Abstract
Adenoid cystic carcinoma of the breast is a rare histologic type of triple-negative breast cancer with an indolent clinical behavior, often driven by the MYB-NFIB fusion gene. Here we sought to define the repertoire of somatic genetic alterations in two adenoid cystic carcinomas associated with high-grade triple-negative breast cancer. The different components of each case were subjected to copy number profiling and massively parallel sequencing targeting all exons and selected regulatory and intronic regions of 488 genes. Reverse transcription PCR and fluorescence in situ hybridization were employed to investigate the presence of the MYB-NFIB translocation. The MYB-NFIB fusion gene was detected in both adenoid cystic carcinomas and their associated high-grade triple-negative breast cancer components. Whilst the distinct components of both cases displayed similar patterns of gene copy number alterations, massively parallel sequencing analysis revealed intra-tumor genetic heterogeneity. In case 1, progression from the trabecular adenoid cystic carcinoma to the high-grade triple-negative breast cancer was found to involve clonal shifts with enrichment of mutations affecting EP300, NOTCH1, ERBB2 and FGFR1 in the high-grade triple-negative breast cancer. In case 2, a clonal KMT2C mutation was present in the cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and high-grade triple-negative breast cancer components, whereas a mutation affecting MYB was present only in the solid and high-grade triple-negative breast cancer areas and additional three mutations targeting STAG2, KDM6A and CDK12 were restricted to the high-grade triple-negative breast cancer. In conclusion, adenoid cystic carcinomas of the breast with high-grade transformation are underpinned by MYB-NFIB fusion gene, and, akin to other forms of cancer, may be constituted by a mosaic of cancer cell clones at diagnosis. The progression from adenoid cystic carcinoma to high-grade triple-negative breast cancer of no special type may involve the selection of neoplastic clones and/ or the acquisition of additional genetic alterations.
Keywords: Adenoid cystic carcinoma, high-grade, triple-negative, breast cancer, tumor progression
INTRODUCTION
Adenoid cystic carcinoma of the breast is a rare special histologic type of breast cancer, accounting for <1% of all cases of invasive disease.1,2 Adenoid cystic carcinomas are characterized by a dual population of neoplastic epithelial and myoepithelial cells, arranged in cribriform, tubular, trabecular or solid patterns.1,3 Like other salivary gland-like tumors occurring in the breast, breast adenoid cystic carcinomas almost invariably display a triple-negative phenotype (i.e. lack of estrogen receptor (ER), progesterone receptor (PR), and HER2 expression). At variance with common forms of triple-negative breast cancer, adenoid cystic carcinomas of the breast generally have an indolent clinical course with a 10-year overall survival rate of about 90%.1,3–5
At the genetic level, adenoid cystic carcinomas are characterized by the t(6;9)(q22–23;p23–24) translocation, resulting in the MYB-NFIB gene fusion,6,7 which is considered an early event in the tumorigenesis of these lesions.7–9 More recently, rearrangements affecting MYBL1 have also been documented in a subset of MYB-NFIB-negative adenoid cystic carcinomas.10,11 In contrast to common forms of triple-negative breast cancer, which are characterized by complex genomes and high mutation rates, breast adenoid cystic carcinomas have been shown to have rather simple genomes with low levels of genetic instability and low mutation rates.7,12,13,14–16 In fact, adenoid cystic carcinoma lack somatic mutations found in common types of triple-negative breast cancer, including TP53 and PIK3CA,16 but display a heterogeneous constellation of non-synonymous somatic mutations affecting cancer-related genes that converge into several functional categories, including chromatin remodeling and cell adhesion.12,13
High-grade transformation of salivary gland adenoid cystic carcinomas has been reported;17,18 this phenomenon, however, has also been documented in breast adenoid cystic carcinomas,19–21 where it presents as adenoid cystic carcinomas with solid architectural patterns and areas with histologic features consistent with those of high-grade triple-negative breast cancers of no special type. The genetic events associated with the progression from conventional breast adenoid cystic carcinoma to high-grade triple-negative breast cancer of no special type remain to be elucidated. The aim of this study was to define whether the high-grade triple-negative breast cancers associated with breast adenoid cystic carcinomas are clonally related and to investigate the pattern of somatic genetic alterations of breast adenoid cystic carcinomas showing progression to high-grade triple-negative breast cancers.
PATIENTS AND METHODS
Clinicopathologic characteristics
Two patients with diagnosis of adenoid cystic carcinoma of the breast with heterogeneous morphology and associated with areas of high-grade triple-negative breast cancer were included in the study. Patient 1 (AdCC1) was a 40-year-old woman with a nodule of 1.9 cm in the upper quadrants of the right breast. No other relevant clinical conditions or family history were documented. An excisional biopsy was performed at the Indiana University Hospital, IN, USA, and a diagnosis of trabecular adenoid cystic carcinoma with a solid component was rendered. Unequivocal areas of high-grade triple-negative breast cancer were not observed. A total mastectomy with sentinel lymph node excision was performed. Histologic examination revealed a multifocal high-grade triple-negative breast cancer of no special type, and no additional areas diagnostic of adenoid cystic carcinoma were present. All sentinel lymph nodes were negative. Immunohistochemical analysis revealed that all lesions displayed a triple-negative phenotype. After 58 months of follow-up, the patient is alive and free of disease.
Case 2 (AdCC2) was a 36 year-old woman who underwent a wide local excision at the Kato Breast Surgery Clinic, Japan, for a 3.5 cm nodule of the upper quadrant of the right breast. Histologic examination revealed a triple-negative breast cancer consistent with adenoid cystic carcinoma but the tumor showed high-grade triple-negative breast cancer areas. All sentinel lymph nodes were negative. Since the lesion was focally present on one surgical margin, the patient underwent adjuvant radiotherapy. A combination of paclitaxel and doxifluridine was also delivered. After 8 years of follow-up, the patient is alive and free of disease.
Samples, histopathologic and immunohistochemical analyses
All diagnostic blocks and slides of both breast adenoid cystic carcinomas were centrally reviewed by four pathologists (NF, EG-R, IOE & JSR-F) and morphologically distinct areas of each tumor were identified. Approval was obtained from the Institutional Review Boards (IRB) of the authors’ institutions, and written consent was obtained according to the IRB-approved protocols. Four µm-thick sections from representative formalin-fixed paraffin-embedded (FFPE) tissues of both cases were subjected to immunohistochemical analysis using antibodies against ER, PR, Ki67, HER2, cytokeratin 7, p63, c-KIT and MYB, following previously described protocols (Supplementary Table 1).22,23 Positive and negative controls were included in each slide run. All immunohistochemical stains were independently analyzed by four of the authors (NF, FCG, EG-R & JSR-F) and the immunohistochemical characteristics of each morphologically distinct area of the tumors were evaluated. ER, PR, and HER2 status were assessed following the American Society of Clinical Oncology (ASCO)/ College of American Pathologists (CAP) guidelines.24,25 The Ki67 index was assessed according to the recommendations of the International Ki67 in Breast Cancer working group.26 For cytokeratin 7 and p63 the percentage of tumor cells within a given component was recorded, while for c-Kit the immunoreactive score (IRS) was calculated according to Remmele and Stegner.27 Discordant results were resolved on a multi-headed microscope.
