Abstract
Comparative analysis of the enediyne biosynthetic gene clusters revealed sets of conserved genes serving as outstanding candidates for the enediyne core. Here we report the crystal structures of SgcJ and its homologue NCS-Orf16, together with gene inactivation and site-directed mutagenesis studies, to gain insight into enediyne core biosynthesis. Gene inactivation in vivo establishes that SgcJ is required for C-1027 production in Streptomyces globisporus. SgcJ and NCS-Orf16 share a common structure with the nuclear transport factor 2-like superfamily of proteins, featuring a putative substrate binding or catalytic active site. Site-directed mutagenesis of the conserved residues lining this site allowed us to propose that SgcJ and its homologues may play a catalytic role in transforming the linear polyene intermediate, along with other enediyne polyketide synthase associated enzymes, into an enzyme-sequestered enediyne core intermediate. These findings will help formulate hypotheses and design experiments to ascertain the function of SgcJ and its homologues in 9-membered enediyne core biosynthesis.
Keywords: biosynthesis, C-1027, enediyne, neocarzinostatin, nuclear transport factor 2-like superfamily
Introduction
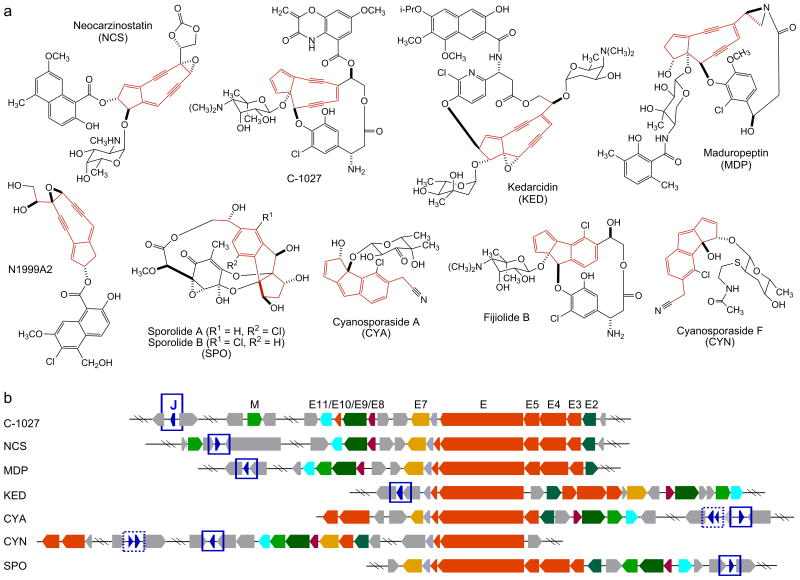
The enediynes represent one of the most fascinating families of natural products for their unprecedented molecular architecture and extraordinary biological activities, and they have had profound impact on modern chemistry, biology, and medicine.1-4 Since the structure of the neocarzinostatin (NCS) chromophore was first elucidated in 1985,5 the enediyne family of natural products has grown steadily with a total of 15 enediynes structurally characterized to date, of which four were isolated in the cycloaromatized form.4 The enediynes are classified into two subcategories according to the size of the enediyne core structures.1,2 Members of the 9-membered enediyne core subcategory included NCS, C-1027, kedarcidin (KED), maduropeptin (MDP), N1999A2, the sporolides (SPO), the cyanosporasides (CYA, CYN), and the fijiolides (Fig. 1a). Members of the 10-membered enediyne core subcategory included the calicheamicins (CAL), esperamicins (ESP), dynemicin (DYN), namenamicin, shishijimicin, and uncialamycin (Fig. S1). The enediynes have provided an outstanding opportunity to decipher the genetic and biochemical basis for the biosynthesis of complex natural products,1-4 to explore ways to make novel analogues by manipulating genes governing their biosynthesis,6-10 and to discover new enediyne natural products by mining microbial genomes for the trademark enediyne biosynthetic machineries.3,4,11,12
Figure 1.
Structures of the 9-membered enediyne natural products and their biosynthetic gene clusters. (a) Structures of the five 9-membered enediyne natural products (NCS, C-1027, KED, MDP, N1999A2) and the four additional natural products (fijiolide, SPO, CYA, CYN) proposed to be derived from 9-membered enediyne precursors after cycloaromatization. The 9-membered enediyne cores or their aromatized products are highlighted in red. (b) Alignment of the seven known 9-membered enediyne biosynthetic gene clusters highlighting the enediyne PKS gene cassette (i.e., E3, E4, E5, E, E10) (shown in red) and the seven additional conserved genes (i.e., E2, E7, E8, E9, E11, M, J) (color-coded). C-1027 biosynthetic gene cluster nomenclature is used. SgcJ and its homologues reported in this study are shown in blue and highlighted with blue boxes. Additional SgcJ homologues were also noted in the CYA and CYN clusters (boxed with dotted blue lines), but they were not included in the current study due to their varying length and significantly lower amino acid sequence homology.
The first set of biosynthetic gene clusters for the 9-membered enediyne C-102713 and the 10-membered enediyne CAL14 was cloned in 2002. Since then, a total of seven biosynthetic gene clusters for 9-membered enediynes (i.e., C-1027,13 NCS,15 MDP,16 KED,17 SPO,18 CYA,19 and CYN19) and three biosynthetic gene clusters for 10-membered enediynes [i.e., CAL,14 ESP (partial),11 and DYN20] have been reported. Comparative analysis of these gene clusters revealed a set of five genes common to both 9- and 10-membered enediynes [i.e., the enediyne polyketide synthase (PKS) cassette consisting of E3/E4/E5/E/E10 (Fig 1b)], characterization of which has unambiguously established (i) the polyketide origin for both 9- and 10-membered enediynes and (ii) a convergent model for enediyne biosynthesis.3,4,21,22 While significant progress has been made toward elucidating the biosynthesis of the peripheral moieties present in enediynes, little is known about the enediyne core biosynthesis. In vivo and in vitro studies have established that the iterative type I PKS enzyme E initiates both 9- and 10-membered enediyne core biosynthesis via an acyl carrier protein (ACP)-tethered linear polyene intermediate, which, in the absence of other enediyne PKS associated enzymes, could be released by the thioesterase (TE) E10 to afford a heptaene.21-25 However, the enzymes and chemistry responsible for converting heptaene, or the nascent ACP-tethered linear polyene intermediate, into the 9- and 10-membered enediyne cores remain elusive. Many of the candidate genes, predicted to be associated with enediyne core biosynthesis, are often annotated to encode proteins of unknown function.3,4 Inactivation of these candidate genes in vivo afforded mutant strains that often failed to accumulate any biosynthetic intermediate, revealing few clues for their function in enediyne core biosynthesis. Lack of functional prediction, together with the unavailability of suitable substrates, essentially forfeits any practical attempt to directly characterize these proteins biochemically in vitro.
