Abstract
Currently, evaluation of sperm quality is primarily based on in vitro measures of sperm function such as motility, viability and/or acrosome reaction. However, results are often poorly correlated with fertility, and alternative diagnostic tools are therefore needed both in veterinary and human medicine. In a recent pilot study, we demonstrated that MS profiles from intact chicken sperm using MALDI-TOF profiles could detect significant differences between fertile/subfertile spermatozoa showing that such profiles could be useful for in vitro male fertility testing. In the present study, we performed larger standardized experimental procedures designed for the development of fertility- predictive mathematical models based on sperm cell MALDI-TOF MS profiles acquired through a fast, automated method. This intact cell MALDI-TOF MS-based method showed high diagnostic accuracy in identifying fertile/subfertile males in a large male population of known fertility from two distinct genetic lineages (meat and egg laying lines). We additionally identified 40% of the m/z peaks observed in sperm MS profiles through a top-down high-resolution protein identification analysis. This revealed that the MALDI-TOF MS spectra obtained from intact sperm cells contained a large proportion of protein degradation products, many implicated in important functional pathways in sperm such as energy metabolism, structure and movement. Proteins identified by our predictive model included diverse and important functional classes providing new insights into sperm function as it relates to fertility differences in this experimental system. Thus, in addition to the chicken model system developed here, with the use of appropriate models these methods should effectively translate to other animal taxa where similar tests for fertility are warranted.
Male fertility can be described as the success by spermatozoa to fertilize oocytes, which is a quite intricate process that relies on the correct performance of different, very specialized sperm cells functions such as motility, structural integrity, viability or acrosomic reaction (1, 2). Each of these functions can be tested in vitro to evaluate sperm quality, but results are often poorly correlated with real male fertility, and so a change in the paradigm of sperm quality testing is needed. A new trend in reproduction biotechnology is the elucidation of male infertility etiology through proteomics. This tactic is justified by the fact that mature spermatozoa are transcriptionally and translationally quiescent, so the functionality of those cells is based largely in early gene expression during spermatogenesis and in the complex protein abundance changes and post-transcriptional modifications occurring during maturation (2, 3). Mass spectrometry (MS)-based proteomics has been therefore used as a tool in search of biomarkers of sperm quality basically by applying discovery approaches (4–8). These usually ends up with long biomarker candidates lists, whose validation for diagnostic purposes requires further verification using large cohorts, and high-throughput analysis through the application of multiplexed assays (9). Clinical proteomics has succeeded in solving the latter through the application of targeted approaches such as multiple reaction monitoring-MS methods. However, the development of such tests is cumbersome and expensive and it is often difficult to detect and/or quantify some of the desired targets (9, 10). Alternatively, recent advances in MS instrumentation in combination with the development of bioinformatics tools have allowed the development of untargeted, much simpler and less expensive clinical proteomic approaches based on MALDI-TOF MS profiling. One of these approaches is Intact Cell MALDI-TOF-Mass Spectrometry (ICM-MS)1, a method that relies in the identification of cell-specific peptide/protein MS fingerprints (11–13). In this context, “intact cell” means that whole cells are subjected directly to MS analysis without any preparatory steps, which is highly convenient in diagnostics. This approach has been proven useful for routine high-throughput bacterial (11, 14) and yeast (15, 16) biotyping in clinical specimens, but it has also been applied in superior eukaryotic cells such as macrophages (17, 18) or neuroglial cells (19). In the past, the progress in the general use of MS-based profiling has been somewhat hampered by the difficulties in confidently identifying diagnostic MS signatures, because traditional bottom-up approaches are certainly not adequate. In this sense, the development of direct identification of the peptide/protein through top-down high resolution mass spectrometry (HRMS) has opened new possibilities. The top-down proteomic approach allows for the acquisition in one analysis of both the complete characterization of endogenous peptide/protein sequences and their post-translational modifications (20, 21). A large number of native, intact biomolecules can be hence directly identified, whose masses can be matched with MALDI m/z peaks. As described in the literature, the combination of ICM-MS and top-down HRMS is therefore a convenient analytical strategy (22, 23). In an initial attempt to test if this analytical approach could be employed to phenotype sperm cells with different fertility abilities, we observed that subfertile sperm cells ICM-MS profiles showed characteristic features that could be useful to discriminate individuals regarding their fertility, and that these could be confidently identified by top-down HRMS (23). In that study (as the present one), chicken was chosen as the animal model because of their several advantages, such as their relative small size (easy to handle and house in large numbers), the non-invasive collection of sperm in sufficient quantity, the use of artificial insemination for accurate fertility determination and the high standardization of egg incubation, which also allows proper tracking of the embryonic development. Plus, the commercial breeds used for meat or egg production are genetically uniform but their genetic background differs between the two production types. Using chicken as an experimental model for male fertility has the added advantage of the direct application of results in order to help developing reproduction strategies for poultry breeders and to better characterize semen for bird genetic biodiversity preservation programs (24). Our previous results indicated that sperm cells ICM-MS profiling coupled with top-down HRMS could be employed as a fertility screening tool. However, before demonstrating this fact, the diagnostic performance of the latter had to be proven in a larger and more representative group of males, and the method adapted for a higher throughput analysis.
Due to the abovementioned, the objectives of the present study were (1) to acquire sperm cells ICM-MS profiles in a standardized and automated way from a large and representative cohort of roosters of known fertility, (2) to employ sperm cells ICM-MS profiles to construct fertility-predictive mathematical models, (3) to test the diagnostic performance of the ICM-MS -based models versus other traditional sperm cell quality tests, and (4) to analyze the native and endogenous peptides/proteins of spermatozoa by μLC-top-down HRMS after two different fractionation modes (gel filtration and reverse phase chromatography), in order to identify represented biomolecules in the ICM-MS spectra.
EXPERIMENTAL PROCEDURES
Chemicals
Unless otherwise indicated, chemicals were purchased by Sigma-Aldrich (Saint Quentin Fallavier, France).
Experimental Design and Statistical Rationale
The study population consisted of 72 roosters including 36 roosters from a grandparent generation of chicken bred for meat production (meat line) and 36 roosters from a grandparent generation of chicken bred for egg laying production (laying line). This number was the maximum of animals that could be housed in the INRA experimental unit UE-PEAT, which was enough to obtain statistically confident results, and included two different genetic lines in order to include some natural variability. The fertility values of those animals were determined as described below and used as reference to classify animals. Three ejaculates were analyzed per rooster through ICM-MS as well as in vitro sperm quality tests to account for day-to-day variability. Each sample was divided in 12 replicates that were analyzed three consecutive times through ICM-MS (to account for read-to-read variability), thus 36 technical replicates were performed per ejaculate. Fertility data were unknown at the moment of the acquisition, which served as a way to randomizing the analysis. Four animals per genetic line were selected randomly among those with fertility values above 70% (fertile) and below 40% (very low fertility) and considered as set of training examples for the construction of fertility-predictive models by using supervised learning algorithms. The best model was then subjected to external validation using all animals as classified using the reference fertility value. The number of animals correctly classified as fertile and subfertile by the ICM-MS data-based model was calculated and employed to estimate the diagnostic accuracy of method by means of calculating tests' sensitivity % (rate of true positives detected), specificity % (rate of true negatives detected), positive and negative predictive values (respectively PPV and NPV, which represent proportions of positive and negative results that are true positive and true negative results), as well as likelihood ratios for positive (sensitivity/(1-specificity) and negative ((1-sensitivity)/specificity) results. Same parameters were calculated for each in vitro sperm quality test and results were compared.
Animal Housing and Classification Depending on Individual Fertility Rates
Animals were housed at the INRA experimental unit UE-PEAT at Nouzilly (France) following the European welfare and the French Direction of Veterinary Services regulations (agreement number C37–175-1). Seventy-two 30 weeks-old males were acquired from a commercial pedigree stock (Hubbard and Novogen, Quintin, France) and housed in individual battery cages under a 14L/10D photoperiod and fed with a standard diet of 12.5 MegaJoules/day. The 360 females employed for artificial insemination were adult ISABROWN hens (ISA, Ploufragan, France) housed in 5 hens rooms under a 14L/10D photoperiod and fed a standard diet of 12.5 MegaJoules/day, supplemented with calcium. Semen was routinely collected twice a week by massage (25) during 5 weeks. Sperm concentrations were immediately determined by light absorption of semen with a photometer (Accucell photometer, IMV Technologies, L'Aigle, France) at a wavelength of 530 nm (26). Individual ejaculates were then diluted 1:1 in Beltsville Poultry Semen Extender (26) and then used directly to analyze semen or inseminate females.
