The mechanisms by which PIWI proteins generate piRNAs to repress expression of transposable elements in germ cells are unclear. Andress et al. examine the dynamic cycling of the RNA helicase Spindle-E with PIWI proteins and cofactors in and out of nuage granules in the Drosophila female germline and suggest that nuage granules are hubs for transient, active protein complexes associated with piRNA amplification.
Abstract
Transposable elements (TEs) are silenced in germ cells by a mechanism in which PIWI proteins generate and use PIWI-interacting ribonucleic acid (piRNA) to repress expression of TE genes. piRNA biogenesis occurs by an amplification cycle in microscopic organelles called nuage granules, which are localized to the outer face of the nuclear envelope. One cofactor required for amplification is the helicase Spindle-E (Spn-E). We found that the Spn-E protein physically associates with the Tudor domain protein Qin and the PIWI proteins Aubergine (Aub) and Argonaute3 (Ago3). Spn-E and Qin proteins are mutually dependent for their exit from nuage granules, whereas Spn-E and both Aub and Ago3 are mutually dependent for their entry or retention in nuage. The result is a dynamic cycling of Spn-E and its associated factors in and out of nuage granules. This implies that nuage granules can be considered to be hubs for active, mobile, and transient complexes. We suggest that this is in some way coupled with the execution of the piRNA amplification cycle.
Introduction
The eukaryotic cell is compartmentalized into microscopic granules where specific RNA–protein biochemical reactions are concentrated and localized. By electron microscopy, these granules appear as compact granulo-fibrillar aggregates with no surrounding membrane. One of the central mysteries about these organelles is how they are established and how their molecular components are shuttled in and out with specificity and integrity. Although many RNP organelles such as the nucleolus are found in virtually all cells, some specialized cells contain RNP organelles with unique structures and functions. One such organelle is called the nuage granule, which is found in the cytoplasm of developing germ cells (Eddy, 1974). Nuage (meaning cloud) granules are closely juxtaposed to the cytoplasmic face of nuclear pores, where they receive sense and antisense RNA transcribed from active transposable elements (TEs) within the nucleus (Zhang et al., 2012). TEs are capable of replicating and moving within genomes, which can lead to heritable mutations and expansion in TE number. Germ cells elicit a molecular mechanism to silence expression of TE genes and suppress their transposition. Nuage granules are sites of active TE silencing and suppression (Voronina et al., 2011), and they have substantial lifetimes when associated with the nuclear envelope, although individual components within them are highly dynamic (Snee and Macdonald, 2004).
The PIWI family of proteins and PIWI-interacting RNAs (piRNAs) are critical for TE silencing (Aravin et al., 2007; Ghildiyal and Zamore, 2009; Saito and Siomi, 2010). Aubergine (Aub) and Argonaute3 (Ago3) are two PIWI family members in Drosophila melanogaster involved in germline piRNA generation (Thomson and Lin, 2009; Khurana and Theurkauf, 2010). They do so by using an amplification loop known as the ping-pong cycle (Brennecke et al., 2007; Gunawardane et al., 2007). Aub is loaded with antisense piRNAs that guide Aub to sense-stranded mRNAs made from active TEs. Subsequent cleavage of the mRNAs produces sense piRNAs, which load into Ago3. Ago3 is then guided to antisense-stranded piRNA cluster transcripts, where it slices these to form antisense piRNAs. These novel piRNAs are loaded into Aub, and the cycle repeats itself. Because Aub and Ago3 presumably act as catalysts, their activities account for the majority of piRNAs present in the Drosophila germline (Li et al., 2009; Malone et al., 2009). The ping-pong signature is mainly confined to Aub–Ago3 pairing, and Aub–Aub pairing to a lesser extent. In the absence of Ago3, Aub–Aub pairing prevails, although total piRNA levels are drastically reduced (Li et al., 2009). In the absence of Aub, the ping-pong signature is almost entirely lost (Li et al., 2009; Malone et al., 2009). Most proteins required for ping-pong amplification, including Aub and Ago3, are enriched in nuage granules (Voronina et al., 2011). Hence, nuage represent the cytoplasmic portion of a piRNA processing compartment that spans the nuclear envelope.
Genetic studies have identified many other factors involved in the germline piRNA pathway (Siomi et al., 2011). However, molecular mechanistic details of their functions are largely lacking. One exception is Vasa, a DEAD-box helicase protein. DEAD-box proteins use ATP to unwind short duplex RNA and remodel RNA–protein complexes, but they can also function as ATP-dependent RNA clamps to provide nucleation centers that establish larger RNA–protein complexes (Linder and Jankowsky, 2011). In the silkworm, handoff of sense-stranded piRNA precursors from Aub to Ago3 is facilitated by Vasa (Xiol et al., 2014; Nishida et al., 2015). An ATPase-defective Vasa generates a futile Ago3–Aub–Vasa complex in which the RNA handoff fails (Xiol et al., 2014; Nishida et al., 2015). Interestingly, all three proteins remain statically bound within nuage, unable to move to the cytosol, as though helicase activity is needed for exit of multiple proteins from nuage granules (Xiol et al., 2014). This suggests that complex disassembly leads to exit of disassembled subunits from the nuage.
A second DEAD-box helicase implicated in piRNA production is Spindle-E (Spn-E). Spn-E is essential for Drosophila germline piRNA production and the ping-pong signature (Gillespie and Berg, 1995; Malone et al., 2009; Czech et al., 2013). In silkworm, Spn-E associates with the Aub orthologue and is essential for processing of primary piRNAs, which initiate the ping-pong cycle of secondary piRNA production (Nishida et al., 2015). Spn-E is localized to nuage (Nishida et al., 2015) and is necessary for proper localization of Ago3 and Aub into Drosophila nuage granules (Lim and Kai, 2007; Malone et al., 2009). However, its physical and dynamic relationships with nuage components are poorly understood. Here, we have studied the dynamics and associations of Spn-E in Drosophila nuage granules. Spn-E associates with Aub and Ago3 in protein–protein complexes, but no evidence of Vasa association with Spn-E was found. We find that Spn-E and the Tudor domain protein Qin are mutually dependent for their exit from the nuage, whereas Spn-E and both Aub and Ago3 are mutually dependent for their entry or retention in nuage. Absence of Qin results in accumulation of Spn-E in nuage. We propose that Qin–Spn-E complexes disassociate from stable nuage scaffolds, whereas Aub–Spn-E and Ago3–Spn-E complexes are retained on such scaffolds. Exchange of Spn-E from one complex to another might be facilitated or passive, but the end result is a dynamic cycling of this RNA helicase.
Results
Qin, Ago3, and Aub physically interact with Spn-E
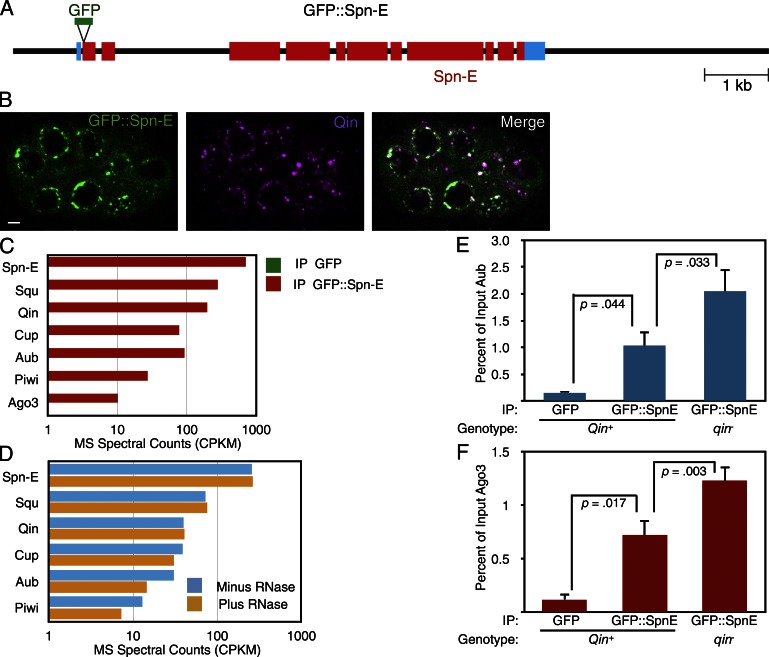
Spn-E plays a critical though mysterious role in piRNA biogenesis. The antibody reagents for Drosophila Spn-E are of variable quality, making its biochemical analysis limited. Therefore, we generated a GFP fusion to the N terminus of Spn-E by bacterial artificial chromosome recombineering (Venken et al., 2006). A recombineered interval of 8 kb of genomic DNA encompassing the modified spn-E gene was inserted into a [Pacman] vector and transformed into Drosophila (Fig. 1 A). The recombineered GFP::Spn-E gene was able to rescue the developmental defects associated with loss of the endogenous spn-E gene. This result argues that the transgene and its products contain most or all of the necessary functions of Spn-E.
