Abstract
Cortico-basal ganglia circuits are critical for speech and language and are implicated in autism spectrum disorder (ASD), in which language function can be severely affected. We demonstrate that in the striatum, the gene, Foxp2, negatively interacts with the synapse suppressor, Mef2C. We present causal evidence that Mef2C inhibition by Foxp2 in neonatal mouse striatum controls synaptogenesis of corticostriatal inputs and vocalization in neonates. Mef2C suppresses corticostriatal synapse formation and striatal spinogenesis, but can, itself, be repressed by Foxp2 through direct DNA binding. Foxp2 deletion de-represses Mef2C, and both intrastriatal and global decrease of Mef2C rescue vocalization and striatal spinogenesis defects of Foxp2-deletion mutants. These findings suggest that Foxp2-Mef2C signaling is critical to corticostriatal circuit formation. If found in humans, such signaling defects could contribute to a range of neurologic and neuropsychiatric disorders.
INTRODUCTION
Speech and language endow human beings with unrivalled refinement and breadth in their social communications. In ASD, these communicative abilities can be severely compromised along with other problems related to social communication and repetitive behavior1. The neural mechanisms underlying language difficulties in ASD individuals present a challenge to neuroscientists2, but neuroimaging studies have detected aberrant neuronal activity and connectivity in the neocortex and striatum of ASD individuals3,4. We were struck by the parallels between these findings in ASD individuals and findings linking mutations of the FOXP2 gene to spoken language disabilities. In both, corticostriatal dysfunction has been found along with the behavioral problems in humans and in mutant mouse models5–9. We therefore sought for signaling pathways within corticostriatal circuits that could link these two functional domains.
Human FOXP2 has been shown to interact with several known ASD-risk genes, including CNTNAP2, MET and uPAR10–12, but what links FOXP2 and these ASD-risk genes to specific neural circuits with defined neurological functions has not been clear. Nor is it known how such interactions could alter the development of language-related circuits in ASD brains. Given the potential involvement of the striatum in both ASD and language dysfunction, we searched for evidence that might link influences of ASD-risk genes and language-linked Foxp2 during postnatal striatal development, a time of particular vulnerability in ASD. We focused on ASD candidate genes associated with synapse formation, at risk in ASD13,14. We chose for study myocyte enhancer factor 2C (MEF2C), suggested to be a possible ASD risk gene15–17, identified as one of 175 FOXP2 targets identified by chromatin immunoprecipitation (ChIP)-chip analysis of human basal ganglia during development18, and known to be a mental retardation risk gene19. Mef2C has been shown to regulate negatively dendritic spines and excitatory synapses in hippocampal neurons and in cultured striatal neurons20–23. Here, we demonstrate that Mef2C also negatively regulates synaptogenesis in the striatum in vivo, but that negative interactions between Foxp2 and Mef2C lead to repression of the effects of Mef2C coinciding spatially and temporally with the development of corticostriatal connections during early postnatal development. We show that Foxp2 can suppress Mef2C through direct DNA binding, and that the negative Foxp2-Mef2C signaling interactions have specific synaptic and behavioral effects including control of ultrasonic vocalizations (USVs) in young mouse pups. We suggest that negative Foxp2-Mef2C interactions could allow cortical fibers to synapse progressively onto striatal neurons, thus building the corticostriatal pathway by de-repression of synaptogenesis in the striatum. Such Foxp2-Mef2C signaling could provide a valuable model for identifying candidate molecular mechanisms underlying developmental defects in corticostriatal circuits.
RESULTS
Progressive dissociation of striatal Foxp2 and Mef2C
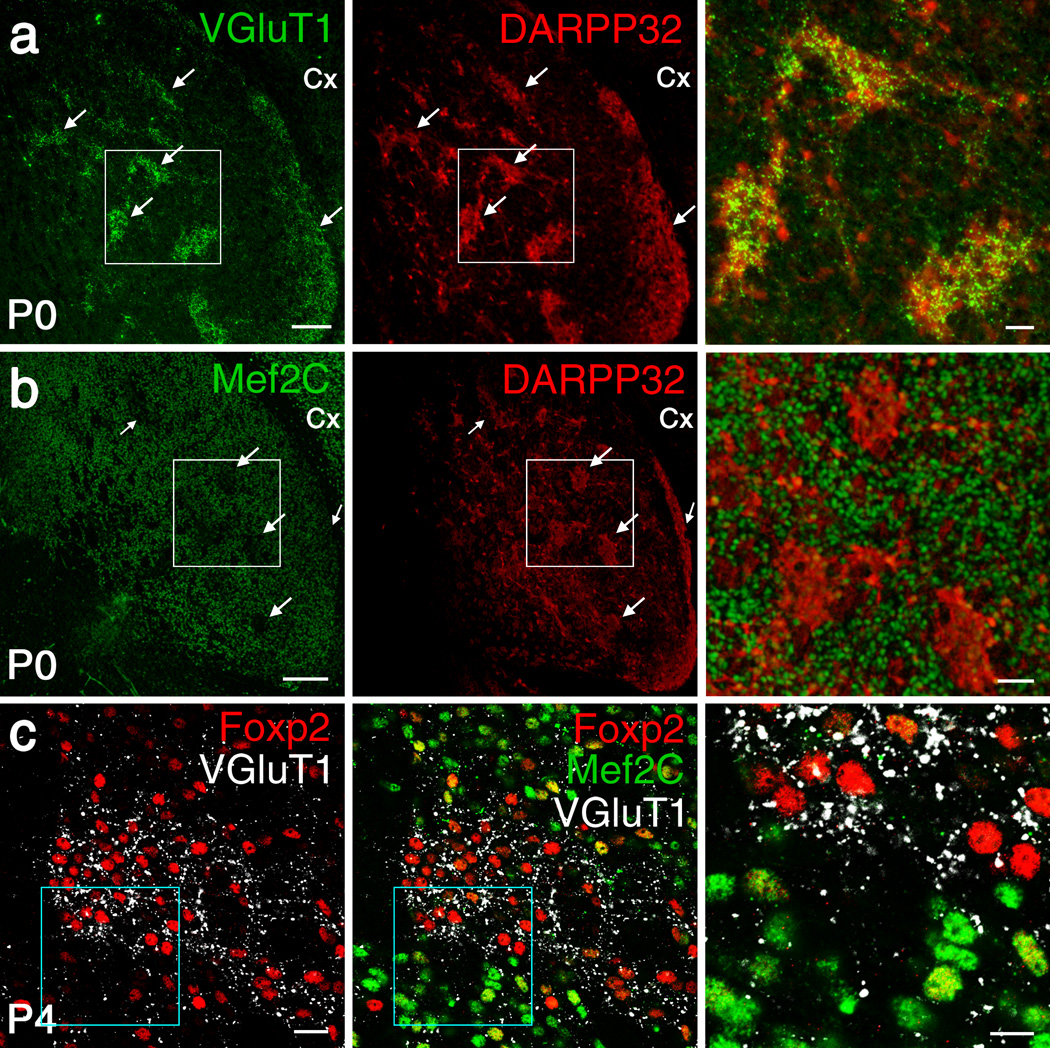
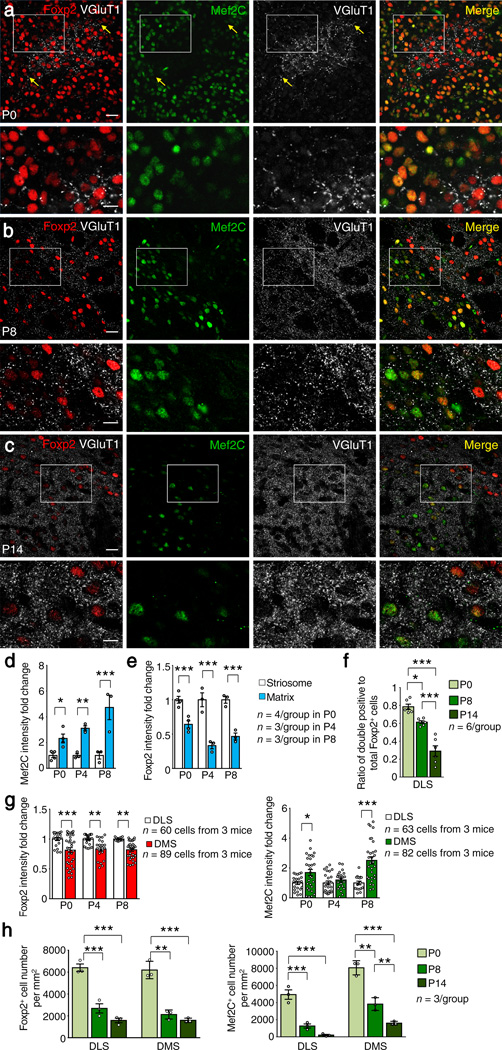
We performed immunostaining for Mef2C and Foxp2 in the mouse brain with antibodies validated by the absence of corresponding immunofluorescent signals in Mef2C and Foxp2 knockout striatum (Supplementary Fig. 1a–f). Mef2C and Foxp2 were co-localized in many individual striatal projection neurons (SPNs) at birth, but their expression patterns soon became complementary and non-overlapping (Figs. 1 and 2). This inverse patterning was first evident in the striosome compartment of the striatum, where we found the earliest incoming corticostriatal terminals as marked by vesicular glutamate transporters 1 (VGluT1; Fig. 1a). During the early postnatal (P) period, P0-P4, striosomes, marked by DARPP-32, were nearly devoid of Mef2C, despite strong Mef2C expression in the surrounding matrix (Figs. 1b and 2d). Strong Foxp2 expression, by contrast, was found in striosomes (Fig. 1). By P8 and P14, the inverse expression patterns of Foxp2 and Mef2C had extended across the striatum (Fig. 2b,c,f), paralleled by a progressive expansion of the corticostriatal innervation from the striosomes to the entire striatum (Fig. 2a–c)24, dorsolateral expansion leading slightly (Fig. 2g). This striking progression from co-localization to dissociation of Mef2C and Foxp2 expression coincided with the postnatal time-period during which corticostriatal axonal collaterals arborize and the dendritic spines of SPNs develop24–26, suggesting that Mef2C and Foxp2 might oppositely regulate corticostriatal synapse formation.
Figure 1.
Dissociation of Foxp2 and Mef2C in SPNs is initiated in VGluT1-positive (VGluT1+) striosomes in neonatal striatum. (a,b) Dual-antibody immunostaining shows VGluT1+ corticostriatal axon terminals in P0 striatum (a, left, arrows) clustered in DARPP-32+ striosomes (a, middle, arrows) that are devoid of Mef2C (b, arrows). Boxed regions in a and b are shown as merged images at higher magnification in the right panels of a and b. (c) In P4 striatum, VGluT1-rich, Mef2C–poor striosomes contain clustered, strongly Foxp2+ cells. Images represent five repeats. Scale bars: 100 µm (in a, b, left and middle), 25 µm (in a, b, right), 20 µm (in c, left and middle) and 10 µm (in c, right).
Figure 2.
Progressive dissociation of Foxp2 and Mef2C in SPNs during postnatal development. (a–c) Triple-antibody immunostaining shows predominant concentration of VGluT1+ corticostriatal axonal puncta in striosomes that are Mef2C–poor but exhibit strong Foxp2 immunostaining in P0 (a) and P8 (b) striatum. By P14 (c), VGluT1+ corticostriatal axonal puncta become widespread and relatively evenly distributed in the striatum, and Foxp2 expression is equally widespread, whereas Mef2C is down-regulated. The boxed regions in a–c are shown at high magnification at bottom rows. Note that strongly Foxp2+ cells express weak Mef2C, and vice versa. Yellow arrows in a mark the boundary of a striosome. Scale bars: 20 µm (top rows) and 10 µm (bottom rows). Images represent five repeats. (d,e) Mef2C (d) and Foxp2 (e) immunofluorescence intensity in P0-P8 wildtype striatum. Data represent at least three mice per group. (f) Ratio of cells positive for both Foxp2 and Mef2C relative to total Foxp2+ cells in the dorsolateral striatum (n = 6 mice/group). (g) Ratio of Foxp2 (left) or Mef2C (right) immunofluorescence intensity in striosomes of dorsomedial striatum (DMS) relative to that in dorsolateral striatum (DLS). Data represent at least 60 cells from three mice per group. (h) The densities of Foxp2+ and Mef2C+ cells in DLS and DMS of wildtype mice (n = 3 mice/group). *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent s.e.m. Two-way ANOVA with Tukey’s honest significant difference (HSD) post hoc test are used in d, F(2, 16) = 4.400, P = 0.033 for interaction, F(1, 16) = 49.072, P = 0.000006 for compartment, F(2, 16) = 4.400, P = 0.033 for age, in e, F(2,16) = 3.456, P = 0.060 for interaction, F(1,16) = 110.852, P = 0.000000 for compartment, F(2,16) = 3.001, P = 0.060 for age, and in g, for Foxp2, F(2, 139) = 0.039, P = 0.962 for interaction, F(1, 139) = 40.321, P = 0.000000 for region, F(2, 139) = 0.039, P = 0.962 for age; for Mef2C, F(2, 141) = 5.838, P = 0.004 for interaction, F(1, 141) = 25.477, P = 0.000001 for region. F(2, 141) = 5.838, P < 0.004 for age. One-way ANOVA with Tukey’s HSD post hoc test is used in f, F(2, 15) = 41.852, P = 0.000001, and in h, for Foxp2, F(2, 6) = 56.571, P = 0.000128 in DLS, F(2, 6) = 67.197, P = 0.000078 in DMS; for Mef2C, F(2, 6) = 50.443, P = 0.000177 in DLS, F(2, 6) = 75.536, P = 0.000056 in DMS.