Fertility (% fertile/incubated eggs) was measured after individual intravaginal artificial insemination (AI) of 10 females/male with a dose of 100 million sperm/female (day 0). Two inseminations per female were made at a 1-week interval for the same male. Eggs were collected and candled from day 2 to 9 post insemination for the first AI and day 2 to 23 post insemination for the second AI. This second AI was made in order to (1) confirm the first AI results and (2) to observe the coherence of the fertile period according to the reproductive profile of the species (28). Hatchability of the eggs was also measured and did not show added differences between male progeny results. A mean of 250 eggs was checked per male.
This test is considered the reference test to define fertility, and values to establish two cohorts of animals, namely fertile and subfertile animals. This was done taking into account that 70% of fertility is usually the lower limit applied for male selection at chicken grandparent flocks. Animals were therefore considered fertile when rates were above 70% and subfertiles when rates were below 70%. We also designated a subgroup within subfertile animals showing fertility rates lower than 40% that we considered as those exhibiting very poor fertility rates.
ICM-MS Analysis
Spermatozoa (SPZ) were separated from seminal plasma by centrifugation (600 × g for 10 min at 20 °C), washed twice in Tris-Sucrose buffer (260 mm sucrose 20 mm Tris-HCl pH 6.8) by centrifugation at 400 × g for 5 min at 4 °C, and resuspended in the same buffer at 109cells/ml. One microliter of cell suspension (106 cells) was overlaid with 2.5 μl of matrix (20 mg/ml sinapinic acid, 2% trifluoroacetic acid, 50% acetonitrile in water) using the dried droplet method, and 12 replicates were spotted onto a MTP Ground Steel 384 MALDI plate (Bruker Daltonics, Germany). The matrix/sample mix was allowed to evaporate at room temperature and kept subsequently under vacuum for 30 min. Spectra were acquired three consecutive times per spot using a Bruker UltrafleXtreme MALDI-TOF instrument (Bruker Daltonics, Germany) equipped with a Smartbeam laser at 2 kHz laser repetition rate following an automated method controlled by FlexControl 3.0 software (Bruker Daltonics, Germany). Spectra were obtained in positive linear ion mode in the m/z 1,000–20,000 range, and collected from each spot as a sum of 1000 laser shots in 5-shot steps (total of 5,000 spectra per spot). The parameters used for spectra acquisition were: ion source 1, 25 kV; ion source 2, 23.55 kV; lens, 7 kV; pulsed ion extraction; and laser parameter set, large. External calibration was followed using a mixture of peptides and proteins (1 μl of calibrant solution plus 1 μl of matrix composed of 20 mg/ml sinapinic acid, 0,1% trifluoroacetic acid, 50% acetonitrile in water) containing Glu1-fibrinopeptide B, ACTH (fragments 18–39), insulin and ubiquitin, both at 1 pmol/μl, 2 pmol/μl cytochrome C, 4 pmol/μl myoglobin and 8 pmol/μl trypsinogen. To increase mass accuracy (mass error < 0.05%), internal calibration was subsequently applied to all spectra. The latter was achieved by performing a lock mass correction using flexAnalysis 4.0 software (Bruker) with the mass of the highest intensity peak, corresponding to the protein phosphoglycerate kinase (7983.457 m/z). ICM-MS spectra were generated from three different sperm samples per male.
The intra-assay precision of the ICM-MS acquisition was determined by calculating the coefficient of variation (% CV) of the normalized peak intensity values of the 36 technical replicates from three different males/genetic line, whereas the inter-assay precision was determined by calculating the % CV of the mean peak normalized values for the three ejaculates of all animals. The mean % CV of the normalized peak intensity and of the mean peak normalized values did not exceed 30 and 33%, respectively.
Spectral processing and analysis was performed with ClinProTools v3.0 software (Bruker Daltonics, Germany). Data analysis began with an automated raw data pre-treatment workflow, comprising baseline subtraction (Top Hat, 10% minimum baseline width), normalization (total ion count), smoothing (Savitzky-Golay algorithm 2 cycle, m/z range = 15) and spectra realignment using prominent peaks (maximal peak shift 1000 ppm, 30% of peaks matching most prominent peaks, exclusion of spectra that could not be recalibrated). Automatic peak detection was applied to the total average spectrum (a weighted average of all experimental spectra at 800 ppm resolution) with a signal/background noise greater than 2.
Model Generation and Validation
ClinProTools allows for the use of machine-learning algorithms that generate models for discriminating between samples belonging to different established classes. We established two model generation cohorts carefully taking into account that they should be representative for each condition (fertility versus subfertility) but also including some natural variability. Individuals were therefore chosen randomly from those meeting the following requisites: Class 1 (subfertile males; n = 4) should show egg fertilization rates below 40% (very poor fertility) whereas class 2 (Fertile males; n = 4) should show average fertilization rates above 70% (fertile males). Spectra from 3 different ejaculates of each of these animals were loaded onto the software and predictive models using 3 different algorithms were generated: genetic algorithm (GA), supervised neural network (SNN), and quick classifier (QC; including parametric Student's t and non-parametric Mann-Whitney u tests). For each model, the default settings were left unaltered. The ability of each model to correctly identify its component spectra (recognition capability) as well as to handle variability among spectra (cross-validation) was estimated. The model showing the best values of these two parameters was selected for each genetic line and its clinical applicability was confirmed through a blinded validation test using spectra from all the animals included in this study. The discrimination power of each model to identify fertile and subfertile males (thus above or below 70% fertility rates) was then quantified by measures of sensitivity % (true positive rate), specificity % (true negative rate), positive and negative predictive values (PPV and NPV, respectively), as well as likelihood ratios for positive (sensitivity/(1-specificity) and negative ((1-sensitivity)/specificity) test results.
The spectra from meat line and laying line roosters used for model construction as detailed above were combined and uploaded in ClinProTools with the objective of producing a model useful for both genetic lines. We expected this to be possible given the high degree of similarity in the spectra composition between genetic lines (all the analyzed spectra in this study were 85.6% qualitatively identical), but the software repeatedly failed to process spectra together. We discovered later that this was because of differences in the spectral total intensities between the two datasets. We also learnt that with the present version of ClinProTools there was unfortunately no way to treat the spectra so we could process them together.
Protein/Peptide Fractionation and Top-Down HRMS Identification
It was observed that the sperm ICM-MS spectra composition was highly constant between males independently of their fertility rates and genetic origin (85.6% of common masses between all spectra). This means that the molecular composition of sperm cells can be considered qualitatively uniform, and that any sample could be employed for top-down protein identification analysis. A fresh sperm sample was thus randomly chosen (200 μl), and protein extraction was performed by sonication in 400 μl of 6 m Urea 50 mm Tris-HCl pH 8.8 buffer containing protease inhibitor mixture (Roche, Switzerland). Samples were centrifuged during 45 min at 13,000 rpm and 4 °C. One milligram of the extracted peptides/proteins contained in supernatants were subjected to fractionation through chromatographic separation on an UltiMate 3000 RSLC system controlled by Chromeleon version 6.80 SR13 software (Thermo Scientific Dionex) using two different chromatographic approaches. First, proteins were separated on the base of their hydrophobicity by reversed phase (RP) HPLC using a Waters XBridge BEH C18 column (Waters). A second approach was based on the biomolecules separation by molecular weight using a Superdex 75 10/300 GL gel filtration (GF) column (GE Healthcare Life Sciences) in 100 mm ammonium bicarbonate buffer. Thus, 35 and 38 fractions were generated by RP and GF liquid chromatography processes, respectively. Samples were immediately vacuum-dried and kept at −80 °C until further analysis.
