Abstract
Objective
Increased type I interferon (IFN-I) and a broad signature of IFN-I-induced gene transcripts are observed in patients with SLE and other systemic autoimmune diseases. To identify disease-relevant triggers of the IFN-I pathway we investigated whether endogenous virus-like genomic repeat elements, normally silent, might be expressed in patients with systemic autoimmune disease, activate an innate immune response and induce IFN-I.
Methods
Expression of IFN-I and long interspersed nuclear element-1 (LINE-1; L1) was studied in kidney tissue from lupus patients and minor salivary gland (MSG) tissue from patients with primary Sjogren’s syndrome (SS) by PCR, western blot and immunohistochemistry. Induction of IFN-I by L1 was investigated by transfection of plasmacytoid dendritic cells (pDCs) or monocytes with an L1-encoding plasmid or L1 RNA. Involvement of innate immune pathways and altered L1 methylation were assessed.
Results
L1 mRNA transcripts were increased in lupus nephritis kidneys and in MSG from SS patients and correlated with IFN-I expression and L1 DNA demethylation. L1 open reading frame 1/p40 protein and IFNβ were expressed in MSG ductal epithelial cells and in lupus kidneys, and IFNα was detected in infiltrating pDCs. Transfection of pDCs or monocytes with L1-encoding DNA or RNA induced IFN-I. Inhibition of TLR7/8 reduced L1 induction of IFNα in pDCs and an inhibitor of IKKε/TBK1 abrogated induction of IFN-I by L1 RNA in monocytes.
Conclusion
L1 genomic repeat elements represent endogenous nucleic acid triggers of the IFN-I pathway in SLE and SS and may contribute to initiation or amplification of autoimmune disease.
Systemic autoimmune diseases, with systemic lupus erythematosus (SLE) the prototype, are characterized by protean immunologic alterations, heterogeneous clinical manifestations, and tissue and organ damage. Recent studies have documented numerous common and several rare genetic variants that are associated with SLE, but the endogenous and exogenous triggers that initiate and perpetuate disease have not yet been defined (1). The presence of elevated serum type I interferon (IFN-I) activity (2) and a broad signature of IFN-I-induced gene (IFIG) transcripts and proteins in blood and tissue of patients with lupus (3–6) and other systemic autoimmune diseases, including primary Sjogren’s syndrome (SS) (7–14), systemic sclerosis (15), and dermatomyositis (16) are consistent with a viral trigger, but available data have not identified an exogenous virus as an etiologic agent in any of these diseases.
Although extensive in vitro data document the capacity of nucleic acid-containing immune complexes to access endosomal Toll-like receptors (TLR) and induce IFNα production by peripheral blood cells, particularly plasmacytoid dendritic cells (pDCs), the genetic and environmental factors that might initiate IFN-I production prior to development of autoantibodies require further investigation. Some first degree relatives of lupus patients show elevated IFN-I activity even in the absence of measureable autoantibodies (17). Moreover, data from individuals with polymorphisms or single gene mutations in components of the TLR-independent nucleic acid-sensing pathway suggest alternative mechanisms of induction of IFN-I that do not require stimulatory immune complexes. In that regard, mutations in genes associated with Aicardi-Goutieres syndrome, such as TREX1, encoding a DNase, or variants in IFIH1, encoding MDA5, an RNA sensor, have been documented in lupus patients and indicate that endogenous cytoplasmic nucleic acids can drive IFN-I production and innate immune system activation (18,19).
Virus-like repetitive elements comprise a high portion of the human genome (roughly 48%). Those sequences originated from retroviruses that integrated into our genome more than 25 million years ago (20). We hypothesized that these virus-like genomic elements might represent an endogenous source of ligands for nucleic acid sensors, induce IFN-I and promote a host microenvironment supportive of immune dysfunction, autoimmunity, and inflammation, similar to the immunopathology characteristic of some chronic virus infections (21,22). Long interspersed nuclear element-1, LINE-1 or L1, is an autonomous family of retroelements that remains active in mammalian genomes. It comprises ~ 17% of genome content, representing about a half-million copies. L1 contains two open reading frames (ORF): ORF1 encoding a 40 kDa RNA binding protein (ORF1/p40) that co-localizes with L1 RNA in cytoplasmic ribonucleoprotein (RNP) particles, and ORF2, encoding a reverse transcriptase and endonuclease (~150 kDa). The majority of L1 inserts are 5’-truncated or mutated and unable to transpose. However a limited number of L1s in the human genome are full-length and capable of generating an RNA transcript with the potential to be translated into proteins. In some cases, an L1 DNA copy is inserted at a new genome location. In view of its potential for genome disruption, L1 expression is stringently regulated, particularly through methylation of CpG motifs in its 5’ regulatory region (23). L1 is predominantly expressed during specific stages of germ cell maturation (24), however recent data document its expression in somatic cells, including endothelial cells of human male gonads, in normal human brain, and in synovial tissue from patients with rheumatoid arthritis (24–27). We hypothesized that inappropriate L1 expression might trigger an innate immune response characterized by production of IFN-I that could promote immune dysfunction. Altered regulation of genome methylation, inadequate expression of viral restriction elements or genetic variants in regulators of nucleic acid integrity or metabolism might represent potential mechanisms leading to L1 transcript expression.
To test our hypothesis, we studied mRNA and protein expression of L1 and IFN-I in affected tissues from patients with SLE, SS, or sicca syndrome in the setting of an IFN-I-associated autoimmune disease. Our data demonstrate increased L1 in kidney tissue from patients with class IV lupus nephritis and salivary gland tissue from patients with SS or other autoimmune disorders that have been associated with elevated IFN-I and implicate L1 RNA-mediated activation of pattern recognition innate immune pathways in the induction of IFN-I. Our data support relative hypomethylation of the L1 promoter region as one potential mechanism that might account for impaired regulation of L1 in these diseases.
