Abstract
Mutations in MECP2 cause the neurodevelopmental disorder Rett syndrome (RTT). The RTT missense MECP2R306C mutation prevents MeCP2 interaction with NCoR/Histone deacetylase 3 (HDAC3); however, the neuronal function of HDAC3 is incompletely understood. We report that neuronal deletion of Hdac3 in mice elicits abnormal locomotor coordination, sociability, and cognition. Transcriptional and chromatin profiling revealed HDAC3 positively regulates a subset of genes and is recruited to active gene promoters via MeCP2. HDAC3-associated promoters are enriched for the FOXO transcription factors, and FOXO acetylation is elevated in Hdac3 KO and Mecp2 KO neurons. Human RTT patient-derived MECP2R306C neural progenitor cells have deficits in HDAC3 and FOXO recruitment and gene expression. Gene editing of MECP2R306C cells to generate isogenic controls rescued HDAC3-FOXO-mediated impairments in gene expression. Our data suggests that HDAC3 interaction with MeCP2 positively regulates a subset of neuronal genes through FOXO deacetylation, and disruption of HDAC3 contributes to cognitive and social impairment.
RTT is a neurodevelopmental disorder that leads to impaired motor and intellectual abilities, hand stereotypies and is often associated with autistic features1. Initial studies indicate that MeCP2 negatively regulates transcription through binding to methylated DNA and recruiting HDAC complexes2,3. However, transcriptional and epigenomic analyses in both mouse brain and human-embryonic-stem-cell-derived (hESC) neurons suggest that MeCP2 may act as a transcriptional activator for a subset of genes4-8. Likewise, emerging evidence indicates that HDACs may regulate dynamic changes in transcription9-12. MeCP2 recruits the Sin3a complex containing HDAC1 and HDAC2, and the NCoR complex with HDAC3. However, the role of individual HDACs in RTT pathology is unknown, which prompted us to investigate HDAC function in RTT. Recent studies show that a cluster of single amino acid missense RTT-causative MECP2 mutations abolish the interaction of MeCP2 with NCoR/HDAC3, whereas binding with the Sin3a complex is unaffected13,14. As HDAC3 is the major enzymatic component of the NCoR complex, this led us to explore the role of HDAC3 in RTT pathology and whether this could provide clarity for the function of MeCP2 in regulating transcription.
Results
Neuronal loss of HDAC3 leads to abnormal locomotor behavior
To investigate whether loss of HDAC3 models RTT-associated behaviors, and to avoid embryonic lethality that occurs following deletion of Hdac3 during development15, we used Hdac3 conditional knockout mice (Hdac3 cKO) that lack Hdac3 expression in forebrain excitatory neurons. Hdac3 cKO mice were generated by crossing Hdac3 flox mice (Hdac3f/f)16 with transgenic αCamKII promoter-driven Cre (CW2) mice17. Neuronal loss of HDAC3 was confirmed by immunostaining in the hippocampus (Supplementary Fig. 1a-d), and by western blot analysis in the hippocampus and the cortex. HDAC3 expression is maintained in the striatum as previously described for the CW2-cre line (Supplementary Fig. 1e-j)17. To assess locomotor activity, Hdac3 cKO mice were compared to control mice in the open field arena and exhibited hyperactivity (Supplementary Fig. 2a-e) and abnormal exploratory behavior (Fig. 1a and Supplementary Fig. 2f). Mecp2 cKO mice generated using the CamKII-Cre93 line have MeCP2 deleted in the forebrain including the striatum and exhibited modest hypolocomotor activity18. Striatal expression of MeCP2 has been shown to regulate locomotor activity19. Normal striatal expression of HDAC3 in the CW2-Cre driven Hdac3 cKO mice (Supplementary Fig. 1i, j) suggests that differences in locomotor activity may be due to regional specificity of the CamKII-Cre lines. Hdac3 cKO mice also displayed impaired motor coordination as assessed by accelerating rota-rod (Fig. 1b), which has been observed in MeCP2 loss-of-function models including the forebrain-specific Mecp2 cKO mice18. In addition to locomotor coordination deficits, Hdac3 cKO mice exhibit hind limb clasping (Fig. 1c). Stereotypic hand-wringing behavior in RTT patients is thought to resemble hind limb clasping in Mecp2 KO mice20. A severe hind limb paralysis was observed in neuron-specific Hdac3 cKO mice, supporting our observation of locomotor impairments15.
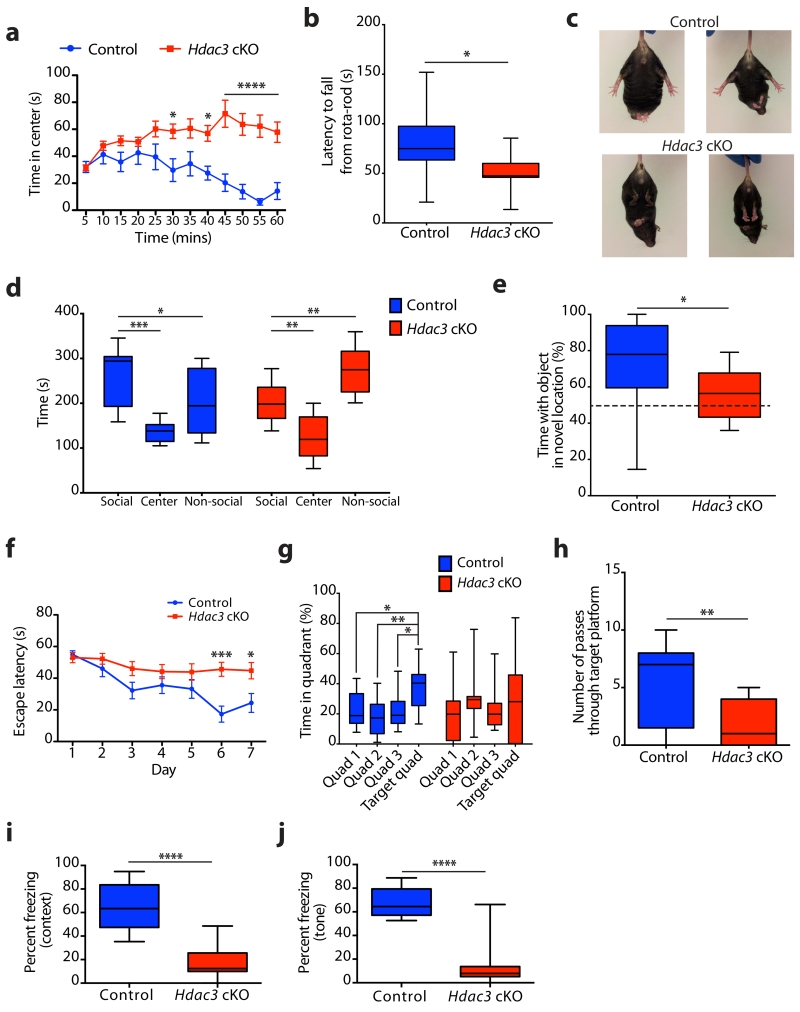
Figure 1. Hdac3 cKO mice exhibit social and cognitive impairments similar to models of RTT.
(a) Control and Hdac3 cKO mice were subjected to the open-field test for 60 minutes and analyzed in 5 minute bins for time spent in the center. Two-way ANOVA, genotype: F(1,44) = 24.82, **** P < 0.0001; time: F(11, 484) = 2.131, * P = 0.0171; Bonferroni post hoc; 30 min bin, * P = 0.0256; 40 min bin, * P = 0.0203; 45 min, 50 min, 55 min, 60 min bins, **** P < 0.0001; n = 22, 24 mice. (b) Latency to fall from an accelerating rota-rod (s), average of two trials per mouse for control and Hdac3 cKO mice; two-tailed t test; t(20) = 2.527; * P = 0.0201; n = 11, 11 mice. (c) Hind limb clasping phenotype was observed in Hdac3 cKO at 12 weeks of age, but not in controls. (d) Time spent in each chamber of the sociability arena (s) during 10 minutes with a novel mouse confined to one chamber (social) for control and Hdac3 cKO mice. Control; one-way ANOVA, F(2,24) = 11.10, *** P = 0.0004; Bonferroni post hoc, social versus non-social, * P = 0.0441; social versus center, *** P = 0.0002; n = 9, 9 mice. Hdac3 cKO; one-way ANOVA, F(2,24) = 21.65, **** P < 0.0001; Bonferroni post hoc, social versus non-social, ** P = 0.0065; social versus center, ** P = 0.0058; n = 9 mice. (e) Control and Hdac3 cKO mice were assessed for object location memory as the percent time spent with the object in the novel location versus time spent with the object in the familiar location; two-tailed t test; t(26) = 2.374; * P = 0.0253; n = 16, 12 mice. (f) The escape latency (s) to reach the hidden platform during the training days of the MWM was scored for control and Hdac3 cKO mice. Two-way ANOVA; genotype, F(1,133) = 23.65, **** P < 0.0001; day, F(6,133) = 5.624, **** P < 0.0001, Bonferroni post hoc; day 1, P > 0.99; day 2, P > 0.99; day 3, P = 0.2874; day 4, P > 0.99; day 5, P = 0.7824; day 6, *** P = 0.0003; day 7, * P = 0.0197; n = 10, 11 mice. (g) During the probe trial (day 8), the percent time spent in the target quadrant, and the 3 control quadrants was scored. Control; one-way ANOVA, F(3,36) = 4.789, ** P = 0.0066; Bonferroni post hoc; quadrant 1 (Q1) versus target quadrant (T), * P = 0.0325; Q2 versus T, ** P = 0.0037; Q3 versus T, * P 0.0225; n = 10 mice. Hdac3 cKO; one-way ANOVA, F(3,40) = 0.6259, P = 0.6025; Bonferroni post hoc; Q1 versus T, P > 0.99; Q2 versus T, P > 0.99; Q3 versus T, P > 0.99; n = 11 mice. (h) During the probe trial (day 8) the number of passes through the location where the hidden platform was positioned during the training (days 1 – 7) was calculated for control and Hdac3 cKO mice; two-tailed t test; t(19) = 3.056, ** P = 0.0065; n = 10, 11 mice. (i) Contextual fear-conditioned memory scored as percent time spent freezing during a 3 minute exposure to the context; two-tailed t test; t(20) = 6.719, **** P < 0.0001; n = 11, 11 mice. (j) Cued fear-conditioned memory scored as percent time spent freezing during a 3 minute exposure to the tone presented in a novel context; two-tailed t test; t(20) = 7.959, **** P < 0.0001; n = 11, 11 mice. a, f report mean value ± s.e.m. b, d, e, g - j, report median, 25th and 75th percentile, min and max value.
Hdac3 cKO mice exhibit social and cognitive deficits
Mice with Mecp2 loss-of-function mutations are suggested to model RTT and display impaired sociability and cognition21-24. Sociability of Hdac3 cKO mice was tested using a three-chamber arena, whereby an initial habituation to an empty arena is followed by exposure to an unfamiliar mouse restricted to one of the lateral chambers. Hdac3 cKO mice displayed similar behavior to controls during the habituation phase (Supplementary Fig. 3a). Upon exposure to an unfamiliar mouse, control mice spent more time in the chamber with the social stimulus (Fig. 1d). However, Hdac3 cKO mice spent more time in the non-social chamber indicating aberrant sociability (Fig. 1d).
To test whether loss of HDAC3 affects cognition we assessed object location memory (OLM), a test for hippocampal-dependent episodic memory based on the premise that rodents will preferentially explore a familiar object that moved to a new location. Training of mice to the position of two identical objects, and later repositioning only one object, resulted in control mice spending more time at the novel location (Fig. 1e). However, Hdac3 cKO mice spent a comparable amount of time at both the familiar and novel location, indicating a deficit in OLM (Fig. 1e). The total time spent with both objects was higher for Hdac3 cKO mice (Supplementary Fig. 3b), reflecting hyperactivity in these mice; and indicate that sociability deficits of Hdac3 cKO mice are not due to diminished exploratory behavior.
