Abstract
Estrogen receptor alpha (ERα)-mediated proliferation of breast cancer cells is facilitated through expression of multiple primary target genes, products of which induce a secondary response to stimulation. To differentiate between the primary and secondary target proteins of ERα signaling, we measured dynamics of protein expression induced by 17β-estradiol in MCF-7 breast cancer cells. Measurement of the global proteomic effects of estradiol by stable isotope labeling by amino acids in cell culture (SILAC) resulted in identification of 103 estrogen-regulated proteins, with only 40 of the corresponding genes having estrogen response elements. Selected reaction monitoring (SRM) assays were used to validate the differential expression of 19 proteins and measure the dynamics of their expression within 72 h after estradiol stimulation, and in the absence or presence of 4-hydroxytamoxifen, to confirm ERα-mediated signaling. Dynamics of protein expression unambiguously revealed early and delayed response proteins and well correlated with presence or absence of estrogen response elements in the corresponding genes. Finally, we quantified dynamics of protein expression in a rarely studied network of transcription factors with a negative feedback loop (ERα-EGR3-NAB2). Because NAB2 protein is a repressor of EGR3-induced transcription, siRNA-mediated silencing of NAB2 resulted in the enhanced expression of the EGR3-induced protein ITGA2. To conclude, we provided a high-quality proteomic resource to supplement genomic and transcriptomic studies of ERα signaling.
Steroid hormone receptors are ubiquitous nuclear receptors with key roles in control of reproduction, fetal development, metabolism, homeostasis, immune response and cognitive functions (1). Estrogen receptor-alpha (ERα)1 belongs to the family of transcription factors which control cell growth and differentiation and regulate the expression of proto-oncogenes. ERα action is exerted through four distinct pathways: direct ligand-dependent transcription through binding to estrogen response elements (EREs), tethered mode through its binding to other transcription factors which interact with their DNA response elements, nongenomic pathway mediated through membrane or cytoplasmic ERα and rapid signaling by protein kinases, and finally, estrogen-independent pathway through growth factor signaling and ERα phosphorylation (2). Although the nongenomic pathways are rapidly exerted in a matter of minutes, ERE-mediated genomic pathways are relatively slow, with protein expression being deployed over the course of hours. Because some ERα target genes include transcription factors and regulatory proteins, a secondary cascade of gene expression is triggered following the initial stimulation of ERα.
Given the fundamental importance of estrogen signaling and its involvement in breast cancer progression, numerous approaches have been undertaken to discover primary ERα target genes and their secondary messengers. Global profiling of estradiol-stimulated ERα-positive cells by ChIP-sequencing and DNA microarrays revealed thousands of EREs (3–6) and hundreds of estrogen-regulated mRNA transcripts (7, 8), respectively. In contrast, the impact of ERα stimulation at the proteomic level was either measured for individual proteins or simply inferred from transcriptomic data. Even though as many as 62% of human genes have EREs (9), the diversity of the estrogen-regulated proteome is yet to be elucidated by proteomic methods. Quantitative proteomic methods would also be indispensable to reveal the protein expression dynamics that cannot be deduced from genomic and transcriptomic data.
Quantitative mass spectrometry recently advanced to the level of reproducible measurements of thousands of proteins in mammalian cells (10, 11). Stable isotope labeling by amino acids in cell culture (SILAC) facilitated global profiling of ligand-induced protein expression whereas targeted proteomic approaches by selected reaction monitoring (SRM) allowed measuring accurate temporal dynamics of protein expression in the presence of receptor agonists and inhibitors (12, 13).
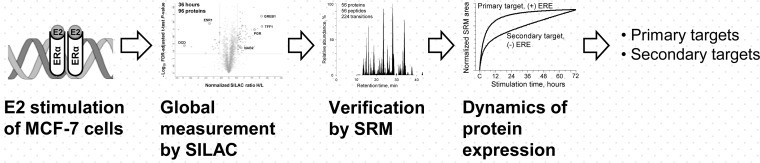
In this work, we have chosen MCF-7 breast cancer cells as a model ERα-positive cell line. We used SILAC to discover estrogen-regulated proteins and SRM to verify candidate proteins and also measure temporal dynamics of their expression. In addition, dynamics of protein expression in the presence and absence of the ERα antagonist 4-hydroxytamoxifen was used to exclude false candidates and confirm the ERα-mediated signaling. We also hypothesized that measurement of the dynamics of protein expression over the period of 72 h would reveal early-response (primary) and late-response (secondary) targets of estrogen stimulation. A brief schematic of our discovery workflow is presented in Fig. 1. Finally, we focused on dynamics of protein expression in a rarely studied network composed of a primary target of ERα signaling (transcription factor EGR3) and its secondary messengers (NAB2 and ITGA2 proteins). We hypothesized that siRNA silencing of NAB2 protein, which is also a repressor of EGR3-induced transcription, would lead to the unrestrained expression of EGR3-regulated genes. To investigate the dynamics of protein expression in the ERα-EGR3-NAB2 network, we relied on quantitative multiplex SRM assays.
Fig. 1.
Experimental workflow to identify and validate estrogen-regulated proteins and measure dynamics of protein expression.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The objective of this study was to identify by SILAC and verify by SRM ERα-regulated proteins. Taking into account our previous measurements of 76 proteins in MCF-7 cells by SRM and the median biological reproducibility of 8% (13), the required sample size to detect a fold change of 1.5 (our suggested cut-off for SILAC-derived candidates) was estimated at 3 (two-tailed t test for matched pairs with 80% power at α = 0.05). With these parameters, the minimal changes of protein expression which can be detected with 80% power in three (SILAC identification), eight (SRM verification) and four (NAB2 silencing) biological replicates were estimated at 1.46, 1.14, and 1.28 (two-tailed t test for matched pairs with α = 0.05 and biological reproducibility of 8%). G*Power (version 3.1.7, Heinrich Heine University Dusseldorf) was used for power calculations.
Cell Culture
MCF-7 breast cancer cell line was purchased from the American Type Culture Collection (Manassas, VA). MCF-7 cells were maintained as a monolayer at 37 °C in a humidified atmosphere containing 5% CO2 in RPMI 1640 medium supplemented with 10% FBS. Dextran-coated charcoal-treated FBS (Thermo Fisher Scientific, Mississauga, ON) was used to minimize effects of endogenous steroids and maximize the effect of stimulation with exogenous 10 nm 17β-estradiol (Sigma-Aldrich, Oakville, ON) (14). With an estimated doubling time of 33 ± 3 h, cell density did not exceed ∼80% confluence during the whole stimulation experiment (0 to 72 h).
Western Blotting
Cell pellets were resuspended in 1% sodium dodecyl sulfate in Tris-buffered saline and sonicated three times for 10 s. Total protein amount in the cell lysate was measured by PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific). Twenty micrograms of total protein from each sample were loaded onto an SDS-PAGE gel (4–15%) and transferred onto PVDF membranes (BioRad, Mississauga, ON). Western blot analysis was performed with trefoil factor 1 (HPA003425, Sigma-Aldrich), SPINT1 (HPA006903, Sigma-Aldrich), NAB2 (PA5–27925, Thermo Fisher Scientific), EGR3 (sc-191, Santa Cruz Biotechnology, Dallas, TX) and beta-actin (ab8227, Abcam, San Francisco, CA) antibodies.
Cell Labeling by SILAC
SILAC media were prepared using customized RPMI 1640 media devoid of l-arginine and l-lysine (Athena ES, Baltimore, MD). “Heavy” media was prepared by addition of l-13C6-arginine (87 mg/L) and l-13C6-,15N2-lysine (54 mg/L) from Cambridge Isotope Laboratories (Tewksbury, MA), whereas “light” media was prepared by addition of l-12C6-arginine and l-12C6-,14N2-lysine (Sigma-Aldrich). Both heavy and light media were supplemented with dialyzed FBS (10% final, Gibco, Thermo Fisher Scientific). Supplemental l-proline (150 mg/L, Sigma-Aldrich) was added to suppress conversion of supplemental arginine to proline during metabolic labeling. Cells were seeded into T25 flasks and grown for ∼ 9 days (six doublings) to ensure near-complete labeling. Labeled cells were detached with a nonenzymatic cell dissociation buffer (Sigma-Aldrich) and seeded in 6-well plates (Cellstar, Greiner Bio One, Monroe, NC) with 106 cells per well, in triplicates. Cells were left for 24 h in the FBS-free RPMI 1640 to induce cell cycle synchronization, washed twice with PBS and then media was changed for RPMI 1640 supplemented with 10% dextran-coated charcoal-treated FBS. Following the 24 h-cell attachment, the media of heavy cells was replaced with RPMI 1640 with 10% dextran-coated charcoal-treated FBS and 10 nm 17β-estradiol (Sigma Aldrich), whereas the media of control light cells contained RPMI 1640, 10% dextran-coated charcoal-treated FBS and 0.1% ethanol. Cells were grown for 6 or 36 h, washed with PBS and centrifuged at 290 g for 10 min. Supernatants were discarded, and cell pellets were kept at −80 °C.
Cell Lysis and Proteomic Sample Preparation
Three biological replicates of ∼3 × 106 cells (500 μg of total protein) were reconstituted in 200 μl of 0.1% RapiGest SF (sodium-3-[(2-methyl-2-undecyl-1,3-dioxolan-4-yl)-methoxyl]-1-propanesulfonate, Waters, Milford, MA) and lysed as previously described (15). All lysates were centrifuged for 20 min at 16,000 × g at 4 °C, even though no debris was observed after lysis. Total protein was measured by the BCA assay (Pierce), and equimolar amounts of heavy- and light-labeled cell lysates were mixed. Trypsin digestion of proteins was performed as previously described (13). Briefly, proteins were denatured at 65 °C, reduced with 10 mm dithiothreitol, alkylated with 20 mm iodoacetamide and digested overnight at 37 °C using sequencing grade modified trypsin (trypsin/total protein ratio 1:30; Promega, Madison, WI). RapiGest SF was cleaved with 1% trifluoroacetic acid and removed by centrifugation.