Fluorescence in situ hybridization (FISH)
FISH was performed on 4 µm-thick FFPE sections using a three-color probe mix consisting of bacterial artificial chromosomes (BACs) for 5′ MYB (RP11-614H6, RP11-104D9; green), 3′ MYB (RP11-323N12, RP11-1060C14; orange) and 3′ NFIB (RP11-413D24, RP11-589C16; red) using validated protocols at the Memorial Sloan Kettering Cancer Center (MSKCC) Molecular Cytogenetics Core as previously described.23,28 For analysis, at least 50 non-overlapping, interphase nuclei of morphologically unequivocal neoplastic cells were analyzed, and components with ≥15% of cells displaying at least one 5’MYB-3’NFIB fusion signal were considered fusion-positive.23
Microdissection and DNA extraction
Eight µm-thick sections of representative FFPE blocks of tumor and normal breast tissue from each case were stained with nuclear fast red. The morphologically distinct components of each case (i.e. AdCC1, trabecular adenoid cystic carcinoma and high-grade triple-negative breast cancer; AdCC2, cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and high-grade triple-negative breast cancer) were microdissected separately with a sterile needle under a stereomicroscope (Olympus SZ61) to ensure >80% of tumor cell content. Furthermore, the normal tissue was microdissected to be devoid of any neoplastic cells as previously described.29,30 Genomic DNA from each tumor component and matched normal tissue was extracted using the DNeasy Blood & Tissue Kit (Qiagen), according to the manufacturer’s instructions, and quantified using the Qubit 2.0 Fluorometer (Invitrogen, Life Technologies).
Reverse transcription PCR
The MYB-NFIB fusion transcript variants, including MYB exon 14 fused to NFIB exons 8 or 9, were defined in each tumor component by reverse transcription PCR (RT-PCR) as previously described.13,23 Total RNA was extracted from the different components of each case from FFPE sections using the RNeasy FFPE Kit (Qiagen) and reverse transcribed (SuperScript III Reverse Transcriptase; Invitrogen), and PCRs performed to detect specific fusion transcripts as previously described (primers see Supplementary Table 2).13,31 A breast adenoid cystic carcinoma known to harbor a MYB-NFIB fusion gene was employed as a positive control.7 All experiments were performed in duplicate.
Targeted massively parallel sequencing and amplicon re-sequencing
Tumor and normal DNA samples were subjected to targeted capture massively parallel sequencing at the MSKCC Integrated Genomics Operation, using the MSK Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) sequencing assay targeting all exons and selected introns of 341 key cancer genes,32,33 as well as a sequencing assay targeting all exons of 254 genes recurrently mutated in breast cancer and related to DNA repair34 (Supplementary Table 3). Of the 595 genes captured, 107 genes were common to both targeted capture sequencing assays (i.e. 488 unique genes; Supplementary Table 3). Targeted sequencing on an Illumina HiSeq2500 was performed as previously described.32,34 Reads were aligned to the reference human genome GRCh37 using the Burrows-Wheeler Aligner,35 and local realignment, duplicate removal and base quality recalibration were performed using the Genome Analysis Toolkit36 and picard (http://broadinstitute.github.io/picard/). Somatic single nucleotide variants were detected by MuTect,37 small insertion and deletions by Varscan 2 and Strelka,38,39 and further curated by manual inspection. Single nucleotide variants and small insertion and deletions located outside of the target regions, with mutant allelic fraction of <1% and/or supported by <5 reads were disregarded.13 We further excluded single nucleotide variants and small insertion and deletions for which the tumor mutant allelic fraction was <5 times that of the matched normal mutant allelic fraction, as well as single nucleotide variants and small insertion and deletions found at >5% global minor allele frequency of dbSNP (build 137).13
The 107 genes common to both targeted capture sequencing assays were used for cross-validation. In addition, selected mutations identified by target capture sequencing present in only one sequencing assay were validated independently in each of the distinct morphological components using amplicon re-sequencing (for primers, see Supplementary Table 4) on an Illumina MiSeq using a 150×150 chemistry in the MSKCC Integrated Genomics Operation as previously described.13 The depth of coverage was 5,230×–32,321×. The overall validation rate of 28 non-synonymous somatic mutations assessed was 96.4% (27/28); a MYB mutation found to be absent by targeted capture massively parallel sequencing in one sample (AdCC2, solid component) was identified to be present by high-depth amplicon re-sequencing (Supplementary Table 5). Only validated mutations were employed for subsequent analyses.
The potential functional effect of each single nucleotide variant was defined using a combination of MutationTaster40 and CHASM41 as previously described.42 Genes affected by non-passenger mutations were further annotated according to their presence in three cancer gene datasets, Kandoth et al.,43 the Cancer Gene Census44 and Lawrence et al.45
Sequencing data have been deposited to the NCBI Sequence Read Archive under accession SRP068515.
Copy number analysis
For whole genome copy number analysis, DNA from each component of each case was hybridized to OncoScan FFPE v3 arrays (Affymetrix) separately as previously described.46 Raw data files were loaded into Nexus Express for OncoScan analysis software (BioDiscovery) and analyzed using ASCAT47 as implemented in the Nexus Express for OncoScan software. Regions of copy number gains/losses, amplifications and homozygous deletions were generated based on the ploidy and purity adjusted modal copy numbers from ABSOLUTE (v1.0.6),48 where segments of modal copy number 0 were considered homozygously deleted, modal copy number >0 and ≤ ploidy−1 considered lost, modal copy number ≥ ploidy considered gained and modal copy number ≥ ploidy considered amplified. For each case, dimension reduction was performed using the BioConductor package CGHregions49 to generate a matrix of collapsed copy number regions. OncoScan array data have been deposited to the NCBI Gene Expression Omnibus under the accession GSE79052.
Cancer cell fraction
The cancer cell fraction of each validated mutation was inferred using the number of reads supporting the reference and the alternate alleles, and the segmented Log ratios from OncoScan arrays as input for ABSOLUTE (v1.0.6).48 Solutions from ABSOLUTE were manually reviewed as recommended.48,50 A mutation was classified as clonal if its probability of being clonal was >50%50 or if the lower bound of the 95% confidence interval of its cancer cell fraction was >90%;29 mutations that did not meet the above criteria were considered subclonal.
