Abstract
Meiotic recombination involves a combination of gene conversion and crossover events that along with mutations produce germline genetic diversity. Here, we report the discovery of 3,176 SNP and 61 indel gene conversions. Our estimate of the non-crossover (NCO) gene conversion rate (G) is 7.0 for SNPs and 5.8 for indels per Mb per generation, and the GC bias is 67.6%. For indels we demonstrate a 65.6% preference for the shorter allele. NCO gene conversions from mothers are longer than those from fathers and G is 2.17 times greater in mothers. Notably, G increases with the age of mothers, but not fathers. A disproportionate number of NCO gene conversions in older mothers occur outside double strand break (DSB) regions and in regions with relatively low GC content. This points to age-related changes in the mechanisms of meiotic gene conversions in oocytes.
Introduction
New combinations of alleles, generated through the shuffling of material between homologous chromosomes during meiosis, contribute to genetic diversity. Meiotic recombination is mostly thought to stem from programmed DNA double-strand breaks (DSBs)1 generated by the evolutionary conserved SPO11 protein2 at hotspot locations determined by the DNA-binding specificity of PRDM93. These DSBs are either resolved through the reciprocal crossing-over of large regions between homologous chromosomes (often referred to as recombination in previous publications) or NCO gene conversion, the non-reciprocal transfer of short DNA segments between homologues. Crossover events, which arise through the resolution of the double Holliday junction (dHj)4, are also frequently accompanied by gene conversions, hereafter referred to as CO gene conversions. NCO gene conversions most commonly arise via the synthesis-dependent strand annealing (SDSA) pathway5, generating short regions of unidirectional homologous exchange6,7, but the resolution of the dHj can also lead to NCO gene conversions8,9. A schematic overview of these DSB repair mechanisms is given in Supplementary Figure 1.
Repair through meiotic gene conversion uses sequence copied from a homologous chromosome. Heteroduplex DNA is formed in the recombination intermediates10 and mismatched nucleotides in the heteroduplex DNA are subsequently corrected. A preference of strong (G/C) over weak (A/T) base-pairs of mismatch repaired nucleotides in converted segments, known as GC bias, has been previously observed11. Consequently, gene conversion plays a key role in shaping the genome's GC content12,13 , has a potential confounding impact on mutation rate inferences and evolutionary divergence time estimates14,15 and may skew allele frequencies increasing the disease burden of recessive alleles16.
In previous publications, we quantified the rate of chromosomal crossover in meiosis, its determinants and genomic distribution in humans17–19. Here we report on the discovery of allelic gene conversions in meiosis inferred from the germline genotypes of probands and their relatives. We adopted a study design similar to that of Williams et al.20, based on probands in three-generation families, where genotypes are available for a proband, its spouse, both its parents, at least two of its siblings and at least one child. This family structure was chosen to limit the impact of genotyping errors that can mimic gene conversion, such that haplotypes carried by the parents can be independently verified in the siblings and the gene conversion can be verified in the child. We searched for contiguous tracts of markers (no longer than 100kb) consistent with non-reciprocal transfer of chromosome fragments in a transmission from parent to proband. This study design is adapted to the detection of NCO gene conversions, which are typically shorter than 100kb and where the converted tract is flanked by sequence from the original recipient chromosome on both sides4,20. Our study design does not allow for the detection of the subset of CO gene conversions, where the converted tract is flanked by sequence from the original recipient chromosome on only one side4,8. However, we are able to detect crossover recombination events that are accompanied by one or more such gene conversion events – hereafter referred to as complex crossover (CCO) gene conversions20–22.
Using this approach, we sought gene conversions in two overlapping datasets. The first consists of 7,219 proband-family sets genotyped on Illumina HumanHap and Omni BeadChip arrays (chip dataset). The second consists of 101 whole genome sequenced (at > 30×) proband-family sets (sequencing dataset), 91 of which are contained in the first dataset (cf. Figure 1).
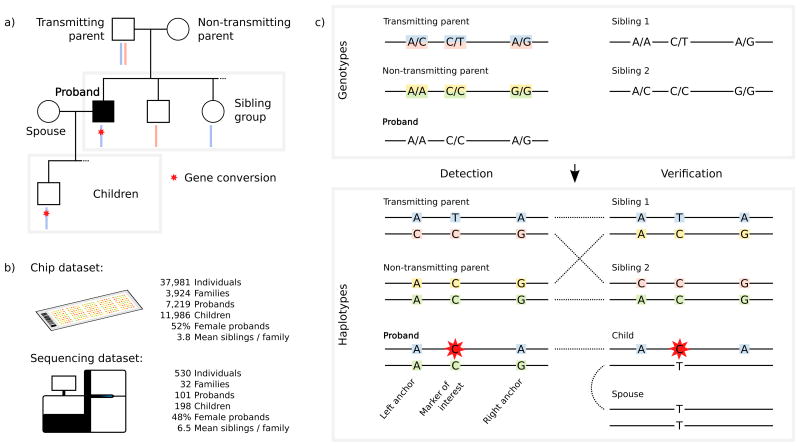
Figure 1.
The study design and method used for detecting gene conversions. Including a) the family structure and b) the two sources of genotypes. c) The genotypes of the siblings are necessary to verify the haplotypes of both parents. We require at least one child of the proband to verify the gene conversion. The method is shown for NCO gene conversion.
Results
As gene conversions can only be detected at polymorphic markers, we restricted our analysis to high quality SNPs and indels with a minor allele frequency over 0.5%. Indels were further restricted to be shorter than 10 bp, as longer indels are rare and yield less reliable genotypes. This restriction left 624,955 SNPs in the chip dataset and 8,195,014 SNPs and 465,054 indels in the sequencing dataset. For each proband-family set, we considered only variants where at least one parent was heterozygous, the haplotypes transmitted by both parents could be verified in a sibling and the gene conversion could be verified in an offspring. Using this approach, we assessed 214,241,663 marker proband pairs (mpps) in the chip dataset, yielding 2,192 mpps involved in a gene conversion (cf. Table 1,Table 2). In the sequencing dataset, 147,368,280 SNP and 8,715,873 indel mpps were assessed, yielding 1,027 and 61 gene converted mpps, respectively.
