Abstract
Over 100 genetic loci harbor schizophrenia associated variants, yet how these variants confer liability is uncertain. The CommonMind Consortium sequenced RNA from dorsolateral prefrontal cortex of schizophrenia cases (N = 258) and control subjects (N = 279), creating a resource of gene expression and its genetic regulation. Using this resource, ~20% of schizophrenia loci have variants that could contribute to altered gene expression and liability. In five loci, only a single gene was involved: FURIN, TSNARE1, CNTN4, CLCN3, or SNAP91. Altering expression of FURIN, TSNARE1, or CNTN4 changes neurodevelopment in zebrafish; knockdown of FURIN in human neural progenitor cells yields abnormal migration. Of 693 genes showing significant case/control differential expression, their fold changes are ≤ 1.33, and an independent cohort yields similar results. Gene co-expression implicates a network relevant for schizophrenia. Our findings show schizophrenia is polygenic and highlight the utility of this resource for mechanistic interpretations of genetic liability for brain diseases.
Keywords: Schizophrenia, dorsolateral prefrontal cortex, postmortem study, gene expression, RNA-seq, case-control study, biomarker, eQTL, functional GWAS, zebrafish, hiPSC
INTRODUCTION
How the human brain dynamically performs its innumerable functions is recognized as one of this century’s “Grand Challenges”. Indeed, seemingly straightforward fundamental information such as which genes are expressed therein and what functions they perform are only partially characterized. To overcome these obstacles, we established the CommonMind Consortium (CMC; www.synapse.org/CMC), a public-private partnership to generate functional genomic data in brain samples obtained from autopsies of cases with and without severe psychiatric disorders. The CMC is the largest existing collection of collaborating brain banks and includes over 1,150 samples. A wide spectrum of data is being generated on these samples, including regional gene expression, epigenomics (cell-type specific histone modifications and open chromatin), whole genome sequencing, and somatic mosaicism.
Schizophrenia (SCZ), affecting roughly 0.7% of adults, is a severe psychiatric disorder characterized by abnormalities in thought and cognition1. Despite a century of evidence establishing its genetic basis, only recently have specific genetic risk factors been conclusively identified, including rare copy number variants2 and >100 common variants3. However, there is not a one-to-one Mendelian mapping between these SCZ risk alleles and diagnosis. Instead, SCZ is truly complex and appears to result from a myriad of genetic variants exerting small effects on disease risk4,5, conforming closely to a classical polygenic model. The available data are incomplete but implicate synaptic components, including calcium channel subunits and post-synaptic elements5–7. A consequence of polygenic inheritance is that the small effect sizes of individual variants complicate characterization of the biological processes they influence, both at the level of particular genes and pathways.
Post-mortem gene expression studies of SCZ cases suggest subtle abnormalities in multiple brain regions including the prefrontal and temporal cortices, hippocampus, and several specific cell types8. More than 50 gene expression studies of SCZ cases and controls have been reported, often of overlapping samples and mostly of modest scale (prior RNA sequencing studies evaluated only 5–31 cases, Supplementary data file 1). Results are often inconsistent and there are few replicated findings. These studies are probably underpowered to detect subtle effects that might be expected to arise as a result of this complex disease and within tightly regulated brain tissue9, among other limitations of existing microarray-based gene expression studies10.
RNA sequencing can accurately detect transcription at the gene and isoform level. We sequenced a cohort of SCZ and control subjects that is an order of magnitude larger than prior RNA sequencing studies. By applying state-of-the-art analytic methods and including genome-wide characterization of common variants, we generated a rich resource of the genetics of gene expression in the brain. This resource can serve as a useful catalogue of regulatory variants underlying the molecular basis of SCZ and other brain disorders. We use this resource to identify: (a) specific effects on gene expression of genetic variants previously implicated in risk; (b) genes showing a significant difference in expression between SCZ cases and controls; and (c) coordinated expression of genes implicated in SCZ. Our results shed light on the subtle effects expected from the polygenic nature of SCZ risk and thus substantially refine our understanding of the neurobiology of SCZ.
RESULTS
Samples and sequencing
We generated RNA sequence data from post-mortem human dorsolateral prefrontal cortex (DLPFC; Brodmann areas 9 and 46) from brain banks at the Icahn School of Medicine at Mount Sinai, the University of Pennsylvania, and the University of Pittsburgh (Supplementary Table 1). To control for batch effects, multiple randomization steps were introduced and DNA and RNA isolation and library preparation were performed at one site (Supplementary Fig. 1A). Samples were genotyped on the Illumina Infinium HumanOmniExpressExome array (958,178 SNPs) and imputed using standard techniques with the 1000 Genomes Project as reference data11. These genotypes were then used to detect SNPs that have an effect on gene expression (eQTLs, expression quantitative trait loci), to estimate ancestry of the samples, and to ensure sample identity across DNA and RNA experiments. Ethnicity was similar between cases and controls (Caucasian 80.7%, African-American 14.7%, Hispanic 7.7%, East Asian 0.6%, Supplementary Figs. 1B, C). As expected3, SCZ cases inherited an increased number of common variant alleles previously associated with SCZ risk (P = 1.6 × 10−8, Supplementary Fig. 1D).
RNA sequencing was performed after depleting ribosomal RNA (rRNA). Following quality control, there were 258 SCZ cases and 279 controls. Fifty-five cases with affective disorder were included to increase power to detect eQTLs. The median number of paired end reads per sample was 41.6 million, with low numbers of rRNA reads (Supplementary Fig. 2). Following data normalization, 16,423 genes (based on Ensembl models) were expressed at levels sufficient for analysis, of which 14,222 were protein coding. Validation using PCR showed high correlation (r > 0.5) with normalized expression from RNA-seq for the majority of genes assessed (Supplementary Fig. 3). Gene expression measurement can be influenced by a number of variables; some are well documented (e.g., RNA integrity (RIN) and post-mortem interval (PMI)), but others may be unknown. We investigated known covariates by standard model selection procedures to find a good statistical model (Supplementary Fig. 4 and 5). Covariates for RIN, library batch, institution (brain bank), diagnosis, age of death, genetic ancestry, PMI, and sex together explained a substantial fraction (0.42) of the average variance of gene expression, and were thus employed to adjust the data for all analyses.
Generation of a brain eQTL resource
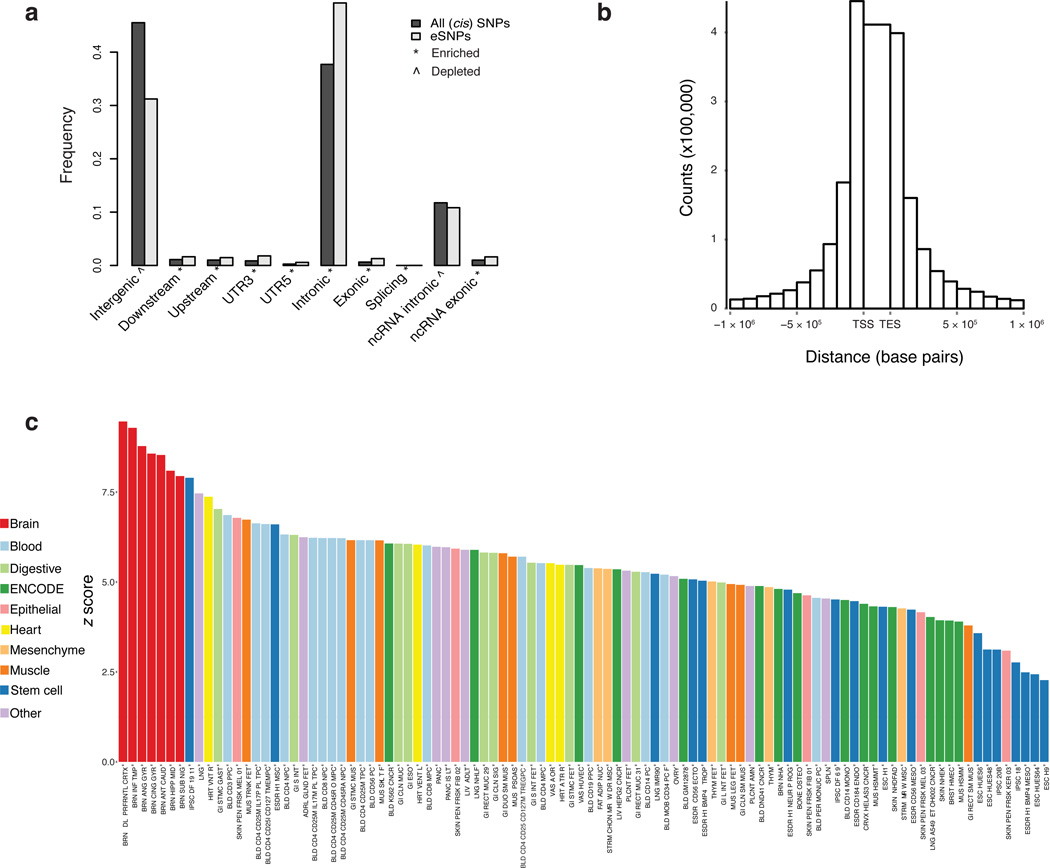
To identify eQTLs, gene expression data from European-ancestry subjects (N = 467) were adjusted for known and hidden variables detected by surrogate variable analysis (SVA) conditional on diagnosis but excluding ancestry (Supplementary Fig. 2 and 4). Adjusted expression levels were then fit to imputed SNP genotypes, covarying for ancestry and diagnosis, using an additive linear model implemented in MatrixEQTL. The model identified 2,154,331 significant cis-eQTLs, (i.e., within 1 Mb of a gene) at a false discovery rate (FDR) ≤ 5%, for 13,137 (80%) of 16,423 genes. Many eQTLs for the same gene were highly correlated, due to linkage disequilibrium, and 32.8% of eQTL SNPs (“eSNPs”) predict expression of more than one gene. Cis-eSNPs were enriched within genic elements and non-coding RNAs, particularly within 100 kb of the transcription start and end sites12, and depleted in intergenic regions (Fig. 1A, B). As defined by GTEx13, an “eGene” is a gene with at least one significant eSNP after strict correction for multiple marker testing for that gene. There were 8,427 eGenes at FDR ≤ 5%, or 18 eGenes discovered per sample, consistent with a prediction from GTEx. We examined the enrichment of max-eQTLs (defined as the most significant eSNP per gene, if any) in predicted enhancer sequences derived from the Roadmap Epigenomics Consortium and ENCODE across 98 human tissues and cell lines14. Cis-eQTLs were enriched for enhancer sequences present in brain tissues (Kolmogorov-Smirnov (KS) test versus non-brain: D = 1, P = 4.5 × 10−6), and the strongest enrichment is observed in DLPFC enhancers (Z = 9.5) (Fig. 1C).
Figure 1. Enrichment of cis-eQTLs in regulatory and other genomic elements.
(a) Enrichments of cis-eQTLs compared to all eQTLs in sequence-defined elements according to the Ensembl annotations implemented in the ANNOVAR (version 2014-07-14) software51. The bars illustrate the proportion of SNPs that belong to each category for significant cis-eQTLs (at FDR 5%) compared to all cis-SNPs (within 1 Mb from expressed genes). These categories are illustrated: exonic (fold change (FC) = 2.14); intronic (FC = 1.3); upstream (1 kb region upstream of transcription start site (TSS); FC = 1.48); downstream (1 kb region downstream of transcription end site (TES); FC = 1.52); UTR3 (3’ untranslated region; FC = 2.10); UTR5 (5’ untranslated region; FC = 2.35); splicing (within 2 bp of a splicing junction; FC = 2.51); ncRNA (transcripts without coding annotation in the gene definition, within either the exonic or intronic region; FC = 1.62 or 0.91, respectively); intergenic (FC = 0.69). (^) and (*) indicate significant (Iadjusted < 0.05) depletion or enrichment of cis-eQTLs compared to all cis-SNPs, respectively. (b) Distribution of cis-eQTL location relative to the gene. (c) Enrichment of “max-cis-eQTLs” (most associated eSNP per gene) within enhancer sequences across 98 human tissues and cell lines. Bars represent the Z score for the overlap of max-cis-eQTLs compared to 1,000 sets of random SNPs matched with respect to allele frequency, gene density, distance from the TSS, and linkage disequilibrium density. Brain (red) shows significantly higher enrichment for eQTLs compared to non-brain tissues and cell lines (P = 4.5 × 10−6) and the strongest enrichment is observed in DLPFC enhancers.
To assess the utility of analyzing a much larger brain dataset, we compared previously reported DLPFC eQTLs to CMC-derived eQTL, estimating the proportion of non-null hypotheses (π1) in CMC and the number of additional eQTL found in CMC that were not detected in the other studies. GTEx v6 is the largest public dataset of eQTLs from DLPFC tissue (n = 92) assayed by RNA-seq; its replication in CMC is π1 = 0.98. Considering microarray-based eQTLs from the Harvard Brain Bank 15, BrainCloud16, NIH17, and the UK Brain Expression Consortium (UKBEC)18, we estimated π1 to be 0.75, 0.70, 0.79, and 0.93, respectively, indicating that our results captured most eQTLs found in other independent samples. Replication was somewhat lower for a recent meta-analysis that included a mix of several distinct brain regions19 (π1 = 0.62), and for eQTLs detected in blood (π1 = 0.54)20. We also derived eQTL for 279 DLPFC samples as part of the NIMH Human Brain Collection Core (HBCC) microarray data and found replication π1 = 0.77. Moreover, concordance of the direction of allelic effect was high, with 93% of eQTL showing the same direction of effect when intersecting CMC eQTL (FDR ≤ 5%) with even a liberally defined set of HBCC eQTL (FDR ≤ 20%). In addition to containing the vast majority of eQTL found in the literature, the CMC sample finds a substantial number of genes with previously undetected eQTL (Table 1).
Table 1.
Overlaps and differences between CMC and other publicly available eQTL resources
| Comparison cohort eQTL genes compared to CMC eQTL |
||||||||
|---|---|---|---|---|---|---|---|---|
| Cohort | Sample Size | Study PMID/GEO ID/dbGaP ID |
Number of cis eQTL |
Proportion of non-null hypotheses (π1) in CMC |
Unique Genes with eQTL |
eQTL Genes Expressed in CMC |
Genes with eQTL in CMC |
Genes w/ eQTL in CMC but not in comparison cohort |
| Blood eQTL | 2494 twins | 24728292 | 9640* | 0.54 | 9533 | 8108 | 6794 | 5052 |
| Brain Cloud | 108 | GSE30272 | 374223 | 0.7 | 6199 | 5386 | 4666 | 7180 |
| Brain Meta- analysis |
424 | 25290266 | 3520** | 0.62 | 3503 | 2806 | 2507 | 9339 |
| GTEx PFC | 92 | 25954002 | 173026 | 0.98 | 1922 | 1326 | 1284 | 11853 |
| HBCC | 279 | phs000979.v1.p1 | 788338 | 0.77 | 7514 | 6785 | 5862 | 7275 |
| HBTRC | 146 | GSE44772 | 531400 | 0.75 | 6473 | 5186 | 4555 | 7291 |
| NIM | 145 | GSE15745 | 105735 | 0.79 | 2127 | 2057 | 1851 | 9995 |
| UKBEC | 134 | 25174004 | 52593 | 0.93 | 808 | 618 | 546 | 11300 |
| UNION | 1573706 | 0.7 | 16568 | 12644 | 10544 | 2593 | ||
Best eQTL per probeset reported
Best eQTL per gene reported
FDR ≤ 5% used to define eQTL in all cohorts. eQTL for Brain Cloud, HBCC, HBTRC, NIH and UKBEC were computed as described in the supplement. eQTL for the Blood cohort, Brain Meta-analysis and GTEx were downloaded from public resources. All eQTL resources represent prefrontal or frontal cortex except the Blood cohort (peripheral blood) and the Brain Meta-analysis (meta-analysis across multiple brain regions). The UNION set was derived by including all unique eQTL from all 8 cohorts.
The patterns of results should be different for “trans-eQTLs”, i.e., SNPs correlated with expression of a gene beyond 1 Mb of its genomic location. Trans-eQTLs incur a greater penalty for multiple testing, require greater power for detection, and are thus more susceptible to false positives and less likely to replicate than cis-eQTL. Nevertheless, the data supported 45,453 significant trans-eQTL at FDR ≤ 5%, of which 20,288 were also cis- eQTL SNPs for local genes, and 34% predicted expression of more than one distant gene. The proportion of trans eQTL in CMC that replicate in HBCC is 18.6% (both FDR ≤ 5%). The proportion of HBCC trans eQTL that replicate in CMC is 29.7%. Enrichment of trans-eQTLs with brain enhancers was not observed (data not shown), though enrichment in genic regions and depletion in intergenic regions was observed, particularly when restricting to trans eQTL ≥ 10 Mb from the gene location. We used similar techniques to derive isoform expression quantitative trait loci (isoQTLs).
eQTL signatures at SCZ risk loci point to specific genes
A hallmark of polygenic inheritance is that individual SNPs confer small effects on risk. For some risk SNPs, perhaps the majority, their impact could be mediated through effects on gene expression. Indeed, GWAS SNPs associated with SCZ risk occur more often than expected by chance in cis-regulatory functional genomic elements, such as enhancers or eQTL SNPs3,21–24. Yet, GWAS loci typically contain many genes, and SNPs therein are often highly correlated via linkage disequilibrium, so that assigning a biological role for a particular risk SNP has been difficult. Here, we leverage CMC-derived eQTL to relate SCZ risk variants to expression of specific genes.
Of the 108 SCZ GWAS loci previously reported3, 73 harbor cis-eQTL SNPs for one or more genes (FDR ≤ 5%). To determine if 73 out of 108 loci were larger than that expected by chance, we conducted an experiment that randomly chose such loci in the genome; it showed that 73 loci with cis-eQTL SNPs is consistent with chance expectation (data not shown). Moreover, the simple presence of an eQTL does not imply disease causality. We used Sherlock 25, a Bayesian approach that prioritizes consistency between disease association and eQTL signatures in GWAS loci, to identify genes likely to contribute to SCZ etiology. While Sherlock evaluated genes across the genome, we only evaluated genes within the 108 SCZ GWAS loci because SNPs in these loci showed genome-wide significant association with SCZ; thus, in essence, we fine mapped these loci. The results suggested that GWAS risk and eQTL association signals co-localized for 84 genes in 30 of these loci (adjusted P < 0.05; Supplementary Fig. 6A, Supplementary data file 2). After removing genes where additional evaluation indicated lack of consistency (Supplementary Fig. 7B), there were 33 genes highlighted in 18 of the 108 GWAS loci (Supplementary data file 2). Genes found to have variants affecting risk for autism are often found enriched for variation affecting risk for SCZ; indeed, compared to other genes with eQTL in the GWAS loci, these 33 genes are more enriched for nonsynonymous de novo mutations in autism (fold enrichment = 2.4, Pcorrected = 0.03), although not for SCZ, intellectual disability, or epilepsy.
Repeating the analyses using isoform-level eQTLs (isoQTL) identified nine genes in eight GWAS loci, with all but three genes already identified in the gene-level analysis (Supplementary data file 2). Combining the gene and isoform data, 20 of 108 GWAS loci (19%) had evidence suggesting that mis-regulated gene expression could, in part, explain the genetic association with schizophrenia: 18 cis-QTL loci (cis-eQTL for 33 genes + 2 genes with cis-isoQTL), one locus implicated only by cis-isoQTL (SNX19), and one trans-eQTL association for IMMP1L at a GWAS locus on chr7. We discuss other genes identified by Sherlock in the Supplement.