Prior to analysis all fractions were reconstituted in 20 μl of Formic acid 1%, desalted using Zip-Tips (Millipore, MA), and then analyzed by on-line micro-liquid chromatography tandem mass spectrometry (μLC-MS/MS) on a dual linear ion trap Fourier Transform Mass Spectrometer (FT-MS) LTQ Orbitrap Velos (Thermo Fisher Scientific, Germany) coupled to an Ultimate® 3000 RSLC Ultra High Pressure Liquid Chromatographer (Dionex, The Netherlands) controlled by Chromeleon Software (version 6.8 SR11; Dionex). Ten microliters of each sample were injected using μl-pickup mode and loaded on a Dionex trap column (Monolithic PS-DVB PepSwift, 200 μm inner diameter × 5 mm long). Mobile phases consisted of (A) 0.1% formic acid, 97.9% water, 2% acetonitrile (v/v/v) and (B) 0.1% formic acid, 15.9% water, 84% acetonitrile (v/v/v). Biomolecules were automatically preconcentrated for 10 min at 10 μl/min with 4% solvent B. The separation was conducted using a Dionex column (Monolithic PS-DVB PepSwift, 200 μm inner diameter x 5 cm long). The gradient consisted of 4–10% B for 1 min, 10–75% B for 60 min, 75 to 99% B for 1 min, constant 99% B 20 min and return to 4% B in 1 min. The column was re-equilibrated for 15 min at 4% B between runs. The flow rate was set to 1 μl/min. The eluate was sprayed using a SilicaTip emitter with 30 μm inner diameter and 360 μm outer diameter (New Objective, Woburn, MA) into a Thermo Finnigan Nanospray Ion Source 1. Standard mass spectrometric conditions for all experiments were spray voltage 1.3 kV, no sheath and auxiliary gas flow; heated capillary temperature, 250 °C; predictive automatic gain control enabled, and an S-lens RF level of 70%. Data were acquired using Xcalibur software (version 2.1; Thermo Fisher Scientific, San Jose, CA). The LTQ Orbitrap Velos instrument was operated in positive mode in data-dependent mode using a high-high strategy, meaning that a FT-MS spectrum using the profile mode was followed by an FT-MS2 spectrum. Target resolution in the Orbitrap was set to r = 100,000. In the scan range of m/z 400–2000, the 10 most intense ions with charge states ≥2 were sequentially isolated and fragmented by HCD (Higher-Energy Collisional Dissociation) with normalized collision energy of 38% and wideband-activation enabled. Ion selection threshold was 500 counts for MS/MS with an isolation width = 3 m/z. The maximum allowed ion accumulation times were 200 ms for full scans (2 microscans) and 500 ms for HCD-MS/MS measurements (3 microscans) in the Orbitrap analyzer. Target ion quantity for FT full MS was 1 × 106 and for MS2 was 5 × 105. Dynamic exclusion was enabled with a repeat count of 3 and exclusion duration of 5000 secondes.
Proteo/peptidoform identification and structural characterization were performed using ProSight PC software v 3.0 SP1 (Thermo Fisher, San Jose, CA). μLC-MS/MS raw files were collected and automatically processed inside ProSightHT using the cRAWler algorithm average related scans to assign precursor and fragment masses and using the THRASH algorithm to convert each precursor/fragmentation scan pair into monoisotopic neutral mass values. All fragmentation data were filtered using the following parameters: signal/noise = 3/1, minimum fragment intensity at 100, retaining only the top 5 most intense neutral fragment masses within a 100 Da window below 2000 Da. Individual ProSightPC experiments were created for each precursor and corresponding fragment masses generating an XML file in ProSight Upload Format (PUF). From PUF files, automated searches were performed using the “Biomarker” search option against a database made via shotgun annotation from the Swiss-Prot Gallus gallus release from the UniProtKnowledgebase release 2012_06 (gallus_gallus_2012_06_top_down_simple.pwf downloaded from ftp://prosightpc.northwestern.edu/). An iterative search tree was designed to begin with high mass accuracy (both 10 ppm at the intact and fragment ion level) for monoisotopic precursors. If the top result matched with a p-score of ≤1 × 10−5 the search engine accepted this result as valid. Two subsequent searches were performed for invalid results using larger intact mass tolerances (average precursors with 3 and 85 Da mass tolerance). For all searches, both delta mass feature and disulfide modifications were deactivated and the N-terminal post-translation modifications (acetylation and initial methionine cleavage) were considered. First hits were considered positively identified with a minimum of five matching fragment ions with E-value ≤1 × 10−5 for monoisotopic precursors and E-value ≤1 × 10−8 for average precursors. However, for average precursors, identifications with E-values between 1 × 10−7 and 1 × 10−6 (considered as low confidence proteoform identifications) were accepted after being manually controlled and independently analyzed using Sequence Gazer.
Raw data derived from top-down HRMS analysis have been deposited to the ProteomeXchange Consortium (29) via the PRIDE partner repository (30) with the dataset identifier PXD002768.
Functional Analysis
In order to estimate which cell compartments/functions/pathways are mainly represented by the peptides/proteins analyzed through ICM-MS, a systems biology analysis was performed. In brief, the UniProtKB accession numbers from all m/z masses matching those detected in the ICM-MS spectra that were confidently identified through top-down HRMS analysis were recovered and listed. From these, the official human gene symbols (HuGO Gene Nomenclature Committee) were retrieved from Gallus gallus annotated proteins, whereas uncharacterized proteins were mapped to the corresponding Homo sapiens orthologs by identifying the reciprocal-best-BLAST hits. When manual annotation was needed (i.e. for deleted entries), this was performed by BLASTp (E value ≤ e-04), together with Gene Ontology (GO) functional annotation. These lists were uploaded in GeneAnalytics (http://geneanalytics.genecards.org/), a commercially available software program based on proprietary, comprehensive and organized databases from the LifeMap suite.
Immunoblotting Analysis
The fertility-predictive models obtained in this study from the acquired ICM-MS spectra employed a defined list of m/z masses for its construction, which corresponded mostly with fragment peptides of bigger proteins that showed a differential abundance between fertile and subfertile males for each genetic line. We tested the hypothesis that relative differences in the intracellular abundance of these fragment peptides could be associated with similar variations in the corresponding whole proteins. We selected three proteins that were employed in the construction of fertility-predictive models, namely heat-shock protein 90-alpha (HSP90A) for meat-line model as well as Tektin 3 (TEKT3) and sperm-associated protein antigen 6 (SPAG6) for laying models, and we established the differences in the relative abundance of the whole protein by immunoblotting in sperm protein extracts from the same animals used to build the models (see above). In brief, 30 μg of total protein extracts (obtained as detailed above) were separated on 4–12% SDS-PAGE minigels under reducing conditions, blotted onto nitrocellulose membranes and blocked for one hour in 5% skimmed milk TBS-Tween 20 buffer. HSP90A, SPAG6, and TEKT3 were detected by commercial antibodies showing cross-reactivity with chicken (references Abcam-ab59459, Antibodies-online-ABIN503491 and Aviva ARP57677_P050, respectively), followed by anti-mouse (HSP90A) or anti-rabbit (SPAG6 and TEKT3) secondary antibodies conjugated with Alexa Fluor® 680 (Thermo-Fisher Scientific). Total protein staining of the membrane with SyproRuby blot stain (Thermo) prior to blocking served as a loading control. Images were obtained by scanning on a Fusion FX (SyproRuby-stained membranes; Vilber-Lourmat) or a Li-Cor Odyssey Infrared Imager (Immunoblots; Li-Cor Biosciences, Lincoln, NE). All the images were digitalized and analyzed by Image Studio Lite Software (Li-Cor Biosciences). For each lane, band intensity values were normalized onto the respective overall protein staining.
In Vitro Semen Quality Tests
We investigated if the diagnostic performance of MS-based method to predict fertility was comparable to that of the different tests routinely employed for sperm quality, namely mass motility, objective measures of motility and viability. Tests were performed in three different ejaculates from the same males as above, using samples collected ± 4 days around the collection dates of sperm samples tested by ICM-MS. Values were averaged for the three ejaculates and objective cut-off values were defined for each test to classify the sperm quality into “fertile” or “subfertile.” The discrimination power of each model to identify fertile and subfertile males was then quantified by measures of sensitivity, specificity, likelihood ratios as well as positive and negative predictive values taking values of in vivo fertility rates as reference as described previously. Results were compared with those obtained by ICM-MS.
Mass motility was measured as a subjective evaluation of the speed of movement of a group of sperm in 20 μl of semen (scale 0 to 9) as previously described (31). On this motility scale, 0 represents a total lack of movement with agglutinates maybe covering the entire slide area and 9 representing whirlwinds covering 30–60% of the area. It was considered that sperm of good quality should show a value of 5 or higher. This measure was performed five times per ejaculate and the mean was calculated.
Objective measurements of motility were evaluated by the computer-assisted sperm analysis (CASA) system with an HTM-IVOS (Hamilton-Thorn Motility Analyzer, IVOS; (31). In this experiment, the parameters retained for the analyses were percentage of motile sperm (%) and average path velocity (VAP, in μm/s). It was considered that good quality sperm should show at minimum values of 50 and 60%, respectively.
