Abstract
The induction of host defense against Candida species is initiated by recognition of the fungi by pattern recognition receptors and activation of downstream pathways that produce inflammatory mediators essential for infection clearance. In the present study, we present complementary evidence based on transcriptome analysis, genetics, and immunological studies in knockout mice and humans, that the cytosolic RIG-I-like receptor MDA5 (IFIHI) has an important role in the host defense against C. albicans. Firstly, IFIH1 expression in macrophages is specifically induced by invasive C. albicans hyphae, and patients suffering from chronic mucocutaneous candidiasis (CMC) express lower levels of MDA5 than healthy controls. Secondly, there is a strong association between missense variants in the IFIH1 gene (rs1990760 and rs3747517) and susceptibility to systemic Candida infections. Thirdly, cells from Mda5 knockout mice and human PBMCs with different IFIH1 genotypes display an altered cytokine response to C. albicans. These data strongly suggest that MDA5 is involved in immune responses to Candida infection. As a receptor for viral RNA, MDA5 receptor until now has been linked to antiviral host defense, but these novel studies show unexpected effects in antifungal immunity as well. Future studies are warranted to explore the potential of MDA5 as novel target for immunotherapeutic strategies.
Keywords: Candida albicans, host defense, candidemia, genetic susceptibility, MDA5, IFIH1
Introduction
Candida species are one of the most common human fungal pathogens. They inhabit skin, mucosa, and the gastrointestinal tract. Normally, Candida colonization does not lead to disease in healthy individuals; however, this peaceful cohabitation can drastically change when the host immune system is compromised. Oropharyngeal and vaginal Candida infections are relatively commonly present in the population, while systemic candidiasis is the fourth most common form of bloodstream infections in the US. It is often life-threatening due to invasion of deep tissues and organs, with mortality rates reaching up to 40% [1-4].
Recent progress has partly elucidated the environmental and genetic risk factors that contribute to the causes and severity of Candida infections [5, 6]. Immunosuppressive medication, treatment with antibiotics, and prolonged hospitalization in intensive care units, are some of the most important risk factors [7, 8]. Several monogenic disorders that result in primary immunodeficiencies increase the susceptibility to Candida infection, as demonstrated for mutations in CARD9 and STAT1 [9-11]. Common genetic variants, for example in pattern recognition receptors (e.g. Dectin-1 and TLR1) and interleukins (e.g. IL-4, IL-10 and IL-12B), also increase the risk of infection by affecting Candida recognition and cytokine signaling [6, 12]. It is likely that combinations of these and other common genetic variants in multiple genes underlie the complex patterns of susceptibility to different forms of disease caused by Candida spp.
The discovery of genetic factors contributing to infection susceptibility has improved our understanding of the molecular pathways involved in the immune response against Candida. However, identifying pathway components that are suitable targets for novel immunotherapeutic approaches requires more insight. Recently, Smeekens et al. discovered that the type I interferon (IFN) pathway plays a central role in host defense against C. albicans [13]. In the present work, we demonstrate that MDA5 (IFIHI), a RIG-I-like receptor, until now described as a receptor of viral RNA that induces a signaling pathway leading to the production of type I interferons, is directly involved in the inflammatory response against Candida infections in humans. To this end, we present complementary evidence based on genetics, gene expression, and functional immunological studies in knockout mice and in healthy humans, as well as in patients suffering from systemic candidiasis or chronic mucocutaneous candidiasis (CMC). This is the first time that a receptor of the RIG-I-like helicase family has been shown to be involved in the antifungal immune response.
Materials and Methods
Macrophage transcriptome analysis after stimulation with wild-type and hgc1-defective Candida albicans
CD14+ monocytes were derived from healthy volunteers and differentiated into macrophages using M-CSF for 7 days. 200000 monocytes per well remained unstimulated or were stimulated for 4 or 24 hours with either wild-type C. albicans (UC820) or a HGC1 null mutant, which is unable to form hyphae. Global gene expression was then profiled as previously described [13]. Gene expression levels in the various conditions were compared to unstimulated macrophages. We identified genes that showed significant differential expression after Benjamini-Hochberg (BH)-correction (P < 0.05 and > 2-fold change in expression) in at least one of the conditions (i.e. stimulation with wild-type or HGC1 null Candida, for 4h or 24h, compared to unstimulated control). From this set, we selected the 62 genes that are exclusively induced after stimulation with wild-type C. albicans for 24 hours. These genes were analyzed for enrichment in KEGG pathways [49] using the DAVID suite [50], with all human genes as background.
Candidemia and control cohorts
In this study we included 227 unrelated adult Caucasian patients suffering from candidemia (described in detail in [6]). Patients were enrolled after giving written informed consent at the Duke University Hospital (DUMC, Durham, North Carolina, USA) as well as the Radboud University Medical Centre (Radboudumc, Nijmegen, The Netherlands). Patient enrollment took place after confirmation of at least one positive blood culture for a Candida species. The study was approved by the Institutional Review Boards of both medical centres, the Institutional Review Board of Duke University (CR4_Pro00006427) in North Carolina (USA) and the ‘Commissie Mensgebonden Onderzoek Arnhem-Nijmegen’ (2001/198) in Nijmegen (The Netherlands). The study was performed in accordance with the international guidelines of the declaration of Helsinki (year 2000) of the World Medical Association adopted by the World Medical Assembly. Enrollment took place between January 2003 and January 2009.
A control cohort of 176 Caucasians was employed. This cohort consists of non-infected (candidiasis free) matched patients from the same medical centres as the patient cohort. Controls were recruited consecutively from the same hospital wards/services as infected patients during the study period, with a similar balance of medical, surgical, and oncology patients in case and control groups.
Genotyping and quality control
The case and control individuals of the candidemia cohort were genotyped on the Illumina Immunochip SNP array platform, which contains ~200,000 SNPs focused on genomic regions known to be involved in immune-mediated diseases [16]. DNA was isolated from whole blood using the Gentra Pure Gene Blood kit (Qiagen, Venlo, The Netherlands), according to the protocol of the manufacturer. Only samples with a SNP call rate above 90% were included. We applied quality control filters to exclude SNPs with: (i) a genotype call rate of less than 90%, (ii) strong deviation from Hardy-Weinberg equilibrium in control samples (Hardy-Weinberg exact test P ≤ 1 × 10−3), and (iii) significant differences in missingness between cases and controls (Fisher's exact P < 1 × 10−2). As our cohort does not have the power to detect associations with rare variants, we only included SNPs with a minor allele frequency (MAF) of greater than 5% in cases and controls together. 843 SNPs in the FAP-IFIH1-GCA-KCNH7 LD region were tested in the case-control association analysis.
Genetic analyses
Linkage disequilibrium (LD) measures were calculated using Haploview version 4.2 [51] for both the candidemia cohort (LD patterns are based on genotypes of control individuals only) and common SNPs (MAF > 5%) in the CEU population (Utah Residents (CEPH) with Northern and Western European ancestry) in HapMap 3, release 2 [52]. The 405 kb FAP-IFIH1-GCA-KCNH7 LD region on chromosome 2 (hg18 coordinates: 162,720 kb – 163125 kb) was defined based on the LD patterns in the larger genomic region (Figure S1). It includes the complete FAP gene and part of the KCNH7 gene.
