Abstract
The glycoproteome remains severely understudied because of significant analytical challenges associated with glycoproteomics, the system-wide analysis of intact glycopeptides. This review introduces important structural aspects of protein N-glycosylation and summarizes the latest technological developments and applications in LC-MS/MS-based qualitative and quantitative N-glycoproteomics. These maturing technologies provide unique structural insights into the N-glycoproteome and its synthesis and regulation by complementing existing methods in glycoscience. Modern glycoproteomics is now sufficiently mature to initiate efforts to capture the molecular complexity displayed by the N-glycoproteome, opening exciting opportunities to increase our understanding of the functional roles of protein N-glycosylation in human health and disease.
Protein glycosylation encompasses a broad class of post-translational modifications involving the covalent attachment of complex carbohydrates (glycans) to specific amino acid residues of polypeptide chains. The human biosynthetic machinery catalyzes diverse types of glycosylation, with the best studied being attachment of glycans to asparagine (N-glycosylation) and serine/threonine (O-glycosylation) residues (1). As with all types of protein glycosylation, N-glycosylation is a template-less modification synthesized by a suite of glycosylation enzymes in the secretory pathway, Fig. 1A (2). Template-less synthesis means that glycosylation is determined by the physiological state of the glycosylation machinery and the nature of the proteins undergoing glycosylation. Jointly, these attributes determine the repertoire of glycans present on synthesized glycoproteins (glycoforms) and create the important features of protein site- and cell-specific glycosylation (3–5). Protein glycosylation is therefore a spatiotemporal dynamic modification that cells can utilize to respond to the constantly changing milieu.
Fig. 1.
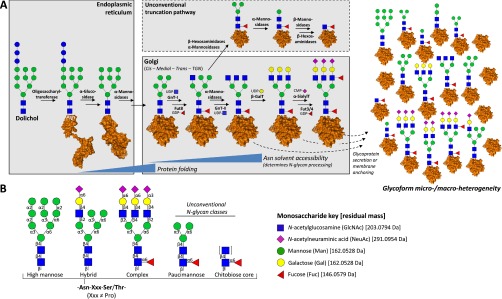
Overview of the biosynthesis and structural classes of mammalian protein N-glycosylation. A, Schematic summary of the biosynthetic machinery of N-glycoproteins. The enzymatic processing, which is initiated while the glycoproteins are still being translated, translocated, and folded, may terminate at any point in the enzymatic sequence depending partially on the Asn solvent accessibility of the maturely folded glycoprotein (3). This generates site-, cell-, and even subcellular-specific glycoform heterogeneity forming one of the functionally most important features of the glycoproteome (5), and also creates substantial analytical challenges. TGN: trans-Golgi network. B, Mammalian N-glycoproteins are typically divided into three main N-glycan classes: high mannose, hybrid, and complex type. Unusual paucimannosidic and chitobiose core type N-glycans arising from unconventional truncation pathways (dashed box) have been reported in specific cell types and physiological conditions (21). Monosaccharide residues are depicted according to the establish nomenclature (180) with residual monoisotopic masses provided.
N-linked glycans are typically present on asparagine residues in AsnXxxSer/Thr, Xxx ≠ Pro consensus sequences (sequons) in humans. This preference is caused by specific recognition of the sequon by the peptide-binding site of an oligosaccharyltransferase (OST)1 (6), the enzyme which catalyzes this reaction. However, it is now clear that mammalian cells also have the ability to rarely glycosylate more relaxed sequons (e.g. AsnXxxCys) (7–13). The use of such nonconsensus sequons seems to be more frequent in rodents, where even glutamine-linked glycosylation has been reported (14). Low efficiency N-glycosylation in these noncanonical sequons is consistent with a role of the canonical sequon in recognition and high-affinity binding to OST to promote glycan transfer (15). Mammalian N-glycans share a common trimannosylchitobiose core comprised of three mannose (Man) and two N-acetylglucosamine (GlcNAc) residues, extended with a variety of monosaccharides including Man, GlcNAc, galactose (Gal), fucose (Fuc), N-acetylgalactosamine (GalNAc) and sialic acids such as N-acetylneuraminic acid (NeuAc) and N-glycolylneuraminic acid (NeuGc). N-glycans can be further modified by noncarbohydrate moieties including phosphorylation, sulfation and acetylation (16, 17). The conserved trimannosylchitobiose core of N-glycans is a remnant of the N-glycan precursor (Glc3Man9GlcNAc2) initially transferred to proteins in mammalian cells. This oligosaccharide structure is built stepwise on a dolichol pyrophosphate carrier embedded in the endoplasmic reticulum (ER) membrane, and is transferred en bloc to nascent polypeptides by OST. The terminal Glc and Man play critical roles in assisting glycoprotein folding and in ensuring glycoprotein quality control in the ER. After glycoproteins are correctly folded, the terminal Glc and Man are generally removed by α-glucosidases and α-mannosidases in the ER and cis-Golgi. N-glycans can then be extended by glycosyltransferases in the multiple Golgi compartments, potentially resulting in an extreme diversity of structures on mature glycoproteins in an organism-, cell-, or regulation-specific manner. The diverse mammalian N-glycans can be crudely classified into three conventional classes: high mannose, hybrid, and complex type, Fig. 1B. However, it is becoming clear that other unconventional N-glycan classes such as paucimannosidic and chitobiose core types decorate some mammalian glycoproteins (see below) (7, 13, 18–21). The structures, biosynthetic pathways, and associated disorders of N-glycosylation have been recently reviewed elsewhere and readers are encouraged to use these resources for a deeper introduction (16, 22).
A substantial proportion of mammalian genomes is dedicated to genes encoding proteins involved in glycosylation pathways, and these are highly conserved. Consistent with this, glycosylation is central to many biological processes. N-glycans are critical for enabling efficient glycoprotein folding and for maintaining the structural and functional integrity of glycoproteins (16). Protein N-glycosylation is also intimately associated with development processes (23, 24), with facilitating or preventing bacterial binding to the host (25) and with sustaining the normal function of individual cells, tissues, organs, and organisms (26). Finally, protein glycosylation is a strictly regulated modification process in healthy cells, and these biochemical processes are dysregulated in various pathologies including, but not restricted to, cancer (27, 28), inflammation (29), Alzheimer's disease (30), multiple sclerosis (31), and cystic fibrosis (25). Disease-associated changes in protein glycosylation may arise from changes in glycoprotein abundance, glycosylation site occupancy (macro-heterogeneity or “glycosylation efficiency”), or glycan micro-heterogeneity at different sites on a glycoprotein (see details below). Changes in the glycoprotein micro-heterogeneity are dictated by the capacity of glycan-processing enzymes (glycosidases and glycosyltransferases) in the glycoprotein biosynthesis pathway, the nature of specific glycoprotein substrates, and other cellular factors (3, 4).
This review summarizes the present analytical tools and technologies capable of performing large-scale (system-wide) analysis of protein N-glycosylation micro-heterogeneity and the unique structural insights that can be derived from such experiments by covering the very latest literature describing recent technological developments and applications in LC-MS/MS-based qualitative and quantitative N-glycoproteomics.
System-wide Structural Analysis of Protein N-glycosylation
Deciphering the glycosylation 'code' has been the ambition of generations of glycobiologists. The ability to accurately characterize the structure of glycoproteins is necessary if we are to succeed in our quest to unravel the diverse functions of glycans and develop the next generations of glycoprotein-based therapeutics (32). However, glycoproteins are challenging to characterize because of the multiple layers of structural diversity that form a spectrum of chemically similar glycofoms. The information needed to unambiguously characterize such heterogeneous glycoproteins is therefore consequently much larger than for unmodified proteins or for proteins with structurally simple modifications such as phosphorylation or methylation. Even the most detailed modern glycoprotein characterization studies usually only capture part of the glycoprotein structure. Some glycoprotein structural features can be inferred or predicted from the biosynthetic constraints of the well-studied glycosylation machinery of mammalian cells (33). Nevertheless, it is important to stress that even incomplete structural information can often be very useful in deciphering the structure/function relationships of glycoproteins, and in identifying alterations in the biosynthetic glycosylation machinery.
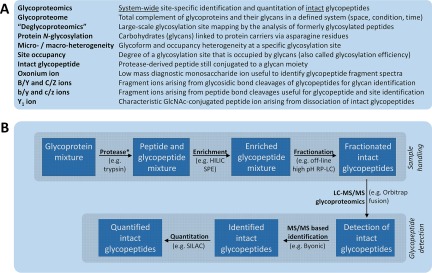
Glycoproteomics is the site-specific analysis of the glycoproteome at the systems level, Fig. 2A. Glycoproteomics experimental workflows are typically initiated with protein extraction from biological samples, denaturation, and protease digestion, Fig. 2B. At this step, isotopic labels assisting in glycopeptide quantitation or enhancing their MS features (e.g. ionization and fragmentation) can be introduced. The resulting peptide mixtures are often extremely complex and glycopeptides are consequently typically enriched and/or prefractionated prior to detection, usually by LC-MS/MS. Glycoproteomics experiments are commonly based on the identification, and less frequently, also the quantitation of intact glycopeptides. Glycoproteomics yields system-wide information on the glycoprotein carriers, the glycan attachment sites, the occupancies of glycosylation at each site, and the structure and heterogeneity of the attached glycans. As showcased in this review by recent examples, glycoproteomics is a powerful technology to map disease-associated alterations in the glycoproteome, Fig. 3. Such glycosylation alterations may originate from multiple tissues, which may be differently regulated during pathogenesis. Typically, in glycoproteomics investigations, intact glycopeptides derived from the total complement of glycoproteins extracted from bodily fluids or complex tissues from healthy and diseased individuals (or other biological scenarios) are qualitatively and quantitatively compared. By also measuring any changes in glycoprotein abundance and site occupancy, the exact mechanism(s) contributing to the observed glycoproteome regulation can be interrogated (see example below).
Fig. 2.
A, Definitions and explanations of commonly used nomenclature in glycoproteomics. B, Generic workflow illustrating important components of a glycoproteomics experiment, which crudely can be divided into segments related to glycopeptide sample preparation (top box) and detection (bottom box). Typical examples of the individual components are provided. *Additional sample handling including glycoprotein derivatization and glycopeptide labeling for quantitation purposes may be introduced at this step.
Fig. 3.
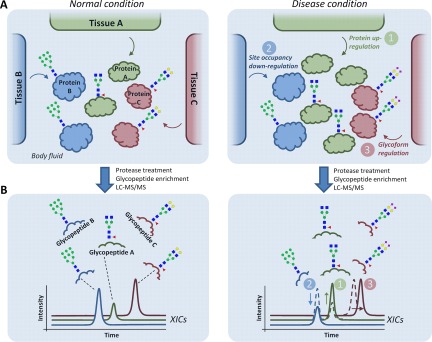
Three fundamental levels of molecular dysregulation of multiple tissues contributing to an altered secreted N-glycoproteome during disease. A, Hypothetical example illustrating three sources of dysregulation: 1) protein level (green), 2) site occupancy (blue), or 3) glycosylation micro-heterogeneity (red) from three separate tissues (Tissue A-C) contributing to a joint secreted N-glycoproteome (Protein A-C) in a body fluid derived from disease (right) and 'normal' healthy (left) condition. B, After proteolysis and enrichment, the altered abundance of the resulting glycopeptides can be detected using LC-MS/MS-based label-free quantitative glycoproteomics as shown by color-coded traces representing extracted ion chromatograms (XICs). However, establishing which of the three mechanisms causes the glycopeptide alterations for the detected glycoproteins may be challenging solely with glycopeptide analysis, especially in glycoproteomes arising from multiple tissues. Parallel quantitative proteomics and “deglycoproteomics” (detection of formerly occupied N-sites) of the same samples can assist in this task.
Many other analytical approaches can be used to characterize aspects of glycoprotein structural diversity. These include site-specific analysis of N-glycoproteins isolated to relative purity rather than in complex mixtures (11, 12, 20, 34–42), N-glycomics analyses of glycans released from glycoproteins (18, 38, 43–47), and identification and quantification of previously glycosylated sites on de-N-glycosylated proteins (“deglycoproteomics”) after removal of the entire glycan or with remnant N-glycan core remaining (48–57). Although these studies per se do not qualify under our definition of glycoproteomics, (site-specific analysis of the glycoproteome at the intact glycopeptide level), they still provide useful information in conjunction with glycoproteomics for the glycobiologist, provided correct experimental design is applied (58).
Key Technologies and Recent Analytical Developments in N-glycoproteomics
In 2014, we reviewed the status of modern LC-MS/MS-based glycoproteomics (59). Other excellent recent reviews are also available (15, 30, 60–68). Thus, this review will highlight the very latest (∼2014–present) technological advances and applications in glycoproteomics, which have been instrumental for the recent performance improvements in detection limits, accuracy of glycopeptide identification and quantitation, and gains in glycoproteome coverage.