Here we report the crystal structures of SgcJ and its homologue NCS-Orf16, together with gene inactivation and site-directed mutagenesis studies, to gain insight into enediyne core biosynthesis. We first closely examined the seven gene clusters that encode 9-membered enediyne biosynthesis and uncovered seven genes (E2, E7, E8, E9, E11, M, and J), in addition to the five genes, i.e., E3/E4/E5/E/E10, encoding the enediyne PKS cassette, that are absolutely conserved but their function could not be predicted on the basis of bioinformatics analysis alone. We then subjected these targets to high-throughput structural biology analysis. This effort resulted in several structures, including SgcJ from the C-1027 and its homologue NCS-Orf16 from the NCS biosynthetic machineries. We next confirmed that SgcJ is absolutely required for C-1027 biosynthesis, inactivation of which in the C-1027 overproducer Streptomyces globisporus SB102210 completely abolished C-1027 production in the resultant ΔsgcJ mutant strain SB1027. We finally showed that SgcJ and NCS-Orf16 share a common structure with the nuclear transport factor 2 (NTF2)-like superfamily of proteins, featuring a hydrophobic pocket in the α+β barrel structure that could constitute as a putative substrate binding or catalytic active site. Site-directed mutagenesis of the conserved residues lining this site abolished C-1027 production, suggesting that SgcJ and its homologues may play a catalytic role in 9-membered enediyne core biosynthesis.
Materials and Methods
Strains, plasmids, and culture conditions
Bacterial strains, plasmids, and primers used in this study are summarized in Tables S1, S2, and S3, respectively. Escherichia coli strains and Micrococcus luteus ATCC 9431 were cultured in lysogeny broth (LB) or grown on LB agar plates. S. globisporus wild-type and recombinant strains were cultivated at 28 °C on ISP Medium 4 (Becton Dickenson, Franklin Lakes, NJ) for sporulation. Antibiotics for selection were used at the following concentrations: 25 μg/mL for apramycin and thiostrepton, and 50 μg/mL for chloramphenicol and kanamycin.
Construction of the ΔsgcJ mutant strain S. globisporus SB1027
The ΔsgcJ mutant strain SB1027 was constructed in the C-1027 overproducer S. globisporus SB102210 by gene replacement via homologous recombination. Briefly, the 1.5-kb kanamycin resistance cassette was amplified by PCR from pJTU4659 with primers sgcJtgtF and sgcJtgtR (Table S3) and used to replace sgcJ in cosmid pBS100526 via λ-RED-mediated PCR targeting mutagenesis27 to generate pBS1143. The ΔsgcJ gene was then excised from pBS1143 as a ∼21 kb XbaI-SpeI fragment and inserted into the XbaI site of pSET151 to afford pBS1144. pBS1144 was finally introduced into S. globisporus SB1022 by E. coli-S. globisporus conjugation.28 Exconjugates resulting from the desired double-crossover homologous recombination were selected on the basis of kanamycin-resistant and thiostrepton-sensitive phenotype, and named SB1027, the genotype of which was confirmed by PCR and Southern analysis (Fig. S3).
Construction of ΔsgcJ complementation strains S. globisporus SB1028 and SB1029
A 0.8-kb fragment bearing oriT was amplified by PCR from plasmid pSET152 with primers oriT152F and oriT152R (Table S3), digested with KpnI, and cloned into the same site of pUWL201pw to generate pBS1145. A 420-bp fragment of sgcJ and a 432-bp fragment of ncs-orf16 were amplified by PCR from cosmids pBS100526 and pBS500715, with primers sgcJ201NdeIF and sgcJ201EcoRIR, and ncs16NdeIF and ncs16HindIIIR, respectively (Table S3). The resultant products were digested with NdeI and EcoRI (for sgcJ), and NdeI and HindIII (ncs-orf16), and cloned into the same sites of pBS1145 to afford pBS1146 (for sgcJ) and pBS1147 (for ncs-orf16), respectively. Both pBS1146 and pBS1147, in which the expressions of sgcJ and ncs-orf16 were under the control of the constitutive ErmE* promoter,28 were finally introduced into the ΔsgcJ mutant strain S. globisporus SB1027 by E. coli-S. globisporus conjugation.28 Exconjugates were selected on the basis of thiostrepton-resistant phenotype as the desired complementation strains, and named SB1028 (i.e., sgcJ expressing) and SB1029 (i.e., ncs-orf16, expressing), respectively.
Site-directed mutagenesis of SgcJ
Plasmids of the sgcJ mutants, pBS1148 (W29A), pBS1149 (F37A), pBS1150 (Y72A), pBS1151 (D111A), pBS1152 (W118A), and pBS1153 (Y132A), were constructed by the QuikChange site-directed mutagenesis method, following the manufacturer's protocol (Agilent Technologies, Santa Clara, CA) and using pBS1146 as a template. The primers used are listed in Table S3. The mutations were verified by DNA sequencing. Each of the mutant constructs was then introduced into the ΔsgcJ mutant strain SB1027 by conjugation, yielding the complementation strains SB1030 (i.e., SB1027/pBS1148), SB1031 (i.e., SB1027/pBS1149), SB1032 (i.e., SB1027/pBS1150), SB1033 (i.e., SB1027/pBS1151), SB1034 (i.e., SB1027/pBS1152), and SB1035 (i.e., SB1027/pBS1153), respectively.
Production, isolation and analysis of C-1027
S. globisporus recombinant strains were cultured following a two-step fermentation procedure reported previously, and both stages utilized the same medium (1% glycerol, 2% dextrin, 1% fish meal, 0.5% peptone, 0.2% (NH4)2SO4, 0.1% MgSO4, 0.2% CaCO3, pH 7.0).9,10,13,26 Briefly, fresh spores of the recombinant strains were inoculated into 250-mL baffled flasks containing 50 mL of medium and incubated at 28 °C and 250 rpm for 48 hr. The resultant seed cultures (2.5 mL) were then inoculated into 250-mL baffled flasks containing 50 mL of the same medium, and fermentation continued at 28 °C and 250 rpm for 7 days. The C-1027 overproducer SB1022 and the ΔsgcJ mutant strain SB1027 were cultured in medium without any antibiotics. All other recombinant strains used in this study were cultured in medium supplemented with 5 μg/L thiostrepton to retain the introduced plasmids.
Isolation and HPLC analysis of the C-1027 chromophore were carried out by following published procedures.9,10,13,26 Briefly, fermentation broth (50 mL) was adjusted to pH 4.0 with 0.1 N HCl and centrifuged to remove any precipitate. To the supernatant, (NH4)2SO4 was then added to 50% saturation, and the precipitated C-1027 chromoprotein was collected by centrifugation and dissolved in 2 mL of 0.1 M potassium phosphate, pH 8.0. The latter was extracted with 2 mL of EtOAc twice, and the combined EtOAc extract was concentrated in vacuo and re-dissolved in CH3OH. HPLC was carried out on a Beckman ultrasphere-ODS dp analytical column (5 μm, 150 × 4.6 mm) (Beckman Coulter, Indianapolis, IN), eluted isocratically with 20 mM potassium phosphate (pH 6.8)/CH3CN (50:50 v/v) at a flow rate of 1.0 mL/min and UV detection at 350 nm on a Varian HPLC system with a Prostar 330 PDA detector (Agilent Technologies). LC-MS analysis of C-1027 was performed on an Agilent 6230 TOF LC-MS instrument (Agilent Technologies).
