Abstract
Recent studies have indicated a metabolic temperature sensitivity in both the arcto-boreal krill species Thysanoessa inermis and Thysanoessa raschii that may determine these species’ abundance and population persistence at lower latitudes (up to 40° N). T. inermis currently dominates the krill community in the Barents Sea and in the high Arctic Kongsfjord. We aimed to increase the knowledge on the upper thermal limit found in the latter species by estimating the CT50 value (19.7 °C) (critical temperature at which 50 % of animals are reactive) and by linking metabolic rate measurements with molecular approaches. Optical oxygen sensors were used to measure respiration rates in steps of 2 °C (from 0 to 16 °C). To follow the temperature-mediated mechanisms of passive response, i.e., as a proxy for molecular stress, molecular chaperone heat shock protein 70 (Hsp70) sequences were extracted from a transcriptome assembly, and the gene expression kinetics were monitored during an acute temperature exposure to 6 or 10 °C with subsequent recovery at 4 °C. Our results showed upregulation of hsp70 genes, especially the structurally constitutive and mitochondrial isoforms. These findings confirmed the temperature sensitivity of T. inermis and showed that the thermal stress took place before reaching the upper temperature limit estimated by respirometry at 12 °C. This study provides a baseline for further investigations into the thermal tolerances of arcto-boreal Thysanoessa spp. and comparisons with other krill species under different climatic regimes, especially Antarctica.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0720-6) contains supplementary material, which is available to authorized users.
Keywords: Krill, Thysanoessa inermis, Heat shock, Metabolic rate, Transcriptome, Hsp70 expression
Introduction
The arcto-boreal krill Thysanoessa inermis appears well adapted to the Arctic marine environment that is characterized by low temperatures, strong seasonality in light conditions, and, hence, primary production (Buchholz et al. 2012; Hop et al. 2006). As a regular expatriate from the Barents Sea, it currently dominates the krill community in West Spitsbergen fjords including the high Arctic Kongsfjord at 79° N (Buchholz et al. 2010; Hop et al. 2006).
The Kongsfjord is mainly influenced by two different water masses, which determine the abiotic conditions (e.g., temperature, salinity, nutrients) within this ecosystem: the cold Arctic current and the warm West Spitsbergen Current. The West Spitsbergen Current is the major transporter of heat from the Atlantic to the Arctic, which intrudes into the fjord during the Arctic summer (Hop et al. 2006; Svendsen et al. 2002).
During the last decade, continuous hydrographic studies have indicated a climatic shift within the Arctic, i.e., a transition to a warmer state attributed to the increasing influence of warm Atlantic water masses (Polyakov et al. 2007; Spielhagen et al. 2011). This has been particularly documented for the Kongsfjord ecosystem over the last 3 years. During this time, the inner part of the Kongsfjord remained ice-free over winter (e.g., in 2011/2012, 2012/2013, and 2013/2014; personal communication AWIPEV station leader Rudolf Denkmann) documented by mean sea surface temperatures >0 °C from October to January (e.g., for 2012/2013 and 2013/2014 see COSYNA ferrybox at Spitzbergen operated by AWI and HZG Geesthacht; http://codm.hzg.de/codm). Furthermore, exceptionally warm water temperatures of up to 8 °C were recorded during the summer of 2014 (July/August; ref. COSYNA ferrybox) which were two degrees higher compared to the mean summer temperatures in 2012 and 2013 but almost double when compared to the maximum values of 3–4 °C reported a decade ago (Svendsen et al. 2002).
Remarkably, population persistence at lower latitudes, i.e., in particular the abundance of the Thysanoessa species, T. inermis and Thysanoessa raschii, was found to be determined by ambient water temperatures (Coyle et al. 2011; Hunt et al. 2011). Furthermore, a recent study of Huenerlage and Buchholz (Huenerlage and Buchholz 2015) highlighted the thermal metabolic sensitivity of both Thysanoessa species as a potential explanation for the species’ restricted biogeographic distribution. At the whole animal level, the authors found that the species were not able to compensate metabolic oxygen demands when ambient temperatures exceeded 12 °C; i.e., the species were not able to maintain cardiac activity at temperatures ≥12 °C possibly due to a limited capacity for oxygen uptake and/or oxygen transport mechanisms. Nevertheless, there is a clear need for further investigations at the molecular level (i.e., induction of molecular chaperones) to understand the underlying constraints at the cellular level that can provide early indicators of chronic stress and long-term survival capacities Accordingly, questions arise about whether the specimens are merely “passively tolerating” increased temperatures or whether the thermal limit of long-term survival may be already reached at temperatures <12 °C.
If the species’ long-term thermal limit is below 12 °C, then the current summer water temperatures of up to 8 °C (see above) may already come close to the true thermal limit of the arcto-boreal Thysanoessa species. Hence, in the future, these species may experience maximum water temperatures close to their thermal limit, which may negatively affect their overall metabolic performance and as a consequence, regional persistence.
In this context, our first aim was to estimate the upper critical temperature limit of T. inermis (CT50 = critical temperature at which 50 % of animals are reactive) and to confirm the Arrhenius break temperature from temperature dependent respiration curves. These data were supplemented by a molecular approach based on the monitoring of expression of HSP70 gene family members, which are considered as traditional markers of thermal shock. This first study of the heat-induced gene expression of the molecular chaperone heat shock protein 70 (Hsp70) may help to understand the thermal adaptive (i.e., survival) capacity of T. inermis within the changing ecosystem of the high Arctic Kongsfjord and increase the knowledge on the adaptive potential of this species with regard to its physiological reaction to sudden temperature exposure.
Material and methods
Sample collection
Krill (T. inermis) were sampled in late summer 2012 (August 17th–28th) on-board the Kings Bay AS workboat MS Teisten in the inner part of the high Arctic Kongsfjord (W-Spitsbergen) at 78.95° N, 12.33° E. A 1-m2 Tucker trawl (1000 μm mesh size and soft cod-end bucket) was deployed at a speed of two knots.
Immediately after being caught, adult T. inermis were transferred to aerated aquaria containing filtered seawater (0.2 μm) and kept at 4 °C in dim light before use in the experiments (respiration measurements and heat shock experiment, see below).
No specific permissions were required for these locations and field activities concerning sampling zooplankton, which includes the named Euphausiid species that are not endangered or protected species.
Illumina sequencing
The sequencing process included total RNA extraction from three whole animals (one control and two treated (6 and 10 °C)) and 60 eyestalks using the SV Total RNA Isolation System (Promega, Madison, WI, USA). Sequencing was conducted by the McGill University and Génome Québec Innovation Centre (Montréal, Québec, Canada) following the manufacturer’s instructions (Illumina, San Diego, CA). These data have been submitted to the SRA-EBI with accession number (SAMN04001594).
RNA-seq data sets
The cDNA library was sequenced to produce 150 bp paired-end reads. Raw reads were filtered with removal of low-quality and low-complexity sequences and trimmed using FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The reads were trimmed and filtered using a quality threshold of 25 (base calling) and a minimal size of 60 bp. Only reads in which more than 75 % of nucleotides had a minimum quality threshold of 20 were retained. Afterwards, rRNA contaminants were removed using ribopicker (Schmieder et al. 2012). Adapter ends were cleaned using cutadapt (version 1.01—(Martin 2011)). Finally, the whole quality control process was checked using fastQC (version 0.10.01 http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/).