Patients and Methods
Patients
Salivary gland biopsies
MSG biopsies were obtained from 31 patients with SS (mean age±SD: 53±14.1), 12 patients presenting with sicca features and an IFN-I-associated autoimmune disease, AD, and 5 non-autoimmune sicca controls, SC (mean age±SD: 48±12.1). SS patients were diagnosed according to the revised American-European Consensus Group Criteria (28). Patients and controls had undergone biopsy of labial MSGs between May 2002 and May 2013 at the Department of Pathophysiology of the School of Medicine, University of Athens, Athens, Greece, as part of the routine diagnostic evaluation for SS. All patients and controls were females between the ages of 15 and 72 at the time of biopsy and provided informed consent. Sicca patients characterized as IFN-I-associated autoimmune patients (AD) included 3 patients with SLE, 4 with Hashimoto thyroiditis, 3 with primary biliary cirrhosis, 1 with morphea and 1 with a positive titer of antinuclear antibodies.
Kidney biopsies
Kidney biopsies from 24 patients with lupus nephritis were obtained from the Department of Pathology at New York Presbyterian Hospital, New York, NY and were classified according to the International Society of Nephrology ISN/RPS 2003 classification criteria (6 class III, 14 class IV, 4 class V) (29). All but two patients were females between the ages of 11 and 55 at the time of biopsy, fulfilled the American College of Rheumatology classification criteria for SLE (30) and provided informed consent. In addition, control renal tissue was obtained commercially from Ambion and from 2 patients who presented with proteinuria/hematuria and whose renal biopsies showed no pathology (male individuals ages 8 and 12 respectively). The study was approved by the Institutional Review Board of Hospital for Special Surgery.
Methods
See Supplemental Methods for detailed description
Cell and tissue preparation and gene expression analysis
RNA was extracted from kidney and MSG tissue and L1, IFN-I and IFIG expression determined by real time-PCR using a Bio-Rad iCycler system with iTaq SYBR Green supermix (Bio-Rad). Primers used for PCR detection are listed in Supplemental Table 1. Data are expressed as relative expression compared to a housekeeping gene control.
Immunohistochemistry and western blot analysis
Expression of L1 ORF-1/p40 protein, IFNα and IFNβ and BDCA-2 were examined in serial sections of MSG and renal tissues. Incubation with mouse anti-IFNα (Chemicon), anti-IFNβ (GeneTex) or anti-BDCA-2 (Pierce) monoclonal antibody, rabbit anti-ORF1/p40 antibody or concentration-matched isotype control antibodies was performed overnight. Tissue sections were subsequently incubated for 60 min with a biotinylated F(ab’)2 fragment of anti-mouse or anti-rabbit immunoglobulin, followed by horseradish peroxidase (HRP)-conjugated streptavidin and were developed with DAB chromogen to the manufacturer’s specifications (DakoCytomation). The specificity of staining was confirmed using matched isotype control antibodies. Western blot analysis was performed using the anti-ORF1/p40 antibody and anti-GAPDH to document the relative level of expression of ORF1/p40 in MSG tissue from SS patients and SC.
Assessment of L1 promoter methylation at CpG sites
DNA was isolated from SS and SC MSG tissue and methylation analysis of the L1 promoter (GenBank accession number U09116) was performed by bisulfite-PCR Pyrosequencing. DNA (1 µg) was bisulfite modified using EZ DNA Methylation kit™, according to the manufacturer’s instructions (Zymo Research). The degree of methylation was expressed as percentage methylation based on the allele quantification of artificial “C/T” SNPs.
Cell cultures
Healthy donor PBMCs, salivary epithelial and kidney cell lines as well as non-neoplastic salivary epithelial cells were cultured with recombinant human IFN-Is and/or TLR ligands. Induction of L1 and IFN-I-related transcripts was assessed by real-time PCR.
Isolation and stimulation of human pDCs and monocytes
PDCs were isolated from human peripheral blood by MACS-based negative selection (Miltenyi Biotech) and EasySep human pDC enrichment kit (STEMCELL™ Technologies). Human CD14+ monocytes were isolated by negative selection with EasySep human monocyte enrichment kit (STEMCELL™ Technologies). Purity of isolated cells was assessed by flow cytometry. Cells were transfected with plasmid DNA encoding a full-length L1 retroelement or the empty vector (pDONR™221, Invitrogen). Alternatively, cells were transfected with in vitro-transcribed RNA molecules. PDCs and monocytes stimulated with empty vehicles served as the negative control. For some experiments pDCs or monocytes were pre-cultured with inhibitors of TLR7/8, TLR9 or TBK1/IKKε prior to transfection with a plasmid containing an L1 element cloned from human chromosome X (AC002980), in vitro-transcribed L1, U1 or Y3 RNA or control plasmid or RNA. Induction of IFN-I-related genes was assessed by real-time PCR, and IFN-I protein activity in culture supernatants was quantified using the WISH reporter cell assay (2).
Statistics
Two-group comparisons of continuous data were assessed using t-tests or the Mann-Whitney test when data were not normally distributed. Correlation between gene expression data and methylation levels was determined using non-parametric Spearman’s test. For in vitro experiments, data are expressed as means±s.e.m., and mean values of relative expression between unstimulated and stimulated cells were compared using paired two-tailed student t-tests. Differences were considered statistically significant for p<0.05.