LC-MS/MS and SILAC Data Analysis
Following protein digestion, mixtures of light and heavy cell lysates were fractionated by strong-cation exchange chromatography, eighteen fractions were collected and analyzed by reverse phase liquid chromatography-tandem mass spectrometry (LTQ-Orbitrap XL,Thermo Fisher Scientific) as previously described (13, 15). For protein identification and data analysis, XCalibur software (v. 2.0.5; Thermo Fisher Scientific) was utilized to generate RAW files. Mass spectra were analyzed using MaxQuant software (version 1.1.1.25), which generated a peak list as well as SILAC- and extracted ion current-based quantitation for SILAC pairs. MaxQuant executed spectral search against a concatenated International Protein Index (IPI) human protein database (version 3.71) and a decoy database. Parameters included: trypsin enzyme specificity, SILAC double measurements of Lys8 and Arg6, 1 missed cleavage, minimum peptide length of 7 amino acids, minimum of 1 unique peptide, top 6 MS/MS peaks per 100 Da, peptide mass tolerance of 20 ppm for precursor ion and MS/MS tolerance of 0.5 Da and fixed modification of cysteines by carbamidomethylation. Variable modifications included oxidation of methionine and acetylation of the protein at N terminus. All entries were filtered using a false positive rate of 1% both at the peptide and protein levels, and false positives were removed. MaxQuant search file proteinGroups.txt was uploaded to Perseus software (version 1.4.1.3). Protein identifications annotated in the columns “Only identified by site”, “Reverse,” and “Contaminant” as well as proteins identified only in a single replicate were filtered out. Heavy-to-light ratios (“Ratio H/L Normalized”) for two or three replicates were log2-transformed and used to calculate average ratio and statistical significance (t test with Benjamini-Hochberg false-discovery rate-adjusted p values) and generate volcano plots. Raw mass spectrometry proteomics data and MaxQuant ouput files with peptide and protein identifications were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD002797. Reviewer account details (http://www.ebi.ac.uk/pride/archive/login): Username: reviewer78691@ebi.ac.uk; Password: ZPNADXAH.
Development of SRM Assays
The Peptide Atlas database (www.peptideatlas.org) was used to select top 5–8 peptides for each of 161 proteins. Fully tryptic peptides with 7–20 amino acids were chosen, and peptides with methionine and N-terminal cysteine residues were avoided, if possible. A list of 805 peptides was uploaded to Pinpoint software, and all possible y-ion transitions (y3 .y[n-1]) were generated in silico for light and corresponding heavy peptides. An equimolar mixture of regular MCF-7 cells and SILAC-labeled heavy MCF-7 cells was used to experimentally test nearly 12,000 transitions within five iterations. The rationale for multiple iterations was to promptly develop SRM methods for high- and medium-abundance peptides and exclude them from the following iterations. That allowed us to focus on low-abundance peptides and test their SRM assays with higher scan times, lower background and more points per peak. If we were in doubt about some proteins, peptides or transitions, we confirmed them using our SILAC data or our MCF-7 proteome (13).
In the first iteration of method development, 13–15 light and heavy peptide pairs and about 200 transitions were included into each of 60 survey SRM methods and analyzed in a nonscheduled mode with 7 ms scan times per transition. In the second iteration, eight peptide pairs and around 110 transitions were included into each of 50 survey SRM methods and run in a nonscheduled mode with 15 ms scan times per transition. In the third iteration, three peptide pairs and around 45 transitions were included into each of 25 survey SRM methods and run in a nonscheduled mode with 40 ms scan times per transition. In the forth iteration, retention times from the previous three steps were used to schedule 109 proteins, 197 peptide pairs and nearly 2700 transitions in 12 methods (∼240 transitions with 25 ms scans per method). In the fifth iteration, peptides were reanalyzed and the following parameters were verified, recorded or optimized, if required: (1) top 3 transitions; (2) retention times of light and heavy peptides and scheduling intervals; (3) heavy-to-light ratios of transitions; (4) selectivity of transitions and possible interferences; (5) scan times. Transitions with fragment m/z higher than precursor m/z were preferable; however, transitions with lower m/z but high signal-to-noise ratio were also used. As a reference to exclude possible interferences, we used corresponding heavy-isotope labeled peptides derived from the digest of heavy SILAC cells. To ensure high selectivity and the correct identity of each SRM peak, we applied the following criteria for each light and heavy peptides: (1) correspondence of LC retention times of light- and heavy-peptide forms; (2) superposition of transitions: light- and heavy-peptide forms should have a minimum of 6–8 overlaid y-ion transitions to ensure correct peptide identity in complex mixtures (16); (3) order of transitions: same order of y-ion transition intensities for light- and heavy-peptide forms, e.g. y1>y2>y3; (4) integrated area of all transitions: area of light and heavy peptides should be equal in the equimolar mixture of lysates. For proteins with multiple peptides, two peptides with the highest SRM area and significantly different retention times were chosen. All peptides were analyzed with the Basic Local Alignment Search Tool at http://blast.ncbi.nlm.nih.gov/Blast.cgi to ensure that peptides were unique to each UniProtKB/Swiss-Prot protein identifier. The exception was made for some high-abundance proteins selected for data normalization, such as tubulins, for which peptides represented several protein isoforms. For peptides which were shared between several members of the same protein family, we applied the “Occam's Razor” principle: within a protein family, we assigned a peptide to that protein isoform which was found differentially expressed in SILAC experiment, was detected with a substantially higher mass spectrometry intensity in the cell lysate, and whose mRNA transcript was identified by RNA sequencing in MCF-7 cells according to the Human Protein Altas (17).
At the final step, 31 candidate proteins and 16 housekeeping and control proteins were scheduled in a single SRM method within 3.2-min (±1.6 min) intervals during a 60 min LC gradient. Two most intense and reproducible transitions were monitored per each light and heavy peptides. Scan times were optimized for each peptide to ensure the measurement of 15–20 points per LC peak per transition.
Cell Culture and Growth Conditions for SRM Quantification
For SRM-based experiments, four biological replicates for each growth condition were used. Approximately 0.5 × 106 cells were seeded individually into 6-well tissue culture plates and left for 1 day for cell attachment. Cells were then transferred to RPMI 1640 culture medium supplemented with 10% dextran-coated charcoal-treated FBS and grown for 24 h. Following this, cells were stimulated with either 10 nm 17β-estradiol in 0.1% ethanol (final concentration) or 0.1% ethanol. Cells were grown as a monolayer for up to 72 h after 17β-estradiol stimulation and then trypsinized and lysed. For ERα antagonist experiment, cells were treated with 10−10 to 10−6 m 4-hydroxytamoxifen (Sigma Aldrich), then immediately treated with 10 nm 17β-estradiol and grown for 36 h.
NAB2 Silencing by siRNA
Increasing concentrations (20–80 nm) of siGENOME human NAB2 siRNA pool (m-020080–01-0005, GE Dharmacon) and vehicle control (20–80 nm) siGENOME nontargeting siRNA pool (d-001206–14-05, GE Dharmacon) were used to define the optimum concentration for NAB2 silencing. MCF-7 cells were transfected for 24 h with the optimum concentration (40 nm) NAB2 siRNA pool or vehicle control. Following transfection, cells were grown for 24 h in RPMI 1640 culture medium supplemented with 10% dextran-coated charcoal-treated FBS and then stimulated with 10 nm 17β-estradiol.
Cell Lysis and Protein Digestion for SRM Quantification
Four biological replicates of ∼2 × 105 cells (∼30 μg of total protein) were reconstituted in 100 μl of 0.1% RapiGest SF (Waters), vortexed, sonicated three times for 30 s and centrifuged for 20 min at 16,000 × g at 4 °C to ensure absence of cell debris and completeness of lysis. Equimolar amounts of total protein derived from the nonstimulated heavy SILAC cells were added to serve as internal standards for the accurate relative quantification. Samples with 60 μg total protein were transferred to the 96-well plate, and proteins were denatured at 65 °C using PCR thermocycler (MasterCycler 5332, Eppendorf, Germany), reduced with 10 mm dithiothreitol and alkylated with 20 mm iodoacetamide. Samples were then digested overnight at 37 °C with sequencing grade modified trypsin (trypsin/total protein ratio 1:30; Promega). RapiGest SF was cleaved with 1% trifluoroacetic acid and samples in the 96-well plate were centrifuged at 290 g. Peptides were extracted with 10 μl OMIX C18 tips (Varian, Lake Forest, CA) using 12-channel pipettes and eluted with 10 μl 64.5% acetonitrile. Recovery yield from Omix C18 microextraction tips was ∼80%, as previously estimated by SRM analysis of BSA peptides. Heavy isotope-labeled peptide LSEPAELTDAVK* peptide was spiked into each digest and used as a quality control for C18 microextraction. Peptides were diluted to 130 μl with 0.1% formic acid in water. The following precautions were taken to minimize possible modifications of peptides (oxidation of methionines and deamidation of asparagines and glutamines) during storage and analysis: (1) supplementation of the protein digest with 0.4 m methionine, (2) storage of tryptic peptides at −20 °C until use; and (3) sealing of 96-well plates with silicone rubber mats and preservation of plates at 7 °C during SRM analysis.
Protein Quantification by SRM
Peptides were separated by 60-min C18 reversed-phase liquid chromatography (EASY-nLC, Thermo Fisher Scientific) and analyzed by a triple-quadrupole mass spectrometer (TSQ Vantage or TSQ Quantiva, Thermo Fisher Scientific) using a nanoelectrospray ionization source, as previously described (18, 19). Reproducibility of SRM signal was ensured by running a solution of 0.25 fmol/μl BSA every 9 runs. Carryover was estimated in the range 0.05–0.2%. Use of heavy-isotope labeled peptides as internal standards ensured stability and reproducibility of SRM analysis. Two technical replicates (40 μl each) and four biological replicates were analyzed for each biological condition. Blocks of four biological replicates were randomized within time-response and antagonist-response experiments. Proteins in the ERα-EGR3 network were quantified by a triple-quadrupole mass spectrometer TSQ Quantiva (Thermo Fisher Scientific) and the identical chromatography setup.