Foxp2 and Mef2C inversely control corticostriatal synaptogenesis
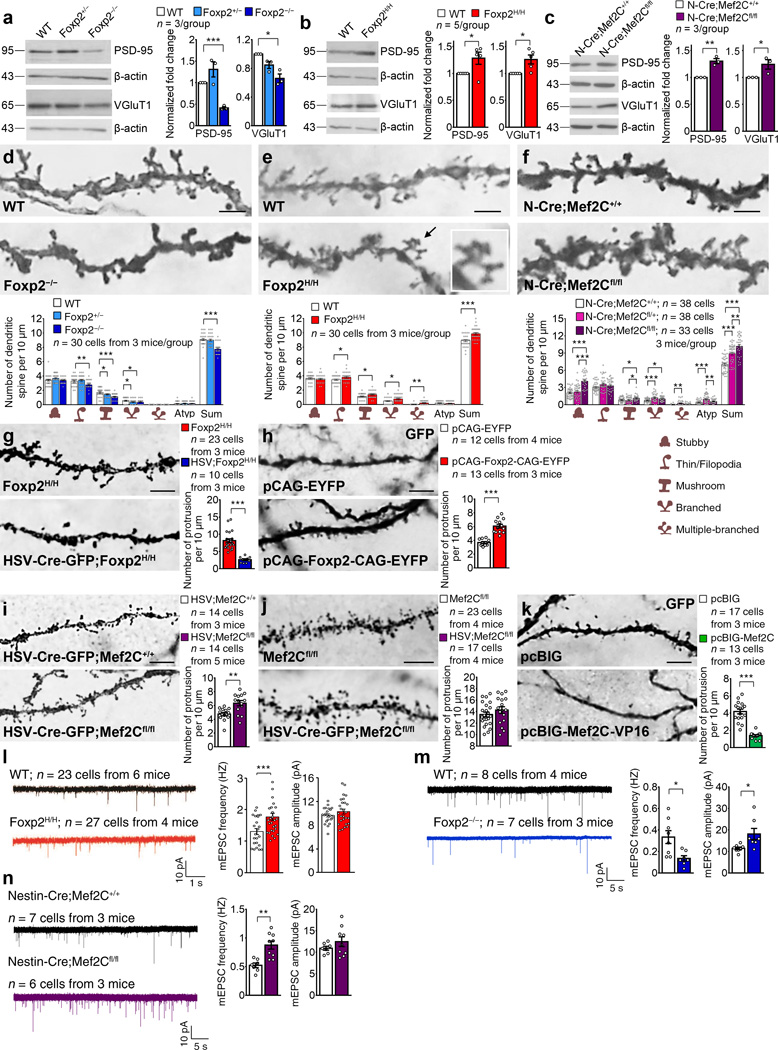
To examine the relationship of Foxp2 and Mef2C expression to the development of the corticostriatal projection system, we analyzed synaptogenesis in Mef2C knockout (Nestin-Cre;Mef2Cfl/fl)21 and Foxp2 knockout (Foxp2−/−)8 mice, and also in Foxp2 humanized (Foxp2H/H) mice8 as a gain-of-function model. We identified corticostriatal synapses with VGluT1 as a pre-synaptic marker27 and postsynaptic density protein 95 (PSD-95) and glutamate receptor 1 (GluR1) as post-synaptic markers (Supplementary Fig. 1g). We focused on P12-P14, by which time Foxp2 was expressed throughout the striatum and Mef2C was nearly absent except far medially (Fig. 2h).
In the Foxp2−/− knockout mice, western blots of striatal VGluT1 and PSD-95 proteins and immunostaining of striatal GluR1 showed that these synaptic markers all were reduced relative to those in wildtype mice (Fig. 3a and Supplementary Fig. 2a–c), but these markers were increased in the humanized Foxp2H/H striatum (Fig. 3b and Supplementary Fig. 2d–f), suggesting that Foxp2 positively regulates striatal synaptogenesis. By contrast, Mef2C negatively regulated striatal synaptogenesis. VGluT1 and PSD-95 proteins and GluR1 immunostaining significantly increased in the striatum of Nestin-Cre;Mef2Cfl/fl conditional Mef2C knockout mice relative to control Nestin-Cre;Mef2C+/+ mice (Fig. 3c and Supplementary Fig. 2g–i). Thus, Foxp2 and Mef2C had opposite effects on synaptogenesis in the developing striatum.
Figure 3.
Synaptic proteins and dendritic spines of SPNs are oppositely regulated by Foxp2 and Mef2C. (a–c) Western blots showing expression of PSD-95 and VGluT1 proteins in the P12 Foxp2−/− striatum (a), P14 Foxp2H/H striatum (b) and P14 Mef2C knockout striatum (c). Data represent at least three mice per genotype. Full-length blots are presented in Supplementary Fig. 10. (d–f) Golgi staining showing dendritic spines in dorsolateral SPNs of Foxp2−/− mice (d), Foxp2H/H (e) and Mef2C knockout (f) mice. Inset in e: multiple-branched spines. Data represent at least 30 cells from three mice per genotype. Atyp: atypical. Scale bars: 2.5 µm. (g) Intrastriatal injection of HSV-Cre-GFP virus in P2 Foxp2H/H mice decreases spine density in GFP-positive SPNs at P8. (h) In utero electroporation of pCAG-EYFP-CAG-Foxp2 at E13.5 increases spines of P14 wildtype SPNs. Scale bar: 5 µm. (i,j) Intrastriatal injection of HSV-Cre-GFP virus in Mef2Cfl/fl mice at P2 increases spines in GFP-positive SPNs at P8 (i), but the same injection at P14-P15 does not affect spine counts at P19-P20 (j). Scale bars: 5 µm. (k) In utero electroporation of pcBIG-Mef2C–VP16 at E12.5 decreases spines of P14 wildtype SPNs. Scale bar: 5 µm. Data in g–k represent at least 10 cells from three mice per group. (l–n) mEPSC recordings of SPNs in Foxp2H/H (l), Foxp2−/− (m), and Mef2C knockout (n) mice. Data represent at least 7 cells from at least three mice per genotype. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent s.e.m. One-way ANOVA with Tukey’s HSD post hoc test are used in a, for VGluT1, F(2, 6) = 14.649, P = 0.005; for PSD-95, F(2, 6) = 19.954, P = 0.002, d”, for stubby, F(2, 87) = 2.992, P = 0.055; for thin/filopodia, F(2, 87) = 8.396, P = 0.000463; for mushroom, F(2, 87) = 12.889, P = 0.000013; for branched, F(2, 87) = 4.169, P = 0.019; for atypical, F(2, 87) = 0.293, P = 0.747; for sum, F(2, 87) = 28.470, P = 0.000000, f”, for stubby, F(2,106) = 34.658, P = 0.000000; for thin/filopodia, F(2,106) = 2.012, P = 0.139; for mushroom, F(2,106) = 5.182, P = 0.007; for branched, F(2,106) = 9.555, P = 0.000153; for multiple branched, F(2,106) = 5.965, P = 0.004 for atypical, F(2,106) = 11.944, P = 0.000021; for sum, F(2,106) = 38.448, P = 0.000000. Student’s t test are used in b, for VGluT1, t(8) = −3.155, P = 0.014; for PSD-95, t(8) = 2.518, P = 0.036, c, for PSD-95, t(4) = −6.442, P = 0.003; for VGluT1, t(4) = −2.795, P = 0.049, e”, for stubby, t(29) = 0.623, P = 0.538; for thin/filopodia, t(29) = −2.249, P = 0.032; for mushroom, t(29) = −2.041, P = 0.050; for branched, t(29) = −2.300, P = 0.029; for multiple branched, t(29) = −3.525, P = 0.001, for atypical, t(29) = 0.769, P = 0.448; for sum, t(29) = −4.216, P = 0.000383, g, t(31) = 9.429, P = 0.000000, h, t(23) = −7.612, P = 0.000001, i, t(26) = −3.509, P = 0.002, j, t(38) = −1.122, P = 0.269, k, t(28) = 9.453, P = 0.000000, l, for frequency, t(13) = 2.890, P = 0.013; for amplitude, t(13) = −2.2505, P = 0.043, and n, for frequency, t(14) = −4.022, P = 0.001, for amplitude, t(14) = −1.131, P = 0.237. Mann-Whitney U test are used in m, U = 167.5, P = 0.009 for frequency and U = 273, P = 0.471 for amplitude.
Markers of striosomes (µ-opioid receptor 1, MOR1) and of the matrix (calcium and diacylglycerol-guanine nucleotide exchange factor I, CalDAG-GEFI) showed that striosome-matrix compartmentation was maintained in the three genotypes (Supplementary Fig. 3a–f). No abnormal cell death was found in P0 or P8 Foxp2−/− knockouts, nor in Nestin-Cre;Mef2Cfl/fl knockout mice at embryonic day (E) 18.5 and P8, as assayed by TUNEL staining and immunostaining for activated caspase3 (Supplementary Fig. 3i–r). The alterations in synaptic markers were thus likely not secondary to loss of compartmentation or to widespread cell death in the mutant mice.
Foxp2 and Mef2C oppositely regulate SPN spinogenesis
The dendritic spines of striatal SPNs are the main sites of corticostriatal synapses. We examined the development of these synapses in the three mouse genotypes (Fig. 3d–f, Supplementary Fig. 4 and Supplementary Table 1–3). Golgi-staining in P12 Foxp2−/− knockout mice showed reductions in the total spine density of SPNs of 14.0% dorsolaterally and 19.1% dorsomedially (Fig. 3d, Supplementary Fig. 4a and Supplementary Table 1). In heterozygous Foxp2+/− mice, only the most mature spine types28 were reduced (Fig. 3d, Supplementary Fig. 4a and Supplementary Table 1), a finding potentially relevant to pathophysiology in KE patients, who carry heterozygous FOXP2R553H missense mutations9. In the positive control mice carrying humanized Foxp2H/H, SPN spine densities were increased both dorsolaterally and dorsomedially relative to those in wildtype littermates (Fig. 3e, Supplementary Fig. 4b and Supplementary Table 2). We microinjected HSV-Cre-GFP viruses into the striatum of P2 Foxp2H/H mice carrying floxed human versions of Foxp2 alleles8. Immunostaining demonstrated a loss of Foxp2 protein expression in HSV-Cre-GFP virus-infected cells (Supplementary Fig. 5d–e), and this postnatal deletion of Foxp2H/H decreased spines by 68% in HSV-Cre-GFP virus-infected SPNs at P8 (Fig. 3g). These findings suggested that Foxp2 positively regulated synaptogenesis, but that Mef2C negatively regulated synaptogenesis, as found in hippocampal neurons20–22.
We used in utero electroporation to over-express mouse Foxp2 gene in wildtype striatal cells. Introduction of Foxp2 plasmid co-expressing enhanced yellow fluorescent protein (EYFP) into the E13.5 striatal primordium increased dendritic spine numbers as tested by EYFP/GFP immunostaining of P14 SPNs (Fig. 3h). Conversely, spine density was significantly increased in striatal SPNs in Nestin-Cre;Mef2Cfl/fl conditional knockouts relative to control Nestin-Cre;Mef2C+/+ mice for all but the least mature subtypes (Fig. 3f, Supplementary Fig. 4c and Supplementary Table 3).
