ABSTRACT
Tumor‐targeted Salmonella VNP20009 preferentially replicate within tumor tissue and partially suppress tumor growth in murine tumor models. These Salmonella have the ability to locally induce apoptosis when they are in direct contact with cancer cells but they lack significant bystander killing, which may correlate with their overall lack of antitumor activity in human clinical studies. In order to compensate for this deficiency without enhancing overall toxicity, we engineered the bacteria to express epidermal growth factor receptor (EGFR)‐targeted cytotoxic proteins that are released into the extracellular milieu. In this study, we demonstrate the ability of the Salmonella strain VNP20009 to produce three different forms of the Pseudomonas exotoxin A (ToxA) chimeric with a tumor growth factor alpha (TGFα) which results in its producing culture supernatants that are cytotoxic and induce apoptosis in EGFR positive cancer cells as measured by the tetrazolium dye reduction, and Rhodamine 123 and JC‐10 mitochondrial depolarization assays. In addition, exchange of the ToxA REDLK endoplasmic reticulum retention signal for KDEL and co‐expression of the ColE3 lysis protein resulted in an overall increased cytotoxicity compared to the wild type toxin. This approach has the potential to significantly enhance the antitumor activity of VNP20009 while maintaining its previously established safety profile. Biotechnol. Bioeng. 2016;113: 2698–2711. © 2016 The Authors. Biotechnology and Bioengineering published by Wiley Periodicals, Inc.
Keywords: Tumor‐targeted Salmonella, VNP20009, msbB, Pseudomonas ToxA, PE38, TGFα, ColE3 lysis protein
Introduction
Cancer remains a major health concern throughout the world and is the second leading cause of death in the United States, with more than 1.5 million new cases and over 500,000 deaths each year (Siegel et al., 2015). Among the various forms of cancer, prostate, breast, lung, and colon are the most common. Treatment of cancer remains complex due to substantial phenotypic variations based on underlying genotypic differences found among cancers of the same tissue type. Breast cancers, for example, are categorized into the groups luminal A and B, basal, claudin‐low, and HER2, each of which have further subtypes with specialized treatment modalities, including ones based on cell surface receptors (Holliday and Speirs, 2011). Cell surface targeted treatments for cancer include transtuzumab (Herceptin®), a monoclonal antibody that is targeted to HER2 and cetuximab (Erbitux®), a monoclonal antibody targeted to epidermal growth factor receptor (EGFR). EGFR is sometimes present in breast cancers lacking progesterone receptor (PR), estrogen receptor (ER), and HER2, which are known as triple negative breast cancers. EGFR is correlated with a more rapid time to progression and lower post‐relapse survival in patients that respond to primary therapy (Tsutsui et al., 2002). Overall, efficacy of antitumor therapies targeted to these surface receptors remains moderate, with chemotherapy and antibody resistances among the factors that limit long‐term effectiveness, indicating that development of alternative therapies are needed.
Historically, there are a number of reports suggesting that natural bacterial infections can have beneficial, even curative, effects on malignant tumors (reviewed by Nauts, 1976). Among the live bacteria now implicated with potentially positive therapeutic effects against cancer is Salmonella, which is currently being studied by a number of investigators for its tumor‐targeted properties (see Forbes, 2010 and Hoffman, 2015 for reviews). Remarkably, attenuated strains of Salmonella preferentially grow in tumor tissue greater than 1000‐fold compared to normal healthy tissue, creating a tumor‐localized infection (Low et al., 1999; Pawelek et al., 1997, 2003). In studies using mice, Salmonella preferentially grow within all major forms of solid tumors, including prostate, breast, lung, and colon tumors, presenting the opportunity to develop a broadly effective cancer treatment. However, in a human clinical study of lipid‐modified Salmonella strain VNP20009 on patients with melanoma (ClinicalTrials.gov Identifier NCT00004988; Toso et al., 2002), there was no antitumor activity, even in patients that were confirmed to have the bacteria in their tumors. Despite the lack of success in humans, VNP20009 was shown to be effective against spontaneous neoplasms in dogs, including four complete responses and an overall response rate of 42% (Thamm et al., 2005).
Tumor inhibition associated with bacterial colonization may arise from multiple factors. The tumor inhibition observed likely includes effects from the immune system, including CD8+ cytotoxic lymphocytes (Avogadri et al., 2005; Saltzman, 2005; Saltzman et al., 1997) and TNFα (Leschner et al., 2009). By comparing the studies in murine and canine systems with those in humans, it now appears that the antitumor effects in mice and dogs either do not occur or are insufficient to suppress tumor growth in humans. Thus in humans, although the bacteria may be at a high concentration within the tumor, few tumor cells are eliminated. Although antitumor efficacy was not demonstrated, the human clinical study did establish that the bacteria can safely localize within tumors, and therefore have the potential to deliver therapeutics such as proteins or nucleic acids. The purpose of this study was to assess the ability of Salmonella strain VNP20009 to heterologously express chimeric bacterial toxins targeted to a receptor over‐expressed on the surface of some forms of malignant cancer cells in order to increase the ability of the bacteria to kill tumor cells.
Pseudomonas exotoxin A (ToxA or PE) is an ADP ribosylating toxin that has undergone extensive evaluation as the effector component of molecularly targeted toxins, with numerous studies by Pastan and co‐workers demonstrating its potential (see Weldon and Pastan, 2011 for a review). The fundamental premise of those studies is the replacement of the naturally occurring mammalian cell‐binding domain of the toxin with a ligand that is specific for an internalizing receptor overexpressed on cancer cells. EGFR (HER1) and its ligand tumor growth factor alpha (TGFα) is one of the most widely studied cancer cell receptor/ligand complexes. Multiple forms of ToxA chimeras have previously been generated that are capable of specifically killing cancer cells overexpressing EGFR. One of the early constructs had the binding domain Ia (DIa; amino acids 1‐252 of the mature peptide as numbered following cleavage of the signal sequence) deleted and substituted with the epidermal growth factor receptor (EGFR) ligand TGFα fused to the remaining domains II, Ib, and III consisting of amino acids 253‐613 and was designated TGFα‐PE40 (Chaudhary et al., 1987; Siegall et al., 1989b). A reduction of a portion of PE40 by removal of amino acids 365‐380 from domain Ib (DIb) was used to generate PE38 (Siegall et al., 1989a), which exhibited increased cytotoxicity to cancer cells. Deletion of DIa and part of domain II (DII; amino acids 253‐279) resulted in the PE35/TGFαKDEL truncation that further increased cytotoxicity (Theuer et al., 1993). In that construct, the TGFα targeting moiety was positioned functionally downstream of PE35 followed by substitution with the consensus amino acid sequence KDEL for the naturally occurring endoplasmic reticulum (ER)/golgi retrieval signal for the C‐terminal REDLK, and constituted among the most potent of the truncated cytotoxins (Seetharam et al., 1991).
ToxA is among the proteins that has been shown to be secreted by attenuated Salmonella in a form capable of killing cancer cells (Swofford et al., 2014). Because Salmonella localize to tumors at substantially greater concentrations than other tissues, expression of a toxin is inherently tumor‐specific by virtue of the tumor‐localized replication of the bacteria (Pawelek et al., 1997). However, Salmonella can still be found in other tissues such as liver, spleen and bone marrow (Clairmont et al., 2000), and thus it is not yet apparent if toxicity to these tissues from a bacterial protein toxin would be significant. An example of an additional, independent tumor localization mechanism for expression by a tumor‐targeted Salmonella is the use of a hypoxic promoter, which has bacterial targeting as a primary localization mechanism and the co‐occurrence of hypoxia as a second targeting mechanism and was used for expression of the cytotoxic protein HlyE (ClyA) (Ryan et al., 2009). A potential alternative improvement to expression of the wildtype toxins that could avoid toxicity to normal tissues would be expression of toxins specifically targeted to tumor cells.
ToxA contains a classical N‐terminal Type II hydrophobic signal sequence that is cleaved in the process of Pseudomonas aeruginosa secreting the mature protein into the extracellular medium. When the wild type gene for ToxA has been expressed by Escherichia coli, various results have been achieved ranging from no secretion (Gray et al., 1984), periplasmic secretion (Lory et al., 1988) to extracellular secretion using an OmpA signal sequence substituted for that of the ToxA signal sequence (Chaudhary et al., 1988). Among the different chimeric proteins, TGFα‐PE40 can be recovered from inclusion bodies, the periplasm, and supernatant when used with the OmpA signal sequence (Kondo et al., 1988), while PE35/TGFαKDEL was only found in inclusion bodies.
These various results in regard to secretion may stem from several different aspects of this heterologous system including the amino acid sequence of the ToxA, the genetic expression system, and the host strain of E. coli. In order for an attenuated Salmonella strain to effectively deliver a TGFα:ToxA chimera to tumor cells and to further achieve a bystander effect, whereby the toxin may contact multiple cancer cells through diffusion, the protein must either be released or secreted through both the inner and outer membranes into the media. Secretion of chimeric forms of ToxA by Salmonella in a biologically active form has not been investigated.
In this study, we cloned and expressed modified forms of three TGFα‐targeted ToxA chimeras based on previous reports (Edwards et al., 1989; Kihara and Pastan, 1994a; Kreitman et al., 1992). Each of these forms includes an in‐frame fusion of TGFα upstream of DIII of ToxA. Components of the three chimeras also includes a Type II hydrophobic secretion signal derived from OmpA, a long flexible linker, and presence or absence of DIb. In addition, we substituted the naturally occurring REDLK endoplasmic reticulum retention signal with the KDEL consensus sequence. We also compared the cytotoxicity of culture supernatants when co‐expressed with the colicin E3 (ColE3) lysis protein that aids in liberation of colicin and other proteins from the periplasm. This report is the first to demonstrate secretion of a chimeric ToxA that selectively kills EGFR‐expressing tumor cells by an attenuated Salmonella and has potential for the treatment of neoplasias overexpressing EGFR such as breast, colon, lung, prostate, bladder, and/or brain cancers (Brendel et al., 1999; Woodburn, 1999).
