Abstract
This study interrogates the antigen-specificity of inflammatory infiltrates in renal biopsies with BK polyomavirus (BKPyV) viremia (BKPyVM) with or without allograft nephropathy (BKPyVN). PBMC from 5 healthy HLA-A0101 subjects were stimulated by peptides derived from the BKPYV proteome or polymorphic regions of HLA. Next generation sequencing (NGS) of the T-cell receptor (TCR) cDNA was performed on peptide stimulated PBMC and 23 biopsies with T-cell mediated rejection (TCMR) or BKPyVN. Biopsies from patients with BKPyVM or BKVPyVN contained 7.7732 times more alloreactive than virus reactive clones. Biopsies with TCMR also contained BKPyV-specific clones, presumably a manifestation of heterologous immunity. The mean cumulative T-cell clonal frequency was 0.1378 for alloreactive clones and 0.0375 for BKPyV reactive clones. Samples with BKPyVN and TCMR clustered separately in dendrograms of V-family and J-gene utilization patterns. Dendrograms also revealed that V-gene, J-gene, and D-gene usage patterns were a function of HLA type. In conclusion, biopsies with BKPyVN contain abundant allospecific clones that exceed the number of virus reactive clones. The T-cell component of tissue injury in viral nephropathy appears to be mediated primarily by an ‘innocent bystander’ mechanism in which the principal element is secondary T-cell influx triggered by both anti-viral and anti-HLA immunity.
INTRODUCTION
Renal allograft biopsies with BK polyomavirus (BKPyV) viremia (BKPyVM) or allograft nephropathy (BKPyVN) typically show T-cell rich mononuclear infiltrates associated with tubular injury(1). The relative proportion of lymphocytes that target viral versus human leukocyte alloantigens (HLA) in these specimens is not known. This leads to a particularly challenging problem following development of T-cell mediated rejection (TCMR), which complicates the course of 5–30% of cases with BKPyV infection (2). Both TCMR and BKPyVN are characterized by inflammation and tubulitis that look similar on routine light microscopy (3). Further characterization of the antigen-specificity of the infiltrating T-cells may allow distinction between these two different types of inflammation. This may assist in (a) the differential diagnosis of graft dysfunction, (b) implementation of appropriate antiviral and/or anti-rejection treatment, and (c) assessing the prognosis in this patient population (4).
T-cells are activated and stimulated to proliferate when they recognize and bind non–self-peptides complexed to HLA on the cell surface of antigen presenting cells. This binding is determined primarily by the amino acid sequence of the hypervariable complementarity-determining region 3 (CDR3) of the T cell receptor (TCR). It follows that one can expect different patterns of TCR Vβ utilization in T-cells depending on whether they are reacting to donor-HLA antigens or viral antigens (5–7). Accordingly, we have explored T-cell repertoire analysis as a tool to define the antigen specificity of inflammatory infiltrates in renal allograft biopsies with TCMR, BKPyVM or BKPyVN.
Conventional techniques to define T-cell repertoire, namely Southern blotting, flow cytometry, and PCR can reliably detect only the most abundant clones in biologic samples, and do not adequately cover the genetic diversity of the TCR (8, 9). Typically, these tests are only informative about broad patterns of TCR Vβ usage. It is now well recognized that T-cell clones utilizing the same TCR Vβ gene and identical CDR3 lengths can differ in TCR sequence. Therefore, we have used next generation sequencing (NGS) as the primary investigational tool in this study (10). Many published studies have used RNA reverse transcribed to complimentary DNA (cDNA) as the starting material for NGS. RNA has the potential advantage of supplying more copies of template material transcribed from TCR genes. However, it has been reported that gDNA is more successful in the detection of rare virus specific clones which can play a very important role in the pathobiology of virus induced tissue injury (4). For this reason the present investigation utilized genomic DNA for NGS. The focus was on analysis of T-cell receptor sequences, since it was not the purpose of this work to address the issue of viral genomic diversity in the pathogenesis of BKPyVN.
MATERIALS AND METHODS
Recruitment of study subjects
Healthy subjects (n=5) and kidney transplant recipients (n= 18) were recruited into the study using protocols approved by the University of Pittsburgh IRB (IRB protocol # 060814 and #000586). Kidney transplant patients belonged to two categories: (a) HLA-A01 positive recipients of HLA-A02 positive donors (RA1DA2)(12 biopsies, 6 at the time of TCMR, and 6 at the time of BKPyVM) or (b) HLA-A02 positive recipients of HLA-A01 positive donors (RA2DA1) (11 biopsies, of which 8 had TCMR, and 3 were in the context of BKPyVM). Donor and recipient serology was not available.
