Abstract
Protein arginylation is a posttranslational modification of both N-terminal amino acids of proteins and sidechain carboxylates and can be crucial for viability and physiology in higher eukaryotes. The lack of arginylation causes severe developmental defects in moss, affects the low oxygen response in Arabidopsis thaliana and is embryo lethal in Drosophila and in mice. Although several studies investigated impact and function of the responsible enzyme, the arginyl-tRNA protein transferase (ATE) in plants, identification of arginylated proteins by mass spectrometry was not hitherto achieved. In the present study, we report the identification of targets and interaction partners of ATE in the model plant Physcomitrella patens by mass spectrometry, employing two different immuno-affinity strategies and a recently established transgenic ATE:GUS reporter line (Schuessele et al., 2016 New Phytol., DOI: 10.1111/nph.13656). Here we use a commercially available antibody against the fused reporter protein (β-glucuronidase) to pull down ATE and its interacting proteins and validate its in vivo interaction with a class I small heatshock protein via Förster resonance energy transfer (FRET). Additionally, we apply and modify a method that already successfully identified arginylated proteins from mouse proteomes by using custom-made antibodies specific for N-terminal arginine. As a result, we identify four arginylated proteins from Physcomitrella patens with high confidence.
Data are available via ProteomeXchange with identifier PXD003228 and PXD003232.
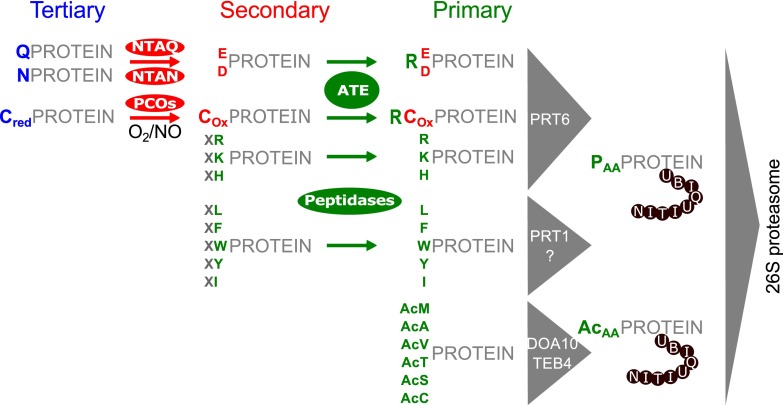
Posttranslational protein arginylation is the process of ribosome-independent incorporation of arginine from charged tRNA to N-terminal amino acids of proteins or sidechains of internal glutamate or aspartate residues and is mediated by arginyl-tRNA protein transferases (ATE)1. Arginylation at the N-terminus of proteins plays a central role in the hierarchically ordered N-end rule degradation pathway that relates the half-life of proteins to the nature of their N-terminus (Fig. 1). Here, arginine, and other N-terminal amino acids (Lys, His, Phe, Leu, Trp, Tyr, Ile), can act as primary destabilizing residues (N-degrons) that serve as recognition site for a certain class of ubiquitin ligases, the N-recognins. Subsequently, poly-ubiquitination of the recognized protein can trigger rapid degradation via the 26S proteasome (1–3). Primary destabilizing residues can become exposed after ATE-mediated arginylation of the secondary destabilizing residues glutamic acid, aspartic acid or oxidized cysteine, as well as after proteolytic cleavage. Further, deamidation of the tertiary destabilizing residues glutamine or asparagine can be mediated by N-terminal amidohydrolases (NTAQ, NTAN) (4), generating glutamic acid and aspartic acid, respectively. N-terminal cysteine can act as secondary destabilizing residue after oxygen- and nitric oxide-dependent oxidation by the action of cysteine oxidases or after nonenzymatic oxidation (5–7). In yeast and mammals, a second branch of the N-end rule pathway, the Ac/N-end rule pathway (8–10), exists where N-terminally acetylated amino acids of certain proteins can function as N-degrons.
Fig. 1.
Schematic overview of the plant N-end rule pathway (according to (2)). The tertiary destabilizing residues glutamine and asparagine can become deamidated by N-terminal amidohydrolases (NTAN/NTAQ) to represent the secondary destabilizing residues glutamic acid or aspartic acid. N-terminal cysteine residues can be oxidized enzymatically (mediated by plant cysteine oxidases (PCOs)) in the presence of oxygen and nitric oxide or non-enzymatically, resulting in the secondary destabilizing oxidized cysteine. Arginyltransferases (ATE) mediate N-terminal arginylation of secondary destabilizing residues (D, E, Cox). Copy numbers for arginyltransferases differ between organisms i.e. A. thaliana harbors 2 genes (ATE1, ATE2) whereas P. patens harbors only one (11, 14–16). Primary destabilizing amino acids (PAA) can occur either via arginylation of secondary destabilizing residues or become exposed after proteolytic cleavages by peptidases. Subsequently, they can be recognized by the ubiquitin ligases PRT1, PRT6 or others, resulting in poly-ubiquitination of the corresponding protein and consequently triggering proteasomal degradation. Positively charged N-terminal residues can be recognized by PRT6 whereas bulky hydrophobic amino acids at the N-terminus can be recognized by PRT1 or yet unidentified ubiquitin ligases (98). In yeast and mammals, a second branch of the N-end rule pathway directly targets certain N-terminally acetylated amino acids (AcAA) via the ubiquitin ligases DOA10 (yeast (8)) or TEB4 (mammals (10)). This branch has not yet been proven for plants but is proposed to exist (78). Unacetylated N-terminal methionine can further act as primary destabilizing residue in certain cases (99) but is not depicted in the present figure.
The protein ATE is conserved among eukaryotes (4, 11). Basal ATE function can be reconstituted in vitro, is independent of co-factors and requires only charged arginyl-tRNA and a substrate (12). Nevertheless, an increase of arginylation efficiency was observed upon addition of cell lysates in the same study, suggesting the presence of interaction partners or supporting co-factors. To date, no such interaction partners have been identified in any eukaryote, except LIAT1 (ligand of ATE1) from mouse (13). LIAT1 is exclusively present in mammals and specific biological functions of this interaction are not yet described.
ATE is encoded by a single copy gene in the moss Physcomitrella patens (11), whereas the vascular plant Arabidopsis thaliana harbors two genes, ATE1 and ATE2 (14–16). Loss of function of one or both ATE genes in A. thaliana results in abnormal shoot and leaf development, delayed senescence and impaired stress- and hormone related responses (14–18). In P. patens, ATE gene knock-out causes altered cell division planes, severe developmental defects and strong starch accumulation (11) demonstrating an important role of protein arginylation in moss. Interestingly, knock-out mutants in yeast are only marginally affected (19) whereas the ATE knock-outs in mice and Drosophila are embryo lethal (20–22). In mouse, ∼100 arginylated proteins were identified, employing an immuno-affinity approach specific for N-terminal arginine (23, 24). Intriguingly, in many cases the destabilizing effect of arginylation on the corresponding target proteins in mouse was lacking and most arginylated N-terminal amino acids were not classical destabilizing residues in the context of the N-end rule pathway (23). In fact, arginylation of mammalian proteins can increase stability and influence oligomerization (25, 26). In contrast, arginylation of glutamic acid residues can also target proteins via p62 (Sequestosome-1) binding to autophagy-mediated degradation in mammalian cell lines (27). Further, ATE-mediated arginylation can also occur on acidic side-chains of non-N-terminal amino acids (28, 29). Although ATE activity was demonstrated quite early in plants (30), the functional investigation of protein arginylation in plants started only recently. Several studies investigated the impact of ATE loss of function on plant physiology and development (14–18), whereas the identification of targets or interacting proteins of ATE remains challenging. To date, arginylation of plant proteins was only indicated via reporter gene fusion for group VII ERF-domain transcription factors in A. thaliana, which act as oxygen and nitric oxide sensors via their N-terminal cysteine residues (17, 18, reviewed in (2)). A few more potential arginylation targets were proposed based on quantitative mass spectrometry including enrichment of N-termini (31, 32). However, corresponding arginylated peptides were not identified in these studies. Whereas the identification of the exact arginylation targets of ATE is pending in plants, the identification of interaction partners of ATE is also necessary to fully understand the function of the N-end rule pathway in eukaryotes. Thus, although several studies have shown the importance of function, no study succeeded in identifying arginylated proteins in plants by mass spectrometry, suggesting that arginylation might be significantly less abundant in plants than in mammals (32).
Physcomitrella patens has become a suitable model organism to study gene function as it offers a versatile toolbox for genetic engineering and a fully sequenced genome (33, 34). Further, the axenic cultivation of different growth states under highly standardized conditions facilitates reproducible high throughput analysis of proteomes, transcriptomes, and metabolites (35–41). Recently, we reported that ATE abundance underlies spatiotemporal patterning in the moss Physcomitrella patens using a translational GUS reporter fusion via knock-in at the endogenous genomic locus (11). In this work we employed the same tagged version of ATE to pull down specifically ATE together with potential interaction partners from tissues where ATE abundance was monitored via histochemical GUS staining. Further, to identify targets of arginylation in P. patens we applied and modified a method using antibodies (23, 42) to pull down specifically proteins bearing N-terminal arginine. Additionally, we incorporated reductive dimethylation of our protein samples into the workflow as artificial dimethylation of peptides facilitates the formation of useful diagnostic reporter ions representing the N-terminal amino acid of a peptide (43).
Here, we present the first identification of N-terminally arginylated proteins from a model plant by mass spectrometry as well as a novel interaction partner of ATE, an Hsp20 class I chaperone. We show that the application of dimethylation can present highly specific reporter ions useful to identify unambiguously N-terminally arginylated proteins. Further, we validate the in vivo interaction of the small heat shock protein with ATE in P. patens using knock-in of fluorescent protein tags and Förster resonance energy transfer (FRET).
On the basis of our findings we distinguish between arginylation targets that interact with ATE to become arginylated and functional interaction partners of ATE that interact with ATE without being targets for arginylation.
EXPERIMENTAL PROCEDURES
Cultivation of Physcomitrella patens
Physcomitrella patens (Hedw.) Bruch & Schimp. ecotype “Gransden 2004” as well as the ATE:GUS line #9 was cultivated in Knop medium supplemented with microelements. The wild type strain as well as the ATE:GUS line is accessible via the International Moss Stock Center (IMSC, www.moss-stock-center.org) under the accession number 40001 and 40965, respectively. Knop medium (pH 5.8) was prepared according to (44) containing 250 mg/l KH2PO4, 250 mg/l KCl, 250 mg/l MgSO4 × 7 H2O, 1000 mg/l Ca(NO3)2 × 4 H2O, and 12.5 mg/l FeSO4 × 7 H2O. Additionally, 10 ml of a microelement (ME) stock solution (309 mg/l H3BO3, 845 mg/l MnSO4 × 1 H2O, 431 mg/l ZnSO4 × 7 H2O, 41.5 mg/l KI, 12.1 mg/l Na2MoO4 × 2 H2O, 1.25 mg/l CoSO4 × 5 H2O, 1.46 mg Co(NO3)2 × 6 H2O) was added per liter of Knop medium as described previously (45, 46). For the cultivation of gametophores on solid medium, 12 g/l agar was added. In order to prevent the formation of gametophores, moss liquid cultures were disrupted weekly with an ULTRA-TURRAX (IKA, Staufen, Germany) at 18,000 rpm for 90 s. If not indicated otherwise, moss was grown under standard light conditions (70 μmol photons/m2 s) at 23 °C in a 16 h/8 h light/dark cycle.