Table 1. Estimates of NCO gene conversion of SNPs.
| Chip dataset | Sequencing dataset | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Paternal | Maternal | Total | Paternal | Maternal | Total | |
| Converted mpps | 641 | 1,393 | 2,034 | 300 | 565 | 865 |
| N mpps considered | 106,740,626 | 106,761,366 | 213,501,992 | 73,919,056 | 73,016,289 | 146,935,345 |
| Observed gene conversion rate per Million bp (95% CI) | 6.0 (5.5–6.5) |
13.0 (12.0–14.1) |
9.5 (9.0–10.1) |
4.1 (3.4–4.8) |
7.7 (5.7–9.9) |
5.9 (4.9–7.0) |
| Corrected gene conversion rate per Million bp (95% CI) | 3.9 (3.5–4.4) |
10.0 (8.5–11.6) |
7.0 (6.0–8.0) |
– | – | – |
| N events | 618 | 1,018 | 1,636 | 244 | 241 | 485 |
| Short | 606 | 849 | 1,455 | 234 | 196 | 430 |
| Long | 12 | 169 | 181 | 10 | 45 | 55 |
| Gene converted mpps / event (SE) | 1.04 (±0.01) | 1.37 (± 0.03) | 1.24 (±0.02) | 1.23 (±0.06) | 2.34(±0.28) | 1.78(±0.15) |
| Short | 1.01 (±0.00) | 1.04 (±0.01) | 1.03 (±0.01) | 1.09 (± 0.02) | 1.17 (±0.04) | 1.13 (±0.02) |
| Long | – | 3.01(±0.13) | 2.96 (±0.12) | – | 7.44 (±1.15) | 6.93 (±0.99) |
| GC bias (95% CI) | 64.4 (60.7–68.3) |
69.1 (66.6–71.6) |
67.6 (65.7–69.8) |
63.1 (57.1–69.1) |
72.5 (69.1–76.1) |
69.3 (65.8–72.3) |
| Short | 64.7 (61.1–68.5) |
69.5 (66.4–72.3) |
67.5 (65.3–69.7) |
65.3 (58.3–72.0) |
73.0 (66.5–79.7) |
68.9 (64.1–74.1) |
| Long | – | 68.6 (64.5–72.3) |
67.9 (64.0–71.7) |
– | 72.2 (67.2–77.4) |
69.8 (64.9–74.5) |
| OR DSB (95% CI) | 42.3 (35.2–50.9) |
5.4 (4.7–6.2) |
10.7 (9.6–11.9) |
45.7 (32.8–67.1) |
7.1 (5.2–9.7) |
14.9 (11.3–19.8) |
| Short | 49.7 (41.2–61.1) |
9.1 (7.9–10.5) |
17.5 (15.9–19.4) |
66.9 (49.4–92.1) |
20.3 (15.7–25.8) |
37.0 (30.2–45.7) |
| Long | – | 1.1 (0.7–1.5) |
1.1 (0.7–1.5) |
– | 1.6 (0.9–2.3) |
2.0 (1.2–3.0) |
| OR crossover recombination hotspots (95% CI) | 8.2 (6.6–10.1) |
4.6 (3.8–5.4) |
– | 12.7 (8.6–18.3) |
8.0 (5.7–11.1) |
– |
| Short | 8.8 (7.1–10.6) |
6.5 (5.4–7.7) |
– | 14.8 (10.7–20.5) |
14.1 (9.8–19.5) |
– |
| Long | – | 2.0 (1.1–3.2) |
– | – | 4.0 (1.5–6.9) |
– |
Corrected gene conversion rate is corrected for crossover recombination rate. This correction and the odds ratio for hotspots are computed using sex specific maps. Short events < 1,000 base-pairs, long events >= 1,000 base-pairs. Results for long NCO events are not presented in paternal transmissions due to small sample size. As crossover maps are sex specific no joint analysis is provided. NCO = non-crossover, OR = odds ratio, DSB = double strand break, mpp = marker proband pair, CI = confidence interval, SE = standard error
Table 2. Estimates of CCO gene conversions of SNPs.
| Chip dataset | Sequencing dataset | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Paternal | Maternal | Total | Paternal | Maternal | Total | |
| Converted mpps | 8 | 150 | 158 | 5 | 157 | 162 |
| N mpps considered | 289,364 | 450,307 | 739,671 | 170,612 | 262,323 | 432,935 |
| Observed gene conversion rate per Million bp (95% CI) | 28 (10–52) |
333 (265–411) |
214 (168–259) |
29 (6–58) |
598 (289–1,011) |
374 (182–624) |
| N events | 7 | 107 | 114 | 5 | 34 | 39 |
| Gene converted mpps / event (SE) | 1.14 | 1.40 (±0.08) | 1.39 (±0.07) | 1 | 4.76(±1.29) | 4.26(±1.16) |
| GC bias (95% CI) |
12.5 |
73.3 (65.2–80.7) |
70.2 (62.5–77.8) |
75 |
70.2 (63.1–78.6) |
70.1 (63.1–78.8) |
| N crossovers considered | 45,025 | 70,079 | 115,104 | 1,624 | 2,490 | 4,114 |
| Complex crossovers, per thousand | 0.16 (0.07–0.28) |
1.5 (1.3–1.8) |
1.0 (0.8–1.2) |
3.1 (0.6–6.0) |
13.3 (8.5–18.2) |
9.2 (6.2–12.5) |
Confidence intervals for CCO gene conversions in paternal transmissions are not reported due to small number of events. CCO = complex crossover, OR = odds ratio, DSB = double strand break, mpp = marker proband pair, CI = confidence interval
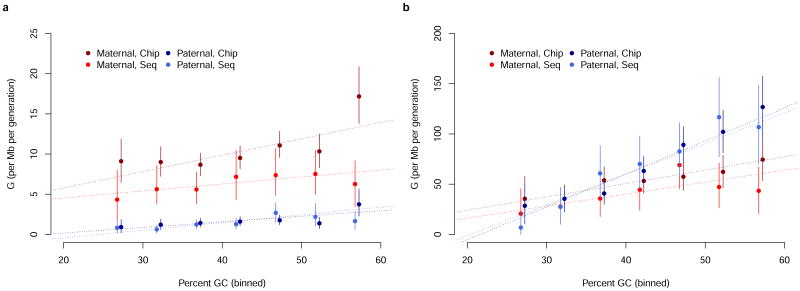
As most sequenced individuals also have chip data, it follows that a subset of 5,233,293 chip data mpps can be informative about the sensitivity and specificity of gene conversion calling in the two sets. From this subset 1,060,127 were omitted from the sequencing dataset due to quality control thresholds, leaving 4,173,166 overlapping mpps that were assessed in both datasets. All yielded concordant results, including an overlap of 43 gene conversions (cf. Supplementary Note 1.1). We further confirmed phased haplotypes using read pairs (Supplementary Note 1.2, Supplementary Table 1) and genotypes using Sanger sequencing (Supplementary Note 1.3) and whole genome sequencing (Supplementary Note 1.4, Supplementary Table 2), which revealed error rates between 0.0% and 1.1%.