Case definitions and clinical follow up
TCMR was defined using Banff 1997 criteria based on the presence of interstitial inflammation and tubulitis (11). PCR tests for BKPyV DNA in the urine and plasma using primers directed against the VP-1 gene as previously published (12). The diagnosis of BKPyVN depended on presence of viral cytopathic effect with confirmation by immunohistochemistry. Graft function was monitored by tracking monthly measurements of serum creatinine, and recording the time of any occurrences of graft loss (defined as graft nephrectomy or return to dialysis).
Flow cytometric measurement of peripheral blood mononuclear cells (PBMC) responses to HLA and BKPYV peptide stimulation in-vitro
Flow cytometry was used to confirm PBMC responses to HLA and BKPYV peptide stimulation in-vitro prior to DNA sequencing (13). For assessment of anti-HLA reactivity, two sets of 14–22 mer HPLC purified peptides (>95% purity) were synthesized from the polymorphic regions of each of the three most common human class I HLA alleles, namely HLA-A0101, HLA- A0201 and HLA-A0301 (Proimmune Limited, Oxford, United Kingdom). The actual amino acid sequences were as follows:
HLA-A1a : EGPEYWDQETRNMKAHSQTDRANLGTLRGYY
HLA-A1b: AQITKRKWEAVHAAEQRRVYLEGRCVDGLRR
HLA-A2a : EGPEYWDGETRKVKAHSQTHRVDLGTLRGYY
HLA-A2b: AQTTKHKWEAAHVAEQLRAYLEGTCVEWLRR
HLA-A3a: EGPEYWDQETRNVKAQSQTDRVDLGTLRGYY
HLA-A3b: AQITKRKWEAAHEAEQLRAYLDGTCVEWLRR
Viral antigen stimulation used a peptide mixture (JPT Technologies, Berlin, Germany) that contained 170 peptides (15 mer each, 11 amino acid overlap) spanning the entire 695 amino acid sequence of the largest BKPyV encoded protein, namely Large T antigen (LTA, SwissProt ID: P14999). This pepmix enables a global assessment of T-cell-mediated immunity directed against BKPyV independent of the subject’s HLA type (14).
Peptide reactivity was monitored by flow cytometric assessment of intra-cellular interferon-γ production in cells pre-treated with Brefeldin A and permeabilized with Perm/Wash buffer (13). Data was collected using a BD LSR II flow cytometer and analyzed with FlowJo software. Signals were collected only on live cells as identified by the Fixable Aqua Dead Cell Stain Kit. PBMC were designated as responsive to an antigen if a minimum of 10 events were captured after correction for background in negative control tubes.
NGS-multiplex assay for determination of T-cell TCR Vβ usage in PBMC and renal allograft biopsy tissue
These assays were run by a commercially available service (Adaptive Biotechnologies, Seattle, WA). Briefly, total genomic DNA (gDNA) was extracted from 5 million PBMC or 10 formalin fixed paraffin embedded tissue sections, each of 25 micron thickness, using the Qiagen DNeasy Blood and Tissue Kit (Cat # 69504). Multiplex PCR amplification of TCRβ CDR3 regions was performed with a DNA input of 3.6 μg for PBMC samples and 63 ng to 2.6 μg for the biopsy samples. A template library was generated using 45 TCR Vβ forward primers, and 13 TCR Jβ reverse primers. Genomic templates were amplified using equimolar pools of the forward and reverse primers and amplicons were submitted to NGS. This assay is sensitive enough to detect T-cell clones at a frequency of 0.001% (1 in 100,000)(4). The information obtained is quantitative and accurate within an overall factor of three over a dynamic range of 100,000 (15).
Data Analysis using ImmunoSEQ analyzer
Raw sequence data was preprocessed to remove technical errors and identify those sequences that had a minimum of a 6-nt match to one of the 45 Vβ gene segments and one of the 13 Jβ gene segments. A nearest neighbor algorithm was used to collapse the data into unique sequences. V- region, D-region and J-regions were identified by the ImMunoGeneTics IMGT/Junction Analysis tool (16). Rearranged CD3 sequences containing substitutions, insertions or deletions that resulted in frameshifts or premature stop codons were be regarded as nonproductive and excluded from analysis. The Illumina GA2 System sequence reads average 54 bp in length. Hence, sequences starting from the Jβ segment tag consistently captured the complete CDR3 region, which has an average nucleotide length 35 ± 3bp. The amplification bias of each V and J segment primer was corrected for using data derived from synthetic peptides. In each sample the total number of unique TCR β CDR3 sequences (unique T-cell clones or clonotypes) were exported as a CSV file to Microsoft Excel, ranked by abundance and expressed in terms of relative frequencies. Clonality index was calculated for all productive clones as the inverse of normalized Shannon’s entropy. This index varies from 0–1 with 1 corresponding to clonal proliferation. Differences in frequency of T-cell clonotypes directed against different HLA or viral peptides were be evaluated by the Student’s t-test using SPSS 22.0 (IBM; Armonk, New York). To study patterns of Vβ family and Jβ gene utilization in biopsy samples, dendrograms of TCR clonotype frequencies were derived by implementing the ‘hcclust’ complete linkage clustering algorithm in R version 3.1.2 (The R Foundation for Statistical Computing). Customized scripts were used to comparing large sequence sets in paired samples for the presence of complete or partial amino acid match.
