Abstract
Dysregulated proteolysis represents a hallmark of numerous diseases. In recent years, increasing number of studies has begun looking at the protein termini in hope to unveil the physiological and pathological functions of proteases in clinical research. However, the availability of cryopreserved tissue specimens is often limited. Alternatively, formalin-fixed, paraffin-embedded (FFPE) tissues offer an invaluable resource for clinical research. Pathologically relevant tissues are often stored as FFPE, which represent the most abundant resource of archived human specimens. In this study, we established a robust workflow to investigate native and protease-generated protein N termini from FFPE specimens. We demonstrate comparable N-terminomes of cryopreserved and formalin-fixed tissue, thereby showing that formalin fixation/paraffin embedment does not proteolytically damage proteins. Accordingly, FFPE specimens are fully amenable to N-terminal analysis. Moreover, we demonstrate feasibility of FFPE-degradomics in a quantitative N-terminomic study of FFPE liver specimens from cathepsin L deficient or wild-type mice. Using a machine learning approach in combination with the previously determined cathepsin L specificity, we successfully identify a number of potential cathepsin L cleavage sites. Our study establishes FFPE specimens as a valuable alternative to cryopreserved tissues for degradomic studies.
Formalin fixation and paraffin embedment are the prevailing methods to preserve tissues for routine clinical diagnostics and archival purposes. As such, formalin-fixed, paraffin-embedded (FFPE)1 specimens represent a large collection of clinically annotated samples that are stored for long periods at room temperature. While many still consider cryopreserved specimens as the gold standard in clinical research, the recruitment of cryopreserved tissues in sufficient numbers for robust study designs is challenging. FFPE tissues offer an attractive alternative for the retrospective analysis of pathological processes.
Proteomic analysis of FFPE tissues has gained increasing interest since it was first presented (1). Studies have successfully demonstrated that FFPE tissues are amenable to all widely applied mass spectrometry (MS) platforms, including reversed-phase liquid tandem MS, matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF), and surface-enhanced laser desorption ionization TOF analyses (2), as well as MALDI imaging (3, 4). Interestingly, protein identification numbers and proteome coverage were found to be equivalent for FFPE and cryopreserved tissue, and FFPE tissues can be analyzed to a depth of up to 10 000 proteins per sample (5). Similarly, FFPE and cryopreserved tissues do not differ with regard to localization and function of identified proteins. Moreover, studies have also shown that identified protein subsets share a substantial overlap (6, 7) and that utilization of different FFPE processes does not impede proteomic analysis (8).
While formaldehyde is known to fix proteins in tissue by reacting with basic amino acids (such as lysine, asparagine, and glutamine (9)) to form methylol adducts or reacting with carbonyl functional groups to form imine adducts between amines and aldehydes, these modifications are rarely detected in FFPE proteomes (10). In fact, it is known that very few carryovers of the formalin fixation process are retained following protein extraction for proteomics analysis. However, a minor shift of the arginine to lysine ratio has been observed, indicating the persistence of yet undefined modifications or cross-links (8, 11, 12). Nevertheless, analysis of common posttranslational modifications such as phosphorylation and glycosylation showed equal preservations in FFPE and cryopreserved tissue specimens (13).
Proteolysis is an irreversible posttranslational modification, often generating stable cleavage products with novel functionality or cell-contextual localization (14). Dysregulated proteolytic processing is a hallmark feature in numerous diseases (14, 15). Thus, it is not surprising that many have turned to proteomics for the elucidation of the precise role of specific protease(s) as well as the identification their physiological substrates. At present, the majority of proteomics-based approaches for the system-wide analysis of proteolytic processing rely on the enrichment and subsequent investigation protein termini with the most widely used techniques focusing on amino termini (14, 15). This is witnessed by the number of established strategies, which have been developed to investigate protein N termini (14). Typically, terminal and side-chain amino groups of full-length proteins are chemically modified, followed by protein digestion using trypsin to generate internal peptides that possess free amino termini. This chemical difference (“free” versus “protected” amino termini) between protein N termini and internal peptides is used to specifically enrich for native N-terminal peptides with subsequent LC-MS/MS analysis. Commonly used enrichment strategies are based on differential chromatography (combined fractional diagonal chromatography (16) and charge-based fractional diagonal chromatography) (17)), charge-reversal enrichment of protein amino termini termini (18), or usage of a high-molecular weight, amine-reactive polymer in combination with ultrafiltration (terminal amine isotopic labeling of substrates (TAILS) (19)).
To date, N-terminomics investigation from FFPE tissues has not yet been probed, perhaps owing to an existing reservation of whether FFPE specimens are amenable to degradomic strategies, as well as skepticism concerning their ability to preserve the “proteolytic signature” of biological specimens. In this study, we have developed a TAILS-based workflow for the degradomic investigation of FFPE specimens. Using corresponding cryopreserved specimens, we show that FFPE processing does not damage protein amino termini and resulting N-terminal peptides do not retain any carryover from the formalin fixation process after N-terminal enrichment. Furthermore, we demonstrate the feasibility of quantitative degradomic studies by comparing liver FFPE specimens from cathepsin L deficient and correspondingo wild-type mice. As a perspective, our study highlights the amenability of FFPE tissues to terminomic profiling and thus enables the potential in harnessing FFPE specimens from the clinical archives as a valuable source for the investigation of disease-associated proteolysis.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
A total of three sample sets of FFPE mice liver tissues (male, 6-months old C57BL/6 strain) were analyzed and described in Results. Each sample set comprised of three biological replicates. Experimental controls from each sample set include wild-type tissues (comparison with knock-out tissues) or cryopreserved tissues (comparison with FFPE tissues) or nonlabeled samples (comparison with 13COD2 formaldehyde-labeled samples). Three biological replicates were investigated in combination with a label-switch between 12COH2 formaldehyde and 13COD2 formaldehyde to provide statistical significance. Statistical analysis using linear models for microarray data (Limma) (20, 21) allows for the use of linear models to assess differential expression in the context of multifactor designed experiments. In addition, Limma has the ability to analyze complex experiments involving comparisons between many peptides simultaneously in a small sample size.
Processing of Tissue Specimens
For formalin fixation and paraffin embedment, whole livers were harvested from male, 6 months old C57BL/6 wild-type mice or male, 6 months old C57BL/6 mice lacking cathepsin L (Ctsl−/−) and fixed in 4% (v/v) formaldehyde solution in phosphate buffered saline for 16 h. After formalin fixation, tissue specimens were processed using a xylene-based STP 120 Spin Tissue Processor (Thermo Scientific, Bremen, Germany) and embedded in standard paraffin blocks. Subsequently, 30 tissue sections at 10 μm thickness were cut from each paraffin block. All FFPE slides were deparaffinized using four times xylene for 5 min, two times with 100% ethanol for 1 min, one time with 96% ethanol for 1 min, one time with 70% ethanol for 1 min, one time with 50% ethanol for 1 min, and one time with distilled water for 5 min. For cryopreservation, fresh livers were withdrawn from mouse and were immediately snap-frozen in liquid nitrogen. Cryopreserved specimens were stored at -80°C.