The resulting assembly was produced using the “de novo” transcriptome assembler Trinity (release 2013-02-25—(Grabherr et al. 2011)). Finally, reads were remapped on the full transcriptome using Bowtie (version 0.12.8—(Langmead et al. 2009)) and relative abundances were estimated using RSEM (version 1.2.0—(Li and Dewey 2011)) to get the FPKM (fragments per kilobase of exon per million fragments mapped) values and thus identify the low coverage contigs (FPKM < 1) and rare isoforms (<1 %) that were excluded later from the analysis (both software programs were launched through the Trinity package Wrapper filter_fasta_by_rsem_values.pl).
Peptide prediction was performed using Transdecoder (Haas et al. 2013). Sequence similarity searches (blastp of the Transdecoder predicted peptides) were conducted against the UniProt-Swiss-Prot database (release 2013-09). Peptide signal prediction was performed using signalP v4.0 (Petersen et al. 2011). Transmembrane peptide detection was performed using TMHMM v2.0c (Krogh et al. 2001). Protein domain searches were conducted using hmmscan from the hmmer v.3.1b1 suite against the Pfam-A database release 27.0 (Finn et al. 2015). Finally, functional annotation was produced using the Trinotate pipeline (http://trinotate.github.io) described in (Haas et al. 2013). GO annotation plotting was carried out using WEGO (Ye et al. 2006) and GO slim analysis mapped against the previously obtained GO annotation against the generic GO slim ontology using custom R and Perl scripts.
Critical temperature estimation
The experimental protocol was identical to that used on the Antarctic species Euphausia crystallorophias and Euphausia superba (Cascella et al. 2015). After acclimation of ~24 h at 4 °C in the main aquarium, actively swimming animals were selected for experiments (n = 41).
The rate of temperature increase in this experiment was 1 °C every 10 min. The animals were maintained in the experimental tank until they were no longer able to respond to tactile stimuli of a probing rod. At this point, it was considered that the critical temperature limit had been reached and the animals were taken from the aquarium and snap frozen in liquid nitrogen. The CT50 was considered as the temperature at which survival of the experimental animals declined to 50 %. This was determined through the non-linear curve fitting option in JMP10 (SAS). The survival curve used was as follows: Survival = c/(1 + (T/CT50)b) where c is the plateau value before the sharp decrease, CT50 is the temperature at which 50 % of mobile animals is reached, and b is a sigmoidicity coefficient. The program explores the different values of these three parameters and calculates a chi-square value. While exploring the different parameter values, the program aims at minimizing chi-square and converges towards a value for each parameter (standard error provided).
Respiration measurements
Twelve hours after capture, T. inermis specimens were randomly chosen for the respiration measurements. In groups of 10 to 20 individuals, the specimens were brought to the final experimental temperatures (0, 2, 6, 8, 10, 12, 14, or 16 °C), at a rate of 1 °C h−1 followed by a 12-h acclimation at the constant experimental temperature. To avoid starvation effects on the metabolic rates, the total exposure times before measurement were not longer than 36 h (e.g., at 16 °C).
After acclimation, the specimens were individually incubated in closed tubular respiration chambers (Perspex; 20 ml) specially designed for measuring routine rates in krill (Huenerlage and Buchholz 2013; Werner et al. 2012). The chambers were filled with filtered seawater at the experimental temperature and stored in a water bath in a temperature-controlled refrigerator. Oxygen consumption (mg O2 L−1) was monitored every 30 s using a 10-channel optode respirometer (Oxy-10 Mini; PreSens Precision Sensing, Germany). This apparatus enabled the measurement of up to eight individuals in parallel. Two chambers were left blank (without a specimen) and served as controls. After the experiments, the specimens were weighed (mg) and measured for size (from the front of the eyes to the tip of the telson to the nearest mm).
Characterization of Hsp70 isoforms and cDNA cloning
To confirm the Illumina transcriptome assemblies and verify the identified Hsp70 contigs, nested PCR and sequencing were performed before evaluating the kinetics of Hsp70 expression in the heat shock experiments.
Ribonucleic acid (RNA) was isolated from the abdominal muscle of individual T. inermis specimens following the RNeasy® protocol (ref. Qiagen N.V., The Netherlands). The concentrations of total RNA were determined photometrically at 260 nm using a Nanodrop® (Thermo Fisher Scientific, USA). RNA purity was checked using the A260/A280 ratio (i.e., absorbance at 260 nm to the absorbance at 280 nm).
Purified RNA (1 μg) was retro-transcribed into single-stranded complementary deoxyribonucleic acid (cDNA) using SKdT primers (Roche, France) and the M-MLV Reverse Transcriptase kit (Affymetrix USB®, USA) according to manufacturer’s instructions. The Hsp70 isoforms were PCR amplified from the cDNA using specific primers that were designed from sequences obtained from the Illumina assembly. Four pairs of PCR primers were used for each gene in order to clone each isoform in overlapping 1000 base pair sections to facilitate full length sequencing of the whole gene. PCR products were gel purified and amplicons were inserted into the pGEM®-T Vector (Promega Corporation, USA). Plasmids were transformed into DH5α bacteria (Escherichia coli; Life Technologies™, USA). Transformed bacteria were selected and positive clones were verified by PCR. Plasmids were then extracted and sequenced with the same primers as before.
Heat shock experiments
The heat shock experiments were performed 24 h after capture. The experiment was started by immediately transferring ~200 T. inermis from the lab maintained at 4 °C to one aquarium (30 L) containing aerated seawater at 6 or 10 °C (“heat shock”). After 3 or 6 h, one set of animals (n = 50) were returned to the control temperature tank at 4 °C for recovery. The recovery lasted 6 h. During the heat shock and recovery time, sub-samples of 10 specimens were taken every 1.5 h or every 2 h, respectively. The individuals were snap frozen in liquid nitrogen and stored at −80 °C until further analysis at the Station Biologique de Roscoff, France.
In parallel to the experiment above, one group of T. inermis was kept at 4 °C and served as the control, i.e., was sub-sampled synchronously with the specimens from the heat shock experiment.
qPCR analysis
The RNA was extracted as described above. Messenger RNA (mRNA) levels of the Hsp70 isoforms were determined by reverse transcription qPCR amplification. Reactions were performed in a 5 μl total volume containing 2.1 μl of diluted reverse transcription product (1:200), 0.4 μmol of each specific primer, and 2.5 μl of SYBR Green I master mix (Roche, France). The amplification was carried out at 95 °C for 15 min, then in 55 cycles at 95 °C for 10 s and at 60 °C for 30 s. A dissociation curve was generated and PCR efficiency was estimated for each primer pair. All primer pairs tested generated a single peak in the dissociation curve and a PCR efficiency of 80–100 %. Data were analyzed with the LightCycler 480 software (Roche, France). The RPL8 gene was chosen as a reference gene using the BestKeeper algorithm (Pfaffl et al. 2004) after testing EF1α, 18S, RPL8, and GAPDH as potential normalizing housekeeping gene. Hsp70 expression was subsequently normalized to this reference. Six to ten animals were used for each point.