Hydroponic Gametophore Culture on Glass Rings
Hydroponic gametophore culture on glass rings was performed as described (41) with some modifications. Gametophores were grown on glass rings (outer diameter 5 cm, height 2.5 cm with four notches of each 1 cm) covered with gauze (PP, 250 μm mesh, 215 μm thread, Zitt Thoma GmbH, Freiburg, Germany) in Magenta®Vessels (Sigma-Aldrich, St. Louis, USA). Knop medium containing microelements was added until reaching the bottom of the gauze. The Knop medium was exchanged every 4 weeks. Each culture was started with a thin layer of protonema that was applied on top of the gauze 1 week after subculturing. Gametophores were harvested after 12–16 weeks.
Treatments
Inhibition of the 26S proteasome was performed using the inhibitor MG132 (47) purchased from Selleckchem (Houston, TX). Gametophores were cultivated in water and a final concentration of 100 μm MG132 and 1% DMSO for 24 h.
Glucose treatments were performed using Knop medium with microelements supplemented with 1% glucose. Gametophores were treated for 24 h.
Histochemical GUS Staining
Histochemical GUS staining was performed as described (48).
Generation of Stable Transgenic Moss Lines
The coding sequence for Citrine with linkers (poly G/A) was obtained from a plasmid from Tian et al. (2004) (49), and mCerulean from the pGEMHE-X-Cerulean vector (BIOSS toolbox University of Freiburg) (50). In the following all primers are listed in 5′->3′ orientation. The C-terminus of ATE was tagged by inserting linker-Citrine-Citrine, whereas the N-terminus of sHSP17.2a was tagged with Cerulean-linker (forward primer GTGAGCAAGGGCGAGGAG, reverse primer AGCTCCACCTCCACCTCCCTTGTACAGCTCGTCCATGCC). Knock-in constructs for the fluorescent reporters at the endogenous loci of sHSP17.2a (Pp1s8_244V6) and ATE (Pp1s333_56V6) were generated using triple template PCR as described (38). Flanking homologous sequences were amplified from genomic DNA using P1/P2 and P3/P4, with P2 and P3 containing overlapping regions to the fluorescent protein and linker coding sequences. sHSP17.2a:
GCTCTTCATTGTGCCACTCCTCATCGTC (P1), CCCGGTGAACAGCTCCTCGCCCTTGCTCACCATGTCTGCCTCGACCTCTC (P2), GAGCTGTACAAGGGAGGTGGAGGTGGAGCTGCTCTATCGCTGTTCGGCG (P3), GCTCTTCACAGGGGGCCAAGATTTGGTG (P4).
The ATE:Citrine knock-in construct was generated by Gibson assembly as described (51), using the Gibson Assembly®Cloning Kit from New England Biolabs (Ipswich, Massachusetts, USA). All parts were amplified using P1-P8 containing overlapping regions to the respective neighboring part, and P1 and P8 additionally containing overlapping parts to the pJet1.2 Vector (Thermo Scientific, Waltham, USA). The amplified flanking homologous sequences additionally contained a BspQI restriction site, whereas the first Citrine contained an additional 18 bp linker sequence at its N-terminus (corresponding to 5xGly,1xAla) (49).
CCGCCAGATCTTCCGGATGGCTCGAGTTTTTCAGCAAGATGCTCTTCCCCGAATGCCTCAGCTGC (P1), TGCTCACAGCTCCACCTCCACCTCCTCTTGGAACCAAAAGCATGCGAGAA (P2), TTCTCGCATGCTTTTGGTTCCAAGAGGAGGTGGAGGTGGAGCTGTGAGCA (P3), TGCTCACAGCTCCACCTCCACCTCCCTTGTACAGCTCGTCCATGCCGAGA (P4), TCTCGGCATGGACGAGCTGTACAAGGGAGGTGGAGGTGGAGCTGTGAGCA (P5), CGAAGTGTTACATTTCTTTCCTAAACTACTTGTACAGCTCGTCCATGCCG (P6), CGGCATGGACGAGCTGTACAAGTAGTTTAGGAAAGAAATGTAACACTTCG (P7), ATGGCAGCTGAGAATATTGTAGGAGATCTTCTAGAAAGATGCTCTTCGTAGATTAAATGTGAGGCCAGGATC (P8).
Two independent ATE:Citrine reporter lines (#49, #101) were generated by transfection into wildtype protoplasts, using a co-transfected neomycin phosphotransferase resistance (nptII) cassette as transient selection marker (pBSNNNEV) (38). The knock-in construct for the generation of Cerulean:sHSP was transfected into the background of a stable ATE:Citrine knock-in line (#49), as well as into the background of a stable ATE:GUS knock-in line (#9) (10), using a co-transfected hygromycin b phosphotransferase (hpt) cassette as transient selection marker (52, 53) (hpt cassette in pJET1.2, Thermo Scientific), selection was performed on Knop-ME medium with 12.5 mg/l hygromycin). Integration of the knock-in construct at the target locus was validated by PCR on genomic DNA of the transgenic lines using primers that annealed to the genomic sequence upstream and downstream of the homologous regions of the knock-in construct, respectively (see supplemental Fig. S2D for a schematic view): sHSP, forward: CTTGCGTTGTGATGCAACTT; reverse: CAACAATATTGATCGTGGTTGG; ATE, forward: ATGGGTCCCTTTTGAAAAAGCA; reverse: CTCGAGATCTCCTTGCCACATGTCAAAG2.
The moss lines used in this study were: ATE:Citrine #49 (IMSC: 40766), ATE:GUS #9 (IMSC: 40965), FRET-lines Cer:sHSP in ATE:Citrine49 #30 (IMSC: 40769) and #36 (IMSC: 40770) and Cer:sHSP in ATE:GUS#9 #16 (IMSC: 40768). Stable transgenic lines were cryopreserved and are available from the International Moss Stock Center (IMSC).
Coimmunoprecipitation (CoIP) of Interaction Partners of ATE
Six grams fresh weight of gametophores from ATE:GUS and wild type were harvested from hydroponic ring cultures after 7 d cultivation in darkness and used for CoIP. For the CoIP, an anti-GUS antibody (C-terminal, Sigma Aldrich, G5545) was covalently coupled to Dynabeads (Dynabeeds M270 Epoxy Co-Immunoprecipitation Kit (Life Technologies, Carlsbad, CA)). The IP-Buffer was further supplemented with 100 mm NaCl, 2 mm MgCl2, 1 mm DTT and 0.1% plant protease inhibitor mixture (Sigma Aldrich, P9599). Three biological CoIP replicates were performed for each CoIP (ATE:GUS and wild type). All steps of the CoIP procedure using Dynabeads were performed according to the manufacturer's instructions employing the cryolysis method. Eluted proteins were precipitated overnight using 5 volumes of cold acetone containing 0.2% DTT at −20 °C. The samples were centrifuged at 20,000 × g for 15 min at 0 °C and the acetone supernatant was discarded. The protein pellet was washed for 1 h in cold acetone without DTT at −20 °C. The samples were again centrifuged and the supernatant was discarded. 20 μl from each eluate were precipitated separately for Western blot analysis. The protein pellets were air-dried and stored at −20 °C until further usage. Protein pellets were dissolved in 50 mm Tris-HCl, pH 7.6, 2% SDS and reduced at 95 °C for 10 min using Reducing Agent (Life Technologies). Alkylation of cysteines was performed at a final concentration of 100 mm iodoacetaminde (IAA) for 20 min at RT. Finally, the samples were mixed with Laemmli buffer (Bio-RAD, Munich, Germany) for SDS-PAGE. Western blot analysis with the sub-fractions of the eluates was performed as described (54) using an anti-rabbit HRP-conjugate secondary antibody (Amersham Biosciences, Buckinghamshire, UK).
Immunoprecipitation (IP) of N-Terminally Arginylated Proteins
Antibodies against N-terminal arginine were ordered from GenScript USA Inc. (Piscataway, NJ) custom-made using synthetic peptides (REHKHANQHMSVC, RDHKHANQHMSVC, sequences were chosen according to (23)). The obtained antibodies were subjected to negative selection against “scrambled” synthetic peptides (HKERDANQHMSVC, HKRREANQHMSVC, HKHANQHMSVC, sequences were chosen according to (23)). For this purpose, a mixture containing equal amounts of the “scrambled” peptides was coupled to Sulfolink® Coupling Resin (Thermo Scientific) according to the manufacturer's instructions. A mixture of equal amounts of both antibodies was applied and fractions of the flow-through, the washing steps as well as elution fractions were collected. The specificity of the antibodies from all fractions was monitored via dot blot analysis using the peptides containing N-terminal arginine as well as the “scrambled” peptides (supplemental Fig. S1). Antibodies from the washing and the flow-through fractions were used for the immunoprecipitation of arginylated proteins. For this, the Dynabeeds M270 Epoxy Co-Immunoprecipitation Kit with the same buffer as before was used. For each experiment of the glucose and MG132 treated samples 8–10 g fresh weight gametophores from liquid culture were used. Immunoprecipitation of arginylated proteins after cultivation in darkness was performed with 5 g (fresh weight) gametophores harvested from hydroponic ring cultures. Eluted proteins were precipitated using acetone as described before. The dimethylation reaction was carried out according to (55) with modifications. Protein pellets were dissolved in 100 mm HEPES-NaOH, pH 7.5, 0.2% SDS and reduced at 95 °C for 10 min using Reducing Agent (Life Technologies). Alkylation of cysteines was performed at a final concentration of 100 mm IAA for 20 min at RT. Dimethylation was performed by adding 2 μl per 100 μl sample of 4% isotope-labeled formaldehyde solution (13C,d2, Sigma Aldrich) and 2 μl of a 500 mm NaCNBH3 solution. The reaction was carried out at 37 °C for 4 h. Then the same amounts of formaldehyde and NaCNBH3 were added and the reaction continued overnight. The reaction was stopped by adding 2 μl of 4% NH4OH solution per 100 μl sample for 1 h at 37 °C. Finally, the dimethylated samples were again precipitated using acetone without DTT as described before. Dry protein pellets were dissolved in 50 mm Tris-HCl pH 7.6, 2% SDS and mixed with Laemmli buffer prior to SDS PAGE.
MS/MS Sample Preparation
After Coomassie staining (PageBlue, Thermo Scientific), 12 gel slices per gel were excised, either at same size or surrounding prominent bands. The slices were chopped to small pieces and destained with 30% ACN (Promochem, Teddington, UK) in 100 mm NH4HCO3 (Sigma-Aldrich) for 10 min. The step was repeated until the gel was completely destained. The supernatant was discarded and the gel was equilibrated in 100 μl 100 mm NH4HCO3. The supernatant was discarded again and the gel was shrunk with 100 μl ACN for 5 min with gentle shaking. Again, the supernatant was discarded and the gel was vacuum-dried. Digests with trypsin (Promega, Madison, USA) or elastase (Promega) were performed overnight at 37 °C in 50 mm NH4HCO3 (pH 8). About 0.1 μg of protease was used for one gel band. Peptides were extracted from the gel slices with 5% formic acid.
Mass Spectrometry (LC-MS/MS)
Nano LC-MS/MS analyses were performed using an LTQ-Orbitrap Velos Pro (Thermo Scientific) equipped with an EASY-Spray Ion Source and coupled to an EASY-nLC 1000 (Thermo Scientific). Peptides were applied on a trapping column (2 cm × 75 μm ID, PepMap C18, 3 μm particles, 100 Å pore size) and separated on an EASY-Spray column (25 cm × 75 μm ID, PepMap C18, 2 μm particles, 100 Å pore size) with a 30 min linear gradient from 3% to 30% acetonitrile and 0.1% formic acid, and 200 nl/min flow rate. MS scans were acquired in the Orbitrap analyzer with a resolution of 30 000 at m/z 400. A TOP5 data-dependent MS/MS method was used and MS/MS scans were acquired in the Orbitrap analyzer with a resolution of 7500 at m/z 400 using HCD fragmentation with 30% normalized collision energy. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 30 s; unassigned and singly charged precursors were excluded from the selection. Minimum signal threshold for precursor selection was set to 50 000. Predictive AGC was used with AGC target value of 5e5 for MS scans and 5e4 for MS/MS scans. Lock mass option was applied for internal calibration using background ions from protonated decamethylcyclopentasiloxane (m/z 371.10124).