Sperm Viability was assessed with SYBR-14/Propidium iodide fluorescent dyes (32). Aliquots from each sample were adjusted to a final concentration of 109 cells/ml. Then, sperm were diluted in Lake 7.1 buffer down to 2 × 106 cells/ml and 5 μl SYBR-14 was added before the solution was incubated for 10 min in darkness at 4 °C. Afterward, 2 μl of propidium iodide was added and the incubation was continued for 5 min in the dark at 4 °C. After incubation, sperm viability was assessed by fluorescence microscopy (Zeiss Axioplan 2; Zeiss Gruppe, Jena, Germany): living cells appeared green and dead ones red. A total of 300 sperm per slide were counted (two slides/sample = 1 replicate) per sample, and minimum of 85% viable sperm cells was set as the criterion to consider high quality. All preparations were analyzed by the same observer.
RESULTS
Animal Classification Regarding Fertility Rates
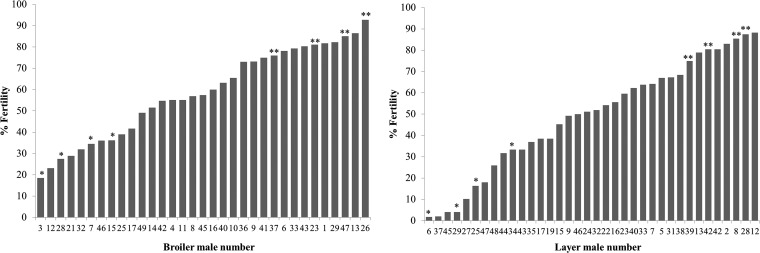
Fertility results ranged from 18 to 90% fertility in meat type line and from 2 to 85% fertility in laying type line (Fig. 1). Three meat line roosters stopped producing sperm during the trial and were therefore eliminated from the study. Fertility rates were employed to classify the roosters, and two classifications were made: the first was used to construct our fertility-predictive mathematical models using ICM-MS spectra and thus 4 males were randomly selected from those with fertility rates higher than 70% (n = 13 for the meat line and n = 8 for the laying line) and lower than 40% (n = 9 for the meat line and n = 14 for the laying line). The second classification was used to perform the clinical validation of both ICM-MS and in vitro sperm quality tests and thus roosters were grouped in those with fertility range over 70% (n = 13 for the meat line and n = 8 for the laying line) and below 70% (n = 20 for the meat line and n = 28 for the laying line).
Fig. 1.
Fertility results obtained with the meat type line (left) and the lay type line (right). Fertility rates reported here were calculated by candling of the eggs laid from days 2 to 9 post insemination. A mean of 10 females were inseminated per male with 100 million sperm cells per insemination dose. Some individuals were randomly selected to construct the fertility-predictive models from those highly subfertile (Fertility rate lower than 40%, indicated with and asterisk) or fertile (Fertility rate higher than 70%, indicated with two asterisks).
Diagnostic Performance of Fertility-predictive ICM-MS-based Model and In Vitro Sperm Quality Tests
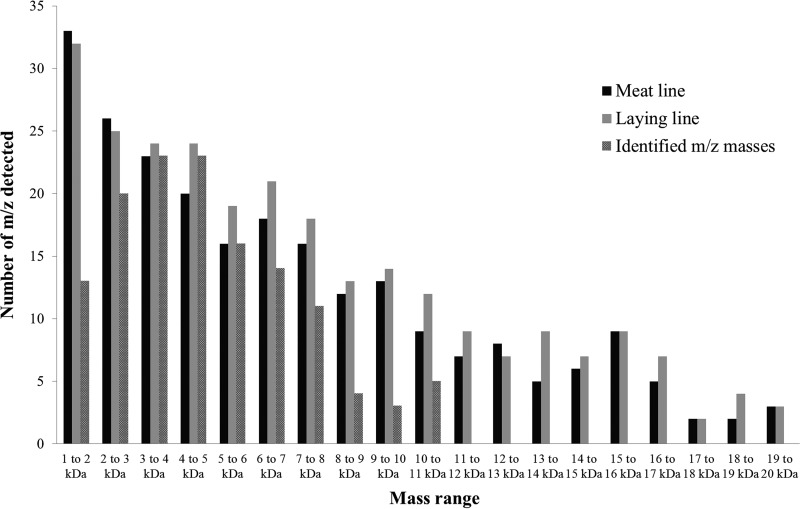
Sperm ICM-MS spectra from meat and laying line roosters included respectively 233 and 259 detected peaks in the mass range of 1–20 kDa. The distribution of detected m/z over the mass range was very similar for both genetic lines (Fig. 2).
Fig. 2.
Distribution of detected m/z peaks over the 1–20 kDa mass range in the average ICM-MS spectra from meat and laying line sperm cells and of species identified by top-down High Resolution Mass Spectrometry.
After comparing the mass lists detected in the ICM-MS spectra from meat and layer chicken sperm cells, we observed that more than 85% of the m/z masses were present in both genetic lines, whereas spectra from fertile and subfertile spermatozoa within each line shared more than 95% m/z masses. This shows that the molecular composition of chicken sperm ICM-MS spectra is quite constant.
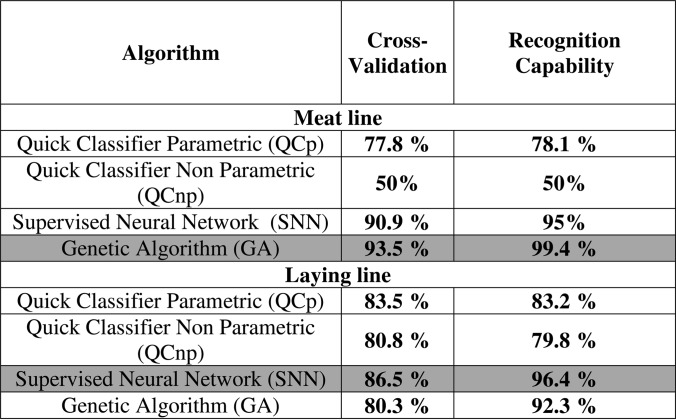
Three different algorithms: genetic algorithm (GA), supervised neural network (SNN), and quick classifier (QCp including parametric Student's T, and QCnp non-parametric Mann-Whitney U tests) were tested for the construction of fertility-predictive models for each genetic line. The best model was chosen based on values of recognition capability (model's ability to correctly identify its component spectra) and cross-validation (the model's ability to handle variability among spectra) for each genetic line (Table I).
Table I. Comparison of cross-validation and recognition capability rates of all the fertility-predictive models constructed for meat and laying line roosters using the available algorithms present in ClinProTools software. The darker background indicates the model showing the best validation parameters.
The model with best values for meat line males was the one based in the use of the GA algorithm which showed 99.4% of recognition capability and 93.48% of cross validation values. This algorithm retained 10 m/z masses for model construction (4777,19; 6539,47; 6695,12; 7035,19; 7164,42; 7340,66; 7487,43; 7633; 8297,38; 11337,16). The best model for laying line males was the one built using the SNN algorithm showing 96.4% of recognition capability and 86.5% of cross validation values, and being built using 20 m/z masses (2188,3; 2400,68; 2574,16; 4012,62; 4537,88; 4984,8; 5226,72; 5951,27; 6538,96; 6615,99; 7341,89; 7487,59; 8569,91; 9026,89; 9543,82; 12443,22; 13328,61; 14060; 14300,17; 18050,71). Three m/z masses were common between those employed to build fertility-discriminating models for meat and laying lines (6539.45, 7341.27, and 7487.51 Da).