The modern discipline of glycoproteomics has deep roots in the protein and carbohydrate analytical chemistry pioneered in the late 1980s and early 1990s with the advent of biomolecular mass spectrometry (69). Impressive analytical strategies using relatively insensitive MS instrumentation were developed e.g. the selective detection of glycopeptides in mixtures using deglycosylation-based mass shifts (70) and selection ion chromatograms (71), and the fundamentals of glycopeptide ionization and fragmentation behavior were accurately described (72, 73). These early studies remain a solid foundation on which many modern glycoanalytical strategies are conceived. It is also clear that glycoproteomics more recently has profited handsomely from technology developments arising from the larger and more mature discipline of proteomics including sample handling, LC-MS/MS acquisition strategies, and data handling and processing (74). In parallel, glycoproteomics has been a beneficiary of the continual performance enhancements of modern mass spectrometers including improved speed, sensitivity, resolution, and accuracy, most notably implemented on the latest Q-TOF (Sciex, Waters, Agilent, Bruker) and on multiple Orbitrap (Thermo) instrument platforms (75). Developments and applications of several key glycoproteomics-specific technologies have additionally been critical for the rapid maturation of glycoproteomics workflows over the past two years, Table I. Specifically, key advances have been made in the enrichment of intact glycopeptides from complex peptide mixtures, in LC-MS/MS-based detection of intact glycopeptides through optimized dissociation and acquisition styles of glycopeptides, and in data handling for more automated, yet still confident, glycopeptide identification and quantitation.
Table I. Latest developments and applications of the key components in N-glycoproteomics.
| Component of glycoproteomics workflow | References | |
|---|---|---|
| Sample preparation of N-glycopeptides | ||
| Enrichment | HILIC SPE | (21, 89, 94, 95, 105–107, 110, 135, 159, 161, 162, 165, 181–184) |
| Synthetic polymer mixed-mode HILIC SPE | (98, 99, 101, 102, 108) | |
| Lectin SPE/chromatography | (7, 9, 13, 40, 60, 88, 104, 107, 135, 138, 140, 165, 182) | |
| Hydrazide chemistry* | (48, 52, 140, 185) | |
| Boronic acid magnetic beads | (80–82) | |
| Acetone precipitation | (86, 103) | |
| Porous graphitized carbon (PGC)-RP SPE | (89, 138) | |
| Size exclusion chromatography | (87, 88) | |
| Metabolic labelling and enrichment | (84, 85) | |
| Separation/fractionation | Neutral/high pH RP-LC | (7, 13, 46, 105, 106, 108, 138, 152) |
| 2D (SCX/RP) -LC (online) | (159) | |
| 2D (HILIC/RP) -LC (online) | (186) | |
| 2D (PGC/RP) -LC (online) | (135) | |
| PGC-LC | (110) | |
| HILIC-LC | (138) | |
| Detection of intact N-glycopeptides using LC-MS/MS | ||
| Dissociation schemes | Beam-type (Q-TOF) CID/HCD | (13, 21, 46, 87, 88, 94, 105, 107, 108, 110, 111, 126, 152, 157, 159, 161, 162, 165, 183, 184, 186, 187) |
| Resonance activation (ion trap) CID | (21, 40, 86, 103–105, 126, 131, 138, 157, 159, 165, 187) | |
| ETD/EThcD/ECD | (7, 13, 21, 105, 114, 126, 157, 159, 187) | |
| Acquisition styles | Data-dependent acquisition (DDA) | (7, 13, 21, 46, 86, 87, 94, 103, 106–108, 152, 157, 159, 162, 165, 183, 184, 186) |
| DDA-PD (m/z 204.086) ETD or ion trap CID/ETD | (105, 125, 126) | |
| Data-independent acquisition (DIA) | (135–138) | |
| Glycopeptide mass defect classifier | (127) | |
| Targeted glycopeptide analysis (MRM) | (134, 140, 141, 143) | |
| Identification strategies | Protein Prospector | (7, 13) |
| Byonic | (21, 46, 105, 106, 135, 150) | |
| GPQuest | (46, 108, 152, 162) | |
| GlycopeptideID | (87, 88, 104) | |
| Sweet-Heart | (126, 154, 155) | |
| GlycoFragWork | (157, 165) | |
| GlycoMod | (40, 138) | |
| Less used programs (i. e. GlycoMaster DB GRIP, MAGIC, GlycoPep Evaluator) | (158–161) | |
| Other (e.g. in-house program/script, manual annotation etc) | (86, 94, 103, 107, 134, 138, 162, 184, 186) | |
| N-Glycome-assisted identification | (21, 88, 105–108, 162) | |
| Parallel proteome- and/or deglycoproteome (formerly occupied glycopeptides) profiling | (21, 46, 88, 94, 103, 105–108, 135, 159, 162) | |
| Quantitation | SILAC | (105) |
| iTRAQ | (46, 108) | |
| Other isotopes | (164, 166) | |
| Label-free quantitation (XIC, precursor ion intensity, spectral count, internal ref.) | (21, 86, 104, 106, 159, 165, 184) | |
* Hydrazide is usually used to capture N-glycopeptides with a subsequent peptide N-glycosidase F release of formerly N-glycosylated peptides for glycosylation site mapping and is, thus, not a generally used tool in glycoproteomics.
Enrichment and Prefractionation Strategies for Glycoproteomics
Because of the substoichiometry of glycopeptides in complex peptide mixtures arising from the extensive glycan micro- and macro-heterogeneity, and inherently poor detectability, glycopeptide enrichment is a critical component of glycoproteomics experiments. Recent advances in glycopeptide enrichment have been fully reviewed elsewhere (76–79). Exciting initiatives in glycopeptide enrichment strategies include the optimized use of boronic acid as a reversible glycopeptide capture method on magnetic graphene (80) and on magnetic microspheres (81). Boronic acid, which reacts with cis-diol-containing monosaccharide residues, has also recently been used to enrich intact glycopeptides (82) and glycoproteins (83), but remains an infrequently used glyco-enrichment method. Solid phase hydrazide-based glycopeptide capture has also seen developments (48) and applications (see Table I), but this approach is most commonly used in conjunction with peptide N-glycosidase F-catalyzed release and analysis of formerly N-glycosylated peptides and does not satisfy our definition of glycoproteomics. Other new enrichment methods of interest include the metabolic incorporation of N-azido sugars into N-glycopeptides to facilitate their specific enrichment and detection (84, 85). The selective precipitation of glycopeptides by acetone (86), use of size exclusion chromatography (87, 88) and the combined use of porous graphitized carbon (PGC) and reversed phase (RP) (89) and titanium dioxide (90) solid phase extraction (SPE) for efficient enrichment of glycopeptides and sialoglycopeptides, respectively, are also promising developments. However, common for most of these proof-of-principle methodology studies is the need for further validation to demonstrate their true potential in glycoproteomics. The frequently used zwitterionic-HILIC SPE-based methods for enrichment and analysis of intact N-glycopeptides (91–93) have been further tested. Usefully, it was found that N-glycopeptides are still efficiently retained when using higher concentrations (0.1%) of surfactants and detergents including SDS and Triton X-100 (94). In addition, other HILIC phases were used to enhance the loading capacity (95) and have been synthetically tweaked by using “click-chemistry” (96, 97) and by introducing mixed mode-HILIC retention mechanisms (98–102). As all N-glycopeptides harbor a minimum degree of localized hydrophilicity arising from a high density of polar hydroxyl groups, HILIC remains the most used and, in our opinion, the most efficient and least biased enrichment method facilitating large-scale analysis of intact and native (nonderivatized) glycopeptides in N-glycoproteomics.
Only few developments in the off-line separation and fractionation of glycopeptides prior to LC-MS/MS detection have recently been published. Some glycoproteomics approaches even by-pass this step because of the increased capacity of modern LC-MS/MS instrumentation to handle the extreme complexity of biologically-relevant glycopeptide mixtures, and perhaps because the multiple LC fractions resulting from off-line separations dramatically increase the required LC-MS/MS instrument time, lower the overall sensitivity of the analysis and complicate the downstream quantitative data analysis (103, 104). However, multiple glycoproteomics strategies, particularly in studies where the amounts of biological sample material are not a limiting factor, still opt to use off-line separation to increase the glycoproteome coverage (46, 105–107). The most exciting new developments in this area include the off-line use of neutral/high pH (pH 7–10) RP-LC (7, 108), which displayed orthogonal glycopeptide retention behavior relative to the conventional acidic (∼pH 2) RP-LC-(online) MS/MS. Importantly in this system, the higher pH RP separation maintained the “clustering effect” by eluting glycopeptide families with the same peptide backbone in narrow bands within single LC fractions enabling their downstream identification and quantitation within a single LC-MS/MS analysis (106).
MS Acquisition Strategies in Glycoproteomics
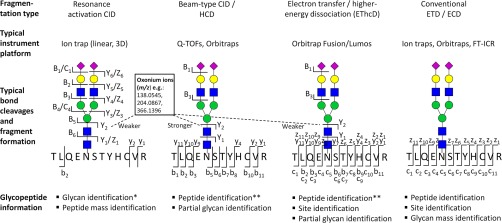
LC-MS/MS-based detection of glycopeptides has seen many exciting advances in the past two years. Jointly, these advances have significantly contributed to the enhanced performance of glycoproteomics workflows. As accurately reviewed (109), much focus has been directed to developing, optimizing and combining informative dissociation methods to enable confident large-scale N-glycopeptide identification. Systematic studies have investigated the glycopeptide (glycan and peptide) sequence coverage as a function of several variables e.g. the adduct formation, proton mobility and (normalized) collision energy in higher-energy collision dissociation (HCD) (110), beam-type (Q-TOF) CID (111–113), and the combined use of resonance activation (ion trap) CID and HCD (77). Together with conventional electron transfer dissociation (ETD), HCD and ion trap CID, which are often activated as “back-to-back” alternating dissociation events of the same isolated precursor ions or alternatively engaged in separate LC-MS/MS runs, form the most popular dissociation techniques because of their complementary nature when applied to N-glycopeptides. Electron capture dissociation (ECD) typically used on Fourier transform ion cyclotron resonance instrumentation is a less used (and less efficient) electron-based dissociation method used in glycopeptide analysis (114, 115). No single dissociation method yields information on the entire structural detail of N-glycopeptides. In general terms, the c/z-ion series formed in ETD-MS/MS yield information on the glycosylation site and the peptide identity, but only reveal the glycan as a “blind” modification with an accurate mass, Fig. 4. HCD produces abundant diagnostic oxonium ions and partial B- and Y-ion series for glycopeptide classification and solid b/y-ion series for confident peptide identification (116, 117), but typically without the Asn-conjugated glycan, which is lost in the fragmentation process. The glycopeptide fragments generated by beam-type CID on Q-TOF instrumentation resemble the glycopeptide fragmentation pattern produced by HCD in the C-trap of Orbitrap mass spectrometers. In contrast, resonance activation CID as generated on linear and 3D (Paul) ion traps produces more complete B- and Y-ion series (and even occasionally C- and Z-ions) as well as some oxonium ions to identify the conjugated glycan, but typically neither the site nor the carrier peptide is confidently identified, and is often left as a “blind” mass. In addition, ion trap CID induces mostly single bond cleavages of glycopeptides and some important oxonium ions may therefore not be formed by this dissociation method. Other limitations of ion traps are the reduced resolution and mass accuracy relative to high resolution mass analyzers and the cut-off in the low-m/z region, which can impede glycopeptide identification by masking the important diagnostic oxonium ions. ETD is valuable for eliminating incorrect assignments from unexpected peptide modifications e.g. carbamidomethylation side-reactions (118) or nonspecific protease activity (119), but may still compromise the identification accuracy in the case of unexpected glycan derivatization e.g. from reactive buffers (120). The “hybrid” dissociation method, electron transfer and higher-energy collision dissociation (EThcD), where a supplemental energy is applied to all fragment ions formed by ETD, including the usually abundant unreacted precursor ions, has shown great potential by the production of richer and more informative spectra consisting of both b/y- and c/z-ion series thereby increasing the sequence coverage of both nonmodified peptides (121) and N-glycopeptides (105, 122). Although engaging multiple dissociation modes is becoming more popular, some researchers still opt for applying single fragmentation techniques i.e. HCD (46) and ETD (7) in glycoproteomics, for example because of the nature of the research question (e.g. only investigating the site-specific glycan monosaccharide composition), limited availability of instruments allowing multiple fragmentation modes or to avoid the longer MS duty cycle associated with approaches using alternating/hybrid dissociation methods.
Fig. 4.
The main dissociation methods in glycoproteomics, typically used in conjunction i.e. resonance activation (ion trap type) CID, beam-type (Q-TOF type) CID or HCD, hybrid-type EThcD, and the conventional ETD (or ECD) fragmentation. The resulting bond cleavages and fragment ion formations when applied to N-glycopeptides are indicated including the presence of diagnostic oxonium ions (insert) (116, 117). *N-Glycan identification is usually restricted to specification of the monosaccharide composition and the partial or complete topology. **The b/y-ions in the beam-type CID (HCD) and EThcD dissociation schemes usually lose the conjugated glycan and therefore do not provide information about the glycosylation site.