SRM Data Analysis
Raw files were analyzed with Pinpoint software (Thermo Fisher Scientific), and areas of each light and heavy SRM transitions were extracted. Analysis of SRM data included normalization of all light endogenous peptides by spiked-in heavy peptides (to account for the variability of mass spectrometry analysis), followed by re-normalization of light peptides of all proteins by a set of high-abundance house-keeping proteins (to account for the variability of sample handling, cell lysis, and total protein analysis). Essentially, protein abundances were normalized to the equal number of cells in each biological condition.
To normalize SRM data, we followed a previously proposed intensity-based approach for pairs of light and heavy peptides (20), rather than a standard ratio-based approach used to analyze SILAC data. In the first step of data normalization, the mean area of two technical injections was calculated for each light and heavy SRM transitions. Because the equal amount of heavy cell lysate was spiked into each biological replicate, in the second step the median area of each heavy SRM transition was used to calculate a unique correlation coefficient for each biological replicate. In the third step, such coefficient was used to calculate the normalized area of light SRM transitions for each biological replicate. In the fourth step, areas of “light” SRM transitions of high-abundance house-keeping proteins (13 proteins and 24 transitions in the verification experiment and 7 proteins and 14 transitions in the ERα-EGR3-NAB2 network analysis) were used to calculate the median correlation coefficient for each biological replicate. Such coefficient was used to calculate the renormalized areas of each light transition in each biological replicate. Intensities of two transitions of each peptide were summed to obtain the area of each light peptide in each biological replicate. We also assumed that the area of each unique peptide represented a proxy of the abundance of the corresponding protein. Finally, the mean abundance of each protein in four biological replicates and the corresponding standard deviations were calculated. Our normalization approach reduced the variability and facilitated an accurate analysis of relative protein abundances.
SRM raw mass spectrometry data and data processed with Pinpoint viewer have been deposited to Peptide Atlas repository with the data set identifier PASS00796. URL: http://www.peptideatlas.org/PASS/PASS00796; Username: PASS00796; Password: NR473vp Full URL: ftp://PASS00796:NR473vp@ftp.peptideatlas.org.
Statistical Analysis of a NAB2 Silencing Experiment
Repeated measures two-way ANOVA was used to compare time courses of stimulation (0–36 h; first factor) and the impact of NAB2 silencing (siRNA-NAB2 versus nontargeting siRNA, second factor). All tests were performed with α value of 0.05. Bonferroni's multiple comparisons post-tests at 95% confidence level were used to test selected combinations of factors. To indentify proteins which expression might be enhanced upon NAB2 silencing, we used the following four criteria which had to be met simultaneously: (1) significant overexpression in 36 versus 0 h in a nontargeting siRNA group (adjusted p < 0.05); (2) significant overexpression in 36 versus 0 h in NAB2-silenced group (adjusted p < 0.05); (3) significant overexpression in 36 h between NAB2-silenced and control groups (adjusted p < 0.05); (4) substantial fold changes (≥1.2; see Experimental design).
RESULTS
Optimization of Estradiol Stimulation
As a model cell line, we selected MCF-7 breast cancer cells which express ERα, but not ERβ (21). MCF-7 cells have been extensively used for identification of estrogen-regulated genes by genomic and transcriptomic platforms (3–6). We previously confirmed by shotgun and SRM mass spectrometry the presence of ERα and absence of ERβ in the MCF-7 cell lysate (13).
To optimize cell culture conditions for estradiol stimulation, we relied on SRM measurement of trefoil factor 1 (TFF1), an estrogen-regulated protein and a well-known marker of estradiol stimulation (13). Using TFF1, we optimized cell culture conditions, estradiol concentration and established time points to monitor the maximum effect of stimulation (supplemental Fig. S1). Starvation of cells in the FBS-free media for 24 h prior to stimulation was used to induce a reversible cell cycle arrest and synchronize cell cycle to G0/G1 (22). Thus, cells progressed to S phase after addition of the 10% FBS-supplemented media. In addition, we compared cell proliferation with continuous stimulation by 10 nm estradiol remaining in the media versus single-point stimulation during which culture media was replaced after 1 h of stimulation by 10 nm estradiol. No significant differences in the proliferation rate of MCF-7 cells were observed.
Based on optimization experiments, we proceeded with 6 and 36 h of stimulation in the SILAC experiment, in order to identify the early- and late-response proteins. These time points were also in agreement with previous transcriptomic approaches, which suggested 4 and 24 h to identify the early- and late-response genes (21). A period of 6 h cell growth after stimulation was chosen based on previously measured mRNA transcription and protein translation rates (23) and the limit of detection of our SRM protocol (∼50,000 protein copies per cell) (13).
Identification and Quantification of ERα-regulated Proteins in MCF-7 Cells Using SILAC
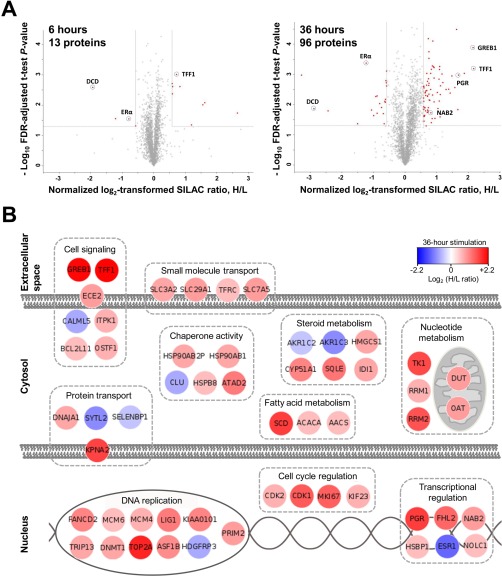
A total of 3105 and 3198 proteins were quantified by mass spectrometry in 3 biological replicates of MCF-7 cells exposed to stimulation with 10 nm 17β-estradiol (heavy SILAC cells) and vehicle control (light SILAC cells) for 6 and 36 h, respectively. As expected, the abundance of most quantified proteins was not affected by the estradiol treatment, resulting in an average normalized heavy-to-light ratio close to one. Statistical analysis (Benjamini-Hochberg-adjusted p values < 0.05) revealed 13 and 96 proteins differentially expressed upon estradiol stimulation in 6 and 36 h of stimulation, respectively (Fig. 2A). Because measurements of small relative changes in protein expression may not be reproducible because of high biological variation, we selected proteins with rather substantial differential fold changes (>1.5 or <0.67) which can be verified experimentally by SRM assays. A high-quality list of estradiol-regulated proteins thus included 103 proteins (supplemental Table S1).
Fig. 2.
Global profiling of estrogen-regulated proteins as measured by SILAC. A, Statistical analysis revealed 13 and 96 proteins differentially expressed upon estradiol stimulation at 6 and 36 h of stimulation, respectively (fold changes >1.50 or <0.67 and FDR-adjusted t test p values <0.05 for 3 biological replicates). Identified proteins are shown as red dots, and fold change and p value cut-offs are presented as vertical and horizontal lines, respectively. See supplemental Table S1 for the full list of estrogen-regulated proteins. B, The search for molecular function and sub-cellular localization using the NextProt database (www.nextprot.org) revealed that the majority of identified proteins were involved in cell proliferation, cell cycle regulation, transcriptional regulation, nucleotide metabolism and DNA replication.
Interestingly, only 40 of 103 corresponding genes had estrogen response elements in their promoter regions, based on the Human ERE Database (9). Trefoil factor 1 (TFF1), a known primary target of ERα, was found to be increased 1.6- and 4.5-fold at 6 and 36 h upon stimulation, respectively (supplemental Fig. S2). In accordance to our previous observations (13), ERα itself was found to be down-regulated 0.6- and 0.4-fold at 6 and 36 h. Other primary targets of estradiol stimulation such as progesterone receptor (PGR) and growth regulation by estrogen in breast cancer 1 protein (GREB1) were found among the top candidates at 36 h stimulation, up-regulated by 4.4- and 3.2-fold, respectively. Reproducibility of fold change ratios between biological replicates of 36 h stimulation for TFF1, GREB1, and PGR were 7, 3, and 7%, respectively. The robust performance of known estrogen-regulated proteins suggested that our list of 103 estrogen-regulated proteins was of a high quality.
It should be noted that identified estrogen-regulated proteins included multiple markers of cellular proliferation such as CDK1, CDK2, TOP2A, MKI67, MCM4, MCM6 and others (24). Some of these markers were secondary-response proteins, such as MKI67 (antigen ki-67), a prognostic breast cancer biomarker and a major determinant of the Oncotype DX score used clinically (25).
A manual search of the differentially expressed proteins in the NextProt database (www.nextprot.org) was performed to examine their molecular function and subcellular localization. Proteins found to be involved in the same molecular function were grouped together and the major groups were depicted in Fig. 2B. As expected, functions related to cell proliferation, cell cycle regulation, transcriptional regulation, nucleotide metabolism and DNA replication were identified.
Development of a Multiplex SRM Assay for Candidate ERα-regulated Proteins
To develop SRM assays, we followed our previously published approach (13). In addition to 103 SILAC candidates which were found significantly over- or under-expressed in three biological replicates, we also included a group of 58 proteins with substantial up-regulation in one or two biological replicates. Briefly, for each of the 161 proteins we selected five peptides based on the Peptide Atlas data (www.peptideatlas.org). We monitored by SRM both light and heavy forms of 805 peptides in a digest of an equimolar mixture of lysates of light and heavy isotope-labeled SILAC MCF-7 cells. Several iterations of experimental testing of SRM assays reduced that list to 56 proteins and 81 proteotypic peptides.