Associations between SNPs and susceptibility to candidemia were assessed using both genotypic and allelic tests. Genotypic association was calculated using the Fisher's exact test, asking whether candidemia cases and controls have significantly different genotype count distributions (H0: genotype counts are the same). We corrected for testing multiple SNPs using the Benjamini-Hochberg procedure. Similarly, allelic association was calculated using the Fisher's exact test, asking whether candidemia cases and controls have significantly different allele count distributions (H0: allele counts are the same). Odds ratios (OR, with 95% confidence intervals) are reported for the allelic association tests and represent the odds of disease for individuals carrying the non-risk allele versus the risk allele. Quality filtering and genetic analyses were performed using PLINK v1.07 [53] and custom R scripts. Regional association plots were created using code adapted from http://www.broadinstitute.org/diabetes/scandinavs/figures.html.
Expression analysis of PBMCs
To assess the expression levels of genes in the FAP-IFIH1-GCA-KCNH7 LD region, blood was collected from healthy volunteers after written informed consent. Isolated peripheral blood mononuclear cells (PBMCs) were obtained using density centrifugation described previously [43]. 0.5 × 106 isolated PBMCs remained unstimulated (RPMI), or were stimulated with either 1 × 106/ml heat killed Borrelia burgdorferi [54], 1 × 106/ml heat killed Candida albicans (UC820) [55], 10 ng/ml Escherichia coli-derived lipopolysaccharide (LPS), or 1 × 107/ml sonicated Mycobacterium tuberculosis (MTB) (Hv37Rv) for either 4 or 24 hours. Gene expression was profiled using Illumina Human HT-12 expression BeadChip following the protocol of the manufacturer [13]. Data generation and processing was performed as described elsewhere [13]. Additionally, we measured gene expression in PBMCs from two patients suffering from chronic mucocutaneous candidiasis (CMC) as well as healthy controls. Both CMC patients carried the STAT1 mutation (Arg274Trp) described earlier [10]. Cells were stimulated with C. albicans for 4 hours and samples subsequently underwent RNA sequencing. RNA isolation as well as enrichment, library preparation and data processing are described elsewhere [13].
PBMC stimulation experiments
After obtaining written informed consent, PBMCs were isolated from blood of healthy volunteers using density centrifugation as described elsewhere [43]. 0.5 × 106 cells were plated in 96 wells round bottom plates and subsequently stimulated with either heat killed C. albicans yeast or hyphae (both UC820, 1 × 106/ml, heat-killed at 100°C for 1 hour) for 24 hours (IL-10) or 7 days (IL-17 and IFN-γ). Additionally, PBMCs were stimulated with Mycobacterium tuberculosis (MTB) (1μg/μl) for 24h. Supernatants were collected and measured for IL-10 and IFN-γ (Sanquin, Amsterdam, The Netherlands), and IL-17 cytokines (R&D Systems, Abingdon, UK) according to the instructions of the manufacturers.
Mda5 knockout mice studies
Mda5−/− mice on C57BL/6J background that have been backcrossed at least ten times (kindly provided by dr. M. Colonna) were housed in the facilities of the University Hospital Bonn, Germany. C57BL/6J wild-type mice were purchased from Charles River Breeding Laboratories. All in vivo mice protocols were approved by the internal animal care committee of the University Hospital Bonn and were performed according to national and European regulations.
Splenocytes were isolated from wild-type and Mda5−/− mice and stimulated with RPMI, Poly I:C, and heat-killed C. albicans yeasts or hyphae for 24 hours. Supernatants were stored at −20°C before cytokine measurement by ELISA. For qPCR, splenocytes from both Mda5 knockout mice and B6 control mice were stimulated for 24 hours with heat-killed C. albicans hyphae. RNA was isolated according to the TRIzol® isolation protocol supplied by the manufacturer (Life Technologies).
Results
Candida germination induces expression of RLR pathway components in macrophages
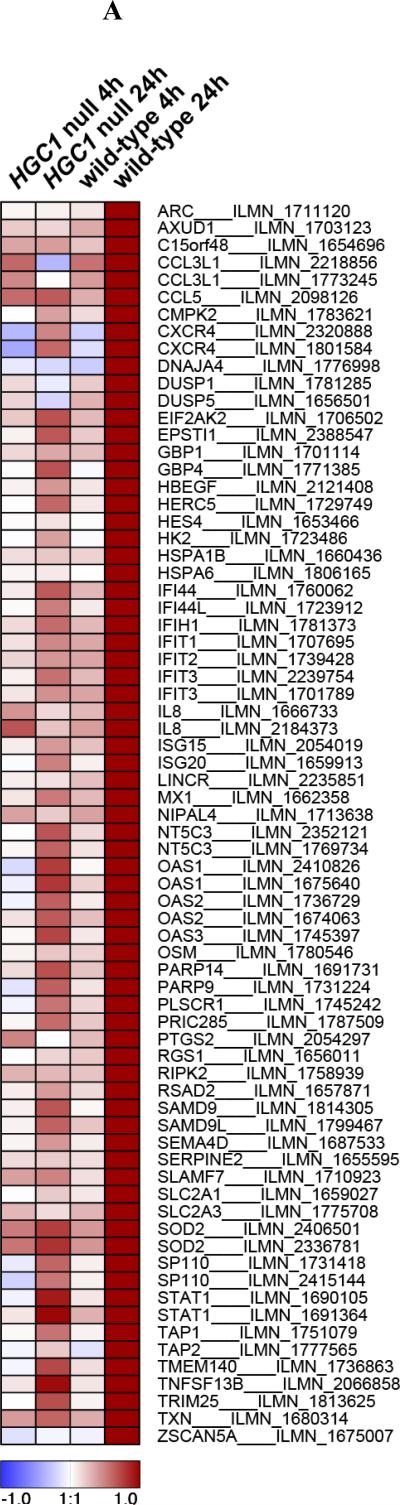
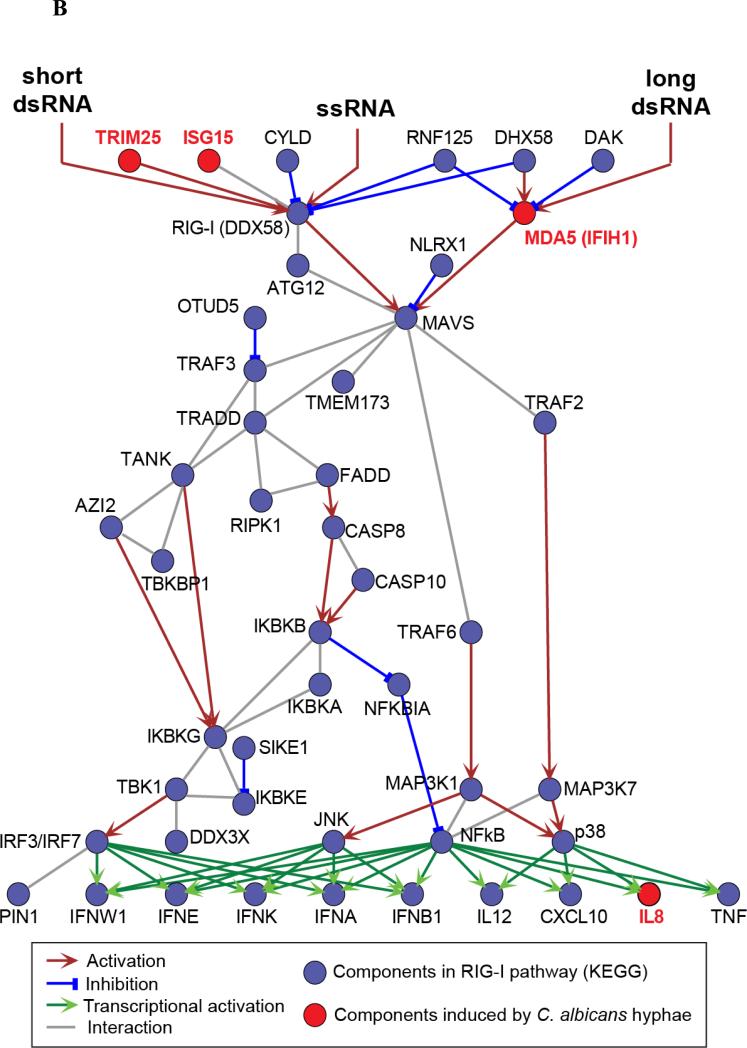
Candida albicans is a dimorphic fungus that exists either in a colonizing yeast form, or as an invasive filamentous form (hyphae). In a first set of experiments we aimed to identify the specific transcription profile induced in human macrophages by C. albicans hyphae. To this end, we profiled gene expression in macrophages stimulated with wild-type and HGC1 null (yeast-locked) strains of C. albicans for either 4 or 24 hours, the latter time point being required for fungal germination into hyphae (Figure 1). HGC1 is required for the yeast-to-hyphae transition and HGC1 null Candida fails to form hyphae, thus remaining in the colonizing yeast form [14]. We identified 62 genes exhibiting significant differential expression specifically in macrophages stimulated with Candida hyphae for 24h, but not for 4h, nor in macrophages stimulated with yeast-locked Candida (Benjamini-Hochberg (BH)-corrected P < 0.05 and > 2-fold change in expression, compared to unstimulated macrophages, Figure 1A). Many of these genes are involved in interferon (IFN) signaling, consistent with a previous study uncovering an important role for type I interferons in the response to Candida [13]. Interestingly, four of the genes induced by Candida hyphae stimulation (IFIH1, ISG15, IL8, and TRIM25) are key components of the RIG-I-like receptor (RLR) signaling pathway, which is significantly more than expected for a random set of genes (P = 4.3 × 10−3, 11.5-fold enrichment, Table 1 and Figure 1B).