The Morris water maze (MWM), a spatial learning task, requires mice to locate a hidden platform in an opaque pool of water using visual cues. Despite the hyperactivity of Hdac3 cKO mice, the swim speed was similar to controls during training days 1-7 (Supplementary Fig. 3c), as observed previously for hyperactive mice in swim tasks25. Acquisition of spatial learning in control mice was observed as reduced latency to reach the hidden platform by days 6 and 7, which did not occur in Hdac3 cKO mice (Fig. 1f). To assess reference memory, a probe trial was performed 24 h after the last training session (day 8) during which the platform was removed. As expected, Hdac3 cKO mice have diminished memory recall, as indicated by reduced time spent in the target quadrant (Fig. 1g), and a low number of passes through the platform location (Fig. 1h).
To further evaluate hippocampal-dependent learning a fear-conditioning paradigm was performed, whereby an auditory cue was paired with a mild aversive foot shock. Both control and Hdac3 cKO mice displayed a similar aversive reaction to the foot shock (Supplementary Fig. 3d). Contextual memory recall to the conditioning chamber as measured by freezing behavior was impaired in Hdac3 cKO mice (mean, 17.16 % ± 4.219) compared to controls (mean, 65.95 % ± 5.908) (Fig. 1i). Likewise, cued memory recall to the auditory tone was impaired in Hdac3 cKO mice (mean, 13.62 % ± 5.581) compared to controls (mean, 66.43 % ± 3.589) (Fig. 1j). Activity suppression is an alternative indicator of fear used for hyperactive mice26, which compares activity during training (before the shock) with activity during testing. Activity suppression is calculated as a ratio; values below 0.5 indicate a fear response and values at 0.5 indicate no fear. Activity suppression in control mice shows a robust fear response during contextual (mean, 0.2899 ± 0.0416) and cued (mean, 0.3131 ± 0.0289) memory recall (Supplementary Fig. 3e, f). Similar to freezing behavior, the activity suppression was impaired in the Hdac3 cKO for contextual (mean, 0.4615 ± 0.0117) and cued (mean, 0.5417 ± 0.0346) memory recall (Supplementary Fig. 3e, f). Collectively, behaviors observed in Hdac3 cKO mice resemble a number of phenotypes observed in forebrain-specific Mecp2 cKO mice, including hind limb clasping, and abnormal motor-coordination, sociability, and cue-dependent memory18.
HDAC3 positively regulates transcription for a subset of genes
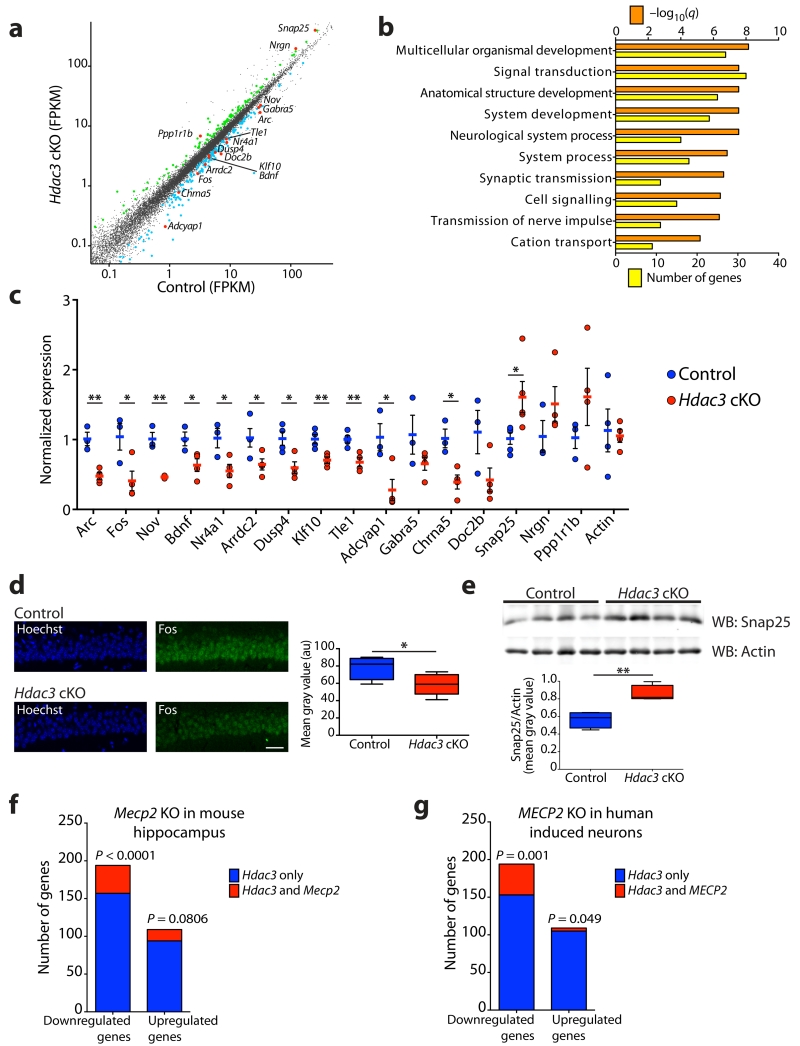
Transcriptional analysis of MeCP2 loss-of-function in the mouse brain and hESC-derived neurons suggests that MeCP2 is a dynamic regulator of transcription5-7, however, gene expression profiling following HDAC3 loss-of-function in the brain remains to be explored. To assess the transcriptional consequence of HDAC3 loss-of-function we performed RNA-seq in the CA1 area of the hippocampus, a region enriched for neurons with deletion of Hdac3 in the Hdac3 cKO mice (Supplementary Fig. 1a, b). Successful excision of exons 11 - 14 in Hdac3 cKO mice was demonstrated by Reverse Transcriptase (RT)-PCR and quantitative (q)RT-PCR analysis, and quantification of exon-specific read number of the Hdac3 gene locus following RNA-seq (Supplementary Fig. 4a-d). Subsequent RNA-seq analysis revealed that 303 genes are differentially regulated in Hdac3 cKO mice compared to controls (Fig. 2a and Supplementary Table 1). Of the 303 genes that exhibited differential expression in the Hdac3 cKO mice, 64.03% of the transcripts were found to be downregulated (194 genes downregulated; 109 genes upregulated). Transcriptional profiling following MeCP2 deletion in the mouse brain and hESC-derived neurons5-7 also revealed that genes are predominantly downregulated. Gene ontology (GO) analysis of HDAC3 dysregulated genes revealed an enrichment of neuron-specific GO groups, including synaptic transmission, neurological system process and transmission of nerve impulse (Fig. 2b). Five immediate-early genes (IEG’s; Arc, Fos, Nov, Bdnf, Nr4a1) were downregulated in the Hdac3 cKO mice, and were validated by qRT-PCR (Fig. 2a, c). Additional neuronal genes identified as downregulated in the Hdac3 cKO mice (Arrdc2, Dusp4, Klf10, Tle1, Adcyap1) were also validated by qRT-PCR (Fig. 2a, c). Genes related to synaptic functions were identified by RNA-seq analysis and confirmed by qRT-PCR as either downregulated (Gabra5, Chrna5, Doc2b) or upregulated (Snap25, Nrgn, Ppp1r1b) in the Hdac3 cKO mice (Fig. 2a, c). Downregulation of Fos protein in the CA1 pyramidal cell layer and upregulation of Snap25 in the hippocampus of Hdac3 cKO mice were validated by immunostaining and immunoblotting, respectively, confirming the transcriptional dysregulation observed in the Hdac3 cKO mice (Fig. 2d, e).
Figure 2. HDAC3 regulates the transcription of a subset of genes in common with MeCP2.
(a) Scatter plot of gene expression using RNA-seq (log10 FPKM values) of the hippocampus CA1 region; 109 genes are upregulated (green), and 194 genes are downregulated (blue) in the Hdac3 cKO mice; n = 2, 2. FPKM, fragments per kilobase of exon per million fragments mapped. (b) GO analysis performed using 303 genes identified in (a) were broadly classified according to biological function using MSigDB. (c) Validation by qRT-PCR of 16 genes identified as differentially expressed between Hdac3 cKO mice compared to controls and a housekeeping gene actin (fold change relative to control, normalized to Gapdh). Two-tailed t test; Arc, t(5) = 5.359, ** P = 0.003; Fos, t(5) = 2.741, * P = 0.0407; Nov, t(5) = 6.857, ** P = 0.001; Bdnf, t(5) = 2.772, * P = 0.0393; Nr4a1, t(5) = 2.909, * P = 0.0334; Arrdc2, t(6) = 2.473, * P = 0.0483; Dusp4, t(6) = 3.273, * P = 0.0170; Klf10, t(6) = 3.726, ** P = 0.0098; Tle1, t(6) = 3.892, ** P = 0.0081; Adcyap1, t(5) = 3.122, * P = 0.0262; Gabra5, t(5) = 1.611, P = 0.1681; Chrna5, t(5) = 3.822, * P = 0.0123; Doc2b, t(5) = 2.084, P = 0.0916; Snap25, t(8) = 2.487, * P = 0.0377; Nrgn, t(5) = 1.330, P = 0.2409; Ppp1r1b, t(5) = 1.166, P = 0.2962; Actin, t(6) = 0.2431, P = 0.8160; n = 3, 4 mice. (d) Coronal brain slices of control and Hdac3 cKO mice at 3-month old were stained using Hoechst (blue) and anti-Fos (green), and images captured within the CA1 using a confocal microscope. Scale bar, 40 μm. Mean gray value of Fos immunofluorescence; two-tailed t test; t(8) = 2.397, * P = 0.0434; n = 4, 6 mice. (e) Western blot analysis of Snap25 and Actin protein levels using hippocampal lysates from control and Hdac3 cKO mice. Ratio of Snap25 and Actin mean gray value; two-tailed t test; t(6) = 4.440, ** P = 0.0044; n = 4, 4 mice. Full-length blots are presented in Supplementary Fig. 10a. (f) Comparison of genes with altered expression in Hdac3 cKO and the hippocampus of Mecp2 KO mice5. Downregulated genes; 194 genes downregulated in Hdac3 cKO of which 37 are downregulated in Mecp2 KO. Upregulated genes; 109 genes upregulated in Hdac3 cKO of which 15 are upregulated in Mecp2 KO. Two-tailed Chi-square test with Yates correction; n = 303 HDAC3 genes; n = 3217 MeCP2 genes, n = 22548 genes in mm9 genome. (g) Comparison of genes with altered expression in Hdac3 cKO mice and human MECP2 loss-of-function neurons7. Downregulated genes; 194 genes downregulated in Hdac3 cKO of which 41 are downregulated in MECP2 loss-of-function neurons. Upregulated genes; 109 genes upregulated in Hdac3 cKO of which 4 are upregulated in MECP2 loss-of-function neurons. Two-tailed Chi-square test with Yates correction; n = 303 HDAC3 genes; n = 7569 MeCP2 genes, n = 22548 genes in mm9 genome. c, report mean value ± s.e.m. d, e, report median, 25th and 75th percentile, min and max value.
To ascertain whether there is a functional link in transcriptional regulation by HDAC3 and MeCP2, the differentially regulated genes in the Hdac3 cKO mice were compared to those identified in the hippocampus of Mecp2 KO mice27. We found a significant overlap in genes downregulated in the absence of HDAC3 and MeCP2 (Fig. 2f and Supplementary Table 2). A similar comparison of HDAC3 dysregulated genes was performed with transcriptional analysis obtained from hESC-derived neurons with TALEN-mediated MECP2 loss-of-function7. We found a significant overlap in dysregulated genes, and these genes were predominantly downregulated in the Hdac3 cKO and human MECP2 loss-of-function neurons (Fig. 2g and Supplementary Table 3). Our transcriptional data suggests that HDAC3, similar to observations for MeCP2, may facilitate transcription of a subset of neuronal genes.
