Abstract
RNA-binding motif proteins (RBMs) belong to RNA-binding proteins that display extraordinary posttranscriptional gene regulation roles in various cellular processes, including development, growth, and stress responses. Nevertheless, only a few examples of the roles of RBMs are known in insects, particularly in Apis cerana cerana. In the present study, we characterized the novel RNA-binding motif protein 11 from Apis cerana cerana, which was named AccRBM11 and whose promoter sequence included abundant potential transcription factor binding sites that are connected to responses to adverse stress and early development. Quantitative PCR results suggested that AccRBM11 was expressed at highest levels in 1-day postemergence worker bees. AccRBM11 mRNA and protein levels were higher in the poison gland and the epidermis than in other tissues. Moreover, levels of AccRBM11 transcription were upregulated upon all the simulation of abiotic stresses. Furthermore, Western blot analysis indicated that AccRBM11 protein expression levels could be induced under some abiotic stressors, a result that did not completely in agree with the qRT-PCR results. It is also noteworthy that the expression of some genes that connected with development or stress responses were remarkably suppressed when AccRBM11 was silenced, which suggested that AccRBM11 might play a similar role in development or stress reactions with the above genes. Taken together, the data presented here provide evidence that AccRBM11 is potentially involved in the regulation of development and some abiotic stress responses. We expect that this study will promote future research on the function of RNA-binding proteins.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0725-1) contains supplementary material, which is available to authorized users.
Keywords: Apis cerana cerana, RNA-binding protein, Abiotic stresses, Expression patterns
Introduction
As an Asian species, the Chinese honeybee brings huge economic benefits to the apiculture industry and plays a critical role in maintaining biodiversity. Compared to western honeybees, Apis cerana cerana has an acute sense of smell, a strong resistance to mites, and can forage a wide range of nectars and pollens; these advantages are irreplaceable (Peng et al. 1987; Cheng 2001; Oldroyd and Wongsiri 2006). However, recently, the population of Apis cerana cerana has severely declined, which can be attributed to the many abiotic stresses that exist in the environment, such as excessive pesticide use, climate changes of extreme heat and cold, and the presence of heavy metals and ultraviolet radiation. Publication of the Apis mellifera genomic sequence in 2006 powerfully facilitated honeybee research (The Honeybee Genome Sequencing Consortium 2006), and the report of the genomic sequence of Apis cerana cerana in 2015 provided a wealth of information for better understanding the natural biology and complex behaviors of the Asian honeybee (Park et al. 2015). However, it remains essential to identify specific genes and their corresponding proteins and to reveal their expression characteristics and related biological functions in stress responses.
Cellular response to environmental stresses is complex. Cells contain multiple regulatory mechanisms that are generally considered to have protective functions. The regulation can cause specific gene regulation or activation as well as posttranscriptional and translational events. With regard to posttranscriptional regulation, diverse RNA-binding proteins (RBPs) are the central posttranscriptional regulators of RNA metabolism. Typical RBPs are characterized by the presence of one or more RNA-recognizing domains (RRMs, also known as CS-RBD, RNP, or RBD domains), which are the largest parts of the protein and are composed of 75–85 amino acids (Norbury 2013). Large-molecular-weight RBPs contain a nuclear localization signal and can combine with nascent mRNAs to be responsible for their export from the nucleus. The structure of RBPs may be related to their function. In recent years, more and more studies have begun to explore the functions of RBPs.
RBPs not only play a role in genome organization, growth, and development but also in stress responses through the regulation of posttranscriptional mechanisms. RBPs were implicated in low oxygen level (Kang et al. 2007) and could respond to H2O2 stress in HeLa cells (Mironova et al. 2014). A glycine-rich RBP could be induced by cold stress and mediate cold-inducible suppression of cell growth (Nishiyama et al. 1997). In the Pashmina goat, RNA-binding motif protein 3 (RBM3) was downregulated under deep hypothermic conditions (Zargar et al. 2015). RBMs belong to RBPs. Guo et al. (2003) proposed that RBM4, RBM7, and RBM11 had strong homology in the RBD family according to GenBank. The role of RBM4 could be modulated by stressful cellular conditions (Lin et al. 2007). RBM7 phosphorylation by the p38(MAPK)/MK2 axis allowed stress-dependent modulations of the noncoding transcriptome (Tiedje et al. 2014). RBM11 displayed dynamic movement between the speckle and the nucleoplasm when cells were exposed to genotoxic and oxidative stresses (Pedrotti et al. 2011).
Although the functions of RBPs have been explored in other species, there is limited knowledge on the role of RBPs in honeybees, particularly in Apis cerana cerana. In this study, to gain insight into the role of the Chinese honeybee RNA-binding motif protein 11 gene (AccRBM11), we characterized the RBM11 gene from Apis cerana cerana and investigated its mRNA levels in different tissues and at different developmental stages. We also simulated common abiotic stress conditions encountered by Apis cerana cerana during its life to examine AccRBM11 mRNA and protein expression profiles. To our knowledge, this is the first report to examine the role of RBPs in stress responses in honeybees.
Experimental procedures
Biological specimens and various treatments
The Chinese honeybees (Apis cerana cerana) used in this study were maintained in artificial beehives at the Shandong Agricultural University (Taian, China). Honeybees in various developmental stages were classified according to the criteria of Michelette and Soares (1993). The egg, larvae, pre-pupal phase pupae, pupae, 1-day postemergence worker bee, non-egg-laying queen bee, and egg-producing queen bee were collected from the hive, whereas 19-day postemergence worker bees and 30-day postemergence worker bees were collected from the hive entrance by marking 1-day postemergence worker bee with paint 19 and 30 days earlier. Drones were also collected from the hive. The 19-day worker bees were collected randomly from the hive and were kept in a thermostat-regulated environment (34 °C) with 70 % relative humidity under a 24 h dark regimen. Then, they were randomly divided into ten groups (n = 60/group) and each group was treated with different stress conditions (Supplementary Table 1), which were simulated and similar to those that Apis cerana cerana are subjected to during its life. All of the control groups (untreated 19-day worker bee) were fed a normal adult diet and were incubated at 34 °C with 70 % relative humidity. The controls for the group that was injected with H2O2 were injected with phosphate-buffered saline (0.5 μl/honeybee). The honeybees were collected at the appropriate time after treatment (Supplementary Table 1). The wing, honey sac, muscle, epidermis, poison gland, midgut, hemolymph, tentacle, and rectum of 19-day worker bees were dissected on the ice to detect tissue-specific expression. All of the samples were frozen in liquid nitrogen and stored at −70 °C until use.
Total RNA extraction, cDNA synthesis, and genomic DNA preparation
Total RNA extraction, cDNA synthesis, and preparation of genomic DNA were performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and EasyScript First-Strand cDNA Synthesis (TransGen Biotech, Beijing, China) and EasyPure Genomic DNA Extraction Kits (TransGen Biotech, Beijing, China), respectively, according to each manufacturer’s instructions.
Acquisition of AccRBM11 cDNA, 5′-flanking region, and genomic DNA sequence
The genome information for the Chinese honeybee was uncovered in 2015 (Park et al. 2015); however, it was not released. Therefore, the open reading frame (ORF), 5′-flanking region (partial sequence of the promoter) and the genomic DNA sequence of AccRBM11 were obtained using specific primers, which are designed based on the RBM11-like sequence from Apis mellifera (a honeybee shares high homology with Apis cerana cerana). The 5′-untranslated region (5′ UTR) and 3′-untranslated region (3′ UTR) of AccRBM11 were cloned by 5′- and 3′- rapid amplification of the cDNA ends (RACE).
Bioinformatics analysis
The AccRBM11 homologous sequences used in this paper were downloaded from the NCBI servers (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The physical and chemical properties of AccRBM11 were predicted with the ProtParam tool (http://www.expasy.ch/tools/protparam.html) and DNAMAN version 5.22 (Lynnon Biosoft, Quebec, Canada). Multiple alignments were performed in DNAMAN version 5.22. The phylogenetic tree was generated with the neighbor-joining method using Molecular Evolutionary Genetics Analysis (MEGA version 4.1). The putative cis-acting elements of the AccRBM11 5′-flanking sequence were predicted by the MatInspector database (http://www.cbrc.jp/research/db/TFSEARCH.html). A PSORT II server was used to predict the nuclear localization of AccRBM11. Putative AccRBM11 antimicrobial peptides were predicted using the following website: http://aps.unmc.edu/AP/design/design_improve.php.
Fluorescent real-time quantitative PCR
Fluorescent real-time quantitative PCR (qRT-PCR) was carried out using the SYBR® PrimeScript™ RT-PCR Kit (TaKaRa, Dalian, China) and a CFX96TM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) to determine the AccRBM11 transcription profile. mRNA expression levels were normalized to the housekeeping gene β-actin (GenBank accession no. XM-640276), which was stably expressed and could be considered as a reference gene (Lourenço et al. 2008; Scharlaken et al. 2008; Li et al. 2012; Umasuthan et al. 2012). A 25-μL amplification reaction volume was used to perform the qRT-PCR reaction, which contained 12.5 μL Takara SYBR® premix Ex Taq™, 9.5 μL double distilled water, 2 μL cDNA, and 0.5 μL of each primer. The following qRT-PCR protocol was performed: (1) 30 s at 95 °C for pre-denaturation, (2) 40 cycles of amplification (5 s at 95 °C for denaturation, 15 s at 54 °C for annealing, and 15 s at 72 °C for extension), and (3) a single melt cycle from 65 to 95 °C. Relative AccRBM11 gene expression was analyzed with the CFX Manager software (version 1.1), using the 2 (−delta delta C(T)) method (Livak and Schmittgen 2001). Statistical Analysis System (SAS) software (version 9.1) was used to determine the statistical significance. All of the experimental conditions were implemented in three independent biological replicates, and the PCR reactions were performed in triplicate.