Materials and Methods
Bacterial Strains and Plasmids
The bacteria used in the study were E. coli cloning strain EC100 (Epicentre, Madison, WI; F‐ mcrA Δ(mrr‐hsdRMS‐mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ‐ rpsL (StrR) nupG), Salmonella enterica serotype Typhimurium restriction (−) methylation (+) strain JR501 (hsdSA29 hsdSB121 hsdL6 metA22 metE551 trpC2 ilv‐452 H1‐b H2‐e,n,x (cured of Fels 2) fla‐66 nml rpsL120 xyl‐404 galE719 (Salmonella Genetic Stock Center, Calgary, Canada, strain 1593; Tsai et al., 1989); attenuated Salmonella enterica serotype Typhimurium strain VNP20009 from the American Type Culture Collection (Manassas, VA; strain 202165, a.k.a. 41.2.9 or YS1646; Low et al., 2004) which include the attenuating mutations msbB and purI and the Suwwan suppressor; Murray et al., 2004). Pseudomonas aeruginosa strain PA103‐LAC was obtained from the ATCC (strain 47086). All strains are stored frozen at −80°C in 15% glycerol. Bacterial media used were LB (Miller, 1992) which contained 1% tryptone, 0.5% yeast extract, and 1% NaCl or LB plates containing 1.5% agar, LB‐0 (LB no NaCl; Murray et al., 2001), and a novel media CTY containing 0.2% casamino acids, 0.2% tryptone, 0.1% yeast, and 0.9% NaCl adjusted to pH 7.4 with 0.2 M NaOH, which we found was less toxic to mammalian cells than LB, yet supported bacterial protein expression. Antibiotics used were 100 μg/mL ampicillin (Amp100) except in CTY which was 50 μg/mL. The expression plasmid pAra99a utilized in these studies is a derivative of pTrc99a (Amann et al., 1988; GenBank U13872.1) where the trc promoter and lacIq were removed with NdeI and NcoI and replaced with the arabinose‐inducible operon consisting of AraBAD, which we derived from plasmid pBAD‐IEE (GenBank AB598835) based on previous studies in Salmonella (Lossner et al., 2007). Bacterial culture supernatants were prepared by innoculating CTY media at 37°C rotated at 15 RPM overnight without arabinose, diluting the culture 1:5 in fresh CTY media and incubating for a 2 h pre‐induction period at 37°C with rotation, and then inducing with 1 mM arabinose at 37°C with rotation for 16 h. The cultures were then centrifuged at 3000g for 10 min at room temperature and then the supernatant was sterilized using a 0.2 μM low binding PES filter (Whatman, Buckinghamshire, UK; Puradisc 25 AS).
Tumor Cells
MDA‐MB‐468 human breast adenocarcinoma and HeLa human cervical adenocarcinoma were obtained from the ATCC (MDA‐MB‐468 was kindly provided by Dr. Jonathan Kelber, California State University, Northridge). H460 human large cell lung carcinoma was obtained from the National Cancer Institute Division of Cancer Treatment and Diagnosis (DCTD; Frederick, MD). Each of the cell lines were authenticated at the University of Arizona Genetics Core (Tucson, AZ). MDA‐MB‐468 has been shown to have high‐level expression of epidermal growth factor receptor (EGFR; Xu et al., 2005) whereas HeLa (Eiblmaier et al., 2008) and H460 (Kirk et al., 1994) have low and undetectable EGFR, respectively. MDA‐MB‐468 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM), HeLa cells were cultured in Minimal Essential Media (MEM), and H460 were cultured in RPMI, each of which was supplemented with heat‐inactivated fetal bovine serum (10% vol/vol; Gibco/Life Technologies, Grand Island, NY) and 1% of penicillin‐streptomycin (10,000 units penicillin and 10 mg/mL streptomycin; Sigma‐Aldrich, St. Louis, MO). Cells were trypsinized (0.5% trypsin 0.2% EDTA, Sigma–Aldrich) for use in passages and seeding cell proliferation assays whereas cells used for immunoblots were removed with a cell scraper.
Cell Proliferation and Cytotoxicity Assays
The methylthiazol diphenyltetrazolum (MTT; Calbiochem, San Diego, CA) viability assay (Mosmann, 1983) was used to quantify the relative number of tumor cells present using a Spectramax M3 (Molecular Devices; Sunnyvale, CA) set at A570 in order to determine the relative cytotoxicity of bacterial culture supernatants. The supernatants (10 μL, with twofold dilutions in phosphate buffered saline (PBS)) were added to 96‐well culture plates in which the tumor cells had been seeded at 2500/well in 100 μL tissue culture media one day prior to treatment. When 24 well plates were used for microscopy, tumor cells were seeded at 25,000 cells per well in 1 mL. The fraction of tumor cells affected was based on the MTT assay of cells treated for 72 h. The absorbance results were normalized using a PBS control (100% survival) and a media only control (0% survival) in Prism 6.0f (Graphpad, La Jolla, CA) based upon Sebaugh (2011). Each assay was performed using triplicate wells, and each experiment was performed at least three times on separate days except where noted. The data from at least three combined experiments was used for statistics. The replicate data for each treatment was then grouped and the log of the volume of active supernatant was then plotted with its respective mean and SD. Where the median inhibition caused by a culture supernatant was greater than 50% but did not approach 100%, the 50% inhibitory volume of culture supernatant was determined using a simple arithmetic ratio based on the highest cell kill achieved and indicated as an approximation (∼). For the determination of the amount of active supernatant required to achieve 50% cell death, the mean of each repeated independent experiment was pooled and the data interpolated using the sigmoidal, 4PL, log function to generate the IC50 curves. LogIC50 values and their standard error were utilized for statistics using a one‐way Anova (P < 0.05) followed by post hoc Tukey's multiple comparison test. Error bars on IC50 bar graph indicate 95% confidence intervals (CI). Actual and logarithmic supernatant volumes are indicated on their respective axes.
PCR and Cloning
Oligonucleotide primers are listed in Table I. PCR reactions were conducted using high‐fidelity polymerases (Phusion Hot Start II, Fermentas/Thermo Fisher, Waltham, MA); Phusion, Finnzymes/Thermo Fisher; KOD Hot Start Master Mix, Novagen/EMD Millipore, Billerica, MA), with the program typically consisting of one cycle of 95 or 98°C for 2 min followed by 33 cycles of 95 or 98°C for 10 s, 58°C for 30 s and 70 or 72° C for 15–30 s/Kb, with a final extension of 70 or 72° C for 5 min; variations in the annealing temperature occurred as indicated in Table I. Overlapping PCRs (Wurch et al., 1998) were conducted using independent PCR reactions for 10 cycles, gel purifying the products using 0.9% agarose gels run in tris‐acetate ethylenediaminetetraacetic acid EDTA (TAE) buffer consisting of 40 mM Tris, 20 mM acetic acid, and 1 mM EDTA with 1:10,000 of a 1% solution of ethidium bromide added to the gel for visualization under ultraviolet light, then extracted from the gel by filtration (Merck Millipore, Cork, Ire., Ultrafree MC 0.45 μM PVDF membrane cartridge). The purified products were precipitated with sodium acetate and ethanol, dried and reconstituted in sterile nanopure water then used as templates with the outer primers for 40 cycles to create the overlapping product. PCR products were digested with the appropriate restriction endonucleases (Fermentas) as indicated in Table I. DNA fragments were ligated into the appropriately digested pAra99a vectors using T4 DNA ligase (Fermentas). Clones were transformed into chemically competent EC100 and grown on LB‐Amp plates. Plasmid minipreps (Fermentas) were then screened with the appropriate restriction endonucleases and gel electrophoresis. The DNA sequence of the cloned PCR products was determined by Sanger DNA sequencing of both strands (Sequetech, Mountain View, CA); DNA sequencing primers are indicated in Table I. DNA sequence analysis and graphic displays utilized 4Peaks (Nucelobytes, Aalsmeer, Netherlands) and Geneious (Biomatters, Aukland, NZ). All DNA constructs were confirmed to be complete and accurate before proceeding to subsequent steps.
Table I.