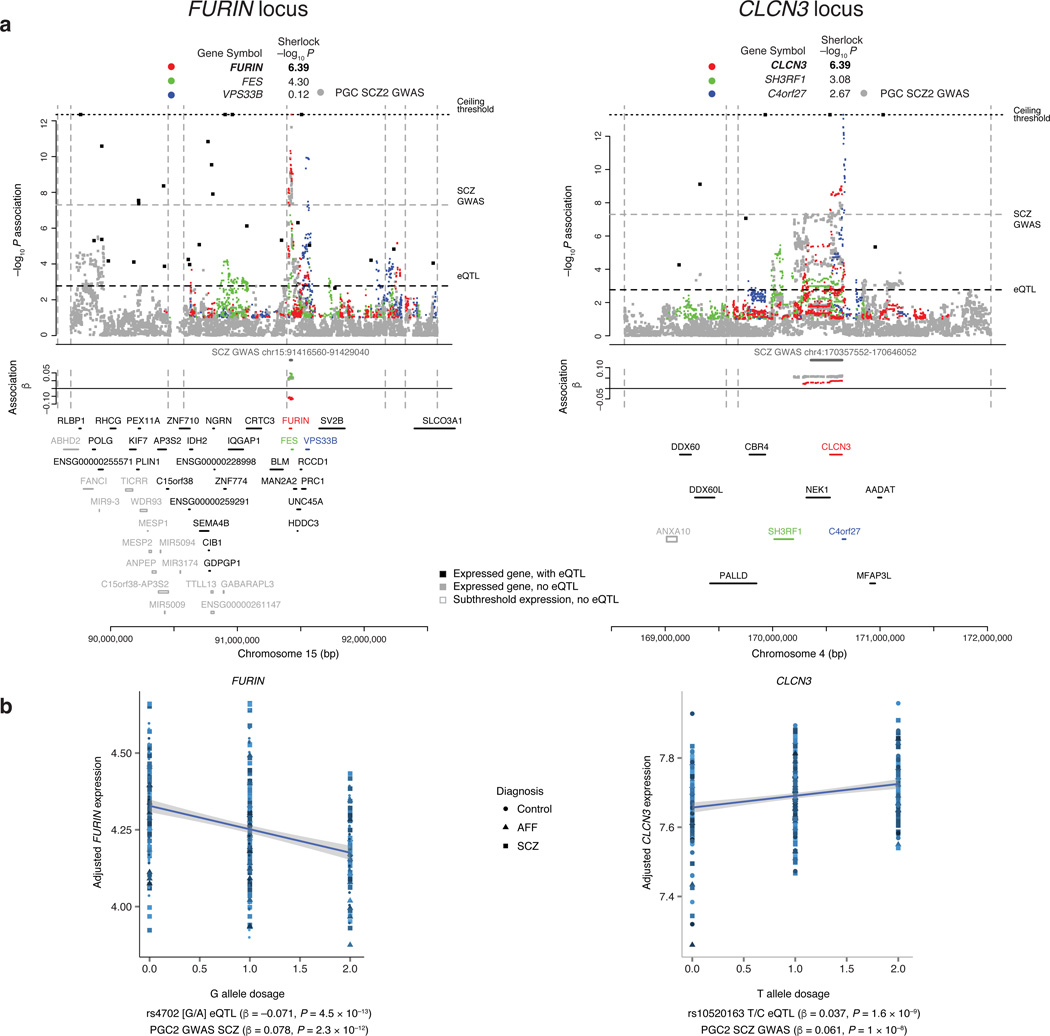
Of the 19 GWAS loci harboring SCZ-associated cis-eQTLs, eight involved only a single gene (i.e., no additional gene with relaxed adjusted Sherlock p < 0.5): furin (FURIN, down-regulated by risk allele), t-SNARE domain containing 1 (TSNARE1, up), contactin 4 (CNTN4, up), voltage-sensitive chloride channel 3 (CLCN3, up), synaptosomal-associated protein of 91 kDa (SNAP91, up), ENSG00000259946 (up), ENSG00000253553 (down), and the ENST00000528555 isoform of sorting nexin 19 (SNX19, down) (Fig. 2 and Supplementary Fig. 6B and 7A). For functional follow-up, we focused on the five single-gene loci encoding known proteins implicated at the gene level. First, we replicated these eQTL in the Religious Orders Study and Memory and Aging Project (ROS/MAP)26, with unpublished human DLPFC RNA sequencing data (N = 461). The most significant GWAS SNP was also a significant eQTL with the same direction of effect as in CMC for FURIN (rs4702: P = 1 × 10−6), CLCN3 (rs10520163: P = 9 × 10−6), and SNAP91 (rs3798869: P = 3 × 10−4); TSNARE1 (rs4129585: P = 0.057) and CNTN4 (rs17194490: P = 0.07) also had alleles in the same direction of effect as in CMC but did not reach significance.
Figure 2. Overlap of GWAS for schizophrenia with eQTL in the DLPFC.
(a) eQTL association profiles across two representative SCZ GWAS loci on chromosomes 15 and 4, respectively. SNP-level associations are plotted for the SCZ GWAS (gray), and cis-eQTL association profiles for genes with Sherlock Pcorrected < 0.5 (or RTC > 0.9) are plotted in colors, with colors and Sherlock P values noted on top of the graphic (P = 4.07 × 10−7 and P = 4.07 × 10−7 for FURIN and CLCN3, respectively). For additional genes in the region with significant eQTL, the single eSNP with minimal eQTL P value (“max-eQTL”) is marked by a black point (corresponding genes names are located above the chromosome marker bar). Locations of regional protein-coding genes and non-coding RNAs without significant eQTL are annotated in gray. Vertical dotted lines mark recombination hotspot boundaries; horizontal dotted lines denote the significance thresholds for eQTL and GWAS, and the ceiling imposed for visualization purposes. Association betas (effect sizes) are plotted for SNP alleles associated with increased SCZ risk, in colors corresponding to genes as above. The red points illustrate the betas for the SCZ risk alleles on expression of the corresponding gene (FURIN and CLCN3, respectively), where values above the 0 line mark up-regulation (CLCN3) and below the line down-regulation (FURIN). (b) The association of expression of FURIN (N = 467, β = −0.071, P = 4.5 × 10−13) and CLCN3 (N = 467, β = 0.037, P = 1.6 × 10−9) with SCZ risk allele at the GWAS index SNP in the respective loci from (a), with shape corresponding to diagnosis.
CLCN3, SNAP91, and TSNARE1 are direct synaptic components, and CNTN4 and FURIN play roles in neurodevelopment. Specifically, CLCN3 (or ClC-3) is a brain-expressed chloride channel, where it appears to control fast excitatory glutamatergic transmission 27. SNAP91 is enriched in the presynaptic terminal of neurons where it regulates clathrin-coated vesicles, the major means of vesicle recycling at the presynaptic membrane. TSNARE1 plays key roles in docking, priming, and fusion of synaptic vesicles with the presynaptic membrane in neurons, thus synchronizing neurotransmitter release into the synaptic cleft. CNTN4 is a member of the contactin extracellular cell matrix protein family responsible for development of neurons including network plasticity28. It plays a key role in olfactory axon guidance29, and there is evidence for association of copy number variants overlapping CNTN4 with autism30. FURIN processes precursor proteins to mature forms, including brain-derived neurotrophic factor (BDNF), a key molecule in brain development whose down-modulation has been hypothesized as related to schizophrenia31, and BDNF and FURIN are up-regulated in astrocytes in response to stress.
The major histocompatibility complex (MHC / human leukocyte antigen / HLA) region is consistently most highly associated with SCZ, but it is a difficult region to dissect for causal variation because of its unusually high linkage disequilibrium and gene density (>200 DLPFC-expressed genes in chr6:25–36 Mb). Nevertheless, only five genes in this locus were ranked highly by Sherlock and passed evaluation for concordance of associations (Supplementary data file 2): C4A, HCG17, VARS2, HLA-DMB, and BRD2. Consistent with recent work identifying structural variation of the C4 genes as partly mediating the genetic MHC association, resulting in higher expression and perhaps driving pathological synapse loss in schizophrenia32, we found a strong correlation between the risk alleles for SCZ and up-regulation of expression of C4A (complement component 4A; Spearman’s ρ = 0.66, P < 10−16).
Functional dissection of genes highlighted
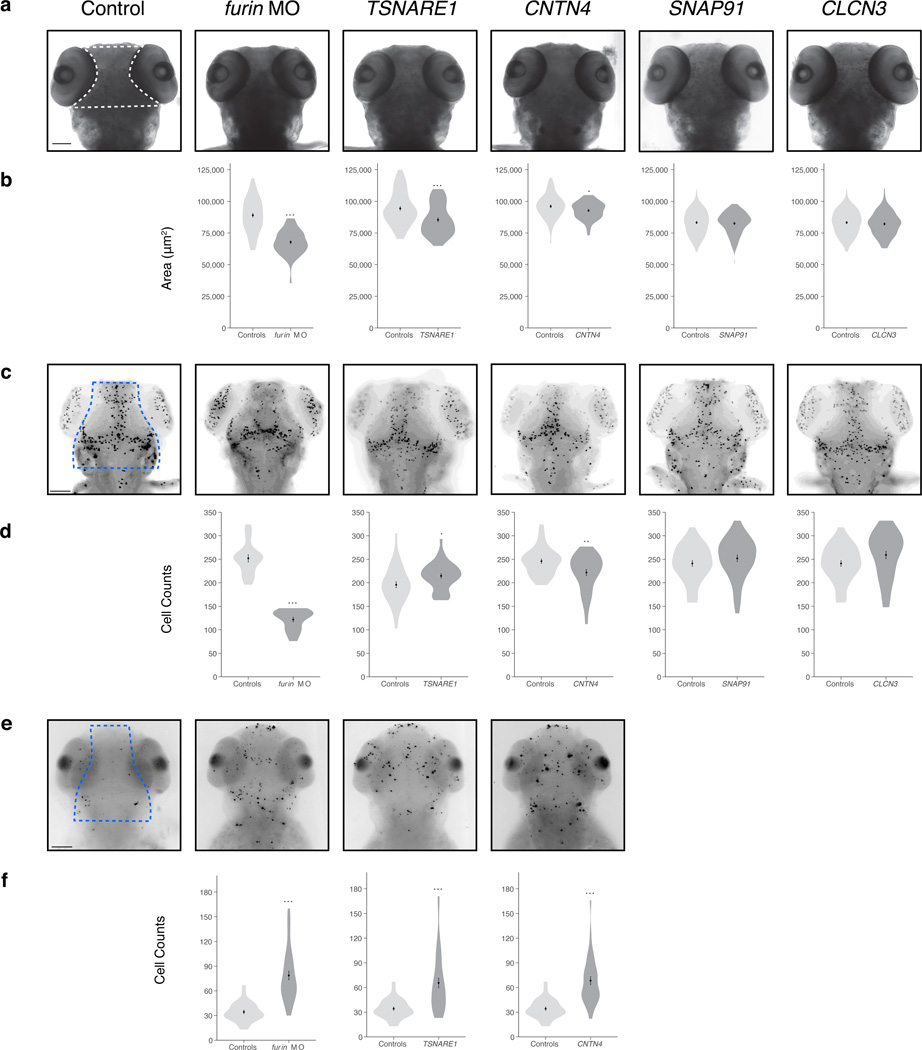
Our results point to a number of genes worthy of follow-up, and we sought an assay that was rapid and amenable to over- and under-expression. Manipulation of zebrafish embryos fits these requirements, especially for evaluation of anatomical phenotypes of early development, such as head and brain size (or area). Perturbing expression of one or more genes in zebrafish has been used to identify genes contributing to neuropsychiatric disorders33–35. Therefore, we asked whether suppression or overexpression of the corresponding gene within each of the five SCZ risk loci could identify key proteins that regulate brain development. To evaluate the four genes up-regulated by risk alleles in the GWAS loci, we injected 200pg of human capped mRNA encoding TSNARE1, CNTN4, SNAP91, or CLCN3 in 1–8 cell stage embryos (N = 60 per experiment, at least two biological replicates performed). At 3 days post-fertilization (dpf), we assessed the area of the head that contains the forebrain and midbrain structures (Fig. 3A, B). Relative to control embryos, overexpression of TSNARE1 or CNTN4 resulted in a significant decrease in head size, 9.5% (P < 0.001) and 3.5% (P = 0.018), respectively, while SNAP91 or CLCN3 showed no statistically significant effect (Fig. 3A, B). Body length and somitic structures were similar across all embryos, suggesting that our observations were unlikely due to gross developmental delay. For FURIN, we sought to mimic the transcriptional down-regulation in human brains associated with SCZ risk. A reciprocal BLAST search of the zebrafish genome revealed a FURIN ortholog with two potential paralogs; both copies were expressed at ~40–60 counts per million reads in mRNA from heads of 3 dpf zebrafish embryos36. We depleted furin_a, the isoform most closely resembling the human ortholog, using a splice blocking morpholino (sbMO) that almost completely extinguished expression of the endogenous message by triggering the inclusion of intron 7 (Supplementary Fig. 8). Suppression of furin_a led to a 24% decrease in head size (Fig. 3A, B); this observation was replicated in CRISPR/Cas9 mutants (Supplementary Fig. 8) and in embryos injected with a second sbMO targeting exon 5 (data not shown) Importantly, expression of human FURIN mRNA could rescue the phenotype induced by either morpholino, providing evidence for specificity (Supplementary Fig. 8).
Figure 3. Neuroanatomical phenotypes upon suppression or overexpression of genes at SCZ risk loci.
(a) Head size phenotype after suppression of furin_a (3ng MO) or overexpression of TSNARE1, CNTN4, SNAP91 or CLCN3 (200ng). Representative head size images per treatment condition are shown, quantified area is depicted by the dashed white lines in the control image. (b) Quantification of head size phenotype in each treatment condition as compared to control embryos for furin MO (Ncontrol = 76, Nfurin MO = 66, P = 5.32 × 10−20), TSNARE1 (Ncontrol = 78, NTSNARE1 = 64, P = 4.69 × 10−5), CNTN4 (Ncontrol = 66, NCNTN4 = 75, P = 0.018), SNAP91 (Ncontrol = 114, NSNAP91 = 106, p = 0.57), CLCN3 (Ncontrol = 114, NCLCN3 = 100, P = 0.40). (c) Representative images of PH3 staining assessing proliferation phenotypes. Dashed blue lines depict the area included in the quantification of cell counts. (d) Quantification of PH3-labeled cells with respect to each treatment condition for furin MO (Ncontrol = 19, Nfurin MO = 20, P = 7.56 × 10−17), TSNARE1 (Ncontrol = 40, NTSNARE1 = 40, P = 0.018), CNTN4 (Ncontrol = 39, NCNTN4 = 38, P = 0.0032), SNAP91 (Ncontrol = 40, NSNAP91 = 40, P = 0.25), CLCN3 (Ncontrol = 40, NCLCN3 = 40, P = 0.07). (e) Representative images of TUNEL staining per condition marking cells undergoing apoptosis. Area quantified is depicted within the dashed blue lines. (f) Cell counts of apoptotic cells in each treatment condition as compared to controls for furin MO (Ncontrol = 33, Nfurin MO = 39, P = 1.10 × 10−10), TSNARE1 (Ncontrol = 33, NTSNARE1 = 38, P = 9.44 × 10−6), CNTN4 (Ncontrol = 33, NCNTN4 = 35, P = 1.98 × 10−8). Error bars are s.e., * P < 0.05, ** P < 0.005, *** P < 0.0005; MO - morpholino. Scale bar = 100 µm. In all cases, t- tests were used to generate P values.
Given a potential role for FURIN, TSNARE1, and CNTN4 during neurogenesis, we asked whether the decrease in head size could be attributed to changes in cell proliferation and/or apoptosis. Overexpression of CNTN4 and suppression of furin_a led to a 9.8% (P = 0.003) and a 29.8% (P < 0.001) decrease, respectively, in proliferating cells marked by phospho-histone3 (PH3), and overexpression of TSNARE1 led to a 9.5% increase (P = 0.018) in proliferating cells (N = 20 per experiment; Fig. 3C, D). Next, we wondered how more proliferating cells nevertheless resulted in a smaller head size phenotype for the case of TSNARE1. To test the possibility that cells exiting cell cycle experience a higher apoptotic index, we performed TUNEL staining on injected embryos, and determined that modulation of all three target genes led to a significant increase in apoptotic cells in the head region corresponding to our head size measurements (N = 20 per experiment; P < 0.001; Fig. 3E, F). Taken together, the data support the hypothesis that changes in FURIN, TSNARE1, and CNTN4 expression levels induce subtle neuroanatomical variation in multiple brain regions.
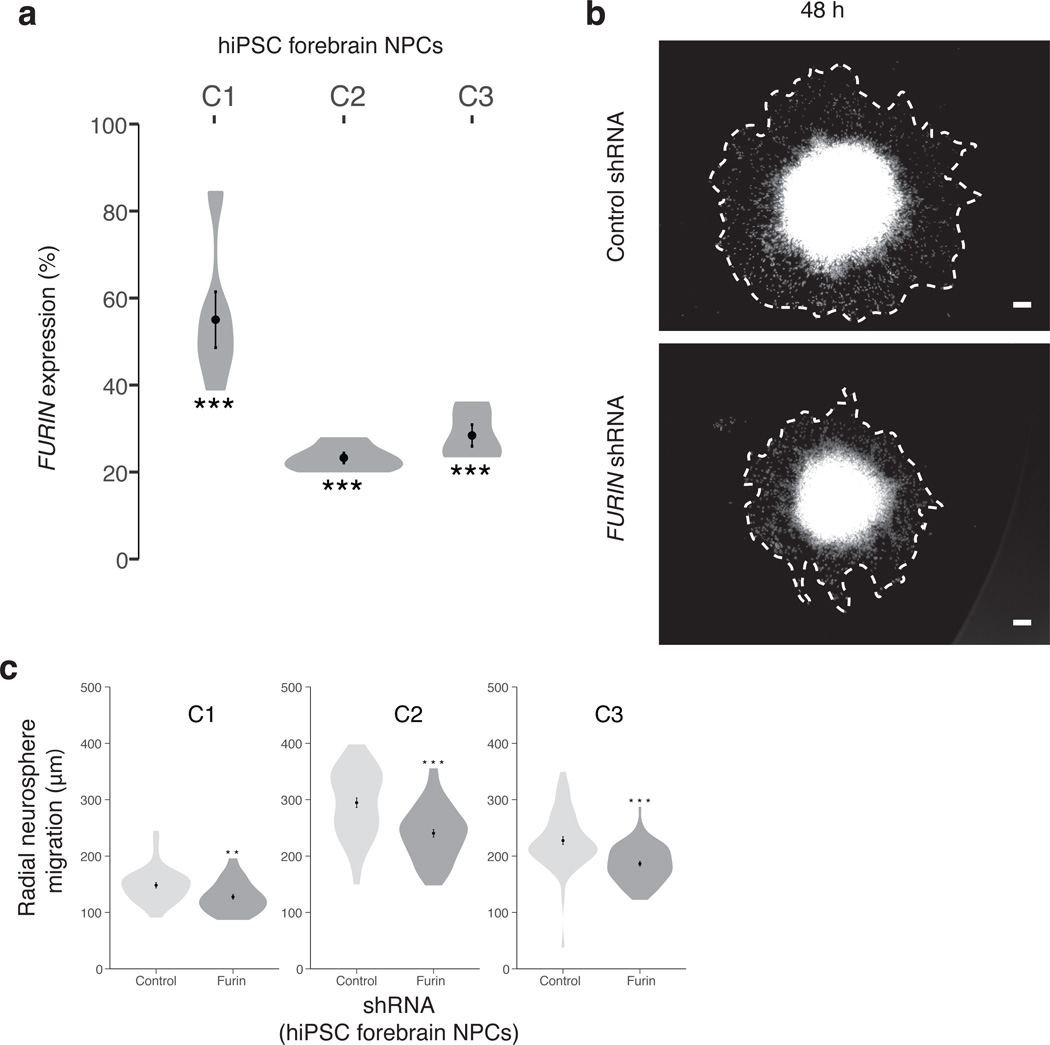
Depletion of furin in our in vivo zebrafish model had the largest impact on head size. Thus we further tested the impact of FURIN knockdown in human neural progenitor cells (NPCs) capable of differentiating into mixed populations of post-mitotic neurons and astrocytes37,38. Neurosphere outgrowth is a well-established neural migration assay measuring the distance NPCs migrate away from the neurosphere. NPCs were differentiated from human induced pluripotent stem cells (hiPSCs) reprogrammed from human fibroblasts using sendai viral vectors39,40. Pairwise isogenic comparisons were conducted in 307 neurospheres from three independent unaffected controls. We measured migration of DAPI-positive nuclei from pLKO.1 non-hairpin-PURO control neurospheres (n = 147) and LV-FURIN shRNA-PURO (shRNA-FURIN) knockdown neurospheres (n = 160). FURIN knockdown in the hiPSC NPCs resulted in significantly decreased total radial migration for all three individuals (C1: 1.16-fold decrease, P < 0.0017; C2: 1.23-fold decrease, P < 3 × 10−6; C3: 1.22-fold decrease, P < 2 × 10−6) (Fig. 4).
Figure 4. Decreasing FURIN expression in human NPCs perturbs neural migration.
(a) FURIN expression reduction achieved by lentiviral (LV)-FURIN shRNA-PURO, relative to LV-non-hairpin-PURO control (C1: N = 6; P = 4.5 × 10−4; C2: N = 6, P = 6.2 × 10−9; C3: N = 5, P = 4.2 × 10−6). (b) Representative images of the hiPSC NPC neurosphere outgrowth assay after 48 hours of migration, following transduction with LV-FURIN shRNA-PURO and LV-non-hairpin-PURO control. The average distance between the radius of the inner neurosphere (dense aggregate of nuclei) and outer circumference of cells (white dashed line) was calculated. DAPI-stained nuclei (blue), scale bar 100 µm. (c) Across hiPSC NPCs generated from three controls (C1: Nvehicle = 42, NshRNA-FURIN = 44, 1.16-fold decrease, P < 0.0017; C2: Nvehicle = 49, NshRNA-FURIN = 53, 1.23-fold decrease, P < 3 × 10−6; C3: Nvehicle = 56, NshRNA-FURIN = 63, 1.22-fold decrease, P < 2 × 10−6), average radial neurosphere migration following transduction with LV-FURIN shRNA-PURO (red bars) or LV-non-hairpin-PURO (gray bars). Error bars are s.e., *P < 0.05, **P < 0.01, ***P < 0.001. In all cases, a t-test was used to generate P values.