Phylogenetic tree construction
Phylogenetic tree construction was performed using somatic mutations and copy number alterations. For somatic mutations, binary presence/absence matrices based on the non-synonymous and synonymous somatic mutations were constructed. For copy number alterations, the matrix of collapsed copy number regions was used. Maximum parsimony trees for the two cases were constructed as previously described.51 A starting tree was constructed using the Neighbor-joining method and Hamming distance and optimized using the parsimony ratchet method52 implemented in the R package Phangorn.53 Trees were rooted at the hypothetical germline where all somatic genetic alterations are absent. Branch lengths were determined according to the ACCTRAN criterion as implemented in the Phangorn package53 and are drawn to scale.
RESULTS
Histologic and immunohistochemical features of adenoid cystic carcinoma and associated high-grade triple-negative breast cancer of no special type
Based on histology and immunohistochemical findings, both cases were classified as bona fide adenoid cystic carcinomas (Figures 1 and 2, Table 1). In addition to areas unequivocally diagnostic of adenoid cystic carcinoma, both cases displayed a minor component of high-grade triple-negative breast cancer of no special type. Case 1 (AdCC1) was an adenoid cystic carcinoma with infiltrative borders characterized by a predominantly trabecular proliferation of a dual population of neoplastic cells: the more abundant small cells with angular dark nucleus and scant cytoplasm, and cuboidal-to-polyhedral cells with more abundant cytoplasm, which often lined the duct-like structures. Both cell types displayed relatively bland and uniform nuclei, devoid of conspicuous pleomorphism. Mitotic figures were infrequent (1 and 6 per 10 high power fields (0.238mm2) in the adenoid cystic carcinoma and high-grade triple-negative breast cancer components, respectively). At the periphery of the lesion, a component of high-grade triple-negative breast cancer of no special type comprising approximately 20% of the lesion was identified (Figure 1). This area was composed of larger, more atypical cells with larger nuclei, more conspicuous nucleoli and scant cytoplasm. The high-grade triple-negative breast cancer displayed a higher proportion of cells displaying an epithelial rather than myoepithelial immunohistochemical profile (Figure 1, Table 1). Consistent with this observation, a decreased cell population with myoepithelial differentiation has been consistently described in high-grade breast adenoid cystic carcinomas and transformed adenoid cystic carcinoma of the salivary glands.17,54
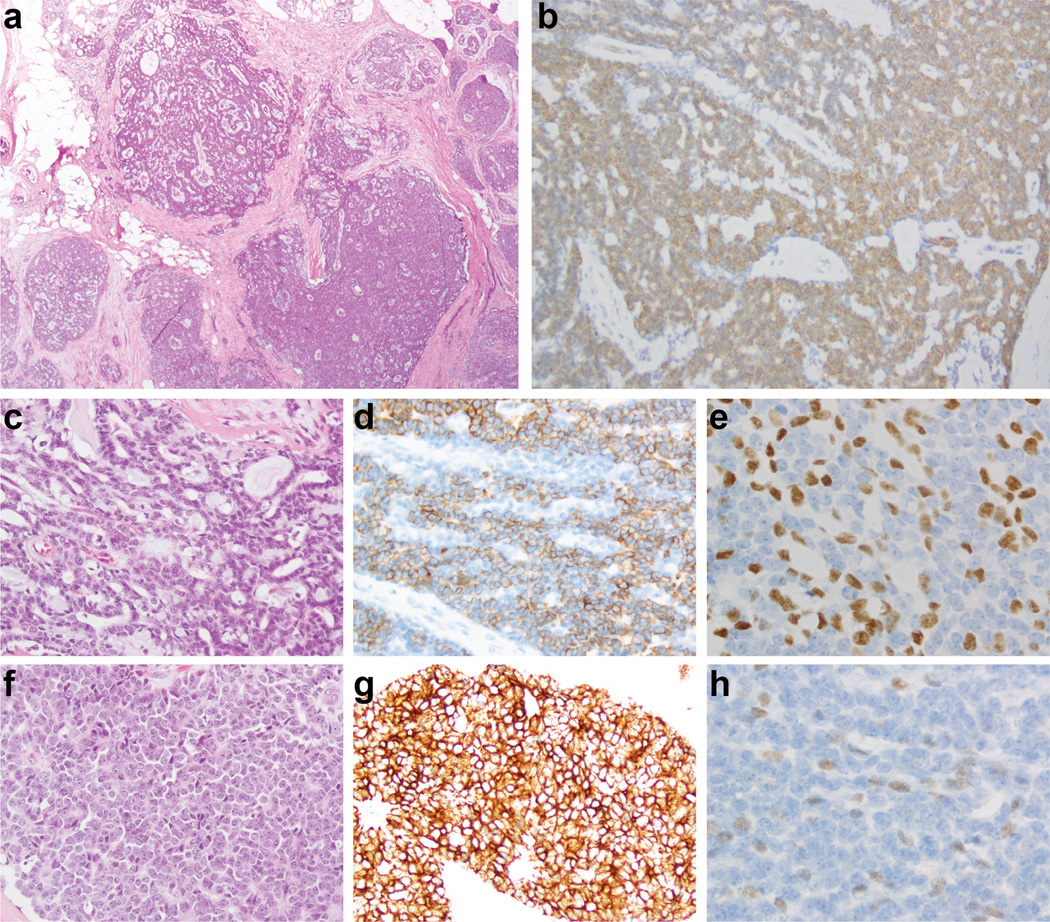
Figure 1. Histologic and immunohistochemical features of an adenoid cystic carcinoma with high-grade transformation (case AdCC1).
Low-power view of an adenoid cystic carcinoma (a) with diffuse c-Kit immunoreactivity (b), which was composed of a predominant trabecular component (c) and a minor component of high-grade triple-negative breast cancer of no special type (f). Cytokeratin 7 (d, g) and p63 (e, h) expression highlights the populations with epithelial and myoepithelial phenotype in the trabecular and high-grade components, respectively.