Results
Expression of L1 retroelement in affected tissues from systemic autoimmune disease patients
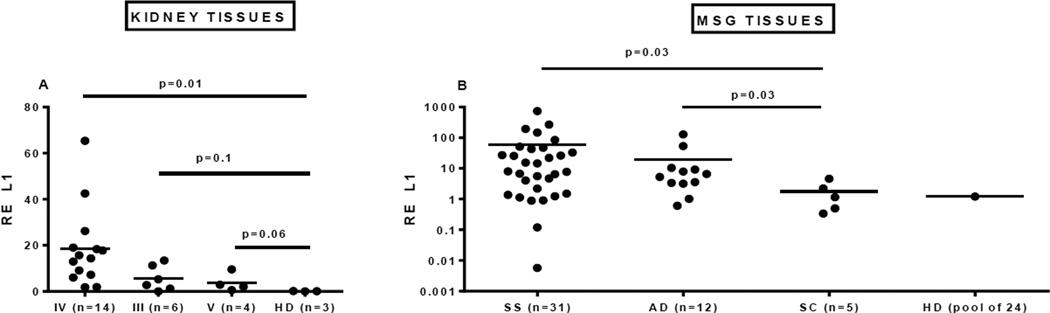
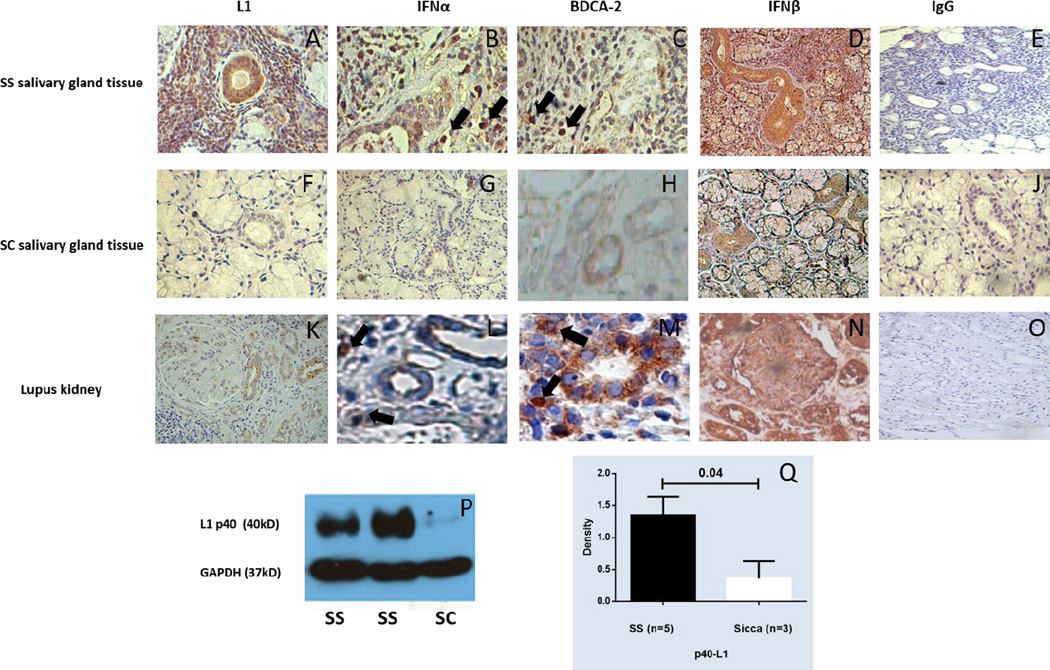
We hypothesized that abnormal expression of L1 retroelements might trigger an innate immune response similar to that induced by exogenous viruses and contribute to activation of the IFN-I pathway in patients with a systemic autoimmune disease. To test this hypothesis we studied expression of L1 transcripts and protein in affected tissues from patients with two related autoimmune diseases, SLE and SS. Kidney tissue from 24 patients with lupus nephritis and MSG tissue from 31 patients with primary SS (not complicated by lymphoma), 12 patients with an IFN-I-associated autoimmune disease (AD), 5 patients with non-autoimmune conditions (SC), and pooled RNA from healthy individuals were analyzed. We found elevated L1 transcripts in renal biopsies from patients with diffuse proliferative lupus glomerulonephritis (class IV) compared to renal tissue from non-autoimmune subjects, with class III and V (membranous) lupus nephritis kidneys showing a similar trend (Fig. 1A). Relative expression of full-length L1 transcripts was also higher in MSG from primary SS patients and those with other autoimmune disorders compared to MSG tissue from non-autoimmune and healthy controls (Fig. 1B). Serial sections of renal and MSG tissues stained with an antibody to ORF1/p40 showed p40 protein in renal tubular cells from SLE kidneys and in MSG ductal epithelial cells and some infiltrating mononuclear cells from SS patients and to a much lesser extent in SC (Fig. 2A, F, K). Ductal epithelial cells also showed striking staining with antibodies specific for IFNβ (Fig. 2D, N) while scattered infiltrating mononuclear cells, many staining with anti-BDCA2 antibody, reflecting pDCs, were stained by the anti-IFNα antibody (Fig. 2B, C, I, M). Expression of ORF1/p40 was confirmed by western blot analysis of protein extracts from MSG tissues, with SS tissues showing increased expression compared to non-autoimmune sicca controls (SC) (Fig. 2P, Q). These results demonstrate that L1 retroelements are transcribed, and at least one of the L1 proteins - ORF1/p40 - is expressed in organs targeted by the immune system in SLE and SS. Moreover, the data demonstrate presence of IFNβ and IFNα in tubular epithelial cells and infiltrating pDCs, respectively, suggesting the possibility that L1 transcript expression might drive IFN-I expression through either TLR-independent or -dependent pathways.
Figure 1. Expression of L1 mRNA in involved tissues from SLE and SS patients.
(A) Total mRNA from kidney tissue was extracted and relative expression of L1 mRNA quantified by RT-PCR, using a primer set that amplifies the 5’UTR of human LRE2. Kidney biopsy tissue was obtained from either healthy individuals (HD, healthy donors) or lupus nephritis patients with different classes (IV, III, V) of renal pathology. (B) Relative expression of L1 mRNA was quantified in human MSG from 31 patients with primary SS; 12 patients with IFN-I-associated autoimmune disease (AD) and sicca symptoms; 5 non-autoimmune sicca controls (SC); and pooled mRNA from MSG of 24 healthy individuals (Clontech laboratories, Inc.) (HD).