Figure 1. Transcriptional changes in macrophages stimulated with Candida albicans.
(A) The heatmap shows differential gene expression after 4h or 24h stimulation of human macrophages with yeast-locked HGC1 null Candida albicans (which are unable to form hyphae), or wild-type invasive Candida albicans (that can form hyphae), compared to expression levels in unstimulated macrophages (control). 62 genes exhibited a significant change in expression level (Benjamini-Hochberg-corrected P < 0.05 and > 2-fold change in expression) specifically after 24h stimulation with wild-type Candida, during which germination into hyphae takes place. Signal-to-noise ratio scaled to the maximum absolute deviation is shown for each probe corresponding to the 62 differentially expressed genes. (B) Candida albicans hyphae-induced genes, IFIH1, TRIM25, ISG15 and IL8 (indicated in red), are key components of the RIG-I-like receptor (RLR) signaling pathway. These genes represent both the MDA5 (IFIH1) and RIG-I (ISG15 and TRIM25) branches, as well as inflammatory cytokines that are produced by activation of the pathway (IL8). Figure based on the KEGG map of the RLR pathway [49].
Table 1.
KEGG pathway enrichment for genes that are specifically induced in macrophages stimulated with wild-type Candida for 24 hours.
| KEGG identifier | Pathway | P value | Fold enrichment | Genes |
|---|---|---|---|---|
| hsa04622 | RIG-I-like receptor signaling pathway | 4.3 × 10−3 | 11.5 | IFIH1, ISG15, IL8, TRIM25 |
| hsa04060 | Cytokine-cytokine receptor interaction | 6.6 × 10−3 | 4.7 | OSM, TNFSF13B, IL8, CXCR4, CCL3L1, CCL5 |
| hsa04612 | Antigen processing and presentation | 6.6 × 10−3 | 9.8 | TAP2, TAP1, HSPA6, HSPA1B |
| hsa04062 | Chemokine signaling pathway | 1.1 × 10−2 | 5.4 | IL8, CXCR4, CCL3L1, CCL5, STAT1 |
| hsa04621 | NOD-like receptor signaling pathway | 3.4 × 10−2 | 9.8 | IL8, RIPK2, CCL5 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 4.0 × 10−2 | 9.0 | IL8, HBEGF, CCL5 |
RIG-I-like receptors are well-known intracellular receptors of viral RNA leading to the production of type I IFNs and proinflammatory cytokines [15]. IFIH1 (interferon induced with helicase C domain 1), with its protein product known as MDA5 (melanoma differentiation-associated protein 5), is a receptor of long double-stranded RNA (dsRNA), which constitutes one branch of the RLR pathway (Figure 1B). ISG15 and TRIM25 are involved in the RIG-I branch of that pathway, which is activated by shorter, 5’ triphosphate-containing dsRNA. The two branches of the pathway converge at the signaling adaptor MAVS (also known as IPS-1). Thus, the invasive form of Candida induces expression of components of two branches of the virus-recognition RLR pathway in macrophages.
Genetic variation linked to IFIH1 modulates susceptibility to candidemia
To validate this unexpected role for components of the virus recognition RLR pathway, IFIH1, ISG15, IL8, and TRIM25, in invasive Candida infection, we investigated whether genetic variation linked to these genes correlates with susceptibility to candidemia. For this, a cohort of 227 Caucasian patients with candidemia and 176 matched controls (described in detail in [6]) was genotyped using the Immunochip single-nucleotide polymorphism (SNP) array, which contains ~200,000 SNPs focused on genomic regions known to be involved in immune-mediated diseases [16]. We checked for the presence of SNPs associated with the four RLR pathway components on the array and tested the loci for association with candidemia. ISG15 and TRIM25 were not covered, and genetic variation linked to IL8 did not reveal any association.