Figure 1.
Spn-E associates with piRNA pathway proteins in the Drosophila germline. (A) A schematic of the GFP::Spn-E transgene. The black line shows 5′ and 3′ flanking regions and introns within the transcription unit. Blue boxes show the 5′ and 3′ UTRs. Red boxes show Spn-E coding regions, and the position of the GFP insertion is also shown. (B) Localization of GFP::Spn-E and Qin proteins in a stage 4 egg chamber. The chamber is composed of 16 interconnected germ cells, some of which are visible in this plane of focus. Both proteins are strongly enriched in perinuclear foci called nuage granules. The proteins are depleted in the nuclei, and GFP::Spn-E in particular is also detected throughout the cytoplasm. Bar, 5 µm. (C) Normalized spectral counts from mass spectrometry of proteins that coimmunoprecipitated with either GFP::Spn-E or GFP alone (signal is below detection in the graph). Although the results of only one experiment are shown, similar results were obtained in two other experiments. (D) Normalized spectral counts from mass spectrometry of proteins that coimmunoprecipitated with GFP::Spn-E. Before mass spectrometry analysis, half of the immunoprecipitate was treated with an RNase cocktail and the other half was mock treated, and dissociated proteins were washed away before mass spectrometry analysis of the precipitate. (E and F) Anti-GFP immunoprecipitation of proteins from wild-type and qin mutant ovary extracts and Western quantitation of Aub (E) and Ago3 (F) proteins that coimmunoprecipitated. As indicated, some ovaries expressed GFP alone and some ovaries expressed GFP::Spn-E. n = 3 replicate experiments. Shown are means with error bars representing SEM. P-values are the results of paired and unpaired two-tailed t tests. IP, immunoprecipitate. MS, mass spectrometry. CPKM, counts per kilodalton per million.
We examined the localization of GFP::Spn-E within the developing ovary. Expression was detected in the germarium as early as the germ stem cells, and it persisted in egg chambers through oogenesis (Fig. S1). As expected, GFP::Spn-E was restricted to the germline and found within both nurse cells and the oocyte (Fig. 1 B). It was localized to germ cell cytoplasm and, to a much lesser extent, the nucleus and was particularly enriched in perinuclear granules (Fig. 1 B). These most likely correspond to nuage granules. To confirm that GFP::Spn-E was enriched in nuage, we performed immunostaining of the Qin protein in GFP::Spn-E ovarioles because Qin is primarily found in nuage granules (Zhang et al., 2011; Anand and Kai, 2012). Indeed, Qin and GFP::Spn-E showed significant colocalization in these perinuclear bodies (Fig. 1 B).
GFP::Spn-E is enriched in nuage granules, where other piRNA pathway components reside. We wondered whether Spn-E formed protein complexes with these factors. Therefore, we immunoprecipitated GFP::Spn-E from ovary lysates and performed multidimensional protein identification technology (MUDPIT) mass spectrometry to identify coimmunoprecipitated proteins. As a control for specificity, we also performed immunoprecipitations with lysate from ovaries expressing GFP in the ovary. SDS-PAGE analysis of immunoprecipitated proteins showed a number of proteins that specifically coimmunoprecipitated with GFP::Spn-E but not GFP (Fig. S2). We then subjected the material to MUDPIT mass spectrometry, which can identify many individual proteins from complex mixtures (Link et al., 1999). As expected, the most abundant signal detected in the GFP::Spn-E immunoprecipitate was Spn-E itself (Fig. 1 C). We also detected the PIWI protein Aub and, to a lesser extent, the other two PIWI-class proteins, Piwi and Ago3 (Fig. 1 C). We furthermore detected Qin that was specifically associated with GFP::Spn-E (Fig. 1 C). Qin contains five Tudor domains (Zhang et al., 2011; Anand and Kai, 2012), which recognize symmetrically dimethylated arginines on proteins (Siomi et al., 2010; Handler et al., 2011). Qin is required for Aub–Ago3 association and ping-pong to occur (Zhang et al., 2011, 2014). Qin physically associates with both Aub and Vasa (Anand and Kai, 2012; Xiol et al., 2014), suggesting that Qin might help Vasa in the handoff of piRNA precursors from Aub to Ago3. In silkworm, Qin is also associated with Spn-E (Nishida et al., 2015).
Squash (Squ), a putative nuclease that localizes to the nuage (Pane et al., 2007), was also identified as an associated protein. Mutations in squ have little effect on piRNA profiles, although expression of some TEs such as I elements is dramatically derepressed (Malone et al., 2009; Haase et al., 2010). For these reasons, it has been suggested that Squ plays a role in the effector phase of the piRNA pathway (Haase et al., 2010). Cup, an eIF4E-binding protein (Richter and Sonenberg, 2005), was also identified as an associated protein with GFP::Spn-E. Cup represses translation of several mRNAs during oogenesis (Richter and Sonenberg, 2005) but appears to have other activities as well. It transiently localizes to the nuclear envelope of germ cells (Keyes and Spradling, 1997), where it physically associates with the Nup154 subunit of the nuclear pore complex (Grimaldi et al., 2007). Cup also interacts with a complex of Trailer Hitch (Tral) and Me31b (Wilhelm et al., 2005), which are localized within nuage granules (Lim et al., 2009).
One potential reason that GFP::Spn-E pulled down all of these proteins was because they were mutually bound to common RNA transcripts rather than bound directly within protein complexes. Therefore, we repeated the immunoprecipitation experiment but treated half of the immunoprecipitate with a cocktail of ribonucleases followed by extensive washing away of released proteins. We used spectral counting as a semiquantitative means to compare protein levels from ribonuclease-treated immunoprecipitates with control samples (Bantscheff et al., 2007). Strikingly, Qin, Cup, and Squ association with GFP::Spn-E was unaffected by ribonuclease treatment, indicating that they are in direct protein complexes with GFP::Spn-E (Fig. 1 D). Aub and Piwi showed slightly reduced levels with GFP::Spn-E from ribonuclease-treated lysate. Thus, Spn-E forms protein complexes with a variety of factors, including Aub and Qin.
It is possible that either Spn-E forms a multisubunit complex composed of all these factors, or it forms distinct complexes with different subunit composition. A common feature of multisubunit complexes is the codependence of complex formation/stability on the presence of subunits (Butland et al., 2005). Conversely, if distinct complexes compete with one another for a limiting common subunit, the complexes often form antagonistically with one another. We genetically removed one interacting partner of Spn-E and measured the associations between Spn-E and the remaining factors. Unfortunately, available anti-Qin antibodies did not work for Western blotting or immunoprecipitation, making it impossible to test whether loss of Ago3 or Aub affected Qin–Spn-E association. Instead, we performed an immunoprecipitation of GFP::Spn-E in a qin mutant and measured the levels of Aub and Ago3 that coimmunoprecipitated with GFP::Spn-E (Fig. 1, E and F). In the qin mutant, the levels of Aub and Ago3 bound to GFP::Spn-E were greater than the levels of bound Aub and Ago3 when Qin was present (Fig. 1, E and F). Thus, Qin inhibits Spn-E association with Aub and Ago3. It might suggest that Qin competes with Aub and Ago3 for Spn-E binding.
