Abstract
Promysalin is a species-specific Pseudomonad metabolite with unique bioactivity. To better understand the mode of action of this natural product, we synthesized 16 analogs utilizing diverted total synthesis (DTS). Our analog studies revealed that the bioactivity of promysalin is sensitive to changes within its hydrogen bond network whereby alteration has drastic biological consequences. The DTS library not only yielded three analogs that retained potency but also provided insights that resulted in the identification of a previously unknown ability of promysalin to bind iron. These findings coupled with previous observations hint at a complex multifaceted role of the natural product within the rhizosphere.
Over the past century, groundbreaking discoveries in the field of antibiotics have resulted in the saving of countless lives. This can be primarily attributed to their broad-spectrum activity that is ideal for the treatment of a wide range of bacterial infections but can also result in significant misuse and unintended side effects.1 For example, recent work from the Blaser lab strongly suggests that prolonged broad-spectrum antibiotic exposure early in life can lead to an increased likelihood of obesity, allergies, and inflammatory diseases.1 These findings highlight the effect that certain antimicrobials have on one’s commensal population, resulting in an altered community dynamic and leading to undesired outcomes lending credence to the recent call for the development of narrow-spectrum therapies.
To date very few options currently exist for pathogen-specific treatments.2 Furthermore, with species-specific compounds in hand one would also have access to tool compounds to aid in deconvoluting the complex multispecies environments present. The recent advances in genetic sequencing, as exemplified by the human microbiome project, would allow for the probing of these environments if the appropriate tools were available.3
Our group has been particularly interested in developing narrow-spectrum agents to combat Pseudomonas aeruginosa (PA) infections. PA is a Gram-negative pathogen that is typically found in people with weakened immune systems and/or hospital settings accounting for an estimated 51,000 infections/year in the United States alone.4 The community, including our own lab,5 has postulated that the rhizosphere, which is defined as the immediate area of soil that is directly influenced by microorganisms and root secretions, is an ideal environment to discover such compounds as the soil is teeming with Pseudomonads competing to establish their own niches. This hypothesis has motivated our interest in the unique and highly specific Pseudomonad natural product promysalin (1, Figure 1).
Figure 1.
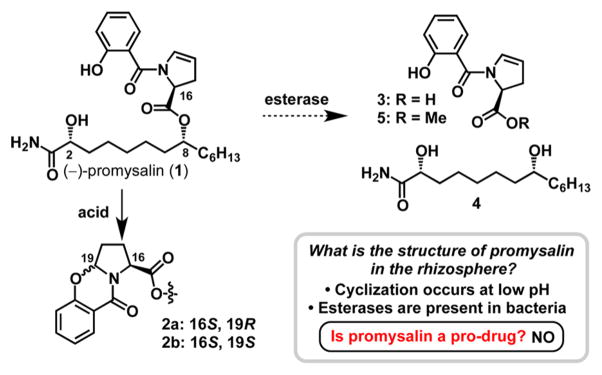
Structure of promysalin (1) and compounds 2–5 involved in our pro-drug hypothesis.
We recently reported the total synthesis and structural elucidation of promysalin, which is produced by Pseudomonas putida (PP), and inhibits the growth of PA at nM concentrations,6 while Gram-positive bacteria show no susceptibility.7 Furthermore, during our biological studies we found that promysalin inhibits the production of pyoverdine (a siderophore) in P. putida KT2440, and as expected, only one stereoisomer elicited this phenotype. Siderophores are essential for the growth of Pseudomonads, as they actively chelate Fe3+ for its end use in enzymatic processes. Intriguingly, siderophore production is closely linked to virulence; therefore, inhibiting such a process may yield novel antivirulence therapies.8 These initial discoveries prompted us to initiate a synthetic campaign to elucidate what chemical moieties are responsible for the species-specific activity of promysalin.
