherent to commonly used shotgun proteomics approaches (16–19) represent major obstacles for proteome-based systems biology. This is in stark contrast to the emerging importance of “network medicine” as a powerful strategy to correct functional misalignments of complex cellular processes (2, 76, 95). Consequently, it might provide promising tools for achieving analgesia as exemplified by studies linking mitochondrial dysfunction (33, 80–82) and ER stress to neuropathic pain (77, 79, 83). However, the underlying molecular changes and altered proteins are only scarcely understood preventing the design of targeted interventions with therapeutic potential. Here, we report highly reproducible DIA-proteome profiling of 2526 proteins, which granted unique insights into significant alterations of candidate proteins belonging to distinct cellular signaling networks. These data are not only in agreement with previous studies implicating mitochondrial (33, 80–82) and ER (77, 79, 83) abnormalities in neuropathic pain, but importantly (i) identify affected members of these protein networks and (ii) provide evidence for their differential involvement also in inflammatory pain. Indeed, we could demonstrate attenuation of fully established chronic inflammatory pain by application of mitochondrial complex I or protein disulfide isomerase inhibitors in vivo. Taken together, we expect the current study to spark further research to decipher the exact mechanisms how the here detected protein changes contribute to pain pathology.
Although we affirm selected candidates by examples of orthogonal validation as well as correlation with known pain biology, we did not detect several previously reported regulations of proteins during chronic pain. For example, we did not observe a regulation of TRPA1 in our study whereas other reports demonstrated its up-regulation in DRG of CFA-injected mice (96). Several technical considerations might underlie these discrepancies. On one hand, we pooled ipsilateral lumbar DRG from different levels in order to obtain sufficient tissue per DIA-MS replicate. Although this is common practice in most -omics studies, it is noteworthy that even neurons within one DRG are highly heterogeneous, as affected and nonaffected neurons are intermingled upon induction of chronic pain in mice (97). Furthermore, the relative proportions of affected and nonaffected neurons in one DRG critically depend on the injection site of agents in respect to the projection sites of individual nerve branches innervating the hind paw (97). Indeed, differences between affected versus nonaffected nerve branches might potentially explain our results showing disparate regulation of Serca in distinct sciatic nerve fascicles. On the other hand, our study differs from previous proteomics studies in respect to biochemical sample preparation. With the goal of deciphering the (patho-) physiologically relevant (4–6, 58–61, 63) but traditionally understudied membrane proteome (64) we used membrane-enriched lysates of DRG. Consequently, the depletion of soluble proteins in our samples might explain the marginal overlap of our results with published proteomics studies using whole-cell lysates (9–14).
In addition, species, tissue and pain model differences have to be considered when interpreting and comparing data among studies (97). For instance, Gemes and colleagues reported elevated activity of plasma membrane calcium-ATPase (PMCA) in the neuropathic pain model of spinal nerve ligation in rats (98). In contrast, we employed the CFA-induced inflammatory pain model and the neuropathic pain model of spared nerve injury in mice and could not detect any alterations of PMCA, neither by DIA-MS profiling nor on the functional level using calcium-imaging in sensory neurons harvested from CFA-injected mice.