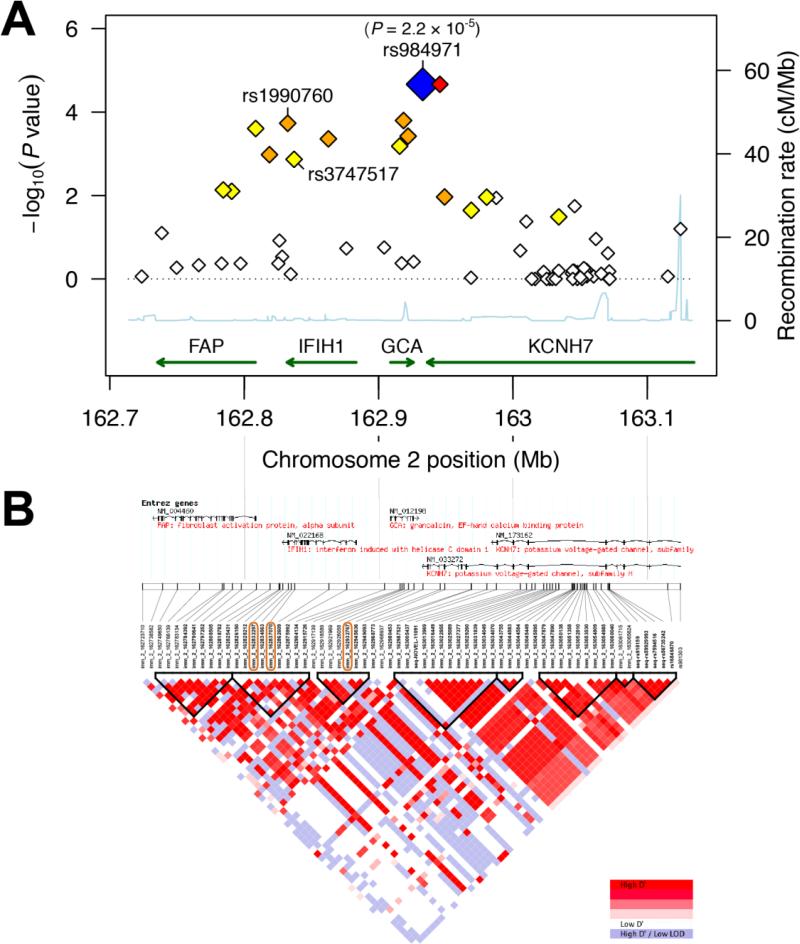
In contrast, analysis of 64 SNPs associated with IFIH1 revealed strong allelic and genotypic association with susceptibility to candidemia (Figure 2A and Table 2). The IFIH1 locus is present in a 405 kb genomic region on chromosome 2 with low recombination rates (Figure 2A) and accompanying strong linkage disequilibrium (LD) in both the candidemia cohort (Figure 2B and Figure S1D) and the HapMap CEU population (Figures S1A-C), as also seen in other studies [17-19]. Besides IFIH1, the LD region contains the genes FAP (fibroblast activation protein), GCA (grancalcin), and part of KCNH7 (potassium voltage-gated channel subfamily H member 7). Of the 64 SNPs in the FAP-IFIH1-GCA-KCNH7 LD region, 15 have genotype count distributions that differ significantly between cases and controls after applying Benjamini-Hochberg multiple testing correction (BH-corrected genotypic P < 0.05, Table S1). The significant SNPs are distributed mainly across the central part of the LD region (Figure 2A). Indeed, the association with candidemia does not extend beyond the LD region (Figure S2). An intergenic SNP between GCA and KCNH7 shows the strongest association with candidemia in the FAP-IFIH1-GCAKCNH7 LD region (rs984971, genotypic P = 2.2 × 10−5, allelic P = 2.2 × 10−4, odds of disease 0.43 – 0.77, Table 2). Interestingly, although the Immunochip covers missense coding variants in all four genes, only IFIH1 harbors significant missense SNPs (rs1990760 – Ala946Thr, and rs3747517 – His843Arg, which are also in strong LD with each other; HapMap CEU: D’ = 1, r2 = 0.42 – candidemia cohort: D’ = 1, r2 = 0.55).
Figure 2. (A) Regional association plot and (B) linkage disequilibrium (LD) map for the FAP-IFIH1-GCA-KCNH7 LD region on chromosome 2.
(A) 64 SNPs with MAF > 5% in 403 Caucasian individuals of the candidemia cohort (cases and controls together) were assessed for genotypic association with candidemia. The resulting - log10(genotypic P values) (left y-axis) are plotted as a function of genomic coordinates (hg18, x-axis). The blue diamond highlights the most significant SNP with its P value (rs984971). rs1990760 and rs3747517 are the only two significant missense SNPs; both are in the coding region of IFIH1. Recombination rates, estimated from the CEU, YRI and JPT+CHB HapMap populations (HapMap 2, Release 22) [56], are plotted to reflect the local LD structure (right y-axis, cyan line). SNPs are colored according to the degree of LD with the most significant SNP, rs984971 (R-squared, calculated across the controls in the candidemia cohort; from strong to weak LD - red: r2≥0.8; orange: 0.5≤r2<0.8; yellow: 0.2≤ r2<0.5; white: r2<0.2). Genes with their direction of transcription are shown at the bottom; KCNH7 is only partly in this region. (B) LD patterns across the 405 kb FAP-IFIH1-GCA-KCNH7 LD region are calculated based on genotypes of control individuals in the candidemia cohort, measured using the Immunochip SNP array. The intersections of the diagonals between pairs of SNPs are colored according to the degree of LD, which is calculated as D' and LOD: SNPs with D' values between 0 and 1 and with LOD ≥ 2 are colored from white to red. Haplotype blocks (triangles with bold black borders) are regions where at least 95% of SNPs are in strong LD, defined by high D' values [57]. Chromosome 2 coordinates (hg18) and Entrez genes are shown at the top. Orange boxes around SNP identifiers indicate the top SNP and two IFIH1 m SNPs significantly associated with susceptibility to candidemia (see Table 2). The corresponding R-squared LD map for the candidemia cohort is depicted in Figure S1D. See Figures S1A-C for R-squared and D’/LOD LD maps calculated based on the HapMap CEU population.
Table 2.
Selection of SNPs in the FAP-IFIH1-GCA-KCNH7 LD region that are significantly associated with susceptibility to candidemia
| SNP | Immunochip | Closest gene(s) | Alleles (dbSNP) | Functional class (AA change) | BH-corrected genotypic P value | |
|---|---|---|---|---|---|---|
| rs984971 | imm_2_162932767 | GCA | KCNH7 | A/G | intergenic | 6.9 × 10−4 | |
| Genotypes | GG | GA | AA | |||
| Controls | 25 (14.2%) | 103 (58.5%) | 48 (27.3%) | P = 2.2 × 10−5 | ||
| Cases | 25 (11.0%) | 89 (39.2%) | 113 (49.8%) | |||
| Alleles | G | A* | ||||
| Controls | 153 (43.5%) | 199 (56.5%) | P = 2.2 × 10−4 | |||
| Cases | 139 (30.6%) | 315 (69.4%) | OR, G vs. A = 0.57 (0.43 – 0.77) | |||
| rs1990760 | imm_2_162832297 | IFIH1 | C/T | missense (Ala946Thr) | 3.0 × 10−3 | |
| Genotypes | CC | CT | TT | |||
| Controls | 31 (17.6%) | 99 (56.3%) | 46 (26.1%) | P = 1.9 × 10−4 | ||
| Cases | 37 (16.3%) | 87 (38.3%) | 103 (45.4%) | |||
| Alleles | C | T* | ||||
| Controls | 161 (45.7%) | 191 (54.3%) | P = 3.7 × 10−3 | |||
| Cases | 161 (35.5%) | 293 (64.5%) | OR, C vs. T = 0.65 (0.49 – 0.87) | |||
| rs3747517 | imm_2_162837070 | IFIH1 | T/C (A/G) | missense (His843Arg) | 8.7 × 10−3 | |
| Genotypes | TT | TC | CC | |||
| Controls | 12 (6.8%) | 88 (50.0%) | 76 (43.2%) | P = 1.4 × 10−3 | ||
| Cases | 20 (8.8%) | 73 (32.2%) | 134 (59.0%) | |||
| Alleles | T | C* | ||||
| Controls | 112 (31.8%) | 240 (68.2%) | P = 3.3 × 10−2 | |||
| Cases | 113 (24.9%) | 341 (75.1%) | OR, T vs. C = 0.71 (0.52 – 0.97) | |||
Genotypic and allelic associations were assessed using the Fisher's exact test. P values are shown next to the corresponding contingency tables. Odds ratios (OR, with 95% confidence intervals) are reported for the allelic association tests and represent the odds of disease for individuals carrying the non-risk allele versus the risk allele. Risk alleles are denoted by an asterisk. BH-corrected genotypic P value: Benjamini-Hochberg correction of the genotypic association test P values for testing multiple SNPs (64 in total). Immunochip: the identifier of the SNP on the Immunochip SNP array. Alleles: the alleles measured with the Immunochip, and the complementary alleles reported by dbSNP (build 138), if they are different. The table shows the top SNP in the LD region, along with the only two significant missense SNPs. All SNPs tested are in Hardy–Weinberg equilibrium in the controls (P > 1 × 10−3). See Table S1 for the full list of significant SNPs.