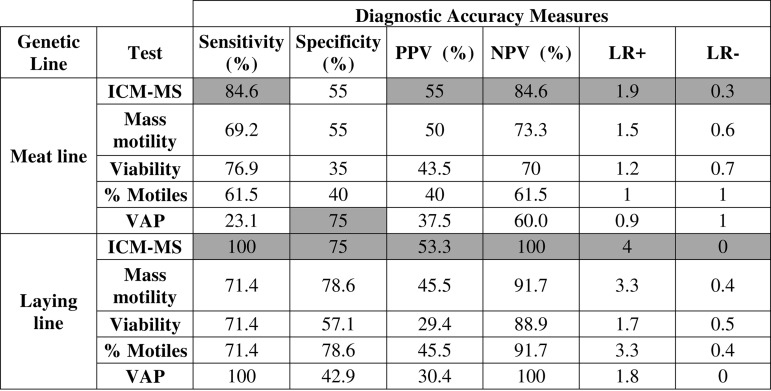
To perform the clinical validation of these models, both male populations were divided into fertile and subfertile males using a fertilization rate cut-off value of 70% and we determined if the models generated for each genetic line classified each male into the correct group. The number of correctly and incorrectly classified males were determined and used to calculate different measurements of diagnostic performance such as sensitivity (true positive rate), specificity (true negative rate), positive and negative predictive values (PPV and NPV, respectively), as well as likelihood ratios for positive (sensitivity/(1-specificity) and negative ((1-sensitivity)/specificity) test results. The same was performed for each in vitro sperm quality test and results were compared (Table II).
Table II. Diagnostic accuracy measures of intact cell MALDI-TOF Mass Spectrometry (ICM-MS) and sperm quality tests for meat and laying line for fertility diagnosis. PPV: Positive Predictive Value; NPV: Negative Predictive Value; LR+: Positive Likelihood Ratio; LR-: Negative Likelihood Ratio; VAP: Average Path Velocity. The darker background highlights the higher values of each parameter for each genetic line.
The ICM-MS method showed the highest trade-off between sensitivity and specificity, as well as between negative and positive predictive values. It also showed the best likelihood ratios for positive and negative testing. The ICM-MS method showed very good sensitivity for both genetic lines, meaning that it performed very well at detecting animals with high fertility, whereas the lower specificity values indicated a still good but lower ability at detecting subfertile males. Likelihood ratios were consistent with this: LR- were very close to zero, indicating that males classified as fertile by the ICM-MS method are very likely to be actually fertile. However, although LR+ values were higher than 1 and thus satisfactory, results suggested that the tests were efficient but little less sensitive when detecting subfertile males. The same can be interpreted from the fact that NPV was better than PPV.
Top-down HRMS Protein Identification
The top-down HRMS proteomics experiment, operated in discovery mode, was carried out by μLC coupled to tandem high resolution mass spectrometry from pre-fractionated sperm protein extracts after RP or GF separations. In all, 796 intact or fragment protein masses were detected in a mass range between 1156.46 and 14,907.14 Da ([M+H]+ theoretical average mass), and were confidently identified by ProSight PC software with E-values ranging from 7 × 10−6 to 1.32 × 10−126. These peptido- and proteoforms derived from 366 unique UniprotKB accession numbers corresponding to 342 unambiguously identified genes. Out of them, 34 biomolecules showed post-translational modifications as N-terminal acetylation including 13 N-acetylations on methionine, one on valine, 12 on alanine, and 8 on serine residues after the cleavage of the N-methionine. At the exception of the N-acetylvaline on FABP7_CHICK and the N-acetylalanine on 1433E_CHICK, all these modifications were newly characterized. All the results are available in supplementary Table S1.
A total of 313 biomolecules were identified in fractions derived from RP chromatography fractionation, with E values ranging between 1 × 10−126 and 6 × 10−6, masses ranging between 1,156.46 Da and 14,907.14 Da, and corresponding to 188 nonredundant accession numbers and 182 unambiguously identified genes. In fractions derived from GF chromatography fractionation, a total of 483 biomolecules were identified with E values ranging between 1 × 10−102 and 7 × 10−6, masses between 687.79 Da and 14,837.86 Da, and corresponding to 240 nonredundant accession numbers and 226 unambiguously identified genes. Between the two strategies, only 62 accession numbers and 66 genes names were shared, showing the complementarity of both fractionation methods. Approximately 72% of the identified biomolecules (580/796) presented C-terminal arginine or C-terminal lysine residues, suggesting that the major part of the biomolecules identified in the mass range < 15 kDa are the products of protease activities from trypsin-like or kallikrein enzymes.
The total peak lists from the average broiler and layer ICM-MS spectra were retrieved from ClinProTools, and after the suppression of replicates (219 m/z masses were common between genetic lines), a list of 256 m/z masses representing all the peaks detected by ICM-MS in all the studied conditions was obtained. From these, the top-down MS proteomic analysis described above was able to identify 102 m/z masses, which corresponded to 83 unique peptido-and proteoforms. In this work, from the 17 reviewed and 70 unreviewed proteins (15 deleted sequences from TrEMBL) from UniProtKB database (accessed in August 2015), five proteins were annotated with a previously characterization at protein level, 21 at transcript level, 21 were inferred from homology and 40 as protein predicted. Out of 102 peaks observed by ICM-MS, only one whole protein was identified (Mitochondrial ubiquinol-cytochrome c reductase 7.2 kDa protein; UniProtKB accession number D0VX27) whereas four biomolecules were identified as N-terminal fragments of proteins, eight as C-terminal fragments and 75 as internal fragments. However, most of them corresponded to tryptic or semi-tryptic sequences very close to the N or C terminus and one of them has even been characterized with an N-acetylation. These results suggest that the biomolecules observed by ICM-MS are essentially products of protein degradation. How these fragments were generated and under which mechanism is unknown. According to the results of the bioinformatics analysis with GeneAnalytics, these biomolecules corresponded to fragments of proteins expressed mostly in testicular tissues, and attributed to the reproductive system phenotype. Most biomolecules were associated with molecular functions such as ATP/GTP-related activities, transmembrane transport and cell structure/movement, whereas the most represented biological processes included different metabolic routes and cell motility/structure. Among the enriched pathways identified in this dataset, the most significant were related with cell movement, centromere activity, energy metabolism and transmembrane ion transport. Complete results as retrieved from GeneAnalytics are included in supplemental Table S2.
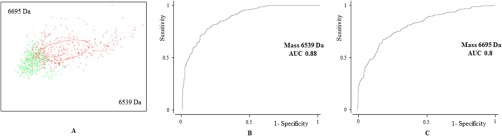
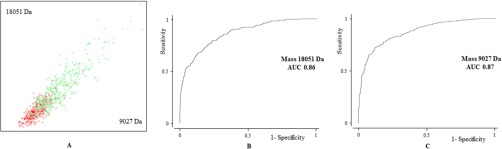
For each dataset (meat line and laying line), ClinProTools automatically displayed the two peaks with the highest ability for class discrimination, which constituted the most interesting peaks for diagnostic purposes. Those peaks corresponded to masses 6539.47 Da and 6695.12 Da in the meat line; 9026.89 Da and 18,050.71 Da in the laying line. The ability of these differentially displayed peaks to segregate the male populations in the 2 studied classes is represented by 2D peak distribution diagrams (Fig. 3a and 4a). As shown in the plot, although there is some overlapping, the combination of the abovementioned peaks can separate fertile from subfertile individuals with a 95% confidence interval. Receiver operating characteristic (ROC) curves were generated for each of these peaks (Figs. 3B, 3C, 4B, and 4C), and results showed a good diagnostic performance, with areas under the curve (AUC) above 0.8 in all cases. We were able to confidently identify masses 6695.12 Da and 6539.47 Da by top-down HRMS, which corresponded with fragments of the proteins heat shock protein 90 alpha and tubulin alpha 3e, respectively. We also tentatively identified mass 18,050.71 Da as vitelline membrane outer layer 1 (VMO1) based on previous data (Labas et al., 2015), although this should be confirmed.
Fig. 3.
Bidimensional peak distribution diagrams (A) and receiver operating characteristic (ROC) curves (B, C) of the two m/z peaks with the highest ability for subfertility discrimination in meat line males.
Fig. 4.
Bidimensional peak distribution diagrams (A) and receiver operating characteristic (ROC) curves (B, C) of the two m/z peaks with the highest ability for subfertility discrimination in laying line males.