MeCP2 affects HDAC3 binding at promoters of transcribed genes
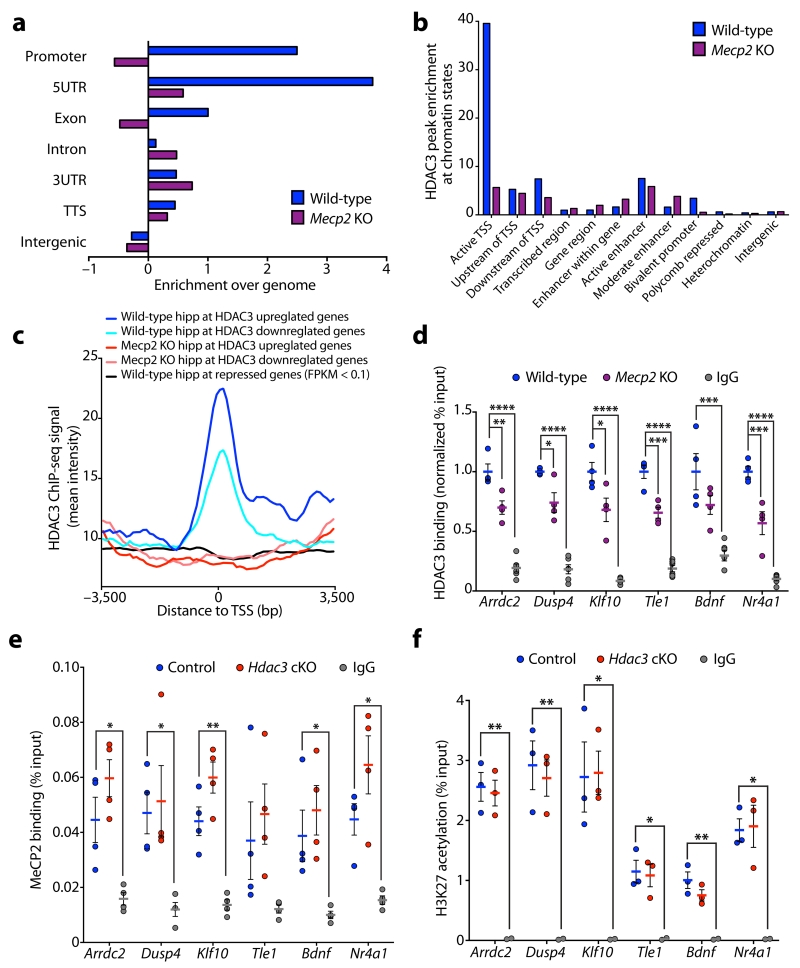
To examine whether HDAC3 binds to the regulatory regions of transcribed genes genome-wide we performed chromatin immunoprecipitation (ChIP) against HDAC3 in the hippocampus of 3-month wild-type mice, followed by next-generation sequencing (ChIP-seq). The DFilter peak-finding algorithm28 was utilized to define HDAC3 binding sites and identified 6149 regions. Downstream analysis revealed that HDAC3 binding is enriched at the promoter and 5’ UTR of genes, indicative of a general role for HDAC3 in regulating gene expression (Supplementary Fig. 5a). A more detailed analysis was performed using ChromHMM software29, to assess HDAC3 binding in relation to chromatin states as defined by histone modifications in the mouse hippocampus30. HDAC3 was found to bind near the transcriptional start site (TSS) of active gene promoters enriched for Histone 3 Lysine 27 acetylation (H3K27ac) and H3K4 trimethylation (H3K4me3). HDAC3 binding was also present at some active enhancer elements as distinguished by H3K27ac and H3K4me1 (Supplementary Fig. 5b). Aggregate plots showing the average HDAC3 binding intensity across the promoters of upregulated and downregulated genes in Hdac3 cKO mice were generated to assess whether HDAC3 directly modulates these genes. The average HDAC3 binding intensity was enriched at the promoter of both up- and down-regulated genes, including the downregulated genes Tle1 and Klf10 (Supplementary Fig. 5c, d). This data implicates that HDAC3 directly regulates the expression of genes identified as either up- and down-regulated in Hdac3 cKO mice. To further test HDAC3 binding at genes downregulated in our RNA-seq we carried out HDAC3 ChIP in the hippocampus of 3-month wild-type mice followed by qPCR analysis for Arrdc2, Dusp4, Klf10, Tle1, Bdnf and Nr4a1. HDAC3 binding was enriched near the TSS or promoters of all six genes (Supplementary Fig. 5e). HDAC3 binding was reduced at these gene regulatory regions in hippocampal CA1 of 3-month Hdac3 cKO mice relative to controls indicating specificity of the HDAC3 antibody (Supplementary Fig. 5f).
As HDAC3 is a binding partner of MeCP213, we assessed whether global chromatin distribution of HDAC3 is perturbed in the absence of MeCP2. HDAC3 ChIP-seq was carried out in the hippocampus of Mecp2 KO mice and wild-type littermates at postnatal day 45 (P45) when Mecp2 KO mice exhibit RTT-like phenotypes20. HDAC3 binding in P45 wild-type hippocampus was enriched at the TSS of active gene promoters genome wide (Fig. 3a, b), and at the TSS of genes upregulated and downregulated in the Hdac3 cKO (Fig. 3c), similar to that of 3-month wild-type hippocampus (Supplementary Fig. 5a-c). In P45 Mecp2 KO hippocampus, HDAC3 binding at active promoters and HDAC3 dysregulated genes was reduced (Fig. 3a-c and Supplementary Fig. 6a). As behavioral and transcriptional effects of Hdac3 cKO mice are similar to that of MeCP2 loss-of-function, we assessed HDAC3 binding at the promoters of genes dysregulated in the hippocampus of Mecp2 KO mice27. HDAC3 binding at the TSS of genes downregulated and upregulated in the hippocampus of Mecp2 KO mice was enriched in P45 wild-type hippocampus, and reduced in the absence of MeCP2 (Supplementary Fig. 6b, c).
Figure 3. MeCP2 modulates HDAC3 localization to the promoters of transcribed genes.
(a) Enrichment of HDAC3 binding peaks in hippocampus of P45 wild-type and Mecp2 KO mice at annotated regions of the genome analyzed using Homer (enrichment over genome). TTS; transcriptional termination site, UTR; untranslated region. (b) Enrichment of HDAC3 binding peaks in hippocampus of P45 wild-type and Mecp2 KO mice at genomic regions analyzed using chromatin state profiling of the hippocampus. TSS; transcriptional start site. (c) Aggregate plots of average HDAC3 ChIP-seq intensity signal in P45 wild-type hippocampus (blue) and Mecp2 KO hippocampus (red). Aggregate plots were generated at the TSS of genes upregulated and downregulated in the CA1 of Hdac3 cKO mice. Aggregate plot of average HDAC3 ChIP-seq intensity signal at the TSS of genes with FPKM values below 0.1 in control hippocampal CA1 (black; negative control). TSS, transcriptional start site. (d) ChIP of HDAC3 and IgG followed by qPCR analysis at the promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in cortex of P45 wild-type and Mecp2 KO mice (normalized % input to wild-type); one-way ANOVA; Arrdc2, F(2,11) = 74.68, **** P < 0.0001; Dusp4, F(2,11) = 75.39, **** P < 0.0001; Klf10, F(2,11) = 62.38, **** P < 0.0001; Tle1, F(2,11) = 106.5, **** P < 0.0001; Bdnf, F(2,11) = 16.68, **** P = 0.0005; Nr4a1, F(2,11) = 75.92, **** P < 0.0001; Bonferroni post hoc; wild-type versus Mecp2 KO: Arrdc2, ** P = 0.0039; Dusp4, * P = 0.0119; Klf10, * P = 0.0112; Tle1, *** P = 0.0003; Bdnf, P = 0.1318; Nr4a1, *** P = 0.0005; wild-type versus IgG: Arrdc2, **** P < 0.0001; Dusp4, **** P < 0.0001; Klf10, **** P < 0.0001; Tle1, **** P < 0.0001; Bdnf, *** P = 0.0003; Nr4a1, **** P < 0.0001; n = 4, 4, 6 mice. (e) ChIP of MeCP2 and IgG followed by qPCR analysis at the promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in hippocampal CA1 region of 3-month control and Hdac3 cKO mice (% input); one-way ANOVA; Arrdc2, F(2,9) = 12.58, ** P = 0.0025; Dusp4, F(2,9) = 6.043, * P = 0.0217; Klf10, F(2,9) = 26.48, *** P = 0.0002; Tle1, F(2,9) = 2.967, P = 0.1024; Bdnf, F(2,9) = 6.930, * P = 0.0151; Nr4a1, F(2,9) = 12.53, ** P = 0.0025; Bonferroni post hoc; control versus Hdac3 cKO: Arrdc2, P = 0.2449; Dusp4, P > 0.9999; Klf10, P = 0.0727; Tle1, P > 0.9999; Bdnf, P = 0.8101; Nr4a1, P = 0.1520; control versus IgG: Arrdc2, * P = 0.0205; Dusp4, * P = 0.0398; Klf10, ** P = 0.0022; Tle1, P = 0.2465; Bdnf, * P = 0.0490; Nr4a1, * P = 0.0314; n = 4, 4, 4 mice. (f) ChIP of H3K27ac and IgG followed by qPCR analysis at the promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in hippocampal CA1 region of 3-month control and Hdac3 cKO mice (% input); one-way ANOVA; Arrdc2, F(2,5) = 37.22, *** P = 0.0010; Dusp4, F(2,5) = 19.15, ** P = 0.0045; Klf10, F(2,5) = 9.955, * P = 0.0181; Tle1, F(2,5) = 10.58, P = 0.0160; Bdnf, F(2,5) = 17.82, ** P = 0.0053; Nr4a1, F(2,5) = 13.43, ** P = 0.0098; Bonferroni post hoc; control versus Hdac3 cKO: Arrdc2, P > 0.9999; Dusp4, P > 0.9999; Klf10, P > 0.9999; Tle1, P > 0.9999; Bdnf, P = 0.3040; Nr4a1, P > 0.9999; control versus IgG: Arrdc2, ** P = 0.0011; Dusp4, ** P = 0.0045; Klf10, * P = 0.0220; Tle1, * P = 0.0165; Bdnf, ** P = 0.0041; Nr4a1, * P = 0.0122; n = 3, 3, 2 mice. d – f, report mean value ± s.e.m.
Our ChIP-seq analysis indicates HDAC3 recruitment to the promoter of active genes is altered in the absence of MeCP2. This was further tested by ChIP qPCR analysis of MeCP2 and HDAC3 binding at the promoter of six genes identified as downregulated in both Hdac3 cKO mice (Fig. 2a) and Mecp2 KO neurons7. MeCP2 binding was enriched at the gene promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in P45 wild-type hippocampus, but was not enriched in Mecp2 KO indicating antibody specificity (Supplementary Fig. 6d). Furthermore, we observed decreased HDAC3 binding at these promoters in the absence of MeCP2 (Fig. 3d), indicating MeCP2 regulates HDAC3 binding at these genes. Conversely, we tested whether DNA binding of MeCP2 is regulated by HDAC3. MeCP2 was enriched at the gene promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in 3-month hippocampal CA1 of both control and Hdac3 cKO mice (Fig. 3e). Our ChIP-seq and qPCR analysis indicates that MeCP2 regulates HDAC3 binding at the promoter region of active genes, including a subset of genes downregulated in Hdac3 cKO mice.