Determination of C-1027 production by bioassay and biochemical induction assays
Determination of C-1027 production by bioassay against M. luteus ATCC 9431 was carried out as described previously. 9,10,13,26 Alternatively, C-1027 production was also followed by the biochemical induction assay (BIA) according to literature procedures,11 which uses the E. coli BR513 strain as an indicator and specifically detects agents with DNA damage activities. Briefly, 10 μL of fermentation supernatant or an agar plug were applied onto agar plates seeded with E. coli BR513 and incubated for 3-4 hr at 37 °C. The plates were then overlaid with soft agar containing 0.7 mg/mL of X-gal and incubated at 37 °C for additional 30-60 min to develop the characteristic blue color, indicative of DNA damage, and thus C-1027 production.
Gene expression and protein purification
PCR amplification of sgcJ from S. globisporus genomic DNA13 and ncs-orf16 from S. carzinostaticus genomic DNA15 by KOD Hot Start DNA polymerase (EMD Millipore, Billerica, MA) followed the manufacturer's protocols using primers sgcJ-F and sgcJ-R and primers orf16-F and orf16-R primers, respectively (Table S3). The amplification buffer was supplemented with betaine to a final concentration of 2.5 M. The PCR products were purified and cloned into pMCSG57, yielding pBS1154 (expressing sgcJ) and pBS1155 (expressing ncs-orf16), by the ligation-independent procedures.29 The expression plasmids were then transformed into E. coli BL21(DE3)-Gold strain (Stratagene, San Diego, CA) for protein production. Production and purification of SeMet-labeled SgcJ and NCS-Orf16 were performed according to standard protocol.30 Briefly, the cell were cultured at 37 °C in 1 L of enriched M9 medium30 until OD600 = 1.0. After air-cooling the culture down at 4 °C for 60 min, inhibitory amino acids (25 mg each/L l-valine, l-isoleucine, l-leucine, l-lysine, l-threonine, and l-phenylalanine), selenomethionine (SeMet), and isopropyl-β-D-thiogalactoside (IPTG) were added. The cells were incubated overnight at 18 °C, harvested and re-suspended in lysis buffer [500 mM NaCl, 5% (v/v) glycerol, 50 mM HEPES pH 8.0, 20 mM imidazole, and 10 mM β-mercaptoethanol]. The SeMet labeled proteins were purified using Ni-NTA affinity chromatography by the AKTAxpress system (GE Healthcare Life Sciences, Marlborough, MA) and digested with recombinant His6-tagged TEV protease to remove the His6-tag. The final pure proteins were concentrated using Amicon Ultra-15 concentrators (Millipore, Bedford, MA) in 20 mM HEPES pH 8.0 buffer, 250 mM NaCl, and 2 mM dithiothreitol (DTT). Protein concentrations were determined based on the absorbance at 280 nm using a molar absorption coefficient (ε280 = 19,480 and 20,970 M−1 cm−1 for SgcJ and NCS-Orf16, respectively).31 The concentrations of SgcJ and NCS-Orf16 used for crystallization were both ∼50 mg/mL. Size-exclusion chromatography was performed using a Superdex 200 16/600 column (GE Healthcare Life Sciences) with an Äkta FPLC chromatographic system (GE Healthcare Life Sciences) at 4 °C. The column was calibrated with a size-exclusion calibration kit (GE Healthcare Life Sciences) and developed with the elution buffer (200 mM NaCl, 100 mM Tris, pH 8.0) at flow rate of 0.5 mL/min with UV detection at 280 nm.
Protein crystallization
Both SgcJ and NCS-Orf16 were screened for crystallization conditions using a Mosquito liquid dispenser (TTP Labtech, Melbourn, UK) and the sitting-drop vapor-diffusion technique in 96-well CrystalQuick plates (Greiner Bio-one, Monroe, NC). For each condition, 0.4 μL of protein (52.8 mg/mL) and 0.4 μL of crystallization formulation were mixed. The mixture was equilibrated against 140 μL of the reservoir in the well. Commercially available crystallization screens were used, including MCSG-1–4 (Microlytic Inc., Burlington, MA) at 24 °C, 16 °C, and 4 °C. For SgcJ, crystals were obtained under several conditions, with the most promising condition being from 0.1 M Na2HPO4 (adjust to pH 4.2 with citric acid) and 40% (v/v) PEG 300 at 16 °C. The crystals grew within one week and reached sizes of approximately 0.100 mm × 0.020 mm × 0.010 mm. For NCS-Orf16, suitable crystals for X-ray diffraction were grown from the condition containing 0.2 M sodium formate and 20% (w/v) PEG 3350 at 16 °C.
Data collection, structure determination, and refinement
Diffraction data were collected at 100 K at the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory.32 A single dataset was taken near the Se K-edge peak anomalous position (0.9792 Å) from a single protein crystal of SgcJ to a resolution of 1.70 Å. The crystal was exposed for 3 s per 1.0° rotation with a distance of 240 mm from crystal to detector. The data were recorded on an ADSC Quantum 315r CCD detector. For NCS-Orf16, data collection was the same except the crystal to detector distance was 327 mm, and three datasets were collected and merged. Data collection strategy, integration, and scaling were performed with the HKL3000 program package.33 A summary of the crystallographic data can be found in Table 1.
Table I. Data collection, phasing, and refinement statistics for the SgcJ and NCS-Orf16 structures.
| SgcJ | NCS-Orf16 | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 72.72, 86.90, 55.27 | 98.32, 52.83, 131.80 |
| α, β, γ (°) | 90.00, 121.6, 90.00 | 90.00, 90.14, 90.00 |
| Wavelength (Å) | 0.9792 | 0.9794 |
| Resolutiona (Å) | 33.90-1.70 (1.73-1.70) | 38.00-2.72 (2.77-2.72) |
| Rsymb or Rmergec (%) | 9.0 (49.8) | 8.4 (95.9) |
| I/σI | 16.1 (3.2) | 21.8 (2.7) |
| Completeness (%) | 99.3 (98.2) | 99.8 (100.0) |
| Redundancy | 3.9 (2.8) | 7.1 (7.2) |
| Refinement | ||
| Resolution (Å) | 31.9-1.70 | 38.0-2.72 |
| No. reflections | 30859 (2046) | 32801 (1011) |
| Rwork/ Rfree | 0.168/0.195 | 0.217/0.256 |
| Ramachandran plot (%)d | ||
| favored | 99.60 | 97.24 |
| outliers | 0.0 | 0.0 |
| Average B-factors (Å2) | 24.2 | 71.8 |
| Wilson B-factor (Å2) | 14.7 | 43.53 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.010 | 0.002 |
| Bond angles (°) | 1.325 | 0.648 |
| Clashscoree | 2.33 | 1.44 |
Numbers in parentheses are values for the highest-resolution bin.
