Abstract
Edible lamellibranch molluscs can be involved in foodborne disease and infections of varying severity. They are filter feeding animals able to retain and concentrate in their organism bacteria, parasites, viruses and biotoxins marine algae present in their external environment. Major shellfish harvesting and relaying areas from different areas in Sardinia region were defined and studied by analysing different physicochemical parameters in the water and the levels of Escherichia coli (E. coli), Norovirus (NoVs) genogroup I (NoVGI), NoVs genogroup II (NoVGII) and hepatitis A virus (HAV) in the shellfish harvested and farmed from 2009 to 2011. During that period the identification of the viral agents was carried out by one step real-time reverse transcriptase-polymerase chain reaction and Escherichia coli according to ISO TS 16649-3:2005 standard method. A total of 1266 shellfish samples were tested for NoVGI, NoVGII, HAV and faecal indicators. Norovirus contamination was found in 337 samples (26.6%); only one sample of mussels was positive for HAV (0.08%); while E. coli prevalence was 3.8% in shellfish. The probability of observing shellfish samples positive for NoVs, HAV and E. coli presence was associated with harvesting, growing and relaying areas, period of sampling, environmental parameters, animal species (P<0.05). Although the higher prevalence rate of human enteropathogenic viruses was found in the winter period, we did not observe a significant relationship between the effect of seawater temperature (seasonality) and NoVs presence all over the study period; in fact, according to statistical analysis, the presence of human enteric viruses does not appear to be related to water temperature.
Key words: Bivalve molluscs, Logistic regression, Food safety
Introduction
The Sardinian shellfish farming is a relevant and traditional economical aspect within the local economy. Most of production is mainly carried out in marine coastal areas with typical and geomorphic features, involving numerous human and manufacturing activities. Therefore, this marine environment becomes a potential source of seawater’s pollution and contamination for shellfish production. Bivalve molluscs may be contaminated by human sewage and they can constitute the main vector in the transmission of enteric diseases favouring the occurrence of situations of human health risk (Vilariño et al., 2009; Maalouf et al., 2010). The percentage of enteric viruses outbreak associated with shellfish is approximately 20% in countries such us United States, but this percentage increases up to 80% in countries in which seafood consumption is greater like Eastern areas or wherever seafood is eaten raw (Lees, 2000). Foodborne diseases linked to the consumption of bivalve molluscs are mainly caused by enteric viruses, especially from enteric human Calicivirus [Norovirus (NoVs)] and hepatitis A virus (HAV) (Davies et al., 2001; Corrain et al., 2007); indeed, epidemiological indications suggest that human enteric viruses are the most common pathogens transmitted by bivalve shellfish (Lees, 2000). These viruses have the capability to persist in shellfish for several days (as far as 130 days in sea water), even if when this animals are placed in clean seawater used for growing or farming or depuration cycles (Croci and Suffredini, 2003). Norovirus genogroup I (NoVGI) and II (NoVGII) have been detected in bivalve molluscan shellfish especially in oyster samples harvested and marketed from farm-pond and bays worldwide (Beuret et al., 2003; Cheng et al., 2005; Gallimore et al., 2005). In respect to shellfish market, the health controls for consumer’s protection is regulated by European Union (EU) legislations (EC Regulations No. 852/04, No. 853/04, No. 854/04, No. 2073/05, No. 2074/05, No. 1881/06 and No. 1441/07; European Commission, 2004a, 2004b, 2004c, 2005a, 2005b, 2006, 2007). Specifically, Regulation of the European Commission (EC) No. 2073/05 (2005a) lays down the microbiological criteria based only on the determination of some bacteriological parameters (Salmonella spp. and Escherichia coli). However, the same regulation makes it clear that determining of faecal indicators is unreliable for demonstrating the presence or absence of viral contamination.
Shellfish from category A classification sites can be taken for direct human consumption. Shellfish from these sites must contain <230 E. coli bacteria per 100 g of flesh. Shellfish from category B classification (<4600 E. coli bacteria per 100 g of flesh for 90% of samples and <46,000 E. coli bacteria per 100 g of flesh for all samples) must be purified by relaying or depuration prior to consumption. Shellfish from category C classification sites (all samples with 46,000 E. coli bacteria per 100 g of flesh) must be subjected to protracted relaying (2 months) or commercial heat treatment prior to consumption (European Commission, 2004c).