NCO gene conversions in DSB regions and recombination hotspots
We first examined the distribution of NCO gene conversions in the genome and their rate, G. As most such events are thought to be due to programmed DSBs, we compared our predicted NCO gene conversions to a map of DSB regions generated from spermatocyte samples of five human males23, which have a mean size of 1,464 bp, s.d. 586bp. Table 1 shows that NCO gene conversions are highly overrepresented in spermatocyte DSB regions (odds ratio > 10). In paternal transmissions, the overrepresentation was 42.3 and 45.7 fold in the chip and sequencing data, respectively. In maternal transmissions, the overrepresentation was only 5.4 fold and 7.1 fold in chip and sequencing datasets, respectively, albeit highly significant (p-value < 0.001, for both datasets). As the locations of crossover recombination hotspots are known to differ between male and female meioses18, and hot spots are largely determined by DSB regions23, it follows that a stronger elevation of maternal G would be expected against a map of DSB regions from oocytes (such a map is currently not available). A previous study20 also showed an overrepresentation of NCO gene conversions in male DSB regions and indications of a sex difference in their localization. Recent research has shown that PRDM9 alleles carried by the parent strongly influences the locations of DSBs23 and consequently the locations of crossovers and NCOs in chromosomes transmitted to the offspring. Our results confirm these findings. The PRDM9 allele of the proband strongly affects the distribution of NCO gene conversion, but not their overrepresentation in DSB regions (cf.Supplementary Note 2, Supplementary Table 3).
We next compared NCO gene conversions to a sex specific map of crossover recombination hotspots24. Crossover recombinations, when estimated in the sexes separately, are enriched 38.0 and 27.6 fold in male and female crossover recombination hotspots, respectively17. Table 1 shows that for males, the enrichment of G in male crossover recombination hotspots is 8.2 and 12.7 fold in chip and sequencing datasets, respectively. For females, the enrichment of G was 4.6 and 8.0 fold in female crossover recombination hotspots.
NCO gene conversion rate
Williams et al.20, estimated G as 5.9/Mb/generation, using a genomewide approach similar to the one presented here (but restricting to events shorter than 5kb). Comparable estimates have been obtained based on sperm genotyping6,7 (not genomewide) and coalescent inferences14,15,25. Our genomewide estimate of G is 9.5/Mb/generation in the chip dataset (unadjusted for SNP ascertainment) and 5.9/Mb/generation in the sequencing dataset (cf. Table 1). While the sequencing dataset is minimally affected by ascertainment bias, the markers on Illumina BeadChip arrays are preferentially selected in genomic regions of low linkage disequilibrium26, i.e. regions with a high rate of crossover recombination (and underlying DSBs). After correcting for this ascertainment bias in the chip dataset, we estimate the genomewide G to be 7.0/Mb/generation (95% CI 6.0–8.0). The difference between this estimate and the one obtained from the sequencing data is not significant (p-value: 0.12 ) and both are consistent with previous estimates.
Genomic distribution and sex differences
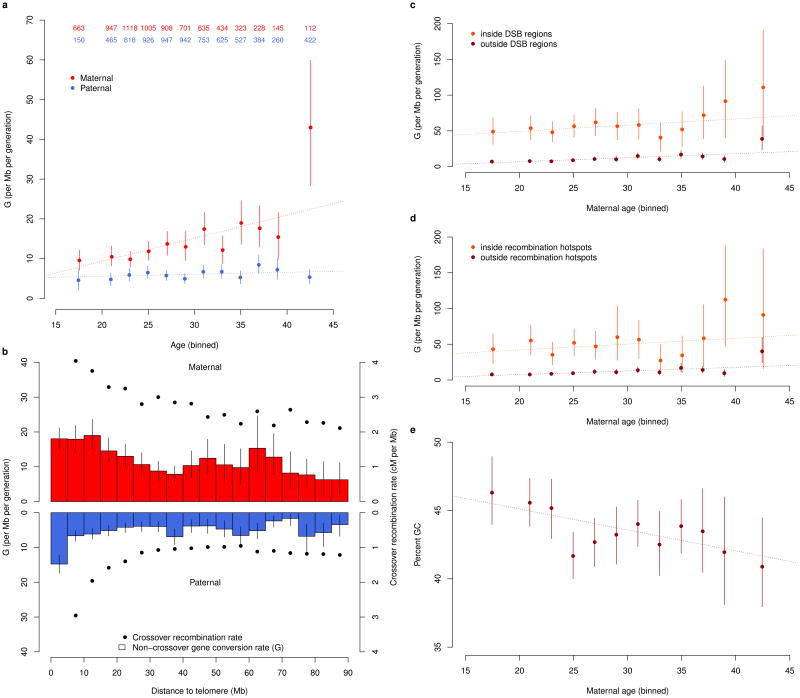
An assessment of the genomic distribution of G in the two sexes (cf. Supplementary Note 3) shows that G is elevated near telomeres and therefore decreases with chromosome length (Figure 2b). This pattern is analogous to that observed for crossover recombinations18,27 and is consistent with a higher stationary GC-content near telomeres28. Moreover, as in the case of crossover recombinations, the proportion of events near telomeres is greater in fathers than in mothers. Overall, G is 2.17 (95% CI 1.94–2.45) and 1.91 (95% CI 1.34–2.58) times higher in maternal transmissions than paternal transmissions when assessed in the chip and sequencing datasets, respectively. This sex difference is very similar to that observed for the crossover recombination rate17, which is 2.03, although the difference in G can largely be attributed to longer events in maternal transmissions.
Figure 2.
NCO gene conversion rate, G. As a function of a) age of parent, b) distance from telomere, c) age of mother stratified by presence in DSB region and d) age of mother stratified by presence in crossover recombination hotspots. e) percent GC content of neighboring 100 bases for gene converted markers outside of double strand break regions as a functionof age of mother. Blue represents paternal and red maternal transmission. Error bars represent 95% confidence intervals. a) Number of parents in each age bin are presented in top of figure. a,c,d,e) Individuals are grouped into 2 year age bins with all individuals younger than 20 grouped together and all individuals 40 years or older grouped together. b) mpps are grouped into 5Mb bins. Crossover recombination rate is computed as the average over all marker proband pairs in a bin of average crossover recombination rate in +/– 5kb interval around a marker. cM = centiMorgan, Mb = 1 million base pairs.
G increases with maternal age
As previous studies have reported an age-related increase in the crossover recombination rate in females that is not seen in males19,29, we examined the impact of age on G. In the chip dataset, there is a marked increase of G with maternal age of 0.58/Mb/year (95% CI 0.38– 0.78, p-value: 1.4·10–8) that is not observed in fathers (Figure 2a, Supplementary Note 4). Based on a very small number of events, the maternal age effect in the sequencing dataset is 0.33/Mb/year (95% CI –0.18 – 0.83). Although this estimate is not significantly different from 0 (p-value: 0.21), this effect is also not significantly different from the chip data estimate (p-value: 0.35).