The abundant formation of the glycopeptide-specific oxonium ions upon beam-type CID (HCD) has been utilized to create more intelligent glycoproteomics acquisition styles that enable more instrument time being spent on fragmenting glycopeptides rather than the usually more abundant nonglycosylated peptides in biological mixtures. The GlcNAc-derived fragments m/z 138.0545 and m/z 204.0867 are particularly useful diagnostic ions for such acquisition styles. In fact, the same oxonium ions are often generated upon beam-type CID (HCD) fragmentation of GalNAc-containing peptides, albeit in different intensities (123), meaning that these acquisition strategies are also useful for O-glycopeptides. The saccharide oxonium ion profile derived from glycopeptide fragmentation can even provide valuable supporting information of the monosaccharide compositions of the attached N-glycans (123, 124). The original product-dependent (PD) method, which triggered ETD events upon recognition of m/z 204.086 in HCD-MS/MS (125), was lately followed up with a more informative approach where alternating ion trap CID and ETD events are triggered upon oxonium ion recognition in HCD-MS/MS spectra (126). In another innovative LC-MS/MS study, N-glycopeptides in nonenriched peptide mixtures were crudely isolated (postacquisition) from their nonglycosylated counterparts based on their accurate mass (127). This type of glycopeptide classification was based on a common mass defect of N-glycopeptides arising from their oxygen-rich elemental compositions. This unique property can be used to filter large LC-MS/MS data sets for the presence of N-glycopeptides.
The growing volume of glycoproteomics literature can be used to identify trends and common methods utilized across multiple laboratories. Convenient examples include the common use of RP-LC as a standardized chromatographic platform to separate glycopeptides in an online-conjugated manner, together with the LC-ESI-MS/MS acquisition style, which commonly utilizes data-dependent acquisition (DDA) to detect native (underivatized) intact glycopeptides at high resolution/high mass accuracy (better than 20 ppm accuracy of precursor and product ions) in positive ion polarity mode. Such approaches are shared by most glycoproteomics researchers, see Table I. In fact, compared with the more established discipline of glycomics that still, based on lab-to-lab or researcher-to-researcher preferences and traditions, utilizes a wide spectrum of analytical approaches to achieve structural information of glycans (128), glycoproteomics strategies appear surprisingly more uniform, likely because of similarities with standard proteomics workflows. However, the literature does harbor recent examples of studies exploring new avenues deviating from these mainstream approaches. For example, ion mobility MS was used to separate and analyze intact ovomucoid glycopeptides (40) and urine glycopeptides from a patient diagnosed with Schindler disease (129). Furthermore, recent studies have explored the feasibility of measuring deprotonated glycopeptides in negative ion mode with (130) and without (131) derivatization. Innovative strategies for the derivatization of sialic acid residues have also been designed to achieve linkage-specific (α2,3 versus α2,6-sialyl) information at the glycopeptide level (132) and to obtain more equal ionization response from neutral and sialylated glycopeptides (133). Furthermore, attempts to generate universal workflows using Pronase digestion and C18-PGC LC-MS/MS instead of the conventional RP-LC-MS/MS of tryptic glycopeptides were described (39, 134). In addition, data-independent acquisition (DIA), referred to as “SWATH-MS” on Sciex MS platforms and “MSE” on Waters MS platforms, has also been trialed to monitor both micro- (135, 136) and macro- (136–138) heterogeneity of protein N-glycosylation using LC-MS/MS instrumentation. However, common for these less used, but still very exciting analytical choices, is the fact that much work is still needed to accurately document their potential in glycoproteomics relative to their already implemented counterparts.
Interesting developments have been described in targeted glycopeptide detection and quantitation strategies, as recently reviewed (139). Several applications of multiple reaction monitoring (MRM)-based glycopeptide analysis from a variety of biological systems and diseases were lately published including the use of MRM-based serum glycoproteomics to study esophagus diseases (140), liver disease (141), and the immunoglobulin subclasses (134, 142, 143). Among other technical challenges unique to MRM-based glycopeptide detection and quantitation, one of the most important considerations is the selection of the precursor-product transitions being monitored. Needless to say, the transitions must be unique for the glycopeptide(s) of interest. The low-mass oxonium ions may not provide sufficient specificity because they can arise from multiple glycopeptides. It is anticipated that the need for targeted glycopeptide analysis will increase in the immediate future as a direct result of the maturation of discovery-type glycoproteomics technologies, yielding more “glycopeptide candidates” to target. MRM-based strategies are ideal for targeted analyses because of their high sensitivity, dynamic range and throughput directly from complex biological matrices requiring minimal sample handling. As recently summarized (15), several recent studies also report large-scale (proteome-wide) quantification of N-glycosylation site occupancy. Novel deglycosylation-based (144) or nondeglycosylation-based (145) analytical LC-MS/MS approaches have been established for the accurate quantitation of glycosylation site occupancy. These analytical platforms were applied to measure site occupancy in various biological settings including in plants (146), human saliva (83), and yeast (137). Recently, an alternative method using differential glycoprotein oxidation and labeling was utilized to map the sialoglycan occupancy on specific protein N-glycosylation sites (145). These methods and the results they produce are highly interesting and of great value to glycobiologists, and emphasize the diverse analytical techniques applicable for glycoprotein analysis.
Glycoinformatic Tools for Glycopeptide Analysis
Following the acquisition step of large-scale glycopeptide data, a bottleneck that has severely limited the field of glycoproteomics is downstream glycopeptide identification. The identification process was, until recently, largely driven by manual expert annotation of the resulting MS/MS spectra. As described in topical reviews (122, 147–149), many glycoinformatics initiatives have been initiated to automate the glycopeptide identification process. Recent commercial and open-source software developments facilitating automated glycopeptide identification or providing assistance in the process include ByonicTM (150), Protein Prospector (151), GPQuest (152), GlycopeptideID (153), Sweet Heart (154, 155), GlycoMaster DB (156), GlycoFragWork (157), GlycoPep Evaluator (158), GRIP (159), MassyTools (160), and MAGIC (161). The proprietary Byonic (Protein Metrics), which recently was integrated as a node into Proteome Discoverer v2.0 (Thermo Scientific), appears to be the most widely used search engine in glycoproteomics because it allows semi-automated identification of N- and O-glycopeptides (and other post-translational modifications) using fine control of the search space in a probability based manner from high resolution MS/MS data (see Table I). The different programs utilize various strategies to identify N-glycopeptides including the use of characteristic Y1 ions, oxonium ions, B/Y- and C/Z- type glycan fragment ion series, and b/y- and c/z-type peptide fragment ions (116, 117). In order to limit the search space and enhance the detailed structural knowledge of the glycans encountered in a given glycoproteome, several studies have performed parallel N-glycome profiling (21, 40, 88, 105, 106, 108, 162). This so-called 'glycomics-assisted glycoproteomics' approach can also be complemented with quantitative proteome and de-N-glycoproteome profiling (mapping of formerly N-glycosylated peptides) (21, 46, 88, 103, 105–108, 135, 159, 162), which reduce the search space even further and provide supporting evidence to pinpoint the exact mechanism(s) driving the observed glycoproteome alterations. At this stage, for most of the available glycopeptide identification software tools it is strongly advised that manual validation of the glycopeptide assignments are still performed to generate sufficient confidence in the reported identifications. This is particular true for a subset of glycopeptides that are known to cause frequent misassignments including 1) peptides with susceptibility for unexpected peptide modifications/derivatizations e.g. Met carbamidomethylation (addition of CH2-CO-NH2) (Δm = 57 Da/41 Da from nonmodified/oxidized Met equating to the mass difference of HexNAc/Fuc and HexNAc/Hex, respectively) (118), 2) peptides carrying isobaric (Fuc1NeuAc1-R versus Hex1NeuGc1-R, Δm = 0 Da) or near-isobaric monosaccharide subcompositions (e.g. Fuc3-R vs. NeuAc1Fuc1-R, Δm = 1 Da), the latter relying crucially on correct monoisotopic peak picking, and 3) unexpected analyte formation e.g. noncovalent glycopeptide dimer formation (163). Analysts should also be aware that peptides may harbor multiple (N- and O-) glycosylation sites and that the established monosaccharide composition may therefore not necessarily be restricted to a single glycan moiety on a single site. Similar to proteomics, it is necessary that glycoproteomics moves toward false discovery rate (FDR)-based identifications, where the accuracy of the reported glycopeptides are statistically assessed by a joint probability score for the entire glycopeptide structure or by multiple probability scores to assess the correctness of the individual components of the identified glycopeptide i.e. the peptide, site and monosaccharide composition. However, assigning appropriate and meaningful FDRs to glycopeptide identifications involves more variables than generating FDRs of nonmodified peptide identifications and has consequently proven challenging and is still in its infancy (158).
Quantitative Glycoproteomics
As discussed in more detail below, the first studies performing isotope-assisted quantitative glycoproteomics using stable isotope labeling with amino acids in cell culture (SILAC) (105) and isobaric tags for relative and absolute quantitation (iTRAQ) (46, 108) have recently been published. This has enabled accurate relative quantitation of single glycoforms on individual protein sites by comparing the precursor (SILAC) or the product (iTRAQ) ion intensities across two or more conditions. However, as we discussed in a recent review (67), comparing the entire glycoform distribution (micro-heterogeneity) at individual glycosylation sites in separate conditions may be more informative in glycobiology. Both isotope-assisted quantitative glycoproteomics (46, 105, 108, 164) and label-free (based on XIC area, precursor intensity or spectral count) quantitative glycoproteomics (21, 104, 106, 159, 165) can generate quantitative information of site micro-heterogeneity. Importantly, if the ambition is to understand the mechanisms driving alterations of the glycoproteome in specific conditions (see Fig. 3) parallel proteome profiling and site occupancy experiments are typically required. Recently, isotope-coded carbamidomethylation was shown to be a simple and inexpensive alternative to SILAC and iTRAQ-based glycopeptide labeling and quantitation, but this strategy suffers from the severe limitation that it is only applicable to cysteine-containing glycopeptides (166). Stable isotope dimethyl and succinic anhydride labeling appear to be more widely applicable alternatives when aiming for a universal glycopeptide analytical workflow (167, 168).
Glycoproteomics Databases
Glycoproteome data deposition and storage are important aspects to consider at the end of the glycoproteomics pipeline. Large glycoproteome data sets often harbor information of great value to glycobiologists pursuing other research questions that can tentatively be probed using the acquired data (62). This naturally requires the data (processed or raw) to be freely available in a data repository (e.g. ProteomeXchange and PRIDE) and the experimental design and conditions explicitly described. In analogy to the MIAPE and MIRAGE guidelines detailing the minimum information required for proteomics (169) and glycomics (170) experiments, respectively, it is desirable that an equivalent set of guidelines are designed and implemented for published glycoproteomics data. The curated data in the form of the identified N-glycan structures at specified protein sites has begun to be captured and stored at established glycoinformatic platforms such as UnicarbKB (http://unicarbkb.org) (171) for easy browsing and relevant connectivity to the larger protein-centric UniProt (http://uniprot.org) knowledge repository and the glycan-centric UnicarbDB (http://unicarb-db.biomedicine.gu.se) MS/MS database. Informatics initiatives aiming at integrating glycoproteomics data and other glyco- and protein-specific data as well as enzyme information at a higher level have also been presented (147). Taken together, it is clear that the recent advances of glycoproteomics strategies are driven by exciting initiatives in all components of the analytical workflow. As described below, the significant aggregate value of these advances is tangible in recent glycoproteomics applications reporting thousands of unique N-glycopeptides from biological protein samples.
Glycoproteomics Facilitates Insight in the Biosynthesis and Regulation of the N-glycoproteome
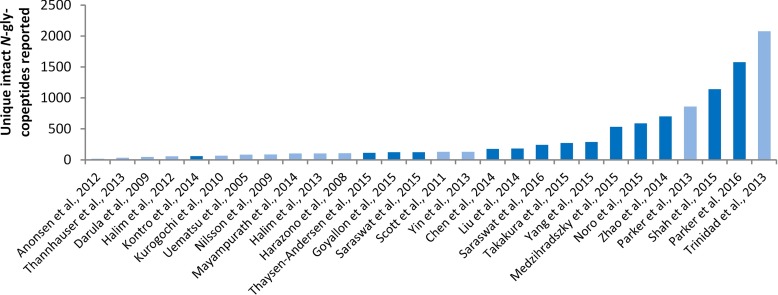
The technological advances described above have facilitated an increasing number of glycoproteomics-centered studies in recent years, Table II. As expected, considering their diverse experimental design (sample origin/amount, sample preparation, LC-MS/MS acquisition, and data handling strategies), these glycoproteomics studies differed markedly in their N-glycoproteome coverage, with some reporting fewer than 100 unique glycopeptides from less than 50 glycoproteins and others reporting thousands of unique glycopeptides from several hundred glycoproteins, Fig. 5. Using this metric to evaluate analytical performance it becomes clear that glycoproteomics is now capable of achieving a relatively deep coverage of the N-glycoproteome, albeit far from the extensive proteome coverage provided by modern ultra-sensitive proteomics technologies (172). This is particularly true when considering the immense size of the glycoproteome that is orders of magnitude larger than proteome. We would like to emphasize that although deep glycoproteome coverage is worth pursuing in our efforts to enhance our understanding of the structures and regulation associated with of the glycoproteome, important glycobiology can be learnt from glycoproteomics experiments without access to the latest high performance MS instrumentation provided an intelligent experimental design and hypothesis is applied.