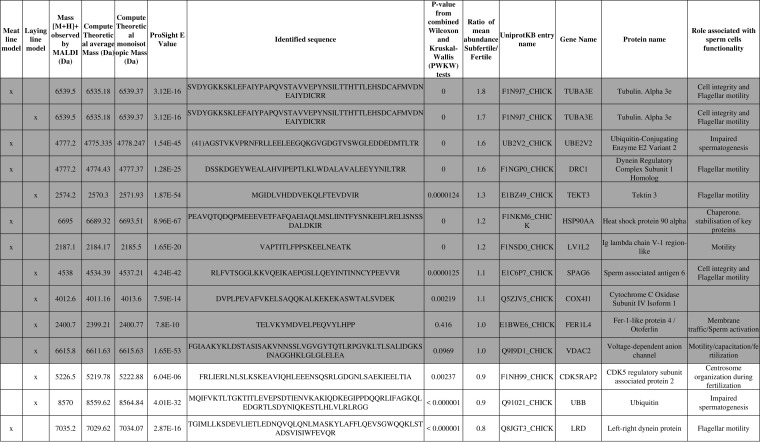
From the m/z masses employed by ClinProTools to elaborate the fertility-discriminating models, 12 were confidently identified, including the mass 6539.45 Da (TUBA3E), which was included in the models from both meat and laying genetic lines. These masses corresponded with fragments of proteins mostly implicated in functions related with cell structure and movement, ATP-related activities and ubiquitin-proteasome dependent proteolysis, and most of them are already known fertility biomarkers in other species (Table III).
Table III. Characteristics of the intact cell MALDI-TOF Mass Spectrometry m/z masses employed to generate ClinProTools fertility predictive models in meat- and laying-type chicken that were confidently identified after top-down High Resolution Mass Spectrometry. Gene names are indicated according with the Hugo Gene Nomenclature Committee-approved gene nomenclature. Grey background indicates peptides that show higher abundance in Subfertile (SubF) males, whereas white background indicates peptides that show higher abundance in Fertile (F) males.
Immunoblotting Analysis
The m/z masses employed by ClinProTools to elaborate the fertility-discriminating models corresponded to fragment peptides of bigger proteins. We investigated if fold-changes in abundance between subfertile and fertile roosters detected at the peptide (obtained from ICM-MS data analysis) and the protein level (obtained by immunoblotting) were comparable. In laying-type males, we tested the proteins SPAG6 and TEKT3, and we observed that fold-changes in relative abundance were identical at the peptide and protein level (Table IV). In meat type males, we studied the protein HSP90A, and results showed that the abundance of this biomolecule was higher in subfertile sperm both at the peptide and the protein level, although differences were slightly more marked at the protein level. Results from Western blot analysis are included in supplemental Table S3.
Table IV. Fold-changes observed in peptide/protein abundance of the proteins HSP90A, SPAG6 and TEKT3 between subfertile and fertile sperm samples in the two genetic lines studied, as calculated after ICM-MS or Immunoblotting (WB) analysis, respectively.
| Protein name | Genetic line tested | Fold-change in abundance in subfertile (n = 4) relative to fertile (n = 4) males |
|
|---|---|---|---|
| ICM-MS (fragment peptide) | Immunoblotting (whole protein) | ||
| HSP90A | Meat line | 1.2 | 1.8 |
| SPAG6 | Laying line | 1.1 | 1.1 |
| TEKT3 | Laying line | 1.3 | 1.3 |
DISCUSSION
In the present study, we were able to show for the first time that fertility-predictive mathematical models can be generated from sperm cells ICM-MS spectra, and that these can be used for male fertility phenotyping with a high degree of diagnostic performance. Characterization of ICM-MS spectra through top-down HRMS showed that they represent most of cellular processes and compartments, hence allowing a global testing of the main sperm cell functions.
Ideally, a diagnostic test should be simple (few preparatory steps), fast, automated and reliable. Fertility testing with the ICM-MS method presented here meets these requirements, because cells are tested after only a simple wash, and the spectral acquisition and evaluation is performed in an automated way in a few minutes. Furthermore, intra- and inter-assay variability were kept under 30%, and mass accuracy was high (error <0.05%), thus showing analytical robustness. MS data were used to construct two male fertility-predictive mathematic models for meat and egg-laying roosters, showing very satisfactory values of class recognition capability and cross-validation, and very good values of diagnostic accuracy. Our initial objective was to obtain a common model for both genetic lines, but each data set presented global intensity differences which prevented their combined processing by the software. We expect that further updates of the software allow for a solution to this problem. However, we observed that even if the molecular composition of chicken sperm ICM-MS spectra was quite constant between genetic lines (ICM-MS spectra from both lines showed more than 85% of m/z masses in common), the m/z masses selected by the software to build each model were very different (only three m/z masses were common to both mass lists of 10 and 20 masses for the meat and laying line models, respectively). This suggests that spite of showing a very similar composition, the relative abundances of each species differed between lines, thus possibly reflecting the marked differences in genetic background.
When evaluating the diagnostic accuracy of the ICM-MS test we observed very good values for all the parameters, noticing that it performed particularly well in terms of sensitivity, negative predictive values and negative likelihood ratio. To interpret these results, we have to bear in mind that, in general, the objective measures of the performance from diagnostic tests are highly influenced by the spectrum of the studied condition (33). When the spectrum is wide (i.e. a disorder that can appear with different clinical pictures), we can expect a loss in specificity, and to improve it we have to rely in specific testing of each feature of the studied condition. Since the spectrum of subfertility is quite wide (the reasons of a sperm cell for not being functional are various), we can expect a relatively wide range of changes happening at the molecular level, whereas functional (thus fertile) sperm cells should be more uniform in their molecular composition. Results show that the ICM-MS-based models are very good at identifying this latter stable phenotype, and that is why they are able to identify the most fertile animals with higher sensitivity whereas the wide spectrum of subfertility is reflected in relatively lower but still satisfactory levels of specificity. Both positive and negative predictive values as well as likelihood ratios for positive and negative results were better for the ICM-MS test than for any other in vitro sperm quality test. Likelihood ratios results suggest that ICM-MS is the most reliable rest for ruling-in and -out a fertility diagnosis, whereas the better predictive values indicate that the probability of being correctly classified through the results of ICM-MS as fertile/subfertile were higher than if using results from other in vitro tests. In view of these results, it can be hence concluded that ICM-MS profiling is more accurate for fertility screening of individual males than any other in vitro sperm quality test.
We were interested in defining the identity of the m/z peaks present in the sperm ICM-MS spectra in order to having a better understanding on the molecular species being detected by this diagnostic tool and therefore possibly explaining some cellular mechanisms implicated in male fertility/infertility. The top-down HRMS analysis employed here allowed us to characterize approximately half of the m/z masses represented in the sperm ICM-MS spectra. This high percentage of identified masses could be achieved mainly thanks to a combination of two fractionation methods based on gel filtration and reverse phase chromatography, which were highly complementary. Our HRMS top-down strategy allowed the identification of endogenous low molecular weight biomolecules (< 15 kDa) directly in their biological context, with several post-translational modifications carried by some of these endogenous peptides. Additionally, we highlighted protein cleavages at specific sites (mainly at R or K) in most of these peptides. Gross degradation of proteins during sample preparation was highly unlikely, as we took measures to avoid proteolysis during sample preparation (use of anti-proteases, preservation of dried samples at −80 °C). The absence of significant protein degradation during sample preparation is also suggested by the fact that the molecular composition of all the sperm samples used in the present study was qualitatively very constant, as mentioned above. Our hypothesis is hence that these masses corresponded mostly to protein degradation products generated by biologically relevant cleavages because of endogenous degradative or targeted proteolysis. Functional analysis showed that the native proteins are implicated in the main sperm functions, energy metabolism, protein degradation, membrane transport and cellular structure and movement. We can therefore suggest that through ICM-MS analysis, we are taking a sort of snapshot of the relative abundance of degradation peptides representing the main sperm cell functions. This “holistic” nature of ICM-MS analysis in juxtaposition with the “reductionist” functional in vitro tests might be also the reason for the better diagnostic performance of the first. This is in line with previous reports that describe that degradomic-peptidomic profiling is highly sensitive to physio-pathological changes and potentially provides unique signatures of possible diagnostic utility (34, 35).
A total of 27 different m/z masses were used for the generation of meat and laying lines fertility predictive models, from which 12 could be identified by top-down HRMS. The identified peptides were more abundant in subfertile sperm cells except CDK5RAP2, LRD and Ubiquitin, which were more abundant in the most fertile cells. Changes in the relative abundance of some of these fragment peptides (HSP90A, TEKT3 and SPAG6) between fertile and subfertile sperm coincided with similar changes observed in corresponding native proteins in previous reports (23). Moreover, the combined evaluation of the 27 m/z list in each mathematical algorithm allowed for the estimation of fertility, and so we considered interesting to investigate their functional role in sperm.
