Abstract
Pluripotent embryonic stem cells (ESCs) self-renew or differentiate into all tissues of the developing embryo and cell-specification factors are necessary to balance gene expression. Here we delineate the function of the PHD-finger protein 5a (Phf5a) in ESC self-renewal and ascribe its role in regulating pluripotency, cellular reprogramming, and myoblast specification. We demonstrate that Phf5a is essential for maintaining pluripotency, since depleted ESCs exhibit hallmarks of differentiation. Mechanistically, we attribute Phf5a function to the stabilization of the Paf1 transcriptional complex and control of RNA polymerase II elongation on pluripotency loci. Apart from an ESC-specific factor, we demonstrate that Phf5a controls differentiation of adult myoblasts. Our findings suggest a potent mode of regulation by the Phf5a in stem cells, which directs their transcriptional program ultimately regulating maintenance of pluripotency and cellular reprogramming.
INTRODUCTION
The remarkable cellular plasticity that defines ESCs is central towards their ability to differentiate into all somatic lineages and the germline1, 2. Their unique identity is governed by regulation on multiple levels, ultimately orchestrating gene expression3. However, ESCs must readily alter their transcriptional program to allow proper cell specification. Therefore, elucidating molecular mechanisms of cellular adaptation is paramount in understanding stem cell function. We have previously characterized several factors necessary for maintaining ESC self-renewal or initiating differentiation4. Using an RNAi-based screen we identified the poorly characterized PHD-finger protein 5a (Phf5a) as a potential modulator of pluripotency4. Phf5a is a small, highly conserved protein (Supplementary-Fig.1a) harboring a characteristic PHD-fold5. Ectopic Phf5a was suggested to localize to the nucleus, postulated to associate with chromatin mediating transcription5. Deletion of Phf5a is lethal in yeast and knockdown in C. elegans results in aberrant organogenesis during early development5, 6, suggesting its importance for embryo formation and tissue morphogenesis6. However, its functional role in transcription regulation in mammals remains unexplored.
Modulation of gene expression is crucial for stem cell self-renewal or cell specification. The Paf1 transcriptional complex (Paf1C) is central to these processes dictating RNA-PolII function and deposition of histone modifications7, 8. Paf1C plays important roles in development and is necessary for differentiation9–11, however details regarding its function in stem cells remain unknown. Here we demonstrate that Phf5a is a potent regulator of Paf1C stability and chromatin binding. Furthermore, we show it is essential for ESC self-renewal, and cellular reprogramming and found that RNA-PolII elongation of pluripotency genes is defective after Phf5a depletion. Beyond its role in ESCs, we found Phf5a to regulate muscle specification suggesting additional functions in adult stem cells. We conclude that Phf5a is crucial regulating RNA elongation of genes controlling pluripotency and cell differentiation.
RESULTS
Phf5a depletion leads to loss of ESC pluripotency and inhibits reprogramming
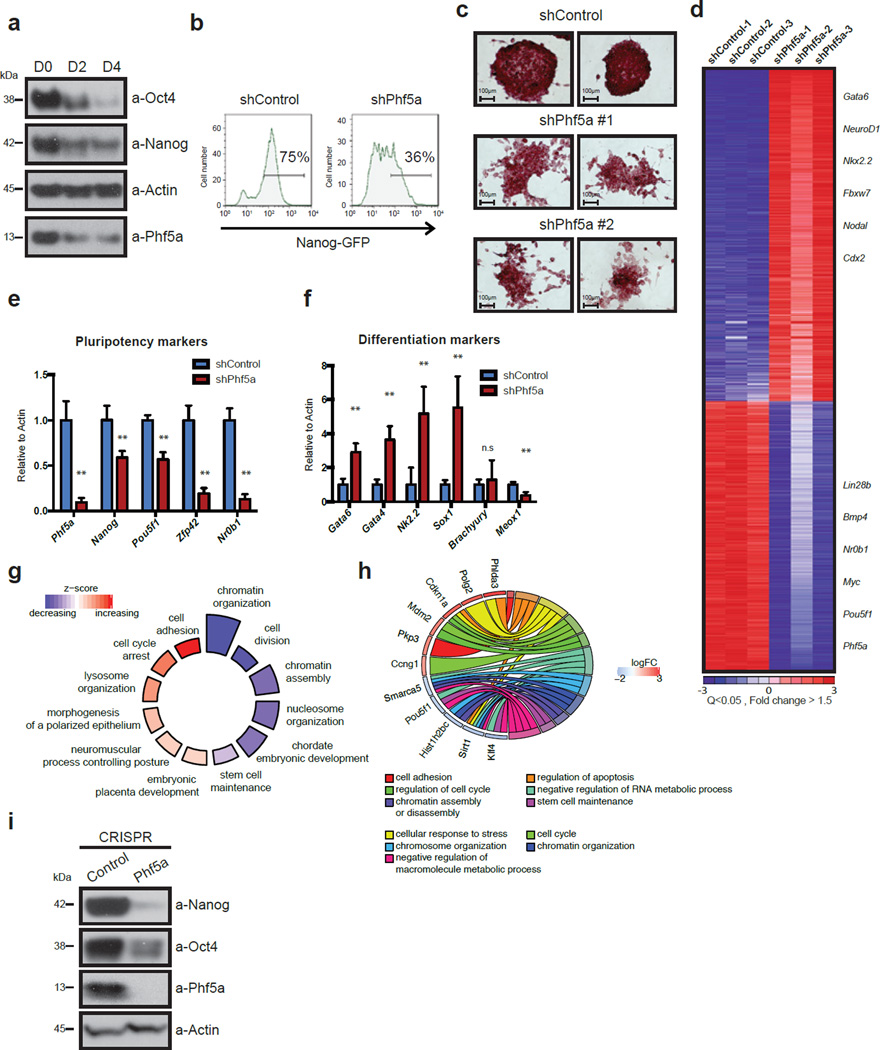
To delineate the role of Phf5a in pluripotency, we investigated its expression levels during mouse ESC differentiation. We utilized the Nanog-GFP (NG) reporter ESC line, a faithful indicator of self-renewal4, 12. Phf5a expression, both at mRNA and protein levels, is high in pluripotent ESCs, but becomes rapidly downregulated upon differentiation (Fig.1a and Supplementary-Fig.1b). Consistent with a possible role in preserving self-renewal, knockdown of Phf5a led to a significant loss of Nanog-GFP fluorescence (Fig.1b). This was accompanied by morphological changes (Supplementary-Fig.1c), and cells exhibited considerably reduced alkaline-phosphatase (AP) staining, an additional marker of pluripotency (Fig.1c). Importantly, we did not observe increased levels of cell death or apoptosis upon Phf5a knockdown (Supplementary-Fig.1d–e), suggesting that its loss leads to differentiation without effects on viability.
Figure 1. Phf5a is required for maintenance of ESC self-renewal.
(a) Western blot analysis of Phf5a and Nanog, Oct4 proteins during ESC differentiation (see Supplementary Figure 7). (b) Histogram FACS plots representing loss of GFP fluorescence in Nanog-GFP transcriptional reporter ESCs following knockdown with shControl or shPhf5a respectively. (c) Alkaline phosphatase (AP) staining of ESCs following knockdown with shControl or shPhf5a (2 different hairpins), respectively. Scale bars, 100 µm. (d) Heatmap of Affymetrix microarrays for differentially expressed genes of Nanog-GFP ESCs following knockdown with shControl or shPhf5a respectively. Red: upregulated genes, blue: downregulated genes. Q-value<0.05 Fold change (log2)>1.5. (e and f) Bar graphs showing expression levels by qRT-PCR of Phf5a, pluripotency markers (e) and differentiation markers (f), respectively, following shPhf5a knockdown in ESCs. n=6 biologically independent replicates (see Supplementary Table 5). Phf5a, Nanog, Pou5f1, Sox2, Zfp42 and Nr0b1: **p=0.0001, respectively. Gata6, Gata4, Nkx2-5, Sox1, and Meox1, **p=0.0001, respectively, Brachyury: n.s: non-significant, p=0.5632, two-sided Student’s t-test, values represent the mean ± s.d.. (g) GO-Circle plot displaying gene-annotation enrichment analysis. Blue and red indicate downregulated or upregulated gene-associated GO Terms, respectively, relative to the z-score of the analysis. (h) GO-Chord plot displaying relationships between several representative downregulated and upregulated GO Terms and associated genes. Distinct categories linked to pluripotent or differentiated cells cluster separately. (i) Western blot analysis of pluripotency factors following CRISPR-Cas9 mediated Phf5a depletion in ESCs. (see Supplementary Figure 7).
We next determined the transcriptional response of Phf5a silencing by gene expression array analysis revealing two sets of genes significantly up- or down-regulated (Fig.1d and Supplementary-Table 1). Changes in expression of selected genes were validated with qRT-PCR. Consistent with ESC differentiation, pluripotency markers were significantly downregulated, whereas lineage markers were upregulated with the exception of several mesoderm markers (Fig.1e–f). To further characterize gene expression profiles of Phf5a-depleted ESCs, we performed RNA-seq followed by Gene Ontology (GO) and gene-annotation enrichment analysis13. Downregulated genes associated with GO terms related to stem cell function, such as stem cell maintenance, chromatin organization and cell division, whereas upregulated genes associated with GO terms related to early embryo development such as cell adhesion, cell cycle arrest, and morphogenesis (Fig.1g–h and Supplementary-Fig.1f). Transcriptional profiling therefore strongly supports the notion that Phf5a loss triggers ESC differentiation.