Interdependence of Spn-E with PIWI proteins and Qin
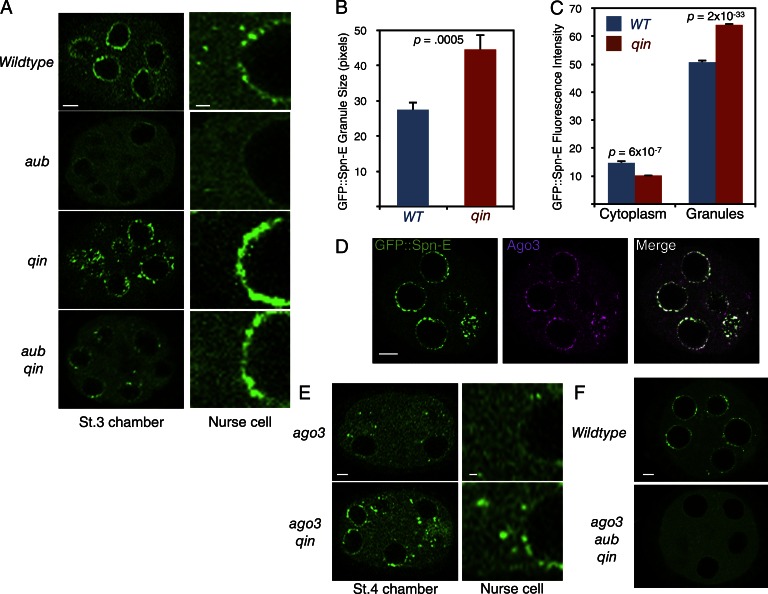
Our biochemical analysis suggests that Qin and PIWI proteins act antagonistically with one another for binding to Spn-E. If they act in such a manner, possibly Spn-E localization is regulated in opposite ways. Therefore, we examined the localization pattern of GFP::Spn-E in aub and qin mutants. GFP::Spn-E localization to nuage granules was greatly reduced in aub mutants, although the protein remained diffuse throughout the cytoplasm (Fig. 2 A). In contrast, qin mutants had GFP::Spn-E levels that were reduced in the cytoplasm and more localized in nuage (Fig. 2, A–C). These effects on GFP::Spn-E were not caused by a large change in overall protein abundance; total GFP::Spn-E levels were not greatly altered in the mutants (Fig. S3). Thus, it appears that aub and qin have opposing effects on GFP::Spn-E localization. To determine whether there was an epistatic relationship between Aub and Qin on Spn-E, we looked at GFP::Spn-E in an aub qin double mutant. We found the localization of GFP::Spn-E was restored to a situation intermediate between the single mutants (Fig. 2 A). It suggests that Qin and Aub act in parallel and antagonistically to affect Spn-E localization.
Figure 2.
GFP::Spn-E localization is regulated by Qin and PIWI proteins. (A) Confocal micrographs of GFP::Spn-E in stage 3 egg chambers of wild type and mutants, as indicated. (Left) One egg chamber section. Bar, 5 µm. (Right) Magnified images of the perinuclear and nuclear regions of one nurse cell. Bar, 1 µm. (B) Relative sizes of nuage granules positive for GFP::Spn-E as measured by pixel number of individual granules. Error bars are SEM, and p-value is the result of a two-tailed t test comparing the difference in sizes between wild type (WT) and qin. n = 22 replicates. (C) Quantification of GFP::Spn-E fluorescence intensity in defined regions of nurse cells at stage 3 of oogenesis. Shown are mean intensities detected in peripheral cytoplasm and in individual nuage granules. Error bars are SEM, and the p-values are results of two-tailed t tests comparing the difference in intensities between wild type and qin. n = 23 replicates. (D) Localization of GFP::Spn-E and Ago3 proteins in a stage 3 egg chamber. Bar, 5 µm. (E) Confocal micrographs of GFP::Spn-E in stage 4 egg chambers of mutants, as indicated. (Left) Section through one egg chamber. Bar, 5 µm. (Right) Magnified images of the perinuclear and nuclear regions of one nurse cell. Bar, 1 µm. (F) GFP::Spn-E localization in wild-type and aub ago3 qin triple mutant stage 4 egg chambers. Bar, 5 µm.
As previously noted, GFP::Spn-E has an almost normal localization pattern in nuage and cytosol when Aub and Qin are both absent. This indicates that there are other factors influencing Spn-E localization as well. We asked whether Ago3 could be one of those factors. Ago3 physically associates with Spn-E, and both proteins colocalize in nuage (Fig. 2 D). Therefore, we examined the localization of GFP::Spn-E in an ago3 mutant and found it to be strongly depleted from nuage (Fig. 2 E). This depletion was partially recovered in an ago3 qin double mutant. These results are consistent with the aub effects on Spn-E and suggest that each PIWI protein promotes Spn-E to nuage granules in parallel and antagonistically with Qin.
If Ago3 and Aub act redundantly to promote nuage localization of Spn-E, we predicted that loss of both PIWI proteins would block Spn-E localization to nuage even in the absence of Qin. To test this prediction, we made an ago3 aub qin triple mutant and observed that GFP::Spn-E completely failed to localize within nuage granules (Fig. 2 F). Therefore, Aub and Ago3 act epistatically to Qin and together are essential for proper Spn-E localization to nuage.
Spn-E regulates Qin and PIWI protein localization
Our evidence thus far suggests that when bound to Qin, Spn-E is more likely to be found in the cytoplasm, whereas when bound to Aub or Ago3, Spn-E is more likely to be found in nuage granules. One mechanism by which Qin and PIWI proteins could affect Spn-E localization is by each acting as a sink located in separate cellular compartments. If this hypothesis is correct, each molecule would be strictly located within a unique compartment—Qin in the cytoplasm and the two PIWI proteins in the nuage. However, Qin is detected in both compartments and is enriched in the nuage (Fig. 1 B). The PIWI proteins have a similar localization pattern (Snee and Macdonald, 2004; Brennecke et al., 2007; Li et al., 2009). Given that their localization patterns resemble one another and resemble that of Spn-E, a more attractive mechanism comes to mind. In this mechanism, Qin and PIWI proteins would shuttle with Spn-E from one compartment to another. If this were the case, we would expect their localization to be affected when we removed Spn-E.
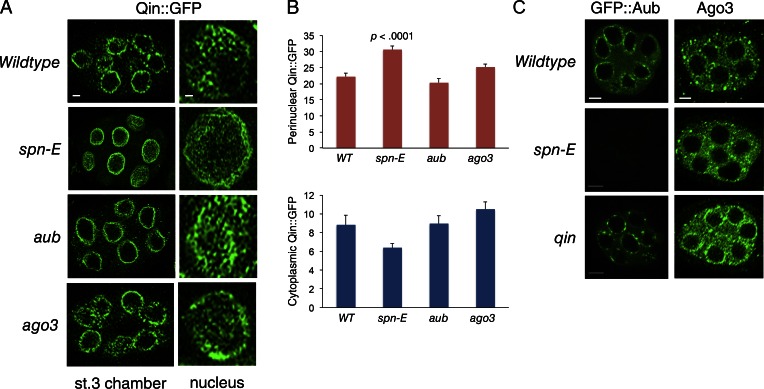
We looked at the localization of GFP-tagged Qin in a spn-E mutant background. As expected, the removal of Spn-E affected the localization of Qin (Fig. 3, A and B). Qin::GFP was more concentrated at the nucleus in spn-E mutant cells. Strikingly, the granular appearance of Qin::GFP in nuage was transformed into a web-like perinuclear localization (Fig. 3 A). We also looked at the effects of Aub and Ago3 on Qin localization and did not observe a change in Qin::GFP localization in aub or ago3 mutant cells (Fig. 3, A and B). Thus, Qin is recruited to perinuclear regions by a mechanism independent of the two PIWI proteins.
Figure 3.
Spn-E regulates localization of Qin and PIWI proteins. (A) Qin::GFP localization in stage 3 egg chambers of wild type and mutants, as indicated. (Left) Section through one egg chamber. Bar, 5 µm. (Right) Tangent planar views of a single nurse cell nucleus from each genotype. This plane images the perinuclear localization of Qin::GFP. Bar, 1 µm. Shown are representative egg chambers from a total number imaged and analyzed of n = 50 (wild type), n = 22 (spn-E), n = 16 (aub), and n = 12 (ago3). (B) Quantification of Qin::GFP fluorescence intensity in defined regions of nurse cells. Shown are mean intensities detected for perinuclear Qin::GFP and peripheral cytoplasmic Qin::GFP. Error bars are SEM; two-tailed t tests were done to compare the difference in intensities between wild type (WT) and each mutant. The result of the only t test returning significance is shown. Replicate number for each analysis (bar) ranged from 12 to 18. (C) GFP::Aub localization in stage 3 egg chambers of wild type and mutants, as indicated. Ago3 protein localization as detected by anti-Ago3 in stage 3 egg chambers of wild type and mutants, as indicated. Bars, 5 µm.