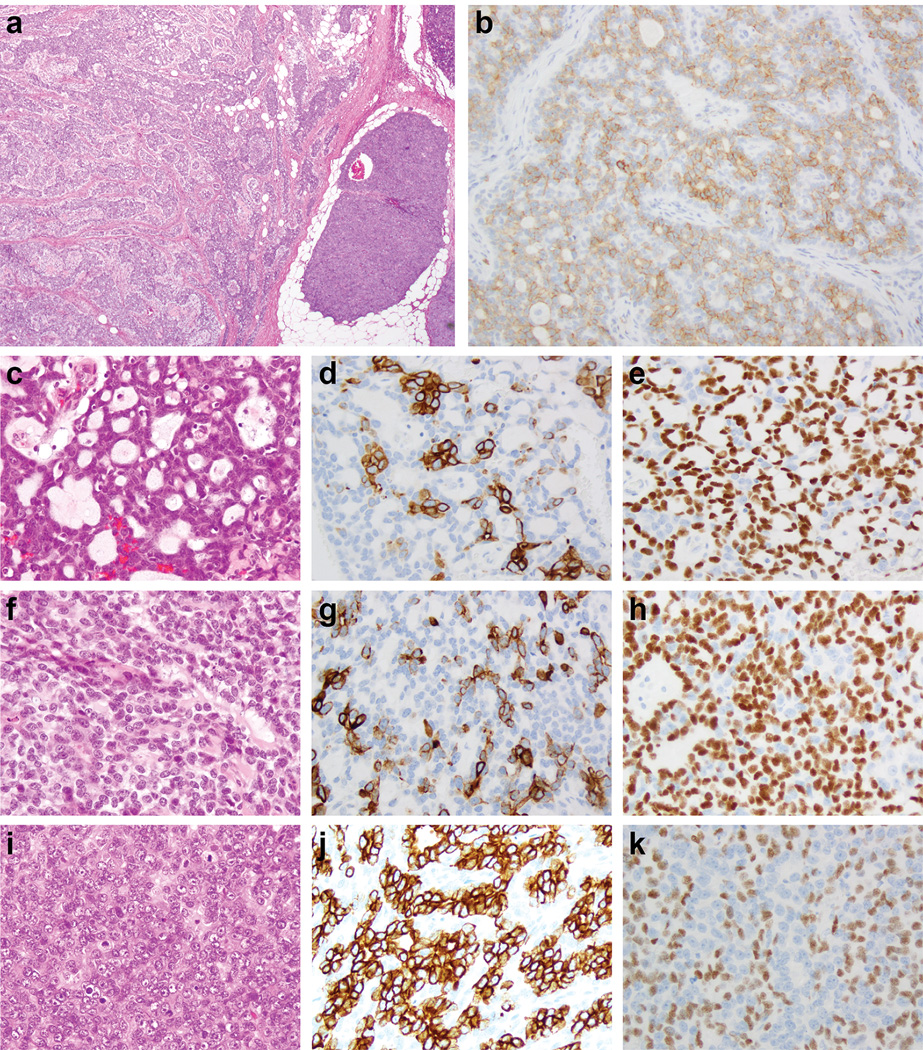
Figure 2. Histologic and immunohistochemical features of an adenoid cystic carcinoma with high-grade transformation (case AdCC2).
Low-power view of an adenoid cystic carcinoma (a) with diffuse c-Kit immunoreactivity (b), which was composed of three distinct components, including a cribriform (c), a solid (f) and a minor component high-grade triple-negative breast cancer of no special type (i). Cytokeratin 7 (d, g, j) and p63 (e, h, k) expression highlights the populations with epithelial and myoepithelial phenotype, respectively.
Table 1.
Immunohistochemical features of the adenoid cystic carcinomas with high-grade transformation included in this study.
| AdCC1 | AdCC2 | ||||
|---|---|---|---|---|---|
| Component/Marker | Trabecular adenoid cystic carcinoma |
High-grade triple- negative breast cancer |
Cribriform adenoid cystic carcinoma |
Solid adenoid cystic carcinoma |
High-grade triple- negative breast cancer |
| Estrogen receptor | negative | negative | negative | negative | negative |
| Progesterone receptor | negative | negative | negative | negative | negative |
| HER2 | negative | negative | negative | negative | negative |
| Ki67 | Not performed | Not performed | 19% | 25% | 45% |
| Cytokeratin 7 | 75% | 90% | 25% | 40% | 45% |
| p63 | 40% | 20% | 60% | 50% | 35% |
| c-Kit (immunoreactive score) | 100 | 120 | 42 | 50 | 60 |
Case 2 (AdCC2) was composed of a single mass with infiltrative borders, encompassing a dual population of cells similar to those found in AdCC1, but arranged in two distinct histologic patterns, solid and cribriform. The solid component comprised approximately 70% of the entire lesion, while the cribriform component accounted for approximately 20%. In both components, a dual population of cells similar to that described in AdCC1 was observed. In continuity with the solid component, an area composed of cells with more abundant cytoplasm, and larger, more pleomorphic nuclei with conspicuous nucleoli was observed. This high-grade area, where the neoplastic cells were arranged in solid sheets and abortive duct-like structures, was classified as high-grade triple-negative breast cancer of no special type (Figure 2, Table 1). Akin to AdCC1, an increased ratio of cells displaying epithelial rather than myoepithelial phenotype was observed in the solid and high-grade triple-negative breast cancer components (Figure 2, Table 1). As expected, Ki67 levels were higher in the high-grade triple-negative breast cancer component as compared to the cribriform and solid components (Table 1).
Adenoid cystic carcinoma and associated high-grade triple-negative breast cancer of no special type harbor the MYB-NFIB fusion gene
To define whether the high-grade triple-negative breast cancer components of AdCC1 and AdCC2 would be related to the bona fide areas of adenoid cystic carcinoma, we investigated the presence of the MYB-NFIB fusion gene in the adenoid cystic carcinoma and high-grade triple-negative breast cancer components of each case by means of FISH and RT-PCR. FISH analysis revealed the presence of the MYB-NFIB fusion gene in both the trabecular and the high-grade triple-negative breast cancer component of AdCC1 (Figure 3). RT-PCR analysis confirmed the expression of the MYB-NFIB transcript in both components of AdCC1, and showed that it involved MYB exon 14 linked to NFIB exon 8c (Supplementary Figure 1). All components of AdCC2, the cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and high-grade triple-negative breast cancer, harbored the MYB-NFIB fusion gene as defined by FISH analysis (Figure 4). Akin to AdCC1, RT-PCR revealed the expression of the corresponding fusion transcript MYB-NFIB in all neoplastic areas of case AdCC2, involving MYB exon 14 linked to NFIB exon 8c (Supplementary Figure 1). These data provide evidence consistent with the notion that the high-grade triple-negative breast cancer components of both cases are likely clonally related to their respective bona fide adenoid cystic carcinomas, and may represent high-grade transformation of the adenoid cystic carcinomas.
