Abstract
Reaction prediction remains one of the major challenges for organic chemistry and is a prerequisite for efficient synthetic planning. It is desirable to develop algorithms that, like humans, “learn” from being exposed to examples of the application of the rules of organic chemistry. We explore the use of neural networks for predicting reaction types, using a new reaction fingerprinting method. We combine this predictor with SMARTS transformations to build a system which, given a set of reagents and reactants, predicts the likely products. We test this method on problems from a popular organic chemistry textbook.
Short abstract
We introduce a neural network machine learning algorithm for the prediction of basic organic chemistry reactions. We explore the performance for two compound types using a novel fingerprint.
Introduction
To develop the intuition and understanding for predicting reactions, a human must take many semesters of organic chemistry and gather insight over several years of lab experience. Over the past 40 years, various algorithms have been developed to assist with synthetic design, reaction prediction, and starting material selection.1,2 LHASA was the first of these algorithms to aid in developing retrosynthetic pathways.3 This algorithm required over a decade of effort to encode the necessary subroutines to account for the various subtleties of retrosynthesis such as functional group identification, polycyclic group handling, relative protecting group reactivity, and functional group based transforms.4−7
In the late 1980s to the early 1990s, new algorithms for synthetic design and reaction prediction were developed. CAMEO,8 a reaction predicting code, used subroutines specialized for each reaction type, expanding to include reaction conditions in its analysis. EROS9 identified leading structures for retrosynthesis by using bond polarity, electronegativity across the molecule, and the resonance effect to identify the most reactive bond. SOPHIA10 was developed to predict reaction outcomes with minimal user input; this algorithm would guess the correct reaction type subroutine to use by identifying important groups in the reactants; once the reactant type was identified, product ratios would be estimated for the resulting products. SOPHIA was followed by the KOSP algorithm and uses the same database to predict retrosynthetic targets.11 Other methods generated rules based on published reactions and use these transformations when designing a retrosynthetic pathway.12,13 Some methods encoded expert rules in the form of electron flow diagrams.14,15 Another group attempted to grasp the diversity of reactions by creating an algorithm that automatically searches for reaction mechanisms using atom mapping and substructure matching.16
While these algorithms have their subtle differences, all require a set of expert rules to predict reaction outcomes. Taking a more general approach, one group has encoded all of the reactions of the Beilstein database, creating a “Network of Organic Chemistry”.2,17 By searching this network, synthetic pathways can be developed for any molecule similar enough to a molecule already in its database of 7 million reactions, identifying both one-pot reactions that do not require time-consuming purification of intermediate products18 and full multistep reactions that account for the cost of the materials, labor, and safety of the reaction.2 Algorithms that use encoded expert rules or databases of published reactions are able to accurately predict chemistry for queries that match reactions in its knowledge base. However, such algorithms do not have the ability of a human organic chemist to predict the outcomes of previously unseen reactions. In order to predict the results of new reactions, the algorithm must have a way of connecting information from reactions that it has been trained upon to reactions that it has yet to encounter.
Another strategy of reaction prediction algorithm draws from principles of physical chemistry and first predicts the energy barrier of a reaction in order to predict its likelihood.19−24 Specific examples of reactions include the development of a nanoreactor for early Earth reactions,20,21 heuristic aided quantum chemistry,23 and ROBIA,25 an algorithm for reaction prediction. While methods that are guided by quantum calculations have the potential to explore a wider range of reactions than the heuristic-based methods, these algorithms would require new calculations for each additional reaction family and will be prohibitively costly over a large set of new reactions.
A third strategy for reaction prediction algorithms uses statistical machine learning. These methods can sometimes generalize or extrapolate to new examples, as in the recent examples of picture and handwriting identification,26,27 playing video games,28 and most recently, playing Go.29 This last example is particularly interesting as Go is a complex board game with a search space of 10170, which is on the order of chemical space for medium sized molecules.30 SYNCHEM was one early effort in the application of machine learning methods to chemical predictions, which relied mostly on clustering similar reactions, and learning when reactions could be applied based on the presence of key functional groups.13
Today, most machine learning approaches in reaction prediction use molecular descriptors to characterize the reactants in order to guess the outcome of the reaction. Such descriptors range from physical descriptors such as molecular weight, number of rings, or partial charge calculations to molecular fingerprints, a vector of bits or floats that represent the properties of the molecule. ReactionPredictor31,32 is an algorithm that first identifies potential electron sources and electron sinks in the reactant molecules based on atom and bond descriptors. Once identified, these sources and sinks are paired to generate possible reaction mechanisms. Finally, neural networks are used to determine the most likely combinations in order to predict the true mechanism. While this approach allows for the prediction of many reactions at the mechanistic level, many of the elementary organic chemistry reactions that are the building blocks of organic synthesis have complicated mechanisms, requiring several steps that would be costly for this algorithm to predict.