In this list, we found fragments of the proteins TEKT3, SPAG6, TUB3E, DRC1, and LRD, which are components of both the sperm flagella axoneme and periaxonemal space, thus responsible for flagellum structure and/or motility. Several references can be found in the literature that relates the differential abundance of these proteins with fertility (36–40). Along with these, we also observed a fragment of the CDK5RAP2, a protein that displays several functions in the control of centrosome and microtubule organization (41, 42). The role of this regulatory protein in spermatozoa has not been investigated, but it is known that it might play a role in keeping correct function of the centrosome (41). An additional member of the models mass lists is voltage-dependent anion channel 2. This pore-forming protein is involved in the membrane transport of ions and small molecules and has been found in sperm of different species, with relevant roles including functions related with motility, capacitation and fertilization (43). COX4I is the terminal enzyme of the respiratory chain and is necessary for ATP synthesis (44), so lower levels of this protein may be associated with defective sperm functions including motility (45) and other energetic metabolic dependent functions. Another interesting member of the mass list is Fer-1-like protein 4 (FER1L4), a protein that belongs to the ferlin family, whose members are implicated in Ca2+ mediated membrane-membrane fusion events (46). The function of FER1L4 is unknown, and no evidences of ferlins implication on vertebrate fertility have been published, but it is known that these proteins are expressed in the gonads and required for fertility in both D. melanogaster and C. elegans (47, 48). Specifically, the single ferlin gene found in D. melanogaster is required for sperm plasma membrane breakdown and completion of sperm activation during fertilization (48). Future studies should be needed to prove the exact function of FER1L4 during fertilization in chicken. A fragment of ubiquitin was also identified as a list member. This is a small protein that serves as a tag for degradation through the ubiquitin-proteasome pathway(49). Differential ubiquitination patterns may be necessary for reproductive success. The involvement of this small molecule in male fertility has been evidenced at different levels such as sperm centrosome reduction, histones replacement by protamines, paternal mitochondria elimination at fertilization, epididymal elimination of defective spermatozoa and zona penetration (Reviewed by 49). The heat shock protein 90 (HSP90) is a molecular chaperone present in many tissues, including the sperm tail. In sperm, HSP90 preferentially interacts with a specific subset of proteins such as tyrosine kinases, serine-threonine kinases and other enzymes, activating and stabilizing them, thus playing a role in mediating sperm function and fertilizing ability (51, 52). A fragment of IgG light chain was also present in the mass list. Levels of immunoglobulins have been found in semen of different species, associated with infertility, because their binding with sperm surface can cause cells to clump together and prevent motility (53, 54).
In conclusion, we designed an automated, high-throughput ICM-MS method efficient for sperm phenotyping. Acquired spectra were successfully used to generate fertility-predictive models showing a better diagnostic performance than any traditional in vitro sperm quality tests. The identification of the peptides/proteins represented in the ICM-MS spectra determined that this method evaluates different sperm functions at once, and helped us identifying particular proteins whose relative abundance might be linked with fertility.
Supplementary Material
Footnotes
Author contributions: E.B. designed research; L.S., V.L., A. Thelie, I.G., and A. Teixeira-Gomes performed research; L.S. and V.L. analyzed data; L.S., V.L., and E.B. wrote the paper.
* This work was supported by the French National Infrastructure of Research CRB anim funded by “Investissements d'avenir”, ANR-11-INBS-0003 and from the French National Institute of Agronomic Research. The high resolution mass spectrometer was financed (SMHART project) by the European Regional Development Fund (ERDF), the Conseil Régional du Centre, the French National Institute for Agricultural Research (INRA) and the French National Institute of Health and Medical Research (Inserm). Laura Soler has received the support of the EU in the framework of the Marie-Curie FP7 COFUND People Programme, through the award of an AgreenSkills fellowship (under grant agreement n° 267196).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ICM
- intact cell MALDI-TOF
- AI
- Artificial Insemination
- MTP
- micro-titer plate
- %CV
- Coefficient of variation
- GA
- Genetic Analyzer
- SNN
- Supervised Neural Network
- QC
- Quick classifier
- QCp
- Quick classifier-parametric
- QCnp
- Quick classifier-non parametric
- HRMS
- High Resolution Mass Spectrometry
- RP
- Reverse Phase
- GF
- Gel Filtration
- HCD
- High Collision Energy
- PUF
- ProSight Upload Format
- CASA
- Computer-Assisted Sperm Snalysis
- HTM
- Hamilton-Thorn Motility Analyzer
- IVOS
- Integrated Visual Optical System
- VAP
- Average Path Velocity
- PPV
- Positive Predictive Value
- NPV
- Negative Predictive Value
- LR+
- Positive Likelihood Ratio
- LR-
- Negative Likelihood Ratio
- ROC
- Receiver Operating Characteristic
- AUC
- Area Under the Curve
- HGNC
- Hugo Gene Nomenclature Committee
- WB
- Western Blotting
- ATP
- Adenosine Triphosphate.
REFERENCES
- 1. Lemoine M., Grasseau I., Brillard J. P., and Blesbois E. (2008) A reappraisal of the factors involved in in vitro initiation of the acrosome reaction in chicken spermatozoa. Reproduction 136, 391–399 [DOI] [PubMed] [Google Scholar]
- 2. Peddinti D., Nanduri B., Kaya A., Feugang J. M., Burgess S. C., and Memili E. (2008) Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst. Biol. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aitken R. J., and Baker M. a. (2008) The role of proteomics in understanding sperm cell biology. Int. J. Androl. 31, 295–302 [DOI] [PubMed] [Google Scholar]
- 4. Oliva R., De Mateo S., and Estanyol J. M. (2009) Sperm cell proteomics. Proteomics 9, 1004–1017 [DOI] [PubMed] [Google Scholar]
- 5. Rahman M. S., Lee J.-S., Kwon W.-S., and Pang M.-G. (2013) Sperm proteomics: road to male fertility and contraception. Int. J. Endocrinol. 2013:360986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Mateo S., Martínez-Heredia J., Estanyol J. M., Domíguez-Fandos D., Vidal-Taboada J. M., Ballescà J. L., and Oliva R. (2007) Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 7, 4264–4277 [DOI] [PubMed] [Google Scholar]
- 7. Park Y. J., Kim J., You Y. A., and Pang M. G. (2013) Proteomic revolution to improve tools for evaluating male fertility in animals. J. Proteome Res. 12, 4738–4747 [DOI] [PubMed] [Google Scholar]
- 8. Dorus S., Busby S. A., Gerike U., Shabanowitz J., Hunt D. F., and Karr T. L. (2006) Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38, 1440–1445 [DOI] [PubMed] [Google Scholar]
- 9. Savaryn J. P., Catherman A. D., Thomas P. M., Abecassis M. M., and Kelleher N. L. (2013) The emergence of top-down proteomics in clinical research. Genome Med. 5, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Picotti P., and Aebersold R. (2012) Selected reaction monitoring–based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 11. Claydon M. A., Davey S. N., Edwards-Jones V., and Gordon D. B. (1996) The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14, 1584–1586 [DOI] [PubMed] [Google Scholar]
- 12. Holland R. D., Wilkes J. G., Rafii F., Sutherland J. B., Persons C. C., Voorhees K. J., and Lay J. O. (1996) Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 13. Krishnamurthy T., and Ross P. L. (1996) Rapid identification of bacteria by direct matrix-assisted laser desorption/ionization mass spectrometric analysis of whole cells. Rapid Commun. Mass Spectrom. 10, 1992–1996 [DOI] [PubMed] [Google Scholar]
- 14. Welker M. (2011) Proteomics for routine identification of microorganisms. Proteomics 11, 3143–3153 [DOI] [PubMed] [Google Scholar]
- 15. Qian J., Cutler J. E., Cole R. B., and Cai Y. (2008) MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal Bioanal Chem. 392, 439–449 [DOI] [PubMed] [Google Scholar]