Protein Extraction and Sample Preparation
Following deparaffinization, FFPE tissue sections were incubated in 100 mm 4–2(2- hydroxyethl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 4% (w/v) sodium dodecyl sulfate (SDS), 50 mm dithiothreitol (DTT) for 1 h at 95°C with gentle agitation. For cryopreserved samples, tissues were homogenized using Ultra-Turrax T8 Homogenizer (IKA-Werke, Wilmington, NC, USA) in 200 mm HEPES, pH 8.0, and 4% (w/v) SDS following by heating at 95 °C for 30 min with gentle agitation. Lysates from cryopreserved tissues were reduced using 10 mm DTT at 60 °C for 30 min. FFPE and cryopreserved protein lysates were cooled and alkylated using 20 mm of iodoacetamide for 30 min in the dark, followed by centrifugation at 14,000 × g for 15 min. Extracted proteins in the supernatant were precipitated using nine volumes of ice cold acetone and one volume of ice cold methanol at –80°C for 2 h. Precipitated proteins were harvested using centrifugation at 4,500 × g for 2 h at 4°C. Resulting protein pellets were washed four times with ice cold methanol and then resolubilized in ice-cold 100 mm NaOH by water-bath ultrasonication at 4°C. The solution was brought to pH 7.5–8.0 by the addition of 200 mm HEPES free acid. Protein concentration was determined using bicinchoninic acid protein assay (Thermo Fisher).
N-Terminal Amino Isotopic Labeling of Substrates
Enrichment of protein N termini using terminal amine isotopic labeling of substrates (TAILS) was conducted as described previously (19). Briefly, extracted proteins from deparaffinized FFPE tissues and cryopreserved tissues were dimethylated using 40 mm 12COH2 formaldehyde or 40 mm 13COD2 formaldehyde in the presence of 40 mm sodium cyanoborohydride at 37 °C for 16 h. Excess formaldehyde was quenched by the addition of 50 mm tris(hydroxymethyl)aminomethane (TRIS). Following the amine protection step, proteins were precipitated using nine volumes of ice-cold acetone and one volume of ice-cold methanol at -80°C for 2 h. Precipitated proteins were harvested using centrifugation at 4,500 × g for 2 h at 4°C. Protein pellets were washed four times with ice-cold methanol and then redissolved in ice-cold 100 mm NaOH by water-bath ultrasonication at 4°C. The solution was brought to pH 7.50–8.0 by the addition of 200 mm HEPES free acid and. Protein concentration was determined using bicinchoninic acid protein assay. Proteins were digested using sequencing-grade trypsin (Worthington Biochemical Corp, Lakewood, NJ) in a 100:1 (w/w) ratio at 37°C pH 7.0 for 16 h. Resulting free neo-N termini generated from the tryptic digestion were captured by hyperbranched polyglycerol-aldehydes (HPG-ALD) polymers in the presence of 40 mm sodium cyanoborohydride at 37°C for 16 h. Following capture, HPG-ALD hyperbranched polymers were saturated using 50 mm glycine for 1 h at room temperature and subsequently removed by ultracentrifugation using 10 kDa MWCO Microcon spin filters (Milipore, Billerica, MA). Collected flow-through fractions containing N-terminal peptides were desalted using C-18 Sep Pak (Waters, Milford, MA) and fractionated on high-performance liquid chromatography (SCX-HPLC) coupled to a strong cation exchange column (PolyLC, Columbia, MD). Buffer A consisted of 5 mm KH2PO4 and 25% (v/v) acetonitrile (pH 2.7), and buffer B consisted of 5 mm KH2PO, 1 m KCl, and 25% acetonitrile (pH 2.7). Peptides were eluted in a linear gradient with increasing concentration of buffer B. Resulting fractions were collected, desalted using self-packed C18 STAGE tips (Empore, St. Paul, MN) (22), and analyzed by LC-MS/MS.
LC-MS/MS and Data Analysis
Samples were analyzed on an Orbitrap XL (Thermo Scientific) or an Orbitrap Q-Exactive plus (Thermo Scientific) mass spectrometer. The Orbitrap XL was coupled to an Ultimate3000 micro pump (Thermo Scientific). Buffer A was 0.5% (v/v) acetic acid, buffer B 0.5% (v/v) acetic acid in 80% acetonitrile (HPLC grade). Liquid phases were applied at a flow rate of 300 nl/min with an increasing gradient of organic solvent for peptide separation. Reprosil-Pur 120 ODS-3 (Dr. Maisch, Ammerbuch-Entringen, Germany) was used to pack column tips of 75 μm inner diameter and 11 cm length. The MS was operated in data-dependent mode, and each MS scan was followed by a maximum of five MS/MS scans. The Q-Exactive plus mass spectrometer was coupled to an Easy nanoLC 1000 (Thermo Scientific) with a flow rate of 300 nl/min. Buffer A was 0.5% (v/v) formic acid, and buffer B was 0.5% (v/v) formic acid in acetonitrile (water and acetonitrile were at least HPLC gradient grade quality). A gradient of increasing organic proportion was used for peptide separation (5–40% (v/v) acetonitrile in 80 min). The analytical column was an Acclaim PepMap column (Thermo Scientific), 2 μm particle size, 100 Å pore size, length 150 mm, inner diameter 50 μm. The mass spectrometer operated in data-dependent acquisition mode with a top 10 MS/MS method at a mass range of 300–2000 Da.
MS data were converted to mzML format (23) using ProteoWizard (24). The complete data analysis was performed with a fully automated workflow within the OpenMS framework (25) (Supplemental Fig. 1). Peptide sequences were identified by MS-GF+ (26) peptide search engine with decoy search strategy. A complete mouse proteome sequence file was downloaded from UniProt (27) on October 16, 2011, comprising 44,819 protein sequences. It was appended to an equal number of randomized sequences, derived from the original mouse proteome entries. Semi Arg-C specificity was used as search parameters with mass tolerance set at 20 ppm for precursor ions. Static modifications applied include cysteine carboxyamidomethylation (+57.02 Da), lysine and N-terminal dimethylation (12COH2 formaldehyde +28.03 Da or 13COD2 formaldehyde +34.06 Da, if applicable), N-terminal monomethylation (12COH2 formaldehyde +14.02 Da or 13COD2 formaldehyde +17.03 Da, if applicable), and N-terminal acetylation (+42.01 Da). The MS-GF+ results were further validated by OpenMS at a confidence level greater than 95%. The relative quantification for each peptide was calculated using the FeatureFinderMultiplex tool (28) (as part of OpenMS). For cleavage events in which peptides were only present in wild-type or Ctsl−/− condition, a ratio of 2−10 or 210 was assigned, respectively. A list of potential cathepsin L substrates has been predicted on the basis of a dataset for cathepsin L cleavage specificity from the MEROPS peptidase database. The method is based on an efficient string kernel implemented in the Explicit Decomposition with Neighborhood library (DOI:10.5281/zenodo.27945). The method uses the notion of k-mers with gaps to enumerate all possible substrings of increasing orders (starting from monomers up to eight-mers), which are used as features in a linear binary classification estimator. The full computational pipeline, which allows a good estimate of the likelihood of cleavage target sites, is available under the Galaxy open, web-based platform for data-intensive biomedical research (https://toolshed.g2.bx.psu.edu).
Data Availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (29) PRoteomics IDEntifications (PRIDE) partner repository with dataset identifier PXD002847 (reviewer account details—username: reviewer38683@ebi.ac.uk password: mIR5jHGS). Search results (pepXML format), along with .raw and mzML files, have been deposited. Annotated spectra are provided via MS-Viewer (30) with URLs being listed in Supplemental Table 1.