The present work was carried out in accordance with the Regional Plan of Surveillance, Vigilance and Sanitary Control of Production and Marketing of Molluscs, and of Periodic Monitoring of Shellfish Farming Areas, in force in Sardinia region, during the period 2008-2011 (Regione Sardegna, 2008). The aims of this work were i) to detect faecal indicators (E. coli) and NoVGI, NoVGII, HAV contamination in shellfish samples collected from harvesting, growing, relaying areas in five production locations in Sardinia region, and ii) to evaluate a correlation between the presence of these contaminants and seasonality. Physicochemical parameters of all shellfish harvesting and relaying areas were measured. The data analysis obtained in this study is highly valuable for improving microbiological and viral contamination control of shellfish, increasing the level of understanding and safety for consumers, and helping aquaculture farming.
Materials and Methods
Site selection and shellfish collection
For this study to be representative of the regional country, samples from 1266 bivalve molluscs consisting of mussels (Mytilus galloprovincialis), oysters (Crassostrea gigas, Ostrea edulis), clams (Tapes decussatus, Tapes philippinarum and Ruditapes philippinarum, autochthonous native species in north Sardinian aquaculture) and Cerastoderma sp. or Cardium spp. were collected and tested for human enteric viruses and E. coli. Samples were collected in Sardinia region from five (Site 1 to 5) geographically separate harvesting, growing and relaying areas, classified from A (product suitable for direct consumption) to C (product suitable for consumption after long depuration treatment) (European Commission, 2004c). Each area was in marine coast and pond locations with considerable urban populations (>50,000) (Figure 1). Shellfish was harvested by local authority sampling officers in accordance with the official monitoring and surveillance programme and in compliance with the Shellfish Regional Plan (Regione Sardegna, 2008). Sampling officers collected shellfish per week/month from each selected site between January 2009 and December 2011 included. Each sample consisted of a variable number of individuals (20-30), based on size and species. Samples were shipped directly to laboratory via cold storage in 12 h and immediately tested for E. coli, while tested on different days for viral contamination (HAV, NoVs).
Figure 1.
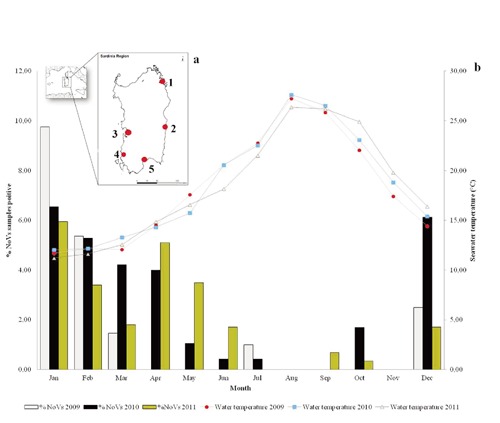
Collection sites (from 1 to 5) of shellfish tested for microbiological and environmental parameters in Sardinia region (a). Monthly distribution of Novovirus positive samples and average seawater temperature corresponding to each site and month from 2009 to 2011 (b).
Escherichia coli analysis
Escherichia coli was isolated using most probable number (MPN) standard methods as described in ISO TS 16649-3:2005 (ISO, 2005b). Calculation of the number of E. coli was performed using the ISO 7251 MPN table (ISO 7251:2005; ISO, 2005a). Processed shellfish was stored at -80±1.0°C and later used for enteric virus detection by real-time reverse transcriptase polymerase chain reaction (rRT-PCR) as described by Croci (2009) and at a later time by Lees and CEN WG6 TAG4 Working Grasp (2010). When workable, environmental parameters, in particular water temperatures, at the sampling site were determined by using a mobile equipment always approved by the competent authority at the time of sampling.
Shellfish processing for virus detection and viral RNA extraction
Shellfish samples were washed under clean running water and opened with sterile equipment. For each sample, the digestive glands and hepatopancreas of 10/20 shellfish were excised, pooled, and then blended by using T 25 basic Ultra-Turrax®. From the original homogenate 2.0±0.1 g of chopped glands were transferred into a clean tube, and inoculated with 1 106 50% tissue culture infective doses feline calicivirus (FCV) (10 µL). Feline calicivirus was chosen as the sample process control virus (Mattison et al., 2007; Di Pasquale et al., 2010). Homogenates were prepared by treating the glands with 100 g/mL proteinase K solution (30 U/mg; 2.0±0.1 mL for each sample) (Promega, Madison, WI, USA). The samples were then incubated at 37±1.0°C with shaking at 300 rpm for 1 h and then incubated at 60±1.0°C for 15±1 min in a water bath. Finally, the sample was centrifuged at 3000 g for 5 min, the soluble portion (homogenate) was retained for testing, and the pellet was discarded. Homogenates were stored at -80±1.0°C prior to testing. Total RNA was extracted from 140 µL of shellfish homogenate by using mini-kits QIAamp® Viral RNeasy (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions (RNA was eluted in 60 µL and was stored at -80±1.0°C until testing).