Gene expression is subtly disrupted in schizophrenia
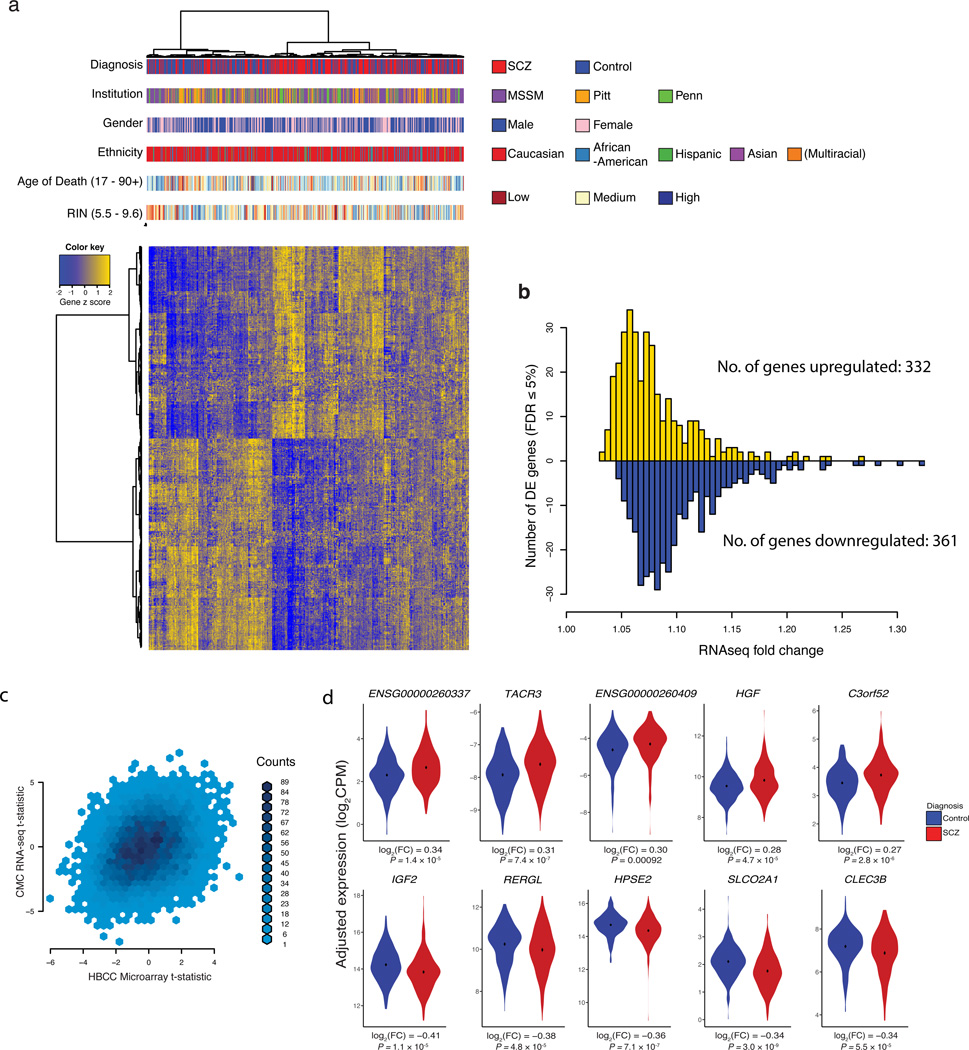
We next evaluated whether SCZ cases versus controls differed in their expression levels per gene. Following normalization of read counts for each gene, a weighted linear regression adjusting for known covariates was performed (Supplementary Figs. 2 and 4). Analysis of the distribution of P values for the 16,423 genes was tested for a mixture of disease-associated and null distributions for 25 cases and 25 controls and suggests that approximately 44% of genes are perturbed in SCZ; this excess of low P values disappears when case and control labels are permuted. While polygenic inheritance, where many genes are affected but to a small degree3, could explain this result, treatment and environmental factors also likely play a role. Without imposing a threshold on the magnitude of fold change in mean expression between SCZ and controls, we find 693 genes to be differentially expressed after correction for multiple testing (FDR ≤ 5%), 332 up-regulated and 361 down-regulated (Fig. 5A, Supplementary data file 3). All had modest fold changes (Fig. 5B), with a mean of 1.09 and range 1.03–1.33 (inverting down-regulated expression ratios). As expected, hierarchical clustering of the differentially expressed genes showed case-control distinctions but were independent of institution, sex, age at death, ethnicity, and RIN (Fig. 5A). We examined differential expression in an independent sample, the NIMH Human Brain Collection Core (HBCC), which generated DLPFC gene expression data using Illumina HumanHT-12_V4 Beadchip microarrays from 131 SCZ cases and 176 controls. Though these arrays differ from RNA-seq in their capture features, there was high correlation of test statistics for differential expression in CMC compared to HBCC for the differentially expressed genes also present in the HBCC data (480 of 693), Pearson correlation r = 0.58 (P < 10−16); the correlation remains high (r = 0.28, P < 10−16) across all 10,928 genes common to both platforms after QC (Fig. 5C).
Figure 5. Differential expression between schizophrenia cases and controls in the DLPFC.
(a) For the N = 693 genes differentially expressed at FDR ≤ 5%, bivariate clustering of individuals (columns) and genes (rows) depicts the case-control differences, as marked by the red-blue horizontal colorbar at top (‘Diagnosis’). An individual’s expression (converted to a z-score per gene) is red for above-average values, and green for below-average values; thus, the top cluster of the plot consists of genes up-regulated in cases versus controls (green in top left; red in top middle), and the bottom cluster of down-regulated genes (red in bottom left; green in bottom middle). In addition to the horizontal colorbar marking case-control status for each sample, additional colorbars denote brain bank (‘Institution’), gender, reported ancestry (‘Ethnicity’), age of death, and RNA quality (‘RIN’), where the latter two use a continuous-values color scale (with low, medium, and high as colored), relative to the range denoted on the figure. (b) Distribution of fold-change of differential expression for 693 differentially expressed genes. Case:control fold-changes for up-regulated genes are plotted in red (N = 332, positive values), and control:case fold-changes for down-regulated genes in green (N = 361, negative values). (c) Binned density scatter plot comparing the t-statistics for case versus control differential expression between the independent HBCC replication cohort assayed on microarrays and the CommonMind RNA-seq data; correlation between the statistics is 0.28 (P < 10−16). (d) For the 10 significantly differentially expressed genes with the largest fold changes (5 up- and 5 down-regulated), the 25 cases and 25 controls of normalized and adjusted gene expression in cases (red) versus controls (blue).
The differential expression observed here is smaller than that reported in earlier studies (Supplementary data file 1), but it is consistent with plausible models for average differential gene expression and the polygenic inheritance of SCZ (Supplementary Text, “Differential gene expression: expectation, variability, and power analyses”). Consider, for example, a gene for which the major determinant of differential expression is the case-control difference in allele frequency at an eQTL SNP. For that gene, the expected magnitude of differential expression fold change will be on the order of the allele frequency differences seen in the recent large Psychiatric Genomic Consortium SCZ genetic association study (~1–2%)3, precisely what is observed in the CMC data. Beyond case-control difference in allele frequency, our modeling can also explain the difference between earlier studies and CMC results (Supplementary Fig. 9); because earlier studies tend to be far smaller in sample size, their larger differential expression is consistent with either the well-known “Winner’s Curse”41 or false positives that may occur in smaller samples. Finally, our results imply a need for thousands of samples to ensure 80% statistical power to observe differential expression between cases and controls for the genes implicated at SCZ-associated eQTL, e.g., the five genes of interest above.
The most highly up-regulated protein-coding gene is tachykinin receptor 3 (TACR3, NK3 receptor, 1.24-fold, Fig. 5D). NK3 antagonists have been tested in SCZ and other CNS diseases42. Moreover, rat and human studies have suggested a role for the NK3 receptor in memory and cognition43, both key impairments of schizophrenia. Insulin-like growth factor 2 (IGF2), the most strongly down-regulated gene (1.33-fold, Fig. 5D), can rescue neurogenesis and cognitive deficits in certain mouse models of schizophrenia44. Also included among the top 100 differentially expressed genes are the alpha 5 subunit of the GABA A receptor (GABRA5) and calbindin (CALB1), genes previously reported as differentially expressed in cortical tissue from schizophrenia patients, suggesting GABAergic interneuron dysfunction45. Available in situ hybridization data from DLPFC suggest that genes identified by DE analysis display various degrees of cell-type specificity, which could affect the estimated fold changes (Supplementary Fig. 10).
We identified 239 isoforms differentially expressed between SCZ cases and controls: 94 up-regulated and 145 down-regulated. These isoforms derive from 223 genes, which are enriched, as expected, for overlap with the 693 differentially expressed genes (P = 2 × 10−131, Fisher’s exact test), and 136 are differentially expressed at both the gene and isoform levels (Supplementary Fig. 11). No obvious unifying biological theme emerges from this set of genes and isoforms on the basis of pathway enrichment analysis (Supplementary data file 4). An assessment of the impact of age at death or cell type proportions suggests that these variables do not explain significant differential expression (Supplementary Fig. 12). Although analyses of experiments performed using either monkeys or rodents indicate that genes whose expression are affected by antipsychotics are often the same as those we find altered in individuals with SCZ, the impact of antipsychotic drugs nevertheless tends to be significantly in the opposite direction of that observed in the SCZ subjects (Supplementary Table 2). Thus, our analyses find that genes highlighted by the contrast of SCZ cases versus control subjects do not largely trace their differential expression to antipsychotic medications, although intriguingly they do suggest a mechanism for the efficacy of these drugs46.
Brain co-expression networks capture SCZ associations
Coordinated expression of genes is critical to brain development and function. One expectation of polygenic inheritance of disease is that this coordination may be subtly altered in individuals with SCZ. To assess this, we applied weighted gene co-expression network analysis (WGCNA) to the matrix of pairwise gene co-expression values. WGCNA recovers a network that consists of nodes (genes) and edges connecting nodes (i.e., the degree of co-expression for a pair of genes, measured as their correlation after transformation by raising the value to a power β that results in an overall scale-free topology). WGCNA divides the network into subnetworks called modules, or clusters of genes with more highly correlated expression.
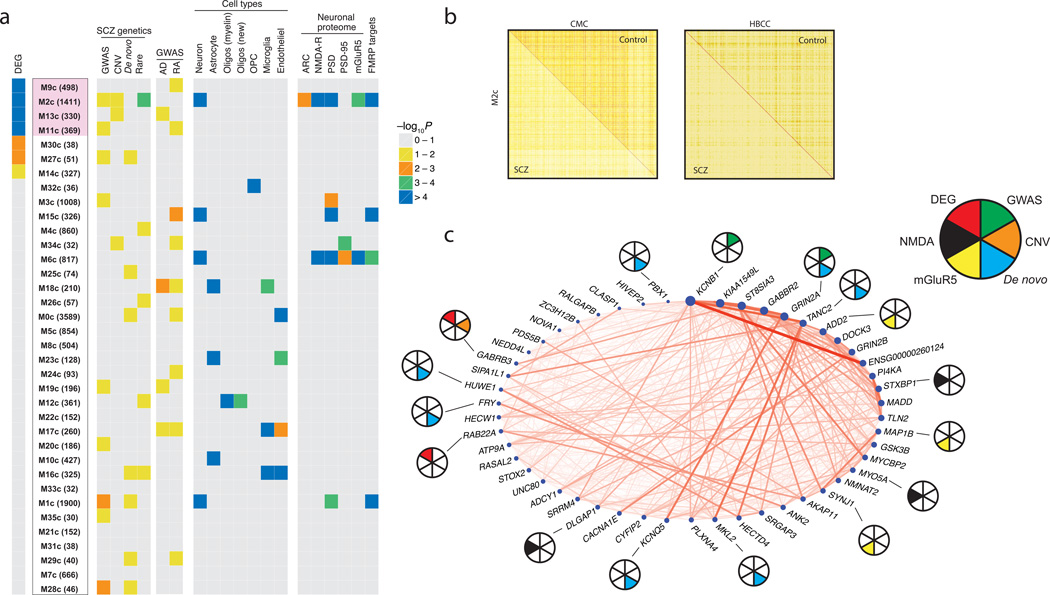
We constructed gene co-expression networks separately from control individuals and SCZ cases (Supplementary data file 5), since we wished to assess disease-dependent changes in co-expression for modules of interest15. The co-expression network generated from the controls consisted of 35 modules each containing between 30 and 1,900 genes, along with ~3,600 unclustered genes (Supplementary data file 5). Four modules stand out in harboring an excess of differentially expressed genes (Fig. 6A, Supplementary data file 6). Of these, however, only one (M2c) shows association with differential expression (OR = 2.3, P = 1 × 10−13) and multiple prior genetic associations with SCZ; the latter encompasses genes in GWAS loci (FE [fold-enrichment] = 1.36, P = 0.04), rare CNV (FE = 1.52, P = 0.051), and rare nonsynonymous variants (FE = 1.18, P = 2 × 10−4) (Supplementary table 3). Given its apparent relevance to SCZ risk, we tested if the co-expression pattern for M2c was perturbed in SCZ samples relative to controls. We used two categories of network-based preservation statistics: (a) testing whether highly connected nodes in a module remain as highly connected (“density”), or (b) testing for differences in the overall connectivity pattern in a module (“connectivity”). The M2c module exhibits a loss of density in the SCZ cases (permutation Z = −1.79, one-tailed P = 0.037, Fig. 6B) but no loss of connectivity. The loss of density replicates in the HBCC cohort (Z = −3.02, P = 0.003), indicating that the regulatory coordination of genes in this module is disrupted in SCZ. The dysregulation of M2c in SCZ is not due to medication effect or clinical and technical confounds.
Figure 6. Co-expression network analysis in control DLPFC samples.
(a) Control-derived modules were ranked by enrichment [estimated based on Fisher’s exact test (FET)] with differentially expressed genes; number of genes in each module is given in parentheses. Among the 4 modules with strongest overlap (marked in blue), only the M2c module genes are strongly enriched for multiples lines of prior genetic evidence: differential expression (FET: OR = 2.3, Bonferroni adjusted P = 1.9 × 10−12), SCZ GWAS loci (tested by INRICH: FE [fold-enrichment] = 1.36, P = 0.04), rare CNV (tested by INRICH: FE = 1.52, P = 0.051), and rare nonsynonymous variants (tested by PLINK/Seq and SMP: FE = 1.18, P = 2 × 10−4). The enrichment of each module with SCZ genetics, cell type-specific markers, neuronal proteome sets (proteins that are localized to the postsynaptic density of neurons), and fragile X mental retardation protein (FMRP) targets is depicted at right. As a control, note the lack of enrichment of M2c with common variants for Alzheimer’s disease (AD) and rheumatoid arthritis (RA). (b) Topological overlap matrix of the differentially connected M2c module in controls (upper right triangle) and SCZ cases (lower left triangle) in the CMC (left) and HBCC (right) cohorts. (c) Circle plot showing connection strengths for the top 50 hub genes of the M2c module, where node size corresponds to intramodular connectivity and nodes are ordered clockwise based on connectivity. Pie chart: SCZ susceptibility genes based on GWAS PGC2-SCZ (green), CNV (orange) or de novo (cyan) studies; Genes that belong in the NMDA (black) or mGluR5 (yellow) signalling pathway; Genes that are differentially expressed in schizophrenia vs. controls at FDR ≤ 5% (red).
Consistent with prior studies of the brain transcriptome15,47–50, we find gene co-expression to be organized into modules of distinct cellular and functional categories (Supplementary data file 7). In particular, the M2c module is enriched for multiple categories, including axon guidance, postsynaptic membrane, transmission across chemical synapses, and voltage-gated potassium channel complexes (Fig. 6C). Gene sets identified in prior genetic studies that highlighted certain neurobiological functions are also enriched in the M2c module, including the activity-regulated cytoskeleton-associated (ARC) protein complex, targets of fragile X mental retardation protein (FMRP), neuronal markers, post-synaptic density (PSD) proteins, and NMDA receptors (Fig. 6A). Overall, our results point to the M2c module of ~1400 genes that possess functions related to synaptic transmission as being enriched for differential expression, overlapping SCZ genetic signal, and with some genes having less dense co-expression in SCZ cases.
DISCUSSION
Deficits in executive functions, especially cognitive function, are key features of SCZ. The roots of these deficits lie in cortical function and integration, at least in part tracing to the DLPFC. Here we have used gene expression derived from this tissue to understand how genetic liability is related to the molecular etiology of SCZ. Our analyses had two fundamental goals: to identify mechanisms that underlie genetic risk and to describe differences in gene expression and co-expression related to disease. By intersecting transcriptomics and genetics, we elucidated important aspects of the genetic control of transcription and found that genetic variants in 20 of the 108 SCZ GWAS risk loci alter expression of one or more genes. Prior analyses using brain eQTL datasets derived from older technologies have pointed to less than a handful of such associations3. In five of the 20 loci for which we observed regulatory potential of GWAS variants the risk variants altered expression of only one gene. Experimental manipulation of three of these genes had an impact on neuroanatomical and developmental attributes in model systems, making these genes excellent candidates for further biological investigation. We also detected replicable differences in gene expression in SCZ that point to subtle but broad disruption in transcription, which is consistent with the polygenic nature of genetic risk underlying SCZ. Finally, we identified a subnetwork of ~1400 genes sub-serving functions related to synaptic transmission that is significantly perturbed in SCZ and is highly enriched for SCZ genetic signal.
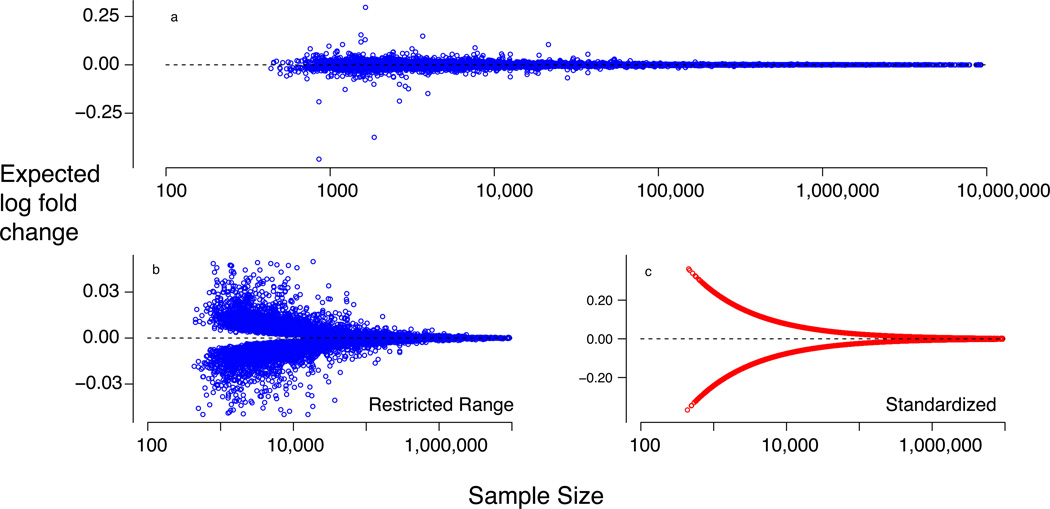
In contrast, we did not find evidence for case-control differential expression among the implicated GWAS risk genes. At first blush this appears to contradict evidence for impact on risk. Yet the magnitude of differential expression will be determined largely by case-control differences in allele frequencies, which we know are small. Modeling the differential in allele frequencies and the predicted effect of alleles on gene expression demonstrates that the distribution of expected differential expression, across genes, is quite similar to the observed distribution from the CMC data (Fig. 7A). Using allele frequencies from the PGC schizophrenia data, we can ask what the number of cases are needed to detect differential expression. For example, 11,784 cases and 11,784 controls would be needed to have 80% power to detect a significant case-control difference in FURIN expression. Genome-wide, the median number of cases and controls needed to obtain 80% power assuming 10,000 genes is ~28,500, well beyond any available dataset (Fig. 7B,C). Our model demonstrates that the distribution of expected differential expression, across genes, is quite similar to the observed distribution from the CMC data (Fig. 7). This calls into question results from smaller studies that report large differential expression. Our analyses show that these studies would have notably larger variability, and because genome-wide surveys test a large number of genes, that variability can translate into large observed differential expression: even when no gene is differentially expressed, studies with only 25 cases and 25 controls can lead to estimates of differential expression exceeding twofold. Notably, this pattern not seen when the N is raised to 250. (See supplementary text for additional scenarios, discussion and modeling.).