RESULTS AND DISCUSSION
Enrichment of N-terminal Peptides from FFPE Specimens
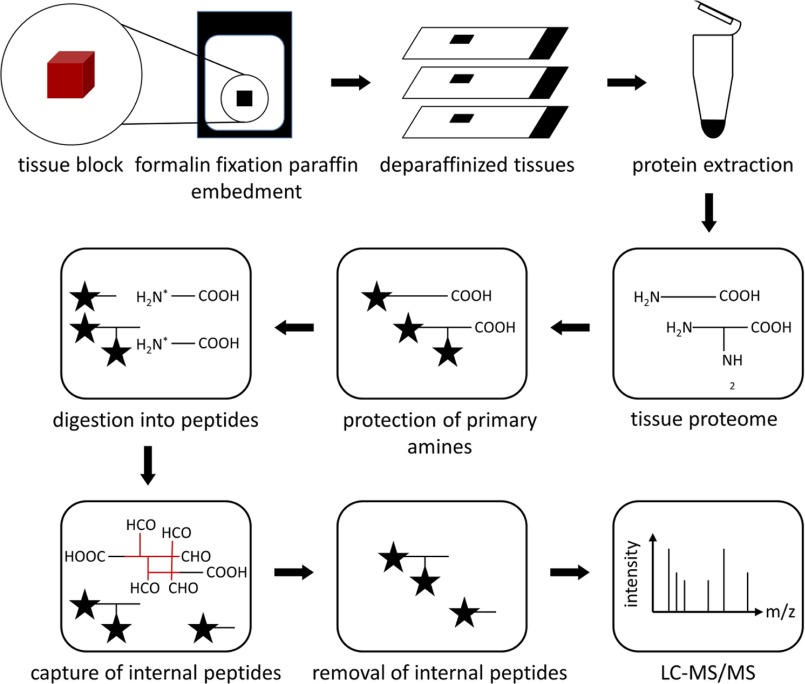
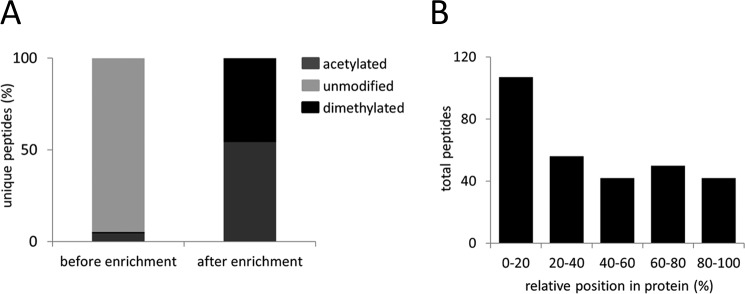
This study aims at the establishment of a protocol for the enrichment of N termini from formalin-fixed, paraffin embedded tissues for mass spectrometry analysis (Fig. 1). Proteins were extracted from deparaffinized tissues, using an extraction buffer containing HEPES as buffering agent, SDS as denaturing agent, and DTT as reducing agent, together with heating at 95 °C for an extended period of time in order to revert the chemical modifications that are formed during formalin fixation. Once proteins were successfully extracted, the enrichment of N termini for mass spectrometry analysis was performed according to the original TAILS workflow (19). The technique depletes internal peptides after tryptic digest thereby enriching for naturally occurring N termini. Prior to the enrichment step, mass spectrometry analysis identified the majority of the N termini as being unmodified, while only few dimethylated and acetylated N termini were detected (Fig. 2A) On the other hand, when the same sample was subjected to N-terminal enrichment using TAILS, unmodified N termini were completely depleted (Fig. 2A). The identified dimethylated peptides were mainly derived from the first 20% of the full length protein chain (Fig. 2B), similarly observed in a previous study in canonical positional profiling of N termini from fresh or cryopreserved tissue or cultured cells (31).
Fig. 1.
Schematic of protein N termini enrichment from formalin-fixed paraffin embedded (FFPE) tissue specimen. FFPE tissues are deparaffinized prior to protein extraction. Protein N termini are enriched using terminal amine isotopic labeling of substrates (TAILS).
Fig. 2.
N termini peptides from FFPE liver tissue of C57BL/6 wild-type mouse. (A) Composition of acetylated, chemically dimethylated (naturally unmodified protein N termini) and unmodified peptides (internal peptides with neo-N termini from protein digestion) before and after N-terminal enrichment. (B) Positional clustering of all identified dimethylated peptides showing relative position in protein.
N-terminal Coverage from Cryopreserved and FFPE Specimens
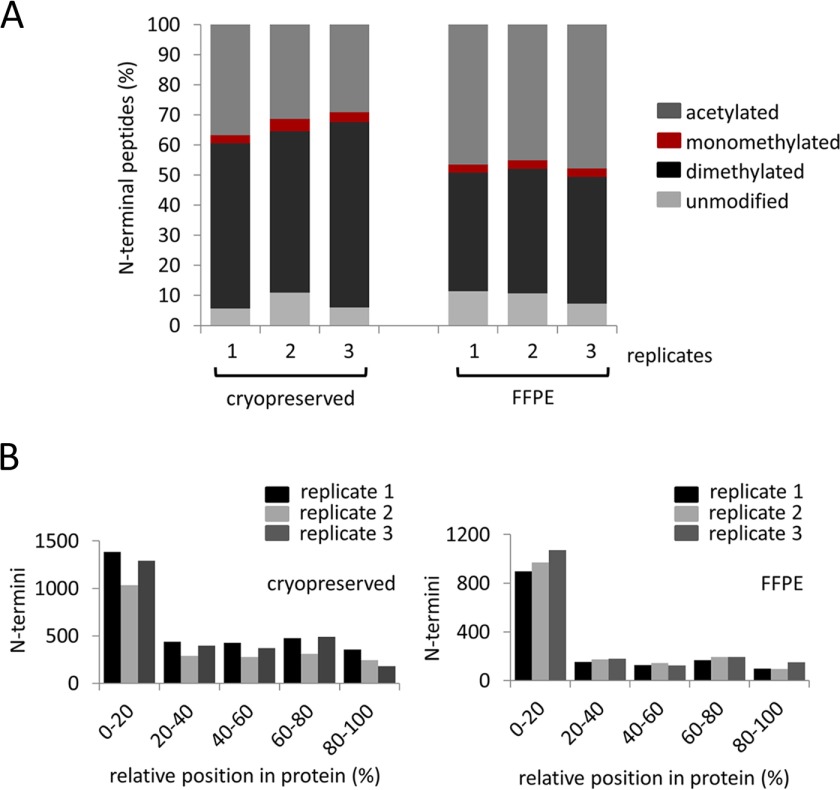
While cryopreserved tissues are immediately snap-frozen upon harvest, formalin fixation/paraffin embedment of tissues involves extended workflows from tissue harvest to paraffin block embedment and histoprocessing. Moreover, FFPE specimens were stored at room temperature for extended periods of time. To gain insight into the differential status of N termini in cryopreserved and formalin-fixed tissues, the N-terminal enrichment procedure was applied to assess N-terminal peptides from the different preservation conditions from liver tissue of C57BL/6 wild-type mouse. LC-MS/MS analysis yielded comparable numbers of N termini identifications (unmodified, acetylated and dimethylated) among the three biological replicates in each of the two differentially processed tissues (Supplemental Tables 2–7). 3,000–3,800 N termini were identified in cryopreserved specimens, while 2,000–2,300 N termini were identified in formalin-fixed tissues. Incomplete overlap between proteomic experiments is an intrinsic characteristic when comparing between different biological replicates (32). In this study, a total of 987 N termini was identified among the three biological replicates of FFPE tissue, while 1,199 overlapping N termini were identified among the cryopreserved counterparts. From these, a total of 486 N termini were shared between both preservation methods (among all replicates of cryopreserved and FFPE tissues, respectively). Previous studies indicate that proteins extracted from FFPE tissues are susceptible to a +12 Da addition at N termini, lysine, tryptophan, tyrosine, serine, and threonine residues, as well as a +30 Da addition at cysteine, histidine, lysine, and arginine residues (20, 33, 34). In both cryopreserved and FFPE samples, the fraction of peptides displaying these modifications remained below the 5% false discovery rate (data not shown).