Oligonucleotide primers for PCR, gene assembly and DNA sequencing utilized in this study
| # | Primer Sequence (5′ – 3′) | Anneal temp for PCR | Primer enzyme site | Vector enzyme site | Structural component or function |
|---|---|---|---|---|---|
| Pseudomonas ToxA | |||||
| 1 | GATCTCATGAAACACCTGATACCCCATTGGATCCCC | 62°C | BspHl | NcoI | Complete ToxA forward |
| 1B | GATCGGCGCCTGGATACCATTCTCGGCTGGCCGCTG | 60°C | EheI | EheI | E553D forward |
| 2 | GATCTCTAGATTACTTCAGGTCCTCGCGCG | 60°C/62°C | XbaI | XbaI | Complete ToxA reverse |
| 3 | ACCTGACGCTTTTTATCGCA | n/a | n/a | n/a | DNA forward sequencing; primes in pAra99a plasmid |
| 4 | CCGCCAGGCAAATTCTGT | n/a | n/a | n/a | DNA reverse sequencing; primes in pAra99a plasmid |
| 5 | CACATGTCGCCGATCTACAC | n/a | n/a | n/a | DNA Sequencing F1 |
| 6 | GCTGGTCGCCCTCTACCT | n/a | n/a | n/a | DNA Sequencing F2 |
| 7 | AGGACCTCGACGCGATCT | n/a | n/a | n/a | DNA Sequencing F3 |
| 8 | CGCCCTGACGAAGAAGGT | n/a | n/a | n/a | DNA Sequencing R1 |
| 9 | AGGTAGAGGGCGACCAGC | n/a | n/a | n/a | DNA Sequencing R2 |
| 10 | AGATCGCGTCGAGGTCCT | n/a | n/a | n/a | DNA Sequencing R3 |
| 11 | GATCTCTAGAAAGGAGTCGTTATGAAAAAAATAACAGG | 55°C | XbaI | XbaI | ColE3 RBS and lysis protein forward |
| 12 | GATCCTGCAGTTACTGCGTTGCTAATCCGGTC | 55°C | PstI | PstI | ColE3 RBS and lysis protein reverse |
| OmpA:TGFα:3GS:DIb:DIIIK (O‐T‐G‐DIbK) | |||||
| 13 | GATCTCATGAAACACCTGATACCCCATTGGATCCCCCTGGTCGCCAGCCTCGGCCTGCTCGCCGGCGGCTCGTCCGCGTCCGCAGCTGTGGTGAGCCATTTTAACGATTGCCC | 58°C | BspHl | NcoI | ToxA signal sequence and TGFα forward |
| 14 | CCGGGTTGCCGGCGAGACCCCGGGCCAGCAGATCCGCATGTTCGC | 58°C | n/a | n/a | TGFα and DII overlap reverse |
| 15 | GCGAACATGCGGATCTGCTGGCCCGGGGTCTCGCCGGCAACCCGG | 58°C | n/a | n/a | DII overlap forward; results in SmaI site |
| 16 | CCGGCCTCGTCGTTGCCGGCCAGACGGGCCTGC | 58°C | n/a | n/a | DII overlap reverse |
| 17 | GCAGGCCCGTCTGGCCGGCAACGACGAGGCCGG | 58°C | n/a | n/a | DIII overlap forward |
| 18 | GATCTCTAGATTACTTCAGGTCCTCGCGCG | 58°C | XbaI | XbaI | DIII reverse |
| 19 | GATCCAGCTGAAGAAGTGGTGAGCCATTTTAACGATTGCC | 55°C | PvuII | PvuII | Glu Glu forward |
| 20 | GATCTCTAGAGTACAGATCTACTCGAGTTACAGTTCATCTTTCGGCGGTTTGCCGGGC | 55°C | XbaI | XbaI | KDEL reverse |
| 21 | GATCGAGCTCGGTACCCAG | 58°C | SacI | SacI | OmpA DNA Works 1 |
| 22 | TGCAATCGCGATAGCTGTCTTTTTAGCCATGGTGAATTCCTCCTGGGTACCGAGCTCGAT | 58°C | n/a | n/a | OmpA DNA Works 2 |
| 23 | GACAGCTATCGCGATTGCAGTGGCACTGGCTGGTTTCGCTACCGTAGCGCAGGCAGCTGG | 58°C | n/a | n/a | OmpA DNA Works 3 |
| 24 | GATCCAGCTGCCTGCGCT | 58°C | PvuII | PvuII | OmpA DNA Works 4 |
| 25 | GATCGATATCAACTAGTGGGCGGTGGTGGCAG | 58°C | EcoRV | SmaI | 3GS linker forward |
| 26 | GATCCCCGGGACCCGCCACCACCAGAGCCGCCGCCACCACTGCCACCACCGCCC | 58°C | SmaI | SmaI | 3GS linker reverse |
| 27 | GATCTCCCGGGGTCTGACCTGCCCGGTCG | 58°C | SmaI | SmaI | DIb‐DIIK forward |
| OmpA:TGFα:3GS:DIb:DIIIR (O‐T‐G‐DIbR) | |||||
| No PCR; this clone was generated by subcloning REDLK from wildtype ToxA into O‐T‐G‐DIbK (AatII and XbaI). | |||||
| OmpA:TGFα:3GS:DIIIK (O‐T‐G‐DIIIK) | |||||
| 28 | GACTCCCGGGGTACTGGCGCGGAGTTCC | SmaI | SmaI | DIIIK forward | |
| 20 | GATCTCTAGAGTACAGATCTACTCGAGTTACAGTTCATCTTTCGGCGGTTTGCCGGGC | 58°C | XbaI | XbaI | KDEL reverse |
| OmpA:TGFα:3GS:DIIIR (O‐T‐G‐DIIIR) | |||||
| No PCR; this clone was generated by subcloning REDLK from wildtype ToxA into O‐T‐G‐DIIIK (via EheI and XbaI). | |||||
| OmpA:TGFα:3GS:PE38K (O‐T‐G‐PE38K) | |||||
| 29 | GATCCCCGGGGTGGCAGCCTGGCCGC | 62°C | SmaI | SmaI | PE38K forward |
| 30 | GTCGCCGCTGTCCGCCGGGGCGTTGGCCGCGCCGGC | 62°C | n/a | n/a | Overlap reverse |
| 31 | GCCGGCGCGGCCAACGCCCCGGCGGACAGCGGCGAC | 58°C | n/a | n/a | Overlap forward |
| 20 | GATCTCTAGAGTACAGATCTACTCGAGTTACAGTTCATCTTTCGGCGGTTTGCCGGGC | 58°C | XbaI | XbaI | KDEL reverse |
| OmpA:TGFα:3GS:PE38R (O‐T‐G‐PE38R) | |||||
| No PCR; this clone was generated by subcloning REDLK from wildtype ToxA into O‐T‐G‐PE38K (via EheI and XbaI). | |||||
| TGFα:PE38K‐ΔOmpA | |||||
| 32 | GATCCCATGGTGGTGAGCCATTTTAACGATTGCC | 60°C | NcoI | NcoI | ΔOmpA forward |
| 26 | GATCCCCGGGACCCGCCACCACCAGAGCCGCCGCCACCACTGCCACCACCGCCC | 60°C | SmaI | SmaI | 3GS linker reverse |
| OmpA:TGFα:PE38K‐Δ3GS | |||||
| No PCR; this clone was generated by subcloning a deletion of 3GS. | |||||
| OmpA:3GS:PE38K‐ΔTGFα | |||||
| 33 | GATCGAGCTCGGTACCCAG | 60°C | SacI | SacI | ΔTGFα forward |
| 34 | GATCACTAGTTGATGCACCACAGCTGCCTGCGCTA | 60°C | SpeI | SpeI | ΔTGFα reverse |
| O‐T‐G‐PE38K‐E553D | |||||
| IB | GATCGGCGCCTGGATACCATTCTCGGCTGGCCGCTG | 60°C | EheI | EheI | E553D forward |
| 20 | GATCTCTAGAGTACAGATCTACTCGAGTTACAGTTCATCTTTCGGCGGTTTGCCGGGC | 60°C | XbaI | XbaI | KDEL reverse |
| Additional universal chimera sequencing primers | |||||
| 35 | GTCAGCTTCAGCACCCGC | n/a | n/a | n/a | Forward DNA sequencing primer in DIII. |
| 36 | GCGGGTGCTGAAGCTGAC | DNA Sequencing; reverse complement of primer 35. | |||
Cloning of the Wild Type ToxA
The parental Pseudomonas ToxA as well as the functional chimeras described below are shown in Figure 1. The Pseudomonas wild type toxin was cloned by PCR using primers 1 and 2. When the complete construct was confirmed by DNA sequencing using primers 3–10, the plasmids were transformed into electroporation competent JR501 (O'Callaghan and Charbit, 1990; Tsai et al., 1989) to provide the appropriate methylation. Plasmid DNA was then prepared from the JR501 clones and then transformed by electroporation into VNP20009. The ColE3 lysis protein, including its ribosomal binding site (RBS) bases 2660–2815 of the complete colicin E3 plasmid (Morales et al., 2014; GenBank KM287568), was amplified via PCR using primers 11 and 12 and cloned into the pAra99a using the enzymes XbaI and PstI to create a pAra99a‐RBS‐ColE 3‐lysis protein construct.
Figure 1.
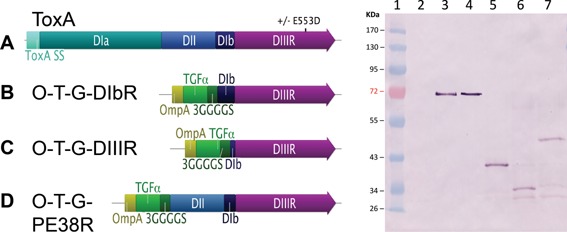
Comparison of the wildtype ToxA and ToxA chimeras. Left panel A) ToxA; the wildtype toxin ± the E553D substitution. B) O‐T‐G‐DIbR, a chimera containing an OmpA secretion signal, TGFα targeting domain, a 3GS linker, ToxA DIb, and DIII followed by REDLK. C) O‐T‐G‐DIIIR, a chimera containing an OmpA secretion signal, TGFα targeting domain, a 3GS linker, truncated DIb, and ToxA DIII followed by REDLK. D) O‐T‐G‐PE38R, a chimera containing an OmpA secretion signal, TGFα targeting domain, a 3GS linker, ToxA DII, partial DIb (Δ365‐380 amino acids) and DIII followed by REDLK. Right panel, an immunoblot using anti‐ToxA. Lane 1, molecular weight standard. Lane 2, pyrogallol red molybdate methanol (PRMM)‐precipitated VNP20009 empty vector culture supernatant (200 μL). Lane 3, ToxA standard (3.5 μg). Lane 4, VNP20009 ToxA culture supernatant (10 μL). Lane 5, VNP20009 O‐T‐G‐DIbR pyrogallol red molybdate methanol (PRMM)‐precipitated culture supernatant (200 μL). Lane 6, VNP20009 O‐T‐G‐DIIIR PRMM‐precipitated culture supernatant (200 μL). Lane 7, VNP20009 O‐T‐G‐PE38R PRMM‐precipitated culture supernatant (200 μL).