Rmerge = ΣhklΣi|Ii (hkl) – Ī(hkl)|/ΣhklΣiIi(hkl), where Ii(hkl) is the ith observation of reflection hkl and Ī(hkl) is the weighted average intensity for all observations i of reflection hkl.
Rmeas = Σhkl[N/(N − 1)1/2]Σi|Ii (hkl) – Ī(hkl)|/ΣhklΣiIi(hkl).
As defined by MolProbity.
As defined by Molprobity.
The crystal structures of SgcJ and NCS-Orf16 were determined by SAD phasing, utilizing the anomalous signal from Se atoms with shelxc/d/e,34 mlphare,35 and dm36 in HKL300033 for SgcJ and SOLVE/RESOLVE37 for NCS-Orf16, and refined to 1.7 Å and 2.72 Å, respectively. For SgcJ, the initial model contains two protein chains consisting of at least 90% of the residues in each chain. For NCS-Orf16, the initial model contains ten protein chains consisting of 67% of whole model with 20% assigned side-chain. Extensive manual model building with COOT38 and the subsequent refinement using phenix.refine39 were performed until R-factors converged to final values of R(Rfree) = 0.168(0.195) and 0.217(0.256) for the structures of SgcJ and NCS-Orf16, respectively. The geometrical properties of the models were assessed using PROCHECK40 and Molprobity.41 The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 4I4K for SgcJ and 4OVM for NCS-Orf16, respectively.
Results and Discussion
SgcJ and homologues are conserved among the 9-membered enediyne biosynthetic gene clusters but their function could not be predicted
Inspired by the enediyne PKS cassette, consisting of E3/E4/E5/E/E10, that is conserved among the seven 9-membered and three 10-membered enediyne biosynthetic gene clusters characterized to date, we recently completed a virtual survey of all bacterial genomes available in public databases using the enediyne PKS cassette as a probe.3,4 This effort resulted in the identification of an additional 77 putative enediyne biosynthetic gene clusters, implying that enediynes are more common than currently appreciated on the basis of structurally characterized enediyne natural products.1-4,11,12 We subsequently constructed an enediyne genome neighborhood network, including both the 10 known and 77 putative enediyne gene clusters, to facilitate cluster annotation and predict 9- and 10-membered enediyne core biosynthesis. The enediyne PKS cassette is present in all 87 gene clusters, suggesting that they may be responsible for biosynthesis of a common intermediate for both 9- and 10-membered enediyne cores. Subsets of genes that are unique to either 9- or 10-membered enediyne gene clusters are also identified, as exemplified by the E2, E7, E8, E9, E11, M, and J genes from the seven known 9-membered enediyne biosynthetic gene clusters (Fig. 1b), and they may play roles in diversifying the common intermediate into the 9- or 10-membered enediyne cores, respectively.3,4
Among this set of genes is SgcJ (Fig. 1b), and its homologues are present in the 34 putative 9-membered enediyne biosynthetic gene clusters (Fig. S2). SgcJ and homologues are comprised of 140-160 amino acids, with amino acid sequence identities ranging from 30% to 66%. According to the BLASTP search result, SgcJ homologues feature a domain of unknown function (DUF4440) and belong to the NTF2-like superfamily, a large group of related proteins that share a common protein fold. The NTF2-like superfamily proteins are widely found in both prokaryotic and eukaryotic organisms and possess versatile functions.42 Proteins in the NTF2-like superfamily are generally defined into two categories, enzymatically active and non-enzymatically active proteins. The former group includes enzymes with varying activities such as the ketosteroid isomerase,43 scytalone dehydrogenase,44 and polyketide cyclase.45,46 The latter group includes proteins that could play roles as diverse as facilitating protein transport into the nucleus47 or mediating multimerization of calcium/calmodulin dependent protein kinase II (CaMKII),48 or may function as a receptor.49 The enediyne variants of SgcJ show less than 18% amino acid sequence identity to functionally characterized NTF2-like proteins. Due to the diverse functions of the NTF2-like superfamily, bioinformatics analysis alone fell short of predicting the function of SgcJ and its homologues in 9-membered enediyne core biosynthesis.
Gene inactivation reveals that sgcJ is necessary for enediyne biosynthesis
To establish a functional linkage of sgcJ and its homologues with enediyne biosynthesis, we inactivated sgcJ in the C-1027 overproducer S. globisporus SB102210 by replacing it with the kanamycin resistance cassette through λ-RED-mediated PCR targeting mutagenesis27 (Fig. S3a). The genotype of the resulting ΔsgcJ mutant strain SB1027 was confirmed by PCR and Southern analysis (Fig. S3b). SB1027 was fermented under the established conditions for C-1027 production with S. globisporus SB1022 as a positive control.9,10,13,26 While C-1027 production by SB1022 was readily confirmed upon both bioassay against M. luteus and BIA, SB1027 completely abolished the production of C-1027, which was unambiguously verified by HPLC and ESI-MS analysis (Fig. 2, panels I and II). The requirement for sgcJ in C-1027 biosynthesis was further supported by the fact that the ΔsgcJ mutation in SB1027 could be complemented by expressing a functional copy of sgcJ in trans, restoring C-1027 production in the complementation strain SB1028 to the level comparable to that of SB1022 (Fig. 2, panels I and III). Taken together, these data clearly established that SgcJ plays a necessary role in C-1027 biosynthesis and, by analogy, the essential role SgcJ homologues play in 9-membered enediyne core biosynthesis. However, SB1027 failed to accumulate any biosynthetic intermediate to sufficient levels for isolation and structural characterization, revealing no clues for its exact function. We therefore opted to solve the structures of SgcJ and its homologues in an attempt to elucidate their function in 9-membered enediyne core biosynthesis.
Figure 2.
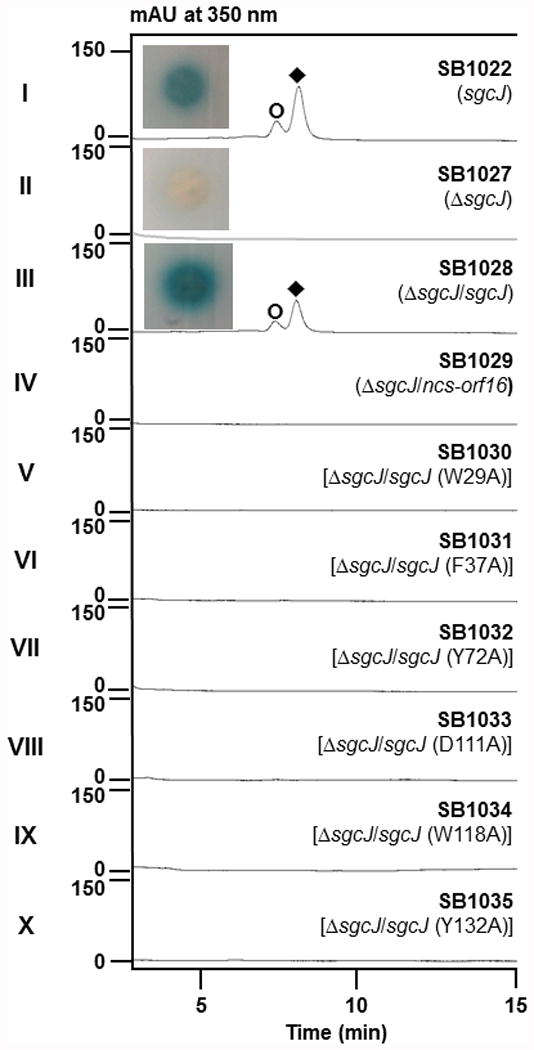
C-1027 production by S. globisporus SB1022 and derived recombinant strains. HPLC analysis for C-1027 production from fermentation of (I) SB1022, (II) SB1027, (III) SB1028, (IV) SB1029, (V) SB1030, (VI) SB1031, (VII) SB1032, (VIII) SB1033, (IX) SB1034, and (x) SB1035. Symbols denote C-1027 chromophore (◆) and aromatized C-1027 (○). BIA (inserts) showed that the production of C-1027 in SB1022, loss of C-1027 production in SB1027, and restored C-1027 production in SB1028.