The here presented spectral library is enriched with fingerprints for transmembrane and membrane-associated proteins found in diverse cellular compartments like the plasma membrane, membranes of the endoplasmic reticulum as well as mitochondrial membranes. For example, TRPA1 and TRPV1 channels are molecular noxious stimulus detectors and critically implicated in nociceptive signaling and pain (4, 99–101). Therefore, they are considered as potential targets for pharmacological interventions (98, 102–104). Moreover, Piezo2 channels confer mechanosensitivity to sensory neurons (105, 106) as well as Merkel cells of the skin (107, 108), and articular cartilage cells (109) highlighting the applicability of our spectral library to different tissues beyond DRG. However, despite successful enrichment of membrane proteins, our spectral library (and therefore our DIA-MS results) is not complete. Although we provide fingerprints for the sodium channels Nav1.8 and Nav1.9, the spectral library lacks information on the sodium channel Nav1.7 known to be involved in several pain states (5). The reasons can be manifold, ranging from relative low expression levels to insufficient solubilization or localization in detergent-resistant microdomains - all factors known to render membrane protein analysis challenging (64). Nonetheless, and compared with published proteomics studies on pain (9–14) or DRG membranes (65), our spectral library constitutes a unique resource for many hundreds of DRG membrane proteins, which has the potential to enormously facilitate research on these proteins in any tissue. Moreover, the nature of DIA-MS profiling allows any experimenter to continuously extend this library, and use it for in-silico analysis of generated data sets without requiring new experiments. Therefore, the depth of protein profiling will steadily improve once experiments in DIA-mode are commonly used by the research community.
Another powerful indication of the value of our data set is the high rate of reproducibility we achieved in our DIA-MS experiments. We reproducibly profiled the same set of 2,526 proteins in all 12 DIA-MS experiments performed, i.e. in three replicates of each of the four conditions. Taking into account that each replicate consisted of independent rounds of surgeries/injections, tissue extraction and biochemical preparation, all being subject to high variability, the observed reproducibility is remarkable and qualitatively superior to shotgun proteomics studies (16–19).
Thus, the (1) enrichment of membrane proteins in our spectral library paired with the (2) high rate of reproducibility, and (3) the possibility for future in-silico examination of generated data sets implement a standardized toolbox for profiling these proteins in any tissue of interest.
Collectively, our findings support the notion that the here generated DRG protein compendium represents a rich and valuable resource for the research community. It provides a framework for comprehensive characterization of particular molecular signatures underlying diverse pathologies, well beyond the here described application in mouse models of chronic pain.
Supplementary Material
Acknowledgments
We thank Sergej Zeiter (MPI of Experimental Medicine, Goettingen, Germany) and Monika Raabe (MPI of Biophysical Chemistry, Goettingen, Germany) for excellent technical assistance, Henning Urlaub and Christof Lenz (UMG and MPI of Biophysical Chemistry, Goettingen, Germany) for initial discussions on technical aspects of SWATH-MS, Michael W. Sereda (UMG and MPI of Experimental Medicine, Goettingen, Germany) for kindly providing access to a dynamic plantar aesthesiometer in the pilot-phase of the project, Luis Pardo, Walter Stuehmer, Klaus-Armin Nave and Nils Brose (all MPI of Experimental Medicine, Goettingen, Germany) for generously providing antibodies. We are grateful to Roland Bruderer (Biognosys AG, Zurich, Switzerland) for fruitful discussions and critically reading the manuscript.
Footnotes
Author contributions: D.G. and M.S. designed research; T.R., J.S., and L.A. performed research; T.R., J.S., L.A., D.G., and M.S. analyzed data; D.G. and M.S. wrote the paper.
* This work was supported by the Emmy Noether-Program of the Deutsche Forschungsgemeinschaft (SCHM 2533/2–1 to MS), a PhD fellowship of the International Max Planck Research School Neurosciences (to JS) and the Max Planck Society. MS received a research award and travel support by the German Pain Society (DGSS), both of which were sponsored by Astellas Pharma GmbH (Germany). DGV received research support from Biognosys AG (Zurich, Switzerland). None of this research support was used for the current study.
 This article contains supplemental material.
This article contains supplemental material.
All authors declare no conflict of interest.