Interestingly, the increase of G with maternal age in the chip is comparable inside and outside of both crossover recombination hotspots and male DSB regions (Figure 2c,d), resulting in a decrease of odds ratios for co-occurence with these genome features (cf. Supplementary Note 5, Supplementary Figure 2). The average female crossover recombination rate of maternally transmitted NCO gene conversions further decreases with age (cf. Supplementary Figure 3). Thus, the NCO gene conversions that accumulate with age appear to be less tied to programmed DSBs than those transmitted by younger mothers.
G increases with local GC content
G increases with local GC content, defined as the GC content of the 100 base pairs surrounding each mpp (cf. Supplementary Note 6), a result consistent with GC-biased NCO gene conversions occurring repeatedly at similar locations in evolutionary history. Local GC content is elevated in DSB regions (cf. Supplementary Figure 4 and Supplementary Table 4), in part due to the GC composition of PRDM9 motif30. We consider the correlation with local GC content inside and outside of DSB regions separately as well as distinguishing between the chip and sequencing datasets and paternal and maternal transmissions. Remarkably, even after we have conditioned the data on DSB region status, in seven out of eight cases G is positively correlated with local GC content (Figure 3 and Supplementary Table 5).
Figure 3.
NCO gene conversion rate G, as a function of GC content of neighboring 100bp. a) G outside of male DSB regions. b) G inside of male DSB regions. Error bars represent 95% confidence intervals. The dotted line represents a linear model. NCO = non-crossover, DSB = double strand break
In the chip dataset, we observe a decrease (p-value: 0.0068) in local GC content with age in maternally transmitted NCO gene conversions. As this result might be attributed to the high GC content of the PRDM9 binding motif, we restricted to mpps outside of male DSB regions, where we also observed a decrease (p-value: 0.0037) (Figure 2e). The result also remained significant after adjustment of local GC content for the HapMap based recombination rate31 (p-value: 0.0029). Again, this suggests that the NCO gene conversions that accrue in aging mothers are different in mechanism from those found in younger mothers.
We investigated whether G was dependent on the SNP type, the base pair composition of the SNP's two alleles, and found no such effect (cf.Supplementary Table 6).
Complex NCO events
We grouped gene converted mpps into events, based on their proximity. In NCO gene conversions, most events contain only a small number of gene converted mpps, with an average of 1.24 mpps per event for the chip dataset and 1.78 gene per event for the sequencing dataset. The smaller number of mpps per event for the chip dataset than the sequencing dataset can in part be attributed to the lower marker density. Interestingly, in NCO gene conversions, maternally transmitted events are tagged by more mpps than paternally transmitted events; 1.37 vs. 1.04 (p-value < 0.001) and 2.34 vs. 1.23 (p-value < 0.001) mpps per event for chip and sequencing data, respectively. However, we did not observe an age dependence in the number of mpps per event (cf. Supplementary Figure 5).
We partitioned the set of NCO gene conversions into short and long NCO events, based on a distance of 1,000 bp (roughly the size of a DSB region), between the first and last gene converted mpp per event. Supplementary Table 7 shows the length distribution of long NCO events by distance between the first and last marker. Due to the denser marker set, the length of the event can be better estimated in the sequencing dataset, hence some events classified as short NCO events in the chip data would be classified as long NCO events, if the denser sequencing data were available (cf. Supplementary Note 7). Table 1 shows that short NCO events are highly overrepresented in male DSB regions and crossover recombinations hotspots, while long NCO events are not overrepresented in male DSB regions, but are overrepresented in crossover recombination hotspots.
The tracts of long NCO events contain both gene converted mpps and non-gene converted mpps (cf. Supplementary Note 7, Supplementary Table 8). A similar pattern has been previously observed20 and are referred to as complex NCO gene conversions. Within complex events both the gene converted and non-gene converted mpps show a GC bias.
In the chip and sequencing datasets we estimate that, respectively, at least 46.1% and 65.3% of all long NCO events are complex (cf. Supplementary Table 9). The true rate of complex events is likely to be higher (cf. Supplementary Note 7), leading us to hypothesize that all long NCO events may be complex. These long, and mostly complex, NCO events are more common in maternal transmissions (p-value < 0.001, for chip and sequencing datasets). A significant increase in G with mother's age is observed both for short and long events (cf. Supplementary Note 7).
Gene conversion of indels
We estimate G for indels as 5.8/Mb/generation (95% CI 4.1–7.9), comparable to that for SNPs. Our results show a bias of 65.6% (95% CI 53.3–76.6, p-value: 0.018, cf.Table 3) toward the shorter allele for indels in allelic gene conversions (cf. Supplementary Note 8). A direct estimate of gene conversions involving indels has to our knowledge not been previously reported. Comparisons between species have yielded conflicting results; a deletion bias has previously been reported for non-allelic gene conversion32, while a bias towards insertion has previously been reported for allelic gene conversion33.
Table 3. Gene conversion of indels.
| Sequencing dataset | |||
|---|---|---|---|
|
|
|||
| Paternal | Maternal | Total | |
| Converted mpps | 23 | 38 | 61 |
| NCO | 22 | 28 | 50 |
| CCO | 1 | 10 | 11 |
| N mpps considered | 4,391,571 | 4,324,302 | 8,715,873 |
| NCO | 4,382,511 | 4,310,148 | 8,692,659 |
| CCO | 9,060 | 14,154 | 23,214 |
| Observed gene conversion rate per Million bp (95% CI) | 5.2 (3.0–8.0) |
8.8 (5.5–12.1) |
7.0 (5.0–9.0) |
| NCO | 5.0 (2.7–7.8) |
6.5 (3.9–9.5) |
5.8 (4.1–7.9) |
| CCO | 110 | 707 | 474 |
| Deletion bias (95% CI) | 55.5% (33.3–77.5) |
71.1% (53.2–84.5) |
65.6% (53.2–77.4) |
| NCO | 54.5 (31.0–75.4) |
67.8 (50.0–82.8) |
62.0 (48.0–74.7) |
| CCO | – |
80 |
81.8 |
| OR NCO in DSB (95% CI) | 35.6 (14.8–86.0) |
4.9 (1.0–11.8) |
13.9 (7.0–24.1) |
| OR NCO in crossover recombination hotspots (95% CI) | 10.4 (2.0–29.0) |
8.4 (0.0–25.4) |
|
OR = odds ratio, DSB = double strand break, mpp = marker proband pair, CI = confidence interval, CCO = complex crossover, NCO = non-crossover.
Complex crossover (CCO) gene conversions
In crossovers we are only able to detect complex events (cf. Supplementary Note 9, Supplementary Figure 6). The rate reported for CCO gene conversions should be interpreted with caution, as it refers to the fraction of mpps within a distance of 100kb from a crossover recombination that show evidence of gene conversion (cf. Supplementary Note 9) and is not a genomewide rate. We observe a greater CCO gene conversion rate in the sequencing dataset, where more events are detectable (cf.Table 2). Due to our inability to detect all complex events the true CCO gene conversion rate is likely to be higher than the estimates in both datasets.