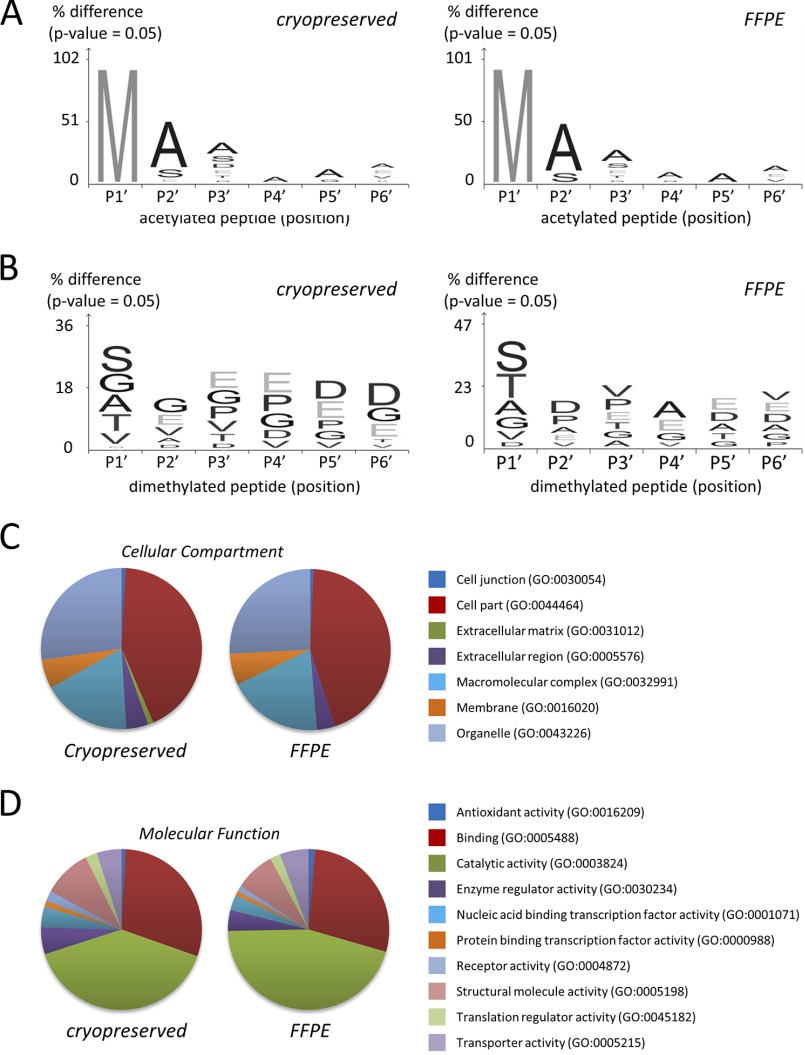
Acetylation is the most prevalent native N-terminal modification, which occurs in a posttranslational manner while N-terminal dimethylation is introduced during the TAILS procedure to protect free N termini that are often generated by endogenous proteolysis (14, 35). The ratio of acetylated to dimethylated N termini differs to a limited extent between cryopreserved and FFPE specimens. In cryopreserved samples, 58.7 ± 4.3% of N termini were chemically dimethylated; this number was 42.1 ± 1.4% for FFPE specimens (Fig. 3A). In both cryopreserved and FFPE specimens, dimethylated N termini mainly map to the first 20% of the full length protein chain (Fig. 3B) in good correspondence with the canonical positional profile of N termini (31). The increased proportion of dimethylated N termini in cryopreserved samples may indicate that formalin fixation and paraffin embedment prevents chemical dimethylation of a limited number of protein termini. On the other hand, the situation may also be indicative of increased proteolysis during cryopreservation. Noteworthy, the elevated number of dimethylated N termini in cryopreserved samples coincides with an increased fraction of termini that map to internal positions (Fig. 3B), thus signaling aberrant cleavage within the protein chain. In another recent degradomic study of mouse embryonic kidney, we also observed a reduced level of dimethylated peptides when compared with acetylated peptides (18). By introducing a novel gel-based enrichment of N termini, we observed a significantly reduced fraction of peptides mapping to internal positions (18). For this reason, we consider it likely that the increased proportion of dimethylated N termini in cryopreserved samples is indicative of increased proteolysis during cryopreservation rather than signifying limited reactivity of N termini from FFPE samples. Further studies have also indicated that proteins are not proteolytically damaged during formalin fixation and histoprocessing (8). Nevertheless, the N-terminal peptides (acetylated and dimethylated) identified in cryopreserved or FFPE tissues show a high degree of similarity. For acetylated N termini, residues in P1′ are predominantly methionine in peptides derived from both cryopreserved and FFPE tissues, along with a consistent alanine fingerprint in position P2′-P6′ (Fig. 4A). On the other hand, for N-terminally dimethylated peptides, residues, such as serine, glycine, valine, alanine, and threonine, are equally observed at position P1′ in both sample types (Fig. 4B). N termini from both FFPE and cryopreserved tissues map to proteins, which represent similar cellular components and molecular functions (Figs. 4B and 4C). The congruence in the proteome analysis of FFPE and cryopreserved samples is in line with an earlier study (8).
Fig. 3.
N termini peptides from cryopreserved and FFPE liver tissues of C57BL/6 wild-type mouse. (A) Composition of acetylated, chemically dimethylated (naturally unmodified protein N termini) and unmodified peptides (internal peptides with neo-N termini from protein digestion) from three biological replicates (n = 3). (B) Positional clustering of all identified dimethylated and acetylated peptides showing relative position in protein from cryopreserved and FFPE tissues.
Fig. 4.
Characterization of N-terminal peptides from cryopreserved and FFPE liver tissues of C57BL/6 wild-type mouse. Visualization of sequence specificity of the (A) N-α acetylated and (B) N-terminally dimethylated peptides consistently identified in all biological replicates of either cryopreserved or FFPE tissues, respectively (n = 3). Sequence logos were generated using iceLogo (60). Gene Ontology database analysis of (C) cellular component and (D) molecular function of N-terminal peptides consistently identified in all biological replicates either cryopreserved or FFPE tissues, respectively (n = 3).
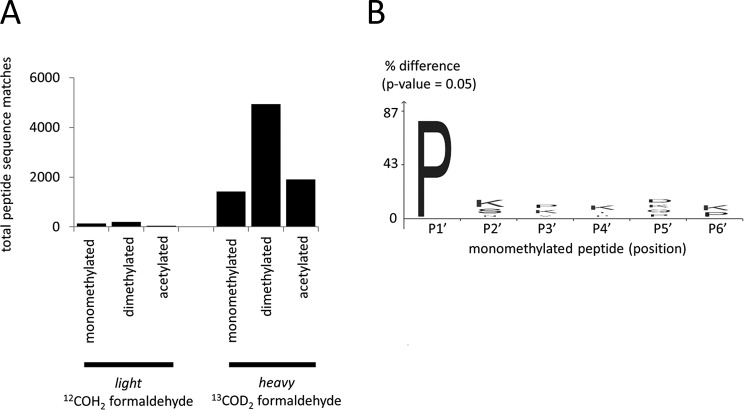
To specifically probe for putative formalin carryover, we further performed the TAILS N-terminomic analysis from FFPE specimens using only “heavy” 13COD2 formaldehyde. This setup clearly distinguishes light, carryover formaldehyde. We detected almost exclusively N termini with the heavy form of formaldehyde labeling (Fig. 5A, Supplemental Table 8), with the large population of heavy dimethylated peptides strongly indicating the absence of significant formalin carryover. Monomethylated N termini were also detected, which is attributed to N-terminal monomethylated proline-starting peptide sequences (Fig. 5B), as reported previously in (36, 37). In total, the very low detection numbers of the 12COH2 formaldehyde “light” counterparts remain within the 5% false discovery rate margin, which we employed for peptide identifications. Evidently, our technique is not biased by carryover of light formaldehyde from formalin fixing of tissues. Therefore, we conclude that FFPE specimens are readily amenable to N-terminal degradomic profiling.
Fig. 5.
