Complex interactions between genetic and environmental factors have long been implicated in the development of autoimmune diseases, including systemic lupus erythematosus (SLE) and Sjögren's syndrome (SS) 1. Endogenous retroviruses (ERVs) have been proposed as molecular links between the human genome and environmental factors, such as viruses and other infectious organisms 2. ERVs may contribute to autoimmunity via 1 of 2 mechanisms: 1) structural molecular mimicry, serving as cross‐reactive antigen targets for antiviral immune responses, and 2) functional interference with immune responses, by directly regulating gene expression and signal transduction 2.
ERVs belong to the larger family of retrotransposable elements that make up as much as 40% of the human genome 2. While most ERVs have only 1–100 copies per haploid genome, the copy numbers of retrotransposable elements are as high as 300,000 for Alu elements and ∼10,000 for long interspersed nucleotide elements (LINEs) 2. Alu elements are transcribed by RNA polymerase III, are not poly‐adenylated, and do not code for proteins. In contrast, LINEs are transcribed by RNA polymerase II, are poly‐adenylated, and can code for proteins. However, due to truncations and accumulated mutations in coding regions, retrotransposable elements and ERVs are rarely expressed on the RNA or protein level. The 3 ′‐terminus of transfer RNA (tRNA) is known to initiate reverse transcription by annealing to an 18‐nucleotide–long primer binding site at the 5 ′ long terminal repeat (LTR) (Figure 1). The LTR regions, which are typically several hundred nucleotides in length, surround the protein‐coding open‐reading frames (ORFs) and harbor transcriptional regulatory sequences, such as a TATA box, a poly‐adenylation site, a tRNA primer binding site, and inverted repeats at typical locations 2.
Figure 1.
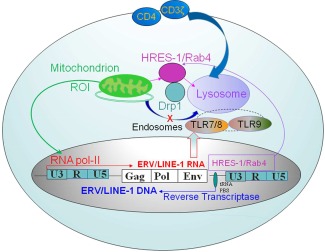
Mechanistic diagram of endogenous retrovirus (ERV)–regulated endosomal recycling in autoimmune diseases. ERVs and other retroelements are integrated into the nuclear DNA. ERVs are transcribed into RNA by RNA polymerase II (RNA pol II). In turn, retroviral transcripts can be reverse transcribed into DNA using transfer RNA (tRNA) as a primer. While most ERVs are transcriptionally silent, as a rare example, human T lymphotropic virus–related endogenous sequence 1 (HRES‐1) is transcribed into RNA and translated into protein, such as HRES‐1/Rab4 (4). This small GTPase promotes the recycling and lysosomal degradation of cell surface receptors, such as CD4 (4), CD3ζ 5, and Drp1 (6). The HRES‐1/Rab4–mediated depletion of dynamin‐related protein 1 (Drp1) inhibits mitochondrial fission and causes the accumulation of mitochondria that results in increased production of reactive oxygen intermediates (ROIs) in lupus T cells 6. Thus, the accumulation of mitochondria may underlie oxidative stress in patients with systemic lupus erythematosus 8. TLR‐7 = Toll‐like receptor 7; LINE‐1 = long interspersed nuclear element 1; PBS = primer binding site.
Human ERVs have generally been found to be defective proviruses with deletions or stop codons in the ORFs, which generally code for the structural proteins Gag and Env and the reverse transcriptase–generated enzyme termed Pol 2. Human T lymphotropic virus–related endogenous sequence 1 (HRES‐1), which is one of the few human ERVs that have been implicated in autoimmunity, is transcribed 3. It encodes a viral Gag‐like, 28‐kd autoantigen and a 24‐kd small GTPase, termed HRES‐1/Rab4, which has been recently redesignated as Rab4A 4. While the HRES‐1/p28 protein is a target of cross‐reactive antiviral antibodies in patients with SLE, HRES‐1/Rab4 controls the recycling of endosomes 4 (Figure 1). It regulates the surface expression of CD4 (4) and CD3ζ, both of which are involved in abnormal signal transduction in lupus T cells 5. The typical LTR promoter sequence of HRES‐1 contains a trans‐activation region, or TAR, similar to the LTR of human immunodeficiency virus type 1 (HIV‐1) 3. Notably, the TAR region of HRES‐1 interacts with HIV‐1 (4). Through the Tat trans‐activator, HIV‐1 stimulates transcription of HRES‐1 (4). In turn, the HRES‐1/Rab4 protein stimulates the endocytic recycling of CD4, and thereby causes the lysosomal degradation of CD4 and prevents reinfection of T cells by HIV‐1 (4).