The CCO gene conversion rate is greater than the NCO gene conversion rate (p-value <0.001 chip dataset, < 0.001 sequencing dataset). This confirms that as a group CCO gene conversions are not independent of crossovers. How CCO gene conversions are related to crossover recombinations remains to be elucidated.
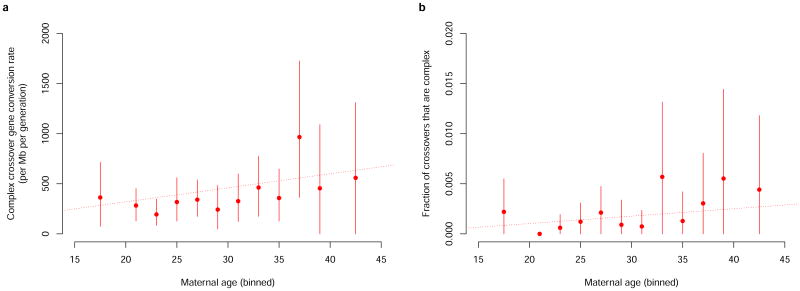
A large majority of the CCO gene conversions we identified are maternal, demonstrating that complex crossovers are more common in maternal transmissions, as is the case for NCO gene conversions. Another similarity is that, we observe an increase in CCO gene conversion rate with maternal age of 14.0/Mb/year (95% CI 0.7– 27.3, p-value: 0.04), in the chip dataset (cf. Figure 4a). Moreover, the fraction of crossovers that are complex increases with maternal age (p-value: 0.02 in the chip dataset) (cf. Supplementary Note 9,Figure 4b). In the sequencing dataset, we observe that 0.31% of all paternally transmitted crossover recombinations are complex. This result is in close agreement with a previous estimate of 0.33% obtained using sperm analysis22.
Figure 4.
Complex crossovers (CCO) and maternal age. a) Complex crossover (CCO) gene conversion rate by maternal age in the chip dataset. b) Fraction of maternally transmitted crossovers in the chip dataset that are complex, as a function of mother's age. Error bars represent 95% confidence intervals. The dotted line represents a linear model.
GC bias
Like previous studies7,20, our results reveal a significant GC bias for NCO gene conversions4, where strong base pairs (G or C) preferentially appear on polymorphic gene converted base pairs (cf.Table 1): the GC bias is 67.6% (95% CI 65.7–69.8) in the chip dataset and 69.3% (95% CI 65.8–72.3) in the sequencing dataset. Short and long NCO events exhibit the same GC bias (cf.Table 1).
Our results (cf. Supplementary Note 10) indicate that the bias is greater in maternal than paternal transmissions (p-value: 0.032 chip, 0.004 sequencing). Further, CpG SNPs show a greater GC-bias than other SNPs (p-value: 0.038 chip, <0.001 sequencing).
The GC bias in CCO gene conversionns is 70.2 (95% CI 62.5-77.8) in the chip dataset and 70.1 (95% CI 63.1-78.8) in the sequencing dataset, which is not significantly different from that observed in NCO gene conversions (p-values were: 0.56 and 0.73 for the chip and sequencing data, respectively).
Discussion
In summary, we have used both SNP chip and whole genome sequencing datasets from three-generation families to search for meiotic gene conversions in humans. Overall, we identified 3,237 mpps involved in gene conversions. Based on the sequencing data, we obtained a sex-averaged estimate of G, as 5.9/Mb/generation. Crucially, our results demonstrate that G varies with both age and sex. Thus, the rate for mothers (7.7/Mb/generation) is 1.91 (95% CI 1.34 – 2.58) times greater than for fathers (4.1/Mb/generation), in the sequencing dataset. Given that the fraction of heterozygous loci (where gene conversions can be detected) in Icelanders34 is 6·10–4, it follows there is an expectation of 7 detectable NCO gene conversions from fathers and 14 from mothers. These numbers are 12 and 23, respectively, based on a worldwide average heterozygosity35 of 1·10–3.
A surprising result was the magnitude of the age-related increase of G in females, where we estimate a 2.42 (95% CI 1.83–3.10) fold increase from the ages of 20 to 40 years. In the case of crossover recombination, the estimated increase between these age points is only 1.042 fold (4.2%), which is thought to be the result of greater viability of eggs with more crossover events19,29. Such selection might account for some of the age-related increase of G in females. However, given the more drastic increase in G, some other age-related factor must be at work. The increased G in older mothers is less biased towards male DSBs, sex specific crossover recombination hotspots (cf. Supplementary Note 5) or regions of high local GC content (cf. Supplementary Note 6). Although our results are not conclusive, they indicate that a large fraction of the increase in crossover recombinations with maternal age are due to complex crossover recombinations (cf. Supplementary Note 9).
Overall, the large fraction of NCO gene conversions in spermatocyte DSB regions and crossover recombination hotspots is consistent with the view that most of them occurred in response to programmed DSBs prior to the meiotic prophase 1 arrest of oocytes in the fetal ovary36. However, an accumulation of programmed DSBs over subsequent decades does not seem a likely source of the age-related increase of NCO gene conversions in females. Other possible sources may be linked to the age-related deterioration of the oocytes across the decades that they are in dictyate arrest, possibly leading to non-disjunction37,38. This deterioration may be due to damage-induced DSBs, deficiencies in checkpoint mechanisms36,39, failure of cohesins to maintain the cohesion of sister-chromatids36,40 or that cohesive linkages are not restored at the same rate as they are lost38. Further research is needed to determine the source of these additional NCO gene conversions in older females and whether they have the same source as the additional CCO gene conversions. It is obviously interesting to note in this context that risk of aneuploidies increases drastically with the age of mothers41 – although there is no direct evidence to link aneuploidies with the age-related increase of gene conversions.
Our results further demonstrate that the control of gene conversions differs between the sexes. While most paternally transmitted NCO events are short and complex crossovers are rare in paternal transmissions, maternally transmitted NCO events tend to be long and complex. Our results suggest that there is a different biological mechanism underlying short and long NCO events and consequently maternal and paternal transmissions. As the definition of an event is based solely on proximity of gene converted mpps, we cannot discern whether the gene converted mpps within the same long event occurred simultaneously in a single process or in several collocated processes. Crossover interference has recently been shown to decrease with maternal age42, possibly leading to double crossover recombination events which in our study design could be detected as long NCO events. The complex nature of long NCO events and their GC bias make it unlikely that crossover interference explains a large fraction of long NCO events (cf. Supplementary Note 7). Paternal NCO gene conversions may be enriched for those derived from the SDSA pathway and maternal NCO gene conversions may be enriched for those resulting from dHj resolution4. The long NCO events, which are complex and mostly maternally transmitted may also arise from a more complex set of underlying biological mechanisms43, including repeated template switching44. Analysis of sperm6,22, oocyte45 or tetrad analysis8 are promising approaches for obtaining a more complete picture of meiotic gene conversion and its mechanisms.