Figure 7. Power to detect differential expression.
Analysis of power to detect differential expression of a gene for case versus control subjects, where differential expression is expressed as expected log-fold change, the sample size is the total number of cases and controls to achieve significance (50:50 cases:controls), and the significance level for 80% power is 5 × 10−6. (a) For each gene in the differential expression analysis, we found the cis-eQTL with the smallest P value (see text for additional restrictions). Expected differential expression to achieve 80% power was computed for 10,094 gene-by- cis-eQTL associated pairs. (b) Increased resolution of (a) by limiting the range of differential expression. (c) Standardized log-fold change (80% power) obtained by dividing estimated log-fold change by its estimated standard deviation.
It is conceivable, indeed probable, that certain cells or cell types (e.g., pyramidal neurons) are more salient for risk than the heterogeneous tissue evaluated here. Depending on the pattern of cell-specific gene expression, this scenario could have little or no impact on differential expression or it could diminish it somewhat. The same is true for detection of eQTLs. We do not expect, however, that the scenario will compromise the bulk of our results, all of which complement the genomic studies of this disease. Alterations of the cellular composition in SCZ versus controls might also introduce a systemic bias in the analysis of differential expression; e.g., if the proportion of neurons were reduced by 2% in SCZ versus controls, multiple neuronal genes might appear to be downregulated in SCZ. Analyses of cell composition, however, do not support global differences in the cellular composition in DLPFC tissue from SCZ versus control subjects.
The findings reported here by the CommonMind Consortium (CMC) represent a unique resource to understand brain function, basic neuroscience, and brain diseases at the molecular level. They include a comprehensive compilation of gene expression patterns, together with intensive evaluation of eQTLs across the genome. The expertise and support to produce and analyze these data required a consortium of brain banks, pharmaceutical companies, a foundation, academic centers, and the NIMH, and this work represents the first phase of our ongoing project. All results are available through the CommonMind Knowledge Portal with a searchable database of eQTLs and other visualizations (https://shiny.synapse.org/users/ssiebert/cmc_eqtl_query/). Both alone, and in combination with other datasets such as GTEx, the CMC data will empower future studies paving the way for connecting genetic influences on cellular function with changes in macroscopic circuits of the brain that may ultimately lead to disease.
ONLINE METHODS
Post-mortem samples
Data generated for this study came from postmortem human brain specimens originating from the tissue collections at the three brain banks described below. All samples were shipped to the Icahn School of Medicine at Mount Sinai (ISMMS) for nucleotide isolation and data generation. See Supplementary Fig. 1A for an overview of the sample collection and aggregation workflow.
Selection criteria
Postmortem tissue from schizophrenia (SCZ) and bipolar or other affective/mood disorder (AFF) cases were included if they met the appropriate diagnostic DSM-IV criteria, as determined in consensus conferences after review of medical records, direct clinical assessments, and interviews of family members or care providers. Cases were excluded if they had neuropathology related to Alzheimer’s disease and/or Parkinson’s disease, acute neurological insults (anoxia, strokes, and/or traumatic brain injury) immediately prior to death, or were on ventilators near the time of death. Three case samples (2 with leukotomies, and 1 with a history of a head injury prior to diagnosis) were included; these were not outliers on any metrics that we used to evaluate our samples (see “RNA-seq outliers” below).
“MSSM” sample - Mount Sinai NIH Brain Bank and Tissue Repository (NBTR) (http://icahn.mssm.edu/research/labs/neuropathology-and-brain-banking)
The Mount Sinai Brain Bank was established in 1985. The NBTR obtains brain specimens from the Pilgrim Psychiatric Center, collaborating nursing homes, Veteran Affairs Medical Centers and the Suffolk County Medical Examiners Office. Diagnoses are made based on DSM-IV criteria and are obtained through direct assessment of subjects using structured interviews and/or through psychological autopsy by extensive review of medical records and informant and caregiver interviews52,53. Informed consent is obtained from the next of kin. The brain bank procedures are approved by the ISMMS IRB and exempted from further IRB review due to the collection and distribution of postmortem specimens. All samples for the study were dissected from the left hemisphere of fresh frozen coronal slabs cut at autopsy from the dorsolateral prefrontal cortex (DLPFC) from Brodmann areas 9/46. Immediately after dissection, samples were cooled to −190°C and dry homogenized to a fine powder using a L-N2 cooled mortar and pestle. Tissue was transferred on dry ice to ISMMS as a dry powder for DNA and RNA extraction.
“Pitt” sample - The University of Pittsburgh Brain Tissue Donation Program
Brain specimens from the University of Pittsburgh Program are obtained during routine autopsies conducted at the Allegheny County Office of the Medical Examiner (Pittsburgh) following the consent of the next of kin 54. An independent committee of experienced research clinicians makes consensus DSM-IV diagnoses for all subjects on the basis of medical records and structured diagnostic interviews conducted with the decedent’s family member 55. All procedures for Pitt samples have been approved by the University of Pittsburgh’s Committee for the Oversight of Research involving the Dead and Institutional Review Board for Biomedical Research. At autopsy, the right hemisphere of each brain is blocked coronally, immediately frozen, and stored at −80°C56. Samples for this study contained only the gray matter of DLPFC, where Brodmann area 9/46 was cut on a cryostat and collected in tubes appropriate for DNA or RNA extraction. The DNA and RNA tubes were shipped on dry ice to ISMMS as homogenized tissue in trizol for RNA extraction and thinly sliced tissue for DNA extraction. Specimens from Pitt were provided as matched case/control pairs. These were perfectly matched for sex, and as closely as possible for age (73% of pairs were matched within 5 years, and 95% within 10 years) and race (71% of pairs were matched for race). Members of a pair were always processed together for RNA-seq. Tissue for 10 of the Pitt controls was extracted in duplicate, once as part of a SCZ pair and once as part of a bipolar pair.
“Penn” sample - University of Pennsylvania Brain Bank of Psychiatric illnesses and Alzheimer’s Disease Core Center (http://www.med.upenn.edu/cndr/biosamples-brainbank.shtml)
Brain specimens are obtained from the Penn prospective collection. Disease diagnoses were made based on DSM-IV criteria and obtained through a clinical interview by psychiatrist and review of medical records. All procedures for Penn are approved by the Committee on Studies Involving Human Beings of the University of Pennsylvania, and the use of control postmortem tissues was considered exempted research in accordance with CFR 46.101 (b), item 65 of Federal regulations and University policy. At autopsy, the right or left hemisphere of each brain is blocked into coronal slabs, which are immediately frozen and stored at −80°C. For this study, Brodmann areas 9/46 were dissected from either the left or right hemisphere and pulverized in liquid nitrogen. The tissue was shipped in tubes appropriate for DNA or RNA extraction to ISMMS as homogenized tissue in trizol for RNA extraction and as dry pulverized tissue for DNA extraction.
Tissue, RNA and DNA preparation
Total RNA was isolated from approximately 50 mg homogenized tissue in Trizol using the RNeasy kit according to manufacturer protocol. Samples were processed in batches of 12, and the Pitt matched case/control pairs were always processed in the same batch. The order of extraction for SCZ-affected and control samples was assigned randomly with respect to brain bank, diagnosis, and all other sample characteristics. Because the affective disorder cases (AFF) and matched controls from Pitt were not available until after the processing of the SCZ and controls was underway, these samples were randomized among the remaining 132 SCZ and control samples still queued for extraction at that time. The mean total RNA yield was 15.3 ug (+/− 5.7). The RNA Integrity Number (RIN) was determined by fractionating RNA samples on the 6000 Nano chip (Agilent Technologies) on the Agilent 2100 Bioanalyzer. 51 samples with RIN < 5.5 were excluded from the study (see Sample QC below). Among the remaining samples, the mean RIN was 7.7 (+/− 0.9), and the mean ratio of 260/280 was 2.0 (+/− 0.02).
DNA was isolated from approximately 10 mg dry homogenized tissue from specimens coming from the MSSM and Penn brain banks. The thinly sliced tissue from Pitt was homogenized before DNA isolation. All DNA isolation was preformed using the Qiagen DNeasy Blood and Tissue Kit according to the manufacturer’s protocol. DNA yield was quantified using Thermo Scientific’s NanoDrop. The mean yield was 12.6 ug (+/− 4.6), the mean ratio of 260/280 was 2.0 (+/− 0.1), and the mean ratio of 260/230 was 1.8 (+/− 0.6).
RNA Library Preparation and Sequencing
Processing order was re-randomized prior to ribosomal RNA (rRNA) depletion, and samples were processed in batches of 8. To expedite sequencing, processing began before extraction was complete and randomization occurred among all available extracted samples in sets of 120 to 226. Briefly, rRNA was depleted from about 1 ug of total RNA using Ribo-Zero Magnetic Gold kit (Illumina/Epicenter Cat # MRZG12324) to enrich for polyadenylated coding RNA and non-coding RNA. The Pitt case/control pairs were batched together in each processing step, including Ribo-Zero depletion, sequence library preparation, and sequencing lane. 10 of the Pitt controls were extracted and sequenced as independent duplicates, once as part of a SCZ pair and once as part of a bipolar pair. The sequencing library was prepared using the TruSeq RNA Sample Preparation Kit v2 (RS-122–2001-48 reactions) in batches of 24 samples. The insert size and DNA concentration of the sequencing library was determined on Agilent Bioanalyzer and Qubit, respectively. A pool of 10 barcoded libraries were layered on a random selection of two of the eight lanes of the Illumina flow cell bridge amplified to ~250 million raw clusters. One-hundred base pair paired end reads were obtained on a HiSeq 2500. The sequence data were processed for primary analysis to generate QC values (reads were mapped to the human reference genome using TopHat; see “Mapping, QC and quantification of Gene Expression” below). Samples with a minimum of 50 million mapped reads (~25 million paired end reads) and less than 5% rRNA-aligned reads were retained for downstream analysis. We attempted a single round of re-sequencing for samples that failed these QC criteria. In the end, a total of 15 samples did not meet these sequencing criteria (see “Sample QC” below) and were discarded.
DNA genotyping, QC, ancestral evaluation and polygenic scoring
Genotyping was preformed on the Illumina Infinium HumanOmniExpressExome 8 v 1.1b chip (Catalog #: WG-351–2301) using the manufacturer’s protocol. Samples for genotyping were aliquoted onto 96 well plates, where each plate had an internal control from the HapMap project (NA12878 - Coriell Institute) in two unique locations. Initial QC was preformed using PLINK 57 to remove markers with: zero alternate alleles, genotyping call rate ≤ 0.98, Hardy-Weinberg P value < 5 × 10−5, and individuals with genotyping call rate < 0.90. This removed 2 samples from the analysis. After QC, 668 individuals genotyped at 767,368 markers were used for imputation. Phasing was performed on each chromosome using ShapeIt v2.r79058, and variants were imputed in 5 Mb segments by Impute v2.3.159 with the 1000 Genomes Phase 1 integrated reference panel11 excluding singleton variants. Note that, in addition to the 22 autosomes, we also included chromosome X, split out into pseudoautosomal (PAR) and non-PAR genomic regions to properly handle male haploidy in the non-PAR regions.
To infer ancestry from genetic data, we identified a set of high quality autosomal SNPs from the pre-imputed data with the following properties: an rs dbSNP database identifier, known physical location in the hg19 reference genome, alleles coded as either A, C, G, or T, call rate ≥ 99.5%, minor allele frequency MAF > 0.05. These criteria yielded 552,351 SNPs. Next, using PLINK57, we performed LD pruning using sliding windows of 50 SNPs, with steps of 5 and a pairwise r2 < 0.04 and found 28,663 SNPs. Ancestry was determined using clusterGem in GemTools (arXiv:1104.116260,61, http://www.wpic.pitt.edu/wpiccompgen/GemTools/GemTools.htm). Gemtools found that 5 dimensions and 7 clusters were sufficient to describe the ancestry space. Because one sample was missing key phenotypic information, 667 subjects were assigned ancestry based on DNA genotypes. Supplementary Fig. 1B, C describe the distribution of nominal ancestry and diagnosis and plot several informative dimensions of genetically-inferred ancestry.
We carried out analyses for polygenic scoring of schizophrenia risk using the largest available schizophrenia association dataset3 as the “discovery” set. Quantitative scores were computed for each subject in this paper based on the set of SNPs with P values less than predefined P value thresholds (pT) in the discovery data set: pT < 0.0001, pT < 0.001, pT < 0.01, pT < 0.05, pT < 0.1, pT < 0.2, pT < 0.3, pT < 0.5, and pT < 1. For each SNP set defined by pT, we calculated the proportion of variance explained (Nagelkerke’s r2, Supplementary Fig. 1D). Throughout this work, we refer to the scores defined at pT < 0.5 simply as “polygenic risk scores” (PRS).
RNA Sample QC
Samples were excluded if RIN < 5.5 or genetic information from the sample was inconsistent with subject descriptors such as sex. Of the 633 samples sent for sequencing (those with RIN ≥ 5.5), 15 samples were removed because they yielded < 50 million total reads (~25 million paired end reads) or had > 5% of reads aligning to rRNA, based on two attempts to produce quality sequence (all samples failing either QC criterion on the first attempt were re-prepped and/or re-sequenced, and those failing twice were removed); calculation of RNA-seq QC metrics is described in “Mapping, QC and Quantification of Gene Expression” below. Of the 10 Pitt control samples that were sequenced twice, only the first sequencing run was included in our analysis. Of the remaining 609 samples, two were removed because their DNA genotypes had high rates of missingness; one sibling pair was identified and the sample with the lower RNA quality (RIN) was removed; 14 samples were removed (see details below) because they were determined to be outliers based of a series of multivariate analyses of the RNA-seq data (N =10), or due to sample contamination/mix-up (N = 4). This left 592 samples for subsequent analyses.
To evaluate discordance between nominal and genetically-inferred sex, we used PLINK 57 to calculate the mean homozygosity rate across X-chromosome markers and to evaluate the presence or absence of Y-chromosome markers. Pairwise comparison of samples across all genotypes was done to identify potentially duplicate samples (duplicate pair defined as having genotypes > 99% concordant) or related individuals, again using PLINK.
RNA-seq outliers were detected using two methods in parallel.
To evaluate the data for outliers, one group of analysts used four approaches to normalization: FPKM (fragments per kilobase per million reads) from Cufflinks; quantile normalization across samples; quantile normalization across genes; and trimmed mean of M values (TMM) from the edgeR package62,63. We applied three different methods of analysis to these normalized data sets: Hierarchical Clustering with average linkage (HC); the number of extreme transcripts (NT: the number of transcripts with expression value outside the 95% confidence interval for the transcript, across individuals); and Principal Component Analysis (PCA). For HC, a sample (or small group of samples) was declared an outlier if it did not cluster with other samples. If NT > 7.6% of total transcripts, it was declared an outlier. Finally, if the PCA revealed a sample or small group of samples represented by a leading PC (largest 5), it was declared an outlier. When combining these results, if a sample was declared an outlier by all three methods, it was labeled an outlier.
Separately, another group of analysts applied two procedures to detect outliers on the TMM-normalized data, namely Inter Array Correlation (IAC49) and “Iterative” PCA (iPCA). IAC computes the pairwise correlation over genes for all pairs of samples, plots the distribution of the resulting correlations, and empirically finds outliers. Here we used 3 standard deviations as a threshold to declare a sample an outlier. Alternatively, for iPCA, the following algorithm was implemented: the first two PCs were computed from the data; samples beyond the 95% confidence envelope were identified and removed; then the first two PC were recomputed, outliers identified and removed; and so on, until no outliers were detected. All of the samples removed were declared outliers. The full set of samples labeled outliers was then the union of the IAC and iPCA sets.
The results from analysis (i) and (ii), were compared for consensus. In total, 10 samples were identified as outliers by both groups and these were eliminated from all subsequent analyses.
We ensured DNA and RNA data were from a single individual by making SNP calls from RNA-seq results using samtools and bcftools 0.1.19, using the author-recommended protocol, which includes the “Bayesian inference” option. Calls were made only for SNP locations that were assayed on the genotyping chip. Raw variant calls were filtered, as recommended, using the vcfutils.pl varFilter (v0.1.18) option with the maximum depth set to 120 (roughly twice the average read depth). SNP calls from the DNA genotyping were converted to reference forward strand using PLINK. PLINK/Seq (https://atgu.mgh.harvard.edu/plinkseq/) was then used to generate a VCF file by running the fix-strand and write-vcf commands.
Pairwise-discordance of SNP calls between RNA-seq and the genotyping chip was assessed for all possible combinations of RNA-seq samples and DNA genotyping samples. Discordance was calculated using the variant tools software64, which reports the fraction of discordant sites out of the total number of sites where both samples report a genotype. The basic approach for calling a match was to plot the discordance values across all samples, for an all-by-all comparison, and look for a bimodal distribution with an obvious cutoff point (consistent with pairs that should match and all other pairs which do not). Indeed, all of the distributions were bimodal with regions of zero frequency in between the two peaks. The distributions of discordance values were different for RNA-RNA vs. RNA-DNA. For RNA-RNA sample matches, we called matches as instances where two samples had less than 15% discordance from each other; for DNA-RNA matches, the cutoff was 25%. We verified RNA-DNA matching within samples. Finally, we predicted gender for each sample based on the fraction of total reads aligning to the Y chromosome; if the log(fraction) was ≤ −7.4, the sample was called female, otherwise male. This called gender was evaluated to ensure it matched the reported gender from the corresponding brain bank manifest. By this process, we identified one sample mix-up (wrong sample sent for RNA-seq), and three samples were likely contaminated with other samples (high degree of genotype matching). These four samples were removed.
The entire QC process yielded 592 high-quality samples for analysis (258 SCZ, 279 control individuals, and 55 AFF [47 bipolar disorder, 6 major depressive disorder, and 2 mood disorder, unspecified]), with demographic breakdown of the cases and controls as described in Supplementary Table 1.
Evaluation of RNA Quality
RIN is a standard measure of RNA quality, but it focuses on the integrity of ribosomal RNA, rather than surveying quality of RNA from genes throughout the genome. A few alternatives to RIN have been proposed, a very recent proposal being the “mRIN” method65, which analyzes read coverage over transcripts and derives statistics related to quality. Here we use the mRIN software to evaluate the RNA quality of the samples. The CMC data were processed using the pipeline described on the mRIN website (http://zhanglab.c2b2.columbia.edu/index.php/MRIN). Parameters were set as suggested in the documentation. Additional filtering based on gene expression values was not performed. Data were analyzed without any QC beyond what is automatically implemented in mRIN.
We computed mRIN on the 537 SCZ case and control samples for 18,338 (17,527 uniquely identified) RefSeq transcripts using the mRIN package by Feng et al. Sample by transcript combinations were required to have an abundance > 2. After this step, 6,072 transcripts with a missing rate > 50% were removed from the analysis. Finally, for transcripts with more than one entry in the dataset the entry with the lowest missing rate was retained. After these edits a total of 12,246 transcripts remained. The mRIN statistics and associated P values were subsequently computed using the formulas from the Feng et al. paper. Samples with extreme negative values for the mRIN statistic should indicate low quality samples. The distribution is centered near zero and has no extreme negative values. There were 17 and 3 samples with P value < 0.05 and < 0.01, respectively. One would expect a total of 29 and 5 samples to have P values of this magnitude by chance alone. We therefore conclude the RNA quality of the samples is adequate.
Mapping, QC and Quantification of Gene Expression
The top panel of Supplementary Fig. 2 gives an overview of the RNA-seq data processing pipeline and QC metrics. In detail, reads were mapped to human reference genome hg19 using TopHat version 2.0.9 and Bowtie version 2.1.0, with the following parameters: 0 mismatches in a 20 bp seed, reference guided against Ensembl genes and isoforms (version 70). For each sample, this produced a coordinate-sorted BAM file of mapped paired end reads including those spanning splice junctions, as well as a BAM file of unmapped reads.
Overall quality control metrics were calculated using RNA-SeQC66 for each sample, including total number of reads (counting twice each fragment sequenced, once for each end in pair), number of mapped reads (again, separately counting each end of a paired end since one may map and not the other), the rates of reads mapping to rRNA, intergenic regions, intragenic regions, introns, exons, and the number of genes and transcripts detected (defined here simply as those with at least 5 exon-mapping reads). UCSC Genome Browser transcripts were used for this quality control (QC) analysis.
Genes
Known Ensembl gene levels were quantified by HTSeq version 0.6.0 in intersection-strict mode (the BAM file was streamed to HTSeq through novosort version 1.0.1, as HTSeq accepts read-name-sorted alignments). This provides an integral count of reads for each gene in each sample to be used in downstream analyses (a sample-by-gene “read count matrix”).