Detection of Norovirus genogroup I and II and hepatitis A virus by realtime TaqMan reverse transcriptase polymerase chain reaction
For NoVGI, primers QNIF4 (da Silva et al., 2007) and NV1LCR (Svraka et al., 2007) and probe NVGG1 (Svraka et al., 2007) were used. For NoVGII primers, QNIF2 (Loisy et al., 2005), COG2R (Kageyama et al., 2003) and probe QNIFS (Loisy et al., 2005) were used. The two probes for NoVGI and NoVGII were labeled 5’ 6-carboxyfluorescein and 3’ 6-carboxytetram-thylrhodamine. Hepatitis A virus assay was performed with primers HAV68, HAV240 and probes HAV150 (Costafreda et al., 2006). Probe labeled 5’ 6-carboxyfluorescein and 3’ MGB (minor groove binder). Feline calicivirus primers and probes were described by Mattison and colleagues (2007) and Ward and colleagues (2009). In the same rRT-PCR reaction viral nucleic acid of FCV was also amplified. Retrotranscription and one step PCR were performed on ABI Prism 7900 and 7700 sequence detector system (Applied Biosystems, Carlsbad, CA, USA). All amplification reactions were carried out using the Ultrasense® one-step qRT-PCR system (Invitrogen, Carlsbad, CA, USA), in a total volume of 25 µL in 96-wells plates (MicroAmp; Applied Biosystems). For all targets the concentrations of forward primer, reverse primer and TaqMan probe were 500, 900 and 250 nM, respectively, and 1×Ultrasense reaction mix, 1×ROX reference dye, 1.25 µL of Ultrasense enzyme mix. The cycling conditions were reverse transcription for 60 min at 55°C followed by 5 min at 95°C and 45 cycles of 15 s at 95°C, 1 min at 60°C and 1 min at 65°C. Each run included extracted RNA from NoVGI, NoVGII, HAV and FCV as positive RT-PCR controls and water as negative control. Fluorescence was measured at the end of each cycle. Each sample was amplified in duplicate in each run and considered positive when Ct was ≤44.0 in at least two replicas (Croci, 2009).
Physicochemical parameters
At the sampling times, when possible, the physicochemical parameters were monitored every month in each shellfish harvesting or growing site by public veterinary service or by the help of companies staff using portable weather instrument. The seawaters parameters considered were temperature (°C), salinity g/L (‰), dissolved oxygen (DO) content (% saturation) and pH. The temperature and the pH were measured at the depth at which shellfish were collected (HI 9024C pH meter kit; Hanna Instruments, Woonsocket, RI, USA). Salinity and DO were measured with a conductivity meter and a mobile potency meter (HI 9143 Microprocessor dissolved oxygen meter; Hanna Instruments), or, as an alternative, with multi-parameter probe (with pH, temperature, salinity and O2 sensors; Hydrolab, Solihull, UK).
Statistical analysis
In order to perform statistical analysis the data set was opportunely transformed in logit function. The link function F (F-1) is used for modeling the relationship between the probability of observing a 1 (0) with the covariates through P(Y=1|,X)=F(X). The hypothesis of interest was to see if the environmental parameter, in particular, seawater temperature, was the cause of the microorganisms presence in shellfish, and if this presence occurred regardless of the impact of anthropogenic and geomorphologic coastal waters used for molluscs production. To verify the existence of a risk factor associated with the presence of microorganisms (HAV, NoVGI, NoVGII and E. coli) a multivariate logistic regression model was developed in which the dependent variable result (presence/absence Y=1; Y=0) was placed in relation to independent variables classified as: year (2009-2011), month (January to December), production area (Site 1 to 5), classification areas, samples (clams, mussels, oysters), microbiological analysis (NoVs, HAV, E. coli determination) and environmental parameters (seawater temperature, pH, salinity, DO). All data analysis were carried out with Statgraphic Centurion (StatPoint Technologies, Warrenton, VA, USA) software, in a Intelcore i5 machine running MS Windows7 Professional for laboratory use only.
Results
The results of environmental monitoring showed that the Sardinian seawater had a stable salinity in all areas investigated. Salinity fluctuated between 27.0±1.0 and 36.0±1.0‰ (average=32.26‰). The seawater temperature during all monitoring varied between 11.0±1.0°C in January and 26.0±1.0°C in August. Dissolved oxygen and pH parameters were substantially equivalent in the five areas under observation during the same monitoring time, showing no significant variation during all three years of analysis.