At the outset, we hypothesized two potential mechanisms of action. First, promysalin may be a “prodrug”, whereby it would be activated by either an enzyme (i.e., an esterase) unique to PA or the physical environment surrounding PA itself. A second alternative is that the chemical architecture of the molecule is exquisitely selective for a target harbored by PA but not present in other bacteria. Our earlier synthetic studies hinted that the exact three-dimensional shape of the molecule was key to its biological activity, as we observed a 100–200-fold decrease in activity by altering either stereocenter on the fatty acid fragment. To address these questions, we constructed a library of 16 hypothesis-driven analogs via diverted total synthesis (DTS).9
Figure 1 depicts the structure of promysalin (1) and the structures involved in our hypotheses. Enamines are quite susceptible to acid hydrolysis, and we were curious about the stability of promysalin in low pH environments. Somewhat surprisingly, we (and others)10 noticed the instability of promysalin under mildly acidic conditions, which we were able to attribute to the formation of cyclized products 2a/b. We wondered if this cyclization event was biologically relevant, whereby the compound would be dispensed by PP and would undergo cyclization in the known acidic environment present around PA.11 However, the cyclized compounds showed no biological activity. Similarly, it is well-known that bacteria possess enzymes capable of hydrolyzing esters. Therefore, we questioned if the hydrolyzed fragments (3 and 4) would be active. We synthesized methyl ester 5 in lieu of the acid, for cell permeability purposes. Methyl ester 5 and diol 4 were both inactive up to concentrations of 250 μM. Additionally, we have also recently shown that appending the acid fragment of promysalin to another myristate-derived natural product, lyngbic acid, yielded compounds devoid of activity against PA.12 Taken in sum, it appears that the cyclization and hydrolysis reactions are synthetic artifacts and not biologically relevant.
Based on these results we next sought to leverage DTS to access a library of analogs to build a structure–activity relationship (SAR) profile and further test the importance of the dehydroproline heterocycle. DTS has proven useful in previous natural product mechanistic studies13 and, in some cases, has provided therapeutically useful analogs, exemplified by the development of fludelone.14 It should be noted that the analogs accessed from DTS presented here are inaccessible by enzymatic or chemical manipulation of the natural product directly, thus showcasing the power of organic synthesis.
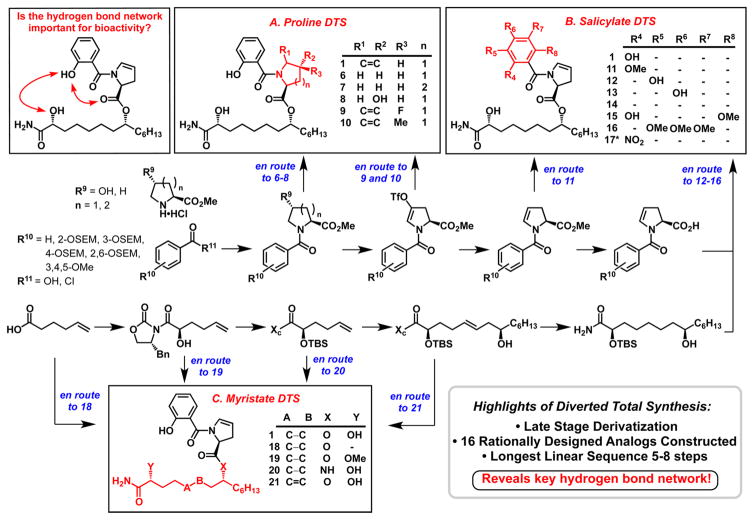
We initially hypothesized that the enamide moiety of promysalin was responsible for the bioactivity wherein it would covalently interact with its biological target; therefore, we postulated that any modification would render the molecule inactive. We utilized DTS to access five specific dehydroproline derivatives (Scheme 1, Box A). The synthesis of the proline, piperidine, and hydroxyproline analogs 6–8 was straightforward using standard coupling reactions and protecting group manipulations. We next exploited the inherent reactivity of our triflate intermediate via Pd-mediated coupling reactions to provide access to analogs 9 and 10.
Scheme 1. Diverted Total Synthesis of Promysalin Analogs To Probe the Hydrogen-Bonding Networka.
aBlue wording depicts the branching points from our previously reported synthesis (for reaction conditions see ref 6 and SI). Dashes in tables denote where hydrogens are present. *Compound 17 was prepared by a different route (see SI).
Preliminary computational models of promysalin highlighted an intricate intramolecular hydrogen-bonding network resulting in a rigidified molecular framework (vide infra). We hypothesized that this network, composed of the phenol, hydroxyl, and ester moieties, were therefore critical for activity. To test this hypothesis we designed analogs varying these key functionalities (Scheme 1, Box B). The salicylate fragment was systematically altered in three specific ways: (1) the position (12 and 13) and presence (14) of the phenol; (2) the substitution of the phenol with either the corresponding methyl ether (11) or nitro group (17); or (3) by increasing the electron density within the ring (15 and 16). In a similar fashion we sought to probe the role of the ester linkage and secondary alcohol (Scheme 1, Box C). For example, we envisioned converting the ester to the corresponding amide (20) to create a hydrolytically more stable analog, albeit one possessing a drastically altered hydrogen-bond network. Alkene analog 21 would add rigidity to the scaffold and potentially stabilize the active conformation. Finally, the importance of the hydroxyl group was probed by either methylation (19) or omission (18). The detailed synthetic route for the analogs library is presented in the Supporting Information (SI) (Schemes S1 and S2).