HDAC-containing chromatin complexes are thought to modulate gene expression through deacetylation of histones, which closely correlates with gene repression. To test whether histone acetylation at promoters is altered in the hippocampal CA1 of Hdac3 cKO mice, we assessed acetylation of histone H3 lysine 27 (H3K27ac), H3K9ac and H4K12ac by ChIP qPCR. H3K27ac is enriched at active promoters and enhancers, H3K9ac at the promoters of transcribed genes, and H4K12ac is associated with learning-induced gene expression31. Both H3K9ac and H4K12ac have been shown to be elevated upon HDAC3 loss-of-function in mouse embryonic fibroblasts and hepatoctyes32,33. We found an enrichment of H3K27ac, H3K9ac and H4K12ac at the promoter of Arrdc2, Dusp4, Klf10, Tle1, Bdnf and Nr4a1 (Fig. 3f and Supplementary Fig. 6e, f). There was no significant change in H3K27ac, H3K9ac or H4K12ac at these promoters in Hdac3 cKO mice compared to controls (Fig. 3f and Supplementary Fig. 6e, f). Together with our RNA-seq analysis (Fig. 2a), this data shows that Arrdc2, Dusp4, Klf10, Tle1, Bdnf and Nr4a1 are actively transcribed genes in the hippocampus, and that downregulated expression of these genes in Hdac3 cKO may be independent of histone acetylation.
HDAC3 promotes localization of FOXO3 at active gene promoters
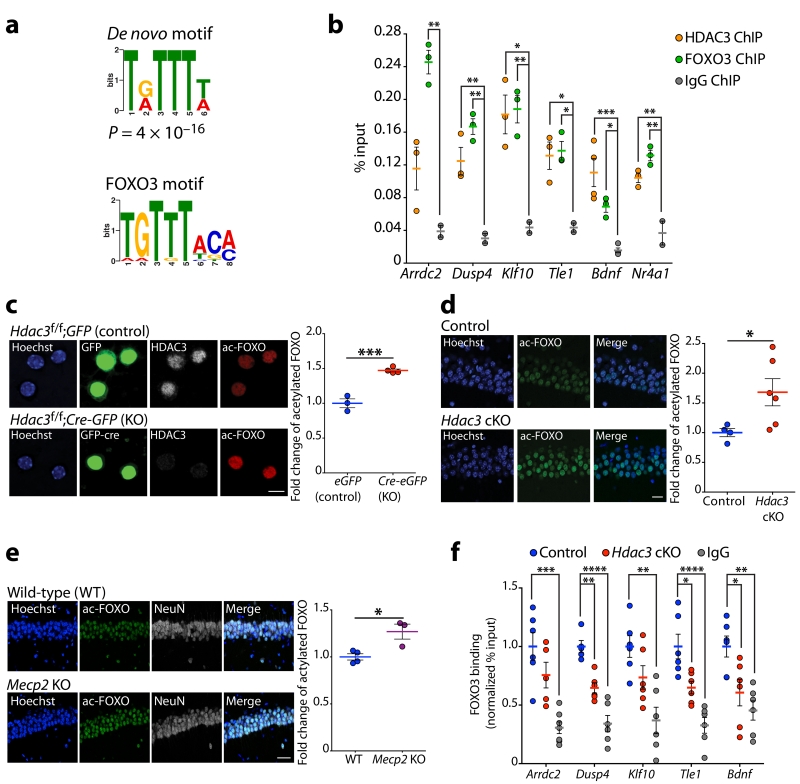
To gain a mechanistic insight into transcriptional regulation by HDAC3 and MeCP2, enrichment of common conserved putative cis-regulatory elements was assessed using the MSigDB motif gene dataset. The most enriched known transcription factor-binding motif identified at genes downregulated in the hippocampus of Hdac3 cKO was the TTGTTT motif for the FOXO transcription factors (Supplementary Fig. 7a). Furthermore, the FOXO motif was highly enriched among genes downregulated in the hippocampus, hypothalamus and cerebellum of Mecp2 KO mice5,6,27 (Supplementary Fig. 7a). We extended our analysis to global HDAC3 binding peaks, as identified by our ChIP-seq data, for enrichment of common de novo motifs using MEME-ChIP software and identified a significant enrichment for a sequence that matches the FOXO binding motif (Fig. 4a and Supplementary 7b).
Figure 4. HDAC3 regulates FOXO3 deacetylation and recruitment to gene promoters in neurons.
(a) De novo motif identified by MEME-ChIP analysis of HDAC3 binding regions resembles FOXO motif; P = 4×10−16. (b) ChIP of HDAC3, FOXO3 and IgG followed by qPCR analysis at the promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in 3-month wild-type hippocampus (% input); one-way ANOVA; Arrdc2, F(2,5) = 25.40, ** P = 0.0024; Dusp4, F(2,5) = 24.67, ** P = 0.0026; Klf10, F(2,5) = 14.58, ** P = 0.0082; Tle1, F(2,5) = 12.37, * P = 0.0116; Bdnf, F(2,5) = 18.50, *** P = 0.0010; Nr4a1, F(2,5) = 30.88, ** P = 0.0015. Bonferroni post hoc; HDAC3 ChIP versus IgG: Arrdc2, P = 0.1036; Dusp4, ** P = 0.0095; Klf10, * P = 0.0105; Tle1, * P = 0.0156; Bdnf, *** P = 0.0006; Nr4a1, ** P = 0.0052; FOXO3 ChIP versus IgG: Arrdc2, ** P = 0.0020; Dusp4, ** P = 0.0019; Klf10, ** P = 0.0087; Tle1, * P = 0.0118; Bdnf, * P = 0.0268; Nr4a1, ** P = 0.0011; n = 3, 3, 2 mice. (c) Hdac3f/f neurons infected with lentivirus expressing either GFP (control) or Cre-GFP (Hdac3 KO) and stained with Hoechst (blue), anti-GFP (green), anti-HDAC3 (white) and anti-acetyl-FOXO (red). Images captured using a confocal microscope. Scale bar, 10 μm. Mean gray value quantification of acetyl-FOXO immunofluorescence in control and Hdac3 KO neurons. Two-tailed t test; t(5) = 8.021, *** P = 0.0005; n = 3, 4 coverslips. (d) Coronal brain slices of 3-month control and Hdac3 cKO mice were stained using Hoechst (blue) and anti-acetyl-FOXO (green). Images of the CA1 using a confocal microscope, scale bar, 20 μm. Mean gray value quantification of acetyl-FOXO immunofluorescence of control and Hdac3 cKO mice. Two-tailed t test; t(8) = 2.343, * P = 0.0472; n = 4, 6 mice. (e) Coronal brain slices of P45 wild-type and Mecp2 KO mice were stained using Hoechst (blue), anti-acetyl-FOXO (green), and anti-NeuN (white). Images of the CA1 using a confocal microscope, scale bar, 40 μm. Mean gray value quantification of acetyl-FOXO immunofluorescence normalized to NeuN in wild-type and Mecp2 KO mice. Two-tailed t test; t(5) = 3.470, * P = 0.0178; n = 4, 3 mice. (f) ChIP of FOXO3 and IgG followed by qPCR at the promoters of Arrdc2, Dusp4, Klf10, Tle1, and Bdnf in hippocampal CA1 region of 3-month control and Hdac3 cKO mice (normalized % input to controls). One-way ANOVA; Arrdc2, F(2,16) = 14.55, *** P = 0.0003; Dusp4, F(2,14) = 32.19, **** P < 0.0001; Klf10, F(2,15) = 9.606, ** P = 0.0021; Tle1, F(2,15) = 18.44, **** P < 0.0001; Bdnf, F(2,15) = 8.623, ** P = 0.0032. Bonferroni post hoc; control versus Hdac3 cKO: Arrdc2, P = 0.5124; Dusp4, ** P = 0.0015; Klf10, P = 0.1759; Tle1, * P = 0.0127; Bdnf, * P = 0.0220; control versus IgG: Arrdc2, *** P = 0.0002; Dusp4, **** P < 0.0001; Klf10, ** P = 0.0011; Tle1, **** P < 0.0001; Bdnf, ** P = 0.0022; n = 6, 6, 6 mice. b - f, report mean value ± s.e.m.
The FOXO family members, FOXO1 and FOXO3, have been identified as direct targets for deacetylation by HDAC3 in the liver34. To test whether HDAC3 and FOXO co-localize at the promoter of transcribed genes we performed ChIP qPCR in 3-month wild-type hippocampus. There are four genes coding for FOXO transcription factors in mouse and human, Foxo1, Foxo3, Foxo4 and Foxo6. Our RNA-seq data shows Foxo3 has the highest expression in the hippocampal CA1 region (Supplementary Fig. 7c), thus we performed ChIP experiments targeting FOXO3. Both HDAC3 and FOXO3 bind at the same promoter regions of six genes downregulated in Hdac3 cKO mice, Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 (Fig. 4b). Collectively, these data show that HDAC3 binding at a subset of genes downregulated in Hdac3 cKO mice is modulated by MeCP2, and that HDAC3 and FOXO3 are co-localized at these same genomic regions.
Acetylation of the FOXO transcription factors has been shown to reduce their binding affinity to DNA and inhibit their ability to activate gene transcription35-37. To determine whether FOXO acetylation is altered in neurons lacking HDAC3, primary neuronal cultures prepared from Hdac3f/f embryonic cortices were infected with lentivirus expressing Cre recombinase tagged with green fluorescent protein (Cre-GFP). Immunostaining with an antibody raised against acetyl-FOXO1 that also detects acetyl-FOXO334, shows that FOXO acetylation was elevated in Hdac3 KO neurons (Hdac3f/f;Cre-GFP) compared to controls (Hdac3f/f;GFP) (Fig. 4c). Furthermore, levels of acetylated FOXO in 3-month Hdac3 cKO mice were markedly elevated in CA1 neurons (Fig. 4d), and were normal in the striatum (Supplementary Fig. 7d). These results indicate that in vivo HDAC3 regulates FOXO acetylation in neurons. Acetylated FOXO was also increased in the hippocampus of P45 Mecp2 KO mice compared to controls (Fig. 4e).
Next, we tested whether FOXO3 directly binds MeCP2 and HDAC3 by co-immunoprecipitation experiments using recombinant human protein. FOXO3 co-immunoprecipitated HDAC3/NCoR, but not MeCP2 (Supplementary Fig. 7e), and HDAC3/NCoR was confirmed to co-immunoprecipitate MeCP2 (Supplementary Fig. 7f)13. FOXO3 binding to chromatin was then assessed at the promoters of Arrdc2, Dusp4, Klf10, Tle1, and Bdnf in hippocampal CA1 of Hdac3 cKO mice by ChIP qPCR, and we found that FOXO3 binding was reduced compared to controls (Fig. 4f). Collectively, these data suggest that DNA binding of FOXO may be compromised upon loss of MeCP2 through decreased recruitment of HDAC3 and increased FOXO acetylation.
HDAC3 and FOXO3 function is impaired in human MECP2R306C cells
To determine whether human RTT-causative mutations of MECP2 affect HDAC3 and FOXO3 binding at gene promoters we obtained induced pluripotent stem cells (iPSCs) derived from a 7-year old RTT patient harboring a heterozygous MECP2R306C point mutation (Coriell: GM23298). The R306C mutation is located within the transcriptional repressor domain of MeCP2, selectively blocking its interaction with NCoR/HDAC313,14. The MECP2R306C derived iPSCs have skewed X-inactivation, and express only the MECP2R306C allele38. Sequencing of genomic (g)DNA from MECP2R306C iPSCs revealed the heterozygous mutation (Supplementary Fig. 8a), whereas sequencing of complementary (c)DNA indicates expression of only the MECP2R306C allele (Supplementary Fig. 8b). To ensure homogeneity of the MECP2 allele expression, neural progenitor cells (NPCs) were generated from single-cell derived MECP2R306C iPSC lines. Sequencing of cDNA derived from two independent MECP2R306C NPC lines (lines 4 and 14) showed exclusive expression of the MECP2R306C mutated allele (Supplementary Fig. 8c). Two independent isogenic control iPSC lines (lines 6 and 36) were generated from the MECP2R306C iPSCs using CRISPR/Cas9-mediated gene editing to correct the R306C mutation (Supplementary Fig. 8d). CRISPR/Cas9 editing was successfully designed to target only the mutant allele (see methods), as indicated by the inclusion of two heterozygous silent mutations, indicating high specificity of genome editing (Supplementary Fig. 8d). Five predicted off-target sites were also verified as unedited by PCR sequencing (Supplementary Fig. 9). NPCs were generated from the isogenic control iPSC lines, and from iPSCs of a healthy individual (C1; ATCC: CRL-2097)39. All NPC lines were validated by immunostaining for the human NPC marker Musashi-1 (MSI1) and Nestin (Supplementary Fig. 7g).