N termini peptides from 13COD2 formaldehyde heavy labeled proteins from FFPE liver tissue of C57BL/6 wild-type mouse. (A) Composition of light and heavy acetylated, chemically dimethylated (naturally unmodified protein N termini) and monomethylated N termini. (B) Visualization of identified monomethylated N termini being predominantly proline residue. Sequence logo was generated using iceLogo (60).
Altered N-Terminal Processing in the Liver of Cathepsin L Deficient Mice
The strength of the TAILS procedure is its suitability for comparative degradomic studies by straightforward incorporation of different formaldehyde isotopes as an integral part of the procedure. To assess compatibility of FFPE specimens with quantitative-comparative N-terminal profiling, we chose FFPE liver samples of cathepsin L deficient and corresponding wild-type mice. We previously showed that cathepsin L deletion in mice (Ctsl−/−) results in a fundamentally perturbed protease network with a large number of downstream and secondary effects (38).
Several quantitative proteomic techniques have been utilized to investigate FFPE tissues, such as trypsin-mediated 18O labeling (2), isobaric tag for relative and absolute quantitation (iTRAQ) (39–41), label-free quantitation (42–46), and chemical dimethylation (47). The successful application of iTRAQ and chemical dimethylation are especially noteworthy, as these labeling techniques target primary amines, which is of importance to N-terminal degradomic analysis.
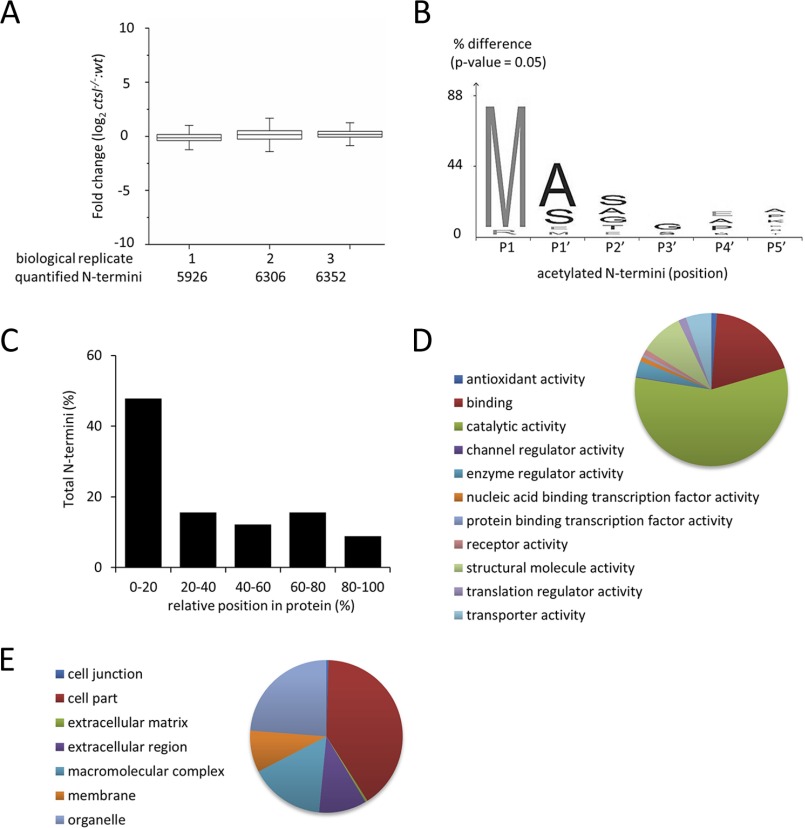
Stable isotopic formaldehyde labeling and TAILS were applied for the quantitative N-terminomic comparison of formalin-fixed liver tissues from wild-type and cathepsin L deficient mice. Three formalin-fixed liver tissues of cathepsin L deficient and wild-type mice were compared, incorporating a label-switch strategy. A total of 8,061 nonredundant N termini (monomethylated, dimethylated, and acetylated) were quantified in all three biological replicates; with 5,926, 6,306, and 6,352 N-terminal peptides identified in individual biological replicates, respectively (Fig. 6A) (Supplemental Tables 9, 10, and 11). The distribution of fold changes of all three replicates show a near normal distribution, with the majority N-terminal peptides being equally abundant in wild-type and Ctsl−/− liver tissues. The characterization on the specificity of N-α acetylation in these FFPE tissues showed that acetylated N-terminal peptides have a preference for alanine, serine, and glutamate residues in P1′ and also to a lesser extent in P2′ (Fig. 6B). These results are in direct agreement with the prototypical profile of N-terminal acetylation and from previous studies on N-terminal acetylation in murine skin (35, 48).
Fig. 6.
Identification of N-terminal peptides from FFPE liver tissues of C57BL/6 wild-type mice and C57BL/6 cathepsin L (Ctsl−/−) knock out mice. (A) Fold change distribution and Shapiro–Wilk normalization test (p value) of acetylated N termini and chemically dimethylated (naturally unmodified) N termini from three biological replicates (n = 3). (B) Visualization of N-α acetylation pattern in cathepsin L deficient tissue. Sequence logo was generated using iceLogo (60). (C) Positional clustering of acetylated and chemically dimethylated N termini from three biological replicates (n = 3). Gene Ontology database analysis of (D) molecular function and (E) cellular components of N-terminal peptides consistently identified all biological replicates (n = 3).
The identified monomethylated, dimethylated, and acetylated peptides were mainly attributed to the first 20% of the full length protein chain (Fig. 6C). A total of 1,720 mono- and dimethylated N termini (matched to 1,713 mouse proteins) were consistently identified in all three biological replicates. Gene ontology annotation for molecular functions classified these peptides to be predominantly involved in binding and catalytic activities (Fig. 6D) while cellular compartmental annotation showed that these peptides are mostly localized within intracellular compartments (Fig. 6E).
We are predominantly interested in cleavage events that depend on the presence of cathepsin L. Statistical analysis of mono- and dimethylated N termini using a moderated t test based on linear model for microarray data (Limma) (20, 21) combined with Benjamini–Hochberg procedure of 5% false discovery rate (n = 3) indicated that 205 peptides showed significant reduction in abundance when comparing Ctsl−/− versus wild-type liver tissues (Supplemental Table 12). Among these peptides, ten N-terminal peptides mapped to the postremoval of initiator methionine, ten stem from the removal of a signal peptide domain, and five stem from the removal of a transit peptide domain while the remaining 187 peptides stem from aberrant cleavage within the protein chain (Fig. 6D). Cathepsin l-mediated cleavage is primarily guided by a strong preference for aromatic and aliphatic residues in P2 with limited prime-site specificity contributions (33). Previous studies using Ctsl−/− mice have shown the involvement of this protease in the regulation in cardiac homeostasis (49–51), immune system (52, 53), hormonal processing (54, 55), and tumorigenesis (56–59). We employed an artificial neural network (machine learning) approach to distinguish significantly downregulated cleavage sites that adhere to the annotated cathepsin L specificity. For the machine learning process, we employed cathepsin L cleavage sites from MEROPS as training data. This approach yielded a list of 23 potential substrates for cathepsin L in the FFPE liver tissues of Ctsl−/− mice (Table I). Cathepsin L dependent proteolytic processing for some of these proteins was previously observed in murine skin (35), namely protein disulfide-isomerase, ATP synthase subunit beta, alpha-enolase, and cytoplasmic actin 1. While these substrate candidates are predominantly localized in the cytoplasm, cathepsin L is a lysosomal protease. We consider it likely that autophagic processes participate in delivering the aforementioned substrate candidates to the endolysosomal system.