Challenges encountered during SRM development revealed that many estrogen-regulated proteins were expressed in MCF-7 cells at very low levels and thus were not amenable to SRM quantification in the unfractionated cell lysate. Even though additional fractionation approaches, such as strong-cation exchange chromatography or off-gel isoelectric focusing, would increase sensitivity of analysis up to several hundred protein copies per cell (26), these additional steps would significantly decrease the throughput and reproducibility of protein quantification. To increase sensitivity of a multiplex SRM assay, we retained a single unique peptide per protein and removed peptides and proteins measured with poor reproducibility. Our final SRM assay included 23 candidate estrogen-regulated proteins identified in three biological replicates, 17 proteins with substantial fold changes only in a single replicate, 13 high-abundance house-keeping proteins for data normalization and three markers of hypoxia. The latter markers were used to detect the adverse effects of growing cells for 72 h and possible starvation and hypoxia which could decrease levels of ERα and thus affect estrogen-regulated proteins, as previously reported (13).
Validation of ERα-regulated Proteins by SRM
Before proceeding to validation of estrogen-regulated proteins and measuring the dynamics of their expression, we measured by SRM the relative expression of trefoil factor 1 after 36 h of stimulation with different combinations of 17β-estradiol and 4-hydroxytamoxifen, an active metabolite of tamoxifen and a potent antagonist of ERα (27). Such experiment provided an estimate of the optimal concentration of 4-hydroxytamoxifen (IC50<100 nm) and facilitated selection of five different concentrations of 4-hydroxytamoxifen within the range 0.1 to 1000 nm, in order to set-up a dose-response study (supplemental Fig. S3).
Following that, we stimulated MCF-7 cells with 10 nm 17β-estradiol or vehicle control and cultured for different time points and in the presence of different concentrations of 4-hydroxytamoxifen, before proceeding to cell lysis. In total, MCF-7 cells were exposed to 14 different biological conditions, with four biological replicates per condition (supplemental Table S2).
Upon cell lysis, equimolar amounts of lysates of nonstimulated SILAC heavy isotope-labeled MCF-7 cells were added to allow for accurate relative quantification of peptides by SRM, and the protein mixture was subjected to the proteomic sample preparation. All lysates were simultaneously subjected to the sample preparation protocol, and the peak areas for each light and heavy peptide were measured with a multiplexed SRM assay (supplemental Table S3). Because the accuracy of colorimetric total protein assays may be affected in the complex mixtures and in the presence of detergents, we measured by SRM 13 high-abundance house-keeping proteins, selected from our previous work (13). Our analysis of relative abundances of proteins thus relied on the stepwise normalization of SRM area using heavy isotope-labeled peptide standards followed by normalization with 13 high-abundance house-keeping proteins. Such stepwise normalization excluded possible analytical issues arising because of the cell lysis, sample loss, protein digestion, LC peptide separation and MS ion suppression, and substantially improved reproducibility of relative quantification.
After data analysis, 9 of 40 proteins were excluded because of their poor reproducibility of biological replicates. Finally, accurate dynamics of expression was measured for 31 proteins (supplemental Tables S4–S5). Such low success rate of SRM development (31/161) can be explained by the low abundance of estrogen-regulated proteins. Dynamic range of SRM analysis was 3.5 orders of magnitude, with the lowest levels for CDK1 and CDK2 and the highest levels for beta-actin. This range correlated well with the previously reported levels of ERα (120,000 copies per MCF-7 cell), progesterone receptor (∼50,000 copies per cell) and beta-actin (∼108 copies per cell) (28, 29).
Estradiol stimulation resulted in the strong over-expression of known estrogen-regulated proteins such as trefoil factor 1 (Table I and Fig. 3). In total, 18 of 19 SILAC candidates identified in three biological replicates responded to estradiol and 4-hydrohytamoxifen, thus leaving a single protein (ACACA) as a false positive. Five control and house-keeping proteins contained EREs, but did not respond to estradiol stimulation or 4-hydroxytamoxifen. Expression of these control and house-keeping proteins may require the recruitment of additional transcription factors or regulatory proteins absent in MCF-7 cells. Interestingly, all 10 proteins identified by SILAC in a single biological replicate did not respond to estradiol and 4-hydrohytamoxifen and were noted as false-positives (supplemental Table S6 and supplemental Fig. S4). Even though SRM results revealed only one false-positive candidate in the group of 19 validated proteins (Table I), we believe it was essential to verify the expression of SILAC-derived candidates by alternative assays, such as SRM, in the unlabeled cells which were grown in their native media. We also concluded that our list of 103 estrogen-regulated proteins was of a high quality.
Table I. Candidate estrogen-regulated proteins verified by SRM assay. 4-OHT, 4-hydroxytamoxifen. *See notes in the supplemental Table S3.
| Gene symbol | Protein symbol | UniProt accession | Protein name | ERE | SILAC fold change 36 h | SRM fold change 36 h | Response to 4-OHT | Category of response |
|---|---|---|---|---|---|---|---|---|
| Candidates identified by SILAC in three biological replicates (adjusted p < 0.05) | ||||||||
| TFF1 | TFF1 | P04155 | Trefoil factor 1 | + | 4.5 | 6.6 | Yes | Primary |
| MCM6 | MCM6 | Q14566 | DNA replication licensing factor MCM6 | + | 1.5 | 2.7 | Yes | Primary |
| KPNA2 | IMA1 | P52292 | Importin subunit alpha-1 | + | 3.1 | 2.5 | Yes | Primary |
| TST | THTR | Q16762 | Thiosulfate sulfurtransferase | + | 1.7 | 1.9 | Yes | Primary |
| BCL2L11 | B2L11 | O43521 | Bcl-2-like protein 11 | + | 1.5 | 1.7 | Yes | Primary |
| ASNS | ASNS | P08243 | Asparagine synthetase [glutamine-hydrolyzing] | + | 1.7 | 1.5 | Yes | Primary |
| CDK2 | CDK2 | P24941 | Cyclin-dependent kinase 2 | + | 1.6 | 1.4 | Yes | Primary |
| RAB31 | RAB31 | Q13636 | Ras-related protein Rab-31 | + | 1.5 | 1.3 | Weak | Transient |
| TPM1 | TPM1 | P09493–3 | Isoform 3 of Tropomyosin alpha-1 chain | + | 1.7 | 1.1 | Weak | Transient |
| DNAJA1 | DNJA1 | P31689 | DnaJ homolog subfamily A member 1 | + | 1.7 | 1.1 | Weak | Transient |
| DUT | DUT | P33316 | Deoxyuridine 5Ȳ-triphosphate nucleotidohydrolase | − | 1.8 | 3.5 | Yes | Secondary |