Data analysis
Metabolic rates were normalized to 1 mg fresh weight (FW) and expressed in micromole per hour (h−1). The Arrhenius break temperatures (ABT) of the temperature dependent respiration curve was estimated from the Arrhenius plots of the corresponding respiration rates (Dahlhoff et al. 1991). A one-way ANOVA with post hoc Dunnett test was performed to test for the temperature influence on the specimens’ respiration rates.
Differences of the mean normalized expression (MNE) of the five Hsp70 isoform genes (B, C1, C2, D, and E) over the time course of the experiment were analyzed using a non-parametric Kruskal–Wallis test. Relative gene expression (fold Hsp70 expression) was calculated from the MNE of test specimens (heat shock at 6 or 10 °C for 3 or 6 h and subsequent recovery at 4 °C) divided by the MNE of control specimens (kept at 4 °C control temperature).
Statistical analyses were carried out using GraphPad Prism 6 (GraphPad Software, Inc., USA). The significance level was set at p < 0.05.
Phylogenetic reconstruction
Phylogenetic reconstructions were carried out on 68 Hsp70 family proteins including Grp78 and mitochondrial isoforms from different crustacean species, using Bayesian Inference (BI) methods. Bayesian analysis was performed using MrBayes 3.1.2 with four chains of 106 generations, trees sampled every 100 generations, and the burning value set to 20 % of the sampled trees. Protein sequences were analyzed with a mixed amino acid model (Ronquist and Huelsenbeck 2003).
Results and discussion
CT50
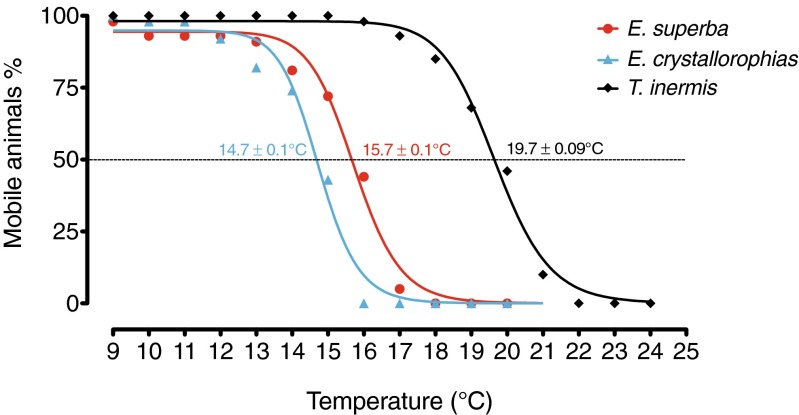
During the temperature challenges, T. inermis did not seem to be affected up to a temperature of 18 °C. The interpretation of the curve, i.e., the loss of mobility as a function of temperature, shows a CT50 value of 19.7 ± 0.09 °C (Fig. 1). However, this value provides little clue in direct relation to the environmental conditions that the animal would encounter. It is nevertheless helpful to compare pelagic species from different climates, for example, different species of krill. This experiment, when conducted in an identical manner on animals from other ecological or climatic backgrounds, enables differential sensitivities to be highlighted, which serve as comparative basis and characterize different resilience capabilities (Peck et al. 2009; Terblanche et al. 2011).
Fig. 1.
Curves representing the loss of mobility of krill populations subjected to gradual temperature increases (0.1 min−1) (E. crystallorophias n = 130; E. superba n = 43 (Cascella et al., 2015); T. inermis n = 41). CT50 is the temperature at which a 50 % loss mobile animal was reached
T. inermis therefore appears to be very thermo-tolerant with this high CT50 value. Indeed, this temperature is higher than those observed under the same experimental conditions in Antarctic Euphausia species (Cascella et al. 2015). However, in the context of this comparison, it is necessary to relate these values to habitat temperature. The starting temperature of the experiment was 4 °C for the boreal species; therefore, this species survived up to an additional 15.7 °C of warming, a value which is very similar to the CT50 of the Antarctic species whose ambient temperature is around 0 °C. Thus, both northern and southern species appear to have similar resiliencies to temperature.
T. inermis transcriptome assembling
A total of 207,011,779 raw sequences with read lengths of 150 bp were generated. After data cleaning to remove adapters and quality control, 205,445,915, high quality reads were obtained. These were used to produce a first assembly of 340,890 transcripts (corresponding to 214,624 Trinity “genes”) from 201 to 19,191 bp with an average length of 904 bp and a median length of 411 bp. The majority (90.7 %) of the cleaned reads were successfully mapped back to the full transcriptome indicating strong support for the assembly. Lowly expressed transcripts (FPKM < 1) and rare isoforms (<1 %) were excluded from the initial assembly leading to a filtered assembly of 54,319 transcripts (corresponding to 34,066 Trinity “genes”) from 201 to 18,966 bp with an average length of 1222 bp and a median length of 723 bp.
Putative functional analysis of the T. inermis transcriptome
20,626 proteins were predicted in the transcriptome. 11,124 of them had blastp matches (with an e value <10−4) against the UniProt-Swiss-Prot database (Online Resources 1A). Particular attention was paid to the GO terms “response to stress” (Online Resources 1B). The ontology GO006950 represented 7.2 % of GO designations and reached 11 % after the GO-Slim annotation. This category included different members of the Hsp family, including the Hsp70s.
Structure of Hsp70 isoforms
To estimate the onset of thermal stress in an organism, it is necessary to analyze the effects of temperature at the molecular scale. The primary effect of high temperature is denaturation of proteins and destabilization of cellular homeostasis, which leads to cell death and finally to the death of the organism. Heat shock proteins (HSP) are well known to counteract these deleterious effects, facilitating the refolding of proteins. There are many HSPs classified according to their weight in kilodaltons (from 10 to 110 kDa). Within this family, the heat shock protein 70 kDa (Hsp70) are the most studied. They are generally highly transcribed in response to heat shock, where they act as chaperone proteins and orchestrate the recruitment of other HSPs. They are, by their activity, largely responsible for the thermal tolerance of an organism. Indeed, the absence of these molecules can significantly decrease tolerance capacity (Bettencourt et al. 2008).
The Hsp70s are traditionally separated into one of two categories; the first contains the inducible Hsp70s called so because their expression is induced during stress. The second comprises the Hsc70s (heat shock cognate), often called constitutive because they are constantly expressed in the cell, at a basal level.