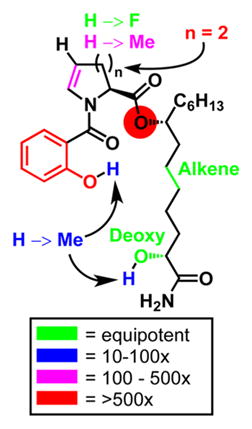
With a library of 16 analogs in hand, we evaluated all of the compounds for inhibitory activity against P. aeruginosa strains PAO1 and PA14. The data for the active analogs (IC50 < 250 μM) are shown in Table 1 along with the inhibitory data for the less potent diastereomers of promysalin (for numbering, see Figure 1). The inhibition data supported our initial hypothesis that the conformation of promysalin is exquisitely linked to its inhibition of PA. Of our modifications to the proline structure, fluorination (9)—being the smallest steric perturbation—was the only compound with equipotency to that of promysalin. Methylation (10) was slightly tolerated, while the piperidine (7) and hydroxyproline (8) derivatives were inactive. To our surprise, proline derivative 6 retained modest activity, which may hint at an inhibitory mechanism that involves both structural recognition and covalent binding.
Table 1.
IC50 Values of Promysalin Diastereomers and Active Compounds in μM (Compounds with IC50 > 250 μM Are Not Shown)a
| PA01 | PA14 | |
|---|---|---|
| 1 (2R,8R) | 4.1 | 0.067 |
| 1b (2R,8S) | 46 | 6.6 |
| 1c (2S,8S) | 90 | 22 |
| 1d (2S,8R) | 33 | 4.3 |
| 6 | 111 | 28 |
| 9 | 7.7 | 0.019 |
| 10 | 32 | 12 |
| 11 | 57 | 6.7 |
| 18 | 5.8 | 0.035 |
| 19 | 38 | 11 |
| 21 | 8.3 | 0.067 |
Values are averages of three independent experiments (see SI for graphs and experimental details). A color-coded SAR is provided depicting the fold decrease in activity (right).
The structure of the salicylate moiety was largely unforgiving in terms of the position (12, 13), substitution (17), or exclusion of the phenol (14). In addition, adding methoxy substituents in the presence (15) or absence (16) of the o-phenol was not tolerated. The only active salicylate analog was the methyl ether (11), albeit with an order of magnitude less potency. In contrast, the side chain analogs all retained some, if not all, activity, with the one exception being the amide (20). Methylation of the hydroxyl group (19) resulted in a decrease in potency on par with the methylated phenol (11). Rigidifying the side chain by including the alkene (21) led to an equipotent analog. However, when the secondary alcohol is removed, providing compound 18, biological activity is fully retained. At first this result was particularly surprising, as we postulated that the alcohol was integral to the hydrogen-bonding network, as evidenced by the difference in activity between 1 and 1b/c (a 10–100-fold decrease in potency). However, when considering our proposed macrocyclic structure, the implications of epimerization at a hydroxyl group can lead to drastic bond angle changes; in contrast, 18 can adopt a similar conformation simply by substituting the amide carbonyl as a Lewis base in place of the alcohol. Furthermore, the synthesis of 18 requires two less overall steps, and is more atom-economical, than that of promysalin, as the use of a protecting group and chiral auxiliary is avoided.