Table II. Overview of recent N-glycoproteomics studies. Important experimental aspects and findings are summarized. N/A, information/data not provided.
| Biological sample | Main finding(s) | LC-MS/MS equipment | Fragmen-tation (acq. style) | Type of glycoproteomics analysis (method) | Comments | Unique N-glycopeptides/sites/proteins | Reference |
|---|---|---|---|---|---|---|---|
| Human serum (healthy, pancreatitis and pancreas cancer) | Regulation of sialylated subset of the N-glyco-proteome in pancreatitis and pancreas cancer | Synapt G2 HDMS Q-TOF | Beam-type CID (N/A) | Qualitative (GlycopeptideID) and quantitative (label-free XIC, int. standard) | Protein concentrations measured separately | 62/27/17 | Kontro et al., 2014 (104) |
| Human embryonic kidney (HEK293T cells) | In-depth N-glycoproteome analysis of the widely used HEK293T cells | LTQ-Orbitrap Velos | HCD (DDA) | Qualitative (manual) | De-N-glycoproteome also profiled | 177/82/N/A | Chen et al., 2014 (94) |
| Human serum | Site-specific analysis of serum N-glycoproteome | LTQ-OrbitrapXL | Ion trap CID and HCD (DDA) | Qualitative (GRIP) and quantitative (label-free spectral count) | De-N-glycoproteome also profiled + serum N-glycome database generated | 183/37/24 | Liu et al., 2014 (159) |
| Cynomolgus monkey plasma | In-depth site-specific analysis of monkey plasma N-glycoproteome | QSTAR XL, TripleTOF 5600 Q-TOF | Beam-type CID (DIA) | Qualitative (Byonic/manual) | Proteome database generated | 705/254/130 | Zhao et al., 2014 (135) |
| Human cystic fibrosis sputum | Discovery of unconventi-onal class of human N-gly-coproteins (paucimannose, Man1–3Fuc0–1-GlcNAc2) | Orbitrap-Elite | Ion trap CID, HCD and ETD (DDA) | Qualitative (Byonic) and quantitative (label-free XIC) | N-glycome and proteome also profiled | 115/36/30 | Thaysen-Andersen et al., 2015 (21) |
| Human cerebrospinal fluid | “Brain-type” N-glycoproteome signature of CSF | Maxis UHR Q-TOF | Beam-type CID (DDA) | Qualitative (GlycoQuest in ProteinScape) | N-glycome, proteome and de-N-glycoproteome also profiled | 124/55/36 | Goyallon et al., 2015 (162) |
| Human urinary exosomes | First site-specific analysis of the urinary exosome N-glycoproteome | Synapt G2 HDMS Q-TOF | Beam-type CID (N/A) | Qualitative (GlycopeptideID) | N-glycome also profiled | 126/51/37 | Saraswat et al., 2015 (88) |
| Human fetal lung fibroblast (TIG-1–20 cells) | In-depth site-specific analysis of the fibroblast membrane N-glycoproteome (comparison to serum) | LTQ-FT | Ion trap CID (DDA) | Qualitative (manual) and quantitative (label-free XIC) | De-N-glycoproteome also profiled | 272/63/44 | Takakura et al., 2015 (103) |
| Mouse heart tissue (WT vs transverse aortic constriction) | Regulation of the N-glycoproteome in response to transverse aortic constriction | Orbitrap Q-Exactive | HCD (DDA) | Qualitative (Proteome Discoverer, GPQuest) and quantitative (iTRAQ) | N-glycome, proteome and de-N-glycoproteome also profiled | 291/N/A/195 | Yang et al., 2015 (108) |
| Mouse liver | Tissue-specific N-glycoproteome signature from mouse liver relative to mouse brain | LTQ-Orbitrap Velos | HCD and ETD (DDA) | Qualitative (Protein Prospector) | Data compared qualitatively to mouse brain N- and O-glycoproteome | 534/168/106 | Medzihradszky et al., 2015 (13) |
| Mouse kidney (WT and FUT9 knock out) | Lewis X regulation of N-glycoproteome in FUT9 knock-out mice | LTQ-Orbitrap | HCD (DDA) | Qualitative (in-house program) | N-glycome and de-N-glycoproteome profiled, glycopeptides desialy-lated prior to profiling | 591/70/48 | Noro et al., 2015 (107) |
| Human prostate cancer cell cultures (LNCaP and PC3 cells) | Androgen-dependent N-glycosylation signatures in prostate cancer | Orbitrap Q-Exactive | HCD (DDA) | Qualitative (Byonic and GPQuest) and quantitative (iTRAQ) | Proteome and de-N-glycoproteome also profiled | 1145/N/A/227 | Shah et al., 2015 (46) |
| Mouse adipocyte cultures (WT and insulin resistant (IR) 3T3-L1 cells) | IR-induced N-glyco-proteome regulation (galactose and sialylation switching) | Orbitrap-Fusion | Ion trap CID, HCD, EThcD (DDA) | Qualitative (Byonic) and quantitative (SILAC) | N-glycome, proteome and de-N-glycoproteome also profiled | 1580/332/154 | Parker et al., 2016 (105) |
| Human seminal plasma | First deeper investigation of the N-glycoproteome of human seminal plasma | Synapt G2 HDMS Q-TOF | Beam-type CID (DDA) | Qualitative (GlycopeptideID) | N/A | 243/73/50 | Saraswat et al., 2016 (87) |
Fig. 5.
Nonchronological summary of the N-glycoproteome coverage as evaluated by the number of reported unique intact N-glycopeptides of glycoproteomics studies published over the past decade. Dark blue bars represent recent glycoproteomics studies (2014–present) covered in this review (see also Table II) and light blue bars represent older studies (2005–2014) covered elsewhere (59).
Glycoproteomics applications can be crudely classified into two types: deep glycoproteome profiling and comparative glycoproteomics. The former seeks to perform deep structural exploration of N-glycoproteomes derived from diverse bodily fluids, tissues/cells or even subcellular compartments thereby providing species-, tissue-, and protein-specific knowledge of the N-glycoproteome produced at a single condition, time and space (13, 21, 88, 103, 135, 159, 162). Although predominantly qualitative and descriptive in their nature, such efforts enhance our fundamental knowledge of glycobiology. This is particularly relevant considering the largely unexplored state of the N-glycoproteome even in many accessible biological tissues and fluids (173). Knowledge obtained from such efforts include a spatiotemporal understanding of protein- and tissue-specific N-glycosylation patterns (13), information that cannot be captured by (deglyco-) proteomics, glycomics or other -omics technologies. As described further below, deep glycoproteome profiling studies may also expand our understanding of the N-glycosylation machinery by identifying novel N-glycan structures that may only be present on a small subset of glycoproteins or restricted to specific tissues or physiological conditions. As such, deep site-specific glycoprotein profiling may ultimately provide new “structure-driven” insight into the capacity and regulatory mechanisms of the biosynthetic machinery (21).
The other class of glycoproteomics studies aims to identify alterations in a specified “N-glycoproteome system” comparing two or more biological conditions (46, 104, 105, 107, 108). Occasionally, such comparative glycoproteomics studies may also aim to pinpoint the underlying mechanism(s) driving the pathway dysregulation causing the altered N-glycosylation signatures (46, 105). In order to specify which of the three levels of regulation is responsible for the observed glycoproteome alterations (see Fig. 3), the proteome and site-specific occupancies are typically mapped in parallel with site-specific glycoform profiling. The molecular mechanisms controlling regulation can be supported by orthogonal techniques at the protein or glycan level, for example by using antibodies or lectins in Western/lectin blotting, protein/lectin arrays, ELISA, immunofluorescence, immunohistochemistry (and qPCR/RNAseq at the transcriptional level) or better at the glycoprotein level e.g. by proximity ligation assays (174) or using antibodies directed to shared glycan and polypeptide epitopes (175). Below, two recently published examples of these approaches are discussed to illustrate the potential of modern glycoproteomics.
Cystic Fibrosis Sputum Glycoproteomics
We previously performed N-glycomics of human sputum and found that a significant proportion of the N-glycome of sputum from inflamed and bacterial-infected cystic fibrosis patients (∼25–45%) was comprised of a new class of N-glycans, the so-called paucimannosidic N-glycans (Fuc0–1Man1–3GlcNAc2, see Fig. 1B) compared with healthy individuals (<5%) (18). By using an established glycoproteomics workflow (106), glycoproteome profiling confirmed the human origin and mapped the identity of the proteins carrying this unconventional modification. Because of limited biological starting material, crude mixtures of glycopeptides enriched solely by HILIC-SPE were profiled (92). Proteomics of de-N-glycosylated peptides together with the sputum N-glycome profiles, significantly limited the search space thereby yielding more confident and faster glycopeptide identifications and provided additional structural insights on the glycosylation of sputum proteins. The N-glycopeptides were analyzed using nano-RP-LC-MS/MS Orbitrap Elite instrumentation employing ion trap CID, HCD, and ETD fragmentation. HCD-MS/MS data were filtered for m/z 204.086 containing diagnostic oxonium ions (see Fig. 4) and used for automated identification of the peptide components of the glycopeptides by Byonic (see above). The ETD and resonance activation (ion trap) CID-MS/MS served as a source to verify the glycosylation sites and the topology of the glycan moieties of the identified glycopeptides, respectively. The glycoform distributions at the individual sites were profiled using XIC-based label-free relative quantitation. From a total of 115 unique N-glycopeptides covering 36 sites distributed across 30 human glycoproteins, paucimannosidic N-glycans were mapped to 35 unique glycopeptides from 23 sites and 18 proteins. The sputum glycoproteome and glycome data suggested that the paucimannosidic structures were carried by proteins located in the azurophilic granules of neutrophils in the (neutrophil-rich) sputum, which was confirmed by colocalization of immunofluorescence using subcellular- and paucimannosidic epitope-specific antibodies (21). A new spatially and temporally regulated biosynthetic pathway based on expression of β-hexosaminidases and α-mannosidases in specialized compartments of maturing neutrophils was suggested to generate paucimannosidic glycoproteins (see Fig 1A for the proposed truncation pathway) (21). Proteins isolated from the azurophilic granule of human neutrophils also carried further truncated epitopes of the chitobiose core type (Fuc0–1GlcNAc1–2, see Fig. 1B) (20). Recently, protein paucimannosylation was detected in other cell and tissue types including human fetal lung fibroblasts (103), saliva (176), epithelial colorectal (44, 177, 178) and breast (179) cancer cells, and in the mouse brain (19) also using glycoproteomics and other -omics technologies. Glycoproteomics was furthermore used to identify core and noncore fucosylated full and truncated chitobiose core type N-glycans from mouse synaptosome (7) and liver (13). Taken together, these findings illustrate the potential of modern glycoproteomics technologies to identify new classes of protein glycosylation hidden in the glycoproteome and to study their biosynthesis and spatial and temporal location.
Comparative Glycoproteomics of TNF-induced Insulin Resistance (IR) in Adipocytes
As discussed in more general terms above, glycoproteomics is a powerful tool to map the regulation of glycoproteomes derived from the same cellular system under different biological conditions. A prime example of this application is a recent study by Parker and colleagues, in which the regulation of the N-glycoproteome during TNF-based induction of IR was investigated in vitro in murine adipocytes (105). Wild-type and IR cells were grown in SILAC media and the N-glycome (87 N-glycans), proteome (7258 proteins), and the de-N-glycoproteome (2187 formerly N-glycosylated sites on 1041 proteins) were first profiled. The N-glycoproteome was then profiled using HILIC-SPE enriched and high-pH RP-LC off-line separated glycopeptides using Orbitrap Fusion LC-MS/MS instrumentation. The ten most abundant precursors in each cycle were selected for HCD fragmentation followed by PD (m/z 138.0545 and 204.0867) re-isolation and fragmentation of glycopeptides using EThcD- and ion trap CID-MS/MS. In total, 1580 unique N-glycopeptides covering 332 unique N-sites on 154 proteins were confidently identified using Byonic and quantified using the ion intensities of the isotopically labeled precursors. The novelty of this study was the ability to accurately detect glycoproteome alterations at the site-specific level and, importantly, to be able to differentiate their regulation at the protein, glycosylation site occupancy, and glycan level (see Fig. 3 and text for details). The vast majority of the N-glycoproteome alterations observed upon IR-induction could be accounted for by protein regulation; more than 200 proteins were significantly changed in abundance. Very limited differences in site occupancy (only three sites) and glycosylation micro-heterogeneity pattern (only 16 N-glycopeptides) were observed when the altered protein levels were adjusted for. Interestingly, the conventional proteomics experiments acquired on the Orbitrap Q-Exactive platform proved to be sufficiently sensitive to identify and accurately quantify 94 low-abundant glycosyltransferases and glycosidases involved in the glycosylation biosynthetic pathway. Together with transcriptomics data of altered expression of competing galactosyl- and sialyltransferases, these data yielded important clues to the enzymatic origin of the altered N-glycosylation signature associated with IR-transformation (terminal galactosylation and sialylation switching).