Figure 2. Immunohistochemical and western blot detection of L1 in salivary gland tissue and kidney tissue.
(A–O) Representative examples of immunohistochemical data are demonstrated. MSG tissue from a SS patient and a SC patient and renal tissue from a patient with class IV lupus nephritis were stained with antibodies specific for ORF1/p40 (L1), BDCA-2 (a specific marker for pDCs), IFNα, IFNβ or an IgG control. L1 and IFNβ staining were particularly strong in SS salivary ductal epithelial cells and lupus renal tubular epithelial cells (A, K, D, N), while positive IFNα stained cells (representative cells indicated by arrow) coincided with anti-BDCA-2-stained cells (pDCs) (B, C, L, M). (P, Q) Western blot analysis of protein extracts from 5 SS and 3SC MSG samples was performed with rabbit anti-ORF1/p40 and mouse anti-GAPDH antibodies. ORF-1/p40 levels were significantly higher in the SS than the SC samples. Representative gels from 2 SS patients and 1 SC are shown.
L1 promoter methylation levels in SS salivary gland tissues
Since methylation of the CpG-rich promoter of L1 retroelements represents one of the major mechanisms of L1 transcriptional regulation, we investigated whether increased L1 expression in MSG tissues might be associated with reduced L1 methylation. L1 methylation was determined in DNA isolated from MSG tissues of 13 SS patients and 4 SC of comparable age included in our initial cohort. A negative correlation of L1 expression with % L1 methylation was documented for the majority of L1 promoter CpG sites tested, suggesting that augmented demethylation processes, or alternatively impaired remethylation, might account for the observed L1 overexpression in SS MSG tissues (Table 1). Supplemental Figure S1 reveals a trend toward reduced % L1 methylation levels in MSG tissues derived from SS patients compared to the MSG control samples tested (mean±SD: 49.9 ± 5.3 vs 54.9 ± 3.0, p=0.12). These data support a role for altered L1 methylation as a potential mechanism of the observed L1 mRNA transcript expression.
Table 1.
Inverse correlation of L1 mRNA expression with L1 promoter methylation levels in salivary gland tissues from SS patients and sicca controls.
| CpG Promoter Site |
Spearman's rho | |
|---|---|---|
| Methylation levels (%) vs. L1 mRNA expression | ||
| r | p-values | |
| CpG pos #1 | −0.674 | 0.003 |
| CpG pos #2 | −0.617 | 0.008 |
| CpG pos #3 | −0.674 | 0.003 |
| CpG pos #4 | −0.621 | 0.008 |
| CpG pos #5 | −0.278 | 0.41 |
| CpG pos #7 | −0.664 | 0.004 |
| CpG pos #8 | −0.496 | 0.09 |
| CpG pos #9 | −0.462 | 0.112 |
| Mean | −0.598 | 0.011 |
r: Spearman’s correlation rho coefficients and corresponding p-values at several CpG sites are shown for 17 MSG samples. No amplification was detected for CpG positions #6 and #10.
Association of L1 mRNA expression with IFN-I in vivo in SLE and SS
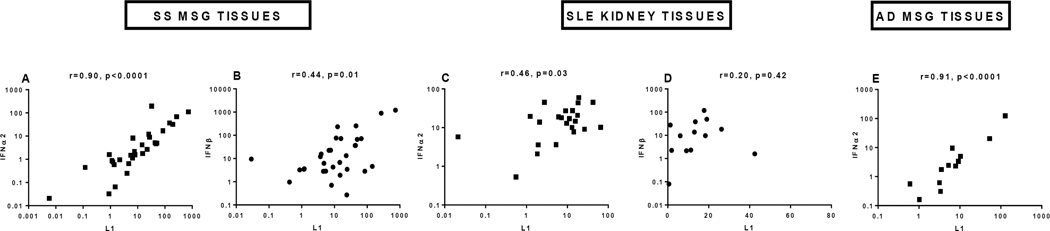
As SLE and SS are characterized by increased production of IFN-I, we explored whether L1 expression might be related to IFN-I synthesis in individual patients. As noted, lupus kidney tissue and SS MSG showed expression of IFN-I protein in a similar distribution as ORF1/p40: in renal tubular cells, MSG ductal epithelial cells, and infiltrating mononuclear cells (Fig. 2). A striking correlation of L1 mRNA expression with IFNα2 was detected in MSG tissue from SS and AD patients (r=0.90, p<0.0001; r=0.91, p<0.0001, respectively) (Fig. 3A and E). Similarly, coordinate expression of L1 and IFNα2 was detected in kidney tissue (r=0.46, p=0.03) from SLE patients (Fig. 3C). Correlation of L1 with IFNβ transcripts was also observed in SS MSG tissues and to a lesser extent in lupus kidney tissues (Fig. 3B and D). Collectively, these results point to a potential functional relationship between L1 transcript expression and IFN-I production in SLE and SS tissues.
Figure 3. Coordinate expression of L1 and type I IFN mRNA in affected tissues from patients with systemic autoimmune disease.
IFNα2 (A, C, E) and IFNβ (B, D) mRNA expression were measured by RT-PCR in MSG tissue from patients with SS (A,B), in lupus nephritis renal tissues (C,D), and in MSG tissue from patients with IFN-I-associated AD and sicca symptoms (E) and related to expression of L1 mRNA. Spearman correlation is shown for L1 with IFNα2 or IFNβ.