Protein expression, purification, and preparation of anti-AccRBM11
The ORF of AccRBM11 was cloned into the BamHI and SacI sites of the expression vector pET-30a(+) (Novagen, Madison, WI). Next, the expression plasmid pET-30a(+)-AccRBM11 was transformed into Transetta (DE3) chemically competent cells (a type of Escherichia coli; TransGen Biotech, Beijing, China). Next, the bacteria were cultured in 500 mL Luria-Bertani (LB) with 250 μL kanamycin (100 mg/mL) until the cell density reached 0.4–0.6 at 600 nm. Next, the cultures were induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Promega) at 37 °C for 12 h. Finally, a HisTrap™ FF column (GE Healthcare, Uppsala, Sweden) was used to purify the recombinant AccRBM11. The target SDS-PAGE albumen glue was added moderate sodium chloride injection (0.9 %) (Cisen Pharmaceutical, Jining, China) and benzylpenicillin sodium for injection (Lukang Pharmaceutical, Jining, China) and ground in a mortar with a pestle. The appropriate amount of ground sample was injected into the body of a white mouse (Taibang, Taian, China). The remaining experimental procedures were carried out as described in Meng et al. (2010). The obtained AccRBM11 antibody was hybridized to a blot containing the overexpressed protein of AccRBM11 in order to detect the specificity of the anti-AccRBM11.
Western blot analysis
Total protein was extracted from Apis cerana cerana following the manufacturer’s instructions that provided by a tissue protein extraction kit (ComWin Biotech, Beijing, China) and quantified with a total protein assay kit (using a standard BCA method; ComWin Biotech, Beijing, China). A 12 % SDS-PAGE was used to separate equal concentration of total protein from each sample of the same treatment. Then, the albumin glue that contained target proteins was subsequently electrotransferred onto a PVDF membrane (ComWin Biotech, Beijing, China) using a wet transfer method. The anti-AccRBM11 serum was used as the primary antibody at a 1:500 (v/v) dilution. Peroxidase-conjugated goat anti-mouse immunoglobulin G (Jingguo Changsheng Biotechnology, Beijing, China) was used as the secondary antibody at a dilution of 1:2000 (v/v). α-Tubulin was used as a control at a dilution of 1:50,000 (v/v); its protein levels is usually not change in the body of an organism (Liu et al. 2009; Zhang et al. 2009; Tang et al. 2010). The tubulin antibody (1:50,000) purchased from Beyotime (China) recognizes the C-terminal region of α-tubulin. The results of the binding reaction were visualized using a SuperSignal™ West Dura Extended Duration Substrate (Thermo Fisher Scientific, Shanghai, China).
RNA interference mediated silencing of the AccRBM11 gene and transcriptional levels of some other genes after AccRBM11 knockdown
RNA interference experiment was used to knock down AccRBM11 transcripts. The non-conserved sequence region in the AccRBM11 ORF was chosen to design the specific primers. The primers with a T7 polymerase promoter at their 5′-end were used to synthesize a linear DNA template. AccRBM11 double-stranded RNA (dsRNA) was produced using RiboMAX T7 large-scale RNA production systems as per the manufacturers’ instructions (Elias-Neto et al. 2010). The green fluorescent protein gene (GFP, synthetic construct, GenBank accession number U87974) was used as a control (Elias-Neto et al. 2010). Apis cerana cerana genes do not share homology with dsRNA-GFP, so an RNAi response will not be triggered by dsRNA-GFP in the body of Apis cerana cerana. The 19-day postemergence worker bees were selected for the RNAi treatments and were divided into three groups (n = 40/group). Using a microsyringe, 6 μg of dsRNA-AccRBM11 or dsRNA-GFP was injected between the first and second abdominal segments into each adult. The last group was untreated. The above three groups were maintained in an incubator (at 34 °C with 60 % relative humidity and under a 24-h dark regimen). The healthy adult bees were sampled each day and stored at −70 °C. qRT-PCR was performed to detect AccsHsp22.6, AccSOD2, AccTpx4, AccAK, AccRPL11, AccTpx1, Accp38b, AccGSTO1, and AccTrx1 expression profiles when AccRBM11 was knocked down. These experiments were carried out in three independent biological replicates.
Disk diffusion assay
Cells overexpressing AccRBM11 were plated onto LB-kanamycin agar plates and incubated at 37 °C for 50 min. Then, filter disks (6-mm diameter) were placed on the surface of the agar plates. The filter disks were soaked with 2.5 μL of different concentrations of cumene hydroperoxide (0, 50, 100, 200, and 400 mM), paraquat (0, 100, 200, and 300 mM), HgCl2 (0, 10, 20, 50, and 70 mg/ml) and CdCl2 (0, 8, 16, 23, and 42 mg/ml). The bacteria were incubated at 37 °C for 10 h, and the killing zones around the disks were measured. Transetta (DE3) chemically competent cells containing the pET30-a (+) vector were used as control.
Primers and amplification conditions
All of the primer pairs and polymerase chain reaction (PCR) amplification procedures used in this paper are presented in Supplementary Tables 2 and 3, separately. An ideal primer pair used for qRT-PCR should have an efficiency (E) value of 90–110 %, a coefficient (R 2) of more than 0.980, and single peak melting curves (Bustin et al. 2009; Yang et al. 2016). The primers used for qRT-PCR in this paper were designed on the basis of the principle of quantitative primer design; the E value, R 2, and numbers of melting curve peaks for each qRT-PCR primer pair are listed in Supplementary Table 4, all of which complied with the qRT-PCR requirements. All of the primers used in this study were synthesized by the Sangon Biotechnological Company (Shanghai, China).
GenBank accession characteristic of the genes used in this study
A lot of genes were used to execute bioinformatics analysis, and their GenBank accession number and species name are listed in Supplementary Table 5.
Results
Characterization of AccRBM11
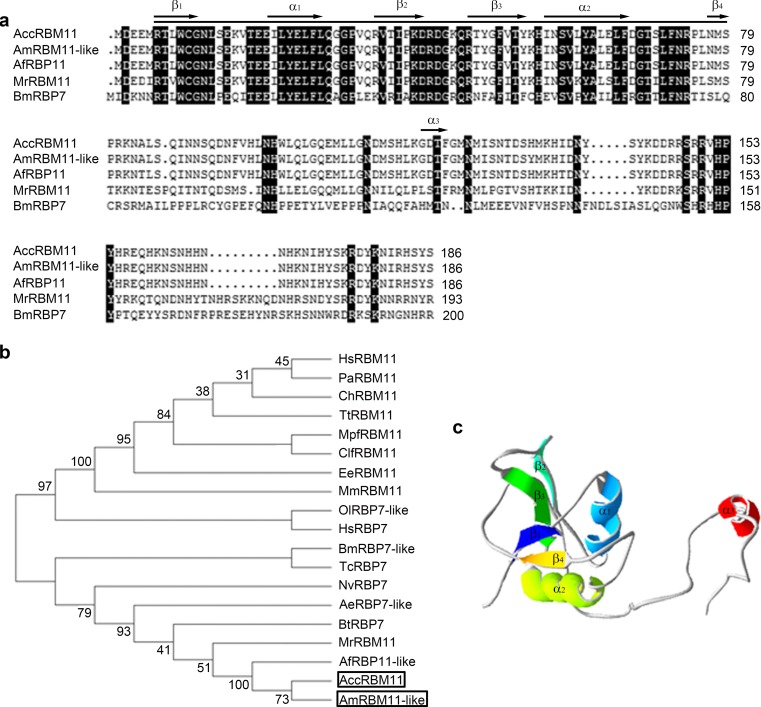
The full-length cDNA of AccRBM11 (GenBank accession number KR827409) is 1205 bp, which contains a 116 bp 5′ UTR, a 561 bp complete ORF, and a 528 bp 3′ UTR (Fig. 1). A typical polyadenylation signal sequence (AATAA) was found in the 3′ UTR (Fig. 1). Figure 2a reveals that AccRBM11 was most homologous to its ortholog protein in Apis mellifera (99.46 %), and the N-terminus was more highly conserved than the C-terminus. The ORF of AccRBM11 encodes a 186 amino acids protein with a predicted molecular mass of 22.115 kDa and a predicted theoretical pI of 9.10. AccRBM11 included an RRM-RBM7-like (RNA recognition motif in the RBP7 and similar proteins) specific hit that contained 74 residues (6–79) (Fig. 2a), which was predicted by the NCBI Conserved Domain Database Search. The secondary structure of AccRBM11 is represented as β1α1β2β3α2β4α3 and the secondary structure of the RRM domain of AccRBM11 as β1α1β2β3α2β4 (Fig. 2a).
Fig. 1.
AccRBM11 nucleotide sequence and deduced amino acid sequence. The cDNA sequence is shown on the top line, and the deduced amino acid sequence is shown on the second line. The boxes mark the ATG (start codon) and the TAA (stop codon), and the asterisk denotes the stop codon. The predicted antimicrobial peptides are underlined. Ellipses show the possible polyadenylation signal. The sequence of AccRBM11 was deposited into GenBank with the accession no. KR827409
Fig. 2.