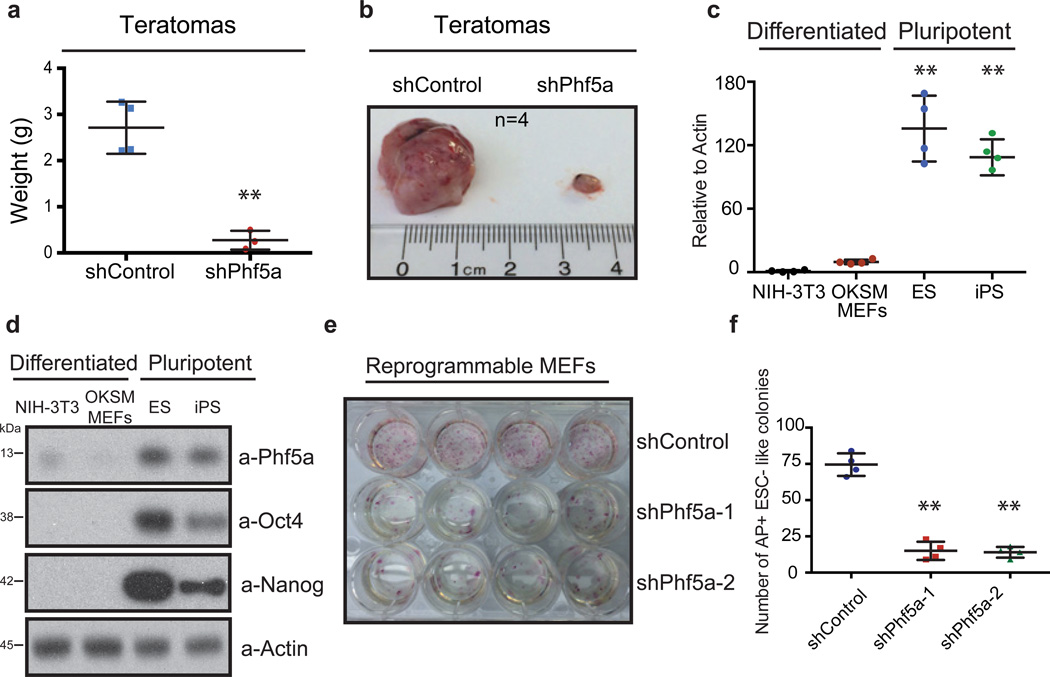
To validate our findings, we first ruled out potential off-target effects. Phf5a silencing using individual shRNAs, siRNAs, as well as CRISPR-Cas9-mediated gRNAs, targeting distinct regions of its transcript, resulted in identical downregulation of pluripotency markers Oct4 and Nanog, as well as decreased AP staining (Fig.1i and Supplementary-Fig.1g–1j). Examination of Phf5a levels in ESCs from different backgrounds revealed no significant differences during differentiation (Supplementary-Fig.1k–l). Finally, we engineered inducible knockdown ESC lines by introducing mir30-shPhf5a cassettes in the Col1a1 locus of cells constitutively expressing the M2rtTA transactivator14, and were able to faithfully reproduce differentiation phenotypes following addition of doxycycline (Supplementary-Fig.2a–d). Using these lines, we also investigated the role of Phf5a in pluripotency in vivo. Injection of inducible ESCs in immuno-deficient mice post-induction with doxycycline significantly inhibited the ability to form teratomas (Fig.2a–b). Examination by hematoxylin and eosin (H&E) stain as well as immunohistochemistry showed reduction of mesoderm markers compared to ectoderm or endoderm (Supplementary-Fig.2e–g). Specifically, we noticed apparent histological differences, including depletion of skeletal muscle formation, which we validated using desmin immunohistochemistry (Supplementary-Fig.2g). To further document defects in mesoderm differentiation we examined in vitro differentiation towards the mesoderm lineage15, 16 using the reporter ESC line Dppa4-RFP/Brachyury-GFP17. We found that shPhf5a depletion results in loss of self-renewal (Dppa4-RFP reduction), however, without an increase of mesoderm differentiation (Brachyury-GFP gain) (Supplementary-Fig.2h). Last, we were unable to detect upregulated Brachyury protein levels or additional mesoderm markers (Supplementary-Fig.2i–j). These results suggest that Phf5a loss results in failure of self-renewal maintenance while affecting lineage skewing, demonstrating its importance in stem cells.
Figure 2. Phf5a regulates ESC pluripotency and cellular reprogramming.
(a and b) Comparison of mass (a) and size (b) of teratomas generated in SCID mice following injection of doxycycline-induced ESCs engineered to express shControl or shPhf5a cassettes from the Col1a1 locus. n=4 biologically independent replicates (see Supplementary Table 5). **p=0.001 two-sided Student’s t-test, values represent the mean ± s.d. (c) Comparison of Phf5a transcript levels between differentiated fibroblasts and pluripotent stem cells by qRT-PCR. n=4 biologically independent replicates (see Supplementary Table 5). ESCs: **p=0.0035, iPSCs: **p=0.0013, two-sided Student’s t-test, values represent the mean ± s.d. (d) Western blot analysis of Phf5a protein in differentiated fibroblasts or pluripotent stem cells. (see Supplementary Figure 7). (e and f) Alkaline phosphatase (AP) staining (e) and comparison of AP-positive ESC-like colony number (f), respectively, of reprogrammable OKSM MEFs on day14 post-initial doxycycline induction following shPhf5a knockdown. n=4 biologically independent replicates (see Supplementary Table 5). **p=0.001, respectively, two-sided Student’s t-test, values represent the mean ± s.d.
These phenotypes prompted us to further investigate its role in pluripotency and cellular reprogramming. We compared Phf5a expression in pluripotent or differentiated cells and found that it correlated with the pluripotent state (Fig.2c–d). Consistent with that, analysis of proliferation profiles in ESCs or primary mouse embryo fibroblasts (MEFs) following shPhf5a silencing resulted only in ESC self-renewal defects (Supplementary-Fig.2k–l). Finally, Phf5a overexpression during ESC differentiation maintained pluripotent marker expression (Supplementary-Fig.2m–n).
We reasoned that Phf5a expression might also regulate induced pluripotent stem cell (iPSC) formation. We silenced Phf5a in reprogrammable primary MEFs, engineered to express the Oct4, Klf4, Sox2 and c-Myc (OKSM) reprogramming casette18–20 and interrogated its effects 14 days post-induction of reprogramming factors. We observed that Phf5a loss resulted in a dramatic reduction of reprogrammed fibroblasts (Fig.2e and Supplementary-Fig.2o). We noticed a significant decrease in the absolute number of AP-positive ESC-like colonies (Fig.2f), suggesting that Phf5a silencing leads to decreased efficiency of reprogramming. Last, we examined transgene-independent expression of endogenous markers of pluripotency18 and found that Phf5a deficiency blocked their upregulation (Supplementary-Fig.2p). Collectively, we conclude that Phf5a is necessary for ESC self-renewal and efficient iPSC generation as its silencing results in aberrant initiation of differentiation and a block to reprogramming.
Phf5a interacts with the Paf1 complex
The impact of Phf5a knockdown on ESCs and iPSCs prompted us to investigate its functional role propagating pluripotency. We first explored its intracellular localization. We engineered inducible Phf5a-expressing or control ESC lines, fractionated cytoplasmic and nuclear extracts following doxycycline induction and confirmed a primarily nuclear localization in ESCs (Supplementary-Fig.3a). Immunofluorescence (IF) confirmed nuclear accumulation of Phf5a (Supplementary-Fig.3b). Given its internal PHD-finger motif, we hypothesized a possible role in transcription and chromatin regulation.
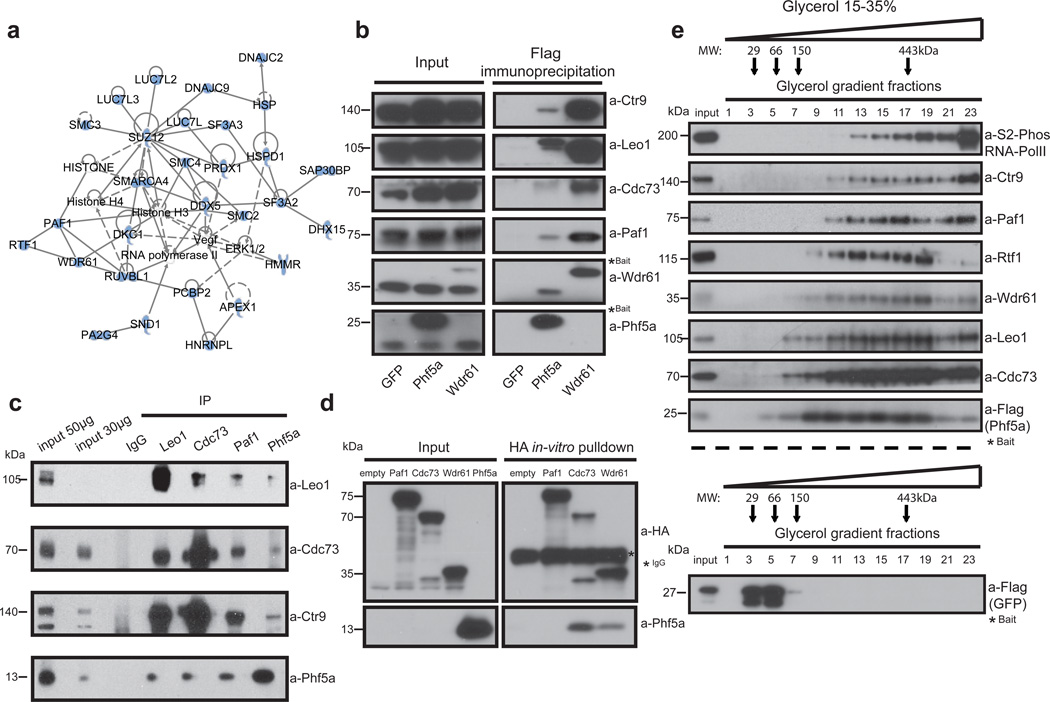
To identify its molecular function we undertook an unbiased approach by purifying Phf5a from ESCs and analyzing its interacting partners by mass-spectrometry (Fig.3a and Supplementary-Table 2). Among the top interacting proteins we found 3 out of 6 subunits of the Paf1 transcriptional complex (Paf1C). The mammalian Paf1C, which consists of the subunits Ctr9, Rtf1, Leo1, Paf1, Cdc73, and Wdr61, has been implicated in transcriptional regulation and deposition of histone modifications7. Since mass spectrometry suggested close association of Paf1C with Phf5a and since Paf1C depletion also results in ESC differentiation21, 22, we decided to further investigate this interaction.