Table I. List of potential cathepsin L substrates generated using artificial machine learning prediction with a training dataset for cathepsin L cleavage specificity from MEROPS peptidase database.
| Non-prime Sequence | Prime Sequence | UniProt | Protein Name | Average log2 (Ctsl−/−/wt) | Position | Length | Machine learning score |
|---|---|---|---|---|---|---|---|
| ADIALVELLY | HVEELDPGVVDNFPLLKALR | P30115 | Glutathione S-transferase A3; | −10.0 | 166 | 221 | 7.98 |
| FEESFQKALR | MCHPSVDGFTPR | Q8C196 | Carbamoyl-phosphate synthase (ammonia), mitochondrial; | −10.0 | 815 | 1500 | 6.85 |
| AEGFKGKILF | IFIDSDHTDNQR | P09103 | Protein disulfide-isomerase; | −10.0 | 291 | 509 | 6.81 |
| KSGENFKLLY | DLADQLHAAVGASR | Q99LC5 | Electron transfer flavoprotein subunit alpha, mitochondrial; | −10.0 | 236 | 333 | 6.13 |
| AMDGTEGLVR | GQKVLDSGAPIKIPVGPETLGR | P56480 | ATP synthase subunit beta, mitochondrial; | −10.0 | 122 | 529 | 5.70 |
| PSPSPSPSLS | STQSAVSKAGAGAVVPKLSHLPR | Q9DC70 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 7, mitochondrial; | −10.0 | 46 | 224 | 5.63 |
| DLYTAKGLFR | AAVPSGASTGIYEALELR | P17182 | Alpha-enolase; | −10.0 | 33 | 434 | 5.58 |
| EVGALAKVLR | LFEENEINLTHIESR | P16331 | Phenylalanine-4-hydroxylase; | −10.0 | 54 | 453 | 4.19 |
| EHPGGEEVLR | EQAGGDATENFEDVGH | P56395 | Cytochrome b5; | −10.0 | 53 | 134 | 3.45 |
| EHPGGEEVLR | EQAGGDATENFEDVGHSTDAR | P56395 | Cytochrome b5; | −10.0 | 53 | 134 | 3.45 |
| CDVDIRKDLY | ANTVLSGGTTMYPGIADR | P60710 | Actin, cytoplasmic 1; | −10.0 | 295 | 375 | 2.55 |
| SLLQQQKTSR | SNMDNMFESYINNLR | P11679 | Keratin, type II cytoskeletal 8; | −10.0 | 140 | 490 | 2.40 |
| PFSQHVRRLR | SSITPGTVLIILTGR | P47911 | 60S ribosomal protein L6; | −10.0 | 150 | 296 | 2.33 |
| DILNMDKTLK | GLNSDSVTEETLR | Q8C196 | Carbamoyl-phosphate synthase (ammonia), mitochondrial; | −0.93 | 893 | 1500 | 2.16 |
| LQDCMSKMQR | MVQESSSGGLLDR | Q571F8 | Glutaminase liver isoform, mitochondrial; | −10.0 | 118 | 602 | 1.69 |
| DTDDTATALR | EAQEEVGLHPH | Q99P30 | Peroxisomal coenzyme A diphosphatase NUDT7; | −10.0 | 92 | 236 | 1.43 |
| TKYPQLLSGIR | GISEETTTGVHNLY | P50247 | Adenosylhomocysteinase; | −10.0 | 152 | 432 | 0.83 |
| FMAILCRGID | HTVVYWLGRR | Q3V0D6 | Protein 4930544L04Rik | −10.0 | 26 | 104 | 0.70 |
| AVSCLWGKVN | SDEVGGEALGR | P02088 | Hemoglobin subunit beta−1; | −1.16 | 21 | 147 | 0.58 |
| AAVAAAREER | GLSPIWAINSPATAEVIR | G3X982 | Aldehyde oxidase 3; | −10.0 | 1291 | 1335 | 0.52 |
| GGFLGQRIVR | MLVQEEELQEIR | Q61694 | 3 beta-hydroxysteroid dehydrogenase type 5; | −10.0 | 22 | 373 | 0.36 |
| KTQDPAKAPN | TPDVLEIEFKKGVPVKVTNIKDGTTR | J3QNG0 | Uncharacterized protein | −10.0 | 219 | 412 | 0.25 |
| KTQDPAKAPN | TPDVLEIEFKKGVPVKVTNIKDGTTR | J3QNG0 | Uncharacterized protein | −10.0 | 219 | 412 | 0.25 |
| NRRRLSELLR | YHTSQSGDEMTSLSEYVSR | P11499 | Heat shock protein HSP 90-beta; | −10.0 | 457 | 724 | 0.17 .te |
Taken together, these data highlight that quantitative degradomic investigation of FFPE is feasible. Moreover, innovative strategies, such as machine learning enable the rapid classification of affected cleavage sites according to protease specificity patterns.
CONCLUSION
Proteolysis as a pivotal posttranslational modification plays a fundamental role in patho-physiological regulation in numerous diseases. While novel “terminomic” approaches have recently enabled the system-wide investigation of native proteolytic processing in various kinds of biological materials, the strategy for FFPE specimens has yet to be brought forward. Given that FFPE specimens are the most abundant resource for clinical and biomedical research, the present study reports, for the first time, the use FFPE specimens for protease research, henceforth opening novel avenues to study the role of proteolysis in a clinical setting.
Supplementary Material
Acknowledgments
We thank Alejandro Gomez-Auli (Spemann Graduate School of Biology and Medicine, University of Freiburg, Freiburg, Germany) for performing Limma statistical analysis using R package, Bjoern Gruening (Department of Computer Science, University of Freiburg) for implementation of machine learning prediction of potential substrates, Christopher S. Hughes (European Molecular Biology Laboratory, Heidelberg, Germany) for performing mass spectrometry measurements on cryopreserved and FFPE liver tissues, and Franz Jehle for mass spectrometry technical support. We thank Thomas Reinheckel for donating murine liver FFPE tissue samples.
Authors declare no competing interests.
Footnotes
Author contributions: O.S. designed the research; Z.W.L., J.W., E.K., M.B., P.B., and O.S. performed the research; M.T., J.N.K., and P.B. contributed new reagents or analytic tools; Z.W.L., L.N., F.C., M.B., and O.S. analyzed data; and Z.W.L. and O.S. wrote the paper.