Characterization of RNA-binding proteins (RBPS) from different species and the tertiary structure of AccRBM11. Protein sequences were acquired from NCBI (Supplementary Table 5). a Multiple alignment of AccRBM11 amino acid sequences with other RNA-binding proteins. Identical regions are shaded in black. The putative RRM domain of AccRBM11 is marked by a straight line, and the predicted secondary structure of AccRBM11 is labeled with arrows. b Phylogenetic analysis of RBPs from various species; AfRBM11 and AccRBM11 are boxed. c The tertiary structure of AccRBM11. The helices, sheets, and coils are marked with different colors
Phylogenetic tree analysis revealed that AccRBM11 was more closely related to AmRBM11-like protein than to other species (Fig. 2b). Figure 2c shows the possible three-dimensional structure of AccRBM11 that might be important for understanding AccRBM11 protein structure. In addition, the NLS prediction showed that AccRBM11 was most likely localized in the nucleus, suggesting that it might play a role in the nucleus.
Analysis of the AccRBM11 genetic sequence structure
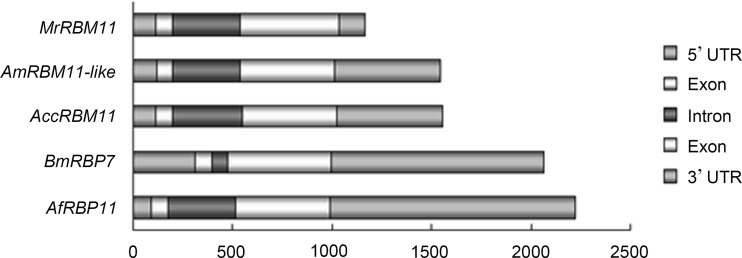
To further study the AccRBM11 genomic locus, a 1552-bp full-length segment of DNA was obtained, which contained one intron and two exons (GenBank accession number KR827410). As shown in Fig. 3 and Supplementary Table 6, the lengths and number of exons had been conserved; however, the sizes of the introns, the 3′ UTR, and the 5′ UTR were not conserved during the evolutionary period. Supplementary Table 6 revealed that the A+T content was richer than the content of G+C and that the A+T contents of the exons were much lower than in the introns, which was consistent with the results from a previous study (Park et al. 2015).
Fig. 3.
Schematic representation of DNA structures. The pattern of untranslated regions, introns, and exons in AccRBM11, AmRBM11-like, AfRBP11, MrRBM11, and BmRBP7 are presented according to the scale shown below. The genomic DNA from the above genes (except AccRBM11) was loaded from the NCBI database, and their GenBank accession numbers are presented in Supplementary Table 5
Putative TFBs on the AccRBM11 promoter
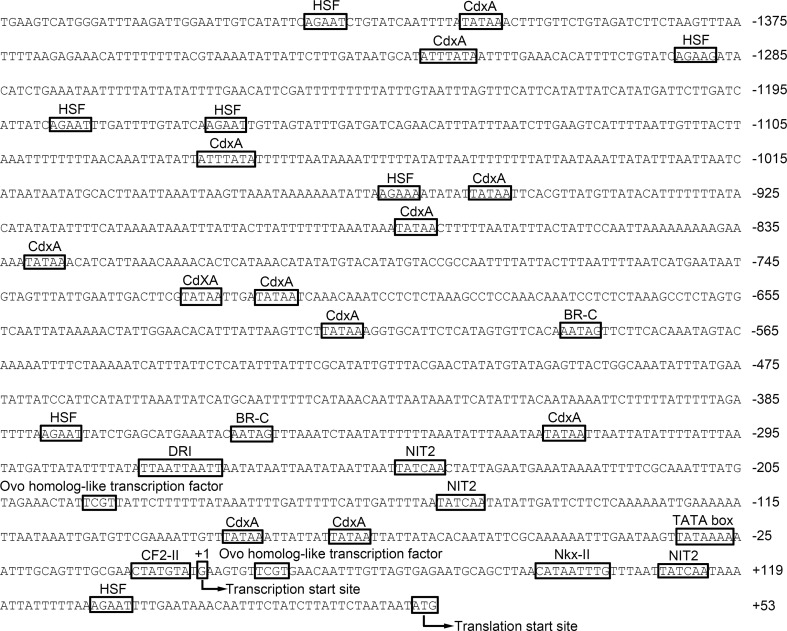
A 1464-bp promoter sequence (GenBank accession number KR827410) was isolated to investigate the transcription regulatory regions of AccRBM11. Many putative transcription factor binding sites (TFBs) were predicted by the software MatInspector in both the sense strand (Fig. 4) and the antisense strand (data not shown). These included sites for heat shock factor (HSF), CdxA, Nkx-II, NIT2, BR-C, DRI, CF2-II, and ovo homolog-like transcription factor. Moreover, many possible TFBs were also detected in the 5′ UTR of AccRBM11. TATA box was also identified in the promoter region of AccRBM11, which can combine with TFII to initiate gene transcription.
Fig. 4.
Partial nucleotide sequences and putative transcription factor binding sites in the promoter of AccRBM11. The transcription start and translation sites are labeled with an arrow, and the putative TFBs are marked with boxes
Expression and characterization of recombinant AccRBM11
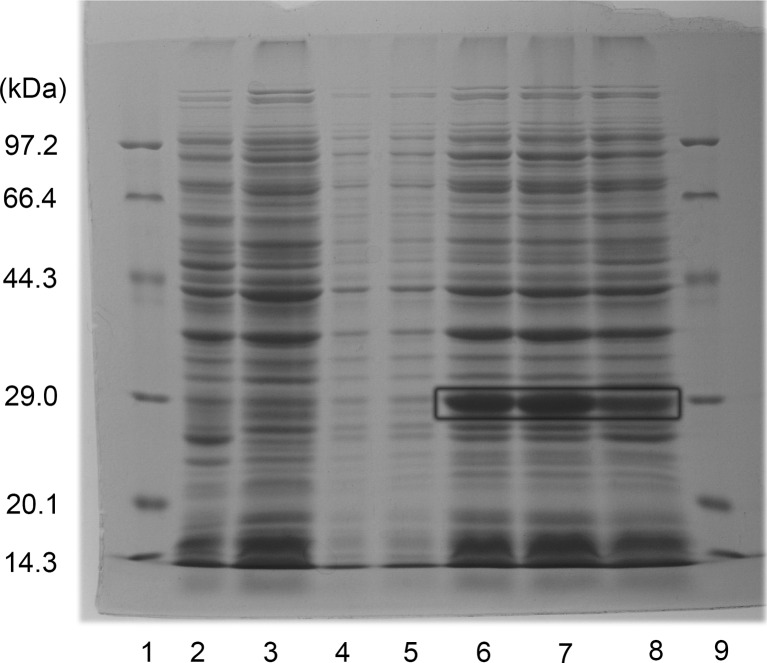
AccRBM11 was overexpressed in Transetta cells as a histidine fusion protein. SDS-PAGE result showed that the recombinant had a molecular mass of less than 29 kDa (Fig. 5), which was a little smaller than the predicted molecular mass of 29.946 kDa (including cleavable C- and N-terminal His-tags of approximately 7.831 kDa). The recombinant protein was presented in the form of an inclusion body in the Transetta cells, and the nearly insoluble recombinant protein could not be purified with a HisTrap™ FF column, even at low temperature (data not shown).
Fig. 5.
The expression of the recombinant AccRBM11 protein. Recombinant AccRBM11 was separated by SDS-PAGE. Lanes 1 and 9, protein molecular weight marker; lane 2, induced overexpression of pET-30a(+) in Transetta cells; lanes 3 and 8, uninduced and induced overexpression of pET-30a(+)-AccRBM11, respectively, in Transetta cells; lanes 4 and 5, suspension of sonicated recombinant AccRBM11; and lanes 6 and 7, pellets from sonicated recombinant AccRBM11. The site of recombinant AccRBM11 is marked with a box
Specific detection of anti-AccRBM11
Total protein from overexpressed AccRBM11 in Transetta cells was used to detect the specificity of the anti-AccRBM11, whereas total protein from uninduced overexpression of pET-30a(+)-AccRBM11 in Transetta cells was used as a control. As shown in Supplementary Fig. 1, only the lane from the cells overexpressing AccRBM11 had a band, and it was a single band, which suggested that the anti-AccRBM11 was specific for overexpressed AccRBM11.
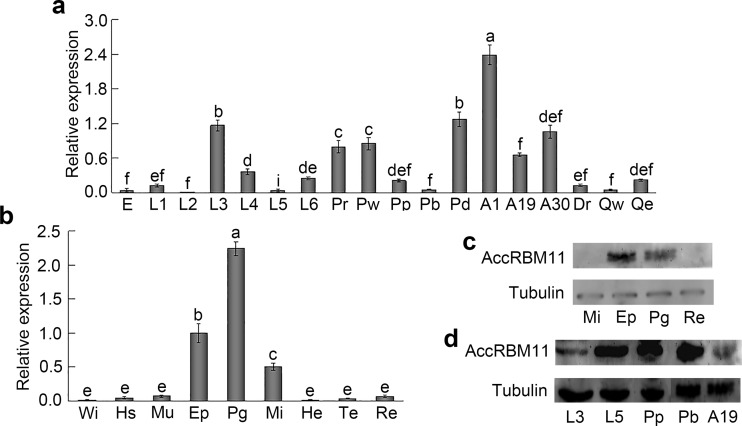
Temporal and spatial AccRBM11 profiles
qRT-PCR was used to examine the expression patterns of AccRBM11 during various developmental stages and in different tissues. As shown in Fig. 6a, AccRBM11 was expressed at all stages and displayed the highest transcriptional levels in the 1-day postemergence worker bees (A1). Among the tissues examined, the most well-defined expression was observed in the poison gland (Pg), followed by the epidermis (Ep) and midgut (Mi) (Fig. 6b). Western blotting was also performed to examine AccRBM11 temporal and spatial protein expression. As shown in Fig. 6c, AccRBM11 expression was higher in the Ep than that in the Pg, whereas AccRBM11 proteins could not be detected in the Mi and Re. The AccRBM11 protein contents were higher in the L5 stage, Pp and Pb, followed by the stage of L3 and A19 (Fig. 6d), which was not completely consistent with the result of qRT-PCR.