Isoforms
Relative isoform abundances (PSI = percent spliced in) of Ensembl genes were estimated using MISO (http://genes.mit.edu/burgelab/miso/; version 0.5.2, run with default parameters 67). We processed the per-sample, per-gene MISO output files to extract the estimated PSI, as well as the standard deviations of the estimated sampled PSI values. We constructed corresponding sample-by-isoform matrices for all subsequent data processing and analysis (see “Isoform-level normalization and analysis” below).
In addition, Cufflinks version 2.1.1 was applied to the BAM files to estimate both gene- and isoform-level FPKM values for all Ensembl genes and isoforms. Separately, Cufflinks was applied to the BAM files to assemble isoforms for each sample. These assembled isoforms were unified across samples using Cuffmerge, resulting in a single GTF file of “merged” genes and isoforms annotated by Ensembl annotations. Cufflinks was then applied to this GTF file to estimate both gene- and isoform-level FPKM values for all merged genes and isoforms.
RAPID RNA-seq pipeline
To robustly facilitate the large-scale nature of the RNA-seq data processing described above for ~600 samples, we utilized RAPiD, an efficient and dependable RNA-seq pipeline manager that automates read alignment, quality control, and quantitative analyses of next-generation sequencing gene expression experiments. By closely integrating with the Apollo framework, RAPiD utilizes high-performance computing clusters and provides pipeline monitoring so that RAPiD runs are automatically tracked, QCd, and visualized on the Apollo Run Console web interface. Of note, RAPiD is designed to be an agile framework that is user-configurable via JSON-formatted “recipes” that define the set of tools and algorithms, and corresponding parameters, for running various pipelines. Thus, in this work, RAPiD easily permitted the addition of alternative splicing analyses by running MISO and custom post-processing of MISO results
Normalization of Gene Expression and Adjustment for Covariates
Gene-level analyses started with the HTSeq-derived sample-by-gene read count matrix. The basic normalization and adjustment pipeline for the expression data matrix (Supplementary Fig. 2, middle and bottom panels) consisted of: a) exploration to determine which known and hidden covariates should be accounted for during analyses; b) voom-based calculation of normalized log(CPM) (read counts per million total reads), along with weights that estimate the precision of each log(CPM) observation estimate68 c) linear regression-based adjustment for the chosen covariates, where linear regression for each gene is performed independently and using the observation weights, so that observations with higher presumed precision will be up-weighted in the linear model fitting process (i.e., weighted least squares regression). We now detail the procedure involved for each of the above steps, where we include both SCZ and AFF cases and controls, and the corresponding diagnosis status (“Dx”) is the primary variable of interest.
Initial normalization of read counts
To define the set of covariates for adjustment, we start by initially normalizing the HTSeq read count matrix for all 56,632 Ensembl genes, using voom without covariates. Next, we filtered out all genes with lower expression in a substantial fraction of the cohort, with 16,423 genes remaining with at least 1 CPM in at least 50% of the individuals; note that only these genes were carried forward into all subsequent analyses. This initially-normalized gene expression matrix was then used to select known covariates (described above). Next, hidden covariates were derived (for use in eQTL analyses only, as is common practice13). These covariates were then included for adjustment in the normalization and adjustment steps.
Normalize observations and estimate confidence of sampling abundance by sequencing
The voom68 normalization scales each sample’s read count for each gene by their total counts across all genes to account for variable sequencing depths across the samples. It then transforms each gene to be more approximately Gaussian by taking the logarithm (base 2) of the counts. Still, as a result of the experimental steps involved in obtaining read counts for genes (PCR, library preparation, sequencing, etc.), the read count for a particular gene will only on average be proportional to the underlying expression level of that gene. Thus, it is critical to model the statistical sampling of gene expression level, since larger log(CPM) typically exhibit lower variance (an example of heteroscedasticity). To this end, voom estimates confidence weights for each normalized observed read count. It does this by residualizing on the covariates (known and surrogate, as applicable), fitting a mean-variance relationship function across all genes, using the fitted function to estimate the variance of a particular read count observation, and then setting the observation weight to be the inverse of the corresponding estimated variance. The normalized observed read counts, along with the corresponding weights, move forward into the next step.
Adjust for covariates
For most analyses, we perform a variant of the following basic linear regression:
where Dx is the disease status of an individual, the gene expression is given in log(CPM), and weighted regression is performed using the voom confidence weights from above. For differential expression, we used the linear regression utilities in the limma package, where regression is performed for each gene separately.
Otherwise, to generate input for the eQTL and network analyses, we directly used the lm() function in R, and the weighted-regression residuals were combined with the estimated effect of the disease status (to preserve the estimated effect of disease on expression); in the main text, we refer to this as expression data that is adjusted for all other covariates “conditional on diagnosis”. This procedure yields a normalized and adjusted gene expression matrix carried forward for eQTL and network analyses.
Technical validation of normalized gene expression levels using qPCR
The voom-normalized log(CPM) levels provide estimates of true gene expression. To determine if these estimates were precise, we compared their values to independent estimates of gene expression. Studies reporting validation of their RNA quantification typically report “technical validation;” i.e., after extraction from a common source, an RNA pool is measured by the primary quantification tool and the same pool is assessed by a secondary quantification tool, such as qPCR. Technical validation often results in excellent fit between the two methods; yet it avoids other sources of experimental variation involved in extracting RNA from tissue. We take a somewhat different approach here. For a selected set of 13 genes that had been previously reported to be altered in this same brain region in 57 SCZ cases relative to 57 matched controls among the Pitt cohort (Supplementary Fig. 3), we compared results from RNA-seq to that of qPCR when these quantifications are taken from different tissue samples, although they were taken from the same subject and roughly the same brain region. Therefore our results also account for possible differences in pathological sampling of brain region and variability in RNA extraction.
Some of these genes showed increased expression and others showed decreased expression between cases and controls in the Pitt cohort, and many have been reported to be similarly altered in other cohorts of SCZ subjects. After selection of uniquely-mapping primers (approximately 20 bp for each of forward and reverse strand), qPCR was performed for each of these 13 genes and mRNA levels were normalized to the expression of ACTB, PPIA, and GAPDH, yielding “expression ratios” calculated using CTs (i.e., the PCR cycle threshold). The Pearson correlation between these expression ratios and the voom-normalized log(CPM) levels for the same subjects was greater than 0.5 for 9 of the 13 genes (Supplementary Fig. 3A); for an additional 3 genes, it was between 0.1 and 0.3, and only for one gene (HIVEP2) was the correlation negative. The correspondence between estimates is notable because of the different measurement methodologies and because, while the samples came from the same subject and brain region, they were drawn independently for the qPCR and RNA-seq experiments. We thus conclude that the genome-wide RNA-seq-based quantification provides good estimates of true gene expression in DLPFC tissue. Voom-normalized log(CPM) are presented by diagnosis and site for GAD1, PVALB, SLC32A1 and SST(Supplementary Fig. 3B).
Evaluation and selection of co-variates
Following basic sample-level normalization and gene-level filtering, we assessed the relationship between known clinical, technical, and experimental sample-level variables and the gene-level expression values in the normalized read count matrix. The purpose of this exploratory analysis was to determine which of these variables should be included as covariates that statistically adjust the gene expression levels for downstream analyses (i.e., eQTL discovery, differential expression, and gene co-expression). The final model, which we call “the covariate model”, included 12 sample variables (Dx [3], Institution [3], Sex [2], AOD, PMI, RIN, RIN2, and 5 ancestry vectors) and 1 experiment variable (clustered LIB [9]), where the number of levels for factor variables is noted here in square brackets. Counting the intercept term, this model accounted for 23 df and yielded an average r2 of 0.42 (For description of the model selection procedure, see Supplementary Text). We use this model in most analyses reported in the manuscript, except where otherwise noted (see Supplementary Fig. 2). We discuss the addition of surrogate variables (Supplementary Fig. 4G, H and Supplementary Text); the fit of the various models to the data is summarized in Supplementary Fig. 4I. Graphical display of the distribution of selected covariates by diagnosis are provided for the CommonMind Consortium (CMC) and Human Brain Cohort Collection (HBCC) data in Supplementary Fig. 5, which demonstrate that cases and controls show roughly the same ranges.
Isoform-level normalization and analyses
Relative isoform abundances were estimated using the MISO software package. The estimates of PSI (percent spliced in; i.e., fraction of each isoform of a gene expressed) and their standard deviations of those estimates, were calculated for a total of 160,305 isoforms. The isoforms were initially filtered to include only those deriving from genes expressed at a CPM > 1 in at least 50% of the samples (the same 16,423 genes used in gene-level analyses). To obtain absolute abundance estimates of isoform expression (“isoform-assigned” CPM), the isoform PSI values were multiplied by their respective effective isoform lengths 67 to control for variable isoform length, re-normalized to sum to 1, and then multiplied by the HTSeq gene-level read counts, which were then converted to isoform-level CPM, and log(CPM), using voom. Next, we retained only isoforms that had sufficient expression for analysis (CPM > 0.5 and PSI > 0.01 in more than 50% of the samples) and sufficiently well-estimated PSI (standard deviation across MISO iterations of PSI estimate < 0.1, and a coefficient of variation on the estimate < 0.5 in more than 50% of samples). After filtering, a total of 43,817 isoforms of 12,329 genes remained for analysis. The covariate model used for gene analyses was used for isoform-level analyses. As a technical assessment of self-consistency, For 85% of the analyzed isoforms, the correlation across samples between the number of unique reads per isoform, arguably, the most direct measure of relative isoform abundance from RNA-seq, and the isoform-level CPM was above 0.2. Analyses for discovery of differential isoform expression and isoform-eQTL association used a strategy analogous to that at the gene level. Of note, we estimated isoform-level voom sampling weights from the isoform log(CPM) data and then used these weights in all linear regression analyses..
eQTL generation and analysis
For the 16,423 genes with above-threshold expression, gene-level eQTL (gene expression quantitative trait loci) were derived using the N = 467 genetically-inferred Caucasian samples (209 SCZ cases, 206 Controls, and 52 AFF cases), across the 6.4 million genotyped and imputed markers with imputation score (INFO) ≥ 0.8 and estimated minor allele frequency (MAF) ≥ 0.05. eQTL were computed using a linear model on the imputed genotype dosages using MatrixEQTL69. The gene expression data were adjusted for the covariate model, although without adjusting for ancestry vectors. In addition, the estimated Dx effect was added back to the residuals because we wanted to allow for an effect of diagnosis on gene expression. The 5 ancestry vectors were included instead in the eQTL model to control for ancestry differences in SNP allele frequencies. Thus, the final regression model for eQTL discovery in the full Caucasian CMC cohort was:
FDR was estimated separately for cis-eQTL (defined as ≤ 1 MB between SNP marker and gene position) and trans-eQTL (> 1 MB between marker and gene), controlling for FDR one chromosome at a time. The regression modeling was performed for SNPs on the X chromosome in the same manner as for those on the autosomes (i.e., with a dosage scaling between 0 and 2 for both males and females); this gender-neutral model was appropriate here since the gene expression was already adjusted for gender.
Additionally, eQTL were generated separately in SCZ cases and controls, and the combination of those samples (excluding AFF cases). However, permutation of disease status indicated that the overlaps between case-derived eQTL and control-derived eQTL were similar to the amount expected for two homogeneous sets of these sample sizes, and there was limited evidence for condition-specific eQTL. Nevertheless, to potentially identify eQTL that differ by disease state, a disease-genotype interaction term was also explicitly tested, but only a handful of such associations were found to be significant after controlling for FDR.
Lastly, per-gene permutations were performed to identify genes with at least one significant eQTL after correcting for multiple marker testing13. 1000 permutations were performed per gene and FDR was estimated on the permutation P values using the qvalue R package (Dabney A and Storey JD. qvalue: Q-value estimation for false discovery rate control. R package version 1.43.0).
Using similar techniques to derive isoform expression quantitative trait loci (isoQTLs), we identified 3,355,111 significant cis-isoQTLs at FDR ≤ 5%, representing 27,691 isoforms of 10,779 genes. IsoQTLs and gene-level eQTLs overlapped substantially; 58% of isoQTLs were cis-eQTLs for the parent gene at FDR ≤ 5%; conversely, 71% of cis-eQTLs for genes with at least one represented isoform were isoQTLs at FDR ≤ 5%. There were, however, 1,584 genes having no cis-eQTL (FDR ≤ 5%) that nevertheless had at least one significant isoQTL. At the isoform level, there were 39,414 significant trans-isoQTLs, representing 964 isoforms (836 genes), of which 61% were also trans-eQTLs for the same gene.
Overlap with other eQTL databases
Since there exist a number of previous brain eQTL studies, we wanted to assess the overlap of the eQTL derived here from CMC with those existing databases. To that end, eQTL for the DLPFC from the (i) Braincloud16 (GEO accession number GSE30272, n samples = 108), (ii) NIH17 (GEO accession number GSE15745, n samples = 145), and (iii) Harvard Brain Tissue Resource Center (HBTRC) / Harvard Brain Bank (HBB)15 (GEO accession number GSE44772, n samples = 146) datasets were generated as previously described21. In addition, eQTL for the frontal cortex from the (iv) UKBEC data18 (GEO accession number GSE46706, n samples = 134) were generated in a similar manner using imputed genotypes obtained directly from the study authors. eQTL for a (v) meta-analysis of brain cortical regions (N = 424) were also obtained from the supplementary materials included with the publication19; note that this meta-analysis included some of the individual studies above. For each of these 5 datasets, an FDR threshold of 5% was used to declare significance of cis-eQTL, and those associated pairs were carried forward for testing. For RNA-seq-based eQTLs from DLPFC (Brodmann area 9, n samples = 92) that are part of the Genotype-Tissue Expression (GTEx) Project13, we utilized those eQTLs significant after permutation (as performed by the GTEx Consortium); these data were downloaded from the GTEx Portal (www.gtexportal.org), corresponding to dbGaP accession number phs000424.v6.p1.
Next, before performing any comparison analyses, the database eQTL were first filtered, removing all eQTL involving: a) array probes that mapped to more than one gene, b) genes not expressed above the minimum threshold in our cohort (and thus would necessarily be missing from our results), c) genes that could not be uniquely mapped to Ensembl (v70) genes, or d) SNPs not included in our analysis.
Then, because the data herein are substantially larger than existing brain eQTL datasets that were therefore more limited in power for eQTL discovery, we focused on testing the sensitivity of our eQTL towards recapitulating publicly available eQTL. To robustly assess this sensitivity, we considered the eQTL P values from our CMC cohort with regards to the eQTL associations described in each public database we had curated. We scored the overlap using π1, the proportion of non-null hypotheses (as estimated by the ‘qvalue’ package in R) among the distribution of CMC P values for the database eQTL SNP-gene association pairs.
For another comparison with genome-wide eQTL, we also used the unpublished, but publicly available HBCC microarray cohort (dbGAP ID: 000979.v1.p1) described below to generate a large set of eQTL better powered for replication of the CMC-derived eQTL. Genotypes were obtained using the HumanHap650Yv3 or Human1MDuov3 chips, and ancestry components were subsequently inferred as above for CMC. Next, eQTL were generated using N = 279 genetically-inferred Caucasian samples (76 controls, 72 SCZ, 43 BP, 88 MDD) in an analogous manner to CMC, adjusting for diagnosis and 5 ancestry components.
Lastly, we employed the eQTL derived from the unpublished ROS/MAP study (https://www.synapse.org/#!Synapse:syn3219045, details on the study given below) in a limited way to replicate the 5 single-gene QTL associations we detected as having strong overlap with GWAS risk variants. The subset of the ROS/MAP cohort currently RNA-sequenced and analyzed (N = 461 DLPFC samples) was used to derive eQTL. To account for non-genetic factors such as batch effects, age, gender, and technical artifacts in the gene expression data, PEER70 was applied. The optimal numbers of PEER factors for association analysis were determined based on the factors that resulted in the maximal number of cis-eQTLs. This procedure identified between 30 and 40 factors in this DLFPC dataset; here, we used 30 PEER factors. We regressed out these factors from the gene expression levels and used the residuals as phenotypes for all eQTL association analyses. The ROS/MAP study 26 takes advantage of data and biological specimens from more than 1000 persons from two prospective, longitudinal clinical-pathologic studies of older subjects that are non-demented at the time of recruitment (the religious order study and the memory and aging project). The subjects have detailed clinical and phenotypic data such as detailed annual cognitive function testing, clinical evaluations for dementia, and a detailed neuropathologic examination.
Religious Orders Study (ROS)
From January 1994 through June of 2010, 1,148 persons agreed to annual detailed clinical evaluation and brain donation at the time of death. Of these, 1,139 have completed their baseline clinical evaluation: 68.9% were women; 88.0% were white, non-Hispanic; their mean age was 75.6 years; and mean education was 18.1 years. To date, there have been 287 cases of incident dementia and 273 cases of incident AD with or without a coexisting condition.
Memory and Aging Project (MAP)
From October 1997 through June 2010, 1,403 persons agreed to annual detailed clinical evaluation and donation of brain, spinal cord, nerve, and muscle at the time of death. Of these, 1,372 have completed their baseline clinical evaluation: 72.7% were women; 86.9% were white, non-Hispanic; their mean age was 80.0 years; and mean education was 14.3 years, with 34.0% with 12 or fewer years of education. To date, there have been 250 cases of incident dementia and 238 cases of incident AD with or without a coexisting condition. At this time, over 900 subjects from either ROS or MAP are deceased and have frozen brain tissue available for data generation. To avoid population stratification artifacts in the genetic analyses, the first 500 subjects were randomly selected from among those subjects that are self-reported to be of white, non-Hispanic ancestry, and have genome-wide genotype data (N = 1,709 for the entire ROS and MAP studies) that confirm this self-reported ancestry.
Overlap of eQTLs with enhancer sequences
To assess how cis- and trans-eQTLs relate to known enhancer sequences, we tested for overlap between eQTLs and enhancer sequences from the Roadmap Epigenomics Consortium71. More specifically, we used chromatin states for enhancer sequences (active, genic, and weak enhancers), derived from a recent joint analysis that the Roadmap Epigenomics Consortium applied in different chromatin immunoprecipitation sequencing (ChIP-seq) data across 98 human tissues and cell lines. We included tissues that were assayed for 6 different chromatin marks (H3K4me1, H3K4me3, H3K27ac, H3K36me3, H3K27me3, and H3K9me3). We tested for enrichment of significant eQTLs at FDR ≤ 5%, using as an index eQTL SNP (eSNP) the most significantly associated SNP per gene (“max-eQTL”), which resulted in 13,137 and 851 cis- and trans-eSNPs, respectively. For each tissue or cell line, we counted the number of index eSNPs that lie within enhancer sequences respectively found in that tissue or cell line. To assess if this overlap is higher than expected by chance, we generated 1,000 sets of random SNPs matched with the index cis- and trans-eSNPs, in terms of allele frequency, gene density, distance from TSS, and density of tagSNPs arising from genomic variability of linkage disequlibrium. Z scores were estimated as:
Where observed is the number of index eSNPs that lie within enhancers, and meannull and SDnull are the mean and standard deviation of the null distribution of overlap, as estimated using the sets of permuted SNPs.
Using genetic association with eQTL - Sherlock
The Sherlock method25 attempts to uncover disease-associated genes (“risk genes”) by using a Bayesian statistical framework to assess overlap between eQTL for a gene and GWA significant SNPs loci for a disease. Its underlying principle is that genetic-driven changes in expression levels of risk genes (discovered as eSNPs) should ultimately also manifest as genetic association of those same SNPs with disease (GWAS SNPs). Specifically, we expect that cis-eQTL and trans-eQTL for a risk gene should be associated with disease (if risk is mediated by expression changes of that gene); note that the converse need not be true (since not all associated SNPs need be related to the function of any single disease gene). Briefly, Sherlock uses a Bayesian model to integrate signal across all statistically independent eQTL loci for a gene, where an independent linkage block is defined as a genomic interval containing one or more eSNPs associated with a gene and having a within-eSNP interval of 500 kb or less. For each such independent block, a single Bayes factor is calculated as the mean of the SNP-level Bayes factors within the block; the SNP-level Bayes factor corresponds to the likelihood of the observed GWAS and eQTL P values under the alternative hypothesis that expression changes in the gene mediate disease risk, relative to the likelihood under the null model where the gene is not related to disease. Bayes factors are multiplied for the independent loci, yielding a single per-gene score. P values for these genic scores are estimated using permuted disease GWAS P values to generate a null distribution of Sherlock Bayes factors across all genes. In this study, we used the eQTL derived from the full cohort of 467 Caucasian-inferred individuals, resulting from the expression-on-SNP regression that included the covariate model with the surrogate variables. The Sherlock method takes as input liberally-defined cis-eQTL associations (P < 10−3) and trans-eQTL associations (P < 10−5). For the trans-eQTL, we used a very strict definition to exclude putatively artifactual associations of SNP and gene expression, requiring, in addition to P < 10−5, that the trans-eQTL association also be present in the 206 controls-only Caucasian cohort, albeit with a P value as high as 10−3. Additionally excluding trans-eQTL where the eSNP was within 10 Mb of the associated gene (since such scenarios are perhaps instances of cis-eQTL for regions with larger LD blocks), yielded a final reduced subset of 13,114 trans-eQTL (~7% of all trans-eQTL SNP-gene pairs at P < 10−5) across 661 genes. This stricter filter increases the replication rate in HBCC to 36% at FDR ≤ 5% in both cohorts. For generating null GWAS P values, we used 100 permutations of random case-control assignments of the 2,504 individuals in the 1000 Genomes Phase 3 genotype data (http://www.1000genomes.org) 72, as suggested by the author of the Sherlock software (Xin He, personal communication). We also slightly modified the Sherlock source code, omitting the exclusion criterion for SNPs (and genes whose expression is associated with those SNPs) that could not be found in the 1000 Genomes data, which encompassed only 49,612 [4.4%] of the 1,127,447 eSNPs also found in the PGC SCZ2 GWAS data 3. Default Sherlock parameters (priors) were used, except for setting the number of individuals in which the eQTL were discovered to N = 467, setting a 1% prevalence for SCZ, and setting the PGC SCZ2 GWAS primary meta-analysis cohort sizes (35,476 cases and 46,839 controls). For input allele frequencies, we used the frequencies estimated from the 46,839 GWAS controls. Also, instead of using the minor allele frequency, we used the “risk” allele frequency at each SNP; i.e., the allele at higher frequency in cases. This ensures that, for significantly associated SNPs, the minor or major allele is appropriately chosen for likelihood calculations based on the direction of risk. Still, for most SNPs in the genome, those not clearly associated with SCZ, the choice of major or minor allele is essentially random and unbiased.