We also microinjected HSV-Cre-GFP virus into the striatum of P2 Mef2Cfl/fl pups and examined their brains at P8. Immunostaining confirmed loss of Mef2C protein expression in HSV-Cre-GFP virus-infected cells (Supplementary Fig. 5a–c) and indicated significant increases in spine counts in the HSV-Cre-GFP virus-infected SPNs (Fig. 3i). Intrastriatal injection of HSV-Cre-GFP viruses in Mef2Cfl/fl mice at P14–15, when Mef2C expression already was low (Fig. 2h), showed no detectable changes in dendritic spines of SPNs at P19-P20 (Fig. 3j). Finally, we over-expressed Mef2C by in utero electroporation of pcBIG-Mef2C–VP16 plasmids into the E12.5 striatal primoridum of wildtypes. This procedure decreased the number of spines in P14 SPNs (Fig. 3k). These findings identify Mef2C as a negative regulator of spine and synapse formation in SPNs of the developing postnatal striatum, consonant with in vitro work on Mef2-class proteins23.
In slice preparations, we analyzed miniature excitatory postsynaptic currents (mEPSCs) of dorsolateral SPNs in Foxp2H/H, Foxp2 knockout, and Nestin-Cre;Mef2Cfl/fl knockout brains relative to their wildtype controls. Whole-cell patch-clamp recordings indicated an increase in mEPSC frequency in 36.2% of SPNs in P14 Foxp2H/H mice without significant change in mEPSC amplitudes (Fig. 3l). In P8 Foxp2−/− knockout mice, mEPSC frequency was decreased in SPNs, but mEPSC amplitude was increased compared to wildtype (Fig. 3m). In P14 Nestin-Cre;Mef2Cfl/fl knockout brains, mEPSC frequency, but not amplitude, was increased in SPNs relative to control Nestin-Cre;Mef2C+/+ mice (Fig. 3n). These findings demonstrated that synaptic activity in striatal SPNs is positively regulated by Foxp2, but negatively regulated by Mef2C, consistent with our anatomical findings (Fig. 3, Supplementary Fig. 4 and Supplementary Tables 1–3).
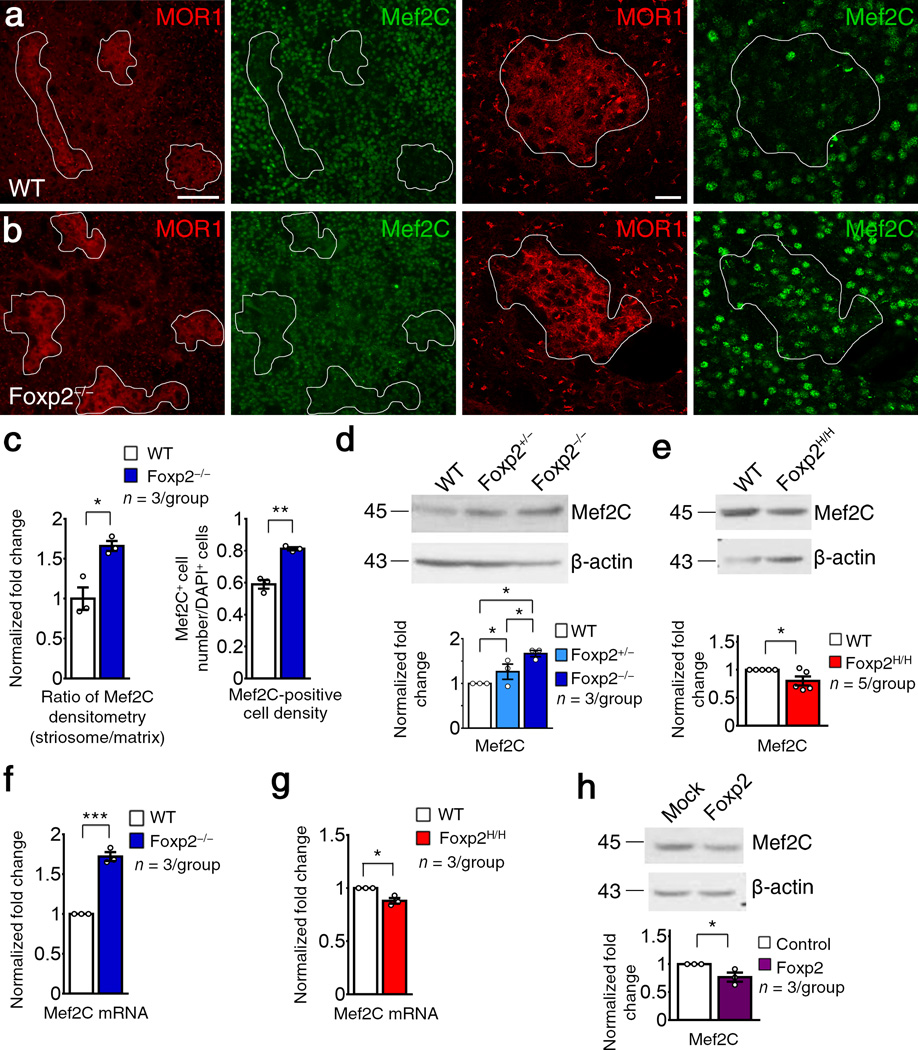
Foxp2 suppresses Mef2C expression in striatal SPNs
The opposing functions of Mef2C and Foxp2 in regulating corticostriatal synapse formation and SPN spinogenesis, and their striking spatiotemporal dissociation during neonatal development, suggested that the positive regulation of spine and synapse formation in striatal SPNs by Foxp2 might depend on its ability to suppress Mef2C. As an initial probe for this potential interaction, we asked whether the near-absence of Mef2C expression in striosomes in the normal neonatal striatum could be reversed by deletion of Foxp2. We found striking de-repression of Mef2C expression in striosomes in the P7 Foxp2−/− striatum (Fig. 4a,b) with the striosome predominance of immunofluorescence intensity of Mef2C–positive cells increased in the Foxp2−/− knockout mice compared to levels in wildtypes (Fig. 4c). In western blots, Mef2C protein levels were increased by 68.3% over control levels in the P12 Foxp2−/− striatum (Fig. 4d) and were decreased in P14 Foxp2H/H mice (Fig. 4e). In quantitative real-time polymerase chain reaction (qRT-PCR) assays, Mef2C mRNA was increased in Foxp2−/− knockout striatum (Fig. 4f), but decreased in the Foxp2H/H striatum (Fig. 4g). In vitro over-expression of Foxp2 in Neuro2A (N2A) cells was sufficient to decrease Mef2C protein by 23.5% (Fig. 4h). These findings were consistent with an antagonism of Mef2C by Foxp2 in the striatum. Mef2C was persistently expressed in neocortex of postnatal wildtype mice (Supplementary Fig. 1h–j). We found no changes in Mef2C protein levels or Mef2C–positive cell numbers in the neocortex of P8 Foxp2−/− knockouts (Supplementary Fig. 6a–d).
Figure 4.
Foxp2 suppresses Mef2C expression in SPNs. (a–b) Dual immunostaining of Mef2C and MOR1 in P7 wildtype (a) and Foxp2−/− (b) striatum. Scale bars: 50 µm (a, two left panels) and 20 µm (a, two right panels). (c) Ratio of immunofluorescence intensity of Mef2C+ SPNs in striosomes over those in the matrix in Foxp2−/− mice compared to wildtype mice (left, Student’s t test; t(4) = −4.317, P = 0.012), and density of Mef2C–positive cells in P7 Foxp2−/− striatum relative to wildtype (right, t(4) = −7.692, P = 0.002). Images and data represent three mice per group. (d,e) Western blots showing expression of Mef2C protein in P12 Foxp2−/− striatum (d; Foxp2+/+ vs. Foxp2+/−: P = 0.022, t(4) = −3.645; Foxp2+/− vs. Foxp2−/−: P = 0.018, t(4) = −3.875; Foxp2+/+ vs. Foxp2−/−: P = 0.016, t(4) = −7.913) and P14 Foxp2H/H striatum (e; t(8) = 2.480, P = 0.038). Data represent at least three mice per genotype. (f,g) qRT-PCR analyses of Mef2C mRNA levels in P12 Foxp2−/− striatum (f; t(4) = −12.987, P < 0.001) and ,P14 Foxp2H/H striatum (g; t(4) = 4.540, P = 0.010). Data represent three mice per genotype. (h) Western blots showing the level of Mef2C protein in N2A cells transfected with Foxp2 expressing plasmid (t(4) = 2.853, P = 0.046). Data represent three repeats. Full-length blots are presented in Supplementary Fig. 10. *P < 0.05, **P < 0.01. Error bars represent s.e.m.
We tracked striatal Mef2C and Foxp2 protein levels by western blot analysis from P4 to P21. For Foxp2 knockout striatum at P4, the levels of Mef2C and the synaptic markers VGluT1, PSD-95 and GluR1 were not different from controls (Supplementary Fig. 6e), but by P8, there was a significant increase of Mef2C and significant decreases of VGluT1, PSD-95 and GluR1 (Supplementary Fig. 6f), patterns maintained at P12 (Supplementary Fig. 6g). In the humanized Foxp2 striatum, Mef2C was decreased, and VGluT1 and GluR1 were increased as early as P4 relative to controls (Supplementary Fig. 6h) and remained elevated at P8 and P21 (Supplementary Fig. 6i,j). These results delineated consistent negative interactions of Foxp2 and Mef2C in their effects on striatal synaptogenesis detectable around P4-P8 and persisting for at least 3 postnatal weeks, suggesting that Foxp2 could act to repress Mef2C expression in the striatum.
Mef2C is a direct downstream target gene of Foxp2
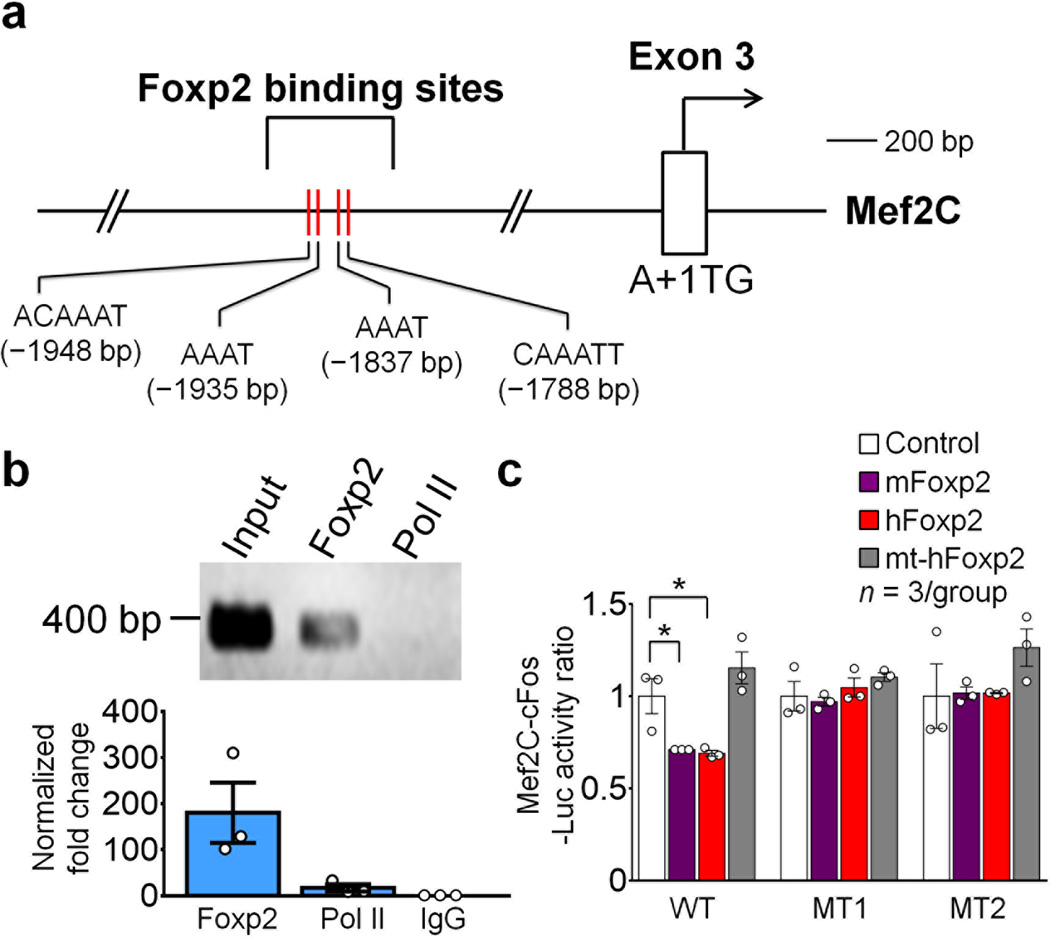
One mechanism by which repression of Mef2C by Foxp2 could occur would be if the Mef2C gene were to contain the canonical FOXP2 binding site “CAAATT” and the binding site core, “AAAT”18. Foxp2 binds to DNA by dimerization29. We found two pairs of putative Foxp2 binding sites in the second intron of the Mef2C gene [ACAAAT (−1948 bp) & AAAT (−1935 bp); and AAAT (−1837 bp) & CAAATT (−1788 bp); translational initiation site A+1TG; Fig. 5a]. With ChIP-quantitative PCR (ChIP-qPCR) assays of mouse striatal cells at E16, we found that Foxp2 antibody, but not control IgG antibody, immunoprecipitated a DNA fragment containing the two pairs of Foxp2 motifs (Fig. 5b). Thus, Foxp2 can bind directly to this region containing the canonical Foxp2 binding motifs in vivo.