Figure 3. Phf5a physically associates with the Paf1 complex.
(a) Ingenuity systems-generated pathway of Phf5a interacting proteins following purification and mass spectrometry in ESCs. Solid or dashed lines illustrate established direct or indirect interactions, respectively. (b) Validation of Phf5a interactions with the Paf1 complex in ESCs using Flag-Phf5a purification. Tagged Phf5a, Wdr61 (positive control) and GFP (negative control) were transiently expressed in engineered Tet-inducible ESC lines following addition of doxycycline. Bait proteins are tagged (marked with a star) and migrate slower than endogenous proteins (see Supplementary Figure 7). (c) Endogenous protein immunoprecipitations for Phf5a and Paf1C subunits in ESCs (see Supplementary Figure 7). (d) Paf1-complex subunits Paf1, Cdc73 and Wdr61 were cloned in HA-tag expressing vectors and subjected into in-vitro transcription and translation. Phf5a protein was expressed and purified from bacteria. In vitro binding of HA-tagged subunits and Phf5a was interrogated by a pull-down assay using HA-immunoprecipitation and western blot analysis (see Supplementary Figure 7). (e) Phf5a interacting proteins from ESCs were subjected to glycerol gradient sedimentation followed by fractionation and western blot analysis resulting in overlapping distributions of Phf5a and Paf1-complex subunits. A control analysis for GFP is shown in the lower panel. (see Supplementary Figure 7).
Although we initially identified specific Paf1C subunits as Phf5a binding partners, we validated interactions with the entire complex (Fig.3b). Similar to Phf5a, Paf1C subunits are downregulated during ESC differentiation (Supplementary-Fig.3c). We engineered inducible knock-in ESC lines expressing the subunits Cdc73 and Wdr61 and confirmed binding with Phf5a (Supplementary-Fig.3d). Furthermore, we validated Phf5a-Paf1C interactions using tandem-affinity purification (Supplementary-Fig.3e), as well as endogenous protein immunoprecipitations in ESCs (Fig.3c). Importantly, we found that DNA or RNA do not mediate this interaction since it persists after extensive nuclease treatment (Supplementary-Fig.3f). These results suggest a robust interaction between Phf5a and Paf1C.
We next attempted to specify whether distinct Paf1C subunits mediate this interaction. We in vitro translated Paf1, Cdc73 and Wrd61 subunits and interrogated binding to purified recombinant Phf5a. We observed interactions with Cdc73 and Wdr61 subunits but not Paf1 (Fig.3d), suggesting that Phf5a binds to a subset of Paf1C subunits directly. Next, we performed glycerol gradient density sedimentation analysis from ESCs and identified that Phf5a forms high-molecular weight complexes and co-fractionates with Paf1C under native conditions (Fig.3e). Furthermore, since Paf1C is implicated in transcriptional elongation, we confirmed Phf5a interaction with RNA-PolII (Supplementary-Fig.3g). Moreover, in density sedimentation analysis the elongating form of RNA-PolII co-sedimented with Phf5a and Paf1C (Fig.3e), indicating active engagement during transcription elongation. In contrast, we were unable to detect interaction with the initiation factor TFIID (Supplementary-Fig.3e), absent in elongating complexes, suggesting Phf5a association with PolII at specific transcription stages. These findings establish interaction of Phf5a with Paf1C and suggest its possible role in transcriptional regulation in ESCs.
Phf5a depletion leads to Paf1C destabilization and loss of binding to its target genes
The direct interaction between Phf5a and Paf1C together with differentiation phenotypes after knockdown, suggested an intimate connection to Paf1C function. To explore this functional association, we first compared gene expression profiles of Phf5a- and Paf1-depleted ESCs and found similar gene expression patterns (Supplementary-Fig.4a and Supplementary-Table 3). Since Phf5a was previously implicated in alternative exon recognition in malignant cells23, we also investigated whether shPhf5a or shPaf1 knockdown result in aberrant alternative splicing in ESCs. We used multivariate analysis of transcript splicing (rMATS)24, and found a small number of splicing differences concluding that their loss do not result in overt changes in splicing patterns (Supplementary-Table 4).
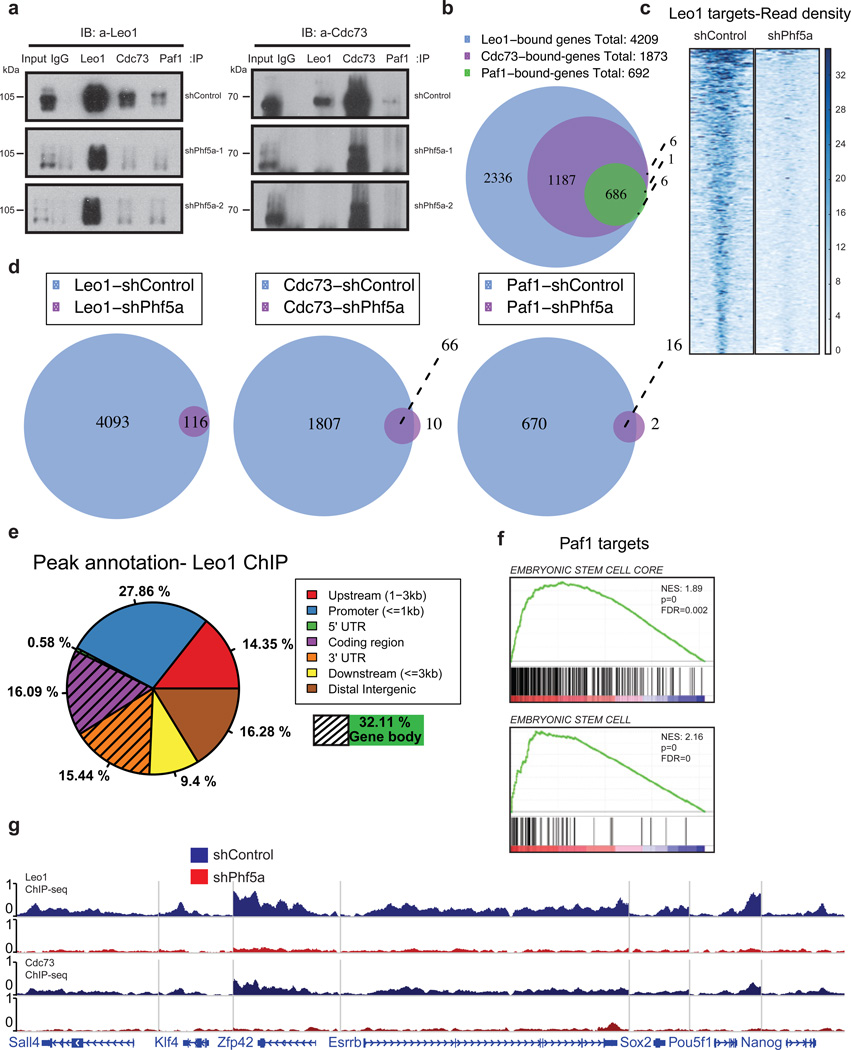
We next studied Paf1C stability upon loss of Phf5a. We re-examined Paf1C composition using density sedimentation analysis in the presence or absence of Phf5a. We found that Phf5a knockdown leads to Paf1C distribution towards lower molecular-weight fractions, indicating destabilization (Supplementary-Fig.4b–e). In contrast other protein complexes, such as Swi/Snf and NELF, remain unaltered (Supplementary-Fig.4f). This suggested loss of interaction among Paf1C subunits and we confirmed significant decrease between subunit associations following Phf5a silencing (Fig.4a).
Figure 4. Phf5a controls interactions among Paf1C subunits and its silencing abrogates Paf1C recruitment on pluripotency genes in ESCs.
(a) Western blot analysis of Paf1C subunit immunoprecipitations in 293T cells following knockdown with shControl or shPhf5a, respectively, showing loss of interactions between different Paf1C members upon Phf5a depletion. Two different shRNA hairpins are shown. Left Panel: Blot for Leo1; Right Panel: blot for Cdc73; IP: immunoprecipitation. IB: immunoblot. (see Supplementary Figure 7). (b) Venn diagram showing number of genes bound by individual Paf1C subunits in ESCs using ChIP-sequencing with antibodies against endogenous Leo1, Cdc73 and Paf1 proteins. (c) Heatmap representations of normalized read density for Leo1 binding in ESCs following shControl or shPhf5a silencing, respectively. (d) Venn diagrams showing the numbers of genes bound by Leo1, Cdc73 and Paf1 in ESCs in the presence or absence of Phf5a, respectively. (e) Binding profiles for genomic distribution of Leo1 peaks (upstream, promoter, coding region, 5'UTR, 3'UTR, downstream and intergenic) in ESCs, showing preferential (32%) binding within gene bodies. (f) Gene set enrichment analysis (GSEA) enrichment plots showing significant enrichment of the top Paf1 targets for genes linked to embryonic stem cell signatures. (g) Snapshots of Leo1 and Cdc73 binding on representative pluripotency gene targets (Sall4, Klf4, Zfp42, Esrrb, Sox2, Pou5f1 and Nanog) in the presence (blue) of absence (red) of Phf5a, respectively.