In this study, 112 samples were taken from sites with a category A classification at the time of sampling, 1139 were taken from sites with a category B classification, and 15 were taken from sites with a category C classification. From a comparison of average E. coli and average NoVs levels in samples collected from the same site showed 30% of a correlation in B areas. The presence of E. coli during the study was 3.8% (49 samples positive/1266 samples analysed) and it most frequently occurred (3.7%; 22 positive samples/588 samples analysed) in the spring season of 2011 (Tables 1 and 2). The studied A class areas showed levels of E. coli higher than 230 MPN/100 g FIL in 6 clams samples. In the studied B class areas 40 shellfish samples showed all values well above of 4600 MPN/100 g FIL with a mussel sample with 30,000 MPN/100 g FIL. Interesting 11 (1.8%) mussels sampled in Site 5 showed E. coli levels far above the European legislation limit (4600 MPN/100 g FIL) (data not shown). With respect to the water temperature a correlation analysis has been performed. At temperatures between 10.0±1.0 and 20.0±1.0°C E. coli presence did not show significant correlation with seawater temperatures during the three years of the study. On the contrary, in 2011 E. coli at temperatures <19.0±1.0°C showed highest counts (Table 2). In respect to sampling shellfish monitoring plan 1266 samples were tested for human enteric viruses and all provided valid results (acceptable extraction and amplification efficiencies). Of these samples, 337 (26.6%) tested positive for NoVs, with 19 (1.5%) being positive for both genogroups, 29 (2.2%) being positive for NoVGI only, and 289 (22.8%) being positive for NoVGII only. The NoVs prevalence showed a high seasonal variability, with a maximum level of positive samples between January and March in 2009, 2010 and 2011 and a minimum of 0.15% positive samples (2/1266) in August 2009 (Table 2; Figure 1). Prevalence of NoVs levels varied markedly by season, with annual peaks occurring between January and March and the lowest levels being recorded between July and August (Table 2). Levels of NoVGII were on average higher than those of NoVGI. For each month of the study, the percentage of samples with total NoVs (GI plus GII summed) in different animal species is shown in Table 1 in which the impact of possible control limits on NoVs levels in production areas at different times of the year is remarkably shown. All five sites sampled during the research returned at least one positive result. Year-by-year NoVs prevalence varied between 20 and 27.9% positive samples in 2010. Site-by-site NoVs prevalence varied from a minimum of 1.2% (6/473 samples tested) up to 16.9% positive samples (80/473 samples tested) in 2010. We observed higher frequency of NoVs in B Class areas (Table 1). The results obtained in relation to a large number of analysis showed interesting analytical data, in fact, hepatitis A virus was found only in a mussel sample collected from the Site 5, during the winter season in 2009 (Tables 1 and 2). Among the NoVs positive samples 2 (0.9%) were collected from a relaying water production area in Site 1 (oysters) in 2009, 4 (0.8%) were collected from a class A water production areas in Site 1 (oysters) and 4 (0.8%) in Site 3 (mussels) in 2010. Only one clam sample was positive for NoVGII from a class C water production area in Site 3 in 2010. Logistic regression showed that there is a risk to find NoVs positive samples, in Site 1, 3.37 times higher than the Site 5 (Site 5 is the reference by algorithm calculation, OR=1) and other areas have not achieved significant results.
Table 1.
Positive shellfish samples tested for microbiological parameters.
| Period (total no. of samples tested) | Site | Samples (no. of positive targets) | Classification | No. of contaminated samples (%) | ||||
|---|---|---|---|---|---|---|---|---|
| E. coli | HAV | NoVGI | NoVGII | NoVGI+NoVGII | ||||
| 2009(205) | 1 | Musel (92) | B | 7(8) | 15(16) | 5(5) | ||
| Oyster (12) | A/relaying | 2(17) | 2(17) | 1(8) | ||||
| 3 | Mussel (35) | B | 1(3) | 4(11) | 1(3) | |||
| Clam (30) | A/B | 6(20) | 1(3) | 1(3) | 1(3) | |||
| 5 | Mussel (11) | B | 1(9) | |||||
| 2010(473) | 1 | Mussel (175) | B | 11(6) | 76(43) | 5(3) | ||
| Oyster (21) | A/B | 1(5) | 4(19) | 1(5) | ||||
| 3 | Mussel (50) | A/B | 4(8) | |||||
| Clam (20) | A/B/C | 2(10) | ||||||
| 5 | Mussel (181) | B | 6(3) | 17(9) | 5(3) | |||
| 2011(588) | 1 | Mussel (161) | B | 6(4) | 59(37) | |||
| Oyster (20) | A/B | 2(10) | 10(50) | |||||
| Clam (15) | B | 1(7) | 8(53) | |||||
| 2 | Mussel (40) | B | 6(15) | 8(20) | ||||
| 3 | Mussel (45) | A/B | 1(2) | 5(11) | ||||
| Clam (43) | A/B/C | 15(35) | 8(17) | |||||
| 5 | Mussel (224) | B | 11(5) | 62(28) | ||||
| Clam (40) | B | 1(3) | 4(10) | |||||
E. coli, Escherichia coli; HAV, hepatitis A virus; NoVGI, Norovirus genogroup I; NoVGII, Norovirus genogroup II; A, product suitable for direct consumption; B, product suitable for consumption after depuration treatment; C, product suitable for consumption after long depuration treatment.