Figure 5.
Mef2C is a direct target gene of Foxp2. (a) Foxp2 binding motifs in the second intron of mouse Mef2C gene. (b) ChIP-qPCR analysis of E16 striatum. Foxp2 antibody, but not the control Pol II antibody, immunoprecipitates a DNA fragment containing the two pairs of Foxp2 motifs. Full-length gel is presented in Supplementary Fig. 10. (c) Transfection of mouse Foxp2 or human FOXP2 plasmids, but not transfection of human FOXP2R553H mutant (mt-hFOXP2) plasmid, represses luciferase activity of Mef2C–pCL3-c-fos-Luc reporter in N2A cells. Repression by mouse Foxp2 or human FOXP2 in wildtypes (WT) is abolished with Mef2C–MT1-c-fos-Luc (MT1) or Mef2C–MT2-c-cfos-Luc (MT2) mutant plasmids (Student’s t test; WT: control vs. mFoxp2, t(4) = 3.017, P = 0.039; control vs. hFoxp2, t(4) = 3.237, P = 0.032). Data represent three repeats. *P < 0.05. Error bars represent s.e.m.
We cloned this DNA region into a pGL3-c-fos-Luciferase (Luc) reporter gene construct and co-transfected into N2A cells the Mef2C–pGL3-c-fos-Luc plasmid with a Foxp2 expression plasmid. Transfection of the mouse Foxp2 gene repressed Luc activity by 29% compared to mock controls (Fig. 5c). Transfection of the human FOXP2 gene, but not the hFOXP2R553H mutant lacking DNA binding capacity, resulted in a similar (31%) decrease of Luc activity (Fig. 5c).
We next mutated the sequences of the ACAAAT and AAAT motifs to ACGGGT & GGGT (MT1), and the sequences of the AAAT & CAAATT motifs to GGGT & CGGGTT (MT2) by site-directed mutagenesis. The repression of Luc reporter gene activity was abolished when either Mef2C–MT1-c-fos-Luc or Mef2C–MT2-c-cfos-Luc mutant plasmid was co-transfected with mouse Foxp2 expression plasmid (Fig. 5c). Loss of repression of Luc activity also occurred with co-transfection of human FOXP2 gene and the MT1 or MT2 mutant reporter gene constructs (Fig. 5c). These results suggest that Foxp2 directly binds to the two canonical Foxp2 binding motifs to repress functionally the capacity for Mef2C gene expression.
We looked for, and identified, putative FOXP2 binding motifs in the promoter region of the human MEF2C gene (Supplementary Fig. 7), a finding potentially linking our mouse work to the suggestion that human MEF2C gene is a putative target gene of FOXP2 in human embryonic basal ganglia tissue18.
Mef2C deletion rescues USVs and SPN spines in Foxp2+/− mice
Foxp2 heterozygous and homozygous knockout mice exhibit defective neonatal isolation-induced USVs30, a form of vocal communication in neonatal rodents. Our finding that Mef2C is normally repressed by Foxp2 suggested that, by reducing abnormally overactive Mef2C in Foxp2 knockouts, we might be able to rescue such defective USV in Foxp2 mutants. If so, this link would constitute functional evidence for an interaction of the two proteins in vocal communication.
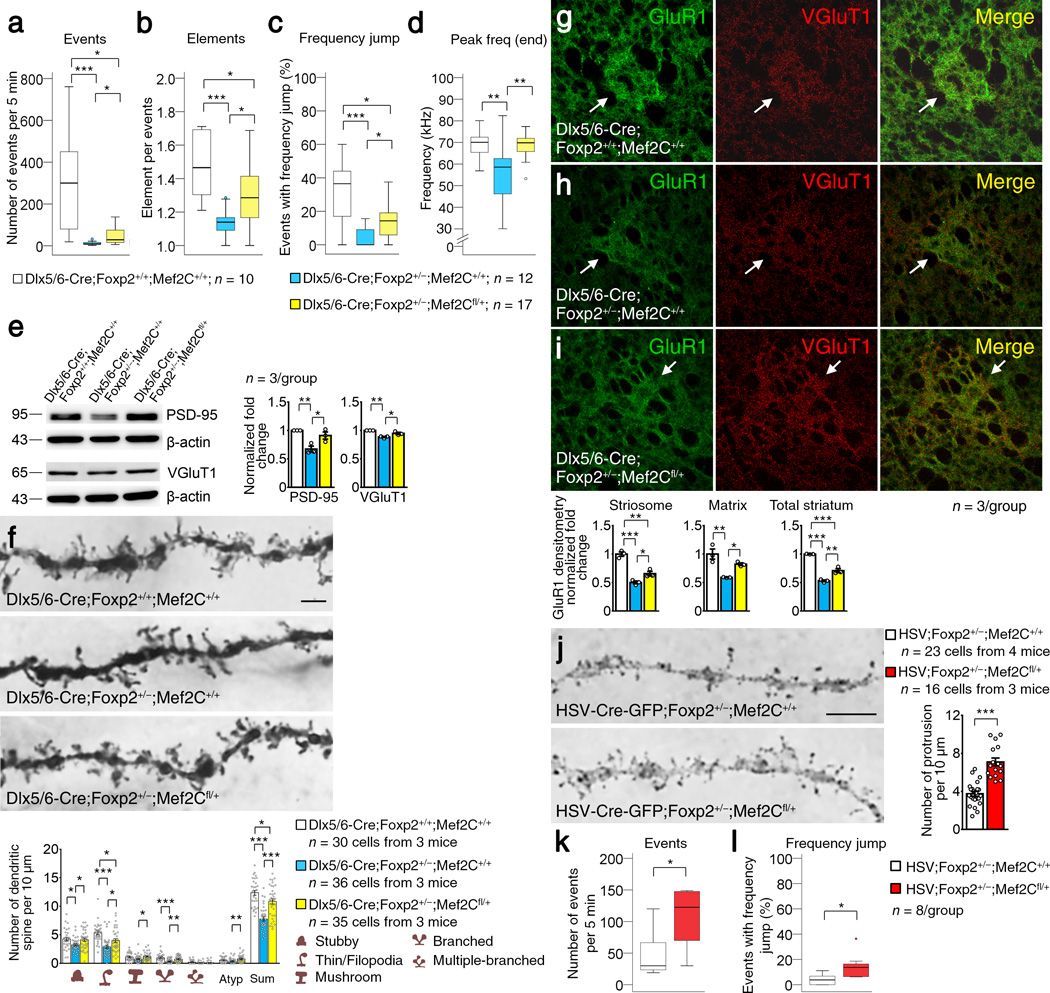
We tested this possibility in Foxp2+/− heterozygotes, mouse models of humans carrying one allele of the FOXP2 mutation. We confirmed that these Foxp2+/− mutants had abnormal USVs (Supplementary Fig. 8a) and then examined the effects of inactivation of one allele in Mef2Cfl/fl, accomplished by genetic introduction of striatum-enriched Dlx5/6-Cre in Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ mice. This partial inactivation of Mef2C significantly rescued many features of the abnormal USVs characteristic of P8 Foxp2+/− mice, as compared to USVs of control P8 Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ mice (Fig. 6a–d and Supplementary Fig. 8b). The fact that we found no marked differences in USV features between Dlx5/6-Cre;Foxp2+/+;Mef2Cfl/+ mice, which had normal Foxp2 but a deleted Mef2C allele, and Dlx5/6-Cre;Foxp2+/+;Mef2C+/+ mice, which had normal Foxp2 and normal Mef2C alleles, ruled out the possibility that the effect of Mef2C reduction was mediated through a pathway independent of Foxp2 (Supplementary Fig. 8c).
Figure 6.
Inactivation of one allele of Mef2C rescues USV, dendritic spines and synaptic protein changes in P8 Foxp2+/− mice. (a–d) Analysis of USVs in P8 Dlx5/6-Cre;Foxp2+/+;Mef2C+/+ (white), Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ (blue) and Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ (yellow) mice. Box plots show the median (horizontal line in the box), range between the 25th and 75th percentiles (box), and 1.5 times this interquartile range (T-bars). Circles show outlying values. Data represent at least ten mice per genotype. (e) Western blots showing increased expression of PSD-95 and VGluT1 protein expression in P8 Dlx5/6-Cre;Foxp2+/+;Mef2C+/+: Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ and Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ striatum. Data represent three mice per genotype. Full-length blots are presented in Supplementary Fig. 10. (f) SPN dendritic spines of P8 Dlx5/6-Cre;Foxp2+/+;Mef2C+/+ (top), Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ (middle) and Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ (bottom) mice. Data represent at least 30 cells from three mice per genotype. Scale bars: 2.5 µm. (g–i) GluR1 immunofluorescence in striosomes and matrix of P8 Dlx5/6-Cre;Foxp2+/+;Mef2C+/+ (g), Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ (h) and Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ (i) striatum. Images and data represent three mice per genotype. Scale bars: 50 µm. (j) Dendritic spines are increased in SPNs of HSV-Cre;Foxp2+/−;Mef2Cfl/+ mice (red) compared to that in control SPNs of HSV-Cre;Foxp2+/−;Mef2C+/+ mice (white). Data represent at least 16 cells from three mice per group. Scale bars: 5 µm. (k,l) The events (k) and frequency jump (l) are increased in HSV-Cre-GFP;Foxp2+/−;Mef2Cfl/+ mice compared to control HSV-Cre-GFP;Foxp2+/−;Mef2C+/+ mice. Data represent 8 mice per group. *P < 0.05, **P < 0.01. ***P < 0.001. Error bars represent s.e.m. Kruskal-Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons test are used in a, test statistic = 21.474, P < 0.001, c, test statistic = 15.832, P < 0.001. One way ANOVA are used in b, F(2, 36) = 10.768, P = 0.000238, d, F(2, 36) = 7.296, P = 0.002, e, for PSD-95, F(2, 6) = 12.645, P = 0.007; for VGluT1, F(2, 6) = 21.487, P = 0.002, f, for stubby, F(2, 98) = 4.521, P = 0.013; for thin/filopodia, F(2, 98) = 13.875, P = 0.000005; for mushroom, F(2, 98) = 3.816, P = 0.025; for branched, F(2, 98) = 11.563, P = 0.000031; for multiple branched, F(2, 98) = 2.023, P = 0.138, for atypical, F(2, 98) = 4.709, P = 0.011; for sum, F(2, 98) = 30.941, P = 0.000000, g-i, for striosome, F(2, 6) = 54.063, P = 0.000145; for matrix, F(2, 6) = 15.055, P = 0.005; for total striatum, F(2, 6) = 117.718, P = 0.000015. Student’s t test is used in j, t(26) = −7.410, P = 0.000000, k, t(14) = −2.789, P = 0.014, and l, t(14) = −2.727, P = 0.016.
The single-allele genetic reduction of Mef2C levels also significantly rescued VGluT1 and PSD-95 protein levels in the striatum of Mef2C–deficient Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ mice compared to those in control Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ mice (Fig. 6e). SPN spine counts were markedly increased dorsolaterally and dorsomedially (Fig. 6f, Supplementary Fig. 4d and Supplementary Table 4). These synaptic rescue effects were evident in both striosome and matrix compartments, judging by significantly increased immunostaining for GluR1 (Fig. 6g–i).
To test the possibility that the spine rescue occurred because of general effects of genetic deletion on development, we restricted deletion of Mef2C specifically in the neonatal striatum by intrastriatal microinjection of HSV-Cre-GFP virus of P2 Foxp2+/−;Mef2Cfl/+ pups and performing GFP immunostaining and Golgi assays at P8. This striatum-specific decrease of Mef2C resulted in significant increases in dendritic spines in HSV-Cre-GFP virus-infected SPNs of Foxp2+/−;Mef2Cfl/+ mice compared to the spines of SPNs in control Foxp2+/−;Mef2C+/+ mice (Fig. 6j). The USV events and frequency-jump scores of the pups were also rescued in the HSV-Cre-GFP;Foxp2+/−;Mef2Cfl/+ pups (Fig. 6k,l and Supplementary Fig. 8d).