HRES‐1/Rab4 is overexpressed in lupus T cells, where it also controls the endocytic recycling of another GTPase, dynamin‐related protein 1 (Drp1), which is essential to initiate mitochondrial fission and mitophagy 6. Due to HRES‐1/Rab4–mediated depletion of Drp1, mitophagy is inhibited and oxidative stress–generating mitochondria accumulate in lupus T cells 6. Interestingly, HIV‐1 also triggers oxidative stress 7, and it is conceivable that a similar virus may underlie the activation of Rab4A and oxidative stress in SLE 8. Although elevated serum levels of type I interferon (IFN) in SLE patients have long been linked to a viral etiology 9, no infectious virus has been convincingly shown to account for the increased IFN production and downstream activation of IFN‐stimulated genes 10. Therefore, significant efforts have been invested in evaluating ERVs as potential triggers of IFN production in autoimmune diseases.
Initial evidence to indicate that ERV‐derived nucleic acids act as inducers of type I IFN production comes from studies of patients with Aicardi‐Goutières syndrome and chilblain lupus 11. Such patients have a genetic deficiency of TREX1 (three prime repair exonuclease 1 gene), a 3 ′‐5 ′ repair exonuclease that metabolizes DNA reverse transcribed from ERVs 11. Consequently, single‐stranded DNA derived from endogenous retroelements accumulates in TREX1‐deficient cells, and these cells exhibit increased production of IFN (Figure 2). Thus, TREX1 is an essential negative regulator of the IFN‐stimulatory DNA (ISD) response 12. In support of the role of TREX1 in autoimmunity, mice lacking TREX1 develop lupus‐like disease, which is remarkably dependent on the nucleic acid sensor cGMP–cAMP synthase (cGAS) 13. During the pathogenesis of lupus in mouse models, the ERV‐induced ISD response also depends on the mitochondrial antiviral signaling (MAVS) protein and retinoic acid–inducible gene 1 (RIG‐1) 14. Thus, cGAS, MAVS, and RIG‐1 have emerged as cardinal transducers of the ISD response (Figure 2).
Figure 2.
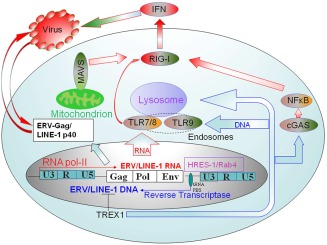
Mechanisms of ERV‐activated interferon (IFN) production in autoimmunity. The human genome harbors retroelements, such as LINE‐1. They can be reverse transcribed into DNA and metabolized by TREX1 (the three prime repair exonuclease 1 gene), which is deficient in patients with Aicardi‐Goutières syndrome and chilblain lupus 11. Increased transcription of LINE‐1 is now recognized as an important trigger of IFN production, which is mediated through RNA sensing, and it involves signal transduction through TLR‐7/TLR‐8, cGMP–cAMP synthase (cGAS), retinoic acid–inducible gene 1 (RIG‐1), and NF‐κB. Alternatively, reverse transcribed ERV DNA is recognized via TLR‐9, which also traffics through endosomes 23. Moreover, mitochondrial oxidative stress activates the mitochondrial antiviral signaling protein (MAVS), which acts as an amplifier of RIG‐1 activation during IFN signaling 14. Oxidative stress also causes DNA hypomethylation 16, a process that underlies increased LINE‐1 expression 15. In addition to promoting nucleic acid–driven IFN production, ERV Gag‐encoded proteins, such as HRES‐1/p28, or the LINE‐1 p40 protein may also contribute to autoimmunity via molecular mimicry 22. See Figure 1 for other definitions.
In this issue of Arthritis & Rheumatology, Mavragani and colleagues demonstrate that the transcription of LINE‐1 is increased in kidney tissue from patients with lupus nephritis and in minor salivary gland (MSG) tissue from patients with SS, and that increased LINE‐1 transcription correlates with greater expression of type I IFN 15. LINE‐1 ORF‐1/p40 protein and IFNβ are also highly expressed in MSG ductal epithelial cells and in lupus kidneys, and IFNα is detected in infiltrating plasmacytoid dendritic cells (PDCs). Moreover, transfection of PDCs or monocytes with LINE‐1–encoding DNA or RNA induces type I IFN. These elegant studies clearly show that LINE‐1 retroelements constitute endogenous nucleic acid triggers of type I IFN production, a process that involves RIG‐1. Therefore, LINE‐1 may contribute to the pathogenesis of inflammation in SLE and SS.
Since Mavragani et al also demonstrated that methylation of CpG sites in the LINE‐1 promoter are negatively correlated with LINE‐1 expression in MSG tissue 15, it is likely that demethylation may play a critical role in the observed overexpression of LINE‐1. Interestingly, LINE‐1 transcription via hypomethylation is readily induced by oxidative stress, and it responds to treatment with the antioxidant N‐acetylcysteine (NAC) 16. Similar to LINE‐1, expression of ERVs is also controlled by epigenetic mechanisms 17, primarily via methylation 18, which appears to also underlie the increased expression of HRES‐1/p28 in B cells from patients with SLE 19. Of note, oxidative stress is a cause of global DNA hypomethylation in SLE 20. Therefore, activation of ERVs, such as increased transcription of LINE‐1 and HRES‐1, may originate from mitochondrial oxidative stress (Figure 2). Importantly, oxidative stress can be moderated by treatment with NAC, which is a potentially safe intervention with promising clinical benefits for patients with SLE 21.