Notably, in the PGC SCZ2 paper detailing the 108 SCZ-associated loci the authors attempted to ask if any eSNPs from brain eQTL databases existing at the time were credibly associated with schizophrenia. Specifically, they tested if the most significant eQTL SNP for any gene is among those SNPs 99% most likely to be credibly causal for SCZ at any locus (assuming only a single causal SNP per locus). This process led to 3 genes based on brain eQTL: 1, TINAGL1, and LIG1. In our CMC eQTL data, there is only overlap between the SCZ association and eQTLs for MLH1, with up-regulation of expression predicted to be associated with the genetic risk variation; however, this overlap is below the genome-wide threshold, Sherlock P = 6 × 10−5, Bonferroni corrected P = 0.69.
Zebrafish functional assays
Morpholino (MO)-mediated depletion and complementation with human mRNA. All zebrafish assays were performed utilizing the wild-type ZDR strain in accordance with standard zebrafish husbandry practices at Duke University. To assess the functional outcome of FURIN down-regulation in a zebrafish model, a splice-blocking morpholino for furin_a targeting the splice site donor region of exon 7 (5’ – CAGTTAAATGCGCCGACTCACCTCC – 3’) was designed from Gene Tools, LLC (Philomath, OR). All eggs were injected with 3ng/µl of the furin_a MO construct at the 1- to 4-cell stage. Embryos were collected at 3 days post-fertilization (dpf), and RT-PCR was performed to validate the efficiency of the MO. The forward RT-PCR primer targets the start of intron 7 (5’ – GTTGTGCTGGAGAGGTTGCT – 3’) with the reverse primer targeting the intronic region bordering exon 8 (5’ – GGTGTGCTCTGTGTGCTGAT – 3’). For mRNA rescue of furin_a MO and the overexpression study of TSNARE1, CNTN4, SNAP91, and CLCN3, human wild-type capped mRNA for each gene was transcribed using the SP6 Message Machine Kit (Ambion). All RNAs were injected at the 1- to 4-cell stage at 200ng concentrations. Immunohistochemistry and phenotyping: For immunostaining purposes, all embryos were collected at 3dpf, dechorionated, and fixed in Dent’s solution (20% DMSO; 80% MeOH) overnight at 4°C. Embryos were rehydrated in a step-wise manner starting with 75% ethanol in 1XPBS, followed by 50%, and 25% ethanol solutions. Embryos were then bleached, post-fixed with 4% PFA, and permeabilized using proteinase-K. Embryos were then washed twice in IF buffer (1% BSA, 0.1% Tween-20 in 1XPBS) and incubated in primary antibodies for anti-α-acetylated tubulin (1:1000, Sigma-Aldrich, T7451) and anti-p-histone H3 (PH3; 1:500, Santa Cruz Biotechnology, sc-8656-R) in blocking solution overnight at room temperature (RT). Following two washes in IF buffer, embryos were placed in secondary antibody solution containing Alexa Fluor 594 goat anti-mouse IgG (1:1000) and Alexa Fluor 488 goat anti-rabbit IgG at 488(1:500; Invitrogen) in blocking solution for 2hrs at RT. Embryos were then washed and stored in IF buffer at 4°C until used for microscopy.
Head size measurements of 3dpf embryos were assessed using brightfield microscopy and quantified using the NIH ImageJ software package. To assess proliferation, PH3-stained embryos, images were taken using fluorescent microscopy along the z-axis and stacked to obtain a focused image spanning the full head. PH3-positive cells from the forebrain to hindbrain (directly behind the cerebellum) were then counted for quantification purposes using ImageJ. TUNEL staining was performed to measure apoptosis using Apoptag Red In Situ Apoptosis Detection Kit (Millipore). TUNEL-stained embryos were then imaged and quantified using the same technique as for proliferation. All experiments were replicated twice and aggregate data was compiled. Statistical differences between controls and treatment conditions for each phenotype were calculated using Student’s t-test.
Human neural progenitor cell (NPC) model of FURIN
Fibroblast biopsies were obtained from healthy controls that were recruited as part of a longitudinal study by Dr. Judith Rapoport (NIMH)73. All participants provided written assent/consent with written informed consent from a parent or legal guardian for minors. Human fibroblasts (HFs) were cultured on plates coated with 0.1% gelatin (in milli-Q water) and grown in HF media (DMEM (Invitrogen), 20% FBS (Gemini)).
hiPSCs were derived as described previously (http://www.nature.com/articles/npjschz201519); replicating but nearly confluent HFs were transfected with Cytotune Sendai virus (Life Technologies). Cells were allowed to recover for at least 3 days, dissociated with TrypleE (Life Technologies) and re-plated onto a 10-cm dish containing 1 million mouse embryonic fibroblasts (mEFs). Cells were switched to HUES media (DMEM/F12 (Invitrogen), 20% KO-Serum Replacement (Invitrogen), 1× Glutamax (Invitrogen), 1× NEAA (Invitrogen), 1× 2-mercaptoethanol (Sigma) and 20 ng/ml FGF2 (Invitrogen)) and fed every 2–3 days. hiPSC colonies were manually picked and clonally plated onto 24-well mEF plates in HUES media. At early passages, hiPSCs were split through manual passaging, but at higher passages, hiPSC could be enzymatically passaged with Collagenase (1mg/ml in DMEM) (Sigma). Karyotyping analysis was performed by Wicell Cytogenetics (Madison WI); only karyotypically normal lines were used for subsequent studies.
hiPSC forebrain NPCs were derived from the three controls as described previously74. These samples were selected irrespective of their genotypes for the FURIN-eQTL SCZ-risk variant at SNP rs4702, with two being heterozygous G/A and the third homozygous risk G/G. Incubation with Collagenase (1 mg/ml in DMEM) at 37°C for 1–2 hours lifted colonies, which were transferred to a nonadherent plate (Corning). Embryoid Bodies (EBs) were grown in suspension with dual-SMAD inhibition (0.1mM LDN193189 (Stemgent) and 10mM SB431542 (Tocris)) N2/B27 media (DMEM/F12-Glutamax (Invitrogen), 1× N2 (Invitrogen), 1X B27 (Invitrogen)). 7-day-old EBs were plated in N2/B27 media with 1 µg/ml Laminin (Invitrogen) onto poly-ornithine/Laminin-coated plates. Neural rosettes were harvested from 14-day-old EBs using Neural Rosette Selection Reagent (STEMdiff™) for 60 minutes at 37°C before being plated in NPC media (DMEM/F12, 1× N2, 1× B27-RA (Invitrogen), 1 µg/ml Laminin and 20 ng/ml FGF2 on poly-ornithine/laminin-coated plates.
hiPSC NPCs were maintained at high density, grown on Matrigel in NPC media (DMEM/F12, 1× N2, 1× B27-RA (Invitrogen), and 20 ng/ml FGF2 (Invitrogen) and split approximately 1:3–1:4 every week with Accutase (Millipore)37. NPCs can be expanded beyond 10 passages. NPC experiments were conducted on passage-matched populations, between passages 9 and 12. Control hiPSC and NPC validation as shown39,75. All hiPSC and NPCs in the laboratory are tested monthly using MycoAlert (Lonza) to ensure they remain mycoplasma free.
Neurosphere migration assay
NPCs were dissociated with accutase and then cultured for 48 hours in nonadherent plates to generate neurospheres. Neurospheres were manually picked and cultured in “Matrigel matrix (0.5 mg Matrigel was plated in cold NPC media on a 96-well plate 1 hour prior to neurosphere plating; following neurosphere picking, an additional 0.5 mg Matrigel was added in cold NPC media per 96-well plate). DAPI-stained neurospheres were imaged at 48 hours. Average radial migration from each neurosphere was measured using NIH ImageJ40,74.
Knockdown of FURIN
pLKO.1 - TRC control was a gift from David Root (Addgene plasmid # 10879)76. A bacterial glycerol stock containing the LV-FURIN-shRNA plasmid was purchased from Sigma (SHCLNG-TRCN0000262167). High-titer lentiviral supernatant was generated by co-transfection of shRNA expression vector together with psPAX2 and pMD2.G to package letivirus particles in HEK-293T cells. psPAX2 (Addgene plasmid # 12260) and pMD2.G (Addgene plasmid # 12259) were gifts from Didier Trono. Lentiviral supernatant was concentrated by centrifugation at 19,300 × g for 2hr at 4°C and resuspended in NPC media. Viral titer was determined using a qPCR lentiviral titration kit (Applied Biological Material Inc. - LV900) and TaqMan® RNA-to-Ct™ 1-Step Kit (ThermoFisher Scientific – 4392938). NPC transduction was performed by addition of lentiviral particals to NPCs at an MOI of 0.5–1 followed by centrifugation of plate at 1,000 × g for 1hr at RT then incubation of NPCs at 37°C for an additional 6 hours. 48hr after infection, transduced cells were selected for with 1µg/mL puromycin for 48hr. FURIN knockdown was validated by qPCR using TaqMan® RNA-to-Ct™ 1-Step Kit (ThermoFisher Scientific – 4392938).
Differential Expression Analyses
Differential expression between SCZ cases and the controls was assessed (Supplementary Fig. 2, bottom panel) utilizing the limma package in R, with the following inputs: the voom-normalized gene expression matrix, the voom precision weights matrix corresponding to the values in the expression matrix, and the final “covariate model”. Note that the expression matrix we utilized contained data for 592 samples in total (SCZ cases, controls, and AFF cases), and 16,423 genes passing the expression-level threshold of > 1 CPM in > 50% of the samples. The rationale for including ~50 AFF, even though they were not analyzed for differential expression, was to: a) increase statistical power during linear modeling of covariates (such as age, RIN, PMI, etc.); and b) place the expression data for the AFF cases on the same scale as for the SCZ and control samples for the sake of simplicity.
For each gene, weighted least-squares linear regression was performed using limma to yield coefficients for the effect on gene expression of each variable on the right-hand side:
Then, for each gene, the SCZ disease status coefficient was statistically tested for being non-zero, implying an estimated effect for SCZ, above and beyond any other effect from the covariates. This test produces a t-statistic (then moderated in a Bayesian fashion) and corresponding P value. P values were then adjusted for multiple hypothesis testing using false discovery rate (FDR) estimation, and the differentially expressed genes were determined as those with an estimated FDR ≤ 5%. FDR was calculated by the limma package, which uses Benjamini-Hochberg from p.adjust() function in R. Significance was also assessed by permuting case-control status for 1,000 experiments. Of these experiments, the average number of significant genes at FDR ≤ 0.05 was 4.3, well below 693 found in our sample. If 5% of the 693 were false, the threshold established of 34.7 genes is exceed in 9 out of 1000 experiments, slightly less than 1%. Differential expression of gene isoforms was performed analogously.
Cross-validation of differential expression
We performed cross-validation of the differential expression by randomly splitting the full cohort into an 80% “discovery” cohort and 20% “replication” cohort (with equal proportions of SCZ cases and controls into the two parts of the split). This splitting process was repeated 20 times. Each time, we chose the t-statistics of the genes considered to be differentially expressed at an FDR < 5% in the discovery cohort and looked up the corresponding statistics in the independent 20% replication cohort. Across the 20 samplings, the median number of FDR < 5% differentially expressed genes was 216 (mean = 315, sd = 261, 25th percentile = 92, 75th percentile = 562). For these FDR < 5% “discovery” differentially expressed genes, the median Pearson correlation of t-statistics with the “replication” cohort was 0.79 (mean = 0.75, sd = 0.16, 25th percentile = 0.67, 75th percentile = 0.88). This strongly supports the robustness of the differential expression results described herein.
In situ hybridization images from the Allen Human Brain Atlas
Given the broader dynamic range of RNASeq and its ability to detect low abundance transcripts – because it does not suffer from hybridization-based limitations associated with microarray such as background noise – we would expect to see large fold-changes, if they existed, even for genes displaying low expression; or expression and differential expression restricted to a specific subset of cells. ISH images for representative genes taken from the same brain region used in our experiment can add extra information when related to RNASeq intensity data. Supplementary Fig. 10 shows in situ hybridization images from Allen Human Brain Atlas for a selected set of genes showing significant differential expression in CMC; the figure shows different cell type specific expression (from high to low specificity). These are from the largest dataset for DLPFC, the Neurotransmitter Study (176 genes across cortical regions and 88 genes across subcortical regions in 4 control cases), 12 of which were in common with our list. The data suggest that genes identified by DE analysis display various degrees of cell-type specificity.
Effect of age on differential expression
To assess the impact of age-at-death on expression differences between SCZ cases and controls, we compared the per-gene differential expression t-statistics derived from various subsets of the entire cohort described here. Specifically, for the 172 cases and controls whose age of death was youngest (mean age 45.5, range 20–60), the t-statistics for differential expression were highly correlated with those from the full cohort (Pearson r = 0.62), yet somewhat lower (though not significantly, P = 0.28) than for 100 random subsets of the same size (mean r = 0.69, standard deviation = 0.13), suggesting that age at death may have only a modest impact among adult cohorts. Furthermore, we explicitly compared the differential expression between the aged 20–60 individuals (172 samples, mean age 45.5) to an analysis of the complementary age 60 or older cohort (362 cases and controls, mean age 77.8) by independently processing the data for each of those sub-cohorts. The differential t-statistics between these independent sub-cohorts were correlated (r = 0.18, P < 2 × 10−16), arguing for some consistency of case-control differences across the lifespan. Still, it is possible that a larger cohort of younger cases and controls would exhibit somewhat different patterns of gene expression changes. It is important to note, however, that the effect of age is not the only possible explanation for a lower correlation; e.g., while Pitt samples compose 26% of all SCZ cases and controls in CMC, 63% of the aged 20–60 CMC samples are from Pitt, and other factors besides age also differ between the Pitt and non-Pitt samples.
Drug effects on differential expression
To examine whether drug treatment effects were responsible for the differential expression observed in SCZ, we examined enrichment of differential expression and directional concordance for drug treatment signatures derived from studies of Rhesus macaque monkeys and rodents. Subjects from a cohort of N = 34 Rhesus macaques born between 1995 and 2004 were randomly selected for four treatment groups: 7 for high doses of haloperidol (4 mg/kg/day), 10 low doses of haloperidol (0.14 mg/kg/day), 9 clozapine (5.2 mg/kg/day), and 8 vehicle. Monkeys were administered the antipsychotic drugs orally for six months, mixed with powdered sugar and given in peanut butter or fruit treats. Monkeys were raised at Wake Forest University and received standard enrichment, including social enrichment, human interaction, variety in diet, and age-appropriate objects as dictated by the Animal Welfare Act and the Emory University and Wake Forest School of Medicine policies for non-human primate environmental enrichment. Animal care procedures strictly followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees of Emory University and Wake Forest School of Medicine. Monkeys were sacrificed and necropsied on average at age 6.2 years (range between 3.6 and 8.2 years old) after the six-month treatment protocol by an overdose of barbiturate and transcardially perfused with ice cold saline. The brains were removed and cut into 4 mm slabs in the coronal plane using a brain matrix (EMS, Fort Washington, PA) and immediately frozen and stored at −80°C. Tissue was dissected from slabs of the right hemisphere that included the basal ganglia from the rostral pole to the beginning of the anterior commissure. The DLPFC was dissected from the dorsal and ventral banks of the principal sulcus (Area 46) and pulverized. The identical RNA-seq protocol (using the RiboZero Gold kit [Illumina]) was followed as for the primary human CMC cohort. Sequencing data were processed similarly as for the human CMC cohort, with reads aligned to the macaque reference genome and transcriptome (mmul1), but with two minor changes: STAR77 was used for efficient alignment, and featureCounts78 was used for gene-level quantification. rRNA rates were all below 1%. RNA expression levels were normalized using voom, and limma differential expression analysis was performed, adjusting for sex and RNA isolation batch, to assess the effects of haloperidol treatment (N = 17 in total, grouped to increase statistical power) and clozapine (N = 9) drug groups, as compared to the baseline untreated group (N = 8). While no genes were considered differentially expressed after multiple test correction, we used a nominal P ≤ 0.01 cutoff to identify signatures for haloperidol and clozapine treatment, which resulted in human-orthologous gene sets of size 237 and 31, respectively.
To assess enrichment of overlapping genes, we performed a one-sided Kolmogorov-Smirnov (KS) test of the P values for the gene signatures versus all genes and assessed significance via resampling. Significant enrichment was observed (Supplementary Table 2A) for the haloperidol signatures, but not the clozapine signature (P = 1 × 10−9 and 0.29, respectively). We also tested whether the direction of effect for drug signature genes was more concordant than expected by chance; this was tested using a hypergeometric test whose null hypothesis assumes that up- and down-regulated drug-mediated genes were randomly sampled from genes either up- or down-regulated (at any P value) in the CMC SCZ differential expression tests. We found the haloperidol signature significantly less concordant than expected by chance (35 out of 237 genes concordant, one-sided P < 1 × 10−4), while the clozapine signature showed significant concordance, despite the lack of enrichment in the previous test (22 of 31 genes concordant, one-sided P = 0.013). For haloperidol, the enrichment of significance but depletion of concordance is perhaps consistent with a scenario in which such antipsychotic drugs target these genes and affect them in the opposite direction of what etiologically occurs in the course of disease (hence their efficacy).
We also performed similar analyses in rodent drug treatment signatures. We had access to unpublished data (PFS) from RNA sequencing performed on mice previously treated with haloperidol. A brief description of data generation is as follows. All experimental procedures were randomized to minimize batch artifacts, including assignment of mice to receive haloperidol (HAL) or placebo (PLA), home cage, order of dissection, RNA extraction, and assay batch. Male C57BL/6J mice were chronically treated with HAL (N = 16) or PLA (N = 12) for 30 days, and the striata were dissected (15–17 mice per treatment group). All individual striatal samples were assayed using RNA-seq. Quality control and analysis for all data types conformed to those developed in our prior publications19. We considered differential expression at FDR q < 0.05 as the criterion for inclusion in the set of genes we considered to be affected by the drug.
Secondly, we culled from the literature rodent drug treatment signatures for experiments that profiled frontal or prefrontal cortex. To ensure higher quality, we required the study to be published after 2005. In total, we curated 11 gene signatures from 7 rodent studies comparing antipsychotic-treated animals to vehicle-treated controls79–85. Nine of the 11 gene signatures were represented by orthologs that were expressed in our study. In total (including the unpublished data described above), 10 rodent gene signatures were tested.
Using the independent rodent datasets, we found overall similar results as that in the monkeys, with some overlaps of genes impacted by drugs and associated with disease, but with opposite directions of effect on gene expression. Specifically, using the KS test for enrichment, none of the gene signatures were strictly significant after Bonferroni correction (Pbonferroni > 0.05), though two signatures were significantly less concordant than expected by chance after multiple test correction (Pbonferroni = 0.01 and 0.01, Supplementary Table 2B). However, we identified a set of 21 genes that appeared in 4 or more of the gene signatures; 16 of these genes were represented by orthologs in our normalized data, and 8 showed concordant direction in all 4 studies. A one-sided test of enrichment for these “most represented” genes showed a non-significant trend towards enrichment (p = 0.061), with one gene from the signature significant in the CMC DLPFC data at FDR ≤ 5% (ATRX, FDR = 4.5%). Sets of genes that appeared in 3 or more studies, or 2 or more studies, also did not reach statistical significance but did show a trend for overlap (P = 0.084 and 0.071, respectively). However, for each of these gene signatures, the direction of effect was significantly less concordant than expected by chance (1 of 8 [P = 0.004], 10 of 55, and 122 of 388 for those in ≥4 studies, ≥3 studies, and ≥2 studies, respectively).