These multiple lines of evidence directly suggest that corticostriatal synaptic protein levels are suppressed by Mef2C and can be co-rescued along with behavioral rescue of USVs by expression of Foxp2. These findings suggest that Mef2C acts downstream of Foxp2 to regulate synaptic wiring of corticostriatal circuits that contribute to vocalization.
USV deficits occur in Mef2C knockout mice
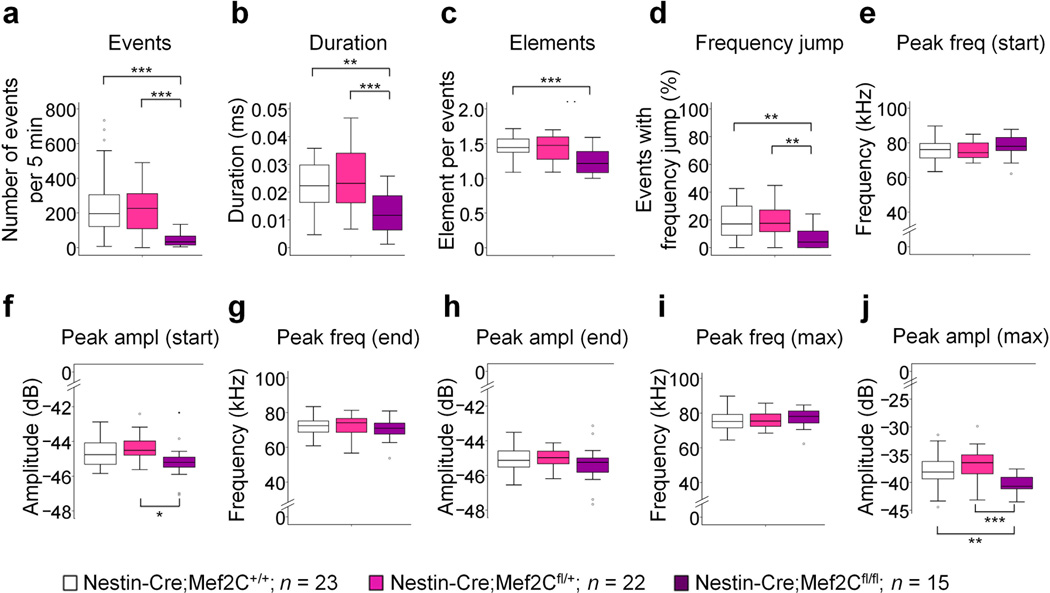
Conditional Mef2C knockout (Nestin-Cre;Mef2Cfl/fl) mice exhibit autistic-like behavioral phenotypes, putatively murine counterparts of Rett syndrome features, including increased anxiety, decreased cognition and paw wringing/clasping stereotypy31. It remained unclear whether such Nestin-Cre;Mef2Cfl/fl mice displayed deficits in vocal communication. Because our rescue experiments suggested that Mef2C acts downstream of Foxp2 to regulate USVs (Fig. 6a–d,k-l), we expected that deletion of Mef2C would alter USVs. We tested this possibility and found that the numbers of emitted USVs were significantly reduced in the Nestin-Cre;Mef2Cfl/fl mice, compared to those in control Nestin-Cre;Mef2C+/+ mice (Fig. 7a), and that the deficit included significant reductions in many USV features (Fig. 7a–d,j), though some USV features were unaffected (Fig. 7e–i). Thus Mef2C deletion can result in a deficit in USVs, a vocalization-like phenotype, adding evidence consistent with the working proposal, made here, that Foxp2-Mef2C signaling could regulate vocalization along with regulating corticostriatal connectivity.
Figure 7.
Reduction of USVs in Nestin-Cre;Mef2C knockout mice at P8. The number of USV calls (a; one-way ANOVA with Tukey’s HSD post hoc test; F(2, 66) = 11.001, P = 0.000196), call durations (b; F(2, 66) = 8.613, P = 0.000475), numbers of elements in each call (c; F(2, 66) = 6.444, P = 0.003), proportion of events with frequency jumps (d; F(2, 66) = 5.723, P = 0.005) and maximum peak amplitudes (j; F(2, 66) = 10.022, P = 0.000371) are decreased in Nestin-Cre;Mef2Cfl/fl mice, compared to those in control Nestin-Cre;Mef2C+/+ mice, whereas peak frequency at start (e; F(2, 66) = 2.497, P = 0.090) and end (g; F(2, 66) = 0.069, P = 0.933) of calls, amplitudes at start (f; F(2, 66) = 4.404, P = 0.016) and end (h; F(2, 66) = 1.325, P = 0.273) of calls, and maximum peak frequency (i; F(2, 66) = 1.218, P = 0.303) are not affected. Data represent at least 15 mice per genotype. Box plots show the median (horizontal line in the box), range between the 25th and 75th percentiles (box), and 1.5 times this interquartile range (T-bars). Outlying values are marked as circles. *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
Our findings provide causal evidence that Mef2C is directly implicated in the conjoint deleterious behavioral and morphological effects of reductions in Foxp2. We demonstrate that Mef2C is a negative regulator of synapse formation in the projection neurons of the developing striatum, and that Mef2C expression is itself a direct target of gene suppression by the Foxp2 gene in these neurons (Supplementary Fig. 9). Thus Foxp2, by negatively regulating Mef2C, can promote the formation of the massive corticostriatal connectivity that is known to be functionally important in behavior.
Our findings suggest that Foxp2 acts in the early postnatal period by coordinately promoting corticostriatal synaptogenesis and retarding the expression of Mef2C in the striatum. Deletion of Foxp2 increased neonatal striatal Mef2C expression and, in parallel, decreased corticostriatal synaptogenesis and spinogenesis of striatal SPNs. Engineered expression of humanized Foxp2 in mouse had opposite effects on the developing corticostriatal circuits and striatal neurons. Our evidence that canonical Foxp2 binding sites are required for the Foxp2 inhibition of Mef2C expression suggests that there are direct molecular interactions between Foxp2 and Mef2C. We found that intrastriatal as well as genome-wide decreases in Mef2C can rescue defects in USVs and striatal spinogenesis otherwise occurring in Foxp2 mutants. Finally, our study of USVs suggests that these Foxp2-Mef2C interactions exert orchestrated control over corticostriatal systems affecting social vocalization in neonates. Our findings are based on mouse models, and so might not be relevant to clinical conditions. Given that MEF2C has been suggested to be an ASD candidate risk gene and is known to be a gene related to mental retardation15–17,19, and that FOXP2 has been identified as a language-associated gene9, a plausible hypothesis raised by our findings is that the Foxp2-Mef2C interactions we demonstrate here, and their profound effects on corticostriatal circuitry, could be relevant to mechanisms underlying spoken language dysfunction in ASD. More generally, such interactions, controlling the development of the corticostriatal system, could be relevant to a range of normal and abnormal functions in which this massive corticostriatal circuitry has been implicated clinically, notably including Huntington’s disease.
Foxp2-Mef2C control of USVs and corticostriatal circuit
The potential behavioral significance of interactions between Foxp2 and Mef2C was demonstrated by assays of USVs in mice that lacked one allele of Foxp2 and one allele of Mef2C (Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ mice). Genetic reduction of Mef2C levels rescued four of the seven USV defects exhibited by Foxp2 heterozygotes and increased VGluT1 and PSD-95 synaptic proteins in the striatum, relative to levels of the corresponding proteins in control Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ mice. The USV and spine formation deficits were also partially rescued by selective early postnatal reduction of Mef2C. While not ruling out other contributions, these findings support the striatal specificity of Foxp2-Mef2C interactions in regulating synaptogenesis in SPNs and vocalization behavior of the pups. We also found a clear correlation between synaptic alterations in Dlx5/6-Cre;Foxp2+/−;Mef2C+/+ mice (Foxp2 heterozygotes) and the USV deficits in these mice, further linking anatomical and functional control by Foxp2-Mef2C signaling.
Foxp2 as a molecular key to unlock Mef2C–induced inhibition
Our findings demonstrate that interactions between Foxp2 and Mef2C are required for establishing the normal synaptic connectivity of corticostriatal circuits. In the hippocampus, Mef2 protein interacts with the ASD-risk genes, FMRP and Pcdh10, to induce degradation of PSD-95 protein for elimination of excitatory synapses22. This work suggests a central role for Mef2 in negative regulation of hippocampal synapses. Our findings identify Foxp2 as a molecular key to unlock the inhibition of striatal synapse formation imposed by Mef2C, thereby allowing the initiation of synaptic connectivity of corticostriatal circuits within the striatum. Our data support a postnatal role, as early as P2, for Foxp2-mediated repression of Mef2C in the regulation of spinogenesis. As spines start to grow in postnatal SPNs, Mef2C, which inhibits spine formation, presumably must be down-regulated to permit the progression of spine formation in SPNs. The gradual decrease in Mef2C in the postnatal striatum and the maintenance of these low levels at least until P14 are consistent with this hypothesis. The evidence of increased spine formation in SPNs by postnatal deletion of Mef2C gene at P2 further supports this working model. Our experiments did not clarify whether Mef2C acts to prune exuberant striatal SPN synapses or to inhibit synapse formation in these SPNs. However, given our observation that Mef2C is strongly expressed in striatal matrix SPNs before these SPNs are innervated by corticostriatal axon terminals in newborn striatum (Fig. 1b), an intriguing possibility is that Mef2C functions as a molecular gate to prevent immature synapse formation unless it is triggered by an appropriate input. This gate-control role for Mef2C would suggest that a rigorously timed and specific molecular mechanism is implemented for developmental control of synaptic wiring of corticostriatal circuits. We suggest that Foxp2 inhibition of Mef2C could provide such a mechanism, potentially through direct DNA binding of Foxp2 to Mef2C, and that defects in this inhibition by Foxp2 could have deleterious effects.
ChIP-chip analysis has identified MEF2D in addition to MEF2C as potential target genes of FOXP2 in embryonic human brains18. For other members of Mef2 family, in contrast to the low levels of Mef2C in the mature striatum, levels of Mef2D and Mef2A are high in the mature mouse striatum, where they have been implicated in the plasticity of dendritic spines in SPNs induced by exposure to cocaine32. In the hippocampus, dendritic spine formation is negatively regulated by Mef2C in the dentate gyrus of hGFAP-Cre;Mef2C mice21, but not in the CA1 region of hGFAP-Cre;Mef2A;Mef2D double-knockout mice or hGFAP-Cre;Mef2A;Mef2C;Mef2D triple-knockout mice, apparently reflecting low Mef2C expression in the CA1 region33. Postnatal deletion of Mef2C also results in increased spines in the dentate gyrus in CaMKII-Cre;Mef2C knockout mice34.
Comparison of Foxp2 in striatal and cortical synaptogenesis
In contrast to the developmental down-regulation of Mef2C in postnatal striatum, Mef2C was persistently expressed in postnatal neocortex of wildtype mice. Our finding that Mef2C protein was not altered in the neocortex of Foxp2 knockout mice suggests that Foxp2-Mef2C interaction does not dominate the development of cortical neurons, but leaves open the possibility of some compensatory mechanism in the neocortex. In cultured neocortical neurons, Foxp2 has been found to regulate negatively excitatory synaptogenesis via inhibition of SPRX2, an epilepsy- and language-associated gene35. The positive action of Foxp2-Mef2C signaling on synaptogenesis in the striatum, shown here, and the reported negative action of Foxp2-SPRX2 signaling on synaptogenesis in cortical neurons, suggest a commanding and highly differentiated role for Foxp2 in developmental control of neural circuits that have been related to language. In the avian model of vocal communication, knocking down songbird FoxP2 in Area X decreases spine density in neurons of Area X and decreases vocal motor learning36,37. Our findings add to evidence for a controlling role for Foxp2, explicitly by introducing its ability to suppress Mef2C as a potential critical mechanism contributing to the control of vocalization.
Potential clinical relevance of Foxp2-Mef2C interactions
Our findings suggest a molecular mechanism by which Foxp2 can regulate excitatory corticostriatal synapses onto striatal projection neurons in mice through suppression of Mef2C, whose progressive down-regulation is synchronized spatially and temporally with the development and synaptogenesis of corticostriatal circuits. Our evidence specifically suggests that Foxp2 transcriptionally suppresses the spinogenesis suppressor, Mef2C, thereby allowing corticostriatal synaptogenesis.