Type I IFN and non-specific stimuli do not induce L1 retroelement expression in vitro
Observing strong correlation between L1 and IFN-I expression in affected tissues, we wanted to exclude the possibility of direct induction of L1 retroelements by IFN-I or innate immune stimuli such as TLR ligands. We stimulated PBMC from healthy individuals with various doses of human recombinant IFNα for 6 hours and analyzed expression of IFIGs IFIT1 or IFIT3 and L1 transcripts. IFNα induced IFIT1 in a dose-related fashion while L1 transcripts were not detected (Fig. S2A). To mimic potential responses of salivary gland and kidney tissue, we used a human embryonic kidney cell line (HEK 293F) and a salivary gland epithelial cell line (HSG) as responder cells. Similar to what was observed with PBMC, stimulation of HEK 293F and HSG cells with various doses of IFNα did not lead to increased expression of L1 mRNA (Fig. S2B and C). Thus, L1 transcription was not induced directly by IFNα. We also ruled out several relevant immune activation pathways as likely contributors to induction of L1 in patients with SLE or SS. Neither the TLR9 agonist CpG DNA (dose range 1–10µg/ml) nor TLR7 agonist CL097 (dose range 1–10µg/ml) increased transcription of L1 in PBMC (Fig. S2D and E). Additionally, stimulation of non-neoplastic salivary epithelial cells derived from SS patients with IFNα or IFNβ did not increase L1 transcription after 6 or 24h (Fig. S2F).
L1 can induce IFN-I production in vitro through TLR-dependent and TLR-independent pathways
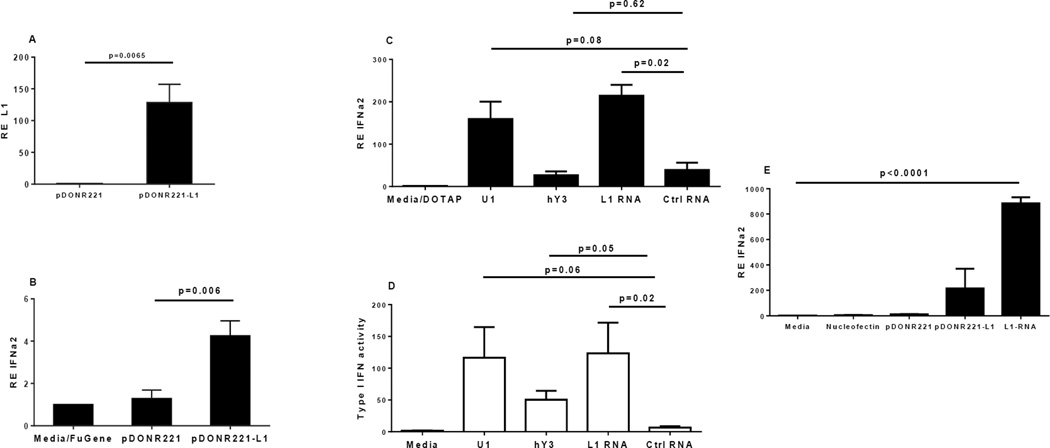
Since IFN-I did not stimulate L1 expression, we investigated whether the strong correlation observed between L1 and IFN-I in SLE and SS tissues could be based on the induction of IFN-I by L1 DNA or RNA. We cloned a full-length L1 retrotransposon together with its natural promoter region from the X chromosome of an SLE patient into a plasmid and then transfected pDC, known to be active producers of IFNα, with either empty or L1-carrying plasmids. L1 transcription from the plasmid (Fig. 4A), as well as modestly increased IFNα2 mRNA (Fig. 4B), were detected in cells transfected with L1-encoding plasmid compared to those receiving an empty vector.
Figure 4. L1 induces IFNα in human plasmacytoid dendritic cells and monocytes.
(A) Expression of L1 mRNA in pDC 6 hours post transfection with an L1-containing plasmid (pDONR221-L1) or empty vector (pDONR221) as measured by RT-PCR. (B) Expression of IFNα2 by pDC 6 hours post-transfection with an L1-containing or control plasmid. (C) Human pDC were transfected directly with a purified L1 RNA fragment, with U1 or hY3 RNA, or with unrelated control RNA using DOTAP transfection reagent. Relative expression of IFNα2 was determined after 6 hours. (D) IFN-I protein activity was assessed in supernatants from the experiments shown in (C) using a reporter cell (WISH) assay. Relative expression of interferon inducible gene MX1 in WISH cells was measured by RT-PCR after 4 hours of incubation with 50% supernatants. (E) Expression of IFNα2 mRNA in human CD14+ monocytes 6 hours after electroporation of an L1-containing or control plasmid or purified L1 RNA.
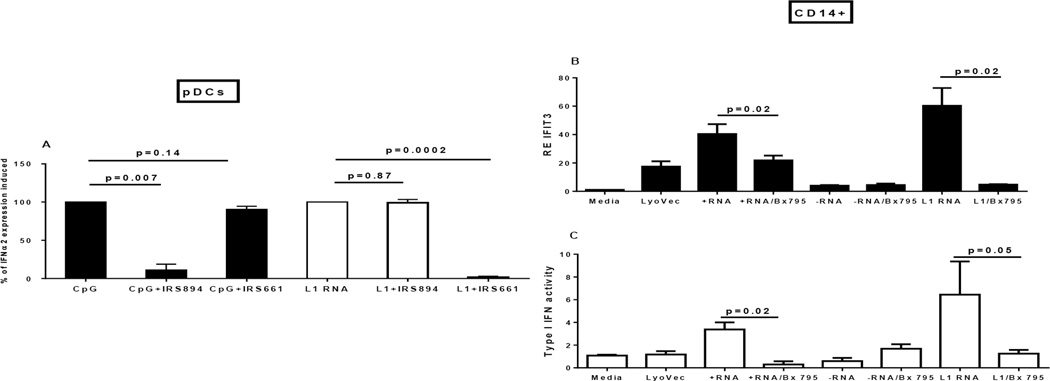
We next generated 5’UTR fragments of L1 mRNA, as 1) we would expect those to be present only in the context of transcription of full-length L1 retroelements and 2) they potentially possess immune stimulatory activity due to their capacity to form secondary structures. We also obtained RNA derived from two classic targets of autoimmunity in SLE and related syndromes, human Y3 RNA (associated with the Ro RNP antigen) and U1 RNA (associated with the Sm/RNP antigen), RNAs with distinct secondary structures. Transfection of human pDC with either L1 or U1 RNA, but not hY3 or an unrelated RNA control, caused robust induction of IFNα mRNA, and IFN-I protein activity (2) was also confirmed in supernatants from the same cultures (Fig. 4C and D). Elevated IFNa2 in the presence of L1 RNA was also observed after electroporating freshly isolated human CD14+ cells with either an L1-containing plasmid or L1 RNA (Fig. 4E). These findings show that L1 and U1 RNA can initiate effectively IFN-I production in human pDCs and monocytes, suggesting their potential role in promoting IFN-related autoimmune diseases.