Table 2.
Physicochemical parameters measured in site production areas and monthly frequency of positive Novovirus and Escherichia coli samples during the study.
| Year | Month | Temperature (°C) | pH | Salinity (‰) | DO (% saturation) | No. of contaminated samples (%) | ||
|---|---|---|---|---|---|---|---|---|
| E. coli | NoVs | HAV | ||||||
| 2009 | Jan-Mar | 11.18±0.50 | 7.39±0.11 | 31.11±2.73 | 96.08±9.39 | 1 (0.5) | 34 (16.5) | |
| Apr-Jun | 16.10±2.00 | 7.98±0.42 | 29.98±2.15 | 109.00±6.33 | 3 (1.5) | |||
| Jul-Sep | 26.11±0.77 | 8.13±0.31 | 32.04±0.67 | 104.43±0.70 | 1 (0.5) | 2(1) | ||
| Oct-Dec | 18.91±2.93 | 8.24±0.16 | 30.95±3.33 | nt | 1 (0.5) | 5 (2.5) | 1 (0.5) | |
| 2010 | Jan-Mar | 12.96±0.97 | nt | 30.91±2.92 | nt | 61(13) | ||
| Apr-Jun | 18.63±2.31 | 8.05±0.11 | 29.85±0.59 | 108.36±6.37 | 52(11) | |||
| Jul-Sep | 24.82±1.28 | 7.96±0.26 | nt | 105.53±1.18 | 10(2) | |||
| Oct-Dec | 17.31±2.40 | 7.55±0.04 | 33.17±1.66 | 99.00±14.03 | 29(6) | |||
| 2011 | Jan-Mar | 13.23±0.76 | 7.93±0.06 | 30.55±2.03 | 94.30±11.42 | 13 (2.2) | 66(11) | |
| Apr-Jun | 19.73±2.21 | 7.82±0.20 | 28.97±0.84 | 102.72±3.99 | 22 (3.7) | 62(10) | ||
| Jul-Sep | 24.99±0.60 | 8.14±0.16 | 31.80±2.75 | nt | 4 (0.7) | 4 (0.7) | ||
| Oct-Dec | 19.86±2.45 | 7.59±0.24 | 33.53±1.68 | 1005.05±9.23 | 4 (0.7) | 12(2) | ||
DO, dissolved oxygen; E coli, Escherichia coli; NoVs, Norovirus; HAV, hepatitis A virus; nt, not tested. Values of temperature, pH, salinity and dissolved oxygen are expressed as mean±standard deviation.
Discussion and Conclusions
In Sardinia region, one of the most important producer in Italy, shellfish production is a significant economic activity, with producing areas distributed along different coasts. Marine areas evaluated in this study support active commercial and recreational shellfish growing and harvesting activities. Most of the shellfish produced are consumed in local trade, especially in the western Sardinian coastal area, and in the North continental Italy (Sardegna Agricoltura/Laore, 2009). Shellfish farmers sell their products directly from the farms to wholesale, national and regional market and retail. During the research we did not observe, like in other countries, human contamination by NoVs (in Sardinia region, at present, clinical cases have not been recorded). Our study evaluated the contamination throughout the three years of the study and observed the probable presence of NoVs levels in shellfish during winter and summer period comparing this data to the environmental parameters (Figure 1; Table 2). The results of our research have shown no correlation between E. coli concentration and NoVs presence. The result clearly showed the absence of correlation between bacterial and viral faecal contaminants. In 2010 and 2011 the frequencies of NoVs positive samples were 26 and 23% in mussels respectively, in winter season, as the data confirm, even though we detected both NoVs genogroups in shellfish that were collected even in summer. The occurrence of frequencies NoVs positive samples in shellfish lead to think of the importance of regional or local conditions, particularly faecal contamination sources. Levels of NoVs contamination were observed at all sampling locations (except in the Site 2 and Site 4 in 2009 and in 2010) in wintertime, especially in Site 2, but this is not a real seasonality. The measures of the environmental parameters in the five harvesting, growing and relaying areas showed regular fluctuations related to seasonal variations, in particular the temperature (Figure 1). Most of NoVs contamination from Site 1 to 5 were detected during the winter season and no samples resulted positive during 2010 for E. coli contamination. We observed the high coexistence of two genogroups (GI, GII) in mussels (10%) in Site 1 showing that, along with all the other data, this area was especially NoVs-sensitive. The risk of finding mussels positive for NoVs in the year 2011 compared to the year 