There is no direct evidence from our work that these interactions occur in the human brain, or that they relate to human conditions including ASD and speech and language. FOXP2 is considered a ‘language-related gene’9, but its precise relation to speech and language is still a matter of study. Similarly, MEF2C has been identified as a candidate ASD gene15–17, but evidence for linkage of MEF2C mutations to ASD is still incomplete. MEF2C mutations have been linked to phenotypes including mental retardation in humans15,19, a matter of current research. It is known, however, that MEF2C is a putative target gene of FOXP2 in the developing human basal ganglia18, and our findings suggest that the human FOXP2 protein is at least as effective as, or more effective than, the mouse Foxp2 protein in promoting synapse and complex spine formation in SPNs in the mouse striatum. A possibility raised by our findings is that the interactions that we have documented in the mouse hold in some form in the human. If so, our findings could be interpreted as relevant to a molecular linkage between the putative ASD candidate gene, MEF2C, and the putative language-associated gene, FOXP2. Notably, Mef2C is regulated by MeCP2, a well-established Rett syndrome gene with broad effects38, and MEF2 is implicated in regulation of the ASD-risk genes, DIA1 and PCDH1039. Thus, our findings could be relevant in the context of multiple ASD-risk genes interacting in signaling networks for neuronal development13,14.
Human patients with MEF2C deletion and mutation have complex neurological syndromes15,16,19. In addition to defective language communication, stereotypic behaviors, potentially linked to striosomes40, have been reported in patients with MEF2C deletion and mutation15,19 and in the Mef2C knockout mouse model of ASD31,41. Our findings indicate that part of the etiology of MEF2C–related ASD symptoms could be rooted in the basal ganglia. We raise the possibility that defective FOXP2-MEF2C signaling affecting developing corticostriatal synaptic circuits could be a previously underestimated pathologic mechanism in conditions characterized by corticostriatal abnormalities.
ONLINE METHODS
Animals
Foxp2+/− mice and humanized Foxp2H/+ mice were generated as previously described8. Mef2Cfl/+ mice (kindly provided by Dr. Eric Olson) were generated as previously described21. All mice were maintained with freely available food and water in a specific pathogen-free room with a constant humidity and 12-hr light-dark cycle (2–3 adult mice/cage; 6–8 pups/cage) in the animal center of National Yang-Ming University. All the experiments on single Foxp2 mutant mice were performed on littermate mice derived from the offspring of crosses between heterozygous Foxp2+/− x Foxp2+/− or Foxp2H/+ x Foxp2H/+ mice. Nestin-Cre mice42 were crossed with Mef2Cfl/+ mice to derive Nestin-Cre;Mef2Cfl/+ mice. Nestin-Cre;Mef2Cfl/+ mice were then crossed with Mef2Cfl/fl mice to generate Nestin-Cre;Mef2Cfl/fl conditional knockout mice. Foxp2+/− mice were crossed with Mef2Cfl/fl mice to derive Foxp2+/−;Mef2Cfl/+ mice. Dlx5/6-Cre mice43 were crossed with Foxp2+/−;Mef2Cfl/+ mice to derive Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ mice. Dlx5/6-Cre;Foxp2+/−;Mef2Cfl/+ mice were then crossed with Foxp2+/−;Mef2Cfl/+ mice. Foxp2+/−, Foxp2H/+, Mef2Cfl/+ and Nestin-Cre mice were maintained in C57Bl/6 genetic background. Dlx5/6-Cre mice were maintained in CD-1 background. We used both genders for experiments. All animal protocols were approved by the Animal Care and Use Committees of National Yang-Ming University.
Genotyping
Genotyping was performed by PCR with the genomic DNA. Tissues for the genotyping were collected, and then were incubated overnight in DNA lysis buffer (10 mM Tris, 10 mM EDTA, 100 mM NaCl, 0.5% SDS, 250 µg/ml protease K) at 55°C. After incubation, lysates were treated with RNase A for 30 min at 37°C, followed by addition of 200-µl protein precipitation solution (Promega), and they were kept on ice for 1 hr to separate proteins and RNA from genomic DNA. Finally, the genomic DNA was extracted by isopropanol (1:1), washed with 500 µl of 70% ethanol and placed overnight in 100 µl of Tris-EDTA buffer at 37°C to dissolve. For PCR genotyping, three primers were used to detect the Foxp2 mice genotypes: Foxp2ko_5425_f (5’-TTCTCTCCTGTCTCCCATTGA-3’), Foxp2ko_8502_f (5’-CACGCCCAGCTACATTTTTA-3’), and Foxp2ko_8729_r (5’-GCAGAAACACTATGGTGGGAAG-3’). The PCR product derived from Foxp2ko_5425_f/Foxp2ko_8729_r was 194 bp in wildtype mice; and that from Foxp2ko_8502_f/Foxp2ko_8729_r was 350 bp in Foxp2−/− knockout mice. The PCR product derived from Foxp2ko_8502_f/Foxp2ko_8729_r was 228 bp in humanized Foxp2H/H mice. The primers used for detecting genotypes of the Mef2Cfl/+, Nestin-Cre and Dlx5/6-Cre mice were as follows:
Mef2C-5’ (5’-GTGATGACCCATATGGGATCTAGAAATCAAGGTCCAGGGTCAG-3’),
Mef2C-3’ (5’-CTACTTGTCCCAAGAAAGGACAGGAAATGCAAAAATGAGGCAG-3’),
Nestin (Dlx5/6)-Cre-5’ (5’-GCTAAACATGCTTCATCGTCGG-3’),
and Nestin (Dlx5/6)-Cre-3’ (5’-GATCTCCGGTATTGAAACTCCAGC-3’).
The PCR genotyping protocols were as follows: 94°C for 5 min, 35 cycles at 94°C for 30 s, 58°C (Foxp2, Nestin-Cre, Dlx5/6-Cre) or 65°C (Mef2C) for 30 s, and 72°C for 30 s. Finally, an additional step was run at 72°C for 5 min before completing the PCR reaction.
In utero electroporation
The pCAG-EYFP-CAG-Foxp2, pCAG-EYFP-CAG, pcBIG-Mef2C–VP16 (Mef2C–VP16 cDNA kindly provided by E. Olson) and pcBIG (vector kindly provided by P. Arlotta) plasmids were prepared using an endotoxin-free kit (Novelgene) and were injected (1.5 μl, 2 μg/μl) into the lateral ventricle of E12.5 (pcBIG-Mef2C–VP16, pcBIG) or E13.5 (pCAG-EYFP-CAG-Foxp2, pCAG-EYFP-CAG) forebrains of wildtype mouse embryos followed by electroporation (7 pulses, 33 V, 30 ms duration, 970 ms interval; BTX ECM 830). The electroporated mice were born and were perfused at P14 for GFP immunostaining.
Virus transduction
ST HSV-Cre-GFP viruses (Viral Core Facility, McGovern Institute for Brain Research, MIT) were diluted 1:1 in D-PBS buffer. Foxp2+/+, Foxp2H/H, Mef2C+/+, Mef2Cfl/fl, Foxp2+/−;Mef2C+/+ and Foxp2+/−;Mef2Cfl/+ mouse pups were anesthetized by hypothermia at P2, and virus solution (0.2 μl) was injected (0.1 μl/min) bilaterally in the rostral (AP: +2.85 mm, ML: ±1.60 mm, DV: −1.80 and −2.00 mm) and middle (AP: +2.55 mm, ML: ±1.51 mm, DV: −1.50 and −1.70 mm) striatum. After injections, the needle was left for 2 min before slow retraction. A few pups received injections only at the middle striatum. The pups were allowed to survive until P8, and following perfusion, the brains were prepared for Golgi staining and immunostaining. In another set of experiments, intrastriatal viral injections were made in Mef2Cfl/fl mice at P14–15 at rostral (AP: + 3.75 mm, ML: ±2.20 mm, DV: −2.5 and −2.8 mm) and middle (AP: +3.33 mm, ML: ±2.35 mm, DV: −3.0 and −3.5 mm) levels. The brains were harvested at P19-P20.
Preparation of brain tissue
For western blotting analysis, P4, P8, P12, P14 and P21 mice were deeply anesthetized by intraperitoneal injection of sodium pentobarbital, and brains were cut into 1 mm coronal slices using acrylic brain slice matrix (Alto). The selected slices containing the regions of interest were then frozen with dry ice and used for analysis. For immunohistochemical analysis, P0, P4, P7, P8, P14 and P30 mice were perfused transcardially with 0.9% saline followed by 4% paraformaldehyde (PFA) in 0.1 M phosphate buffered saline (PBS, pH 7.4). The perfused brains were post-fixed in 4% PFA at 4°C overnight and were then cryoprotected with 30% sucrose in 0.1 M PBS. Brains were cut in the coronal plane on a cryostat at 20 μm (Lecia).
Golgi staining
P12 and P14 brain tissues were prepared and processed for Golgi staining using the FD Rapid GolgiStain Kit (FD NeuroTechnologies) according to the manufacturer’s instruction. Transverse sections were cut at 80 μm on a cryostat (Lecia). P8 brains infected with HSV-Cre-EGFP viruses were processed using the sliceGolgi Kit (Bioenno Tech) for double Golgi staining and GFP immunostaining with DAB substrate.
Immunohistochemistry
Immunohistochemistry was performed as previously described44. Primary antibodies were as follows: rabbit anti-Foxp2 (1:2,000; #ab16046, Abcam), rabbit anti-MOR1 (1:10,000; #24216, ImmunoStar), rabbit anti-CalDAG-GEFI (1:10,000)40, rabbit anti-GluR1 (1:2,000; #ab1504, Millipore), rabbit anti-activated caspase3 (1;1,000, #9661, Cell Signaling), rabbit anti-PSD95 (1:10; #20665-1-AP, Proteintech), goat anti-Mef2C (1:1,000; #sc-13266, Santa Cruz), guinea pig anti-VGluT1 (1:5,000; #ab5905, Millipore), mouse anti-DARPP-32 (1:10,000, kindly provided by Dr. H.C. Hemmings) and chicken anti-GFP (1:500; #ab13970, Abcam). For triple immunostaining of Foxp2, Mef2C and VGluT1, Alexa555-conjugated goat anti-rabbit antibody and Alexa647-conjugated goat anti-guinea pig antibody were used to detect Foxp2 and VGluT1 signals. For detecting Mef2C signals, sections were incubated sequentially in biotin-conjugated donkey anti-goat antibody, ABC complex (ABC kit, Vector Laboratories) and tyramide-FITC (1:2,000, Invitrogen) with several rinses in between. For double immunostaining of DARPP-32 and VGluT1, Alexa660-conjugated goat anti-mouse and Alexa488-conjugated goat anti-guinea pig antibody were used. For double immunostaining of DARPP-32 and Mef2C, Alexa555-conjugated goat anti-mouse and biotin-conjugated donkey anti-goat antibody were used followed by tyramide-FITC amplification as described above. For double Golgi staining and GFP immunostaining, after Golgi staining, the brain sections were processed for GFP immunoistaining using the ABC method with DAB substrate.
TUNEL staining
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining to identify apoptotic cells was performed using In Situ Cell Death Detection Kit, POD (#11684817910, Roche) with instructions from the manufacturer.
Western blotting
Western blotting was performed as previously described45. Brain tissue lysates containing denatured proteins were resolved by 7.5% Tricine-SDS-PAGE and wet-transferred with transfer buffer (25 mM Tris, 192 mM Glycine and 20% Methanol in distilled H2O) to polyvinyl difluoride (PVDF) membranes (Hybond-P, Amersham Biosciences) for 90 min. The protein-bound membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with Tween-20 (TBST; 150 mM NaCl, 25 mM Tris and 0.05% Tween-20, pH 7.4) at room temperature for 1 hr with agitation, and then were incubated with appropriately diluted primary antibody. The dilutions of the primary antibodies were as follows: goat anti-Mef2C (1:1,000; #sc-13266, Santa Cruz), mouse anti-PSD95 (1:4,000; #7E3-1B8, Affinity BioReagents), guinea pig anti-VGluT1 (1:50,000; #ab5905, Millipore), rabbit anti-GluR1 (1:2,000; #ab1504, Millipore), mouse anti-β-actin (1:10,000; #A5441, Sigma), and mouse anti-alpha-tubulin (1:10,000; #T9026, Sigma). The primary antibodies were diluted in TBST containing 1% nonfat dry milk at 4°C overnight with gentle shaking. After being washed with TBST for three times, the membranes were incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (goat anti-mouse or goat anti-guinea pig, 1:10,000) diluted in TBST containing 1% nonfat dry milk at room temperature for 1 hr, and then washed with TBST for three times. For Mef2C detection, biotinylated rabbit anti-goat IgG (1:10,000) was used with 30 min incubations. After being washed with TBST for three times, the membranes were incubated with avidin-biotin-peroxidase complex (Elite kit, Vector Laboratories) for 20 min. The membranes were then developed with enhanced chemiluminescence (ECL) plus (Millipore). For western blots presented in Figs. 3 and 4, the ECL signals were visualized on X-ray films. Otherwise, the ECL signals were detected using Fujifilm LAS-4000 Image System. The immunoblotting proteins in the gel were normalized with the internal controls of β-actin or α-tubulin proteins in the same lane.