The overall structure of SgcJ and NCS-Orf16 reveals structural similarity to NTF2-like superfamily proteins
The crystals of SgcJ were obtained in the monoclinic space group C2 with unit cell parameters a = 72.7, b = 86.9, and c = 55.3 Å and α = γ = 90.0°, and β = 121.6°. The asymmetric unit contained two peptide chains, corresponding to a solvent content of 50.9%. The asymmetric unit also contained molecules of citric acid, glycerol, phosphate, pentaethlene glycol, and tetraethylene glycol, which were present in the crystallization condition. The final model of SgcJ was refined to a resolution of 1.7 Å with an R factor of 16.9% and an Rfree factor of 19.5%. Ramachandran analysis reveals that 99.6% of the residues were in the favored region with none in disallowed regions. Electron density map was well defined for residues Ser3-Asp140 and Ala10-Asp140 for the two polypeptide chains in the asymmetric unit. Data collection and refinement statistics are summarized in Table 1.
The NCS-Orf16 crystals were obtained in the monoclinic space group P21 with unit cell parameters a = 98.3, b = 52.8, and c = 131.8 Å and α = γ = 90.0°, and β = 90.1°. The asymmetric unit contained ten peptide chains, corresponding to a solvent content of 45.0%. The final model of NCS-Orf16 was refined to a resolution of 2.72 Å with an R factor of 21.6% and an Rfree factor of 25.6%. Ramachandran analysis reveals that 97.2% of the residues were in the favored region with none in disallowed regions. Electron density map was well-defined for residues Thr19-Arg142 for each peptide chain in an asymmetric unit. Data collection and refinement statistics are summarized in Table 1.
SgcJ and its homologues show high amino acid sequence homology (Fig. 3a), with SgcJ and NCS-Orf16 sharing 45% amino acid sequence identity. The crystal structures of SgcJ and NCS-Orf16 feature a common three-dimensional structural fold (Fig. 3b). The structure of SgcJ superimposed well with NCS-Orf16 with a root-mean-square deviation (rmsd) of 0.83 Å for the Cα atoms. The overall structures of SgcJ and NCS-Orf16 form a cone-like α+β barrel structure, which are both comprised of a long N-terminal α-helix (α1-α2) passing though the curved six-stranded antiparallel β-sheet (β1-β6), with two additional shorter α-helices (α3 and α4) neighbor upon the α1-α2 helix (Fig. 2b). The β-sheet packs against the three α-helices to form a hydrophobic core within the α+β barrel (Fig. 3b). The crystal structures of SgcJ and NCS-Orf16 are packed as homodimers in an asymmetric unit, which are generated via non-crystallographic twofold axes (Fig. 3c). The dimer interface is formed via a hydrogen-bonding network and salt bridges between the flat-face of the β-sheet from each monomer. Both SgcJ and NCS-Orf16 were indeed found to be homodimers in solution upon size exclusion chromatography (Fig. S4).
Figure 3.
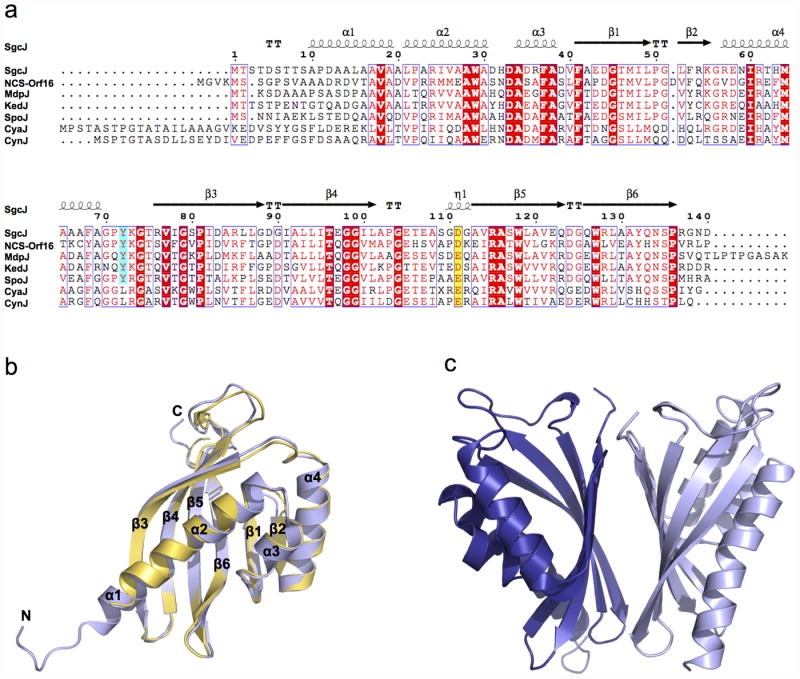
Sequence alignment of SgcJ and its homologues and overall structure of SgcJ and NCS-Orf16. (a) Sequence alignment of SgcJ and its homologues from the seven known 9-membered enediyne biosynthetic gene clusters. Aligned residues are colored on the basis of the level of conservation. Yellow background with red character and cyan background with red character show the putative general base and acid, respectively. Red background with white character shows strict identity, red character similarity and blue frame similarity across groups. The alignment was created with MUSCLE54 and rendered with ESPript 3.0.55 (b) Ribbon diagram and structural alignment of the SgcJ and NCS-Orf16 monomers. SgcJ and NCS-Orf16 are shown in light blue and yellow, respectively. (c) Ribbon diagram of the SgcJ dimer. The two chains are colored in blue and light blue, respectively. The NCBI accession numbers for each of the proteins are: SgcJ (ALU98438), NCS-Orf16 (AAM77985), MdpJ (ABY66022), KedJ (AFV52149), SpoJ (WP028564083), CyaJ (AG0972300), and CynJ (AG097162).