These findings suggested decreased Paf1C binding to target genes after Phf5a loss. We directly interrogated Paf1C occupancy in ESCs using Leo1, Cdc73 and Paf1 ChIP-sequencing in the presence or absence of Phf5a. We identified ~4200 high-stringency targets of Paf1C in ESCs (Fig.4b) with Cdc73- and Paf1-bound genes constituting subsets of Leo1-bound genes. We observed that Paf1C peaks virtually disappear upon Phf5a silencing, supporting its critical role in complex function (Fig.4c–d). We dissected peak localization and determined occupancy among promoters, UTRs, coding and intergenic regions. Consistent with regulation of active gene expression25, we noticed that Leo1 peaks fall mostly within gene bodies and promoters (Fig.4e). Further dissection revealed that downregulated genes engage Leo1 mostly within gene bodies (42%), whereas upregulated genes utilize Leo1 mostly on promoters (47%) (Supplementary-Fig.4g). This suggests that Paf1C sub-complexes may differentially localize on their targets to control gene expression. However, using gene set enrichment analysis (GSEA) we found that Paf1 targets are enriched in pluripotency genes and correlate with ESC signatures (Fig.4f). These Paf1C targets include Pou5f1, Esrrb, Sall4, Prdm14 and other well-described pluripotency genes (Fig.4g). Finally, to test direct Phf5a binding to self-renewal network we engineered a Tet-inducible HA-tagged Phf5a line and performed ChIP-seq using HA-epitope after doxycycline induction. Consistent with previous findings we identified Phf5a binding to pluripotency genes such as Nanog, Pou5f1, Fbxo15, Esrrb, Tcf3, Prdm14, Sall4 and others (Supplementary-Fig.4h).
Loss of Phf5a leads to RNA-PolII promoter-proximal pausing
The Paf1C facilitates transcriptional elongation26 and is required for maximal levels of Ser-2-P-PolII27, 28. Since our studies suggest that Phf5a controls Paf1C binding on self-renewal genes, we hypothesized that its loss might lead to their aberrant elongation. We first investigated Ser-2-P-PolII levels following Phf5a loss and found them significantly lower (Fig.5a and Supplementary-Fig.5a). We next interrogated effects on RNA-PolII stalling and regulation of elongation. Pausing of activated RNA-PolII constitutes a rate-limiting step in gene expression and pause-release is important for embryonic development29, 30 and somatic cell reprogramming31. We profiled nascent RNAs using global run-on sequencing (GRO-seq)32 and calculated the elongation pausing index in ESCs following shPhf5a knockdown. We initially observed that downregulated genes were paused (Fig.5b–c) in contrast to upregulated genes (Fig.5d) signifying a difference between the two sets. We further dissected pausing ratios for multiple GO subsets. Downregulated categories exhibited significantly elevated promoter-proximal pausing compared to upregulated ones (Fig.5e). In addition, downregulated gene read density shows significant decrease within gene bodies (Fig.5f). Similar to shPhf5a, shPaf1 knockdown leads to profound RNA-PolII pausing on downregulated genes (Fig.5g) analogous to flavopiridol treatment33. In contrast, ESCs differentiated in the absence of LIF, did not exhibit increased RNA-PolII stalling (Supplementary-Fig.5b–d), suggesting specific roles for Paf1C/Phf5a in maintenance of pluripotency. Indeed, we found that ~50% of downregulated genes are direct ChIP-seq Paf1C targets and show promoter-proximal pausing after shPhf5a knockdown, compared to upregulated targets (Supplementary-Fig.5e).
Figure 5. Phf5a controls transcriptional elongation and RNA-PolII pause-release of pluripotency genes in ESCs.
(a) Western blot analysis of total PolII and Ser-2 phosphorylated RNA-PolII in ESC following shControl or shPhf5a knockdown or ESCs differentiated in the absence of LIF, respectively. (see Supplementary Figure 7). (b and c) Scatter plot representing pausing indices of downregulated (b) or upregulated genes (c), respectively, 72h following shControl or shPhf5a knockdown using GRO-seq analysis. Read density of 500bp downstream of promoters (5'density) was normalized to read density in the rest of the gene bodies (3'density). Pausing Index= 5’density/3’density. Gray: All genes; Blue: Downregulated genes; Red: Upregulated genes. (d) Box plot showing pausing index ratios after GRO-seq analysis for downregulated (blue) or upregulated (red) genes, respectively, following shPhf5a knockdown. Only downregulated genes exhibit significant promoter-proximal pausing after shPhf5a depletion. n=3 biologically independent replicates, Wilcoxon signed rank test non-parametric. (e) Box plot showing pausing index ratios after GRO-seq analysis for specific GO Terms. Blue: Downregulated and Red: Upregulated GO Term categories, respectively. Only downregulated GO Terms exhibit significant promoter-proximal pausing after shPhf5a depletion. n=3 biologically independent replicates, Wilcoxon signed rank test non-parametric. (f) Comparison of GRO-seq read density profiles of genes 72h following shControl or shPhf5a knockdown, respectively, in ESCs. RPKM: Reads Per Kilobase per Million total reads. (g) Box plot representing comparison of log2 pausing index for downregulated genes, 72h following shControl, shPhf5a, shPaf1 knockdown, or flavopiridol-treated ESCs, respectively, using GRO-seq analysis. Flavopiridol treatment is used as a positive control of pause-release block. n=3 biologically independent replicates, Wilcoxon signed rank test non-parametric. (h) Scatter plot representing RNA-PolII pausing index for Paf1C targets and pluripotency genes based on normalized Ser5 (on TSSs)/Ser2 (on gene bodies) read density ratio of RNA-PolII ChIP-Seq in ESCs following shControl of shPhf5a silencing, respectively. In box plots (d, e and g) the central mark is the median, and the edges of the box are the first and third quartiles. Whiskers extend to the most extreme non-outlier data points.
To further study elongation dynamics we performed ChIP-seq for initiating (Ser5-phosphorylated) and elongating (Ser2-phosphorylated) RNA-PolII in the presence or absence of Phf5a. We calculated the RNA-PolII pausing index25 and found that Paf1C targets and self-renewal genes, such as Nanog Pou5f1, Sox2, Klf4 Fbxo15, Myc, Esrrb and others, exhibit significant stalling (Fig.5h). We also found elevated levels of Ser5-RNA-PolII near gene promoters for GO terms, in contrast to decreased Ser2-RNA-PolII in their gene bodies (Supplementary-Fig.5f–g).
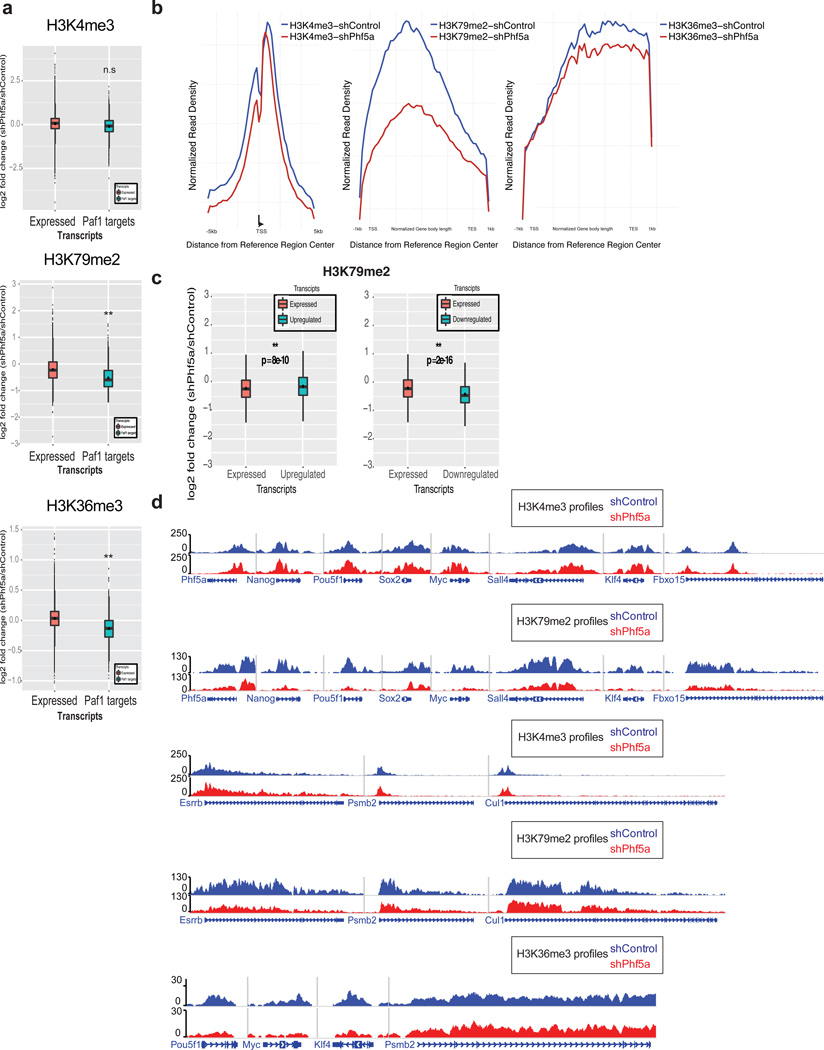
Finally, we interrogated whether Phf5a affects histone modifications associated with Paf1C function7. We performed ChIP-sequencing for H3K4me3, H3K79me2 and H3K36me3 in the presence or absence of Phf5a. We found its depletion negatively affected elongation-associated histone modifications H3K79me2 and H3K36me3 on Paf1C targets and self-renewal genes, but not the promoter-associated mark H3K4me3 (Fig.6a). H3K79me2 and H3K36me3 profiles revealed a pronounced loss from gene bodies after shPhf5a knockdown (Fig.6b). Dissection of H3K79me2 revealed that Paf1C targets have similar profiles to downregulated genes, compared to upregulated ones (Fig.6c). Last, we examined occupancy on pluripotency genes and found diminished H3K79me2 and H3K36me3 levels in their gene bodies, but not H3K4me3 levels on their promoters, compared to housekeeping genes (Fig.6d). These data suggest that Phf5a loss affects Paf1C functions including regulation of RNA-PolII elongation and histone mark occupancy.