* This study was funded by a Marie Curie Fellowship for Career Development (PIIF-GA-2012-329622 GlycoMarker to Z. W. L.), Deutsche Forschungsgemeinschaft (SCHI 871/2 and SCHI 871/5, SCHI 871/6, GR 1748/6, INST 39/900-1, and SFB850-Project B8 to O. S.), European Research Council (ERC-2011-StG 282111-ProteaSys to O.S.), and the Excellence Initiative of the German Federal and State Governments (EXC 294, BIOSS to O.S.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- FFPE
- formalin-fixed, paraffin-embedded
- TAILS
- terminal amine isotopic labeling of substrates
- iTRAQ
- isobaric tag for relative and absolute quantitation
REFERENCES
- 1. Hood B. L., Darfler M. M., Guiel T. G., Furusato B., Lucas D. A., Ringeisen B. R., Sesterhenn I. A., Conrads T. P., Veenstra T. D., and Krizman D. B. (2005) Proteomic analysis of formalin-fixed prostate cancer tissue. Mol. Cell. Proteomics 4, 1741–1753 [DOI] [PubMed] [Google Scholar]
- 2. Prieto D. A., Hood B. L., Darfler M. M., Guiel T. G., Lucas D. A., Conrads T. P., Veenstra T. D., and Krizman D. B. (2005) Liquid Tissue: Proteomic profiling of formalin-fixed tissues. BioTechniques 38, 32–35 [DOI] [PubMed] [Google Scholar]
- 3. Stauber J., MacAleese L., Franck J., Claude E., Snel M., Kaletas B. K., Wiel I. M. V. D., Wisztorski M., Fournier I., and Heeren R. M. A. (2010) On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J. Am. Soc. Mass. Spectr. 21, 338–347 [DOI] [PubMed] [Google Scholar]
- 4. De Sio G., Smith A. J., Galli M., Garancini M., Chinello C., Bono F., Pagni F., and Magni F. (2015) A MALDI-mass spectrometry imaging method applicable to different formalin-fixed paraffin-embedded human tissues. Molecular bioSystems 11, 1507–1514 [DOI] [PubMed] [Google Scholar]
- 5. Wisniewski J. R., Dus K., and Mann M. (2013) Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10 000 proteins. Proteom. Clin. Appl. 7, 225–233 [DOI] [PubMed] [Google Scholar]
- 6. Guo K., Ji C., and Li L. (2007) Stable-isotope dimethylation labeling combined with LC-ESI MS for quantification of amine-containing metabolites in biological samples. Anal. Chem. 79, 8631–8638 [DOI] [PubMed] [Google Scholar]
- 7. Kojima K., Bowersock G. J., Kojima C., Klug C. A., Grizzle W. E., and Mobley J. A. (2012) Validation of a robust proteomic analysis carried out on formalin-fixed paraffin-embedded tissues of the pancreas obtained from mouse and human. Proteomics 12, 3393–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bronsert P., Weisser J., Biniossek M. L., Kuehs M., Mayer B., Drendel V., Timme S., Shahinian H., Kusters S., Wellner U. F., Lassmann S., Werner M., and Schilling O. (2014) Impact of routinely employed procedures for tissue processing on the proteomic analysis of formalin-fixed paraffin-embedded tissue. Proteom. Clin. Appl. 8, 796–804 [DOI] [PubMed] [Google Scholar]
- 9. Fox C. H., Johnson F. B., Whiting J., and Roller P. P. (1985) Formaldehyde fixation. J Histochem. Cytochem. 33, 845–853 [DOI] [PubMed] [Google Scholar]
- 10. Hatakeyama K., Wakabayashi-Nakao K., Aoki Y., Ogura S., Yamaguchi K., Nakajima T., Sato T. A., Mochizuki T., and Hayashi I. (2012) Novel protein extraction approach using micro-sized chamber for evaluation of proteins eluted from formalin-fixed paraffin-embedded tissue sections. Proteome Sci. 10, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sprung R. W. Jr., Brock J. W., Tanksley J. P., Li M., Washington M. K., Slebos R. J., and Liebler D. C. (2009) Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol. Cell. Proteomics 8, 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azimzadeh O., Barjaktarovic Z., Aubele M., Calzada-Wack J., Sarioglu H., Atkinson M. J., and Tapio S. (2010) Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J. Proteome Res. 9, 4710–4720 [DOI] [PubMed] [Google Scholar]
- 13. Ostasiewicz P., Zielinska D. F., Mann M., and Wiśniewski J. R. (2010) Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res 9, 3688–3700 [DOI] [PubMed] [Google Scholar]
- 14. Lai Z. W., Petrera A., and Schilling O. (2015) Protein amino-terminal modifications and proteomic approaches for N-terminal profiling. Curr. Opin. Chem. Biol. 24, 71–79 [DOI] [PubMed] [Google Scholar]
- 15. Shahinian H., Tholen S., and Schilling O. (2013) Proteomic identification of protease cleavage sites: Cell-biological and biomedical applications. Expert Rev. Proteomics 10, 421–433 [DOI] [PubMed] [Google Scholar]
- 16. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., and Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 17. Venne A. S., Vögtle F. N., Meisinger C., Sickmann A., and Zahedi R. P. (2013) Novel highly sensitive, specific, and straightforward strategy for comprehensive N-terminal proteomics reveals unknown substrates of the mitochondrial peptidase Icp55. J. Proteome Res. 12, 3823–3830 [DOI] [PubMed] [Google Scholar]
- 18. Lai Z. W., Gomez-Auli A., Keller E., Mayer B., Biniossek M., and Schilling O. (2015) Enrichment of protein N-termini by charge-reversal of internal peptides. Proteomics 15, 2470–2478 [DOI] [PubMed] [Google Scholar]
- 19. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., and Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 20. Toews J., Rogalski J. C., Clark T. J., and Kast J. (2008) Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal. Chim. Acta 618, 168–183 [DOI] [PubMed] [Google Scholar]
- 21. Liu J. S., Wang C., Lu J. T., Liu H., Fan T., Wang K. J., and Wang H. (2004) Morphological dependence of power spectra from two-dimensional passive random media. Phys. Lett. A 333, 395–398 [Google Scholar]
- 22. Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 23. Deutsch E. (2008) mzML: A single, unifying data format for mass spectrometer output. Proteomics 8, 2776–2777 [DOI] [PubMed] [Google Scholar]
- 24. Kessner D., Chambers M., Burke R., Agus D., and Mallick P. (2008) ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sturm M., Bertsch A., Gropl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., and Kohlbacher O. (2008) OpenMS-An open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S., Gupta N., and Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Apweiler R., Martin M. J., O'Donovan C., Magrane M., Alam-Faruque Y., Antunes R., Barrell D., Bely B., Bingley M., Binns D., Bower L., Browne P., Chan W. M., Dimmer E., Eberhardt R., Fedotov A., Foulger R., Garavelli J., Huntley R., Jacobsen J., Kleen M., Laiho K., Leinonen R., Legge D., Lin Q., Liu W. D., Luo J., Orchard S., Patient S., Poggioli D., Pruess M., Corbett M., di Martino G., Donnelly M., van Rensburg P., Bairoch A., Bougueleret L., Xenarios I., Altairac S., Auchincloss A., Argoud-Puy G., Axelsen K., Baratin D., Blatter M. C., Boeckmann B., Bolleman J., Bollondi L., Boutet E., Quintaje S. B., Breuza L., Bridge A., deCastro E., Ciapina L., Coral D., Coudert E., Cusin I., Delbard G., Doche M., Dornevil D., Roggli P. D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gehant S., Farriol-Mathis N., Ferro S., Gasteiger E., Gateau A., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hulo N., James J., Jimenez S., Jungo F., Kappler T., Keller G., Lachaize C., Lane-Guermonprez L., Langendijk-Genevaux P., Lara V., Lemercier P., Lieberherr D., Lima T. D., Mangold V., Martin X., Masson P., Moinat M., Morgat A., Mottaz A., Paesano S., Pedruzzi I., Pilbout S., Pillet V., Poux S., Pozzato M., Redaschi N., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stanley E., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A. L., Yip L. N., Zuletta L., Wu C., Arighi C., Arminski L., Barker W., Chen C. M., Chen Y. X., Hu Z. Z., Huang H. Z., Mazumder R., McGarvey P., Natale D. A., Nchoutmboube J., Petrova N., Subramanian N., Suzek B. E., Ugochukwu U., Vasudevan S., Vinayaka C. R., Yeh L. S., Zhang J., and Consortium U. (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nilse L., Sigloch F. C., Biniossek M. L., and Schilling O. (2015) Toward improved peptide feature detection in quantitative proteomics using stable isotope labeling. Proteom. Clin. Appl. 9, 706–714 [DOI] [PubMed] [Google Scholar]