2010 was significantly higher (Table 3), while there was no statistically significant difference in the type of samples (clams, mussels, oysters). The analysis of the variable month confirms the above data with high risks, statistically significance, and a positive outcome between November and March, with a peak between January and February. Regarding the temperature, the greatest risk is evident in the range between 11.0±1.0 and 15.0±1.0°C but it was not statistically significant. The data analysis showed that the concentration of NoVs is greater in winter highlighting accordingly that the presence of this organism shows a certain seasonality (Tables 2 and 3), in agreement with a study conducted in 2002 in Northern Italy from human cases in children (Medici et al., 2006). This seasonality, according to our data, would not seem related to the temperature of seawater. Finally, this study showed comprehensible winter seasonality for NoVs contamination in all areas and in all shellfish species investigated, but specifically, this observation was not supported by statistical data analysis in relationship with seawater temperatures. Epidemiological data have reported that the major NoVs illness occur precisely in wintertime, often, after shellfish consumption in many countries (Boxman et al., 2006). Several reports in fact have been published describing outbreaks of acute gastroenteritis in humans related to the consumption of raw shellfish (oysters) (Svraka et al. 2007). One reason could be that the shellfish metabolism changes from winter to summer and in this season there is more animal faecal contamination in shellfish growing areas (Lowther et al., 2012).
Table 3.
Odds ratio values and their confidence intervals (95%) defined by logistic regression.
| Variable | SE | OR | 95% CI, lower limit | 95% CI, upper limit |
|---|---|---|---|---|
| Year 2009 | 0.29909 | 0.447854 | -0.13836 | 1.03407 |
| Year 2010 | 0.171356 | 0.396681 | 0.060823 | 0.732539 |
| Site 1 | 0.196746 | 3.37382 | 2.988198 | 3.759442 |
| Site 2 | 0.356512 | 0.963479 | 0.264715 | 1.662243 |
| Site 3 | 0.270447 | 0.939788 | 0.409712 | 1.469864 |
| Site 4 | 123.389 | 1.1877E-05 | -241.842 | 241.8425 |
| E. coli | 0.193196 | 0.322087 | -0.05658 | 0.700751 |
| January | 1.1615 | 294.58 | 292.3035 | 296.8565 |
| February | 1.16329 | 864.347 | 862.067 | 866.627 |
| March | 1.14057 | 324.475 | 322.2395 | 326.7105 |
| April | 1.08608 | 113.121 | 110.9923 | 115.2497 |
| May | 1.03065 | 60.0302 | 58.01013 | 62.05027 |
| June | 0.987417 | 52.7993 | 50.86396 | 54.73464 |
| July | 0.61474 | 1.11993 | -0.08496 | 2.32482 |
| August | 0.728953 | 0.554168 | -0.87458 | 1.982916 |
| October | 0.952403 | 10.9379 | 9.07119 | 12.80461 |
| November | 1.09492 | 53.13441 | 50.98806 | 55.28014 |
| December | 1.11818 | 103.255 | 101.0634 | 105.4466 |
| 11.00-14.90°C | 1.3498 | 0.0097962 | -2.63581 | 2.655404 |
| 15.00-18.90°C | 1.32932 | 0.0342304 | -2.57124 | 2.639698 |
| 19.00-22.90°C | 1.26068 | 0.0224851 | -2.44845 | 2.493418 |
| 23.00-26.90°C | 0.857982 | 0.154801 | -1.52684 | 1.836446 |
| Clam | 197.9 | 0.000107933 | -387.884 | 387.8841 |
| Mussel | 0.267597 | 0.622397 | 0.097907 | 1.146887 |
| Oyster | 0.437898 | 0.313003 | -0.54528 | 1.171283 |
SE, standard error; OR, odds ratio; CI, confidence interval.
The data obtained in this study could contribute to the introduction of explicit shellfish monitoring programmes, which could be improved with acquisition of all environmental parameters data and real-time qRT-PCR tests on cultured areas. Optimisation of monitoring plans could lead to better management of the shellfish harvest and market.
Acknowledgments
The authors would like to thank Dr. Stefano Congiu (Health, Hygiene and Social Welfare Local Government, Sardinia Region) for his sharing the results and the development of the regional plans, and Dr. Elisabetta Suffredini (Italian National Health Institute) for her contribution in technical protocol specification.