Many algorithms that predict properties of organic molecules use various types of fingerprints as the descriptor. Morgan fingerprints and extended circular fingerprints33,34 have been used to predict molecular properties such as HOMO–LUMO gaps,35 protein–ligand binding affinity,36 and drug toxicity levels37 and even to predict synthetic accessibility.38 Recently Duvenavud et al. applied graph neural networks39 to generate continuous molecular fingerprints directly from molecular graphs. This approach generalizes fingerprinting methods such as the ECFP by parametrizing the fingerprint generation method. These parameters can then be optimized for each prediction task, producing fingerprint features that are relevant for the task. Other fingerprinting methods that have been developed use the Coulomb matrix,40 radial distribution functions,41 and atom pair descriptors.42 For classifying reactions, one group developed a fingerprint to represent a reaction by taking the difference between the sum of the fingerprints of the products and the sum of the fingerprints of the reactants.43 A variety of fingerprinting methods were tested for the constituent fingerprints of the molecules.
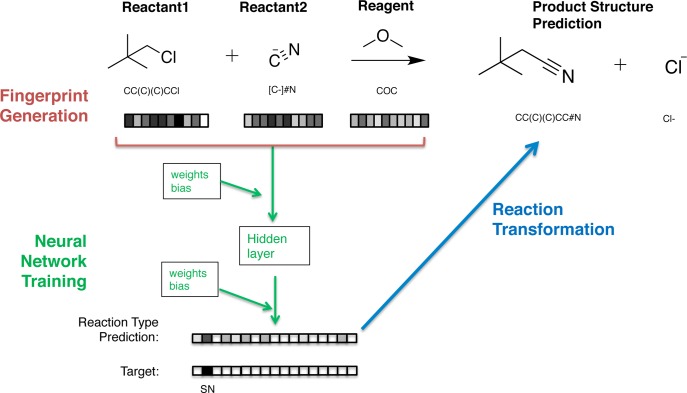
In this work, we apply fingerprinting methods, including neural molecular fingerprints, to predict organic chemistry reactions. Our algorithm predicts the most likely reaction type for a given set of reactants and reagents, using what it has learned from training examples. These input molecules are described by concatenating the fingerprints of the reactants and the reagents; this concatenated fingerprint is then used as the input for a neural network to classify the reaction type. With information about the reaction type, we can make predictions about the product molecules. One simple approach for predicting product molecules from the reactant molecules, which we use in this work, is to apply a SMARTS transformation that describes the predicted reaction. Previously, sets of SMARTS transformations have been applied to produce large libraries of synthetically accessible compounds in the areas of molecular discovery,44 metabolic networks,45 drug discovery,46 and discovery of one-pot reactions.47 In our algorithm, we use SMARTS transformation for targeted prediction of product molecules from reactants. However, this method can be replaced by any method that generates product molecule graphs from reactant molecule graphs. An overview of our method can be found in Figure 1 and is explained in further detail in Prediction Methods.
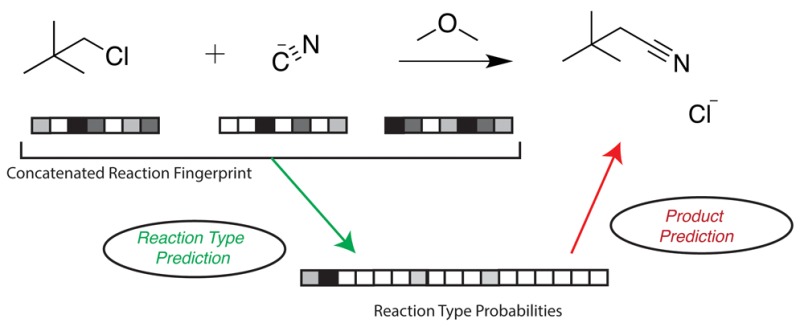
Figure 1.
An overview of our method for predicting reaction type and products. A reaction fingerprint, made from concatenating the fingerprints of reactant and reagent molecules, is the input for a neural network that predicts the probability of 17 different reaction types, represented as a reaction type probability vector. The algorithm then predicts a product by applying to the reactants a transformation that corresponds to the most probable reaction type. In this work, we use a SMARTS transformation for the final step.
We show the results of our prediction method on 16 basic reactions of alkyl halides and alkenes, some of the first reactions taught to organic chemistry students in many textbooks.48 The training and validation reactions were generated by applying simple SMARTS transformations to alkenes and alkyl halides. While we limit our initial exploration to aliphatic, nonstereospecific molecules, our method can easily be applied a wider span of organic chemical space with enough example reactions. The algorithm can also be expanded to include experimental conditions such as reaction temperature and time. With additional adjustments and a larger library of training data, our algorithm will be able to predict multistep reactions and, eventually, become a module in a larger machine-learning system for suggesting retrosynthetic pathways for complex molecules.
Results and Discussion
Performance on Cross-Validation Set
We created a data set of reactions of four alkyl halide reactions and 12 alkene reactions; further details on the construction of the data set can be found in Methods. Our training set consisted of 3400 reactions from this data set, and the test set consisted of 17,000 reactions; both the training set and the test set were balanced across reaction types. During optimization on the training set, k-fold cross-validation was used to help tune the parameters of the neural net. Table 1 reports the cross-entropy score and the accuracy of the baseline and fingerprinting methods on this test set. Here the accuracy is defined by the percentage of matching indices of maximum values in the predicted probability vector and the target probability vector for each reaction.