Contruction of OmpA:TGFα:3GS:DIb:DIIIR (O‐T‐G‐DlbR) and OmpA:TGFα:3GS:DIb:DIIIK (O‐T‐G‐DlbK)
Chimeras were cloned by an initial set of three overlapping PCRs using primers 13 and 14, 15 and 16, and 17 and 18 as independent PCRs whose purified products were then mixed for 10 PCR cycles followed by 40 cycles with the outer primers 13 and 18, which generates an in‐frame ToxA signal sequence (SS), TGFα, DII with a 25 amino acid deletion (Δ25 aa), DIb and DIII (ToxA SS:TGFα‐DII (Δ25 aa):DIb:DIII). A subsequent PCR using primers 19 and 20 which incorporates two additional glutamates after the ToxA SS and substitutes KDEL for REDLK at the end of DIII was cloned into the PvuII and XbaI sites of an intermediate vector and then used for a subsequent cloning step. An OmpA SS was substituted for the ToxA SS using primers 21–24 designed using the online DNA Works (Hoover and Lubkowski, 2002) tool with a first round using all four primers and a second round using primers 21 and 24. An AatII and XbaI fragment was cut out of the cloned PCR 19 and 20 product and substituted for the same fragment from the OmpA SS:TGFα‐DII (Δ25 aa):DIb:DIII construct, which results in substituting the KDEL sequence from the other KDEL‐containing clone (the product of primers 19 and 20). A flexible linker composed of the amino acids GGGGS repeated three times (3GGGGS or 3GS) was generated with overlapping primers 25 and 26 and cloned into the SmaI site immediately after TGFα. Finally, the DII (Δ25 aa):DIb:DIII KDEL was replaced with a DIb:DIII KDEL PCR product of primers 20 and 27 to generate the complete construct designated O‐T‐G‐DIbK. The O‐T‐G‐DIbR chimera was constructed by cloning the AatII/XbaI fragment containing REDLK from ToxA into O‐T‐G‐DIbK.
Construction of OmpA:TGFα:3GS:DIIIR (O‐T‐G‐DIIIR) and OmpA:TGFα:3GS:DIIIK (O‐T‐G‐DIIIK)
Chimeras were generated using primers 20 and 28 and cloned into the DIbK described above by removing the SmaI and XbaI fragment and replacing it with the 20 and 28 PCR product digested with SmaI and XbaI. The O‐T‐G‐DIIIR chimera was constructed by cloning the EheI/XbaI fragment containing REDLK from ToxA into O‐T‐G‐DIIIK.
Construction of OmpA:TGFα:3GS:PE38 K (O‐T‐G‐PE38K) and OmpA:TGFα:3GS:PE38 R (O‐T‐G‐PE38R)
The O‐T‐G‐PE38K chimera was generated by overlapping PCR using primers 29 and 30 and 31 and 20 whose purified products were then amplified with the outer primers 29 and 20, which generates an in‐frame PE38K that was cloned into the DIb‐DIIIK described above by removing the SmaI and XbaI fragment and replacing it with the 29 and 20 PCR product digested with SmaI and XbaI, resulting in O‐T‐G‐PE38K. The O‐T‐G‐PE38R chimera was constructed by cloning the EheI/XbaI fragment containing REDLK from ToxA into O‐T‐G‐PE38K.
Deletion Analysis of O‐T‐G‐PE38 K
The deletion analysis of O‐T‐G‐PE38K is diagramed in Figure 6. Construction of the ΔOmpA construct was performed by cloning the PCR product generated by primers 32 and 26 and digested with NcoI/SmaI into O‐T‐G‐PE38K, whereby the forward primer provides an in‐frame initiating methionine. Construction of Δ3GS construct was performed by digesting O‐T‐G‐PE38K with NcoI and SmaI and replacing it with the NcoI and SmaI fragment containing OmpA‐TGFα without the linker from the OmpA:TGFα‐DII (Δ25 aa):DIb:DIII construct described above. Deletion of TGFα from O‐T‐G‐PE38K to generate the ΔTGFα construct was performed by digesting the PCR product generated by primers 33 and 34 with SacI and SpeI and cloning it into O‐T‐G‐PE38K. Disruption of the ADP ribosyation active site was accomplished by changing the glutamic acid of the active site to aspartic acid (E553D) by cutting the PCR product generated by primers 1B and 20 with EheI and XbaI and cloning it into O‐T‐G‐PE38K which had had its EheI/XbaI fragment removed, which resulted in the construct designated E553D.
Figure 6.

Relative expression levels of epidermal growth factor receptor (EGFR) in different tumor cell lines and the relative sensitivities of those cell lines to culture supernatants of VNP20009 expressing either wild type ToxA or O‐T‐G‐PE38K. A) Immunoblot of whole cancer cell lysates using antibodies against either human EGFR or human GAPDH. B) Dose‐response curve of ToxA against H460 (●), HeLa (▪), and MDA‐MB‐468 (▴) cancer cells. C) Dose‐response curve of O‐T‐G‐PE38K against H460 (○), HeLa (□), and MDA‐MB‐468 (△) cancer cells. The experimental error is expressed as the SD.
Protein Detection and Quantification
The amount of the wild type ToxA protein produced by VNP20009 was determined from immunoblots using a range of known quantities of purified Pseudomonas ToxA (Sigma–Aldrich) as standards defined by NanoDrop 2000 s reading (NanoDrop Products/Thermo Fisher) at A280 using an extinction coefficient of E1% = 11.9, and a rabbit anti‐Pseudomonas ToxA primary antibody (Sigma–Aldrich) with a goat anti‐rabbit alkaline phosphatase secondary antibody (Jackson Laboratories, West Grove, PA) detected with nitro‐blue tetrazolium and 5‐bromo‐4‐chloro‐3′‐indolyphosphate (NBT/BCIP). The bands were quantified by densitometry using Image J (Schneider et al., 2012) in order to construct a standard curve, with the VNP20009 produced ToxA analyzed by linear regression using Excel. In order to normalize the detection of ToxA chimeras, which possess varying amounts of the wild type ToxA antigen and potentially results in different number of epitopes recognized by the polyclonal antibody, quantification was based on the TGFα portion of the chimera using a dilution series of purified human recombinant TGFα (hrTGFα; R&D Systems, Minneapolis, MN) quantified by CBQCA protein assay (Molecular Probes, Eugene, OR) as standards. The amount of the ToxA chimeric proteins produced by the bacteria was determined by linear regression from a representative immunoblot with a mouse anti‐TGFα monoclonal antibody (Clone MF9; Abnova, Taipei, Taiwan) and a secondary goat anti‐mouse alkaline phosphatase antibody (Jackson Laboratories). Preliminary assays of VNP20009 wild type ToxA and ToxA chimeras showed that the chimeric ToxA supernatants required 200 μL of supernatant concentrated by pyrogallol red molybdate methanol (PRMM) precipitation (Caldwell and Lattemann, 2004) in order to be detected, whereas 10 μL the wild type ToxA supernatant could be detected without concentration, and thus these volumes were utilized. The relative amount of epidermal growth factor receptor (EGFR) was determined using a polyclonal rabbit anti‐EGFR antibody (Rockland, Gilbertsville, PA) against 20 μg each of the respective tumor cell lysate, where the lysate total protein concentration was determined by the BCA assay (Pierce/Thermo Fisher) and the relative quantity of EGFR determined by immunoblot. A duplicate blot was probed with an anti‐glyceraldehyde 3‐phosphate dehydrogenase (GAPDH antibody (Thermo Fisher) as a loading control. The EGFR and GAPDH blots utilized a horseradish peroxidase conjugated goat anti‐rabbit (Jackson Laboratories) as a secondary antibody detected by enhanced chemiluminescence (Thermo‐Fisher).
Apoptosis Assay
Induction of apoptosis was determined by fluorescent microscopy using Rhodamine 123 (Acros Organics/ThermoFisher) as a vital stain for mitochondrial membrane polarity (Ferlini and Scambia, 2007). For microscopy, 25,000 live cells in 1 mL of tissue culture media were grown in 24 well optical bottom culture dishes (Eppendorf North America, Hauppauge, NY) overnight, and were then treated with VNP20009 empty vector (no toxin), ToxA or O‐T‐G‐PE38K culture supernatants and stained with 1.0 μg/mL of Rhodamine 123 for 10 min, counterstained with 1.0 μg/mL of Hoecsht 33342 (Molecular Probes) for 10 min in order to stain nuclei, washed three times with PBS and then observed for fluorescence using excitation at 488 nm using a Zeiss Axio Observer inverted microscope with differential interference contrast (DIC) optics. Digital images were captured by a Zeiss AxioCam MRc camera, and the Rhodamine and Hoechst images digitally merged using Zeiss Efficient Navigation (ZEN) software. For quantifying total mitochondrial polarity of the treatments, JC‐10 (Enzo Life Sciences, Farmingdale, NY) was used (Patel et al., 2016) rather than Rhodamine 123 because Rhodamine 123 gave a high background due to adherence to the sides of the wells. Five thousand cells were seeded per well in an optical bottom black 96 well tissue culture plate (Corning, Corning, NY), allowed to adhere overnight, and were then treated with 10 μL VNP20009 empty vector (no toxin), ToxA or O‐T‐G‐PE38K culture supernatants. The average mitochondrial fluorescence per cell was determined by staining the cells with 2 μM JC‐10 for 15 min in cell culture media followed by staining with 1 μg/mL Hoechst for 10 min in PBS, and then determining total relative JC‐10 fluorescence units at 560 nm, which was divided by the total cell number of cells as determined by total Hoechst (nuclear) fluorescence staining at 510 nm.