It could be speculated that glycomics or glycoepitope (e.g. lectin blotting)-centric analytical methods may have incorrectly assigned these protein abundance changes to quantitative glycosylation changes. It should also be recognized that other physiological conditions and tissues consisting of multiple cell types (in contrast to isolated homogeneous cell cultures investigate here) most likely would show much more complex glycoproteome regulation at all three molecular levels (glycan, occupancy, and protein level), supporting the relevance of performing this detailed level of molecular mapping. Similar impressive glycoproteomics efforts have recently been published in other biological systems using iTRAQ labeling and quantitation of glycopeptides (46, 108).
Understanding the exact mechanism(s) contributing to altered glycoproteomes in specific diseases is very important to advance our molecular knowledge of the underlying pathogenesis and when developing accurate biomarkers and targeted therapeutics toward the investigated pathologies. Thus, with the availability of the maturing glycoproteomics technologies, and their potential to provide unique structure-based insight into glycobiology, it is anticipated that similar comparative glycoproteomics strategies will be applied in the immediate future to investigate the glycoproteome regulation in a wide spectrum of biological systems.
CONCLUSIONS
In summary, acknowledging the wealth of biological and chemical information stored in the relatively unexplored N-glycoproteome, the past decade, and the recent two years in particular, have seen tremendous efforts to develop and implement efficient strategies for large-scale site-specific analysis of protein N-glycosylation. An increasing volume of tangible examples are now available in the literature demonstrating the enormous potential of glycoproteomics. The maturation of glycoproteomics has given glycoscientists a new powerful tool which importantly complements existing structural and functional glycobiological techniques and can serve as both initial discovery-type experiments for hypothesis generation or as a confirmatory tool for hypothesis testing. At present, glycoproteomics is still limited to relatively specialized research groups. However, with the recent advances and maturation of workflows, we anticipate that glycoproteomics will rapidly transition to become an accessible tool for the wider research community, in particular to established proteomics scientists. Ultimately this will allow glycoscientists and more general biomolecular scientists alike to explore the intriguing information stored in the spatiotemporally regulated glycoproteome.
Acknowledgments
We thank Dr. Benjamin L. Parker for stimulating glycoproteomics-centric discussions.
Footnotes
Author contribution: M.T.-A., N.H.P. and B.L.S. wrote the review.
* M.T.-A. was supported by an Early Career Fellowship from the Cancer Institute, NSW, Australia and by a Macquarie University Research Development Grant (MQRDG). B.L.S. was supported by a National Health and Medical Research Council R.D. Wright Biomedical Career Development Fellowship Level 2 (APP1087975). N.P. acknowledges the funding of the Australian Research Council Centre of Excellence in Nanoscale Biophotonics (CE140100003).
1 The abbreviations used are:
- OST
- oligosaccharyltransferase
- DDA
- data-dependent acquisition
- DIA
- data-independent acquisition
- ECD
- electron capture dissociation
- ER
- endoplasmic reticulum
- ETD
- electron transfer dissociation
- EThcD
- electron transfer and higher-energy collision dissociation
- FDR
- false discovery rate
- Fuc
- fucose
- GalNAc
- N-acetylgalactosamine
- GlcNAc
- N-acetylglucosamine
- HCD
- higher-energy collision dissociation
- HILIC
- hydrophilic interaction liquid chromatography
- IR
- insulin resistance
- iTRAQ
- isobaric tag for relative and absolute quantitation
- Man
- mannose
- MRM
- multiple reaction monitoring
- NeuAc
- N-acetylneuraminic acid
- NeuGc
- N-glycolylneuraminic acid
- PGC
- porous graphitized carbon
- RP
- reversed phase
- SILAC
- stable isotope labelling with amino acids in cell culture
- SPE
- solid phase extraction
- XIC
- extracted ion chromatogram.
REFERENCES
- 1. Varki A., and Sharon N. (2009) Historical Background and Overview. In: Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E., eds. Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor (NY) [PubMed] [Google Scholar]
- 2. Shrimal S., Cherepanova N. A., and Gilmore R. (2015) Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 41, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thaysen-Andersen M., and Packer N. H. (2012) Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 22, 1440–1452 [DOI] [PubMed] [Google Scholar]
- 4. Rudd P. M., and Dwek R. A. (1997) Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 32, 1–100 [DOI] [PubMed] [Google Scholar]
- 5. Lee L. Y., Lin C. H., Fanayan S., Packer N. H., and Thaysen-Andersen M. (2014) Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 5, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lizak C., Gerber S., Numao S., Aebi M., and Locher K. P. (2011) X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 7. Trinidad J. C., Schoepfer R., Burlingame A. L., and Medzihradszky K. F. (2013) N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 12, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 9. Tan Z., Yin H., Nie S., Lin Z., Zhu J., Ruffin M. T., Anderson M. A., Simeone D. M., and Lubman D. M. (2015) Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J. Proteome Res. 14, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asperger A., Marx K., Albers C., Molin L., and Pinato O. (2015) Low abundant N-linked glycosylation in hen egg white lysozyme is localized at nonconsensus sites. J. Proteome Res. 14, 2633–2641 [DOI] [PubMed] [Google Scholar]
- 11. Faid V., Denguir N., Chapuis V., Bihoreau N., and Chevreux G. (2014) Site-specific N-glycosylation analysis of human factor XI: Identification of a noncanonical NXC glycosite. Proteomics 14, 2460–2470 [DOI] [PubMed] [Google Scholar]
- 12. Chandler K. B., Brnakova Z., Sanda M., Wang S., Stalnaker S. H., Bridger R., Zhao P., Wells L., Edwards N. J., and Goldman R. (2014) Site-specific glycan microheterogeneity of inter-alpha-trypsin inhibitor heavy chain H4. J. Proteome Res. 13, 3314–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Medzihradszky K. F., Kaasik K., and Chalkley R. J. (2015) Tissue-Specific Glycosylation at the Glycopeptide Level. Mol. Cell. Proteomics 14, 2103–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valliere-Douglass J. F., Eakin C. M., Wallace A., Ketchem R. R., Wang W., Treuheit M. J., and Balland A. (2010) Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 285, 16012–16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zacchi L. F., and Schulz B. L. (2015) N-glycoprotein macroheterogeneity: biological implications and proteomic characterization. Glycoconj. J. In Press [DOI] [PubMed] [Google Scholar]
- 16. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 18. Venkatakrishnan V., Thaysen-Andersen M., Chen S. C., Nevalainen H., and Packer N. H. (2015) Cystic fibrosis and bacterial colonization define the sputum N-glycosylation phenotype. Glycobiology 25, 88–100 [DOI] [PubMed] [Google Scholar]
- 19. Dahmen A. C., Fergen M. T., Laurini C., Schmitz B., Loke I., Thaysen-Andersen M., and Diestel S. (2015) Paucimannosidic glycoepitopes are functionally involved in proliferation of neural progenitor cells in the subventricular zone. Glycobiology 25, 869–880 [DOI] [PubMed] [Google Scholar]
- 20. Loke I., Packer N. H., and Thaysen-Andersen M. (2015) Complementary LC-MS/MS-based N-glycan, N-glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil cathepsin G. Biomolecules 5, 1832–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thaysen-Andersen M., Venkatakrishnan V., Loke I., Laurini C., Diestel S., Parker B. L., and Packer N. H. (2015) Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 290, 8789–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeze H. H. (2013) Understanding human glycosylation disorders: biochemistry leads the charge. J. Biol. Chem. 288, 6936–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haltiwanger R. S., and Lowe J. B. (2004) Role of glycosylation in development. Annu. Rev. Biochem. 73, 491–537 [DOI] [PubMed] [Google Scholar]
- 24. Tran D. T., and Ten Hagen K. G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venkatakrishnan V., Packer N. H., and Thaysen-Andersen M. (2013) Host mucin glycosylation plays a role in bacterial adhesion in lungs of individuals with cystic fibrosis. Expert Rev. Respir. Med. 7, 553–576 [DOI] [PubMed] [Google Scholar]
- 26. Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christiansen M. N., Chik J., Lee L., Anugraham M., Abrahams J. L., and Packer N. H. (2013) Cell surface protein glycosylation in cancer. Proteomics 14, 525–546 [DOI] [PubMed] [Google Scholar]
- 28. Stuchlová Horynová M., Raška M., Clausen H., Novak J. (2013) Aberrant O-glycosylation and anti-glycan antibodies in an autoimmune disease IgA nephropathy and breast adenocarcinoma. Cell Mol. Life Sci. 70, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scott D. W., and Patel R. P. (2013) Endothelial heterogeneity and adhesion molecules N-glycosylation: implications in leukocyte trafficking in inflammation. Glycobiology 23, 622–633 [DOI] [PubMed] [Google Scholar]
- 30. Schedin-Weiss S., Winblad B., and Tjernberg L. O. (2014) The role of protein glycosylation in Alzheimer disease. FEBS J. 281, 46–62 [DOI] [PubMed] [Google Scholar]
- 31. Grigorian A., Mkhikian H., Li C. F., Newton B. L., Zhou R. W., and Demetriou M. (2012) Pathogenesis of multiple sclerosis via environmental and genetic dysregulation of N-glycosylation. Semin. Immunopathol. 34, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dalziel M., Crispin M., Scanlan C. N., Zitzmann N., and Dwek R. A. (2014) Emerging principles for the therapeutic exploitation of glycosylation. Science 343, 1235681. [DOI] [PubMed] [Google Scholar]
- 33. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 34. Song E., Hu Y., Hussein A., Yu C. Y., Tang H., and Mechref Y. (2015) Characterization of the glycosylation site of human PSA prompted by missense mutation using LC-MS/MS. J. Proteome Res. 14, 2872–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vinther L., Lademann U., Andersen E. V., Højrup P., Thaysen-Andersen M., Krogh B. O., Viuff B., Brünner N., Stenvang J., Moreira J. M. (2014) Purification and characterization of bioactive his6-tagged recombinant human tissue inhibitor of metalloproteinases-1 (TIMP-1) protein expressed at high yields in mammalian cells. Protein Expr. Purif. 101, 157–164 [DOI] [PubMed] [Google Scholar]
- 36. Ye H., Hill J., Gucinski A. C., Boyne M. T. 2nd, and Buhse L. F. (2015) Direct site-specific glycoform identification and quantitative comparison of glycoprotein therapeutics: imiglucerase and velaglucerase alfa. AAPS J. 17, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klapoetke S. (2014) N-glycosylation characterization by liquid chromatography with mass spectrometry. Methods Mol. Biol. 1131, 513–524 [DOI] [PubMed] [Google Scholar]
- 38. Thaysen-Andersen M., Chertova E., Bergamaschi C., Moh E. S., Chertov O., Roser J., Sowder R., Bear J., Lifson J., Packer N. H., Felber B. K., and Pavlakis G. N. (2015) Recombinant human heterodimeric IL-15 complex displays extensive and reproducible N- and O-linked glycosylation. Glycoconj J. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stavenhagen K., Plomp R., and Wuhrer M. (2015) Site-specific protein N- and O-glycosylation analysis by a C18-porous graphitized carbon-liquid chromatography-electrospray ionization mass spectrometry approach using pronase treated glycopeptides. Anal. Chem. 87, 11691–11699 [DOI] [PubMed] [Google Scholar]