- 29. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baker P. R., and Chalkley R. J. (2014) MS-viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lange P. F., and Overall C. M. (2011) TopFIND, a knowledgebase linking protein termini with function. Nat. Methods 8, 703–704 [DOI] [PubMed] [Google Scholar]
- 32. Liu H., Sadygov R. G., and Yates J. R. Jr. (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 33. Metz B., Kersten G. F. A., Baart G. J. E., de Jong A., Meiring H., ten Hove J., van Steenbergen M. J., Hennink W. E., Crommelin D. J. A., and Jiskoot W. (2006) Identification of formaldehyde-induced modifications in proteins: Reactions with insulin. Bioconjugate Chem. 17, 815–822 [DOI] [PubMed] [Google Scholar]
- 34. Tanca A., Abbondio M., Pisanu S., Pagnozzi D., Uzzau S., and Addis M. F. (2014) Critical comparison of sample preparation strategies for shotgun proteomic analysis of formalin-fixed, paraffin-embedded samples: Insights from liver tissue. Clin. Proteomics 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tholen S., Biniossek M. L., Gansz M., Gomez-Auli A., Bengsch F., Noel A., Kizhakkedathu J. N., Boerries M., Busch H., Reinheckel T., and Schilling O. (2013) Deletion of cysteine cathepsins B or L yields differential impacts on murine skin proteome and degradome. Mol. Cell. Proteomics 12, 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen P. T., Hsu J. L., and Chen S. H. (2007) Dimethyl isotope-coded affinity selection for the analysis of free and blocked N-termini of proteins using LC-MS/MS. Anal. Chem. 79, 9520–9530 [DOI] [PubMed] [Google Scholar]
- 37. Mommen G. P., van de Waterbeemd B., Meiring H. D., Kersten G., Heck A. J., and de Jong A. P. (2012) Unbiased selective isolation of protein N-terminal peptides from complex proteome samples using phospho tagging (PTAG) and TiO(2)-based depletion. Mol. Cell. Proteomics 11, 832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tholen S., Biniossek M. L., Gansz M., Ahrens T. D., Schlimpert M., Kizhakkedathu J. N., Reinheckel T., and Schilling O. (2014) Double deficiency of cathepsins B and L results in massive secretome alterations and suggests a degradative cathepsin-MMP axis. Cell. Mol. Life Sci. 71, 899–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jain M. R., Liu T., Hu J., Darfler M., Fitzhugh V., Rinaggio J., and Li H. (2008) Quantitative proteomic analysis of formalin fixed paraffin embedded oral HPV lesions from HIV patients. Open Proteomics J. 1, 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao Z., Li G., Chen Y., Li M., Peng F., Li C., Li F., Yu Y., Ouyang Y., Xiao Z., and Chen Z. (2010) Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. J. Histochem. Cytochem. 58, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ly L., Barnett M. H., Zheng Y. Z., Gulati T., Prineas J. W., and Crossett B. (2011) Comprehensive tissue processing strategy for quantitative proteomics of formalin-fixed multiple sclerosis lesions. J. Proteome Res. 10, 4855–4868 [DOI] [PubMed] [Google Scholar]
- 42. Negishi A., Masuda M., Ono M., Honda K., Shitashige M., Satow R., Sakuma T., Kuwabara H., Nakanishi Y., Kanai Y., Omura K., Hirohashi S., and Yamada T. (2009) Quantitative proteomics using formalin-fixed paraffin-embedded tissues of oral squamous cell carcinoma. Cancer Sci. 100, 1605–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Byrum S., Avaritt N. L., Mackintosh S. G., Munkberg J. M., Badgwell B. D., Cheung W. L., and Tackett A. J. (2011) A quantitative proteomic analysis of FFPE melanoma. J. Cutan. Pathol. 38, 933–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perroud B., Ishimaru T., Borowsky A. D., and Weiss R. H. (2009) Grade-dependent proteomics characterization of kidney cancer. Mol. Cell. Proteomics 8, 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nirmalan N. J., Hughes C., Peng J., McKenna T., Langridge J., Cairns D. A., Harnden P., Selby P. J., and Banks R. E. (2011) Initial development and validation of a novel extraction method for quantitative mining of the formalin-fixed, paraffin-embedded tissue proteome for biomarker investigations. J. Proteome Res. 10, 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Craven R. A., Cairns D. A., Zougman A., Harnden P., Selby P. J., and Banks R. E. (2013) Proteomic analysis of formalin-fixed paraffin-embedded renal tissue samples by label-free MS: Assessment of overall technical variability and the impact of block age. Proteom. Clin. Appl. 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 47. Weißer J., Lai Z. W., Bronsert P., Kuehs M., Drendel V., Timme S., Kuesters S., Jilg C. A., Wellner U. F., Lassmann S., Werner M., Biniossek M. L., and Schilling O. (2015) Quantitative proteomic analysis of formalin-fixed, paraffin-embedded clear cell renal cell carcinoma tissue using stable isotopic dimethylation of primary amines. BMC Genomics 16, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Van Damme P., Arnesen T., and Gevaert K. (2011) Protein alpha-N-acetylation studied by N-terminomics. FEB J. 278, 3822–3834 [DOI] [PubMed] [Google Scholar]
- 49. Liu H. H., Bodvarsson G. S., Lu S. L., and Molz F. J. (2004) A corrected and generalized successive random additions algorithm for simulating fractional levy motions. Math. Geol. 36, 361–378 [Google Scholar]
- 50. Spira D., Stypmann J., Tobin D. J., Petermann I., Mayer C., Hagemann S., Vasiljeva O., Günther T., Schüle R., Peters C., and Reinheckel T. (2007) Cell type-specific functions of the lysosomal protease cathepsin L in the heart. J. Biol. Chem. 282, 37045–37052 [DOI] [PubMed] [Google Scholar]
- 51. Tang Q. Z., Cai J., Shen D., Bian Z., Yan L., Wang Y. X., Lan J., Zhuang G. Q., Ma W. Z., and Wang W. (2009) Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3 beta signaling. J. Mol. Med. 87, 249–260 [DOI] [PubMed] [Google Scholar]
- 52. Honey K., Nakagawa T., Peters C., and Rudensky A. (2002) Cathepsin L regulates CD4(+) T cell selection independently of its effect on invariant chain: A role in the generation of positively selecting peptide ligands. J. Exp. Med. 195, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakagawa T., Roth W., Wong P., Nelson A., Farr A., Deussing J., Villadangos J. A., Ploegh H., Peters C., and Rudensky A. Y. (1998) Cathepsin L: Critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280, 450–453 [DOI] [PubMed] [Google Scholar]
- 54. Friedrichs B., Tepel C., Reinheckel T., Deussing J., von Figura K., Herzog V., Peters C., Saftig P., and Brix K. (2003) Thyroid functions of mouse cathepsins B, K, and L. J. Clin. Invest. 111, 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Funkelstein L., Toneff T., Mosier C., Hwang S. R., Beuschlein F., Lichtenauer U. D., Reinheckel T., Peters C., and Hook V. (2008) Major role of cathepsin L for producing the peptide hormones ACTH, beta-endorphin, and alpha-MSH, illustrated by protease gene knockout and expression. J. Biol. Chem. 283, 35652–35659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gocheva V., Zeng W., Ke D., Klimstra D., Reinheckel T., Peters C., Hanahan D., and Joyce J. A. (2006) Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Gene Dev. 20, 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gocheva V., Wang H. W., Gadea B. B., Shree T., Hunter K. E., Garfall A. L., Berman T., and Joyce J. A. (2010) IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Gene Dev. 24, 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Benavides F., Perez C., Blando J., Contreras O., Shen J. J., Coussens L. M., Fischer S. M., Kusewitt D. F., DiGiovanni J., and Conti C. J. (2012) Protective role of cathepsin L in mouse skin carcinogenesis. Mol. Carcinogen 51, 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dennemärker J., Lohmüller T., Mayerle J., Tacke M., Lerch M. M., Coussens L. M., Peters C., and Reinheckel T. (2010) Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene 29, 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (29) PRoteomics IDEntifications (PRIDE) partner repository with dataset identifier PXD002847 (reviewer account details—username: reviewer38683@ebi.ac.uk password: mIR5jHGS). Search results (pepXML format), along with .raw and mzML files, have been deposited. Annotated spectra are provided via MS-Viewer (30) with URLs being listed in Supplemental Table 1.