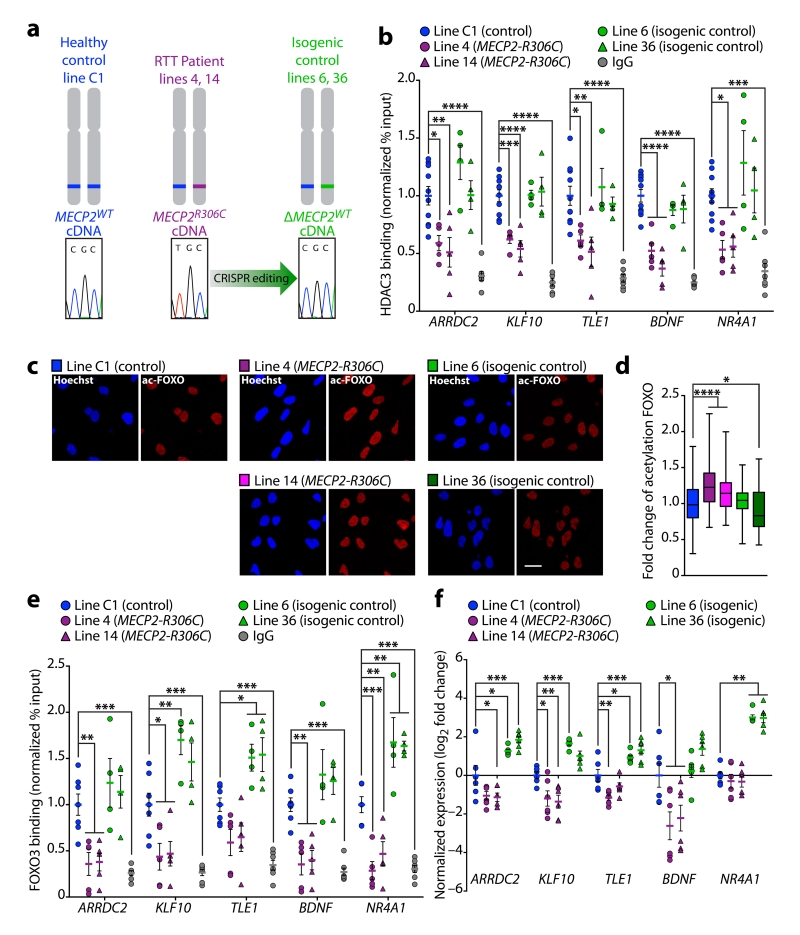
To test whether the RTT-causative point mutation of MeCP2 affects HDAC3 recruitment to the promoters of genes downregulated in the Hdac3 cKO mice, we performed ChIP qPCR experiments using NPCs from the healthy individual (C1), two MECP2R306C lines, and two isogenic control lines (Fig. 5a). We found that HDAC3 binding was reduced at the promoters of ARRDC2, KLF10, TLE1, BDNF, and NR4A1 in MECP2R306C NPCs compared to the C1 healthy control and isogenic control NPCs (Fig. 5b), consistent with reports that the interaction of mutant MeCP2R306C with NCoR/HDAC3 is compromised13,14. To test whether FOXO function is altered in the MECP2R306C NPC lines, we assessed FOXO3 binding at these same promoter regions and FOXO acetylation levels. We found that acetylation of FOXO was elevated in the MECP2R306C NPC lines compared to the C1 healthy control and isogenic control lines (Fig. 5c, d). Furthermore, FOXO3 binding was compromised in the MECP2R306C NPC lines compared to the C1 healthy control and isogenic control NPCs (Fig. 5e). These data indicate that the RTT-causative MeCP2 point mutation located outside of the methyl DNA binding domain reduces the recruitment of HDAC3 and FOXO3 to gene promoters. Lastly, gene expression of ARRDC2, KLF10, TLE1, and BDNF was decreased in MECP2R306C NPCs compared to the C1 healthy control, and was rescued in the isogenic control NPCs to levels that surpassed the C1 healthy control (Fig. 5f). Together, our data indicates that MeCP2, in concert with HDAC3, can regulate gene transcription through modulating FOXO deacetylation and binding to DNA.
Figure 5. FOXO3 and HDAC3 recruitment to gene regulatory regions is abrogated in NPCs harboring a patient-derived MECP2 mutation.
(a) NPC lines were generated using iPSCs derived from a healthy control (C1) and an RTT patient with a heterozygous MECP2R306C mutation (line 4 and 14). To generate isogenic control NPCs (lines 6 and 36), the R306C mutation in the RTT patient-derived line was changed to the wild-type sequence by CRISPR/Cas9 gene editing. (b) ChIP of HDAC3 and IgG followed by qPCR at the promoters of ARRDC2, KLF10, TLE1, BDNF and NR4A1 in healthy (C1), MECP2R306C (line 4, line 14), and isogenic control NPCs (line 6, line 36) (normalized % input to C1). One-way ANOVA; ARRDC2, F(5,29) = 14.79, **** P < 0.0001; KLF10, F(5,29) = 31.08, **** P < 0.0001; TLE1, F(5,29) = 12.65, **** P < 0.0001; BDNF, F(5,29) = 28.76, **** P < 0.0001; NR4A1, F(5,29) = 9.570, **** P < 0.0001. Bonferroni post hoc; C1 versus line 4: ARRDC2, * P = 0.0132; KLF10, *** P = 0.0002; TLE1, * P = 0.0154; BDNF, **** P < 0.0001; NR4A1, * P = 0.0221; C1 versus line 14: ARRDC2, ** P = 0.0024; KLF10, **** P < 0.0001; TLE1, ** P = 0.0018; BDNF, **** P < 0.0001; NR4A1, * P = 0.0345; C1 versus line 6: ARRDC2, P = 0.2027; KLF10, P > 0.9999; TLE1, P > 0.9999; BDNF, P = 0.8822; NR4A1, P = 0.4572; C1 versus line 36: ARRDC2, P > 0.9999; KLF10, P > 0.9999; TLE1, P > 0.9999; BDNF, P > 0.9999; NR4A1, P > 0.9999; C1 versus IgG: ARRDC2, **** P < 0.0001; KLF10, **** P < 0.0001; TLE1, **** P < 0.0001; BDNF, **** P < 0.0001; NR4A1, *** P = 0.0002; n = 10, 5, 5, 4, 4, 7 cell pellets from 4 cultures. (c) Immunostaining of C1 (control), line 4 and line 14 (MECP2R306C), and line 6 and 36 (isogenic control) NPCs using Hoechst (blue) and anti-acFOXO (red). Imaged using confocal microscope; Scale bar, 20 μm; n = 3 coverslips per line. (d) Quantitation of acFoxo immunostaining in NPCs (c). One-way ANOVA; F(4,795) = 39.15, **** P < 0.0001. Bonferroni post hoc; C1 versus line 4: **** P < 0.0001, C1 versus line 14: **** P < 0.0001, C1 versus line 6: P = 0.4427, C1 versus line 36: * P = 0.0174; n = 160 nuclei per line. (e) ChIP of FOXO3 and IgG followed by qPCR at the promoters of ARRDC2, KLF10, TLE1, BDNF and NR4A1 in healthy (C1), MECP2R306C (line 4, line 14), and isogenic control NPCs (line 6, line 36) (normalized % input to C1). One-way ANOVA; ARRDC2, F(5,26) = 11.38, **** P < 0.0001; KLF10, F(5,26) = 19.39, **** P < 0.0001; TLE1, F(5,26) = 16.96, **** P < 0.0001; BDNF, F(5,26) = 15.59, **** P < 0.0001; NR4A1, F(5,26) = 27.79, **** P < 0.0001. Bonferroni post hoc; C1 versus line 4: ARRDC2, ** P = 0.0052; KLF10, * P = 0.0158; TLE1, P = 0.0746; BDNF, ** P = 0.0020; NR4A1, *** P = 0.0005; C1 versus line 14: ARRDC2, ** P = 0.0071; KLF10, * P = 0.0249; TLE1, P = 0.1697; BDNF, ** P = 0.0042; NR4A1, ** P = 0.0098; C1 versus line 6: ARRDC2, P > 0.9999; KLF10, ** P = 0.0042; TLE1, * P = 0.0279; BDNF, P = 0.3387; NR4A1, ** P = 0.0020; C1 versus line 36: ARRDC2, P > 0.9999; KLF10, P = 0.0961; TLE1, * P = 0.0173; BDNF, P = 0.7238; NR4A1, ** P = 0.0037; C1 versus IgG: ARRDC2, *** P = 0.0004; KLF10, *** P = 0.0004; TLE1, *** P = 0.0005; BDNF, *** P = 0.0002; NR4A1, *** P = 0.0002; n = 7, 5, 5, 4, 4, 7 cell pellets from 4 cultures. (f) qRT-PCR analysis of RNA from healthy control (C1), MECP2R306C (line 4, line 14), and isogenic control (line 6, line 36) NPCs for ARRDC2, KLF10, TLE1, BDNF and NR4A1 (Log2 fold change to C1, normalized to GAPDH). One-way ANOVA; ARRDC2, F(4,25) = 22.57, **** P < 0.0001; KLF10, F(4,25) = 23.57, **** P < 0.0001; TLE1, F(4,25) = 23.62, **** P < 0.0001; BDNF, F(4,25) = 9.01, **** P < 0.0001; NR4A1, F(2,25) = 48.89, **** P < 0.0001. Bonferroni post hoc; C1 versus line 4: ARRDC2, P = 0.0582; KLF10, * P = 0.0192; TLE1, ** P = 0.0026; BDNF, * P = 0.0118; NR4A1, P > 0.9999. C1 versus line 14: ARRDC2, * P = 0.0358; KLF10, ** P = 0.0067; TLE1, P = 0.2614; BDNF, * P = 0.0396; NR4A1, P > 0.9999; C1 versus line 6: ARRDC2, * P = 0.0.165; KLF10, *** P = 0.0009; TLE1, * P = 0.0189; BDNF, P > 0.9999; NR4A1, **** P < 0.0001; C1 versus line 36: ARRDC2, *** P = 0.0004; KLF10, P = 0.0611; TLE1, *** P = 0.0007; BDNF, P = 0.3875; NR4A1, **** P < 0.0001; n = 6, 6, 6, 6, 6 cell pellets from 2 cultures. b, e, f, report mean value ± s.e.m. d, report median, 25th and 75th percentile, min and max value.
Discussion
We demonstrated that neuronal deletion of HDAC3 causes behavioral phenotypes observed in Mecp2 loss-of-function mouse models of RTT, including impaired locomotor coordination, sociability and cognition. The genetic manipulations we utilized examine HDAC3 loss over a prolonged period and likely reflect disease conditions, however, previous work suggests that an acute reduction in HDAC3 activity may be beneficial for aspects of cognition40. Similar to models of RTT we show that HDAC3 positively regulates the transcription of a subset of neuronal genes, and that MeCP2 regulates global recruitment of HDAC3 to the promoter of active genes. At a subset of gene promoters, loss of HDAC3 impairs recruitment of FOXO3 and correlates with elevated levels of FOXO acetylation, a modification that likely affects its DNA binding. The lack of changes in histone acetylation at the promoters of downregulated genes in the Hdac3 cKO mice suggests a departure of the conventional role for HDAC3 in regulating histone acetylation, at least for certain genes regulating neuronal functions. Rather, HDAC3 appears to adopt a unique role in regulating acetylation of transcriptional factors in neuronal lineages. Importantly, NPCs derived from an RTT patient harboring an MECP2 mutation that prevents MeCP2 interaction with NCoR/HDAC313, impairs HDAC3 and FOXO3 localization to gene promoters and negatively affects gene expression. Collectively, our data supports a role for MeCP2 and HDAC3 in regulating transcription factor recruitment and creating an environment permissive for gene expression, which is dysregulated in RTT patient-derived NPCs.