Quantitative PCR
RNA was extracted from the dissected brain tissue and then was reverse transcribed into cDNA. The cDNA was used as the template for SYBR® Green-based qRT-PCR (ABI) reaction that was performed by the National Yang-Ming University Genome Center. The Mef2C forward (5’-GGATGAGCGTAACAGACAGGT-3’) and reverse primers (5’-ATCAGTGCAATCTCACAGTCG-3’) were used for detecting expression levels of Mef2C gene (NM_025282). The PCR program was as follows: 95°C for 60 s, 45 cycles at 95°C for 15 s, 60°C for 15 s, and 72°C for 45 s. The Ct value (cycle of threshold) was used for the calculation of gene fold change with the formula, 2−[Mutant(Gene Ct – GAPDH Ct) – Wildtype(Gene Ct – GAPDH Ct)].
Cell culture and transfection
Primary striatal cells from E16 mouse brains were cultured with 5% FCS and 5% horse serum in Dulbecco’s Modified Eagle Medium (DMEM), and were used for ChIP assays. N2A cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and maintained in a humidified incubator with 5% CO2 at 37°C. N2A cells were seeded in 6-well plates one day before transfection at a density that reached 50% confluency on the following day. For over-expression studies of Foxp2, pCAG-EYFP-CAG-mFoxp2 or pCAG-EYFP-CAG plasmids were transfected into N2A cells with Lipofectamine 2000.
DNA cloning and site-directed mutagenesis
The nucleotides of mouse Mef2C gene (−2131 to −1718, translational initiation site +1; TAAGCGCTTAGGCAGGCATAGTCCGGTAAGCCGATAGTGCCACAGGTCTCAGCTAAGAGAAAGGAAGGGAAGATGCTCCCATATACTTTGACTTGAGATTCATGGATTGTTTTTGTTTGTTTGTTTGTTTGTTTTTTGTGGTGGAGAAAGCCTTCTTGTCTCATTGGGGTATTACAAATGCATGGGAAATACAAGTCTGGGCTGCTGCTTGCTTCTTGGGTAAGACTATAGTAAATGCAGAAAAGATTCCCACTTGTATGCTGTTTAAGACCATTTCTAAGACTAAATGGGTAGACTATTTAACAATGTTGGATACATCATGTGTGTGCTTTGCAAATTCTTTATCTATATGATCTTGGTGTCTTTATAGTAGTGTCTTTGCCTTGTGAGTGTCTTAGTGTGTGTTGTTCATAT) containing ACAAAT (−1948) & AAAT (−1935) and AAAT (−1837) & CAAATT (−1788) Foxp2 binding motifs were custom-made by DNA synthesis (Neogene Biomedicals Taiwan/Genscript), and then were cloned into the Kpn1 and SacI sites of pGL3-c-fos-Luc plasmid (kindly provided by Dr. O. Marin). To produce the mutant reporter gene plasmids, the sequences of the ACAAAT & AAAT motifs were mutated to ACGGGT & GGGT (MT1) to generate a Mef2C–MT1-c-fos-Luc plasmid by site-directed mutagenesis (Neogene Biomedicals Taiwan/Genscript). The sequences of the AAAT & CAAATT motifs were mutated to GGGT & CGGGTT (MT2) to generate a Mef2C–MT2-c-fos-Luc plasmid. For construction of the human MEF2C reporter gene plasmid, the nucleotides of the 5’ flanking region of human MEF2C gene (−872 to 261 bp, transcription initiation site: +1) were cloned by PCR into the KpnI and NheI sites of pGL3-c-fos-Luc plasmid using genomic DNA of SH-SY5Y cell.
Reporter gene assay
Mouse Mef2C–pGL3-c-fos-Luc or pGL3-c-fos-Luc plasmids were co-transfected along with pCAG-EYFP-CAG-mFoxp2, pCAG-EYFP-CAG-hFOXP2 or pCAG-EYFP-CAG-hFOXP2R553H and pGL4-Renilla plasmids into N2A cells. For the mutant reporter gene assay, Mef2C–MT1-c-fos-Luc or Mef2C–MT2-c-fos-Luc plasmids were co-transfected along with pCAG-EYFP-CAG-mFoxp2, pCAG-EYFP-CAG-hFOXP2 or pCAG-EYFP-CAG-hFOXP2R553H and pGL4-Renilla plasmids into N2A cells. For the human MEF2C reporter gene assay, human MEF2C–pGL3-c-fos-Luc or pGL3-c-fos-Luc plasmids were co-transfected along with pCAG-EYFP-CAG-hFOXP2 or pCAG-EYFP-CAG-hFOXP2R553H and pGL4-Renilla plasmids into SH-SY5Y cells.
Chromatin immunoprecipitation
Cultured striatal cells were fixed and processed for chromatin immunoprecipitation assay using the ChIP-IT® Express Chromatin Immunoprecipitation Kit (Active Motif) according to the manufacturer’s instruction. An affinity-purified polyclonal rabbit anti-Foxp2 antibody (H. Takahashi) was used to immunoprecipitate DNA that was then used as templates for PCR to detect the 5’ flanking region of mouse Mef2C gene (−1718 to −2131 bp, translational initiation site +1) using forward (5’-TGTCTTAGTGTGTGTTGTTCATAT-3’) and reverse (5’-TAAGCGCTTAGGCAGGCATAGTCC-3’) primers. The PCR product size was 414 bp. For ChIP-qPCR, the DNA products of ChIP assays were analyzed by quantitative PCR (Applied Biosystems StepOne system). Briefly, the PCR reaction solution containing 200-fold diluted DNAs and 2X TaqMan® Fast Universal PCR Master Mix (Applied Biosystems®) and specific primer pair (Mef2C promoter F: 5’-AGGCAGGCATAGTCCGGTAA-3’, R: 5’-CATCTTCCCTTCCTTTCTCTTAGCT-3’). Each qRT-PCR reaction was performed in triplicate with the following conditions: 95°C for 10 min followed by 40 cycles at 95°C for 10 s, and 60°C for 1 min. The ChIP-qPCR signals from Foxp2 antibody and RNA Pol II antibody reactions were normalized with that of IgG control.
Electrophysiology
Brains of male P14 Foxp2H/H and Foxp2+/+ mice were placed into ice-cold sucrose-based cutting solution (85 mM sucrose, 60 mM NaCl, 3.5 mM KCl, 6 mM MgCl2, 0.5 mM CaCl2, 38 mM NaHCO3, 1.25 mM NaH2PO4, 10 mM HEPES, and 25 mM glucose), and coronal slices (250 µm) were cut (Vibroslice 7000smz, Campden Instruments, UK), incubated in artificial cerebrospinal fluid (aCSF; 120 mM NaCl, 3.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 30 mM NaHCO3, 1.25 mM NaH2PO4, and 15 mM glucose) supplemented with 5 mM HEPES, 1 mM MgCl2 for 30 min at 35°C, and allowed to recover at room temperature for at least 40 min. Whole-cell patch clamp recordings of SPNs were performed in the dorsolateral striatum (Foxp2H/H: n = 27 cells from 6 mice; Foxp2+/+: n = 23 cells from 4 mice) to measure mEPSCs. SPNs were identified by their size, morphology and characteristic electrophysiological properties including negative resting membrane potentials (< −80 mV), slow capacitance transients and low input resistances46. The mean resting membrane potentials were −87.5 ± 0.46 mV in wildtype mice and −84.7 ± 0.67 mV in Foxp2H/H mice; the input resistances were 176.8 ± 14.6 MΩ in wildtype mice and 211.0 ± 13.8 MΩ in Foxp2H/H mice; the cell capacitance levels were 47.7 ± 2.2 pF in wildtype mice and 44.8 ± 2.9 pF in Foxp2H/H mice; and series resistances during recordings were 21.5 ± 1 MΩ in wild-type mice and 18.7 ± 0.7 MΩ in Foxp2H/H mice. Glass electrodes (5–8 MΩ) were filled with an internal solution containing K gluconate (150 mM), NaCl (10 mM), Mg-ATP (4 mM), GTP (0.5 mM), HEPES (10 mM) and EGTA (0.05 mM) adjusted to pH = 7.3 and 310 mOsm. The liquid junction potential (15 mV) was corrected online and the cells were clamped to a membrane potential of −85 mV. Slices were perfused (2–3 ml/min, aCSF, 21–23°C) in the presence the GABAA receptor antagonist, SR-95531 (10 µM, Sigma), and tetrodotoxin (TTX, 0.1 µM, Tocris). All solutions were continuously bubbled with carbogen (95% O2 and 5% CO2). Signals were low-pass filtered at 2.5 kHz and sampled at 20 kHz (EPC-10 amplifier, HEKA, Lambrecht, Germany) using Patchmaster (HEKA). Traces were analyzed with Mini Analysis software (V 6.0.7, Synaptosoft). For electrophysiological recordings in Foxp2+/+ and Foxp2−/− mice at P8, and in Nestin-Cre;Mef2C+/+ and Nestin-Cre;Mef2Cfl/fl mice at P14, the brains were placed in ice-cold aCSF (119 mM NaCl, 2.5 mM KCl, 26.2 mM NaHCO3, 1 mM NaH2PO4·H2O, 1.3 mM MgSO4, 11 mM glucose, and 2.5 CaCl2) oxygenated with 95% O2 and 5% CO, and were then cut in the coronal plane at 300 µm with a vibrotome. After recovering for 2 hr at 25°C, the brain slices were perfused with oxygenated aCSF containing picrotoxin (100 µM, Tocris Bioscience) and tetrodotoxin (1 µM, Tocris Bioscience). Whole-cell patch clamp recordings of SPNs were performed in the striatum (Foxp2+/+: n = 8 cells from 4 mice; Foxp2−/−: n = 7 cells from 3 mice; Nestin-Cre;Mef2C+/+: n = 7 cells from 3 mice; and Nestin-Cre;Mef2Cflfl: n = 9 cells from 3 mice) with glass pipettes filled with internal solution (131 mM potassium gluconate, 10 mM HEPES, 2 mM EGTA, 20 mM KCl, 8 mM NaCl, 2 mM MgATP, 0.3 mM Na3GTP, pH7.2, 300–310 mOsm). Membrane potentials were held at −70 mV by voltage clamp. mEPSCs were recorded with Axon MultiClamp 700B (AutoMate Scientific, USA) for ~10 min. The recorded currents were analyzed with MiniAnalysis (Synaptosoft, USA). The investigator who performed electrophysiological recordings of Foxp2 knockout and Mef2C knockout mice was blind to the genotype of the mice.
USV analysis
The USV recordings were performed with mouse genotypes blinded to the experimenter. USVs were recorded in pups on P8 during the daylight period of light/dark cycle. After a 30-min habituation period in a soundproof room, each pup was removed from the cage containing its mother and littermates, and was placed in a glass beaker (diameter: 6 cm) in a sound-attenuating plastic chamber. Within the beaker, a microphone (condenser ultrasound microphone Avisoft-Bioacoustics CM16) was hung 6 cm above the pup. After ~1 min of habituation, recordings began and were continued for 5 min (Avisoft UltraSoundGate 116, Avisoft-RECORDER software). The recorded data were transferred to Avisoft-SASLab Pro (Version 5.2) to analyze spectrograms of vocalizations with settings of 50% overlapping FlatTop windows, 100% frame size and 256 points fast Fourier transform (FFT) length. After de-noising, the automatic single threshold was set to 2 dB above the baseline. The post-filter was set to a maximal entropy of 100. Emitted events with frequency between 40–100 kHz were included, and the following measures were recorded and averaged for each group: events (number of calls), elements (discontinuous signals separated by more than 5-ms duration within a single event); peak frequency at start (the peak frequency at the beginning of each event); peak frequency at end (the peak frequency at the end point of each event); peak amplitude at start (the peak amplitude at the beginning of each event); peak amplitude at end (the peak amplitude at the end point of each event); peak frequency at max (the peak frequency at the point of the maximum amplitude of the entire event); peak amplitude at max (the peak amplitude of the entire event); and frequency jump (abrupt upward increase in frequency exceeding 25 kHz within a single event).