| CDK1 | CDK1 | P06493 | Cyclin-dependent kinase 1 | − | n/a | 2.8 | Yes | Secondary |
| NAB2 | NAB2 | Q15742 | NGFI-A-binding protein 2 | − | 1.8 | 1.9 | Yes | Secondary |
| TOP2A | TOP2A | P11388 | DNA topoisomerase 2-alpha | − | 3.7 | 1.5 | Yes | Secondary |
| HSP90AB1* | HS90B | P08238 | Heat shock protein HSP 90-beta | − | 1.7 | 1.3 | Yes | Secondary |
| ESR1 | ESR1 | P03372 | Estrogen receptor | − | 0.4 | 0.7 | Yes | Secondary |
| LXN | LXN | Q9BS40 | Latexin | − | 1.5 | 0.5 | Yes | Secondary |
| SLC9A3R1 | NHRF1 | O14745 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | − | 1.6 | 1.3 | No | Indirect |
| ACACA | ACACA | Q13085 | Acetyl-CoA carboxylase 1 | − | 1.5 | 0.9 | Weak | False |
| Controls for hypoxia | ||||||||
| SLC2A1 | GTR1 | P11166 | Solute carrier family 2, facilitated glucose transporter member 1 | + | 1.4 | 1.8 | Weak | |
| ALDOA | ALDOA | P04075 | Fructose-bisphosphate aldolase A | 23 | 1.1 | 1.1 | No | |
| ALDOC | ALDOC | P09972 | Fructose-bisphosphate aldolase C | 23 | 1.2 | 1.2 | No | |
| Housekeeping proteins used for normalization of SRM data | ||||||||
| ACTB | ACTB | P60709 | Actin, cytoplasmic 1 | + | 1.0 | 1.0 | No | |
| CS | CISY | O75390 | Citrate synthase, mitochondrial | + | 1.1 | 1.0 | No | |
| IDH2 | IDHP | P48735 | Isocitrate dehydrogenase [NADP], mitochondrial | + | 1.0 | 1.1 | No | |
| TALDO1 | TALDO | P37837 | Transaldolase | + | 0.9 | 1.1 | No | |
| ENO1 | ENOA | P06733 | Alpha-enolase | − | 0.9 | 0.8 | No | |
| GAPDH | G3P | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | − | 1.1 | 0.9 | No | |
| PKM | KPYM | P14618 | Pyruvate kinase PKM | − | 1.2 | 1.0 | No | |
| PKM/PKLR | KPYM/KPYR | P14618/P30613 | Pyruvate kinase PKM/Pyruvate kinase PKLR | − | 1.2 | 1.0 | No | |
| RPL27A | RL27A | P46776 | 60S ribosomal protein L27a | − | 1.1 | 1.1 | No | |
| RPL30 | RL30 | P62888 | 60S ribosomal protein L30 | − | 0.9 | 1.2 | Weak | |
| RPS3 | RS3 | P23396 | 40S ribosomal protein S3 | − | 1.2 | 1.1 | No | |
| TUBB | TBB5 | P07437 | Tubulin beta chain | − | 1.2 | 1.2 | Weak | |
| UBB | UB | P0CG47 | Polyubiquitin-B | − | 1.0 | 1.1 | No |
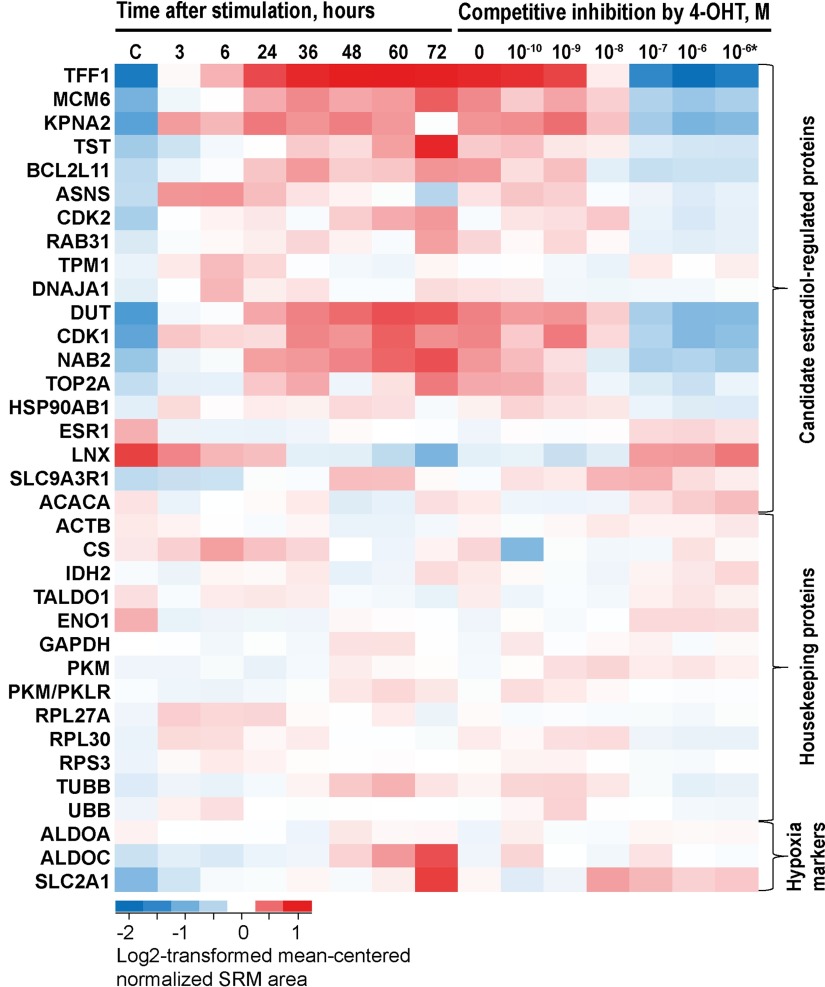
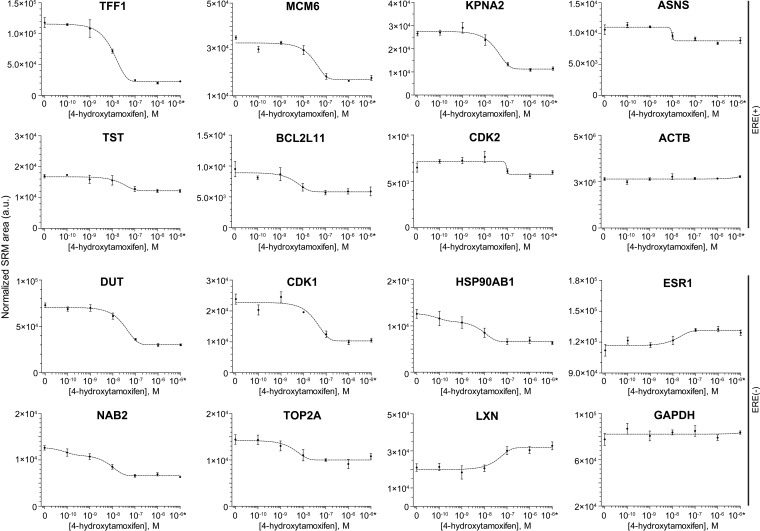
Fig. 3.
Verification of true ERα-regulated proteins through the dynamics of protein expression and competitive inhibition by 4-hydroxytamoxifen. Four biological replicates of MCF-7 cells were stimulated with 10 nm 17β-estradiol and grown for 3–72 h, or stimulated with 10 nm 17β-estradiol in the presence of increasing concentrations of 4-hydroxytamoxifen (4-OHT) and grown for 36 h. Relative abundances of estrogen-regulated, house-keeping and control proteins were measured by a multiplex SRM assay. C, control: Cells treated with vehicle control (0.1% ethanol) and grown for 36 h. *Cells treated with vehicle control (0.1% ethanol) and 10−6 m 4-hydroxytamoxifen.
It should also be noted that ALDOA, ALDOC and SLC2A1 markers revealed possible hypoxia and starvation at 72 h of cell growth in the 6-well tissue culture plates. According to our previous findings (13), hypoxia may result in the reduced expression of estrogen-regulated proteins, because of degradation of ERα. Thus, measured expression of some proteins at 72 h may be biased. We would recommend continuous monitoring of ALDOA, ALDOC, or SLC2A1 proteins in the future studies on ERα signaling to avoid the negative impact of hypoxia and starvation.
Comparison of SILAC and SRM Data
Our data showed that temporal dynamics of protein expression in the SILAC-labeled cells may be delayed and even distorted. For example, expression of trefoil factor 1 at 6 h was noticeably weaker in the SILAC experiment (1.6-fold increase) relative to the SRM experiment (3.7-fold). Overall, 6-hour SILAC experiment provided only 14 candidate proteins and failed to identify many robust estrogen-regulated proteins, such as CDK1, KPNA2 and MCM6 which were later verified by SRM. It is likely that during growth in the SILAC media, which are supplemented with dialyzed but not standard FBS, cells undergo high stress which affects the dynamics of ERα signaling. Our preliminary experiments with SILAC labeling also showed that metabolically labeled MCF-7 cells might even completely fail to respond to estradiol stimulation. For example, we previously observed a subset of SILAC-labeled MCF-7 cells which had normal phenotypical features and expressed ERα, but had a substantially increased population doubling time (72 versus 36 h) and did not respond to estradiol, as measured by the expression of trefoil factor 1. Thus, in order to validate SILAC-derived candidates and measure the accurate dynamics of protein expression by SRM, we relied on MCF-7 cells grown in their native media (RPMI 1640 supplemented with 10% FBS).
Measuring the Accurate Dynamics of Expression of ERα-regulated Proteins
Measurement of protein expression at multiple time points after estradiol stimulation allowed us to observe accurate dynamics of primary targets and secondary response proteins, as defined by the presence and absence of EREs, respectively. We observed a rapid response for primary targets and delayed response for secondary targets (Table I and Fig. 4). Using increasing doses of 4-hydroxytamoxifen during estradiol stimulation (Fig. 5), we also confirmed that the observed response was mediated through ERα.
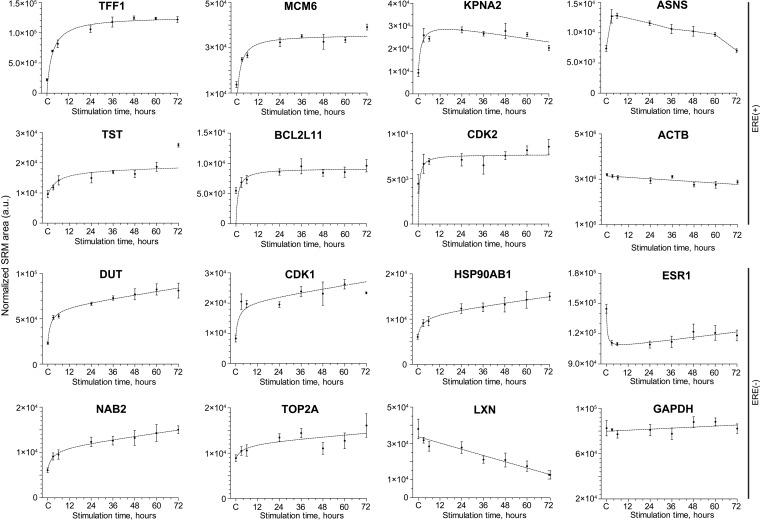
Fig. 4.
Dynamics of temporal expression of selected primary target ERE(+) proteins and secondary response ERE (-) proteins. Four biological replicates of MCF-7 cells were stimulated with 10 nm 17β-estradiol, grown for 3–72 h and analyzed by SRM. Primary targets (such as TFF1) revealed a sharp increase of expression at 3 and 6 h after stimulation, whereas secondary response proteins (such as NAB2) showed a slower and delayed expression. Levels of two house-keeping proteins ACTB and GAPDH are presented for comparison. C, control: Cells treated with 0.1% ethanol and grown for 36 h. a.u., arbitrary units.
Fig. 5.
Dynamics of expression of selected groups of primary target ERE(+) proteins and secondary response ERE(-) proteins in the presence of increasing concentrations of 4-hydroxytamoxifen. Four biological replicates of MCF-7 cells were stimulated with 10 nm 17β-estradiol in the presence of increasing concentrations of 4-hydroxytamoxifen, grown for 36 h and analyzed by SRM. Expression of true primary targets and secondary response proteins was inhibited at 100 nm concentration of 4-hydroxytamoxifen. IC50 of 4-hydroxytamoxifen was estimated at 15 nm. Levels of two house-keeping proteins ACTB and GAPDH are presented for comparison. *Cells treated with vehicle control (0.1% ethanol) and 10−6 m 4-hydroxytamoxifen. a.u., arbitrary units.