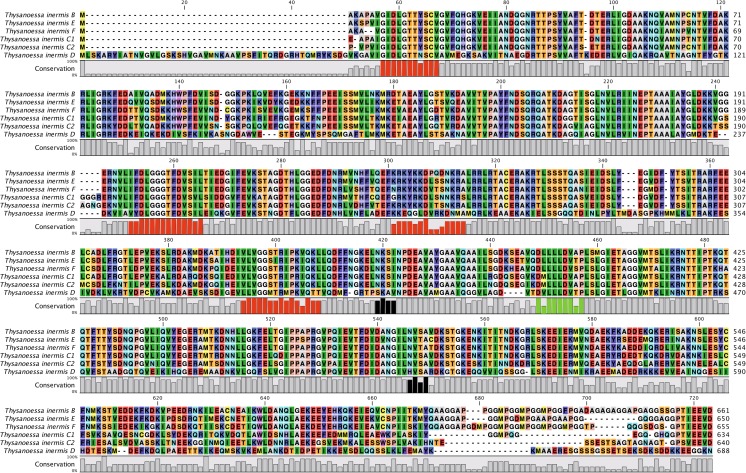
Six isoforms of Hsp70 were extracted from transcriptomic data. They were named B, C1, C2, D, E, and F according to their sequence similarities and in agreement with orthologous comparisons with the Southern Ocean species (Cascella et al. 2015) (Fig. 2). Unlike the Southern Ocean species, the A form was not identified in the transcriptome data, while two isoforms C (C1 and C2) have been characterized in T. inermis. The F isoform was subsequently extracted from the transcriptome assembly, but has not been confirmed by RT-PCR and no expression studies have been carried out to date. However, the orthologous sequence has been extracted from a new E. superba transcriptome (pers. data, unpublished), validating the identity of this sequence and the identification of six krill Hsp70 paralogues to date.
Fig. 2.
Alignment of six Hsp70 isoforms from T. inermis. B, E, and F isoforms represent the potential constitutive forms, C1 and C2 the potential inducible ones, and D the mitochondrial. In red: Hsp70 diagnostic motifs; in black: possible glycosylation sites; in green: hydrophobic linker between nucleotide-binding domain and substrate-binding domain. Alignment was realized with CLC MainWorkbench 7
The different isoforms were generally very similar to each other in terms of their primary sequence with high percentage identities ranging from 71 to more than 85 % at the amino acid level. The exception was the D form with 42–44 % identity, which was putatively designated as the mitochondrial form Hsp74 (Cascella et al. 2015). These isoforms clearly associated with potentially orthologous sequences previously identified in E. superba (ThiHsp70B/EusHsp70B = 96 %; ThiHsp70C1-C2/EusHsp70C = 77–85 %; ThiHsp70D/EusHsp70D = 90 %; ThiHsp70E/EusHsp70E = 91 %; ThiHsp70F/EusHsp70F = 93 %). The B, E, and F isoforms were structurally related to constitutive isoforms because of the tetrapeptide repeat motif (GGMP), which was present 3, 1, and 4 times, respectively. The C1 and C2 isoforms were equivalent to the inducible forms due to the presence of additional tetrapeptide sequences (residues 191–194); however, the C1 form carried a final GGMP, indicating a potential hybrid molecule. All these isoforms had the terminal I/VEEVD as signature motifs indicating to their cytoplasmic localisation.
The E and F isoforms were equally represented in the T. inermis transcriptome and were structurally designated as constitutive and cytoplasmic. They were present in similar quantities as the mitochondrial D form. This observation overlaps with the FPKM data obtained in E. crystallorophias for the D form and partial E form, which was not fully characterized (Cascella et al. 2015). The E form has only been found in one other species with a unique potential orthologous sequence in the crab Portunus trituberculatus (ACZ02405.1) (Cui et al. 2010). The latter is annotated as Hsp70 cognate-4 and is also found in various insects where it has been identified from genomic data. The F forms have not yet been identified in any other species.
There are two other key differences in the comparison with the Antarctic krill species: the lack of the A form and the presence of two inducible forms. In Euphausia, the two A and B forms, which were characterized as structurally constitutive, were the most highly expressed Hsp70s. In contrast, in T. inermis, only the B form was present. It also is the most highly represented form in the mRNA population of the transcriptome sampled (Table 1). The potential inducible forms in T. inermis were not equally represented. The C2 isoform was more represented in the transcriptome than the C1 isoform suggesting different functions. This low level of C1 may also be an explanation for the lack of characterization in Euphausia and in other crustaceans. In T. raschii, a short sequence potentially related to a C1 form has also been identified (pers. obs. data, unpublished).
Table 1.
T . inermis Hsp70 isoform expression values from the transcriptome and associated Blast matches
| Size (aa) | Size (pb) | FPKM | BLAST matches (E value) and accession number | |
|---|---|---|---|---|
| ThiHsp70B | 661 | 1983 | 942.9 | Hsp70B Euphausia crystallorophias (0.00) AIR72266 |
| ThiHsp70C1 | 634 | 1902 | 33.7 | Hsp70C Euphausia crystallorophias (0.00) AIR72268 |
| ThiHsp70C2 | 640 | 1920 | 197.1 | Hsp70C Euphausia crystallorophias (0.00) AIR72268 |
| ThiHsp70D | 687 | 2061 | 65.5 | Hsp70D Euphausia superba (0.00) AIR72274 |
| ThiHsp70E | 650 | 1950 | 54.6 | Hsp70E Euphausia crystallorophias (0.00) AIR72267 |
| ThiHsp70F | 655 | 1955 | 54.1 | Hsp70E Euphausia crystallorophias (0.00) AIR72267 |
Size (aa): deduced coding sequences. Size (pb): corresponding sizes in pair bases
FPKM fragments per kilobase of exon per million fragments mapped
These qualitatively and quantitatively differential representations of Hsp70 isoforms in boreal and Antarctic krill species may be associated with various response strategies. These could be potentially associated with different environmental stresses or different temperature ranges and stabilities, via the action of selection on gene duplication events and the subsequent sub-functionalization that ensures either their maintenance within the genome or their disappearance (Prince and Pickett 2002).
Molecular phylogeny of the Hsp70 family
To confirm and establish the phylogenetic relationships in crustaceans between the different isoforms of Hsp/Hsc70, 68 sequences of Hsp70 largo sensu (Grp78 and mitochondrial isoforms were included) were aligned. A tree was produced using the Bayesian inference method. This tree confirmed the positions of the Euphausia isoforms (Cascella et al. 2015) and the designations assigned to different isoforms of T. inermis previously assigned solely on the basis of their sequence similarities (Fig. 3).
Fig. 3.
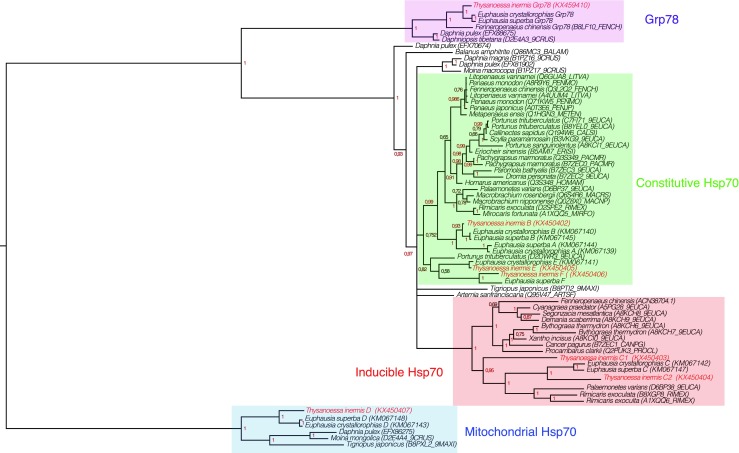
Phylogeny of the Hsp70 family including Grp78 in Eucrustacea based on a Bayesian analysis of the amino acid data set. Numbers above branches are posterior probabilities. T. inermis isoforms are in bold. Figure was created with FigTree v1.3.1
The positions of the E and F forms were confirmed and they constitute a sister group to a cluster classically considered as the constitutive isoforms, which includes the Euphausiid A and B forms.