Our results and others12,13,20 show that gene conversions are biased towards GC base-pairs, while mutations are biased towards AT base-pairs and increase with age in both sexes46,47, but more strongly with father's age48. Now it is clear that gene conversions increase with mothers' age. On average, the number of gene conversions per generation is comparable to that of mutations. Intriguingly, this means that the nucleotide composition of the human genome represents an equilibrium that is maintained by an unwitting battle between the sexes, where male driven AT-biased mutations48 are offset by female driven GC-biased gene conversion events.
Online methods
In order to detect gene conversions, we use three-generation pedigrees, where a proband as well as both of its parents, at least two of the proband's siblings, a child of the proband and the proband's spouse are all genotyped (cf. Figure 1).
Data descriptions
We use two datasets of Icelandic samples collected as a part of disease association efforts at deCODE genetics34: Chip data consisting of 7,219 probands genotyped on Illumina HumanHap and Omni BeadChip arrays and sequencing data consisting of 101 whole genome sequenced probands.
The study was approved by the Icelandic Data Protection Authority (ref. 2004120649) and the National Bioethics Committee, Iceland (ref. VSN 13-028). All participating subjects who donated blood signed informed consent. Personal identities of the participants and biological samples were encrypted by a third party system approved and monitored by the Icelandic Data Protection Authority.
A description of how families were selected and datasets were proprocessed is given in Supplementary Note 11. A list of the chip types used in the chip dataset is given in Supplementary Table 10.
Identifying gene conversion mpps in pedigrees
Gene conversions appear in the proband's haplotype as short tracts of mpps from one of the parent's haplotype on the background of its other haplotype, i.e. the markers in question are inherited from one haplotype while nearby markers on both sides are inherited from the other haplotype. We can detect them if the two haplotypes of a parent and the haplotype transmitted from the parent to the proband are known.
To determine the haplotypes of the parents we phase their nuclear family (the parents, the proband and its siblings). This is done in three steps; In the first step we construct a set of mpps, where we are confident of the inheritance pattern from a given parent of the sibling group. We refer to these mpps as anchors. In the second step, we phase the remaining mpps by minimizing the discrepancy between the genotypes of the mpps compared to the inheritance pattern observed at neighboring anchors. In the third step, we determine the location of crossover recombinations in order to phase the proband.
Finally, we identify gene conversions as tracts of inconsistencies between genotypes of the proband and the phased inheritance pattern at each mpp. If the tracts of inconsistencies indicate a gene conversion we attempt to verify them in the proband's children.
A schematic overview of the algorithm is given in Supplementary figure 7.
Determining anchor mpps
An anchor is an mpp where one parent is heterozygous, the other is homozygous and the genotypes of the proband, its parents and all of its siblings meet the accuracy thresholds defined above. With respect to the heterozygous parent, the phase of the sibling group (the proband and its siblings) is unambiguous at anchors and the sibling group can be partitioned into two sets, determined by which allele was inherited from the heterozygous parent. Two adjacent anchors, with the same heterozygous parent, induce the same partition in the sibling group, unless either anchor is genotyped incorrectly or a gene conversion or crossover recombination occurred between them.
Given an mpp and a parent we define a left anchor as the closest anchor with a lower numerical coordinate where the parent is heterozygous. A right anchor is defined analogously with a higher numerical coordinate.
Unless a genotyping error occurred we can be confident in the inheritance pattern for all anchor mpps. We now remove mpps that appear to be the result of a genotyping error from the set of anchor markers. When removing these we may also remove mpps that are the result of a gene conversion. In a later step we determine whether the mpp is the result of a gene conversion or a genotyping error.
Mpps whose partition does not agree with neighboring anchors are removed. To formally delineate which markers are removed from the set of anchors, we define the discrepancy between two anchors as the minimum number of individuals that need to be moved between the sibling group partitions of the anchors such that the partitions become identical. We compute a local discrepancy score for an anchor, A, as the sum of the discrepancy between A and its two closest anchors to the left and two closest anchors to the right. The anchor A is removed if, when doing so, the sum of the discrepancy scores of all other anchors is reduced.
Phasing the parents
At a given mpp the sibling group can be split into four inheritance groups based on which of the two haplotypes they inherit from each parent. The inheritance groups are not known but when there are no crossover recombinations, gene conversions or genotyping errors, the haplotypes will agree with the haplotypes of both parents inherited at the neighbouring anchors. We define two inheritance groupings, left and right. The left inheritance grouping is determined by the left anchors of both parents and the right inheritance grouping is based on the right anchors of both parents. Both the left and right inheritance groupings should be identical to the inheritance grouping at the given mpp unless there has been a crossover recombination for either parent in the region or the mpp being examined is gene converted in the proband or one of its siblings. A genotyping error in one of the siblings or either parent may also occur, causing the genotypes not to agree with the left and right inheritance groupings even if they are identical to the true inheritance grouping at the mpp.
Given the genotypes of the individuals in the nuclear family and the inheritance groupings we assign alleles to the parents' haplotypes. For binary mpps, there are a total of 24 = 16 possible assignments of the two alleles to the four haplotypes. For each such assignment we infer genotypes of both parents and the siblings according to the left inheritance grouping and compare them to observed genotypes. We define left phasing discrepancies in the nuclear family as the combined number of mismatches between observed and inferred genotypes. Right phasing discrepancies are defined analogously from the right inheritance groups. For each assignment of alleles to haplotypes we define the number of phasing discrepancies as the smaller of the left and right phasing discrepancies. A phasing discrepancy can be explained with a crossover recombination, a gene conversion or genotyping error.
If there exists exactly one assignment of the alleles to the parents' haplotypes with fewer than two phasing discrepancies, the mpp is considered phased by the assignment. All other Mpps are removed from further consideration as candidates when searching for gene conversions. When the assignment has no phasing discrepancy either there is no gene conversion or the mpp is part of a gene conversion tract that includes neighboring anchors. Assignments with a single phasing discrepancy are further candidates for where a gene conversion may have taken place. Not all single phasing discrepancies will represent a gene conversion as they may also represent a genotyping error or a non-gene converted mpp in a long gene conversion tract including both neighboring anchors.
When there are more than one assignments that have fewer than two phasing discrepancies we cannot reliably determine which assignment is the correct one, since all of them can arise from a single genotyping error or gene converted mpp. An example of when an mpp has multiple assignments with fewer than two phasing discrepancies is when one of the parents' haplotypes is not carried by any members of the sibling group. If all individuals are correctly genotyped and there is no gene conversion then the correct assignment of haplotypes leads to zero phasing discrepancies, while switching the assignment of the haplotype not carried by any members of the sibling group leads to one phasing discrepancy.