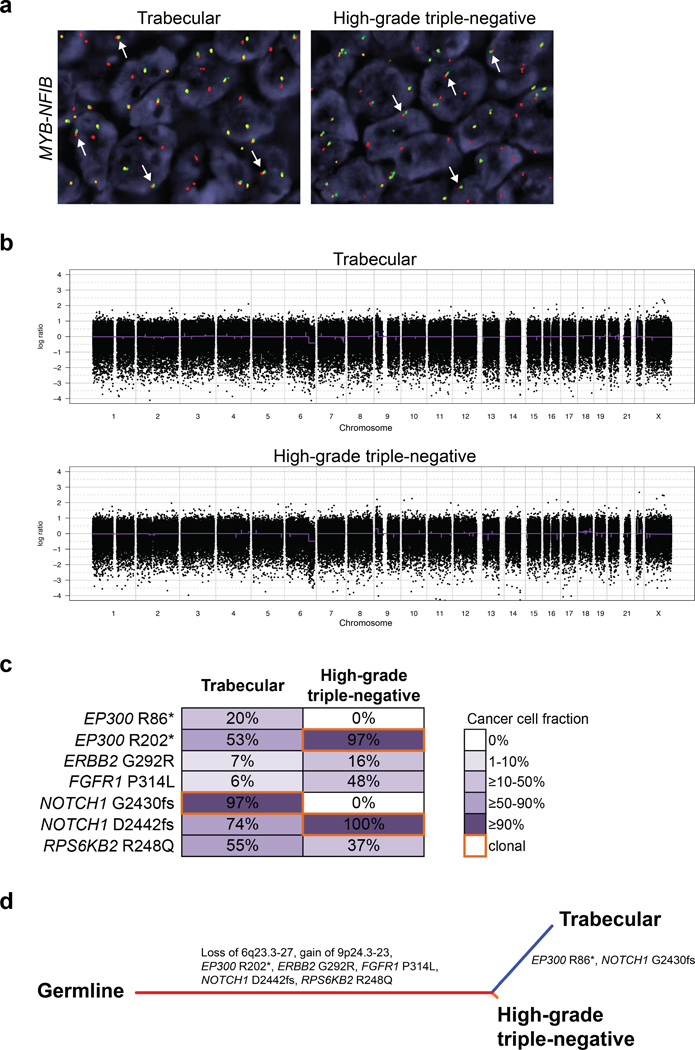
Figure 3. Genomic profiling of an adenoid cystic carcinoma with high-grade transformation (case AdCC1).
(a) FISH analysis using a three-color MYB–NFIB probe, with 5’ MYB (green), 3’ MYB (orange) and 3’ NFIB (red), showing the presence of the fusion gene in the trabecular adenoid cystic carcinoma and high-grade triple-negative breast cancer components (white arrows). (b) The gene copy number profiles of the trabecular and high-grade triple-negative breast cancer components are highly similar. In the genome plots, Log2 ratios were plotted on the y-axis according to their genomic positions indicated on the x-axis. (c) Diagram depicting the somatic mutations identified in the trabecular and high-grade triple-negative breast cancer components. Cancer cell fractions are shown, which are the estimated percentage of cancer cells harboring a given somatic mutation, and are color-coded according to the legend. Clonal somatic mutations are marked by an orange border. In the progression from the trabecular adenoid cystic carcinoma to the high-grade triple-negative breast cancer, the subclonal mutations affecting EP300 and NOTCH1 became clonal. (d) Phylogenetic tree depicting the evolution of the trabecular adenoid cystic carcinoma and the high-grade triple-negative components, where the colored branches represent each of the subclones identified. The length of the branches is representative of the number of mutations and copy number alterations that distinguishes a given clone from its ancestral clone;51 somatic genetic alterations that define a given subclone are illustrated along the branches.
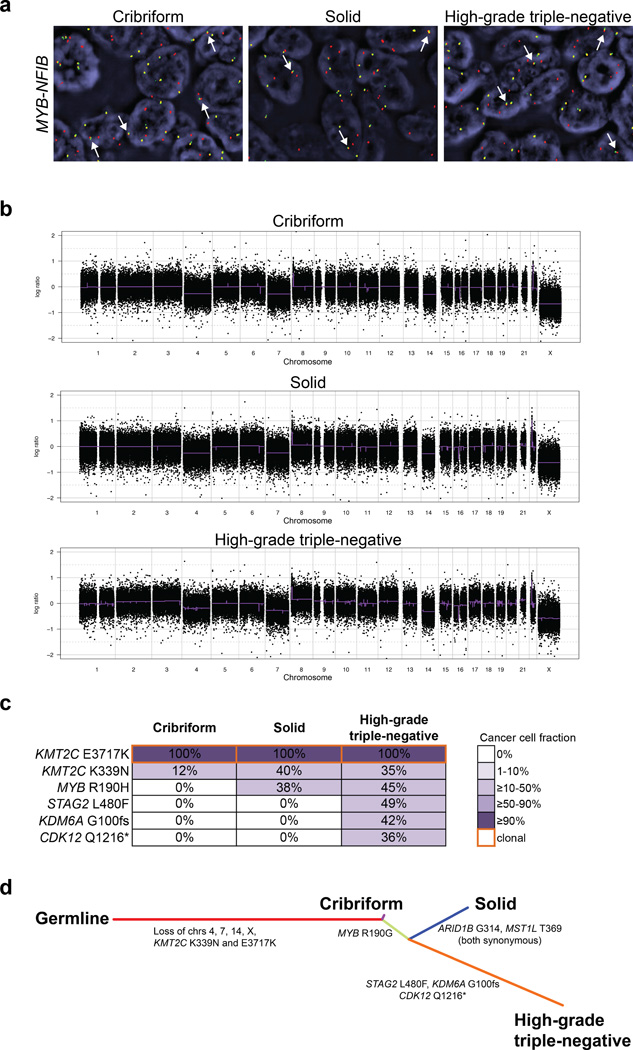
Figure 4. Genomic profiling of an adenoid cystic carcinoma with high-grade transformation (case AdCC2).
(a) FISH analysis using a three-color MYB–NFIB probe, with 5’ MYB (green), 3’ MYB (orange) and 3’ NFIB (red), showing the presence of the fusion gene in the cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and high-grade triple-negative breast cancer components (white arrows). (b) The gene copy number profiles of the cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and high-grade triple-negative breast cancer are highly similar. In the genome plots, Log2 ratios were plotted on the y-axis according to their genomic positions indicated on the x-axis. (c) Diagram depicting the somatic mutations identified in the cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and high-grade triple-negative breast cancer components. Cancer cell fractions are shown, which are the estimated percentage of cancer cells harboring a given somatic mutation, and are color-coded according to the legend. Clonal somatic mutations are marked by an orange border. In the progression from the cribriform adenoid cystic carcinoma to the high-grade triple negative breast cancer, additional subclonal mutations were acquired. (d) Phylogenetic tree depicting the evolution of the cribriform adenoid cystic carcinoma, solid adenoid cystic carcinoma and the high-grade triple-negative components, where the colored branches represent each of the subclones identified. The length of the branches is representative of the number of mutations and copy number alterations that distinguishes a given clone from its ancestral clone;51 somatic genetic alterations that define a given subclone are illustrated along the branches.
Adenoid cystic carcinoma and associated high-grade triple-negative breast cancer of no special type display similar patterns of gene copy number alterations
Consistent with the presence of the MYB-NFIB fusion gene in the bona fide adenoid cystic carcinoma and the high-grade triple-negative breast cancer components of both adenoid cystic carcinomas studied here, gene copy number analysis by OncoScan molecular inversion probe arrays revealed the lack of complex gene copy number alterations in the high-grade triple-negative breast cancer components and low levels of genomic instability. In fact, the gene copy number profiles of the high-grade triple-negative breast cancer components were highly similar to those of the respective adenoid cystic carcinoma components in both cases (Figures 3b and 4b).