The T. inermis C isoforms were positioned in a second set, grouping the inducible Hsp70s. The branch lengths of this set attest to a faster rate of evolution of these isoforms compared to the Hsc70s. Although only one C form was characterized in Euphausia, two are present in the transcriptome of T. inermis. The two sequences are not, as in the case of Rimicaris exoculata, present on the same branch of the tree. Indeed, the C1 isoform is positioned at the base of the cluster indicating that it is potentially close to an ancestral form.
It is clear from the phylogeny that the group of the inducible forms is numerically less represented than that of the constitutive forms. However, it may simply be that the sampling is not representative. Most of these data arise from transcriptome studies and referring to the FPKM data in the current study the C1 form is very poorly represented compared to the C2. This very low abundance could explain the absence of orthologous forms in the other species. These may well be discovered with more intensive transcriptome sampling or the production of draft genomes in the crustacean in the future.
Metabolic rates
In total, 122 adult T. inermis were sampled for the respiration measurements. Of these, 46 % were determined as females, 38 % as males, and 16 % were determined as neuter due to sexual regression which did not allow for a clear sex determination. The specimens had an average fresh weight of 115.74 ± 4.2 mg and an average size of 24.5 ± 0.2 mm. There was no significant size difference between the sexes. Furthermore, the respiration rates did not differ between sexes. Therefore, the data were pooled for the comparison of experimental temperature effects (Table 2, Fig. 4).
Table 2.
Mean respiration rates of adult T. inermis (n = 4–40) in relation to experimental temperatures
| Experimental temperature (°C) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | |
| n | 8 | 8 | 32 | 8 | 8 | 40 | 6 | 8 | 4 |
| Respiration rate ± SEM (μmol O2h−1gFW−1) | 3.3 ± 0.4 | 3.8 ± 0.4 | 4.7 ± 0.2 | 6.3 ± 0.5 | 7.8* ± 0.3 | 8.6* ± 0.4 | 10.2* ± 0.9 | 9.5* ± 0.9 | 7.3 ± 2.1 |
Values are given as means ± SEM, n = number of individuals used in the temperature experiments
*Significant difference to 4 °C control temperature
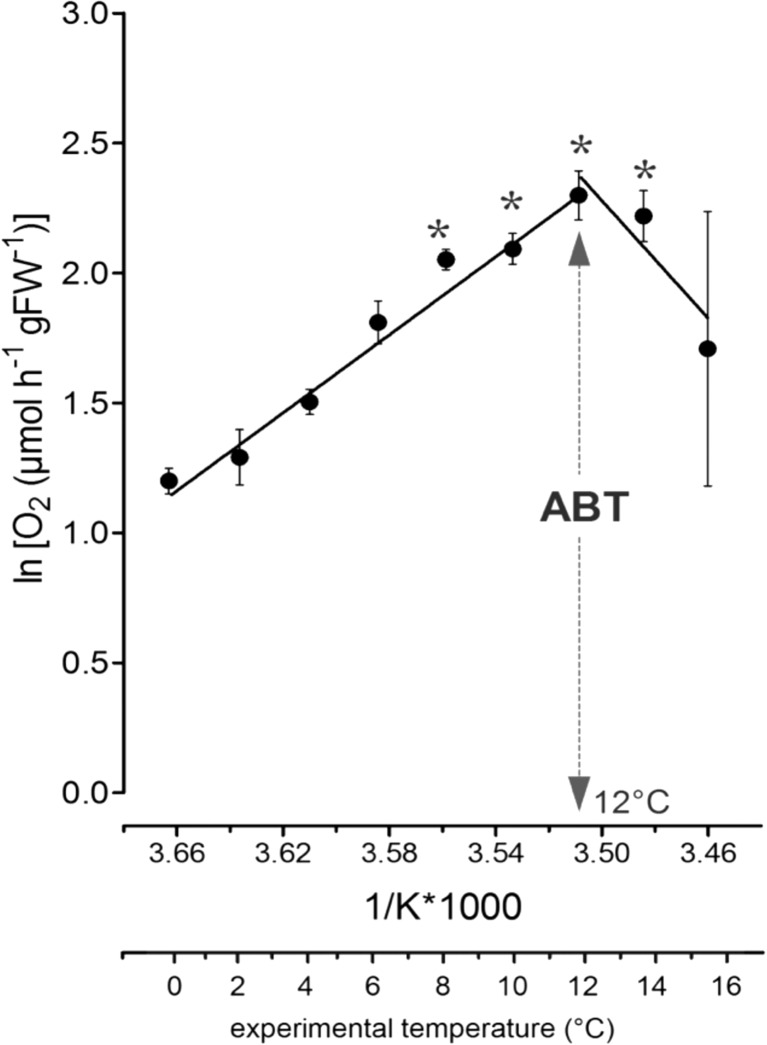
Fig. 4.
The Arrhenius plot of normalized respiration rates of adult T. inermis (n = 4–40) in relation to experimental temperatures. Lines show linear regressions. ABT Arrhenius breakpoint temperature defined by a significant change in slope. Values are given as means ± SEM, n = number of individuals used in the temperature experiments. (Asterisk) significant difference to 4 °C control temperature
The normalized respiratory performance over the experimental temperatures could be divided into two phases (Fig. 4). In the first temperature increment (0–12 °C), respiration rates increased exponentially. In reference to the 4 °C control temperature, the increase was significant from 8 °C (4.7 ± 0.2 μmol O2h−1gFW−1 at 4 °C vs. 7.8 ± 0.3 μmol O2h−1gFW−1 at 8 °C; Table 1, Fig. 1; p < 0.0001, F = 15.2, one-way ANOVA with Dunnett’s multiple comparison test against 4 °C control temperature). However, at experimental temperatures beyond 12 °C, mean oxygen consumption decreased from 10.2 ± 0.9 μmol O2h−1gFW−1 at 12 °C over 9.5 ± 0.9 μmol O2h−1gFW−1 at 14 °C to 7.3 ± 2.1 μmol O2h−1gFW−1 at 16 °C. The tipping point was depicted by the Arrhenius plot and showed the respiratory Arrhenius breakpoint temperature at 12 °C indicated by a sharp change in the slope of linear regression (=ABT; Fig. 4).
The respiratory response of adult T. inermis to experimental temperature change was the same as previously determined (Huenerlage and Buchholz 2015): increasing temperatures resulted in remarkable metabolic disturbance at temperatures exceeding 12 °C. This tipping point was determined by the Arrhenius breakpoint temperature (ABT) and consequently, characterized the upper limit of temperature-induced oxygen demand, i.e., the upper pejus temperature limit (TpII) (Frederich and Pörtner 2000) after which metabolism changes from aerobic to anaerobic (Pörtner 2012).
However, the respiratory response does not provide a measure for temperature-induced stress at the cellular level. In marine ectotherms, increased ambient water temperature is one of the major factors causing cellular damage due to the denaturation of proteins (Feder and Hofmann 1999; Kültz 2005). Accordingly, species have evolved responses to environmental stressors using molecular chaperones that help to prevent protein degradation and hence, enable them to survive during the periods when the species are exposed to unfavorable abiotic conditions.