When all assignments of alleles to the parents' haplotypes yield at least two phasing discrepancies, then the mpps genotypes are not consistent with the left and right inheritance groupings, even when allowing for a single individual to be either carrier of a gene conversion or having a genotyping error. This can occur due to multiple recombinations, a structural variant at the locus, a misplacement of the marker in the assembly or repeated genotyping errors at the marker.
Determining crossover recombinations
For each proband we locate crossover recombinations from each parent separately. We refer to an mpp as informative if the phase of the proband can be determined directly at the mpp without any assumption about the phase inherited from the other parent. Thus, the set of informative mpps includes anchor mpps as well as mpps where the parent of interest is heterozygous and either the proband or the other parent are homozygous. In particular, the set includes mpps where some of the siblings of the proband do not meet genotyping accuracy thresholds.
In order to distinguish candidate gene conversions from crossover recombinations, we look for all inheritance tract changes in the proband. Initially, we assign inheritance tract changes to all regions between two adjacent informative mpps where the proband inherits alleles from different haplotypes of the parent of interest. The region of an inheritance tract change can be further narrowed if the parent of interest is heterozygous for additional mpps between the two informative markers, if the haplotype of the other parent is known, and if the two informative mpps agree on a haplotype for the other parent. In this case, we assume that the proband inherited the same haplotype for the whole region from the other parent and we can determine the proband's phase at the all mpps in the region where the parent of interest is heterozygous. If the two informative mpps do not agree on a haplotype for the other parent, the inheritance tract change region is excluded from the search for gene conversions. Once the inheritance tract change region has been narrowed, we assign an inheritance tract change to the center of the region.
A crossover recombination is assigned to all tract changes where no other tract change occurs within 100 kb. All other tract changes are candidate gene conversions. Additionally, a crossover recombination is assigned if there is an odd number of multiple consecutive tract changes within 100 kb of each other; more precisely, the crossover recombination is assigned to the leftmost or rightmost tract change depending on which induces fewer gene converted mpps (see below). In this case a crossover recombination has occurred along with possible gene conversions.
Determining gene conversion mpps
Having assigned crossover recombinations and phased the haplotypes of the parents, we search in the proband for mismatches in observed genotypes compared to the genotypes defined by the haplotypes inherited from the parents.
We search for gene conversion from each parent separately for all mpps in the phased mpp set after applying the quality filters described in Supplementary Note 12.
Mpps passing these filters are counted towards the denominator in our rate computations. If there is a mismatch between the proband's genotype and its phase-determined haplotypes, we examine whether this mismatch can be due to a gene conversion.
If the mismatch can be explained with the other haplotype of the parent of interest, we attempt to verify the gene conversion in the proband's children. We verify the genotype in one of two ways; If a child is homozygous, we verify the gene converted haplotype without requiring the proband's spouse to be genotyped. Otherwise, if the proband's spouse is genotyped and homozygous we use that together with the child's genotype to verify the gene converted haplotype. We may be unable to verify the haplotype due to inconsistencies such as structural variants, a misplacement in the assembly or repeated genotyping errors.
In order to verify a putative gene conversion in the proband's child, we first determine whether the child carries the gene converted haplotype. We search for the closest mpps to the left (of lower numerical order) and to the right (of higher numerical order) where the proband is heterozygous and the spouse is homozygous, ignoring mpps at the end of chromosomes where there is no such left or right mpp. At these mpps we can determine which haplotype the child inherited from the proband. If the child inherited the same haplotype from the proband at both of these mpps and the distance between these mpps is less than 1 Mb we assume that the child is carrying the corresponding haplotype. The gene conversion can be verified in the child if this is the gene conversion haplotype.
If a marker shows evidence of being part of a structural variant, being misplaced in the assembly or having more than one genotyping error, the marker is flagged as problematic in the family. Specifically, a marker is flagged as problematic either if all of the assignments of alleles to the parents' phases produces two or more phasing discrepancies or if we fail to verify a gene conversion in one of the proband's children. Markers that are flagged as problematic in more than one family are removed from the set of quality markers input to the algorithm.
Crossover regions and CCO vs. NCO gene conversions
For each crossover recombination we defined a crossover region as the adjacent 100 kb in each direction. If a gene converted mpp was found within the crossover region the region was iteratively extended so that it contained all mpps within 100 kb of the crossover recombination and the gene converted mpps found.
Mpps determined to be within and outside a crossover region were used in the computation of CCO gene conversion rate and NCO gene conversion rate, respectively.
Determining gene conversion events
Gene conversion events are determined once all gene converted mpps have been determined. While searching for gene converted mpps we restrict our search to contiguous tracts of mpps where the length of the tract is 100kb. Gene conversion events may however contain both gene converted mpps and non-gene converted mpps.
Within non-crossovers, we arranged mpp positions from a parent-proband pair into distinct gene conversion events by traversing the chromosome in numerical order. We considered the first gene converted mpp found on a chromosome to be part of a new event and iteratively extended the event if a gene converted mpp was found within 100 kb of the previous gene converted mpp. Consequently, gene conversion events may be longer than 100kb.
Within crossovers, mpps neighbouring the same crossover recombination were considered a part of the same event.
Crossover recombination map data
We use a dataset of crossover recombinations24 preprocessed as described in Supplementary Note 13.
Computation of rates, confidence intervals and p-values
We compute the observed gene conversion rate as the number of mpps where a gene conversion occurs divided by the number of mpps that were tested for a gene conversion.
For two events, A and B, we compute the odds ratio as (N11*N22)/(N12*N21), where N11 represents the number of mpps that are part of event A and B, N12 the number of mpps that are part of A and not B, N21 the number of mpps that are part of B and not A and N22 the number of mpps that are part of neither event. Odds ratios for crossover recombination hotspots are computed considering only those markers where the crossover recombination rate has been estimated.
All confidence intervals presented are 95% confidence intervals and all p-values are two-sided. Confidence intervals for G, odds ratios, number of mpps per event, rate of increase in G between age of 20 and 40 and GC bias, are computed using a bootstrap method49. For the chip dataset 1,000 sets of 7,219 individuals are sampled with replacement from the set of 7,219 probands. For the sequencing dataset 1,000 sets of 101 individuals are sampled with replacement from the set of 101 probands. The statistic in question is computed within each set, creating a list of 1,000 statistics. Following the sorting of this list, the lower bound of the confidence interval is computed as the mean of entries 25 and 26 and the upper bound is computed as the mean of entries 975 and 976.
Age of parent effects are determined using a weighted linear regression using the function lm in R50. To determine age effect of G we first compute G for each proband-parent pair separately. The final model can be expressed as: lm( G ∼ ParentAge, weights = sqrt(N)), where N is the number of mpps considered for the proband. To determine age effect on other statistics we compute for each proband-parent pair separately the statistic, S, in question. The final model can be expressed as: lm( S ∼ ParentAge, weights = sqrt(N)).