Figure 6. Phf5a regulates the deposition of histone marks characteristic of transcriptional elongation in pluripotency gene loci.
(a) Box plots representing log2 fold change of normalized read density for H3K4me3, H3K79me2 and H3K36me3 ChIP-seq in ESCs following shControl or shPhf5a silencing. Plots represent comparisons of all expressed transcripts in ESCs with direct Paf1 targets around transcription start sites (TSSs) (H3K4me3) or gene bodies (H3K79me2 and H3K36me3). n=3 biologically independent replicates, Wilcoxon signed rank test non-parametric. (b) Normalized read density profiles around TSSs (H3K4me3) or gene bodies (H3K79me2 and H3K36me3) on Paf1C targets and pluripotency genes in ESCs in the presence (blue) or absence (red) of Phf5a. (c) Box plots showing log2 fold change H3K79me2 occupancy on gene bodies of target genes. H3K79me2 occupancy is increased in upregulated genes, however, H3K79me2 occupancy is decreased in downregulated genes compared to all expressed genes. n=3 biologically independent replicates, Wilcoxon signed rank test non-parametric. (d) Snapshots of representative H3K4me3 H3K79me2 and H3K36me3 density tracks on pluripotency genes or control loci under conditions of shControl (blue) or shPhf5a silencing (red). In box plots (a and c) the central mark is the median, and the edges of the box are the first and third quartiles. Whiskers extend to the most extreme non-outlier data points.
Phf5a regulates myogenic differentiation
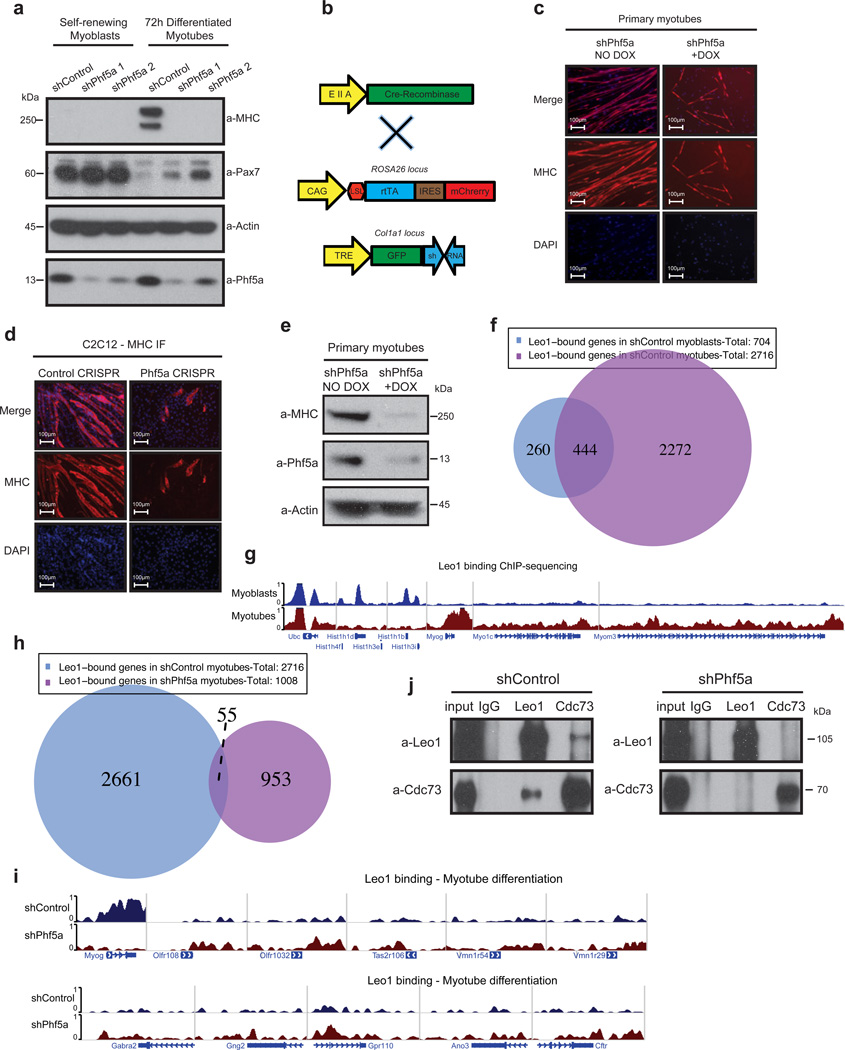
We next asked whether Phf5a functions specifically in ESCs or it controls differentiation in additional systems. Since Phf5a depletion results in aberrant mesoderm differentiation and muscle formation in teratomas, and Paf1 is also implicated in cardiomyocyte specification in zebrafish10, we decided to study Phf5a function in muscle. Myoblasts self-renew, however, in differentiation conditions commence myogenic programs and fuse forming elongated, multi-nucleated myotubes (Supplementary-Fig.6a). We initially depleted Paf1C subunits in C2C12 myoblasts and verified its role in myotube differentiation (Supplementary-Fig.6b). We then depleted Phf5a and found it also compromises their ability to differentiate. We observed that myoblasts fail to upregulate the differentiation marker myosin-heavy chain (MHC) (Supplementary-Fig.6c–d). Additionally, Phf5a silencing results in maintenance of Pax7, a marker of myoblast self-renewal (Fig.7a). To support our findings in C2C12 cells we generated a Tet-inducible RNAi mouse model by knocking-in individual shPhf5a hairpins in the Col1a1 locus of ESCs (Fig.7b). We generated Rosa26rtTACol1a1TREshPhf5a mice and crossed them to EIIA-Cre mice to drive hairpin expression (Fig.7b). We generated primary mouse myoblasts from these animals, and observed defects in myotube differentiation upon Phf5a silencing (Fig.7c). We observed upregulation of Phf5a and Paf1C during primary myoblast differentiation (Supplementary-Fig.6e) and were also able to mimic effects of RNAi depletion on myotube differentiation using CRISPR-Cas9 strategies (Fig.7d–e and Supplementary-Fig.6f–g). These results demonstrate that loss of Phf5a blocks myogenic differentiation.
Figure 7. Phf5a loss leads to Paf1C destabilization and inhibits myogenic differentiation.
(a) Western blot analysis of myoblast self-renewal and myotube differentiation markers (myosin heavy chain and Pax7, respectively) following shControl or shPhf5a knockdown (see Supplementary Figure 7). (b) Schematic of Tet-inducible Rosa26rtTACol1a1TREshRNA animals for the derivation of primary myoblasts. Addition of doxycycline drives expression of shPhf5a from the Col1a1 locus. LSL: LoxP-stop-LoxP cassette. (c and d) Myosin heavy chain (MHC) immunofluorescence on primary myotubes purified from Rosa26rtTACol1a1TREshRNA animals (c) and CRISPR-Cas9-mediated Phf5a silencing on C2C12 cells (d), respectively, depicting suppression of myoblast differentiation. Scale bars, 100 µm. (e) Western blot analysis of Phf5a on primary myotubes purified from Rosa26rtTACol1a1TREshRNA animals. Addition of Doxycyclin induces shRNA hairpin expression and the silencing of Phf5a (see Supplementary Figure 7). (f) Venn diagram of Leo1-bound genes in myoblasts and myotubes following ChIP-sequencing. (g) Genome browser tracks showing peaks of Leo1 ChIP-sequencing for representative genes in myoblasts and myotubes. Histone-1 cluster genes, Myog, Myo1c, Myom3 and Ubc are shown as examples. (h) Venn diagram of Leo1 bound genes in myotube differentiation in the presence or absence of Phf5a following ChIP-sequencing. (i) Genome browser tracks showing peaks of Leo1 ChIP-sequncing for representative genes in shControl and shPhf5a conditions, respectively. Myog, and several olfactory, taste and smell receptors and G-protein coupled receptors, ion channels and neurotransmitter receptors are shown as examples. (j) Western blot analysis of Paf1C subunit composition using immunoprecipitations in C2C12 cells differentiated for 72h following knockdown with shControl or shPhf5a, respectively. A significant loss among Paf1C subunit interactions is observed upon Phf5a silencing (see Supplementary Figure 7).
To further investigate how Paf1C/Phf5a control mechanisms of muscle differentiation we performed ChIP-sequencing for Leo1 in myoblasts and myotubes. We found an increased number of Leo1-bound genes during muscle differentiation from 700 genes in myoblasts, to more than 2700 in myotubes (Fig.7f). GO analysis of Leo1-bound genes identified multiple chromatin- and transcription-associated GO terms in myoblasts, such as chromatin assembly, nucleosome organization and others, compared to muscle-specific GO terms in differentiated myotubes, such as actin organization, muscle development, muscle organization and myofibril assembly (Supplementary-Fig.6h–i), Examples for Leo1 binding include Hist1 cluster genes in myoblasts, Myog, Myo1c and Myom3 in myotubes and many others (Fig.7g).