We also tested whether PIWI protein localization was influenced by Spn-E. GFP::Aub is present in germ cell cytoplasm and highly enriched in perinuclear nuage granules (Fig. 3 C). Loss of Spn-E caused almost complete loss of GFP::Aub in egg chambers. Ago3 protein is normally concentrated in heterogeneously sized granules, some of which are perinuclear (Fig. 3 C). Loss of Spn-E did not abolish Ago3 as it did for Aub; rather, perinuclear localization of Ago3 granules was less pronounced (Fig. 3 C). We also looked at the effect of qin mutants on Aub and Ago3. The GFP::Aub nuage was less homogeneous and more punctate, reminiscent of Spn-E distribution in the qin mutant (Fig. 3 C). Likewise, Ago3 appeared to be more concentrated in the perinuclear nuage of qin mutants (Fig. 3 C).
Spn-E dynamics are regulated by Qin
Nuage granules are stable structures that maintain size, seldom detach from the nuclear envelope, and exchange their protein components with the cytosol (Snee and Macdonald, 2004). To measure the dynamic movement of Spn-E in nuage granules, we performed FRAP of GFP::Spn-E. FRAP is used to study the kinetic properties of fluorescent proteins by measuring the fluorescence recovery rate in a bleached area (Reits and Neefjes, 2001; Snapp et al., 2003). Unbleached molecules enter into the bleached area from the outside, and the fluorescence intensity is recorded by time-lapse microscopy. The recovery curve provides information about the diffusion and binding dynamics of the protein.
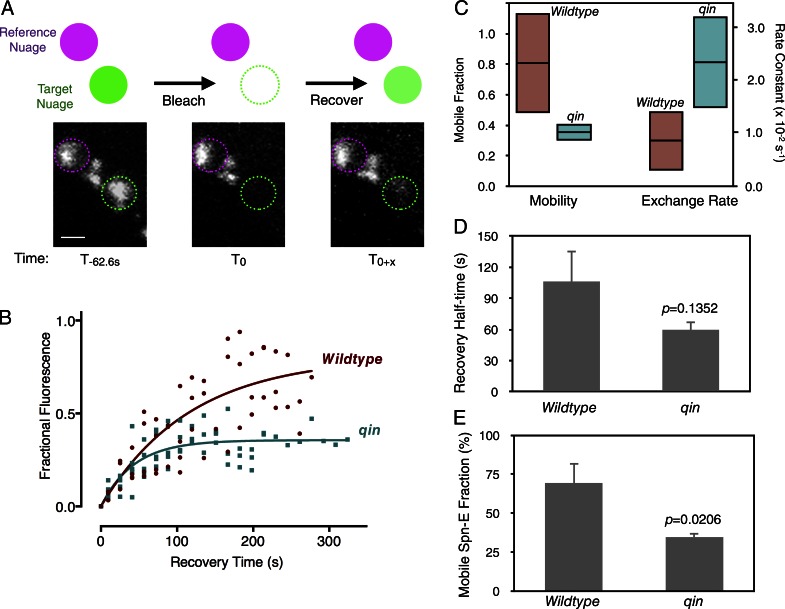
We dissected stage 3 egg chambers from GFP::Spn-E ovaries and cultured them in vitro in a confocal microscopy chamber (Video 1). One or two nuage granules were then photobleached in a 1-μm2 region of interest (ROI), and the reappearance of GFP::Spn-E fluorescence in the ROI was monitored (Fig. 4 A). A nearby unbleached nuage granule was also monitored over time and used as a reference to normalize for the relative recovery of the bleached granule. New GFP::Spn-E fluorescence reappeared at the same position as the original nuage granule, suggesting that there is exchange of GFP::Spn-E between cytosolic pools and stable nuage (Video 2). For each time point, we calculated the level of recovering GFP::Spn-E fluorescence as a fraction of the prebleach fluorescence intensity. This recovery response is shown in Fig. 4 B, which presents data from five biological replicates each for wild-type and qin mutants.
Figure 4.
GFP::Spn-E dynamics in nuage is controlled by Qin. (A) A schematic of the FRAP experiment. After prebleach measurement of GFP::Spn-E fluorescence in nuage, one granule was bleached for 62.6 s; thereafter, fluorescence in both reference and bleached nuage was followed over time. Below are representative confocal images in nuage before and after bleaching. Bar, 0.25 µm. (B) Kinetics of GFP::Spn-E fluorescence recovery expressed as a normalized fraction of the prebleach level. Values measured from all replicates are plotted. The fitted curves are single-phase exponential functions. (C) Exponential model parameters estimated from B. (Left) The percentage of prebleached GFP::Spn-E that is mobile within the nuage. Shown are point estimates (horizontal lines) and 99% confidence intervals (shaded boxes) for wild-type and qin mutant mobile fractions. (Right) The recovery rate constants of GFP::Spn-E within the nuage. Point estimates (horizontal lines) and 95% confidence intervals (shaded boxes) are shown. (D) Mean recovery half-time for the mobile fraction of GFP::Spn-E in wild-type and qin mutant nuage. (E) Mean mobile fraction of GFP::Spn-E in bleached nuage of wild-type and qin mutant germline. n = 5 replicates. Error bars are SEM, and p-values are the results of two-tailed t tests.
Using nonlinear regression analysis, we found that single-phase exponential functions best fit the data for both genotypes (see the Analysis of FRAP experiments section within Materials and methods). Using the exponential models, we estimated the rate constant for recovery of fluorescence (k) in each condition (Fig. 4 B). If the bleached molecules have no effect on the dynamics of unbleached molecules, k is determined by the unbleached molecule’s diffusion coefficient. This relationship is k = 4D × ln(2)/r2, where r is the bleach spot radius and D is the diffusion coefficient. The observed k for GFP::Spn-E in wild-type nuage translates to a diffusion coefficient of ∼0.001 µm2/s. Given that GFP and its fusion proteins exhibit diffusion coefficients in cytoplasm ranging between 0.5 and 25 µm2/s (Snapp et al., 2003), this large discrepancy indicates that GFP::Spn-E recovery is not limited by monomolecular diffusion. Instead, it describes a recovery in which bleached molecules are bound to static molecules and must dissociate before unbleached molecules can occupy the bleached area (Snapp et al., 2003). Consequently, the recovery rate constant is determined by the dissociation rate constant of binding to the static partners. For GFP::Spn-E, this would translate to an approximate binding half-life of 1.5 min. When comparing the rate constants for GFP::Spn-E between wild-type and qin nuage, there was a small difference (Fig. 4 C). The mutant exhibited a slightly higher rate constant that was weakly significant (P < 0.05 by confidence interval).
The exponential model also defined the fraction of bleached molecules that can freely dissociate from their static partners (mobile) and the fraction of molecules that are permanently bound (immobile; Snapp et al., 2003). The relative levels of mobile and immobile fractions can be seen by the fitted curves approaching asymptotes that are less than prebleached levels (Fig. 4 B). The fitted parameter for the size of the GFP::Spn-E mobile fraction greatly differed between wild-type and qin mutant nuage (Fig. 4 C). Approximately 80% of GFP::Spn-E in wild-type nuage was mobile, whereas only 35% of GFP::Spn-E in qin mutant nuage was mobile, and this difference between wild type and mutant was significant (P < 0.01 by confidence interval).