Loss of MeCP2 and HDAC3 also leads to increased expression of a number of genes, which are likely suppressed by this complex. A recent analysis of gene expression datasets from Mecp2 mutant mice, including Mecp2R306C mice, found that MeCP2 loss-of-function leads to increased expression of long genes41. Specifically, MeCP2-mediated repression of long genes correlates with MeCP2 binding to methylated CA sites within the gene body41,42. MeCP2 binds broadly to chromatin in neurons and its genomic localization likely affects its function as a transcriptional regulator. A possibility is that MeCP2 binding within the body of long genes suppresses transcription, whereas MeCP2 recruitment of HDAC3 to certain gene promoters positively regulates transcription. Whether MeCP2 interacts with the NCoR/HDAC3 complex within long genes remains to be determined. However, the molecular targets for HDAC3-mediated deacetylation within gene bodies likely differ from those located at promoter regions.
Many transcription factors and chromatin regulators are known to be acetylated43, therefore, HDAC3-mediated deacetylation could be applicable to multiple chromatin factors. In this regard, binding motifs for TCF/LEF, mediators of Wnt signaling, were also enriched among genes downregulated in Hdac3 cKO and Mecp2 KO mice. It will be of interest to test whether Wnt signaling, a pathway implicated in the autism spectrum disorders44, is regulated by MeCP2 and HDAC3. Network analysis of gene expression and protein interaction profiles implicate the NCoR/HDAC3 complex with autism and intellectual impairment45. Human mutations in two key components of the NCoR/HDAC3 complex, TBL1XR1 and TBL1X, are associated with sporadic autism46,47. TBL1XR1 human mutations are also linked to intellectual disability48, and to a patient diagnosed with West syndrome displaying RTT-like features49. Taken together with our observations that HDAC3 loss leads to social and cognitive impairments, the transcriptional function of NCoR/HDAC3 could be more widely applicable to the autism spectrum disorders and intellectual disability.
Methods
Animals
Hdac3 conditional knockout mice (Hdac3 cKO) were generated by crossing Hdac3 floxed mice (Control)16 with the transgenic αCamKII promoter-driven Cre (CW2) line17. Hdac3 floxed mice and CW2 Cre mice were backcrossed to C57BL/6 a minimum of nine generations prior to generating Hdac3 cKO mice. Mecp2 KO mouse strain was obtained from The Jackson Laboratory (B6.129P2(C)-Mecp2tm1.1Bird/J)20. Mecp2 KO males were generated through crossing Mecp2 heterozygous females with C57BL/6J males. Mice were housed in groups of 3 – 5 on a standard 12h light / 12 h dark cycle, and all behavioral experiments were performed during the light cycle. Food and water were provided ad libitum. All animal work was approved by the Committee for Animal Care of the Division of Comparative Medicine at the Massachusetts Institute of Technology. No animals were excluded from the study and randomization of experimental groups was not required. Experimenter was blind to animal genotypes during behavioral testing, and data analysis was automated for all experiments apart from Novel Object Location testing. Prior to all behavioral tests, mice were habituated to behavior rooms for 60 min. Hdac3 cKO experimental mice were male and aged 3 – 4 months. Mecp2 KO experimental mice were male and aged P45. Mice that were tested on multiple behavioral paradigms were given a minimum of 1 week resting period between experiments. Mice used for all experiments were male. A summary of published datasets analyzed in this manuscript from Mecp2 KO models can be found in Supplementary Table 4.
Cell Lines
C1 control iPSCs were gifted from Guo-li Ming, and were previously generated using commercially available fibroblasts (ATCC, CRL-2097)39. Karyotyping analysis of C1 iPSCs had previously been performed39. MECP2R306C iPSCs were commercially obtained (Coriell, GM23298), and were derived from fibroblasts of a 7-year old RTT patient with a heterozygous MECP2R306C point mutation. MECP2R306C iPSCs were authenticated by Coriell, including karyotyping analysis, and mycoplasma testing, and pluripotency was assessed by embryoid body formation and in vivo teratoma formation. Directed induction of NPCs from human iPSCs was as previously described50, aside from minor alterations in medium composition. Neural Maintenance Medium consisted of N2 medium and B27 medium at a 1:1 ratio. N2 medium consisted of DMEM/F-12 GlutaMAX (Life Technologies, 10565-018), 1 × N2 supplement (Life Technologies, 17502-048), 5 μg ml−1 insulin (Sigma, I9278), 1 mM L-Glutamine (Life Technologies, 25030-024), 100 μM non-essential amino acid solution (Life Technologies, 11140-050), 100 μM 2-mercaptoethanol (Sigma, M7522), 50 U ml−1 penicillin and 50 mg ml−1 streptomycin (Life Technologies, 15140-122). B27 medium consisted of Neurobasal (Life Technologies, 12348-017), 1 × B27 (Life Technologies, 17504-044), 200 mM L-Glutamine, 50 U ml−1 penicillin and 50 mg ml−1 streptomycin. Neural Induction Medium consisted of Neural Maintenance Medium supplemented with 1 μM Dorsomorphin (Tocris Bioscience, 3093) and 10 μm SB431542 (Tocris Bioscience, 1614). An absence of mycoplasma in all cell lines was routinely assessed by Hoechst staining.
CRISPR/Cas9 gene editing in MECP2R306C iPSCs
Short guide (sg)RNA sequences were generated by software at http://crispr.mit.edu. The selected sgRNA sequence covered the R306C locus and was modified to match the sequence of the mutant allele (CGGGTCTTGCGCTTCTTGAT; antisense). The sgRNA was introduced into the pSpCas9(BB)-2A-GFP plasmid (Addgene, PX458) by BbsI mediated cloning, and confirmed by sequencing. Sense single-strand oligodeoxynucleotide (ssODN) was designed to correct the R306C missense mutation and to introduce two silent mutations within the sgRNA targeting site (5’GGTGGCAGCCGCTGCCGCCGAGGCCAAAAAGAAAGCCGTGAAGGAGTCTTCTATCCGATCTGTGCAGGAGACCGTACTCCCCATtAAaAAGCGCAAGACCCGGGAGACGGTCAGCATCGAGGTCAAGGAAGTGGTGAAGCCCCTGCTGGTGTCCACCCTCGGTGAGAAGAGCGGGAAAGGACTG3’)
The two silent point mutations were to prevent further cutting after gene editing has occurred. iPSCs (5M cells) were electroporated with an Amaza Nucleofector using the Human Stem Cell Nucleofector Kit 1 reagents (Lonza, VPH-5012) according to the manufactures protocol. Briefly, 5M iPSCs were resuspended in 100 µl Lonza Reaction Buffer supplemented with ssODN (15 µg) and pSpCas9(BB)-2A-GFP-sgRNA plasmid (7.5 µg) and electroporated using Program A-23. Electroporated iPSCs were resuspended in hES medium and seeded onto 1× 6-well plate. Two days later, iPSCs were dissociated (accutase), washed and resuspended with DPBS. A single-cell suspension (Falcon, 352235) was prepared followed by fluorescence-activated cell sorting to collect GFP-positive cells. GFP-positive iPSCs (50,000 cells) were plate onto 1 × 6-well plate, and single colonies were collected 10 days later and maintained in single wells (24-well plates). Successful gene editing was determined by PCR-amplification of the R306C locus followed by sequencing (R306Csurveyor_F AGTCCTGGGAAGCTCCTTGT; R306Csurveyor_R CTTTGGGGACTCTGAGTGGT).
Immunohistochemistry
Adult male mice (control and Hdac3 cKO at 3-months; wild-type and Mecp2 KO at P45) were perfused with 10% formaldehyde under deep anesthesia and brains were post-fixed overnight in 10% formaldehyde. Brains were sectioned at 40 μm using a vibratome (Leica). Sections were permeabilized and blocked in PBS containing 0.3% Triton X-100 and 10% normal donkey serum at room temperature for 1 h. Sections were incubated overnight at 4 °C in primary antibody diluted 1:200 in PBS with 0.3% Triton X-100 and 10% normal donkey serum. Primary antibodies used were anti-HDAC3 (Cell Signaling Technology; 3949), anti-GFAP (Cell Signaling Technology; 12389), anti-Parvalbumin (Swant; PV-25), anti-Fos (Santa Cruz, sc-52), anti-GFP (Aves Labs, GFP-1020), anti-Acetyl-FOXO (Santa Cruz, sc-49437), anti-NeuN (SySY, 266-004), anti-Musashi-1 (Millipore, AB5977). Primary antibodies were visualized with Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 antibodies and nuclei were visualized with Hoechst 33342, all diluted 1:500 in PBS and incubated at room temperature for 90 min. Sections were mounted on slides with Fluoromount G (Electron Microscopy Sciences) overnight at room temperature and stored at 4 °C . Images were acquired using an LSM 710 Zeiss confocal microscope. Quantitation of nuclear immunofluorescence levels of Fos, HDAC3, acFOXO3 and NeuN was performed using ImageJ 1.46a software by generating an ROI of the nucleus using the Hoechst channel and measuring the mean gray value (MGV) in the channel of interest. Analysis of primary neurons was quantitated as the mean MGVs of 21 nuclei per coverslip (Fig. 4c). Analysis of neurons in the CA1 pyramidal cell layer was quantitated as the mean nuclear MGVs per mouse, as follows: 40 nuclei per mouse for acFOXO in HDAC3 cKO (Fig. 4d), 80 nuclei per mouse for acFOXO in MeCP2 KO mice (Fig. 4e), and 30 nuclei per mouse for HDAC3 in Hdac3 cKO (Supplementary Fig. 1a). Neurons in the striatum were identified by NeuN, followed by quantitation of acFOXO as the mean MGVs of 40 nuclei per mouse (Supplementary Fig. 7d).
Recombinant Protein Binding Assay
Binding reactions were carried out using 1 μg of human recombinant FOXO3 (OriGene, TP302894), HDAC3/NCoR(DAD) (Enzo, BML-SE15-0050), and MeCP2 (Abnova, H00004204-P01). Recombinant proteins (1 μg) were combined as indicated in a final volume of 30 μl supplemented with Binding Buffer (50 mM Tris HCl pH 8.0, 0.1 mM EDTA, 1 mM DTT, 10% glycerol, 1 mM PMSF), and incubated at 30 °C for 60 min. Antibodies (2 μg) for immunoprecipitation were added and reactions were adjusted to a final volume of 120 μl with Binding Buffer and incubated at room temperature for 60 min. Reactions were supplemented with 30 μl Protein A Sepharose beads (50% slurry), and rotated at 4 °C for 2 h. Beads were washed five times with 1 ml Binding Buffer (3,000 rcf centrifugation), and eluted with 30 μl 2× Laemmli sample buffer at 95 °C for 10 min prior to western blot analysis.