Linear regression, its confidence intervals and p-values, for distance to the telomere and length of chromosome, were computed using the lm function in R, using a matrix containing all mpps where a gene conversion event could be ascertained. All other linear regressions were implemented using Python51.
All other p-values, not previously discussed, were computed using bootstrapping. 1,000 simulations are used analogously to the description above and a p-value is computed by counting the number of times the single-sided event of interest occurred and dividing by the number of simulations. The single-sided p-value was then multiplied by 2, in order to obtain a double-sided p-value. In cases when the event of interest did not occur in 1,000 simulations the p-value was reported as < 0.001.
In order to compute G corrected for crossover recombination, a linear regression was performed with gene conversion as a response and local sex specific crossover recombination rate as an explanatory variable. All marker proband pairs where an NCO gene conversion could be ascertained and the crossover recombination rate had been determined were used. A corrected G was computed by inserting the genomic average crossover recombination rate24, of 1.572 cM/Mb for maternal transmissions and 0.772 cM/Mb for paternal transmissions, into the regression formula. Confidence intervals were computed using the predict.lm function in R.
Supplementary Material
Acknowledgments
This work was supported in part by NIH (NIDA) (R01–DA017932).
Footnotes
Author contributions: BVH, DFG and KS designed the experiments. BVH wrote the first draft of the paper. BVH, MTH, BK, US, PS, AH, AK, DFG and KS reviewed and contributed to subsequent drafts of the paper. BVH, MTH and AG implemented the methodology. BVH, MTH and BK prepared tables and figures. AsJ and AdJ performed the Sanger sequencing. UT oversaw the operations of the genotyping facility. BVH, MTH, FZ, GT, AG and GM processed the data. BVH and MTH analyzed the data. All authors contributed to the final version of the manuscript.
Competing financial interests: All authors are employees of deCODE genetics/Amgen.
References for main text
- 1.Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 2.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol. 2015;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haber J. Genome stability. Garland Science; 2013. [Google Scholar]
- 5.McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–6. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 7.Odenthal-Hesse L, Berg IL, Veselis A, Jeffreys AJ, May CA. Transmission distortion affecting human noncrossover but not crossover recombination: a hidden source of meiotic drive. PLoS Genet. 2014;10:e1004106. doi: 10.1371/journal.pgen.1004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole F, et al. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet. 2014;46:1072–80. doi: 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allers T, Lichten M. Differential Timing and Control of Noncrossover and Crossover Recombination during Meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 10.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 11.Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-Content Evolution in Mammalian Genomes: The Biased Gene Conversion Hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glemin S, et al. Quantification of GC-biased gene conversion in the human genome. Genome Res. 2015;25:1215–1228. doi: 10.1101/gr.185488.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 14.Narasimhan VM, et al. bioRxiv. Cold Spring Harbor Labs Journals; 2016. A direct multi-generational estimate of the human mutation rate from autozygous segments seen in thousands of parentally related individuals. [DOI] [Google Scholar]
- 15.Palamara PF, et al. Leveraging Distant Relatedness to Quantify Human Mutation and Gene-Conversion Rates. Am J Hum Genet. 2015;97:775–789. doi: 10.1016/j.ajhg.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachance J, Tishkoff SA. Biased gene conversion skews allele frequencies in human populations, increasing the disease burden of recessive alleles. Am J Hum Genet. 2014;95:408–420. doi: 10.1016/j.ajhg.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 18.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 19.Kong A, et al. Recombination rate and reproductive success in humans. Nat Genet. 2004;36:1203–1206. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- 20.Williams AL, et al. Non-crossover gene conversions show strong GC bias and unexpected clustering in humans. Elife. 2015;4:e04637. doi: 10.7554/eLife.04637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20:563–73. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Webb AJ, Berg IL, Jeffreys A. Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proc Natl Acad Sci U S A. 2008;105:10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratto F, et al. Recombination initiation maps of individual human genomes. Science. 2014;346:1–9. doi: 10.1126/science.1256442. 1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong A, et al. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet. 2014;46:11–16. doi: 10.1038/ng.2833. [DOI] [PubMed] [Google Scholar]
- 25.Padhukasahasram B, Rannala B. Meiotic gene-conversion rate and tract length variation in the human genome. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.30. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–62. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 27.Pardo-Manuel de Villena F, Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm Genome. 2001;12:318–22. doi: 10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- 28.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin HC, et al. Multicohort analysis of the maternal age effect on recombination. Nat Commun. 2015;6:7846. doi: 10.1038/ncomms8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 31.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assis R, Kondrashov AS. A strong deletion bias in nonallelic gene conversion. PLoS Genet. 2012;8:e1002508. doi: 10.1371/journal.pgen.1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leushkin E. V & Bazykin, G. A Short indels are subject to insertion-biased gene conversion Evolution. 2013;67:2604–2613. doi: 10.1111/evo.12129. [DOI] [PubMed] [Google Scholar]
- 34.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 35.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian V. V & Bickel, S. E Aging predisposes oocytes to meiotic nondisjunction when the cohesin subunit SMC1 is reduced PLoS Genet. 2008;4:e1000263. doi: 10.1371/journal.pgen.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng KA, Jeffreys CA, Bickel SE. Rejuvenation of meiotic cohesion in oocytes during prophase I is required for chiasma maintenance and accurate chromosome segregation. PLoS Genet. 2014;10:e1004607. doi: 10.1371/journal.pgen.1004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leland S, et al. Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc Natl Acad Sci U S A. 2009;106:12776–12781. doi: 10.1073/pnas.0903075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- 41.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell CL, Furlotte NA, Eriksson N, Hinds D, Auton A. Escape from crossover interference increases with maternal age. Nat Commun. 2015;6:6260. doi: 10.1038/ncomms7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martini E, et al. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011;7:e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsaponina O, Haber JE. Frequent Interchromosomal Template Switches during Gene Conversion in S. cerevisiae Mol Cell. 2014;55:615–25. doi: 10.1016/j.molcel.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Boer E, Jasin M, Keeney S. bioRxiv. Cold Spring Harbor Labs Journals; 2015. Local and sex-specific biases in crossover vs noncrossover outcomes at meiotic recombination hotspots in mouse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong WSW, et al. New observations on maternal age effect on germline de novo mutations. Nat Commun. 2016;7:10486. doi: 10.1038/ncomms10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldmann JM, et al. Parent-of-origin-specific signatures of de novo mutations. Nat Genet. 2016;48:935–939. doi: 10.1038/ng.3597. [DOI] [PubMed] [Google Scholar]
- 48.Kong A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 49.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. CRC Press; 1994. [Google Scholar]
- 50.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 51.van Rossum G, Drake FL. PYTHON Reference Manual. Centrum voor Wiskunde en Informatica; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.