1 The abbreviations used are:
- DRG
- dorsal root ganglia
- LC
- liquid chromatography
- GO
- gene ontology
- DTT
- dithiothreitol
- HEPES
- 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid
- lDRG
- lumbar DRG
- MF
- molecular function
- min
- minutes
- MPI
- Max Planck Institute
- mRNA
- messenger RNA
- MS/MS
- tandem mass spectrometry
- Ndufs4
- NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
- Ndufv2
- NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
- NGF
- Nerve growth factor
- NS
- not significant
- NT-3/4
- Neurotrophin-3/4
- PACMA31
- propynoic acid carbamoyl methyl amide 31
- PBS
- phosphate-buffered saline
- PDI
- protein disulfide isomerase
- PFA
- Paraformaldehyde
- PMCA
- plasma membrane calcium ATPase
- PVDF
- Polyvinylidene fluoride
- RNA
- Ribonucleic acid
- RNA-Seq
- RNA-sequencing
- Rot
- Rotenone
- rpm
- revolutions per min
- RT
- room temperature
- Serca
- sarco/endoplasmic reticulum calcium-ATPase
- SNI
- spared nerve injury
- STRING
- Search Tool for the Retrieval of Interacting Genes/Proteins
- SWATH
- sequential window acquisition of all theoretical fragment ion spectra
- TG
- thapsigargin
- TRPA1
- Transient receptor potential cation channel, subfamily A, member 1
- TRPV1
- transient receptor potential cation channel, subfamily V, member 1
- Veh
- vehicle.
REFERENCES
- 1. Costigan M., Scholz J., and Woolf C. J. (2009) Neuropathic pain: a maladaptive response of the nervous system to damage. Annu. Rev. Neurosci. 32, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borsook D., Hargreaves R., Bountra C., and Porreca F. (2014) Lost but making progress–Where will new analgesic drugs come from? Sci. Transl. Med. 6, 249sr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basbaum A. I., Bautista D. M., Scherrer G., and Julius D. (2009) Cellular and molecular mechanisms of pain. Cell 139, 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patapoutian A., Tate S., and Woolf C. J. (2009) Transient receptor potential channels: targeting pain at the source. Nature Rev. Drug discovery 8, 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raouf R., Quick K., and Wood J. N. (2010) Pain as a channelopathy. J. Clin. Invest. 120, 3745–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bennett D. L., and Woods C. G. (2014) Painful and painless channelopathies. Lancet Neurol. 13, 587–599 [DOI] [PubMed] [Google Scholar]
- 7. Manteniotis S., Lehmann R., Flegel C., Vogel F., Hofreuter A., Schreiner B. S., Altmuller J., Becker C., Schobel N., Hatt H., and Gisselmann G. (2013) Comprehensive RNA-Seq expression analysis of sensory ganglia with a focus on ion channels and GPCRs in Trigeminal ganglia. PloS One 8, e79523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Usoskin D., Furlan A., Islam S., Abdo H., Lonnerberg P., Lou D., Hjerling-Leffler J., Haeggstrom J., Kharchenko O., Kharchenko P. V., Linnarsson S., and Ernfors P. (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 [DOI] [PubMed] [Google Scholar]
- 9. Zou W., Zhan X., Li M., Song Z., Liu C., Peng F., and Guo Q. (2012) Identification of differentially expressed proteins in the spinal cord of neuropathic pain models with PKCgamma silence by proteomic analysis. Brain Res. 1440, 34–46 [DOI] [PubMed] [Google Scholar]
- 10. Niederberger E., and Geisslinger G. (2008) Proteomics in neuropathic pain research. Anesthesiology 108, 314–323 [DOI] [PubMed] [Google Scholar]
- 11. Vacca V., Marinelli S., Pieroni L., Urbani A., Luvisetto S., and Pavone F. (2014) Higher pain perception and lack of recovery from neuropathic pain in females: a behavioural, immunohistochemical, and proteomic investigation on sex-related differences in mice. Pain 155, 388–402 [DOI] [PubMed] [Google Scholar]