RESULTS
Clinical and pathology parameters
Patients varied in age from 20–73 years (mean ± SD, 48.89 ± 15.32) with a male:female ratio of 3.7:1. The biopsies had been performed 97–7610 days post-transplant (1418.77 ± 1797.25).
There were 5 Banff type 1A and 1-Banff Type 1B rejections amongst 6 patients in the RA1DA2 groups. The 8 patients belonging to the RA2DA1 group experienced 7 rejection episodes graded as 1A and one as grade 1B. Amongst 9 patients with viremia 3/6 in RA1DA2 and 2/3 in RA2DA1 group satisfied histologic criteria of BKPyV nephropathy (stage B1, B2, B2 and C in 1/5, 1/5, 2/5 and 1/5 biopsies respectively). The remaining patients showed predominantly mononuclear inflammation and tubulitis but no viral cytopathic effect or antigens in the tissue available for analysis. In the setting of viremia, it was presumed that the biopsy had not sampled the site of active infection in the allograft. Therefore, these biopsies were assigned to the BKPYV infection group. In the rejection biopsies, the Banff scores for inflammation were as follows: i = 1.0 +/− 0.7, total i = 2.5 +/− 0.7, i-IFTA 2.0 +/− 0.8, and t = 2.2 +/− 0.9. These were not statistically different from the corresponding scores in patients with BKV infection: i = 1.0 +/− 0.4, total i = 2.1 +/− 0.9, i-IFTA 1.7 +/− 1.0. and t = 1.7 +/− 1.5. Two of nine patients with viremia/nephropathy had preceding but no follow up biopsies with TCMR. In 3 patients rejection episodes occurred for the first time following the diagnosis of BKPyV viremia or nephropathy. In the remaining patients rejection were documented both before and after the index biopsy,
The serum creatinine was higher in patients with BKPyVM or BKPyVN than rejection at all time points and the difference was statistically significant 3m, 6m,12m and 24m post-biopsy, and at last available follow up (2.57 ± 0.79 versus 0.82 mg/dl, 3.67 ± 2.43 versus 2.09 ± 0.54, 4.79 ± 2.99 versus 2.48 ± 1.16, 4.15 ± 3.58 versus 3.22 ± 3.51) respectively.
Flow cytometric assays to confirm antigenicity of HLA and BKPyV peptides
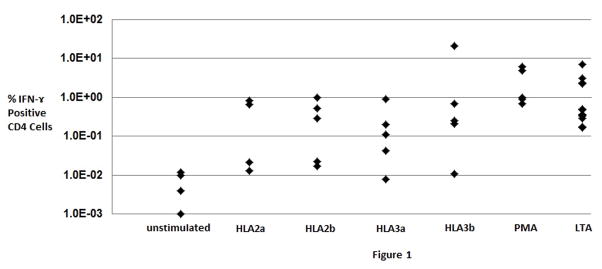
PBMC from 5 healthy BKPyV seropositive HLA-A0101 subjects were stimulated by HLA peptides 2a, 2b, 3a, and 3b with phorbol myristate acetate (PMA) as a positive control. For each peptide tested, at least 4/5 subjects tested gave signals that exceeded the unstimulated (no peptide) negative control (0.02 ± 0.07%, mean ± 2sd, n=5, Fig. 1). The frequency of circulating peptide responsive CD4 positive T-cells was 1.46 ± 4.11 % for BKPyV LTA pepmix, 0.38 ± 0.85% for peptide HLA2a, 0.37 ± 0.83% for peptide HLA2b, 0.26 ± 0.76 for peptide HLA3a, and 4.47± 18.71 for peptide HLA3b. A single highly responsive subject accounts for the high standard deviation associated with HLA3b stimulation. The specificity of these responses was shown in parallel experiments where responses to HIV peptides and actin were found to be up to 100 fold lower and equivalent to the background signals.
Fig. 1.
Response of healthy subject peripheral blood mononuclear cells to stimulation by HLA-A02, HLA-A03, or virus large T antigen (LTA) peptides as measured by flow cytometric assessment of % CD4 positive T-cells producing interferon-γ. CD8 positive T-cells reactive to viral and HLA peptides could not be consistently detected with the cell numbers analyzed. The polymorphic region of each HLA allele studied was represented by two peptides (sub-labeled a and b). Phorbol myristate acetate (PMA) was used as a positive control for stimulation, while unstimulated cells were used as a negative control to assess the background signal in the assay. Note the log scale on the Y-axis indicating considerable subject-to-subject differences in response. Each diamond represents a separate healthy subject (n= 4–5 for most conditions, n=12 for LTA peptides).