- 40. Zhu F., Trinidad J. C., and Clemmer D. E. (2015) Glycopeptide site heterogeneity and structural diversity determined by combined lectin affinity chromatography/IMS/CID/MS techniques. J. Am. Soc. Mass Spectrom. 26, 1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pabst M., Küster S. K., Wahl F., Krismer J., Dittrich P. S., and Zenobi R. (2015) A microarray-matrix-assisted laser desorption/ionization-mass spectrometry approach for site-specific protein N-glycosylation analysis, as demonstrated for human serum immunoglobulin M (IgM). Mol. Cell. Proteomics 14, 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu G., Hitchen P. G., Panico M., North S. J., Barbouche M. R., Binet D., Morris H. R., Dell A., and Haslam S. M. (2015) Glycoproteomic studies of IgE from a novel hyper IgE syndrome linked to PGM3 mutation. Glycoconj J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meitei N. S., Apte A., Snovida S. I., Rogers J. C., and Saba J. (2015) Automating mass spectrometry-based quantitative glycomics using aminoxy tandem mass tag reagents with SimGlycan. J. Proteomics 127, 211–222 [DOI] [PubMed] [Google Scholar]
- 44. Sethi M. K., Kim H., Park C. K., Baker M. S., Paik Y. K., Packer N. H., Hancock W. S., Fanayan S., and Thaysen-Andersen M. (2015) In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology 25, 1064–1078 [DOI] [PubMed] [Google Scholar]
- 45. Stavenhagen K., Kolarich D., and Wuhrer M. (2015) Clinical glycomics employing graphitized carbon liquid chromatography-mass spectrometry. Chromatographia 78, 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shah P., Wang X., Yang W., Toghi Eshghi S., Sun S., Hoti N., Chen L., Yang S., Pasay J., Rubin A., Zhang H. (2015) Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveals glycoprotein alteration in protein abundance and glycosylation. Mol. Cell. Proteomics 14, 2753–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hyakumura M., Walsh R., Thaysen-Andersen M., Kingston N. J., La M., Lu L., Lovrecz G., Packer N. H., Locarnini S., and Netter H. J. (2015) Modification of asparagine-linked glycan density for the design of Hepatitis B virus virus-like particles with enhanced immunogenicity. J. Virol. 89, 11312–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang J., Qin H., Sun Z., Huang G., Mao J., Cheng K., Zhang Z., Wan H., Yao Y., Dong J., Zhu J., Wang F., Ye M., and Zou H. (2015) A peptide N-terminal protection strategy for comprehensive glycoproteome analysis using hydrazide chemistry based method. Sci. Rep. 5, 10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DeCoux A., Tian Y., DeLeon-Pennell K. Y., Nguyen N. T., de Castro Brás L. E., Flynn E. R., Cannon P. L., Griswold M. E., Jin Y. F., Puskarich M. A., Jones A. E., Lindsey M. L. (2015) Plasma glycoproteomics reveals sepsis outcomes linked to distinct proteins in common pathways. Crit. Care Med. 43, 2049–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sugahara D., Tomioka A., Sato T., Narimatsu H., and Kaji H. (2015) Large-scale identification of secretome glycoproteins recognized by Wisteria floribunda agglutinin: A glycoproteomic approach to biomarker discovery. Proteomics 15, 2921–2933 [DOI] [PubMed] [Google Scholar]
- 51. Pan S. (2014) Quantitative glycoproteomics for N-glycoproteome profiling. Methods Mol Biol 1156, 379–388 [DOI] [PubMed] [Google Scholar]
- 52. Hill J. J., Tremblay T. L., Fauteux F., Li J., Wang E., Aguilar-Mahecha A., Basik M., and O'Connor-McCourt M. (2015) Glycoproteomic comparison of clinical triple-negative and luminal breast tumors. J. Proteome Res. 14, 1376–1388 [DOI] [PubMed] [Google Scholar]
- 53. Sok Hwee Cheow E., Hwan Sim K., de Kleijn D., Neng Lee C., Sorokin V., and Sze S. K. (2015) Simultaneous enrichment of plasma soluble and extracellular vesicular glycoproteins using prolonged ultracentrifugation-electrostatic repulsion-hydrophilic interaction chromatography (PUC-ERLIC) approach. Mol. Cell. Proteomics 14, 1657–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng M., Fang Y., Han B., Xu X., Fan P., Hao Y., Qi Y., Hu H., Huo X., Meng L., Wu B., and Li J. (2015) In-depth N-glycosylation reveals species-specific modifications and functions of the royal jelly protein from Western (Apis mellifera) and Eastern Honeybees (Apis cerana). J. Proteome Res. 14, 5327–5340 [DOI] [PubMed] [Google Scholar]
- 55. DeLeon-Pennell K. Y., Tian Y., Zhang B., Cates C. A., Padmanabhan Iyer R., Cannon P., Shah P., Aiyetan P., Halade G. V., Ma Y., Flynn E., Zhang Z., Jin Y. F., Zhang H., and Lindsey M. L. (2016) CD36 is a matrix metalloproteinase-9 substrate that stimulates neutrophil apoptosis and removal during cardiac remodeling. Cardiovasc Res. 110, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang J., Wan H., Yao Y., Li J., Cheng K., Mao J., Chen J., Wang Y., Qin H., Zhang W., Ye M., and Zou H. (2015) Highly efficient release of glycopeptides from hydrazide beads by hydroxylamine assisted PNGase F deglycosylation for N-glycoproteome analysis. Anal. Chem. 87, 10199–10204 [DOI] [PubMed] [Google Scholar]
- 57. Clark D. J., Mei Y., Sun S., Zhang H., Yang A. J., and Mao L. (2016) Glycoproteomic approach identifies KRAS as a positive regulator of CREG1 in non-small cell lung cancer cells. Theranostics 6, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Palmisano G., Melo-Braga M. N., Engholm-Keller K., Parker B. L., and Larsen M. R. (2012) Chemical deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 59. Thaysen-Andersen M., and Packer N. H. (2014) Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 1844, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 60. Pagel O., Loroch S., Sickmann A., and Zahedi R. P. (2015) Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev. Proteomics 12, 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kolli V., Schumacher K. N., and Dodds E. D. (2015) Engaging challenges in glycoproteomics: recent advances in MS-based glycopeptide analysis. Bioanalysis 7, 113–131 [DOI] [PubMed] [Google Scholar]
- 62. Baycin Hizal D., Wolozny D., Colao J., Jacobson E., Tian Y., Krag S. S., Betenbaugh M. J., and Zhang H. (2014) Glycoproteomic and glycomic databases. Clin. Proteomics 11, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhu Z., and Desaire H. (2015) Carbohydrates on proteins: site-specific glycosylation analysis by mass spectrometry. Annu. Rev. Anal. Chem. 8, 463–483 [DOI] [PubMed] [Google Scholar]
- 64. Lazar I. M., Deng J., Ikenishi F., and Lazar A. C. (2015) Exploring the glycoproteomics landscape with advanced MS technologies. Electrophoresis 36, 225–237 [DOI] [PubMed] [Google Scholar]
- 65. Thaysen-Andersen M., Larsen M. R., Packer N. H., and Palmisano G. (2013) Structural analysis of glycoprotein sialylation - Part I: pre-LC-MS analytical strategies. Rsc. Advances 3, 22683–22705 [Google Scholar]
- 66. Palmisano G., Larsen M. R., Packer N. H., and Thaysen-Andersen M. (2013) Structural analysis of glycoprotein sialylation -part II: LC-MS based detection. Rsc. Advances 3, 22706–22726 [Google Scholar]
- 67. Moh E. S., Thaysen-Andersen M., and Packer N. H. (2015) Relative versus absolute quantitation in disease glycomics. Proteomics Clin. Appl. 9, 368–382 [DOI] [PubMed] [Google Scholar]
- 68. Miura Y., and Endo T. (2016) Glycomics and glycoproteomics focused on aging and age-related diseases - Glycans as a potential biomarker for physiological alterations. Biochim. Biophys. Acta. S0304–4165(16)00022–2 [DOI] [PubMed] [Google Scholar]
- 69. Burlingame A. L. (1996) Characterization of protein glycosylation by mass spectrometry. Curr. Opin. Biotechnol. 7, 4–10 [DOI] [PubMed] [Google Scholar]
- 70. Carr S. A., and Roberts G. D. (1986) Carbohydrate mapping by mass spectrometry: a novel method for identifying attachment sites of Asn-linked sugars in glycoproteins. Anal. Biochem. 157, 396–406 [DOI] [PubMed] [Google Scholar]
- 71. Huddleston M. J., Bean M. F., and Carr S. A. (1993) Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal. Chem. 65, 877–884 [DOI] [PubMed] [Google Scholar]
- 72. Küster B., Naven T. J., and Harvey D. J. (1996) Effect of the reducing-terminal substituents on the high energy collision-induced dissociation matrix-assisted laser desorption/ionization mass spectra of oligosaccharides. Rapid Commun. Mass Spectrom. 10, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 73. Townsend R. R., Heller D. N., Fenselau C. C., and Lee Y. C. (1984) Determination of the sialylation pattern of human fibrinogen glycopeptides with fast atom bombardment. Biochemistry 23, 6389–6392 [DOI] [PubMed] [Google Scholar]
- 74. Richards A. L., Merrill A. E., and Coon J. J. (2015) Proteome sequencing goes deep. Curr. Opin. Chem. Biol. 24, 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Peng W. P., Chou S. W., and Patil A. A. (2014) Measuring masses of large biomolecules and bioparticles using mass spectrometric techniques. Analyst 139, 3507–3523 [DOI] [PubMed] [Google Scholar]
- 76. Song E., and Mechref Y. (2015) Defining glycoprotein cancer biomarkers by MS in conjunction with glycoprotein enrichment. Biomark. Med. 9, 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Scott N. E., and Cordwell S. J. (2015) Enrichment and identification of bacterial glycopeptides by mass spectrometry. Methods Mol. Bio.l 1295, 355–368 [DOI] [PubMed] [Google Scholar]
- 78. Chen C. C., Su W. C., Huang B. Y., Chen Y. J., Tai H. C., and Obena R. P. (2014) Interaction modes and approaches to glycopeptide and glycoprotein enrichment. Analyst 139, 688–704 [DOI] [PubMed] [Google Scholar]
- 79. Zhao H., Li Y., and Hu Y. (2014) Nanotechnologies in glycoproteomics. Clin. Proteomics 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang J., Wang Y., Gao M., Zhang X., and Yang P. (2015) Multilayer hydrophilic poly(phenol-formaldehyde resin)-coated magnetic graphene for boronic acid immobilization as a novel matrix for glycoproteome analysis. ACS Appl. Mater. Interfaces. 7, 16011–16017 [DOI] [PubMed] [Google Scholar]
- 81. Wang M., Zhang X., and Deng C. (2015) Facile synthesis of magnetic poly(styrene-co-4-vinylbenzene-boronic acid) microspheres for selective enrichment of glycopeptides. Proteomics 15, 2158–2165 [DOI] [PubMed] [Google Scholar]
- 82. Chen W., Smeekens J. M., and Wu R. (2014) A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS). Mol. Cell. Proteomics 13, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xu Y., Bailey U. M., Punyadeera C., and Schulz B. L. (2014) Identification of salivary N-glycoproteins and measurement of glycosylation site occupancy by boronate glycoprotein enrichment and liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 28, 471–482 [DOI] [PubMed] [Google Scholar]
- 84. Woo C. M., Iavarone A. T., Spiciarich D. R., Palaniappan K. K., and Bertozzi C. R. (2015) Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Smeekens J. M., Chen W., and Wu R. (2015) Mass spectrometric analysis of the cell surface N-glycoproteome by combining metabolic labeling and click chemistry. J. Am. Soc. Mass Spectrom. 26, 604–614 [DOI] [PubMed] [Google Scholar]
- 86. Takakura D., Harazono A., Hashii N., and Kawasaki N. (2014) Selective glycopeptide profiling by acetone enrichment and LC/MS. J. Proteomics 101, 17–30 [DOI] [PubMed] [Google Scholar]
- 87. Saraswat M., Joenväärä S., Tomar A. K., Singh S., Yadav S., Renkonen R. J.. Proteome Res. 15, 991–1001 (2016) N-glycoproteomics of human seminal plasma glycoproteins. J. Proteome Res. [DOI] [PubMed] [Google Scholar]
- 88. Saraswat M., Joenvaara S., Musante L., Peltoniemi H., Holthofer H., and Renkonen R. (2015) N-linked (N-) glycoproteomics of urimary exosomes. Mol. Cell. Proteomics 14, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu J., Wang F., Zhu J., Mao J., Liu Z., Cheng K., Qin H., and Zou H. (2014) Highly efficient N-glycoproteomic sample preparation by combining C(18) and graphitized carbon adsorbents. Anal. Bioanal. Chem. 406, 3103–3109 [DOI] [PubMed] [Google Scholar]
- 90. Melo-Braga M. N., Schulz M., Liu Q., Swistowski A., Palmisano G., Engholm-Keller K., Jakobsen L., Zeng X., and Larsen M. R. (2014) Comprehensive quantitative comparison of the membrane proteome, phosphoproteome, and sialiome of human embryonic and neural stem cells. Mol. Cell. Proteomics 13, 311–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Thaysen-Andersen M., Thøgersen I. B., Nielsen H. J., Lademann U., Brünner N., Enghild J. J., Højrup P. (2007) Rapid and individual-specific glycoprofiling of the low abundance N-glycosylated protein tissue inhibitor of metalloproteinases-1. Mol. Cell. Proteomics 6, 638–647 [DOI] [PubMed] [Google Scholar]
- 92. Mysling S., Palmisano G., Højrup P., Thaysen-Andersen M. (2010) Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82, 5598–5609 [DOI] [PubMed] [Google Scholar]
- 93. Hägglund P., Bunkenborg J., Elortza F., Jensen O. N., Roepstorff P. (2004) A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3, 556–566 [DOI] [PubMed] [Google Scholar]