Results
Relative Toxicity of ToxA Chimera Culture Supernatants
Schematics of the constructs generated and the anti‐ToxA immunoblot are shown in Figure 1. Using the ToxA standards and anti‐ToxA antibody, the quantity of ToxA in the supernatant produced by VNP20009 was determined to be 39.9 ng/μL (not μ shown). The predicted molecular weights of the products are 66 kDa for ToxA, 33 kDa for O‐T‐G‐DIbR, 31 kDa for O‐T‐G‐DIIIR, and 44 kDa for O‐T‐G‐PE38R. All of the constructs ran 3–5 kDa higher than predicted, which may have been due to the use of pre‐stained molecular weight standards. The relative supernatant toxicity of the initial cohort of ToxA chimeras containing the native endoplasmic reticulum retention signal (C‐terminal REDLK) are shown in Figure 2. These data demonstrate that the VNP20009 supernatant with the wild type ToxA exhibited a high degree of relative toxicity, and that the single conservative amino acid substitution in ToxA‐E553D completely abolished detectable cytotoxicity. These data also demonstrate the relative lack of cytotoxicity from VNP20009 culture supernatants and underscores the potential for expression of a targeted toxin to increase the antitumor activity of VNP20009. Among the ToxA chimeras, the O‐T‐G‐PE38R had the highest degree of cell killing, followed by O‐T‐G‐DIbR, and then O‐T‐G‐DIIIR, which only showed cell toxicity at the highest concentration.
Figure 2.

Dose‐response curves of ToxA and ToxA chimeras. Dose responses were determined as relative percent survival of MDA‐MB‐468 breast adenocarcinoma cells following exposure to twofold dilutions of the respective VNP20009 culture supernatants, with the error expressed as the SD. Panel A: ToxA wild type toxin (●) and ToxA with the E553D substitution (▪). Panel B: The chimeric toxins O‐T‐G‐DIbR (□); O‐T‐G‐DIIIR (▴); and O‐T‐G‐PE38 R (○).
Relative Quantities of the ToxA Chimeras and Their Potency
Although O‐T‐G‐PE38R had the strongest cytotoxicity among the chimeras, differences in the chimera supernatant cell killing ability could have been due to either production of greater amounts of the protein or that there were actual differences in the potency of the different proteins, or both. Because the preliminary data suggested that VNP20009 produced higher concentrations of the wild type ToxA than it produced for the chimeras, the supernatants were concentrated by PRMM. We used TGFα standards and an antibody against TGFα as a means to normalize the antibody binding activity among the chimeras in order to determine the amount of the chimeric proteins in the supernatants (Fig. 3A and B). The chimeric proteins in ng/μL of supernatant were multiplied by the IC50 in μL of culture supernatant to determine the IC50 in ng/well, and molarity (µM). Using this analysis, O‐T‐G‐PE38R was the most potent on a molar basis, followed by O‐T‐G‐DIIIR and O‐T‐G‐DIbR. Because the cell killing ability of an intratumoral Salmonella will likely be dictated by the activity of the extracellular toxin produced, we chose O‐T‐G‐PE38R for further analysis.
Figure 3.
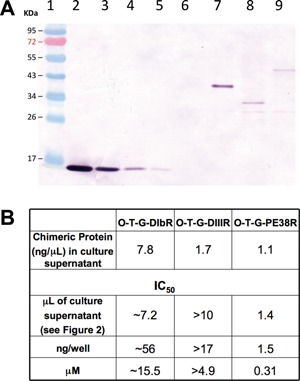
Quantities of the chimeric proteins present in culture supernatants and their relative cytotoxicities. Top Panel: Anti‐TGFα immunoblot comparison of hrTGFα standards and PRMM‐precipitated culture supernatants of O‐T‐G‐DIbR, O‐T‐G‐DIIIR, and O‐T‐G‐PE38R. Lane 1, molecular weight standard. Lane 2, 2.0 μg hrTGFα. Lane 3, 0.67 μg hrTGFα. Lane 4, 0.20 μg hrTGFα. Lane 5, 0.067 μg hrTGFα. Lane 6, VNP20009 empty vector culture supernatant. Lane 7, O‐T‐G‐DIbR. Lane 8, O‐T‐G‐DIIIR. Lane 9, O‐T‐G‐PE38R. Lower Panel: IC50 values of the TGFα:ToxA fusions present in the culture supernatants. The quantities of the fusion proteins present in the culture supernatants were calculated based on a standard curve of the hrTGFα bands in the top panel determined by densitometry using Image J. The volume of culture supernatants in μL/well required to achieve the 50% inhibitory values shown in Figure 2B were calculated from the interpolated dose response curves, which allowed the calculation of the IC50 values in ng/well and nM.
Effect of KDEL Substitution and ColE3 Lysis Protein Co‐Expression on Relative Cell Killing by O‐T‐G‐PE38R Supernatants
Based upon previous studies that have shown that the REDLK C‐terminus of ToxA acts as a endoplasmic reticulum retention signal (Kihara and Pastan, 1994b; Seetharam et al., 1991), and other studies which have shown colicin‐based lysis proteins might enhance the ability of tumor‐targeted Salmonella effector genes (Bermudes et al., 2001a), we assessed these features alone and in combination. As shown in Figure 4 there was a significant loss of activity when ToxA was modified into O‐T‐G‐PE38R (P = 0.04). However, the KDEL substitution for REDLK enhanced the cell‐killing ability of the O‐T‐G‐PE38R by greater than twofold (P = 0.017). The KDEL substitution for REDLK also increased the toxicity of O‐T‐G‐DIbR and O‐T‐G‐DIIIR constructs (data not shown). Furthermore, the addition of ColE3 lysis with KDEL showed an even greater ability of the cell culture supernatant to kill MDA‐MB‐468 cancer cells (P = 0.049), with a statistically significant (P < 0.021) combined increase of O‐T‐G‐PE38R‐ColE3 Lysis compared to the wild type toxin, confirming that the cell‐killing ability of chimeric toxins can be improved by these genetic modifications in Salmonella.
Figure 4.
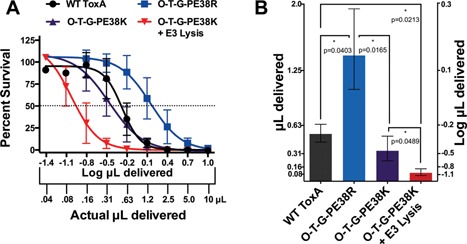
Relative culture supernatant toxicities of wild type ToxA and chimeric variants. A) The individual dose response curves for the wild type toxA, O‐T‐G‐PE38R, O‐T‐G‐PE38K, and O‐T‐G‐PE38K with the ColE3 lysis. Error bars represent SD Actual and logarithmic supernatant volumes are indicated on the x‐axis. B). Comparison by volume (actual and log) of the supernatants required to achieve an IC50 against MDA‐MB‐468 tumor cells. The wild type ToxA, O‐T‐G‐PE38R, O‐T‐G‐PE38K, and O‐T‐G‐PE38K with the ColE3 lysis, were compared by the quantity (in μL) of supernatant required to achieve an IC50 against MDA‐MB‐468 tumor cells. Error bars indicate 95% CI. Brackets indicate comparisons that were made and the level of significance (asterisks) as well as the actual P‐value indicated below the bracket.
Deletion Analysis of O‐T‐G‐PE38K
As the secretion of ToxA remains incompletely understood and there is some potential for growth suppression from TGFα in the MDA‐MB‐468 cell line despite the fact it is normally mitogenic (Wang et al., 1997), we sought to determine the relative contribution of each of the three N‐terminal components. Deletion analysis included ΔOmpA, Δ3GS, and ΔTGFα constructs, and the dose response curves were determined for each of the culture supernatants Figure 5. These results demonstrate substantial contributions from the OmpA and TGFα, with a smaller contribution from the flexible linker. Similar to the ToxA‐E553D construct shown in Figure 1, the O‐T‐G‐PE38K‐E533D lost all detectable activity, while the co‐expression of the ColE3 lysis protein increased the relative cytotoxicity of the O‐T‐G‐PE38K supernatant (also see Fig. 4 for the effect of the ColE3 lysis protein).
Figure 5.
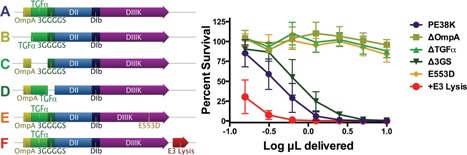
Deletion analysis of the O‐T‐G‐PE38K N‐terminal components and comparisons with the E553D substitution and the coexpression of the ColE3 lysis protein. Left Panel: Diagram of the parent O‐T‐G‐PE38K and variants. Right Panel: Dose responses from the constructs diagramed in the left panel were determined as relative percent survival of MDA‐MB‐468 breast adenocarcinoma cells following exposure to culture supernatants from VNP20009 expressing A) O‐T‐G‐PE38K, B) ΔOmpA (O‐T‐G‐PE38K with a deletion in OmpA), C) ΔTGFα (O‐T‐G‐PE38K with a deletion in TGFα), D) Δ3GS (O‐T‐G‐PE38K with a deletion of the 3GS linker), E) E553D (O‐T‐G‐PE38K with a glutamic acid to aspartic acid substitution at the amino acid corresponding to number 553 of the wild type ToxA), n = 2, and F) +ColE3 Lysis (O‐T‐G‐PE38K coexpressed with the ColE3 lysis protein). The experimental error is expressed as the SD.