Funding Statement
Funding: the work was supported by a local public grant for the Regional Plan of surveillance, vigilance and sanitary control of production and marketing of molluscs, and of periodic monitoring of shellfish farming areas (Sardinia region, 2009/2011 and 2011/2014).
References
- Beuret C, Baumgartner A, Schluep J, 2003. Virus-contaminated oysters: a 3-month monitoring of oysters imported to Switzerland. Appl Environ Microb 69:2292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxman ILA, Tilburg JJHC, Te Loeke NAJM, Vennema H, Jonker H, de Boer E, Koopmans M, 2006. Detection of noroviruses in shellfish in the Netherlands. Int J Food Microbiol 108:391-6. [DOI] [PubMed] [Google Scholar]
- Cheng PKC, Wong DKK, Chung TWH, Lim WWL, 2005. Norovirus contamination found in oysters worldwide. J Med Virol 76:593-7. [DOI] [PubMed] [Google Scholar]
- Corrain C, Arcangeli G, Fasolato L, Manfrin A, Rossetti E, Piazzi E, Mioni R, Pavoni E, Losio N, Sanavio G, Suffredini E, Croci L, 2007. [Influenze climatico ambientali sulla presenza di virus enterici in molluschi bivalvi]. [Article in Italian]. Ind Aliment 46:277-83. [Google Scholar]
- Costafreda MI, Bosch A, Pintó RM, 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microb 72:3846-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci L, 2009. Determinazione di Norovirus ed HAV in molluschi bivalvi mediante Real Time PCR. Available from: http://www.iss.it/binary/spva/cont/Determinazione_NoV_e_HAV_Integrazioni_Correzioni_Dichiarazione_validazione_Tabella_comparativa_POMIAC01.002.pdf [Google Scholar]
- Croci L, Suffredini E, 2003. [Rischio microbiologico associato al consumo di prodotti ittici]. Available from: http://www.iss.it/publ/anna/2003/1/39135.pdf [PubMed] [Google Scholar]
- da Silva AK, Le Saux JC, Parnaudeau S, Pommepuy M, Elimelech M, Le Guyader FS, 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl Environ Microb 73:7891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AR, Capell C, Jehanno D, Nychas GJE, 2001. Incidence of foodborne pathogens on European fish. Food Control 12:67-71. [Google Scholar]
- Di Pasquale S, Paniconi M, De Medici D, Suffredini E, Croci L, 2010. Duplex Real Time PCR for the detection of hepatitis A virus in shellfish using Feline Calicivirus as a process control. J Virol Methods 163:96-100. [DOI] [PubMed] [Google Scholar]
- European Commission, 2004a. [Regolamento del Parlamento Europeo e del Consiglio del 29 aprile 2004 sull’igiene dei prodotti alimentari, 852/2004/CE]. [Regulation in Italian]. In: Official Journal, L 139/1, 30/04/2004. [Google Scholar]
- European Commission, 2004b. [Regolamento del Parlamento Europeo e del Consiglio del 29 aprile 2004 che stabilisce norme specifiche in materia di igiene per gli alimenti di origine animale, 853/2004/CE]. [Regulation in Italian]. In: Official Journal, L 139/55, 30/04/2004. [Google Scholar]
- European Commission, 2004c. Commission regulation of 29 April 2004 laying down specific rules for the organization of official controls on products of animal origin intended for human consumption, 854/2004/EC. In: Official Journal, L 139, 25/06/2004. [Google Scholar]
- European Commission, 2005a. [Regolamento della Commissione del 15 novembre 2005 sui criteri microbiologici applicabili ai prodotti alimentari, 2073/2005/CE]. [Regulation in Italian]. In: Official Journal, L 338/1, 22/12/2005. [Google Scholar]
- European Commission, 2005b. [Regolamento della Commissione del 5 dicembre 2005 recante modalità di attuazione relative a taluni prodotti di cui al regolamento (CE) n. 853/2004 del Parlamento europeo e del Consiglio e all’organizzazione di controlli ufficiali a norma dei regolamenti del Parlamento europeo e del Consiglio (CE) n. 854/2004 e (CE) n. 882/2004, deroga al regolamento (CE) n. 852/2004 del Parlamento europeo e del Consiglio e modifica dei regolamenti (CE) n. 853/2004 e (CE) n. 854/2004, 2074/2005/CE]. [Regulation in Italian]. In: Official Journal, L 338/27, 22/12/2005. [Google Scholar]
- European Commission, 2006. [Regolamento della Commissione del 19 dicembre 2006 che definisce i tenori massimi di alcuni contaminanti nei prodotti alimentari, 1881/2006/CE]. [Regulation in Italian]. In: Official Journal, L 364/5, 20/12/2006. [Google Scholar]