Raw Data Processing and Database Search for Identification of Arginylated Proteins
Raw data processing was performed using Mascot Distiller V2.5.1.0 (Matrix Science, Boston, MA). Database searches on the processed raw data was performed using Mascot Daemon V2.4 (Matrix Science) against the Physcomitrella patens database containing all version 1.6 protein models (56), as well as their reversed sequences used as decoys, and simultaneously against an in-house database containing all sequences of known typical contaminants (e.g. human Keratins, trypsin, 267 entries total, available on request). The fixed modifications were carbamidomethyl (C) +57.021464 Da and 13C,d2 dimethyl (K) +34.063117 Da. Variable modifications used for the database search were Gln->pyro-Glu (N-term Q) −17.026549 Da, Oxidation (M) +15.994915 Da, Acetyl (N-term) +42.010565 Da, 13C,d2 dimethyl (N-term) +34.063117 Da and phospho (ST) +79.966331 Da. Additionally, the mass shifts of an N-terminally dimethylated arginine (+190.164228) was set as modification of any possible N-terminal amino acid including neo-N-termini after proteolytic cleavage of the protein, whereas the mass shift of a dimethylated arginine at glutamine residues that were converted into glutamate residues (+191.148243) was restricted to N-terminal glutamine residues. For all searches the peptide mass tolerance was ± 8 ppm and the fragment mass tolerance was set to ± 0.02 Da. The enzyme specificity was set to semitryptic with a total of 3 missed cleavage sites in the case of the trypsin digests. For samples digested with elastase specificity was set to none.
All Mascot searches were loaded into Scaffold 4 (Version 4.3.4, Proteome Software Inc., Portland, OR) and an additional database search against the Physcomitrella patens database using the same settings was performed using X!Tandem (57) implemented in the Scaffold 4 software. All loaded data was analyzed using the legacy PeptideProphet scoring (high mass accuracy) with independent sample protein grouping. Results were filtered using the Protein- and PeptideProphetTM (58, 59) implemented in Scaffold 4 software.
Data Analysis for the Identification of Interaction Partners
Raw MS data files were analyzed with MaxQuant version 1.4.1.12 (60). Database search was performed with Andromeda, which is integrated in MaxQuant. In addition to these sequences, the search was performed against a database containing common contaminants; a target-decoy database was generated on the fly in MaxQuant by reverse concatenation.
Protein identification was under control of the false-discovery rate (<1% FDR on protein and peptide level). In addition to MaxQuant default settings (e.g. at least 1 razor/unique peptide for identification, two allowed missed cleavages) the search was performed against following variable modifications: Protein N-terminal acetylation, Gln to pyro-Glu formation and oxidation (on Met). Carbamidomethylation of cysteine residues was set as fixed modification. For protein quantitation, the LFQ intensities (61) were used. Proteins with less than two identified razor/unique peptides were dismissed as well as proteins with intensities in only one of the three anti-ATE:GUS-CoIP experiments. Missing LFQ intensities in the control samples were imputed with values close to the baseline if intensities in the corresponding anti-ATE:GUS-CoIP samples were present. Data imputation was performed with values from a standard normal distribution with a mean of the 5% quantile of the combined LFQ intensities and a standard deviation of 0.1.
For each of the three replicates, ratios of the ATE:GUS-CoIP protein intensities to the corresponding control intensities were calculated. The log2 transformed ratios were normalized to the mode of the distributions and averaged (mean with standard deviation) for each protein. Adjusted p values for differential abundance were calculated with the limma package (62) from R/Bioconductor.
Confocal Microscopy
All confocal images were taken on a Zeiss LSM 510 Live DUO (inverted) microscope using the spectral META detector (12 channels, from 465–593 nm, bandwidth of 10 nm) and a 63× objective with water immersion (LCI-Plan Neofluar 63x/1.3 DIC Imm corr.). Channels were recorded simultaneously using the 458 nm line of an Argon laser (intensity 6%, pixel dwell time 3.21 μs, pinhole 2AU, pixel size 0.101 μm, averaging 4). Reference spectra for Cerulean and Citrine were recorded in stable transgenic moss lines expressing only one fusion protein, respectively (Cerulean:sHSP in ATE:GUS background #16, or ATE:Citrine:Citrine in WT background #49). Background signals were recorded in wild type moss cells with the same settings. Acceptor photobleaching of Citrine was started after three excitation cycles with 458 nm, by using the 514 nm line of the Argon laser (50% intensity, 50 iterations) in a region of interest encompassing the nucleus. Bleaching was repeated after each 458 nm cycle until a total of 10 bleaching cycles was reached. Linear unmixing and subsequent extraction of intensity values from bleached regions for Cerulean and Citrine was realized with the Zen 2010 (black) software, using the reference spectra for Cerulean and Citrine, and the WT spectrum as background. The percentage change of donor (Cerulean) and acceptor (Citrine) fluorescence of the FRET pair was quantified in several independent experiments. For every single experiment the maximum fluorescence signal for each channel after linear unmixing was set to 100%. The correlation between the decrease of the Citrine fluorescence and the increase of the Cerulean fluorescence was calculated according to (63) using R (http://www.r-project.org/).
Phylogeny
Protein sequences containing the combination of an N-terminal DUF domain (Pfam DUF3523) and a C-terminal AAA domain (Pfam PF00004) according to the PFAM database (64) where retrieved from the respective genome browsers for P. patens (www.cosmoss.org), A. thaliana (www.arabidopsis.org) and Oryza sativa (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/), and from Uniprot (www.uniprot.org) for Selaginella moellendorffii, Chlorella variabilis, Volvox carteri, Caenorhabditis elegans, Mus musculus and Homo sapiens using the protein sequence of Pp1s106_174V6.1 as a BLAST query. Protein sequences were aligned with Jalview (65) using the Muscle algorithm (default settings). The multiple sequence alignment was subsequently checked and adapted manually, and trimmed at the N- and the C-terminus (Ser88-Leu676 of Pp1s106_174V6.1). A phylogenetic tree was constructed employing Mr. Bayes (V3.1.2) software (66) with the following settings: aamodel = mixed, ngen = 200000, samplefreq = 100, burnins 25%. The deviation of split frequencies was well below 0.01 after 200000 generations. The FigTree software v1.2.3 (http://tree.bio.ed.ac.uk/software/_gtree/) was used to generate figures.
RESULTS AND DISCUSSION
Recently, we demonstrated that ATE abundance was increased in moss (Physcomitrella patens) leafy gametophores after application of the stress hormone abscisic acid (ABA), in darkness or in red light, using a translational ATE:GUS fusion at the endogenous locus (11). Here, we employed this transgenic ATE:GUS line to pull down specifically ATE together with potential interaction partners as well as arginylation targets from tissues where ATE abundance was monitored via histochemical GUS staining. Of the previously employed treatments, cultivation in darkness resulted in the strongest and concurrently most consistent distribution of the GUS-staining across the whole gametophore, and thus was chosen as treatment for the present work. We also performed additional new treatments where GUS-staining increased after cultivation for 1 day in liquid medium supplemented with 1% glucose. Further, inhibition of the proteasome using MG132 was performed to prevent arginylated proteins from proteasomal degradation.
Co-immunoprecipitation of Putative ATE Interaction Partners
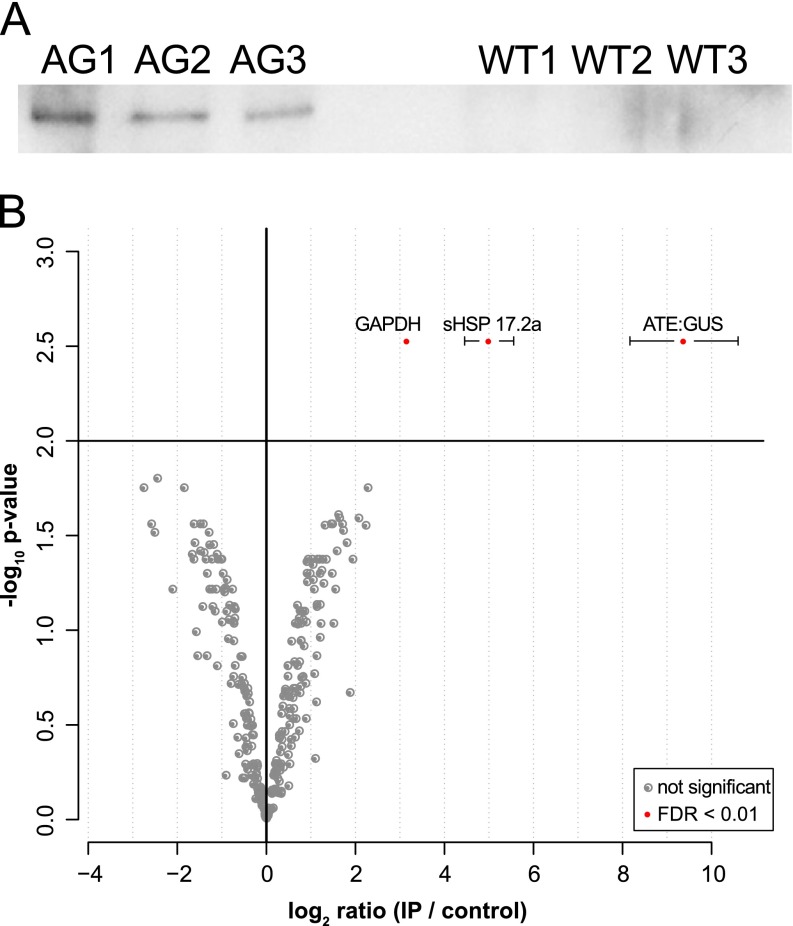
Gametophores of the ATE:GUS line and wild type were cultivated for 8 days in darkness in analogy to (11). For the CoIP of potential interaction partners of ATE, an anti-GUS antibody was used. Its performance was validated via Western blot and a specific signal was exclusively detected in the ATE:GUS samples (Fig. 2A). We identified a total of 820 protein isoforms across the CoIPs of the ATE:GUS and the WT line with an FDR less than 1%. Label free quantitation (LFQ) intensities were used to calculate statistically significant differential abundances of proteins in the ATE:GUS and the wild type control samples. In addition to the ATE:GUS fusion protein, two proteins were significantly overrepresented in the ATE:GUS samples according to our statistical analysis (adjusted p value < 0.01, ratioIP/control > 1, Fig. 2B), namely a glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Pp1s3_238V6.1) and a small heat shock protein (sHSP17.2a, Pp1s8_244V6.1). In mouse, a GAPDH (NCBI: XP_979290.1) with high sequence similarity (68% identity) to the GAPDH identified in the present experiment was identified as a target of arginylation (23). However, we did not detect any arginylated peptide for GAPDH in our data. For the sHSP17.2a, the sequence coverage in the ATE:GUS samples was more than 91% (supplemental Table S1). As the acetylated N-terminus and the C-terminus of sHSP17.2a were identified, sHSP17.2a is a candidate interaction partner of ATE and most likely not a target for arginylation. Additional database searches including N-terminal arginylation (data not shown) did not reveal any arginylated peptide for sHSP17.2a or any other protein within these measurements. In order to validate the protein-protein interaction between ATE and sHSP17.2a in vivo, we used Förster resonance energy transfer (FRET), employing knock-in of fluorescent tags at the endogenous loci of ATE and sHSP17.2a.