Both the trabecular adenoid cystic carcinoma and high-grade triple-negative breast cancer components of AdCC1 had few gene copy number alterations. A loss at 6q23.3-6q27 was found in both components (Figure 3b), and this genomic locus is consistent with the regions previously described as recurrently lost in breast adenoid cystic carcinomas7 and in salivary gland adenoid cystic carcinomas with high-grade transformation.54 All components of AdCC2 displayed losses of chromosomes 4, 7, 14 and X (Figure 4b). The high-grade triple-negative breast cancer component of AdCC2 also harbored a subclonal gain of chromosome 21. These findings provide circumstantial evidence for the clonal relatedness of the different adenoid cystic carcinoma and high-grade triple-negative breast cancer components in the two breast adenoid cystic carcinomas with high-grade transformation studied here, and confirm the existence a subset of high-grade triple-negative breast cancers harboring few copy number alterations.55
Progression from adenoid cystic carcinoma to high-grade triple-negative breast cancer of no special type involves the selection of clones and the acquisition of additional genetic alterations
To define the landscape of somatic mutations of the two adenoid cystic carcinomas and their associated high-grade triple-negative breast cancers, we subjected the DNA extracted from the separately microdissected adenoid cystic carcinoma and high-grade triple-negative breast cancer components to massively parallel sequencing targeting all exons of 488 genes, including clinically actionable genes, the most frequently mutated genes in breast cancer and DNA repair-related genes (see Methods). The tumor samples were sequenced to a median depth of 258× (205×–352×).
Common forms of high-grade triple-negative breast cancer and basal-like breast cancer have been shown to have high mutation rates, and to harbor mutations in TP53 in >80% of cases, as well as mutations in PIK3CA, and DNA repair-related genes.14–16 None of the adenoid cystic carcinomas and their respective high-grade triple-negative breast cancer components studied here harbored somatic mutations affecting TP53, PIK3CA and/ or DNA repair genes (Figures 3c and 4c; Supplementary Table 5). This repertoire of somatic mutations is consistent with those of pure adenoid cystic carcinomas of the breast, which have been shown to be characterized by low mutation rates and lack of mutations affecting TP53, PIK3CA and DNA repair genes.13 These findings are consistent also with the observations made by Shah et al., who reported on a subset of triple-negative breast cancers lacking TP53 mutations and displaying a limited number of somatic mutations.15
In AdCC1, seven and five somatic mutations were identified in the trabecular adenoid cystic carcinomas and high-grade triple-negative breast cancer components, respectively (Figure 3c, Supplementary Table 5). Notably, mutations including those affecting bona fide cancer genes such as ERBB2 and FGFR1 were subclonal, indicating the presence of intra-tumor genetic heterogeneity. Four of the mutations identified targeted two genes, namely EP300 and NOTCH1. A nonsense mutation in EP300 (R86*) and a clonal frameshift mutation in NOTCH1 (G2430fs) were restricted to the trabecular adenoid cystic carcinoma component, with cancer cell fractions of 53% and 97%, respectively, whereas the additional EP300 R202* and NOTCH1 D2442fs mutations were detected in both components (Figure 3c), providing evidence suggestive of a convergent phenotype. Interestingly, the subclonal EP300 R202* and the NOTCH1 D2442fs mutations in the trabecular adenoid cystic carcinoma became clonal in the high-grade triple-negative breast cancer component. In addition, there was enrichment in the cancer cell fractions of the FGFR1 and ERBB2 mutations from 6% and 7% in the trabecular adenoid cystic carcinoma to 48% and 16% in the high-grade triple-negative breast cancer, respectively. These data suggest that a clonal shift, and potentially clonal selection, occurred in the progression from the adenoid cystic carcinoma to high-grade triple-negative breast cancer in this case (Figure 3d).
In AdCC2, only two KMT2C mutations (E3717K and K339N) were found in the cribriform adenoid cystic carcinoma component. In fact, the KMT2C E3717K mutation was identified as the founder genetic event, being clonal and present in all cancer cells of the cribriform adenoid cystic carcinoma, the solid adenoid cystic carcinoma and high-grade triple-negative breast cancer components. Akin to AdCC1, a second but subclonal missense mutation affecting the same gene (KMT2C K339N) was found in all components, with cancer cell fractions ranging from 12% to 40%, indicating the presence of intra-tumor genetic heterogeneity also in this case. In addition, a validated subclonal MYB mutation was restricted to the solid adenoid cystic carcinoma and the high-grade triple-negative breast cancer components. Moreover, subclonal mutations in STAG2, KDM6A and CDK12 were found to be restricted to the high-grade triple-negative breast cancer, with cancer cell fractions of 36–49%. These data suggest that in this AdCC2 case progression from adenoid cystic carcinoma to high-grade triple-negative breast cancer may have occurred through the acquisition of additional genetic alterations (Figure 4c and 4d).
Taken together, our findings demonstrate that in both cases, the high-grade triple-negative breast cancer components were clonally related to the bona fide adenoid cystic carcinomas, providing support to the notion that morphologically unequivocal adenoid cystic carcinomas associated with high-grade triple-negative breast cancers may display intra-tumor genetic heterogeneity, and that in the progression from adenoid cystic carcinoma to high-grade triple-negative breast cancer, clonal shifts, and potentially clonal selection, may take place.
DISCUSSION
Adenoid cystic carcinomas of the breast are rare neoplasms that despite being of triple-negative phenotype usually have an indolent clinical behavior. High-grade transformation can occur and is associated with aggressive clinical behavior and poor response to systemic therapies,19,20 however guidelines for the management of this particular subset of patients have yet to be fully implemented.
Here we performed a genetic analysis of two breast adenoid cystic carcinomas with high-grade triple-negative breast cancer of no special type components and found that adenoid cystic carcinomas associated with high-grade triple-negative breast cancer may be composed of multiple clones at diagnosis, consistent with previous observations by our group,13 and that progression to high-grade triple-negative breast cancer may occur through clonal selection and/ or the acquisition of additional genetic events. Furthermore, in this progression from a low-grade trabecular or cribriform adenoid cystic carcinoma to a high-grade triple-negative breast cancer of no special type, we documented the presence of the MYB-NFIB fusion gene not only in the adenoid cystic carcinoma components, but also in the high-grade triple-negative breast cancer areas of both cases.
The prevalence of the MYB-NFIB fusion gene in breast adenoid cystic carcinomas has been reported to range from 23% to 100%.6,7,12,13,21,56 Recently, D’Alfonso et al.21 reported on the solid variant of breast adenoid cystic carcinomas with basaloid features and confirmed that this high-grade variant may harbor MYB rearrangements, although at lower frequencies (2/16; 12.5%). Here we demonstrate that high-grade triple-negative breast cancer of no special type associated with bona fide adenoid cystic carcinomas may also be underpinned by the MYB-NFIB fusion gene. Further studies analyzing larger numbers of breast adenoid cystic carcinomas are warranted to determine the prevalence of MYB-NFIB fusion gene in this histologic context.