To compare dynamics of mRNA and protein expression, we analyzed publicly available microarray gene expression profiling of MCF-7 cells treated with estradiol and cultured for 0, 3, 6 and 12 h after treatment (3). Comparison of 95 genes discovered by SILAC at 36 h and corresponding mRNAs at 12 h revealed the following trends: (1) 44 genes had the same direction of over- or under-expression, with substantial protein and mRNA fold changes >1.5 or <0.67; (2) 23 genes had the same direction of over- or under-expression, but mRNA fold changes were not substantial <1.5 or >0.67; (3) 26 genes had different directions of over- or under-expression, but mRNA fold changes were not substantial <1.5 or >0.67 and (4) three genes had different directions of over- or under-expression, with substantial protein and mRNA fold changes >1.5 or <0.67. Thus, 44/95 mRNA/proteins were correlated, 49/95 mRNA/proteins were inconclusive and 3 mRNA/proteins revealed the opposite trends.
In general, relative changes of mRNA expression (1.2-fold median) were notably smaller than changes in protein expression (1.7-fold median), if we consider seven ERE(+) estrogen-regulated proteins measured by microarray and SRM at 6 h (supplemental Table S7). In addition, secondary response genes and their corresponding proteins were marginally elevated at the mRNA level, but substantially over-expressed at the protein level. Our results demonstrated that EREs and mRNA expression may predict estrogen-regulated genes, but fail to predict accurate dynamics of protein expression.
Measuring the Dynamics of Protein Expression in the Network of Secondary Messengers Regulated by Transcription Factor EGR3, a Primary Target of ERα Signaling
Our SRM data revealed a strong but delayed expression of several proteins whose genes did not have EREs and thus were defined as secondary response proteins (Table I). One of such proteins was NGFI-A binding protein 2 (NAB2), a primary target of EGR3 transcription factor, which in its turn is a primary target of ERα in MCF-7 cells. Unlike other ERα-regulated networks (for example, well-studied ERα-E2F1-CDK1 network), ERα-EGR3-NAB2 network and dynamics of its signaling was not previously investigated in detail and thus captured our attention.
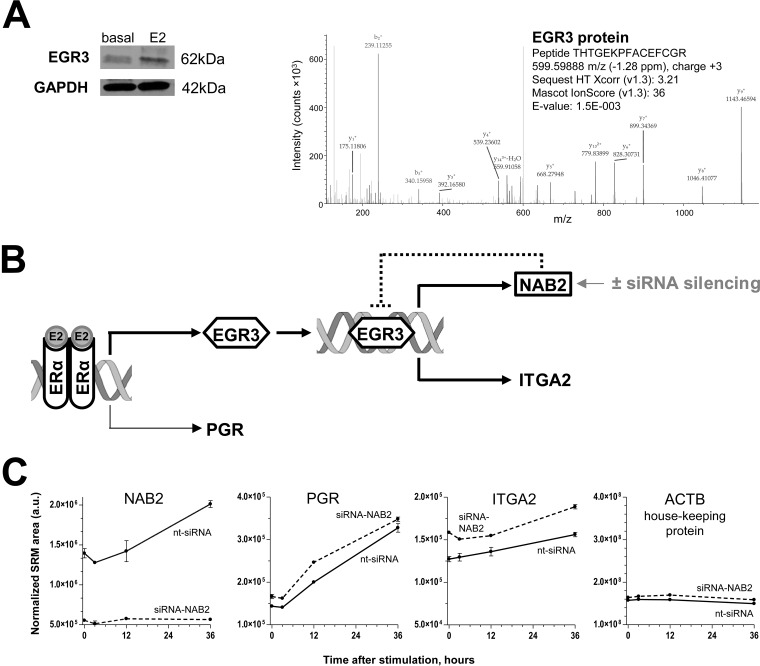
Because of its low expression levels in MCF-7 cells, EGR3 transcription factor was not identified by SILAC. However, the increased expression of EGR3 protein upon estradiol stimulation was confirmed by Western blot (Fig. 6A). We also confirmed by deep proteomic analysis of cell lysate the expression of EGR3 in MCF-7 cells and the absence of EGR1, EGR2 and EGR4 (supplemental Tables S8, S9 and Fig. 6A).
Fig. 6.
Dynamics of protein expression in the ERα-EGR3 network. A, Expression of EGR3 in MCF-7 cells as measured by Western blotting (basal expression and expression at 36 h after stimulation) or by the deep proteomic profiling. Isoform 2 of Early growth response protein 3 (Q06889–2) was identified with 13 peptides, 3 unique peptides and the sequence coverage of 42%. EGR1, EGR2 and EGR4 proteins were not identified. B, SiRNA silencing of NAB2, the repressor of EGR3, may alter the dynamics of protein expression in the ERα-EGR3-NAB2 network. C, Dynamics of protein expression in the ERα-EGR3-NAB2 network upon NAB2 silencing, as measured by SRM. Very low levels of NAB2 protein in siRNA-NAB2 transfected cells, a significant increase of PGR and NAB2 levels upon estradiol stimulation in the control cells transfected with nontargeting siRNA (nt-siRNA), and no effect on house-keeping proteins were observed. Levels of putative EGR3-regulated proteins DCLK1, CDC5L, TPM1, and SPINT1 were not affected either by siRNA-NAB2 transfection or estradiol stimulation (supplemental Fig. S5). Expression of alpha 2 integrin protein (ITGA2) was not only estradiol-dependent, but also increased upon siRNA-NAB2 silencing. E2, 17β-estradiol; a.u., arbitrary units.
Because NAB2 protein is a repressor of EGR3-mediated transcription and acts as a dynamic brake in the ERα-EGR3 network (Fig. 6B), we hypothesized that expression of EGR3-regulated proteins will be enhanced upon siRNA silencing of NAB2. First, we selected from the literature four putative EGR3-regulated transcipts ITGA2, DCLK1, CDC5L, SPINT1, and TPM1 (30). Note that ITGA2, DCLK1, CDC5L and SPINT1 were found in our MCF-7 proteome, but not selected as candidates in the SILAC experiment because changes of their expression were not significant (<1.5-fold, Benjamini-Hochberg-adjusted t test p > 0.05). For example, the expression of ITGA2 in the three replicates of 36 h-stimulated cells increased 1.30, 1.15 and 0.94-fold (adjusted t test p = 0.36). Such small changes might be expected for EGR3-regulated proteins because their expression over time might be suppressed by the increasing amounts of NAB2.
Second, we developed a multiplex SRM assay for ERα-regulated proteins (PGR and NAB2), putative EGR3-regulated proteins (ITGA2, DCLK1, CDC5L, TPM1 and SPINT1) and seven house-keeping proteins (supplemental Table S10). Following optimization of NAB2 silencing by siRNA (supplemental Fig. S5), MCF-7 cells were transfected with siRNA-NAB2 or nontargeting siRNA for 24 h, stimulated with 10 nm estradiol and grown for additional 0, 3, 12, and 36 h in duplicates.
Finally, we measured by SRM the dynamics of estradiol-induced protein expression in the ERα-EGR3 network with and without NAB2 silencing (Fig. 6B). As it was expected, estradiol stimulation resulted in a significant increase of PGR (two-way ANOVA with Bonferroni's multiple comparisons post-tests, α = 0.05, adjusted p < 0.0001) and NAB2 (adjusted p = 0.002) expression at 36 h (Fig. 6C). Levels of NAB2 protein in siRNA-NAB2 transfected cells decreased dramatically (mean fold change 3.6; adjusted p = 0.0002). The 36-hour effect of siRNA-NAB2 on PGR was not significant (fold change 1.06; adjusted p = 0.05).
Impact of siRNA-NAB2 transfection or estradiol stimulation on the levels of putative EGR3-regulated proteins DCLK1, CDC5L, TPM1, and SPINT1 was not significant (supplemental Table S11 and supplemental Fig. S6). Interestingly, only alpha 2 integrin (ITGA2) was significantly up-regulated by estradiol stimulation with and without NAB2 silencing (36 versus 0 h; adjusted p = 0.008 and 0.009). ITGA2 expression also significantly increased at 36 h upon NAB2 silencing (fold change 1.2; adjusted p = 0.008) (Fig. 6C). We thus concluded that expression of ITGA2 protein was enhanced upon NAB2 silencing.
Rational reprogramming of cellular responses and engineering of synthetic gene circuits (31) may require tools to manipulate the dynamics and intensity of response to receptor stimulation. Even though it is too preliminary to make any broad conclusions based on the silencing of a single repressor NAB2 in the present work, such approach may be investigated in future for multiple transcription factors and their repressors or activators.
DISCUSSION
Estrogen-regulated genes were widely studied by genomic and transcriptomic approaches (3). Chromatin immunoprecipitation experiments and bioinformatic algorithms predicted hundreds of potential ERα-binding sites in the human genome (5, 32–34). Transcriptomic studies provided expression levels of estrogen-regulated genes and revealed primary and secondary targets in combination with EREs (35–37). Estrogen-regulated proteins were traditionally studied by antibody-based techniques such as Western blots, whose semiquantitative capabilities did not allow for the accurate measurement of the dynamics of protein expression (38, 39). In this work, we employed two different quantitative proteomic methods to identify and validate estrogen-regulated proteins. The presented approach offers supplementary knowledge on protein abundance and dynamics of expression which cannot be predicted using genomic and transcriptomic data (23).
Our global SILAC approach quantified more than 3100 proteins and revealed 103 putative primary and secondary targets of ERα signaling. Targeted proteomics by SRM verified 19 of those proteins. During SILAC labeling, MCF-7 cells were closely monitored to ensure that their phenotypical characteristics, proliferation rate and active ERα signaling were not altered. However, accurate comparison of both approaches showed that the dynamics of protein expression might be distorted or delayed in the SILAC-labeled cells. This observation might be related to the extended cell culture (six doublings to reach 99% labeling) in the SILAC media supplemented with dialyzed versus native FBS. Recognizing possible shortcomings of SILAC labeling, the differential expression of candidate proteins was verified by SRM in the unlabeled MCF-7 cells grown for a limited number of passages. Even though the SILAC approach revolutionized global quantification of the cellular proteomes, we would thus suggest that SILAC results need to be independently verified.