Fig. 6.
AccRBM11 expression profile in different developmental stages and various tissues. a AccRBM11 expression levels at diverse developmental stages including egg (E), 1- to 6-day larvae (L1–L6), pre-pupal phase (Pr), white-eyed (Pw), pink-eyed (Pp), brown-eyed (Pb) and dark-eyed (Pd) pupae, 1-day postemergence worker bee (A1), 19-day postemergence worker bee (A19), 30-day postemergence worker bee (A30), drone (Dr), the non-egg-laying queen bee (Qw), and the egg-producing queen bee (Qe). b AccRBM11 transcript levels in the wing (Wi), honey sac (Hs), muscle (Mu), epidermis (Ep), poison gland (Pg), midgut (Mi), hemolymph (He), tentacle (Te), and rectum (Re). The β-actin gene is shown for comparison. Vertical bars and letters above vertical bars represent the mean ± SEM of three different samples and significant differences (P < 0.0001), respectively. c The expression level of AccRBM11 protein at different tissue. Tubulin was used as an internal control. d The amount of AccRBM11 protein in different development stage. Tubulin was chosen as an internal control
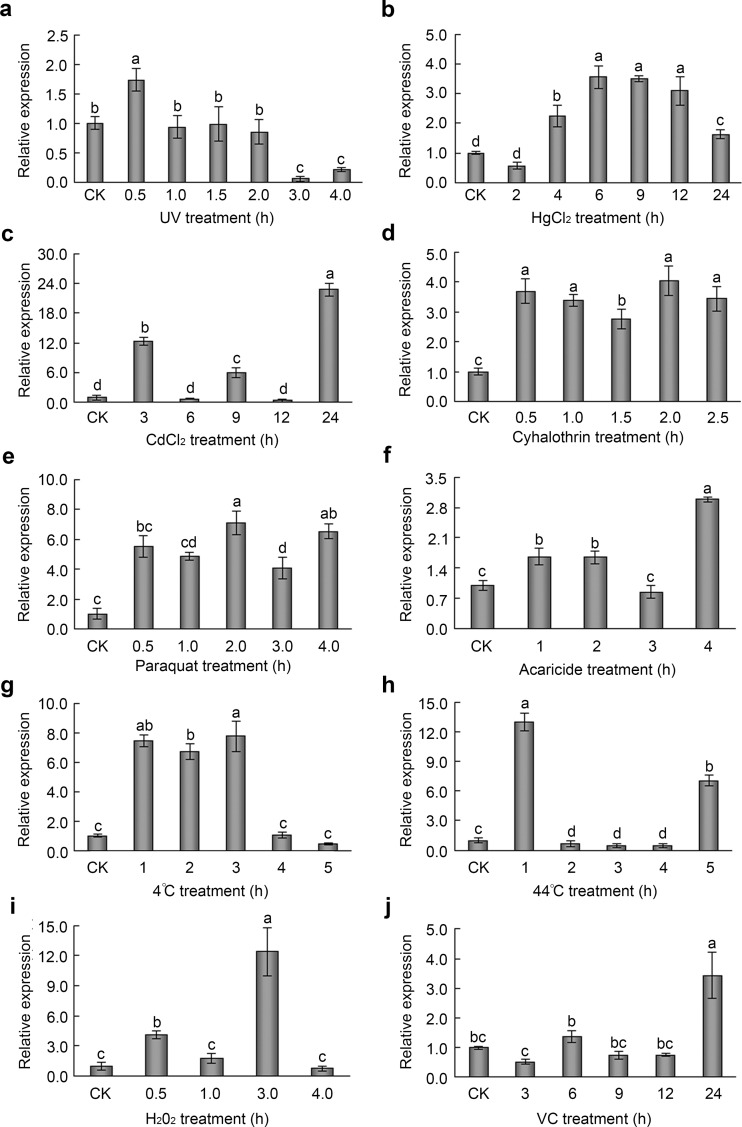
AccRBM11 expression profiles under different types of abiotic stress
Though the mRNA and protein levels in the 19-day postemergence worker bee were not high. Older than 18-day postemergence worker bees are usually responsible for foraging nectar, pollen, and water, so they were most likely to be subject to environmental stresses. Thus, the 19-day postemergence worker bee was chosen to be treated with different stresses to explore the possible response of AccRBM11 at transcript and protein levels. qRT-PCR was used to characterize the mRNA levels of AccRBM11 after exposure to UV, H2O2, VC, heavy metals (HgCl2 and CdCl2), pesticides (acaricide, cyhalothrin, and paraquat), and extreme temperatures (4 and 44 °C). Expression levels were normalized to those in the control adult bees. As shown in Fig. 7a, the transcript level of AccRBM11 was upregulated under UV treatment and peaked at 0.5 h. The mRNA level of AccRBM11 was increased 2.26-fold at 4 h and peaked at 6 h following HgCl2 treatment (Fig. 7b). AccRBM11 level climbed 22.77-fold compared with the control group after treatment with CdCl2 (Fig. 7c). Cyhalothrin, paraquat, and acaricide also upregulated AccRBM11 expression (Fig. 7d–f); however, there were differences in the degrees of the increase and in the transcriptional patterns. Experiments involving exposure to different temperatures are presented in Fig. 7g, h, which indicated that AccRBM11 expression levels were markedly upregulated following exposure to 4 and 44 °C temperatures. According to the qRT-PCR analysis, AccRBM11 levels were upregulated after treatment with H2O2 (Fig. 7i) and VC (Fig. 7j) and reached maximums at 3 and 24 h, respectively.
Fig. 7.
Expression profile of AccRBM11 under various abiotic stress conditions. These stresses included UV (a), HgCl2 (b), CdCl2 (c), cyhalothrin (d), paraquat (e), acaricide (f), 4 °C (g), 44 °C (h), H2O2 (i), and VC (j). The β-actin gene was used as an internal control. Control check is abbreviated CK. The data represent the mean ± SE of independent experiments (n = 3). The letters above the vertical bars indicate significant differences (P < 0.0001) by Duncan’s multiple range tests
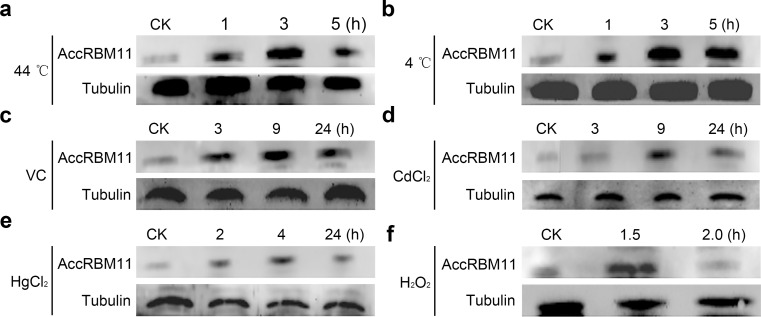
Western blot analysis
Western blot analysis was performed to further explore AccRBM11 expression patterns under adverse abiotic stress conditions. AccRBM11 was detected using an anti-AccRBM11, which was specific to AccRBM11 (Supplemental Fig. 1). After exposure to 44 °C (Fig. 8a) and 4 °C (Fig. 8b) for 1, 3, and 5 h, AccRBM11 expression was increased several fold. VC treatment caused clear increases in AccRBM11 expression levels (Fig. 8c). As shown in Fig. 8d, e, the amount of AccRBM11 was highest at 9 and 4 h of CdCl2 and HgCl2 treatment, respectively. Following exposure to H2O2, AccRBM11 protein levels rose gradually from 0 to 1.5 h (Fig. 8f). In contrast, AccRBM11 expression patterns did not display any significant changes when treated with acaricide (Supplemental Fig. 2a), paraquat (Supplemental Fig. 2b), cyhalothrin (Supplemental Fig. 2c), or UV (Supplemental Fig. 2d).
Fig. 8.
Western blot analysis of AccRBM11. Nineteen-day-old adult bees were treated with 44 °C (a), 4 °C (b), VC (c), CdCl2 (d), HgCl2 (e), and H2O2 (f). An equivalent concentration of extracted protein from the same treatment at different time was loaded in different lane and anti-AccRBM11 was used to immunoblot the targeted protein. Tubulin was used as an internal control
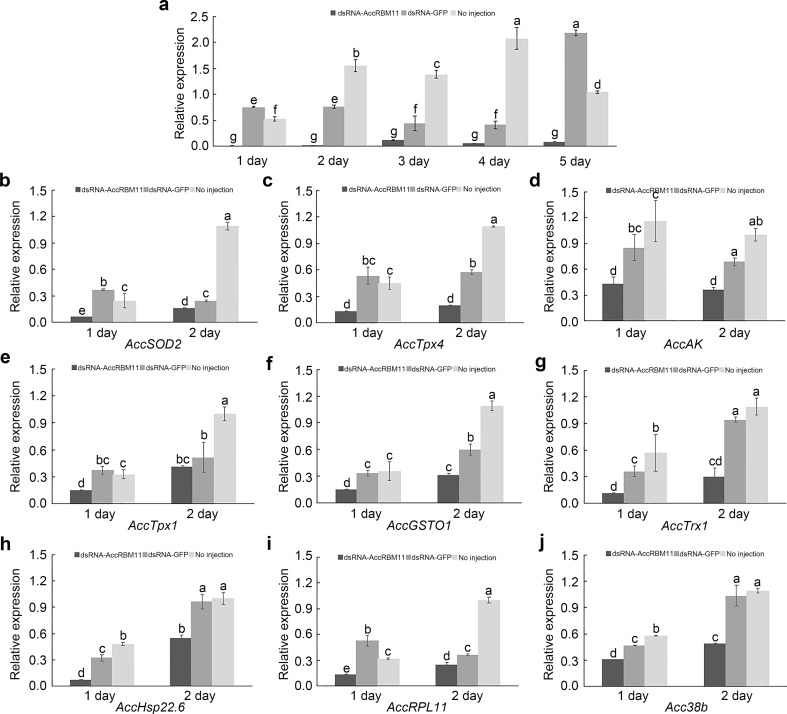
AccRBM11 transcript knockdown and the expression profiles of other genes after AccRBM11 silencing
RNAi experiment was carried out to explore the role of AccRBM11 in development and stress responses. As shown in Fig. 9a, the AccRBM11 gene was successfully silenced compared with the control groups, and the lowest AccRBM11 transcript levels were discovered at 1 and 2 days postinjection of dsRNA-AccRBM11. The qRT-PCR results showed that AccSOD2, AccTpx4, AccAK, AccTpx1, AccGSTO1, AccTrx1, AccsHsp22.6, AccRPL11, and Accp38b were all suppressed when AccRBM11 was silenced (Fig. 9b–j).