HBCC replication microarray cohort
Microarray-based gene expression data were available from the DLPFC samples of the Human Brain Collection Core (HBCC, http://www.nimh.nih.gov/labs-at-nimh/research-areas/research-support-services/hbcc/human-brain-collection-core-hbcc.shtml). These samples were prepared by extraction of RNA using Qiagen RNAeasy kits, generation of biotin-labeled cRNAs using Affymetrix 3’IVT express kits, and hybridization of cRNAs to Illumina HumanHT-12_V4 Beadchips. Expression values were extracted with GenomeStudioV2011.1. After preprocessing and quality control to check for outliers and sex mismatches, there remained 131 SCZ and 176 control samples (as well as 43 BP and 88 MDD samples, where these latter samples were used only for eQTL derivation).
Gene expression data for samples passing quality control were normalized by first aligning data within each batch, then by addressing batch effects. In detail, within-batch normalization included: (i) background correction using negative control probes; (ii) quantile normalization; and (iii) log2 transformation86. We used the Inter-Array Connectivity (IAC) to identify outliers as those samples with values 3 standard deviations lower than the mean in their respective batches; samples identified as outliers were then removed from the batch and preprocessing was repeated. After the within-batch normalization, probes were considered as robustly expressed only if the detection P value was < 0.01 for at least half of the samples in the dataset. Next, systematic batch effects across the entire dataset were addressed, by application of ComBat87 (http://statistics.byu.edu/johnson/ComBat/), a parametric empirical Bayes framework, to achieve cross-batch normalization.
To maximize comparability with the CMC data, we designed an analysis pipeline analogous to that which we used for the CMC RNA-seq processing. We remapped probes to genomic locations of genes using the sequence of the probe (using the same reference genome and Ensembl transcriptome as for the CMC RNA-seq data). For transcripts with more than one probe, we chose the probe with the maximum intensity for each sample (this choice had only minimal impact on results). We retained samples with genotype data so that we could include ancestry as a covariate.We selected covariates based on variance explained in data. The following covariates were used in the differential expression (and eQTL) analysis: Dx, Age of death, sex, PMI, pH, RIN, clustered processing batch, and ancestry markers. We performed differential expression analysis with adjustment for covariates, using linear regression models in limma and identified 2,288 differentially expressed genes at FDR 5%, among which 1,166 and 1,122 were up-regulated and down-regulated in SCZ, respectively.
Overlaps with genetic associations
To assess how genetic risk for schizophrenia relates to brain function in the DLPFC at the molecular level, we tested for overlap between genes found in genetic loci previously associated with SCZ and the genes exhibiting expression differences between SCZ cases and controls in this study. To this end, we curated genetic associations with schizophrenia from the literature, including those derived from: a) 108 loci discovered in a common variant genome-wide association (GWAS) meta-analysis study of 36,989 SCZ cases and 113,075 controls3, b) a literature consensus of 12 copy number variant (CNV) regions collated from numerous rare CNV studies88, c) 756 nonsynonymous (NS; mostly missense, but also including 114 loss-of-function [LoF; nonsense, essential splice site, or frameshifting indels]) de novo mutations discovered from exome-sequencing across 1,024 schizophrenia trios [probands and their parents]7,89–92 and uniformly re-annotated using PLINK/Seq (http://atgu.mgh.harvard.edu/plinkseq), and d) rare variants in an exome-sequencing study of 2,536 SCZ cases and 2,543 matched controls from Sweden5.
For category c (de novo mutations), in addition to the data from SCZ studies, we also collated information from studies of autism (3,446 NS mutations, 579 LoF, from 3,985 trios)93–95, intellectual disability (259 NS, 67 LoF, 192 trios)96–98, and epilepsy (341 NS, 58 LoF, 356 trios)99. However, these additional datasets were only used to test enrichment of genes in GWAS loci that were prioritized by eQTL), not for overlap with differential expression.
To statistically assess the overlap between the genetic and the mRNA expression associations with schizophrenia, we integrated the overlap individually found for each of the four classes of SCZ genetic variation using Fisher’s method for combining P values. For each class of genetic variation and corresponding disease association data, we tested the associations for enrichment in a gene set consisting of the 693 genes found to be significantly (FDR ≤ 5%) differentially expressed between SCZ cases and controls (up- or down-regulated). To control for the fact that some genetically-associated genes may not be brain-expressed, we conditioned all enrichment tests on the background set of 16,423 genes with above-threshold expression that we had included in differential expression analysis (and were thus candidates for being labeled as differential in the first place). Note that all genetic variants and regions were annotated using RefSeq transcripts as downloaded from the UCSC Genome Browser in April 2013; see references 5 and 7 for more details.
In detail, we used the following tests for the four classes of genetic variation: a) GWAS loci: INRICH100 was used to assess if the 108 SCZ-associated PGC SCZ2 GWAS loci (with a 20 kb window added both upstream and downstream) tended to hit the 693 differentially expressed genes (DEG) more than expected by chance loci. These random loci were generated by permutation of the associated loci within the genome, but matched to the associated loci in terms of the number of SNPs, SNP density, and the number of overlapping genes; background SNPs for matching were taken from the full imputed list of 9.4 million SNPs tested for SCZ association. After intersecting with DLPFC-expressed genes from this paper, there were 87 loci spanning one or more genes, encompassing a total of 489 genes. 10,000 permutations were performed. b) CNV regions: INRICH was also utilized to test if the 12 SCZ-associated CNV regions (without any additional genomic window) tended to hit the differentially expressed genes more than expected by randomly generated regions in the genome matched to the associated regions in terms of the number of overlapping genes. After conditioning on DLPFC expression, the 12 regions spanned 127 genes. 10,000 permutations were performed. c) De novo mutations: DNENRICH7 (https://psychgen.u.hpc.mssm.edu/dnenrich) was employed to measure if the 756 nonsynonymous (114 loss-of-function) SCZ mutations affected the differentially expressed genes more than expected by randomly generated de novo mutations matched to the observed mutations for their trinucleotide base context and functional consequence and then placed in the genome uniformly at random to account for gene size (e.g., larger genes tend to have more mutations). Conditioning on DLPFC expression, there were 103 loss-of-function mutations in 101 genes, and a total of 638 nonsynonymous mutations across 605 genes. 50,000 permutations were employed for each test. The two tests, for nonsynonymous and loss-of-function mutations, were combined by taking the minimum P value after Bonferroni correction for the 2 tests. d) Rare variants: PLINK/Seq and SMP5 (http://atgu.mgh.harvard.edu/plinkseq) were used to assess whether the exome-sequenced SCZ cases exhibited a burden of rare singleton variants (observed just once in the entire cohort of ~5,000 individuals) in the differentially expressed genes, as compared to controls. Enrichment statistics for the differentially expressed set (the sum of gene burden statistics) were calculated via permutation that controlled for any exome-wide case-control differences, residual linkage disequilibrium among rare variants in nearby genes, and differences between cases and controls arising from ancestry (based on exome-wide identity-by-state [IBS]), experimental batch, and gender. Case burden in the differentially expressed genes was tested for either nonsynonymous variants (comprised of loss-of-function variants and missense variants predicted in silico as deleterious by each of five different algorithms5), or just the smaller set of loss-of-function variants. Looking only at differentially expressed DLPFC genes, there were 236 genes with one or more singleton loss-of-function variants and a total of 440 genes harboring singleton damaging nonsynonymous variants. 10,000 permutations were used for each test. Again, the two tests were combined by choosing the minimal P value after Bonferroni correction.
Overlap of differential expression with polygenic common variant risk for SCZ
Since the CMC cases bear an aggregate common polygenic schizophrenia risk burden, we subsequently performed an independent controls-only analysis (using limma) of the effect of polygenic risk scores on expression of each gene. While no single gene was found to be significantly associated with PRS after correction for multiple testing using an FDR approach (cutoff of 5%), there was inflation of the P value distribution101 consistent with a non-uniform distribution (π1 = 0.22). Moreover, there was a significantly positive, but small, correlation (Pearson r = 0.095, p < 10−16) between the independent t-statistics for the effect of PRS on expression in controls and those we found for case-control expression differences in the full cohort, consistent with at least some of the SCZ case-control differences in CMC perhaps being driven by underlying genetic differences between the SCZ cases and controls.
Generation of gene sets for enrichment analyses of differential expression
To further attempt to interpret the list of differentially expressed genes and isoforms, we also conducted a series of structured tests to evaluate their functional enrichment, including evaluating primary hypotheses previously implicated by genetic findings in schizophrenia research (e.g., targets of regulation by FMRP, fragile X mental retardation protein), and performing exploratory analyses of a large number of gene sets (such as those obtained from Gene Ontology). In brief, we found no convincing patterns to the primary or exploratory hypotheses, after correction for multiple set testing, confounders such as gene size, and after combining results over multiple enrichment tests.
We started by curating two classes of gene sets for analyzing the differential expression data: 1) a small group of pathways and gene sets previously implicated in genome-wide genetic studies of schizophrenia (“hypothesis-driven”), and 2) a collection of thousands of “hypothesis-free” gene sets from large databases that would allow us to potentially characterize novel biology arising in brain expression related to schizophrenia. We considered each of these classes independently for multiple test correction owing to their dissimilar goals.
Hypothesis-driven: This collection consisted of 12 sets of genes previously implicated in the literature of schizophrenia genetics, including: a) all genes within 20 kb of 108 GWAS loci3, b) genes sitting under rare SCZ-associated CNV88, and c) nonsynonymous and loss-of-function de novo mutations discovered from exome-sequencing of schizophrenia probands and their parents7,89–92; note that these correspond to the data described above, where all genes in associated regions are simply lumped together as a single gene set (losing the important distinction that some loci bear many more genes than others). In addition, we added gene sets previously shown to be enriched for genetic variation associated with schizophrenia102, including genes regulated by FMRP (fragile X mental retardation protein) targeting103, predicted targets of miR-137 (filtered to include those with a total context score ≤ 0.3 or an aggregate PCT (probability of conserved targeting) ≥ 0.9 in TargetScan version 6.2)104, voltage-gated calcium channels102, and 5 related subsets of genes whose protein products are localized to the postsynaptic density of neurons, including those involved in glutamatergic neurotransmission105.
Hypothesis-free: These gene sets were derived from three widely-used databases for functional gene classification: curated GO (Gene Ontology) sets of molecular functions (MF), biological processes (BP), and cellular components (CC) (http://www.geneontology.org)106; the curated Reactome database of pathways and reactions in human biology (http://www.reactome.org)107; and HGNC (HUGO Gene Nomenclature Committee) gene families (http://www.genenames.org)108.
We sought to retain sets that were relevant to the DLPFC brain expression we observed here, as well as address overlap between the 3 databases, using the following strategy. We only retained a gene set in which at least 10% of the genes are expressed in DLPFC (that is, are among the 16,423 genes passing the expression-level threshold. For each set, we filtered out any genes not expressed in DLPFC. We then retained only sets with a final number of genes between 10 and 1,000. For adding the latter two databases, we did not include any set with a Jaccard overlap index > 0.5 to a GO set already included (since in such cases, a substantial portion of the genes were already included in the GO set and the added test would likely be redundant). This procedure yielded 2,902 gene sets in total: 1,938 sets from GO, 824 from Reactome, and 140 gene families.
Geneset enrichment for differential expression
Enrichment methodologies for differential gene expression between cases and controls can be broadly classified into two categories: gene permutation and subject permutation109. In gene permutation methods, such as a hypergeometric test, the null distribution of the overlap statistic is derived by (either analytically or empirically) permuting the genes found in the set being tested. In the subject sampling methods, such as GSEA110, case control labels are (either analytically or empirically) permuted to generate the null distribution of the overlap statistic. Since these methods differ in their statistical assumptions and thus appropriateness for a particular dataset and gene set, which subsequently affects their performance, here we used a combination of methods and then merged the results. Note that for these subject permutation tests, only the expression at the level of genes, but not isoforms, was incorporated.
For the gene permutation test category, we used the Fisher’s exact, hypergeometric, and GOSeq111 tests. For these tests, genes were separated into two classes depending on whether they met FDR criteria for differential expression at the gene or isoform levels (estimated FDR ≤ 5% for either genes or isoforms), or not; this set of differentially expressed genes was then evaluated for overlap versus non-overlap with the gene set being evaluated for enrichment (i.e., a 2 × 2 table was constructed). Compared to the hypergeometric and Fisher’s tests, GOSeq has an advantage for RNA-seq data in that it explicitly accounts for the detection bias of long and highly expressed transcripts. For the subject permutation category of tests, we used GSVA112, ssGSEA110, PLAGE113, and zScore114, all implemented in the gsva package of bioconductor115. To combine the results of these tests, within each of the two primary categories, we used Fisher’s method for combining P values with Brown’s correction, which is an extension of Fisher’s method that accounts for correlation between the different enrichment test statistics 116. Then, within category, P values were Bonferroni corrected across all gene sets tested, yielding two P values for each gene set. Lastly, these two P values arising from the two categories of tests (gene and subject sampling) were again Bonferroni-corrected to adjust for the twofold testing, and the minimum of the two was reported (Supplementary data file 4).
Weighted gene co-expression network analysis (WGCNA)
We constructed gene co-expression networks using the WCGNA and coexpp packages in R (https://bitbucket.org/multiscale/coexpp), starting with the normalized expression data for 16,423 genes. To ensure a more robust correlation-based co-expression analysis, we first removed 5 samples as outliers based on IAC analysis, specifically those more than 4 standard deviations from the mean), leaving a final cohort for co-expression analysis consisting of 278 control samples and 254 cases with schizophrenia. We constructed gene co-expression networks separately in control individuals and SCZ cases117.
The connectivity metric between a pair of genes i and j, or kij, is a transformed correlation between their expression profiles, with the matrix A = (kij) known as the unsigned adjacency matrix. kij is defined as |rij|β, using the absolute value of rij, the Pearson correlation coefficient between the profiles of genes i and j, and β is the parameter of a power function. β is selected using the fitting index proposed by Zhang et al. 117, i.e., to maximize the scale-free topology model fitting index r2 of the linear model that regresses log(p(k)) on log(k), where k is connectivity and p(k) is the frequency distribution of connectivity. For the current data, we used an R2 cutoff of 0.8, which corresponded to a selection of β = 6.5 and β = 9 for the control and schizophrenia networks, respectively.
To explore the modular structures of the co-expression network, the adjacency matrix is further transformed into a topological overlap matrix118. Use of the topological overlap metric leads to more cohesive and biologically meaningful modules, since it not only represents the direct correlation between two genes but also incorporates their indirect interactions through other genes in the network 117,118. Next, to identify discrete modules of highly coregulated genes (either correlated or anti-correlated), average linkage hierarchical clustering of the genes is performed, followed by a dynamic tree-cut algorithm to dynamically cut clustering dendrogram branches into discrete subsets of gene modules119. Ordered from largest (the module containing the most genes) to smallest, each module is sequentially assigned: 1) a unique number (with higher numbers indicating smaller modules), 2) a color, and 3) a label of “c” or “s” for control or schizophrenia modules, respectively. The less well-connected genes are arbitrarily grouped in the “M0” module (grey color in the WGCNA package).
Prioritization of modules for association with SCZ
We aggregated the outcome of the overlap of modules with differentially expressed genes and genetic associations with SCZ, as follows. 1. Overlaps with differentially expressed genes: The genes in each module were used to define a gene set, and each such gene set was tested for overlap with the gene set of differentially expressed genes for schizophrenia (from our CMC data). Briefly, we assess the overlap with genes in each module using Fisher’s exact test, and Bonferroni correction is applied across all modules. Overlaps with genetic associations: The genes in each module were used to define a gene set, and each such gene set was tested for overlap with genetic associations for schizophrenia as described above in the section on differential expression. Briefly, for each module, we consider the genetic overlap for each of the four classes of genetic variation tested (GWAS, CNV, de novo mutations, rare variants), where overlaps within each class of variation are combined by choosing the minimal P value after Bonferroni correction. In Supplementary table 3, we report nominal P values without correction for multiple testing of all modules, since we use this only as a secondary filter for choosing modules of interest.
In addition, we explored the specificity of the enrichment for common SCZ variants by testing the enrichment of each module with common variants for Alzheimer’s disease (AD)120, a neurodegenerative brain disorder, and rheumatoid arthritis (RA)121. Summary statistics were downloaded from publically available datasets for AD (http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php) and RA (http://plaza.umin.ac.jp/~yokada/datasource/software.htm). For each GWAS dataset, SNPs were ‘clumped’ using Plink 1.9 (https://www.cog-genomics.org/plink2) and samples of European ancestry from the 1000 genomes project phase 3, using the following settings: threshold of significance for disease-associated SNPs P value = 5 × 10−8, r2 = 0.6, and a window of 500 kb. Enrichment of modules with AD and RA loci was tested using INRICH as described in the “Overlaps with genetic associations” section.
Module Preservation Analysis
We quantified the preservation (or lack thereof) of within-module topology between schizophrenia and control co-expression networks by calculating network-based preservation statistics. Our analysis is based on previously published methods implemented in the WGCNA package122, which requires a list of genes assigned to modules in a reference network, as well as adjacency matrices for both the reference and a test network. We thus ran two separate analyses, once with the controls-based network as the reference and the SCZ-derived network as the test, and vice versa
We compared networks using various preservation statistics that can be grouped in two main categories:
Density-based preservation statistics are used to determine whether the genes in a reference module remain highly connected in the test network.
Connectivity-based preservation statistics assess whether the overall connectivity pattern between genes in a reference module is similar in the reference and test networks.
Network statistics used to assess preservation of density and connectivity are described in the supplementary text. Within each category (density or connectivity), composite module preservation statistics are constructed to summarize changes in module preservation. In detail, the comparison of network preservation in the reference and test networks is based on a permutation-based approach. The permutation approach implemented in the WGCNA package (module label permutation in the test network) shows a strong dependency on module size in our cohort (fitting index r2 > 0.95 based on a quadratic model). Thus, as an alternative, we performed 1,000 permutations of disease status for the final cohort analyzed for co-expression (278 control and 254 schizophrenia samples), followed by generation of gene co-expression networks and estimation of network preservation statistics. In the permuted sets, we again observed large differences of the network statistics with module size. Therefore, we estimated module size-dependent distributions of null statistics, based on the permuted network statistics for various ranges of binned module sizes: 30–60, 61–125, 125–250, 251–500, 501–1500, and 1501–3000.
For each module q and module preservation statistic α, the z-score , where is the observed value for the statistic α regarding module q, and µα and σα are the mean and standard deviation of the empirical distribution of permuted values for the size bin corresponding to the number of genes in module q.
We define the following composite statistics:
- permuted Z density statistics:
- permuted Z connectivity statistics:
Lower (negative) values of z-scores indicate larger relative non-preservation of the reference module in the test network. Empirical z-scores are then converted into empirical P values using the normal cumulative distribution function.
As a replication of significant findings for non-preserved modules we used the microarray gene expression data from the HBCC cohort, which included 131 SCZ and 176 control samples. We used similar approaches as the ones described above to: (1) generate the null distribution of network preservation statistics in the HBCC cohort and (2) test the non-preservation of CMC significant modules in the HBCC SCZ cases vs. controls.
For the differential expression analysis, we curated two classes of gene sets to characterize the modules:
-
Hypothesis-driven: This collection consisted of the hypothesis-driven sets dpreviously described with additional gene sets derived from previous cell type or region-specific studies or co-expression analyses.
-
Cell type- or compartment-specific annotations:
a) Cell type markers based on in situ hybridization in mouse brain tissue (abbreviated as ABA for Allen Brain Atlas123.
b) Definite (10+ fold) enrichment for seven brain cell types, estimated based on FPKM for the given cell type vs. the average FPKM in the remaining types (abbreviated as Zhang124. For each cell type, only genes with FPKM > 1 were considered.
c) Markers for different organelles and cellular compartments (markers of organelles, or MO)125–128
d) Mitochondrial genes from the somatic vs. synaptic fraction of mouse cells (MitochondrialType) 129.
-
Brain region-specific annotations: We used three categories of markers130:
a) top 200 global marker genes for 22 large brain structures [globalMarker(top200)]. Genes were ranked based on fold change enrichment (expression in region vs. expression in rest of brain).
b) top 200 local marker genes for 90 large brain structures [localMarker(top200)]. Same as a, except that fold change is defined as expression in region vs. expression in larger region (For example, enrichment of CA1 region relative to other subcompartments of the hippocampus).
c) same as b, but only local marker genes with fold change > 2 were included [localMarker(FC>2)]. Regions with < 10 marker genes were omitted.