Consistent with the BLASTP search result, a search of the PDB databank using the DALI server50 revealed that SgcJ and NCS-Orf16 belong to the NTF2-like superfamily. This versatile superfamily is a classic example of divergent evolution wherein the proteins have similar overall structures but diverge greatly in their functions.42,51 Several crystal structures for NTF2-like superfamily proteins were reported, of which the functions have been characterized, including the association domain of CaMKII from mouse (PDB entry 1HKX),48 NTF2 from rat (PDB entry 1OUN),52 ketosteroid isomerase (KSI) from Pseudomonas putida (PDB entry 1OPY),43 scytalone dehydatase (mgSD) from Magnaporthe grisea (PDB entry 1STD),44 and polyketide cyclases SnoaL from Streptomyces avidinii (PDB entry 1SJW)45 and Tcm ARO/CYC from Streptomyces glaucescens (PDB entry 2RER).46 Despite low amino acid sequence identities, ranging from 9.8% to 17.5%, SgcJ was found to share similar folds with each of the NTF2-like superfamily proteins listed, with rmsds of 3.0, 2.6, 2.7, 2.7, 3.1, and 3.0 Å for the Cα atoms, respectively (Fig. 4a). SgcJ, NTF2, KSI, and SnoaL form homodimers, while mgSD and CaMKII form a trimer and tetradecamer, respectively. Most importantly, all these NTF2-like superfamily proteins contain a hydrophobic pocket in the α+β barrel structure (Fig. 4a), which forms a cavity that could be adapted to create an enzyme active site or a small molecule/peptide binding site, thereby serving the versatile functions.
Figure 4.
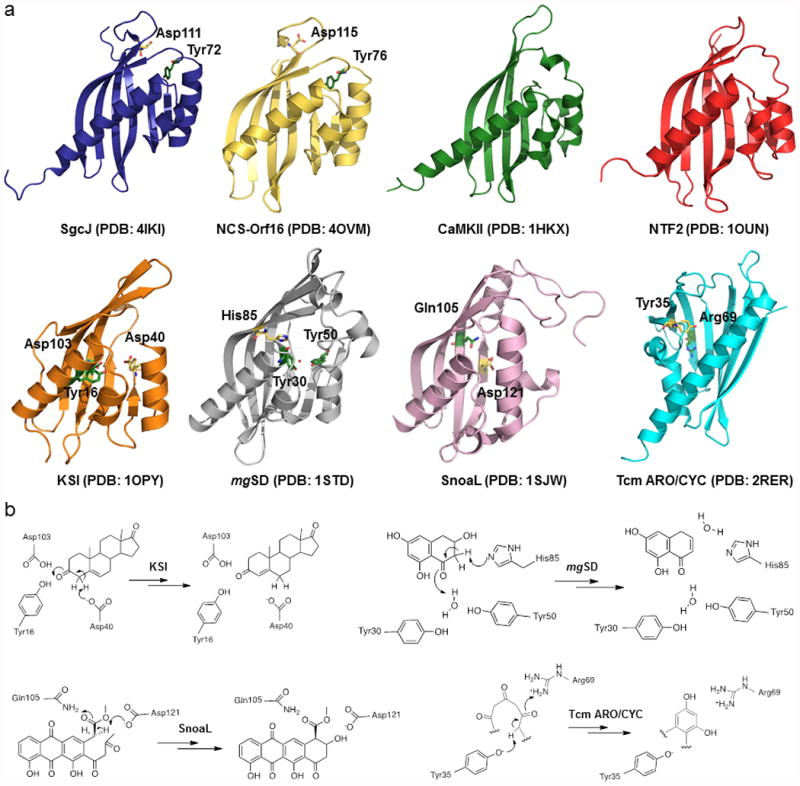
Structure comparison between SgcJ and selected homologous in the NTF2-like superfamily. (a) Each structure shows a curved antiparallel β-sheet wall with a group of α-helices on one side of the wall to form the cone-like shapes. The putative general base and acid are shown as yellow and green sticks, respectively. The water molecule in the structure of mgSD is depicted by a red dot. Given in parentheses are PDB accession codes for each of the structures. (b) The proposed mechanisms for KSI, mgSD, SnoaL, and Tcm ARO/CYC, featuring the conserved general acid-base pairs to catalyze the initial steps of reactions.
Putative substrate binding cavity and catalytic residues of SgcJ and its homologues
KSI (isomerase),43 mgSD (dehydratase),44 SnoaL (cyclase),45 and Tcm ARO/CYC (cyclase)46 are enzymatically active proteins within the NTF2-like superfamily. Although their functions are different, they share a common catalytic mechanism: (i) a general base abstracts a proton from Cα of a carbonyl group to form an enolate intermediate, which is stabilized by a general acid; (ii) the enolate intermediate tautomerizes back to the carbonyl group followed by double bond rearrangement or nucleophilic attack (Fig. 4b). In KSI, mgSD, SnoaL, and Tcm ARO/CYC, the general acid-base pairs that initiate the reactions are Asp40-Tyr16/Asp103, His85-a water bound by Tyr30 and Tyr50, Asp121-Gln105, and Tyr35-Arg69, respectively (Fig. 4b). Interestingly, the crystal structures of SgcJ and NCS-Orf16 reveal conserved Asp111-Tyr72 and Asp115-Tyr76 pairs located at the entrance of the pocket, respectively (Fig. 4a). Since these amino acids are known to act as the general acid-base pair in catalysis, it is tempting to speculate that SgcJ may play a similar catalytic role in transforming the nascent linear polyene intermediate, along with other enediyne PKS associated enzymes, into the 9-membered enediyne core.
Additionally, both SgcJ and NCS-Orf16 form a hydrophobic cavity within their pockets. Despite less than 50% amino acid sequence identity between SgcJ and NCS-Orf16, the amino acids lining the cavities are conserved: Trp29, Phe37, Tyr72, Trp118, and Tyr132 in SgcJ versus Trp32, Phe40, Tyr76, Trp122, and Tyr136 in NCS-Orf16 (Fig. 3a and Fig. 5). Intriguingly, in the crystal structure of SgcJ, a molecule of pentaethylene glycol (1PE in chain A) and one of tetraethylene glycol (PG4 in chain B) were found bound in the cavity and surrounded by the conserved amino acid residues (Fig. 4a). These polyethylene glycol molecules may mimic the binding of the linear polyene intermediate, which is sequestered and stabilized by the conserved aromatic residues lining the cavity during biosynthesis of the otherwise unstable 9-membered enediyne core intermediates.
Figure 5.