Table 1. Accuracy and Negative Log Likelihood (NLL) Error of Fingerprint and Baseline Methods.
| fingerprint method | fingerprint length | train NLL | train accuracy (%) | test NLL | test accuracy (%) |
|---|---|---|---|---|---|
| baseline | 51 | 0.2727 | 78.8 | 2.5573 | 24.7 |
| Morgan | 891 | 0.0971 | 86.0 | 0.1792 | 84.5 |
| neural | 181 | 0.0976 | 86.0 | 0.1340 | 85.7 |
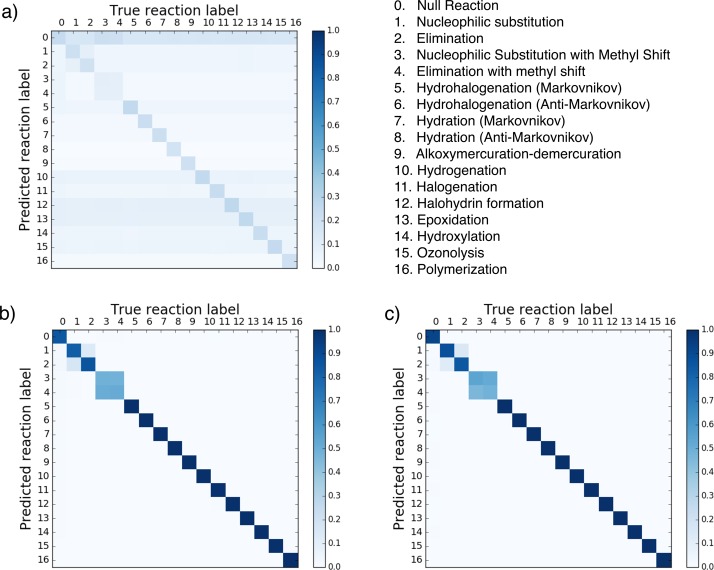
Figure 2 shows the confusion matrices for the baseline, neural, and Morgan fingerprinting methods, respectively. The confusion matrices for the Morgan and neural fingerprints show that the predicted reaction type and the true reaction type correspond almost perfectly, with few mismatches. The only exceptions are in the predictions for reaction types 3 and 4, corresponding to nucleophilic substitution reaction with a methyl shift and the elimination reaction with a methyl shift. As described in Methods, these reactions are assumed to occur together, so they are each assigned probabilities of 50% in the training set. As a result, the algorithm cannot distinguish these reaction types and the result on the confusion matrix is a 2 × 2 square. For the baseline method, the first reaction type, the “NR” classification, is often overpredicted, with some additional overgeneralization of some other reaction type as shown by the horizontal bands.
Figure 2.
Cross validation results for (a) baseline fingerprint, (b) Morgan reaction fingerprint, and (c) neural reaction fingerprint. A confusion matrix shows the average predicted probability for each reaction type. In these confusion matrices, the predicted reaction type is represented on the vertical axis, and the correct reaction type is represented on the horizontal axis. These figures were generated on the basis of code from Schneider et al.43
Performance on Predicting Reaction Type of Exam Questions
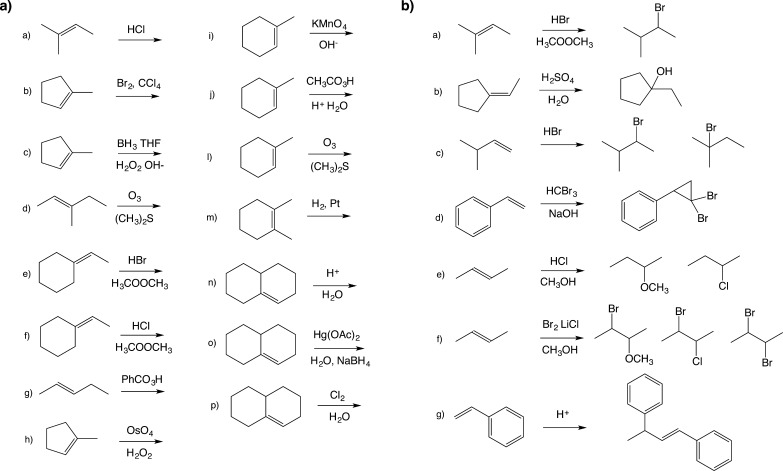
Kayala et al.31 had previously employed organic textbook questions both as the training set and as the validation set for their algorithm, reporting 95.7% accuracy on their training set. We similarly decided to test our algorithm on a set of textbook questions. We selected problems 8-47 and 8-48 from the Wade sixth edition organic chemistry textbook shown in Figure 3.48 The reagents listed in each problem were assigned as secondary reactants or reagents so that they matched the training set. For all prediction methods, our networks were first trained on the training set of generated reactions, using the same hyperparameters found by the cross-validation search. The similarity of the exam questions to the training set was determined by measuring the Tanimoto49 distance of the fingerprints of the reactant and reagent molecules in each reactant set. The average Tanimoto score between the training set reactants and reagents and the exam set reactants and reagents is 0.433, and the highest Tanimoto score oberved between exam questions and training questions was 1.00 on 8-48c and 0.941 on 8-47a. This indicates that 8-48c was one of the training set examples. Table SI.1 show more detailed results for this Tanimoto analysis.