In our work, SRM assays allowed for the simultaneous measurement of dynamics of dozens of estrogen-regulated proteins. Although reviewing the dynamics of protein expression, we noted a sharp increase in the expression of ERE(+) primary response proteins at 3 h and a plateau at 24 h (TFF1), whereas the expression of ERE(-) secondary response proteins was moderate at 3 h and continued to increase until 72 h. Some ERE(+) and ERE(-) genes, for example TST and TOP2A, exhibited little difference for their early and late responses. Such little difference might be because of either the impact of factors which repress transcription of TST and TOP2A or because of the less precise measurements of expression of these proteins by SRM.
It should be mentioned that estradiol-mediated expression of ERE(-) genes may be executed through several indirect mechanisms:
induction of expression of ERE(-) genes by the primary products of ERα stimulation. Such products may include transcription factors (EGR3, PGR, E2F1), transcriptional activators and repressors (FLH2), and growth factors or receptor ligands (OSTF1 and HDGFRP3);
binding of the estradiol-activated ERα to other transcription factors (AP-1, STATs, ATF-2, c-Jun, Sp1 and NF-κB) which then trigger transcription through their cognate DNA binding sites (40);
nongenomic rapid signaling through the membrane or cytoplasmic G protein-coupled estrogen receptor GPR30;
nongenomic rapid signaling through activation of protein-kinase cascades (MAPK, Src, AKT and others).
Even though in this work we focused on the slow genomic signaling exerted through ERα within hours, we cannot completely exclude expression of some ERE(-) proteins through the rapid nongenomic signaling mechanisms. It should be also noted that in other breast cancer cell lines different combinations of transcriptional co-factors, activators and repressors may modulate the response of ERα and trigger the expression of additional proteins.
Overall, ERα is a master regulator of gene expression, and its signaling involves cross-talks with numerous nongenomic and genomic pathways including EGFR and IGFR pathways (41–43). Cross-talks with genomic signaling pathways are exerted through estrogen-regulated expression of multiple transcription factors including PGR, E2F1, GATA3, and others (3–6, 44). Even though these low-abundance transcription factors cannot be directly measured by mass spectrometry as yet, their activity can be inferred through the measurement of their secondary response targets. In this work, we measured secondary messengers of ERα-E2F1 network (CDK1, DUT and TOP2A) and ERα-EGR3 network (NAB2 and ITGA2) (45, 46).
EGR family of transcription factors includes immediate-early growth response genes induced by mitogenic stimulation (47). It was demonstrated that EGR factors digitalize mitogenic stimuli of the epidermal growth factor through MAPK/ERK pathway and ensure that cells undergo proliferation only upon consistent stimulation and do not respond to the single events of exposure to growth factors (48). Expression of EGR3 factor is induced by estradiol and activates the secondary wave of transcriptional events which are modulated by the family of transcriptional co-repressors NAB1 and NAB2. NGFI-A binding protein NAB2 represses the activity of promoters upstream of the DNA binding domains of EGR1, EGR2 and EGR3 (49, 50). NAB2 is thus acting as a brake: it stops transcription of EGR3-regulated genes at a certain period of stimulation, thus preventing an unrestrained activity of EGR3-regulated proteins (51). Because NAB2 is also a direct target of EGR3 in breast tissues and MCF-7 cells (52), such mechanism provides a negative feedback loop in the EGR3-regulated network (53).
Loss of NAB2 co-repressor was previously found in the primary prostate carcinoma and resulted in high transcriptional activity of EGR1 (54, 55). Similarly, association between breast cancer and EGR3 signaling was suggested (56). Investigation of NAB2 loss, EGR3 hyperactivation and unrestrained proliferation of breast cancer cells may provide future directions for studies of ERα-EGR3 network.
Measurement of the dynamics of estrogen-regulated proteins is also important for the translational research. Circulating levels of estradiol in blood of premenopausal women change between 100 to 700 pmol/L within a month (57), resulting in the alternating expression of estrogen-regulated proteins in tissues, blood and urine (58). Because estrogen-regulated proteins are often identified as disease biomarkers (59), dynamic profile of such proteins in biological fluids may eliminate some false biomarkers (60).
To conclude, we identified by SILAC 103 estrogen-regulated proteins and verified by SRM 19 of these candidate proteins. We also measured accurate temporal dynamics of expression of these proteins and differentiated between the primary and secondary target genes of ERα signaling in MCF-7 cells. Finally, we measured the dynamics of protein expression in the subnetwork of primary and secondary targets of ERα. Our work demonstrated the power of quantitative proteomics for the elucidation of primary and secondary effects of hormonal stimulation and accurate measurements of the dynamics of protein expression in the networks of transcription factors with negative feedback loops. Experimental measurement of the dynamics of protein expression in such networks may find numerous applications in systems biology, reprogramming of the cellular responses and cell engineering.
Supplementary Material
Acknowledgments
We thank A. Dimitromanolakis for help with data normalization and L. Dimitrakopoulos for providing access to the unpublished deep MCF-7 proteome.
Footnotes
Author contributions: A.P.D. and M.P. performed SILAC and SRM experiments; C.S. performed siRNA knockdown; A.P.D. designed the research project and wrote the manuscript, and all authors contributed to the revision of the manuscript.
* This work was supported by the Canadian Institutes of Health Research. A.P.D was supported by CIHR and the Mount Sinai Hospital Foundation Norm Hollend post-doctoral fellowships. M.P.P. is a recipient of the Ontario Graduate Scholarship.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- ERα
- Estrogen receptor alpha
- EGR3
- Early growth response protein 3
- ERE
- Estrogen response element
- FBS
- Fetal bovine serum
- FPKM
- Fragments per kilobase of exon per million fragments mapped
- FWHM
- Full width at half maximum
- ITGA2
- Integrin alpha-2
- LC-MS/MS
- Liquid chromatography-tandem mass spectrometry
- NAB2
- NGFI-A-binding protein 2
- SILAC
- Stable isotope labeling by amino acids in cell culture
- SRM
- Selected reaction monitoring.
REFERENCES
- 1. Norman A. W., Mizwicki M. T., and Norman D. P. (2004) Steroid-hormone rapid actions, membrane receptors and a conformational ensemble model. Nat. Rev. Drug Discov. 3, 27–41 [DOI] [PubMed] [Google Scholar]
- 2. Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Strom A., Treuter E., Warner M., and Gustafsson J. A. (2007) Estrogen receptors: how do they signal and what are their targets. Physiol. Rev. 87, 905–931 [DOI] [PubMed] [Google Scholar]
- 3. Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., and Brown M. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 4. Kininis M., Chen B. S., Diehl A. G., Isaacs G. D., Zhang T., Siepel A. C., Clark A. G., and Kraus W. L. (2007) Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 27, 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin C. Y., Vega V. B., Thomsen J. S., Zhang T., Kong S. L., Xie M., Chiu K. P., Lipovich L., Barnett D. H., Stossi F., Yeo A., George J., Kuznetsov V. A., Lee Y. K., Charn T. H., Palanisamy N., Miller L. D., Cheung E., Katzenellenbogen B. S., Ruan Y., Bourque G., Wei C. L., and Liu E. T. (2007) Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kwon Y. S., Garcia-Bassets I., Hutt K. R., Cheng C. S., Jin M., Liu D., Benner C., Wang D., Ye Z., Bibikova M., Fan J. B., Duan L., Glass C. K., Rosenfeld M. G., and Fu X. D. (2007) Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc. Natl. Acad. Sci. U.S.A. 104, 4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charpentier A. H., Bednarek A. K., Daniel R. L., Hawkins K. A., Laflin K. J., Gaddis S., MacLeod M. C., and Aldaz C. M. (2000) Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 60, 5977–5983 [PubMed] [Google Scholar]
- 8. Coser K. R., Chesnes J., Hur J., Ray S., Isselbacher K. J., and Shioda T. (2003) Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc. Natl. Acad. Sci. U.S.A. 100, 13994–13999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bourdeau V., Deschenes J., Metivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J. H., and Mader S. (2004) Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 18, 1411–1427 [DOI] [PubMed] [Google Scholar]
- 10. Geiger T., Wehner A., Schaab C., Cox J., and Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., and Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meissner F., Scheltema R. A., Mollenkopf H. J., and Mann M. (2013) Direct proteomic quantification of the secretome of activated immune cells. Science 340, 475–478 [DOI] [PubMed] [Google Scholar]
- 13. Drabovich A. P., Pavlou M. P., Dimitromanolakis A., and Diamandis E. P. (2012) Quantitative analysis of energy metabolic pathways in MCF-7 breast cancer cells by selected reaction monitoring assay. Mol. Cell. Proteomics 11, 422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiese T. E., Kral L. G., Dennis K. E., Butler W. B., and Brooks S. C. (1992) Optimization of estrogen growth response in MCF-7 cells. In Vitro Cell Dev. Biol. 28A, 595–602 [DOI] [PubMed] [Google Scholar]
- 15. Saraon P., Cretu D., Musrap N., Karagiannis G. S., Batruch I., Drabovich A. P., van der Kwast T., Mizokami A., Morrissey C., Jarvi K., and Diamandis E. P. (2013) Quantitative proteomics reveals that enzymes of the ketogenic pathway are associated with prostate cancer progression. Mol. Cell. Proteomics 12, 1589–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., and Maccoss M. J. (2009) Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J. Proteome Res 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Ponten F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 18. Drabovich A. P., Jarvi K., and Diamandis E. P. (2011) Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell. Proteomics 10, M110.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drabovich A. P., Dimitromanolakis A., Saraon P., Soosaipillai A., Batruch I., Mullen B., Jarvi K., and Diamandis E. P. (2013) Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci. Transl. Med. 5, 212ra160. [DOI] [PubMed] [Google Scholar]