In order to gain insight into the mechanism of IFN-I induction by L1 RNA we used specific inhibitors of two RNA detection pathways: IRS 661, which inhibits endosomal TLR7 and TLR8, and Bx795, an inhibitor of TBK1/IKKε, kinases that are activated downstream of cytoplasmic nucleic acid sensors (including RIG-I and MDA5). Incubation of pDC with the TLR inhibitor or monocytes with the TBK1/IKKε inhibitor significantly reduced IFN-I induction by L1 RNA (Fig. 5A–C), pointing to the capacity of L1 RNA to signal through both TLR-dependent and TLR-independent pathways.
Figure 5. Inhibition of TLR7/8 and TBK1/IKKε pathways.
(A) Human pDC were pre-treated for 1 hour with either TLR9 inhibitor IRS 894 or TLR7/8 inhibitor IRS 661 followed by stimulation for 4 hours with either TLR9 ligand CpG or with L1 RNA. Relative expression of IFNα2 was measured by RT-PCR, and the percent of the level induced by either CpG or L1 RNA is shown. (B) Human CD14+ monocytes were pre-treated for 30 min with TBK1/IKKε inhibitor Bx795 that blocks signaling downstream of the RIG-I-like receptor (RLR) pathway followed by transfection with either positive or negative RNA controls (+RNA/−RNA) or L1 RNA. After 4 hours of incubation relative expression of interferon inducible gene IFIT3 was measured by RT-PCR. (C) Supernatants from the CD14+ cells cultured as shown in (B) were incubated with reporter (WISH) cells to assess IFN-I protein activity. The level of interferon inducible gene MX1 in WISH cells after 4 hours of stimulation with 50% supernatants is shown.
Discussion
We hypothesized that transcriptionally active L1 elements might provide an innate immune trigger similar to that induced by exogenous viruses and contribute to activation of the IFN-I pathway in patients with systemic autoimmune diseases. To pursue this possibility, we studied affected tissues from patients with two related autoimmune diseases, SLE and SS, both characterized by an IFN-I signature. We found that L1 mRNA and L1 ORF1/p40 protein were expressed in renal and MSG tissues from patients with lupus nephritis or primary SS. Similarly, L1 transcripts were detected in MSG tissue from patients with autoimmune diseases that are associated with increased IFN-I. As methylation of CpG sites in the L1 promoter was negatively correlated with L1 expression in MSG tissues, alterations in mechanisms that either demethylate or restore methylation might contribute to the observed L1 overexpression. Moreover, L1 transcript expression strongly correlated with both IFNβ and IFNα expression in the same tissues. While we cannot definitively attribute IFNα or IFNβ expression to a particular cell type, a striking co-localization of L1 ORF1/p40 protein and IFNβ was notable in MSG ductal and renal tubular epithelial cells. After excluding L1 induction by IFNα or -β as well as several non-specific innate immune stimuli, we demonstrated that transfection of pDCs or monocytes with an L1 carrying plasmid or L1 RNA induced IFN-I production by those cells. Additionally, incubation of pDC with an inhibitor of TLR7/8, but not TLR9, and inhibition of RIG-I and MDA5 signaling in monocytes significantly reduced IFN-I induced by L1, pointing to L1 RNA as a stimulus for IFN-I through both TLR-dependent and TLR-independent mechanisms.
A potential role for virus infection in the pathogenesis of autoimmune disease has long been an attractive hypothesis, in part based on clinical manifestations that mimic acute or chronic virus infection (31, 32). Although production of IFN-I is a key component of anti-viral host defense, with the possible exception of Epstein Barr virus (33) there are scant data to directly implicate an exogenous virus in the IFN-I pathway activation observed in many lupus patients. Attention has turned to endogenous triggers of immune activation (21, 34), and a significant advance relates to the role of self-nucleic acid-containing immune complexes as stimuli for endosomal TLRs and IFN-I production (35, 36). Additional support for nucleic acids as inducers of IFN-I production comes from investigation of rare clinical syndromes, such as Aicardi-Goutieres syndrome, characterized by elevated IFN-I, lupus-type autoantibodies, skin rash, and neurologic manifestations (18, 37–39). The molecular basis of these syndromes is informed by mutations in enzymes that cleave or modify DNA or RNA that gains access to the cytoplasm or is otherwise potentially immunostimulatory (18, 37–39). Among these, TREX1 mutations have been extensively studied, and it is proposed that the nucleic acid targets of the TREX1-encoded DNase are enriched in genomic retrotransposon elements (40). Together with our data reported here, the association of mutations in TREX1, RNASEH2, SAMHD1, ADAR1 and IFIH1 draw attention to the potential pathogenic role of cytoplasmic nucleic acids and genomic viral-like elements in human disease (18, 21, 37–40).