We also took a different approach for FRAP data analysis, in which each replicate was analyzed independent of the other replicates, and estimated parameters were then averaged for each genotype. This allowed us to estimate the recovery half-life, which is the time required for 50% of the mobile fraction to recover (Snapp et al., 2003). The mean recovery half-life of mobile GFP::Spn-E in qin nuage was not significantly different from the recovery half-life in wild-type nuage (Fig. 4 D; P = 0.1352; two-tailed t test). Because the recovery half-life is inversely proportional to the dissociation rate constant k (t1/2 = ln(2)/k), it indicates that the GFP::Spn-E dissociation rate constant is not significantly affected by the qin mutant. This is what had also been observed in our single-phase exponential modeling. Also consistent with exponential modeling, the mean mobile fraction of GFP::Spn-E was reduced twofold in qin mutant nuage using the alternative analytic approach (Fig. 4 E). Thus, Qin positively regulates the size of the GFP::Spn-E pool in nuage that can freely exchange with cytoplasmic GFP::Spn-E.
Discussion
The nuage granule is a hallmark of germline cytoplasm in diverse animal species, ranging from nematodes to mammals (Voronina et al., 2011). It is a site for TE RNA degradation, which is essential for inhibiting TE transposition in the germline genome. Nuage granules in Drosophila reside on the cytoplasmic face of certain nuclear pores, adjacent to nuclear foci composed of Rhino and UAP56 proteins plus TE and cluster RNAs (Zhang et al., 2012). This intimate connection between nuclear foci and nuage ensures the efficient transfer of piRNA precursors to the ping-pong apparatus.
A hierarchical model was proposed for Drosophila nuage assembly in which the RNA helicase Vasa sits at the apex (Lim and Kai, 2007; Patil and Kai, 2010). Vasa was proposed to recruit piRNA pathway components in a defined order: first by directly recruiting Tudor domain proteins, which thereafter recruit PIWI proteins by binding to their symmetrically dimethylated arginines. A more nuanced view of Vasa has emerged in which it nucleates assembly of a transient complex within the nuage granule (Xiol et al., 2014; Nishida et al., 2015). There, Vasa’s helicase clamps sense-stranded TE RNAs and assembles PIWI proteins that cleave the RNAs for Ago3 loading. Strikingly, if Vasa cannot unclamp RNA, the complex fails to disassemble, and complex subunits overaccumulate in nuage granules. This strongly hints that the formation of RNA–protein complexes is coupled with movement of complex components between nuage granules and the rest of the cytoplasm. The nuage can be considered to be a hub for active and transient complexes.
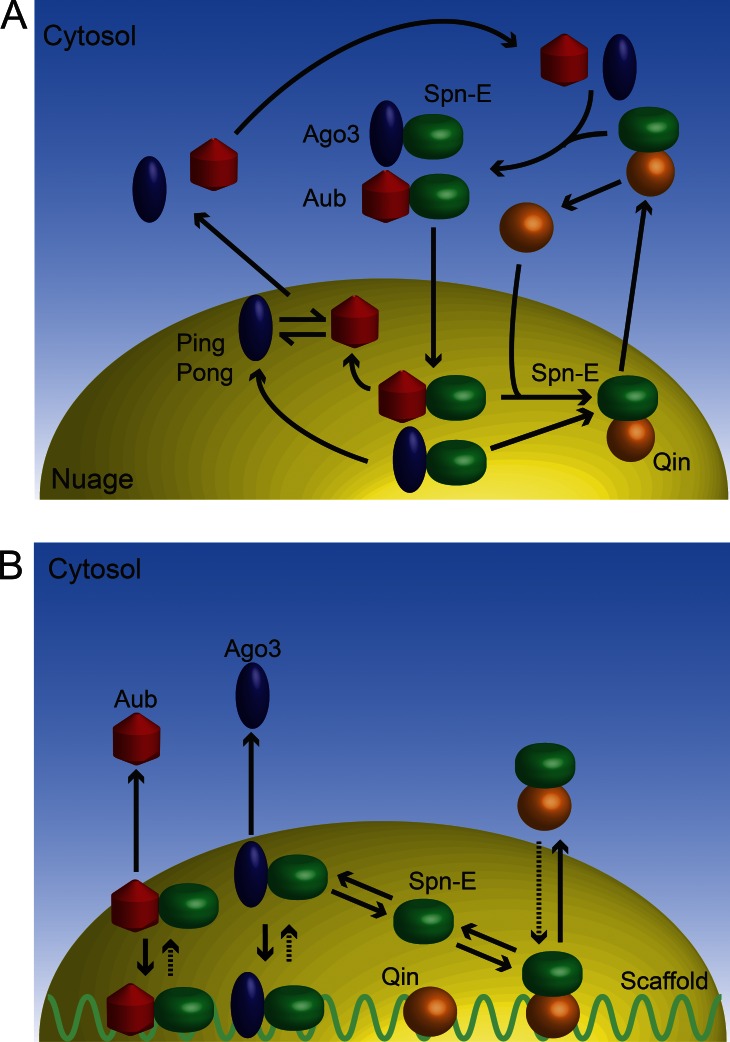
Our studies of Spn-E confirm and expand upon this concept. Spn-E is another RNA helicase essential for piRNA biogenesis, and Spn-E exhibits distinct patterns of localization within the germ cells. Our results allow us to propose a model in which Spn-E cycles between nuage granules and the cytoplasm (Fig. 5). In one version of the model, the cycle is coupled to a defined series of transient complexes composed of Spn-E and other proteins (Fig. 5 A). Within the nuage, Spn-E would bind to Qin, and this complex would exit from the nuage. Exit would be followed by complex dissociation and reassociation of Spn-E with Ago3 and Aub proteins. These complexes would then enter the nuage, where they would remain until they dissociate, whereupon Spn-E and Qin complex formation could be renewed. In another version of the model, cycling would be driven by the relative affinities of different Spn-E complexes for an immobile scaffold within nuage (Fig. 5 B). In this model, Spn-E could form complexes with either Qin or PIWI proteins indiscriminately in both nuage and cytoplasm. However, in the nuage, these complexes interact with an immobile scaffold for which they possess differential binding affinities. PIWI–Spn-E complexes would have strong affinity for the scaffold becoming immobile, whereas Qin–Spn-E complexes would have weaker affinity for the scaffold and be more mobile. If this model is correct, what might the scaffold be? One intriguing possibility is that the scaffold is part of the nuclear envelope localized to the pores.
Figure 5.
Models for Spn-E cycling between cytoplasmic compartments. (A) One model proposes that Spn-E forms transient complexes in different compartments, and this follows a defined series of steps. (B) A second model proposes that Spn-E forms transient complexes in all compartments at steady state, and different complexes have different affinities for a nuage scaffold.
If the scaffold model is correct, the Spn-E complexes at steady state might potentially compete with one another. Indeed, Aub and Ago3 compete with Qin for binding to Spn-E, suggesting that Spn-E is a limiting subunit for these different complexes. The scaffold model would also explain the Spn-E FRAP results. In the absence of Qin, more Spn-E is immobile within nuage granules, presumably because more of it is tightly bound to the scaffold. Because Spn-E is associated with more PIWI proteins under these conditions, it suggests that Aub–Spn-E and Ago3–Spn-E complexes are strongly associated with the scaffold, as the model predicts. The minor fraction of Spn-E in qin mutant nuage that is mobile might be free protein or protein associated in other complexes that have weak affinity for the scaffold, making them highly mobile. The model predicts that a greater fraction of Spn-E would be competed out of high affinity complexes into low affinity complexes when Qin is present, reducing the fraction of Spn-E that is immobile. Indeed, this is precisely what was observed: Qin caused a twofold decrease in the immobile fraction of Spn-E in the nuage.
The dynamic cycling of Spn-E and associated factors is intriguing, and it is worth speculating on a possible function for the cycling. Spn-E and these factors act in a biochemical cycle of piRNA processing called ping-pong. Perhaps the dynamic cycling of Spn-E and associated factors is in some way coupled with the execution of the ping-pong cycle. Transient complexes might form and immobilize in the nuage until one step in the ping-pong cycle is executed, thereafter releasing the complex subunits. These could then diffuse elsewhere and allow new complexes to form that execute other steps in the ping-pong cycle. Hence, complexes would not be physically coupled to one another in executing one complete round of ping-pong, but would move between one another and even between nuage granules to create a highly connected network of RNA processing reactions. Recent evidence that Vasa complexes act in this manner (Xiol et al., 2014) lends credence to the notion that various steps of the ping-pong reaction might be spatially and temporally uncoupled.