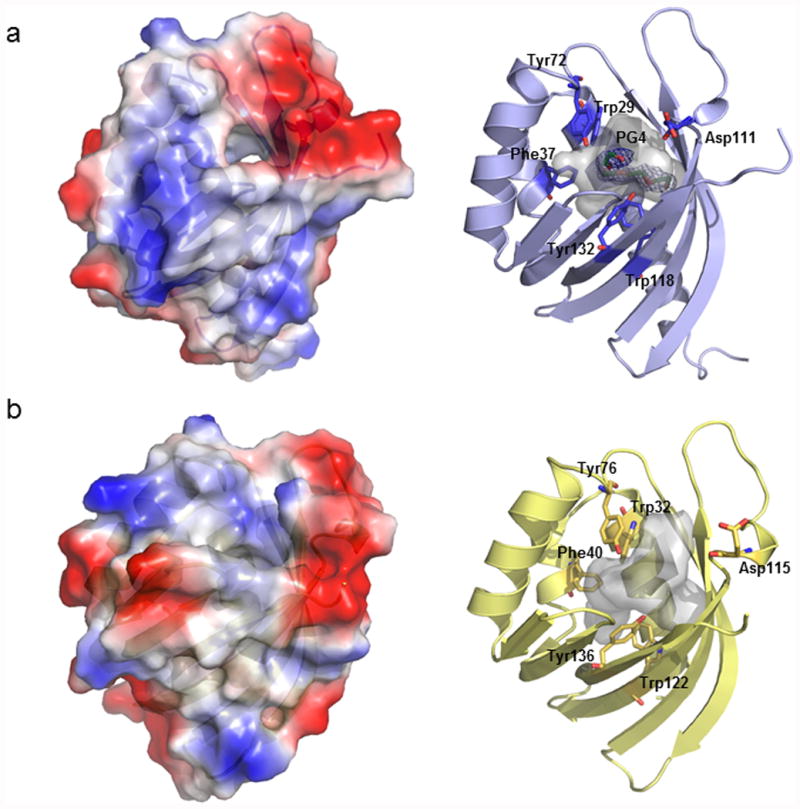
Structure comparison between SgcJ and NCS-Orf16. The electrostatics diagrams (left) and the conserved residues around the cavities (right) of (a) SgcJ and (b) NCS-Orf16. The cavities are shown by grey transparency. The ligand tetraethylene glycol (PG4) molecule is colored in green. The 2Fo – Fc electron density map is contoured at 1.0.
The putative general acid-base catalytic pair and the amino acids lining the cavity are partially conserved among the SgcJ homologues in the seven known (Fig. 3a) and 34 putative (Fig. S2) 9-membered enediyne biosynthetic gene clusters. To provide additional experimental data to support the catalytic role SgcJ and its homologues may play in enediyne core biosynthesis, we mutated each of the six conserved residues in SgcJ (i.e., D111A and Y72A acting as the general acid-base pair, and W29A, F37A, W118A, and Y132A lining the cavity) by site-directed mutagenesis. The expression constructs (pBS1148 to pBS1153) for the mutant variants of sgcJ were identical to pBS1146, in which the expression of sgcJ or its mutant variants was under the control of the constitutive ErmE* promoter. Introduction of pBS1148-pBS1153 individually into SB1027 afforded SB1030-SB1035, respectively, which were fermented, with SB1028 as a positive control, to examine if they could complement the ΔsgcJ mutation in SB1027. Gratifyingly, none of six mutants restored C-1027 production (Fig. 1, panels III and V-X), consistent with the proposal that these conserved residues are involved in substrate recognition, catalysis, or both. Taken together, we now propose that SgcJ plays a catalytic role in transforming the linear polyene intermediate, along with other enediyne PKS associated enzymes, into an enzyme-sequestered 9-membered enediyne core intermediate.
SgcJ and its homologues are pathway specific for enediyne biosynthesis
We have previously demonstrated that PKSEs and TEs from different 9- and 10-membered enediyne machineries are freely interchangeable and 9- versus 10-membered enediyne core biosynthetic divergence occurs beyond the PKSE-TE chemistry.21,22 Given the sequence homology among SgcJ and its homologues (Fig. 3a and Fig. S2) and the structural similarity as exemplified by SgcJ and NCS-Orf16 (Fig. 3b, Fig. 4a, and Fig. 5), as well as the common catalytic role proposed for SgcJ and homologues in 9-membered enediyne core biosynthesis, we finally asked if SgcJ and its homologues are pathway specific. An expression vector pBS1147 for ncs-orf16 was similarly constructed as pBS1146 for sgcJ, in which the expression of ncs-orf16 was under control of the constitutive promoter ErmE*. Introduction of pBS1147 into the ΔsgcJ mutant strain SB1027 afforded SB1029, which offered the opportunity to examine if ncs-orf16 could cross-complement the ΔsgcJ mutation in SB1027. SB1029 was fermented with SB1028 as a positive control. Cross-complementation was not observed as evidenced upon HPLC analysis that showed no C-1027 production in SB1029 (Fig. 1, panels III and IV). This result would suggest that SgcJ and its homologues are pathway specific for 9-membered enediyne core biosynthesis. Close comparison of the SgcJ and NCS-Orf16 structures indeed showed subtle differences in protein surface electrostatics and the shape of the putative cavities (Fig. 5), which may account for unique protein-protein interaction or accommodate varying enediyne core intermediates for different 9-membered enediyne biosynthetic machineries.
Conclusions
The enediynes have served as an outstanding model to study the biosynthesis of complex natural products. Since cloning of the first set of enediyne biosynthetic gene clusters nearly 15 years ago,13,14 significant progress has been made toward elucidating the biosynthesis of the peripheral moieties present in enediynes, but biosynthesis of the eneidyne cores remains elusive.1-4 Comparative analysis of the enediyne gene clusters clearly revealed sets of genes that are highly conserved among the 9-membered, 10-membered, or both enediynes, serving as outstanding candidates to study enediyne core biosynthesis.3,4 Many of these candidate genes, however, are often annotated to encode proteins of unknown function, inactivation of which in vivo afforded mutant strains that often failed to accumulate any biosynthetic intermediates, thereby revealing few clues for their function in enediyne core biosynthesis. As a result, in spite of the great progress made in the past decade in characterizing the enediyne PKS enzyme E and its cognate TE, cumulating to the discovery of a linear heptaene and its variants as the earliest possible intermediates or shunt metabolites for enediyne core biosynthesis,21-25 the exact nature of the nascent linear polyketide intermediates and their subsequent transformation to 9- and 10-membered enediyne cores remain unknown.
Recent technological advance in X-ray crystallography has made it possible to apply high-throughput structural biology as a practical tool to functionally characterize genes with deduced products that show little sequence homology to proteins of known function.53 While the current study fell short of establishing the exact function for SgcJ and its homologues, the structures of SgcJ and NCS-Orf16 and comparison to the NTF2-like superfamily of proteins allowed us to (i) define a putative substrate binding or catalytic active site, (ii) correlate the function of SgcJ to C-1027 biosynthesis by site-directed mutagenesis of the conserved residues lining this site, and (iii) propose that SgcJ and its homologues may play a catalytic role, along with other enediyne PKS associated enzymes, in transforming the linear polyene intermediate into an enzyme-sequestered 9-membered enediyne core intermediate. These findings will surely help formulate hypotheses and design experiments to ascertain the function of SgcJ and its homologues in 9-membered enediyne core biosynthesis in the future.