- 12. Huang H. L., Cendan C. M., Roza C., Okuse K., Cramer R., Timms J. F., and Wood J. N. (2008) Proteomic profiling of neuromas reveals alterations in protein composition and local protein synthesis in hyper-excitable nerves. Mol. Pain 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melemedjian O. K., Yassine H. N., Shy A., and Price T. J. (2013) Proteomic and functional annotation analysis of injured peripheral nerves reveals ApoE as a protein upregulated by injury that is modulated by metformin treatment. Mol. Pain 9, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michaelevski I., Medzihradszky K. F., Lynn A., Burlingame A. L., and Fainzilber M. (2010) Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol. Cell Proteomics 9, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 16. Michalski A., Cox J., and Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 17. Gingras A. C., and Raught B. (2012) Beyond hairballs: The use of quantitative mass spectrometry data to understand protein-protein interactions. FEBS Lett. 586, 2723–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Domon B., and Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 19. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., and White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., and Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillet L. C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sajic T., Liu Y., and Aebersold R. (2014) Using data-independent, high resolution mass spectrometry in protein biomarker research: Perspectives and clinical applications. Proteomics Clin. Appl. 9, 307–321 [DOI] [PubMed] [Google Scholar]
- 23. Bruderer R., Bernhardt O. M., Gandhi T., Miladinovic S. M., Cheng L. Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., and Reiter L. (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell Proteomics 14, 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shao S., Guo T., and Aebersold R. (2015) Mass spectrometry-based proteomic quest for diabetes biomarkers. Biochim. Biophys. Acta 1854, 519–527 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y., Chen J., Sethi A., Li Q. K., Chen L., Collins B., Gillet L. C., Wollscheid B., Zhang H., and Aebersold R. (2014) Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol. Cell Proteomics 13, 1753–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minett M. S., Quick K., and Wood J. N. (2011) Behavioral Measures of Pain Thresholds. Current Protocols in Mouse Biology, John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- 27. Butler R. K., and Finn D. P. (2009) Stress-induced analgesia. Prog. Neurobiol. 88, 184–202 [DOI] [PubMed] [Google Scholar]
- 28. Sorge R. E., Martin L. J., Isbester K. A., Sotocinal S. G., Rosen S., Tuttle A. H., Wieskopf J. S., Acland E. L., Dokova A., Kadoura B., Leger P., Mapplebeck J. C., McPhail M., Delaney A., Wigerblad G., Schumann A. P., Quinn T., Frasnelli J., Svensson C. I., Sternberg W. F., and Mogil J. S. (2014) Olfactory exposure to males, including men, causes stress and related analgesia in rodents. Nat. Methods 11, 629–632 [DOI] [PubMed] [Google Scholar]
- 29. Simonetti M., Hagenston A. M., Vardeh D., Freitag H. E., Mauceri D., Lu J., Satagopam V. P., Schneider R., Costigan M., Bading H., and Kuner R. (2013) Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron 77, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Decosterd I., and Woolf C. J. (2000) Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain 87, 149–158 [DOI] [PubMed] [Google Scholar]
- 31. Xu S., Butkevich A. N., Yamada R., Zhou Y., Debnath B., Duncan R., Zandi E., Petasis N. A., and Neamati N. (2012) Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc. Natl. Acad. Sci. U.S.A. 109, 16348–16353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joseph E. K., and Levine J. D. (2006) Mitochondrial electron transport in models of neuropathic and inflammatory pain. Pain 121, 105–114 [DOI] [PubMed] [Google Scholar]