Fig. 9.
RNA interference on AccRBM11 transcript levels in adults and the effect of AccRBM11 silencing on other genes in Apis cerana cerana. a Each 19-day postemergence worker bee was injected with 6 μg of dsRNA-AccRBM11. An equivalent volume of dsRNA-GFP was also injected for the control group. Untreated adults were also chosen as the controls. b–j Expression profiles of other development or stress response genes when AccRBM11 was knocked down. The β-actin gene was used as an internal control. All of the data are presented as the mean ± SE of three independent experiments. Various letters above the bars indicate significant differences (P < 0.0001) using Duncan’s multiple range tests
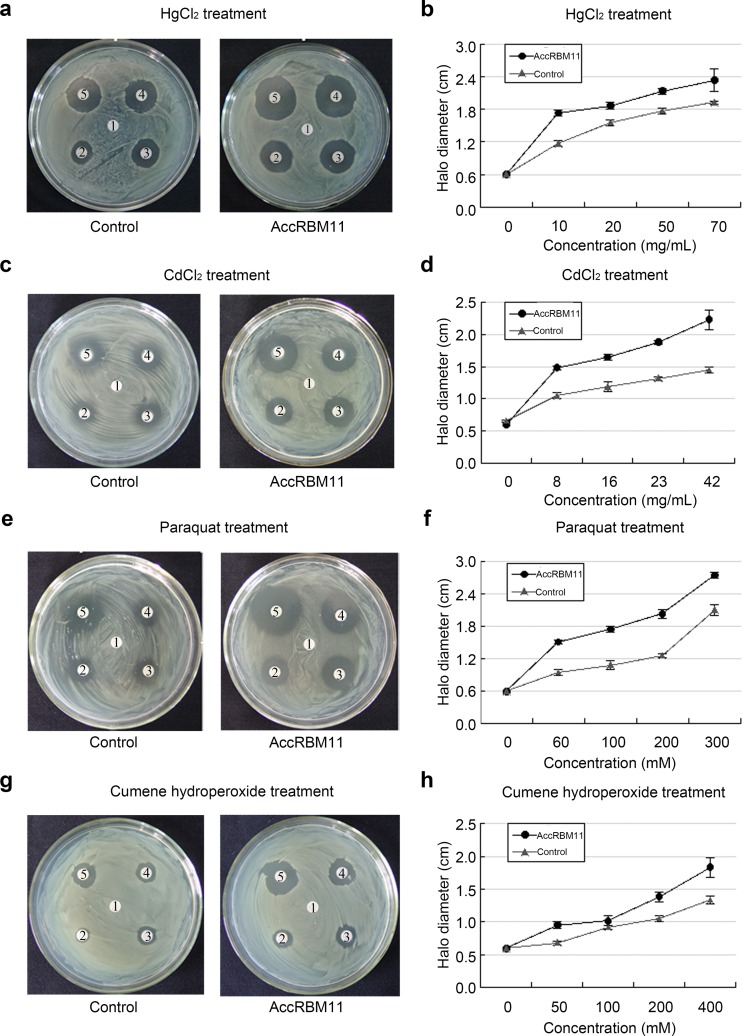
Disk fusion assay of recombinant AccRBM11in response to diverse stressors
combinant AccRBM11 contained a His-tag and AccRBM11. Interestingly, it required much more time for the Escherichia coli (E. coli) cells with AccRBM11 to reach the same optical density at 600 nm compared with that of the E. coil cells without AccRBM11. After exposure to different concentrations of reagents (HgCl2, CdCl2, paraquat, and cumene hydroperoxide), the halo diameters around the filters on agar plates that cultured the bacteria containing overexpression AccRBM11 were larger than around the controls, even though the death zones were distinctly different under the different treatment condition (Fig. 10). The online database APD2 was used to explore the mechanism indicated by the experimental results. At least two antimicrobial peptides were identified in the AccRBM11 amino acid sequence (Fig. 1). The peptides have neutral charges and may display antimicrobial activities.
Fig. 10.
Resistance of bacteria cells overexpressing AccRBM11 to various stressors. A total of 5 × 108 cells were inoculated onto LB agar plates. AccRBM11 was overexpressed in bacterial cells, and the E. coli cells that were transfected with pET30a(+) were used as a control. Killing zone diameters were compared in the line chart. The data are the mean ± SE of three replicates. a, b The HgCl2 concentrations on filter disks 1–5 were 0, 10, 20, 50, and 70 mg/mL, respectively. c, d The CdCl2 concentrations on filter disks 1–5 were 0, 8, 16, 23, and 42 mg/mL, respectively. e, f The paraquat concentrations on filter disks 1–5 were 0, 60, 100, 200, and 300 mM, respectively. g, h The cumene hydroperoxide concentrations on filter disks were 0, 50, 100, 200, and 400 mM, respectively
Discussion
RNA-binding proteins are well known for posttranscriptionally regulation of RNA metabolism. Recently, an increasing number of studies have begun to investigate the functions of RBPs in response to different stresses in plants (Lorković 2009). Studies have also indicated a pivotal role for RBPs in animal under adverse types of stress, such as in Homo sapiens and Mus musculus (Nishiyama et al. 1997; Yang and Carrier 2001; Aoki et al. 2003; Mironova et al. 2014; Zargar et al. 2015). However, the roles of RBPs in stress response are poorly understood in insects. The present study was undertaken to characterize a novel RBP gene (AccRBM11) identified in Apis cerana cerana and to investigate its response to different abiotic stresses. Our findings suggested that AccRBM11 could be induced by some stress agents.
AccRBM11 has a typical RRM domain, which specifically belongs to the RRM-RBM7-like subfamily. This subfamily is characterized by the RRM in RBM7, RBM11, and their eukaryotic homologs. An AccRBM11 phylogenetic tree was generated to analyze the evolutionary relationships among different species, and the result showed that AccRBM11 shared a high degree of homology with RBP11 and RBP7 from other species; however, its greatest homology was to RBM11-like from Apis mellifera (Fig. 2b), which was consistent with the results predicted by multiple protein alignment. We therefore named the newly acquired gene from the Chinese honeybee AccRBM11. Kenan et al. (1991) deemed that RRM domain could form a β1α1β2β3α2β4 secondary structure and that the direction of the two α helixes was perpendicular to the direction of the β-sheet. It is possible that the AccRBM11 RRM domain has this β1α1β2β3α2β4 structure (Fig. 2a). Thus, the secondary structure characteristics of AcRBM11 might be involved in AccRBM11 functions.
To better understand the potential roles of AccRBM11, a 1464-bp 5′-flanking region that contained many TFBs connected with environmental stress and development was cloned (Fig. 4). Interestingly, the AccRBM11 5′ UTR also included some TFBs (Fig. 4), which indicated that the 5′ UTR might also participate in the transcriptional regulation of AccRBM11. Among these, CF2-II is known to be related to development and growth (Štanojević et al. 1989; Gogos et al. 1992). BR-C can regulate the development of the third instar and early pre-pupal larvae (Spokony and Restifo 2007). NIT2 has been described as the major nitrogen regulatory gene (Fu and Marzluf 1987). A previous study suggested that RBPs could be developmental regulators (Bandziulis et al. 1989). It showed that AccRBM11 could be expressed in all Apis cerana cerana stages and its expression displayed a distinct increase during the transition from egg to larva, larva to pupa, and pupa to adult (Fig. 6a). The Western blot results were not completely consistent with the results of qRT-PCR (Fig. 6d). Recent studies have shown that the transitions between developmental stages are often followed by immense changes in gene expression and that the expression of definite qualitative and quantitative proteins that implement specific functions are required in each stage (Gala et al. 2013). This finding suggested that AccRBM11 might be necessary for the transitions in Apis cerana cerana between developmental stages.
CdxA, Nkx-2, DRI, and Ovo homolog-like transcription factor binding sites were also found in the 5′-flanking region of AccRBM11. CdxA participates in tissue-specific expression (Ericsson et al. 2006). Nkx-2 is related to organogenesis (Briscoe et al. 1999). DRI determines the development of neural and gut cells (Gregory et al. 1996), and Ovo homolog-like transcription factor is necessary for epidermal and germline differentiation (Delon et al. 2003). Tissue-specific analysis showed that the highest expression of AccRBM11 appeared in the poison gland, followed by the epidermis and midgut (Fig. 6b), which did not agree with the Western blotting findings (Fig. 6c). This particular expression of AccRBM11 in different tissues most likely reflects the specific demand of that tissue. Moreover, the poison gland plays a key role in self-defense. The epidermis plays an important role in imparting physical stability and in taking part in responses to environmental stresses. The midgut is associated with the detoxification of exogenous substances and protection from oxidative stress (Marionnet et al. 2003; Enayati 2005). These findings indicated that AccRBM11 might be involved in protecting Apis cerana cerana against harm from environmental stresses.