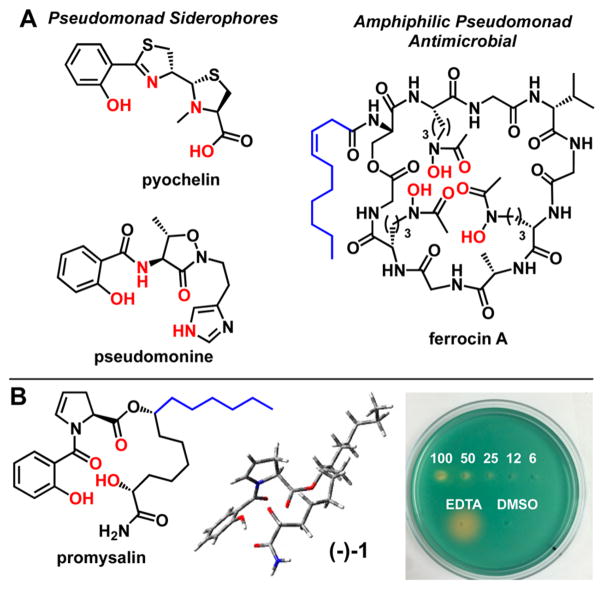
As mentioned earlier, Pseudomonads are well-known to produce a variety of siderophores in the rhizosphere. A cursory look at the literature reveals two specific siderophores with known salicyamide iron-binding motifs, reminiscent of promysalin: pyochelin15 and pseudomonine16 (Figure 2A). PQS, the Pseudomonas aeruginosa 4-Quinolone Signaling Molecule, which regulates virulence in PA, has also been shown to bind iron.17 Intriguingly, ferrocin,18 another Pseudomonad siderophore that bears a fatty acid moiety, has been shown to possess antimicrobial activity specifically against Gram-negative bacteria, and in particular PA. Based on these findings, and in light of our bioactivity and modeling results, we speculated that promysalin might be capable of binding iron. This hypothesis was reinforced by earlier findings demonstrating that promysalin is capable of promoting swarming in PP and inhibiting pyoverdine production. With these questions in mind, we tested the ability of promysalin to bind iron. Indeed, promysalin tested positive for Fe3+ chelation on CAS agar at concentrations ranging from 6 to 100 mM, albeit with reduced affinity when compared to the known iron chelator EDTA (Figure 2B). Furthermore, Fe3+ chelation was confirmed down to 1 mM using the solution-based CAS assay (see Supporting Information Figure S6).19 These results hint at the ability of promysalin to act as a siderophore in various Pseudomonads; however, additional studies are needed to confirm this role and are currently ongoing in our laboratory.
Figure 2.
(A) Structures of PA siderophores pyochelin and pseudomonine, and antimicrobial siderophore ferrocin. Putative iron-binding atoms are colored red, and fatty acid tails, in blue. (B) Structure of promysalin with proposed iron contacts shown in red (left), minimized calculated structure (middle), and CAS agar plate (right). Captions indicate the concentration of promysalin (mM) in 10 μL innoculations on CAS agar, with EDTA (6 mM) and DMSO (10% v/v) as controls.
These findings may allude to how the natural product elicits its species-specific activity against PA, as well as its enhanced potency against PA14 versus PAO1. One possible mechanism of action of promysalin is through the inhibition of siderophore transport pathways thereby severely limiting or inhibiting the organism’s ability to acquire iron. This particular mechanism is especially intriguing since PA14 significantly differs from PAO1 in its acquisition of iron. Rahme and co-workers have recently shown that ybtQ, a yersiniabactin ABC-transporter (yersiniabactin is a phenolate-thiazoline siderophore that shares structural features with pyochelin), is found in PA14 and is primarily responsible for its virulence.20
Chemists, the Miller group in particular, have exploited siderophore transport pathways with great success using “sideromimetic” compounds in which antibiotic moieties are tethered to known siderophores for uptake.21 Conversely, there are only a few reports of iron-binding natural products which also possess antibacterial activity, though none of these have been conclusively shown to inhibit siderophore transport systems. However, it seems increasingly likely that this type of process is indeed taking place in the rhizosphere, resulting in narrow-spectrum Gram-negative agents. As discussed in Figure 2, ferrocin is a compound produced by the soil-dwelling bacteria Pseudomonas fluorescens YK-310 and has been shown to inhibit Gram-negative bacteria. In addition to their affinities for iron, both compounds possess amphiphilic features that may play a role in their potent activities. Nevertheless, the promise of highly potent and selective agents against Pseudomonads warrants further investigation.
In conclusion, we have leveraged the power of DTS to access a 16-membered library of rationally designed synthetic promysalin analogs. This structural diversity, which is inaccessible by semisynthesis, has shed light on the key structural features responsible for bioactivity and highlights the importance of the key functionalities within the hydrogen-bonding network (which are presumably also critical for binding iron). Furthermore, these findings have led to the discovery of the iron-binding ability of promysalin, hinting at a secondary role for the natural product as a rhizosphere siderophore. In light of these results, a potential mechanism of action via the inhibition of siderophore transport seems feasible. Current work in our laboratory is now focused on determining if promysalin is a bona fide siderophore, and the unequivocal identification of its biological target will be reported in due course.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under CHE-1454116 and Temple University. We thank B. A. Buttaro (Temple University) for assistance with biological experiments, G. A. O’Toole (Dartmouth Medical School) for the generous donation of strains, and Materia, Inc. for olefin metathesis catalysts.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b03373.