Supplementary Material
Acknowledgments
This work is supported in part by a fellowship of Academia Sinica-The Scripps Research Institute Postdoctoral Talent Development Program (to C.-Y.C.), a German Research Foundation postdoctoral fellowship (to I.C.), U.S. National Institute of General Medical Science Protein Structure Initiative Grants GM094585 (to A.J.) and GM098248 (to G.N.P.), and U.S. National Institutes of Health Grants GM109456 (to G.N.P.), CA078747 (to B.S.) and GM115575 (to B.S.). The use of Structural Biology Center beamlines at the Advanced Photon Source was supported in part by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Dedicated to Professor David E. Cane, Brown University, for his distinguished contributions to natural product biosynthesis and engineering.
Footnotes
Supplementary Information: Bacterial strains (Table S1), plasmids and cosmids (Table S2), primers (Table S3) used in this study, structures of the six-known 10-membered enediyne natural products (Fig. S1), multiple sequence alignment of SgcJ and its homologues from the seven known and 34 putative 9-membered enediyne biosynthetic gene clusters (Fig. S2), construction of the ΔsgcJ mutant strain S. globisporus SB1027 (Fig. S3), and molecular weight estimations of SgcJ and NCS-Orf16 in solution by size exclusion chromatography (Fig. S4). Supplementary information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja).
The authors declare no competing financial interest.
References
- 1.Van Lanen SG, Shen B. Biosynthesis of enediyne antitumor antibiotics. Curr Top Med Chem. 2008;8:448–459. doi: 10.2174/156802608783955656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang ZX. Complexity and simplicity in the biosynthesis of enediyne natural products. Nat Prod Rep. 2010;27:499–528. doi: 10.1039/b908165h. [DOI] [PubMed] [Google Scholar]
- 3.Shen B, et al. Enediynes: Exploration of microbial genomics to discover new anticancer drug leads. Bioorg Med Chem Lett. 2015;25:9–15. doi: 10.1016/j.bmcl.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudolf JD, Yan X, Shen B. Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J Ind Microbiol Biotechnol. 2016;43:261–276. doi: 10.1007/s10295-015-1671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edo K, et al. The structure of neocarzinostatin chromophore possessing a novel bicyclo [7,3,0]dodecadiyne system. Tetrahedron Lett. 1985;26:331–340. [Google Scholar]
- 6.Kennedy DR, et al. Single chemical modifications of the C-1027 enediyne core, a radiomimetic antitumor drug, affect both drug potency and the role of ataxia-telangiectasia mutated in cellular responses to DNA double-strand breaks. Cancer Res. 2007;67:773–781. doi: 10.1158/0008-5472.CAN-06-2893. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy DR, Ju J, Shen B, Beerman TA. Designer enediynes generate DNA breaks, interstrand cross-links, or both, with concomitant changes in the regulation of DNA damage responses. Proc Natl Acad Sci U S A. 2007;104:17632–17637. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerman TA, Gawron LS, Shin S, Shen B, McHugh MM. C-1027, a radiomimetic enediyne anticancer drug, preferentially targets hypoxic cells. Cancer Res. 2009;69:593–598. doi: 10.1158/0008-5472.CAN-08-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Yin M, Horsman GP, Huang S, Shen B. Manipulation of pathway regulation in Streptomyces globisporus for overproduction of the enediyne antitumor antibiotic C-1027. J Antibiot. 2010;63:482–485. doi: 10.1038/ja.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Yin M, Horsman GP, Shen B. Improvement of the enediyne antitumor antibiotic C-1027 production by manipulating its biosynthetic pathway regulation in Streptomyces globisporus. J Nat Prod. 2011;74:420–424. doi: 10.1021/np100825y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zazopoulos E, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, et al. Rapid PCR amplification of minimal enediyne polyketide synthase cassettes leads to a predictive familial classification model. Proc Natl Acad Sci U S A. 2003;100:11959–11963. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 14.Ahlert J, et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, et al. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohman JR, et al. Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics. Mol BioSyst. 2013;9:478–491. doi: 10.1039/c3mb25523a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGlinchey RP, Nett M, Moore BS. Unraveling the biosynthesis of the sporolide cyclohexenone building block. J Am Chem Soc. 2008;130:2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- 19.Lane AL, et al. Structures and comparative characterization of biosynthetic gene clusters for cyanosporasides, enediyne-derived natural products from marine actinomycetes. J Am Chem Soc. 2013;135:4171–4174. doi: 10.1021/ja311065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q, Thorson JS. The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710. FEMS Microbiol Lett. 2008;282:105–114. doi: 10.1111/j.1574-6968.2008.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, et al. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc Natl Acad Sci U S A. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horsman GP, Chen Y, Thorson JS, Shen B. Polyketide synthase chemistry does not direct biosynthetic divergence between 9- and 10-membered enediynes. Proc Natl Acad Sci U S A. 2010;107:11331–11335. doi: 10.1073/pnas.1003442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belecki K, Crawford JM, Townsend CA. Production of octaketide polyenes by the calicheamicin polyketide synthase CalE8: implications for the biosynthesis of enediyne core structures. J Am Chem Soc. 2009;131:12564–12566. doi: 10.1021/ja904391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belecki K, Townsend CA. Environmental control of the calicheamicin polyketide synthase leads to detection of a programmed octaketide and a proposal for enediyne biosynthesis. Angew Chem Int Ed. 2013;51:11316–11319. doi: 10.1002/anie.201206462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belecki K, Townsend CA. Biochemical determination of enzyme-nound metabolites: preferential accumulation of a programmed octaketide on the enediyne polyketide synthase CalE8. J Am Chem Soc. 2013;135:14339–14348. doi: 10.1021/ja406697t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Shen B. Genes for production of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus are clustered with the cagA gene that encodes the C-1027 apoprotein. Antimicrob Agents Chemother. 2000;44:382–392. doi: 10.1128/aac.44.2.382-392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, United Kingdom: 2000. [Google Scholar]
- 29.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, et al. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Rosenbaum G, et al. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Cryst. 2006;D62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst. 2010;D66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z. Isomorphous replacement and anomalous scattering. Proceedings of Daresbury Study Weekend. 1991:80–85. [Google Scholar]
- 36.Cowtan K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 37.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 41.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberhardt RY, et al. Filling out the structural map of the NTF2-like superfamily. BMC Bioinformatics. 2013;14:327. doi: 10.1186/1471-2105-14-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cha HJ, et al. Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase. Mol Cells. 2013;36:39–46. doi: 10.1007/s10059-013-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundqvist T, et al. Crystal structure of scytalone dehydratase--a disease determinant of the rice pathogen, Magnaporthe grisea. Structure. 1994;2:937–944. doi: 10.1016/s0969-2126(94)00095-6. [DOI] [PubMed] [Google Scholar]
- 45.Sultana A, et al. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 2004;23:1911–1921. doi: 10.1038/sj.emboj.7600201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ames BD, et al. Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. Proc Natl Acad Sci U S A. 2008;105:5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 49.Ott M, et al. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bullock TL, Clarkson WD, Kent HM, Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2) J Mol Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 53.Jacobson MP, Kalyanaraman C, Zhao S, Tian B. Leveraging structure for enzyme function prediction: methods, opportunities, and challenges. Trends Biochem Sci. 2014;39:363–371. doi: 10.1016/j.tibs.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.