In addition to the TFBs mentioned above, a higher amount of HSF, which is involved in heat-induced transcriptional activation (Santoro et al. 1998), was found in the 5′-flanking sequence of AccRBM11. High temperature may be an important environmental factor that influences development because it can induce physiological changes (An and Choi 2010). The behavior of the honeybee can be influenced by temperature changes, and Apis cerana cerana are more sensitive to heat stress (Tautz et al. 2003). qRT-PCR was performed to detect whether AccRBM11 responded to heat. As shown in Fig. 7h, the mRNA level of AccRBM11 reached their highest levels at 1 h, which suggested that AccRBM11 might play a role in heat stress. In organisms, cold pressure suppresses protein synthesis rate, changes cellular membrane lipid composition, and ultimately inhibits cell growth (Rao and Engelberg 1965; Burdon 1986). RBPs have been known to play important roles in the cellular response under cold stress and can be induced by low temperature (Bae et al. 1997; Nishiyama et al. 1997; Zargar et al. 2015). The qRT-PCR results indicated that AccRBM11 was increased several fold (Fig. 7g) under 4 °C treatment. The data suggested that AccRBM11 might play a role in avoiding cold lesions.
In other species, many previous studies have demonstrated that RBPs could respond to environmental stress (Mazan-Mamczarz et al. 2003; Kang et al. 2007; Abdelmohsen et al. 2008; Lorković 2009; Mitobe et al. 2009; Tanaka et al. 2009). Therefore, we also detected AccRBM11 expression levels when Apis cerana cerana was subjected to other abiotic stresses, such as pesticides, heavy metals, UV, H2O2, and VC. Pesticides are important factors that lead to environmental pollution. These include herbicides, insecticides, fungicides, and others, which damage the physiological and biochemical functions of lymphocytes and erythrocytes and result in the lesions to lipid biomembranes (Narendra et al. 2007). The transcript levels of AccRBM11 were all induced by cyhalothrin (Fig. 7d), paraquat (Fig. 7e), and acaricide (Fig. 7f) treatment, suggesting a role for AccRBM11 in protecting honeybees against pesticides. Heavy metals can influence the normal development of insects. Mercury (Hg) and cadmium (Cd) are well known to be the most poisonous heavy metals in the natural world (Rashed 2001) and can inactivate the function of proteins by directly binding to enzyme metal ion sites. Worker honeybees may contact heavy metals when they forage for pollen and nectar outside. The expression levels of AccRBM11 were enhanced by HgCl2 (Fig. 7b) and CdCl2 (Fig. 7c) treatment by 3.6- and 22.7-fold, respectively, indicating that AccRBM11 might function to avoid injury under HgCl2 and CdCl2 stresses. As an environmental stress, UV light influences insect habits and living (Mazza et al. 2002). qRT-PCR findings revealed that UV induced the mRNA level of AccRBM11 (Fig. 7a). The UV-inducible RBP A18 has been shown to protect human cells against genotoxic stress by translocating to the cytosol and stabilizing specific transcripts related to cell survival (Yang and Carrier 2001). AccRBM11 might function similarly to avoid UV injury. Hydrogen peroxide (H2O2) is a main type of oxidant that can cause oxidative damage. We detected high AccRBM11 expression levels after 3 h of H2O2 treatment (Fig. 7i). Recent studies had indicated that RBPs could be involved in oxidative responses (Brégeon and Sarasin 2005; Abdelmohsen et al. 2008; Mironova et al. 2014). AccRBM11 might play a role in oxidative stress. Figure 7j showed that AccRBM11 was upregulated by VC. VC is a typical antioxidant. However, VC can also induce the decomposition of lipid hydroperoxide to endogenous genotoxins and can lead to DNA oxidative damage (Lee et al. 2001). Thus, in this study, the dose of VC might have been sufficient to induce AccRBM11 to take part in the response to oxidative injury.
Moreover, Western blotting was carried out to investigate AccRBM11 protein levels when Chinese honeybees were subjected to various abiotic stresses. When treated with 44 °C (Fig. 8a), 4 °C (Fig. 8b), VC (Fig. 8c), CdCl2 (Fig. 8d), HgCl2 (Fig. 8e), and H2O2 (Fig. 8f), the expression level of AccRBM11 protein increased in some capacity, even though they were upregulated to various degrees and at different times. However, no clear response of AccRBM11 protein levels was observed after treatment with acaricide (Supplemental Fig. 2a), paraquat (Supplemental Fig. 2b), cyhalothrin (Supplemental Fig. 2c), and UV (Supplemental Fig. 2d). It is worth noting that the upregulated degree and time points for AccRBM11 showed distinct differences in the transcript and protein levels. The mRNA and its corresponding protein for a gene are both present in a living body; however, usually only the protein is functional. Changes in AccRBM11 protein levels when subjected to abiotic stresses in this study indicated that AccRBM11 might be implicated in responses to some abiotic pressure.
Regarding the transcriptional patterns of AccRBM11 that were not completely consistent with its protein levels, the following explanations should be taken into account. First, stress treatments may be enough to induce gene transcription, but not enough to necessarily affect its translation. Second, the accumulation and degradation of protein existed in organisms. For instance, when the transcription of a gene is inhibited, increased protein expression levels could be due to protein accumulation. Third, the transcription and translation of a gene could be regulated by various signal transduction pathways under environmental pressure, such as posttranscriptional regulation. Recently, Mitobe et al. (2009) reported that invE mRNA levels were easily detectable, though translation of the protein was tightly inhibited, which is similar for posttranscriptional regulation through RBP Hfq. Last but not least, several RNAs can participate in the transcription and translation of mRNAs, such as circRNAs and miRNAs. Previous studies had shown that circRNAs are implicated in regulating transcription, posttranscription, splicing, and protein activation (Memczak et al. 2013; Ashwal-Fluss et al. 2014). A recent study revealed that miRNAs were involved in regulating many pivotal processes during amelogenesis by influencing translation and mRNA stability in rat incisors (Yin et al. 2014). The differences in the expression patterns of AccRBM11 and its protein may be a result of its regulation by circRNAs and miRNAs.
RNAi experiment was performed to further investigate the function of AccRBM11. RNA silencing works through posttranscriptional gene regulation mechanisms (Ding 2010; Goic et al. 2013). RNAi-mediated knockdown of endogenous target gene expression has become a popular strategy determining gene function, as transgenesis is hard to achieve in some species, especially in animals (Huvenne and Smagghe 2010; Goic et al. 2013). This experimental method also has been used in honeybees, and studies have proven that gene knockdown is important for studying of gene function (Wilson et al. 2014; Yao et al. 2014). Here, AccRBM11 mRNA levels were successfully knocked down by RNAi, especially at 1 and 2 days after injection of dsRNA-AccRBM11 (Fig. 9a). Compared with the controls, AccRBM11 silencing markedly downregulated AccSOD2, AccTpx4, AccAK, AccTpx1, AccGSTO1, AccTrx1, AccsHsp22.6, AccRPL11, and Accp38b transcripts to different degrees (Fig. 9b–j). Among these genes, AccSOD2, AccTpx4, AccAK, AccTpx1, AccGSTO1, and AccTrx1 had been demonstrated to be involved in different environment stress responses (Yu et al. 2011; Jia et al. 2014; Meng et al. 2014; Yao et al. 2014; Chen et al. 2015; Huaxia et al. 2015). AccsHsp22.6, AccRPL11, and Accp38b could not only function in stress defence but also in development (Meng et al. 2012; Zhang et al. 2012; Zhang et al. 2014). Thus, AccRBM11 might have the similar functions in development and stress responses in Apis cerana cerana with the above genes.
We also used a disk diffusion assay to examine the ability of AccRBM11 to resist abiotic stresses. Recent evidence had shown that the human YB-1 protein can bind to RNA and act as an RNA chaperone to play a role in cell survival under environmental stress. When introduced into E. coli, the YB-1 gene conferred high resistance to bacterial cells to environmental stress (Li et al. 2006). However, after exposure to four different treatment conditions, AccRBM11 overexpression in E. coli cells displayed low resistance to these disparate stresses (Fig. 10). This might be because of potential antimicrobial peptides that were identified in the AccRBM11 amino acid sequence (Chen et al. 2015), which might inhibit the growth of bacteria (Xu et al. 2009). The inhibitory effect is relatively weak (not enough to completely restrain the growth of E. coli cells), and the cells still can express recombinant AccRBM11 protein (Yu et al. 2007; Guo et al. 2013).
In conclusion, our study indicated that the expression of AccRBM11 could be enhanced by some abiotic stresses both at the transcriptional and protein levels. The increased expression pattern possibly relates to the changes in ROS levels. AccRBM11 might play an important role in the development of Apis cerana cerana and in stress response challenges and might cause the elevated resistance of honeybees to environmental stresses. These findings may contribute to further inquiry into the function of RNA-binding proteins in honeybees and other insects.