Specificity of the O‐T‐G‐PE38K Construct for EGFR‐Expressing Cell Lines
Immunoblots using anti‐EGFR and anti‐GAPDH as a loading control confirmed that our cell lines had the expected expression levels of EGFR. As shown in the upper panel of Figure 6A, EGFR was highly expressed in MDA‐MB‐468, with a low level of expression by HeLa, and no detectable expression by H460. The anti‐GAPDH loading control blot showed constant levels of GAPDH among the cell lines (Fig. 6A lower panel). When culture supernatant from a Salmonella expressing ToxA was exposed to the different cell lines, all of them were sensitive and exhibited a high degree of cell killing (Fig. 6, center panel). However, when culture supernatant from a Salmonella expressing O‐T‐G‐PE38K was exposed to the same cell lines, those producing little or no EGFR (HeLa and H460, respectively) were less sensitive to O‐T‐G‐PE38K, while the MDA‐MB‐468 cell line that expresses high levels of EGFR had increased sensitivity (Fig. 6, lower panel). These results were further confirmed by analysis of cell number and mitochondrial polarity using Rhodamine 123 combined with Hoechst (Fig. 7). When MDA‐MB‐468 or HeLa cells were exposed to culture supernatant from a Salmonella expressing ToxA, a substantial loss of cells and of Rhodamine 123 staining occurred for both cell lines. However, when MDA‐MB‐468 or HeLa cells were exposed to culture supernatant from a Salmonella expressing O‐T‐G‐PE38K, MDA‐MB‐468 showed a high degree of sensitivity (Fig. 7), whereas the treated HeLa cells were similar to those treated with a culture supernatant from a Salmonella expressing an empty vector (no toxin) control. Quantification of the overall mitochondrial staining per cell using JC‐10 and Hoechst in the HeLa cell line at 18 h showed that ToxA resulted in a 55% reduction of mitochondrial signal (P = 0.0087) while there was no measurable loss in the cells treated with O‐T‐G‐PE38K. Although we could microscopically observe MDA‐MB‐468 cells with reduced mitochondrial signal due to ToxA and O‐T‐G‐PE38K treatments at 18 h using Rhodamine 123 (Fig. 7), there was no statistical significance in the JC‐10 staining, possibly due to the fact that many of the cells were already eliminated at that time and the remainder were a mixture of cells that had not yet responded with those that were loosing their mitochondrial signal. However, significant values were obtained at 22 h, where ToxA resulted in a 25% reduction (P = 0.0014) and O‐T‐G‐PE38K resulted in a 55% reduction (P = 0.0001) relative to the no toxin control. Overall, these results are consistent with ToxA exhibiting relatively non‐selective toxicity compared with O‐T‐G‐PE38K, which was selectively toxic to EGFR positive cells as previously established using purified toxin (Siegall et al., 1989a).
Figure 7.
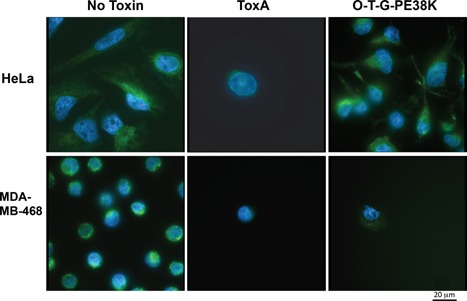
Analysis of apoptosis induction in low‐level EGFR expressing (HeLa) and high‐level EGFR expressing (MDA‐MB‐468) cancer cells using Rhodamine 123 counterstained with Hoechst. Top Panel: HeLa cells treated with either no toxin (empty vector control), wild type ToxA (ToxA), or the O‐T‐G‐PE38K chimera. Bottom Panel: MDA‐MB‐468 cells with either no toxin, wild type ToxA, or the O‐T‐G‐PE38K chimera.
Discussion
Attenuated Salmonella strain VNP20009 has a remarkable ability to colonize solid tumors and suppress tumor growth in mice, yet in humans there is no antitumor activity associated with colonization of melanoma tumors by the same bacteria (Toso et al., 2002), although it is possible there may be antitumor activity in other tumor types. In vitro, this attenuated Salmonella strain is able to locally induce apoptosis of tumor cells (Ganai et al., 2011) but its culture supernatant exhibits little ability to kill cancer cells (Swofford et al., 2014). Augmenting the ability to kill cancer cells by a secreted protein targeted to the tumor cells themselves might compensate for the lack of antitumor activity in humans. We anticipate that this approach will enhance the therapeutic index by lowering the effective dose while retaining the tolerability of the parent vector.
Many of the studies by Pastan and co‐workers utilized protein harvested from bacterial inclusion bodies that were then denatured and refolded (Siegall et al., 1989a). In those studies, comparison of naturally folded protein derived from the periplasm with refolded protein derived from inclusion bodies showed that the refolded protein is approximately 10‐fold more potent (Siegall et al., 1989a). In the context of a live tumor‐targeting Salmonella system, the process of achieving functionality of the proteins expressed must be carried out by the bacteria themselves. The ToxA chimeras that we constructed were selected from among a large range of forms previously investigated outside the context of a bacterial delivery vector (Weldon and Pastan, 2011), and modified as to accommodate their expression and release into the extracellular milieu by Salmonella, which we determined by assessing the cytotoxicity of culture supernatants.
The results of our study are consistent with the previous study by Swofford et al. (2014) that demonstrated VNP20009's ability to express an active form of the Pseudomonas ToxA capable of killing cancer cells (Figs. 1 and 2). Our results further extend that study to include chimeric forms of ToxA. Of the three different forms we initially investigated, the culture supernatant of VNP20009 expressing O‐T‐G‐PE38R was the most active as determined by cytotoxicity (Fig. 2) and also the most potent based on the relative quantity of the toxin present in the culture supernatant (Fig. 3). The O‐T‐G‐DIbR construct resulted in the next highest degree of cell killing (Fig. 2), but this is likely due to the greater amount of protein produced (Fig. 3). Because of the superior potency of the O‐T‐G‐PE38R, we further investigated modulation of its toxicity. Substitution of the consensus endoplasmic reticulum retention signal KDEL for the naturally occurring REDLK is another previously explored modification (Kihara and Pastan, 1994b). This modification also enhanced the cytotoxicity of the VNP20009‐expressed chimeras (Fig. 4). N‐terminus deletion analysis of O‐T‐G‐PE38K that established the OmpA signal sequence and the TGFα ligand were necessary for toxicity, and that the flexible linker slightly enhanced activity (Fig. 5). This result is also consistent with the previous study that showed improved activity from a short flexible linker (Kihara and Pastan, 1994b). Toxicity was further enhanced by co‐expression of the ColE3 lysis protein. When expressed in VNP20009, these modifications result in a supernatant that is more active than the supernatant containing the wild type toxin.
The difference between an in vitro reconstitution of protein activity compared to in vivo delivery of active protein is underscored by the requirement for the OmpA signal sequence, since the OmpA sequence is not required for activity of the refolded protein but was completely necessary for activity to be obtained from a VNP20009 culture supernatant. Our initial studies also represent variation in the presence or absence of DIb in the O‐T‐G‐DlbR and O‐T‐G‐DIIIR constructs, neither of which was as active as the O‐T‐G‐PE38R, but were increased in cytotoxicity when REDLK was replaced with KDEL (data not shown).
In order to assess whether the Salmonella‐expressed O‐T‐G‐PE38K remained specific for EGFR‐expressing cell lines, we compared the relative toxicity of O‐T‐G‐PE38K and ToxA culture supernatants against cell lines that are known to either express little or no EGFR (HeLa and H460) compared with a high level EGFR expressing cell line (MDA‐MB‐468). Our results showed a correlation of cytotoxicity with the presence of EGFR for O‐T‐G‐PE38K, but not for ToxA (Fig. 6), consistent with the original studies using TGFα fusions (Siegall et al., 1989b). Staining of mitochondria with Rhodamine 123 was also consistent with selectivity for EGFR‐expressing cells (Fig. 7). Quantification of total mitochondrial signal per cell using JC‐10 and Hoechst were also consistent with general toxicity of Salmonella‐expressed ToxA, and relative selectivity of O‐T‐G‐PE38K for the EGFR positive cell line MDA‐MB‐468.
Use of live replication competent bacteria as experimental cancer therapeutics is an area of growing interest. Live Salmonella (ClinicalTrials.gov Identifier NCT01099631), Listeria (ClinicalTrials.gov Identifier NCT01598792; ClinicalTrials.gov Identifier NCT01675765; ClinicalTrials.gov Identifier NCT02002182), and Clostridium (ClinicalTrials.gov Identifier NCT01118819) all have current or recent FDA‐sanctioned human clinical studies in the cancer treatment setting. Results of the Clostridium trial have recently been published (Roberts et al., 2014) and demonstrated an anti‐tumor response. The preclinical studies of Clostridium also showed antitumor activity in a veterinary trial with pet dogs (Roberts et al., 2014) and an earlier study using Salmonella VNP20009 also showed significant antitumor activity in dogs with spontaneous neoplasms (Thamm et al., 2005). Continued success is likely to sustain interest in these novel therapeutic approaches.
Targeted toxins have also shown promise in the clinic (Kreitman et al., 2005, 2009). Limitations that have been identified include biodistribution properties and chimeric toxin neutralizing antibodies (Weldon and Pastan, 2011). Modifications that have been investigated to overcome these limitations has included releasable PEGylation (Filpula et al., 2007), which substantially increases the biodistribution. Tumor‐targeted bacteria offer alternative solutions to this and other problems as these bacteria readily travel through the blood, with a portion of them avoiding the innate immune system and taking up residence within the tumor where they then persist despite a subsequent systemic immune response. The immunoprivileged nature of tumors is thought to contribute to the persistence and tumor‐selective growth of the bacteria within the tumor (Bermudes et al., 2001b; Darveau, 1999; Low et al., 1999), including providing partial protection from antibody responses, and thus the effect of neutralizing antibodies against the chimeras within the tumors should also be diminished. Furthermore, targeted proteins delivered systemically have limitations for delivery deep inside of tumors, whereas the bacteria localize even within the necrotic regions and can produce therapeutic proteins directly in situ (Soghomonyan et al., 2005), which could overcome that limitation.