Fig. 2.
Results of the CoIP of total protein extracts from ATE:GUS #9 using anti-GUS antibodies. A, Western blot with anti-GUS (C-term) of the performed CoIP triplicates for ATE:GUS samples (AG1–3) and wild type samples (WT1–3). B, Distribution of protein ratios and p values. Ratios are averaged -log2-ratios of normalized protein intensities from three independent experiments (IP against ATE versus control IP, n > = 2); significantly enriched proteins are marked with filled red circles (adjusted p value <0.01, corresponding to <1% FDR). Error bars represent standard deviation of the mean for significant hits.
Validation of Interaction between ATE and sHSP17.2a using FRET
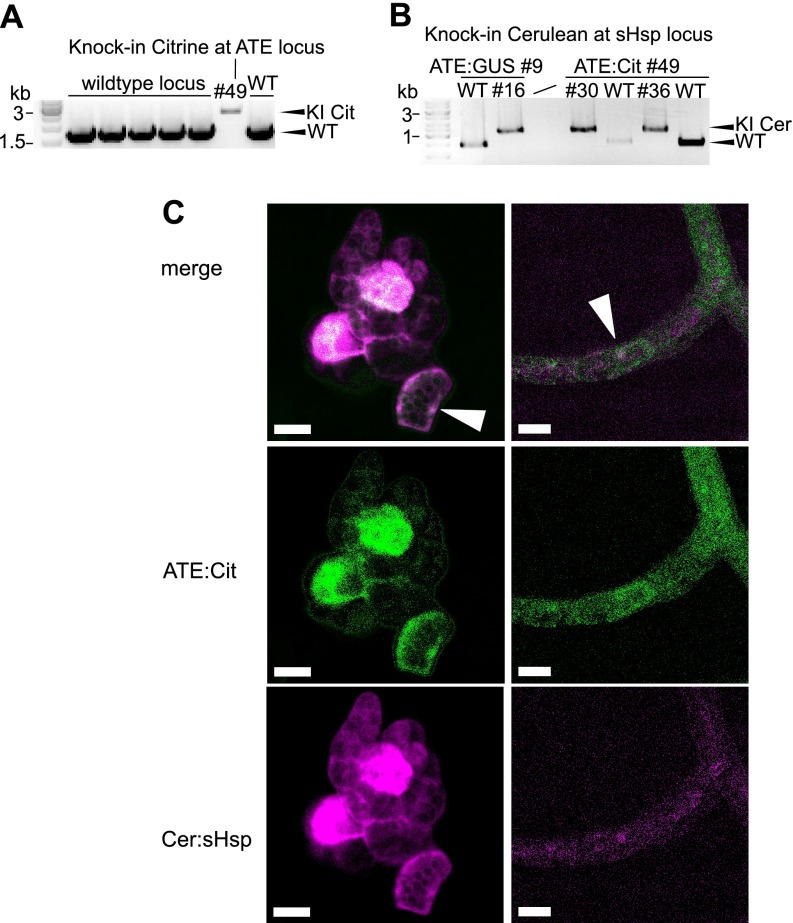
We used the amenability of P. patens to precise gene targeting in order to validate the interaction of sHSP17.2a and ATE in stable transgenic lines, expressing fluorescent fusion proteins of ATE and sHSP17.2a from the endogenous genomic locus of each gene. To achieve this, knock-in constructs were designed to integrate a linker-Citrine-Citrine sequence before the stop codon of ATE (Pp1s333_56V6), and two independent stable lines with a single integration at the target locus were identified (Fig. 3A and supplemental Fig. S2A). In the background of the ATE:Citrine line #49, we generated double transgenic lines by transformation with a Cerulean-linker sequence targeting the sHSP17.2a locus (Pp1s8_244V6) after the start codon (Fig. 3B and supplemental Fig. S2). Tagging both potential interaction partners with fluorophores enables the in vivo analysis of the protein-protein interaction via FRET, in our case using the expression from the endogenous genomic loci. The ATE:Citrine and Cerulean:sHSP17.2a fluorescence signals co-localized, both in cells of the filamentous growth stage of moss, the protonema, and in young gametophores (Fig. 3C), which exhibit three-dimensional growth from a single apical meristematic cell (67). ATE:Citrine abundance varied from cell to cell and was most pronounced in the nucleus (supplemental Fig. S2B), in concordance with previous results from a transient localization assay (11). Cerulean:sHsp17.2a fluorescence was present to different levels in cells and localized to the nucleus and the cytoplasm (supplemental Fig. S2C). In young gametophores fluorescence of Citrine and Cerulean was stronger in shoot meristematic cells. This increased abundance of ATE:Citrine in meristematic cells is in agreement with our previous results from GUS staining in P. patens gametophores (11).
Fig. 3.
Generation of FRET moss (Physcomitrella patens) reporter lines via knock-in. Fluorescent protein tags were introduced into the endogenous ATE gene and sHsp17.2a gene via homologous recombination. A, PCR using genomic DNA of wildtype (WT) and transgenic lines, validating the single integration of Citrine knock-in (KI) construct at the ATE locus using primer flanking the homologous regions used for targeted knock-in (see supplemental Fig. S2 and Material and Methods for additional information). B, PCR using genomic DNA, validating the integration of the Cerulean knock-in construct at the sHsp17.2a locus in two different transgenic background lines. Stable transgenic lines for both the ATE:Citrine and the Cerulean:sHsp17.2a construct were used for FRET analyses, whereas lines with a single fluorophore-tagged protein served as reference lines for spectral imaging and linear unmixing of fluorophore channels using a confocal microscope. C, Exemplary confocal images of the FRET line #36 after linear unmixing of the Citrine and Cerulean signals (see Material and Methods). ATE:Citrine and Cerulean:sHsp17.2a co-localize in several protonema cells, with a low abundance in the cytosol and a higher abundance in the nucleus (arrows). In young gametophores, fluorescence signal was strongest in the single meristematic cell of moss shoots. Scale bars: 20 μm.
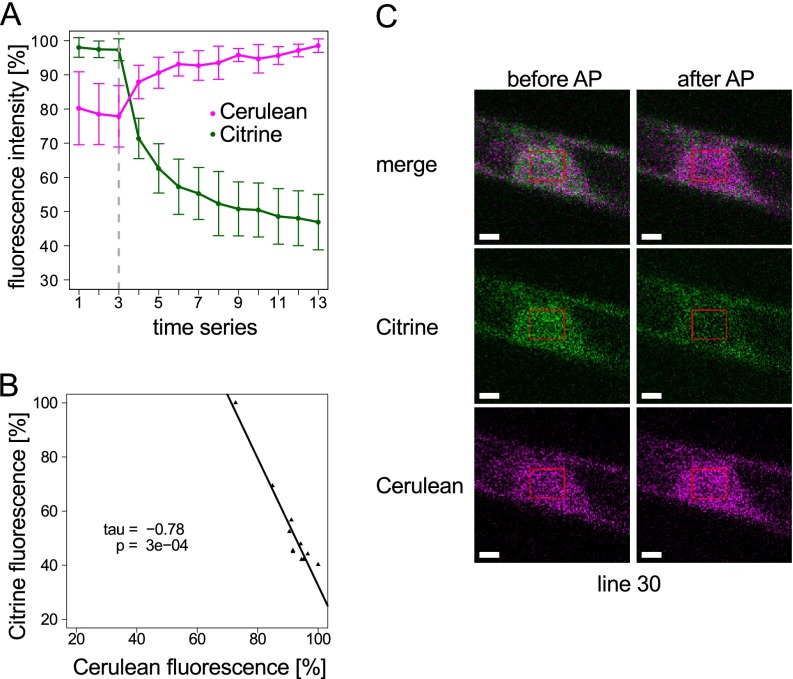
Protein interaction between ATE:Citrine and Cerulean:sHsp17.2 was tested in FRET lines #30 and #36 using the acceptor photobleaching method (68–70), in which the acceptor of a FRET pair (here: Citrine) is bleached, and the change in the fluorescence emission of the donor of the FRET pair (here: Cerulean) is monitored. If FRET is present, the bleaching of the acceptor leads to a proportional increase in the donor fluorescence, as seen in Fig. 4A (for additional FRET line #36 see supplemental Fig. S4). Although Citrine and Cerulean were observed in the cytosol and in the nucleus (Fig. 3C), the ATE:Citrine intensity was stronger in the nucleus and hence, FRET was measured in the nucleus (Fig. 4C). Here we observed a statistically significant (p = 0.0003) and strong negative correlation (Kendall's tau = −0.76) between the fluorescence intensity of Citrine and Cerulean (Fig. 4A, 4B). Thus, we found an in vivo interaction of P. patens ATE with the small heat shock protein sHSP17.2a using two different approaches, namely CoIP and FRET.
Fig. 4.
FRET between Cerulean:sHSP and ATE:Citrine in stable transgenic moss (Physcomitrella patens) lines. FRET was analyzed using acceptor (Citrine) photobleaching and monitoring of the increase in donor (Cerulean) fluorescence. A, Fluorescence intensities of Citrine and Cerulean were monitored using 458 nm excitation before and after acceptor photobleaching (AP) with 514 nm excitation (the gray dashed line indicates start of bleaching, bleaching was repeated after every step). The graph shows average values (± standard deviation) from n = 8 independent experiments. B, Negative correlation between the decrease of the Citrine fluorescence and the increase of the Cerulean fluorescence is strong (Kendall′s tau = −0.78) and statistically significant (p value = 3e−04), indicating FRET. C, Exemplary confocal microscope images showing start (before AP) and end (after AP) of a bleaching experiment in the nucleus of the stable double transgenic Cerulean:sHSP/ATE:Citrine knock-in line #30. Scale bars: 2 μm. See supplemental Fig. S3 for Citrine bleaching test in the ATE:Citrine #49 background line and supplemental Fig. S4 for data of a second independent stable FRET line.
Interestingly, knock-in lines for ATE:Citrine showed a wildtype phenotype, whereas knock-in lines for Cerulean:sHsp17.2a showed a retarded growth of gametophores (supplemental Fig. S5). As the knock-out of ATE leads to severe developmental defects in moss, including an extreme reduction in gametophore formation (11) we suggest that the phenotype observed in our current study could be due to a negative effect of the tagging on sHsp17.2a function. However, this effect is much weaker than in the ATE knockout lines, and FRET lines still exhibit the ATE/sHsp17.2 interaction and form gametophores.
Based on the high sequence coverage and the absence of any detectable arginylation we suggest that sHSP17.2a is a functional interaction partner rather than a target for arginylation in P. patens. As the two interacting proteins (ATE and sHSP17.2a) were identified in a CoIP employing gametophores cultivated in darkness for several days, but FRET was also observed in untreated protonema cells under the control of the endogenous promoters, we conclude that the interaction between ATE and sHSP17.2a is not specific to a treatment but occurs during normal moss development in several cell types and growth stages.
Small heat shock proteins (sHSPs) are present in all domains of life and are subdivided into 11 subfamilies in plants with distinct functionalities and subcellular localizations (71). They are involved in stress responses, but are also present in unstressed cells, preventing protein misfolding and aggregation. Small HSPs harbor a conserved C-terminal alpha-crystalline domain, a variable N-terminal domain and are organized in different oligomeric states composed of up to 24 subunits (71–73).