Antigen-specificity of TCR sequences in PBMC
A TCR DNA sequence does not by itself provide any information about the antigenic epitope being recognized by the corresponding TCR. In an attempt to identify at least a subset of the T-cell repertoire that is stimulated by HLA and BKPyV antigens, we performed NGS on PBMC from a HLA-A01 positive subject stimulated with HLA-A02a or LTA peptides. As a control for reactivity to autoantigens and various components of the tissue culture medium (particularly fetal calf serum derived proteins), HLA-A01a peptide stimulated PBMC were run in parallel, and the results used to correct data obtained after stimulation with allopeptides or BKPyV peptides. NGS on these samples yielded a total of 5,286,621 ± 2,406,354 (mean ± SD) sequences, of which 105,425 ± 39,833 were unique, and 86,452 ± 32,674 were productive (as well as unique). The clonality index of these sequences was 0.1589±0.0127. A total of 1346 T-cell clones expanded at least 5 fold, and 524 more than 10 fold in response to HLA-A02a peptide. By comparison, BKPyV LTA stimulation elicited 5 and 10 fold expansion in 619 and 230 TCR sequences respectively. The 5 most clonally expanding HLA-A02 reactive and BKPyV LTA reactive sequences that showed the greatest fold expansion are listed in Table 1. The clonal frequencies of these CDR3 sequences ranges from 5.02% to 29.9%.
Table 1.
Comparison of the Five Most Rapidly Expanding TCR Sequences in HLA-A01+ PBMC Stimulated by HLA-A02 or BKV LTA peptides.
| A02 Stimulation | LTA Stimulation | ||||
|---|---|---|---|---|---|
|
| |||||
| CDR3 sequence | % Clone Frequency | Fold Change | CDR3 sequence | % Clone Frequency | Fold Change |
| CASSSQGDRVQETQYF* | 0.0502 | 2006.5 | CASSSQGDRVQETQYF* | 0.0539 | 1235.0 |
| CASSSGTGDQPQHF | 0.2990 | 1849.1 | CSASQRQSSNQPQHF | 0.1869 | 711.6 |
| CASSLGLSGGQETQYF | 0.2066 | 1283.9 | CASSPTRYEQYF | 0.0796 | 524.9 |
| CASSPTWTGGPYNEQFF | 0.0775 | 688.7 | CASSPWGAHSGNTIYF | 0.1124 | 428.3 |
| CASSRTVSGANVLTF | 0.0566 | 451.2 | CSVDGPGQGAQYF | 0.1081 | 379.8 |
The most abundant sequence (first row) is common to both stimulation conditions and indicates that a single T-cell clone can potentially cross recognize HLA-A02 and BKV LTA antigens. The remaining sequences are unique and of potential utility in distinguishing between alloimmune and anti-viral immune responses.
Antigen-specificity of TCR sequences in renal allograft tissue
The amount of DNA extracted from ten tissue sections each 25 micron in thickness was 2409.08 ± 5515.39 ng (mean ± SD; range 97.75—26267.80 ng). The mean template inputs for DNA sequencing was 787.33 ± 612.34 ng (range 62.72—2666.56 ng). DNA sequencing yielded a total of 130,101 ± 164,436 sequences, of which 103,445 ± 139,030 were productive, and 2,582 ± 2,594 were unique. The clonal frequencies of the 5 most abundant HLA-A2 reactive clones found in one or more RA1DA2 biopsies with TCMR varied from 0.0011–0.0177. All these biopsies with TCMR had multiple BKPyV and HLA-A2 reactive clones with cumulative frequencies (mean, SD) of 0.0375 ± 0.0431 and 0.1378 ± 0.0682 respectively. The mean alloreactive to BKPYV reactive peptide ratio was 3.6711. By comparison, in the biopsies with BKPyVN/BKPyVM the clonal frequencies of 5 most abundant BKPYV reactive clones in one or more RA1DA2 BKPyVN biopsies varied from 0.0025–0.0824. The cumulative BKPyV reactive and alloreactive frequencies were 0.0387 ± 0.0520 and 0.3004 ± 0.2815 respectively, with mean alloreactive to BKPyV reactive peptide ratio of 7.7732 (Table 3). Differences between biopsies with rejection and BKPyVM or BKPyVN were not statistically significant although the number of patients analyzed is very small.
Table 3.