Finally, we performed ChIP-sequencing in myotube differentiation after shPhf5a knockdown. We found that Leo1 binding was abolished from its myotube targets (Fig.7h and Supplementary-Fig.6j). Interestingly, we identified ~1000 Leo1 targets after shPhf5a knockdown, however almost none of them is associated with myogenic processes (Fig.7h). In addition, we performed GO analysis of Leo1 targets after shPhf5a knockdown and found genes associated with neurogenesis, instead of myogenesis (Supplementary-Fig.6j). Specifically, we identified neurological system process, cognition, sensory perception of smell, and sensory perception of chemical stimulus GO terms after shPhf5a knockdown (Supplementary-Fig.6j). We observed loss of Leo1 binding in specific myogenic genes, but gain in neurogenic ones such as olfactory, taste and neurotransmitter receptors, G-protein-coupled receptors, ion channels and many others (Fig.7i). Finally, to investigate whether Phf5a loss affects Paf1C stability we directly interrogated interactions among Paf1C subunits after its knockdown and found dissociation of the core subunits Leo1 and Cdc73, resulting in complex disruption (Fig.7f). In conclusion, these studies confirm that Phf5a is an essential regulator of myoblast differentiation and suggest that it stabilizes Paf1C in chromatin promoting myogenic programs.
DISCUSSION
Since their early characterization, PHD-finger proteins were recognized as tethering molecules recruiting or stabilizing protein complexes34–36 leading to tight regulation of gene expression37. Here we characterize in detail the functions of Phf5a in ESCs, iPSCs and myoblasts and attribute its requirement for self-renewal to the binding and stabilization of the Paf1C. We found that Phf5a depletion resulted in ESC differentiation and inhibition of cellular reprogramming. Phf5a and Paf1C associate strongly, are recruited on actively transcribed pluripotency genes and positively-regulate RNA-PolII elongation.
Previous efforts to study Paf1C functions focused entirely on promoter occupancy using ChIP-on-chip assays21, or in relation to DNA methylation38. Our studies characterize how Phf5a affects elongation dynamics in self-renewing ESCs and we integrated a combination of approaches to elucidate its functions. First, we characterized promoter-proximal pausing ratios using nascent RNA profiling following Phf5a depletion. Second, utilizing RNA-PolII ChIP-sequencing we elucidate traveling ratios on pluripotency genes and Paf1C targets. Last, we determine how elongation-specific histone marks change in response to Paf1C alterations. Our results suggest that Phf5a directly regulates Paf1C stability, facilitating pause release and productive elongation of the self-renewal network. Elongation of upregulated genes in the absence of Paf1C is due to indirect effects. Direct Paf1C/Phf5a targets include master regulators of pluripotency, and it would be intriguing to further study mechanisms of Paf1C/Phf5a recruitment and possible cooperation with transcription factors22.
Furthermore, we found that Phf5a functions are not limited to ESCs. Using in vivo and in vitro models we demonstrated that Phf5a is essential for differentiation of myoblasts to myotubes. These functions are Paf1C-dependent as loss of Phf5a expression leads to significant decrease of Paf1C occupancy at myogenic genes and de novo targeting at neurogenic ones. These findings are consistent with Paf1C regulating cardiac specification and heart morphogenesis in zebrafish10 and deregulation on muscle organ formation in Phf5a-depleted C. elegans6. Collectively, we conclude that Phf5a mediates Paf1C functions to orchestrate myogenic differentiation.
Processes that balance self-renewal or cell specification can be deregulated in cancer. Paf1C can also act as an oncogene and its amplification or overexpression is implicated in tumor formation8, 39. Although Paf1C stimulates transcriptional elongation in vitro and in vivo7, 26, 40, it was recently suggested to also suppress RNA-PolII transcription in cancer41. Our data suggest that Paf1C is active in ESCs, directly promoting elongation of pluripotency networks. However, Paf1C may function in a cell-type specific manner influenced by chromatin accessibility.
Recent studies in human malignancies, implicated Phf5a in endometrial cancer and glioblastoma23, 42. Although Phf5a is suggested to interact with ATP-dependent helicases and the U2 snRNP spliceosome23, 43, its silencing surprisingly only affects exon recognition and splicing in glioblastoma stem cells (GSCs) but not their normal counterpart neural stem cells (NSCs)23. Despite extensive sequencing studies, we did not find significant splicing defects in Phf5a-depleted ESCs, enforcing the notion that these are cell-type specific. It would be intriguing to speculate that splicing phenotypes are absent from non-malignant cells (ESCs and NSCs) but present in glioblastoma or other malignancies, opening the way to investigate this distinction as a potential vulnerability in cancer. It is interesting to interrogate whether aberrant Phf5a expression correlates with de-regulated Paf1C functions in human disease, and since Phf5a loss inhibits proliferation of cancer cells44, its targeting might be an alternative therapeutic option.
Methods
Mouse ESC and iPSC culture and OKSM MEF reprogramming
Mouse ESCs, iPSCs and “reprogrammable” OKSM MEFs45 were cultured under standard conditions as described previously46, 47 using recombinant LIF, on gelatin-coated plates or feeder-MEFs, respectively. For shRNA viral transductions ESCs and OKSM MEFs were transduced with pLKO.1-puro backbone lentiviruses and selected with puromycin. For reprogramming experiments ESC-like colonies were enumerated 14 days post-initial induction with doxycycline.
Culture of C2C12 myoblasts, myotube differentiation
C2C12 myoblast cells were cultured as described previously48. For differentiation C2C12 cells were grown to confluence followed by culturing in differentiation media (DMEM supplemented with 2% horse serum) for 72h or up to 120h before analysis.
Immunofluorescence
ESC expressing Flag-Phf5a or C2C12 cells were fixed with 4% formaldehyde in PBS for 20 min at room temperature, washed once with PBS and permeabilized for 20 min at room temperature using Block Solution (5% normal goat serum, 0.1% Triton X-100 in PBS). Cells were incubated overnight at 4°C with Flag antibody (1:500), Desmin (1:500) or MHC serum (1:500) in Block Solution, washed 3 times with PBS, incubated for 1h at room temperature with 1:1000 secondary rabbit anti-mouse Alexa594-conjugated antibody in Block Solution and DAPI.
Animal experiments
Female C57BL/6 mice (6–8 weeks old) were obtained from the National Cancer Institute. For teratoma assays nonobese diabetic/severe combined immunodeficient NOD/MrkBomTac-Prkdcscid (NOD-SCID) mice were obtained from Taconic. 106 KH2 ESCs engineered to express inducible shControl or shPhf5a hairpins were injected subcutaneously into NOD-SCID mice, were under 0.1mg doxycycline diet throughout the course of the assay and were euthanized 3 weeks after injection when tumors were collected. Tumors were fixed in formalin followed by imbedding in paraffin, sectioned and stained for histological analysis with hematoxylin and eosin or immunohistochemistry stains following standard procedures. For generation of Tet-inducible knockdown animals Rosa26rtTACol1a1TREshPhf5a targeted ESCs were purchased from Mirimus Inc. Mice were generated by injection into tetraploid blastocysts at the NYU Medical Center Rodent Genetic Engineering Core. Engineered mice were crossed to EIIA-Cre recombinase mice to drive Rosa26rtTA expression. For in vivo Col1a1TREshPhf5a cassette expression mice were placed on 0.1mg doxycycline diet. Isolation and growth of primary myoblasts was performed as described previously49. Briefly, neonatal mice were sacrificed and limb muscle was dissected from skin and bones. Primary myoblasts were isolated in mincing and incubation of muscle in collagenase/dispase/CaCl2 solution followed by incubation in 5% CO2 incubator using F-10-based primary myoblast growth medium. Mice were housed in specific pathogen-free conditions at the Skirball Institute animal facility. For animal experiments, no statistical method was used to predetermine sample size. Furthermore, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. All animal experiments were performed in accordance with protocols approved by the New York University Institutional Animal Care and Use Committee.
CRISPR-Cas9 editing
For CRISPR-Cas9 editing Control- and Phf5a-targeting and repair template vectors were purchased from SantaCruz (sc-418922, sc-427066, sc-427066-HDR) and were transfected in ESCs or C2C12 cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s manual.
Engineering of inducible ESCs expressing tagged proteins or shRNAs
For inducible expression, cDNA was prepared (High-Capacity RNA-to-cDNA Kit, Applied Biosystems). Open reading frames (ORF) were cloned into the Tet-operated vector pINTA, bearing N-terminal Flag/Strep-TagII (F/S) tandem tags50 (kind gift of Dr. R. Bonasio, University of Pennsylvania). Vectors were nucleoporatated (Amaxa) into KH2 ESCs51, 52. ESCs were selected with 50 µg/ml Zeocin (Invitrogen) for 7 days.