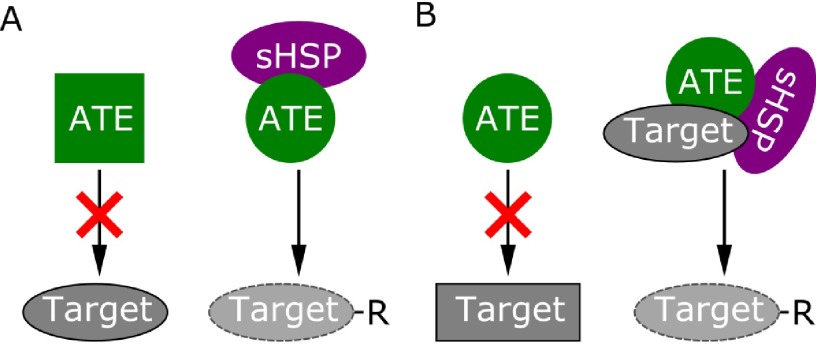
The sHSP17.2a identified in this study belongs to the cytosolic class I sHSPs, according to existing phylogenetic analyses (74, 75) as well as a BLASTP (76) search against recently characterized class I sHSP members of Nicotiana tabacum (77). Small HSPs occur in large oligomers composed of homodimer building blocks with variable stoichiometries of sHSP-substrate ratios (73). Yet, the most thoroughly investigated function of sHSPs is assistance in refolding of proteins either in an ATP-independent manner (77) or via interaction with HSP70 chaperons requiring ATP (73). In addition, other functions such as interaction with proteases acting on sHSP substrates are proposed but far less well investigated (73). Consequently, we consider two possible models for the interaction of ATE with sHSP17.2a (Fig. 5): (1) The interaction might be maintaining and/or enhancing ATE function (Fig. 5A) by supporting correct folding of ATE itself; or (2) sHSP17.2a might act on target proteins of ATE, arranging their folding to present an accessible N-terminus for arginylation (Fig. 5B). Although both hypotheses need experimental verification, the interaction of ATE with a small HSP emphasizes the fact that an integral feature of protein stability within the N-end rule pathway may not solely be the N-terminal amino acid of a protein, but also its accessibility (31). This is already evident in the second branch of the N-end rule pathway, the Ac/N-end rule pathway, which is present in yeast and mammals but is also proposed to exist in plants (8–10, 78). Here, proteins become destabilized because of unfolding or subunit dissociation, resulting in steric unshielding of their N-terminal acetylated amino acid, that is rapidly recognized by the ubiquitin ligases TEB4 (mammals (10)) or DOA10 (yeast (8)). Thus, accessibility is a prerequisite for the recognition of an N-terminus by the components of the N-end rule pathway, namely ubiquitin ligases and ATE. In this context, the sHSP17.2a identified here may represent a candidate to recruit misfolded or unfolded proteins not only for refolding, but also for degradation via the N-end rule pathway. This hypothesis is in line with the function of human sHSP27 that facilitates SUMOylation of the mutant cystic fibrosis transmembrane conductance regulator (CFTR) and thus triggers proteasomal degradation (79).
Fig. 5.
Model of possible modes of interaction between sHSP17.2a and ATE. A, sHSP17.2a may be necessary to support correct folding of ATE to maintain or mediate functionality on target proteins. B, sHSP17.2a might be required to unfold target proteins for arginylation and to present their N-terminus to ATE.
LIAT1 is the first interaction partner of ATE1 identified in mouse (13). Biochemical characterization of the interaction between ATE1 and LIAT1 revealed an increase of ATE1 arginylation capacity on a substrate protein while no arginine incorporation on LIAT1 itself was detectable (13). These findings are in line with the results of (12) who demonstrated cofactor-independent arginylation capacity of ATE that could be increased upon supplementation with cell extracts. Although highly conserved in mammals (13), LIAT1 is completely absent in plants and fungi. Whether LIAT1 represents a general enhancer of the arginylation reaction in mammals remains to be shown. The same is true for the question whether small heat shock proteins are involved in refolding target proteins for arginylation in moss and/or other species or support the maintenance of the functional folding of ATE itself. These challenges remain to be addressed by biochemical characterization of this interaction in subsequent experiments.
Immunoprecipitation of Arginylated Proteins
We also employed P. patens gametophores from the ATE:GUS reporter line (11) to identify arginylation targets in this plant. In analogy to the identification of arginylated proteins in mouse (23), we performed immunoprecipitation (IP) of arginylated proteins from moss, employing antibodies raised against synthetic peptides bearing N-terminal arginine. Further enhancement of the specificity toward N-terminal arginine was achieved via negative selection towards epitopes mimicking N-terminal arginine, resulting in the depletion of such epitopes (supplemental Fig. S1A). Antibodies from the flow-through and the wash fractions that were highly specific for N-terminal arginine (supplemental Fig. S1B) were then used to pull down arginylated proteins.
To increase the amount of putative arginylated proteins, the ATE:GUS reporter line cultivated either in liquid medium or in hydroponic cultures was subjected to different treatments that either increase ATE abundance in moss (darkness, 1% glucose) or block the proteasome (MG132). GUS staining revealed a general increase of stained gametophores after 1 day of cultivation in liquid medium supplemented with 1% glucose (supplemental Fig. S6A, S6B). Cultivation in darkness for 7 days resulted in an increase of the GUS-staining across the whole gametophore (supplemental Fig. S6C, S6D), which is in line with our previous findings (11). We also tested ATE:GUS abundance after inhibition of the 26S proteasome for 24 h using MG132 and did not observe a remarkable increase or decrease of stained gametophores in response to the treatment (Fig. S6E, S6F).
When searching for N-terminal modifications, the application of different proteases can provide alternative evidence for a nontryptic N-terminus of a protein. It also enables the identification of N-termini that have unfavorable peptide lengths related to their fragmentation and identification after tryptic digestion (80). Hence, we conducted two parallel immunoprecipitations of arginylated proteins from MG132-treated samples and digested one sample with trypsin and the other with elastase. Finally, four samples of potentially arginylated proteins (1% glucose, 7 d darkness, 100 μm MG132 elastase and trypsin digested) were analyzed by mass spectrometry.
Across the four samples we identified a total of 1,453 protein isoforms with 0.3% decoy FDR at the protein level and 0.05% decoy FDR at the peptide level using 20,279 spectra (protein threshold 99% ProteinProphetTM, peptide threshold: 90% PeptideProphetTM, minimum number of peptides: 2).
Identification of N-terminal arginylation is a complex task that requires careful verification of the database search engine results because several combinations of amino acids or amino acids with selected modifications can have almost isobaric masses to arginine (42), and thus appear as false positive identifications in database searches. Hence, we employed reductive dimethylation, as this artificial modification results in an increase in a1 ion intensity (43) in the corresponding MS2 spectra of CID or HCD spectra. The a1 ion is the immonium ion of the N-terminal amino acid in a peptide. In consequence, the immonium ion mass of a dimethylated amino acid represent a highly reliable diagnostic reporter ion for the N-terminal amino acid of the identified peptide. Unfortunately, arginine itself is a weak source of immonium ions (81). Hence spectra of false positively identified peptides, where the N-terminal arginine was derived from the genomic sequence were inspected manually to check whether dimethylated N-terminal arginine leads to the formation of a traceable a1 ion. In fact, the presence of corresponding a1 ions of dimethylated arginine (163.1771) was traceable in this study (Fig. S7).
Additionally, all spectra of potentially arginylated peptides were manually inspected regarding isotope peak errors or preceding amino acids from the corresponding protein model that lead to mass ambiguities. We also inspected error distributions of the b- and y-ion series of all arginylation candidates. We observed that the error distribution of the b- and the y-ion series differed markedly if the peptide was falsely annotated to be arginylated based on mass combinations of amino acids that were almost isobaric to the mass of an arginine residue (supplemental Fig. S8). Thus, peptide spectra with imbalanced error series were suspected to be false positive identifications and were discarded from further analysis.
Using the above mentioned quality criteria, arginylated peptides were reliably identified for three different proteins (Table I) namely Acylamino-acid releasing enzyme (PpAARE, Pp1s619_3V6.1), an uncharacterized protein (UP, Pp1s68_62V6.1) and a putative AAA-type ATPase (PpATAD3.1, Pp1s106_174V6.1). All inspected spectra for the arginylated peptides were of high quality (supplemental Fig. S9–S12). However, the a1 ion of a dimethylated arginine could be proven only in the spectra for one of these proteins (Pp1s619_3V6.1, supplemental Fig. S9 and S10). Additionally, using slightly less stringent filter criteria (protein threshold 99% ProteinProphetTM, peptide threshold: 0.5% FDR, minimum number of peptides: 2, resulting in 0.9% decoy FDR at the protein level and 0.15% decoy FDR at the peptide level (min 5% PeptideProphetTM)) one additional protein, an ABC transporter family protein (PpABCB20, Pp1s29_108V6.1, Table I), was identified to be arginylated and the a1 ion was traceable in the corresponding spectrum (supplemental Fig. S13). Further arginylated peptides were not reliably identified, even with considerably less stringent filter settings.
Table I. Overview of identified proteins with validated arginylated peptides.
| Protein Id | Protein Name | AANterma | a1 Ionb | Nr of spectrac | Lengthd |
|---|---|---|---|---|---|
| Pp1s619_3V6.1 | Acylamino-acid-releasing enzyme (PpAARE) | Asp2 | yes | 54 | 791 |
| Pp1s68_62V6.1 | Uncharacterized protein (UP) | Glu3 | no | 1 | 418 |
| Pp1s106_174V6.1 | Putative AAA-type ATPase (PpATAD3.1) | Gln30 | no | 2 | 690 |
| Pp1s29_108V6.1 | ATP-binding cassette transporter (PpABCB20) | Glu529 | yes | 1 | 1422 |
a AANterm: Identified N-terminal arginylated or additionally modified (Gln30) amino acid, the superscript number indicates the position number of the amino acid from the corresponding protein model.
b a1 ion: a1 ion of an N-terminally dimethylated arginine is traceable in assigned spectra.
c Nr of spectra: total number of spectra for the arginylated N-terminus across all experiments.
d Length of the corresponding protein model (number of amino acids).
The number of spectra for the arginylated peptides differed substantially between the different experiments (Table II). Interestingly, for two proteins (Pp1s68_62V6.1, Pp106_174V6.1) spectra were acquired across almost all experiments, whereas arginylated peptides were only monitored in a single experiment. In contrast, spectra for arginylated peptides were monitored in all four experiments for PpAARE (Pp1s619_3V6.1) whereas PpABCB20 (Pp1s29_108V6.1) was only identified in a single experiment.
Table II. Distribution of total spectral counts per experiment for arginylated proteins and number of spectral counts for identified arginylated peptides across the four conducted IPs.
| Protein Id | 1% Glucose |
7 d Darkness |
100 μm MG132 (Trypsin) |
100 μm MG132 (Elastase) |
||||
|---|---|---|---|---|---|---|---|---|
| Total | Arginylated | Total | Arginylated | Total | Arginylated | Total | Arginylated | |
| Pp1s619_3V6.1 | 432 | 1 | 36 | 1 | 750 | 18 | 1157 | 34 |
| Pp1s68_62V6.1 | 1 | 0 | 0 | 0 | 15 | 0 | 13 | 1 |
| Pp1s106_174V6.1 | 63 | 0 | 9 | 2 | 51 | 0 | 36 | 0 |
| Pp1s29_108V6.1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 |
As far as we are aware, this is the first study to identify arginylated proteins from a plant model organism by mass spectrometry. Although more than 1400 proteins were identified in the IP experiments employing arginylation-specific antibodies, arginylated peptides were only reliably identified for four proteins. As even the application of an additional protease (elastase) did not significantly enhance the detection of arginylated peptides from different proteins, one may exclude that their identification suffered from unfavorable peptides based on a single trypsin digest. This further confirms that trypsin is not interfering with the identification of arginylated peptides by cleaving N-terminal arginine residues, in agreement with all arginylated peptides identified in (23) being exclusively tryptic peptides and the fact that trypsin is obviously unable to cleave N-terminal arginine or does so at least at a very low efficiency (82).