The strong LD across the region makes that our genetic data cannot definitively assert which SNP(s), and hence which gene(s), are responsible for the observed association with candidemia. However, it has previously been shown that rs1990760 and rs3747517, among other IFIH1-linked SNPs, are quantitative trait loci (QTLs) to IFIH1 expression, with the candidemia risk alleles correlating with higher IFIH1 expression in PBMCs [18, 20]. Thus, the presence of (missense) SNPs in the IFIH1 locus showing significant association with the infection, together with the observed IFIH1 upregulation in macrophages responding to Candida hyphae, strongly suggest the involvement of IFIH1 in the host defense against candidemia.
IFIH1 is strongly upregulated upon Candida stimulation of PBMCs, while FAP, GCA, and KCNH7 are not
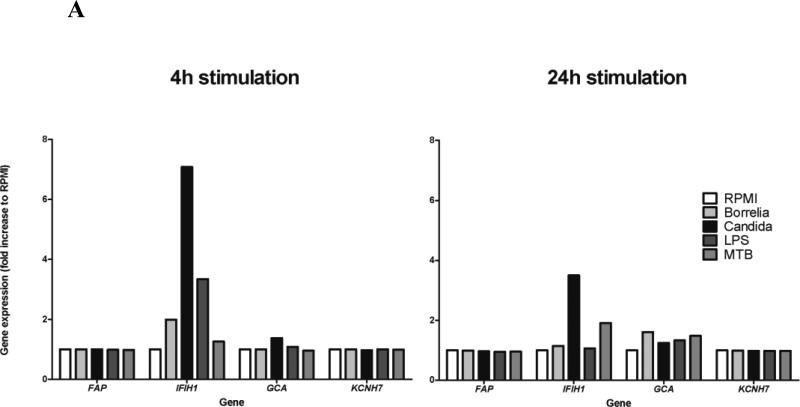
To provide additional evidence regarding which genes in the FAP-IFIH1-GCA-KCNH7 LD region are important for the host response to Candida, we measured gene expression in peripheral blood mononuclear cells (PBMCs) from healthy volunteers in the presence of C. albicans and various other stimuli for 4 or 24 hours. 4h stimulation with bacterial stimuli such as Borrelia burgdorferi and Escherichia coli-derived lipopolysaccharide (LPS) resulted in increased expression of IFIH1, but not of the other LD region genes (Figure 3A). Importantly, stimulation with C. albicans resulted in an even higher IFIH1 expression. Of the other genes, only GCA was also induced by C. albicans, but this expression was negligible compared to IFIH1. Similar trends were apparent after 24h of stimulation: C. albicans induced high expression of IFIH1 (though lower than at 4h), and moderate expression of GCA, while FAP and KCNH7 show no effect (Figure 3A). Heat-killed Mycobacterium tuberculosis also induced expression of IFIH1 at 24h, although to a lesser extent than C. albicans.
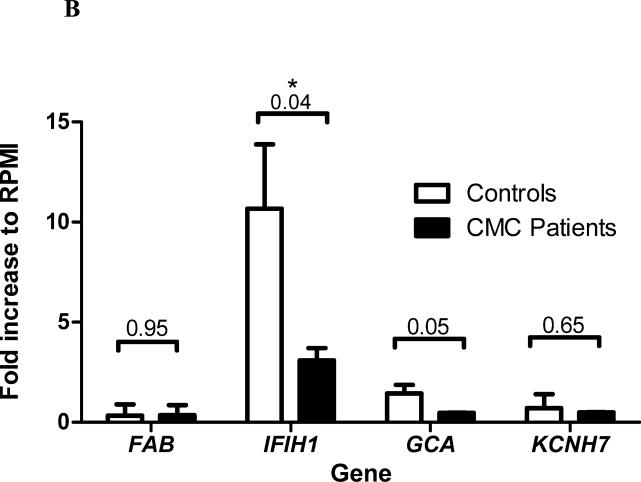
Figure 3. Transcriptional response of genes in the FAP-IFIH1-GCA-KCNH7 LD region to various microbial stimuli.
(A) Peripheral blood mononuclear cells (PBMCs) from healthy volunteers were stimulated for either 4 or 24 hours with Borrelia burgdorferi, Candida albicans, Escherichia coli-derived lipopolysaccharide (LPS), or Mycobacterium tuberculosis (MTB). Gene expression was measured using microarrays and normalized to the control RPMI condition (untreated). (B) Gene expression (Mean ± SD) in PBMCs of healthy controls (n=3) and patients suffering from chronic mucocutaneous candidiasis (CMC) (n=2) were stimulated with C. albicans for 4 hours. P values were calculated using the Welch-corrected t test.
To improve support for a likely involvement of IFIH1 induction in antifungal host defense, we compared the expression patterns of IFIH1 in cells from three healthy individuals with cells isolated from two patients suffering from chronic mucocutaneous candidiasis (CMC) due to a deleterious STAT1 mutation [10]. For this, PBMCs were isolated and stimulated with C. albicans for 4 hours, after which gene expression was measured using RNA sequencing. CMC patients expressed significantly lower levels of IFIH1 after stimulation with C. albicans than healthy individuals (P = 0.04, Welch-corrected t test, Figure 3B). The differences in expression were largely specific for IFIH1, although we also observed minor differences in the expression of GCA (P = 0.05, Welch-corrected t test, Figure 3B). Together, the observations in healthy individuals and CMC patients indicate that expression of IFIH1, and not expression of the other genes in the FAP-IFIH1-GCA-KCNH7 LD region, is specifically induced by stimulation with C. albicans but not by bacterial stimuli. This suggests that IFIH1 plays an important role in the antifungal host defense.
Genetic variants in IFIH1 are associated with an altered cytokine profile in response to Candida
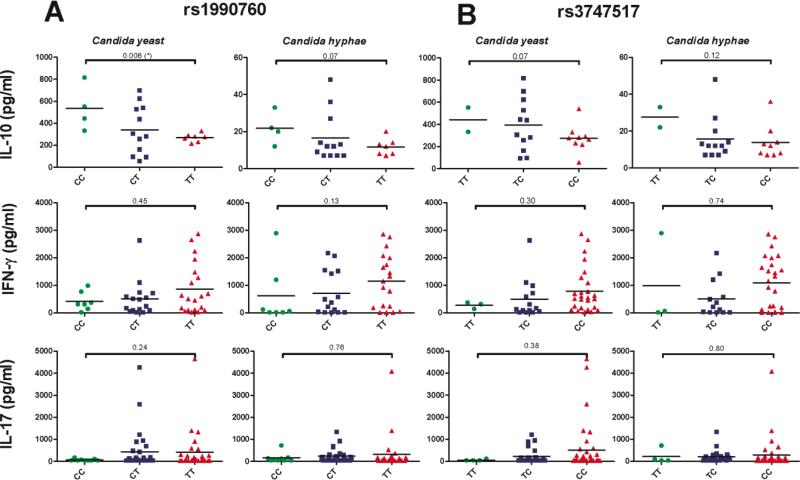
To investigate the functional consequences of genetic variants associated with IFIH1 that predispose individuals to candidemia (rs984971, rs1990760 and rs3747517, Table 2), we correlated the genotypes of these SNPs with in vitro cytokines levels upon Candida stimulation in healthy volunteers. IL-10, IFN-γ, and IL-17 levels produced by PBMCs were measured after stimulation with either C. albicans yeasts or hyphae. Interestingly, we observed a trend towards an increased capacity to release the proinflammatory cytokines IFN-γ and IL-17 in cells isolated from individuals homozygous for the risk allele for both IFIH1 missense polymorphisms (TT for rs1990760; CC for rs3747517) (Figure 4). In contrast, levels of the anti-inflammatory IL-10 tended to be lower in individuals carrying the risk allele for these SNPs (Figure 4). This decrease was consistent between stimulation with the C. albicans yeast and hyphal forms. The top SNP associated with candidemia (intergenic rs984971) did not reveal the same general trends as the two missense SNPs (Figure S3). Furthermore, stimulation with Mycobacterium tuberculosis did not reveal clear correlations between cytokine levels and IFIH1 missense SNP genotypes (Figure S4), strengthening the previous observation that the involvement of IFIH1 under the stimuli studied here is specific for Candida. Thus, genetic variation in IFIH1 influences anti-Candida cytokine profiles in vitro.