For inducible knockdown, miR-30 hairpins were cloned into a modified pColTGM vector53 targeting the Col1a1 locus. Vectors were electroporated with pCAGS-FlpE recombinase in KH2 ESCs and were selected with hygromycin 140µg/mL for 10 days.
shControl: 5’TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGAA3’
shPhf5a: 5’TGCTGTTGACAGTGAGCGACTATCGGAAGACTGTGTGAAATAGTGAAGCCACAGATGTATTTCACACAGTCTTCCGATAGCTGCCTACTGCCTCGGA3’
Antibody dilutions, western blotting and immunoprecipitations
Antibodies were used according to manufacturer’s specifications. For western blots, cells lysed in Lysis Buffer (100mM Tris-HCl pH8.0, 150mM NaCl, 0.1% Triton X-100) supplemented with Complete Mini protease inhibitors (Roche), 10mM NaF (Sigma) and 1mM Na3VO4 (Sigma). For immunoprecipitation (IP) 1mg of pre-cleared cell lysate was incubated overnight with each antibody or IgG control (sc-2027 Santa Cruz), and bound to Protein-A Sepharose beads (Invitrogen) at 4°C. Beads were washed 4 times with 1mL Lysis Buffer. Antibodies used were the following: Nanog (1:10000 A300-397A, Bethyl), Oct-3/4 (1:5000 sc-5279, Santa Cruz), Actin (1:10000 C4, Millipore), Flag (1:10000 M2, Sigma), HA (1:5000 ab-9110, Abcam) Phf5a (1:500 15554-1-AP, ProteinTech), Paf1 (1:1000 A300-173A, Bethyl for IP, A300-172A, Bethyl for WB and purified rabbit antibody for ChIP54), Cdc73 (1:1000 A300-701, Bethyl), Ctr9 (1:1000 A301-395A, Bethyl), Leo1 (1:1000 A300-175A, Bethyl), Rtf1 (1:1000 A300-179A, Bethyl), Wdr61 (Ski8) (1:500 pAB-012-150, Diagenode), Caspase-3 (1:1000 9662, Cell Signalling), GFP (1:5000 sc-9996, Santa Cruz), RNA-PolII (1:1000 sc-899, Santa Cruz), Ser2-phospho-RNA-PolII (1:1000 clone 3E10, Active Motif), Ser5-phospho-RNA-PolII (1:1000 ab5131, Abcam), TFIID (1:1000 sc273 X, Santa Cruz), a-Tubulin (1:10000 sc-53029, Santa Cruz), Lamin B (1:1000 sc-6217, Santa Cruz), Pax7 (1:500 sc-81975, Santa Cruz), NELF-A (1:1000 A301-910A, Bethyl), Brachyury (1:500 ab20680, Abcam), Desmin (1:1000 ab32362, Abcam), Nestin (1:1000 MAB353, clone rat-401, EMD Millipore), H3K4me3 (1:1000 07-473, Millipore), H3K79me2 (1:1000 39143, Active Motif), H3K36me3 (1:1000 ab9050, Abcam). Serum against Myocin Heavy Chain (MHC, kind gift from Dr. B. Dynlacht, NYU School of Medicine) was used 1:10000. For western blots antibodies were used in 5% milk in TBS-T, unless otherwise noted. Secondary horseradish peroxidase (HRP)-conjugated antibodies (GE Healthcare) were used in 5% milk in TBS-T. For immunoprecipitations Rabbit IgG Trueblot (1:5000 18-8816-31, Rockland) or Mouse IgG Veriblot (1:5000 ab131368, Abcam) secondary HRP-conjugated antibodies were used.
Purification of tagged proteins and mass spectrometry
5*107 targeted ESCs were induced with 2µg/mL doxycycline (Sigma) for 3 days and treated with 10µM MG132 (Peptides International) for 3h. Pellets were resuspended in Lysis Buffer (100mM Tris-HCl pH7.5, 150mM NaCl, 1% Triton-X100, 1mM EDTA, 2mM MgCl2, supplemented with Complete Mini protease inhibitors (Roche), 10mM N-elthylmaleimide (Sigma), 10mM NaF (Sigma), 1mM Na3VO4 (Sigma) and 250 units Benzonase nuclease (Novagen), and passed 8 times through a 25 & 5/8 gauge syringe. Protein purification was performed as described before50. Beads were eluted with 5mL Elution Buffer (Buffer E, IBA) and concentrated with Amicon Ultra centrifugal filter units, 10,000 MW cutoff, (Millipore) to 200µL. For tandem affinity purification elutions were bound to 50µL Flag magnetic beads, washed with 1× Buffer E (IBA), and boiled in SDS-loading buffer. For cytoplasmic and nuclear fractionation 1*107 KH2 ESCs were lysed as above and cytoplasmic and nuclear extracts were prepared as described previously using a glass homogenizer55. For mass spectrometry peptides were analyzed by LC-MS/MS on Orbitrap Velos MS. The MS/MS spectra were searched against NCBI database using a local MASCOT search engine (V.2.3). At least two peptides were identified for each protein (false discovery rate <0.01%) with a confidence interval no less than 95%.
Glycerol gradient density sedimentation analysis
5*107 targeted ESCs were induced, lysed and tagged proteins were purified as above and concentrated to final volume of 500µL. 15-35% glycerol gradients were prepared using BIOCOMP Gradient Master in 4.5mL open-top tubes (Beckman) and fraction preparations as described before using TCA precipitation56.
Phf5a purification from bacteria and in vitro pull-down
Phf5a ORF was cloned into the pGEX-6P-1 vector (GE Healthcare) (kind gift from Dr. K.J. Armache, NYU School of Medicine) and transformed into BL21 Star (DE3) bacteria. 12L of liquid cultures were induced overnight with 0.1mM IPTG at 18°C. Bacteria were lysed in 20mM Tris-HCl pH8.0, 200mM NaCl, 1mM DTT passing though a pressure homogenizer. Soluble fraction was bound to glutathione agarose beads (Pierce) for 1h at 4°C, beads were washed with 200 column volumes (CV) Lysis Buffer and GST-Phf5a protein was eluted using reduced glutathione. GST tag was cleaved by cleavage with PreScission Protease (kind gift from Dr. K.J. Armache, NYU School of Medicine) and Phf5a was further purified by ion exchange and size exclusion FPLC chromatography. Paf1, Cdc73 and Wdr61 were cloned in Flag/HA-modified pCDNA3.1 vector (Invitrogen) and in vitro translated using TNT Coupled Wheat Germ Extract System (L4140, Promega). 5µg of purified Phf5a protein was added, mixtures were bound overnight at 4°C and HA-tagged proteins were pulled-down using HA affinity gel beads (Invitrogen) for 4h at 4°C. Beads were washed 4 times with 1mL Lysis Buffer and interactions were visualized by WB analysis.
Benzonase treatment assay
1*107 KH2 ESCs were lysed as above and 50 units Benzonase nuclease (Novagen) was added. Lysate was incubated at 4°C for a total of 12h and 100µL of material was removed and snap-frozen every 3h. Nucleic acids were phenol/chloroform extracted, ethanol precipitated and visualized in a 2% agarose gel.
Flow Cytometry Analysis
Apoptosis and cell death was detected using Annexiv-V APC-conjugated detection kit (BD Biosciences) along with 7-AAD following manufacturers protocol on a BD LSRFortessa (BD Biosciences) flow cytometer.
Real-time quantitative reverse transcription PCR (qRT-PCR) and microarray analysis
Total RNA was harvested from cells using the Qiagen RNeasy Kit (Qiagen) and 2µg of was used for cDNA synthesis using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). qRT-PCR was carried out using LightCycler SYBR green mix (Roche) using a LightCycler 480 II (Roche). For microarray analysis RNA samples were hybridized to Affymetrix MoGene2.0.st Chips and scanned at the NYU Genome Technology Center. CEL files were loaded into GeneSpring (Agilent). Feature intensities for each probe set were condensed into a single intensity value.
GRO-seq and library preparation
Analysis of nascent RNAs using global-run-on experiments were performed as described previously57. Briefly, nuclei were isolated in swelling buffer (10mM Tris-HCl pH7.5, 2mM MgCl2, 3mM CaCl2), lysed twice in lysis buffer (10mM Tris-HCl pH7.5, 2mM MgCl2, 3mM CaCl2, 10% glycerol, 0.5% NP-40) and snap-frozen in freezing buffer (50mM Tris pH8.0, 40% glycerol, 5mM MgCl2, 0.1mM EDTA), For run-on reaction, an equal volume of reaction buffer was added to thawed nuclei (10mM Tris pH8.0, 5mM MgCl2, 300mM KCl, 500uM ATP, 500µM GTP, 5µM CTP, 500µM BrUTP, 1mM DTT, 100U/mL SuperaseIN, 1% Sarcosyl), mixed and incubated at 30°C for 5min. The reaction was stopped with Trizol reagent and RNA was phenol/chloroform extracted and ethanol precipitatated. RNA was heated in Fragmentation buffer (40mM Tris pH8.0, 100mM KCl, 6.25mM MgCl2, 1mM DTT), DNAse treated and purified using Zymo RNA Clean & Concentrator (Zymo Research) using the >17nt protocol. Run-on RNA was immunoprecipitated using BSA-blocked BrDU beads (Santa Cruz) in Binding buffer (SSPE 0.5X, 1mM EDTA, 0.05% Tween-20) for 1h at 4°C, washed and eluted in Elution buffer (5mM Tris pH7.5, 300mM NaCl, 20mM DTT, 1mM EDTA, 1% SDS) at 65°C for 20min. Nascent RNA was further phenol/chloroform extracted and sequencing libraries were prepared.
Data sources and computational pipelines
The samples were run using Illumina HiSeq2000. Raw reads were aligned against the mouse genome assembly mm10/GRCm38. Alignments were performed using Bowtie v.1.0.058. MACS 1.4.259 or MACS 2.0 was used for peak-calling in the case of ChIP-Seq data, while the suite GenomicTools version 2.8.160 was used for genome binning, genomic annotations and the construction of occupancy profiles, both in the case of polymerases and histone marks. For plotting, R version 3.2.0 was used (R Core Team (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria) (https://www.R-project.org), along with the VennDiagram package61 for the generation of Venn diagrams and ggplot262 for the generation of boxplots, density profiles and scatterplots with ggrepel (https://CRAN.R-project.org/package=ggrepel). For the generation of heatmaps depicting the binding profiles of Pac1C components, deepTools was used63.
ChIP-seq and library preparation
ChIP experiments were performed as described previously60, 64. Antibodies coupled to magnetic beads were added to precleared chromatin and incubated for 12–16 h. Beads were washed and eluted according to protocol. ChIP-Seq libraries for Paf1C components, PolII samples and histone marks were prepared using the Illumina TruSeq system, including end repair, A-tailing, adaptor ligation and PCR amplification. AMPure XP beads (Beckman Coulter, A63880) were used for DNA cleaning in each step of the process. Raw images generated by Illumina HiSeq2000 using default parameters, were processed by CASAVA to remove the first and last bases and then they were used to generate sequence reads in fastq format. Reads were aligned to mm10 mouse genome using Bowtie with the standard parameters (except for –m 1 in order to report only unique alignments). MACS version 1.4.259 was used to perform peak calling using the parameter values listed below:
In the case of ChIP-Seq experiments for Paf1C components and Phf5a, MACS v1.4.2 was used and the parameter values were: (a) --nomodel, (b) --shiftsize=120, (c) --slocal 5000, (d) --llocal 50000 (e) –p 1e-4.