Western Blot
Hippocampal whole cell lysates were prepared using tissue from control and Hdac3 cKO male mice (3-month). Tissue was homogenized in 1 ml RIPA (50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) buffer with a hand homogeniser (Sigma), incubated on ice for 15 min, and rotated at 4 °C for 30 min. Cell debris was isolated and discarded by centrifugation at 14,000 rpm for 10 minutes. Lysates were quantitated using a nanodrop and 25 μg protein was loaded on a 10 % acrylamide gels. Protein was transferred from acrylamide gels to PVDF membranes (Invitrogen) at 100 V for 90 min. Membranes were blocked using bovine serum albumin (5% w/v) diluted in TBS:Tween. Membranes were incubated in primary antibodies overnight at 4 °C and secondary antibodies at room temperature for 90 min. Western blots were imaged using the Odyssey Imaging System (LI-COR Biosciences) and analyzed with ImageJ 1.46a software.
Open field Test
Male mice (3 month) were placed into an arena (40 cm width × 40 cm length × 30 cm height) with orthogonal lasers to track position and locomotor activity for 60 min using VersaMax software, version 4.12.1AFE (AccuScan Instruments, OH). Activity was measured as movement across a grid of infrared light beams and analysis using 5 min intervals was automated using the Versamax software.
Rotarod Test
Motor coordination was measured using an accelerating rotarod (Ugo Basile model 47600) consisting of 5 beams (5 cm length 3 cm diameter) divided by round plates. Each mouse performed two consecutive 5 min trials with an inter-trial interval of 10 min. The rotarod was set at a starting speed of 4 rpm, accelerating to a maximum speed of 60 rpm. Assessment of the time at which the mouse falls was automated and an average of two trials is used to calculate the latency.
Sociability Test
Control and Hdac3 cKO mice (3-month) were habituated for 10 min to an empty arena consisting of 3 chambers, each chamber is 20 cm (width) × 40.5 cm (length) × 22 cm (height). A stimulus mouse (3-month) is then confined to one of the lateral chambers using a circular wire enclosure (11 cm high, 10.5 cm diameter, 1 cm spaced bars). An empty wire enclosure is included in the opposing lateral chamber as a non-social cue. The test mouse is placed in the central chamber and was allowed to explore the arena for 10 min. The movement of the test mouse within the 3-chamber arena is recorded during both the habituation and sociability phase using a ceiling mounted camera. The choice of lateral chamber for the stimulus mouse is alternated between trials, and the stimulus mouse is habituated to the wire enclosures for 30 min the previous day. Sociability is measured as time spent in each of the three chambers (social, central and non-social) and was automated using Ethovision XT software (Noldus Information Technology).
Object location memory test
Male mice (3-month) were habituated to the arena (Allentown rat cage) without objects for 2 days (10 min per day). To provide adequate spatial discrimination, the outside walls were marked using colored tape. On day three, mice were habituated to the cages for an additional 10 min. After 1 h, mice were trained two times (with a 1 h inter-trial interval) for 10 min in an arena with two identical objects placed in two corners of the arena. After 90 min, the mice are tested by using the same two objects, one object placed in a familiar position, and the second object placed in a new ‘novel’ corner of the arena. Mice were recorded for 5 min during the test phase. Scoring of mice was blinded to the experimenter. Mice were scored as exploring the objects when climbing, sniffing or observing (within 2 cm) of the object. Time spent on top of the object, without simultaneously directing attention to the object, was not recorded.
Morris Water Maze
Spatial memory was performed using a circular tank (1.2 m in diameter) filled with opaque water at 22 °C. The walls contained spatial reference cues, and inside the tank was a fixed hidden platform (10 cm in diameter) in a target quadrant. Male mice (4-month) were placed into the maze at random locations and allowed to search for the platform for 60 s. Two trials a day were conducted with a 60 min inter-trial interval. On day 8, the hidden platform was removed and a probe trial was conducted for 60 s to assess spatial learning. Ethovision XT software (Noldus Information Technology) was used for automated recording and analysis of swim speed and escape latency during training (day 1-7), time spent in quadrants and platform crossings during the probe trial (day 8).
Fear-conditioned learning
Contextual and cued fear conditioning was conducted over 3 days using male mice (4 months). On day 1, mice were placed in a fear conditioning apparatus chamber for 3 min and then presented with a 30 s 75 dB tone followed by a 2 s constant foot shock at 0.8 mA (TSE Systems, Germany). Mice remained in the context for a further 15 s after the footshock and were then returned to their homecage. 24 h after training (day 2), the mice were tested for contextual learning by quantitating freezing behavior for 3 min in the context used during training. On day 3, the mice were tested for cue learning by placing them in a novel context and providing the 75 dB tone for 3 min, during which freezing behavior is quantitated. Prior to each trial, each context was cleaned with 70% ethanol except for on day 3, when it was cleaned with isopropanol. Freezing behavior analysis was automated using software provided by TSE Systems. Activity suppression was calculated as a Suppression Ratio defined as ActivityTesting/(ActivityTraining + ActivityTesting)51. Activity values (au) were automated using software provided by TSE Systems for the 3 min habituation on day 1 (ActivityTraining) and the 3 min testing on day 2 and day 3 (ActivityTesting). Suppression ratio values below 0.5 indicate a fear response; suppression ratio values at or greater than 0.5 indicate no fear response or may indicate conditioned safety.
RNA isolation and reverse transcription
The CA1 was isolated from the hippocampus of male mice (3-month). Tissue was rapidly frozen using liquid nitrogen and stored at −80 °C, and RNA extracted using Trizol according to manufacturers protocol (Invitrogen). RNA (3 μg) was DNase I treated (4 U, Worthington Biochemical Corporation), purified using RNA Clean and Concentrator-5 Kit (Zymo Research) according to manufacturers’ instructions and eluted with 14 μl DEPC-treated water. For each sample, 1 μg RNA was reverse transcribed in a 20 μl reaction volume containing random hexamer mix and Superscript III reverse transcriptase (50 U, Invitrogen) at 50 °C for 1 h. First strand cDNAs were diluted 1:10 and 1 μl were used for qRT-PCR amplification in a 20 μl reaction (SsoFast EvaGreen Supermix, Bio-Rad) containing primers (0.2 μM). Relative changes in gene expression were assessed using the 2−ΔΔCt method52.
Mouse primer sequences: Hdac3exon10_F GCTGTGATCGATTAGGCTGC, (Hdac3exon11_R) GGCAACATTTCGGACAGTGT; Arrdc2_F CTTCCAGCTGCCTATCTCC, Arrdc2_R CCACAGGTTCAATGACAGTG; Dusp4_F TTACCAGTACAAGTGCATCC, Dusp4_R TTACTGCGTCGATGTACTC; Klf10_F GACCTTCAGACAGTCCCAG, Klf10_R GAGGGTTCGAAGTCAGAGG; Tle1_F CTCAGGAACATCAACAACAGG, Tle1_R CGATGATGGCATTCAACTCTG; Arc_F ATGACACCAGGTCTCAAGG, Arc_R ATGTAGGCAGCTTCAGGAG; Fos_F GAACGGAATAAGATGGCTGC, Fos_R TTGATCTGTCTCCGCTTGG; Bdnf_F AATGGTGTCGTAAAGTTCCAC, Bdnf_R GCAACCGAAGTATGAAATAACC; Nov_F AGGAAGTAACAGACAAGAAAGG, Nov_R AATTCTCGAACTGTAGGTGG; Nr4a1_F CTCATCACTGATCGACACG, Nr4a1_R CTCCTTCAGACAGCTAGCA; Chrna5_F TCGGAATACTTTGGAGGCC, Chrna5_R AATCTTCAACAACCTCGCG; Doc2b_F CCAATGATTTCATCGGTGGT, Doc2b_R TCTTCAAGCAGTCAAACCAG; Adcyap1_F GGTGTATGGGATAATAATGCATAGC, Adcyap1_R GTCTTCTGGTCTGATCCCAG; Ppp1r1b_F GGACCGCAAGAAGATTCAG, Ppp1r1b_R AGAGGTTCTCTGATGTGGAG; Snap25_F CATATGGCTCTAGACATGGG, Snap25_R GTTGGAATCAGCCTTCTCC; Nrgn_F TTTAGAAGTTCCAGAGGAGAGTC, Nrgn_R TAGGGAAGTCTTGTCACTGC; Gabra5_F CCAGCACAGGTGAATATACG, Gabra5_R GGAAGGTAGGTCTGGATGAC; Gapdh_F TCCTTTAGGATTTGGCCGT, Gapdh_R TTGATGGCAACAATCTCCAC; Hprt_F TACCTAATCATTATGCCGAGGA, Hprt_R GAGCAAGTCTTTCAGTCCTG.
Human primer sequences: ARRDC2_F CTCCTGAGTACTCGGAGGT, ARRDC2_R GGTTTGGATCCTCCTCAGAG; KLF10_F CTGCGGAGGAAAGAATGGA, KLF10_R GACATAAGTGCTTCTACAGCT; TLE1_F AGTTCACTATCCCGGAGTC, TLE1_R CCAATTTAAGGCTGTGATACTG; BDNF_F AGAGGAATGGTTCCACCAG, BDNF_R GATGTTTGCTTCTTTCATGGG; NR4A1_F ACTGGACTACTCCAAGTTCC, NR4A1_R TCGTAGAACTGCTGTACATCC; GAPDH_F CCTCCTGTTCGACAGTCAG, GAPDH_R CATACGACTGCAAAGACCC.
Chromatin Immunoprecipitation protocol
Tissue from control and Hdac3 cKO mice (3-month) and wild-type and Mecp2 KO mice (P45) were dissected on ice, snap frozen in liquid nitrogen and stored at −80 °C. Frozen tissue and cell pellets were briefly homogenized and cross fixed with 2 mM disuccinimidyl glutarate in PBS for 35 min followed by 1% formaldehyde for 10 min, and then quenched with 125 mM glycine for 5 min. Fixed homogenate was washed twice (0.5% triton X-100, 0.1 M sucrose, 5 mM MgCl2, 1 mM EDTA, 10 mM Tris-HCl pH 8.0) and douce-homogenized. Fixed nuclei were pelleted and resuspend in Lysis Buffer (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8.0, 0.5% sarcosyl) and chromatin sheared using a BioruptorPlus (Diagenode Inc, NJ) set to high for 40 cycles of 30 s on, 30 s off. Chromatin was quantitated using 6.25% of the sample, and equivalent amounts of chromatin were used for each ChIP (roughly 3 μg per tissue sample, 8 μg per cell pellet sample). Chromatin was precleared using an adjusted Lysis Buffer (Final: 1% Triton, 0.1% sodium deoxycholate, 5 mM EDTA, 1 mM PMSF, 2 μg ml−1 leupeptin and aprotinin). ChIP was performed using the following antibodies (antibodies reported for ChIP-sequencing are referenced): 5 μg HDAC3 (Abcam, ab7030)53, 5 μg FOXO3 H-144 (Santa Cruz, sc-11351)54, 5 μg MeCP2 (Sigma, M6818), 1 μg H3K9ac (Abcam, ab4441)55, 1 μg H4K12ac (Millipore, 07-595) or normal rabbit IgG (Millipore, 12-370) antibody incubated overnight, followed by enrichment using protein A sepharose beads for 4 h. Beads were washed 4 times with RIPA buffer (50 mM Hepes pH 7.6, 10 mM EDTA, 0.7% sodium deoxycholate, 1% NP-40, 0.5 M LiCl), and once with TE (50 mM Tris HCl, 10 mM EDTA). Chromatin was eluted by agitation at 65 °C for 20 min in TES (TE plus 1% SDS), and reverse crosslinked overnight at 65 °C . Chromatin was subjected to RNase and proteinase K treatment, followed by DNA purification by phenol chloroform extraction and ethanol precipitation. DNA pellets were resuspended in 10 mM Tris and subjected to qPCR or ChIP-seq analysis.