Quantification of dendritic spine density
Dendritic spines were counted in Golgi-stained material with the aid of light microscopy using a 100X oil immersion objective (Nikon Eclipse E800M; Olympus BX63). Coronal sections of three pairs of brains were used for analysis. For brains of each Foxp2 genotype, we counted 10 neurons in the dorsomedial striatum and 10 in the dorsolateral striatum. For each Nestin-Cre;Mef2C genotype, we counted 13–19 neurons in the dorsomedial striatum and 15–22 neurons in the dorsolateral striatum. For each Dlx5/6-Cre;Foxp2;Mef2C genotype, we counted 10–12 neurons in the dorsomedial striatum and 11–17 neurons in the dorsolateral striatum. The number of spines in the proximal parts of secondary dendrites was counted for each neuron. Spine densities were quantified by tracing dendritic segments in which spines were counted. The data were presented as averaged spine number/10 µm. The dendritic spines were categorized into six types (stubby, thin/filopodia, mushroom, branched, multiple-branched and atypical) according to Harris et al.28 with slight modifications. Thin spine and filopodia were included in the same group as spine/filopodia, because it was difficult to distinguish them with light microscopy. Atypical spines were those that could not be assigned to a specific group. Spines were categorized as ‘stubby’ if the diameter of the neck was similar to the total length of the spine. Spines were categorized as ‘thin/filopodia’ if the length was longer than the neck diameter, and the diameters of the head and neck were similar or the head-neck ratio was less than two. Spines were categorized as ‘mushroom’ if the diameter of the head was at least 2.5 fold the diameter of the neck. Spines were categorized as ‘branched’ spines if they had two heads and as ‘multiple-branched’ if they had more than two heads. For photomicrographic illustration of dendritic spines (Fig. 3d–f and Supplementary Fig. 4a–c), the images were processed with the AutoQuant de-convolution software (Media Cybernetics) to de-blur the images caused by spines oriented in different depths of focal planes.
Microscopic image analyses
Photomicrographs were acquired using Olympus BX51, BX63 fluorescence microscopes and a Zeiss LSM700 confocal microscope. Quantification of cell number was performed with the aid of Image J software. For comparison between the brains with different genotypes, we analyzed photomicrographs at matched anatomical levels. Fluorescence intensity was measured for single cells (Fig. 2g) or in the region of interest (Figs. 2d,e, 4c, Fig. 6g–i and Supplementary Fig. 2) using the RGB measure plugins of ImageJ. For quantification of striatal compartmentation (Supplementary Fig. 3a–h), the sum of MOR1-positive striosome areas or CalDAG-GEFI-poor matrix areas was normalized with the total striatal area. For quantification of Foxp2- or Mef2C–positive cells (Fig. 2h and Supplementary Fig. 3), Foxp2- or Mef2C–positive cells were counted in six 0.05-mm2 squares that were placed evenly in the striatum of each section, and the cell numbers averaged over 3 sections, from rostral to caudal striatal levels, were analyzed with statistical tests described below.
Statistics
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those generally employed in the field. For small number of samples, we assumed normal distribution because the number of samples was too small for normality tests. Statistical analyses for the data of cell counts, dendritic spine counts, and biochemical and immunochemical measurements of protein levels were done with Student’s two-tailed t-test or analysis of variance (ANOVA) followed by Tukey’s post hoc test. Statistical analysis for electrophysiological data was done with Origin (V 8.6, OriginLab; Northampton, USA), by Mann-Whitney U test and Student’s two-tailed t-test. Statistical analysis for the USV data was performed with SPSS (IBM, version 21). The USV data were first assessed by the Kolmogorov-Smirnov test for normal distribution exploration. Normally distributed data were processed with one-way ANOVA followed by Tukey’s test for post hoc comparisons between two groups. For the data sets without normal distributions, the data were analyzed with Kruskal-Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons test between two groups. All statistical data were presented as mean ± s.e.m. A supplementary methods checklist is available.
Data collection and analysis
Animals/samples were assigned to various experimental groups according to their genotypes. Data collection and analysis were not performed blind to the conditions of the experiments, unless specifically specified. No data points were excluded from analyses for any reason.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank P. Arrollta of Harvard University, K. Campbell of Cincinnati Children’s Hospital Medical Center, H.C. Hemmings of Rockefeller University, O. Marin of King’s College London and E. Olson of University of Texas Southwestern Medical Center for providing transgenic mice and reagents, M. Bear, M. Sur, M.-M. Poo and D. Homma for insightful discussion, and Y. Kubota for help with illustrations and manuscript preparation. This work was supported by NIH/NICHD grant R37 HD028341 (A.M.G.), the Nancy Lurie Marks Family Foundation (A.M.G.), the Simons Center for the Social Brain at MIT (A.M.G.), the Paul G. Allen Family Foundation (S.P.), National Science Council grants NSC97-2321-B-010-006, NSC98-2321-B-010-002, NSC99-2321-B-010-002, NSC100-2321-B-010-002, NSC101-2321-B-010-021, NSC102-2321-B-010-018, NSC102-2911-I-010-506 (F.-C.L.), Ministry of Science and Technology grant MOST103-2321-B-010-009, MOST104-2321-B-010-022 (F.-C.L.), National Health Research Institutes grant NHRI-EX104-10429NI, NHRI-EX105-10429NI (F.-C.L.) and by an Aiming for Top University grant, Ministry of Education, Taiwan (F.-C.L.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
F.-C.L. conceived and supervised the project. Y.-C.Chen, H.-Y.K., H.T., S.-Y.C., K.-M.L., C.-T.C., U.B., W.H., W.E., S.P., A.M.G. and F.-C.L. designed the experiments. Y.-C.Chen, H.-Y.K., U.B., W.H., S.-Y.C., H.-Y.Y., G.-M.C., Y.-H. L. and S.-J.C. performed experiments on Foxp2 and Mef2C mutant mice; K.-M.L., J.-R.L. and Y.-C.Chou performed Foxp2 and Mef2C over-expression experiments; H.T., Y.-C.Chen and S.-Y.C. characterized Foxp2 binding sites; W.E., S.P. provided Foxp2 mutant mice and interpretation of data and discussion. Y.-C.Chen., H.-Y.K., S.-Y.C., K.-M.L., H.T., U.B., W.H., W.E., S.P., A.M.G. and F.-C.L. analyzed the data. A.M.G. and F.-C.L. wrote the paper with inputs from all authors.
COMPETING FINANCIAL INTERESTS
We declare no competing financial interests.
REFERENCES
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mody M, Belliveau JW. Speech and language impairments in autism: insights from behavior and neuroimaging. N. Am. J. Med. Sci. (Boston) 2013;5:157–161. doi: 10.7156/v5i3p157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollander E, et al. Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol. Psychiatry. 2005;58:226–232. doi: 10.1016/j.biopsych.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 4.Di Martino A, et al. Aberrant striatal functional connectivity in children with autism. Biol. Psychiatry. 2011;69:847–856. doi: 10.1016/j.biopsych.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins KE, et al. MRI analysis of an inherited speech and language disorder: structural brain abnormalities. Brain. 2002;125:465–478. doi: 10.1093/brain/awf057. [DOI] [PubMed] [Google Scholar]
- 6.Vargha-Khadem F, et al. Neural basis of an inherited speech and language disorder. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12695–12700. doi: 10.1073/pnas.95.21.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groszer M, et al. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr. Biol. 2008;18:354–362. doi: 10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enard W, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 9.Graham SA, Fisher SE. Decoding the genetics of speech and language. Curr. Opin. Neurobiol. 2013;23:43–51. doi: 10.1016/j.conb.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Mukamel Z, et al. Regulation of MET by FOXP2, genes implicated in higher cognitive dysfunction and autism risk. J. Neurosci. 2011;31:11437–11442. doi: 10.1523/JNEUROSCI.0181-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernes SC, et al. A functional genetic link between distinct developmental language disorders. New Engl. J. Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roll P, et al. Molecular networks implicated in speech-related disorders: FOXP2 regulates the SRPX2/uPAR complex. Hum. Mol. Genet. 2010;19:4848–4860. doi: 10.1093/hmg/ddq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 2012;4:a009886. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novara F, et al. Refining the phenotype associated with MEF2C haploinsufficiency. Clin. Genet. 2010;78:471–477. doi: 10.1111/j.1399-0004.2010.01413.x. [DOI] [PubMed] [Google Scholar]
- 16.Rauch A, Bartholdi D, Rueegger CM, Zweier M, Zweier C, Bijlsma EK, van Haeringen A, Reardon W, Zollino M, Baumer A. MEF2C mutations are a frequent cause of Rett- or Angelman syndrome like neurodevelopmental disorders; Montreal, Canada. Presented at the 12th International Congress of Human Genetics/61st Annual Meeting of The American Society of Human Genetics; 2011. October 12. Program Number: 1026T. [Google Scholar]
- 17.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiteri E, et al. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am. J. Hum. Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Meur N, et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J. Med. Genet. 2010;47:22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flavell SW, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 21.Barbosa AC, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai NP, et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian X, Kai L, Hockberger PE, Wokosin DL, Surmeier DJ. MEF-2 regulates activity-dependent spine loss in striatopallidal medium spiny neurons. Mol. Cell Neurosci. 2010;44:94–108. doi: 10.1016/j.mcn.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisenbaum LK, Webster SM, Chang SL, McQueeney KD, LoTurco JJ. Early patterning of prelimbic cortical axons to the striatal patch compartment in the neonatal mouse. Dev. Neurosci. 1998;20:113–124. doi: 10.1159/000017307. [DOI] [PubMed] [Google Scholar]
- 25.Somogyi P, Bolam JP, Smith AD. Monosynaptic cortical input local axon collaterals of identified striatonogral neurons A light and electron microscopic study using the golgi-peroxidase transport degeneration procedure. J. Comp. Neurol. 1981;195:567–584. doi: 10.1002/cne.901950403. [DOI] [PubMed] [Google Scholar]
- 26.Sheth AN, McKee ML, Bhide PG. The sequence of formation and development of corticostriate connections in mice. Dev. Neurosci. 1998;20:98–112. doi: 10.1159/000017306. [DOI] [PubMed] [Google Scholar]
- 27.Fremeau RT, Jr, Voglmaier S, Seal RP, Edwards RH. VGLUTs define subsets of excitatory neurons and suggest novel roles for glutamate. Trends Neurosci. 2004;27:98–103. doi: 10.1016/j.tins.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu W, et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulipparacharuvil S, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar MW, et al. vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS ONE. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adachi M, Lin PY, Pranav H, Monteggia LM. Postnatal Loss of Mef2c Results in Dissociation of Effects on Synapse Number and Learning and Memory. Biol. Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.09.018. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sia GM, Clem RL, Huganir RL. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science. 2013;342:987–991. doi: 10.1126/science.1245079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulz SB, Haesler S, Scharff C, Rochefort C. Knockdown of FoxP2 alters spine density in Area X of the zebra finch. Genes Brain Behav. 2010;9:732–740. doi: 10.1111/j.1601-183X.2010.00607.x. [DOI] [PubMed] [Google Scholar]
- 37.Haesler S, et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crittenden JR, Graybiel AM. Basal Ganglia disorders associated with imbalances in the striatal striosome and matrix compartments. Front. Neuroanat. 2011;5:59. doi: 10.3389/fnana.2011.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipton SA, et al. Autistic phenotype from MEF2C knockout cells. Science. 2009;323:208. doi: 10.1126/science.323.5911.208b. [DOI] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES FOR ONLINE METHODS
- 42.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 43.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J. Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu KM, Evans SM, Hirano S, Liu FC. Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E168–E177. doi: 10.1073/pnas.1319138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang SL, et al. Ectopic expression of nolz-1 in neural progenitors promotes cell cycle exit/premature neuronal differentiation accompanying with abnormal apoptosis in the developing mouse telencephalon. PLoS ONE. 2013;8:e74975. doi: 10.1371/journal.pone.0074975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawlak V, Kerr JN. Dopamine receptor activation is required for corticostriatal spike-timing-dependent plasticity. J. Neurosci. 2008;28:2435–2446. doi: 10.1523/JNEUROSCI.4402-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.