- 94. Chen R., Seebun D., Ye M., Zou H., and Figeys D. (2014) Site-specific characterization of cell membrane N-glycosylation with integrated hydrophilic interaction chromatography solid phase extraction and LC-MS/MS. J. Proteomics 103, 194–203 [DOI] [PubMed] [Google Scholar]
- 95. Dedvisitsakul P., Jacobsen S., Svensson B., Bunkenborg J., Finnie C., Hägglund P. (2014) Glycopeptide enrichment using a combination of ZIC-HILIC and cotton wool for exploring the glycoproteome of wheat flour albumins. J. Proteome Res. 13, 2696–2703 [DOI] [PubMed] [Google Scholar]
- 96. Dedvisitsakul P., Jacobsen S., Svensson B., Bunkenborg J., Finnie C., Hägglund P. (2014) Sample preparation for mass spectrometric analysis of human serum N-glycans using hydrophilic interaction chromatography-based solid phase extraction. Analyst 139, 4538–4546 [DOI] [PubMed] [Google Scholar]
- 97. Cao L., Yu L., Guo Z., Shen A., Guo Y., and Liang X. (2014) N-Glycosylation site analysis of proteins from Saccharomyces cerevisiae by using hydrophilic interaction liquid chromatography-based enrichment, parallel deglycosylation, and mass spectrometry. J. Proteome Res. 13, 1485–1493 [DOI] [PubMed] [Google Scholar]
- 98. Pan Y., Ma C., Tong W., Fan C., Zhang Q., Zhang W., Tian F., Peng B., Qin W., and Qian X. (2015) Preparation of sequence-controlled triblock copolymer-grafted silica microparticles by sequential-ATRP for highly efficient glycopeptides enrichment. Anal. Chem. 87, 656–662 [DOI] [PubMed] [Google Scholar]
- 99. Huang G., Xiong Z., Qin H., Zhu J., Sun Z., Zhang Y., Peng X., ou J., and Zou H. (2014) Synthesis of zwitterionic polymer brushes hybrid silica nanoparticles via controlled polymerization for highly efficient enrichment of glycopeptides. Anal. Chim. Acta 809, 61–68 [DOI] [PubMed] [Google Scholar]
- 100. Chen Y., Xiong Z., Zhang L., Zhao J., Zhang Q., Peng L., Zhang W., Ye M., and Zou H. (2015) Facile synthesis of zwitterionic polymer-coated core-shell magnetic nanoparticles for highly specific capture of N-linked glycopeptides. Nanoscale 7, 3100–3108 [DOI] [PubMed] [Google Scholar]
- 101. Bodnar E. D., and Perreault H. (2015) Synthesis and evaluation of carboxymethyl chitosan for glycopeptide enrichment. Anal. Chim. Acta 891, 179–189 [DOI] [PubMed] [Google Scholar]
- 102. Zheng J., Xiao Y., Wang L., Lin Z., Yang H., Zhang L., and Chen G. (2014) Click synthesis of glucose-functionalized hydrophilic magnetic mesoporous nanoparticles for highly selective enrichment of glycopeptides and glycans. J. Chromatogr. A 1358, 29–38 [DOI] [PubMed] [Google Scholar]
- 103. Takakura D., Tada M., and Kawasaki N. (2016) Membrane glycoproteomics of fetal lung fibroblasts using LC/MS. Proteomics 16, 47–59 [DOI] [PubMed] [Google Scholar]
- 104. Kontro H., Joenväärä S., Haglund C., Renkonen R. (2014) Comparison of sialylated N-glycopeptide levels in serum of pancreatic cancer patients, acute pancreatitis patients, and healthy controls. Proteomics 14, 1713–1723 [DOI] [PubMed] [Google Scholar]
- 105. Parker B. L., Thaysen-Andersen M., Fazakerley D. J., Holliday M., Packer N. H., and James D. E. (2016) Terminal galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes. Mol. Cell. Proteomics 15, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Parker B. L., Thaysen-Andersen M., Solis N., Scott N. E., Larsen M. R., Graham M. E., Packer N. H., and Cordwell S. J. (2013) Site-specific glycan-Peptide analysis for determination of N-glycoproteome heterogeneity. J. Proteome Res. 12, 5791–5800 [DOI] [PubMed] [Google Scholar]
- 107. Noro E., Togayachi A., Sato T., Tomioka A., Fujita M., Sukegawa M., Suzuki N., Kaji H., and Narimatsu H. (2015) Large-scale identification of N-glycan glycoproteins carrying lewis x and site-specific N-glycan alterations in Fut9 Knockout mice. J. Proteome Res. 14, 3823–3834 [DOI] [PubMed] [Google Scholar]
- 108. Yang S., Mishra S., Chen L., Zhou J. Y., Chan D. W., Chatterjee S., and Zhang H. (2015) Integrated glycoprotein immobilization method for glycopeptide and glycan analysis of cardiac hypertrophy. Anal. Chem. 87, 9671–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nilsson J. (2016) Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides. Glycoconj J. In Press [DOI] [PubMed] [Google Scholar]
- 110. Cao L., Tolić N., Qu Y., Meng D., Zhao R., Zhang Q., Moore R. J., Zink E. M., Lipton M. S., Paša-Tolić L., Wu S. (2014) Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation. Anal. Biochem. 452, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Aboufazeli F., Kolli V., and Dodds E. D. (2015) A comparison of energy-resolved vibrational activation/dissociation characteristics of protonated and sodiated high mannose N-glycopeptides. J. Am. Soc. Mass Spectrom. 26, 587–595 [DOI] [PubMed] [Google Scholar]
- 112. Hinneburg H., Stavenhagen K., Schweiger-Hufnagel U., Pengelley S., Jabs W., Seeberger P. H., Silva D. V., Wuhrer M., and Kolarich D. (2016) The art of destruction: optimizing collision energies in quadrupole-time of flight (Q-TOF) instruments for glycopeptide-based glycoproteomics. J. Am. Soc. Mass Spectrom. 27, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kolli V., Roth H. A., De La Cruz G., Fernando G. S., and Dodds E. D. (2015) The role of proton mobility in determining the energy-resolved vibrational activation/dissociation channels of N-glycopeptide ions. Anal. Chim. Acta 896, 85–92 [DOI] [PubMed] [Google Scholar]
- 114. Manri N., Satake H., Kaneko A., Hirabayashi A., Baba T., and Sakamoto T. (2013) Glycopeptide identification using liquid-chromatography-compatible hot electron capture dissociation in a radio-frequency-quadrupole ion trap. Anal. Chem. 85, 2056–2063 [DOI] [PubMed] [Google Scholar]
- 115. Håkansson K., Cooper H. J., Emmett M. R., Costello C. E., Marshall A. G., Nilsson C. L. (2001) Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal. Chem. 73, 4530–4536 [DOI] [PubMed] [Google Scholar]
- 116. Domon B., and Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5, 397–409 [Google Scholar]
- 117. Biemann K. (1990) Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods Enzymol. 193, 886–887 [DOI] [PubMed] [Google Scholar]
- 118. Darula Z., and Medzihradszky K. F. (2015) Carbamidomethylation side reactions may lead to glycan misassignments in glycopeptide analysis. Anal. Chem. 87, 6297–6302 [DOI] [PubMed] [Google Scholar]
- 119. Fang P., Liu M., Xue Y., Yao J., Zhang Y., Shen H., and Yang P. (2015) Controlling nonspecific trypsin cleavages in LC-MS/MS-based shotgun proteomics using optimized experimental conditions. Analyst 140, 7613–7621 [DOI] [PubMed] [Google Scholar]
- 120. Darula Z., and Medzihradszky K. F. (2014) Glycan side reaction may compromise ETD-based glycopeptide identification. J. Am. Soc. Mass Spectrom. 25, 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mommen G. P., Frese C. K., Meiring H. D., van Gaans-van den Brink J., de Jong A. P., van Els C. A., and Heck A. J. (2014) Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc. Natl. Acad. Sci. U.S.A. 111, 4507–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hu H., Khatri K., Klein J., Leymarie N., and Zaia J. (2015) A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconj J. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Halim A., Westerlind U., Pett C., Schorlemer M., Rüetschi U., Brinkmalm G., Sihlbom C., Lengqvist J., Larson G., Nilsson J. (2014) Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 [DOI] [PubMed] [Google Scholar]
- 124. Yu J., Schorlemer M., Gomez Toledo A., Pett C., Sihlbom C., Larson G., Westerlind U., and Nilsson J. (2016) Distinctive MS/MS Fragmentation Pathways of Glycopeptide-Generated Oxonium Ions Provide Evidence of the Glycan Structure. Chemistry 22, 1114–1124 [DOI] [PubMed] [Google Scholar]
- 125. Saba J., Dutta S., Hemenway E., and Viner R.. Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. Int. J. Proteomics 2012:560391, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wu S. W., Pu T. H., Viner R., and Khoo K. H. (2014) Novel LC-MS(2) product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal. Chem. 86, 5478–5486 [DOI] [PubMed] [Google Scholar]
- 127. Froehlich J. W., Dodds E. D., Wilhelm M., Serang O., Steen J. A., and Lee R. S. (2013) A classifier based on accurate mass measurements to aid large scale, unbiased glycoproteomics. Mol. Cell. Proteomics 12, 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ito H., Kaji H., Togayachi A., Azadi P., Ishihara M., Geyer R., Galuska C., Geyer H., Kakehi K., Kinoshita M., Karlsson N. G., Jin C., Kato K., Yagi H., Kondo S., Kawasaki N., Hashii N., Kolarich D., Stavenhagen K., Packer N. H., Thaysen-Andersen M., Nakano M., Taniguchi N., Kurimoto A., Wada Y., Tajiri M., Yang P., Cao W., Li H., Rudd P. M., and Narimatsu H. (2015) Comparison of analytical methods for profiling N- and O-linked glycans from cultured cell lines : HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycoconj J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sarbu M., Zhu F., Peter-Katalinić J., Clemmer D. E., Zamfir A. D. (2015) Application of ion mobility tandem mass spectrometry to compositional and structural analysis of glycopeptides extracted from the urine of a patient diagnosed with Schindler disease. Rapid Commun. Mass Spectrom. 29, 1929–1937 [DOI] [PubMed] [Google Scholar]
- 130. Nishikaze T., Kawabata S., and Tanaka K. (2014) In-depth structural characterization of N-linked glycopeptides using complete derivatization for carboxyl groups followed by positive- and negative-ion tandem mass spectrometry. Anal. Chem. 86, 5360–5369 [DOI] [PubMed] [Google Scholar]
- 131. Nishikaze T., Kawabata S., and Tanaka K. (2014) Fragmentation characteristics of deprotonated N-linked glycopeptides: influences of amino acid composition and sequence. J. Am. Soc. Mass Spectrom. 25, 988–998 [DOI] [PubMed] [Google Scholar]
- 132. de Haan N., Reiding K. R., Haberger M., Reusch D., Falck D., and Wuhrer M. (2015) Linkage-specific sialic acid derivatization for MALDI-TOF-MS profiling of IgG glycopeptides. Anal. Chem. 87, 8284–8291 [DOI] [PubMed] [Google Scholar]
- 133. Gomes de Oliveira A. G., Roy R., Raymond C., Bodnar E. D., Tayi V. S., Butler M., Durocher Y., and Perreault H. (2015) A systematic study of glycopeptide esterification for the semi-quantitative determination of sialylation in antibodies. Rapid Commun. Mass Spectrom. 29, 1817–1826 [DOI] [PubMed] [Google Scholar]
- 134. Hong Q., Ruhaak L. R., Stroble C., Parker E., Huang J., Maverakis E., and Lebrilla C. B. (2015) A method for comprehensive glycosite-mapping and direct quantitation of serum glycoproteins. J. Proteome Re.s 14, 5179–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhao Y., Szeto S. S., Kong R. P., Law C. H., Li G., Quan Q., Zhang Z., Wang Y., and Chu I. K. (2014) Online two-dimensional porous graphitic carbon/reversed phase liquid chromatography platform applied to shotgun proteomics and glycoproteomics. Anal. Chem. 86, 12172–12179 [DOI] [PubMed] [Google Scholar]
- 136. An Y., McCullers J. A., Alymova I., Parsons L. M., and Cipollo J. F. (2015) Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J. Proteome Res. 14, 3957–3969 [DOI] [PubMed] [Google Scholar]
- 137. Xu Y., Bailey U. M., and Schulz B. L. (2015) Automated measurement of site-specific N-glycosylation occupancy with SWATH-MS. Proteomics 15, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 138. Ruiz-May E., Hucko S., Howe K. J., Zhang S., Sherwood R. W., Thannhauser T. W., and Rose J. K. (2014) A comparative study of lectin affinity based plant N-glycoproteome profiling using tomato fruit as a model. Mol. Cell. Proteomics 13, 566–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Goldman R., and Sanda M. (2015) Targeted methods for quantitative analysis of protein glycosylation. Proteomics Clin. Appl. 9, 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Song E., Zhu R., Hammoud Z. T., and Mechref Y. (2014) LC-MS/MS quantitation of esophagus disease blood serum glycoproteins by enrichment with hydrazide chemistry and lectin affinity chromatography. J. Proteome Res. 13, 4808–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Benicky J., Sanda M., Pompach P., Wu J., and Goldman R. (2014) Quantification of fucosylated hemopexin and complement factor H in plasma of patients with liver disease. Anal. Chem. 86, 10716–10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Yuan W., Sanda M., Wu J., Koomen J., and Goldman R. (2015) Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LC-MS-MRM in liver disease. J. Proteomics 116, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hong Q., Lebrilla C. B., Miyamoto S., and Ruhaak L. R. (2013) Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Anal. Chem. 85, 8585–8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang B., Tsybovsky Y., Palczewski K., and Chance M. R. (2014) Reliable determination of site-specific in vivo protein N-glycosylation based on collision-induced MS/MS and chromatographic retention time. J. Am. Soc. Mass Spectrom. 25, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhang Z., Sun Z., Zhu J., Liu J., Huang G., Ye M., and Zou H. (2014) High-throughput determination of the site-specific N-sialoglycan occupancy rates by differential oxidation of glycoproteins followed with quantitative glycoproteomics analysis. Anal. Chem. 86, 9830–9837 [DOI] [PubMed] [Google Scholar]