EGFR is a major target in cancer therapeutics because of its over‐expression in a number of cancer cell types, including breast, colon lung, head and neck, bladder, and brain cancers. Approved EGFR targeting cancer therapeutics include cetuximab, an antibody that is used in the treatment of metastatic colorectal cancer which binds EGFR and blocks EGFR signaling (Chu and DeVita, 2014) and gefitinib and erlotinib which are used in the treatment of non‐small cell lung cancer (NSCLC) and block EGFR signaling by specifically inhibiting EGFR tyrosine kinase activity. Targeting of EGFR by another bacterial anticancer agent, antibody targeted bacterial minicells, is a recently developed approach to treatment of EGFR‐expressing cancers (MacDiarmid et al., 2009). Unlike bacterial minicells however, live replication competent bacteria have the potential to persist in tumors for weeks, continuously producing an anticancer agent or stimulating immune responses, which may represent a significant therapeutic advantage. Furthermore, some Salmonella strains such as Salmonella saintpaul isolate (SstpNPG) can reach the brain and have been proposed as a vector for treating brain tumors (Mlynarczyk et al., 2014). This approach may be of special use since some brain tumors overexpress EGFR and may, therefore, be amenable to Salmonella expressing EGFR‐targeted therapeutics.
Our future investigations are expected to include efficacy studies of murine tumor models overexpressing EGFR treated with VNP20009 expressing O‐T‐G‐PE38K and other targeted constructs. It will be of interest to evaluate the effect that the presence of this targeted toxin has on the therapeutic index. New chimeric proteins are also of interest for their potential to expand the therapeutic index and/or to target other receptors or achieve tumor cell specificity through other means. By targeting more than one weakness of tumor cells, development of resistance could be minimized. The capacity of bacterial vectors to express more than one effector gene makes this possibility of particular importance.
This work was supported by NIH Grant SC3GM098207 to DB. LV was supported by the NIH Research Initiative for Scientific Enhancement (RISE) Grant GM063787. The sponsors did not have any influence on the project design or interpretation. We wish to thank Jonathan Kelber for providing the MDA‐MB‐468 cell line and helpful advice. DB has financial interest in Aviex Technologies and Magna Therapeutics, and receives royalties from Yale University. DB and DQ have proportional rights to royalties from a patent application filed by California State University, Northridge.
References
- Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli . Gene 69:301–315. [DOI] [PubMed] [Google Scholar]
- Avogadri F, Martinoli C, Petrovska L, Chiodoni C, Transidico P, Bronte V, Longhi R, Colombo MP, Dougan G, Rescigno M. 2005. Cancer immunotherapy based on killing of Salmonella‐infected tumor cells. Cancer Res 65:3920–3927. [DOI] [PubMed] [Google Scholar]
- Bermudes D, Low KB, Pawelek J, Feng M, Belcourt M, Zheng L‐M, King I. 2001a. Tumor‐specific Salmonella‐based cancer therapy. Biotechnol Genet Eng Rev 18:219–233. [DOI] [PubMed] [Google Scholar]
- Bermudes D, King I, Clairmont CA, Lin SL, Belcourt M. 2001b. Compositions and methods for tumor‐targeted delivery of effector molecules, WO/2001/025397.
- Brendel M, Pollack IF, Hamilton RL, James CD. 1999. Epidermal growth factor receptor expression and gene amplification in high‐grad non‐brainstem gliomas of childhood. Clin Cancer Res 5:1786–1792. [PubMed] [Google Scholar]
- Caldwell RB, Lattemann CT. 2004. Simple and reliable method to precipitate proteins from bacterial culture supernatant. Appl Environ Microbiol 70:610–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary VK, FitzGerald DJ, Adhya S, Pastan I. 1987. Activity of a recombinant fusion protein between transforming growth factor type α and Pseudomonas toxin. Proc Natl Acad Sci USA 84:4538–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary V, Wu Y‐H, FitzGerald D, Sankar A, Pastan I. 1988. Role of domain II of Pseudomonas exotoxin in the secretion of proteins into the periplam and medium by Escherichia coli . Proc Natl Acad Sci U S A 85:2934–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, DeVita VT Jr. 2014. Physicians’ cancer chemotherapy drug manual. Burlington, MA: Jones and Bartlett; p 664. [Google Scholar]
- Clairmont C, Lee KC, Pike J, Ittensohn M, Low KB, Pawelek J, Bermudes D, Brecher SM, Margitich D, Turnier J, Li Z, Luo X, King I, Zheng L‐M. 2000. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium . J Infect Dis 181:1996–2002. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov Identifier: NCT00004988, Treatment of patients with cancer with genetically modified Salmonella typhimurium bacteria.
- ClinicalTrials.gov Identifier: NCT01099631, IL‐2 expressing, attenuated Salmonella typhimurium in unresectable hepatic spread.
- ClinicalTrials.gov Identifier: NCT01118819, Safety study of Clostridium novyi‐NT spores to treat patients with solid tumors that have not responded to standard therapies.
- ClinicalTrials.gov Identifier: NCT01598792, Safety study of recombinant Listeria monocytogenes (Lm) based vaccine virus vaccine to treat oropharyngeal Cancer (REALISTIC).
- ClinicalTrials.gov Identifier: NCT01675765, CRS‐207 Cancer vaccine in combination with chemotherapy as front‐line treatment for malignant pleural mesothelioma.
- ClinicalTrials.gov Identifier: NCT02002182, ADXS 11‐001 Vaccination prior to robotic surgery, HPV‐positive oropharyngeal cancer.
- Darveau R. 1999. Infection, inflammation and cancer. Nat Biotechnol 17:19. [DOI] [PubMed] [Google Scholar]
- Edwards GM, DeFeo‐Jones D, Tai JY, Vuocolo GA, Patrick DR, Heimbrook DC, Oliff A. 1989. Epidermal growth factor receptor binding is affected by structural determinants in the toxin domain of transforming growth factor‐alpha‐Pseudomonas exotoxin fusion proteins. Mol Cell Biol 9:2860–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiblmaier M, Meyer LA, Watson MA, Francasso PM, Pike LJ, Anderson CJ. 2008. Correlating EGFR expression with receptor‐binding properties and internalization of 64Cu‐DOTA‐Cetuximab in 5 cervical cancer lines. J Nuclear Med 49:1472–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlini C, Scambia G. 2007. Assay for apoptosis using the mitochondrial probes, Rhodamine123 and 10‐N‐nonyl acridine orange. Nat Protoc 2:3111–3114. [DOI] [PubMed] [Google Scholar]
- Filpula D, Yang K, Basu A, Hassan R, Xiang L, Zhang Z, Wang M, Wang QC, Ho M, Beers R, Zhao H, Peng P, Zhou J, Li X, Petti G, Janjua A, Liu J, Wu D, Yu D, Zhang Z, Longley C, FitzGerald D, Kreitman RJ, Pastan I. 2007. Releasable PEGylation of mesothelin targeted immunotoxin SS1 P achieves single dosage complete regression of a human carcinoma in mice. Bioconjug Chem 18:773–784. [DOI] [PubMed] [Google Scholar]
- Forbes NS. 2010. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer 10:785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. 2011. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther 18:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GD, Smith S, Baldridge J, Markins R, Vasil M, Chen E, Heyneker M. 1984. Cloning, nucleotide sequence, and expression in Escherichia coli of the exotoxin A structural gene of Pseudomonas aeruginosa . Proc Natl Acad Sci U S A 81:2645–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RM. 2015. Back to the future: Are tumor‐targeting bacteria the next‐generation cancer therapy? Methods Mol Biol 1317:239–260. [DOI] [PubMed] [Google Scholar]
- Holliday DL, Speirs V. 2011. Choosing the right cell line for breast cancer research. Breast Cancer Res 13:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J. 2002. DNAWorks: An automated method for designing oligonucleotides for PCR‐based gene synthesis. Nucleic Acids Res 30(10):e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Pastan I. 1994a. Small chimeric toxins containing only transforming growth factor α and domain III of Pseudomonas exotoxin with good antitumor activity in mice. Cancer Res 54:5154–5159. [PubMed] [Google Scholar]
- Kihara A, Pastan I. 1994b. Analysis of sequences required for the cytotoxic action of a chimeric toxin composed of Pseudomonas exotoxin and transforming growth factor alpha. Bioconjug Chem 5:532–538. [DOI] [PubMed] [Google Scholar]
- Kirk J, Carmichael J, Stratford IJ, Harris AL. 1994. Selective toxicity of TGFα‐PE40 to EGFR‐positive cell lines: selective protection of low EGFR‐expressing cell lines by EGF. Br J Cancer 69:988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, FitzGerald D, Chaudhary VK, Adhya S, Pastan I. 1988. Activity of immunotoxin constructed with modified Pseudomonas exotoxin A lacking the cell recognition domain. J Biol Chem 263:9470–9475. [PubMed] [Google Scholar]
- Kreitman RJ, Siegall CB, Chaudhary VK, Fitzgerald DJ, Pastan I. 1992. Properties of chimeric toxins with two recognition domains: Interleukin 6 and transforming growth factor α at different locations in Pseudomonas exotoxin. Bioconjugate Chem 3:63–68. [DOI] [PubMed] [Google Scholar]