-
Previous WGCNA studies in brain tissue:
-
Previous modules associated with schizophrenia:
a) modules (modules 1, 2, 7, 16, and 21) that are significantly enriched in genes differentially expressed in DLPFC between subjects with schizophrenia (N = 47) versus control (n = 54) subjects (Torkamani48).
b) modules (M1A and M3A) that are significantly affected in the parietal cortex of subjects with schizophrenia (n = 50) versus control (n = 50) subjects (Chen133).
c) a module (tan module) that is affected in peripheral blood of cases with schizophrenia (deJong134).
-
Hypothesis-free: The same hypothesis-free gene sets described above were used here.
The genes in each module were tested for overlap with each hypothesis-driven and hypothesis-free gene set using Fisher’s exact test. For each class of gene sets (hypothesis-driven and hypothesis-free), Bonferroni correction was applied across all modules and all gene sets tested.
Cross-validation of module reproducibility
Using the same 20 sets of 80%-20% splits used to evaluate differential expression (see “Cross-validation of differential expression”), we estimated the module reproducibility. We generated modules in the controls and SCZ using the 80% split and then examined the reproducibility of connectivity in the independent 20% replication cohort. The connectivity is estimated based on adjacency matrix using the same power (beta = 6) across all comparisons. The median Pearson correlation of connectivity values among the “discovery” and “replication” cohorts was 0.77 (mean = 0.76, sd = 0.06, 25th percentile = 0.73, 75th percentile = 0.78) and 0.80 (mean = 0.78, sd = 0.07, 25th percentile = 0.70, 75th percentile = 0.84) for cases with SCZ and controls, respectively. This strongly supports the robustness of the gene-gene correlation structure, since this replication process occurs in a completely independent sub-cohort of 20% of the brain samples.
Effect of genetic risk variants on M2c hub genes
We examined whether genes implicated in genetic studies are more likely to affect hub nodes (genes with higher number of connections) in the M2c module. For each gene in the M2c module, we estimated the intramodular connectivity (connectivity of nodes to other nodes within the M2c module). We then examined whether genes that have association for common GWAS variants (PGC SCZ2 GWAS loci), CNVs or de novo mutations have higher intramodular connectivity compared to genes that are not genetically associated with SCZ. We found a significant effect for PGC SCZ2 GWAS loci (T test: t = 2.6; P = 0.013) and de novo mutations (T test: t = 5.1; P = 2.9 × 10−6) but no CNVs (T test: t = 0.88; P = 0.4), where genes associated with SCZ have higher intramodular connectivity. Nodes from the top 50 hub genes that have been associated with SCZ are illustrated in Figure 6C.
Effect of medication exposure on genetic risk variants on M2c hub genes
In theory drug treatment could have a strong effect on the abundance of specific transcripts in cases with SCZ and thereby induce a subset of genes to cluster together and have different co-expression patterns compared to controls. To explore this hypothesis, we performed enrichment analysis of drug gene expression signatures (see “Drug effects on differential expression” section), and identified an overlap for 3 out of 18 drug signature datasets with M2c. While the overlap was significant after correcting for multiple testing, this is not surprising because M2c contains multiple receptor subunits and genes underlying synaptic neurotransmission, including direct targets of different neuroleptics. We then explored the hypothesis that genes affected by medications (or belonging to a drug signature) are differentially expressed between cases with SCZ and controls, which subsequently leads to loss of density in SCZ modules. To explore this hypothesis, we focused on genes that cluster within the M2c module and examined whether the distribution of the differentially expressed genes significance (estimated as −log10 P value) is different for genes with (“Drug”) and without (“NonDrug”) a drug signature. We did not find a significant difference in the distribution of −log10 P values for genes that have or do not have drug signature (drug versus non-drug: Kolmogorov–Smirnov test: P = 0.54). Therefore, our results do not support the hypothesis that drugs drive the loss of density through alteration in the transcript abundance of target genes. We also explored whether “Drug” versus “NonDrug” signatures within the M2c module show a different effect for loss or gain of connectivity in controls compared to SCZ. We did not observe any significant effect (Kolmogorov–Smirnov test: P = 0.054). This analysis provides additional evidence that the density loss in SCZ is not driven by medication effects.
Effect of covariates on networks
We examine the correlation of clinical/technical covariates, including: Institution, Gender, Age of death, PMI, RIN, Library batch and Ancestry with the Module Eigengene (ME) values from the control and SCZ networks. There was no significant association at FDR < 20% (range of Pearson’s r: −0.16 to 0.21). At nominal P value < 0.05 we found an association of M0c, M16c, M6c, M26c, M28c, M32c, M7s and M12s MEs with Institution, RIN or Library batch. We found no association of the blue (M2c) module with any covariate at P < 0.1, indicating that our differential co-regulated results are not biased from clinical or technical covariates.
A supplementary reporting checklist is available.
Supplementary Material
Acknowledgments
We thank the patients and families who donated material for these studies. We thank T. Lehner for his early and inspirational ideas about this project, as well as organizational and intellectual support. We thank X. He for helpful discussions regarding Sherlock, J. Scarpa for help running and interpreting WGCNA, L. Essioux for support in establishing and managing interactions with the Consortium, and Alessandro Bertolino and Anirvan Ghosh for continuous encouragement. Data and results were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and grants R01MH093725-02S1 (JB), P50MH066392 (JB), R01MH097276 (PS, ES), R01MH075916 (CGH), P50MH096891 (CGH), P50MH084053-S1 (DAL), R37MH057881 (BD) and R37MH057881S1 (BD), R01MH085542-S1 (PS), U01MH096296-S2 (PS), HHSN271201300031C (VH), VA VISN3 MIRECC (VH), P50MH066392 (JDB), NIMH Intramural program (BKL), R01MH101454 (KJB), R01MH109677 (PR), R01AG050986 (PR), VA Merit BX002395 (PR), R01 AG036836 (PDH) New York Stem Cell Foundation (KJB), the Silvio O Conte Center grant P50MH094268 (NK), NARSAD (ECO), NARSAD Young Investigator (DMR, PR, EAS), and the Stanley Medical Research Foundation and NIMH-R01MH074313 (SEH). Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh Brain Tissue Donation Program, the NIMH Human Brain Collection Core, and Wake Forest University. CMC Leadership: P. Sklar, J. Buxbaum (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), R. Gur, C.. Hahn (University of Pennsylvania), K. Hirai, H.Toyoshiba (Takeda Pharmaceuticals Company Limited), E. Domenici, L. Essioux (F. Hoffman-La Roche Ltd), L. Mangravite, M. Peters (Sage Bionetworks), T. Lehner, B. Lipska (NIMH).
COMPETING FINANCIAL INTEREST STATEMENT
E Domenici was an employee of F.Hoffmann-La Roche for the first portion of the study and later served as a consultant to Roche in the area of genetic biomarkers. H Toyoshiba and K Hirai are employees of Takeda Pharmaceutical Company Limited and L Shinobu is a former employee. DAL currently receives investigator-initiated research support from Pfizer and in 2012–2014 served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals, and Sunovion. M Fromer was an employee of Mount Sinai until March 2016, he is now an employee of Google Verily.
Footnotes
AUTHOR CONTRIBUTIONS:
P Roussos, J Johnson, K Talbot, R Gur, C Hahn, D Lewis, V Haroutunian, B Lipska and J Buxbaum contributed to sample collection. S Hemby contributed monkey brain tissue, P Sullivan contributed mouse data.
M Fromer, P Roussos, S Sieberts, D Kavanagh, T Perumal, D Ruderfer, K Dang, E Oh, A Topol, T Chess, M Peters, E Domenici, B Devlin and P Sklar contributed to the writing of this manuscript.
M Fromer, P Roussos, S Sieberts, H Shah, D Ruderfer, K Dang, M Mahajan, J Dudley, A Chess, S Purcell, L Shinobu, L Mangravite, H Toyoshiba, D Lewis, M Peters, J Buxbaum, E Schadt, K Hirai, K Brennand, N Katsanis, B Devlin and P Sklar contributed to experimental and study design and planning analytical strategies.
L Shinobu, H Toyoshiba, D Lewis, B Lipska, J Buxbaum, E Schadt, K Hirai, E Domenici, B Devlin and P Sklar contributed the funding of this work.
M Fromer, P Roussos, S Sieberts, J Johnson, D Kavanagh, T Perumal, D Ruderfer, H Shah, L Klei, R Kramer, D Pinto, Z Gumus, A Chess, K Dang, A Browne, C Lu, B Readhead, E Stahl, T Hamamsy, J Fullard, Y Wang, J Derry, B Logsdon, T Raj, J Zhu, B Zhang, P Sullivan, S Purcell, E Schadt, E Domenici, B Devlin and P Sklar contributed to data analyses.
E Oh, A Topol, M Parvizi, K Brennand and N Katsanis contributed to the model system experiments.
T Raj, D Bennett, P De Jager contributed the ROS/MAP data.
A Chess, L Shinobu, L Mangravite, H Toyoshiba, R Gur, C Hahn, D Lewis, M Peters, B Lipska, J Buxbaum, K Hirai, E Schadt, E Domenici, B Devlin and P Sklar contributed to the management and leadership of phase 1 of the CommonMind Consortium.
DATA AVAILABILITY
The CommonMind investigators are committed to the release of data and analysis results, with the anticipation that data sharing in a rapid and transparent manner will speed the pace of research to the benefit of the greater research community. Data and analytical results generated through the CommonMind Consortium are available through the “CommonMind Consortium Knowledge Portal”: doi:10.7303/syn2759792.
URLS:
Human Brain Collection Core (HBCC): http://www.nimh.nih.gov/labs-at-nimh/research-areas/research-support-services/hbcc/human-brain-collection-core-hbcc.shtml)
CommonMind Consortium (CMC): www.synapse.org/CMC
Gemtools: http://www.wpic.pitt.edu/wpiccompgen/GemTools/GemTools.htm
GTEx Portal: www.gtexportal.org, dbGaP accession number phs000424.v6.p1
ROSMAP Study: https://www.synapse.org/#!Synapse:syn3219045
1000 Genomes Phase 3 genotype data: http://www.1000genomes.org
GO (Gene Ontology): http://www.geneontology.org
Reactome database: (http://www.reactome.org)
HGNC (HUGO Gene Nomenclature Committee) gene families: http://www.genenames.org
Plink 1.9: https://www.cog-genomics.org/plink2
ComBat: http://statistics.byu.edu/johnson/ComBat/
PLINK/Seq: https://atgu.mgh.harvard.edu/plinkseq/
MRIN: http://zhanglab.c2b2.columbia.edu/index.php/MRIN
MISO (http://genes.mit.edu/burgelab/miso/
GEO accession numbers for datasets - Braincloud: GSE30272; NIH: GSE15745, HBTRC: GSE44772; UKBEC: GSE46706;
HBCC microarray: dbGAP study_id=phs000979.v1.p1
DNENRICH: https://psychgen.u.hpc.mssm.edu/dnenrich
Alzheimer’s GWAS data: http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php)
Rheumatoid arthritis GWAS data: http://plaza.umin.ac.jp/~yokada/datasource/software.htm
Code Availability
Code for gene expression normalization and differential expression are provided in a public repository: https://bitbucket.org/commonmind/commonmind/src/PIPELINE-FOR-PUBLIC-FREEZE-1.0/scripts/phaseI
eQTL and coexpression network analyses utilized standard software packages.
REFERENCES
- 1.McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 2.Kirov G. CNVs in neuropsychiatric disorders. Hum Mol Genet. 2015;24:R45–R49. doi: 10.1093/hmg/ddv253. [DOI] [PubMed] [Google Scholar]
- 3.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 7.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath S, Janka Z, Mirnics K. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry. 2011;69:157–162. doi: 10.1016/j.biopsych.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mistry M, Gillis J, Pavlidis P. Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls. BMC Neurosci. 2013;14:105. doi: 10.1186/1471-2202-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitzemann R, et al. Introduction to sequencing the brain transcriptome. Int Rev Neurobiol. 2014;116:1–19. doi: 10.1016/B978-0-12-801105-8.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs JR, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramasamy A, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y, et al. A meta-analysis of gene expression quantitative trait loci in brain. Transl Psychiatry. 2014;4:e459. doi: 10.1038/tp.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright FA, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roussos P, et al. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards AL, et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharadwaj R, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84:997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Jager PL, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman RE, Alekov AK, Filippov M, Hegermann J, Fahlke C. Involvement of ClC-3 chloride/proton exchangers in controlling glutamatergic synaptic strength in cultured hippocampal neurons. Front Cell Neurosci. 2014;8:143. doi: 10.3389/fncel.2014.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimoda Y, Watanabe K. Contactins: emerging key roles in the development and function of the nervous system. Cell Adh Migr. 2009;3:64–70. doi: 10.4161/cam.3.1.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57:834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan Q, et al. Regulation of Brain-Derived Neurotrophic Factor Exocytosis and Gamma-Aminobutyric Acidergic Interneuron Synapse by the Schizophrenia Susceptibility Gene Dysbindin-1. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 32.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra-Gorur K, et al. Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron. 2014;84:1226–1239. doi: 10.1016/j.neuron.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golzio C, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho CM, et al. Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. Am J Hum Genet. 2014;95:565–578. doi: 10.1016/j.ajhg.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borck G, et al. BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome Res. 2015;25:155–166. doi: 10.1101/gr.176925.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topol A, et al. Altered WNT Signaling in Human Induced Pluripotent Stem Cell Neural Progenitor Cells Derived from Four Schizophrenia Patients. Biol Psychiatry. 2015;78:e29–e34. doi: 10.1016/j.biopsych.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee IS, et al. Characterization of molecular and cellular phenotypes associated with a heterozygous CNTNAP2 deletion using patient-derived hiPSC neural cells. NPJ Schizophr. 2015;1 doi: 10.1038/npjschz.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaloy C, Gao FB. A new role for microRNA-9 in human neural progenitor cells. Cell Cycle. 2010;9:2913–2914. doi: 10.4161/cc.9.15.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao R, Boehnke M. Quantifying and correcting for the winner’s curse in genetic association studies. Genet Epidemiol. 2009;33:453–462. doi: 10.1002/gepi.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson LA, Porter RA. Progress in the development of neurokinin 3 modulators for the treatment of schizophrenia: molecule development and clinical progress. Future Med Chem. 2013;5:1525–1546. doi: 10.4155/fmc.13.122. [DOI] [PubMed] [Google Scholar]
- 43.de Souza Silva MA, et al. Neurokinin3 receptor as a target to predict and improve learning and memory in the aged organism. Proc Natl Acad Sci U S A. 2013;110:15097–15102. doi: 10.1073/pnas.1306884110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouchi Y, et al. Reduced adult hippocampal neurogenesis and working memory deficits in the Dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J Neurosci. 2013;33:9408–9419. doi: 10.1523/JNEUROSCI.2700-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai T, et al. Changes in density of calcium-binding-protein-immunoreactive GABAergic neurons in prefrontal cortex in schizophrenia and bipolar disorder. Neuropathology. 2008;28:143–150. doi: 10.1111/j.1440-1789.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- 46.Carboni L, Domenici E. Proteome effects of antipsychotic drugs: Learning from preclinical models. Proteomics Clin Appl. 2015 doi: 10.1002/prca.201500087. [DOI] [PubMed] [Google Scholar]
- 47.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torkamani A, Dean B, Schork NJ, Thomas EA. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res. 2010;20:403–412. doi: 10.1101/gr.101956.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian V. A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen Psychiatry. 2012;69:1205–1213. doi: 10.1001/archgenpsychiatry.2012.704. [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 51.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powchik P, et al. Postmortem studies in schizophrenia. Schizophr Bull. 1998;24:325–341. doi: 10.1093/oxfordjournals.schbul.a033330. [DOI] [PubMed] [Google Scholar]
- 53.Purohit DP, et al. Alzheimer disease and related neurodegenerative diseases in elderly patients with schizophrenia: a postmortem neuropathologic study of 100 cases. Arch Gen Psychiatry. 1998;55:205–211. doi: 10.1001/archpsyc.55.3.205. [DOI] [PubMed] [Google Scholar]
- 54.Kimoto S, Bazmi HH, Lewis DA. Lower expression of glutamic acid decarboxylase 67 in the prefrontal cortex in schizophrenia: contribution of altered regulation by Zif268. Am J Psychiatry. 2014;171:969–978. doi: 10.1176/appi.ajp.2014.14010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 56.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 57.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Connell J, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee AB, Luca D, Klei L, Devlin B, Roeder K. Discovering genetic ancestry using spectral graph theory. Genet Epidemiol. 2010;34:51–59. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luca D, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.San Lucas FA, Wang G, Scheet P, Peng B. Integrated annotation and analysis of genetic variants from next-generation sequencing studies with variant tools. Bioinformatics. 2012;28:421–422. doi: 10.1093/bioinformatics/btr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng H, Zhang X, Zhang C. mRIN for direct assessment of genome-wide and gene-specific mRNA integrity from large-scale RNA-sequencing data. Nat Commun. 2015;6:7816. doi: 10.1038/ncomms8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang T, Cai YD. An information-theoretic machine learning approach to expression QTL analysis. PLoS One. 2013;8:e67899. doi: 10.1371/journal.pone.0067899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Topol A, Tran NN, Brennand KJ. A guide to generating and using hiPSC derived NPCs for the study of neurological diseases. J Vis Exp. 2015:e52495. doi: 10.3791/52495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Topol A, et al. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep. 2016;15:1024–1036. doi: 10.1016/j.celrep.2016.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 77.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 79.Cheng MC, et al. Chronic treatment with aripiprazole induces differential gene expression in the rat frontal cortex. Int J Neuropsychopharmacol. 2008;11:207–216. doi: 10.1017/S1461145707008048. [DOI] [PubMed] [Google Scholar]
- 80.Orsetti M, Di Brisco F, Rinaldi M, Dallorto D, Ghi P. Some molecular effectors of antidepressant action of quetiapine revealed by DNA microarray in the frontal cortex of anhedonic rats. Pharmacogenet Genomics. 2009;19:600–612. doi: 10.1097/FPC.0b013e32832ee573. [DOI] [PubMed] [Google Scholar]
- 81.Ikeda M, et al. Identification of novel candidate genes for treatment response to risperidone and susceptibility for schizophrenia: integrated analysis among pharmacogenomics, mouse expression, and genetic case-control association approaches. Biol Psychiatry. 2010;67:263–269. doi: 10.1016/j.biopsych.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 82.Fatemi SH, Folsom TD, Reutiman TJ, Novak J, Engel RH. Comparative gene expression study of the chronic exposure to clozapine and haloperidol in rat frontal cortex. Schizophr Res. 2012;134:211–218. doi: 10.1016/j.schres.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 83.Rizig MA, et al. A gene expression and systems pathway analysis of the effects of clozapine compared to haloperidol in the mouse brain implicates susceptibility genes for schizophrenia. J Psychopharmacol. 2012;26:1218–1230. doi: 10.1177/0269881112450780. [DOI] [PubMed] [Google Scholar]
- 84.Kondo MA, et al. Unique pharmacological actions of atypical neuroleptic quetiapine: possible role in cell cycle/fate control. Transl Psychiatry. 2013;3:e243. doi: 10.1038/tp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Santoro ML, et al. Effect of antipsychotic drugs on gene expression in the prefrontal cortex and nucleus accumbens in the spontaneously hypertensive rat (SHR) Schizophr Res. 2014;157:163–168. doi: 10.1016/j.schres.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 86.Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 2010;38:e204. doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 88.Kirov G, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014;75:378–385. doi: 10.1016/j.biopsych.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 90.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012 doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gulsuner S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCarthy SE, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang YH, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 97.Hamdan FF, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 99.De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tansey KE, Owen MJ, O’Donovan MC. Schizophrenia genetics: building the foundations of the future. Schizophr Bull. 2015;41:15–19. doi: 10.1093/schbul/sbu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashburner M, et al. Gene ontology: tool for the unification of biology The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 110.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tomfohr J, Lu J, Kepler TB. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinformatics. 2005;6:225. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brown MB. 400: A method for combining non-independent, one-sides tests of significance. Biometrics. 1975;31:987–992. [Google Scholar]
- 117.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 118.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 119.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 124.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bachoo RM, et al. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci U S A. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Foster LJ, et al. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 127.Morciano M, et al. Immunoisolation of two synaptic vesicle pools from synaptosomes: a proteomics analysis. J Neurochem. 2005;95:1732–1745. doi: 10.1111/j.1471-4159.2005.03506.x. [DOI] [PubMed] [Google Scholar]
- 128.Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 129.Winden KD, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen C, et al. Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry. 2013;18:1308–1314. doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Jong S, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS One. 2012;7:e39498. doi: 10.1371/journal.pone.0039498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.