Several characteristics of L1 raised our interest in investigating those elements as candidate triggers of an innate immune response and autoimmunity (20, 21). First, L1 uses both RNA and DNA intermediates for retrotransposition, with each present in forms that could be deleterious to the host. Single-stranded DNA, generated in the process of retrotransposition, is atypical in eukaryotic cells, and bicistronic forms of full-length L1 mRNA have been described. Second, there is differential regulation of L1 expression during meiosis in females and males, suggesting mechanisms relevant to the female gender bias in systemic autoimmune diseases (41). Third, accumulation of L1 in lymphoid organs of lupus-prone mice and in inflamed RA synovium points to a potential direct role of this retroelement in the induction of aberrant immune responses (27, 42). Fourth, plasma DNA from lupus patients is enriched in repetitive sequences and is characterized by high levels of CpG-dinucleotides sharing homologies with human retroviruses (43, 44). Additionally, recent data demonstrate strong similarities between the gene expression profile of HIV-1-infected CD4+ T cells, including notable representation of L1 and human endogenous retroviral transcripts, and the gene expression profile of lupus T cells, further supporting investigation of a role for L1 in the immunopathology of systemic autoimmune diseases (45).
Altered epigenetic regulation of gene expression in association with autoimmunity also draws attention to a potential role for L1. Methylation is regarded as the predominant mechanism suppressing expression of L1 retroelements under physiological conditions, and changes in methylation status have been increasingly appreciated in association with autoimmune disorders such as SLE (46, 47). The inverse correlation between L1 expression and L1 promoter methylation levels in MSG tissues in our study suggests epigenetic alterations as significant underlying contributors to the inappropriate overexpression of L1 endogenous retroelements in systemic autoimmune disorders.
Since L1 RNA possesses characteristics of viral RNA and is present in the cytoplasm and as RNP particles during its life cycle, we hypothesized that it might be recognized by RNA sensors and potentiate an innate immune response. The RNA component of some RNPs acts as an endogenous adjuvant and plays a role in the pathogenesis of SLE (35,36,48), and autoantibodies targeting RNA-containing particles are strongly associated with expression of IFIGs in lupus PBMC (49). Among the pattern recognition receptors that initiate immune responses, TLRs recognize nucleic acids – double stranded (ds)RNA (TLR3), ssRNA (TLR7/8) and unmethylated CpG-rich DNA (TLR9) – and are localized in endosomal compartments. Another class of nucleic acid receptors - the RIG-I-like receptors – are located in the cytoplasm and recognize modified ds- and ssRNA and DNA. While demethylated L1 DNA fragments of apoptotic cells in vivo or a plasmid in vitro (our experiment) might signal through TLR9 or cytoplasmic DNA sensors and are relevant candidate stimuli for IFN-I, we emphasize the role of the L1 RNA transcript in IFN-I induction. L1 mRNA synthesis precedes formation of reverse-transcribed L1 ssDNA. Additionally, while reverse transcription occurs in the nucleus (it requires a primer in the form of a 3'-hydroxyl generated by ORF2 nicks in the host genome), L1 mRNA is localized to the cytoplasm where it can be recognized by cytosolic RNA sensors or form stable RNA-protein complexes that could be exposed to the immune system after apoptosis or necrosis. In view of these features, we hypothesized that L1 ssRNA stimulates IFN-I production, either through TLR7 or cytosolic RNA sensors, or both. While delivery to TLRs of L1 RNA in RNP-containing immune complexes is one potential mechanism of induction of IFN-I, we highlight recognition of L1 RNA mediated by cytosolic helicases, such as RIG-I and MDA5. Staining data from SS MSG and lupus kidney tissue suggest co-localization of L1 p40 protein and IFNβ, a major target of signaling initiated by cytoplasmic nucleic acids, and we observed the strongest induction of IFN-I by in vitro-transcribed L1 RNA. Thus we propose that L1 RNA can trigger an innate immune response through RNA-sensing receptors, resulting in significant synthesis of IFN-I and the protean immunologic alterations attributable to IFN-I.
A potential role for retroviruses or retroviral-like elements as pathogenic agents in autoimmune diseases has been a focus of interest over several decades (10, 20, 21, 34, 40, 43, 44). While limited by the availability of sufficient tissue for full analysis of the cell populations expressing L1 transcripts or protein and the precise mechanisms through which L1 transcripts interact with and stimulate nucleic acid sensors, our data support the hypothesis that L1, perhaps along with other virus-derived genomic elements, might be a common denominator in autoimmune disorders characterized by activation of the IFN-I pathway. Investigation of the genetic and environmental factors that alter the usual stringent regulation of L1 elements and increase availability of these endogenous immunostimulatory factors should suggest new avenues for therapeutic intervention in these diseases. More than thirty years ago, Barbara McClintock viewed the activation of endogenous transposons as a genomic defense mechanism against potential environmental threats, contributing to genetic diversity and the possibility of an effective adaptation (50). In this context, we speculate that autoimmune disease is the price we pay for the genomic flexibility that permits adaptation and survival of our species.
Supplementary Material
Acknowledgments
Supported by a Stavros Niarchos Fellowship grant through the Arthritis Foundation, New York Chapter, to CPM; NIH T32 training grants AI007621and AR007517 to IS; a Stavros Niarchos grant to Department of Physiology, Medica; and NIH R01AI059893, R21AR050673, a Novel Research Grant from the Lupus Research Institute, a Target Identification in Lupus grant from the Alliance for Lupus Research, and the Mary Kirkland Center for Lupus Research to MKC.
In the past twelve months Dr. Crow has served as a consultant to AstraZeneca, Bristol-Myers Squibb, Eli Lilly and Company, GSK, Neovacs, and Novartis related to development of lupus therapeutics.
We would like to acknowledge Evi Poulou, B.Sc, and Effie Papageorgiou, PhD, for assistance in immunohistochemical stainings and western blot analysis.