Electronic supplementary material
The specific detection of anti-AccRBM11. Total protein of overexpression AccRBM11, induced overexpression of pET-30a(+)-AccRBM11 in Transetta cells were loaded in different lanes and the targeted protein was identified using an anti-AccRBM11 (GIF 3 kb)
Western blot analysis of AccRBM11. Western blot analysis of AccRBM11 changes after acaricide (a), paraquat (b), cyhalothrin (c), or UV (d) treatments. All of the lanes were loaded with an equivalent concentration of extracted protein from the same treatment at different times. Tubulin was selected as an internal control (GIF 31 kb)
Abiotic stress conditions for each experimental group (DOC 32 kb)
Primer sequences used in this study (DOC 82 kb)
Procedures used in this paper (DOC 28 kb)
Efficiency, coefficient, and number of melting curve peaks for each primer pair used for qRT-PCR (DOC 34 kb)
Characterization of the genes used in this paper (DOC 59 kb)
Size of the introns and exons and the A + T content in AccRBM11, AmRBM11-like, AfRBP11, MrRBM11 and BmRBP7 are shown (DOC 44 kb)
Acknowledgments
This work was financially supported by the earmarked fund for the China Agriculture Research System (No. CARS-45), the National Natural Science Foundation of China (No. 31172275), and Shandong Province Fine Varieties Breeding Projects (2014-2016).
Abbreviations
- 5′ UTR
5′ Untranslated region
- A1
One-day postemergence worker bee
- A19
Nineteen-day postemergence worker bee
- A30
Thirty-day postemergence worker bee
- AccRBM11
Apis cerana cerana RNA-binding motif protein 11
- An
Antennae
- CK
Control check
- Dr
Drone
- dsRNA
Double-stranded RNA
- E. coli
Escherichia coli
- E
Egg
- Ep
Epidermis
- GFP
Green fluorescent protein gene
- H2O2
Hydrogen peroxide
- He
Hemolymph
- HSF
Heat shock factor
- L1
One-day larval instar
- Pr
Pre-pupal
- Mi
Midgut
- Mu
Muscle
- ORF
Open reading frame
- Pb
Brown-eyed pupae
- Pd
Dark-eyed pupae
- Pg
Poison gland
- Pp
Pink-eyed pupae
- Pw
White-eyed pupae
- Qe
The egg-producing queen bee
- qRT-PCR
Fluorescent real-time quantitative PCR
- Qw
The non-egg-laying queen bee
- RBM4
RNA-binding motif protein 4
- RBM7
RNA-binding motif protein 7
- RBMs
RNA-binding motif proteins
- RBPs
RNA-binding proteins
- Re
Rectum
- RRM-RBM7-like
RNA recognition motif in the RNA-binding protein 7 and similar proteins
- RRMs
RNA-recognizing domains
- RNAi
RNA interference
- TFBs
Transcription factor binding sites
- Wi
Wing
- Hs
Honey sac
Contributor Information
Qinghua Sun, Phone: +86-538-8242656, Email: qhsun@sdau.edu.cn.
Baohua Xu, Phone: +86-538-8242656, Email: bhxu@sdau.edu.cn.
References
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389(3):243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An MI, Choi CY. Activity of antioxidant enzymes and physiological responses in ark shell, Scapharca broughtonii, exposed to thermal and osmotic stress: effects on hemolymph and biochemical parameters. Comp Biochem Physiol B Biochem Mol Biol. 2010;155(1):34–42. doi: 10.1016/j.cbpb.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Aoki K, Matsumoto K, Tsujimoto M. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J Biol Chem. 2003;278(48):48491–48497. doi: 10.1074/jbc.M308328200. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Bae W, Jones PG, Inouye M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol. 1997;179(22):7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis RJ, Swanson MS, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Brégeon D, Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat Res. 2005;577(1):293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein J, et al. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398(6728):622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Burdon RH. Temperature and animal cell protein synthesis. Symp Soc Exp Biol. 1986;41:113–133. [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The miqe guidelines: minimum information for publication of quantitative real-time pcr experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen X, Yao P, Chu X, Hao L, Guo X, Xu B. Isolation of arginine kinase from Apis cerana cerana and its possible involvement in response to adverse stress. Cell Stress Chaperones. 2015;20(1):169–183. doi: 10.1007/s12192-014-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SL. The apicultural science in China. Beijing: China Agriculture Press; 2001. [Google Scholar]
- Delon I, Chanut-Delalande H, Payre F. The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech Dev. 2003;120(7):747–758. doi: 10.1016/S0925-4773(03)00081-9. [DOI] [PubMed] [Google Scholar]
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Elias-Neto M, Soares MPM, Simões ZLP, Hartfelder K, Bitondi MMG. Developmental characterization, function and regulation of a laccase2 encoding gene in the honey bee, Apis mellifera, (hymenoptera, apinae) Insect Biochem Mol Biol. 2010;40(3):241–251. doi: 10.1016/j.ibmb.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Enayati AA, Ranson H, Hemingway J (2005) Insect glutathione transferases and insecticide resistance. Insect Mol Biol 14(1):3–8 [DOI] [PubMed]
- Ericsson A, Kotarsky K, Svensson M, Sigvardsson M, Agace W. Functional characterization of the CCL25 promoter in small intestinal epithelial cells suggests a regulatory role for caudal-related homeobox (Cdx) transcription factors. J Immunol. 2006;176(6):3642–3651. doi: 10.4049/jimmunol.176.6.3642. [DOI] [PubMed] [Google Scholar]
- Fu YH, Marzluf GA. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol Cell Biol. 1987;7(5):1691–1696. doi: 10.1128/MCB.7.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gala A, Fang Y, Woltedji D, Zhang L, Han B, Feng M, et al. Changes of proteome and phosphoproteome trigger embryo-larva transition of honeybee worker (Apis mellifera ligustica) J Proteome. 2013;78:428–446. doi: 10.1016/j.jprot.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Gogos JA, Hsu T, Bolton J, Kafatos FC. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992;257(5078):1951–1955. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, et al. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model drosophila. Nat Immunol. 2013;14(4):396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- Gregory SL, Kortschak RD, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol Cell Biol. 1996;16(3):792–799. doi: 10.1128/MCB.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Jin X, Li S, Yu A, Wu M, Tan S, et al. A novel C-type lectin from Eriocheir sinensis functions as a pattern recognition receptor with antibacterial activity. Fish Shellfish Immunol. 2013;35(5):1554–1565. doi: 10.1016/j.fsi.2013.08.021. [DOI] [PubMed] [Google Scholar]
- Guo TB, Boros LG, Chan KC, Hikim APS, Hudson AP, Swerdloff RS, et al. Spermatogenetic expression of RNA-binding motif protein 7, a protein that interacts with splicing factors. J Androl. 2003;24(2):204–214. doi: 10.1002/j.1939-4640.2003.tb02664.x. [DOI] [PubMed] [Google Scholar]
- Huaxia Y, Fang W, Yan Y, Feng L, Wang H, Guo X, et al. A novel 1-Cys thioredoxin peroxidase gene in Apis cerana cerana: characterization of AccTpx4 and its role in oxidative stresses. Cell Stress Chaperones. 2015;20(4):1–10. doi: 10.1007/s12192-015-0594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of rnai for pest control: a review. J Insect Physiol. 2010;56(3):227–235. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Jia H, Sun R, Shi W, Yan Y, Han L, Guo X, et al. Characterization of a mitochondrial manganese superoxide dismutase gene from Apis cerana cerana, and its role in oxidative stress. J Insect Physiol. 2014;60(1):68–79. doi: 10.1016/j.jinsphys.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Kang WH, Park YD, Hwang JS, Park HM. RNA-binding protein Csx1 is phosphorylated by LAMMER kinase, Lkh1, in response to oxidative stress in Schizosaccharomyces pombe. FEBS Lett. 2007;581(18):3473–3478. doi: 10.1016/j.febslet.2007.06.053. [DOI] [PubMed] [Google Scholar]
- Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-D. [DOI] [PubMed] [Google Scholar]
- Lee SH, Oe T, Blair IA. Vitamin C-induced decomposition of lipid hydroperoxides to endogenous genotoxins. Science. 2001;292(5524):2083–2086. doi: 10.1126/science.1059501. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang L, Kang M, Guo X, Xu B. AccERK2, a map kinase gene from Apis cerana cerana, plays roles in stress responses, developmental processes, and the nervous system. Arch Insect Biochem Physiol. 2012;79(3):121–134. doi: 10.1002/arch.21011. [DOI] [PubMed] [Google Scholar]
- Li Z, Wu J, DeLeo CJ. RNA damage and surveillance under oxidative stress. IUBMB life. 2006;58(10):581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- Lin JC, Hsu M, Tarn WY. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci U S A. 2007;104(7):2235–2240. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhao Y, Dai Z, Chen H, Yang W. Formation of diapause cyst shell in brine shrimp, Artemia parthenogenetica, and its resistance role in environmental stresses. J Biol Chem. 2009;284(25):16931–16938. doi: 10.1074/jbc.M109.004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorković ZJ. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009;14(4):229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Lourenço AP, Mackert A, dos Santos Cristino A, Simões ZLP. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie. 2008;39(3):372–385. doi: 10.1051/apido:2008015. [DOI] [Google Scholar]