Figure 4. IFIH1 missense SNP genotypes correlate with Candida-induced cytokine levels.
PBMCs from healthy volunteers with different genotypes for (A) rs1990760 (candidemia risk allele T) and (B) rs3747517 (candidemia risk allele C) were stimulated in vitro with either C. albicans yeast or hyphae. Cytokine levels (scatterplots with mean indicated) were measured after 24 hours (IL-10) or 7 days (IL-17 and IFN-γ) by enzyme linked immunosorbent assay (ELISA). P values were calculated using the Mann-Whitney U test comparing cytokine levels of the two homozygous genotypes.
Missense SNPs could affect MDA5 protein function
We next sought to gain insight into the possible consequences of having alternative alleles at the IFIH1 missense SNPs on MDA5 protein function. MDA5 arose from a duplication of the MDA5/LGP2 gene in the ancestor of jawed vertebrates [21]. Analysis of MDA5 orthologs in 59 jawed vertebrates reveals that both amino acids of rs1990760 (Ala946Thr) are common (alanine: 36/59=61%, threonine: 10/59=17%), while for rs3747517 (His843Arg) the type sequence of human is the only sequence that encodes a histidine (similar data in [22]). The occurrence of alternative alleles at both SNPs suggests that both lead to a functional protein, although there may be functional differences.
Residue 946 is part of an intrinsically disordered loop [23] in the C-terminal domain (CTD) of MDA5. The equivalent loop is rigid in RIG-I, and this differential flexibility contributes to the different RNA binding preferences: displacement of the loop upon dsRNA binding allows MDA5 to recognizes long dsRNA, while the loop in RIG-I specifically caps shorter dsRNA [24]. The human MDA5 crystal structure has an arginine at position 843, which interacts with the negatively charged RNA backbone (Figure S5). Histidine would weaken this electrostatic interaction because it is less often positively charged at physiological pH than arginine. Furthermore, position 843 is close to the interface likely involved in interactions between MDA5 monomers (Figure S5) [24]. The formation of MDA5 filaments along the RNA is critical for downstream activation of MAVS [25]. Indeed, mutation of nearby residues 841 and 842 disrupts filament formation and signaling [24]. Thus, the Ala946Thr and His843Arg substitutions could alter dsRNA binding selectivity and affinity, and the latter might also affect signaling activity. Furthermore, as noted above, these and other SNPs also correlate with IFIH1 gene expression.
Mda5 knockout mice have reduced cytokine production in response to C. albicans
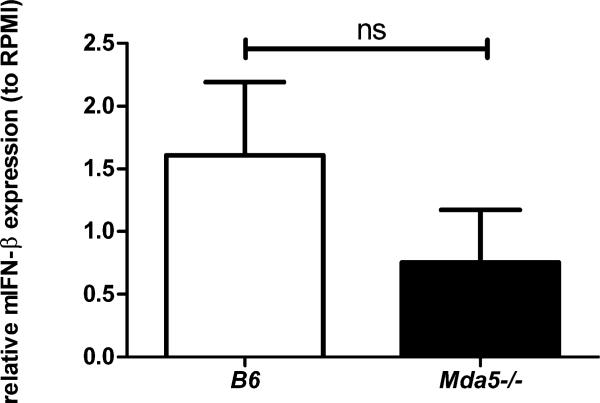
To provide an additional argument for the role of MDA5 in the anti-Candida response, we stimulated splenocytes from Mda5 knockout mice and B6 control mice (C57BL/6J) with C. albicans yeasts or hyphae. Quantitative PCR analysis showed a defective production of interferon β induced by Candida in the cells isolated from mice deficient in Mda5 compared to controls, comparable to the positive control stimulus poly I:C (Figure 5). Similarly, the IL-6 and IL-10 cytokine responses were also lower under Candida stimulation of cells from Mda5 knockout mice, and these differences were more pronounced when cells were stimulated with hyphae compared to the yeast form (Figure S6).
Figure 5.
Relative gene expression (mean ± SEM) of mouse interferon β (mIFN-β) in splenocytes isolated from control B6 control mice (C57BL/6J) and Mda5 knockout mice, upon stimulation with C. albicans hyphae (106/ml) (n=5/group). The P value was not significant at α < 0.05 (calculated using the Welch-corrected t test).
Discussion
In the present study we propose that the pattern recognition receptor MDA5, which belongs to the RIG-I-like receptor (RLR) family and is known for its role in antiviral immunity, is also involved in antifungal host defense. Through a combined approach involving transcriptomics, genetic susceptibility studies and immunological validation, these experiments demonstrate that MDA5 modulates cytokine production induced in human leukocytes by C. albicans, while genetic variants in the IFIH1 gene that encodes MDA5 influence susceptibility to disseminated candidiasis. Based on these data and the known role of MDA5 in interferon production, it is most likely that this effect is mediated through induction of type I interferons (IFN).
MDA5 is composed of two N-terminal CARD domains, responsible for the signaling activity, coupled to a central DExD/H box helicase domain and a C-terminal domain (CTD), which together recognize the long dsRNA often associated with replicating viruses [15]. RNA binding causes MDA5 to interact with MAVS in a CARD-dependent manner, leading to downstream signaling and subsequent activation of transcription factors such as IRF3, IRF7, and NF-κB. These factors induce the expression of type I interferons that are a mainstay of antiviral host defense [26]. Surprisingly, transcriptome analysis aiming to identify the immunological programs induced in human macrophages by Candida identified the MDA5/RIG-I signaling pathway as the top target. C. albicans is a dimorphic fungus, and germination from yeasts to hyphae is a central process for the invasion of tissues, often at the level of the mucosa. We therefore aimed to identify the mechanisms through which human macrophages respond specifically to the germination process. We approached this by stimulating the macrophages either with wild-type C. albicans that germinates during the 24h culture period, or with the hgc1-defective strain that is unable to form hyphae. Pathway analysis identified the MDA5 pathway as essential among the macrophage genes that were specifically induced by hyphae. The hypothesis that MDA5 is important for host defense against Candida is strengthened by the observation that MDA5 induction was significantly reduced in cells isolated from patients suffering from chronic mucocutaneous candidiasis.