The treatments that either increase ATE abundance (1% glucose, 7 d darkness, supplemental Fig. S6A–S6D) or block the proteasome (MG132, supplemental Fig. S6E, S6F) to prevent arginylated proteins from degradation did also not significantly increase the overall detection of arginylated proteins (Table II). However, at least for PpAARE (Pp1s619_3V6.1) the number of identified spectra for the arginylated peptides was remarkably increased in MG132 treated samples (Table II). As the application of a proteasome inhibitor (here MG132) was beneficial for their identification, the identified arginylated proteins may indeed undergo degradation following the N-end rule resulting in short half-lives hampering their identification under steady state conditions.
Among the arginylated proteins, we identified a putative AAA-type ATPase (PpATAD3.1, Pp1s106_174V6.1) where an N-terminal glutamine residue was deamidated to glutamic acid, which subsequently underwent arginylation (Table I, supplemental Fig. S12). This is evidence for a stepwise transformation of a tertiary destabilizing residue into a primary destabilizing residue in a plant, and thus reflects all known hierarchical orders of the N-end rule pathway (Fig. 1). We did not observe any similar transformation on N-terminal asparagine residues in our database searches (data not shown), which supports previous findings (4, 11) that P. patens possesses the ability to deamidate N-terminal glutamine residues (NTAQ, Pp1s114_102V6.1) but not to deamidate N-terminal asparagine residues (NTAN). Although the a1 ion of a dimethylated arginine was not detected in the corresponding spectra, we exclude the possibility that the identified arginylation may represent a sidechain arginylation for several reasons. First, although sidechain arginylation was detected in mouse applying the same experimental setup (29), there is to our knowledge no report on sidechain arginylation that was preceded by deamidation of a glutamine or asparagine residue into a glutamic acid or an aspartic acid residue, respectively. Second, the modified Gln30 was the most N-terminal amino acid identified across all measurements suggesting that it was the apparent N-terminus of PpATAD3.1. As the molecular mechanism of N-terminal arginylation most likely depends on a free α-amino group of the target (29), we suggest that the present arginylation is linked via a peptide bond to the N-terminus rather than via an isopeptide bond to the sidechain.
Interestingly, PpATAD3.1 (Pp1s106_174V6.1) was previously identified in the mitochondrial proteome of P. patens (38). This localization is evolutionary conserved, as homologous proteins in A. thaliana (AT2G18330, AT5G16930; according to (56)) localized as well to mitochondria (83). However, a cleavable N-terminal mitochondrial targeting peptide for PpATAD3.1 was not predicted, neither with TargetP (84) nor with MitoProt II (85) although the position of the arginylated residue (Gln30) might indicate the cleavage of a targeting peptide or other proteolytic processing. Notably, the N-end rule pathway mediated degradation of mitochondrial proteins has already been shown in mammals for PINK1 (PTEN-induced putative kinase 1) (86). Depending on the membrane potential, PINK1 can insert into the inner mitochondrial membrane or remain at the outer membrane where it triggers mitophagy (87). In healthy mitochondria, PINK1 undergoes additional proteolytic processing mediated by PARL (Presenilins-associated rhomboid-like protein) after import and cleavage of its mitochondrial presequence leading to the presentation of primary destabilizing residue according to the N-end rule (86). Subsequently, PINK1 is re-translocated to the cytosol where it is recognized by ubiquitin ligases of the N-end rule pathway. Based on the position of the identified arginylated residue and the potential mitochondrial localization of PpATAD3.1, a similar mechanism may occur in plants. However, proteins that undergo arginylation of their N-terminal secondary destabilizing residue after re-translocation from subcellular compartments can also follow an alternative degradation route, at least in mammals (27). Here, the ER chaperone BiP (GRP78) is re-translocated to the cytosol upon specific stress induction, becomes arginylated and is subsequently bound to p62 triggering autophagy-mediated degradation. Further, PpATAD3.1 is a homolog of the human ATAD3A (Q9NVI7) and ATAD3B (Q5T9A4) according to our phylogenetic analysis (supplemental Fig. S14). The N-terminal region harboring the identified arginylated Gln30 is not well conserved between species, although several homologous proteins exhibit destabilizing residues in the context of the N-end rule pathway in this region (supplemental Fig. S15). The C-terminal domain of human ATAD3A localizes to the mitochondrial matrix whereas the N-terminal domain faces the cytosol (88). This may provide an alternative mechanism for a mitochondria-targeted protein to undergo N-terminal arginylation. Moreover, Caenorhabditis elegans mutants with RNAi suppressed ATAD3 are impaired in fat metabolism (89). This is especially interesting because ATE function is affecting energy homeostasis in plants (11). Here, the arginylation of the putative AAA-type ATPase observed in our study may indicate a yet undescribed link between the N-end rule pathway and the degradation of mitochondrial proteins in plants.
In comparison to the positions of the arginylated residues of PpATAD3.1 (Pp1s106_174V6.1, Gln30), PpAARE (Pp1s619_3V6.1, Asp2) and UP (Pp1s68_62V6.1, Glu3), a more internally positioned residue (Glu529, Table I) was identified for PpABCB20 (Pp1s29_108V6.1). PpABCB20 is a member of the large ATP-binding cassette transporter family in P. patens comprising 121 members that are membrane-bound and transport various substances such as lipids, hormones or other secondary metabolites (34). The identified arginylated residue lies within one of two ABC transporter domains (PF00005.23, 441–589) according to a PFAM domain (64) search. Catalytic cleavages within functional domains with subsequent arginylation of resulting N-terminal amino acids have already been proposed (23), but although crystal structures of this domain are available (90, 91) they do not indicate any functional cleavage within this domain. Thus, the identified residue (Glu529) may be the result of a proteolytic event that deactivated the transporter domain whereas clearance of the proteolytic fragment is possibly mediated via the arginylation branch of the N-end rule pathway. This is further supported by the fact that arginylation was only identified in the samples treated with MG132 (Table II).
Intriguingly, a multiple sequence alignment including homologous proteins from Arabidopsis thaliana, Selaginella. moellendorffii, Populus trichocarpa and Oryza sativa revealed that the identified arginylated residue (Glu529, Pp1s29_108V6.1) lies within a conserved domain (supplemental Fig. S16). Although the glutamic acid is not conserved in all investigated species, all residues at this position represent either tertiary or secondary destabilizing residues in the context of the N-end rule pathway.
Here, we exclude the possibility that the identified arginylation may represent a sidechain arginylation as the a1 ion of a dimethylated arginine was present in the corresponding HCD spectrum (supplemental Fig. S13). As a sidechain arginylation would be connected via an isopeptide bond of the α-amino group of the arginine to the γ-carboxyl group of the glutamate residue, a dimethylated arginine cannot be present in that case. Additionally, spectra for this protein were only monitored in the gel slice excised on the bottom of the gel (low molecular weight range) indicating that the protein was not of full size anymore (theoretical molecular weight 157 kDa) and underwent proteolytic processing.
For two proteins the identified arginylated amino acid was in close proximity to the annotated N-terminus according to the corresponding gene model (Pp1s619_3V6.1: Asp2; Pp1s68_62V6.1: Glu3; Table I). Although UP (Pp1s68_62V6.1) is conserved among several plants (56), there are no functional annotations or predicted functional domains available. The N-terminus and the identified Glu3 of UP related homologous proteins are not conserved (supplemental Fig. S17). Here, we cannot fully exclude a potential sidechain arginylation of the identified arginylated Glu3 of UP as no a1 ion of a dimethylated arginine was observed in the corresponding HCD spectrum. The preceding amino acids of the arginylated Glu3 from the corresponding protein model are methionine followed by glycine. The initiator methionine of proteins is cleaved cotranslationally by methionine aminopeptidase (MetAPs) if the subsequent amino acid has a small sidechain, such as glycine, alanine or serine (92, 93) including subsequent co-translational acetylation of the resulting N-terminal amino acid. In consequence, this would result in an N-terminal acetylated glycine residue of UP. However, we did not observe the arginylated peptide preceded by glycine suggesting that the glutamic acid may have been the N-terminal amino acid. We consider a cleavage of the N-terminal glycine residue by the used elastase as unlikely because elastase belongs to the serine endopeptidases like trypsin (Peptidase_S1, according to MEROPS, https://merops.sanger.ac.uk). Notably, as the arginylated peptide was only observed in the proteasome-inhibited sample, we propose that its arginylation might be linked to proteasomal degradation via the arginylation branch of the N-end rule pathway.
PpAARE (Pp1s619_3V6.1) was among the most abundant proteins in all performed measurements (supplemental Table S2). Its identified arginylated Asp2 represents an exceptional N-terminal amino acid as cleavage of the initiator methionine by MetAPs are likely unusual to occur when aspartic acid is the subsequent amino acid (92, 93). In consequence, this may indicate the presence of a yet undescribed proteolytic processing event, at least in moss. Interestingly, selected homologous proteins from other plant species harbor secondary destabilizing residues following their initiator methionine, whereas the N-terminus in general is far less well conserved between the homologous proteins (supplemental Fig. S18). The question whether the observed cleavages may occur in other plant species as well and if this represents a yet undescribed mechanism to generate N-end rule pathway substrates has to be challenged by future work.
The overall high abundance of PpAARE in the performed experiments may have several reasons. First, across the dynamic protein abundance range in which arginylated proteins are likely to occur, PpAARE might have been the most abundant in all our experimental conditions. Second, the antibodies might be biased toward the N-terminus of PpAARE compared with other proteins.
The identified moss PpAARE is a homolog of Arabidopsis thaliana AtAARE (AT4G14570.1) based on an all-versus-all protein homology clustering (56). AtAARE forms a bifunctional tetrameric protease complex composed of four identical subunits that cleaves oxidized and glycated proteins on the one hand and N-terminally acetylated amino acids from short peptides on the other (94, 95). Glycation of proteins in plants can be caused by degradation products of ascorbic acid or reducing sugars (96). An increase of total spectra after glucose treatment compared with the dark treatment is evident in our data but spectral counts for the arginylated peptide did not increase (Table II). Moreover, the spectral counts for the arginylated N-terminus strongly increased after blocking the proteasome with MG132. In consequence, we propose that the dynamic equilibrium of PpAARE is controlled by the arginylation branch of the N-end rule pathway and that the observed arginylation was not a specific response to the plant treatment with glucose or darkness.
All arginylated N-terminal residues identified in the present work correspond to destabilizing residues according to the N-end rule. Consequently, it is tempting to speculate that all corresponding proteins may undergo proteasomal degradation triggered by the arginylation branch of the N-end rule pathway. In contrast to (23), we did not reliably identify any arginylated N-terminal amino acid that does not correspond to the hierarchical order of the N-end rule pathway in the moss P. patens. This is astonishing, although we cannot fully exclude that the antibodies used in this study resulted in a biased sampling of arginylated proteins due to their specificity to N-terminal RE or RD instead of being only specific for N-terminal Arg, similar to those used in (23). Additionally, it remains unknown why other proteomic approaches (31, 32) failed to detect arginylation events in plants that did not correspond to the hierarchical order of the N-end rule pathway. Thus, we suggest that other functions of arginylation besides targeting for degradation may not be present in moss or at least not to the extent observed in mammals.
CONCLUSION
In the present study we employed a transgenic ATE:GUS moss reporter line to identify targets of arginylation and a novel interaction partner of P. patens ATE. Although the exact role of the protein-protein interaction between sHSP17.2a and ATE needs further elucidation, we exclude this small heat shock protein as a target of arginylation based on our MS data. Further, we suggest a yet undescribed link between sHSP function and the N-end rule pathway, namely to modulate the accessibility of N-termini.