- European Commission, 2007. [Regolamento della Commissione del 5 dicembre 2007 che modifica il regolamento (CE) n. 2073/2005 sui criteri microbiologici applicabili ai prodotti alimentari, 1441/2007/CE]. [Regulation in Italian]. In: Official Journal, L 322/12, 07/12/2007. [Google Scholar]
- Gallimore CI, Cheesbrough JS, Lamden K, Bingham C, Gray JJ, 2005. Multiple norovirus genotypes characterised from an oyster-associated outbreak of gastroenteritis. Int J Food Microbiol 103:323-30. [DOI] [PubMed] [Google Scholar]
- ISO, 2005a. Microbiology of food and animal feeding stuffs. Horizontal method for the detection and enumeration of presumptive Escherichia coli. Most probable number technique. ISO Norm 7251:2005. International Standardization Organization ed., Geneva, Switzerland. [Google Scholar]
- ISO, 2005b. Microbiology of food and animal feeding stuffs. Horizontal method for the enumeration of beta-glucuronidase-positive Escherichia coli - Part 3: most probable number technique using 5-bromo-4-chloro-3-indolyl-beta-D-glucuronide. ISO Norm 16649:2005. International Standardization Organization ed., Geneva, Switzerland. [Google Scholar]
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K, 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol 41:1548-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees DN, 2000. Viruses and bivalve shellfish. Int J Food Microbiol 59:117-26. [DOI] [PubMed] [Google Scholar]
- Lees DN, CEN WG6 TAG4 Working Grasp , 2010. International standardization of a method for detection of human pathogenic viruses in molluscan shellfish. Food Environ Virol 2:146-55. [Google Scholar]
- Loisy F, Atmar RL, Guillon P, Le Cann P, Pommepuy M, Le Guyader FS, 2005. Real time RT-PCR for norovirus screening in shellfish. J Virol Methods 123:1-7. [DOI] [PubMed] [Google Scholar]
- Lowther JA, Gustar NE, Powell AL, Hartnell RE, Lees DN, 2012. Two-year systematic study to assess Norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Appl Environ Microb 78:5812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maalouf H, Zakhour M, Le Pendu J, Le Saux JC, Atmar RL, Le Guyader FS, 2010. Distribution in tissue and seasonal variation of Norovirus genogroup I and II ligands in oysters. Appl Environ Microb 76:5621-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Brassard J, Houde A, Simard C, Pagotto F, Jones T, Trottier Y, Octobre L, 2007. The feline calicivirus (FCV) as an internal control for the detection of RNA viruses from foods. Federal Food Safety and Nutrition Research Meeting Publ., Winnipeg, Manitoba, Canada. [Google Scholar]
- Medici MC, Martinelli M, Abelli LA, Ruggeri FM, Di Bartolo I, Arcangeli MC, 2006. Molecular epidemiology of Norovirus infections in sporadic cases of viral gastroenteritis among children in northern Italy. J Med Virol 78:1486-92. [DOI] [PubMed] [Google Scholar]
- Regione Sardegna, 2008. [Piano regionale per la vigilanza ed il controllo sanitario della produzione e commercializzazione dei molluschi bivalvi vivi e per il monitoraggio periodico delle zone di produzione e di stabulazione di molluschi bivalvi vivi, anno 2008-2011]. [Regulation in Italian]. Regione Sardegna Publ., Cagliari, Italy. [Google Scholar]
- Sardegna Agricoltura/Laore, 2009. [Il comparto dell’acquacoltura in Sardegna alla luce dei risultati dell’indagine conoscitiva]. [Article in Italian]. Available from: http://www.sardegnaagricoltura.it/documenti/14_43_20091201175044.pdf [Google Scholar]
- Svraka S, Duizer E, Vennema H, de Bruni E, van der Veer B, Dorresteijn B, Koopmans M, 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J Clin Microbiol 45:1389-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño ML, Le Guyader FS, Polo D, Schaeffer J, Kröl J, Romaldel JL, 2009. Assessment of human enteric viruses in cultured and wild bivalve molluscs. Int Microbiol 12:145-51. [PubMed] [Google Scholar]
- Ward P, Poitras E, Leblanc D, Letellier A, Brassard J, Plante D, Houde A, 2009. Comparative analysis of different TaqMan real-time RT-PCR assays for the detection of swine Hepatitis E virus and integration of Feline calicivirus as internal control. J Appl Microbiol 106:1360-9. [DOI] [PubMed] [Google Scholar]