Additional evidence that MDA5 is indeed important for human antifungal defense was provided by the assessment of genetic variation predisposing to candidemia. In a cohort of candidemia patients we found a strong association between the disease and genetic variation in the linkage disequilibrium (LD) region on chromosome 2 that contains IFIH1. While genetic variation in IFIH1 has previously been shown to influence susceptibility to several autoimmune diseases such as type I diabetes, Graves’ disease, and multiple sclerosis [18-20, 27-32], this is the first report of polymorphisms in IFIH1 linked to a fungal infection. These data are in line with recent studies showing that polymorphisms in other pattern recognition receptors such as TLRs [6, 33-35], or components of the IFN pathway such as STAT1 or IRF1 [13], also influence susceptibility to systemic fungal infections.
It is important to point out that the candidemia-associated LD region contains several genes: FAP (fibroblast activation protein), IFIH1 (interferon induced with helicase C domain 1), GCA (grancalcin) and KCNH7 (potassium voltage-gated channel subfamily H member 7). IFIH1 and grancalcin are the strongest a priori candidates for causing the susceptibility to candidemia because, in contrast to FAP and KCNH7, these genes have known functions in immunity. Grancalcin is abundant in macrophages and neutrophils, particularly those recovered from sites of bacterial infection [36], and is thought to mediate leukocyte adhesion and migration [37]. To obtain direct evidence for a role of genes in the LD region in candidemia susceptibility, we assessed their gene expression in PBMCs stimulated with Candida. IFIH1 was strongly induced and GCA to lesser extent, while the other genes did not show any expression changes. IFIH1 and GCA are divergently transcribed neighboring genes with ~25kb separating their transcription start sites. As such neighboring genes tend to be co-expressed more often than random genes [38, 39], the moderate upregulation of GCA in response to Candida stimulation could be a by-effect of the strong induction of IFIH1, although the genes are still relatively far apart.Importantly, grancalcin-deficient (Gca−/−, homozygous mutant) mice do not have an increased susceptibility to candidiasis compared to wild-type mice [40], which strongly argues against an important role for GCA in the immune response against C. albicans. Therefore, we concluded that genetic variants acting at IFIH1 are the most likely cause of the association of the FAP-IFIH1-GCA-KCNH7 LD region with candidemia.
The candidemia risk alleles of the IFIH1-linked SNPs identified in our study have previously been shown to lead to higher expression of IFIH1 in PBMCs [18, 20]. Furthermore, our protein structure analyses indicate a possibly stronger RNA binding by MDA5 in the presence of an arginine at position 843, which is encoded by the risk allele C of rs3747517. To gain insight into the downstream immunological effects of IFIH1 variants, we measured cytokine levels produced by PBMCs from donors with different genotypes for the two IFIH1 missense SNPs (rs1990760 and rs3747517). These data indicate that cells from individuals bearing the candidemia risk alleles produce more proinflammatory cytokines (IFN-γ and IL-17) and less anti-inflammatory IL-10 in response to C. albicans yeast and hyphal forms than cells bearing the protective alleles.
These observations bring into discussion the nature of the involvement of MDA5 in the host defense against Candida. MDA5 activates the RLR pathway leading to the production of type I interferons during viral infections [26]. A similar biological activity during Candida stimulation was shown by our data from Mda5 knockout mouse splenocytes, which displayed a decreased capacity to induce IFN-β compared to control cells. A role for type I IFNs in antifungal immunity has been recently proposed [13], and MDA5 is likely the receptor that is at least partially responsible for the type I IFN induction during C. albicans infection.
A recent study showed that rare IFIH1 variants causing the type I interferon-mediated pathology of the Aicardi-Goutières syndrome are gain of function mutations that lead to stronger RNA binding, and thereby increase baseline and RNA-induced IFN signaling [41]. Therefore, other mutations leading to inherently increased expression or activity of MDA5, such as those associated with candidemia as identified in this study, are likely to increase IFN production in a similar manner. Aberrant production of type I IFNs in turn might be cause imbalances in the immune response that are reflected in the observed alterations in the levels of other cytokines.
The apparent deleterious effect of MDA5 hyperactivity on the anti-Candida host defense is consistent with observations that type I IFNs could be harmful for this response as well: mice defective in type I interferon receptors (Ifnar1−/− mice) are actually more resistant to systemic Candida infections [42]. This is also in line with our findings that PBMCs with the candidemia risk genotype in IFIH1 tend to release more inflammatory cytokines. The hypothesis that MDA5 has a negative effect on the anti-Candida immune response has been proven by a very recent elegant study demonstrating that Mda5−/− mice are more resistant to disseminated candidiasis (Malireddi and Kanneganti, personal communication).
It is currently unclear which ligands cause activation of MDA5 in Candida infection. Candida is mainly recognized by cell surface pattern recognition receptors such as TLRs and C-type lectin receptors (CLRs), after which the fungus is internalized and subsequently digested in the phagolysosome [43, 44]. It is conceivable that, during this process, Candida-derived structures may leak from these organelles and enter the cytoplasm, a process earlier described for the recognition of mycobacterial peptidoglycans by the cytoplasmic receptor NOD2 [45-47]. Interestingly, a recent study has suggested that NOD2 is also important for the recognition of Candida chitin [48]. Nevertheless, we currently have no experimental evidence to support that either the wild-type form or a variant form of MDA5 has other ligands than the described RNAs, and future studies should investigate this aspect.
In conclusion, this study demonstrates that the viral receptor MDA5 has an important role in modulating the innate immune response against the fungal pathogen C. albicans. Genetic variation in this gene is associated with candidemia, which is likely mediated by a shift in cytokine responses. Future research should aim to shed light on the exact mechanisms through which MDA5 participates in the immune response against the fungus. Nevertheless, the possible deleterious effects of MDA5-dependent stimulation during systemic candidiasis shown by our data suggest its potential usefulness as a novel therapeutic target.
Supplementary Material
Acknowledgments
MJ and MGN were supported by a European Research Council Consolidator Grant (nr. 310372 to MGN). RvdL and MAH were supported by the Virgo consortium, funded by the Dutch government project number FES0908, and by the Netherlands Genomics Initiative (NGI) project number 050-060-452. CW was supported by the European Research Council Advanced Grant, ERC-671274. We thank Martin Oti for helpful discussions.