Several studies including small cohorts of patients with salivary gland adenoid cystic carcinomas54,57,58 suggested that TP53 inactivation17,54,57 and/ or amplification of 8q24.12-q24.13 encompassing MYC57 may play a role in high-grade transformation of these lesions. Neither breast adenoid cystic carcinomas analyzed here harbored TP53 mutations and/ or MYC amplification. Our previous analyses of breast adenoid cystic carcinomas revealed that these lesions harbor somatic mutations that affect cancer-related genes, but lack TP53 and PIK3CA mutations found in common forms of high-grade triple-negative breast cancer and that the genes mutated in breast adenoid cystic carcinomas converge into several functional categories including chromatin remodeling and signaling pathway genes amongst others.13 Consistent with these findings, we found that the adenoid cystic carcinomas studied here harbored mutations in chromatin remodeling genes (i.e. EP300, AdCC1; KMT2C, KDM6A, AdCC2) and in signaling pathway genes (i.e. ERRB2, FGFR1 and RPS6KB2, AdCC1), as well as in MYB itself (AdCC2). Interestingly, whilst salivary gland adenoid cystic carcinomas have been reported to harbor recurrent mutations affecting NOTCH signaling pathway genes, including NOTCH1, NOTCH2 and SPEN,28,59 these genes were not found to be altered in 12 classical tubular/ cribriform breast adenoid cystic carcinomas previously studied.13 In contrast, in one of the adenoid cystic carcinomas with high-grade transformation (AdCC1) analyzed here, two distinct NOTCH1 frameshift mutations were identified, one of which was restricted to the trabecular adenoid cystic carcinoma component (G2430fs) whereas the second NOTCH1 mutation (D2442fs) was subclonal in the trabecular adenoid cystic carcinoma component and became clonal in the high-grade triple-negative breast cancer of no special type. This clonal shift was also observed for mutations affecting the histone acetyltransferase EP300 in AdCC1: the EP300 R86* mutation was restricted to the trabecular adenoid cystic carcinoma component while the EP300 R202* mutation was subclonal in the adenoid cystic carcinoma component but clonal in the high-grade triple-negative breast cancer component. In AdCC2, high-grade transformation was likely driven by the acquisition of additional mutations including a CDK12 frameshift mutation or by the selection of a clone harboring these alterations but not sampled in our extensive sampling of the distinct components of the adenoid cystic carcinoma. A suclonal MYB R190H somatic mutation was detected in the solid adenoid cystic carcinoma and in the high-grade triple-negative carcinoma components of AdCC2. One could posit that this somatic mutation may have conferred a growth and/ or survival advantage; it should be noted, however, that this mutation was present in a minor subclone of the solid adenoid cystic carcinoma and high-grade triple-negative carcinoma components, was considered by multiple mutation function predictors as a passenger/ non-deleterious mutation, and has not been previously documented in the Catalog of Somatic Mutations in Cancer44 and the cBioPortal database (www.cBioPortal.org; accessed on May 30th 2016)60. Our findings provide evidence to suggest that the genetic events driving progression of breast adenoid cystic carcinomas are heterogeneous; however as these involve mutations in potentially actionable cancer genes, comprehensive sequencing analyses are warranted to identify therapeutic targets in adenoid cystic carcinomas with high-grade transformation.
Triple-negative breast cancers have been shown to be heterogeneous at the genetic level with some tumors harboring only a few non-synonymous somatic mutations.14–16 Based on the findings of our study, one may hypothesize that the high-grade transformation of low-grade triple-negative breast cancers such as adenoid cystic carcinomas may account for a subset of triple-negative breast cancers with low levels of copy number alterations and few somatic mutations.15,55 Further studies are warranted to characterize genetically stable triple-negative breast cancers thought to be of common type.
To define whether the MYB-NFIB fusion gene would be present in a subset of common-type triple-negative breast cancers, we searched for the presence of breast cancers in The Cancer Genome Atlas (TCGA) data set harboring the MYB-NFIB fusion using the TCGA Fusion gene Data Portal (http://54.84.12.177/PanCanFusV2/). We identified one invasive breast cancer from the TCGA dataset (case TCGA-A1-A0SB-01A), which according to the pathology report provided in the cBioPortal (www.cBioPortal.org)60 is a breast adenoid cystic carcinoma.
This study has several limitations. First, owing to the rarity of breast adenoid cystic carcinomas with progression to high-grade triple-negative breast cancer, our sample size is small. Second, given the limited amount of DNA extracted from each component of each case, we were unable to retrieve sufficient DNA for whole exome or whole genome sequencing; however, the use of two independent targeted capture massively parallel sequencing assays allowed for a validation of somatic mutations affecting 107 genes, in addition to the targeted amplicon validation of the mutations identified in AdCC1 and AdCC2. Third, owing to the fact that these cases were obtained from distinct institutions, we were unable to ascertain accurately the clinical behavior of the high-grade triple-negative breast cancer components of these cases. Despite these limitations, here we demonstrate that breast adenoid cystic carcinomas with high-grade transformation may be underpinned by the MYB-NFIB fusion gene and display intra-tumor genetic heterogeneity. Progression from conventional adenoid cystic carcinoma to high-grade triple-negative breast cancer of no special type involves the selection of specific clones and/ or acquisition of mutations in bona fide cancer genes. This morphologic and genetic heterogeneity should be taken into account when performing histologic analyses of breast biopsies, given that the recognition of transformed areas of high-grade triple-negative breast cancer of no special type within an adenoid cystic carcinoma of the breast might allow for a more precise prognostication and the identification of potentially actionable genetic alterations.