Figure 3.
Wade problems (a) 8-47 and (b) 8-48.48
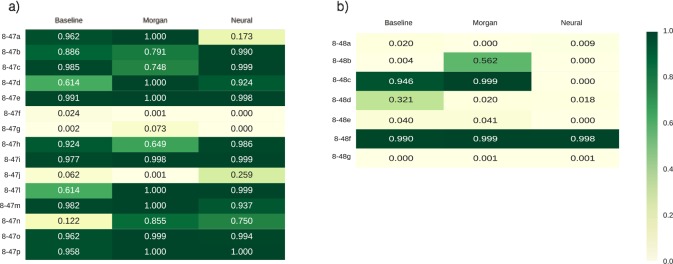
For each problem, the algorithm determined the reaction type in our set that best matched the answer. If the reaction in the answer key did not match any of our reaction types, the algorithm designated the reaction as a null reaction. The higher the probability the algorithm assigned for each reaction type, the more certainty the algorithm has in its prediction. These probabilities are reported in Figure 4, color-coded with green for higher probability and yellow/white for low probability.
Figure 4.
Prediction results for (a) Wade problem 8-47 and (b) Wade problem 8-48, as displayed by estimated probability of correct reaction type. Darker (greener) colors represent a higher predicted probability. Note the large amount of correct predictions in 8-47.
In problem 8-47, the Morgan fingerprint algorithm had the best performance with 12 of the 15 correct answers, followed by the neural fingerprint algorithm and the baseline method, both of which had 11 out of 15 correct answers. Both the Morgan fingerprint algorithm and the neural fingerprint algorithm predicted the correct answers with higher probability than the baseline method. Several of the problems contained rings, which were not included in the original training set. Many of these reactions were predicted correctly by the Morgan and neural fingerprint algorithms, but not by the baseline algorithm. This suggests that both Morgan and neural fingerprint algorithms were able to extrapolate the correct reactivity patterns to reactants with rings.
In problem 8-48, students are asked to suggest mechanisms for reactions given both the reactants and the products. To match the input format of our algorithm, we did not provide the algorithm any information about the products even though it disadvantaged our algorithm. All methods had much greater difficulty with this set of problems possibly because these problems introduced aromatic rings, which the algorithm may have had difficulty distinguishing from double bonds.
Performance on Product Prediction
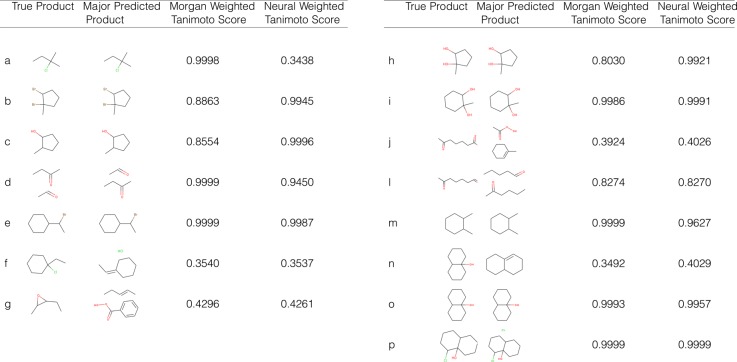
Once a reaction type has been assigned for a given problem by our algorithm, we can use the information to help us predict our products. In this study, we chose to naively use this information by applying a SMARTS transformation that matched the predicted reaction type to generate products from reactants. Figure 5 shows the results of this product prediction method using Morgan reaction fingerprints and neural reaction fingerprints on problem 8-47 of the Wade textbook, analyzed in the previous section. For all suggested reaction types, the SMARTS transformation was applied to the reactants given by the problem. If the SMARTS transformation for that reaction type was unable to proceed due to a mismatch between the given reactants and the template of the SMARTS transformation, then the reactants were returned as the predicted product instead.
Figure 5.
Product predictions for Wade 8-47 questions, with Tanimoto score. The true product is the product as defined by the answer key. The major predicted product shows the product of the reaction type with the highest probability according to the Morgan fingerprint algorithm’s result. The Morgan weighted score and the neural weighted score are calculated by taking an average of the Tanimoto scores over all the predicted products weighted by the probability of that reaction type which generated that product.
A product prediction score was also assigned for each prediction method. For each reaction, the Tanimoto score49 was calculated between the Morgan fingerprint of the true product and the Morgan fingerprint of the predicted product for each reaction type, following the same applicability rules described above. The overall product prediction score is defined as the average of these Tanimoto scores for each reaction type, weighted by the probability of each reaction type as given by the probability vector. The scores for each question are given in Figure 5.
The Morgan fingerprint algorithm is able to predict 8 of the 15 products correctly, and the neural fingerprint algorithm is able to predict 7 of the 15 products correctly. The average Tanimoto score for the products predicted by the Morgan fingerprint algorithm compared to the true products was 0.793, and the average Tanimoto score between the true products and the neural fingerprint algorithm products was 0.776. In general, if the algorithm predicted the reaction type correctly with high certainty, the product was also predicted correctly and the weighted Tanimoto score was high, however, this was not the case for all problems correctly predicted by the algorithm.