Cumulative BKV Reactive and Alloreactive T-cell Clonotype Frequencies in HLA-A01+ Patients Receiving Kidneys from HLA-A02+ Donors
| Rejection (n=6) | BKV Viremia/Nephropathy (n=6) | |
|---|---|---|
| Allo-reactive (HLA-A02) | 0.1378 ± 0.0682 * | 0.3004 ± 0.2815 |
| BKV Reactive | 0.0375 ± 0.0431 * | 0.0387 ± 0.0520 |
| Allo/BKV Ratio | 3.6711 | 7.7732 |
: p<0.05; Student’s t-test
Tracking of circulating T-cell clones in allograft tissue
HLA-A02 reactive T-cell clones identified in PBMC could be tracked in 6/6 biopsies with TCMR and 5/6 biopsies with BKPyVN. The number of clonotypes varied from 1–18, and was subject dependent. Each kidney transplant patient had a preference for specific clones, although three rejection biopsies had one or two shared TCR sequences. Likewise, BKPyV reactive T-cell clones identified in PBMC could be tracked in 3/6 biopsies each with TCMR or BKPyVN. The number of clonotypes varied from 1–7, and three rejection biopsies had one or two shared TCR sequences. None of the tracked clones showed sequence identity with clonotypes found in TCMR.
V-family and J-family Utilization patterns
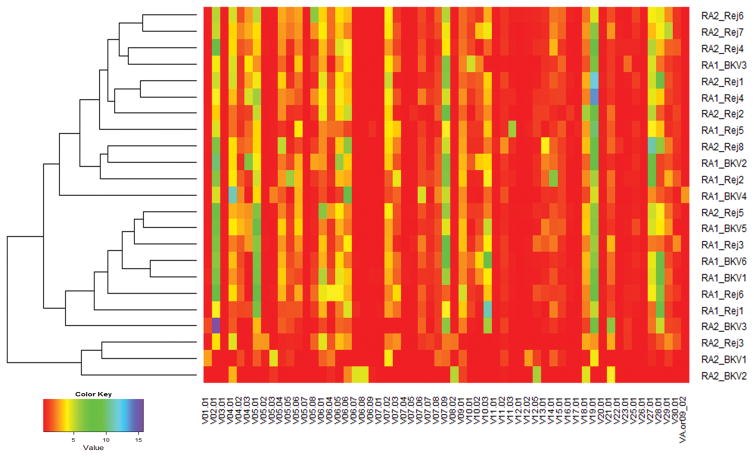
Analyses were performed to ascertain if histopathologic diagnoses are associated with specific V-family and J-family utilization. Amongst BKPyVN patients, RA1DA2 subjects showed greater utilization of select V-families (V-04, V-05. V-06, V-07, V-09 and V-27) and J-family genes (J01–04, J01–05, J02-01, 02–03 and 02–07) compared to RA2DA1 subjects (Tables 4 and 5). Thus, TCR β V-family and J-gene utilization in response to viral infection is HLA-dependent. The question was then asked if TCR β usage patterns can distinguish rejection from BKPyVN. No differences in V-family or J-gene usage were seen amongst RA1D02 patients. However, when RA2DA1 patients were analyzed, biopsies with BKPyVN showed lower utilization of TCRB J-family genes J01–04, J01–05, J02-01 and J02–07, compared to biopsies with TCMR. The same pattern was observed with respect to usage of TCRB V-families V-04, V-05, V-06, V-07, V-15, V-16, V-20, V-25 and V-27. Thus, TCR usage is modulated by recipient as well as donor HLA type. In a dendrogram based on the complete linkage algorithm, biopsies with BKPyVN and TCMR tended to cluster separately by virtue of their distinctive V-family usage (Figure 2).
Table 4.
TCR V-Family Usage Patterns Distinguish BKV Viremia or Nephropathy form TCMR
| TCRBV04 | TCRBV05 | TCRBV06 | TCRBV07 | TCRBV09 | TCRBV20 | TCRBV27 | |
|---|---|---|---|---|---|---|---|
| RA2DA1 BKV Viremia/Nephropathy | 1.21±0.62 * Δ | 3.20±2.42 * Δ | 4.94±2.76 * Δ | 3.74±3.10 * Δ | 1.01±0.37 Δ | 0.68±1.18 * | 0.52±0.65 * Δ |
| RA2DA1 Rejection | 4.56±1.25 * | 7.81±2.39 * | 12.06±2.29 * | 8.28±1.44 * | 1.16±0.52 * | 3.80±1.14 * | 3.31±2.02 * |
| RA1DA2 BKV Viremia/Nephropathy | 5.70±2.60 Δ | 8.38±1.88 Δ | 10.63±3.00 Δ | 9.47±1.28 Δ | 1.69±0.28 Δ | 1.84±0.81 | 3.30±1.45 Δ |
| RA1DA2 Rejection | 3.69±1.25 | 8.95±2.23 | 10.22±3.32 | 8.83±1.56 | 2.15±0.58 * | 2.56±1.23 | 2.79±1.73 |
: p<0.05 in the same v subfamily on comparison of BKV versus rejection in RA2DA1 biopsies; Student’s t-test
: p<0.05 in the same v subfamily on comparison of BKV versus rejection in RA2DA1 biopsies; Student’s t-test
Table 5.