- Marionnet C, Bernerd F, Dumas A, Verrecchia F, Mollier K, Compan D, et al. Modulation of gene expression induced in human epidermis by environmental stress in vivo. J Invest Dermatol. 2003;121(6):1447–1458. doi: 10.1111/j.1523-1747.2003.12629.x. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galbán S, de Silanes IL, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100(14):8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza CA, Izaguirre MM, Zavala J, Scopel AL, Ballaré CL. Insect perception of ambient ultraviolet-B radiation. Ecol Lett. 2002;5(6):722–726. doi: 10.1046/j.1461-0248.2002.00379.x. [DOI] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Meng F, Lu W, Yu F, Kang M, Guo X, Xu B. Ribosomal protein l11 is related to brain maturation during the adult phase in Apis cerana cerana, (hymenoptera, apidae) Naturwissenschaften. 2012;99(5):343–352. doi: 10.1007/s00114-012-0905-5. [DOI] [PubMed] [Google Scholar]
- Meng F, Zhang L, Kang M, Guo X, Xu B. Molecular characterization, immunohistochemical localization and expression of a ribosomal protein L17 gene from Apis cerana cerana. Arch Insect Biochem Physiol. 2010;75(2):121–138. doi: 10.1002/arch.20386. [DOI] [PubMed] [Google Scholar]
- Meng F, Zhang Y, Liu F, Guo X, Xu B. Characterization and mutational analysis of omega-class GST (GSTO1) from Apis cerana cerana, a gene involved in response to oxidative stress. PLoS One. 2014;9(3):e93100. doi: 10.1371/journal.pone.0093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelette EDF, Soares AEE. Characterization of preimaginal developmental stages in Africanized honey bee workers (Apis mellifera L) Apidologie. 1993;24(4):431–440. doi: 10.1051/apido:19930410. [DOI] [Google Scholar]
- Mironova AA, Barykina NV, Zatsepina OV. Cytological analysis of the response of nucleolar RNA and RNA-binding proteins to oxidative stress in HeLa cells. Tsitologiia. 2014;8(6):461–472. [PubMed] [Google Scholar]
- Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H. Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 2009;9(1):110. doi: 10.1186/1471-2180-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra M, Bhatracharyulu NC, Padmavathi P, Varadacharyulu NC. Prallethrin induced biochemical changes in erythrocyte membrane and red cell osmotic haemolysis in human volunteers. Chemosphere. 2007;67(6):1065–1071. doi: 10.1016/j.chemosphere.2006.11.064. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137(4):899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol. 2013;14(10):643–653. doi: 10.1038/nrm3645. [DOI] [PubMed] [Google Scholar]
- Oldroyd B, Wongsiri S. Asian honey bees: biology, conservation, and human interactions. Cambridge: Harvard University Press; 2006. [Google Scholar]
- Park D, Jung JW, Choi BS, Jayakodi M, Lee J, Lim J, et al. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics. 2015;16(1):1. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti S, Busa R, Compagnucci C, Sette C. The RNA recognition motif protein RBM11 is a novel tissue-specific splicing regulator. Nucleic Acids Res. 2011;40:1021–1032. doi: 10.1093/nar/gkr819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YS, Fang Y, Xu S, Ge L. The resistance mechanism of the Asian honey bee, Apis cerana Fabr., to an ectoparasitic mite, Varroa jacobsoni Oudemans. J Invertebr Pathol. 1987;49(1):54–60. doi: 10.1016/0022-2011(87)90125-X. [DOI] [Google Scholar]
- Rao PN, Engelberg J. HeLa cells: effects of temperature on the life cycle. Science. 1965;148(3673):1092–1094. doi: 10.1126/science.148.3673.1092. [DOI] [PubMed] [Google Scholar]
- Rashed MN. Monitoring of environmental heavy metals in fish from Nasser Lake. Environ Int. 2001;27(1):27–33. doi: 10.1016/S0160-4120(01)00050-2. [DOI] [PubMed] [Google Scholar]
- Santoro N, Johansson N, Thiele DJ. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol. 1998;18(11):6340–6352. doi: 10.1128/MCB.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharlaken B, de Graaf DC, Goossens K, Peelman LJ, Jacobs FJ. Differential gene expression in the honeybee head after a bacterial challenge. Dev Comp Immunol. 2008;32(8):883–889. doi: 10.1016/j.dci.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Restifo LL. Anciently duplicated broad complex exons have distinct temporal functions during tissue morphogenesis. Dev Genes Evol. 2007;217(7):499–513. doi: 10.1007/s00427-007-0159-y. [DOI] [PubMed] [Google Scholar]
- Štanojević D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. 1989;341(6240):331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sasaki K, Kamata R, Hoshino Y, Yanagihara K, Sakai R. A novel RNA-binding protein, Ossa/C9orf10, regulates activity of Src kinases to protect cells from oxidative stress-induced apoptosis. Mol Cell Biol. 2009;29(2):402–413. doi: 10.1128/MCB.01035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wang L, Xu X, Zhou Y, Jin W, Wang H, et al. Autocrine/paracrine action of vitamin d on fgf23 expression in cultured rat osteoblasts. Calcif Tissue Int. 2010;86(5):404–410. doi: 10.1007/s00223-010-9355-2. [DOI] [PubMed] [Google Scholar]
- Tautz J, Maier S, Groh C, Rössler W, Brockmann A. Behavioral performance in adult honey bees is influenced by the temperature experienced during their pupal development. Proc Natl Acad Sci U S A. 2003;100(12):7343–7347. doi: 10.1073/pnas.1232346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443(7114):931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C, Lubas M, Tehrani M, Menon MB, Ronkina N, Rousseau S, et al. p38MAPK/MK2-mediated phosphorylation of RBM7 regulates the human nuclear exosome targeting complex. Rna (New York, NY) 2014;21(2):262–278. doi: 10.1261/rna.048090.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umasuthan N, Revathy KS, Lee Y, Whang I, Choi CY, Lee J. A novel molluscan sigma-like glutathione S-transferase from Manila clam, Ruditapes philippinarum: cloning, characterization and transcriptional profiling. Comp Biochem Physiol C Toxicol Pharmacol. 2012;155(4):539–550. doi: 10.1016/j.cbpc.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Kenny NJ, Dearden PK. Components of the dorsal-ventral pathway also contribute to anterior-posterior patterning in honeybee embryos ( Apis mellifera ) EvoDevo. 2014;5(1):1419–1419. doi: 10.1186/2041-9139-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Shi M, Chen XX. Antimicrobial peptide evolution in the Asiatic honey bee Apis cerana. PLoS One. 2009;4(1):4–4239. doi: 10.1371/journal.pone.0004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kemps-Mols B, Spruyt-Gerritse M, Anholts J, Claas F, Eikmans M. The source of SYBI green master mix determines outcome of nucleic acid amplification reactions. BMC Res Notes. 2016;9(1):292. doi: 10.1186/s13104-016-2093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J Biol Chem. 2001;276(50):47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- Yao P, Chen X, Yan Y, Feng L, Zhang Y, Guo X, et al. Glutaredoxin 1, glutaredoxin 2, thioredoxin 1, and thioredoxin peroxidase 3 play important roles in antioxidant defense in Apis cerana cerana. Free Radic Biol Med. 2014;68(3):335–346. doi: 10.1016/j.freeradbiomed.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Yin K, Hacia JG, Zhong Z, Paine ML. Genome-wide analysis of miRNA and mRNA transcriptomes during amelogenesis. BMC Genomics. 2014;15(1):998. doi: 10.1186/1471-2164-15-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Kang M, Meng F, Guo X, Xu B. Molecular cloning and characterization of a thioredoxin peroxidase gene from Apis cerana cerana. Insect Mol Biol. 2011;20(3):367–378. doi: 10.1111/j.1365-2583.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- Yu Y, Yu. Y, Huang H, Feng K, Pan M, Yuan S, et al. A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J Immunol. 2007;179(12):8425–8434. doi: 10.4049/jimmunol.179.12.8425. [DOI] [PubMed] [Google Scholar]
- Zargar, R., Urwat, U., Malik, F., Shah, R. A., Bhat, M. H., Naykoo, N. A. et al. (2015) Molecular characterization of RNA binding motif protein 3 (RBM3) gene from Pashmina goat. Res Vet Sci 98: 51–58. [DOI] [PubMed]
- Zhang Z, Wang Q, Li P, Zhou Y, Li S, Yi W, et al. Overexpression of the interferon regulatory factor 4-binding protein in human colorectal cancer and its clinical significance. Cancer Epidemiol. 2009;33(2):130–136. doi: 10.1016/j.canep.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Zhang L, Meng F, Li Y, Kang M, Guo X, Xu B. Molecular characterization and immunohistochemical localization of a mitogen-activated protein kinase, Acc38b, from Apis cerana cerana. BMB Rep. 2012;45(5):293–298. doi: 10.5483/BMBRep.2012.45.5.293. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Guo X, Li Y, Gao H, Guo X, et al. sHsp22.6, an intronless small heat shock protein gene, is involved in stress defence and development in Apis cerana cerana. Insect Biochem Mol Biol. 2014;53(2):160–165. doi: 10.1016/j.ibmb.2014.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The specific detection of anti-AccRBM11. Total protein of overexpression AccRBM11, induced overexpression of pET-30a(+)-AccRBM11 in Transetta cells were loaded in different lanes and the targeted protein was identified using an anti-AccRBM11 (GIF 3 kb)
Western blot analysis of AccRBM11. Western blot analysis of AccRBM11 changes after acaricide (a), paraquat (b), cyhalothrin (c), or UV (d) treatments. All of the lanes were loaded with an equivalent concentration of extracted protein from the same treatment at different times. Tubulin was selected as an internal control (GIF 31 kb)
Abiotic stress conditions for each experimental group (DOC 32 kb)
Primer sequences used in this study (DOC 82 kb)
Procedures used in this paper (DOC 28 kb)
Efficiency, coefficient, and number of melting curve peaks for each primer pair used for qRT-PCR (DOC 34 kb)
Characterization of the genes used in this paper (DOC 59 kb)
Size of the introns and exons and the A + T content in AccRBM11, AmRBM11-like, AfRBP11, MrRBM11 and BmRBP7 are shown (DOC 44 kb)