Mouse primer sequences: Arrdc2_F AAAAGAGATCGGCCAGGTG, Arrdc2_R CCGCTTGTGTGTGTACGTAG; Dusp4_F AGCCCTCTCTCGTAAACACA, Dusp4_R ATAGCAGTCCCAGCCTTCTC; Klf10_F CTCTGTCAGTGGAGCGTGTA, Klf10_R AGGACTGAAGGCTAGGGTTG; Tle1_F CTTCTGCAAACTTCAACCCC, Tle1_R GCCGAGCTGTCAATCAAAGT; Bdnf_F GCGGTGTAGGCTGGAATAGA, Bdnf_R GCGGTGTAGGCTGGAATAGA; Nr4a1_F TCAACGACGATTTGCATGCT, Nr4a1_R GCCAGGATTCCATTACATCACC.
Human primer sequences: ARRDC2_F CCGAGGATGGCAAAGTCAAC, ARRDC2_R ACTTCCTGGTCCTCTGCATC; KLF10_F GAGCGTGTACACAATCCCC, KLF10_R GCGTCACTCAATCAGGTGG; TLE1_F GACGCCAAAACCAGCCAAT, TLE1_R ACTTTGATTGACAGCCCAGC; BDNF_F TTCTTTGCGGCTTACACCAC, BDNF_R CCGGGTTGGTATACTGGGTT; NR4A1_F AACGAATCCAGAGCCTGTGA, NR4A1_R TCTGATAACGAGTCCCAGCC.
Library Preparation for RNA-seq and ChIP-seq
Libraries were prepared and sequenced as previously described30. RNA-seq of the CA1 region of the hippocampus was performed using two replicates for control and Hdac3 cKO samples. RNA-seq reads were aligned to the mouse mm9 genome using TopHat. The mean yield per sample was 34.33 million 36-bp single-end reads, of which 31.17 million reads were aligned (90.8%).
HDAC3 ChIP-seq of the 3-month wild-type hippocampus (C57BL/6J), P45 wild-type hippocampus and P45 Mecp2 KO hippocampus was performed using 3 replicates. For 3-month wild-type hippocampus, after filtering, a total of 94 million unique reads (roughly 30 million reads per replicate) were obtained for the HDAC3 ChIP, and 78.8 million reads were obtained for total input. For P45 wild-type hippocampus, after filtering, a total of 10 million unique reads were obtained for the HDAC3 ChIP, and 16 million reads were obtained for total input. For P45 Mecp2 KO hippocampus, after filtering, a total of 13 million unique reads were obtained for the HDAC3 ChIP, and 16 million reads were obtained for total input. Sequencing reads were mapped to the mm9 mouse genome using BWA aligner (samse option). Duplicate reads were marked and removed using SAM tools.
RNA-seq and ChIP-seq analysis
For RNA-seq analysis, aligned reads were mapped to the RefSeq database and counted (HTSeq). Differential expression analysis was performed using DESeq (Bioconductor) followed by Student t test to model the experimental and gene-specific dispersion, respectively. Genes were considered differentially expressed if P ≤ 0.05. Ontological analyses of differentially expressed genes were performed using Gene Set Enrichment Analysis (GSEA) for GO biological process (MSigDB, Broad Institute). Transcription factor-binding motif analysis of RNA-seq data was performed using Gene Set Enrichment Analysis (GSEA) for transcription factor targets (MSigDB, Broad Institute). To compare differentially expressed genes (DEG) in the Hdac3 cKO with DEG from human-derived Mecp2 KO neurons7, mouse gene names were converted to human homologs using MGI annotation database (www.informatics.jax.org/homology.shtml).
For ChIP-seq analysis, ChIP reads were normalized and presented as a ratio over input reads using dFilter software28. Bigwig files and HDAC3 peaks were generated using dFilter software and visualized using the UCSC genome browser. Enrichment of HDAC3 binding peaks over genome (Fig. 3a) was assessed using HOMER software. Enrichment of HDAC3 binding peaks over chromatin states (Fig. 3b) was assessed with ChromHMM software56, using chromatin states obtained from the hippocampus of adult mice30. Aggregation plots of normalized ChIP-seq intensity were generated using deepTools Galaxy with ComputeMatrix set to 3,500 bp upstream and downstream of the TSS. De novo motif analysis was assessed with MEME-ChIP using nucleotide sequences (FASTA format) identified as HDAC3 binding peaks. Data deposited under accession number GSE72196. For HDAC3 ChIP-Seq data generated from wild-type and Mecp2 KO mice, the raw data were mapped to mouse mm9 reference genome using BWA aligner. After filtering out duplicate reads (PCR artifact), the reads from biological replicates were concatenated for peak calling using dFilter software (threshold P value = 1×10−5, bin size = 25 bp, kernel size = 50 bp). Enrichment of HDAC3 binding peaks at different genomic regions were compared for WT and Mecp2 KO mice.
In vitro FOXO3 binding assay
Human recombinant proteins were purchased: MeCP2 (Abnova, H00004204-P01), HDAC3/NCOR1 (Enzo Life Sciences, BML-SE15-0050) and FOXO3 (OriGene, TP302894). Binding assays were carried out in a final volume of 30 μl in binding buffer (50 mM Tris HCl pH 8.0, 0.1 mM EDTA, 1 mM DTT, 10% glycerol, 1 mM PMSF) containing 1 μg of each recombinant protein. Samples were incubated at 30 °C for 60 min, then supplemented in 80 μl binding buffer and 10 μl (2 μg) FOXO3 H-144 antibody (Santa Cruz, sc-11351) and incubated at room temperature for 60 min. Protein A sepharose beads (30 μl) were added to each sample and rotated at 4 °C for 2 h, followed by five washes (5 min each) in 1 ml binding buffer. Beads were eluted in 2× Laemmli buffer followed by western blot analysis.
Statistics
Results are presented as mean ± s.e.m. All statistical analysis was performed using Prism GraphPad software. Data distribution was presumed to be normal with equal variance between groups, however, this was not formally tested. Comparison data consisting of two groups was analyzed by two-tailed unpaired t tests. Comparison of data consisting of three or more groups was analyzed by one-way ANOVA followed by Bonferroni post hoc test. Comparison of two or more factors across multiple groups was analyzed by two-way ANOVA followed by Bonferroni post hoc test. The statistical test, exact P values, sample size (n), t values, ANOVA F values, and degrees of freedom for each experiment is specified in the figure legend. No statistical method was used to estimate sample size, but is consistent with previous publications. Molecular and biochemical analysis was performed using a minimum of three biological replicates per condition. Behavioral experiments require larger data sets due to increased variability. All behavioral experiments consist of a minimum of 9 animals per group.
Data Availability
Sequencing data are available from the NCBI Gene Expression Omnibus (GEO) database under accession number GSE72196. Additional data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank Dr. Eric N. Olson (UT Southwestern Medical Center) for kindly providing the HDAC3f/f mice. We thank Dr. G.-L. Ming (Johns Hopkins University) for kindly providing the C1 iPSC cell line. We thank R. Madabhushi, A. Watson, and J. Penney for comments on the manuscript. We thank E. Demmons for help with mouse colony maintenance. This work was supported by the National Institutes of Health grants (MH102690 and NS079625) and Rettsyndrome.org to P.J., and NIH grant NS78839 and the JPB Foundation to L.-H. T.
Footnotes
Accession Codes
Sequencing data are available from the NCBI Gene Expression Omnibus (GEO) database under accession number GSE72196.
Author contributions
A.N. and L.-H.T. designed this study and L.-H.T. directed and coordinated this study. A.N initiated, planned and performed experimental work. J.C. helped with ChIP experiments. F.G. and J.M. contributed to bioinformatic analysis. P.M. helped with some behavioral experiments. A.V.Z. helped with validation of the RNA-seq. E.G. prepared sequencing libraries for the ChIP-seq experiment. T.K. derived the NPC lines. T.K. and Y.-T.L. helped maintain the iPSC. F.Z. and P.J. provided Mecp2 KO tissue. A.N. and L.-H.T. wrote the manuscript with critical input from all the authors.
Competing Financial Interest Statement
The authors declare no competing financial interests.
References
- 1.Pohodich AE, Zoghbi HY. Rett syndrome: disruption of epigenetic control of postnatal neurological functions. Hum Mol Genet. 2015;24:R10–6. doi: 10.1093/hmg/ddv217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 3.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 4.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasui DH, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J, et al. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell. 2014;53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Research. 2013;41:8072–8084. doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zupkovitz G, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyst MJ, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert DH, et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. doi: 10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norwood J, Franklin JM, Sharma D, D’Mello SR. Histone deacetylase-3 is necessary for proper brain development. J Biol Chem. 2014 doi: 10.1074/jbc.M114.576397. doi:10.1074/jbc.M114.576397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng H, et al. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–629. doi: 10.1016/s0092-8674(01)00585-2. [DOI] [PubMed] [Google Scholar]
- 18.Gemelli T, et al. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry. 2006;59:468–476. doi: 10.1016/j.biopsych.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 19.Su S-H, Kao F-C, Huang Y-B, Liao W. MeCP2 in the rostral striatum maintains local dopamine content critical for psychomotor control. J Neurosci. 2015;35:6209–6220. doi: 10.1523/JNEUROSCI.4624-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 21.Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- 22.Yasui DH, et al. Mice with an isoform-ablating Mecp2 exon 1 mutation recapitulate the neurologic deficits of Rett syndrome. Hum Mol Genet. 2014;23:2447–2458. doi: 10.1093/hmg/ddt640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelka GJ, et al. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain. 2006;129:887–898. doi: 10.1093/brain/awl022. [DOI] [PubMed] [Google Scholar]
- 24.Moretti P, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyakawa T, Yamada M, Duttaroy A, Wess J. Hyperactivity and intact hippocampus-dependent learning in mice lacking the M1 muscarinic acetylcholine receptor. J Neurosci. 2001;21:5239–5250. doi: 10.1523/JNEUROSCI.21-14-05239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankland PW, O’Brien C, Ohno M, Kirkwood A, Silva AJ. Alpha-CaMKII-dependent plasticity in the cortex is required for permanent memory. Nature. 2001;411:309–313. doi: 10.1038/35077089. [DOI] [PubMed] [Google Scholar]
- 27.Baker SA, et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V, et al. Uniform, optimal signal processing of mapped deep-sequencing data. Nat Biotechnol. 2013;31:615–622. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 29.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peleg S, et al. Altered Histone Acetylation Is Associated with Age-Dependent Memory Impairment in Mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 32.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhaskara S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihaylova MM, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatta M, Liu F, Cirillo LA. Biochemical and Biophysical Research Communications. Biochem Biophys Res Commun. 2009;379:1005–1008. doi: 10.1016/j.bbrc.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzaki H, et al. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung AYL, et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabel HW, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci USA. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 44.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends in Neurosciences. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hormozdiari F, Penn O, Borenstein E, Eichler E. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2014 doi: 10.1101/gr.178855.114. doi:10.1101/gr.178855.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung R-H, et al. An X chromosome-wide association study in autism families identifies TBL1X as a novel autism spectrum disorder candidate gene in males. Mol Autism. 2011;2:18. doi: 10.1186/2040-2392-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pons L, et al. A new syndrome of intellectual disability with dysmorphism due to TBL1XR1 deletion. Am. J. Med. Genet. A. 2015;167A:164–168. doi: 10.1002/ajmg.a.36759. [DOI] [PubMed] [Google Scholar]
- 49.Saitsu H, et al. A girl with West syndrome and autistic features harboring a de novo TBL1XR1 mutation. J. Hum. Genet. 2014;59:581–583. doi: 10.1038/jhg.2014.71. [DOI] [PubMed] [Google Scholar]
- 50.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 51.Anagnostaras SG, Josselyn SA, Frankland PW, Silva AJ. Computer-assisted behavioral assessment of Pavlovian fear conditioning in mice. Learn Mem. 2000;7:58–72. doi: 10.1101/lm.7.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eijkelenboom A, et al. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Molecular Systems Biology. 2013;9:1–15. doi: 10.1038/msb.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available from the NCBI Gene Expression Omnibus (GEO) database under accession number GSE72196. Additional data that support the findings of this study are available from the corresponding author upon request.