Materials and methods
Spn-E recombineering
To insert eGFP into BACR06B08, which contains Spn-E genomic DNA, a 635-bp left homology arm (LA) was generated from the spn-E locus such that its 3′ endpoint corresponded to the nucleotide upstream of the spn-E start codon. The LA fragment was ligated to an EGFP fragment containing the complete ORF and start codon but no stop codon. A 696-bp right homology arm (RA) was generated from the spn-E locus such that its 5′ endpoint corresponded to the second codon of spn-E. The RA fragment was ligated to the 3′ end of the LA-eGFP fragment. The LA-eGFP-RA linear fragment was transformed into DY380 containing BACR06B08, and recombineered eGFP insertions were identified. To construct a FLAG-tagged version of Spn-E, we used a modified tandem affinity purification tag that had been developed for Drosophila proteins (Yang et al., 2006). This tri tag was a gift from H. Krause (University of Toronto, Toronto, Canada), and it consists of N-3 × FLAG-TEV-Strep-6 × His-C, where three copies of the FLAG epitope are upstream of a recognition site for TEV protease, a Strep tag, and a polyhistidine tag. The tri tag was inserted into BACR06B08 by recombineering to create an N-terminal fusion between the tag and Spn-E polypeptides.
LAs and RAs corresponding to fragments of the spn-E locus were inserted into the multicloning site of attB-[Pacman] (Venken et al., 2006). The endpoint of the 5′-most arm corresponded to 3R:15,836,621, and the endpoint of the 3′-most arm corresponded to 3R:15,844,693. The vector was linearized and transformed into DY380 containing the eGFP- or tri tag–modified BACR06B08. Through recombineered gap repair, an 8,072-bp region from 3R:15,836,621 to 3R:15,844,693 was incorporated into attB-[Pacman] to generate GFP::Spn-E–attB-[Pacman] and FLAG::Spn-E–attB-[Pacman]. The 5′ endpoint resides in the first intron of the ND23 gene on the 5′ side of spn-E, whereas the 3′ endpoint resides in the final exon of the cv-d gene on the 3′ side of spn-E. GFP::Spn-E–attB-[Pacman] and FLAG::Spn-E–attB-[Pacman] were transformed by ψC31-mediated recombination into the VK22 attP landing site near the distal end of 2R (Venken et al., 2006). The functionality of the GFP::Spn-E and FLAG::Spn-E transgenes was tested by recombining them with spn-E1 and spn-EΔ125 mutations. One copy of the transgenes completely rescued the sterility phenotype of spn-E1/spn-EΔ125 trans-heterozygous mutants. All genome positions described were taken from Release 6 (Drosophila Genome Project).
Genetics
All Drosophila stocks were maintained on cornmeal-molasses food between 23 and 25°C. qin1, Df(3R)Exel6180, Df(2L)BSC187, aubHN2, aubQC42, ago3t2, ago3t3, spn-E1, spn-EΔ125, nanos-Gal4, nanos-Gal4::VP16, and tub>GFP were obtained from the Bloomington Stock Center. Qin::GFP was a gift from P. Zamore (University of Massachusetts Medical School, Worcester, MA). UASp-GFP::Aub was a gift from P. MacDonald (University of Texas, Austin, TX). qinkumo was a gift from T. Kai (Temasek Research Laboratories, Singapore). The following trans-heterozygous allelic combinations were generated for all experimental analyses described: aubHN2/aubQC42 (aub), ago3t2/ago3t3 (ago3), spn-E1/spn-EΔ125 (spn-E), qin1/Df(3R)Exel6180 (qin), and qinkumo/Df(3R)Exel6180 (qinkumo). Trans-heterozygous mutants were used to minimize the effects of second-site mutations on the relevant chromosomes.
Immunohistochemistry and microscopy
1- or 3-d-old ovaries were dissected in cold PBS and fixed for 30 min in 4% paraformaldehyde. Permeabilization was performed in PBS + 0.3% Triton X-100 (PBST). PBST was supplemented with 5% goat serum and incubated with fixed tissue for 30 min as a preblock. Ovaries were incubated with primary antibodies for 2 h at room temperature or overnight at 4°C at the appropriate concentrations. After washing with PBST four times, ovaries were incubated with secondary Alexa Fluor 488 or Alexa Fluor 546 (Invitrogen) at a 1:200–1:500 dilution in PBST. TO-PRO-3 diluted 1:1,500 (Thermo Fisher Scientific) was added to the incubation for counterstaining nuclei. Ovaries were washed four times in PBST and mounted with Vectashield (Vector Laboratories). Rabbit anti-Ago3 and rabbit anti-Aub were gifts from G. Hannon (University of Cambridge, Cambridge, England, UK) and were used at a 1:500 dilution. Rabbit anti-Vasa was a gift from P. Lasko (McGill University, Montreal, Canada) and was used at a 1:2,000 dilution. Rabbit anti-Qin was a gift from T. Kai and used at a 1:1,000 dilution. Guinea pig anti-Coracle was a gift from R. Fehon (University of Chicago, Chicago, IL) and used at a 1:200 dilution. Monoclonal anti-adducin 1B1 was obtained from the Developmental Studies Hybridoma Bank and used at a 1:20 dilution. All samples were imaged at room temperature using a confocal microscope (510 Meta; ZEISS) with an oil objective (20×/NA 0.5 or 100×/NA 1.45; ZEISS). All images were collected in Z stacks with sequential channel collection. Acquisition software (ZEISS) was used.
Quantification of protein localization
For imaging of GFP protein localization in wild-type and mutant egg chambers, we were careful to minimize intersample measurement error. Mutant and control females were collected together and aged for an identical length of time. Ovaries were dissected and fixed at the same time in identical buffers. Ovarioles were cleared and mounted on slides at the same time; some genotypes were mounted on the same slide. All samples were imaged by microscopy at the same time, and microscope/imaging settings were kept constant. Imaging parameters were first set to a nonrecorded sample with the greatest fluorescence so that fluorescence measurements did not saturate for any sampled specimens. High magnification Z sections were imaged using laser intensity and pinhole settings such that 8-bit pixel values did not exceed half-maximal level. Only nurse cells within stage 3 or 4 egg chambers were considered. Finally, experiments were repeated with new specimens on different days using different buffers and microscopy sessions. Three or more of these replicates were performed per experiment.
Image analysis was performed using Fiji (National Center for Biotechnology Information). Nonnuage fluorescence was hand segmented such that a minimum of 100 pixels was analyzed for each sampled segment. The mean pixel value per segment was recorded, and the mean of 12–24 segmented means per genotype was taken as cytoplasmic fluorescence intensity. Perinuclear nuage fluorescence was hand segmented over tangential sections through the nuage. Mean pixel value was recorded, and the mean of 12–50 segmented means per genotype was taken as perinuclear fluorescence intensity. To measure nuage granule size and fluorescence intensity, the Fiji freeform tool was used to segment individual granules. Pixel number and mean pixel values for each segment were recorded. The mean pixel values of 150–200 individually segmented objects per genotype were used to derive mean granule fluorescence intensity. The mean pixel number of these objects was taken as granule size. Western blot experiments indicated that total levels of GFP::Spn-E did not significantly vary with genotype (Fig. S3). To confirm this observation using fluorescence microscopic images, we segmented the entire germline of stage 3 egg chambers under study and measured mean pixel values for each. Wild-type chambers averaged 16.06 ± 2.36 (SEM) U/pixel, and qin mutant chambers averaged 15.78 ± 1.72 (SEM) U/pixel. Total GFP::Spn-E fluorescence was not significantly different between genotypes (P = 0.92 by two-tailed t test).