Supplementary Material
Acknowledgments
FUNDING
AMS was supported by “Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung” and SP in part by a Susan G Komen Postdoctoral Fellowship Grant (PDF14298348). Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DISCLOSURE/ CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Sapino A, Sneige N, Eusebi V. Adenoid cystic carcinoma. In: Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO classification of tumours of the breast. 4th. Lyon: IARC press; 2012. pp. 56–57. [Google Scholar]
- 2.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchio C, Weigelt B, Reis-Filho JS. Adenoid cystic carcinomas of the breast and salivary glands (or 'The strange case of Dr Jekyll and Mr Hyde' of exocrine gland carcinomas) J Clin Pathol. 2010;63:220–228. doi: 10.1136/jcp.2009.073908. [DOI] [PubMed] [Google Scholar]
- 4.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 5.Ghabach B, Anderson WF, Curtis RE, et al. Adenoid cystic carcinoma of the breast in the United States (1977 to 2006): a population-based cohort study. Breast Cancer Res. 2010;12:R54. doi: 10.1186/bcr2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persson M, Andren Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wetterskog D, Lopez-Garcia MA, Lambros MB, et al. Adenoid cystic carcinomas constitute a genomically distinct subgroup of triple-negative and basal-like breast cancers. J Pathol. 2012;226:84–96. doi: 10.1002/path.2974. [DOI] [PubMed] [Google Scholar]
- 8.Costa AF, Altemani A, Garcia-Inclan C, et al. Analysis of MYB oncogene in transformed adenoid cystic carcinomas reveals distinct pathways of tumor progression. Lab Invest. 2014;94:692–702. doi: 10.1038/labinvest.2014.59. [DOI] [PubMed] [Google Scholar]
- 9.Fusco N, Guerini-Rocco E, Schultheis AM, et al. The birth of an adenoid cystic carcinoma. Int J Surg Pathol. 2015;23:26–27. doi: 10.1177/1066896914548795. [DOI] [PubMed] [Google Scholar]
- 10.Brayer KJ, Frerich CA, Kang H, Ness SA. Recurrent Fusions in MYB and MYBL1 Define a Common, Transcription Factor-Driven Oncogenic Pathway in Salivary Gland Adenoid Cystic Carcinoma. Cancer Discov. 2016;6:176–187. doi: 10.1158/2159-8290.CD-15-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitani Y, Liu B, Rao PH, et al. Novel MYBL1 Gene Rearrangements with Recurrent MYBL1-NFIB Fusions in Salivary Adenoid Cystic Carcinomas Lacking t(6;9) Translocations. Clin Cancer Res. 2016;22:725–733. doi: 10.1158/1078-0432.CCR-15-2867-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetterskog D, Wilkerson PM, Rodrigues DN, et al. Mutation profiling of adenoid cystic carcinomas from multiple anatomical sites identifies mutations in the RAS pathway, but no KIT mutations. Histopathology. 2013;62:543–550. doi: 10.1111/his.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martelotto LG, De Filippo MR, Ng CK, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng CK, Schultheis AM, Bidard FC, Weigelt B, Reis-Filho JS. Breast cancer genomics from microarrays to massively parallel sequencing: paradigms and new insights. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv015. [DOI] [PubMed] [Google Scholar]
- 17.Seethala RR, Hunt JL, Baloch ZW, Livolsi VA, Leon Barnes E. Adenoid cystic carcinoma with high-grade transformation: a report of 11 cases and a review of the literature. Am J Surg Pathol. 2007;31:1683–1694. doi: 10.1097/PAS.0b013e3180dc928c. [DOI] [PubMed] [Google Scholar]
- 18.Nagao T. "Dedifferentiation" and high-grade transformation in salivary gland carcinomas. Head Neck Pathol. 2013;7(Suppl 1):S37–S47. doi: 10.1007/s12105-013-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin SJ, Rosen PP. Solid variant of mammary adenoid cystic carcinoma with basaloid features: a study of nine cases. Am J Surg Pathol. 2002;26:413–420. doi: 10.1097/00000478-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Silva I, Tome V, Oliveira J. Adenoid cystic carcinoma of the breast with cerebral metastisation: a clinical novelty. BMJ Case Rep. 2011;2011 doi: 10.1136/bcr.08.2011.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Alfonso TM, Mosquera JM, MacDonald TY, et al. MYB-NFIB gene fusion in adenoid cystic carcinoma of the breast with special focus paid to the solid variant with basaloid features. Hum Pathol. 2014;45:2270–2280. doi: 10.1016/j.humpath.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Weigelt B, Horlings HM, Kreike B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–150. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 23.Fusco N, Colombo PE, Martelotto LG, et al. Resolving quandaries: basaloid adenoid cystic carcinoma or breast cylindroma? The role of massively parallel sequencing. Histopathology. 2016;68:262–271. doi: 10.1111/his.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 26.Dowsett M, Nielsen TO, A'Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 28.Ho AS, Kannan K, Roy DM, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerini-Rocco E, Hodi Z, Piscuoglio S, et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015;237:166–178. doi: 10.1002/path.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez L, Wilkerson PM, Lambros MB, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol. 2012;227:42–52. doi: 10.1002/path.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fehr A, Kovacs A, Loning T, et al. The MYB-NFIB gene fusion-a novel genetic link between adenoid cystic carcinoma and dermal cylindroma. J Pathol. 2011;224:322–327. doi: 10.1002/path.2909. [DOI] [PubMed] [Google Scholar]
- 32.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piscuoglio S, Ng CK, Murray M, et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J Pathol. 2016;238:508–518. doi: 10.1002/path.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natrajan R, Wilkerson PM, Marchio C, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol. 2014;232:553–565. doi: 10.1002/path.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 41.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martelotto LG, Ng CK, De Filippo MR, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 2014;15:484. doi: 10.1186/s13059-014-0484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bidard FC, Ng CK, Cottu P, et al. Response to dual HER2 blockade in a patient with HER3-mutant metastatic breast cancer. Ann Oncol. 2015;26:1704–1709. doi: 10.1093/annonc/mdv217. [DOI] [PubMed] [Google Scholar]
- 47.Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van de Wiel MA, Wieringen WN. CGHregions: dimension reduction for array CGH data with minimal information loss. Cancer Inform. 2007;3:55–63. [PMC free article] [PubMed] [Google Scholar]
- 50.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultheis AM, Ng CK, De Filippo MR, et al. Massively Parallel Sequencing-Based Clonality Analysis of Synchronous Endometrioid Endometrial and Ovarian Carcinomas. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nixon KC. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics-the International Journal of the Willi Hennig Society. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 53.Schliep KP. Phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa AF, Altemani A, Vekony H, et al. Genetic profile of adenoid cystic carcinomas (ACC) with high-grade transformation versus solid type. Cell Oncol (Dordr) 2011;34:369–379. doi: 10.1007/s13402-011-0037-5. [DOI] [PubMed] [Google Scholar]
- 55.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brill LB, 2nd, Kanner WA, Fehr A, et al. Analysis of MYB expression and MYB-NFIB gene fusions in adenoid cystic carcinoma and other salivary neoplasms. Mod Pathol. 2011;24:1169–1176. doi: 10.1038/modpathol.2011.86. [DOI] [PubMed] [Google Scholar]
- 57.Seethala RR, Cieply K, Barnes EL, Dacic S. Progressive genetic alterations of adenoid cystic carcinoma with high-grade transformation. Arch Pathol Lab Med. 2011;135:123–130. doi: 10.5858/2010-0048-OAR.1. [DOI] [PubMed] [Google Scholar]
- 58.Frierson HF, Jr, Moskaluk CA. Mutation signature of adenoid cystic carcinoma: evidence for transcriptional and epigenetic reprogramming. J Clin Invest. 2013;123:2783–2785. doi: 10.1172/JCI69070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephens PJ, Davies HR, Mitani Y, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest. 2013;123:2965–2968. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.