- 16. Sendid B., Ducoroy P., François N., Lucchi G., Spinali S., Vagner O., Damiens S., Bonnin A., Poulain D., and Dalle F. (2013) Evaluation of MALDI-TOF mass spectrometry for the identification of medically-important yeasts in the clinical laboratories of Dijon and Lille hospitals. Med. Mycol. 51, 25–32 [DOI] [PubMed] [Google Scholar]
- 17. Ouedraogo R., Flaudrops C., Ben Amara A., Capo C., Raoult D., and Mege J.-L. (2010) Global analysis of circulating immune cells by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS ONE 5, e13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ouedraogo R., Daumas A., Ghigo E., Capo C., Mege J.-L., and Textoris J. (2012) Whole-cell MALDI-TOF MS: a new tool to assess the multifaceted activation of macrophages. J. Proteomics 75, 5523–5532 [DOI] [PubMed] [Google Scholar]
- 19. Hanrieder J., Wicher G., Bergquist J., Andersson M., and Fex-Svenningsen A. (2011) MALDI mass spectrometry based molecular phenotyping of CNS glial cells for prediction in mammalian brain tissue. Anal. Bioanal. Chem. 401, 135–147 [DOI] [PubMed] [Google Scholar]
- 20. Parks B. a, Jiang L., Thomas P. M., Wenger C. D., Roth M. J., Boyne M. T., Burke P. V., Kwast K. E., and Kelleher N. L. (2007) Top-down proteomics on a chromatographic time scale using linear ion trap fourier transform hybrid mass spectrometers. Anal. Chem. 79, 7984–7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahlf D. R., Thomas P. M., and Kelleher N. L. (2013) Developing top down proteomics to maximize proteome and sequence coverage from cells and tissues. Curr. Opin. Chem. Biol. 17, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Labas V., Spina L., Belleannee C., Teixeira-Gomes A.-P., Gargaros A., Dacheux F., and Dacheux J.-L. (2015) Analysis of epididymal sperm maturation by MALDI profiling and top-down mass spectrometry. J. Proteomics 113, 226–243 [DOI] [PubMed] [Google Scholar]
- 23. Labas V., Grasseau I., Cahier K., Gargaros A., Harichaux G., Teixeira-Gomes A.-P., Alves S., Bourin M., Gérard N., and Blesbois E. (2015) Qualitative and quantitative peptidomic and proteomic approaches to phenotyping chicken semen. J. Proteomics 112, 313–335 [DOI] [PubMed] [Google Scholar]
- 24. Blesbois E. (2012) Biological Features of the Avian Male Gamete and their Application to Biotechnology of Conservation. J. Poult. Sci. 49, 141–149 [Google Scholar]
- 25. Burrows W. H., and Quinn J. P. (1937) The Collection of Spermatozoa from the Domestic Fowl and Turkey. Poult. Sci. 16, 19–24 [Google Scholar]
- 26. Brillard J. P., and McDaniel G. R. (1985) The reliability and efficiency of various methods for estimating spermatozoa concentration. Poult. Sci. 64, 155–158 [DOI] [PubMed] [Google Scholar]
- 27. Sexton T. J., Jacobs L. A., and McDaniel G. R. (1980) New poultry semen extender. 4. Effect of antibacterials in control of bacterial contamination in chicken semen. Poult. Sci. 59, 274–281 [DOI] [PubMed] [Google Scholar]
- 28. Blesbois E., and Brillard J. P. (2007) Specific features of in vivo and in vitro sperm storage in birds. Animal 1, 1472–1481 [DOI] [PubMed] [Google Scholar]
- 29. Vizcaíno J., Deutsch E., and Wang R. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. …. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. a., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2013) The Proteomics Identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41, 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blesbois E., Grasseau I., Seigneurin F., Mignon-Grasteau S., Saint Jalme M., and Mialon-Richard M. M. (2008) Predictors of success of semen cryopreservation in chickens. Theriogenology 69, 252–261 [DOI] [PubMed] [Google Scholar]
- 32. Chalah T., and Brillard J. P. (1998) Comparison of assessment of fowl sperm viability by eosin-nigrosin and dual fluorescence (SYBR-14/PI). Theriogenology 50, 487–493 [DOI] [PubMed] [Google Scholar]
- 33. Eusebi P. (2013) Diagnostic accuracy measures. Cerebrovasc. Dis. 36, 267–272 [DOI] [PubMed] [Google Scholar]
- 34. Shen Y., Tolić N., Liu T., Zhao R., Petritis B. O., Gritsenko M. a, Camp D. G., Moore R. J., Purvine S. O., Esteva F. J., and Smith R. D. (2010) Blood peptidome-degradome profile of breast cancer. PLoS ONE 5, e13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Z., Wu C., Xie F., Slysz G. W., Tolic N., Monroe M. E., Petyuk V. a, Payne S. H., Fujimoto G. M., Moore R. J., Fillmore T. L., Schepmoes A. a, Levine D. a, Townsend R. R., Davies S. R., Li S., Ellis M., Boja E., Rivers R., Rodriguez H., Rodland K. D., Liu T., and Smith R. D. (2014) Comprehensive Quantitative Analysis of Ovarian and Breast Cancer Tumor Peptidomes. J. Proteome Res. 10.1021/pr500840w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Donnell L., and O'Bryan M. K. (2014) Microtubules and spermatogenesis. Semin. Cell Dev. Biol. 30, 45–54 [DOI] [PubMed] [Google Scholar]
- 37. Takiguchi H., Murayama E., Kaneko T., Kurio H., Toshimori K., and Iida H. (2011) Characterization and subcellular localization of Tektin 3 in rat spermatozoa. Mol. Reprod. Dev. 78, 611–620 [DOI] [PubMed] [Google Scholar]
- 38. Sapiro R., Kostetskii I., Olds-Clarke P., Gerton G., Radice G., and Strauss J. III (2002) Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 22, 6298–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wirschell M., Olbrich H., Werner C., Tritschler D., Bower R., Sale W. S., Loges N. T., Pennekamp P., Lindberg S., Stenram U., Carlén B., Horak E., Köhler G., Nürnberg P., Nürnberg G., Porter M. E., and Omran H. (2013) The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat. Genet. 45, 262–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pazour G. J., Agrin N., Walker B. L., and Witman G. B. (2006) Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J. Med. Genet. 43, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fong K.-W., Choi Y.-K., Rattner J. B., and Qi R. Z. (2008) CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fong K.-W., Hau S.-Y., Kho Y.-S., Jia Y., He L., and Qi R. Z. (2009) Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol. Biol. Cell 20, 3660–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwon W.-S., Park Y.-J., Mohamed E.-S. a, and Pang M.-G. (2013) Voltage-dependent anion channels are a key factor of male fertility. Fertil. Steril. 99, 354–61 [DOI] [PubMed] [Google Scholar]
- 44. Li Y., Park J.-S., Deng J.-H., and Bai Y. (2006) Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J. Bioenerg. Biomembr. 38, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Binder N. K., Sheedy J. R., Hannan N. J., and Gardner D. K. (2015) Male obesity is associated with changed spermatozoa Cox4i1 mRNA level and altered seminal vesicle fluid composition in a mouse model. Mol. Hum. Reprod. 21, 424–434 [DOI] [PubMed] [Google Scholar]
- 46. Bansal D., and Campbell K. P. (2004) Dysferlin and the plasma membrane repair in muscular dystrophy. Trends Cell Biol. 14, 206–213 [DOI] [PubMed] [Google Scholar]
- 47. Washington N. L., and Ward S. (2005) FER-1 regulates membrane fusion during spermiogenesis. Int. Worm Meet. [DOI] [PubMed] [Google Scholar]
- 48. Smith M. K., and Wakimoto B. T. (2007) Complex regulation and multiple developmental functions of misfire, the Drosophila melanogaster ferlin gene. BMC Dev. Biol. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shabek N., and Ciechanover A. (2010) Degradation of ubiquitin: The fate of the cellular reaper. Cell Cycle 9, 523–530 [DOI] [PubMed] [Google Scholar]
- 50. Sutovsky P. (2003) Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: Killing three birds with one stone. Microsc. Res. Tech. 61, 88–102 [DOI] [PubMed] [Google Scholar]
- 51. Jha K. N., Coleman A. R., Wong L., Salicioni A. M., Howcroft E., and Johnson G. R. (2013) Heat shock protein 90 functions to stabilize and activate the testis-specific serine/threonine kinases, a family of kinases essential for male fertility. J. Biol. Chem. 288, 16308–16320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Purandhar K., Jena P. K., Prajapati B., Rajput P., and Seshadri S.. Understanding the Role of Heat Shock Protein Isoforms in Male Fertility, Aging and Apoptosis. World J. Mens. Health 32, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu D. Y., Clarke G. N., and Baker H. W. (1991) Inhibition of human sperm-zona pellucida and sperm-oolemma binding by antisperm antibodies. Fertil. Steril. 55, 440–442 [PubMed] [Google Scholar]
- 54. Bandoh R., Yamano S., Kamada M., Daitoh T., and Aono T. (1992) Effect of sperm-immobilizing antibodies on the acrosome reaction of human spermatozoa. Fertil. Steril. 57, 387–392 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.