References
- 1.Gudlaugsson O, Gillespie S, Lee K, et al. Attributable mortality of nosocomial candidemia, revisited. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2003 Nov 1;37(9):1172–7. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2004 Aug 1;39(3):309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Miller LG, Hajjeh RA, Edwards JE., Jr. Estimating the cost of nosocomial candidemia in the united states. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2001 Apr 1;32(7):1110. doi: 10.1086/319613. [DOI] [PubMed] [Google Scholar]
- 4.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2005 Nov 1;41(9):1232–9. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 5.Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. Genetic susceptibility to Candida infections. EMBO molecular medicine. 2013 Jun;5(6):805–13. doi: 10.1002/emmm.201201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plantinga TS, Johnson MD, Scott WK, et al. Toll-like receptor 1 polymorphisms increase susceptibility to candidemia. The Journal of infectious diseases. 2012 Mar 15;205(6):934–43. doi: 10.1093/infdis/jir867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullberg BJ, Verweij PE, Akova M, et al. European expert opinion on the management of invasive candidiasis in adults. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011 Sep;17(Suppl 5):1–12. doi: 10.1111/j.1469-0691.2011.03615.x. [DOI] [PubMed] [Google Scholar]
- 8.Guery BP, Arendrup MC, Auzinger G, et al. Management of invasive candidiasis and candidemia in adult non-neutropenic intensive care unit patients: Part II. Treatment. Intensive care medicine. 2009 Feb;35(2):206–14. doi: 10.1007/s00134-008-1339-6. [DOI] [PubMed] [Google Scholar]
- 9.Glocker EO, Hennigs A, Nabavi M, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. The New England journal of medicine. 2009 Oct 29;361(18):1727–35. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Veerdonk FL, Plantinga TS, Hoischen A, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. The New England journal of medicine. 2011 Jul 7;365(1):54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Okada S, Kong XF, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. The Journal of experimental medicine. 2011 Aug 1;208(8):1635–48. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babula O, Lazdane G, Kroica J, Linhares IM, Ledger WJ, Witkin SS. Frequency of interleukin-4 (IL-4) -589 gene polymorphism and vaginal concentrations of IL-4, nitric oxide, and mannose-binding lectin in women with recurrent vulvovaginal candidiasis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2005 May 1;40(9):1258–62. doi: 10.1086/429246. [DOI] [PubMed] [Google Scholar]
- 13.Smeekens SP, Ng A, Kumar V, et al. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nature communications. 2013;4:1342. doi: 10.1038/ncomms2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Wang Y, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. The EMBO journal. 2004 Apr 21;23(8):1845–56. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010 Mar 19;140(6):805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Trynka G, Hunt KA, Bockett NA, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nature genetics. 2011 Dec;43(12):1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu HQ, Marchand L, Grabs R, Polychronakos C. The association between the IFIH1 locus and type 1 diabetes. Diabetologia. 2008 Mar;51(3):473–5. doi: 10.1007/s00125-007-0895-6. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Wang H, Jin Y, et al. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Human molecular genetics. 2009 Jan 15;18(2):358–65. doi: 10.1093/hmg/ddn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth DJ, Cooper JD, Bailey R, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature genetics. 2006 Jun;38(6):617–9. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 20.Downes K, Pekalski M, Angus KL, et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PloS one. 2010;5(9) doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee K, Korithoski B, Kolaczkowski B. Ancient origins of vertebrate-specific innate antiviral immunity. Molecular biology and evolution. 2014 Jan;31(1):140–53. doi: 10.1093/molbev/mst184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molineros JE, Maiti AK, Sun C, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS genetics. 2013;9(2):e1003222. doi: 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Lee R, Buljan M, Lang B, et al. Classification of Intrinsically Disordered Regions and Proteins. Chemical reviews. 2014 Apr 29; doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu B, Peisley A, Richards C, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013 Jan 17;152(1-2):276–89. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 25.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011 Aug 5;146(3):448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006 May 4;441(7089):101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Zhou D, Zhang Z, et al. Genetic variants in IFIH1 play opposite roles in the pathogenesis of psoriasis and chronic periodontitis. International journal of immunogenetics. 2012 Apr;39(2):137–43. doi: 10.1111/j.1744-313X.2011.01068.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Wang Z, Xu K, et al. IFIH1 gene polymorphisms in type 1 diabetes: genetic association analysis and genotype-phenotype correlation in Chinese Han population. Autoimmunity. 2012 May;45(3):226–32. doi: 10.3109/08916934.2011.633134. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland A, Davies J, Owen CJ, et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves' disease susceptibility. The Journal of clinical endocrinology and metabolism. 2007 Aug;92(8):3338–41. doi: 10.1210/jc.2007-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cen H, Wang W, Leng RX, et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013 Nov;46(7):455–62. doi: 10.3109/08916934.2013.796937. [DOI] [PubMed] [Google Scholar]
- 31.Martinez A, Santiago JL, Cenit MC, et al. IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. European journal of human genetics : EJHG. 2008 Jul;16(7):861–4. doi: 10.1038/ejhg.2008.16. [DOI] [PubMed] [Google Scholar]
- 32.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009 Apr 17;324(5925):387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Graaf C, Kullberg BJ, Joosten L, et al. Functional consequences of the Asp299Gly Toll-like receptor-4 polymorphism. Cytokine. 2005 Jun 7;30(5):264–8. doi: 10.1016/j.cyto.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 34.van der Graaf CA, Netea MG, Drenth IP, te Morsche RH, van der Meer JW, Kullberg BJ. Candida-specific interferon-gamma deficiency and toll-like receptor polymorphisms in patients with chronic mucocutaneous candidiasis. The Netherlands journal of medicine. 2003 Nov;61(11):365–9. [PubMed] [Google Scholar]
- 35.Van der Graaf CA, Netea MG, Morre SA, et al. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. European cytokine network. 2006 Mar;17(1):29–34. [PubMed] [Google Scholar]
- 36.Liu F, Shinomiya H, Kirikae T, Hirata H, Asano Y. C haracterization of murine grancalcin specifically expressed in leukocytes and its possible role in host defense against bacterial infection. Bioscience, biotechnology, and biochemistry. 2004 Apr;68(4):894–902. doi: 10.1271/bbb.68.894. [DOI] [PubMed] [Google Scholar]
- 37.Lollike K, Johnsen AH, Durussel I, Borregaard N, Cox JA. Biochemical characterization of the penta-EF-hand protein grancalcin and identification of L-plastin as a binding partner. The Journal of biological chemistry. 2001 May 25;276(21):17762–9. doi: 10.1074/jbc.M100965200. [DOI] [PubMed] [Google Scholar]
- 38.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome research. 2004 Jan;14(1):62–6. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008 Mar;91(3):243–8. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Roes J, Choi BK, Power D, Xu P, Segal AW. Granulocyte function in grancalcin-deficient mice. Molecular and cellular biology. 2003 Feb;23(3):826–30. doi: 10.1128/MCB.23.3.826-830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice GI, del Toro Duany Y, Jenkinson EM, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature genetics. 2014 May;46(5):503–9. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majer O, Bourgeois C, Zwolanek F, et al. Type I interferons promote fatal immunopathology by regulating inflammatory monocytes and neutrophils during Candida infections. PLoS pathogens. 2012;8(7):e1002811. doi: 10.1371/journal.ppat.1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Netea MG, Gow NA, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. The Journal of clinical investigation. 2006 Jun;116(6):1642–50. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Djeu JY. Role of tumor necrosis factor and colony-stimulating factors in phagocyte function against Candida albicans. Diagnostic microbiology and infectious disease. 1990 Sep-Oct;13(5):383–6. doi: 10.1016/0732-8893(90)90007-i. [DOI] [PubMed] [Google Scholar]
- 45.Ferwerda G, Girardin SE, Kullberg BJ, et al. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS pathogens. 2005 Nov;1(3):279–85. doi: 10.1371/journal.ppat.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahman A, Sobia P, Gupta N, Kaer LV, Das G. Mycobacterium tuberculosis subverts the TLR-2-MyD88 pathway to facilitate its translocation into the cytosol. PloS one. 2014;9(1):e86886. doi: 10.1371/journal.pone.0086886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamm LM, Morisaki JH, Gao LY, et al. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. The Journal of experimental medicine. 2003 Nov 3;198(9):1361–8. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagener J, Malireddi RK, Lenardon MD, et al. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS pathogens. 2014 Apr;10(4):e1004050. doi: 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000 Jan 1;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 52.International HapMap Consortium. Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010 Sep 2;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oosting M, Ter Hofstede H, Sturm P, et al. TLR1/TLR2 heterodimers play an important role in the recognition of Borrelia spirochetes. PloS one. 2011;6(10):e25998. doi: 10.1371/journal.pone.0025998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehrer RI, Cline MJ. Interaction of Candida albicans with human leukocytes and serum. Journal of bacteriology. 1969 Jun;98(3):996–1004. doi: 10.1128/jb.98.3.996-1004.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.International HapMap Consortium. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007 Oct 18;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002 Jun 21;296(5576):2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.