J-Gene Utilization in Patients Classified by Diagnosis and HLA Type
| TCRBJ01–04 | TCRBJ01–05 | TCRBJ02-01 | TCRBJ02–03 | TCRBJ02–07 | |
|---|---|---|---|---|---|
| RA2DA1 BKV Viremia/Nephropathy | 0.89±1.53 * Δ | 2.04±1.81 * Δ | 2.26±1.53 * Δ | 1.95±3.04 Δ | 4.60±3.80 * Δ |
| RA2DA1 Rejection | 4.59±2.02 * | 4.79±1.37 * | 8.88±3.23 * | 7.29±0.92 | 10.71±2.22 * |
| RA1DA2 BKV Viremia/Nephropathy | 3.59±1.03 Δ | 6.15±1.78 Δ | 9.38±1.25 Δ | 6.19±1.74 Δ | 13.19±2.64 Δ |
| RA1DA2 Rejection | 3.96±2.69 | 5.23±1.18 | 9.94±2.80 | 8.14±3.15 | 13.56±3.18 |
: p<0.05 in the same J subfamily on comparison of BKV versus rejection in RA2DA1 biopsies; Student’s t-test
: p<0.05 in the same J subfamily on comparison of BKV versus rejection in RA2DA1 biopsies; Student’s t-test
Fig. 2.
Dendrogram of V-family Utilization in RA1DA2 & RA2DA1 Biopsies with BKPyVM/BKPyVN (BKV) or T-cell mediated rejection (Rej). The X-axis lists all the 59 V-families evaluated. The individual biopsies on the Y-axis are designated as RA1 if they represent an HLA-A01+ patient with an HLA-A02+ graft or RA2 if they represent an HLA-A02+ patient with an HLA-A01+ graft. V-family usage in each sample is color coded from least abundant (red) to most abundant (purple). It can be appreciated that biopsy samples with rejection tend to cluster at the top portion of the diagram (9 of the first 11 rows), while those with viral infection tend to cluster in the lower half (6 of the last 11 rows), although the separation is not perfect.
DISCUSSION
The primary goal of this study was to define the antigen specificity of T-cell infiltrates in renal allograft biopsies with BKPyVM/BKPyVN or TCMR. The approach taken was to use NGS and define the TCR repertoire in these specimens. The relationship between specific TCR sequences and the antigenic epitope being recognized was determined empirically by defining the T-cell clones that expanded in-vitro when PBMC were stimulated by viral or HLA peptides. These circulating T-cell clones were then tracked and quantitated in biopsy specimens. The average frequency of virus reactive T-cell clones in biopsies with viremia/nephropathy was 0.0387 which compared with a frequency of 0.3004 for alloreactive clones in the same tissue (Table 3). Thus, as one would expect, the presence of BKPyV infection does not interfere with the development of an immune response to the graft. Instead, both processes appear to proceed in parallel. Steroid therapy in these patients may dampen the inflammatory response directed at either alloantigens or viral antigens, but carries with it the risk of viral replication, which may result in a rebound of the viral load and worsening of viral nephropathy. Conversely, it is surprising that even biopsies with rejection had on average a frequency of 0.0375 for virus reactive cells. We speculate that this reflects heterologous cell mediated immunity directed at viral antigens, as has been noted by other investigators (17–20). Additionally, it is clear that even the combined total of BKPyV reactive and alloreactive cells accounts for only a small proportion of the total T-cell infiltrate in the biopsy. Thus, the T-cell component of tissue injury in viral nephropathy appears to be mediated primarily by an ‘innocent bystander’ mechanism in which the principal player is secondary T-cell influx, which amplifies initial tissue injury mediated anti-viral and anti-HLA T-cell clones, antibodies directed at viral antigens, innate immune effector mechanisms, and viral cytopathic effect. This study is not able to investigate the contribution of granulocytes as this method is targeting T-cells only. The role of innate immunity in controlling viral infection is not yet completely understood.
NGS also demonstrated that BKPyVN and TCMR are characterized by diagnosis and subject-specific TCR β utilization patterns. Thus, in biopsies from RA2 DA1 subjects, BKPyVN was associated with lower utilization of genes J01–04, J01–05, J02-01, J02–07, V-04, V-05, V-06, V-07, V-15, V-16, V-20, V-25 and V-27 compared to biopsies with TCMR. These differences were not seen in RA1DA2 subjects. TCR-BV06 usage was associated with alloreactivity while TCRBV18 and TCRBV27 were associated with T-cell response to BKPyV infection. Samples with BKPyVM/BKPyVN and TCMR clustered separately in dendrograms. Thus, TCR β usage patterns can potentially help determine the relative contribution of alloimmune mechanism and anti-viral immunity in the pathogenesis of tissue injury.