- 20. Chang C. Y., Picotti P., Huttenhain R., Heinzelmann-Schwarz V., Jovanovic M., Aebersold R., and Vitek O. (2012) Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol. Cell. Proteomics 11, M111.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Madak-Erdogan Z., Charn T. H., Jiang Y., Liu E. T., Katzenellenbogen J. A., and Katzenellenbogen B. S. (2013) Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta, and their coregulators. Mol. Syst. Biol. 9, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davis P. L., Miron A., Andersen L. M., Iglehart J. D., and Marks J. R. (1999) Isolation and initial characterization of the BRCA2 promoter. Oncogene 18, 6000–6012 [DOI] [PubMed] [Google Scholar]
- 23. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 24. Whitfield M. L., George L. K., Grant G. D., and Perou C. M. (2006) Common markers of proliferation. Nat. Rev. Cancer 6, 99–106 [DOI] [PubMed] [Google Scholar]
- 25. Sahebjam S., Aloyz R., Pilavdzic D., Brisson M. L., Ferrario C., Bouganim N., Cohen V., Miller W. H. Jr., and Panasci L. C. (2011) Ki 67 is a major, but not the sole determinant of Oncotype Dx recurrence score. Br. J. Cancer 105, 1342–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Picotti P., Bodenmiller B., Mueller L. N., Domon B., and Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reddel R. R., Murphy L. C., and Sutherland R. L. (1983) Effects of biologically active metabolites of tamoxifen on the proliferation kinetics of MCF-7 human breast cancer cells in vitro. Cancer Res. 43, 4618–4624 [PubMed] [Google Scholar]
- 28. Brunner N., Boulay V., Fojo A., Freter C. E., Lippman M. E., and Clarke R. (1993) Acquisition of hormone-independent growth in MCF-7 cells is accompanied by increased expression of estrogen-regulated genes but without detectable DNA amplifications. Cancer Res. 53, 283–290 [PubMed] [Google Scholar]
- 29. Zuo X., Lee K., and Speicher DW (2004) Electrophoretic prefractionation for comprehensive analysis of proteomes. In: Speicher D. W., ed. Proteome Analysis: Interpreting the Genome, pp. 93–115, Elsevier Science, New York [Google Scholar]
- 30. Baron V. T., Pio R., Jia Z., and Mercola D. (2015) Early Growth Response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br. J. Cancer 112, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bacchus W., Aubel D., and Fussenegger M. (2013) Biomedically relevant circuit-design strategies in mammalian synthetic biology. Mol. Syst. Biol. 9, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin C. Y., Strom A., Vega V. B., Kong S. L., Yeo A. L., Thomsen J. S., Chan W. C., Doray B., Bangarusamy D. K., Ramasamy A., Vergara L. A., Tang S., Chong A., Bajic V. B., Miller L. D., Gustafsson J. A., and Liu E. T. (2004) Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 5, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welboren W. J., van Driel M. A., Janssen-Megens E. M., van Heeringen S. J., Sweep F. C., Span P. N., and Stunnenberg H. G. (2009) ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joseph R., Orlov Y. L., Huss M., Sun W., Kong S. L., Ukil L., Pan Y. F., Li G., Lim M., Thomsen J. S., Ruan Y., Clarke N. D., Prabhakar S., Cheung E., and Liu E. T. (2010) Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol. Syst. Biol. 6, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tullai J. W., Schaffer M. E., Mullenbrock S., Sholder G., Kasif S., and Cooper G. M. (2007) Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J. Biol. Chem. 282, 23981–23995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bourdeau V., Deschenes J., Laperriere D., Aid M., White J. H., and Mader S. (2008) Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 36, 76–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hah N., Danko C. G., Core L., Waterfall J. J., Siepel A., Lis J. T., and Kraus W. L. (2011) A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Louie M. C., McClellan A., Siewit C., and Kawabata L. (2010) Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol. Cancer Res. 8, 343–352 [DOI] [PubMed] [Google Scholar]
- 39. Caldon C. E., Sergio C. M., Schutte J., Boersma M. N., Sutherland R. L., Carroll J. S., and Musgrove E. A. (2009) Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol. Cell. Biol. 29, 4623–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bjornstrom L., and Sjoberg M. (2005) Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 19, 833–842 [DOI] [PubMed] [Google Scholar]
- 41. Song R. X., Barnes C. J., Zhang Z., Bao Y., Kumar R., and Santen R. J. (2004) The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor alpha to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 101, 2076–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jin K., Kong X., Shah T., Penet M. F., Wildes F., Sgroi D. C., Ma X. J., Huang Y., Kallioniemi A., Landberg G., Bieche I., Wu X., Lobie P. E., Davidson N. E., Bhujwalla Z. M., Zhu T., and Sukumar S. (2012) The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 2736–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osborne C. K., Shou J., Massarweh S., and Schiff R. (2005) Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clin. Cancer Res. 11, 865s–870s [PubMed] [Google Scholar]
- 44. Mohammed H., Russell I. A., Stark R., Rueda O. M., Hickey T. E., Tarulli G. A., Serandour A. A., Birrell S. N., Bruna A., Saadi A., Menon S., Hadfield J., Pugh M., Raj G. V., Brown G. D., D'Santos C., Robinson J. L., Silva G., Launchbury R., Perou C. M., Stingl J., Caldas C., Tilley W. D., and Carroll J. S. (2015) Progesterone receptor modulates ERalpha action in breast cancer. Nature 523, 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ngwenya S., and Safe S. (2003) Cell context-dependent differences in the induction of E2F-1 gene expression by 17 beta-estradiol in MCF-7 and ZR-75 cells. Endocrinology 144, 1675–1685 [DOI] [PubMed] [Google Scholar]
- 46. Ji J.-Y., and Dyson N. (2010) Interplay Between Cyclin-Dependent Kinases and E2F-Dependent Transcription. In: Enders G. H., ed. Cell Cycle Deregulation in Cancer, pp. 23–41, Springer; New York [Google Scholar]
- 47. Patwardhan S., Gashler A., Siegel M. G., Chang L. C., Joseph L. J., Shows T. B., Le Beau M. M., and Sukhatme V. P. (1991) EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene 6, 917–928 [PubMed] [Google Scholar]
- 48. Zwang Y., Sas-Chen A., Drier Y., Shay T., Avraham R., Lauriola M., Shema E., Lidor-Nili E., Jacob-Hirsch J., Amariglio N., Lu Y., Mills G. B., Rechavi G., Oren M., Domany E., and Yarden Y. (2011) Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Mol. Cell 42, 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Russo M. W., Sevetson B. R., and Milbrandt J. (1995) Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 92, 6873–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svaren J., Sevetson B. R., Apel E. D., Zimonjic D. B., Popescu N. C., and Milbrandt J. (1996) NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol. Cell. Biol. 16, 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Silverman E. S., Khachigian L. M., Santiago F. S., Williams A. J., Lindner V., and Collins T. (1999) Vascular smooth muscle cells express the transcriptional corepressor NAB2 in response to injury. Am. J. Pathol. 155, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inoue A., Omoto Y., Yamaguchi Y., Kiyama R., and Hayashi S. I. (2004) Transcription factor EGR3 is involved in the estrogen-signaling pathway in breast cancer cells. J Mol. Endocrinol. 32, 649–661 [DOI] [PubMed] [Google Scholar]
- 53. Kumbrink J., Kirsch K. H., and Johnson J. P. (2010) EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J. Cell. Biochem. 111, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Abdulkadir S. A., Carbone J. M., Naughton C. K., Humphrey P. A., Catalona W. J., and Milbrandt J. (2001) Frequent and early loss of the EGR1 corepressor NAB2 in human prostate carcinoma. Hum. Pathol. 32, 935–939 [DOI] [PubMed] [Google Scholar]
- 55. Qu Z., Wolfraim L. A., Svaren J., Ehrengruber M. U., Davidson N., and Milbrandt J. (1998) The transcriptional corepressor NAB2 inhibits NGF-induced differentiation of PC12 cells. J. Cell Biol. 142, 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suzuki T., Inoue A., Miki Y., Moriya T., Akahira J., Ishida T., Hirakawa H., Yamaguchi Y., Hayashi S., and Sasano H. (2007) Early growth responsive gene 3 in human breast carcinoma: a regulator of estrogen-meditated invasion and a potent prognostic factor. Endocr. Relat. Cancer 14, 279–292 [DOI] [PubMed] [Google Scholar]
- 57. Stricker R., Eberhart R., Chevailler M. C., Quinn F. A., Bischof P., and Stricker R. (2006) Establishment of detailed reference values for luteinizing hormone, follicle stimulating hormone, estradiol, and progesterone during different phases of the menstrual cycle on the Abbott ARCHITECT analyzer. Clin. Chem. Lab. Med. 44, 883–887 [DOI] [PubMed] [Google Scholar]
- 58. Haynes B. P., Viale G., Galimberti V., Rotmensz N., Gibelli B., A'Hern R., Smith I. E., and Dowsett M. (2013) Expression of key oestrogen-regulated genes differs substantially across the menstrual cycle in oestrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat 138, 157–165 [DOI] [PubMed] [Google Scholar]
- 59. Radon T. P., Massat N. J., Jones R., Alrawashdeh W., Dumartin L., Ennis D., Duffy S. W., Kocher H. M., Pereira S. P., Guarner Posthumous L., Murta-Nascimento C., Real F. X., Malats N., Neoptolemos J., Costello E., Greenhalf W., Lemoine N. R., and Crnogorac-Jurcevic T. (2015) Identification of a Three-Biomarker Panel in Urine for Early Detection of Pancreatic Adenocarcinoma. Clin. Cancer Res. 21, 3512–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drabovich A. P., Martinez-Morillo E., and Diamandis E. P. (2015) Toward an integrated pipeline for protein biomarker development. Biochim. Biophys. Acta 1854, 677–686 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.