References
- 1.Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol. 2014;192:5459–5468. doi: 10.4049/jimmunol.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54:1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 3.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow MK, Wohlgemuth J. Microarray analysis of gene expression in lupus. Arthritis Res Ther. 2003;5:279–287. doi: 10.1186/ar1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 7.Bave U, Nordmark G, Lovgren T, Ronnelid J, Cajander S, Eloranta ML, et al. Activation of the type I interferon system in primary Sjogren's syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52:1185–1195. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 8.Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjogren's syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–1544. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 9.Mavragani CP, Niewold TB, Moutsopoulos NM, Pillemer SR, Wahl SM, Crow MK. Augmented interferon-alpha pathway activation in patients with Sjogren's syndrome treated with etanercept. Arthritis Rheum. 2007;56:3995–4004. doi: 10.1002/art.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavragani CP, Crow MK. Activation of the type I interferon pathway in primary Sjogren's syndrome. J Autoimmun. 2010;35:225–231. doi: 10.1016/j.jaut.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren's syndrome. Proc Natl Acad Sci U S A. 2006;103:2770–2775. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall JC, Baer AN, Shah AA, Criswell LA, Shiboski CH, Rosen A, et al. Molecular subsetting of interferon pathways in Sjogren's Syndrome. Arthritis Rheumatol. 2015;67:2437–2446. doi: 10.1002/art.39204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brkic Z, Maria NI, van Helden-Meeuwsen CG, van de Merwe JP, van Daele PL, Dalm VA, et al. Prevalence of interferon type I signature in CD14 monocytes of patients with Sjogren's syndrome and association with disease activity and BAFF gene expression. Ann Rheum Dis. 2013;72:728–735. doi: 10.1136/annrheumdis-2012-201381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nezos A, Gravani F, Tassidou A, Kapsogeorgou EK, Voulgarelis M, Koutsilieris M, et al. Type I and II interferon signatures in Sjogren's syndrome pathogenesis: Contributions in distinct clinical phenotypes and Sjogren's related lymphomagenesis. J Autoimmun. 2015;63:47–58. doi: 10.1016/j.jaut.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57:664–678. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- 17.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, et al. Mutations in the gene encoding the 3'–5' DNA exonuclease TREX1 are associated with systemic lupus erythematosis. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 19.Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crow MK. Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity. 2010;43:7–16. doi: 10.3109/08916930903374865. [DOI] [PubMed] [Google Scholar]
- 22.Crow MK, Olferiev M, Kirou KA. Targeting of type I interferon in systemic autoimmune diseases. Transl Res. 2015;165:296–305. doi: 10.1016/j.trsl.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 24.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 25.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–3922. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhart M, Rethage J, Kuchen S, Kunzler P, Crowl RM, Billingham ME, et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 2000;43:2634–2647. doi: 10.1002/1529-0131(200012)43:12<2634::AID-ANR3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 28.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61:554–558. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weening JJ, D'Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol. 2004;15:241–250. doi: 10.1097/01.asn.0000108969.21691.5d. [DOI] [PubMed] [Google Scholar]
- 30.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 31.Phillips PE, Christian CL. Myxovirus antibody increases in human connective tissue disease. Science. 1970;168:982–984. doi: 10.1126/science.168.3934.982. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz RS. Viruses and systemic lupus erythematosus. N Engl J Med. 1975;295:132–136. doi: 10.1056/NEJM197507172930308. [DOI] [PubMed] [Google Scholar]
- 33.James JA, Neas BR, Moser KL, Hall T, Bruner GR, Sestak AL, et al. Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum. 2001;44:1122–1126. doi: 10.1002/1529-0131(200105)44:5<1122::AID-ANR193>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Perl A, Fernandez D, Telarico T, Phillips PE. Endogenous retroviral pathogenesis in lupus. Curr Opin Rheumatol. 2010;22:483–492. doi: 10.1097/BOR.0b013e32833c6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovgren T, Eloranta ML, Kastner B, Wahren-Herlenius M, Alm GV, Ronnblom L. Induction of interferon-alpha by immune complexes or liposomes containing systemic lupus erythematosus autoantigen- and Sjogren's syndrome autoantigen-associated RNA. Arthritis Rheum. 2006;54:1917–1927. doi: 10.1002/art.21893. [DOI] [PubMed] [Google Scholar]
- 37.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 38.Fye JM, Orebaugh CD, Coffin SR, Hollis T, Perrino FW. Dominant mutation of the TREX1 exonuclease gene in lupus and Aicardi-Goutieres syndrome. J Biol Chem. 2011;286:32373–32382. doi: 10.1074/jbc.M111.276287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 42.Benihoud K, Bonardelle D, Soual-Hoebeke E, Durand-Gasselin I, Emilie D, Kiger N, et al. Unusual expression of LINE-1 transposable element in the MRL autoimmune lymphoproliferative syndrome-prone strain. Oncogene. 2002;21:5593–5600. doi: 10.1038/sj.onc.1205730. [DOI] [PubMed] [Google Scholar]
- 43.Li JZ, Steinman CR. Plasma DNA in systemic lupus erythematosus. Characterization of cloned base sequences. Arthritis Rheum. 1989;32:726–733. doi: 10.1002/anr.1780320610. [DOI] [PubMed] [Google Scholar]
- 44.Krapf FE, Herrmann M, Leitmann W, Kalden JR. Are retroviruses involved in the pathogenesis of SLE? Evidence demonstrated by molecular analysis of nucleic acids from SLE patients' plasma. Rheumatol Int. 1989;9:115–121. doi: 10.1007/BF00271867. [DOI] [PubMed] [Google Scholar]
- 45.Sherrill-Mix S, Ocwieja KE, Bushman FD. Gene activity in primary T cells infected with HIV89.6: intron retention and induction of genomic repeats. Retrovirology. 2015;12:79. doi: 10.1186/s12977-015-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, et al. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altorok N, Sawalha AH. Epigenetics in the pathogenesis of systemic lupus erythematosus. Curr Opin Rheumatol. 2013;25:569–576. doi: 10.1097/BOR.0b013e328364206f. [DOI] [PubMed] [Google Scholar]
- 48.Koh YT, Scatizzi JC, Gahan JD, Lawson BR, Baccala R, Pollard KM, et al. Role of nucleic acid-sensing TLRs in diverse autoantibody specificities and anti-nuclear antibody-producing B cells. J Immunol. 2013;190:4982–4990. doi: 10.4049/jimmunol.1202986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 50.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.