It was further noted that individual CDR3 sequences in T-cell clones sensitized to HLA-antigens were often distinct from those seen in BKPyV sensitized T-cells (Table 2). Quantitation of these individual sequences could be potentially used to serially monitor anti-viral immunity or to detect evolving rejection in a subclinical phase. Implementing such an approach would require subject-specific identification of clones directed at defined target epitopes. This could be accomplished by routinely obtaining a baseline PBMC sample from all patients early post-transplant, and performing in-vitro peptide stimulation followed by NGS. Subsequently, when the patient undergoes a biopsy for graft dysfunction in the setting of BKPyV infection or rejection, the frequency of virus and HLA directed T-cell clones could be determined in kidney tissue. The same clones could also be tracked again in PBMC. In one study, circulating alloreactive T-cells correlated with graft function in longstanding renal transplant recipients (7). However, it is also known that T-cell clonal frequencies in the peripheral blood can differ substantially from those in specific organs (21, 22). Clonal amplification thresholds corresponding to specific biopsy diagnoses such as TCMR or BKPyVN will have to be established in a prospective clinical trial.
Table 2.
Comparison of the Five Most Abundant TCR Sequences in BKVN Renal Alloreactive Biopsies with BKV Viremia or Nephropathy from HLA-A01 + Kidney Transplant Patients who Received HLA-A02+ Donor Kidneys.
| HLA-A02 Reactive T-cell Clones in BKV Viremia/Nephropathy | LTA-Reactive T-cell Clones in BKV Viremia/Nephropathy | ||||
|---|---|---|---|---|---|
|
| |||||
| CDR3 sequence | % Clone Frequency | Fold Change | CDR3 sequence | % Clone Frequency | Fold Change |
| CASSSPSGANVLTF | 0.0046 | 52.7 | CASSLGGELFF | 0.0824 | 118.9 |
| CASSLEGAGNTIYF | 0.0177 | 36.2 | CASSLGGYTEAFF | 0.0297 | 24.7 |
| CASSGTDTQYF | 0.0012 | 33.0 | CASSPDRNTEAFF | 0.0030 | 15.1 |
| CASSEQMNTEAFF | 0.0064 | 28.2 | CASSLRGSSYEQYF | 0.0025 | 7.7 |
| CASSFTGNQETQYF | 0.0011 | 28.0 | CASSLGWEQYF | 0.0019 | 6.0 |
In conclusion, this study has shown that biopsies from patients with BKPyV viremia/nephropathy contain abundant alloreactive T-cell clones that exceed the number of virus reactive clones. This observation highlights the complex pathogenesis of tissue injury in viral nephropathy, and provides a mechanistic basis for the increased risk of acute rejection as these patients. Our data also confirms and extends a prior report which suggested that NGS in formalin fixed allograft biopsies can assist in the differential diagnosis of graft dysfunction, and thereby enhance the clinical care of kidney transplant patients (4).
Our study has limitations that need to be taken into account while designing future investigations. Firstly, the analysis is restricted to the TCR α-β chains and not all known TCR genes, HLA antigens or viral epitopes were evaluated. Secondly, in-vitro peptide stimulations were performed in the context of antigen presentation by autologous cells, which corresponds to the indirect antigen presentation pathway. Determination of the complete T-cell repertoire also needs attention to the direct antigen presentation, which is particularly important in the first few weeks of transplantation. Third, we have made no attempt to phenotype the T-cell clones that expanded following antigenic stimulation. Finally, analysis of small biopsy samples also has inherent limitations that stem from the large size of the T-cell repertoire, which is estimated to be of the order of 1016 with 107 unique TCRs in the periphery.
Acknowledgments
The research was supported by NIH grant R21 AI117644, The Thomas E Starzl Transplantation Institute, Stephen A Hamill Charitable Fund, and The Robert Weis Family Foundation, University of Pittsburgh.