FRAP experiments
Stage 3 egg chambers were dissected from 1-d-old ovaries in Schneider’s media (Invitrogen) supplemented with 15% fetal bovine serum (Gibco), 0.6× penicillin–streptomycin (Invitrogen), and 200 µg/ml bovine insulin (Sigma-Aldrich). Samples were imaged at room temperature on glass-bottom microwell dishes (MatTek Corporation) with a moist Kimwipe to maintain humidity. The oil objective (100×/NA 1.45) was used on a microscope (510 Meta). Prebleach and postbleach scans were performed such that each pixel in captured images had 0.09 × 0.09–µm dimensions. Photobleaching was performed by focusing the 488-nm laser on a ROI with a dimension of 1 × 1 µm that contained a stable nuage granule. Bleaching was performed at maximum laser intensity for 62.6 s, with bleaching iterations of 0.21 s. Up to 120 postbleach scans were then obtained for each specimen, with each image acquired every 3.2 s at a high scanning speed and minimal laser intensity. The first postbleach scan was performed immediately at the termination of the bleach and was taken as the zero time point.
Analysis of FRAP experiments
Acquisition software (ZEISS) was used for image capture. ImageJ (National Institutes of Health) was used to quantitate the image data. The prebleach image was overlaid with the image from each postbleach time point to correct for drift of the sample over time and to rigorously identify the ROI and reference at each time point. A defined field of 52 pixels was used to quantitate ROI pixel intensity, and the field was positioned such that the inner half of a bleached ROI was quantitated. A similar sized field was used to quantitate pixel intensity in reference nuage for each image. Total pixel intensity in each user-defined field was used as the measure of fluorescence intensity. ROI fluorescence intensity at each postbleach time point was normalized to the reference fluorescence intensity by
where Fn(t) is normalized ROI fluorescence intensity, Rx(tp) is prebleach reference fluorescence intensity, Rx(t) is reference fluorescence intensity at time t, and Fx(t) is ROI fluorescence intensity at time t. Normalized fluorescence intensities were then converted to recovered fractional fluorescence, FF(t), by
where Fn(tp) is prebleach normalized ROI fluorescence intensity and Fn(t0) is normalized ROI fluorescence intensity at t = 0. For each genotype studied, a total of five biological replicates were analyzed for fractional fluorescence. All data per genotype were pooled and analyzed by nonlinear regression analysis (least squares) in Prism (GraphPad Software). A wide variety of linear and nonlinear models were tested, and these were compared with one another by Akaike’s information criterion (AICc; Burnham and Anderson, 2002). The top two models based on relative AICc scores were a one-phase exponential model and a second-order polynomial (quadratic) model. The total difference in AICc scores was 10.72 in favor of the one-phase exponential model. Therefore, we selected this model: Y = (Y0 − AS) × ekx + AS, where Y0 is the Y value when x = 0, AS is the Y value when x = ∞, and k is the rate constant, expressed in units that are the inverse of units for x.
We also compared the one-phase exponential model with nested models and found that the AICc score for the one-phase model was superior. We further tested the one-phase model by constraining each parameter and comparing their AICc scores with each other and the unconstrained model. In doing so, we found that a model in which Y0 is constrained by setting it to zero was optimal and gave particularly strong AICc scores relative to other nonlinear models. When this model was applied to determine the parameter estimates, we also obtained the 95% and 99% confidence intervals for the unconstrained parameters.
Alternative analyses on the FRAP data were also performed. The mobile fraction of GFP::Spn-E was calculated as
where Rx(t∞) is the asymptote of reference fluorescence intensity, Fx(t∞) is the asymptote of the ROI fluorescence intensity, Fx(t0) is the ROI fluorescence intensity at t = 0, and Fx(tp) is the prebleach ROI fluorescence intensity. The half-time of recovery for the mobile GFP::Spn-E fraction was estimated as described previously (Snapp et al., 2003). First, we transformed the normalized ROI fluorescence intensity, Fn(t), to a percentage of mobile GFP::Spn-E intensity, MF(t):
where Fn(t∞) is the asymptote of normalized ROI fluorescence intensity. We then plotted MF(t) versus an adjusted time, tt, in which tt = t + 31.3 s, where 31.3 s is the half-time of the bleach. The reasons for using an adjusted time are explained elsewhere (Snapp et al., 2003). The time at which MF(t) = 50% was taken as the recovery half-life.. Replicate estimates of Mf and the recovery half-life were averaged for each genotype, and two-tailed t tests were performed in Prism to determine the significance in differences between means.
Immunoprecipitations and Western blots
For immunoprecipitation experiments, roughly 100 ovaries from females expressing GFP::Spn-E or GFP were dissected per sample in cold PBS and flash frozen before lysis. Ovaries were homogenized in lysis buffer (10 mM Tris/Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0.5% NP-40) supplemented with protease inhibitors (MG-132, trypsin, aprotinin, and pepstatin) with a hand homogenizer for 2 min. Samples were centrifuged at 16,100 g for 15 min at 4°C. Lysates were diluted to the desired concentration with lysis buffer (without NP-40). Lysates were incubated with GFP-Trap_A (ChromoTek) nanobody-agarose beads overnight at 4°C. Beads were washed three times with dilution buffer. Proteins were eluted from the beads by heating at 95°C in SDS loading buffer and electrophoresed in a 7% SDS-polyacrylamide gel. Proteins were transferred onto polyvinylidene fluoride, and blots were probed with mouse anti-Aub at a 1:1,000 dilution (a gift from H. Siomi, Keio University, Tokyo, Japan), mouse anti-Ago3 at a 1:500 dilution (a gift from H. Siomi), rabbit anti-GFP at a dilution of 1:1,000 (Thermo Fisher Scientific), and mouse anti–α-tubulin at a dilution of 1:50,000 (Developmental Studies Hybridoma Bank). Goat anti–mouse IgG HRP (Bio-Rad Laboratories) and goat anti–rabbit HRP (GE Healthcare) were diluted to 1:1,000 and used as negative controls.
Immunoprecipitated proteins were quantitated using Fiji and normalized to the input proteins present in the starting extracts. Four biological replicates were performed for coimmunoprecipitation experiments, and the mean and interval estimates were derived. Student’s t tests were used to test for significance.
Immunoprecipitation and mass spectrometry
Ovaries from females containing the GFP::Spn-E–[Pacman] transgene were homogenized in lysis buffer supplemented with protease inhibitors (50 mM Hepes-KOH, pH 7.5, 150 mM KCl, 1.5 mM MgCl2, 1.5 mM EGTA, 10% glycerol, and 0.2% NP-40). As a negative control, ovaries from tub>GFP females were separately homogenized for analysis. Samples were centrifuged at 16,100 g for 10 min at 4°C. Supernatant volume was adjusted to bring protein concentration to 5 mg/ml, and the supernatant was then precleared with washed protein G magnetic beads (Invitrogen) for 1 h. Monoclonal anti-GFP antibody (mFX73; Wako Pure Chemical Industries) was added to precleared samples and incubated overnight at 4°C. Protein G magnetic beads were then added to samples and incubated for 2 h at 4°C. Beads were washed with lysis buffer and then with wash buffer (50 mM Hepes, pH 7.5, 200 mM KCl, and 0.2% NP-40) before a final wash in water. Treatment of immunoprecipitations with RNase to determine coimmunoprecipitation resistance to RNase was performed essentially as described previously (Landthaler et al., 2004). Immunoprecipitates bound to protein G beads were washed with lysis buffer and then with buffer R (50 mM Hepes, pH 7.5, 200 mM KCl, and 0.2 mM DTT). Beads were incubated in buffer R + 50 µg/ml RNase A (Sigma-Aldrich) + 10,000 U/ml RNase T1 (Sigma-Aldrich) at 15°C overnight. RNase minus control precipitates were treated identically except RNase was left out of the incubation. Beads were then washed extensively with buffer R before a final wash in water.
Protein pellets were dissolved in digestion buffer (2 M urea and 100 mM Tris, pH 8.5), reduced with TCEP (Tris[2-carboxyethyl]-phosphine hydrochloride), alkylated with iodoacetamide, and digested with trypsin. Digested peptides were analyzed by tandem liquid chromatography/mass spectrometryusing a mass spectrometer (LTQ; Thermo Fisher Scientific). Multidimensional chromatography was performed online with four salt steps (MacCoss et al., 2002). Tandem mass spectra were collected in a data-dependent manner with up to 5-ms2 scans performed for each initial scan (m/z range of 400–2,000). The search program ProLuCID (Xu et al., 2006) was used to match data to a Drosophila protein database. Peptide identifications were filtered using DTASelect (Tabb et al., 2002).