Hsp70 gene expression
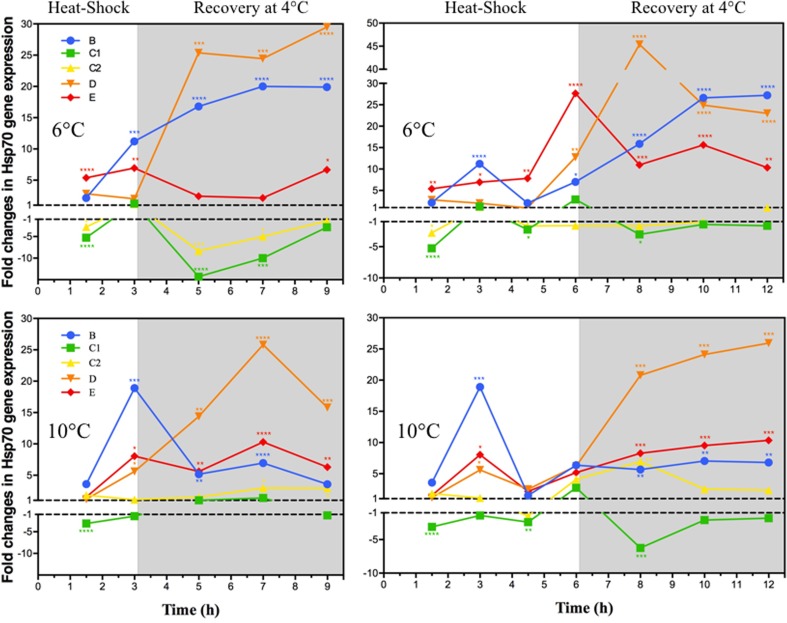
In this study, the molecular chaperones “heat shock protein 70” (Hsp70), which are well known as characteristic indicators of cellular stress, were investigated. The expression kinetics of five of the six isoforms were established according to the intensity of the heat shock (6 or 10 °C) and the duration (3 or 6 h). The responses after a return to the starting temperature (recovery) were also measured every 2 h for a total of 6 h (Fig. 5).
Fig. 5.
Mean normalized expression ratios of five Hsp70 isoforms in the muscle tissue of adult T. inermis (n = 6–10) during 3 h and 6 h continuous heat shock at 6 or 10 °C followed by 6 h of recovery at control temperature (4 °C; shaded). Values relate to the control group (specimens continuously kept at 4 °C). Asterisk: significant MNE difference to 4 °C control temperature evaluated with a Kruskal–Wallis test. Figure was created using GraphPad Prism 6.0 h
The kinetics showed clearly that the various isoforms did not have identical responses to the heat shocks. Furthermore, the notions of inducibility and constitutivity were not met as designated by sequence similarity and identification of signature motifs. Indeed, the forms assigned as constitutive (B, D, E) showed the most significant fold changes in expression whilst the designated inducible forms (C) appeared as the least expressed and even repressed (Fig. 5, Online Resource 2).
The B isoform did not show any significant increase before 3 h of heat shock, regardless of its amplitude, 6 or 10 °C. The kinetics of expression of this isoform showed peaks that suggested regulation by a feedback control as classically expected for an inducible Hsp70. The height of the expression peaks after 3 h shock was greater at 10 °C than at 6 °C suggesting a relationship between shock intensity and response amplitude. The expression could be further modulated as shown by the different successive peaks. In contrast, the amplitude of the response during the post shock, i.e., after a return to 4 °C was higher after a shock to 6 °C rather than 10 °C. The answer after 3 h of shock might be insufficient to counter the effects of stress and might explain this important secondary response. It is also interesting to note that the maximum fold increases in expression levels were similar whether during the shock and post-shock. Furthermore, the delay of the response could be explained by a constitutively large concentration of Hsp proteins present in the tissues.
The response kinetics of the mitochondrial D form also depended on the intensity and length of the thermal shock with a significant upregulation after 3 h at 10 °C and after 6 h at 6 °C. Whatever the shock time, elevated expression was most important during recovery, indicating a metabolic disturbance in the mitochondria. Little is known about the behavior of this type of HSP, but this observation would demonstrate that the heat shock disrupted the functioning of the mitochondria requiring extensive repair when returning to normal. Indeed, high temperature increases the metabolism and makes the mitochondria less efficient. More free radicals are produced. If Hsp70 cannot directly repair free radical damage, they might provide stability to mitochondrial and cellular antioxidant enzymes, which may be less efficient at high temperatures as well. The early response to 10 °C is consistent with the results obtained from the respirometry experiments highlighting an increased metabolism via the proxy of oxygen consumption and a much greater involvement of the mitochondrial respiratory chain in response to the acute stress.
Paradoxically, for the isoforms considered as inducible (C1 and C2), the trend of the response to thermal shock appeared stable or even negative. This decrease could be attributed to a massive recruitment of mRNAs, originally present before the heat shock, for protein synthesis. Much of the response would then consist in replacing the mRNAs involved in translation. On the other hand, the absence of elevated expression may be related to the existence of a base rate quantitatively close to the maximum limit of production of mRNAs encoding these isoforms. Indeed, the quantities measured by qPCR showed, for the C2 form especially, values close to those observed for the B isoform in the controls.
The HSP response characterizes a state of stress at the cellular level in an organism (Colson-Proch et al. 2010). However, the response is not the same depending on the species and the stress applied. A few organisms like the Antarctic Notothenioid fish have lost the inducible heat shock response (Hofmann et al. 2000). Whilst others, such as the starfish Odonaster validus or the amphipod Paraceradocus gibber (Clark et al. 2008) may have also lost these mechanisms during evolution, as an effective response of HSP70s to warming has yet to be shown in these species. The lack of a typical HSP response for this type of organism cannot be considered as an absence of stress, but rather as an inability to overcome damage caused by a factor, which denatures proteins, such as temperature. This absence is evidenced by a special sensitivity to an increase in their habitat temperature. However, many of these studies evaluated a restricted number of HSP70 genes, and it may be that other family members remain to be discovered (Clark et al. 2016). This will become more apparent with the increase in NGS studies, as the krill work is demonstrating with six paralogous HSP70 gene family members identified to date (Cascella et al. 2015). The environmental and the evolutionary histories of organisms have a direct impact on the level of activation of the transcription of Hsp70. We can distinguish between those organisms living in thermally variable environments, such as the temperate pelagic zone and intertidal rocky shores, and thermally stable environments, such as the tropical and polar zones (Tomanek 2010). Thus, the HSP response will vary depending on the type of thermal environment: the more stable is the environment, the weaker is the HSP response, with activation caused by a temperature close to the thermal optimum of the species studied. In contrast, in a more variable environment, the HSP70 response will be more intense and occurs when the individual is subjected to a temperature exceeding its thermal optimum and also across a wider range of temperatures. In this context, the HSP response is a critical molecular mechanism in terms of the thermo-tolerance capacity of individuals and contributes considerably to survival in case of thermal stress. Animals exposed for several million years to varying thermal conditions will have experienced positive selection for a very active heat shock response in contrast to organisms living in stable environments, where there has been less evolutionary pressure on this specialized biochemical pathway and upregulation of HSP70 genes in response to a stress is far more muted.