In addition, our data indicate that arginylation in plants is in fact a very low abundant modification, which was found on proteins of unknown function and putative functions in protein quality control and transport. Thus, an identification of arginylated proteins was only possible by choosing cells with a measurable ATE level and by using specific immuno-enrichment of the modified proteins. The low number of identified arginylated peptides compared with the high number of identified proteins shows that additional improvements of the method may increase the number of reliable identifications. Such improvements may include the addition of proteasome inhibitors into the protein extraction buffer (31), the use of several different proteases or the selective enrichment of N-terminal peptides subsequent to the immuno-purification. Our present data indicate the beneficial effects of MG132 treatments for the identification of arginylated proteins in plants, hinting at a low abundance due to a low half-life of arginylated proteins in plants. However, the exact effects of arginylation on the corresponding target proteins have to be investigated in further studies. The approach used in this study paves the way for further identification of arginylation targets in plants and thus to unravel the exact mechanisms of the physiological and developmental roles of the N-end rule pathway in plants.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (97) via the PRIDE partner repository with the data set identifier PXD003228, PXD003232, and 10.6019/PXD003232.
Supplementary Material
Acknowledgments
We thank the staff of the Life Imaging Center (LIC) in the Center for Biological System Analysis (ZBSA) of the University of Freiburg for help with their confocal microscope resources. We thank Christine Glockner for technical support and Anne Katrin Prowse for proof reading the manuscript.
Footnotes
Author contributions: R.R., A.S., and G.L.I. designed the research. S.N.W.H., S.J.M., K.F., M.S. and C.S. performed experiments. S.N.W.H., J.T.V., S.J.M., D.L. and R.N. performed data analysis. S.N.W.H., S.J.M. and R.R. wrote the manuscript. All authors discussed and approved the final version of the manuscript.
* This work was funded by Deutsche Forschungsgemeinschaft (DFG IG9/8-1 to G. L. I., A. S. and R. R.) and the Excellence Initiative of the German Federal and State Governments (EXC294 to R. R. and R. N.) is gratefully acknowledged.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ATE
- arginyl-tRNA protein transferase
- AARE
- acylamino-acid releasing enzyme
- ABA
- abscisic acid
- AcAA
- N-terminally acetylated amino acid
- AGC
- automatic gain control
- AP
- acceptor photobleaching
- BLAST
- basic local alignment search tool
- CID
- collision induced dissociation
- CoIP
- Co-immunoprecipitation
- FDR
- false discovery rate
- FRET
- Förster resonance energy transfer
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GUS
- β-glucuronidase
- HCD
- higher-energy collisional dissociation
- HRP
- horseradish peroxidase
- IAA
- iodoacetamide
- IMSC
- International Moss Stock Center
- IP
- immunoprecipitation
- LFQ
- label free quantitation
- LIAT1
- ligand of ATE1
- MS/MS
- tandem mass spectrometry
- NTAQ
- N-terminal glutamine amidohydrolase
- NTAN
- N-terminal asparagine amidohydrolase
- PAA
- primary destabilizing amino acid
- PRT1/6
- proteolysis 1/6
- sHSP
- small heat shock protein
- SUMO
- small ubiquitin-like modifier
- UP
- uncharacterized protein
- ATAD
- ATPase family AAA domain
- WT
- wild type.
REFERENCES
- 1. Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U.S.A. 93, 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gibbs D. J., Bacardit J., Bachmair A., and Holdsworth M. J. (2014) The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24, 603–611 [DOI] [PubMed] [Google Scholar]
- 3. Ciechanover A., and Stanhill A. (2014) The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 1843, 86–96 [DOI] [PubMed] [Google Scholar]
- 4. Graciet E., Mesiti F., and Wellmer F. (2010) Structure and evolutionary conservation of the plant N-end rule pathway. Plant J. 61, 741–751 [DOI] [PubMed] [Google Scholar]
- 5. Hu R. G., Sheng J., Qi X., Xu Z., Takahashi T. T., and Varshavsky A. (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 [DOI] [PubMed] [Google Scholar]
- 6. Gibbs D. J., Isa N. M., Movahedi M., Lozano-Juste J., Mendiondo G. M., Berckhan S., Marín-de la Rosa N., Conde J. V., Correia C. S., Pearce S. P., Bassel G. W., Hamali B., Talloji P., Tomé D. F. A., Coego A., Beynon J., Alabadí D., Bachmair A., León J., Gray J. E., Theodoulou F. L., and Holdsworth M. J. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53, 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weits D. A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H.-M., Riegler H., Hoefgen R., Perata P., van Dongen J. T., and Licausi F. (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hwang C.-S., Shemorry A., and Varshavsky A. (2010) N-Terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shemorry A., Hwang C.-S., and Varshavsky A. (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park S.-E., Kim J.-M., Seok O.-H., Cho H., Wadas B., Kim S.-Y., Varshavsky A., and Hwang C.-S. (2015) Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 347, 1249–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schuessele C., Hoernstein S. N. W., Mueller S. J., Rodriguez-Franco M., Lorenz T., Lang D., Igloi G. L., and Reski R. (2016) Spatio-temporal patterning of arginyl-tRNA protein transferase (ATE) contributes to gametophytic development in a moss. New Phytol. 209, 1014–1027 [DOI] [PubMed] [Google Scholar]
- 12. Wang J., Han X., Saha S., Xu T., Rai R., Zhang F., Wolf Y. I., Wolfson A., Yates J. R., and Kashina A. (2011) Arginyltransferase is an ATP-independent self-regulating enzyme that forms distinct functional complexes in vivo. Chem. Biol. 18, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brower C. S., Rosen C. E., Jones R. H., Wadas B. C., Piatkov K. I., and Varshavsky A. (2014) Liat1, an arginyltransferase-binding protein whose evolution among primates involved changes in the numbers of its 10-residue repeats. Proc. Natl. Acad. Sci. U.S.A. 111, E4936–E4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida S., Ito M., Callis J., Nishida I., and Watanabe A. (2002) A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 32, 129–137 [DOI] [PubMed] [Google Scholar]
- 15. Graciet E., Walter F., Maoiléidigh D. Ó., Pollmann S., Meyerowitz E. M., Varshavsky A., and Wellmer F. (2009) The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. U.S.A. 106, 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holman T. J., Jones P. D., Russell L., Medhurst A., Tomás S. Ú., Talloji P., Marquez J., Schmuths H., Tung S.-A., Taylor I., Footitt S., Bachmair A., Theodoulou F. L., and Holdsworth M. J. (2009) The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4549–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Licausi F., Kosmacz M., Weits D. A., Giuntoli B., Giorgi F. M., Voesenek L. A. C. J., Perata P., and van Dongen J. T. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 [DOI] [PubMed] [Google Scholar]
- 18. Gibbs D. J., Lee S. C., Isa N. M., Gramuglia S., Fukao T., Bassel G. W., Correia C. S., Corbineau F., Theodoulou F. L., Bailey-Serres J., and Holdsworth M. J. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature, 479, 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balzi E., Choder M., Chen W. N., Varshavsky A., and Goffeau A. (1990) Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J. Biol. Chem. 265, 7464–7471 [PubMed] [Google Scholar]
- 20. Kwon Y. T., Kashina A. S., Davydov I. V., Hu R.-G., An J. Y., Seo J. W., Du F., and Varshavsky A. (2002) An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 [DOI] [PubMed] [Google Scholar]
- 21. Lee M. J., Kim D. E., Zakrzewska A., Yoo Y. D., Kim S.-H., Kim S. T., Seo J. W., Lee Y. S., Dorn G. W., Oh U., Kim B. Y., and Kwon Y. T. (2012) Characterization of arginylation branch of N-end rule pathway in G-protein-mediated proliferation and signaling of cardiomyocytes. J. Biol. Chem. 287, 24043–24052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., Misra S., and Rubin G. M. (1999) The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong C. C. L., Xu T., Rai R., Bailey A. O., Yates J. R., Wolf Y. I., Zebroski H., and Kashina A. (2007) Global analysis of posttranslational protein arginylation. PLos Biol. 5, e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saha S., Wong C. C. L., Xu T., Namgoong S., Zebroski H., Yates J. R., and Kashina A. (2011) Arginylation and methylation double up to regulate nuclear proteins and nuclear architecture in vivo. Chem. Biol. 18, 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karakozova M., Kozak M., Wong C. C. L., Bailey A. O., Yates J. R., Mogilner A., Zebroski H., and Kashina A. (2006) Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196 [DOI] [PubMed] [Google Scholar]
- 26. Saha S., Mundia M. M., Zhang F., Demers R. W., Korobova F., Svitkina T., Perieteanu A. A., Dawson J. F., and Kashina A. (2010) Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol. Biol. Cell 21, 1350–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cha-Molstad H., Sung K. S., Hwang J., Kim K. A., Yu J. E., Yoo Y. D., Jang J. M., Han D. H., Molstad M., Kim J. G., Lee Y. J., Zakrzewska A., Kim S.-H., Kim S. T., Kim S. Y., Lee H. G., Soung N. K., Ahn J. S., Ciechanover A., Kim B. Y., and Kwon Y. T. (2015) Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 17, 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eriste E., Norberg A., Nepomuceno D., Kuei C., Kamme F., Tran D.-T., Strupat K., Jörnvall H., Liu C., Lovenberg T. W., and Sillard R. (2005) A novel form of neurotensin post-translationally modified by arginylation. J. Biol. Chem. 280, 35089–35097 [DOI] [PubMed] [Google Scholar]
- 29. Wang J., Han X., Wong C. C. L., Cheng H., Aslanian A., Xu T., Leavis P., Roder H., Hedstrom L., Yates J. R., and Kashina A. (2014) Arginyltransferase ATE1 catalyzes midchain arginylation of proteins at side chain carboxylates in vivo. Chem. Biol. 21, 331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manahan C. O., and App A. A. (1973) An arginyl-transfer ribonucleic acid protein transferase from cereal embryos. Plant Physiol. 52, 13–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Majovsky P., Naumann C., Lee C.-W., Lassowskat I., Trujillo M., Dissmeyer N., and Hoehenwarter W. (2014) Targeted proteomics analysis of protein degradation in plant signaling on an LTQ-Orbitrap mass spectrometer. J. Proteome Res. 13, 4246–4258 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H., Deery M. J., Gannon L., Powers S. J., Lilley K. S., and Theodoulou F. L. (2015) Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots. Proteomics, 15, 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reski R., Parsons J., and Decker E. L. (2015) Moss-made pharmaceuticals: from bench to bedside. Plant Biotechnol. J., 13, 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P.-F., Lindquist E. A., Kamisugi Y., Tanahashi T., Sakakibara K., Fujita T., Oishi K., Shin-I T., Kuroki Y., Toyoda A., Suzuki Y., Hashimoto S., Yamaguchi K., Sugano S., Kohara Y., Fujiyama A., Anterola A., Aoki S., Ashton N., Barbazuk W. B., Barker E., Bennetzen J. L., Blankenship R., Cho S. H., Dutcher S. K., Estelle M., Fawcett J. A., Gundlach H., Hanada K., Heyl A., Hicks K. A., Hughes J., Lohr M., Mayer K., Melkozernov A., Murata T., Nelson D. R., Pils B., Prigge M., Reiss B., Renner T., Rombauts S., Rushton P. J., Sanderfoot A., Schween G., Shiu S.-H., Stueber K., Theodoulou F. L., Tu H., Van de Peer Y., Verrier P. J., Waters E., Wood A., Yang L., Cove D., Cuming A. C., Hasebe M., Lucas S., Mishler B. D., Reski R., Grigoriev I. V., Quatrano R. S., and Boore J. L. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 [DOI] [PubMed] [Google Scholar]
- 35. Sarnighausen E., Wurtz V., Heintz D., Van Dorsselaer A., and Reski R. (2004) Mapping of the Physcomitrella patens proteome. Phytochemistry 65, 1589–1607 [DOI] [PubMed] [Google Scholar]