- 33. Joseph E. K., and Levine J. D. (2009) Comparison of oxaliplatin- and cisplatin-induced painful peripheral neuropathy in the rat. J. Pain 10, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu A., Wisniewski J. R., and Mann M. (2009) Comparative proteomic profiling of membrane proteins in rat cerebellum, spinal cord, and sciatic nerve. J. Proteome Res. 8, 2418–2425 [DOI] [PubMed] [Google Scholar]
- 35. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 36. Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M. J., and Rinner O. (2012) Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 38. Storey J. D. (2002) A direct approach to false discovery rates. J. Roy. Stat. Soc. B 64, 479–498 [Google Scholar]
- 39. Choi M., Chang C. Y., Clough T., Broudy D., Killeen T., MacLean B., and Vitek O. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 [DOI] [PubMed] [Google Scholar]
- 40. Perkins J. R., Lees J., Antunes-Martins A., Diboun I., McMahon S. B., Bennett D. L., and Orengo C. (2013) PainNetworks: a web-based resource for the visualisation of pain-related genes in the context of their network associations. Pain 154, 2586.et12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrari L. F., Chum A., Bogen O., Reichling D. B., and Levine J. D. (2011) Role of Drp1, a key mitochondrial fission protein, in neuropathic pain. J. Neurosci. 31, 11404–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., and Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genetics 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 44. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bauder A. R., and Ferguson T. A. (2012) Reproducible mouse sciatic nerve crush and subsequent assessment of regeneration by whole mount muscle analysis. J. Vis. Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Avenali L., Narayanan P., Rouwette T., Cervellini I., Sereda M., Gomez-Varela D., and Schmidt M. (2014) Annexin A2 regulates TRPA1-dependent nociception. J. Neurosci. 34, 14506–14516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Duman J. G., Chen L., and Hille B. (2008) Calcium transport mechanisms of PC12 cells. J. Gen. Physiol. 131, 307–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shmigol A., Kostyuk P., and Verkhratsky A. (1995) Dual action of thapsigargin on calcium mobilization in sensory neurons: inhibition of Ca2+ uptake by caffeine-sensitive pools and blockade of plasmalemmal Ca2+ channels. Neuroscience 65, 1109–1118 [DOI] [PubMed] [Google Scholar]
- 51. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., and Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 52. Collins B. C., Gillet L. C., Rosenberger G., Rost H. L., Vichalkovski A., Gstaiger M., and Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14–3-3 system. Nat. Methods 10, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 53. Mogil J. S., Davis K. D., and Derbyshire S. W. (2010) The necessity of animal models in pain research. Pain 151, 12–17 [DOI] [PubMed] [Google Scholar]
- 54. Koltzenburg M. (1998) Painful neuropathies. Current Opinion Neurol. 11, 515–521 [DOI] [PubMed] [Google Scholar]
- 55. Woolf C. J., and Mannion R. J. (1999) Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet 353, 1959–1964 [DOI] [PubMed] [Google Scholar]
- 56. Delmas P. (2008) SnapShot: ion channels and pain. Cell 134, 366–366 e361 [DOI] [PubMed] [Google Scholar]
- 57. Ren K., and Dubner R. (2010) Interactions between the immune and nervous systems in pain. Nat. Med. 16, 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hucho T., and Levine J. D. (2007) Signaling pathways in sensitization: toward a nociceptor cell biology. Neuron 55, 365–376 [DOI] [PubMed] [Google Scholar]
- 59. Bourinet E., Altier C., Hildebrand M. E., Trang T., Salter M. W., and Zamponi G. W. (2014) Calcium-permeable ion channels in pain signaling. Physiol. Rev. 94, 81–140 [DOI] [PubMed] [Google Scholar]