This study on the arcto-boreal krill T. inermis, besides improving the knowledge of the physiology of the species, also takes a comparative approach aimed at highlighting and understanding the strategies chosen in response to changing temperatures by phylogenetically closely related species living in more or less variable cold environments.
Similar to the results from the Antarctic krill species, the structural designations of the different genes as either inducible or constitutive did not fit with the experimental results. The potentially constitutive isoforms were the most involved in the responses to heat shocks. However, unlike the Southern Ocean species, the observed upregulation in gene expression generally occurred earlier in the time frame of the experiment and the levels were more elevated. This is particularly the case when these results are compared to those of E. crystallorophias, which is the most sensitive austral species examined to date using exactly the same regime of thermal shocks, whereas E. superba remained largely unreactive. This last result was particularly interesting because the two species T. inermis and E. superba exhibited similar capacities of thermal tolerance, according to the values of the corrected CT50, i.e., both can cope with a temperature increase of around 16 °C. It seems clear that the strategies adopted by both T. inermis and E. superba, and to a lesser extent E. crystallorophias, are different and related to the characteristics of the environments in which they live. The constitutive forms were particularly involved in these responses. As observed in E. crystallorophias, an upregulation of both isoforms (A and B), which were also the most represented quantitatively in terms of mRNA, represents an effective strategy for economically responding to thermal stress. Indeed, the existence of a significant basal level of these isoforms assures a rapid response to stress by allowing immediate translation of a high amount of protein. Although in T. inermis only the B isoform was found, its behavior was very similar to that of the isoforms of E. crystallorophias. The duplication of these isoforms in Euphausia could constitute a significant advantage in response to either a heat or cold shock. The hypothesis that ectothermic animals living in cold environments favor the accumulation of Hsps to correct the damage directly linked to the problems of protein folding at low temperatures could be an explanation for this physiological behavior (Place and Hofmann 2005). A supplementary argument that might support this hypothesis is that Hsp70A mRNA amounts were 10 times less in E. superba fished in the warmer waters of South Georgia than in animals from the East coast living at around 0 °C (Tremblay et al. submitted). Thus, the lack or the delay of gene expression could find its origin in the constant presence of large amounts of Hsp proteins in animals, quantities that would be sufficient to manage the immediate damages caused by a moderate or an acute increase of temperature early on. When the shock is prolonged and quantities and/or degradation of available Hsp70s become critical to correct cell damage caused by a thermal shock, the available mRNAs could be recruited via translation and at that moment new mRNAs would be transcribed. As a consequence, the higher the amounts of Hsp as protein or mRNA within the cell under normal conditions, the more delayed the heat shock response would be, thus highlighting the importance of the numbers of copies available of the A and B forms. Therefore, monitoring quantitative changes in translated Hsp70 in response to thermal shock will be the next step towards a more complete understanding of response strategies among different species of krill.
Conclusions
In the current study, we found that the 6 °C experimental temperature was sufficient to induce gene expression of the structurally defined constitutive Hsp70 isoform. Upregulation of expression occurred during direct heat exposure as well as during recovery, thus contradicting the structural definition of this isoform as “constitutive.” In addition, the high level of gene expression during recovery (e.g., up to 25–45-fold expression in Hsp70B and D, respectively, compared to the control) was indicative of intense molecular repair activities taking place, even at this relatively low temperature. Isoforms C1 or C2, which are defined structurally as inducible, showed no significant response at either 3 or 6 h after shock or during the recovery phase but in fact showed a reduction in available mRNAs.
The expression kinetics of these different isoforms are not similar to the response expected in a temperate species. They are also different from the observed responses in other species of krill from other cold environments such as Antarctica (Cascella et al. 2015). However, comparing the kinetics of expression between the polar species also revealed similarities between constitutive isoform behaviors. Thus, we developed the hypothesis that maintenance of a high constitutive protein level is associated with a large stock of mRNAs and that this organization enables an instant or continuous response to heat stress. This may be a primitive response to life in the cold, but also alter the threshold of transcriptional induction of new mRNAs, as these are costly to produce. On this basis, T. inermis could be considered as an intermediate type between very cold and stable environments and temperate conditions.
However, even if the CT50 of T. inermis was around 20 °C, the current findings imply that this species experienced molecular damage during exposure at 6–10 °C, which is even lower than their thermal respiratory limit at 12 °C as found by the Arrhenius breakpoint temperature. Therefore, the long-term thermal limit of overall species performance may be <12 °C at least under experimental conditions and will almost certainly be lower under chronic temperature challenges (Peck et al. 2009).
In order to more accurately predict the warming effects on the overall performance and population persistence of the arcto-boreal T. inermis, further research is needed. The data presented here are preliminary, but provide potential directions for further investigations.
Electronic supplementary material
(PDF 3.36 mb)
(PDF 630 kb)
Acknowledgments
KC received a PhD grant from the Emergence-UPMC 2011 research program and the “Région Bretagne.” JYT benefited from funding provided by Institut Paul Emile Victor (IPEV) (KREVET program) and also from the “Région Bretagne” (SAD-1—DRAKAR program).
This work was supported by the French-German AWIPEV project KOP 124, RIS ID 3451. We thank the captains of the Kings Bay AS workboat MS Teisten. We are grateful for the professional support from the lab leaders of the Kings Bay Marine Lab, the AWIPEV station leaders and (logistic) engineers, Ny-Ålesund, Spitsbergen. The data from the COSYNA data web portal were originally provided by Prof. Dr. Philipp Fischer (AWI), Dr. Gisbert Breitbach (HZG), Prof. Dr. Burkard Baschek (HZG), and Dr. Friedhelm Schroeder (HZG). We also thank Dr. Melody Clark for critical reading of the manuscript and Gildas Le Corguillé for GOslim analysis.
Contributor Information
Kim Huenerlage, Email: kim.huenerlage@awi.de.
Jean-Yves Toullec, Email: jean-yves.toullec@sb-roscoff.fr.