- 36. Heintz D., Erxleben A., High A. A., Wurtz V., Reski R., Van Dorsselaer A., and Sarnighausen E. (2006) Rapid alteration of the phosphoproteome in the moss Physcomitrella patens after cytokinin treatment. J. Proteome Res. 5, 2283–2293 [DOI] [PubMed] [Google Scholar]
- 37. Cui S., Hu J., Guo S., Wang J., Cheng Y., Dang X., Wu L., and He Y. (2011) Proteome analysis of Physcomitrella patens exposed to progressive dehydration and rehydration. J. Exp. Bot. 63, 711–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mueller S. J., Lang D., Hoernstein S. N. W., Lang E. G. E., Schuessele C., Schmidt A., Fluck M., Leisibach D., Niegl C., Zimmer A. D., Schlosser A., and Reski R. (2014) Quantitative analysis of the mitochondrial and plastid proteomes of the moss Physcomitrella patens reveals protein macrocompartmentation and microcompartmentation. Plant Physiol. 164, 2081–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lehtonen M. T., Takikawa Y., Rönnholm G., Akita M., Kalkkinen N., Ahola-Iivarinen E., Somervuo P., Varjosalo M., and Valkonen J. P. T. (2014) Protein secretome of moss plants (Physcomitrella patens) with emphasis on changes induced by a fungal elicitor. J. Proteome Res. 13, 447–459 [DOI] [PubMed] [Google Scholar]
- 40. Hiss M., Laule O., Meskauskiene R. M., Arif M. A., Decker E. L., Erxleben A., Frank W., Hanke S. T., Lang D., Martin A., Neu C., Reski R., Richardt S., Schallenberg-Rüdinger M., Szövényi P., Tiko T., Wiedemann G., Wolf L., Zimmermann P., and Rensing S. A. (2014) Large-scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J. 79, 530–539 [DOI] [PubMed] [Google Scholar]
- 41. Erxleben A., Gessler A., Vervliet-Scheebaum M., and Reski R. (2012) Metabolite profiling of the moss Physcomitrella patens reveals evolutionary conservation of osmoprotective substances. Plant Cell Rep. 31, 427–436 [DOI] [PubMed] [Google Scholar]
- 42. Xu T., Wong C. C. L., Kashina A., and Yates J. R. (2009) Identification of N-terminally arginylated proteins and peptides by mass spectrometry. Nat. Protoc. 4, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsu J.-L., Huang S.-Y., Shiea J.-T., Huang W.-Y., and Chen S.-H. (2005) Beyond quantitative proteomics: signal enhancement of the a1 ion as a mass tag for peptide sequencing using dimethyl labeling. J. Proteome Res. 4, 101–108 [DOI] [PubMed] [Google Scholar]
- 44. Reski R., and Abel W. O. (1985) Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165, 354–358 [DOI] [PubMed] [Google Scholar]
- 45. Egener T., Granado J., Guitton M.-C., Hohe A., Holtorf H., Lucht J. M., Rensing S. A., Schlink K., Schulte J., Schween G., Zimmermann S., Duwenig E., Rak B., and Reski R. (2002) High frequency of phenotypic deviations in Physcomitrella patens plants transformed with a gene-disruption library. BMC Plant Biol. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schween G., Hohe A., Koprivova A., and Reski R. (2003) Effects of nutrients, cell density and culture techniques on protoplast regeneration and early protonema development in a moss, Physcomitrella patens. J. Plant Physiol. 160, 209–212 [DOI] [PubMed] [Google Scholar]
- 47. Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., and Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 48. Bierfreund N. M., Reski R., and Decker E. L. (2003) Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep. 21, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 49. Tian G.-W., Mohanty A., Chary S. N., Li S., Paap B., Drakakaki G., Kopec C. D., Li J., Ehrhardt D., Jackson D., Rhee S. Y., Raikhel N. V., and Citovsky V. (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135, 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rizzo M. A., Springer G. H., Granada B., and Piston D. W. (2004) An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- 51. Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 52. Huether C. M., Lienhart O., Baur A., Stemmer C., Gorr G., Reski R., and Decker E. L. (2005) Glyco-engineering of moss lacking plant-specific sugar residues. Plant Biol. 7, 292–299 [DOI] [PubMed] [Google Scholar]
- 53. Büttner-Mainik A., Parsons J., Jérôme H., Hartmann A., Lamer S., Schaaf A., Schlosser A., Zipfel P. F., Reski R., and Decker E. L. (2011) Production of biologically active recombinant human factor H in Physcomitrella. Plant Biotechnol. J. 9, 373–383 [DOI] [PubMed] [Google Scholar]
- 54. Lang E. G. E., Mueller S. J., Hoernstein S. N. W., Porankiewicz-Asplund J., Vervliet-Scheebaum M., and Reski R. (2011) Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 30, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., and Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 56. Zimmer A. D., Lang D., Buchta K., Rombauts S., Nishiyama T., Hasebe M., Van de Peer Y., Rensing S. A., and Reski R. (2013) Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics 14, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Craig R., and Beavis R. C. (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310–2316 [DOI] [PubMed] [Google Scholar]
- 58. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 59. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 60. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 61. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kendall M. G. (1938) A new measure of rank correlation. Biometrika 30, 81–93 [Google Scholar]
- 64. Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L. L., Tate J., and Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., and Barton G. J. (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ronquist F., and Huelsenbeck J. P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 67. Harrison C. J., Roeder A. H. K., Meyerowitz E. M., and Langdale J. A. (2009) Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 19, 461–471 [DOI] [PubMed] [Google Scholar]
- 68. Bastiaens P. I., and Jovin T. M. (1996) Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta I. Proc. Natl. Acad. Sci. U.S.A. 93, 8407–8412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bastiaens P. I., Majoul I. V., Verveer P. J., Söling H.-D., and Jovin T. M. (1996) Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15, 4246–4253 [PMC free article] [PubMed] [Google Scholar]
- 70. Kottgen M., Buchholz B., Garcia-Gonzalez M. A., Kotsis F., Fu X., Doerken M., Boehlke C., Steffl D., Tauber R., Wegierski T., Nitschke R., Suzuki M., Kramer-Zucker A., Germino G. G., Watnick T., Prenen J., Nilius B., Kuehn E. W., and Walz G. (2008) TRPP2 and TRPV4 form a polymodal sensory channel complex. J. Cell Biol. 182, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Waters E. R. (2013) The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403 [DOI] [PubMed] [Google Scholar]
- 72. Haslbeck M., Franzmann T., Weinfurtner D., and Buchner J. (2005) Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 73. Basha E., O'Neill H., and Vierling E. (2012) Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 37, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ruibal C., Castro A., Carballo V., Szabados L., and Vidal S. (2013) Recovery from heat, salt and osmotic stress in Physcomitrella patens requires a functional small heat shock protein PpHsp16.4. BMC Plant Biol. 13, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu L., Carrie C., Law S. R., Murcha M. W., and Whelan J. (2013) Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol. 161, 644–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Park S. M., Kim K. P., Joe M. K., Lee M. O., Koo H. J., and Hong C. B. (2015) Tobacco class I cytosolic small heat shock proteins are under transcriptional and translational regulations in expression and heterocomplex prevails under the high-temperature stress condition in vitro. Plant Cell Environ. 38, 767–776 [DOI] [PubMed] [Google Scholar]
- 78. Gibbs D. J. (2015) Emerging functions for N-terminal protein acetylation in plants. Trends Plant Sci. 20, 599–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ahner A., Gong X., Schmidt B. Z., Peters K. W., Rabeh W. M., Thibodeau P. H., Lukacs G. L., and Frizzell R. A. (2013) Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier-dependent pathway. Mol. Biol. Cell 24, 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lange P. F., Huesgen P. F., Nguyen K., and Overall C. M. (2014) Annotating N-termini for the human proteome project: N-termini and Nα-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome. J. Proteome Res. 13, 2028–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Falick A., Hines W., Medzihradszky K., Baldwin M., and Gibson B. (1993) Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 4, 882–893 [DOI] [PubMed] [Google Scholar]
- 82. Jensen O. N., Vorm O., and Mann M. (1996) Sequence patterns produced by incomplete enzymatic digestion or one-step Edman degradation of peptide mixtures as probes for protein database searches. Electrophoresis 17, 938–944 [DOI] [PubMed] [Google Scholar]
- 83. Nikolovski N., Rubtsov D., Segura M. P., Miles G. P., Stevens T. J., Dunkley T. P. J., Munro S., Lilley K. S., and Dupree P. (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160, 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Emanuelsson O., Nielsen H., Brunak S., and von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 85. Claros M. G., and Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 86. Yamano K., and Youle R. J. (2013) PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., and Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hubstenberger A., Merle N., Charton R., Brandolin G., and Rousseau D. (2010) Topological analysis of ATAD3A insertion in purified human mitochondria. J. Bioenerg. Biomembr. 42, 143–150 [DOI] [PubMed] [Google Scholar]
- 89. Hoffmann M., Bellance N., Rossignol R., Koopman W. J. H., Willems P. H. G. M., Mayatepek E., Bossinger O., and Distelmaier F. (2009) C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS ONE 4, e7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hung L. W., Wang I. X., Nikaido K., Liu P. Q., Ames G. F., and Kim S. H. (1998) Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 396, 703–707 [DOI] [PubMed] [Google Scholar]
- 91. Hollenstein K., Dawson R. J. P., and Locher K. P. (2007) Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 [DOI] [PubMed] [Google Scholar]
- 92. Kendall R. L., and Bradshaw R. A. (1992) Isolation and characterization of the methionine aminopeptidase from porcine liver responsible for the co-translational processing of proteins. J. Biol. Chem. 267, 20667–20673 [PubMed] [Google Scholar]
- 93. Chang Y. H., Teichert U., and Smith J. A. (1990) Purification and characterization of a methionine aminopeptidase from Saccharomyces cerevisiae. J. Biol. Chem. 265, 19892–19897 [PubMed] [Google Scholar]
- 94. Yamauchi Y., Ejiri Y., Toyoda Y., and Tanaka K. (2003) Identification and biochemical characterization of plant acylamino acid-releasing enzyme. J. Biochem. (Tokyo) 134, 251–257 [DOI] [PubMed] [Google Scholar]
- 95. Nakai A., Yamauchi Y., Sumi S., and Tanaka K. (2012) Role of acylamino acid-releasing enzyme/oxidized protein hydrolase in sustaining homeostasis of the cytoplasmic antioxidative system. Planta 236, 427–436 [DOI] [PubMed] [Google Scholar]
- 96. Yamauchi Y., Ejiri Y., and Tanaka K. (2002) Glycation by ascorbic acid causes loss of activity of ribulose-1,5-bisphosphate carboxylase/oxygenase and its increased susceptibility to proteases. Plant Cell Physiol. 43, 1334–1341 [DOI] [PubMed] [Google Scholar]
- 97. Vizcaíno J. A., Deutsch E. W., Wang R, Csordas A, Reisinger F, Ríos D, Dianes J. A., Sun Z, Farrah T, Bandeira N, Binz P. A., Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S, Apweiler R, Omenn G. S., Martens L, Jones A. R., and Hermjakob H (2014) ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nature Biotechnol. 30, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Graciet E., and Wellmer F. (2010) The plant N-end rule pathway: structure and functions. Trends Plant Sci. 15, 447–453 [DOI] [PubMed] [Google Scholar]
- 99. Kim H. K., Kim R. R., Oh J. H., Cho H., Varshavsky A., and Hwang C. S. (2014) The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.