- 60. Garcia-Caballero A., Gadotti V. M., Stemkowski P., Weiss N., Souza I. A., Hodgkinson V., Bladen C., Chen L., Hamid J., Pizzoccaro A., Deage M., Francois A., Bourinet E., and Zamponi G. W. (2014) The deubiquitinating enzyme USP5 modulates neuropathic and inflammatory pain by enhancing Cav3.2 channel activity. Neuron 83, 1144–1158 [DOI] [PubMed] [Google Scholar]
- 61. Rouwette T., Avenali L., Sondermann J., Narayanan P., Gomez-Varela D., and Schmidt M. (2015) Modulation of nociceptive ion channels and receptors via protein-protein interactions: implications for pain relief. Channels 9, 175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gilron I., Jensen T. S., and Dickenson A. H. (2013) Combination pharmacotherapy for management of chronic pain: from bench to bedside. Lancet Neurol. 12, 1084–1095 [DOI] [PubMed] [Google Scholar]
- 63. Gold M. S. (2006) Ion channels: recent advances and clinical applications. Proceedings of the 11th World Congress on Pain, 10 [Google Scholar]
- 64. Tan S., Tan H. T., and Chung M. C. (2008) Membrane proteins and membrane proteomics. Proteomics 8, 3924–3932 [DOI] [PubMed] [Google Scholar]
- 65. Xiong X., Huang S., Zhang H., Li J., Shen J., Xiong J., Lin Y., Jiang L., Wang X., and Liang S. (2009) Enrichment and proteomic analysis of plasma membrane from rat dorsal root ganglions. Proteome Sci. 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deutsch E. W., Lam H., and Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. Embo. Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Callister S. J., Barry R. C., Adkins J. N., Johnson E. T., Qian W. J., Webb-Robertson B. J., Smith R. D., and Lipton M. S. (2006) Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 5, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rost H. L., Rosenberger G., Navarro P., Gillet L., Miladinovic S. M., Schubert O. T., Wolski W., Collins B. C., Malmstrom J., Malmstrom L., and Aebersold R. (2014) OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 [DOI] [PubMed] [Google Scholar]
- 69. Busek P., Stremenova J., Krepela E., and Sedo A. (2008) Modulation of substance P signaling by dipeptidyl peptidase-IV enzymatic activity in human glioma cell lines. Physiol. Res. 57, 443–449 [DOI] [PubMed] [Google Scholar]
- 70. Guieu R., Fenouillet E., Devaux C., Fajloun Z., Carrega L., Sabatier J. M., Sauze N., and Marguet D. (2006) CD26 modulates nociception in mice via its dipeptidyl-peptidase IV activity. Behav. Brain Res. 166, 230–235 [DOI] [PubMed] [Google Scholar]
- 71. Bianchi R., Cervellini I., Porretta-Serapiglia C., Oggioni N., Burkey B., Ghezzi P., Cavaletti G., and Lauria G. (2012) Beneficial effects of PKF275–055, a novel, selective, orally bioavailable, long-acting dipeptidyl peptidase IV inhibitor in streptozotocin-induced diabetic peripheral neuropathy. J. Pharmacol. Exp. Ther 340, 64–72 [DOI] [PubMed] [Google Scholar]
- 72. Urban M. J., Pan P., Farmer K. L., Zhao H., Blagg B. S., and Dobrowsky R. T. (2012) Modulating molecular chaperones improves sensory fiber recovery and mitochondrial function in diabetic peripheral neuropathy. Exp. Neurol. 235, 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Urban M. J., Li C., Yu C., Lu Y., Krise J. M., McIntosh M. P., Rajewski R. A., Blagg B. S., and Dobrowsky R. T. (2010) Inhibiting heat-shock protein 90 reverses sensory hypoalgesia in diabetic mice. ASN Neuro. 2, e00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. De Fusco M., Marconi R., Silvestri L., Atorino L., Rampoldi L., Morgante L., Ballabio A., Aridon P., and Casari G. (2003) Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat. Genetics 33, 192–196 [DOI] [PubMed] [Google Scholar]
- 75. Baxter R. V., Ben Othmane K., Rochelle J. M., Stajich J. E., Hulette C., Dew-Knight S., Hentati F., Ben Hamida M., Bel S., Stenger J. E., Gilbert J. R., Pericak-Vance M. A., and Vance J. M. (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat. Gen. 30, 21–22 [DOI] [PubMed] [Google Scholar]