References
- Bettencourt BR, Hogan CC, Nimali M, Drohan BW. Inducible and constitutive heat shock gene expression responds to modification of Hsp70 copy number in Drosophila melanogaster but does not compensate for loss of thermotolerance in Hsp70 null flies. BMC Biol. 2008;6:5. doi: 10.1186/1741-7007-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Buchholz C, Weslawski JM. Ten years after: krill as indicator of changes in the macro-zooplankton communities of two Arctic fjords. Polar Biol. 2010;33:101–113. doi: 10.1007/s00300-009-0688-0. [DOI] [Google Scholar]
- Buchholz F, Werner T, Buchholz C. First observation of krill spawning in the high Arctic Kongsfjorden, west Spitsbergen. Polar Biol. 2012;35:1273–1279. doi: 10.1007/s00300-012-1186-3. [DOI] [Google Scholar]
- Cascella K, Jollivet D, Papot C, Leger N, Corre E, Ravaux J, Clark MS, Toullec JY. Diversification, evolution and sub-functionalization of 70 kDa heat-shock proteins in two sister species of antarctic krill: differences in thermal habitats, responses and implications under climate change. PLoS One. 2015;10:1–23. doi: 10.1371/journal.pone.0121642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MS, Fraser KPP, Peck LS. Lack of an HSP70 heat shock response in two Antarctic marine invertebrates. Polar Biol. 2008;31:1059–1065. doi: 10.1007/s00300-008-0447-7. [DOI] [Google Scholar]
- Clark MS, Sommer U, Sihra JK, Thorne MA, Morley SA, King M, Viant MR, Peck LS. Biodiversity in marine invertebrate responses to acute warming revealed by a comparative multi-omics approach. Glob Chang Biol. 2016 doi: 10.1111/gcb.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson-Proch C, Morales A, Hervant F, Konecny L, Moulin C, Douady CJ. First cellular approach of the effects of global warming on groundwater organisms: a study of the HSP70 gene expression. Cell Stress Chaperon. 2010;15:259–270. doi: 10.1007/s12192-009-0139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle KO, Eisner LB, Mueter FJ, Pinchuk AI, Janout MA, Cieciel KD, Farley EV, Andrews AG. Climate change in the southeastern Bering Sea: impacts on pollock stocks and implications for the oscillating control hypothesis. Fish Oceanogr. 2011;20:139–156. doi: 10.1111/j.1365-2419.2011.00574.x. [DOI] [Google Scholar]
- Cui Z, Liu Y, Luan W, Li Q, Wu D, Wang S. Molecular cloning and characterization of a heat shock protein 70 gene in swimming crab (Portunus trituberculatus) Fish Shellfish Immunol. 2010;28:56–64. doi: 10.1016/j.fsi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Dahlhoff E, O’Brien J, Somero GN, Vetter RD. Temperature effects on mitochondria from hydrothermal vent invertebrates: evidence for adaptation to elevated and variable habitat temperatures. Physiol Zool. 1991;64:1490–1508. doi: 10.1086/physzool.64.6.30158226. [DOI] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. HMMER web server: 2015 update. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederich M, Pörtner HO. Oxygen limitation of thermal tolerance defined by cardiac and ventilatory performance in spider crab, Maja squinado. Am J Physiol Regulatory Integrative Comp Physiol. 2000;279:R1531–R1538. doi: 10.1152/ajpregu.2000.279.5.R1531. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann GE, Buckley BA, Airaksinen S, Keen JE, Somero GN. Heat-shock protein expression is absent in the Antarctic fish Trematomus bernacchii (family Nototheniidae) J Exp Biol. 2000;203:2331–2339. doi: 10.1242/jeb.203.15.2331. [DOI] [PubMed] [Google Scholar]
- Hop H, Falk-Petersen S, Svendsen H, Kwasniewski S, Pavlov V, Pavlova O, Søreide JE. Physical and biological characteristics of the pelagic system across Fram Strait to Kongsfjorden. Prog Oceanogr. 2006;71:182–231. doi: 10.1016/j.pocean.2006.09.007. [DOI] [Google Scholar]
- Huenerlage K, Buchholz F. Krill of the northern Benguela current and the Angola-Benguela frontal zone compared: physiological performance and short-term starvation in Euphausia hanseni. J Plankton Res. 2013;35:337–351. doi: 10.1093/plankt/fbs086. [DOI] [Google Scholar]
- Huenerlage K, Buchholz F. Thermal limits of krill species from the high Arctic Kongsfjord (Spitsbergen) Mar Ecol Prog Ser doi:DOI. 2015 [Google Scholar]
- Hunt GL, Coyle KO, Eisner LB, Farley EV, Heintz RA, Mueter F, Napp JM, Overland JE, Ressler PH, Salo S, Stabeno PJ. Climate impacts on eastern Bering Sea foodwebs: a synthesis of new data and an assessment of the oscillating control hypothesis. ICES J Mar Sci. 2011;68:1230–1243. doi: 10.1093/icesjms/fsr036. [DOI] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. Bmc Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17
- Peck LS, Clark MS, Morley SA, Massey A, Rossetti H. Animal temperature limits and ecological relevance: effects of size, activity and rates of change. Funct Ecol. 2009;23:248–256. doi: 10.1111/j.1365-2435.2008.01537.x. [DOI] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Place SP, Hofmann GE. Comparison of Hsc70 orthologs from polar and temperate notothenioid fishes: differences in prevention of aggregation and refolding of denatured proteins. Am J Physiol-Reg I. 2005;288:R1195–R1202. doi: 10.1152/ajpregu.00660.2004. [DOI] [PubMed] [Google Scholar]
- Polyakov I, Timokhov L, Dmitrenko I, Ivanov V, Simmons H, Beszczynska-Möller A, Dickson R, Fahrbach E, Fortier L, Gascard JC. Observational program tracks Arctic Ocean transition to a warmer state. Eos, Trans Amer Geophys Union. 2007;88:398–399. doi: 10.1029/2007EO400002. [DOI] [Google Scholar]
- Pörtner HO. Integrating climate-related stressor effects on marine organisms: unifying principles linking molecule to ecosystem-level changes. Mar Ecol Prog Ser. 2012;470:273–290. doi: 10.3354/meps10123. [DOI] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Lim YW, Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics. 2012;28:433–435. doi: 10.1093/bioinformatics/btr669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielhagen RF, Werner K, Sørensen SA, Zamelczyk K, Kandiano E, Budeus G, Husum K, Marchitto TM, Hald M. Enhanced modern heat transfer to the Arctic by warm Atlantic water. Science. 2011;331:450–453. doi: 10.1126/science.1197397. [DOI] [PubMed] [Google Scholar]
- Svendsen H, Beszczynska-Møller A, Hagen JO, Lefauconnier B, Tverberg V, Gerland S, Ørbøk JB, Bischof K, Papucci C, Zajaczkowski M, Azzolini R, Bruland O, Wiencke C, Winther J-G, Dallmann W. The physical environment of Kongsfjorden-Kossfjorden, an Arctic fjord system in Svalbard. Polar Res. 2002;21:133–166. doi: 10.1111/j.1751-8369.2002.tb00072.x. [DOI] [Google Scholar]
- Terblanche JS, Hoffmann AA, Mitchell KA, Rako L, le Roux PC, Chown SL. Ecologically relevant measures of tolerance to potentially lethal temperatures. J Exp Biol. 2011;214:3713–3725. doi: 10.1242/jeb.061283. [DOI] [PubMed] [Google Scholar]
- Tomanek L. Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs. J Exp Biol. 2010;213:971–979. doi: 10.1242/jeb.038034. [DOI] [PubMed] [Google Scholar]
- Tremblay N, Cascella K, Toullec J-Y, Held C, Fielding F, Tarling GA, Abele D (submitted) Evaluating the hypoxic tolerance of Antarctic krill (Euphausia superba) at its range-edge
- Werner T, Hünerlage K, Verheye H, Buchholz F. Thermal constraints on the respiration and excretion rates of krill, Euphausia hanseni and Nematoscelis megalops, in the northern Benguela upwelling system off Namibia. Afr J Mar Sci. 2012;34:391–399. doi: 10.2989/1814232X.2012.689620. [DOI] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 3.36 mb)
(PDF 630 kb)