ABBREVIATIONS
- BKPyV
BK polyomavirus
- BKPyVM
BK polyomavirus viremia
- BKPyVN
BK polyomavirus nephropathy
- NGS
Next generation sequencing
- TCR
T-cell receptor
- TCMR
T-cell mediated rejection
- HLA
leukocyte alloantigens
- cDNA
complimentary DNA
- CDR3
complementarity-determining region 3
Footnotes
DISCLOSURE: The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.Hirsch HH, Randhawa P. BK polyomavirus in solid organ transplantation. Am J Transplant. 2013;13(Suppl 4):179–88. doi: 10.1111/ajt.12110. [DOI] [PubMed] [Google Scholar]
- 2.Schaub S, Hirsch HH, Dickenmann M, Steiger J, Mihatsch MJ, Hopfer H, et al. Reducing immunosuppression preserves allograft function in presumptive and definitive polyomavirus-associated nephropathy. Am J Transplant. 2010;10(12):2615–23. doi: 10.1111/j.1600-6143.2010.03310.x. [DOI] [PubMed] [Google Scholar]
- 3.Menter T, Mayr M, Schaub S, Mihatsch MJ, Hirsch HH, Hopfer H. Pathology of resolving polyomavirus-associated nephropathy. Am J Transplant. 2013;13(6):1474–83. doi: 10.1111/ajt.12218. [DOI] [PubMed] [Google Scholar]
- 4.Dziubianau M, Hecht J, Kuchenbecker L, Sattler A, Stervbo U, Rodelsperger C, et al. TCR repertoire analysis by next generation sequencing allows complex differential diagnosis of T cell-related pathology. Am J Transplant. 2013;13(11):2842–54. doi: 10.1111/ajt.12431. [DOI] [PubMed] [Google Scholar]
- 5.Naesens M, Sarwal MM. Harnessing the diversity of the human T-cell repertoire: a monitoring tool for transplantation tolerance? Eur J Immunol. 2010;40(11):2986–9. doi: 10.1002/eji.201041047. [DOI] [PubMed] [Google Scholar]
- 6.Morris H, DeWolf S, Robins H, Sprangers B, LoCascio SA, Shonts BA, et al. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015;7(272):272ra10. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miqueu P, Degauque N, Guillet M, Giral M, Ruiz C, Pallier A, et al. Analysis of the peripheral T-cell repertoire in kidney transplant patients. Eur J Immunol. 2010;40(11):3280–90. doi: 10.1002/eji.201040301. [DOI] [PubMed] [Google Scholar]
- 8.Ciupe SM, Devlin BH, Markert ML, Kepler TB. Quantification of total T-cell receptor diversity by flow cytometry and spectratyping. BMC Immunol. 2013;14:35. doi: 10.1186/1471-2172-14-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei S, Concannon P. Identification of a novel human T-cell receptor V beta subfamily by genomic cloning. Hum Immunol. 1994;41(3):201–6. doi: 10.1016/0198-8859(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 10.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alpha beta T cells. Blood. 2009;114(19):4099–107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55(2):713–23. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 12.Randhawa P, Ho A, Shapiro R, Vats A, Swalsky P, Finkelstein S, et al. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J Clin Microbiol. 2004;42(3):1176–80. doi: 10.1128/JCM.42.3.1176-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramaswami B, Popescu I, Macedo C, Luo C, Shapiro R, Metes D, et al. The polyomavirus BK large T-antigen-derived peptide elicits an HLA-DR promiscuous and polyfunctional CD4+ T-cell response. Clin Vaccine Immunol. 2011;18(5):815–24. doi: 10.1128/CVI.00487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer MH, Brestrich G, Andree H, Engelmann E, Rosenberger C, Tillmann H, et al. HLA type-independent method to monitor polyoma BK virus-specific CD4(+) and CD8(+) T-cell immunity. Am J Transplant. 2006;6:625–31. doi: 10.1111/j.1600-6143.2005.01221.x. [DOI] [PubMed] [Google Scholar]
- 15.Robins H, Desmarais C, Matthis J, Livingston R, Andriesen J, Reijonen H, et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods. 2012;375(1–2):14–9. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giudicelli V, Chaume D, Lefranc MP. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic acids research. 2004;32(Web Server issue):W435–40. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amir AL, D’Orsogna LJ, Roelen DL, van Loenen MM, Hagedoorn RS, de Boer R, et al. Allo-HLA reactivity of virus-specific memory T cells is common. Blood. 2010;115(15):3146–57. doi: 10.1182/blood-2009-07-234906. [DOI] [PubMed] [Google Scholar]
- 18.Adams AB, Williams MA, Jones TR, Shirasugi N, Durham MM, Kaech SM, et al. Heterologous immunity provides a potent barrier to transplantation tolerance. The Journal of clinical investigation. 2003;111(12):1887–95. doi: 10.1172/JCI17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie S, Lin SJ, Kim SK, Welsh RM, Selin LK. Pathological features of heterologous immunity are regulated by the private specificities of the immune repertoire. Am J Pathol. 2010;176(5):2107–12. doi: 10.2353/ajpath.2010.090656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford ML, Larsen CP. Overcoming the memory barrier in tolerance induction: molecular mimicry and functional heterogeneity among pathogen-specific T-cell populations. Curr Opin Organ Transplant. 2010;15(4):405–10. doi: 10.1097/MOT.0b013e32833b7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HS, Kim BS. Predominant clonal accumulation of CD8+ T cells with moderate avidity in the central nervous systems of Theiler’s virus-infected C57BL/6 mice. J Virol. 2010;84(6):2774–86. doi: 10.1128/JVI.01948-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramalingam RK, Meyer-Olson D, Shoukry NH, Bowen DG, Walker CM, Kalams SA. Kinetic analysis by real-time PCR of hepatitis C virus (HCV)-specific T cells in peripheral blood and liver after challenge with HCV. J Virol. 2008;82(21):10487–92. doi: 10.1128/JVI.00588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]