The main limitation in the algorithm’s ability to predict products despite predicting the reaction type correctly is the capability of the SMARTS transformation to accurately describe the transformation of the reaction type for all input reactants. While some special measures were taken in the code of these reactions to handle some common regiochemistry considerations, such as Markovnikov orientation, it was not enough to account for all of the variations of transformations seen in the sampled textbook questions. Future versions of this algorithm will require an algorithm better than encoded SMARTS transformations to generate the products from the reactant molecules.
Conclusion
Using our fingerprint-based neural network algorithm, we were able to identify the correct reaction type for most reactions in our scope of alkene and alkyl halide reactions, given only the reactants and reagents as inputs. We achieved an accuracy of 85% of our test reactions and 80% of selected textbook questions. With this prediction of the reaction type, the algorithm was further able to guess the structure of the product for a little more than half of the problems. The main limitation in the prediction of the product structure was due to the limitations of the SMARTS transformation to describe the mechanism of the reaction type completely.
While previously developed machine learning algorithms are also able to predict the products of these reactions with similar or better accuracy,31 the structure of our algorithm allows for greater flexibillity. Our algorithm is able to learn the probabilities of a range of reaction types. To expand the scope of our algorithm to new reaction types, we would not need to encode new rules, nor would we need to account for the varying number of steps in the mechanism of the reaction; we would just need to add the additional reactions to the training set. The simplicity of our reaction fingerprinting algorithm allows for rapid expansion of our predictive capabilities given a larger data set of well-curated reactions.2,12 Using data sets of experimentally published reactions, we can also expand our algorithm to account for the reaction conditions in its predictions and, later, predict the correct reaction conditions.
This paper represents a step toward the goal of developing a machine learning algorithm for automatic synthesis planning of organic molecules. Once we have an algorithm that can predict the reactions that are possible from its starting materials, we can begin to use the algorithm to string these reactions together to develop a multistep synthetic pathway. This pathway prediction can be further optimized to account for reaction conditions, cost of materials, fewest number of reaction steps, and other factors to find the ideal synthetic pathway. Using neural networks helps the algorithm to identify important features from the reactant molecules’ structure in order to classify new reaction types.
Methods
Data Set Generation
The data set of reactions was developed as follows: A library of all alkanes containing 10 carbon atoms or fewer was constructed. To each alkane, a single functional group was added, either a double bond or a halide (Br, I, Cl). Duplicates were removed from this set to make the substrate library. Sixteen different reactions were considered, 4 reactions for alkyl halides and 12 reactions for alkenes. Reactions resulting in methyl shifts or resulting in Markovnikov or anti-Markovnikov product were considered as separate reaction types. Each reaction is associated with a list of secondary reactants and reagents, as well as a SMARTS transformation to generate the product structures from the reactants.
To generate the reactions, every substrate in the library was combined with every possible set of secondary reactants and reagents. Those combinations that matched the reaction conditions set by our expert rules were assigned a reaction type. If none of the reaction conditions were met, the reaction was designated a “null reaction” or NR for short. We generated a target probability vector to reflect this reaction type assignment with a one-hot encoding; that is, the index in the probability vector that matches the assigned reaction type had a probability of 1, and all other reaction types had a probability of 0. The notable exception to this rule was for the elimination and substitution reactions involving methyl shifts for bulky alkyl halides; these reactions were assumed to occur together, and so 50% was assigned to each index corresponding to these reactions. Products were generated using the SMARTS transformation associated with the reaction type with the two reactants as inputs. Substrates that did not match the reaction conditions were designated “null reactions” (NR), indicating that the final result of the reaction is unknown. RDKit50 was used to handle the requirements and the SMARTS transformation. A total of 1,277,329 alkyl halide and alkene reactions were generated. A target reaction probability vector was generated for each reaction.
Prediction Methods
As outlined in Figure 1, to predict the reaction outcomes of a given query, we first predict the probability of each reaction type in our data set occurring, and then we apply SMARTS transformations associated with each reaction. The reaction probability vector, i.e., the vector encoding the probability of all reactions, was predicted using a neural network with reaction fingerprints as the inputs. This reaction fingerprint was formed as a concatenation of the molecular fingerprints of the substrate (Reactant1), the secondary reactant (Reactant2), and the reagent. Both the Morgan fingerprint method, in particular the extended-connectivity circular fingerprint (ECFP), and the neural fingerprint method were tested for generating the molecular fingerprints. A Morgan circular fingerprint hashes the features of a molecule for each atom at each layer into a bit vector. Each layer considers atoms in the neighborhood of the starting atom that are at less than the maximum distance assigned for that layer. Information from previous layers is incorporated into later layers, until the highest layer, e.g., the maximum bond length radius, is reached.34 A neural fingerprint also records atomic features at all neighborhood layers but, instead of using a hash function to record features, uses a convolutional neural network, thus creating a fingerprint with differentiable weights. Further discussion about circular fingerprints and neural fingerprints can be found in Duvenaud et al.39 The circular fingerprints were generated with RDKit, and the neural fingerprints were generated with code from Duvenaud et al.39 The neural network used for prediction had one hidden layer of 100 units. Hyperopt51 in conjunction with Scikit-learn52 was used to optimize the learning rate, the initial scale, and the fingerprint length for each of the molecules.