- 146. Song W., Mentink R. A., Henquet M. G., Cordewener J. H., van Dijk A. D., Bosch D., America A. H., and van der Krol A. R. (2013) N-glycan occupancy of Arabidopsis N-glycoproteins. J. Proteomics 93, 343–355 [DOI] [PubMed] [Google Scholar]
- 147. Liu G., and Neelamegham S. (2015) Integration of systems glycobiology with bioinformatics toolboxes, glycoinformatics resources, and glycoproteomics data. Wiley Interdiscip. Rev. Syst. Biol. Med. 7, 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hu H., Khatri K., and Zaia J. (2016) Algorithms and design strategies towards automated glycoproteomics analysis. Mass Spectrom. Rev. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tang H., Mayampurath A., Yu C. Y., and Mechref Y. (2014) Bioinformatics protocols in glycomics and glycoproteomics. Curr. Protoc. Protein Sci. 76, 1–7 [DOI] [PubMed] [Google Scholar]
- 150. Bern M., Kil Y. J., and Becker C. (2012) Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics Chapter 13, Unit13 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Medzihradszky K. F., Kaasik K., and Chalkley R. J. (2015) Characterizing sialic acid variants at the glycopeptide level. Anal. Chem. 87, 3064–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Toghi Eshghi S., Shah P., Yang W., Li X., and Zhang H. (2015) GPQuest: a spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Anal. Chem. 87, 5181–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Peltoniemi H., Natunen S., Ritamo I., Valmu L., Räbinä J. (2013) Novel data analysis tool for semiquantitative LC-MS-MS2 profiling of N-glycans. Glycoconj. J. 30, 159–170 [DOI] [PubMed] [Google Scholar]
- 154. Wu S. W., Liang S. Y., Pu T. H., Chang F. Y., and Khoo K. H. (2013) Sweet-Heart - an integrated suite of enabling computational tools for automated MS2/MS3 sequencing and identification of glycopeptides. J. Proteomics 84, 1–16 [DOI] [PubMed] [Google Scholar]
- 155. Liang S. Y., Wu S. W., Pu T. H., Chang F. Y., and Khoo K. H. (2014) An adaptive workflow coupled with Random Forest algorithm to identify intact N-glycopeptides detected from mass spectrometry. Bioinformatics 30, 1908–1916 [DOI] [PubMed] [Google Scholar]
- 156. He L., Xin L., Shan B., Lajoie G. A., and Ma B. (2014) GlycoMaster DB: software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. J. Proteome Res. 13, 3881–3895 [DOI] [PubMed] [Google Scholar]
- 157. Mayampurath A., Yu C. Y., Song E., Balan J., Mechref Y., and Tang H. (2014) Computational framework for identification of intact glycopeptides in complex samples. Anal. Chem. 86, 453–463 [DOI] [PubMed] [Google Scholar]
- 158. Zhu Z., Su X., Go E. P., and Desaire H. (2014) New glycoproteomics software, GlycoPep Evaluator, generates decoy glycopeptides de novo and enables accurate false discovery rate analysis for small data sets. Anal. Chem. 86, 9212–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Liu M., Zhang Y., Chen Y., Yan G., Shen C., Cao J., Zhou X., Liu X., Zhang L., Shen H., Lu H., He F., and Yang P. (2014) Efficient and accurate glycopeptide identification pipeline for high-throughput site-specific N-glycosylation analysis. J. Proteome Res. 13, 3121–3129 [DOI] [PubMed] [Google Scholar]
- 160. Jansen B. C., Reiding K. R., Bondt A., Hipgrave Ederveen A. L., Palmblad M., Falck D., and Wuhrer M. (2015) MassyTools: A high-throughput targeted data processing tool for relative quantitation and quality control developed for glycomic and glycoproteomic MALDI-MS. J. Proteome Res. 14, 5088–5098 [DOI] [PubMed] [Google Scholar]
- 161. Lynn K. S., Chen C. C., Lih T. M., Cheng C. W., Su W. C., Chang C. H., Cheng C. Y., Hsu W. L., Chen Y. J., and Sung T. Y. (2015) MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS(2) approach. Anal. Chem. 87, 2466–2473 [DOI] [PubMed] [Google Scholar]
- 162. Goyallon A., Cholet S., Chapelle M., Junot C., and Fenaille F. (2015) Evaluation of a combined glycomics and glycoproteomics approach for studying the major glycoproteins present in biofluids: Application to cerebrospinal fluid. Rapid Commun. Mass Spectrom. 29, 461–473 [DOI] [PubMed] [Google Scholar]
- 163. Medzihradszky K. F. (2014) Noncovalent dimer formation in liquid chromatography-mass spectrometry analysis. Anal. Chem. 86, 8906–8909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kurogochi M., and Amano J. (2014) Relative quantitation of glycopeptides based on stable isotope labeling using MALDI-TOF MS. Molecules 19, 9944–9961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Mayampurath A., Song E., Mathur A., Yu C. Y., Hammoud Z., Mechref Y., and Tang H. (2014) Label-free glycopeptide quantification for biomarker discovery in human sera. J. Proteome Res. 13, 4821–4832 [DOI] [PubMed] [Google Scholar]
- 166. Kim J. Y., Oh D., Kim S. K., Kang D., and Moon M. H. (2014) Isotope-coded carbamidomethylation for quantification of N-glycoproteins with online microbore hollow fiber enzyme reactor-nanoflow liquid chromatography-tandem mass spectrometry. Anal. Chem. 86, 7650–7657 [DOI] [PubMed] [Google Scholar]
- 167. Kovanich D., Cappadona S., Raijmakers R., Mohammed S., Scholten A., and Heck A. J. (2012) Applications of stable isotope dimethyl labeling in quantitative proteomics. Anal. Bioanal. Chem. 404, 991–1009 [DOI] [PubMed] [Google Scholar]
- 168. Pabst M., Benešová I., Fagerer S. R., Jacobsen M., Eyer K., Schmidt G., Steinhoff R., Krismer J., Wahl F., Preisler J., Zenobi R. (2016) Differential isotope labeling of glycopeptides for accurate determination of differences in site-specific glycosylation. J. Proteome Res. 15, 326–331 [DOI] [PubMed] [Google Scholar]
- 169. Martínez-Bartolomé S., Deutsch E. W., Binz P. A., Jones A. R., Eisenacher M., Mayer G., Campos A., Canals F., Bech-Serra J. J., Carrascal M., Gay M., Paradela A., Navajas R., Marcilla M., Hernáez M. L., Gutiérrez-Blázquez M. D., Velarde L. F., Aloria K., Beaskoetxea J., Medina-Aunon J. A., Albar J. P. (2013) Guidelines for reporting quantitative mass spectrometry based experiments in proteomics. J. Proteomics 95, 84–88 [DOI] [PubMed] [Google Scholar]
- 170. York W. S., Agravat S., Aoki-Kinoshita K. F., McBride R., Campbell M. P., Costello C. E., Dell A., Feizi T., Haslam S. M., Karlsson N., Khoo K. H., Kolarich D., Liu Y., Novotny M., Packer N. H., Paulson J. C., Rapp E., Ranzinger R., Rudd P. M., Smith D. F., Struwe W. B., Tiemeyer M., Wells L., Zaia J., and Kettner C. (2014) MIRAGE: the minimum information required for a glycomics experiment. Glycobiology 24, 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Campbell M. P., Peterson R., Mariethoz J., Gasteiger E., Akune Y., Aoki-Kinoshita K. F., Lisacek F., and Packer N. H. (2014) UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 42, D215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Beck S., Michalski A., Raether O., Lubeck M., Kaspar S., Goedecke N., Baessmann C., Hornburg D., Meier F., Paron I., Kulak N. A., Cox J., and Mann M. (2015) The Impact II, a Very High-Resolution Quadrupole Time-of-Flight Instrument (QTOF) for Deep Shotgun Proteomics. Mol. Cell. Proteomics 14, 2014–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Clerc F., Reiding K. R., Jansen B. C., Kammeijer G. S., Bondt A., and Wuhrer M. (2015) Human plasma protein N-glycosylation. Glycoconj J. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Greenwood C., Johnson G., Dhillon H. S., and Bustin S. (2015) Recent progress in developing proximity ligation assays for pathogen detection. Expert Rev. Mol. Diagn. 15, 861–867 [DOI] [PubMed] [Google Scholar]
- 175. Chen W., and Bahl O. P. (1992) Polyclonal antibodies against the polypeptide and carbohydrate epitopes of recombinant human choriogonadotropin beta-subunit. Mol. Cell. Endocrinol. 86, 57–66 [DOI] [PubMed] [Google Scholar]
- 176. Everest-Dass A. V., Jin D., Thaysen-Andersen M., Nevalainen H., Kolarich D., and Packer N. H. (2012) Comparative structural analysis of the glycosylation of salivary and buccal cell proteins: innate protection against infection by Candida albicans. Glycobiology 22, 1465–1479 [DOI] [PubMed] [Google Scholar]
- 177. Sethi M. K., Thaysen-Andersen M., Smith J. T., Baker M. S., Packer N. H., Hancock W. S., and Fanayan S. (2014) Comparative N-glycan profiling of colorectal cancer cell lines reveals unique bisecting GlcNAc and alpha-2,3-linked sialic acid determinants are associated with membrane proteins of the more metastatic/aggressive cell lines. J. Proteome Res. 13, 277–288 [DOI] [PubMed] [Google Scholar]
- 178. Balog C. I., Stavenhagen K., Fung W. L., Koeleman C. A., McDonnell L. A., Verhoeven A., Mesker W. E., Tollenaar R. A., Deelder A. M., and Wuhrer M. (2012) N-glycosylation of colorectal cancer tissues: a liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics 11, 571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lee L. Y., Thaysen-Andersen M., Baker M. S., Packer N. H., Hancock W. S., and Fanayan S. (2014) Comprehensive N-glycome profiling of cultured human epithelial breast cells identifies unique secretome N-glycosylation signatures enabling tumorigenic subtype classification. J. Proteome Res. 13, 4783–4795 [DOI] [PubMed] [Google Scholar]
- 180. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., Etzler M. E., Frank M., Vliegenthart J. F., Lütteke T., Perez S., Bolton E., Rudd P., Paulson J., Kanehisa M., Toukach P., Aoki-Kinoshita K. F., Dell A., Narimatsu H., York W., Taniguchi N., Kornfeld S. (2015) Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ma C., Zhang Q., Qu J., Zhao X., Li X., Liu Y., and Wang P. G. (2015) A precise approach in large scale core-fucosylated glycoprotein identification with low- and high-normalized collision energy. J. Proteomics 114, 61–70 [DOI] [PubMed] [Google Scholar]
- 182. Cao Q., Zhao X., Zhao Q., Lv X., Ma C., Li X., Zhao Y., Peng B., Ying W., and Qian X. (2014) Strategy integrating stepped fragmentation and glycan diagnostic ion-based spectrum refinement for the identification of core fucosylated glycoproteome using mass spectrometry. Anal. Chem. 86, 6804–6811 [DOI] [PubMed] [Google Scholar]
- 183. Cheng K., Chen R., Seebun D., Ye M., Figeys D., and Zou H. (2014) Large-scale characterization of intact N-glycopeptides using an automated glycoproteomic method. J. Proteomics 110, 145–154 [DOI] [PubMed] [Google Scholar]
- 184. Scott N. E., Marzook N. B., Cain J. A., Solis N., Thaysen-Andersen M., Djordjevic S. P., Packer N. H., Larsen M. R., and Cordwell S. J. (2014) Comparative proteomics and glycoproteomics reveal increased N-linked glycosylation and relaxed sequon specificity in Campylobacter jejuni NCTC11168 O. J. Proteome Res. 13, 5136–5150 [DOI] [PubMed] [Google Scholar]
- 185. Cao Q., Ma C., Bai H., Li X., Yan H., Zhao Y., Ying W., and Qian X. (2014) Multivalent hydrazide-functionalized magnetic nanoparticles for glycopeptide enrichment and identification. Analyst 139, 603–609 [DOI] [PubMed] [Google Scholar]
- 186. Khatri K., Staples G. O., Leymarie N., Leon D. R., Turiák L., Huang Y., Yip S., Hu H., Heckendorf C. F., Zaia J. (2014) Confident assignment of site-specific glycosylation in complex glycoproteins in a single step. J. Proteome Res. 13, 4347–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ford K. L., Zeng W., Heazlewood J. L., and Bacic A. (2015) Characterization of protein N-glycosylation by tandem mass spectrometry using complementary fragmentation techniques. Front. Plant Sci. 6, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]