- 76. Schapira A. H. (2012) Mitochondrial diseases. Lancet 379, 1825–1834 [DOI] [PubMed] [Google Scholar]
- 77. Inceoglu B., Bettaieb A., Trindade da Silva C. A., Lee K. S., Haj F. G., and Hammock B. D. (2015) Endoplasmic reticulum stress in the peripheral nervous system is a significant driver of neuropathic pain. Proc. Natl. Acad. Sci. U.S.A. 112, 9082–9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Simmen T., Lynes E. M., Gesson K., and Thomas G. (2010) Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 1798, 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lupachyk S., Watcho P., Stavniichuk R., Shevalye H., and Obrosova I. G. (2013) Endoplasmic reticulum stress plays a key role in the pathogenesis of diabetic peripheral neuropathy. Diabetes 62, 944–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Baloh R. H. (2008) Mitochondrial dynamics and peripheral neuropathy. Neuroscientist 14, 12–18 [DOI] [PubMed] [Google Scholar]
- 81. Fernyhough P., Roy Chowdhury S. K., and Schmidt R. E. (2010) Mitochondrial stress and the pathogenesis of diabetic neuropathy. Expert Rev. Endocrinol. Metabolism 5, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Osio M., Muscia F., Zampini L., Nascimbene C., Mailland E., Cargnel A., and Mariani C. (2006) Acetyl-l-carnitine in the treatment of painful antiretroviral toxic neuropathy in human immunodeficiency virus patients: an open label study. J. Peripher. Nerv. Syst. 11, 72–76 [DOI] [PubMed] [Google Scholar]
- 83. Zhang E., Yi M. H., Shin N., Baek H., Kim S., Kim E., Kwon K., Lee S., Kim H. W., Chul Bae Y., Kim Y., Kwon O. Y., Lee W. H., and Kim D. W. (2015) Endoplasmic reticulum stress impairment in the spinal dorsal horn of a neuropathic pain model. Scientific Reports 5, 11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim H. K., Park S. K., Zhou J. L., Taglialatela G., Chung K., Coggeshall R. E., and Chung J. M. (2004) Reactive oxygen species (ROS) play an important role in a rat model of neuropathic pain. Pain 111, 116–124 [DOI] [PubMed] [Google Scholar]
- 85. Shishkin V., Potapenko E., Kostyuk E., Girnyk O., Voitenko N., and Kostyuk P. (2002) Role of mitochondria in intracellular calcium signaling in primary and secondary sensory neurones of rats. Cell Calcium 32, 121–130 [DOI] [PubMed] [Google Scholar]
- 86. Richards N., and McMahon S. B. (2013) Targeting novel peripheral mediators for the treatment of chronic pain. Br. J. Anaesthesia 111, 46–51 [DOI] [PubMed] [Google Scholar]
- 87. Krames E. S. (2014) The role of the dorsal root ganglion in the development of neuropathic pain. Pain Med. 15, 1669–1685 [DOI] [PubMed] [Google Scholar]
- 88. Fischer M. J., Soller K. J., Sauer S. K., Kalucka J., Veglia G., and Reeh P. W. (2015) Formalin evokes calcium transients from the endoplasmatic reticulum. PloS One 10, e0123762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Usachev Y. M., Marsh A. J., Johanns T. M., Lemke M. M., and Thayer S. A. (2006) Activation of protein kinase C in sensory neurons accelerates Ca2+ uptake into the endoplasmic reticulum. J. Neurosci. 26, 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Duncan C., Mueller S., Simon E., Renger J. J., Uebele V. N., Hogan Q. H., and Wu H. E. (2013) Painful nerve injury decreases sarco-endoplasmic reticulum Ca(2)(+)-ATPase activity in axotomized sensory neurons. Neuroscience 231, 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Antunes-Martins A., Perkins J. R., Lees J., Hildebrandt T., Orengo C., and Bennett D. L. H. (2013) Systems biology approaches to finding novel pain mediators. Wires Syst. Biol. Med. 5, 11–35 [DOI] [PubMed] [Google Scholar]
- 92. Antunes-Martins A., Perkins J. R., Lees J., Hildebrandt T., Orengo C., and Bennett D. L. (2013) Systems biology approaches to finding novel pain mediators. Wiley interdisciplinary reviews. Systems biology and medicine 5, 11–35 [DOI] [PubMed] [Google Scholar]