For some reaction types, certain reagents or secondary reactants are required for that reaction. Thus, it is possible that the algorithm may learn to simply associate these components in the reaction with the corresponding reaction type. As a baseline test to measure the impact of the secondary reactant and the reagent on the prediction, we also performed the prediction with a modified fingerprint. For the baseline metric, the fingerprint representing the reaction was a one-hot vector representation for the 20 most common secondary reactants and the 30 most common reagents. That is, if one of the 20 most common secondary reactants or one of the 30 most common reagents was found in the reaction, the corresponding bits in the baseline fingerprint were turned on; if one of the secondary reactants or reagents was not in these lists, then a bit designated for “other” reactants or reagents was turned on. This combined one-hot representation of the secondary reactants and the reagents formed our baseline fingerprint.
Once a reaction type has been predicted by the algorithm, the SMARTS transformation associated with the reaction type is applied to the reactants. If the input reactants meet the requirements of the SMARTS transformation, the product molecules generated by the transformation are the predicted structures of the products. If the reactants do not match the requirements of the SMARTS transformation, the algorithm instead guesses the structures of the reactants instead, i.e., it is assumed that no reaction occurs.
Acknowledgments
The authors thank Rafael Gomez Bombarelli, Jacob Sanders, Steven Lopez, and Matthew Kayala for useful discussions. J.N.W. acknowledges support from the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1144152. D.D. and A.A.-G. thank the Samsung Advanced Institute of Technology for their research support. The authors thank FAS Research computing for their computing support and computer time in the Harvard Odyssey computer cluster. All opinions, findings, and conclusions expressed in this paper are the authors’ and do not necessarily reflect the policies and views of NSF or SAIT.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.6b00219.
Supplementary Table (PDF)
The code and full training data sets will be made available at https://github.com/jnwei/neural_reaction_fingerprint.git.
The authors declare no competing financial interest.
Supplementary Material
References
- Todd M. H. Computer-aided organic synthesis. Chem. Soc. Rev. 2005, 34, 247. 10.1039/b104620a. [DOI] [PubMed] [Google Scholar]
- Szymkuć S.; Gajewska E. P.; Klucznik T.; Molga K.; Dittwald P.; Startek M.; Bajczyk M.; Grzybowski B. A. Computer-Assisted Synthetic Planning: The End of the Beginning. Angew. Chem., Int. Ed. 2016, 55, 5904–5937. 10.1002/anie.201506101. [DOI] [PubMed] [Google Scholar]
- Corey E. J. Centenary lecture. Computer-assisted analysis of complex synthetic problems. Q. Rev., Chem. Soc. 1971, 25, 455–482. 10.1039/qr9712500455. [DOI] [Google Scholar]
- Corey E.; Wipke W. T.; Cramer R. D. III; Howe W. J. Techniques for perception by a computer of synthetically significant structural features in complex molecules. J. Am. Chem. Soc. 1972, 94, 431–439. 10.1021/ja00757a021. [DOI] [Google Scholar]
- Corey E.; Howe W. J.; Orf H.; Pensak D. A.; Petersson G. General methods of synthetic analysis. Strategic bond disconnections for bridged polycyclic structures. J. Am. Chem. Soc. 1975, 97, 6116–6124. 10.1021/ja00854a026. [DOI] [Google Scholar]
- Corey E.; Long A. K.; Greene T. W.; Miller J. W. Computer-assisted synthetic analysis. Selection of protective groups for multistep organic syntheses. J. Org. Chem. 1985, 50, 1920–1927. 10.1021/jo00211a027. [DOI] [Google Scholar]
- Corey E.; Wipke W. T.; Cramer R. D. III; Howe W. J. Computer-assisted synthetic analysis. Facile man-machine communication of chemical structure by interactive computer graphics. J. Am. Chem. Soc. 1972, 94, 421–430. 10.1021/ja00757a020. [DOI] [Google Scholar]
- Jorgensen W. L.; Laird E. R.; Gushurst A. J.; Fleischer J. M.; Gothe S. A.; Helson H. E.; Paderes G. D.; Sinclair S. CAMEO: a program for the logical prediction of the products of organic reactions. Pure Appl. Chem. 1990, 62, 1921–1932. 10.1351/pac199062101921. [DOI] [Google Scholar]
- Gasteiger J.; Hutchings M. G.; Christoph B.; Gann L.; Hiller C.; Löw P.; Marsili M.; Saller H.; Yuki K.. Organic Synthesis, Reactions and Mechanisms; Springer: Berlin, Heidelberg, 1987; pp 19–73. [Google Scholar]
- Satoh H.; Funatsu K. Further Development of a Reaction Generator in the SOPHIA System for Organic Reaction Prediction. Knowledge-Guided Addition of Suitable Atoms and/or Atomic Groups to Product Skeleton. J. Chem. Inf. Comput. Sci. 1996, 36, 173–184. 10.1021/ci950058a. [DOI] [Google Scholar]
- Satoh K.; Funatsu K. A Novel Approach to Retrosynthetic Analysis Using Knowledge Bases Derived from Reaction Databases. J. Chem. Inf. Comput. Sci. 1999, 39, 316–325. 10.1021/ci980147y. [DOI] [Google Scholar]
- ChemPlanner. http://www.chemplanner.com/.