Spectral counts for a given protein were tabulated and normalized to total sample spectral counts to give counts per million. Because spectral counts for a given protein are proportional in number to the protein’s size (number of peptides), we normalized counts per million to the protein mass in kilodaltons to give counts per kilodalton per million.
Online supplemental material
Fig. S1 shows a confocal micrograph of a single ovariole from a female containing the GFP::Spn-E transgene. Fig. S2 shows a silver-stained SDS-PAGE of GFP immunoprecipitations for MUDPIT analysis. Fig. S3 shows a Western blot of GFP::Spn-E protein from wild-type and mutant ovaries. Video 1 shows a time lapse of GFP::Spn-E fluorescence. Video 2 shows a representative FRAP experiment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201411076/DC1.
Supplementary Material
Acknowledgments
We thank G. Hannon, H. Siomi, R. Fehon, P. MacDonald, H. Krause, P. Lasko, P. Zamore, and T. Kai for their gifts of reagents. We acknowledge the service support provided by the Developmental Hybridoma Studies Bank, the Bloomington Stock Center, and FlyBase.
This work was supported by the National Institutes of Health (R01GM068743).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AICc
- Akaike’s information criterion
- MUDPIT
- multidimensional protein identification technology
- piRNA
- PIWI-interacting RNA
- ROI
- region of interest
- TE
- transposable element
References
- Anand A., and Kai T.. 2012. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 31:870–882. 10.1038/emboj.2011.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Hannon G.J., and Brennecke J.. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 318:761–764. 10.1126/science.1146484 [DOI] [PubMed] [Google Scholar]
- Bantscheff M., Schirle M., Sweetman G., Rick J., and Kuster B.. 2007. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389:1017–1031. 10.1007/s00216-007-1486-6 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., and Hannon G.J.. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 128:1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Burnham K.P., and Anderson D.R.. 2002. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer-Verlag, New York. 488 pp. [Google Scholar]
- Butland G., Peregrín-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., et al. . 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 433:531–537. 10.1038/nature03239 [DOI] [PubMed] [Google Scholar]
- Czech B., Preall J.B., McGinn J., and Hannon G.J.. 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell. 50:749–761. 10.1016/j.molcel.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy E.M. 1974. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat. Rec. 178:731–757. 10.1002/ar.1091780406 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., and Zamore P.D.. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94–108. 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D.E., and Berg C.A.. 1995. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 9:2495–2508. 10.1101/gad.9.20.2495 [DOI] [PubMed] [Google Scholar]
- Grimaldi M.R., Cozzolino L., Malva C., Graziani F., and Gigliotti S.. 2007. nup154 genetically interacts with cup and plays a cell-type-specific function during Drosophila melanogaster egg-chamber development. Genetics. 175:1751–1759. 10.1534/genetics.106.062844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., and Siomi M.C.. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 315:1587–1590. 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- Haase A.D., Fenoglio S., Muerdter F., Guzzardo P.M., Czech B., Pappin D.J., Chen C., Gordon A., and Hannon G.J.. 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 24:2499–2504. 10.1101/gad.1968110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D., Olivieri D., Novatchkova M., Gruber F.S., Meixner K., Mechtler K., Stark A., Sachidanandam R., and Brennecke J.. 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 30:3977–3993. 10.1038/emboj.2011.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes L.N., and Spradling A.C.. 1997. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development. 124:1419–1431. [DOI] [PubMed] [Google Scholar]
- Khurana J.S., and Theurkauf W.. 2010. piRNAs, transposon silencing, and Drosophila germline development. J. Cell Biol. 191:905–913. 10.1083/jcb.201006034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M., Yalcin A., and Tuschl T.. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14:2162–2167. 10.1016/j.cub.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Li C., Vagin V.V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M.D., Syrzycka M., Honda B.M., et al. . 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 137:509–521. 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.K., and Kai T.. 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 104:6714–6719. 10.1073/pnas.0701920104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.K., Tao L., and Kai T.. 2009. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186:333–342. 10.1083/jcb.200904063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., and Jankowsky E.. 2011. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12:505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Link A.J., Eng J., Schieltz D.M., Carmack E., Mize G.J., Morris D.R., Garvik B.M., and Yates J.R. III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676–682. 10.1038/10890 [DOI] [PubMed] [Google Scholar]
- MacCoss M.J., McDonald W.H., Saraf A., Sadygov R., Clark J.M., Tasto J.J., Gould K.L., Wolters D., Washburn M., Weiss A., et al. . 2002. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA. 99:7900–7905. 10.1073/pnas.122231399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., and Hannon G.J.. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 137:522–535. 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K.M., Iwasaki Y.W., Murota Y., Nagao A., Mannen T., Kato Y., Siomi H., and Siomi M.C.. 2015. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 10:193–203. 10.1016/j.celrep.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Pane A., Wehr K., and Schüpbach T.. 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell. 12:851–862. 10.1016/j.devcel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.S., and Kai T.. 2010. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 20:724–730. 10.1016/j.cub.2010.02.046 [DOI] [PubMed] [Google Scholar]
- Reits E.A., and Neefjes J.J.. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145–E147. 10.1038/35078615 [DOI] [PubMed] [Google Scholar]
- Richter J.D., and Sonenberg N.. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 433:477–480. 10.1038/nature03205 [DOI] [PubMed] [Google Scholar]
- Saito K., and Siomi M.C.. 2010. Small RNA-mediated quiescence of transposable elements in animals. Dev. Cell. 19:687–697. 10.1016/j.devcel.2010.10.011 [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Mannen T., and Siomi H.. 2010. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 24:636–646. 10.1101/gad.1899210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M.C., Sato K., Pezic D., and Aravin A.A.. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12:246–258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Snapp E.L., Altan N., and Lippincott-Schwartz J.. 2003. Measuring protein mobility by photobleaching GFP chimeras in living cells. In Current Protocols in Cell Biology. John Wiley and Sons Inc., New York: 21.1.1–21.1.24. 10.1002/0471143030.cb2101s19 [DOI] [PubMed] [Google Scholar]
- Snee M.J., and Macdonald P.M.. 2004. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 117:2109–2120. 10.1242/jcs.01059 [DOI] [PubMed] [Google Scholar]
- Tabb D.L., McDonald W.H., and Yates J.R. III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26. 10.1021/pr015504q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T., and Lin H.. 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25:355–376. 10.1146/annurev.cellbio.24.110707.175327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J., He Y., Hoskins R.A., and Bellen H.J.. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 314:1747–1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Voronina E., Seydoux G., Sassone-Corsi P., and Nagamori I.. 2011. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3:a002774 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J.E., Buszczak M., and Sayles S.. 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell. 9:675–685. 10.1016/j.devcel.2005.09.015 [DOI] [PubMed] [Google Scholar]
- Xiol J., Spinelli P., Laussmann M.A., Homolka D., Yang Z., Cora E., Couté Y., Conn S., Kadlec J., Sachidanandam R., et al. . 2014. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 157:1698–1711. 10.1016/j.cell.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Xu T., Venable J.D., Park S.K., Cociorva D., Lu B., Liao L., Wohlschlegel J., Hewel J., and Yates J.R. III. 2006. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics. 5:S174. [Google Scholar]
- Yang P., Sampson H.M., and Krause H.M.. 2006. A modified tandem affinity purification strategy identifies cofactors of the Drosophila nuclear receptor dHNF4. Proteomics. 6:927–935. 10.1002/pmic.200500230 [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang J., Xu J., Zhang Z., Koppetsch B.S., Schultz N., Vreven T., Meignin C., Davis I., Zamore P.D., et al. . 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 151:871–884. 10.1016/j.cell.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu J., Koppetsch B.S., Wang J., Tipping C., Ma S., Weng Z., Theurkauf W.E., and Zamore P.D.. 2011. Heterotypic piRNA ping-pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell. 44:572–584. 10.1016/j.molcel.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Koppetsch B.S., Wang J., Tipping C., Weng Z., Theurkauf W.E., and Zamore P.D.. 2014. Antisense piRNA amplification, but not piRNA production or nuage assembly, requires the Tudor-domain protein Qin. EMBO J. 33:536–539. 10.1002/embj.201384895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.