- 93. Jamieson D. G., Moss A., Kennedy M., Jones S., Nenadic G., Robertson D. L., and Sidders B. (2014) The pain interactome: connecting pain-specific protein interactions. Pain 155, 2243–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Costigan M. (2012) Pain's peptide signature. Pain 153, 509–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Barabasi A. L., Gulbahce N., and Loscalzo J. (2011) Network medicine: a network-based approach to human disease. Nat. Rev. Genetics 12, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. da Costa D. S., Meotti F. C., Andrade E. L., Leal P. C., Motta E. M., and Calixto J. B. (2010) The involvement of the transient receptor potential A1 (TRPA1) in the maintenance of mechanical and cold hyperalgesia in persistent inflammation. Pain 148, 431–437 [DOI] [PubMed] [Google Scholar]
- 97. Laedermann C. J., Pertin M., Suter M. R., and Decosterd I. (2014) Voltage-gated sodium channel expression in mouse DRG after SNI leads to re-evaluation of projections of injured fibers. Mol. Pain 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gemes G., Oyster K. D., Pan B., Wu H. E., Bangaru M. L., Tang Q., and Hogan Q. H. (2012) Painful nerve injury increases plasma membrane Ca2+-ATPase activity in axotomized sensory neurons. Mol. Pain 8, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Petrus M., Peier A. M., Bandell M., Hwang S. W., Huynh T., Olney N., Jegla T., and Patapoutian A. (2007) A role of TRPA1 in mechanical hyperalgesia is revealed by pharmacological inhibition. Mol. Pain 3, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schmidt M., Dubin A. E., Petrus M. J., Earley T. J., and Patapoutian A. (2009) Nociceptive signals induce trafficking of TRPA1 to the plasma membrane. Neuron 64, 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Story G. M., Peier A. M., Reeve A. J., Eid S. R., Mosbacher J., Hricik T. R., Earley T. J., Hergarden A. C., Andersson D. A., Hwang S. W., McIntyre P., Jegla T., Bevan S., and Patapoutian A. (2003) ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 112, 819–829 [DOI] [PubMed] [Google Scholar]
- 102. Garrison S. R., and Stucky C. L. (2011) The dynamic TRPA1 channel: a suitable pharmacological pain target? Current Pharmaceut. Biotechnol. 12, 1689–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Viana F., and Ferrer-Montiel A. (2009) TRPA1 modulators in preclinical development. Expert Opinion Therap. Patents 19, 1787–1799 [DOI] [PubMed] [Google Scholar]
- 104. Lunn C. A. (2010) Membrane proteins as drug targets. Preface. Prog. Mol. Biol. Transl. Sci. 91, xi. [DOI] [PubMed] [Google Scholar]
- 105. Coste B., Mathur J., Schmidt M., Earley T. J., Ranade S., Petrus M. J., Dubin A. E., and Patapoutian A. (2010) Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ranade S. S., Woo S. H., Dubin A. E., Moshourab R. A., Wetzel C., Petrus M., Mathur J., Begay V., Coste B., Mainquist J., Wilson A. J., Francisco A. G., Reddy K., Qiu Z., Wood J. N., Lewin G. R., and Patapoutian A. (2014) Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Woo S. H., Ranade S., Weyer A. D., Dubin A. E., Baba Y., Qiu Z., Petrus M., Miyamoto T., Reddy K., Lumpkin E. A., Stucky C. L., and Patapoutian A. (2014) Piezo2 is required for Merkel-cell mechanotransduction. Nature 509, 622–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Maksimovic S., Nakatani M., Baba Y., Nelson A. M., Marshall K. L., Wellnitz S. A., Firozi P., Woo S. H., Ranade S., Patapoutian A., and Lumpkin E. A. (2014) Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature 509, 617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lee W., Leddy H. A., Chen Y., Lee S. H., Zelenski N. A., McNulty A. L., Wu J., Beicker K. N., Coles J., Zauscher S., Grandl J., Sachs F., Guilak F., and Liedtke W. B. (2014) Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. Proc. Natl. Acad. Sci. U.S.A. 111, E5114–E5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.