- Gelernter H.; Rose J. R.; Chen C. Building and refining a knowledge base for synthetic organic chemistry via the methodology of inductive and deductive machine learning. J. Chem. Inf. Model. 1990, 30, 492–504. 10.1021/ci00068a023. [DOI] [Google Scholar]
- Chen J. H.; Baldi P. No Electron Left Behind: A Rule-Based Expert System To Predict Chemical Reactions and Reaction Mechanisms. J. Chem. Inf. Model. 2009, 49, 2034–2043. 10.1021/ci900157k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H.; Baldi P. Synthesis Explorer: A Chemical Reaction Tutorial System for Organic Synthesis Design and Mechanism Prediction. J. Chem. Educ. 2008, 85, 1699. 10.1021/ed085p1699. [DOI] [Google Scholar]
- Law J.; Zsoldos Z.; Simon A.; Reid D.; Liu Y.; Khew S. Y.; Johnson A. P.; Major S.; Wade R. A.; Ando H. Y. Route Designer: A Retrosynthetic Analysis Tool Utilizing Automated Retrosynthetic Rule Generation. J. Chem. Inf. Model. 2009, 49, 593–602. 10.1021/ci800228y. [DOI] [PubMed] [Google Scholar]
- Gothard C. M.; Soh S.; Gothard N. A.; Kowalczyk B.; Wei Y.; Baytekin B.; Grzybowski B. A. Rewiring Chemistry: Algorithmic Discovery and Experimental Validation of One-Pot Reactions in the Network of Organic Chemistry. Angew. Chem. 2012, 124, 8046–8051. 10.1002/ange.201202155. [DOI] [PubMed] [Google Scholar]
- Grzybowski B. A.; Bishop K. J. M.; Kowalczyk B.; Wilmer C. E. The ’wired’ universe of organic chemistry. Nat. Chem. 2009, 1, 31–36. 10.1038/nchem.136. [DOI] [PubMed] [Google Scholar]
- Zimmerman P. M. Automated discovery of chemically reasonable elementary reaction steps. J. Comput. Chem. 2013, 34, 1385–1392. 10.1002/jcc.23271. [DOI] [PubMed] [Google Scholar]
- Wang L.-P.; Titov A.; McGibbon R.; Liu F.; Pande V. S.; Martínez T. J. Discovering chemistry with an ab initio nanoreactor. Nat. Chem. 2014, 6, 1044–1048. 10.1038/nchem.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-P.; McGibbon R. T.; Pande V. S.; Martinez T. J. Automated Discovery and Refinement of Reactive Molecular Dynamics Pathways. J. Chem. Theory Comput. 2016, 12, 638–649. 10.1021/acs.jctc.5b00830. [DOI] [PubMed] [Google Scholar]
- Xu L.; Doubleday C. E.; Houk K. Dynamics of 1, 3-Dipolar Cycloaddition Reactions of Diazonium Betaines to Acetylene and Ethylene: Bending Vibrations Facilitate Reaction. Angew. Chem. 2009, 121, 2784–2786. 10.1002/ange.200805906. [DOI] [PubMed] [Google Scholar]
- Rappoport D.; Galvin C. J.; Zubarev D. Y.; Aspuru-Guzik A. Complex Chemical Reaction Networks from Heuristics-Aided Quantum Chemistry. J. Chem. Theory Comput. 2014, 10, 897–907. 10.1021/ct401004r. [DOI] [PubMed] [Google Scholar]
- Socorro I. M.; Taylor K.; Goodman J. M. ROBIA: A Reaction Prediction Program. Org. Lett. 2005, 7, 3541–3544. 10.1021/ol0512738. [DOI] [PubMed] [Google Scholar]
- Socorro I. M.; Taylor K.; Goodman J. M. ROBIA: a reaction prediction program. Org. Lett. 2005, 7, 3541–3544. 10.1021/ol0512738. [DOI] [PubMed] [Google Scholar]
- He K.; Zhang X.; Ren S.; Sun J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. 2015 IEEE Int. Conf. Comput. Vision (ICCV) 2015, 1026–1034. 10.1109/ICCV.2015.123. [DOI] [Google Scholar]
- Krizhevsky A.; Sutskever I.; Hinton G. E. In Adv. Neural. Inf. Process. Syst. 25; Pereira F., Burges C. J. C., Bottou L., Weinberger K. Q., Eds.; Curran Associates, Inc.: 2012; pp 1097–1105. [Google Scholar]
- Mnih V.; et al. Human-level control through deep reinforcement learning. Nature 2015, 518, 529–533. 10.1038/nature14236. [DOI] [PubMed] [Google Scholar]
- Silver D.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- Reymond J.-L.; Van Deursen R.; Blum L. C.; Ruddigkeit L. Chemical space as a source for new drugs. MedChemComm 2010, 1, 30–38. 10.1039/c0md00020e. [DOI] [Google Scholar]
- Kayala M. A.; Baldi P. ReactionPredictor: Prediction of Complex Chemical Reactions at the Mechanistic Level Using Machine Learning. J. Chem. Inf. Model. 2012, 52, 2526–2540. 10.1021/ci3003039. [DOI] [PubMed] [Google Scholar]
- Kayala M. A.; Azencott C.-A.; Chen J. H.; Baldi P. Learning to Predict Chemical Reactions. J. Chem. Inf. Model. 2011, 51, 2209–2222. 10.1021/ci200207y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H. L. The Generation of a Unique Machine Description for Chemical Structures-A Technique Developed at Chemical Abstracts Service. J. Chem. Doc. 1965, 5, 107–113. 10.1021/c160017a018. [DOI] [Google Scholar]