Although increased production of IFN is generally considered to be a pathogenic factor in SLE, it may reflect a protective measure against viral infections 22. Ongoing clinical trials aimed at neutralizing IFN signaling are expected to address this important question in the near future.
AUTHOR CONTRIBUTIONS
Dr. Perl was involved in drafting the article and revising it critically for important intellectual content. Dr. Perl approved the final version to be published.
Supported in part by the NIH (National Institute of Allergy and Infectious Diseases grants AI‐48079, AI‐072648, and AI‐122176).
REFERENCES
- 1. Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus [review]. Genes Immun 2009;10:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Perl A, Fernandez D, Telarico T, Phillips PE. Endogenous retroviral pathogenesis in lupus. Curr Opin Rheumatol 2010;22:483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perl A, Rosenblatt JD, Chen IS, DiVincenzo JP, Bever R, Poiesz BJ, et al. Detection and cloning of new HTLV‐related endogenous sequences in man. Nucl Acids Res 1989;17:6841–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagy G, Ward J, Mosser DD, Koncz A, Gergely P, Stancato C, et al. Regulation of CD4 expression via recycling by HRES‐1/RAB4 controls susceptibility to HIV infection. J Biol Chem 2006;281:34574–91. [DOI] [PubMed] [Google Scholar]
- 5. Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, et al. Activation of mTOR controls the loss of TCR in lupus T cells through HRES‐1/Rab4‐regulated lysosomal degradation. J Immunol 2009;182:2063–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caza TN, Fernandez D, Talaber G, Oaks Z, Haas M, Madaio MP, et al. HRES‐1/RAB4‐mediated depletion of DRP1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann Rheum Dis 2014;73:1887–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banki K, Hutter E, Gonchoroff NJ, Perl A. Molecular ordering in HIV‐induced apoptosis: oxidative stress, activation of caspases, and cell survival are regulated by transaldolase. J Biol Chem 1998;273:11944–53. [DOI] [PubMed] [Google Scholar]
- 8. Gergely PJ, Grossman C, Niland B, Puskas F, Neupane H, Allam F, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum 2002;46:175–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rich SA. Human lupus inclusions and interferon. Science 1981;213:772–5. [DOI] [PubMed] [Google Scholar]
- 10. Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, et al. Coordinate overexpression of interferon‐α–induced genes in systemic lupus erythematosus. Arthritis Rheum 2004;50:3958–67. [DOI] [PubMed] [Google Scholar]
- 11. Lee‐Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, et al. Mutations in the gene encoding the 3′ 29‐5 ′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet 2007;39:1065–7. [DOI] [PubMed] [Google Scholar]
- 12. Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell‐intrinsic initiation of autoimmunity. Cell 2008;134:587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1‐deficient mouse model of Aicardi‐Goutières syndrome. J Immunol 2015;195:1939–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeng M, Hu Z, Shi X, Li X, Zhan X, Li XD, et al. MAVS, cGAS, and endogenous retroviruses in T‐independent B cell responses. Science 2014;346:1486–92. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Mavragani CP, Sagalovskiy I, Guo Q, Nezos A, Kapsogeorgou EK, Lu P, et al. Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease. Arthritis Rheumatol 2016;68:2686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kloypan C, Srisa‐art M, Mutirangura A, Boonla C. LINE‐1 hypomethylation induced by reactive oxygen species is mediated via depletion of S‐adenosylmethionine. Cell Biochem Funct 2015;33:375–85. [DOI] [PubMed] [Google Scholar]
- 17. Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral‐like elements in check: the epigenetic perspective [review]. Cell Mol Life Sci 2008;65:3329–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010;463:237–40. [DOI] [PubMed] [Google Scholar]
- 19. Falli T, le Dantec C, Thabet Y, Jousse S, Hanrotel C, Brooks WH, et al. DNA methylation modulates HRES1/p28 expression in B cells from patients with lupus. Autoimmunity 2014;47:265–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Gorelik G, Strickland FM, Richardson BC. Oxidative stress, T cell DNA methylation and lupus. Arthritis Rheumatol 2014;66:1574–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lai ZW, Hanczko R, Bonilla E, Caza TN, Clair B, Bartos A, et al. N‐acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double‐blind, placebo‐controlled trial. Arthritis Rheum 2012;64:2937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caza T, Oaks Z, Perl A. Interplay of infections, autoimmunity, and immunosuppression in systemic lupus erythematosus. Int Rev Immunol 2014;33:330–63. [DOI] [PubMed] [Google Scholar]
- 23. Okuya K, Tamura Y, Saito K, Kutomi G, Torigoe T, Hirata K, et al. Spatiotemporal regulation of heat shock protein 90‐chaperoned self‐DNA and CpG‐oligodeoxynucleotide for type I IFN induction via targeting to static early endosome. J Immunol 2010;184:7092–9. [DOI] [PubMed] [Google Scholar]