Experimental procedures, characterization data, NMR spectra, and supporting figures (PDF)
References
- 1.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Rodriguez JGZ, Rogers AB, Robine N, Loke P, Blaser MJ. Cell. 2014;158:705. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxson T, Mitchell DA. Tetrahedron. 2015 doi: 10.1016/j.tet.2015.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Human Microbiome Project Consortium. Huttenhower C. Nature. 2012;486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pseudomonas aeruginosa in Healthcare Settings. 2014 Retrieved March 19, 2016, from http://www.cdc.gov/hai/organisms/pseudomonas.html.
- 5.Keohane CE, Steele AD, Wuest WM. Synlett. 2015;26:2739. [Google Scholar]
- 6.Steele AD, Knouse KW, Keohane CE, Wuest WM. J Am Chem Soc. 2015;137:7314. doi: 10.1021/jacs.5b04767. [DOI] [PubMed] [Google Scholar]
- 7.Li W, Santos PE, Matthijs S, Xie G, Busson R, Cornelis P, Rozenski J, De Mot R. Chem Biol. 2011;18:1320. doi: 10.1016/j.chembiol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Overhage J, Bains M, Brazas MD, Hancock REW. J Bacteriol. 2008;190:2671. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson RM, Danishefsky SJ. J Org Chem. 2006;71:8329. doi: 10.1021/jo0610053. [DOI] [PubMed] [Google Scholar]
- 10.Kaduskar RD, Dhavan AA, Dallavalle S, Scaglioni L, Musso L. Tetrahedron. 2016;72:2034. doi: 10.3762/bjoc.12.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter RC, Beveridge TJ. Appl Environ Microbiol. 2005;71:2501. doi: 10.1128/AEM.71.5.2501-2510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knouse KW, Wuest WM. J Antibiot. 2016 doi: 10.1038/ja.2016.4. ASAP. [DOI] [PubMed] [Google Scholar]
- 13.Szpilman AM, Carreira EM. Angew Chem, Int Ed. 2010;49:9592. doi: 10.1002/anie.200904761. [DOI] [PubMed] [Google Scholar]
- 14.Rivkin A, Chou T, Danishefsky SJ. Angew Chem, Int Ed. 2005;44:2838. doi: 10.1002/anie.200461751. [DOI] [PubMed] [Google Scholar]
- 15.(a) Cox CD, Rinehart KL, Jr, Moore ML, Cook JC., Jr Proc Natl Acad Sci U S A. 1981;78:4256. doi: 10.1073/pnas.78.7.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schlegel K, Lex J, Taraz K, Budzikiewicz H. Z Naturforsch C. 2006;61:263. doi: 10.1515/znc-2006-3-418. [DOI] [PubMed] [Google Scholar]
- 16.(a) Anthoni U, Christophersen C, Nielsen PH, Gram L, Petersen BO. J Nat Prod. 1995;58:1786. [Google Scholar]; (b) Sattely ES, Walsh CT. J Am Chem Soc. 2008;130:12282. doi: 10.1021/ja804499r. [DOI] [PubMed] [Google Scholar]; (c) Wuest WM, Sattely ES, Walsh CT. J Am Chem Soc. 2009;131:5056. doi: 10.1021/ja900815w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Pesci EC, Milbank JBJ, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Proc Natl Acad Sci U S A. 1999;96:11229. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. Environ Microbiol. 2006;8:1318. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 18.Katayama N, Nozaki Y, Okonogi K, Harada S, Ono H. J Antibiot. 1993;46:65. doi: 10.7164/antibiotics.46.65. [DOI] [PubMed] [Google Scholar]
- 19.Schwyn B, Neilands JB. Anal Biochem. 1987;160:47. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 20.(a) Haag H, Hantke K, Drechsel H, Stojiljkovic I, Jung G, Zähner H. J Gen Microbiol. 1993;139:2159. doi: 10.1099/00221287-139-9-2159. [DOI] [PubMed] [Google Scholar]; (b) Choi JY, Sifri CD, Goumnerov BC, Rahme LG, Ausubel FM, Calderwood SB. J Bacteriol. 2002;184:952. doi: 10.1128/jb.184.4.952-961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Wencewicz TA, Moellmann U, Long TE, Miller MJ. BioMetals. 2009;22:633. doi: 10.1007/s10534-009-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Starr J, Brown MF, Aschenbrenner L, Caspers N, Che Y, Gerstenberger BS, Huband M, Knafels JD, Lemmon MM, Li C, McCurdy SP, McElroy E, Rauckhorst MR, Tomaras AP, Young JA, Zaniewski RP, Shanmugasundaram V, Han S. J Med Chem. 2014;57:3845. doi: 10.1021/jm500219c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.