- Rogers D.; Hahn M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Pyzer-Knapp E. O.; Li K.; Aspuru-Guzik A. Learning from the Harvard Clean Energy Project: The Use of Neural Networks to Accelerate Materials Discovery. Adv. Funct. Mater. 2015, 25, 6495–6502. 10.1002/adfm.201501919. [DOI] [Google Scholar]
- Ballester P. J.; Mitchell J. B. O. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 2010, 26, 1169–1175. 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Golbraikh A.; Oloff S.; Kohn H.; Tropsha A. A Novel Automated Lazy Learning QSAR (ALL-QSAR) Approach: Method Development Applications, and Virtual Screening of Chemical Databases Using Validated ALL-QSAR Models. J. Chem. Inf. Model. 2006, 46, 1984–1995. 10.1021/ci060132x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolyan Y.; Walters M. A.; Karypis G. Assessing Synthetic Accessibility of Chemical Compounds Using Machine Learning Methods. J. Chem. Inf. Model. 2010, 50, 979–991. 10.1021/ci900301v. [DOI] [PubMed] [Google Scholar]
- Duvenaud D. K.; Maclaurin D.; Iparraguirre J.; Bombarelli R.; Hirzel T.; Aspuru-Guzik A.; Adams R. P. Convolutional Networks on Graphs for Learning Molecular Fingerprints. NIPS’15 Proc. 28th Int. Conf. Neural Inf. Process. Syst. 2015, 2224–2232. [Google Scholar]
- Montavon G.; Hansen K.; Fazli S.; Rupp M.; Biegler F.; Ziehe A.; Tkatchenko A.; Lilienfeld A. V.; Müller K.-R. Learning invariant representations of molecules for atomization energy prediction. Adv. Neural Inf. Process. Syst. 2012, 440–448. [Google Scholar]
- von Lilienfeld O. A.; Ramakrishnan R.; Rupp M.; Knoll A. Fourier series of atomic radial distribution functions: A molecular fingerprint for machine learning models of quantum chemical properties. Int. J. Quantum Chem. 2015, 115, 1084–1093. 10.1002/qua.24912. [DOI] [Google Scholar]
- Carhart R. E.; Smith D. H.; Venkataraghavan R. Atom pairs as molecular features in structure-activity studies: definition and applications. J. Chem. Inf. Model. 1985, 25, 64–73. 10.1021/ci00046a002. [DOI] [Google Scholar]
- Schneider N.; Lowe D. M.; Sayle R. A.; Landrum G. A. Development of a Novel Fingerprint for Chemical Reactions and Its Application to Large-Scale Reaction Classification and Similarity. J. Chem. Inf. Model. 2015, 55, 39–53. 10.1021/ci5006614. [DOI] [PubMed] [Google Scholar]
- Virshup A. M.; Contreras-García J.; Wipf P.; Yang W.; Beratan D. N. Stochastic Voyages into Uncharted Chemical Space Produce a Representative Library of All Possible Drug-Like Compounds. J. Am. Chem. Soc. 2013, 135, 7296–7303. 10.1021/ja401184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M.; Ekker H.; Flamm C.. The graph grammar library—a generic framework for chemical graph rewrite systems. In International Conference on Theory and Practice of Model Transformations; 2013; pp 52–53. [Google Scholar]
- Schurer S. C.; Tyagi P.; Muskal S. M. Prospective Exploration of Synthetically Feasible, Medicinally Relevant Chemical Space. J. Chem. Inf. Model. 2005, 45, 239–248. 10.1021/ci0496853. [DOI] [PubMed] [Google Scholar]
- Gothard C. M.; Soh S.; Gothard N. A.; Kowalczyk B.; Wei Y.; Baytekin B.; Grzybowski B. A. Rewiring Chemistry: Algorithmic Discovery and Experimental Validation of One-Pot Reactions in the Network of Organic Chemistry. Angew. Chem. 2012, 124, 8046–8051. 10.1002/ange.201202155. [DOI] [PubMed] [Google Scholar]
- Wade L. G.Organic chemistry, 6th ed.; Pearson: Upper Saddle River, NJ, USA, 2013; p 377. [Google Scholar]
- Bajusz D.; Rácz A.; Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations?. J. Cheminf. 2015, 7, 1–13. 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RDKit: Open-source cheminformatics; http://www.rdkit.org.
- Bergstra J.; Komer B.; Eliasmith C.; Yamins D.; Cox D. D. Hyperopt: a Python library for model selection and hyperparameter optimization. Comput. Sci. Discovery 2015, 8, 014008. 10.1088/1749-4699/8/1/014008. [DOI] [Google Scholar]
- Pedregosa F.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.