- Kreitman RJ, Squires DR, Stetler‐Stevenson M, Noel P, FitzGerald DJ, Wilson WH. Pastan I. 2005. Phase I trial of recombinant immunotoxin RFB4(dsFv)‐PE38 (BL22) in patients with B‐cell malignancies. J Clin Oncol 2005(23):6719–6729. [DOI] [PubMed] [Google Scholar]
- Kreitman RJ, Stetler‐Stevenson M, Margulies I, Noel P, Fitzgerald DJ, Wilson WH, Pastan I. 2009. Phase II trial of recombinant immunotoxin RFB4(dsFv PE38 (BL22) in patients with hairy cell leukemia. J Clin Oncol 27:2983–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschner S, Westphal K, Dietrich N, Viegas N, Jablonska J, Lyszkiewics M, Lienenklaus S, Falk W, Gekara N, Loessner H, Weiss S. 2009. Tumor invasion of Salmonella enterica serotype Typhimurium is accompanied by strong hemorrhage promoted by TNF‐alpha. PLoS ONE 4:e6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lory S, Strom MS, Johnson K. 1988. Expression and secretion of the cloned Pseudomonas aeruginosa exotoxin A by Escherichia coli . J Bacteriol 170:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossner H, Endmann A, Leschner S, Westphal K, Rohde M, Miloud T, Hämmerling G, Neuhaus K, Weiss S. 2007. Remote control of tumour‐targeted Salmonella enterica serovar Typhimurium by the use of L‐arabinose as inducer of bacterial gene expression in vivo . Cell Microbiol 9:1529–1537. [DOI] [PubMed] [Google Scholar]
- Low KB, Ittensohn M, Luo X, Zheng L‐M, King I, Pawelek JM, Bermudes D. 2004. Construction of VNP20009, a novel, genetically stable antibiotic sensitive strain of tumor‐targeting Salmonella for parenteral administration in humans. Methods Mol Med 90:47–60. [PubMed] [Google Scholar]
- Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fisher J, Lin SL, Luo X, Miller SI, Zheng L, King I, Pawelek JM, Bermudes D. 1999. Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor‐targeting in vivo. Nat Biotechnol 17:37–41. [DOI] [PubMed] [Google Scholar]
- MacDiarmid JA, Amaro‐Mugridge NB, Madrid‐Weiss J, Sedliarou I, Wetzel S, Kochar K, Brahmbhatt VN, Phillips L, Pattison ST, Petti C, Stillman B, Graham RM, Brahmbhatt H. 2009. Sequential treatment of drug‐resistant tumors with targeted minicells containing siRNA or a sequential treatment of drug‐resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat Biotechnol 27:643–651. [DOI] [PubMed] [Google Scholar]
- Miller JH. 1992. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Mlynarczyk GSA, Berg CA, Withrock IC, Fick ME, Anderson SJ, Laboissonniere LA, Jefferson MG, Brewer MT, Stock ML, Lange JK, Luna KC, Acharya S, Kanuri S, Sharma S, Kondru NC, McCormack GR, Carlson SA. 2014. Salmonella as a biological “Trojan horse” for neoplasia: Future possibilities including brain cancer. Med Hypothesis 83:343–345. [DOI] [PubMed] [Google Scholar]
- Morales M, Attai H, Troy K, Bermudes D. 2014. Accumulation of single‐stranded DNA in Escherichia coli carrying the colicin plasmid pColE3‐CA38. Plasmid 77:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. [DOI] [PubMed] [Google Scholar]
- Murray SR, Bermudes D, de Felipe KS, Low KB. 2001. Extragenic suppressors of growth defects in msbB Salmonella . J Bacteriol 183:5554–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SR, Suwwan de Felipe K, Obuchowski PL, Pike J, Bermudes D, Low KB. 2004. Hot spot for a large deletion in the 18‐19 Cs region confers a multiple phenotype in Salmonella enterica serovar Typhimurium strain ATCC 14028. J Bacteriol 186:8516–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauts HC. 1976. Immunotherapy of cancer by bacterial vaccines In: Nieburgs HE, editor. Cancer detection and prevention, vol. 1. New York: Marcel Dekker; p 589–605. [Google Scholar]
- O'Callaghan D, Charbit A. 1990. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol Gen Genet 223:156–158. [DOI] [PubMed] [Google Scholar]
- Patel H, Chen J, and, Kavdia M. 2016. Induced peroxidase and crtoprotective enzyme expressions support adaptation of HUVECs to sustain subsequent H2O2 exposure. Microvas Res 103:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek JM, Low KB, Bermudes D. 1997. Tumor‐targeted Salmonella as a novel anticancer vector. Cancer Res 57:4537–4544. [PubMed] [Google Scholar]
- Pawelek J, Low KB, Bermudes D. 2003. Bacteria as tumour‐targeting vectors. Lancet Oncol Rev 4:548–556. [DOI] [PubMed] [Google Scholar]
- Roberts NJ, Zhang L, Janku F, Collins A, Bai R‐Y, Staedtke V, Rusk AW, Tung D, Miller M, Roix J, Khanna KV, Murthy R, Benjamin RS, Helgason T, Szvala AD, Bird JE, Roy‐Chowdhuri S, Zang HH, Qiao Y, Kari McDaniel BJ, Elpiner A, Sahora A, Lachowicz J, Phillips B, Turner A, Klein MK, Post G, Diaz LA Jr, Riggins GJ, Papadopoulos N, Vogelstein B, Bettegowda C, Huso DL, Varerasian M, Saha S, Zhou S. 2014. Intratumoral injection of Clostridium novyi‐NT spores induces antitumor responses. Sci Trans Med 6(Issue 249):249ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RM, Green J, Williams PJ, Tazzyman S, Hunt S, Harmey JH, Kehoe SC, Lewis CE. 2009. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther 16:329–339. [DOI] [PubMed] [Google Scholar]
- Saltzman DA. 2005. Cancer immunotherapy based on killing of Salmonella typhimurium‐infected tumor cells. Exp Opin Biol Ther 5:443–449. [DOI] [PubMed] [Google Scholar]
- Saltzman DA, Katsanis E, Heise CP, Hasz DE, Vigdorovich V, Kelly SM, Curtiss R 3rd, Leonard AS, Anderson PM. 1997. Antitumor mechanisms of attenuated Salmonella typhimurium containing the gene for human interleukin‐2: A novel antitumor agent? J Pediatr Surg 32:301–306. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS and, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaugh JL. 2011. Guidelines for accurate EC50/IC50 estimation. Parmaceut Statist 10:128–134. [DOI] [PubMed] [Google Scholar]
- Seetharam S, Chaudhary VK, FitzGerald D, Pastan I. 1991. Increased cytotoxic activity of Pseudomonas exotoxin and two chimeric toxins ending in KDEL. J Biol Chem 266:17376–17381. [PubMed] [Google Scholar]
- Siegall CB, Chaudhary VK, FitzGerald DJ, Pastan I. 1989a. Functional analysis of domains II, Ib, and III of Pseudomonas exotoxin. J Biol Chem 264:14256–14261. [PubMed] [Google Scholar]
- Siegall CB, Xu Y‐H, Chaudhary VK, Adhya S, FitzGerald D, Pastan I. 1989b. Cytotoxic activities of a fusion protein comprised of TGF alpha and Pseudomonas exotoxin. FASEB J 3:2647–2652. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2015. Cancer statistics, 2015. CA Cancer J Clin 65:5–29. [DOI] [PubMed] [Google Scholar]
- Soghomonyan SA, Doubrovin M, Pike J, Luo X, Ittensohn M, Runyan JD, Balatoni J, Finn R, Gelovani‐Tjuvajev J, Blasberg R, Bermudes D. 2005. Positron emission tomography (PET) imaging of tumor‐localized Salmonella expressing HSV1‐TK. Cancer Gene Ther 12:101–108. [DOI] [PubMed] [Google Scholar]
- Swofford CA, St. Jean AT, Panteli JT, Brentzel ZJ, Forbes NS. 2014. Identification of Staphylococcus aureus α‐hemolysin as a protein drug that is secreted by anticancer bacteria and rapidly kills cancer cells. Biotechnol Bioeng 111:1233–1245. [DOI] [PubMed] [Google Scholar]
- Thamm DH, Kurzman ID, King I, Li Z, Sznol M, Dubielzig RR, Vail DM, MacEwen EG. 2005. Systemic administration of an attenuated tumor‐targeting Salmonella typhimurium to dogs with spontaneous neoplasia: Phase I evaluation. Clin Cancer Res 11:4827–4834. [DOI] [PubMed] [Google Scholar]
- Theuer CP, FitzGerald DJ, Pastan I. 1993. A recombinant form of Pseudomonase exotoxin A containing transforming growth factor alpha near its carboxy terminus for the treatment of bladder cancer. J Urol 149:1626–1632. [DOI] [PubMed] [Google Scholar]
- Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, Freezer LJ, Morton KE, Seipp C, Haworth L, Mavroukakis S, White D, MacDonald S, Mao J, Sznol M, Rosenberg SA. 2002. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol 20:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SP, Hartin RJ, Ryu J‐I. 1989. Transformation in restriction‐deficient Salmonella typhimurium LT2. J Gen Microbiol 135:2561–2567. [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Kataoka A, Ohno S, Murakami S, Kinoshita J, Hachitanda Y. 2002. Prognostic and predictive value of epidermal growth factor receptor in recurrent breast cancer. Clin Cancer Res 8:3458–3460. [PubMed] [Google Scholar]
- Wang WL, Porter W, Burghardt R and, Safe SH. 1997. Mechanisms of inhibition of MDA‐MB‐468 breast cancer cell growth by 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin. Carcinogenesis 18:925–933. [DOI] [PubMed] [Google Scholar]
- Weldon JE, Pastan I. 2011. A guide to taming a toxin—Recombinant immunotoxins constructed from Pseudomonas exotoxin A for the treatment of cancer. FEBS J 278:4683–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodburn JR. 1999. The epidermal growth factor receptor and its inhibition in cancer therapy. Pharmacol Ther 82:241–250. [DOI] [PubMed] [Google Scholar]
- Wurch T, Lestienne F, Pauwels PJ. 1998. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol Tech 12:653–657. [Google Scholar]
- Xu Y‐H, Marciniak D, Rishi AK, Sakar FH, Kucuk O, and, Majumdar APN. 2005. Epidermal growth factor receptor (EGFR)‐related protein inhibits multiple members of the EGFR family in colon and breast cancer cells. Mol Cancer Ther 4:435–442. [DOI] [PubMed] [Google Scholar]