In the case of polymerase and histone ChIP-Seq experiments, MACS v.2.0 was used with the following parameter values: (a) --nomodel, (b) --broad, (c) --shiftsize=200, (d) –q 0.05.
All PolII and histone ChIP-Seq experiments were performed in triplicates and peaks present at least in two out of the three triplicates were used for downstream analysis. The files with the aligned reads were converted to wig format using GenomicTools60 and then to bigwig format using the corresponding UCSC tool.
Peak characterization
Peaks were assigned to the following categories based on their genome-wide distribution: (a) Upstream: this category includes all peaks that fall within 1-3kb upstream of the transcription start site (TSS), (b) Promoter: it includes all peaks that fall within 1kb upstream of the TSS, (c) Gene body: it includes all peaks that fall within the 5’ UTR, the coding region of genes and the 3’ UTR, (d) Downstream: it includes all peaks that fall within 3kb from the transcription end site (TES), (e) Distal Intergenic: All the peaks that fall within the remaining genomic loci. The peak characterization was performed using ChIPSeeker65 and custom in-house scripts.
Calculation of PolII Pausing Index
The calculation of PollII Pausing index was performed as described previously66. Specifically, the PolII densities were initially calculated as reads per kilobase per million (RPKMs)67 in order to normalize for region length and number of reads. The initiating region was defined as the area between 30bp upstream of the transcription start site (TSS) to 300bp after the TSS, while the elongating region was from +300bp to the end of the gene. We calculated the PolII pausing index by dividing the PolII densities for the initiating region vs. the elongating region. We used Wilcoxon’s non-parametric test to compare the distributions of the fold-changes (log2 scale) in the pausing index in control (shControl) and the samples, where either Phf5a (shPhf5a) or Paf1 (shPaf1) were down-regulated.
Gene-set enrichment analysis
For GSEA analysis the online version of the GSEA tool was used68. The peaks that were identified as significant after peak-calling, were ranked according to peak score (from highest to lowest) and they were used as input for GSEA. MSigDB v5.0 (updated in April 2015) was used for GSEA.
Gene ontology
Gene ontology (GO) analysis was performed based on ChIP-Seq data and gene expression data by using the DAVID tool v6.7 (https://david.ncifcrf.gov/). Visualization of the GO results was performed using the R package GOplot69. The reported z-score was calculated as described in GOplot and shows the trend (increasing/decreasing) of the corresponding GO category in terms of gene expression.
Statistics and reproducibility
Sample sizes and reproducibility for each figure are denoted in the figure legends. Unless otherwise noted, data are representative of at least three biologically independent experiments. For mouse experiments, no statistical method was used to predetermine sample size. Furthermore, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. Statistical significance between conditions was assessed by two-tailed Student’s t-tests. Error bars represent s.d., and significance between conditions is denoted. Raw data from independent replicate experiments can be found in the Statistics Source Data (Supplementary Table 5).
Cell lines
No cell lines used in this study were found in the databases of commonly misidentified cell lines that are maintained by ICLAC and NCBI Biosample. The following source of cell lines was used: KH2 ESCs, OKSM MEFs and iPSCs: Kind gift from Dr. Konrad Hochedlinger, Harvard Stem Cell Institute; CCE and Nanog-GFP: Kind gift from Dr. Ihor Lemischka, Mount Sinai School of Medicine; MK6: Kind gift from Dr. Sang Yong Kim, Rodent Genetic Engineering Core, NYU School of Medicine; Dppa4-RFP/Brachyury-GFP ESCs: Kind gift from Dr. H.J Fehling, University Clinics Ulm, Germany; C2C12 Kind gift from Dr. Brian Dynlacht, NYU School of Medicine. The cell lines were not authenticated. The cell lines were routinely tested for mycoplasma contamination.
Primary accessions
Gene Expression Omnibus (GEO) GSE63974
Data availability
The next-generation sequencing data that support the findings of this study in Figures 1, 4, 5, 6, and 7 have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE63974. Statistics source data have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank all members of the Aifantis laboratory for useful comments and discussions throughout the duration of this project; Z. Gao and P. Voigt for experimental help with glycerol gradients and helpful discussions; K.J. Armache and P. De Ioannes Fernandez for help with protein purification, helpful discussions and manuscript preparation; H-J Fehling for the Dppa4-RFP/Brachyury-GFP reporter ESC line. A. Heguy and the NYU Genome Technology Center (supported in part by National Institutes of Health (NIH)/National Cancer Institute (NCI) grant P30CA016087-30) for expertise with sequencing experiments; the NYU Histology Core (5P30CA16087-31) for assistance; C. Loomis and L. Chiriboga for immunohistochemistry experiments; H.Li and T. Liu at the Center for Advanced Proteomics Research, New Jersey School of Medicine for mass spectrometry. This work has used computing resources at the High Performance Computing Facility of the Center of Health Informatics and Bioinformatics at the NYU Medical Center. A.S. is supported by the NYSTEM institutional NYU Stem Cell Training Grant (C026880). I.A. is supported by the NIH (RO1CA133379, RO1CA105129, RO1CA149655, 5RO1CA173636, 1RO1CA194923) and the NYSTEM program of the New York State Health Department (NYSTEM-N11G-255).
Footnotes
AUTHOR CONTRIBUTIONS
I.A and A.S designed the experiments and wrote the manuscript. A.S performed the experiments. A.T C.L. T.T and I.D. designed and performed the analysis of genome-wide data. P.N. provided expertise in sequencing experiments and contributed in manuscript preparation. A.L.G.N performed histological examination of teratomas. S.B contributed ideas. M.S. Y.Y, B.D, S.R. and B.S. provided materials and tips related to this study, helped with ideas and concepts and contributed to manuscript preparation.
REFERENCES
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley SM, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trappe R, et al. Identification and characterization of a novel murine multigene family containing a PHD-finger-like motif. Biochem Biophys Res Commun. 2002;293:816–826. doi: 10.1016/S0006-291X(02)00277-2. [DOI] [PubMed] [Google Scholar]
- 6.Trappe R, Schulze E, Rzymski T, Frode S, Engel W. The Caenorhabditis elegans ortholog of human PHF5a shows a muscle-specific expression domain and is essential for C. elegans morphogenetic development. Biochem Biophys Res Commun. 2002;297:1049–1057. doi: 10.1016/s0006-291x(02)02276-3. [DOI] [PubMed] [Google Scholar]
- 7.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta. 2013;1829:116–126. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akanuma T, Koshida S, Kawamura A, Kishimoto Y, Takada S. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO reports. 2007;8:858–863. doi: 10.1038/sj.embor.7401045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langenbacher AD, et al. The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol. 2011;353:19–28. doi: 10.1016/j.ydbio.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen CT, Langenbacher A, Hsieh M, Chen JN. The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Dev Biol. 2010;341:167–175. doi: 10.1016/j.ydbio.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaniel C, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 14.Premsrirut PK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelton M, et al. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Reports. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madan B, et al. The pluripotency-associated gene Dppa4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol Cell Biol. 2009;29:3186–3203. doi: 10.1128/MCB.01970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 20.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Ponnusamy MP, et al. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem cells. 2009;27:3001–3011. doi: 10.1002/stem.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubert CG, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen S, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Molecular cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 28.Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith E, Shilatifard A. Transcriptional elongation checkpoint control in development and disease. Genes Dev. 2013;27:1079–1088. doi: 10.1101/gad.215137.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, et al. Transcriptional Pause Release Is a Rate-Limiting Step for Somatic Cell Reprogramming. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigbolt KT, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Science signaling. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- 40.Yu M, et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350:1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen FX, et al. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell. 2015;162:1003–1015. doi: 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falck E, Klinga-Levan K. Expression patterns of Phf5a/PHF5A and Gja1/GJA1 in rat and human endometrial cancer. Cancer Cell Int. 2013;13:43. doi: 10.1186/1475-2867-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rzymski T, Grzmil P, Meinhardt A, Wolf S, Burfeind P. PHF5A represents a bridge protein between splicing proteins and ATP-dependent helicases and is differentially expressed during mouse spermatogenesis. Cytogenet Genome Res. 2008;121:232–244. doi: 10.1159/000138890. [DOI] [PubMed] [Google Scholar]
- 44.Nijhawan D, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 45.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckley SM, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reavie L, et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nature immunology. 2010;11:207–215. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. The Journal of cell biology. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Springer ML, Rando TA, Blau HM. Gene delivery to muscle. Current protocols in human genetics/editorial board, Jonathan L. Haines … [et al.] 2002;Chapter 13(Unit13):14. doi: 10.1002/0471142905.hg1304s31. [DOI] [PubMed] [Google Scholar]
- 50.Gloeckner CJ, Boldt K, Ueffing M. Strep/FLAG tandem affinity purification (SF-TAP) to study protein interactions. Current protocols in protein science/editorial board, John E. Coligan … [et al.] 2009;Chapter 19(Unit19):20. doi: 10.1002/0471140864.ps1920s57. [DOI] [PubMed] [Google Scholar]
- 51.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 52.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 53.Premsrirut PK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu B, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic acids research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Molecular cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Use R. 2009:1–212. [Google Scholar]
- 63.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic acids research. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 66.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 68.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The next-generation sequencing data that support the findings of this study in Figures 1, 4, 5, 6, and 7 have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE63974. Statistics source data have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.